Abstract
Background
Thousands of flowering plant species attract pollinators without offering rewards, but the evolution of this deceit is poorly understood. Rewardless flowers of the orchid Erycina pusilla have an enlarged median sepal and incised median petal (‘lip’) to attract oil-collecting bees. These bees also forage on similar looking but rewarding Malpighiaceae flowers that have five unequally sized petals and gland-carrying sepals. The lip of E. pusilla has a ‘callus’ that, together with winged ‘stelidia’, mimics these glands. Different hypotheses exist about the evolutionary origin of the median sepal, callus and stelidia of orchid flowers.
Results
The evolutionary origin of these organs was investigated using a combination of morphological, molecular and phylogenetic techniques to a developmental series of floral buds of E. pusilla. The vascular bundle of the median sepal indicates it is a first whorl organ but its convex epidermal cells reflect convergence of petaloid features. Expression of AGL6 EpMADS4 and APETALA3 EpMADS14 is low in the median sepal, possibly correlating with its petaloid appearance. A vascular bundle indicating second whorl derivation leads to the lip. AGL6 EpMADS5 and APETALA3 EpMADS13 are most highly expressed in lip and callus, consistent with current models for lip identity. Six vascular bundles, indicating a stamen-derived origin, lead to the callus, stelidia and stamen. AGAMOUS is not expressed in the callus, consistent with its sterilization. Out of three copies of AGAMOUS and four copies of SEPALLATA, EpMADS22 and EpMADS6 are most highly expressed in the stamen. Another copy of AGAMOUS, EpMADS20, and the single copy of SEEDSTICK, EpMADS23, are most highly expressed in the stelidia, suggesting EpMADS22 may be required for fertile stamens.
Conclusions
The median sepal, callus and stelidia of E. pusilla appear to be derived from a sepal, a stamen that gained petal identity, and stamens, respectively. Duplications, diversifying selection and changes in spatial expression of different MADS-box genes shaped these organs, enabling the rewardless flowers of E. pusilla to mimic an unrelated rewarding flower for pollinator attraction. These genetic changes are not incorporated in current models and urge for a rethinking of the evolution of deceptive flowers.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-017-0938-7) contains supplementary material, which is available to authorized users.
Keywords: Deceptive pollination, Floral development, MADS-box genes, Mimicry, Vascular bundles
Background
Flowering plants interact with a wide range of other organisms including pollinators. Pollinators can either receive nectar, oil, pollen or shelter in return for pollen transfer in a rewarding relationship, or nothing at all in a deceptive relationship [1]. One of the deceptive strategies is mimicry, defined as the close resemblance of one living organism, ‘the mimic’, to another, ‘the model’, leading to misidentification by a third organism, ‘the operator’. Essential for mimicry is the production of a false signal (visual, olfactory and/or tactile) that is used to mislead the operator, resulting in a gain in fitness of the mimic [1]. Mimicry in plants generally serves the purpose of attraction of pollinators to facilitate fertilization. In these cases, an unrewarding plant species mimics traits typical for co-flowering models, such as a specific floral shape, coloration, and presence of nectar guides, glands, trichomes or spurs. In this way, pollinators, that are unable to distinguish the two types of flowers from each other, are fooled [1, 2]. Despite the fact that deceptive pollination evolved in thousands of plant species, most notably orchids [3], the mechanisms by which this deceit evolved are still poorly understood.
Flowers are the main attractors of the majority of angiosperms to gain attention of pollinators. The outer first whorl of a flower is usually made up of sepals that generally serve as protection covering the other floral parts until anthesis. The outer second whorl consists of often-showy petals mainly involved in pollinator attraction. The sepals and petals together enfold the male and female reproductive organs in the inner floral whorls. Over the past decades, evolutionary developmental (evo-devo) studies have yielded many new insights in the role of duplication and neo-functionalization of developmental genes in floral diversification and the evolution of sepals, petals and male and female reproductive organs. These studies helped redefine the evolutionary origin of such organs [4].
Theoretically, an orchid flower can be considered to consist of five whorls of floral organs. Three sepals and three petals are present in the outer two whorls. Three external and three internal stamens and three carpels are present in the three inner whorls (Fig. 5a). Studies of the genetic plant model species Arabidopsis thaliana have shown that genes only associated with petals in A. thaliana are also expressed in the first floral whorl of petaloid monocots including orchids. Expression of these genes in the first whorl of petaloid monocots plays an important role in the similarity of sepals and petals in lilies, gingers and orchids [5–7]. From an evolutionary perspective, retention of expression of genes associated with petals in the outer floral whorl is considered an ancestral character for angiosperms [8]. In orchid flowers, the median petal, or ‘lip’, is often enlarged and ornamented with a wart-like structure, or ‘callus’. The lip mostly functions as main attractor and landing platform for pollinators. Many hypotheses have been put forward about the evolutionary origin of the lip and its ornaments [9]. Hsu et al. [10] showed that the lip is homologous with true petals but gained an additional function possibly due to the duplication of a complex of modified developmental genes that gained novel expression domains.
Fig. 5.
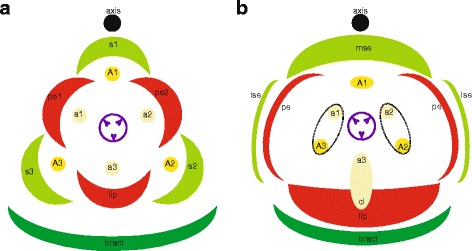
Floral diagrams. a A typical monocot flower. b A resupinate flower of E. pusilla. Abbreviations: s1–3 = sepals; p1–3 = petals; A1–3 = anther in outer floral whorl; a1–3 = anther in inner floral whorl; lse = lateral sepal; mse = median sepal; pe = petal; cl = callus. Color codes: black interrupted = stelidia and callus on lip; purple = gynoecium [Illustrations by Erik-Jan Bosch]
A stamen usually consists of a filament and an anther where the pollen are produced. Many lineages in plant families such as buttercups, orchids, penstemons and witch-hazels, not only have fertile stamens but also rudimentary, sterile or abortive stamen-like structures. These structures are generally called staminodes and are often positioned between the fertile stamens and carpels, although they can also occur in other positions [11]. Multiple hypotheses exist about the function of the morphologically very diverse staminodes. In Aquilegia, staminodes play a role in protecting the early developing fruits as they usually remain present after pollination long after the other organs have abscised [12]. In other plant genera, staminodes are assumed to mediate pollination. Comparative gene expression and silencing studies showed that staminode identity in Aquilegia evolved from a pre-existing stamen identity program. Of the genes involved, one lineage duplicated and one paralog became primarily expressed in the staminodia [11, 12].
Characteristic for orchids is that the male and female reproductive organs are incorporated in a so-called ‘gynostemium’. This structure is thought to result from a fusion of a maximum of six fertile to (partly) sterile stamens and parts of the pistil, in particular the style and stigma. It is a complex organ and the evolutionary origin of its different parts is not yet clear [9, 13, 14]. During the evolution of the orchids over the past 100 million years a reduction in the number of fertile stamens and fusion with the carpels occurred [15–17]. Six fertile stamens, positioned in floral whorls three and four, are commonly present in the closest relatives of the orchids in Asparagales. In the Apostasioideae, the earliest diverging of the five subfamilies of orchids, the number of fertile stamens is reduced to three in the genus Neuwiedia, one in floral whorl three and two in whorl four. In the genus Apostasia, a staminode develops in floral whorl three or nothing resulting in two fertile stamens [18]. In subfamily Cypripedioideae only two fertile stamens are present. A further reduction into a single fertile stamen in floral whorl three evolved in subfamilies Vanilloideae, Orchidoideae and Epidendroideae [13]. Since the two subfamilies with either three or two fertile stamens are the least diverse, reduction to a single fertile stamen may have contributed to species diversification. The sterile stamens have evolved into many other structures. In the majority of the Epidendroid orchids with a single fertile stamen, the mature gynostemium evolved appendages projecting to the front or side, clearly differentiating from broadened or flattened tissue at the base, that help pollinators to position themselves in the correct way to remove or deposit pollinia, which ensures pollination. The shapes of these appendages differ greatly and different terms are used to describe them, e.g. column wings or ‘stelidia’ [19–21]. The oldest hypothesis postulates that the stelidia are remnants of male reproductive tissue [22, 23] and following this hypothesis, stelidia are interpreted as vestiges of the lateral stamens of the third and fourth floral whorls [24].
Current models explaining floral organ development
The genetic basis of floral organ formation can be explained with various genetic models of MADS-box transcription factors. The core eudicot ‘ABCDE model’ included the A-class gene APETALA1 (AP1), B-class genes APETALA3 (AP3) and PISTILLATA (PI), C-class gene AGAMOUS (AG), D-class gene SEEDSTICK (STK) and E-class gene SEPALLATA (SEP). This model has been revised for the monocots to reflect two key differences: (i) there are no AP1 orthologs outside the core eudicots so FRUITFULL (FUL)-like genes are the closest homologs, and (ii) many monocots have entirely petaloid perianths. Class A + B + E genes specify petaloid sepals, A + B + E control petals, B + C + E determine stamens, C + E specify carpels, and D + E are necessary for ovule development [25–27] (Fig. 1a). As in the core eudicots, these genetic combinations are thought to function as protein complexes, as proposed by Theissen and Saedler [27] in the now well accepted ‘floral quartet model’ (Fig. 1b). For the highly specialized flowers of most orchid lineages, further elaborations have been proposed, including the ‘orchid code’ [28, 29], ‘Homeotic Orchid Tepal’ (HOT) model [30] and ‘Perianth code’ (P-code) [10].
Fig. 1.
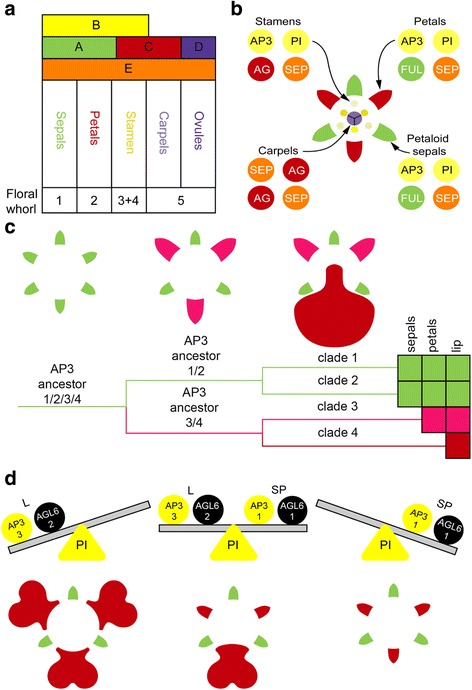
Current models explaining floral organ development. a ABCDE model of floral development in petaloid monocots. b Floral quartet model. c Orchid code and HOT model. d Perianth code model [Illustrations by Bas Blankevoort]
The orchid code and HOT model (Fig. 1c) postulate that the four AP3 lineages in orchids have experienced sub- and neo-functionalization to give rise to distinct petal and lip identity programs. In addition to original MADS-box genes incorporated in the ABCDE model, several AGAMOUS-LIKE-6 (AGL6) gene copies were recently found to play an important role in orchid flower formation. According to the P-code model (Fig. 1d), there are two MADS-box protein complexes active in orchid flowers, one consisting of a set of AP3/AGL6/PI copies, specific for sepal/petal formation, and one consisting of another set of AP3/AGL6/PI copies, specific for the formation of the lip. When the ratio of these two complexes is skewed towards the latter, the lip is large. When the ratio is skewed towards the former, intermediate lip-structures are formed [10]. The P-code model has been functionally validated for wild-type Oncidium and Phalaenopsis, and also for Oncidium peloric mutants, in which the two petals are lip-like. The P-code model was also validated in orchids from other subfamilies than the Epidendroideae, to which Oncidium and Phalaenopsis belong, i.e. Cypripedioideae, Orchidoideae and Vanilloideae, and used to detect gene expression profiles in species with intermediate lip formation [10].
Erycina pusilla as an emergent orchid model: current resources and terminology
MADS-box genes have now been identified for several commercially important orchid genera (e.g. Cymbidium, Dendrobium, Oncidium and Phalaenopsis) [30–32] but long life cycles, large chromosome numbers and complex genomes of these genera hamper functional studies. DNA-mediated transformation can be used to study the function of orchid genes and E. pusilla, with its relatively short life cycle, functions as an emergent orchid model species for such studies [33, 34].
Erycina pusilla belongs to the Oncidiinae, which is a highly diverse subtribe of meso- and south-American epiphytic orchids in subfamily Epidendroideae [35]. It is a rapidly growing orchid species with a low chromosome number (n = 6) and a, for orchids, relatively small sized diploid genome of 1.475 Gb [36, 37]. It can be grown from seed to flowering stage in less than a year [33, 34] and plantlets can be grown without mycorrhizae in test tubes. Flowers develop in a few days in which five distinct floral developmental stages can be observed (Fig. 2a). The species produces deceptive flowers that are self-compatible but incapable of spontaneous self-pollination.
Fig. 2.
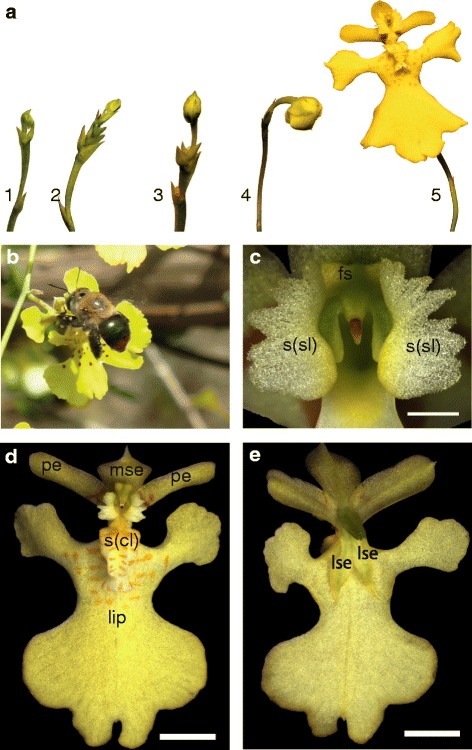
General overview of E. pusilla flowers, pollinator and floral parts. a Five floral stages of E. pusilla [Photo by Rogier van Vugt]. b A female Centris poecila bee pollinating a flower of Tolumnia guibertiana, a close relative of E. pusilla, in Cuba [Photo by Angel Vale], showing the function of the stelidia and callus in freshly opened flowers of these orchids, i.e. attraction and providing a holdfast for the pollinator. c Frontal view of fully developed stelidia. d Adaxial side (with respect to the floral axis) of a flower. e Abaxial side (with respect to the floral axis). Abbreviations: s(cl) = callus; lse = lateral sepal; mse = median sepal; pe = petal; s(sl) = stelidium; fs = fertile stamen
Oil-collecting Centris bees are the main pollinators [38]. The lateral sepals of E. pusilla are small and green. The median sepal is larger and more colorful than the lateral sepals. The lip is the largest part of the flower and very different in shape compared to the lateral petals and sepals. On the basal part of the lip or ‘hypochile’, a callus is present that guides pollinators towards the stamen and stigma to either remove or deposit pollinia effectively. The gynostemium is enveloped on both sides by two large, wing-shaped structures that we further refer to as stelidia. During floral visits, Centris bees cling to these stelidia and the callus with their forelegs while searching for oils (Fig. 2b). In E. pusilla however, these bees are fooled because the flowers employ food deception by Batesian mimicry by resembling flowers of rewarding species of the unrelated Malpighiaceae [38–40]. Flowers of this family have five clawed petals that are often unequal in size. The sepals carry oil glands. It is generally assumed that the enlarged median sepal, incised lip, callus and stelidia of Oncidiinae evolved to mimick the shape of the petals and oil glands of rewarding flowers of Malpighiaceae (Figs. 2b–d and 3) in order to attract oil-collecting bees for pollination [35, 38, 40, 41].
Fig. 3.
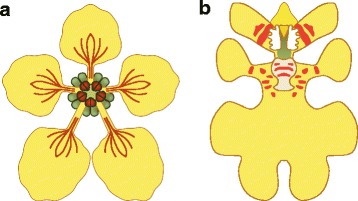
Graphical representation of a flower belonging to (a) Malpigiaceae and (b) Oncidiinae [Illustrations by Bas Blankevoort]
Agrobacterium-mediated genetic transformation was recently developed for E. pusilla [33] and knockdown of genes is currently being optimized. It is expected that the entire genome will have been analyzed using a combination of next-generation sequencing techniques within the following years. Furthermore, transcriptome data of E. pusilla are included in the Orchidstra database [31]. Twenty-eight MADS-box genes from E. pusilla have been identified thus far including the most important floral developmental ones [34]. These resources make E. pusilla an ideal orchid model for evo-devo studies. Lin et al. [34] published expression data of MADS-box genes isolated from sepals, petals, lip, column and ovary of flowers of E. pusilla after anthesis together with a basic phenetic gene lineage analysis.
In this study, we employed a combination of micro-, macromorphological, molecular and phylogenetic techniques to assess the evolutionary origin of the median sepal, callus and stelidia of the flowers of E. pusilla. To accomplish this goal, we investigated early and late floral developmental stages with scanning electron microscopy (SEM), light microscopy (LM), 3D-Xray microscopy (micro-CT) and expression (RT-qPCR) of MADS-box genes belonging to six different lineages. In addition, we investigated gene duplication and putative neo-functionalization as indicated by inferred episodes of diversifying selection. Our aim was to test the hypotheses that the median sepal, callus and stelidia are derived from sepals, petals and stamens, respectively, to unravel the genetic basis of the evolution of deceptive flowers.
Methods
Plant material and growth conditions
A more than 15 year old inbred line of E. pusilla originally collected in Surinam was grown in climate rooms under controlled conditions (7.00–23.00 h light regime), at a temperature of 20 °C and a relative humidity of 50%. The orchids were cultured in vitro under sterile conditions on Phytamax orchid medium with charcoal and banana powder (Sigma-Aldrich) mixed with 4 g/L Gelrite™ (Duchefa) culture medium. Pollinia of flowers from different plants were placed on each other’s stigma after which ovaries developed into fruits. After 18–22 weeks, seeds were ripe and sown into containers with sterile fresh nutrient culture medium. The seeds developed into a new E. pusilla flowering plant within 20 weeks.
Fixation for micromorphology
Flowers and flower buds were fixed with standard formalin-aceto-alcohol (FAA: absolute ethanol, 90%; glacial acetic acid, 5%, formalin; 5% acetic acid) for one hour under vacuum pressure at room temperature and for 16 h at 4 °C on a rotating platform. They were washed once and stored in 70% ethanol until further use.
SEM
Floral buds at different developmental stages were dissected in 70% ethanol under a Wild M3 stereo microscope (Leica Microsystems AG, Wetzlar, Germany) equipped with a cold-light source (Schott KL1500; Schott-Fostec LLC, Auburn, New York, USA). Subsequently, the material was washed with 70% ethanol and then placed in a mixture (1:1) of 70% ethanol and DMM (dimethoxymethane) for five minutes for dehydration. The material was then transferred to 100% DMM for 20 min and critical point dried using liquid CO2 with a Leica EM CPD300 critical point dryer (Leica Microsystems, Wetzlar Germany). The dried samples were mounted on aluminium stubs using Leit-C carbon cement or double-sided carbon tape and coated with Platina-Palladium with a Quorum Q150TS sputtercoater (Quorum Technologies, Laughton, East Sussex, UK). Images were obtained with a JEOL JSM-7600 F Field Emission Scanning Electron Microscope (JEOL Ltd., Tokyo, Japan).
For the images presented in Fig. 4, fixed floral buds were critical point dried using liquid CO2 with a CPD 030 critical point dryer (BAL-TEC AG, Balzers, Lichtenstein) and coated with gold with a SPI-ModuleTM Sputter Coater (SPI Supplies, West-Chester, Pennsylvania, USA). Scanning electron microscope (SEM) images were obtained with a Jeol JSM-6360 (JEOL Ltd., Tokyo) at the Laboratory of Plant Conservation and Population Biology (KU Leuven, Belgium).
Fig. 4.
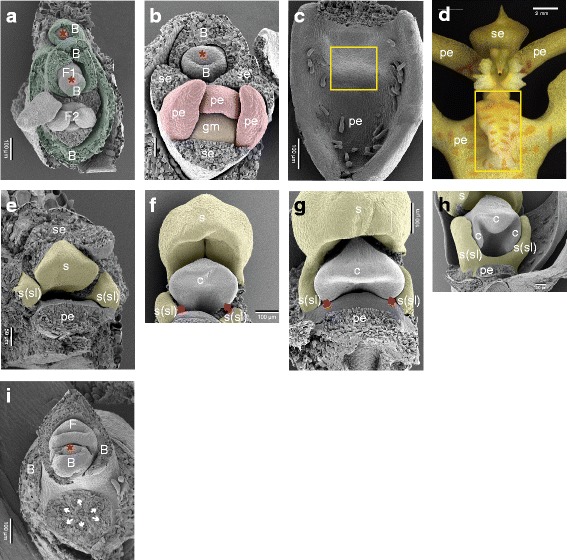
Developing inflorescence of E. pusilla. a Apical view of a young developing inflorescence. A central meristem is present and below it two flowers are visible, each subtended by a bract. The distal flower (F1) is primordial and the next flower (F2) is somewhat more developed. b Apical view of a developing flower in an early developmental stage. The scars of the three removed sepals are visible, two are adaxially (lateral sepals) and one is abaxially (median sepal) situated. More central in the flower, two abaxial-lateral petals and one adaxial developing petal (lip) are present. Most central in the flower is the primordium of the gynostemium. c–d Developing adaxial petal (lip) with callus (boxed). e–h Successive stages of the development of the gynostemium with the developing fertile stamen central and stelidia laterally. In (e), the scar of the removed abaxial sepal is visible. Below the fertile stamen, the scar of the adaxial petal (lip) can be seen. In between the fertile stamen and the adaxial petal (lip), the stigmatic cavity is present. In (f and g), the two adaxial (lateral) carpels are visible (arrowed). In (h), the abaxial carpel is incorporated in the stigmatic cavity. i Apical view of an inflorescence axis with a removed developing flower. In the upper half of the micrograph, the apex of the axis is visible as well as a flower at very early developmental stage, subtended by a bract. In the lower half, in the scar of the removed developing flower, six vascular bundles are visible (arrowed). Abbreviations: Red asterisk = apical meristem; B = bract; F = flower (primordium); c = carpel; gm = gynostemium; pe = petal; se = sepal; s = fertile stamen; s(sl) = stelidium. Color codes: dark green = bract; red = petals; orange = gynostemium; yellow = androecium
3D-Xray microscopy
Fully grown flowers were infiltrated with 1% phosphotungstic acid (PTA) in 70% ethanol for 7 days in order to increase the contrast [42]. The PTA solution was changed every 1–2 days. The flowers were embedded in 1% low melting point agarose (Promega) prior to scanning. The scans were performed on a Zeiss Xradia 510 Versa 3D X-ray with a Sealed transmission 30–160 kV, max 10 W x-ray sources. Scanning was performed using the following settings: acceleration voltage/power 40 kV/3 W; source current 75 μA; exposure time 2 s; picture per sample 3201; camera binning 2; optical magnification 4 ×, with a pixel size of 3.5 μm. The total exposure time was approximately 3, 2 h. 3D images were stacked and processed with Avizo 3D software version 8.1.
RNA extraction
For organ dissection, floral buds of E. pusilla were collected from floral stages 2 and 4 (Fig. 2a). The earliest floral stage to dissect the different flower parts was at floral stage 2. The lateral sepals, median sepal, petals, lip, callus, stamen and the remaining part of the gynostemium with stelidia but excluding the ovary were dissected (Fig. 2c–e) and collected in individual tubes and immediately frozen on dry ice and stored at –80 °C until RNA extraction. Total RNA was extracted from seven different floral organs of E. pusilla using the RNeasy Plant Mini Kit (QIAGEN), following the manufacturer’s protocol. A maximum of 100 mg plant material was placed in a 2.2 ml micro centrifuge tube with 7 mm glass bead. The TissueLyser II (QIAGEN) was used to grind the plant material. The amount of RNA was measured using the NanoVue Plus™ (GE Healthcare Life Sciences) and its integrity was assessed on an Agilent 2100 Bioanalyzer using the Plant RNA nano protocol. RNA samples with an RNA Integrity Number (RIN) < 7 were discarded. RNA was stored at −80 °C until further use. Extracted RNA was treated with DNase I, Amp Grade (Invitrogen 1U/μl) to digest single- and double-stranded DNA following the manufacturer’s protocol.
cDNA synthesis
cDNA was synthesized with up to 1 μg of DNase-treated RNA using iScript™ cDNA Synthesis Kit (Bio-Rad Laboratories) following the manufacturer’s protocol. A reaction mixture was prepared by addition of 1 μg of RNA, 4 μl 5x iScript reaction mix, 1 μl iScript reverse transcriptase to nuclease-free water up to a total volume of 20 μl. The reaction mixture was incubated at 25 °C for 5 min, 42 °C for 30 min and 85 °C for 5 min using a C1000 Touch™ thermal cycler machine (Bio-Rad). During this reaction, a positive control (CTRL) and no reverse transcriptase (NRT) control were included.
Primer design
DNA sequences were downloaded from NCBI Genbank and Orchidstra (http://orchidstra2.abrc.sinica.edu.tw). For the MADS-box genes primers were designed on the C-terminal of the DNA sequences to avoid cross –amplification. Beacon Designer™ (Premier Biosoft, http://www.oligoarchitect.com) software was used to design primers (Additional file 1: Tables S1–S2). All primer pairs were screened for their specificity against the Orchidstra database and in a gradient PCR reaction. The reaction mixture (25 μl) contained: 2.5 ng cDNA, 0.2 μM of each primer, 0.1 mM dNTP’s and 0.6 U Taq DNA polymerase (QIAGEN) in 1x Coral Load Buffer (QIAGEN). The amplification protocol was as follows: initial denaturation step of 5 min 94 °C followed by 40 cycles of [20 s 94 °C, 20 s <55–65 > °C, 20 s 72 °C], one final amplification step of 7 min 72 °C and ∞ 15 °C. Based on the results of the gradient PCR, the annealing temperature was set to 61.3 °C for the Quantitative Real-time PCR as this value gave the best results. Only when a specific product was detected was the primer pair used for subsequent quantification.
Reference genes and quantitative real-time PCR
Experimental and computational analyses with LinRegPCR (http://www.hartfaalcentrum.nl, v2015.1) [43, 44] indicate that E. pusilla Ubiquitin-2, Actin, and F-box were stably expressed in the tissues of interest and these genes were chosen as reference genes for the expression assay. Expression of all MADS-box genes was normalized to the geometric mean of these three reference genes.
Quantitative real-time PCR was performed using the CFX384 Touch Real-Time PCR system (Bio-Rad Laboratories). The assays were performed using the iQ™ SYBR® Green Supermix (Bio-Rad Laboratories). The reaction mixture (7 μl) contained: 1x iQ™ SYBR® Green Supermix, 0.2 μM of each primer, 1 ng cDNA template from a specific floral organ (biological triplicate reactions) for each target gene and floral organ for two sets of isolated RNA (six reactions in total). All reactions were performed in Hard-Shell® Thin-Wall 384-Well Skirted PCR Plates (Bio-Rad Laboratories). For each amplicon group, a positive control was included (=CTRL, flower buds from floral stage 1 to 4), a negative control (=NTC, reaction mixture without cDNA) and a no reverse transcriptase treated sample (=NRT, control sample during the cDNA synthesis). For all the qPCR reactions, the amplification protocol was as follows: initial denaturation of 5 min 95 °C followed by; 20 s 95 °C; 30 s 61.3 °C; 30 s 72 °C; plate read, for 50 cycles; then followed by a melting curve analysis of 5 s, 65 °C to 95 °C with steps of 0.2 °C to confirm single amplified products (Additional file 2: Figure S2).
Normalization, data analysis and statistical analysis
The non-baseline corrected data were exported from the Bio-Rad CFX Manager™ (v3.1) to a spreadsheet. Quantification Amplification results (QAR) were used for analysis with LinRegPCR (v2015.1, dr. J.M. Ruijter). The calculated N0-values represented the starting concentration of a sample in fluorescence units. Removal of between-run variation in the multi-plate qPCR experiments was done using Factor qPCR© (v2015.0) [45, 46]. Geometric means of the corrected N0-values were calculated from the six samples together, i.e. two biological and three technical replicates. GraphPad Prism version 7.00 (http://www.graphpad.com) was used to perform a Two-Way ANOVA with Sidak’s multiple comparison test to calculate significant differences between the two floral stages 2 and 4, and graphed with Standard Error of Measurement (SEM) error bars. Tukey’s multiple comparisons test was used to compare the means between the floral organs. Variation for the two biological replicates was assessed by tests in triplicate.
Phylogenetic analyses
Nucleotide sequences of floral developmental genes were downloaded from NCBI GenBank® (Additional file 1: Table S1) and separate data sets were constructed for MADS-box gene classes FUL-, AP3-, PI-, AG-, STK-, SEP- and AGL6-like. For each gene class, protein-guided codon alignments were constructed by first performing multiple sequence alignments of the protein translations using MAFFT v.7.245 (with the algorithm most suited for proteins with multiple conserved domains, E-INS-I or “oldgenafpair” for backward compatibility), with a maximum of 1000 iterations [47] and then reconciling the nucleotide sequences with their aligned protein translations.
Gene trees were inferred from the codon alignments using PhyML v3.0_360-500M [48] under a GTR + G + I model with six rate classes and with base frequencies, proportion of invariant sites, and γ-shape parameter α estimated using maximum likelihood. Optimal topologies were selected from results obtained by traversing tree space with both nearest neighbor interchange (NNI) and subtree prune and regraft (SPR) branch swap algorithms, ie. PhyML’s “BEST” option. Support values for nodes were computed using approximate likelihood ratio tests (SH-like aLRT, [49]).
To infer where on the gene trees duplications may have occurred the GSDI algorithm [50] was used as implemented in forester V1.038 (https://sites.google.com/site/cmzmasek/home/software/forester). Fully resolved species trees for GSDI testing were constructed based on the current understanding of the phylogeny of the species under study (Additional file 3: Figure S4).
Lastly, to detect lineage-specific excesses of non-synonymous substitutions, BranchSiteREL [51] analyses were performed as implemented in HyPhy [52] on the Datamonkey (http://datamonkey.org) cluster.
Results
Ontogeny, macro- and micromorphology of flowers of E. pusilla
Floral ontogeny in E. pusilla can be divided into two main phases: early and late. Early ontogeny starts from floral initiation (floral stage 1) up to the three-carpel-apex stage (floral stage 2) and late ontogeny starts from the three-carpel-apex stage (floral stage 2) until anthesis (floral stages 3, 4 and 5, Fig. 2a) [53].
The inflorescence of E. pusilla is branched and multiple flowers develop in succession (Fig. 2a). Up to floral stage 1, the perianth is formed following a classic monocot developmental pattern (Fig. 5a) [54] in which the sepals are among the first organs to become visible, followed by the petals. The position of the two abaxial petals is slightly shifted laterally (Fig. 4a). Stamen and carpel primordial are not visible in the course of the early phase, but instead a single massive primordium is present from which the gynostemium will develop (Fig. 4b).
On the hypochile of the lip a callus is formed from floral stage 2 onwards (Fig. 4c–d). The fertile stamen differentiates after floral stage 1. The stelidia appear at each side of the gynostemium (Fig. 4e–h) from where they elongate and start forming wing-like appendices (Fig. 2e). The abaxial carpel is incorporated in the stigmatic cavity, which forms a compound structure with the fertile stamen (Fig. 4h). The three-carpel-apex stage is clearly visible in floral stage 2. At this stage the six staminal vascular bundles can also be observed just above the inferior ovary (Fig. 4i). In floral stage 3, no new organs are formed, but in floral stage 4 (Fig. 2a) the mature flower becomes resupinate (Fig. 5b). The terms adaxial and abaxial are used here to indicate the position of the distinct floral parts with respect to the inflorescence axis (Fig. 4a–b), thereby taking the position of the primordia of the floral organs as a reference. For example, with respect to the inflorescence axis, the lip is the adaxial petal, which by resupination becomes the lowermost part of the flower.
Using micro-CT scanning, vascular bundles were observed in a fully-grown floral stage 5 flower (Fig. 6a–f and Additional file 4: Movie S1). In the inferior ovary six vascular bundles could be discerned, indicated in purple. Three of these vascular bundles, indicated in green, run to the adaxial (median) sepal and abaxial (lateral) sepals, respectively. Three main groups of vascular bundles, indicated in red, run towards the petals including the lip, where they split up. Four vascular bundles (indicated in yellow) are present; one bundle, already split into two at the base, runs to the fertile stamen, where it splits up further towards the two pollinia (Fig. 6a–e); two vascular bundles, originated from two pairs, run up into the stelidia (Fig. 6b–c; e–f) and one vascular bundle runs all the way up into the callus of the lip (Fig. 6b; e–f). When following the yellow vascular bundles downwards, they connect in a plexus situated on top of the inferior ovary with the rest of the vascular system of the flower.
Fig. 6.
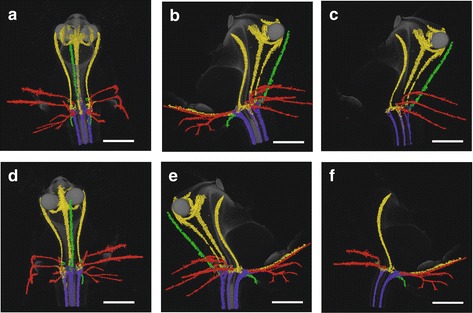
Vascular bundle patterns of E. pusilla. a Frontal view of a 3D X-ray macroscopical reconstruction of the vascular bundle patterns in a mature flower of E. pusilla. b Successive clockwise turn of 45°. c Simplified version of (b). d Successive clockwise turn of 90°. e Successive clockwise turn of 135°. f Simplified version of (e). Color codes: green = vascular bundles in sepals; red = vascular bundles in petals; purple = vascular bundles in gynoecium; yellow = vascular bundles in androecium. Scale bar = 1 mm
Additional file 4: Movie S1. Animation of the 3D visualization as depicted in Fig. 6. (MPG 60302 kb)
Throughout late ontogeny, epidermal cells in all floral organs remained relatively undifferentiated and only expanded in size. Epidermal cells on the abaxial side of floral organs were mostly similar to the cells on the adaxial side, but more convex shaped (Additional file 5: Figure S1). Epidermal cells of the lateral sepals were irregular, flattened and rectangular shaped and longitudinally orientated from the base to the apex (Fig. 7a–c). Epidermal cells of the median sepal, as well as of the petals and the lip, develop from irregularly flattened shaped cells at floral stage 2, to a more convex shape in floral stage 5 (Fig. 7d–l). Epidermal cells of the callus develop from convex shaped cells in floral stage 2 to cells with a more conical shape in floral stage 5 (Fig. 7m–o). Epidermal cells of the stelidia become convex shaped during floral stage 2 and develop papillae on their apices during floral stage 5 (Fig. 7p–r).
Fig. 7.
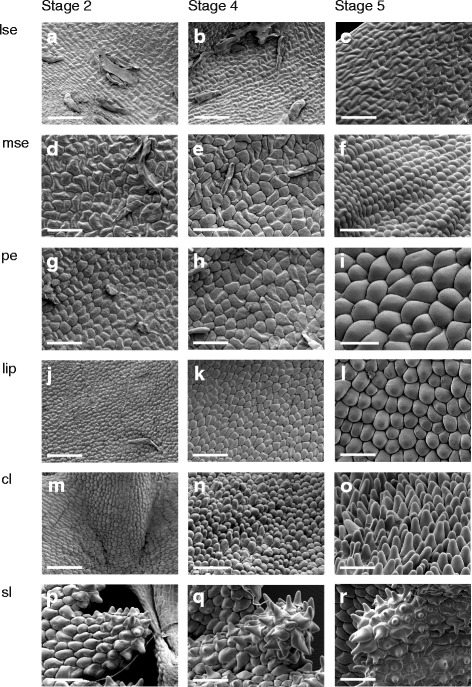
Micromorphology of the epidermal cells on the adaxial side of a flower of E. pusilla. The three columns represent, from left to right, floral stage 2, 4 and 5 of the floral organs. Epidermal cells of (a–c) lateral sepal, (d–f) median sepal, (g–i) petal, (j–l) lip, (m–o) callus on lip and (p–r) stelidia. Scale bar = 100 μm. Abbreviations: lse = lateral sepal; mse = median sepal; pe = petal; cl = callus; sl = stelidia
Duplications, diversifying evolution and expression of eighteen MADS-box genes in selected floral organs of E. pusilla in two developmental stages
FUL-, SEP- and AGL6-like genes
The closest homologs of the Arabidopsis A class gene APETALA1 in E. pusilla are the three FUL-like genes copies EpMADS10, 11 and 12. Our phylogenetic analyses reconstructed three orchid clades of FUL-like genes, containing the three copies present in the genome of E. pusilla (Additional file 6: Figure S5a), which was consistent with previous studies [55]. Diversifying selection was detected along the branch following the gene duplication leading to EpMADS10. The three FUL-like gene copies were expressed in all floral organs of E. pusilla but at low levels only (Additional file 7: Figure S3). During development, expression generally decreased in most floral organs for EpMADS10 and 11 whereas it generally increased for the majority of floral organs for EpMADS12 (Additional file 7: Figure S3 and Additional file 1: Table S3).
Four SEP-like orchid clades were retrieved (Additional file 6: Figure S5f), encompassing the four copies of E. pusilla, consistent with previous studies [55, 56]. The branch leading to the duplication that gave rise to EpMADS6 and EpMADS7 shows evidence of diversifying selection. EpMADS6, 7, 8 and 9 were expressed in all floral organs at varying levels. EpMADS6 was mainly expressed in the fertile stamen, a statistically significant difference as compared to the other six floral organs (Additional file 7: Figure S3 and Additional file 1: Table S3).
Three AGL6 orchid clades, also found by Hsu et al. [10] were retrieved, containing the three different copies present in the E. pusilla genome (Additional file 6: Figure S5g). Evidence for a moderate degree of diversifying selection could be detected on the branch leading to EpMADS4. The three different copies of AGL6-genes were not expressed in all floral organs and the level of expression also varied. EpMADS3 was most highly expressed in the sepals and petals. EpMADS4 was more highly expressed in the lateral sepals as compared with the median sepal, petals and lip. EpMADS5 was mainly expressed in the lip and callus (Fig. 8).
Fig. 8.
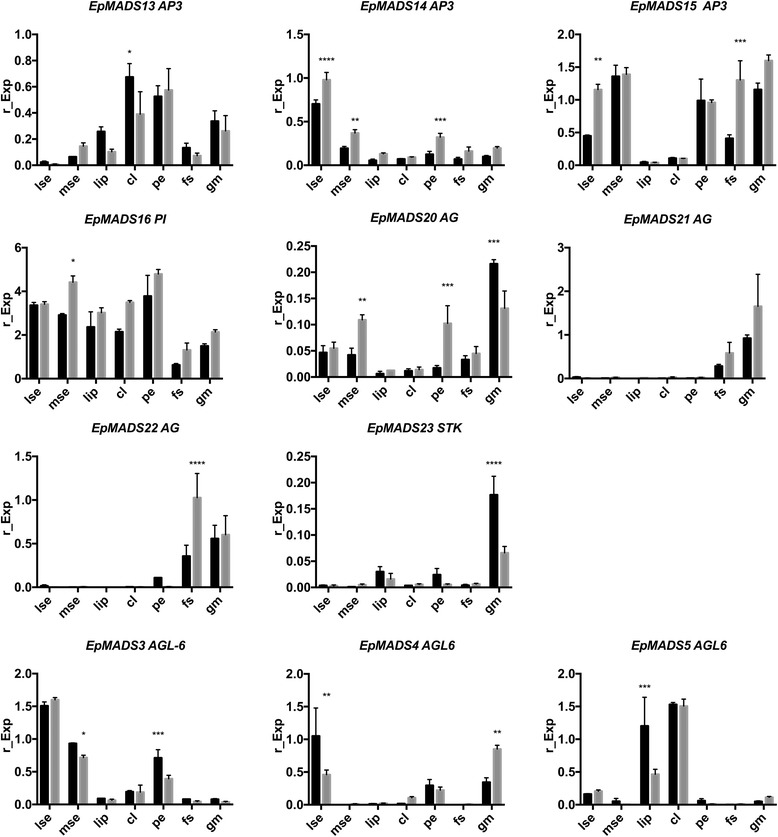
Floral organ specific expression levels of selected MADS-box gene copies in E. pusilla. AP3 (top row), PI (second row), AG (second and third row), STK (second row), ALG6 (third row). RNA was extracted from seven different floral organs during two stages of development of E. pusilla and used for cDNA synthesis. Expression of the MADS-box genes was normalized to the geometric mean of three reference genes Actin, UBI2 and Fbox. Each column shows the relative expression of 20 floral organs in two cDNA pools (10 floral organs per isolation), both tested in triplicate. Abbreviations: lse = lateral sepal; mse = median sepal; cl = callus; pe = petal; fs = fertile stamen; gm = gynostemium. Dark grey = floral stage 2 and light grey = floral stage 4. Y-axis: relative gene expression. The error bars represent the Standard Error of Mean. P-value style: GP: >0.05 (ns), <0.05 (*), <0.01 (**), <0.001 (***), <0.0001 (****)
AP3-like and PI-like genes
Initial phylogenetic analyses reconstructed the main duplication between the AP3 and PI genes also found in many other studies [10, 30, 57] so two separate gene trees were retrieved for each lineage (Additional file 6: Figure S5b–c). Four orchid AP3-clades and three PI-clades were identified in these analyses. The three copies of AP3 and a single copy of PI present in the genome of E. pusilla were placed in AP3-clades 1, 2 and 3 and PI-clade 2, respectively. No evidence for diversifying selection could be detected along the branches leading to the PI-clade containing EpMADS16 but evidence for diversifying selection along the branch in the AP3-1 clade encompassing EpMADS15 was found. AP3-like gene copy EpMADS14 was most highly expressed in the lateral sepals. AP3-like gene copy EpMADS13 was more highly expressed in the lip and callus than in the sepals and petals (Fig. 8). The PI-like gene EpMADS16 was more highly expressed in the first four floral whorls in both floral stages (Figs. 8 and 9).
Fig. 9.
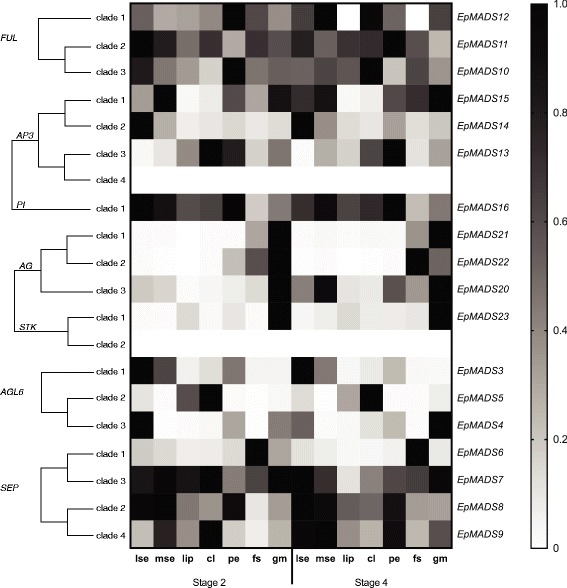
Heat map representation of MADS-box gene expression in E. pusilla. The FUL-, AP3-, PI-, AG-, STK-, SEP- and ALG6- like copies were retrieved from different gene lineage clades during two stages of floral development. Expression of the MADS-box genes was normalised to the geometric mean of three reference genes Actin, UBI2 and Fbox. The relative gene expression was normalised with the CTRL sample (= flower buds from floral stages 1-4). The scales for each gene and developmental stage are independent of each other and set to 1 for the highest value. Abbreviations: lse = lateral sepal; mse = median sepal; cl = callus; pe = petal; fs = fertile stamen; gm = gynostemium
AG- and STK-like genes
Three orchid AG-clades and two STK-clades were identified in the phylogenetic analyses (Additional file 6: Figure S5d–e). EpMADS20, 21 and 22 were placed in AG-clades 3, 1 and 2, respectively, and EpMADS23 was placed in STK-clade 1, as also found by Lin et al. [34]. No evidence for diversifying selection in the branches supporting the three orchid AG-clades and STK-clade containing copies present in the genome of E. pusilla could be detected. AG-like gene copy EpMADS20 was most highly expressed in the stelidia, whereas EpMADS22 was most highly expressed in the stamen as compared with all other floral organs analyzed (Fig. 8). No expression of AG-like genes could be detected in the callus. STK-like gene copy EpMADS23 was most highly expressed in the stelidia as compared with all other floral organs analyzed (Figs. 8 and 9).
Discussion
Homology of the median sepal of E. pusilla
The floral ontogenetic observations and vascularization patterns indicate that the median sepal is derived from the first floral whorl. In contrast, the presence of convex epidermal cells suggests a petaloid origin [58]. The AGL6 and AP3 copies EpMADS3 and EpMADS15, members of the sepal/petal-complex of the P-code model, were most highly expressed in the median sepal, lateral sepal and petal. A possible correlation between expression and petaloidy was found for AGL6 and AP3 copies EpMADS4 and EpMADS14. These two genes were lowly expressed in the median sepal, lip and petal as compared with the lateral sepal. Additional functional studies are needed to show whether loss of function of EpMADS4 and EpMADS14 is linked to sepal morphology in E. pusilla and other species that also possess a petaloid median sepal. The AGL6 gene copy EpMADS4 copy showed evidence of diversifying evolution. Lin et al. [34] identified fifteen motifs in the MIKC-type MADS-box proteins of E. pusilla. Two differences can be noticed within the K-region and C-terminal-region of AP3 and AGL6 genes of E. pusilla: (i) AP3 EpMADS14 is missing motif 11, while the other B-class genes all contain motif 11. AGL6 EpMADS4 also contains motif 11, while the other AGL6 gene copies lack this motif; (ii) AGL6 EpMADS4 is missing motif 6 whereas all the other AGL6 gene copies contain motif 6. The differences found may contribute to the morphological differences between the median and lateral sepals of E. pusilla.
Homology of the lip and callus of E. pusilla
The convex shaped epidermal cells on the lip and conical shaped epidermal cells on the callus are indicative of a petaloid function [58]. The FUL-like gene copy EpMADS12, AP3-like EpMADS13 and AGL6-like EpMADS5 are most highly expressed in lip and callus, further confirming a lip identity based on the ABCDE, floral quartet and P-code models, that dictate joint expression of A, B, E and AGL6-like genes in the petals and lip, respectively. According to these models, B, C and E class genes should be expressed in stamens but no evidence of expression of C class genes was found in the lip or callus. Notwithstanding, the possible staminal origin of the callus is supported by multiple lines of evidence. First of all, the ontogeny and function of the lip of E. pusilla are very different as compared with the ontogeny and function of the callus. The lip is formed from floral stage 1 onwards, mainly acts as a long distance attraction and functions as a soft landing platform for pollinating bees. The callus is formed from floral stage 2 onwards and functions as short distance attraction by offering a sturdy holdfast to pollinators. This is in line with Carlquist [59], who states that different vascularization patterns are driven by different functional needs. Many Oncidiinae have a callus on the lip and in some of these species, the callus produces oil, making the functions of the lip and the callus even more distinct. Flowers with an oil-producing callus evolved twice in unrelated clades from species with non-rewarding flowers according to the molecular phylogeny of the Oncidiinae as presented in Pridgeon et al. [38]. One of the two rewarding clades, i.e. the one containing the genus Gomesa, is the sister group of the Erycina clade, showing that changes between an oil-producing and a non-rewarding callus occur quite easily in this group of orchids. This suggests that evolution towards oil production is correlated with increased venation as also stated by Carlquist [59]. We argue, however, that the venation in the callus is not only driven by functional needs but that the venation pattern is also informative regarding the evolutionary origin of the callus, as the callus of E. pusilla is connected with only one of the six original staminal bundles, physically distinct from the two adjacent vascular bundles leading to the lip. We consider this indicative of a possible staminal origin of the callus because of the occasional appearance of an infertile staminodial structure at this particular position, the inner adaxial stamen (a3), in teratologous orchid flowers [60]. Terata of monandrous orchids with both stelidia carrying an additional anther on their tip next to the anther on the apex of the gynostemium, such as Bulbophyllum triandrum and Prostechea cochleata var. triandrum, are commonly seen as support for a staminal origin of stelidia. Similarly, mutants in Dactylorhiza with a staminodial structure on their lip [60] could be interpreted as support for a staminal origin of the callus. Alternatively, these phenotypes could be caused by ectopic C gene expression that is transforming petal into stamen tissue. Homeotic transformation is not necessarily indicative of derivation. According to Carlquist [59] data from teratology are therefore not useful for studying the evolution of flowers. This publication was written at a time that experimental mutants could not yet be made though. Ongoing work on B- and C- class homeotic mutants in the established plant models Arabidopsis, Antirrhinum and Petunia shows how much can be gained from teratology. We hope that these mutants can be created in emerging orchid models such as E. pusilla in the future to provide more evidence for the evolutionary origin of the callus on the lip.
Homology of the stamen and stelidia of E. pusilla
Five vascular bundles, indicating a stamen-derived origin, lead to the stamen and stelidia. Our observations concur with those of Swamy [24] who showed that the ovary is traversed by multiple vascular bundles in monandrous orchids. He visualized ‘compound’ bundles of staminal origin in the ovary of a species of Dendrobium and discovered vascularizing bundles in the stelidia. In several other plant families, e.g. Brassicaceae (Arabidopsis), Commelinaceae (Tradescantia), and Cyperaceae (Cyperus), it has been shown that vascular bundles of different organs originate in the developing organs and grow towards the stele rather than being branched from the stele [61–64]. Based on Fig. 6 and Additional file 4: Movie S1, we hypothesize that especially the staminal vascular bundles are connected in a similar way to the rest of the vascular system. Of the three copies of AG and four copies of SEP, EpMADS22 and EpMADS6 were found to be highest expressed in the stamen. Another copy of AG, EpMADS20, and the single copy of STK, EpMADS23, were found to be most highly expressed in the stelidia, suggesting that EpMADS23 expression may be correlated with sterility.
Implications for current floral models
The ABCDE, orchid code, HOT and P-code models do not explain the morphological difference between median and lateral sepals as present in orchid species such as E. pusilla. Our results show that a differentiation between the sepaloid lateral sepals and petaloid median sepal of E. pusilla is correlated with a significant reduction of expression of AP3-like EpMADS14 and ALG6-like EpMADS4 in all petaloid organs (Fig. 10a).
Fig. 10.
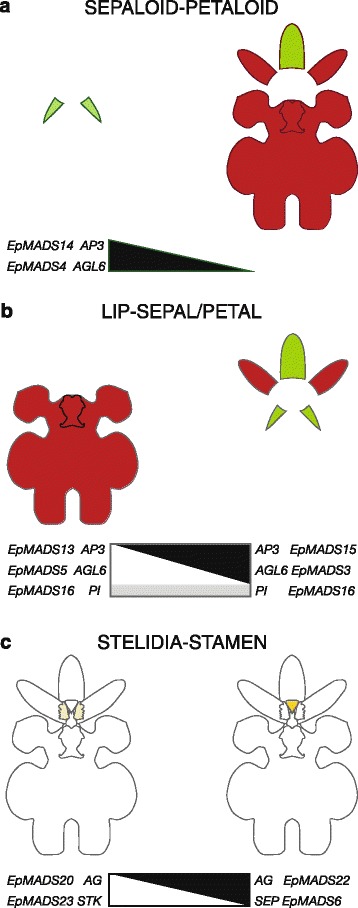
Summary of expression of MADS-box genes involved in the differentiation of selected floral organs of E. pusilla. a Expression of EpMADS4/14 (in black) correlating with a sepaloid-petaloid identity is high in the lateral sepals (left side) but low in the remainder of the perianth (right side), b Expression of the lip complex EpMADS5/13/16 (in white/grey)) correlating with a lip identity is high in in the lip and callus (left side) but low in the remainder of the perianth (right side). Expression of the sepal/petal-complex EpMADS3/15/16 (in black/grey) correlating with a sepal and petal identity is low in the lip (left side) but high in the sepals and petals (right side), c Expression of EpMADS20/23 (in white) correlating with a stelidia-stamen identity is high in the stelidia (left side) but low in the stamen (right side). Expression of EpMADS6/22 (in black) is low in the stelidia (left side) but high in the stamen (right side)
The P-code model explains the development of the lip of E. pusilla as the SP-complex (AP3-like EpMADS15/AGL6-like EpMADS3/PI-like EpMADS16) was found to be most highly expressed in the sepals and petals, whereas the L-complex (AP3-like EpMADS13/AGL6-like EpMADS5/PI-like EpMADS16) was found to be most highly expressed in the lip (Fig. 10b). However, the model does not yet account for the development of the callus and the high expression of AGL6-like EpMADS5 in this particular organ. To incorporate all new evidence found for the evolution and development of first and second floral whorl organs, we propose an Oncidiinae model (Fig. 11), summarizing the gene expression data presented in this study for E. pusilla and earlier studies carried out on Oncidium Gower Ramsey [10] [Illustrations by Bas Blankevoort].
Fig. 11.
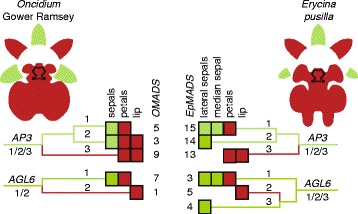
Oncidiinae model summarizing expression of MADS-box genes involved in the differentiation of the perianth of Oncidium Gower Ramsey (left) and E. pusilla (right). Clade 1 AP3-like OMADS5 and EpMADS15 and clade 1 AGL6-like genes OMADS7 and EpMADS3 are expressed in the sepals and petals of both species. Clade 2 AP3-like OMADS3 is expressed in the entire perianth of O. Gower Ramsey whereas EpMADS14 is only expressed in the lateral sepals of E. pusilla. Clade 2 AGL6-like genes OMADS1 and EpMADS5 are expressed in the lip only of both species. Clade 3 AP3-like OMADS9 and EpMADS13 are expressed in the petals and lip of both species. Clade 3 AGL6-like gene EpMADS4 is only expressed in the lateral sepals of E. pusilla [Illustrations by Bas Blankevoort]
All four MADS-box B class gene copies were found to be expressed in the fertile stamen of E. pusilla. In addition, AG-like EpMADS22 and SEP-like EpMADS6 were most highly expressed in this floral organ, confirming a stamen identity as predicted by the ABCDE model. The high expression of AG-like EpMADS20 and STK-like EpMADS23 in the stelidia cannot be explained with the ABCDE model. All current orchid floral models only describe evolution and development of the first and second whorl floral organs. We found evidence for differential gene expression in organs in the third and fourth floral whorl, i.e. the stamen and stelidia (Fig. 10c), and this argues for the development of additional models.
Conclusions
After examining vascularization, macro- and micromorpology, gene duplications, diversifying evolution and expression of different MADS-box genes in selected floral organs in two developmental stages, it can be concluded that: (i) the median sepal obtained a petal-identity, thus representing a particular character state of the character ‘sepal’, (ii) that the lip was derived from a petal but the callus from a stamen that gained petal identity, and (iii) the stelidia evolved from stamens. Duplications, diversifying selection and changes in spatial expressions of AP3 EpMADS14 and AGL6 EpMADS4 may have contributed to an increase of petaloidy of the median sepal. The same can be applied to AP3 EpMADS13 and AGL6 EpMADS5 in the lip and callus. Differential expression of AG copies EpMADS20 and EpMADS22, STK copy EpMADS23 and SEP copy EpMADS6 appear to be associated with the evolution of the stamen and stelidia, respectively.
The evolutionary origin of the median sepal, callus and stelidia of E. pusilla cannot be explained with any of the currently existing floral developmental models. Therefore, new models, like our Oncidiinae model, need to be developed to summarize MADS-box gene expression in more complex floral organs. Such models need validation by functional analyses. The genetic mechanisms discovered in this study ultimately contributed to the evolution of a deceptive orchid flower mimicking the morphologies of rewarding Malpighiaceae flowers. This mimicry enabled flowers of E. pusilla, and many other species in the highly diverse Oncidiinae, to successfully attract Centris bees for pollination, often, as is the case for E. pusilla, without offering a reward. Pollination by deceit is one of the most striking adaptations of orchids to pollinators. It is estimated that approximately a third of all orchid species employ deceit pollination, and that food mimicry is the most common type. Deceptive pollination is hypothesized to be correlated with species diversification as subtle changes in floral morphology can attract different pollinators and eventually lead to reproductive isolation. It was recently discovered that deceptive pollination augmented orchid diversity, not by accelerating speciation but by adding more species at roughly the same rate through time [17]. Ongoing research on the genomics of E. pusilla and other emergent plant models will shed more light on the role that key developmental genes played in the evolution of deceptive flowers.
Additional files
List of sequences used in the alignments and phylogenetic analyses. Table S2. Transcript primer sequences and amplicon characteristics used for quantitative real-time PCR validation of the expression profiles of eighteen MADS-box transcripts following MIQE guidelines [65]. Table S3. Difference in MADS-box gene expression between floral organs; variance analysis of measures using Tukey multicomparisons test. P-value style: GP: >0.05 (ns), <0.05 (*), <0.01 (**), <0.001 (***), <0.0001 (****). Abbreviations: lse = lateral sepal, mse = median sepal, cl = callus, pe = petal, fs = fertile stamen and gm = gynostemium. (DOCX 110 kb)
Melting curve analysis of all primer pairs used in this study performed at the end of the PCR cycles to confirm the specificity of primer annealing. (PDF 9997 kb)
Species phylogeny compiled based on Topik et al. [66], Biswal et al. [67], Takamiya et al. [68] and Chase et al. [69] for (a) FUL-, (b) AP3-, (c) PI- (d) AG- and STK-, (e) SEP- and (f) AGL6-like MADS-box gene lineage trees. (ZIP 584 kb)
Scanning electron micrographs of epidermal cells on the abaxial side of an E. pusilla flower. The three columns represent, from left to right, stage 2, 4 and 5 floral organs. Epidermal cells of (a–c) lateral sepal, (d–f) median sepal, (g–i) petal and (j–l) lip. Scale bar = 100 μm. Abbreviations: lse = lateral sepal; mse = median sepal; pe = petal. (PDF 19591 kb)
MADS-box gene lineage trees. (a) FUL-, (b) AP3-, (c) PI-, (d) AG-, (e) STK-, (f) SEP- and (g) AGL6-like trees. Color codes: green node = speciation event; red node = duplication event. Branches are colored along a gradient between blue and red, in proportion to the value of omega (dN/dS) for the third (i.e. the highest) rate class in the BranchSiteREL analysis. Hence, blue and red branches may be interpreted as suggesting, respectively, stabilizing and diversifying selection. Purple branches implicate a moderate level of diversifying selection. (ZIP 1121 kb)
Floral organ specific expression levels of FUL EpMADS10, EpMADS11 and EpMADS12 and SEP EpMADS6, EpMADS7, EpMADS8 and EpMADS9. RNA was extracted from seven different floral organs during two stages of development of E. pusilla and used for cDNA synthesis. Expression of the MADS-box genes was normalized to the geometric mean of three reference genes Actin, UBI2 and Fbox. Each column shows the relative expression of 20 floral organs in two cDNA pools (10 floral organs per isolation), both tested in triplicate. Abbreviations: lse = lateral sepal; mse = median sepal; cl = callus; pe = petal; fs = fertile stamen; gm = gynostemium. Dark grey = floral stage 2 and light grey = floral stage 4. Y-axis: relative gene expression. The error bars represent the Standard Error of Mean. P-value style: GP: >0.05 (ns), <0.05 (*), <0.01 (**), <0.001 (***), <0.0001 (****). (TIFF 3032 kb)
Acknowledgements
We thank Johan Keus for the culturing of E. pusilla, Bas Blankevoort, Erik-Jan Bosch, Rogier van Vugt (Hortus botanicus Leiden) and Angel Vale for the illustrations and photographs, Pieter van der Velden (LUMC), Stef Janson and Jan M. de Ruijter (UvA) for their support and input with the qPCR, Anneke de Wolf for support with the SEM, Øyvind Hammer for help with the Zeiss X radia and Marcel Lombaerts and Jan Oliehoek for help with the construction of the DNA sequence alignments.
Funding
This work was supported by grant 023.003.015 from the Netherlands Organization for Scientific Research (NWO) to AD and a Fulbright grant to BG.
Availability of data and materials
Scripts and alignments can be found at https://zenodo.org/record/44533#.VpPCP5PhCCR.
Authors’ contributions
ADM and BG designed the gene expression study and KvK, PvS and LK collected the expression data. RAV carried out the phylogenetic analyses with help of JWW. RvdB and SD collected the anatomical and micro-CT data. AV collected the electron microscope data and helped with the interpretation of the floral ontogeny. RB assisted with plant breeding. All authors contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- AG
AGAMOUS
- AGL6
AGAMOUS-LIKE-6
- AP3
APETALA3
- cl
callus
- fs
fertile stamen
- FUL
FRUITFULL
- gm
gynostemium
- L-complex
Lip complex
- LM
Light microscopy
- lse
lateral sepal
- micro-CT
3D-Xray microscopy
- mse
median sepal
- P-code
Perianth code
- pe
petal
- PI
PISTILLATA
- SEM
Scanning electron microscopy
- SEP
SEPALLATA
- SP-complex
Sepal/petal-complex
- STK
SEEDSTICK
Contributor Information
Anita Dirks-Mulder, Email: anita.dirks@naturalis.nl.
Roland Butôt, Email: roland.butot@naturalis.nl.
Peter van Schaik, Email: schaik.van.peter@gmail.com.
Jan Willem P. M. Wijnands, Email: janwillem.wijnands@gmail.com
Roel van den Berg, Email: rca.vd.berg@outlook.com.
Louie Krol, Email: louie.krol.jr@gmail.com.
Sadhana Doebar, Email: sadhanadoebar@gmail.com.
Kelly van Kooperen, Email: k.vankooperen@gmail.com.
Hugo de Boer, Email: h.d.boer@nhm.uio.no.
Elena M. Kramer, Email: ekramer@oeb.harvard.edu
Erik F. Smets, Email: erik.smets@naturalis.nl
Rutger A. Vos, Email: rutger.vos@naturalis.nl
Alexander Vrijdaghs, Email: alexander.vrijdaghs@bio.kuleuven.be.
Barbara Gravendeel, Email: Barbara.Gravendeel@naturalis.nl.
References
- 1.Cho SC, Jang SH, Chae SJ, Chung KM, Moon YH, An GH, Jang SK. Analysis of the C-terminal region of Arabidopsis thaliana APETALA1 as a transcription activation domain. Plant Mol Biol. 1999;40(3):419–429. doi: 10.1023/A:1006273127067. [DOI] [PubMed] [Google Scholar]
- 2.Roy BA, Widmer A. Floral mimicry: a fascinating yet poorly understood phenomenon. Trends Plant Sci. 1999;4(8):325–330. doi: 10.1016/S1360-1385(99)01445-4. [DOI] [PubMed] [Google Scholar]
- 3.Ackerman JD, Cuevas AA, Hof D. Are deception-pollinated species more variable than those offering a reward? Plant Syst Evol. 2011;293(1–4):91–99. doi: 10.1007/s00606-011-0430-6. [DOI] [Google Scholar]
- 4.Preston JC, Hileman LC, Cubas P. Reduce, reuse, and recycle: developmental evolution of trait diversification. Am J Bot. 2011;98(3):397–403. doi: 10.3732/ajb.1000279. [DOI] [PubMed] [Google Scholar]
- 5.Kanno A, Nakada M, Akita Y, Hirai M. Class B Gene Expression and the Modified ABC Model in Nongrass Monocots. Sci World J. 2007;7:268–279. doi: 10.1100/tsw.2007.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanno A, Saeki H, Kameya T, Saedler H, Theissen G. Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana) Plant Mol Biol. 2003;52(4):831–841. doi: 10.1023/A:1025070827979. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura T, Fukuda T, Nakano M, Hasebe M, Kameya T, Kanno A. The modified ABC model explains the development of the petaloid perianth of Agapanthus praecox ssp. orientalis (Agapanthaceae) flowers. Plant Mol Biol. 2005;58(3):435–445. doi: 10.1007/s11103-005-5218-z. [DOI] [PubMed] [Google Scholar]
- 8.Soltis DE, Chanderbali AS, Kim S, Buzgo M, Soltis PS. The ABC model and its applicability to basal angiosperms. Ann Bot. 2007;100(2):155–163. doi: 10.1093/aob/mcm117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endress PK. Development and evolution of extreme synorganization in angiosperm flowers and diversity: a comparison of Apocynaceae and Orchidaceae. Ann Bot. 2016;117(5):749–767. doi: 10.1093/aob/mcv119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu HF, Hsu WH, Lee YI, Mao WT, Yang JY, Li JY, Yang CH. Model for perianth formation in orchids. Nat Plants. 2015;1(5):15046. doi: 10.1038/nplants.2015.46. [DOI] [Google Scholar]
- 11.Decraene LPR, Smets EF. Staminodes: Their morphological and evolutionary significance. Bot Rev. 2001;67(3):351–402. doi: 10.1007/BF02858099. [DOI] [Google Scholar]
- 12.Kramer EM, Holappa L, Gould B, Jaramillo MA, Setnikov D, Santiago PM. Elaboration of B gene function to include the identity of novel floral organs in the lower eudicot Aquilegia. Plant Cell. 2007;19(3):750–766. doi: 10.1105/tpc.107.050385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudall PJ, Bateman RM. Roles of synorganisation, zygomorphy and heterotopy in floral evolution: the gynostemium and labellum of orchids and other lilioid monocots. Biol Rev Camb Philos Soc. 2002;77(3):403–441. doi: 10.1017/S1464793102005936. [DOI] [PubMed] [Google Scholar]
- 14.Rudall PJ, Perl CD, Bateman RM. Organ homologies in orchid flowers re-interpreted using the Musk Orchid as a model. PeerJ. 2013;1:e26. doi: 10.7717/peerj.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKnight TD, Shippen DE. Plant telomere biology. Plant Cell. 2004;16(4):794–803. doi: 10.1105/tpc.160470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramirez SR, Gravendeel B, Singer RB, Marshall CR, Pierce NE. Dating the origin of the Orchidaceae from a fossil orchid with its pollinator. Nature. 2007;448(7157):1042–1045. doi: 10.1038/nature06039. [DOI] [PubMed] [Google Scholar]
- 17.Givnish TJ, Spalink D, Ames M, Lyon SP, Hunter SJ, Zuluaga A, Iles WJ, Clements MA, Arroyo MT, Leebens-Mack J, et al. Orchid phylogenomics and multiple drivers of their extraordinary diversification. In: Proceedings Biological sciences/The Royal Society; 2015, 282(1814). [DOI] [PMC free article] [PubMed]
- 18.Kocyan A, Endress PK. Floral structure and development of Apostasia and Neuwiedia (Apostasioideae) and their relationships to other Orchidaceae. Int J Plant Sci. 2001;162(4):847–867. doi: 10.1086/320781. [DOI] [Google Scholar]
- 19.Vermeulen P. The Different Structure of the Rostellum in Ophrydeae and Neottieae. Acta Bot Neerl. 1959;8(3):338–355. doi: 10.1111/j.1438-8677.1959.tb00544.x. [DOI] [Google Scholar]
- 20.Kurzweil H. Developmental studies in orchid flowers I: Epidendroid and vandoid species. Nord J Bot. 1987;7(4):427–442. doi: 10.1111/j.1756-1051.1987.tb00964.x. [DOI] [Google Scholar]
- 21.Kurzweil H, Kocyan A. Orchid Biology: Reviews and Perspectives, Viii. 2002. Ontogeny of orchid flowers; pp. 83–138. [Google Scholar]
- 22.Brown R, Nees von Esenbeck CGD. Prodromus florae Novae Hollandiae et Insulae Van-Diemen : exhibens characteres plantarum. Norimbergae: Sumtibus L. Schrag; 1827. [Google Scholar]
- 23.Darwin C. The various contrivances by which orchids are fertilised by insects. London: John Murray; 1877. [Google Scholar]
- 24.Swamy BGL. Vascular anatomy of orchid flowers. Bot Mus Leafl Harv Univ. 1948;13(4):61–95. [Google Scholar]
- 25.Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353(6339):31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 26.Theissen G. Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol. 2001;4(1):75–85. doi: 10.1016/S1369-5266(00)00139-4. [DOI] [PubMed] [Google Scholar]
- 27.Theissen G, Saedler H. Plant biology. Floral quartets. Nature. 2001;409(6819):469–471. doi: 10.1038/35054172. [DOI] [PubMed] [Google Scholar]
- 28.Mondragon-Palomino M, Theissen G. Why are orchid flowers so diverse? Reduction of evolutionary constraints by paralogues of class B floral homeotic genes. Ann Bot. 2009;104(3):583–594. doi: 10.1093/aob/mcn258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mondragon-Palomino M, Theissen G. Conserved differential expression of paralogous DEFICIENS- and GLOBOSA-like MADS-box genes in the flowers of Orchidaceae: refining the ‘orchid code’. Plant J. 2011;66(6):1008–1019. doi: 10.1111/j.1365-313X.2011.04560.x. [DOI] [PubMed] [Google Scholar]
- 30.Pan ZJ, Cheng CC, Tsai WC, Chung MC, Chen WH, Hu JM, Chen HH. The duplicated B-class MADS-box genes display dualistic characters in orchid floral organ identity and growth. Plant Cell Physiol. 2011;52(9):1515–1531. doi: 10.1093/pcp/pcr092. [DOI] [PubMed] [Google Scholar]
- 31.Su CL, Chao YT, Yen SH, Chen CY, Chen WC, Chang YC, Shih MC. Orchidstra: an integrated orchid functional genomics database. Plant Cell Physiol. 2013;54(2):e11. doi: 10.1093/pcp/pct004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai J, Liu X, Vanneste K, Proost S, Tsai W-C, Liu K-W, Chen L-J, He Y, Xu Q, Bian C, et al. The genome sequence of the orchid Phalaenopsis equestris. Nat Genet. 2015;47(1):65–72. doi: 10.1038/ng.3149. [DOI] [PubMed] [Google Scholar]
- 33.Lee SH, Li CW, Liau CH, Chang PY, Liao LJ, Lin CS, Chan MT. Establishment of an Agrobacterium-mediated genetic transformation procedure for the experimental model orchid Erycina pusilla. Plant Cell Tissue Organ Cult. 2015;120(1):211–220. doi: 10.1007/s11240-014-0596-z. [DOI] [Google Scholar]
- 34.Lin CS, Hsu CT, Liao DC, Chang WJ, Chou ML, Huang YT, Chen JJ, Ko SS, Chan MT, Shih MC. Transcriptome-wide analysis of the MADS-box gene family in the orchid Erycina pusilla. Plant Biotechnol J. 2016;14(1):284–298. doi: 10.1111/pbi.12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neubig KM, Whitten WM, Williams NH, Blanco MA, Endara L, Burleigh JG, Silvera K, Cushman JC, Chase MW. Generic recircumscriptions of Oncidiinae (Orchidaceae: Cymbidieae) based on maximum likelihood analysis of combined DNA datasets. Bot J Linn Soc. 2012;168(2):117–146. doi: 10.1111/j.1095-8339.2011.01194.x. [DOI] [Google Scholar]
- 36.Felix LP, Guerra M. Chromosome analysis in Psygmorchis pusilla (L.) Dodson & Dressler: the smallest chromosome number known in Orchidaceae. Caryologia. 1999;52(3–4):165–168. doi: 10.1080/00087114.1998.10589169. [DOI] [Google Scholar]
- 37.Chase MW, Hanson L, Albert VA, Whitten WM, Williams NH. Life history evolution and genome size in subtribe Oncidiinae (Orchidaceae) Ann Bot. 2005;95(1):191–199. doi: 10.1093/aob/mci012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pridgeon A. Genera Orchidacearum/Vol. 5, Epidendroideae (part two)/ed. by Alec M. Pridgeon … [et al.]. Oxford: Oxford University Press; 2009.
- 39.Vale A, Navarro L, Rojas D, Alvarez JC. Breeding system and pollination by mimicry of the orchid Tolumnia guibertiana in Western Cuba. Plant Species Biol. 2011;26(2):163–173. doi: 10.1111/j.1442-1984.2011.00322.x. [DOI] [Google Scholar]
- 40.Papadopulos AS, Powell MP, Pupulin F, Warner J, Hawkins JA, Salamin N, Chittka L, Williams NH, Whitten WM, Loader D, et al. Convergent evolution of floral signals underlies the success of Neotropical orchids. In: Proceedings Biological sciences/The Royal Society; 2013, 280(1765):20130960. [DOI] [PMC free article] [PubMed]
- 41.Carmona-Díaz G, García-Franco JG. Reproductive success in the Mexican rewardless Oncidium cosymbephorum (Orchidaceae) facilitated by the oil-rewarding Malpighia glabra (Malpighiaceae) Plant Ecol. 2008;203(2):253–261. doi: 10.1007/s11258-008-9543-6. [DOI] [Google Scholar]
- 42.Staedler YM, Masson D, Schonenberger J. Plant tissues in 3D via X-ray tomography: simple contrasting methods allow high resolution imaging. PloS One. 2013;8(9):e75295. doi: 10.1371/journal.pone.0075295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37(6):e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuomi JM, Voorbraak F, Jones DL, Ruijter JM. Bias in the Cq value observed with hydrolysis probe based quantitative PCR can be corrected with the estimated PCR efficiency value. Methods. 2010;50(4):313–322. doi: 10.1016/j.ymeth.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Ruijter JM, Thygesen HH, Schoneveld OJ, Das AT, Berkhout B, Lamers WH. Factor correction as a tool to eliminate between-session variation in replicate experiments: application to molecular biology and retrovirology. Retrovirology. 2006;3:2. doi: 10.1186/1742-4690-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruijter JM, Ruiz Villalba A, Hellemans J, Untergasser A, van den Hoff MJ. Removal of between-run variation in a multi-plate qPCR experiment. Biomol Detect Quantif. 2015;5:10–14. doi: 10.1016/j.bdq.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 49.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55(4):539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 50.Zmasek CM, Eddy SR. A simple algorithm to infer gene duplication and speciation events on a gene tree. Bioinformatics. 2001;17(9):821–828. doi: 10.1093/bioinformatics/17.9.821. [DOI] [PubMed] [Google Scholar]
- 51.Kosakovsky Pond SL, Murrell B, Fourment M, Frost SD, Delport W, Scheffler K. A random effects branch-site model for detecting episodic diversifying selection. Mol Biol Evol. 2011;28(11):3033–3043. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21(5):676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 53.Kull T, Arditti J. Orchid Biology VIII: Reviews and Perspectives. Netherlands: Springer; 2013. [Google Scholar]
- 54.Rudall PJ, Bateman RM. Evolution of zygomorphy in monocot flowers: iterative patterns and developmental constraints. New Phytol. 2004;162(1):25–44. doi: 10.1111/j.1469-8137.2004.01032.x. [DOI] [Google Scholar]
- 55.Acri-Nunes-Miranda R, Mondragon-Palomino M. Expression of paralogous SEP-, FUL-, AG- and STK-like MADS-box genes in wild-type and peloric Phalaenopsis flowers. Front Plant Sci. 2014;5:76. doi: 10.3389/fpls.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan ZJ, Chen YY, Du JS, Chen YY, Chung MC, Tsai WC, Wang CN, Chen HH. Flower development of Phalaenopsis orchid involves functionally divergent SEPALLATA-like genes. New Phytol. 2014;202(3):1024–1042. doi: 10.1111/nph.12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mondragon-Palomino M, Hiese L, Harter A, Koch MA, Theissen G. Positive selection and ancient duplications in the evolution of class B floral homeotic genes of orchids and grasses. BMC Evol Biol. 2009;9:81. doi: 10.1186/1471-2148-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitney HM, Bennett KM, Dorling M, Sandbach L, Prince D, Chittka L, Glover BJ. Why do so many petals have conical epidermal cells? Ann Bot. 2011;108(4):609–616. doi: 10.1093/aob/mcr065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carlquist S. Toward acceptable evolutionary interpretations of floral anatomy. Phytomorphology. 1969;19(4):332–362. [Google Scholar]
- 60.Bateman RM, Rudall PJ. The Good, the Bad, and the Ugly: using naturally occurring terata to distinguish the possible from the impossible in orchid floral evolution. Aliso. 2006;22:481–496. [Google Scholar]
- 61.Endress PK. Diversity and Evolutionary Biology of Tropical Flowers. Cambridge: Cambridge University Press; 1996. [Google Scholar]
- 62.Reynders M, Vrijdaghs A, Larridon I, Huygh W, Leroux O, Muasya AM, Goetghebeur P. Gynoecial anatomy and development in Cyperoideae (Cyperaceae, Poales): congenital fusion of carpels facilitates evolutionary modifications in pistil structure. Plant Ecol Evol. 2012;145(1):96–125. doi: 10.5091/plecevo.2012.675. [DOI] [Google Scholar]
- 63.Pizzolato TD. Procambial initiation for the vascular system in the aerial shoot of Costus (Costaceae, Zingiberales) Int J Plant Sci. 2007;168(4):393–413. doi: 10.1086/512041. [DOI] [Google Scholar]
- 64.Scarpella E, Marcos D, Friml J, Berleth T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 2006;20(8):1015–1027. doi: 10.1101/gad.1402406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 66.Topik H, Yukawa T, Ito M. Molecular phylogenetics of subtribe Aeridinae (Orchidaceae): insights from plastid matK and nuclear ribosomal ITS sequences. J Plant Res. 2005;118(4):271–284. doi: 10.1007/s10265-005-0217-3. [DOI] [PubMed] [Google Scholar]
- 67.Biswal DK, Marbaniang JV, Tandon P. Age estimation for Asian Cymbidium (Orchidaceae: Epidendroideae) with implementation of fossil data calibration using molecular markers (ITS2 & matK) implying phylogeographic inference. PeerJ PrePrints. 2013;1:e94v91. doi: 10.1142/S0219720016600015. [DOI] [PubMed] [Google Scholar]
- 68.Takamiya T, Wongsawad P, Sathapattayanon A, Tajima N, Suzuki S, Kitamura S, Shioda N, Handa T, Kitanaka S, Iijima H, et al. Molecular phylogenetics and character evolution of morphologically diverse groups, Dendrobium section Dendrobium and allies. Aob Plants. 2014;6:plu045. doi: 10.1093/aobpla/plu045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chase MW, Cameron KM, Freudenstein JV, Pridgeon AM, Salazar G, Van den Berg C, Schuiteman A. An updated classification of Orchidaceae. Bot J Linn Soc. 2015;177(2):151–174. doi: 10.1111/boj.12234. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of sequences used in the alignments and phylogenetic analyses. Table S2. Transcript primer sequences and amplicon characteristics used for quantitative real-time PCR validation of the expression profiles of eighteen MADS-box transcripts following MIQE guidelines [65]. Table S3. Difference in MADS-box gene expression between floral organs; variance analysis of measures using Tukey multicomparisons test. P-value style: GP: >0.05 (ns), <0.05 (*), <0.01 (**), <0.001 (***), <0.0001 (****). Abbreviations: lse = lateral sepal, mse = median sepal, cl = callus, pe = petal, fs = fertile stamen and gm = gynostemium. (DOCX 110 kb)
Melting curve analysis of all primer pairs used in this study performed at the end of the PCR cycles to confirm the specificity of primer annealing. (PDF 9997 kb)
Species phylogeny compiled based on Topik et al. [66], Biswal et al. [67], Takamiya et al. [68] and Chase et al. [69] for (a) FUL-, (b) AP3-, (c) PI- (d) AG- and STK-, (e) SEP- and (f) AGL6-like MADS-box gene lineage trees. (ZIP 584 kb)
Scanning electron micrographs of epidermal cells on the abaxial side of an E. pusilla flower. The three columns represent, from left to right, stage 2, 4 and 5 floral organs. Epidermal cells of (a–c) lateral sepal, (d–f) median sepal, (g–i) petal and (j–l) lip. Scale bar = 100 μm. Abbreviations: lse = lateral sepal; mse = median sepal; pe = petal. (PDF 19591 kb)
MADS-box gene lineage trees. (a) FUL-, (b) AP3-, (c) PI-, (d) AG-, (e) STK-, (f) SEP- and (g) AGL6-like trees. Color codes: green node = speciation event; red node = duplication event. Branches are colored along a gradient between blue and red, in proportion to the value of omega (dN/dS) for the third (i.e. the highest) rate class in the BranchSiteREL analysis. Hence, blue and red branches may be interpreted as suggesting, respectively, stabilizing and diversifying selection. Purple branches implicate a moderate level of diversifying selection. (ZIP 1121 kb)
Floral organ specific expression levels of FUL EpMADS10, EpMADS11 and EpMADS12 and SEP EpMADS6, EpMADS7, EpMADS8 and EpMADS9. RNA was extracted from seven different floral organs during two stages of development of E. pusilla and used for cDNA synthesis. Expression of the MADS-box genes was normalized to the geometric mean of three reference genes Actin, UBI2 and Fbox. Each column shows the relative expression of 20 floral organs in two cDNA pools (10 floral organs per isolation), both tested in triplicate. Abbreviations: lse = lateral sepal; mse = median sepal; cl = callus; pe = petal; fs = fertile stamen; gm = gynostemium. Dark grey = floral stage 2 and light grey = floral stage 4. Y-axis: relative gene expression. The error bars represent the Standard Error of Mean. P-value style: GP: >0.05 (ns), <0.05 (*), <0.01 (**), <0.001 (***), <0.0001 (****). (TIFF 3032 kb)
Data Availability Statement
Scripts and alignments can be found at https://zenodo.org/record/44533#.VpPCP5PhCCR.


