Abstract
A subset of basal cell carcinomas (BCCs) are directly derived from hair follicles (HFs). In some respects, HFs can be defined as ‘ordered’ skin appendage growths, while BCCs can be regarded as ‘disordered’ skin appendage growths. The aim of the present study was to examine HFs and BCCs to define the expression of common and unique signaling pathways in each skin appendage. Human nodular BCCs, along with HFs and non-follicular skin epithelium from normal individuals, were examined using microarrays, qPCR, and immunohistochemistry. Subsequently, BCC cells and root sheath keratinocyte cells from HFs were cultured and treated with Notch signaling peptide Jagged1 (JAG1). Gene expression, protein levels, and cell apoptosis susceptibility were assessed using qPCR, immunoblotting, and flow cytometry, respectively. Specific molecular mechanisms were found to be involved in the process of cell self-renewal in the HFs and BCCs, including Notch and Hedgehog signaling pathways. However, several key Notch signaling factors showed significant differential expression in BCCs compared with HFs. Stimulating Notch signaling with JAG1 induced apoptosis of BCC cells by increasing Fas ligand expression and downstream caspase-8 activation. The present study showed that Notch signaling pathway activity is suppressed in BCCs, and is highly expressed in HFs. Elements of the Notch pathway could, therefore, represent targets for the treatment of BCCs and potentially in hair follicle engineering.
Keywords: basal cell carcinoma, hair follicle, microarray, apoptosis, Notch signaling pathways
Introduction
The skin is the body's first line of defense, providing protection from dehydration, injury, and infection. It comprises the epidermis and its adjoining structures, including the hair follicle (HF) and its associated sebaceous gland; together comprising the pilosebaceous unit. Hair follicles are self-renewing structures that continuously generate new epithelial cells to replenish the skin and pilosebaceous unit in response to injury (1). Skin homeostasis and wound repair requires the presence of epithelial stem cells as the primary source for regenerative cells. Multipotent stem cells that reside within the epidermis and in the bulge region of HFs (2) can give rise to a variety of cell types, including those forming HFs, interfollicular epidermis, and associated epithelial glands (3). Alterations in either proliferation or differentiation have the potential to disrupt normal skin homeostasis.
Certain disorders of the skin, such as cancer, chronic wounds, skin atrophy, skin fragility, hirsutism, and alopecia, can be, fundamentally, viewed as disorders of skin stem cells (4). It has been hypothesized that tumor formation is the result of inappropriate activation of signaling pathways activating these stem cells or their immediate multipotent progeny (5). Consistent with this view is the observation that several types of skin cancers can be derived from HFs, based on observations of histological presentation and the presence of specific molecular markers common to HFs and skin neoplasias (6). Understanding the molecular mechanisms by which proliferation and differentiation are regulated in skin appendages may provide a useful insight into the molecular basis of disease, and may also identify potential targets for treatment intervention.
Previous findings suggest that a significant subset of basal cell carcinomas (BCCs) are directly HF-derived (7–9). The stem cells of the HF bulge region and adjacent cells are a potential primary source of BCCs derived from HFs (8,10). In some regard, HFs and BCCs can be defined as ‘ordered’ and ‘disordered’ skin appendage growths, respectively. The primary mechanism, by which most BCCs develop, a constitutive activation of the Hedgehog pathway, is a principal regulatory mechanism in HF development (11). As such, all BCCs, HF-derived or not, may express similar key growth mechanisms to those involved in HF growth and cycling.
An important property that is shared by BCCs and HFs is the ability of cells to repeatedly proliferate, a mechanism that is responsible for maintaining a tumor mass or normal hair fiber production, respectively. If BCCs utilize HF growth mechanisms, growth factors and regulatory networks fundamental to HF growth would also likely be key mediators of BCC growth and may have the capacity to induce BCC growth and invasion. Several specific molecular mechanisms involved in this process of self-renewal, including the sonic hedgehog (Shh), Notch and Wingless-related integration site (Wnt) signaling pathways, have been found to be active in normal HFs and in BCCs (12–16). However, the roles of these signaling pathways in BCC growth, particularly Notch signaling, remain poorly understood.
The present study examined the potential molecular relationships between nodular BCCs and HFs using microarray profiling, reverse transcription-quantitative polymerase chain reaction (RT-qPCR), and immunohistochemistry. It was anticipated that BCCs and HFs would both exhibit activation of common signaling pathways involved in skin appendage formation (genes and networks commonly involved in ordered skin appendage growth). Specific molecular pathway components that code for ‘hair follicleness’ were also anticipated to be missing or over-represented in BCCs (candidate genes that regulate the networks important in ordered appendage growth that have failed in disordered BCC development). By distinguishing between common pathways and unique pathway components expressed in each type of skin appendage, the aim of the present study was to characterize those components important for BCC growth (genes and networks differentially represented in BCCs not commonly found in healthy skin epithelium or hair follicle appendages) and phenotype presentation, and to identify specific components important for appropriately regulated HF formation.
Materials and methods
Basal cell carcinomas, hair follicles, non-follicular tissues, and clinical information
All the samples were provided through the Department of Surgery and the Department of Dermatology and Skin Science, University of British Columbia, with approval from the University Clinical Research Ethics Board. Samples of human HFs were collected from scalp biopsies of normal individuals undergoing cosmetic procedures, while nodular BCCs and normal skin were obtained from patients undergoing surgical resection. All the nodular BCC samples and normal skin epithelium were taken from the facial area of donors. Only tissue from patients that had not been treated with preoperative chemotherapy or other therapeutic approaches was selected for analysis. BCC morphological subtypes were described and clinically classified during Mohs surgery and initial diagnoses were subsequently confirmed by formalin-fixed, paraffin-embedded histological assessment of the tumors.
Hair follicles (n=10-20/subject) were microdissected to remove the sebaceous gland and upper HF infundibulum and the lower one third, including the hair bulb. The dermal sheath was also removed, leaving the inner and outer root sheaths, including the bulge region, for analysis. Normal skin samples were microdissected to isolate skin epithelium from the dermal component. Samples collected for microarray/qPCR were immediately stored in an RNA stabilization reagent (Qiagen Inc., Toronto, ON, Canada).
RNA isolation
Total RNA was isolated from microdissected tissue or cultured cells with an RNeasy Fibrous Tissue Midi kit (Qiagen Inc.) according to the manufacturer's protocols. The quantity and quality of the RNA was measured using the Agilent 2100 bioanalyzer and RNA 6000 NANO kit (Agilent Technologies, Inc., Santa Clara, CA, USA), and the quantity was measured with a NanoDrop ND-100 spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE, USA).
Microarray production
Human Operon v.2.1 (21K) glass arrays were produced (based on human 70mers from Eurofins MWG Operon Inc., Huntsville, AL, USA) by the Microarray Facility of the Prostate Centre at Vancouver General Hospital (Vancouver, BC, Canada) (17,18). RNAs were amplified using the SenseAmp Plus kit (Genisphere LLC, Hatfield, PA, USA). The 260/280 absorbance ratio was used to determine the appropriate amount of sense RNA for labeling. Total RNA from test samples and universal human reference RNA (Agilent Technologies, Inc.) were respectively labeled with cyanine (Cy) 5 and Cy3, using the 3DNA array detection 350 kit (Genisphere LLC) and cohybridized to cDNA microarrays. Following overnight hybridization and washing, the arrays were imaged using a ScanArray Express scanner (PerkinElmer, Inc., Waltham, MA, USA).
Microarray data processing and analysis
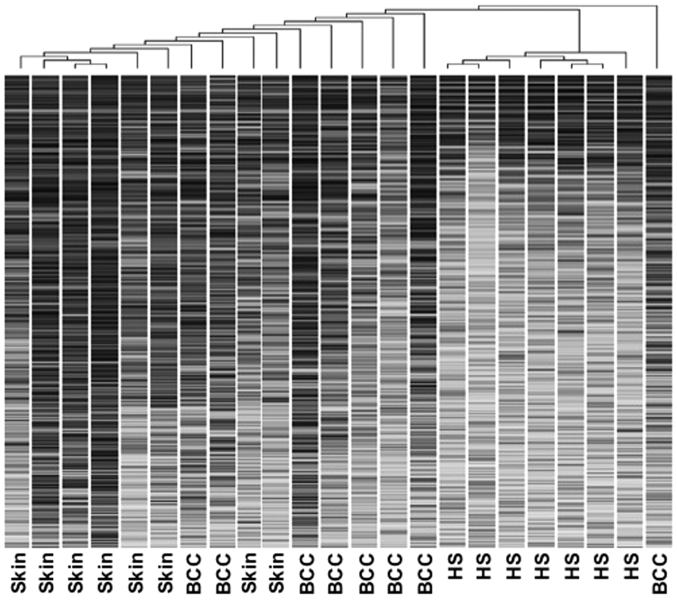
Arrays were scanned at excitation wavelengths of 532 and 635 nm to detect the Cy3 and Cy5 dyes, respectively. Image analysis and quantification were conducted with Imagene 6.0 commercial software (BioDiscovery Inc, El Segundo, CA, USA). The raw signal and background medians were used as the input for the Genespring 7.2 program (Agilent Technologies, Inc.). GeneSpring allows normalization and multiple filter comparisons of data from different experiments, thus generating restriction lists and the functional classification of differentially expressed genes. Raw data were background corrected and normalized using ‘per chip and per spot normalization’, which is an intensity-dependent normalization (non-linear or LOWESS normalization) (19). The expression of each gene is reported as the ratio of the value obtained for each sample relative to the universal reference RNA. Data were subsequently filtered using the raw signal strength value of both channels. Measurements with higher signal strength value are relatively more precise than measurements with lower signal strength. Genes that did not reach this value were discarded (100 out of 65,536). A condition tree was generated using hierarchical clustering of unfiltered data from each sample based on the similarity of their expression data. Similarity was measured using Pearson's correlation, and distances between clusters were calculated via average linkage (Fig. 1). The raw data from the arrays have been entered into the publicly accessible Gene Expression Omnibus (GEO) database in MIAME compliant format (http://www.ncbi.nlm.nih.gov/geo/). The raw data sets are encompassed by a series record number (GSE12542).
Figure 1.
Hierarchical clustering of unfiltered raw microarray data from each sample of hair follicle root sheaths, BCCs and normal skin epithelium. The degree of similarity in gene expression profiles was measured by Pearson's correlation, and distances between clusters were calculated via average linkage. Dendrogram results indicate well-defined cluster groups of cases. HS, hair follicle root sheaths; BCCs, basal cell carcinomas.
Analysis of gene expression differences and similarities
The comparison analyses were conducted by the ‘significance analysis of microarrays’ (SAM) method (20). Gene expression associated with nodular BCCs (n=8 subjects) and HF root sheaths (n=7 subjects) was first evaluated and contrasted (Tables IA and B, and II). The differences analysis between BCCs and HF root sheaths was conducted by the SAM method with a cut-off q-value of 14% and a 2-fold cut-off (Table IA and B). The 2-fold cut-off was employed to reduce the incidence of false-positive results, which can occur when using t-tests (replicates can have similar results by chance), but the probability of which is decreased at higher fold changes. Subsequently, as a prelude to defining the degree of similarity in gene expression between BCCs and HF root sheaths, the cut-off q-value was set to be >40% and the fold change <1.5 (Table II). The cut-off level of <1.5 helped to reduce the selection of genes that were not necessarily regulated, but had a sizable error between their replicate values. A comparison analysis between HF root sheaths and skin (n=8 subjects) was conducted by the SAM method (data not shown). SAM was also used to identify genes differentially regulated between BCC and the normal skin samples as demonstrated in a previous study (21).
Table I.
Gene transcripts.
| A, Top 20 gene transcripts with the highest magnitude fold-change in gene expression upregulation in nodular basal cell carcinomas vs. hair shafts, sorted by the false discovery rate (q-value) | ||||
|---|---|---|---|---|
| Gene product description | Gene name | GenBank accession number | Fold-change | q-value (%) |
| Dipeptidylpeptidase IV (CD26, adenosine | DPP4 | NM_001935 | 11.13121 | 0 |
| Rag D protein deaminase complexing protein 2) | RAGD | AL137502 | 8.835887 | 0 |
| DKFZP564O0463 protein | DKFZP564O0463 | AK001693 | 2.87002 | 0 |
| Propionyl coenzyme A carboxylase, β polypeptide | PCCB | NM_000532 | 37.27286 | 0.326689 |
| DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide, Y chromosome | DBY | NM_004660 | 18.25926 | 0.326689 |
| Ras homolog gene family, member A | ARHA | NM_001664 | 7.591441 | 0.326689 |
| Ubiquitin-like 3 | UBL3 | NM_007106 | 7.215993 | 0.326689 |
| Baculoviral IAP repeat-containing 3 | BIRC3 | AF070674 | 7.08567 | 0.326689 |
| Small nuclear ribonucleoprotein polypeptide E | SNRPE | NM_003094 | 5.495296 | 0.326689 |
| Splicing factor, arginine/serine-rich 10 (transformer 2 homolog, Drosophila) | SFRS10 | NM_004593 | 5.367942 | 0.326689 |
| Proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 | PSMD5 | BC014478 | 4.612349 | 0.326689 |
| Ubiquitously transcribed tetratricopeptide repeat gene, X chromosome | UTX | NM_021140 | 4.392788 | 0.326689 |
| Methionyl aminopeptidase 2 | METAP2 | NM_006838 | 4.388392 | 0.326689 |
| Actin related protein 2/3 complex, subunit 5 (16 kDa) | ARPC5 | NM_005717 | 3.719303 | 0.326689 |
| Sin3-associated polypeptide, 18 kDa | SAP18 | NM_005870 | 2.477771 | 0.326689 |
| F-box and leucine-rich repeat protein 3A | FBXL3A | NM_012158 | 6.425926 | 0.523782 |
| β-2-microglobulin | B2M | AK026463 | 5.708535 | 0.523782 |
| A disintegrin and metalloproteinase domain 9 (meltrin γ) | ADAM9 | NM_003816 | 4.995793 | 0.523782 |
| Lactate dehydrogenase A | LDHA | NM_005566 | 4.698842 | 0.523782 |
| Protein kinase C, ζ | PRKCZ | NM_002744 | 4.646151 | 0.523782 |
| B, Top 20 gene transcripts with the highest magnitude fold-change in gene expression downregulation in nodular basal cell carcinomas vs. hair shaft, sorted by the false discovery rate (q-value) | ||||
| Gene product description | Gene name | GenBank accession number | Fold-change | q-value (%) |
| Keratin-associated protein 4.14 | KAP4.14 | NM_033059 | 0.014887 | 0 |
| Keratin-associated protein 4.10 | KAP4.10 | NM_033060 | 0.018098 | 0 |
| Keratin-associated protein 3.2 | KRTAP3.2 | NM_031959 | 0.026814 | 0 |
| Keratin-associated protein 4.8 | KRTAP4.8 | AJ406940 | 0.055245 | 0 |
| Chromosome 20 open reading frame 28 | C20orf28 | NM_015417 | 0.061437 | 0 |
| Lymphotoxin α (TNF superfamily, member 1) | LTA | NM_000595 | 0.079272 | 0 |
| Desmoplakin (DPI, DPII) | DSP | NM_004415 | 0.087855 | 0 |
| Suppressor of Ty 5 homolog (S. cerevisiae) | SUPT5H | NM_003169 | 0.089178 | 0 |
| Neuronal protein 17.3 | P17.3 | NM_019056 | 0.095102 | 0 |
| Leucine zipper protein 1 | LUZP1 | BC002428 | 0.097131 | 0 |
| Breast carcinoma amplified sequence 1 | BCAS1 | NM_003657 | 0.097779 | 0 |
| Ubiquitin fusion degradation 1-like | UFD1L | NM_005659 | 0.098738 | 0 |
| D component of complement (adipsin) | DF | NM_001928 | 0.11378 | 0 |
| Ribosomal protein S8 | RPS8 | AK023362 | 0.138395 | 0 |
| LIM and cysteine-rich domains 1 | LMCD1 | NM_014583 | 0.149755 | 0 |
| Glutamate receptor, metabotropic 2 | GRM2 | NM_000839 | 0.153922 | 0 |
| Ephrin-B2 | EFNB2 | NM_004093 | 0.157336 | 0 |
| Ribosomal protein L38 | RPL38 | NM_000999 | 0.157418 | 0 |
| Loss of heterozygosity, 12, chromosomal region 1 | LOH12CR1 | NM_058169 | 0.169164 | 0 |
Table II.
Top 20 gene transcripts with the fold-change most consistently close to 0 between nodular basal cell carcinomas and hair shafts, sorted by the false discovery rate (q-value).
| Gene product description | Gene name | GenBank accession number | Fold-change | q-value (%) |
|---|---|---|---|---|
| Proteasome (prosome, macropain) subunit, α type, 4 | PSMA4 | AK055714 | 1.001521 | 38.71807 |
| Calcium channel, voltage-dependent, β 1 subunit | CACNB1 | M92303 | 1.001613 | 38.71807 |
| Leucine-zipper-like transcriptional regulator, 1 | LZTR1 | NM_006767 | 1.001924 | 38.71807 |
| F-box only protein 21 | FBXO21 | NM_033624 | 1.002044 | 38.71807 |
| Carnitine deficiency-associated gene expressed in ventricle 1 | CDV-1 | NM_014055 | 1.002094 | 38.71807 |
| Neurofibromin 1 (neurofibromatosis, von Recklinghausen disease, Watson disease) | NF1 | NM_000267 | 1.002262 | 38.71807 |
| Suppression of tumorigenicity 14 (colon carcinoma, matriptase, epithin) | ST14 | NM_021978 | 1.002397 | 38.71807 |
| Ecotropic viral integration site 5 | EVI5 | AF008915 | 1.002636 | 38.71807 |
| Otoraplin | OTOR | NM_020157 | 1.00287 | 38.71807 |
| HEMK homolog 7kb | HEMK | NM_016173 | 1.002877 | 38.71807 |
| Complement component 9 | C9 | NM_001737 | 1.003119 | 38.71807 |
| RAB11A, member RAS oncogene family | RAB11A | NM_004663 | 1.003131 | 38.71807 |
| ß-actin | ACTB | NA | 0.999976 | 48.18056 |
| Homeobox A6 | HOXA6 | NM_024014 | 0.99973 | 48.18056 |
| Pituitary tumor-transforming 1 interacting protein | PTTG1IP | NM_004339 | 0.99972 | 48.18056 |
| Peroxisome biogenesis factor 10 | PEX10 | NM_002617 | 0.999621 | 48.18056 |
| Bombesin-like receptor 3 | BRS3 | NM_001727 | 0.999605 | 48.18056 |
| Doublesex and mab-3 related transcription factor 2 | DMRT2 | NM_006557 | 0.999424 | 48.18056 |
| High-mobility group (nonhistone chromosomal) protein 4-like | HMG4L | AL049709 | 0.998903 | 48.18056 |
| Microfibril-associated glycoprotein-2 | MAGP2 | NM_003480 | 0.99872 | 48.18056 |
Gene ontology analysis
Functional classification of genes was performed using DAVID software 6.70 (22,23), based on the Gene Ontology (GO) database (www.geneontology.org), to allow the identification of ‘enriched’ or ‘depleted’ gene function categories in assigned biological processes, molecular functions, and cellular components (24,25). This program was used to identify genes belonging to different GO categories (derived from Tables I and II). Benjamini-corrected P<0.05 was used as the cut-off for determination of significant gene enrichment in each defined category. P≤0.05 indicated that the applied genes listed were specifically associated (enriched) in a category, as opposed to random chance, and were selected. For 25–40% of the genes defined, no GO annotation was given and their function was unknown. Ontological analyses were performed at biological process category level 3. The number of genes identified in major categories were normalized by the number of genes annotated in each list and were expressed as percentages (Tables III and IV).
Table III.
Most commonly identified categories with differences in GO categorization analysis between nodular basal cell carcinomas and hair shafts at GO level 3.
| GO category | Number of genes in category | Percentage of total genes in gene set | P-value | Benjamini |
|---|---|---|---|---|
| Regulation of cellular process | 966 | 24.8 | 3.50E-13 | 2.90E-10 |
| Negative regulation of cellular process | 297 | 7.6 | 5.20E-12 | 1.50E-09 |
| Negative regulation of biological process | 308 | 7.9 | 4.30E-12 | 1.80E-09 |
| System development | 417 | 10.7 | 1.00E-09 | 2.10E-07 |
| Regulation of programmed cell death | 147 | 3.8 | 3.30E-07 | 5.60E-05 |
| Positive regulation of cellular process | 243 | 6.2 | 5.20E-07 | 6.30E-05 |
| Positive regulation of biological process | 266 | 6.8 | 5.00E-07 | 7.00E-05 |
| Biopolymer metabolic process | 1090 | 27.9 | 8.30E-07 | 7.80E-05 |
| Organ development | 301 | 7.7 | 8.00E-07 | 8.40E-05 |
| Anatomical structure morphogenesis | 269 | 6.9 | 1.30E-06 | 1.10E-04 |
| Cell death | 206 | 5.3 | 1.70E-06 | 1.30E-04 |
| Cell cycle process | 187 | 4.8 | 2.70E-06 | 1.90E-04 |
| Organ morphogenesis | 114 | 2.9 | 3.40E-06 | 2.20E-04 |
| Regulation of metabolic process | 628 | 16.1 | 6.60E-06 | 4.00E-04 |
| Regulation of cell cycle | 137 | 3.5 | 9.80E-06 | 5.50E-04 |
| Regulation of cellular metabolic process | 604 | 15.5 | 2.00E-05 | 1.00E-03 |
| Regulation of gene expression | 582 | 14.9 | 2.10E-05 | 1.00E-03 |
| Cell differentiation | 399 | 10.2 | 4.20E-05 | 2.00E-03 |
| Regulation of protein metabolic process | 85 | 2.2 | 4.90E-05 | 2.20E-03 |
| Regulation of cell proliferation | 126 | 3.2 | 7.80E-05 | 3.30E-03 |
Top twenty categories identified are shown. GO, gene ontology.
Table IV.
Most commonly identified categories with similarities in GO categorization between nodular basal cell carcinomas and hair shafts at GO level 3.
| GO category | Number of genes in category | Percentage of total genes in gene set | P-value | Benjamini |
|---|---|---|---|---|
| Cellular catabolic process | 317 | 6.5 | 1.50E-05 | 1.70E-02 |
| Cellular nitrogen compound metabolic process | 1015 | 20.7 | 6.40E-05 | 3.40E-02 |
| Cellular macromolecule metabolic process | 1412 | 28.8 | 8.20E-05 | 2.90E-02 |
| Regulation of localization | 194 | 4 | 1.70E-04 | 4.50E-02 |
| Sterol metabolic process | 43 | 0.9 | 2.30E-04 | 4.90E-02 |
| Regulation of cell motion | 72 | 1.5 | 2.40E-04 | 4.20E-02 |
| Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 938 | 19.1 | 3.10E-04 | 4.70E-02 |
| Regulation of cell migration | 63 | 1.3 | 6.20E-04 | 8.00E-02 |
| Regulation of multicellular organismal process | 279 | 5.7 | 8.70E-04 | 1.00E-01 |
| Regulation of locomotion | 69 | 1.4 | 1.10E-03 | 1.10E-01 |
| Regulation of cell communication | 304 | 6.2 | 1.60E-03 | 1.50E-01 |
| Lipid transport | 54 | 1.1 | 1.60E-03 | 1.40E-01 |
| Response to organic nitrogen | 28 | 0.6 | 1.70E-03 | 1.30E-01 |
| Cellular amino acid and derivative metabolic process | 114 | 2.3 | 2.10E-03 | 1.50E-01 |
| Positive regulation of cellular process | 518 | 10.5 | 2.20E-03 | 1.40E-01 |
| Regulation of myeloid cell differentiation | 30 | 0.6 | 2.20E-03 | 1.40E-01 |
| Microtubule organizing center organization | 18 | 0.4 | 2.40E-03 | 1.40E-01 |
| Positive regulation of biological process | 566 | 11.5 | 2.50E-03 | 1.40E-01 |
| Regulation of cellular component organization | 143 | 2.9 | 2.80E-03 | 1.50E-01 |
| Peptide metabolic process | 24 | 0.5 | 3.00E-03 | 1.50E-01 |
GO, gene ontology.
RT-qPCR
Total RNA from each sample (1 µg) was reverse-transcribed into first-strand cDNA according to the protocol of the Superscript III first-strand cDNA synthesis system (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The primers used for SYBR Green qPCR were designed using the Prime Time qPCR Primer Design Software (Integrated DNA Technologies, Inc., Coralville, IA, USA; Table V) and tested with the intron-spanning assay. Assays are defined as intron-spanning if at least one exon/exon border is either directly covered by one primer or contained between the primer binding sites. Compared to the median spanned intron size (2.1 kb), the median Real Time ready assay amplicon size (75 bp) is approximately 30-times shorter. In conjunction with a short amplification time, this size difference can be exploited to gain specificity for mRNA-derived cDNA template vs. template-derived from residual genomic DNA. qPCR was performed using the Applied Biosystems StepOnePlus Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The thermal cycling conditions consisted of 2 min at 50°C and 2 min at 95°C, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. Data were analyzed by the comparative threshold cycle method (26) with normalization to human glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Changes in gene expression following treatments were recorded as fold differences from values in untreated controls.
Table V.
Primer sequences for defined genes.
| Gene name | Genbank accession no. | Forward primer (5′-3′) | Reverse primer (5′-3′) | Amplicon size |
|---|---|---|---|---|
| NOTCH1 | NM_017617 | GAGGCGTGGCAGACTATGC | CTTGTACTCCGTCAGCGTGA | 140 |
| NOTCH2 | NM_024408 | TATTGATGACTGCCCTAACCACA | ATAGCCTCCATTGCGGTTGG | 187 |
| NOTCH4 | NM_004557 | GGGTGAGACGTGCCAGTTTC | CTGGGTGTCAATGGAGAGGGA | 126 |
| DTX1 | NM_004416 | GGTGTGGGAGTGTCTGAATGA | CCTGGCGAAACTGGTGCAT | 176 |
| DTX2 | NM_020892 | GCAAGCCTTTCAGATTTGCCC | GCTGGCACAAACTGTCCCTT | 121 |
| LNX1 | NM_032622 | TTGGCTCAGTCCTGCTAACTA | GGAAGGCACCTTTACAGAGTTCT | 80 |
| DVL2 | NM_004422 | GAGGAAGAGACTCCCTACCTG | CGGGCGTTGTCATCTGAAAT | 167 |
| NUMB | NM_001005743 | CTACCTTCCAAGGGACCGAGT | AGCCCGGACGTTTTTAGACAC | 131 |
| JAG1 | NM_000214 | TCGGGTCAGTTCGAGTTGGA | AGGCACACTTTGAAGTATGTGTC | 143 |
| JAG2 | NM_002226 | AGCTGGACGCCAATGAGTG | GTCGTTGACGTTGATATGGCA | 131 |
| LFNG | NM_001040167 | GGGTCAGCGAGAACAAGGTG | GATCCGCTCAGCCGTATTCAT | 140 |
| KRT17 | NM_000422 | GGTGGGTGGTGAGATCAATGT | CGCGGTTCAGTTCCTCTGTC | 158 |
| HES1 | NM_005524 | ATGGAGAAAAATTCCTCGTCCC | TTCAGAGCATCCAAAATCAGTGT | 182 |
| HES7 | NM_032580 | CGGGATCGAGCTGAGAATAGG | GCGAACTCCAATATCTCCGCTT | 176 |
| HR | NM_018411 | AGGAGGCCATGCTTACCCAT | CACTATGCTCAGGCATCAGGG | 84 |
| RBPSUHL | NM_014276 | CAGTGCCTCCCAATCCTTTGA | CCTCCCCTCAGAATGGTGGT | 139 |
| DAAM1 | NM_014992 | GGTGGACGAGGTATTTCATTCAT | TCTGAAGCGCAAAGTTGCTATC | 100 |
| MAPK8 | NM_002750 | AGCAAGCGTGACAACAATTTTT | GAAATGGTCGGCTTAGCTTCT | 175 |
| GLI1 | NM_005269 | GGCACCATGAGCCCATCTC | ATCACCTTCCAAGGGTTCCTC | 216 |
| GLI2 | NM_005270 | CCCACTCCAACGAGAAACCC | GGACCGTTTTCACATGCTTCC | 96 |
| RAC1 | NM_198829 | ATCCGCAAACAGATGTGTTCT | CGCACCTCAGGATACCACT | 91 |
| ROCK2 | NM_004850 | TTGGTTCGTCACAAGGCATC | AGGGGCTATTGGCAAAGGC | 130 |
| FASL | NM_000639 | AAAGGAGCTGAGGAAAGTGG | CATAGGTGTCTTCCCATTCCAG | 80 |
Immunohistochemistry
Protein expression coded by Notch homolog 1 (NOTCH1), Jagged 2 (JAG2), Disheveled 2 (DVL2), and Hairy and Enhancer of Split 7 (HES7) was assessed by immunohistochemistry on formalin-fixed, paraffin-embedded biopsies (tissues from 4 individuals per group). Biopsy samples were incubated at room temperature for 1 h with the following primary antibodies: Anti-NOTCH1 (cat. no. ab526271; dilution, 100; Abcam, Cambridge, MA, USA), anti-JAG2 (cat. no. ab60041; dilution, 1:50; Abcam), anti-DVL2 (cat no. sc-13974, dilution, 1:100; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and anti-HES7 (cat. no. ARP37926_P05; dilution, 1:100; Aviva Systems Biology Co., San Diego, CA, USA). Following incubation, Tris-buffered saline (0.05 mol/l; Dako North America, Inc.) was used to wash the membranes. The antibody expression was then characterized using the Universal Dako Cytomation Labeled Streptavidin-Biotin kit (Dako North America, Inc., Carpinteria, CA, USA), according to the manufacturer's instructions, with 3,3′-diaminobenzidine as the development substrate. For the negative controls, the primary antibody was replaced with mouse immunoglobulin G at the appropriate dilution. The sections were counterstained with Harris' hematoxylin at room temperature for 30 sec and mounted in Permount (Thermo Fisher Scientific, Inc.). Five visual fields were randomly selected per tissue section for assessment at 400x magnification using a standard pathology microscope (BX40; Olympus Corporation, Richmond Hill, ON, Canada).
Human root sheath cell (HRSC) and BCC cell culture and treatment
HF units were obtained by removing the fat layer; hair bulb and dermal sheath were subsequently removed to expose the outer and inner root sheaths of the HFs. Each tissue was then treated with TrypLE Express (Invitrogen; Thermo Fisher Scientific, Inc.) for 15 min to dissociate the HF keratinocytes [termed ‘human root sheath cells’ (HRSCs)] into a single-cell suspension for culturing. Following digestion, the cells from groups of five HFs were combined and cultured in BioCoat collagen-I coated 24-well plates (BD Biosciences, Franklin Lakes, NJ, USA) in 50% Defined Keratinocyte-serum free medium (Invitrogen; Thermo Fisher Scientific, Inc.) combined with 50% EpiLife (Invitrogen; Thermo Fisher Scientific, Inc.) with a Human Keratinocyte Growth Supplement (HKGS) kit (Invitrogen; Thermo Fisher Scientific, Inc.). Colonies of HRSCs formed in ~1 week, during which fresh media were added every 2 days to replenish the culture, and the cells were passaged into T25 collagen-I-coated flasks (BD Biosciences). Human BCCs were isolated from nodular BCC samples and cultured in base medium M154 (Invitrogen; Thermo Fisher Scientific, Inc.) with HKGS, as described (27).
Each time the culture reached 90% confluence, HRSCs or BCCs were passaged into a T25 flask. At the end of passage 3, HRSCs or BCCs were transferred to BioCoat collagen-I-coated 6-well plates (Corning Incorporated, Corning, NY, USA) with a density of 60,000 cells/ml per well for one day. The next day, the media specific for HRSCs or BCCs were replenished and the cells were incubated with 4 mM recombinant JAG1 or scrambled JAG1 (both from AnaSpec Inc., Fremont, CA, USA) for 3 days. At the end of treatment, the cells were collected for further investigations.
Western blot analysis
Following treatment, the cells were washed twice with ice-cold phosphate-buffered saline and lysed in an ice-cold 1X radioimmunoprecipitation assay buffer [10 mmol/l Tris-HCl (pH 8.0), 140 mmol/l NaCl, 1 mmol/l ethylenediaminetetraacetic acid (pH 8.0), 0.5 mmol/l ethylene glycol-bis (β-aminoethyl ether)-N,N,N',N'-tetraacetic acid, 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), 0.1% sodium deoxycholate] containing protease inhibitors (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany). The extract was centrifuged at 16,400 × g for 15 min at 4°C to remove cellular debris, and protein concentrations were determined by Bradford assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Protein samples (20 µg) were then separated by 10% SDS-polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes (Bio-Rad Laboratories, Inc.). The membranes were blocked at room temperature for 1 h in Tris-buffered saline containing 0.01% Tween 20 with 5% non-fat dried milk, and incubated overnight at 4°C with anti-Fas ligand (cat. no. ab68338; dilution, 1:1,000; Abcam) or anti-caspase-8 (cat. no. 9746S; dilution, 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA). The membranes were washed, and incubated with 1:15,000-diluted IRDye 680LT anti-rabbit (cat. no. P/N 925-68021) or IRDye 800CW anti-mouse (cat. no. P/N 925–32210) secondary antibodies (LI-COR Biosciences, Lincoln, NE, USA) for 1 h at room temperature. Signals were detected with the Odyssey infrared imaging system (LI-COR Biosciences). Antiserum to total β-actin (cat. no. sc-1616; dilution, 1:5,000; Santa Cruz Biotechnology, Inc.) was used as the internal control. Scion Image Analysis software (version 4.0.3.2; Scion Co., Frederick, MD, USA) was used to determine protein density levels.
Flow cytometric analysis
Apoptosis of JAG1-treated HRSCs and BCCs was quantified using an Annexin V-Propidium Iodide Apoptosis Detection Kit (cat. no. 88-8005-72; eBioscience, Inc., San Diego, CA, USA) according to the manufacturer's instructions. The samples were analyzed following appropriate fluorescence compensation and gating strategies with a FACSCanto-II flow cytometer (BD Biosciences), and analyzed using FlowJo software (version 9.0; FlowJo, LLC, Ashland, OR, USA).
Statistical analysis
Data were presented as the mean ± standard deviation from at least three sets of experiments (each from a separate subject). qPCR samples were assayed in triplicate for each experiment. Data were analyzed by one-way analysis of variance, followed by Tukey's multiple comparison tests if the overall P-values were significant, using the computer software PRISM (version 6.0c; GraphPad Software, Inc., San Diego, CA). P<0.05 was considered to indicate a statistically significant difference.
Results
Comparison of sample expression profiles by hierarchical clustering
Unsupervised hierarchical clustering analysis (average linkage method) was applied to the unfiltered datasets from BCCs and normal skin samples from patients undergoing surgical resection, and HF root sheaths from normal individuals undergoing cosmetic procedures. This produced a dendrogram with well-defined cluster groups of cases (Fig. 1). The HF root sheath data were clearly separated from BCC and normal skin data sets with distinct gene expression profiles, but exhibited close similarity within the group. Normal skin samples also yielded relatively distinctive expression profiles. Compared with the HF root sheath and normal skin, BCCs were less readily identified by computer analysis as a distinct entity. Overall, the segregation of data at the molecular level was consistent with the histological distinction of BCCs, HF root sheaths, and normal skin (Fig. 1).
Analysis of gene expression differences and similarities between basal cell carcinoma and hair follicle root sheaths
Gene expression associated with HF root sheaths and BCCs was evaluated, and the two data sets were directly compared. The results indicated that 4329 genes were differentially expressed in BCCs compared with the HF root sheaths with statistical significance (P<0.05). Of these, 3562 genes were upregulated and 767 were downregulated in BCCs compared with the HF root sheaths. Tables IA and B show the top 20 genes exhibiting the highest magnitude of fold-change, with those upregulated in Table IA and those downregulated in Table IB. In addition, the top 20 genes with the fold-change most consistently close to 0 (no change) are listed in Table II.
Similarities and differences in Gene Ontology categorization analysis between basal cell carcinoma and hair follicle root sheaths
Tables I and II were functionally annotated using GO terms, providing a controlled vocabulary to describe genes/gene product attributes to evaluate the potential significance in gene expression functions. The 20 most commonly identified, statistically significant, GO term results (adjusted P<0.05) for genes significantly differentially or similarly expressed in BCCs compared with the HF root sheaths are listed in Tables III and IV, respectively.
The GO analysis revealed that the differentially expressed genes (Table I) and similarly expressed genes (Table II) in BCCs and HF root sheaths were significantly enriched in the designated functional category of ‘developmental process’. For example, significantly differently expressed genes were enriched in ‘embryonic morphogenesis development’, a subcategory of ‘developmental process’. Two-hundred and ninety-seven significantly similarly expressed genes were observed to be associated with ‘regulation of multicellular organismal process’ and 518 similarly expressed genes were associated with ‘positive regulation of cellular process’, which are also subcategories under ‘developmental process’ and ‘multicellular organismal development’.
Enrichment in several gene function categories was commonly identified within significantly differentially expressed genes, including ‘cell cycle phase’, ‘cell cycle process’, ‘cell death’, ‘cell differentiation’, ‘cell maturation’, ‘G1/S transition of mitotic cell cycle’, ‘cell motility’ and ‘cell-cell signaling’, under the primary category of ‘cellular process’ (Table III). In addition, gene enrichment of some subcategories under the term of ‘regulation of cellular process’ was observed, including ‘regulation of cell cycle’, ‘regulation of cell proliferation’, ‘regulation of gene expression’, ‘epigenetic’, ‘regulation of signal transduction’, ‘regulation of transcription’ and ‘regulation of translation’ (Table III).
In parallel, of the genes similarly expressed in BCCs and HF root sheaths, numerous genes with a similar expression were observed in gene function subcategories of the term ‘cellular process’, including ‘positive regulation of cellular process’, ‘regulation of cell communication’, ‘regulation of multicellular organismal process’, ‘regulation of cell migration’ and ‘regulation of localization’ (Table IV). Subcategories such as ‘cellular amino acid and derivative metabolic process’, ‘nucleobase, nucleoside, nucleotide and nucleic acid metabolic process’ and ‘sterol metabolic process’, were also found enriched with similarly expressed genes within the category ‘metabolic process’ (Table IV). The GO analysis also indicated that a number of genes, both similarly and differentially expressed between BCCs and HF root sheaths, are involved in categories under the term of ‘positive regulation of cellular process’. The data suggest patterns of gene activity consistent with skin appendages, though with significant distinctions in individual expression of genes between BCCs and HFs.
Biological network and pathway analysis of basal cell carcinoma and hair shafts
To assess which signaling pathways were affected during early gene regulation, the genes in Tables I and II were classified and grouped into pathways, based on pathway information imported from the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/). Pathways that may be involved in both hair morphogenesis and tumorigenesis were the primary focus, based on published knowledge: The Shh, Notch, and Wnt signaling pathways. Based on the gene set that included genes differentially expressed between the HF root sheaths and nodular BCCs (Table I), 6 genes involved in the Shh signaling pathway were identified, 23 genes in the Wnt signaling pathway, and 4 genes in the Notch signaling pathway. Genes with similar trends of expression in BCCs and HF root sheaths were also analyzed (Table II): 11 genes were involved in the Shh signaling pathway, 37 genes in the Wnt signaling pathway and 9 genes were identified in the Notch signaling pathway. The identified pathway-specific genes in the corresponding pathway maps derived from the KEGG database are presented in Fig. 2. Since Notch signaling i) promotes HF differentiation into sebaceous gland and interfollicular epidermal lineages, ii) is known to act as an epidermal tumor suppressor, and iii) genes in this pathway exhibited the most extreme changes in expression/inhibition by microarray, the present study subsequently focused on the Notch signaling pathway.
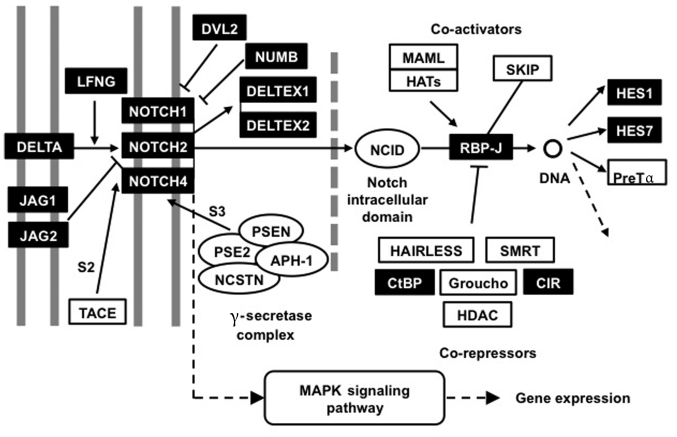
Figure 2.
Notch gene signaling pathway interactions. The Notch signaling pathway network was significantly differentially activated in hair follicle root sheaths compared with basal cell carcinomas. Genes and gene sets identified as significantly differentially expressed by microarray are presented with a dark background. JAG, jagged; LFNG, LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase; TACE, tumor necrosis factor-ACa disintegrin and metalloproteinase metalloprotease converting enzyme; DVL2, dishevelled2; NUMB, NUMB endocytic adaptor protein; PSEN, presenilin; NCSTN, nicastrin; APH, acylaminoacyl-peptide hydrolase; MAPK, mitogen-activated protein kinase; NCID, Notch intracellular cytoplasmic domain; MAML, mastermind-like protein; HAT, histone acetyltransferase; SKIP, Ski-interacting protein; RBP-J, Recombining binding protein suppressor of Hairless; SMRT, Silencing Mediator for Retinoic acid and Thyroid hormone receptor; CtBP, C-terminal binding protein; CIR, CBF1 interacting corepressor; HDAC, histone deacetylase; HES, hairy and enhancer of split.
Validation of the expression of selected genes
Having completed the microarray analysis of BCCs and HFs, 22 genes with known functional significance associated with Notch signaling were selected for evaluation by RT-qPCR (Table VI). Differential Shh pathway gene expression (GLI1, GLI2) was also reconfirmed to validate the study. Although the magnitude of change in expression defined by qPCR was different from that observed by microarray, the trends, whether for increased or decreased gene expression, were generally consistent with microarray data (Table VI).
Table VI.
Selected gene validation results by RT-qPCR with corresponding microarray results for comparison.
| Hair follicle vs. skin | Basal cell carcinomas vs. skin | |||||||
|---|---|---|---|---|---|---|---|---|
| Microarray | RT-qPCR | Microarray | RT-qPCR | |||||
| Gene name | Fold change | q-value | Fold change | P-value | Fold change | q-value | Fold change | P-value |
| NOTCH1 | 4.07350142 | 8.64548474 | 89.7796173 | 3.0878E-08 | 2.25963492 | 55.1681869 | 3.2667374 | 0.01731453 |
| NOTCH2 | 0.49022594 | 0.39254973 | 94.051046 | 2.4841E-06 | 0.46324991 | 2.15276566 | 0.87297527 | 0.84563107 |
| NOTCH4 | 2.69005573 | 0.24322855 | 19.2601231 | 0.00128485 | 1.71844966 | 55.1681869 | 2.67459359 | 0.14313619 |
| DTX1 | 1.17722862 | 48.8237978 | 16.3788042 | 0.03308844 | 2.4295169 | 55.1681869 | 0.28089744 | 0.22707567 |
| DTX2 | 0.99256711 | 40.6530129 | 18.9813992 | 0.00157923 | 0.47607531 | 4.07129257 | 0.43629239 | 0.08540183 |
| LNX1 | – | – | 198.293295 | 3.6156E-08 | 1.72295346 | 55.1681869 | 2.072439 | 0.28247621 |
| DVL2 | 13.8816522 | 3.95106797 | 16.104213 | 0.00293444 | 1.55096741 | 55.1681869 | 2.94822536 | 0.03581873 |
| NUMB | 3.28845002 | 0.48722077 | 182.325561 | 4.9233E-06 | 1.30366339 | 55.1681869 | 1.33412783 | 0.63444821 |
| JAG1 | 0.34149885 | 0.18592759 | 365.28094 | 2.3222E-08 | 1.63466178 | 55.1681869 | 1.83632659 | 0.35967923 |
| JAG2 | 1.5839265 | 41.2725397 | 179.447584 | 8.3358E-05 | 1.33391274 | 55.1681869 | 0.31170539 | 0.23588105 |
| LFNG | 2.07190237 | 6.92879545 | 1889.72022 | 4.5482E-05 | 2.33909215 | 55.1681869 | 1.52408184 | 0.61264104 |
| KRT17 | 3.021 | 0.00846 | 27272.867 | 2.2916E-09 | 7.21676553 | 55.1681869 | 51.4001016 | 0.00100685 |
| HES1 | – | – | 1067.11032 | 1.053E-15 | 0.76047244 | 67.4104045 | 2.91116399 | 0.0400455 |
| HES7 | 1.19000588 | 48.8237978 | 3262.24091 | 1.5035E-08 | 4.16958086 | 29.7263863 | 0.29241452 | 0.13032618 |
| HR | 2.16394516 | 2.23319493 | 226.55094 | 1.0638E-09 | 1.09510072 | 58.338051 | 1.19521471 | 0.76942586 |
| RBPSUHL | 3.11300222 | 41.2725397 | 255.196596 | 7.7485E-10 | 1.22074356 | 55.1681869 | 0.16526889 | 0.06136225 |
| DAAM1 | 0.20611879 | 0.21537795 | 574.06641 | 1.5066E-07 | 0.54865054 | 18.0863137 | 1.63754261 | 0.55059198 |
| MAPK8 | 1.5501382 | 42.0929177 | 199.824645 | 1.1302E-06 | 0.85527761 | 67.7038487 | 0.96727335 | 0.97161104 |
| GLI1 | – | – | 52.4313289 | 0.00072822 | – | – | 1.87408166 | 0.51779197 |
| GLI2 | 2.52604018 | 22.3203103 | 319.860846 | 8.9511E-07 | 2.00283366 | 55.1681869 | 12.2544249 | 0.00422916 |
| RAC1 | 0.26859543 | 1.21189849 | 535.384925 | 2.0661E-11 | 0.65505096 | 61.6040693 | 1.31400567 | 0.66627414 |
| ROCK2 | 0.18494564 | 2.62913482 | 575.373988 | 8.4351E-07 | 0.82910219 | 67.7038487 | 1.38128144 | 0.6297566 |
RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
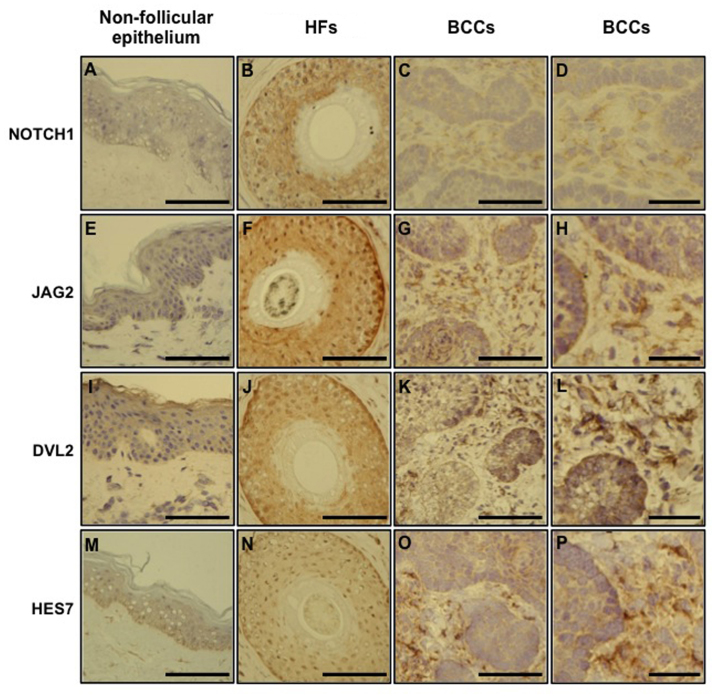
NOTCH1, JAG2, DVL2 and HES7 protein expression in the pathology specimens from BCC patients, scalp/terminal HF biopsies or non-scalp skin (non-follicular epithelium) from normal patients was defined using immunohistochemical analysis. All of the primary antibodies exhibited greater intensity of labeling in terminal HFs compared with nodular BCCs and non-scalp skin when tissues were processed in parallel (Fig. 3). A weak expression of NOTCH1 was observed in non-follicular epithelium (Fig. 3A). By contrast, there was a strong NOTCH1 presence in the outer root sheath (ORS) layer of HFs (Fig. 3B). NOTCH1 showed a more limited expression in the tumor mass compared with the peritumoral stroma in BCCs (Fig. 3C and D). JAG2 had a limited distribution in the inter-follicular epidermis (Fig. 3E), but exhibited a strong expression in HFs, including the ORS and inner root sheath (IRS; Fig. 3F). JAG2 was expressed more weakly in the BCC mass than in the peritumoral stroma (Fig. 3G and H). Compared with the inter-follicular epidermis (Fig. 3I), DVL2 was strongly expressed in the ORS of HFs (Fig. 3J). BCC tumor cells demonstrated a strong immunoreactivity to DVL2 (Fig. 3K and L). The basal layer of epidermis (Fig. 3M) and the ORS of HFs (Fig. 3N) were strongly positive for HES7. The peritumoral stroma of BCCs also exhibited a stronger expression of HES7 than the BCC mass (Fig. 3O and P). A stronger expression of NOTCH1, JAG2, DVL2 and HES7 was observed in the surface layer of the tumor structure (Fig. 3C, G, K and O) than in the internal tumor mass (Fig. 3D, H, L, P). The immunohistochemistry results were consistent with the microarray and qPCR findings.
Figure 3.
NOTCH1, JAG2, DVL2 and HES7 expression in the normal non-follicular epithelium, hair follicle root sheaths and basal cell carcinomas. Immunohistology was conducted to define the expression of NOTCH1, JAG2, DVL2 and HES7 in pathology specimens of normal non-scalp skin, normal scalp/terminal HF biopsies, and BCCs. All of the primary antibodies exhibited a greater intensity of labeling in terminal HFs compared with nodular BCCs and non-scalp skin when tissues were processed in parallel. Scale bar, 100 µm (except D, H, L and P, where the scale bar is 40 µm). JAG2, Jagged2; DVL2, Dishevelled2; HES7, hairy and enhancer of split 7; HF, hair follicle; BCCs, basal cell carcinomas.
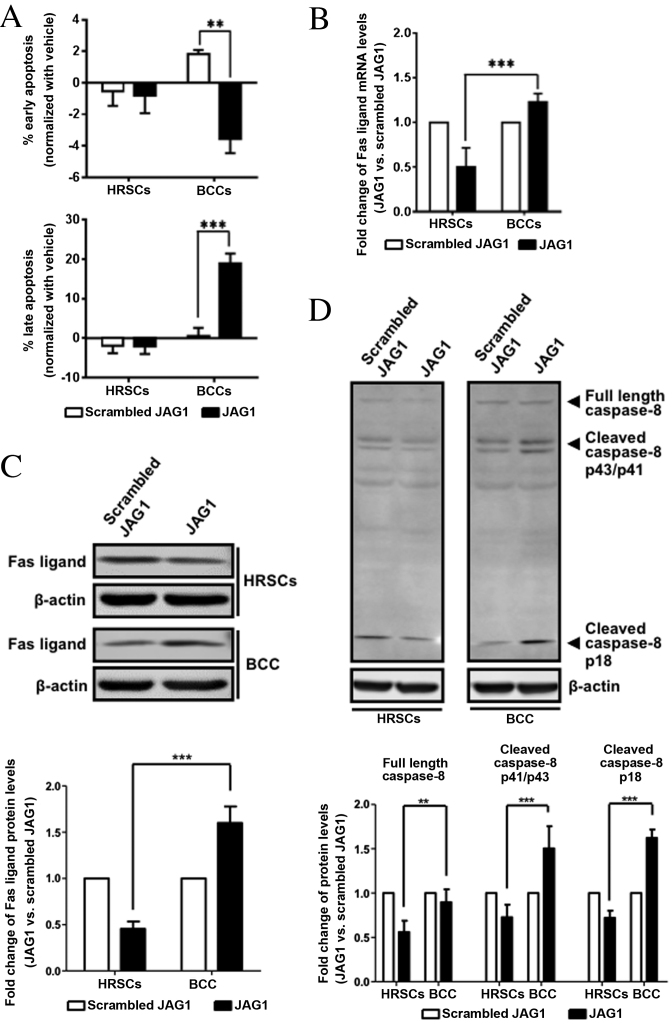
JAG1 treatment induced apoptosis of BCC cells via Fas ligand
Since JAG1 can activate NOTCH1 (28), BCC cell apoptosis was examined in cells induced with recombinant JAG1. A significantly higher percentage of late apoptotic cells was found in JAG1-treated BCC cell cultures compared with cells treated with scrambled JAG1 (0.5 vs. 18.95% Annexin V+PI+ cells, P=0.0004; Fig. 4A bottom channel). By contrast, JAG1 treatment did not induce late apoptosis in HRSCs (P>0.05; Fig. 4A bottom channel). To identify the specific pathway involved in JAG1-induced apoptosis, a human apoptosis PCR array was performed on treated cells, which revealed that Fas ligand mRNA expression was significantly increased in JAG1-treated BCCs, while the level of Fas mRNA did not change (data not shown).
Figure 4.
JAG1 treatment induced BCC cell apoptosis via Fas ligand. HRSCs or BCCs were treated with 4 mM JAG1 or scrambled peptide for 3 days. (A) Percentage of apoptotic cells was quantified by Annexin V-PI labeling and flow cytometric analysis. (B) Fas ligand mRNA expression levels were measured by qPCR. Representative data from three independent experiments are shown. Protein expression levels of (C) Fas ligand and (D) caspase-8 in treated HRSCs or BCCs were measured by western blot analysis. β-actin was used as the internal control. Levels of Fas ligand or caspase-8 proteins levels were normalized to the levels of β-actin protein, and are expressed as fold change relative to control (scrambled JAG1). Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 and ***P<0.001, comparisons indicated by brackets. HRSCs, human hair follicle root sheath cells; BCCs, basal cell carcinomas; JAG1, Jagged1.
qPCR specific for the Fas ligand gene was used to further confirm PCR array data (Fig. 4B). Fas ligand mRNA expression was downregulated in HRSCs following JAG1 treatment compared with the cells treated with scrambled JAG1, and was upregulated in BCCs following JAG1 treatment compared with the cells treated with scrambled JAG1 (Fig. 4B). A statistically significant difference in expression following JAG1 treatment was observed between the two cell types (0.50-fold in HRSCs vs. 1.23-fold in BCCs, P=0.0003; Fig. 4B). Differences in Fas ligand protein expression levels (Fig. 4C) followed the same pattern as the corresponding mRNA expression levels (Fig. 4B).
Caspase-8, is a downstream target of Fas ligand signaling and an initiator for the extrinsic apoptotic pathway (29). JAG1 treatment slightly decreased the protein expression levels of cleaved caspase-8 in HRSCs, but induced a significant increase in cleaved caspase-8 in BCCs (Fig. 4D), suggesting the caspase-dependent pathway was involved in BCC cell apoptosis induced by JAG1.
Discussion
The present study considered the potential relationships between HFs and BCCs, with reference to the similarities/differences in signaling pathway activation. Gene functions and the specific involvement of the Notch and Shh signaling pathways were surveyed to define how these regulatory pathways may control HF and BCC growth (30). Defining gene expression patterns and pathways in BCCs that are distinct from HF growth and cycling may lead to a better understanding of the abnormal proliferation that these cells undergo in the development of skin cancer.
Genes similarly expressed in HFs and BCCs were found under the same functional GO categories, including ‘positive regulation of cellular process’ and ‘regulation of multicellular organismal process’. This suggested that common regulatory genes may be important for the morphogenesis of both skin appendages. By contrast, a number of genes were uniquely expressed in HFs or BCCs only. These genes may serve as stage-specific signatures of appendage formation, either for maintaining and regenerating normal HFs, or for the formation of tumor masses. Commonly and differentially expressed genes related to morphogenesis are consistent with our hypothesis that BCC tumors may follow an abnormal appendage development process that exhibits elements of consistent order, morphogenesis and patterning as observed in HFs.
Shh signaling is required for the proliferation and normal cycling of HF epithelium. Modifications of Shh signaling can lead to tumor development in tissues of different origins (13). Hyperactivation of the Shh signaling pathway is found in several HF derived tumors and in BCCs (31,32). Overexpression of GLI1 and GLI2 products are also common features of BCCs, and suggests increased Shh signaling (31,33). Consistent with previous findings (31,33), the expression of GLI1 and GLI2 was enhanced in BCCs and HF root sheaths, compared with normal skin. However, the HF root sheaths exhibited significantly higher expression levels of GLI1 and GLI2 compared with BCCs. Shh signaling pathway genes may influence the same progenitor cells in BCCs and HFs, but the different gene activation levels in the two tissue types may contribute to the divergent patterns of growth.
Differential expression of the Notch signaling pathway was also revealed in BCCs and HFs compared with each other and with normal skin. The Notch pathway, with its family of four mammalian Notch receptors and their numerous ligands of the Delta, Jagged, and Serrate groups, is important for cell fate determination and organogenesis during embryonic development (34,35). Studies on embryonic mice and rats have demonstrated that the Notch/recombining binding protein suppressor of hairless (RBP-J) signaling pathway promotes epidermal differentiation (12) and cutaneous appendage patterning (36). Aberrant Notch signaling has been linked to a wide variety of tumors, but Notch can either suppress or promote tumors depending on the cell type and context (37,38). Jayaraman et al (14) demonstrated that NOTCH1 and NOTCH2 were significantly mutated in BCCs. The present study indicated that selected Notch pathway genes were differentially activated and inhibited in BCCs, which may be due to positive feedback, and reciprocal negative feedback, from differences in Delta and Notch cell surface expression, or the irregular activation of downstream Notch signaling pathway genes.
Examination of downstream components of the Notch pathway revealed more interesting results: The transcription factor RBP-J and downstream target genes of the Hes and Deltex families, exhibited a high expression in hair shafts compared with BCCs and normal skin. By contrast, two genes that affect the co-repression of RBP-J, CTBP1 and CREBBP, were observed to have a significantly lower expression in HFs compared with BCCs (data not shown). Deletion of RBP-J from follicular stem cells results in an aberrant cell fate switch that leads to the establishment of epidermal progenitors and basal cells (39). This result, therefore, demonstrated that the Notch/RBP-J signaling pathway is strongly activated in HFs. Since the Notch signaling pathway promotes a stem cell phenotype in skin (36), the degree of Notch signaling pathway activation may be important for HF stem cell proliferation and differentiation. The high level of Notch/RBP-J signaling pathway activation may be required for the formation and maintenance of follicles.
The present study suggests a lack of downstream gene expression in the Notch/RBP-J signaling pathway in BCCs. This may allow basal cells to escape from the normal regulation of proliferation that is normally found in the absence of Notch signaling, as observed in mammary epithelium cell lineages (40). Loss of RBP-J action in BCCs may promote cells towards a more stem, or progenitor, cell-like status, enabling basal cell tumor growth. Notch signaling via Notch receptor intracellular domain translocation into the nucleus, and subsequent binding to the transcription factor RBP-J, may be an important stage in the regulation of BCC development. As such, RBP-J signaling may be a focus for the development of new BCC therapies, as has been suggested for other types of cancer (41–44).
Fas ligand is a type II transmembrane protein that can induce apoptosis upon binding to Fas. Compared with normal skin, Fas ligand mRNA expression levels have previously been demonstrated to be lower in BCC specimens and immunostainings were weak or undetected (45). The present findings have shown that activation of the Notch signaling pathway by adding exogenous JAG1 into BCC cell culture resulted in increased Fas ligand mRNA and protein expression. This further activated downstream caspase-8 to initiate BCC cell apoptosis. However, some tumors can decrease Fas expression to resist Fas ligand-mediated T-cell cytotoxicity, and simultaneously upregulate the expression of Fas ligand to induce apoptosis in Fas-expressing T cells (46). BCCs strongly express Fas ligand, which may help prevent attack from surrounding immune effector cells, while also lacking Fas, potentially to make the tumor cells resistant to apoptosis (47). Further investigation is required to characterize the exact role of elevated Fas ligand expression induced by Notch signaling activation by in vivo experiments.
Notch signaling pathway genes are important in HF formation and BCC neoplasia. Hair follicles can develop from skin stem cells or their progeny, in a patterning program that is controlled by multiple signaling pathways, function processes and other components. Skin stem cells that are regulated by multiple signaling pathways, normal function processes and normal components, may allow BCCs to develop through abnormal activation order, irregular cell cycle timing and aberrant activation of multiple signaling pathways.
In summary, the present study suggests that controlling the Notch/RBP-J signaling pathway may stop the dysregulation of cell proliferation and differentiation to BCC skin cancer. The degree of Notch signaling pathway activation may also be important in HF formation. Elements of the Notch pathway are potentially worthwhile targets for future treatment of BCCs and, as a corollary, in HF engineering.
Acknowledgements
The present study was financially supported by the Canadian Dermatology Foundation and the Canadian Institutes of Health Research (CIHR; grant nos. MUS-94025 and MSH-95328). We would like to thank the staff of the Microarray Facility of The Prostate Centre at Vancouver General Hospital for their technical assistance and advice. We would also like to extend thanks to Drs. H. Lui and S. Le Bihan for their assistance and advice.
Glossary
Abbreviations
- BCCs
basal cell carcinomas
- HFs
hair follicles
- JAG1
Jagged1
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- NOTCH
Notch homolog 1
- JAG2
Jagged 2
- DVL2
Disheveled 2
- HES7
Hairy and Enhancer of Split 7
- HRSC
human root sheath cell
- HKGS
Human Keratinocyte Growth Supplement
- ORS
outer root sheath
- IRS
inner root sheath
References
- 1.Tiede S, Paus R. Lhx2-decisive role in epithelial stem cell maintenance, or just the ‘tip of the iceberg’? Bioessays. 2006;28:1157–1160. doi: 10.1002/bies.20506. [DOI] [PubMed] [Google Scholar]
- 2.Alonso L, Fuchs E. Stem cells of the skin epithelium. Proc Natl Acad Sci USA. 2003;100:(Suppl 1) 11830–11835. doi: 10.1073/pnas.1734203100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honeycutt KA, Koster MI, Roop DR. Genes involved in stem cell fate decisions and commitment to differentiation play a role in skin disease. J Investig Dermatol Symp Proc. 2004;9:261–268. doi: 10.1111/j.1087-0024.2004.09312.x. [DOI] [PubMed] [Google Scholar]
- 4.Najafzadeh N, Esmaeilzade B, Dastan Imcheh M. Hair follicle stem cells: In vitro and in vivo neural differentiation. World J Stem Cells. 2015;7:866–872. doi: 10.4252/wjsc.v7.i5.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burkert J, Wright NA, Alison MR. Stem cells and cancer: An intimate relationship. J Pathol. 2006;209:287–297. doi: 10.1002/path.2016. [DOI] [PubMed] [Google Scholar]
- 6.Jahoda C, Reynolds A. Skin stem cells-a hairy issue. Nat Med. 2000;6:1095–1097. doi: 10.1038/80418. [DOI] [PubMed] [Google Scholar]
- 7.Crowson AN. Basal cell carcinoma: Biology, morphology and clinical implications. Mod Pathol. 2006;19:(Suppl 2) S127–S147. doi: 10.1038/modpathol.3800512. [DOI] [PubMed] [Google Scholar]
- 8.Grachtchouk M, Pero J, Yang SH, Ermilov AN, Michael LE, Wang A, Wilbert D, Patel RM, Ferris J, Diener J, et al. Basal cell carcinomas in mice arise from hair follicle stem cells and multiple epithelial progenitor populations. J Clin Invest. 2011;121:1768–1781. doi: 10.1172/JCI46307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez-Takegami JC, Wolter M, Löser C, Maiweg C, Jones M, Metze D, Böer-Auer A. Classification of cysts with follicular germinative differentiation. J Cutan Pathol. 2016;43:191–199. doi: 10.1111/cup.12619. [DOI] [PubMed] [Google Scholar]
- 10.Peterson SC, Eberl M, Vagnozzi AN, Belkadi A, Veniaminova NA, Verhaegen ME, Bichakjian CK, Ward NL, Dlugosz AA, Wong SY. Basal cell carcinoma preferentially arises from stem cells within hair follicle and mechanosensory niches. Cell Stem Cell. 2015;16:400–412. doi: 10.1016/j.stem.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutchin ME, Kariapper MS, Grachtchouk M, Wang A, Wei L, Cummings D, Liu J, Michael LE, Glick A, Dlugosz AA. Sustained Hedgehog signaling is required for basal cell carcinoma proliferation and survival: Conditional skin tumorigenesis recapitulates the hair growth cycle. Genes Dev. 2005;19:214–223. doi: 10.1101/gad.1258705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thélu J, Viallet JP, Dhouailly D. Differential expression pattern of the three Fringe genes is associated with epidermal differentiation. J Invest Dermatol. 1998;111:903–906. doi: 10.1046/j.1523-1747.1998.00372.x. [DOI] [PubMed] [Google Scholar]
- 13.McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/S0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- 14.Jayaraman SS, Rayhan DJ, Hazany S, Kolodney MS. Mutational landscape of basal cell carcinomas by whole-exome sequencing. J Invest Dermatol. 2014;134:213–220. doi: 10.1038/jid.2013.276. [DOI] [PubMed] [Google Scholar]
- 15.Wuest M, Dummer R, Urosevic M. Induction of the members of Notch pathway in superficial basal cell carcinomas treated with imiquimod. Arch Dermatol Res. 2007;299:493–498. doi: 10.1007/s00403-007-0785-2. [DOI] [PubMed] [Google Scholar]
- 16.Rishikaysh P, Dev K, Diaz D, Qureshi WM, Filip S, Mokry J. Signaling involved in hair follicle morphogenesis and development. Int J Mol Sci. 2014;15:1647–1670. doi: 10.3390/ijms15011647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyons P. Advances in spotted microarray resources for expression profiling. Brief Funct Genomic Proteomic. 2003;2:21–30. doi: 10.1093/bfgp/2.1.21. [DOI] [PubMed] [Google Scholar]
- 18.Nelson CC, Hoffart D, Gleave ME, Rennie PS. Application of gene microarrays in the study of prostate cancer. Methods Mol Med. 2003;81:299–320. doi: 10.1385/1-59259-372-0:299. [DOI] [PubMed] [Google Scholar]
- 19.Mungamuri SK, Yang X, Thor AD, Somasundaram K. Survival signaling by Notch1: Mammalian target of rapamycin (mTOR)-dependent inhibition of p53. Cancer Res. 2006;66:4715–4724. doi: 10.1158/0008-5472.CAN-05-3830. [DOI] [PubMed] [Google Scholar]
- 20.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizzo P, Miao H, D'Souza G, Osipo C, Song LL, Yun J, Zhao H, Mascarenhas J, Wyatt D, Antico G, et al. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008;68:5226–5235. doi: 10.1158/0008-5472.CAN-07-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.da W Huang, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.da W Huang, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Lo BK, Yu M, Zloty D, Cowan B, Shapiro J, McElwee KJ. CXCR3/ligands are significantly involved in the tumorigenesis of basal cell carcinomas. Am J Pathol. 2010;176:2435–2446. doi: 10.2353/ajpath.2010.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rooman I, De Medts N, Baeyens L, Lardon J, De Breuck S, Heimberg H, Bouwens L. Expression of the Notch signaling pathway and effect on exocrine cell proliferation in adult rat pancreas. Am J Pathol. 2006;169:1206–1214. doi: 10.2353/ajpath.2006.050926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holtzman MJ, Green JM, Jayaraman S, Arch RH. Regulation of T cell apoptosis. Apoptosis. 2000;5:459–471. doi: 10.1023/A:1009657321461. [DOI] [PubMed] [Google Scholar]
- 30.Katoh M, Katoh M. Notch ligand, JAG1, is evolutionarily conserved target of canonical WNT signaling pathway in progenitor cells. Int J Mol Med. 2006;17:681–685. [PubMed] [Google Scholar]
- 31.Dahmane N, Lee J, Robins P, Heller P, i Altaba A Ruiz. Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature. 1997;389:876–881. doi: 10.1038/39918. [DOI] [PubMed] [Google Scholar]
- 32.Oro AE, Higgins KM, Hu Z, Bonifas JM, Epstein EH, Jr, Scott MP. Basal cell carcinomas in mice overexpressing sonic hedgehog. Science. 1997;276:817–821. doi: 10.1126/science.276.5313.817. [DOI] [PubMed] [Google Scholar]
- 33.Grachtchouk M, Mo R, Yu S, Zhang X, Sasaki H, Hui CC, Dlugosz AA. Basal cell carcinomas in mice overexpressing Gli2 in skin. Nat Genet. 2000;24:216–217. doi: 10.1038/73417. [DOI] [PubMed] [Google Scholar]
- 34.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 35.Ayaz F, Osborne BA. Non-canonical notch signaling in cancer and immunity. Front Oncol. 2014;4:345. doi: 10.3389/fonc.2014.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowell S, Jones P, Le Roux I, Dunne J, Watt FM. Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr Biol. 2000;10:491–500. doi: 10.1016/S0960-9822(00)00451-6. [DOI] [PubMed] [Google Scholar]
- 37.Chappell WH, Green TD, Spengeman JD, McCubrey JA, Akula SM, Bertrand FE. Increased protein expression of the PTEN tumor suppressor in the presence of constitutively active Notch-1. Cell Cycle. 2005;4:1389–1395. doi: 10.4161/cc.4.10.2028. [DOI] [PubMed] [Google Scholar]
- 38.Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, Hui CC, Clevers H, Dotto GP, Radtke F. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto N, Tanigaki K, Han H, Hiai H, Honjo T. Notch/RBP-J signaling regulates epidermis/hair fate determination of hair follicular stem cells. Curr Biol. 2003;13:333–338. doi: 10.1016/S0960-9822(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 40.Buono KD, Robinson GW, Martin C, Shi S, Stanley P, Tanigaki K, Honjo T, Hennighausen L. The canonical Notch/RBP-J signaling pathway controls the balance of cell lineages in mammary epithelium during pregnancy. Dev Biol. 2006;293:565–580. doi: 10.1016/j.ydbio.2006.02.043. [DOI] [PubMed] [Google Scholar]
- 41.Garber K. Notch emerges as new cancer drug target. J Natl Cancer Inst. 2007;99:1284–1285. doi: 10.1093/jnci/djm148. [DOI] [PubMed] [Google Scholar]
- 42.Purow B. Notch inhibition as a promising new approach to cancer therapy. Adv Exp Med Biol. 2012;727:305–319. doi: 10.1007/978-1-4614-0899-4_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yabuuchi S, Pai SG, Campbell NR, De Wilde RF, De Oliveira E, Korangath P, Streppel MM, Rasheed ZA, Hidalgo M, Maitra A, Rajeshkumar NV. Notch signaling pathway targeted therapy suppresses tumor progression and metastatic spread in pancreatic cancer. Cancer Lett. 2013;335:41–51. doi: 10.1016/j.canlet.2013.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan B, Liu L, Zhao Y, Xiu LJ, Sun DZ, Liu X, Lu Y, Shi J, Zhang YC, Li YJ, et al. Xiaotan Sanjie decoction attenuates tumor angiogenesis by manipulating Notch-1-regulated proliferation of gastric cancer stem-like cells. World J Gastroenterol. 2014;20:13105–13118. doi: 10.3748/wjg.v20.i36.13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang XY, Zhang R, Lian S. Aberrant expression of Fas and FasL pro-apoptotic proteins in basal cell and squamous cell carcinomas. Clin Exp Dermatol. 2011;36:69–76. doi: 10.1111/j.1365-2230.2010.03860.x. [DOI] [PubMed] [Google Scholar]
- 46.Satchell AC, Barnetson RS, Halliday GM. Increased Fas ligand expression by T cells and tumour cells in the progression of actinic keratosis to squamous cell carcinoma. Br J Dermatol. 2004;151:42–49. doi: 10.1111/j.1365-2133.2004.05974.x. [DOI] [PubMed] [Google Scholar]
- 47.Erb P, Ji J, Wernli M, Kump E, Glaser A, Buchner SA. Role of apoptosis in basal cell and squamous cell carcinoma formation. Immunol Lett. 2005;100:68–72. doi: 10.1016/j.imlet.2005.06.008. [DOI] [PubMed] [Google Scholar]