ABSTRACT
Myeloid progenitor-derived suppressor cells (MDSCs) arise from myeloid progenitors and suppress both innate and adaptive immunity. MDSCs expand during the later phases of sepsis in mice, promote immunosuppression, and reduce survival. Here, we report that the myeloid differentiation-related transcription factor nuclear factor I-A (NFI-A) controls MDSC expansion during sepsis and impacts survival. Unlike MDSCs, myeloid cells with conditional deletion of the Nfia gene normally differentiated into effector cells during sepsis, cleared infecting bacteria, and did not express immunosuppressive mediators. In contrast, ectopic expression of NFI-A in myeloid progenitors from NFI-A myeloid cell-deficient mice impeded myeloid cell maturation and promoted immune repressor function. Importantly, surviving septic mice with conditionally deficient NFI-A myeloid cells were able to respond to challenge with bacterial endotoxin by mounting an acute inflammatory response. Together, these results support the concept of NFI-A as a master molecular transcriptome switch that controls myeloid cell differentiation and maturation and that malfunction of this switch during sepsis promotes MDSC expansion that adversely impacts sepsis outcome.
KEYWORDS: inflammation, sepsis immunosuppression, MDSCs, NFI-A, sepsis
INTRODUCTION
Myeloid progenitor-derived suppressor cells (MDSCs) represent a heterogenous population of immature myeloid cells that includes progenitors and precursors of monocytes/macrophages, granulocytes, and dendritic cells (1–3). MDSCs are generated under a variety of inflammatory and infection conditions (1, 2) and are best characterized by their immunosuppressive functions (4–6). They may also promote persistent inflammation and chronic infection with catabolism during chronic sepsis (7). MDSCs suppress both innate and adaptive immunity via production of immunosuppressive mediators and inhibition of T cell proliferation and activation (3, 4, 8). Phenotypically, murine MDSCs coexpress the myeloid differentiation markers Gr1 and CD11b, similarly to the Gr1+ CD11b+ myeloid progenitors that arise under normal physiological conditions (2, 9). Unlike the immunosuppressive Gr1+ CD11b+ cells (i.e., MDSCs), normal Gr1+ CD11b+ cells can differentiate into competent innate immune cells (3). Elimination of MDSCs in tumor-bearing mice enhances antitumor immunity (10). Since MDSCs are “immature” cells that deviate from the standard path of differentiation, it has been suggested that arrested myeloid cell differentiation and maturation may be responsible for MDSC generation and immunorepression (2, 6). How immature myeloid cells lose their ability to differentiate and instead generate MDSCs remains unclear. Some studies, however, have suggested that MDSCs retain their potential to differentiate and mature but are trapped in a MDSC phenotype in the environmental milieu of chronic inflammation or growing tumors (1). In support of this, Kusmartsev et al. (10) reported that MDSCs from tumor-bearing mice could differentiate into macrophages and dendritic cells when adoptively transferred into healthy hosts in the presence of all trans-retinoic acid. In contrast, Cuenca et al. (1) were unable to differentiate MDSCs with all trans-retinoic acid in the presence of sepsis. Thus, the molecular events that arrest Gr1+ CD11b+ cell differentiation and promote MDSC expansion in sepsis are unclear.
Sepsis-induced immunosuppression attenuates both innate and adaptive immunity, which disrupts microbial clearance of the original infection and increases the risk of secondary infections (11). Immunosuppression is present during multiple-organ dysfunction, and together, such dysfunctions increase mortality and morbidity in mice and humans with sepsis (12, 13). Using a murine model of polymicrobial sepsis, which, as a consequence of administration of antibiotics, briefly develops into an early proinflammatory phase followed a late, protracted anti-inflammatory/immunosuppressive phase (14), we (15) and others (16) observed massive expansion of MDSCs during the late phase of sepsis.
We discovered that MDSCs isolated from the bone marrow of late-sepsis mice express increased levels of nuclear factor I-A (NFI-A) protein (17, 18). We further showed that ex vivo knockdown of NFI-A in MDSCs restored their differentiation/maturation into macrophages and dendritic cells and diminished their suppressive functions (18), supporting the concept that NFI-A attenuates myeloid cell differentiation (19, 20). Recent studies suggested that NFI-A arrests development of myeloid progenitors in an undifferentiated state. In support of this concept, ectopic expression of NFI-A in human hematopoietic progenitors counteracts monocytic differentiation (20), whereas inhibiting its expression promotes granulocytic differentiation (21).
Here, we used a conditional genetic approach to study NFI-A contributions to MDSC generation during sepsis. Since global deletion of the murine Nfia gene causes perinatal lethality (22), we created a conditional knockout mouse model where the Nfia allele is deleted only in the myeloid-lineage cells. Using this myeloid cell-restricted gene ablation approach, we investigated how NFI-A affects myeloid cell development in a mouse model of polymicrobial sepsis induced by cecal ligation and puncture. NFI-A conditional knockout mice, in contrast to wild-type mice, which suffer profound immunosuppression (15), did not generate MDSCs or develop immunosuppression, and they survived sepsis. We conclude that increased NFI-A expression contributes to lethal sepsis and may be a novel target for sepsis intervention.
RESULTS
NFI-A conditional knockout mice do not modify immune cell numbers.
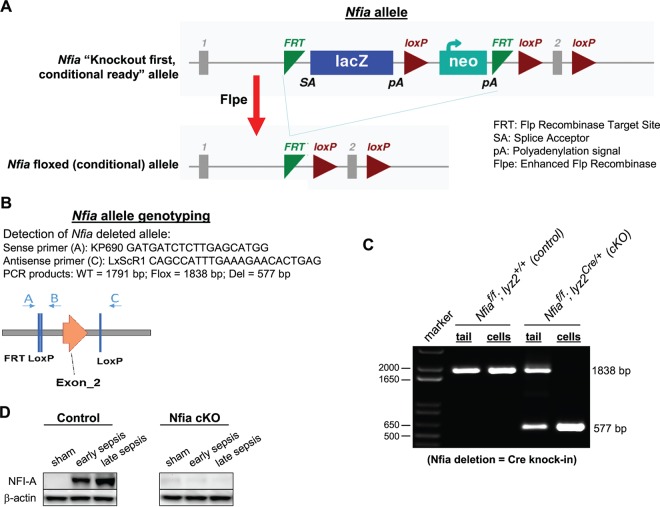
NFI-A protein levels are increased in septic Gr1+ CD11b+ MDSCs, which, unlike Gr1+ CD11b+ cells from naive mice, cannot differentiate into mature innate immune cells (18). Our previous studies implicated NFI-A in the generation and expansion of Gr1+ CD11b+ cells because the knockdown of NFI-A in the Gr1+ CD11b+ cell enhanced their differentiation and maturation into macrophages and dendritic cells (18). To investigate the in vivo effects of NFI-A on the Gr1+ CD11b+ cell development, we generated an NFI-A conditional knockout mouse model in which the Nfia gene was flanked by loxP sites (see Fig. 1). To achieve myeloid cell-specific (conditional) deletion, the Nfia floxed allele was crossed to the Lyz2Cre deletion strain. Mice deficient for the Nfia allele in the myeloid cells did not display any gross phenotypic abnormalities in terms of development, growth, or survival. Because NFI-A is expressed in the Gr1+ CD11b+ cell only after sepsis initiation, we first determined NFI-A protein levels in Gr1+ CD11b+ cells isolated from the bone marrow of the control (Nfiaflox/flox;Lyz2+/+) and knockout (Nfiaflox/flox;Lyz2cre/+) mice undergoing sepsis. NFI-A conditional knockout resulted in a complete loss of the NFI-A protein expression in Gr1+ CD11b+ cells (Fig. 1D).
FIG 1.
Generation of myeloid cell-specific NFI-A knockout model. (A) Schematic representation of gene targeting method used in the generation of the myeloid cell-specific NFI-A knockout mice. (B) The locations and sequences of the PCR primers used to confirm the gene targeting and sizes of the expected PCR products are shown. WT, wild type. (C) Nfia genotyping. The DNA was isolated from tail samples or Gr1+ CD11b+ cells and analyzed by PCR using the primers shown in panel B. PCR of the floxed (wild-type) allele produced a 405-bp product from tail DNA samples or Gr1+ CD11b+ cells, whereas PCR of the deleted allele produced a 577-bp product from Gr1+ CD11b+ cell DNA from the conditional knockout only, confirming successful deletion of Nfia in the myeloid lineage. f, flox. (D) NFI-A protein levels. Gr1+ CD11b+ cells were isolated from the bone marrow of the control (Nfiaflox/flox;Lyz2+/+) and NFI-A-deficient (Nfiaflox/flox;Lyz2cre/+) mice undergoing sepsis, and levels of the NFI-A protein were measured by Western blotting. The results are representative of two immunoblots from two independent experiments. cKO, knockout; Flox, floxed allele; Del, deleted allele.
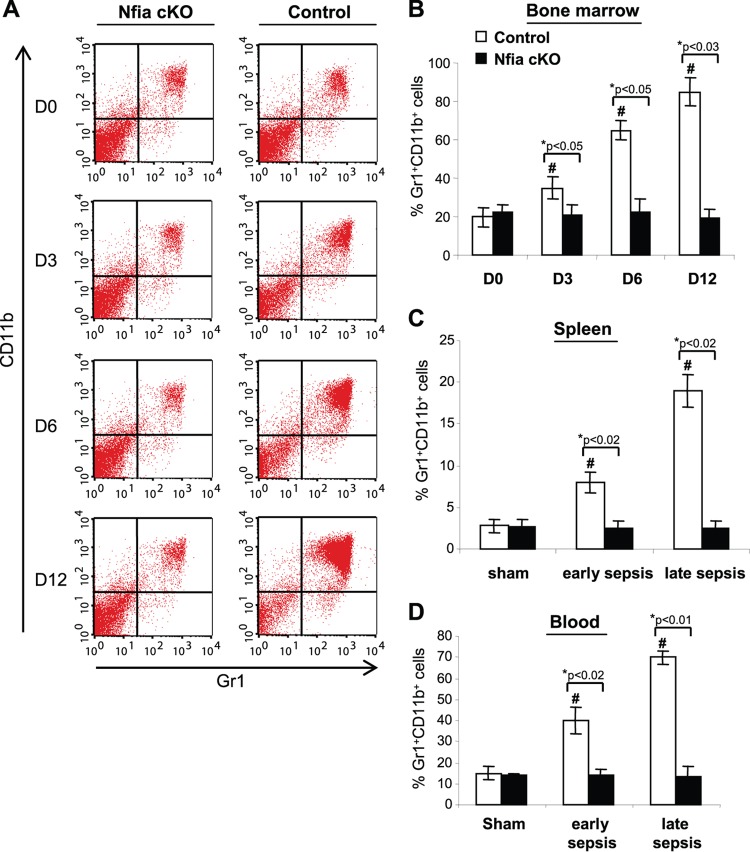
Next, we determined whether the Nfia deletion had any effect on the development of the immune system by measuring numbers of dendritic cells, T and B cells, and Gr1+ CD11b+ cells under steady-state conditions (i.e., in naive mice). Flow cytometry analysis of the bone marrow cells revealed that numbers of dendritic cells and Gr1+ CD11b+ cells were similar in control and NFI-A knockout mice (Fig. 2), suggesting that NFI-A is dispensable for steady-state myelopoiesis. Analysis of splenocytes also revealed normal numbers of T and B cells in NFI-A conditional knockout mice (Fig. 2). These results support the concept that NFI-A deficiency in the myeloid compartment does not affect immune cell numbers under normal conditions.
FIG 2.
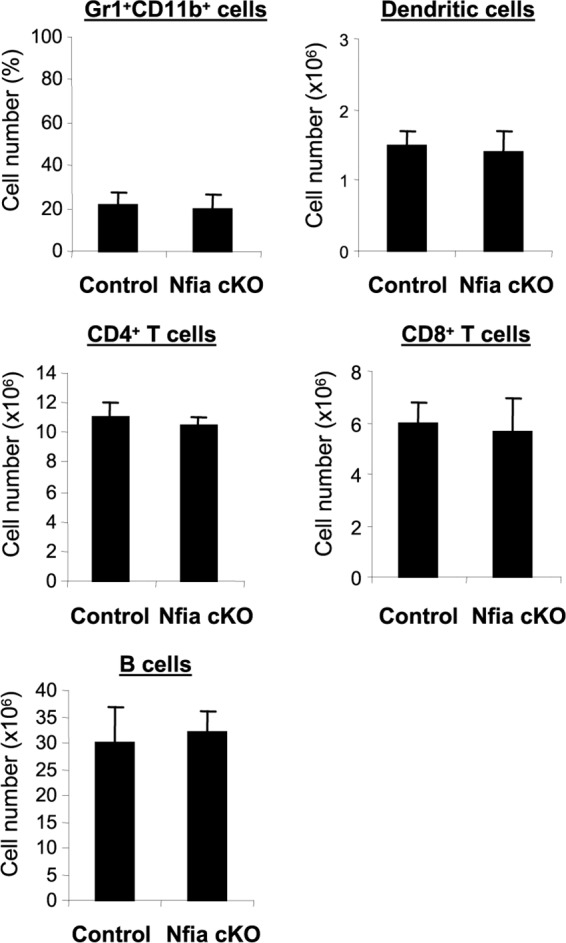
NFI-A conditional knockout does not affect immune cell numbers. Bone marrow cells were stained for Gr1+ CD11b+ cells, and splenocytes were stained for T cells, B cells, and dendritic cells and analyzed by flow cytometry. Data are expressed as means ± SD of results from 5 mice per group and represent results from two experiments. cKO, conditional knockout.
NFI-A conditional knockout mice survive sepsis and do not develop immunosuppression.
Using our sensitive model of polymicrobial sepsis, which develops into early and late sepsis phases (14), with fluid resuscitation and limited antibiotic treatment, mortality was higher during the late/chronic phase and caused immunosuppression concomitant with expansion and accumulation of Gr1+ CD11b+ MDSCs (15). As an initial approach to determine the effect of NFI-A deficiency on sepsis-induced immunosuppression, sepsis was induced by cecal ligation and puncture (CLP), and survival was followed for 4 weeks. Mice moribund during early sepsis (defined as the first 5 days after CLP) or late sepsis (after day 6) (14) were euthanized and analyzed. A corresponding number of mice that appeared healthy were also sacrificed at the same time but are reported as “survivors.” As shown in Fig. 3A, nearly 69% of the control knockout mice died during late sepsis, and none of the mice survived for 4 weeks. In striking contrast, 78% of NFI-A conditional knockout mice survived late sepsis.
FIG 3.
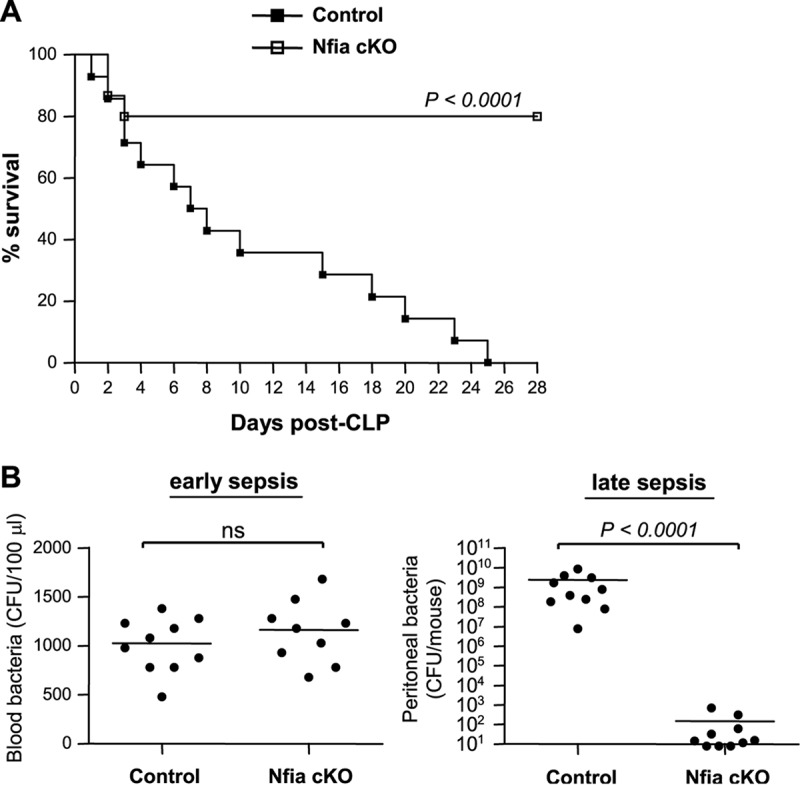
NFI-A conditional knockout mice survive late sepsis. Sepsis was induced by cecal ligation and puncture (CLP) using a 23-gauge needle, and mice were given antibiotics (imipenem) with fluid resuscitation. With this injury and treatment, sepsis develops into an early phase (defined as days 1 to 5) and a late phase (day 6 and thereafter). (A) Kaplan-Meier survival curve. Mortality was monitored for 28 days, and the death rate (moribundity) data were separated into two categories: early deaths (those occurring on days 1 to 5) and late deaths (those occurring on days 6 to 28). All moribund mice suffered significant (>30%) weight loss, hypothermia (<34°C), and loss of righting reflex. The data were collected from 24 mice per group, pooled from three independent experiments. (B) Diminished levels of peritoneal bacteria in late-sepsis NFI-A conditional knockout mice. Mice that were moribund during early or late sepsis were subjected to peritoneal lavage immediately after killing. Blood was collected via cardiac puncture. A corresponding number of surviving mice that appeared healthy (Nfia cKO group) were sacrificed and analyzed at the same time and are reported here as “surviving.” The blood or lavage bacteria were cultured on Trypticase soy agar plates, and the CFU counts were determined 24 h later. Data in panel B were analyzed by GraphPad Prism 5 and are expressed as means ± SD of results from 9 or 10 mice per group pooled from three experiments. cKO, conditional knockout.
The acute phase of sepsis is characterized by blood bacteremia and elevated levels of proinflammatory cytokines, whereas the late (immunosuppressive) phase is associated with localized bacterial growth and production of immunosuppressive cytokines (15). Thus, we measured numbers of bacteria and circulating cytokine levels in control and NFI-A conditional knockout mice during early and late sepsis. Blood bacteremia in NFI-A conditional knockout mice was not significantly different from that seen with the control mice during early sepsis (Fig. 3B). In sharp contrast, local (peritoneal) bacterial growth was diminished in NFI-A conditional knockout mice during late sepsis but was significantly higher in control mice. In addition, plasma levels of the proinflammatory cytokine tumor necrosis factor alpha (TNF-α) were increased in both control and NFI-A conditional knockout mice during early sepsis, but levels were significantly higher in control mice (Fig. 4). In late sepsis, TNF-α levels decreased in both control and NFI-A knockout mice. On the other hand, production of the immunosuppressive cytokine interleukin-10 (IL-10) was slightly increased in control and NFI-A conditional knockout mice during early sepsis. During late sepsis, production of IL-10 was further increased in control mice but was diminished in NFI-A conditional knockout mice (Fig. 4). These results show that NFI-A deficiency attenuates immunosuppressive cytokine production and enhances bacterial clearance in late-sepsis mice and improves late-sepsis survival, and these findings support the concept that NFI-A expression in myeloid cells plays a critical role in late-sepsis outcomes.
FIG 4.
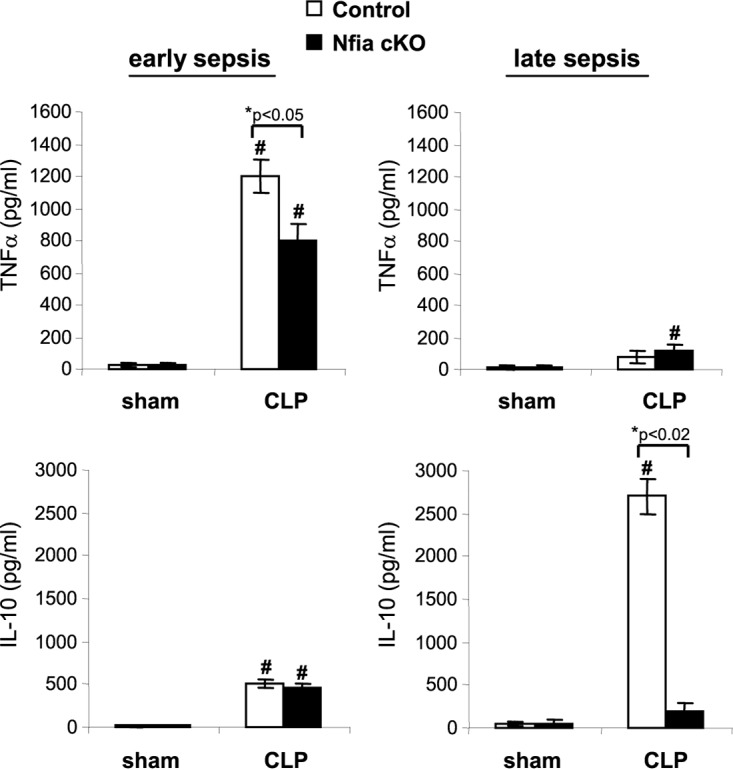
Plasma levels of TNF-α and IL-10. Blood was collected and plasma was recovered from moribund mice that were killed at days 1 to 4 (representing early sepsis) and days 8 to 25 (representing late sepsis) after CLP. TNF-α and IL-10 levels were measured by ELISA. Data are expressed as means ± SD of results from 5 to 7 mice per group pooled from three experiments. *, P value (control versus cKO, Student's t test); #, P < 0.05 (sham treatment group versus CLP group, ANOVA). cKO, conditional knockout.
NFI-A conditional knockout mice do not generate MDSCs during sepsis.
We predicted that the NFI-A deficiency would reduce or prevent Gr1+ CD11b+ MDSC generation and accumulation in late-sepsis mice, because our previous studies showed that late-sepsis Gr1+ CD11b+ cells were unable to differentiate ex vivo (15) and that small interfering RNA (siRNA)-mediated knockdown of NFI-A enhanced their differentiation and maturation (18). Next, we measured numbers of Gr1+ CD11b+ cells in the bone marrow after sepsis induction at intervals chosen to represent early sepsis (day 3) and late sepsis (days 6 to 12). Flow cytometry analysis revealed that numbers of the Gr1+ CD11b+ cells in naive mice (represented by day 0) were approximately 20% of those of the bone marrow cells in the control mice and 22% in the NFI-A conditional knockout mice (Fig. 5A and B). During sepsis, the numbers of Gr1+ CD11b+ cells in control mice increased exponentially from 35% at day 3 (i.e., early sepsis) and reached 85% at day 12 (i.e., late sepsis). In contrast, Gr1+ CD11b+ cell levels did not change significantly in the NFI-A conditional knockout mice during sepsis (Fig. 5B). Since MDSCs migrate systemically, we measured Gr1+ CD11b+ cell numbers in spleens and blood and observed marked increases in the spleens of control mice but not NFI-A conditional knockout mice during sepsis; increases were higher in late-sepsis mice (Fig. 5C). These results show that NFI-A expression is needed to prevent Gr1+ CD11b+ cell generation and accumulation during sepsis.
FIG 5.
Sepsis-induced expansion of Gr1+ CD11b+ cells is diminished in NFI-A conditional knockout mice. Bone marrow cells were harvested and stained with anti-Gr1 and anti-CD11b antibodies (Abs). (A) Example of flow cytometry of bone marrow cells harvested at intervals after sepsis induction and gated by Gr1+ CD11b+ staining. Numbers denote doubly positive cells within the gated cell population and represent their percentages in bone marrow. (B) Quantitative analysis of Gr1+ CD11b+ cell percentages. Data are from 5 mice per time point. (C) Percentages of Gr1+ CD11b+ in spleens harvested from septic mice that were moribund and killed on day 1 to day 4 (early sepsis) or after day 6 (late sepsis). (D) Percentages of Gr1+ CD11b+ cells in blood of sham treatment and septic mice. Data are expressed as means ± SD of results from 5 mice per group and represent one of two experiments. *, P value (control versus cKO, Student's t test); #, P < 0.05 (day 0 versus other time points or sham treatment group versus sepsis group, ANOVA). Day 0 (D0) represents sham treatment mice. cKO, conditional knockout.
NFI-A-deficient Gr1+ CD11b+ cells from septic mice are not immunosuppressive.
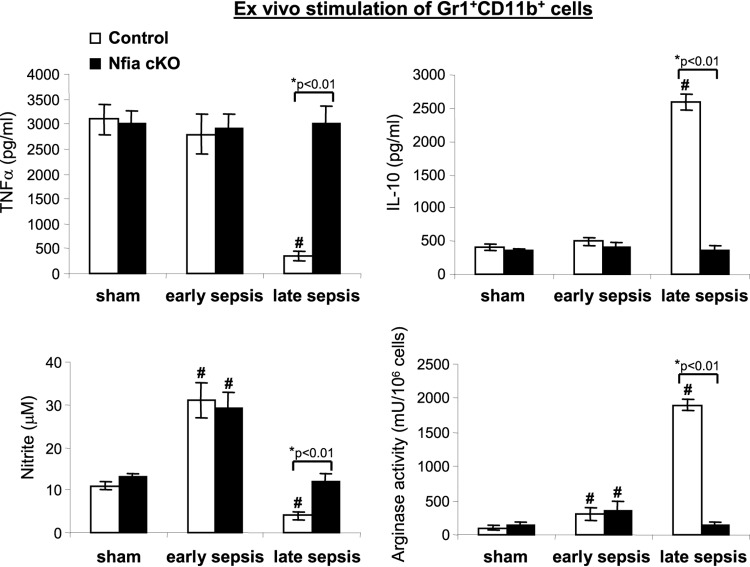
The numbers of Gr1+ CD11b+ cells were not increased in NFI-A conditional knockout mice (Fig. 5), and Gr1+ CD11b+ levels were similar to those seen with naive/sham treatment mice (i.e., at the steady-state level). Gr1+ CD11b+ cells not only increase in number during late sepsis but, unlike cells from naive mice, are immunosuppressive, as they produce immunosuppressive mediators such as IL-10 and overexpress arginase (15). In addition, Gr1+ CD11b+ cells generated during early sepsis are not immunosuppressive, on the basis of their proinflammatory mediators such as TNF-α and nitric oxide (NO), upon ex vivo stimulation with the bacterial lipopolysaccharide (LPS) (15). Therefore, we assessed whether the NFI-A-deficient Gr1+ CD11b+ cells generated during sepsis are immunosuppressive. Bone marrow Gr1+ CD11b+ cells were isolated from sham treatment and septic mice and stimulated with LPS for 24 h. Levels of TNF-α and IL-10 in the culture supernatants were measured by enzyme-linked immunosorbent assay (ELISA), and NO production (measured as nitrite) and arginase levels were measured in cell lysates. TNF-α and NO are proinflammatory mediators whose levels are increased during the early/acute phase of sepsis—to activate immune cells—but their production is decreased during late sepsis (15). These mediators are also produced upon stimulation of normal, immunocompetent immune cells (23, 24).
As shown in Fig. 6, both control and NFI-A-deficient Gr1+ CD11b+ cells from mice undergoing early sepsis produced increased amounts of TNF-α, similarly to cells from sham treatment mice. Importantly, TNF-α production by control cells from late-sepsis mice was diminished but remained significantly higher in the NFI-A-deficient cells. A similar pattern was observed for NO production, except that NO production by early-sepsis cells was much higher (Fig. 6). In contrast, sham treatment and early-sepsis Gr1+ CD11b+ cells from control or NFI-A conditional knockout mice produced small amounts of IL-10. In late sepsis, IL-10 production was significantly increased in control cells but remained at low levels in NFI-A-deficient cells (Fig. 6). In addition, arginase expression (measured by arginase activity) in early-sepsis cells was increased slightly in control and NFI-A-deficient Gr1+ CD11b+ cells, but its levels were significantly increased in late-sepsis control cells compared with NFI-A-deficient cells. These results support the concept that NFI-A-deficient Gr1+ CD11b+ cells generated during sepsis, unlike control cells, produce proinflammatory but not immunosuppressive mediators.
FIG 6.
Gr1+ CD11b+ cells from late-sepsis NFI-A conditional knockout mice exhibit proinflammatory phenotypes. Gr1+ CD11b+ MDSCs were isolated from the bone marrow of sham treatment and septic mice. Cells (1 × 106) were cultured with 1 μg/ml LPS for 24 h. Supernatants were collected and analyzed for TNF-α, IL-10, and NO production (measured as nitrite). The cells were harvested, lysed, and analyzed for arginase activity. Data are expressed as means ± SD of results from 5 mice per group and represent one of two experiments. *, P value (control versus cKO, Student's t test); #, P < 0.05 (sham treatment group versus sepsis group, ANOVA). cKO, conditional knockout.
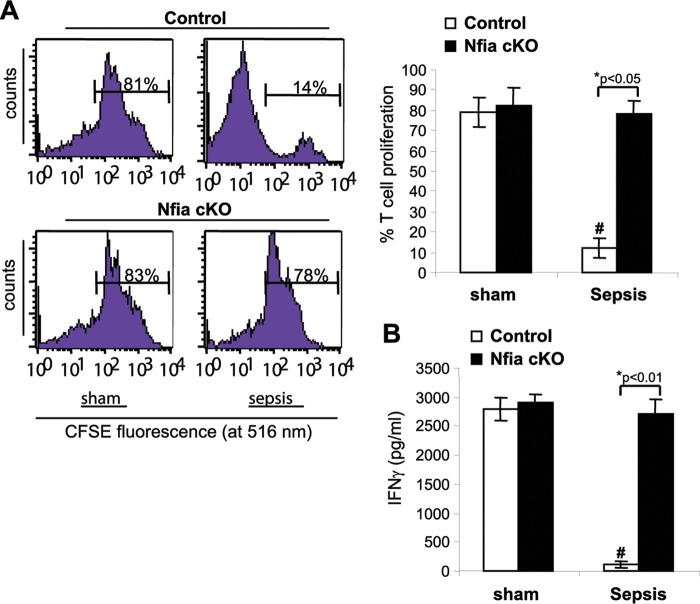
One of the important immunosuppressive functions of the Gr1+ CD11b+ MDSCs is inhibition of T cell activation/function and proliferation (5). Accordingly, we examined the effects of Gr1+ CD11b+ cells on antigen-driven proliferation and activation (measured by gamma interferon [IFN-γ] production) of T cells. Sham treatment or late-sepsis Gr1+ CD11b+ cells from control or NFI-A conditional knockout mice were cocultured with CD4+ T cells isolated from spleens of naive, wild-type mice. T cells were then activated by cross-linking with anti-CD3 plus anti-CD28 antibodies for 3 days. CD4+ T cell proliferation was reduced significantly with the coculture of control Gr1+ CD11b+ cells compared with NFI-A-deficient cells (Fig. 7A). Activated CD4+ T cells produced similar amounts of IFN-γ in coculture with control or NFI-A-deficient Gr1+ CD11b+ cells from sham treatment mice (Fig. 7B). Interestingly, when late-sepsis Gr1+ CD11b+ cells from control mice were used, IFN-γ production was significantly decreased compared with that seen with cells from NFI-A conditional knockout mice, whose production levels were increased similarly to cells from sham treatment mice. Together, the results presented in Fig. 6 and 7 show that Gr1+ CD11b+ cells generated during the course of sepsis in the absence of NFI-A were unable to repress IFN-γ expression in CD4+ T cells and therefore were not immunosuppressive.
FIG 7.
Gr1+ CD11b+ cells from late-sepsis NFI-A conditional knockout mice do not suppress T cell activation or proliferation. A coculture of Gr1+ CD11b+ cells and T cells was used to assess the immunosuppressive function of Gr1+ CD11b+ cells. Spleen CD4+ T cells were isolated from naive wild-type mice and labeled with the fluorescent dye CFSE for 10 min at room temperature. Bone marrow Gr1+ CD11b+ cells isolated from sham treatment or late-sepsis mice were then cocultured (at 1:1 ratio) with CD4+ T cells, and the culture was stimulated with anti-CD3 plus anti-CD28 antibodies (1 μg/ml each). (A) After 3 days, CD4+ T cell proliferation was determined by the stepwise dilution of CFSE dye in dividing CD4+ cells by flow cytometry. Histograms of gated T cells (left) and quantitative analysis of cell proliferation (right) are shown. Percentages of cell proliferation were calculated as follows: percent cell proliferation = 100 × (count from CD4+ T cell + Gr1+ CD11b+ cell culture/count from CD4+ T cell culture alone). (B) The culture supernatants were analyzed for IFN-γ production by ELISA. Data are expressed as means ± SD of results from 5 mice per group pooled from three experiments. *, P value (control versus cKO, Student's t test); #, P < 0.05 (sham treatment group versus sepsis group, ANOVA). cKO, conditional knockout.
NFI-A-deficient Gr1+ CD11b+ cells from septic mice can differentiate ex vivo into immune-responsive macrophages and dendritic cells, and exogenous NFI-A represses this differentiation.
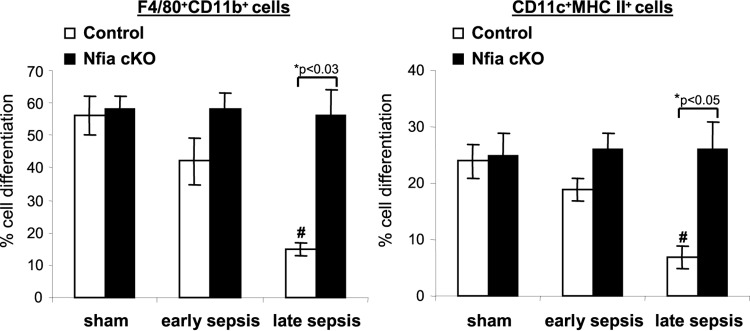
The inflammation-driven generation and accumulation of the MDSCs are caused by attenuated differentiation and maturation of the Gr1+ CD11b+ cell population into innate immune cells, and we have shown that this process becomes more prominent as sepsis progresses from the early phase to the late phase (15). To directly assess the effects of Nfia deletion on MDSC expansion in sepsis, we measured Gr1+ CD11b+ cell ex vivo differentiation and maturation into macrophages and dendritic cells. Bone marrow Gr1+ CD11b+ cells were cultured for 6 days in the presence of macrophage colony-stimulating factor (M-CSF) plus IL-4, and differentiated cells were phenotyped by flow cytometry. Gr1+ CD11b+ cells from control septic mice exhibited decreased differentiation into macrophages and dendritic cells versus sham treatment mice, and such decreases were significant for cells from late-sepsis mice (Fig. 8). Gr1+ CD11b+ cells from septic NFI-A conditional knockout mice produced more macrophages and dendritic cells, similarly to cells from sham treatment mice. This suggests that NFI-A-deficient Gr1+ CD11b+ cells generated during sepsis do not acquire the MDSC phenotype.
FIG 8.
Normal differentiation of septic Gr1+ CD11b+ cells from NFI-A conditional knockout mice. Bone marrow Gr1+ CD11b+ cells were isolated from sham treatment and septic mice. Cells were differentiated for 6 days with M-CSF plus rIL-4 (10 ng/ml each). Percentages of differentiated macrophages (F4/80+ CD11b+) and dendritic cells (CD11c+ MHC II+) were determined by flow cytometry. Data are expressed as means ± SD of results from 5 mice per group and represent one of two experiments. *, P value (control versus cKO, Student's t test); #, P < 0.05 (sham treatment group versus sepsis group, ANOVA). cKO, conditional knockout.
Expression of Nfia is highest in Gr1+ CD11b+ cells during late sepsis (17), a time when differentiation is limited (15). To confirm ex vivo the negative effects of NFI-A on Gr1+ CD11b+ cell differentiation and maturation, we transfected Gr1+ CD11b+ cells from late-sepsis, NFI-A conditional knockout mice with an NFI-A expression construct and confirmed NFI-A protein expression by Western blotting (Fig. 9A). The introduction of Nfia into the NFI-A-deficient Gr1+ CD11b+ cells (which were able to differentiate) significantly decreased their differentiation and maturation into macrophages and dendritic cells compared with cells transfected with an empty vector (Fig. 9B). Not only did reconstitution of Gr1+ CD11b+ cells with NFI-A diminish their differentiation capacity, but these cells also acquired an immunosuppressive phenotype (Fig. 9C), as they produced increased amounts of IL-10 in response to stimulation with LPS (compare with Fig. 4). Together, the results presented in Fig. 8 and 9 support the concept that increased NFI-A expression limits Gr1+ CD11b+ cell differentiation and maturation.
FIG 9.
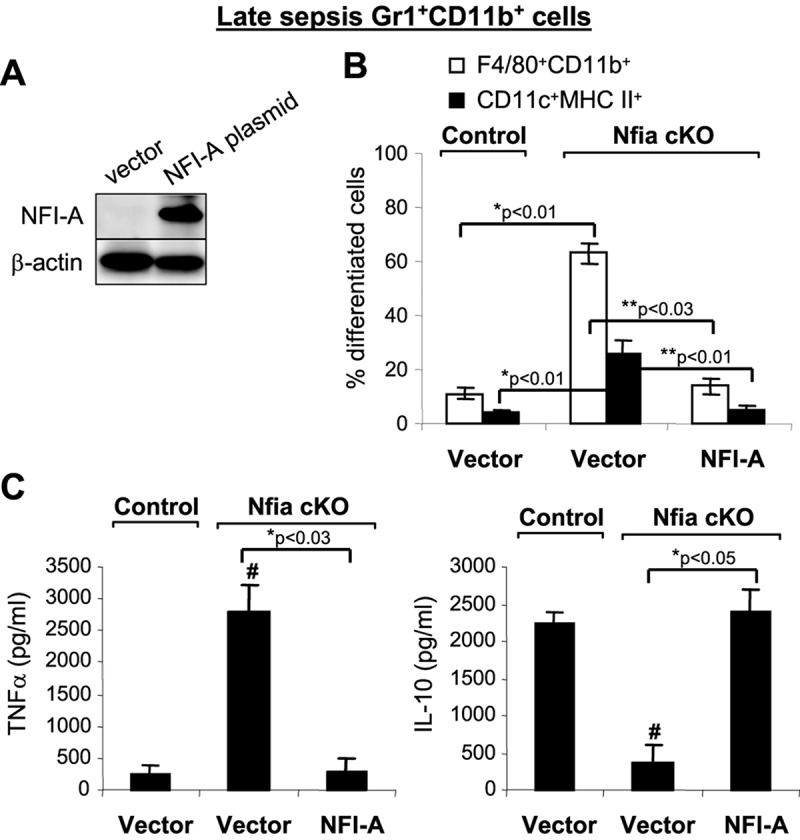
Ectopic expression of NFI-A in septic Gr1+ CD11b+ cells from NFI-A conditional knockout mice blocks their differentiation and switches them to an immunosuppressive phenotype. Bone marrow Gr1+ CD11b+ cells were isolated from late-sepsis mice (days 8 to 22 after sepsis induction) and transfected with an NFI-A expression plasmid or an empty vector. After 36 h, cells were differentiated for 6 days with M-CSF plus rIL-4 (10 ng/ml each). (A) NFI-A expression was confirmed by Western blotting using cells pooled from 3 mice in each group. Results are representative of two Western blottings. (B) Flow cytometry analysis of the differentiated cells gated by F4/80+ CD11b+ or CD11c+ MHC II+ staining. Data are expressed as means ± SD of results from 5 mice per group and represent one of three experiments. *, P value (control versus vector Nfia cKO); **, P value (vector Nfia cKO versus NFI-A, Student's t test). (C) Differentiated cells were stimulated for 12 h with 1 μg/ml LPS (E. coli serotype 0111:B4), and levels of TNF-α and IL-10 in the supernatants were determined by ELISA. Data are expressed as means ± SD of results from 5 mice per group and represent one of three experiments. *, P value (vector versus NFI-A, Student's t test); #, P < 0.05 (control versus Nfia cKO groups, ANOVA).
NFI-A conditional knockout mice survive sepsis and are LPS responsive.
The results described above showed that the NFI-A conditional knockout mouse survivors do not develop immunosuppression and that Gr1+ CD11b+ cells can differentiate into immunocompetent macrophages and dendritic cells and likely have immunocompetent CD4+ T lymphocytes. Sepsis without infection can be partially mimicked by systematic administration of Gram-negative bacterial LPS, which, in mice, nonhuman primates, and humans (25), rapidly activates innate immune cells to generate proinflammatory cytokines such as TNF-α and IL-6. After LPS stimulation in vivo, cells and organs rapidly enter a state of tolerance of endotoxin, with combined organ failure and immunosuppression (26, 27).
To investigate the effect of NFI-A deficiency in the myeloid lineage on the immune competency of innate immune cells in acute inflammation, we challenged naive control (NFI-A-expressing) and NFI-A conditional knockout mice that had recovered from sepsis with LPS. To do this, we intraperitoneally (i.p.) injected both groups with a sublethal dose (25 μg) of LPS 2 weeks after presumed sepsis recovery and measured plasma levels of TNF-α and IL-6 24 h after LPS challenge using ELISA. LPS challenge of the recovered, NFI-A conditional knockout mice increased TNF-α and IL-6 production to levels similar to those seen with the control, naive mice (Fig. 10), suggesting that NFI-A deficiency has no negative effects on innate immunity without sepsis.
FIG 10.
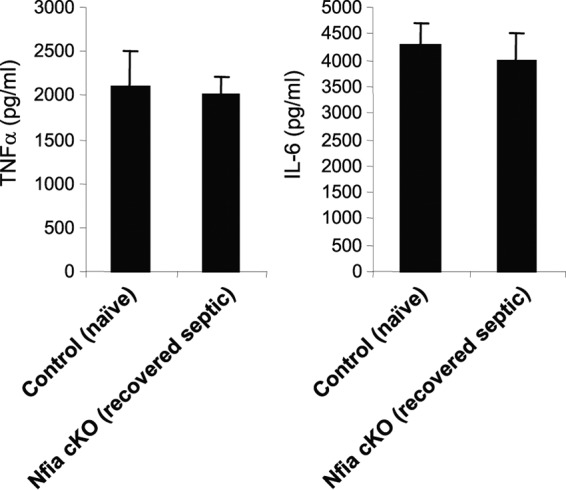
NFI-A conditional knockout mice that had recovered from sepsis mount a normal inflammatory response to LPS challenge. A number of the NFI-A conditional knockout mice that had recovered from sepsis were challenged (i.p.) with a sublethal dose (25 μg) of the Gram-negative bacterial endotoxin lipopolysaccharide (LPS) (from Escherichia coli serotype 0111:B4; Sigma-Aldrich, St. Louis, MO). Mice were subjected to the LPS challenge 2 weeks after the end of the 4-week sepsis time course. A group of naive, control mice were also injected with LPS and served as a positive control. Blood was collected 24 h later, and plasma levels of TNF-α and IL-6 were determined by ELISA. Data are expressed as means ± SD of results from 5 mice per group and represent one of two ELISAs. cKO, conditional knockout.
DISCUSSION
We previously reported that sepsis-induced generation and accumulation of Gr1+ CD11b+ MDSCs are due mainly to arrested differentiation and maturation of the Gr1+ CD11b+ myeloid progenitor population (15). In the present report, we provide genetic support for the concept that sepsis-induced MDSC generation depends on NFI-A expression, since conditional Nfia genetic deletion in myeloid cells prevented late-sepsis MDSC accumulation in bone marrow and spleen and immunosuppression and improved survival. These new findings are compatible with the notion that NFI-A is an essential regulator of the innate and adaptive immune repressor state of sepsis and contributes to poor outcome in murine polymicrobial sepsis.
NFI-A protein is not expressed in the naive Gr1+ CD11b+ cells, but its expression is induced during polymicrobial sepsis in mice (17). Gr1+ CD11b+ cells are intermediates of the myeloid-lineage cell repertoire that arise under normal physiological conditions and differentiate into competent innate immune cells (5). However, during sepsis (15) and under other inflammatory conditions (1, 3), Gr1+ CD11b+ cell numbers expand and are typified by the MDSC phenotype. The increase in Gr1+ CD11b+ cell numbers and suppressive activities has an impact on the later phases of sepsis, during which suppression of both innate and adaptive immunity becomes profound (15). The physiological importance of these findings is highlighted by the observation that the Gr1+ CD11b+ cells from NFI-A conditional knockout mice were able to differentiate normally and were not immunosuppressive. Moreover, NFI-A conditional knockout mice had normal numbers of both T and B cells, supporting the concept that NFI-A may have a global adaptive and innate immunologic impact during murine polymicrobial sepsis.
Through the repression of the cyclin-dependent kinase (cdk) inhibitor p21, the NFI-A protein can disrupt Gr1+ CD11b+ cell differentiation and maturation (18). Previous studies have shown that p21 acts through the cdk's to arrest cell cycle progression in human fibroblasts and breast cancer cell lines (28, 29). Deletion of the p21 gene in mouse increases levels of the myeloid progenitors by attenuating their differentiation (30). We have previously shown that p21 expression is lost in Gr1+ CD11b+ cells during murine sepsis due to the induction of Nfia expression (18). We found that the lack of p21 expression in septic Gr1+ CD11b+ cells enhanced formation of the cyclin D1-cdk4 protein complex, which led to activation of NF-κB p65 (18). The outcome was reduced differentiation of the Gr1+ CD11b+ cells. Silencing of the Nfia gene in Gr1+ CD11b+ cells ex vivo restores p21 expression and enhances their differentiation and maturation (18). In the NFI-A-deficient Gr1+ CD11b+ cells, we observed increased activation of NF-κB, as measured by the increases in NF-κB protein levels (data not shown). Persistent activation of NF-κB has been linked to myeloproliferative disorders and accumulation of Gr1+ CD11b+ cells in mouse (31).
The immunosuppressive activities of Gr1+ CD11b+ MDSCs are associated with different inflammatory conditions and cancer (2, 3). MDSCs from tumor-bearing animals and cancer patients produce increased amounts of the immunosuppressive cytokines IL-10 and transforming growth factor beta (TGF-β) and also overexpress arginase, which inhibits T cell activation and proliferation (2, 32). We have shown that the early/acute-sepsis Gr1+ CD11b+ cells from early/acute-sepsis mice are, like naive cells, proinflammatory, as they produce increased amounts of TNF-α, IL-6, and NO, which activate immune cells (15). However, late-sepsis Gr1+ CD11b+ MDSCs are reprogrammed to produce immunosuppressive IL-10 and arginase (15). Here, we showed that IL-10 and arginase levels did not increase in the NFI-A conditional knockout mice but did increase in control septic mice. NFI-A conditional knockout mice recovering from sepsis mounted an inflammatory response, similarly to naive mice when challenged with bacterial endotoxin. These findings demonstrate that NFI-A deficiency in the myeloid-lineage cells prevents expansion of Gr1+ CD11b+ MDSCs during sepsis.
We also showed that levels of Gr1+ CD11b+ MDSCs decrease in the spleens of NFI-A conditional knockout mice during sepsis. Gr1+ CD11b+ MDSCs accumulate in the spleen under different inflammatory conditions in mice (8, 16), and a small postmortem study showed that spleen harvested at bedside from humans who died from sepsis had increased numbers of Gr1+ CD11b+ MDSCs (33). The source of splenic MDSCs is not clear, but some studies support the concept that MDSCs originate in the bone marrow and then are trafficked to the spleen during injury or inflammation (34, 35). Others support the concept that splenic MDSCs may originate in the spleen as a result of extramedullary myelopoiesis, which is needed to replace myeloid cells depleted from bone marrow (1). This report provides strong evidence that splenic Gr1+ CD11b+ MDSCs in septic mice derive from bone marrow, since they were not detected in the spleen in the NFI-A conditional knockout mouse during sepsis.
Finally, emerging data support the concept that sepsis-associated immunosuppression of adaptive and innate immunity contributes to high mortality and morbidity during persistent sepsis (13, 36). For example, septic patients who die show evidence of unresolved primary infection and acquisition of secondary, opportunistic infections, including reactivation of latent viruses (11, 13). Studies of newer approaches to immunoenhancement treatment of sepsis are under way (37). Our findings showing that sepsis-induced expansion of MDSCs promotes and maintains sepsis immunosuppression are relevant to these newer sepsis treatment strategies, since MDSCs suppress both innate and adaptive immunity (32, 38), and MDSC numbers increase in blood of patients with sepsis (36, 39) and correlate with either early mortality or prolonged hospitalization (36). Moreover, elimination of MDSCs in tumor-bearing animals with chronic immunosuppression improves immune responses and may be an adjuvant to cytotoxic cancer treatment (10).
This report, by identifying a control or checkpoint point, potentially informs a new treatment strategy of targeting NFI-A to control immunosuppressive MDSC expansion.
MATERIALS AND METHODS
Production of BALB/cJ Nfia floxed mice.
The BALB/cJ Nfia floxed allele was created by gene targeting in BALB/cJ embryonic stem (ES) cells (PRX-Balb/cJ 9; Primogenix, Laurie, MO, USA) with a “knockout first, conditional ready” gene targeting vector (vector PGRS0001-B-F10) from the EUCOMM project [see http://www.mousephenotype.org/data/alleles/MGI:108056/tm1a(EUCOMM)Hmgu]. The resulting Nfia knockout first allele incorporated a FLP recombination target (FRT)-flanked splice-acceptor-LacZ-Neo cassette and a loxP site in intron 1 and another loxP site in intron 2, thus flanking Nfia exon 2 with loxP sites (see Fig. 1). Targeted ES cells were initially identified by long-range PCR with one primer outside the vector homology arm and a second primer in the inserted sequence. PCR-positive clones were validated by Southern blotting using 5′ and 3′ probes external to the vector homology arms and with a neomycin internal probe. A single correctly targeted clone (3E) was electroporated with an Flpe expression plasmid, and subclones were screened by PCR for removal of the “knockout first” element. Three subclones were injected into C57BL/6 blastocysts to generate chimeras, which were mated to BALB/cJ females for germ line transmission of the Nfia floxed allele (see Fig. 1).
Production of BALB/cJ Lyz2-Cre knock-in mice.
The structure of the BALB/cJ-Lyz2Cre knock-in allele was similar to that of the allele described by Clausen et al. (40). The allele was constructed as follows. The coding sequence for nucleus-localized Cre recombinase (NLS-Cre) was inserted precisely at the start codon of the Lyz2 gene by clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9-stimulated gene targeting in BALB/cJ ES cells. The targeting vector contained the following: (i) a 1,101-bp 5′ homology arm comprising sequences immediately 5′ of the ATG start codon of the Lyz2 gene; (ii) the NLS-Cre coding sequence (without polyadenylation signal) followed by a FRT-flanked neomycin selection cassette; and (iii) an 860-bp 3′ homology arm comprising sequences immediately 3′ of the Lyz2 ATG start codon. The circular targeting vector was electroporated into BALB/cJ ES cells (PRX-Balb/cJ 9; Primogenix, Laurie, MO, USA) together with a plasmid encoding the Cas9-D10A nickase and two plasmids for U6-promoter-driven expression of guide RNAs targeting Cas9 nickase to the Lyz2 start codon region. The guide RNAs and their target sequences were as follows: Lyz2-g1B (5′-CAGAGTCAGGAGAGTCTTCATGG-3′) and Lyz2-g36T (5′-CCTGCTTTCTGTCACTGCTCAGG-3′). Electroporated ES cells were selected on G418, and the surviving clones were screened by PCR to identify clones with homologous integration of the Cre and Neo cassette region at the Lyz2 locus. PCR-positive clones were analyzed by Southern blotting with 5′ and 3′ probes external to the targeting vector and a neomycin probe. A correctly targeted clone (2G9) was subsequently electroporated with an Flpe expression plasmid, and subclones were screened by PCR for removal of the neomycin cassette. Three correct subclones were injected into C57BL/6 blastocysts to generate chimeras. Chimeras were mated to BALB/cJ females for germ line transmission of the Lyz2Cre knock-in allele (see Fig. S1 in the supplemental material).
Generation of BALB/cJ Nfia conditional knockout mice.
The myeloid cell-specific NFI-A knockout mice were generated by breeding the Nfia floxed mice described above with the Lyz2Cre knock-in mice. Nfiaflox/flox;Lyz2Cre/+ mice were crossed to Nfiaflox/flox;Lyz2+/+ mice to generate Nfiaflox/flox mice with and without Cre. Genotypes were verified for all mice by PCR. Western blotting was also used to confirm that the myeloid cells were negative for Nfia expression. The Nfiaflox/flox;Lyz2Cre/+ mice, where the expression of the Cre recombinase under the control of the Lyz2 gene promoter inactivates the floxed Nfia allele in the myeloid-lineage cells, served as our myeloid cell-specific knockout. The Nfiaflox/flox;Lyz2+/+ mice, which do not express the Cre recombinase (and thus, the floxed Nfia allele is still expressed in the myeloid-lineage cells), served as controls.
The mice were bred and housed in a pathogen-free facility in the Division of Laboratory Animal Resources of East Tennessee State University. Wild-type male BALB/cJ littermates (8 to 10 weeks old) were purchased from the Jackson Laboratory (Bar Harbor, ME) and were acclimated to the new environment for a week before surgery. All experiments were conducted in accordance with National Institutes of Health guidelines and were approved by the East Tennessee State University Animal Care and Use Committee.
Sepsis model.
Polymicrobial sepsis was induced by cecal ligation and puncture (CLP) as described previously (14). Briefly, mice were anesthetized via inhalation with 2.5% isoflurane (Abbott Laboratories, Abbott Park, IL). A midline abdominal incision was made, and the cecum was exteriorized, ligated distal to the ileocecal valve, and then punctured twice with a 21-gauge needle. A small amount of feces was extruded into the abdominal cavity. The abdominal wall and skin were sutured in layers with 3-0 silk. Sham treatment mice were treated identically except that the cecum was not ligated or punctured. Mice received 1 ml lactated Ringers plus 5% dextrose for fluid resuscitation intraperitoneally (i.p.). This level of injury creates a prolonged infection with 100% mortality over 4 weeks. To generate late sepsis, mice were subcutaneously administered antibiotic (imipenem; 25 mg/kg body weight) or an equivalent volume of 0.9% saline solution. To establish intra-abdominal infection and approximate the clinical situation of early human sepsis, when there often is a delay between the onset of sepsis and the delivery of therapy (41), injections of imipenem were given at 8 and 16 h after CLP, a treatment which results in high (∼70%) mortality during the late/chronic phase (14). The presence of early sepsis was confirmed by transient systemic bacteremia and elevated cytokine levels in the first 5 days after CLP. Late/chronic sepsis (after day 5) was confirmed by the presence of enhanced peritoneal bacterial overgrowth and reduced levels of circulating proinflammatory cytokines.
Isolation of Gr1+ CD11b+ cells.
Gr1+ CD11b+ cells were isolated from the bone marrow immediately after mice were euthanized by use of magnetically assisted cell sorting according to the manufacturer's protocol (Miltenyi Biotech, Auburn, CA). The bone marrow cells were flushed out of the femurs with RPMI 1640 medium (without serum) under aseptic conditions (14). A single-cell suspension of the bone marrow was made by pipetting up and down and filtering through a 70-μm-pore-size nylon strainer, followed by incubation with erythrocyte lysis buffer. After washing, total Gr1+ CD11b+ cells were purified by subjecting the single-cell suspension to positive selection of the Gr1+ CD11b+ cells by incubation with biotin-coupled mouse anti-Gr1 antibody (clone RB6-8C5; eBioscience, San Diego, CA) for 15 min at 4°C. Cells were then incubated with antibiotin magnetic beads for 20 min at 4°C and subsequently passed over a mass spectrometry (MS) column. Purified Gr1+ CD11b+ cells were then washed and resuspended in sterile saline solution. For blood Gr1+ CD11b+ cells, blood was collected via cardiac puncture, and red blood cells were removed by lysis. After washing, cells were phenotyped by flow cytometry or subjected to positive selection of the Gr1+ CD11b+ cells as described above. The cell purity was determined by flow cytometry. Typically, ∼90% Gr1+ CD11b+ cells were obtained by this procedure.
Cell culture.
Gr1+ CD11b+ cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine (all from HyClone Laboratories, Logan, UT), and 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) at 37°C and 5% CO2.
Flow cytometry.
Total or differentiated Gr1+ CD11b+ cells were stained by incubation for 30 min on ice in staining buffer (phosphate-buffered saline [PBS], 2% fetal bovine serum [FBS]) with the following antibodies: anti-Gr1 conjugated to fluorescein isothiocyanate (FITC), anti-CD11b conjugated to phycoerythrin (PE), anti-F4/80 conjugated to allophycocyanin (APC), anti-CD11c conjugated to PE, and anti-major histocompatibility complex (MHC) II conjugated to FITC. CD4+ T cells were stained with anti-CD4 antibody conjugated to PE (all antibodies were from eBioscience, San Diego, CA). An appropriate isotype-matched control was used for each antibody. After washing, the samples were analyzed by the use of a FACSCaliber flow cytometer (BD Biosciences, Sparks, MD). About 25,000 events were acquired and analyzed using CellQuest Pro software (BD Biosciences).
Ex vivo differentiation of Gr1+ CD11b+ cells.
Gr1+ CD11b+ cells were cultured for 6 days with complete RPMI 1640 medium in the presence of 10 ng/ml of M-CSF (PeproTech Inc., Rocky Hill, NJ) and 10 ng/ml recombinant IL-4 (rIL-4) (eBioscience, San Diego, CA). The cell phenotypes were analyzed by flow cytometry. In some experiments, a portion of differentiated cells was washed and stimulated for 12 h with 1 μg/ml of LPS, and culture supernatants were used for cytokine measurements by ELISA.
ELISA.
Cytokine concentrations were determined using specific enzyme-linked immunosorbent assay (ELISA) kits (eBioscience) according to the instructions of the manufacturer. Each sample was run in duplicate.
T cell suppression assay.
Gr1+ CD11b+ cells were cultured with CD4+ T cells to determine their effects on T cell proliferation and IFN-γ production. Briefly, spleen CD4+ T cells from naive wild-type mice were isolated by positive selection using biotinylated anti-CD4 magnetic beads (Miltenyi). Cells were fluorescently labeled with carboxy-fluorosceindiacetate, succinimidyl ester (CFSE) dye using a Vybrant 2′,7′-dichlorodihydrofluorescein diacetate (CFDA) SE Cell Tracer kit (Invitrogen/Molecular Probes, Eugene, OR). Cells were incubated for 10 min at room temperature with 10 μM CFSE dye and then cocultured (at a 1:1 ratio) with Gr1+ CD11b+ cells. T cell proliferation was induced by stimulation with an anti-CD3 antibody plus an anti-CD28 antibody (R&D Systems, Minneapolis, MN) (1 μg/ml each). After 3 days, cells were harvested and CD4+ T cell proliferation was determined by stepwise dilution of CFSE dye in dividing, CD3-gated CD4+ T cells using flow cytometry. Culture supernatants were collected for the IFN-γ measurement.
NFI-A construct and transfection.
Full-length mouse Nfia cDNA was cloned in a pEZ-M07 plasmid expression vector downstream of the cytomegalovirus (CMV) promoter, and NFI-A protein expression was verified by Western blotting. An empty pEZ-M07 vector served as a negative control. Plasmid DNA was suspended in HiPerFect reagent (Qiagen, Valencia, CA) (final concentration, 0.5 μg/ml) and transfected into Gr1+ CD11b+ cells, using a Gene Pulser MXCell system (Bio-Rad, Hercules, CA). After 24 h, cells were differentiated for 6 days with M-CSF plus rIL-4 (as described above). In some experiments, differentiated cells were stimulated for 12 h with LPS.
Western blotting.
Equal amounts of protein extracts were mixed with 5× Laemmli sample buffer, separated by the use of an SDS-10% polyacrylamide gel (Bio-Rad), and subsequently transferred to nitrocellulose membranes (Thermo Fisher Scientific, Waltham, MA). After blocking with 5% milk–Tris-buffered saline–Tween 20 was performed for 1 h at room temperature, membranes were probed overnight at 4°C with mouse anti-NFI-A antibody (Santa Cruz Biotechnology, Dallas, TX). After washing, blots were incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (Life Technologies, Grand Island, NY) for 2 h at room temperature. Proteins were detected with an enhanced chemiluminescence detection system (Thermo Fisher Scientific). The developed bands were visualized using a ChemiDoc XRS system (Bio-Rad), and the images were captured with Image Lab software V3.0. Membranes were stripped and reprobed with β-actin antibody (Sigma-Aldrich) as a loading control.
Blood and peritoneal bacterial culture.
Immediately after mice were euthanized, the peritoneal cavity was subjected to lavage performed with 5 ml PBS. The lavage fluid was cleared by centrifugation and diluted 6- to 8-fold. Blood was collected via cardiac puncture in heparinized tubes and diluted 5-fold. Diluted lavage fluid or blood was plated on Trypticase soy agar base (BD Biosciences, Sparks, MD). The plates were incubated for 24 h at 37°C under aerobic conditions. The plates were read by a microbiologist, and the CFU counts were determined and multiplied by the dilution factor.
Nitric oxide production.
Gr1+ CD11b+ cells were cultured in RPMI 1640 medium and stimulated with 1 μg/ml LPS (Escherichia coli [0111:B4]; Sigma, St. Louis, MO) for 24 h. Nitric oxide production (which reflects inducible nitric oxide synthase [iNOS] activity) was determined by analysis of nitrite concentrations in the culture supernatants using Griess reagent according to the protocol of the manufacturer (Molecular Probes, Eugene, OR). Briefly, Griess reagent was made immediately before use by mixing equal volumes of 0.1% N-1-naphthylethylenediamine dihydrochloride and 1% sulfanilamide in 5% phosphoric acid. Samples and sodium nitrite standards (150 μl) were mixed with 20 μl Griess reagent and 130 μl double-distilled water (dH2O) and then incubated for 30 min at room temperature in a 96-well microplate. Absorbance was read at 548 nm using a spectrophotometer. Nitrite concentrations were calculated from the NaNO2 standard curve.
Arginase activity assay.
After the culture supernatants were collected for the NO production assay, Gr1+ CD11b+ cells were collected and arginase 1 activity was determined by measuring urea concentration (a by-product of arginase 1 activity) in the cell lysates using an arginase assay kit (Abnova, Walnut, CA). Briefly, ∼1 × 106 cells were lysed with 100 μl of 10 mM Tris-HCl (pH 7.4) containing 1× protease inhibitor cocktail and 0.4% Triton X-100. Lysates were cleared by centrifugation for 10 min at 15,000 rpm. Arginine hydrolysis by arginase was conducted by incubating 40 μl of lysate with 10 μl of 5× substrate buffer (containing l-arginine) in a 96-well plate at 37°C for 2 h. The reaction was stopped by adding 200 μl of urea reagent to all wells, including those containing the urea standard. The plate was then incubated at room temperature for 20 min, and the urea concentration was measured at 520 nm. One unit of arginase 1 converts 1 μmol of l-arginine to ornithine and urea per minute at pH 9.5 and 37°C.
Statistical analysis.
The Kaplan-Meier survival curve was plotted by the use of GraphPad Prism version 5.0 (GraphPad Software, La Jolla, CA), and survival significance was determined by a log-rank test. All other data were analyzed by the use of Microsoft Excel, V3.0, and are presented as means ± standard deviations (SD). Differences between 2 groups were analyzed by an unpaired Student's t test. One-way analysis of variance (ANOVA) was used to analyze differences in comparisons of 3 or more groups. P values of <0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01GM103887 (to M.E.G.) and C06RR0306551 (to the East Tennessee State University College of Medicine).
We thank Dale Cowley and colleagues (TransViragen, Chapel Hill, NC) for providing gene targeting services to generate the NFI-A conditional knockout and Lyz2-Cre strains. We also thank Michelle Duffourc and Rhesa Dykes (ETSU Molecular Biology Core) for assisting with the mouse genotyping.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00066-17.
REFERENCES
- 1.Cuenca AG, Delano MJ, Kelly-Scumpia KM, Moreno C, Scumpia PO, Laface DM, Heyworth PG, Efron PA, Moldawer LL. 2011. A paradoxical role for myeloid-derived suppressor cells in sepsis and trauma. Mol Med 17:281–292. doi: 10.1007/s00894-010-0723-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gabrilovich DI, Nagaraj S. 2009. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostrand-Rosenberg S, Sinha P. 2009. Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol 182:4499–4506. doi: 10.4049/jimmunol.0802740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lees JR, Azimzadeh AM, Bromberg JS. 2011. Myeloid derived suppressor cells in transplantation. Curr Opin Immunol 23:692–697. doi: 10.1016/j.coi.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Nagaraj S, Youn JI, Gabrilovich DI. 2013. Reciprocal relationship between myeloid-derived suppressor cells and T cells. J Immunol 191:17–23. doi: 10.4049/jimmunol.1300654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ostrand-Rosenberg S. 2010. Myeloid-derived suppressor cells: more mechanisms for inhibiting antitumor immunity. Cancer Immunol Immunother 59:1593–1600. doi: 10.1007/s00262-010-0855-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mira JC, Gentile LF, Mathias BJ, Efron PA, Brakenridge SC, Mohr AM, Moore FA, Moldawer LL. 2017. Sepsis pathophysiology, chronic critical illness, and persistent inflammation-immunosuppression and catabolism syndrome. Crit Care Med 45:253–262. doi: 10.1097/CCM.0000000000002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong YY, Fuchsberger M, Xiang SD, Apostolopoulos V, Plebanski M. 2013. Myeloid derived suppressor cells and their role in diseases. Curr Med Chem 20:1437–1444. doi: 10.2174/0929867311320110006. [DOI] [PubMed] [Google Scholar]
- 9.Condamine T, Gabrilovich DI. 2011. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol 32:19–25. doi: 10.1016/j.it.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kusmartsev S, Cheng F, Yu B, Nefedova Y, Sotomayor E, Lush R, Gabrilovich D. 2003. All-trans-retinoic acid eliminates immature myeloid cells from tumor-bearing mice and improves the effect of vaccination. Cancer Res 63:4441–4449. [PubMed] [Google Scholar]
- 11.Hotchkiss RS, Monneret G, Payen D. 2013. Immunosuppression in sepsis: a novel understanding of the disorder and a new therapeutic approach. Lancet Infect Dis 13:260–268. doi: 10.1016/S1473-3099(13)70001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shubin NJ, Monaghan SF, Ayala A. 2011. Anti-inflammatory mechanisms of sepsis. Contrib Microbiol 17:108–124. doi: 10.1159/000324024. [DOI] [PubMed] [Google Scholar]
- 13.Hotchkiss RS, Monneret G, Payen D. 2013. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol 13:862–874. doi: 10.1038/nri3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brudecki L, Ferguson DA, Yin D, Lesage GD, McCall CE, El Gazzar M. 2012. Hematopoietic stem-progenitor cells restore immunoreactivity and improve survival in late sepsis. Infect Immun 80:602–611. doi: 10.1128/IAI.05480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brudecki L, Ferguson DA, McCall CE, El Gazzar M. 2012. Myeloid-derived suppressor cells evolve during sepsis and can enhance or attenuate the systemic inflammatory response. Infect Immun 80:2026–2034. doi: 10.1128/IAI.00239-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delano MJ, Scumpia PO, Weinstein JS, Coco D, Nagaraj S, Kelly-Scumpia KM, O'Malley KA, Wynn JL, Antonenko S, Al-Quran SZ, Swan R, Chung CS, Atkinson MA, Ramphal R, Gabrilovich DI, Reeves WH, Ayala A, Phillips J, Laface D, Heyworth PG, Clare-Salzler M, Moldawer LL. 2007. MyD88-dependent expansion of an immature GR-1(+)CD11b(+) population induces T cell suppression and Th2 polarization in sepsis. J Exp Med 204:1463–1474. doi: 10.1084/jem.20062602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClure C, Brudecki L, Ferguson DA, Yao ZQ, Moorman JP, McCall CE, El Gazzar M. 2014. MicroRNA 21 (miR-21) and miR-181b couple with NFI-A to generate myeloid-derived suppressor cells and promote immunosuppression in late sepsis. Infect Immun 82:3816–3825. doi: 10.1128/IAI.01495-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClure C, Ali E, Youssef D, Yao ZQ, McCall CE, El Gazzar M. 2016. NFI-A disrupts myeloid cell differentiation and maturation in septic mice. J Leukoc Biol 99:201–211. doi: 10.1189/jlb.4A0415-171RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. 2005. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell 123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 20.Rosa A, Ballarino M, Sorrentino A, Sthandier O, De Angelis FG, Marchioni M, Masella B, Guarini A, Fatica A, Peschle C, Bozzoni I. 2007. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci U S A 104:19849–19854. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zardo G, Ciolfi A, Vian L, Starnes LM, Billi M, Racanicchi S, Maresca C, Fazi F, Travaglini L, Noguera N, Mancini M, Nanni M, Cimino G, Lo-Coco F, Grignani F, Nervi C. 2012. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood 119:4034–4046. doi: 10.1182/blood-2011-08-371344. [DOI] [PubMed] [Google Scholar]
- 22.das Neves L, Duchala CS, Tolentino-Silva F, Haxhiu MA, Colmenares C, Macklin WB, Campbell CE, Butz KG, Gronostajski RM. 1999. Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc Natl Acad Sci U S A 96:11946–11951. doi: 10.1073/pnas.96.21.11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munder M, Eichmann K, Modolell M. 1998. Alternative metabolic states in murine macrophages reflected by the nitric oxide synthase/arginase balance: competitive regulation by CD4+ T cells correlates with Th1/Th2 phenotype. J Immunol 160:5347–5354. [PubMed] [Google Scholar]
- 24.Williams MA, Withington S, Newland AC, Kelsey SM. 1998. Monocyte anergy in septic shock is associated with a predilection to apoptosis and is reversed by granulocyte-macrophage colony-stimulating factor ex vivo. J Infect Dis 178:1421–1433. doi: 10.1086/314447. [DOI] [PubMed] [Google Scholar]
- 25.Kopanakis K, Tzepi IM, Pistiki A, Carrer DP, Netea MG, Georgitsi M, Lymperi M, Droggiti DI, Liakakos T, Machairas A, Giamarellos-Bourboulis EJ. 2013. Pre-treatment with low-dose endotoxin prolongs survival from experimental lethal endotoxic shock: benefit for lethal peritonitis by Escherichia coli. Cytokine 62:382–388. doi: 10.1016/j.cyto.2013.03.028. [DOI] [PubMed] [Google Scholar]
- 26.Brudecki L, Ferguson DA, McCall CE, El Gazzar M. 2012. Adoptive transfer of CD34(+) cells during murine sepsis rebalances macrophage lipopolysaccharide responses. Immunol Cell Biol 90:925–934. doi: 10.1038/icb.2012.32. [DOI] [PubMed] [Google Scholar]
- 27.Remick DG, Newcomb DE, Bolgos GL, Call DR. 2000. Comparison of the mortality and inflammatory response of two models of sepsis: lipopolysaccharide vs. cecal ligation and puncture. Shock 13:110–116. doi: 10.1097/00024382-200013020-00004. [DOI] [PubMed] [Google Scholar]
- 28.Dulić V, Kaufmann WK, Wilson SJ, Tlsty TD, Lees E, Harper JW, Elledge SJ, Reed SI. 1994. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell 76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 29.Rashidian J, Iyirhiaro GO, Park DS. 2007. Cell cycle machinery and stroke. Biochim Biophys Acta 1772:484–493. doi: 10.1016/j.bbadis.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 30.Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, Scadden DT. 2000. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 31.Zhao JL, Rao DS, Boldin MP, Taganov KD, O'Connell RM, Baltimore D. 2011. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc Natl Acad Sci U S A 108:9184–9189. doi: 10.1073/pnas.1105398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eruslanov E, Daurkin I, Ortiz J, Vieweg J, Kusmartsev S. 2010. Pivotal advance: tumor-mediated induction of myeloid-derived suppressor cells and M2-polarized macrophages by altering intracellular PGE(2) catabolism in myeloid cells. J Leukoc Biol 88:839–848. doi: 10.1189/jlb.1209821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boomer JS, To K, Chang KC, Takasu O, Osborne DF, Walton AH, Bricker TL, Jarman SD, Kreisel D, Krupnick AS, Srivastava A, Swanson PE, Green JM, Hotchkiss RS. 2011. Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA 306:2594–2605. doi: 10.1001/jama.2011.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bronte V, Apolloni E, Cabrelle A, Ronca R, Serafini P, Zamboni P, Restifo NP, Zanovello P. 2000. Identification of a CD11b(+)/Gr-1(+)/CD31(+) myeloid progenitor capable of activating or suppressing CD8(+) T cells. Blood 96:3838–3846. [PMC free article] [PubMed] [Google Scholar]
- 35.Noel JG, Guo X, Wells-Byrum D, Schwemberger S, Caldwell CC, Ogle CK. 2005. Effect of thermal injury on splenic myelopoiesis. Shock 23:115–122. doi: 10.1097/01.shk.0000154239.00887.18. [DOI] [PubMed] [Google Scholar]
- 36.Mathias B, Delmas AL, Ozrazgat-Baslanti T, Vanzant EL, Szpila BE, Mohr AM, Moore FA, Brakenridge SC, Brumback BA, Moldawer LL, Efron PA; and the Sepsis, Critical Illness Research Center Investigators. 9 May 2016. Human myeloid-derived suppressor cells are associated with chronic immune suppression after severe sepsis/septic shock. Ann Surg doi: 10.1097/SLA.0000000000001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutchins NA, Unsinger J, Hotchkiss RS, Ayala A. 2014. The new normal: immunomodulatory agents against sepsis immune suppression. Trends Mol Med 20:224–233. doi: 10.1016/j.molmed.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vuk-Pavlović S, Bulur PA, Lin Y, Qin R, Szumlanski CL, Zhao X, Dietz AB. 2010. Immunosuppressive CD14+HLA-DRlow/− monocytes in prostate cancer. Prostate 70:443–455. doi: 10.1002/pros.21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janols H, Bergenfelz C, Allaoui R, Larsson AM, Ryden L, Bjornsson S, Janciauskiene S, Wullt M, Bredberg A, Leandersson K. 2014. A high frequency of MDSCs in sepsis patients, with the granulocytic subtype dominating in gram-positive cases. J Leukoc Biol 96:685–693. doi: 10.1189/jlb.5HI0214-074R. [DOI] [PubMed] [Google Scholar]
- 40.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 8:265–277. doi: 10.1023/A:1008942828960. [DOI] [PubMed] [Google Scholar]
- 41.Mazuski JE, Sawyer RG, Nathens AB, DiPiro JT, Schein M, Kudsk KA, Yowler C; Therapeutic Agents Committee of the Surgical Infections Society. 2002. The Surgical Infection Society guidelines on antimicrobial therapy for intra-abdominal infections: an executive summary. Surg Infect (Larchmt) 3:161–173. doi: 10.1089/109629602761624171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.