Abstract
In addition to natural competence, some Thermus thermophilus strains show a high rate of DNA transfer via direct cell-to-cell contact. The process is bidirectional and follows a two-step model where the donor cell actively pushes out DNA and the recipient cell employs the natural competence system to take up the DNA, in a hybrid transformation-dependent conjugation process (transjugation). While the DNA uptake machinery is well known as in other bacterial species that undergo transformation, the pushing step of transjugation remains to be characterized. Here we have searched for hypothetical DNA translocases putatively involved in the pushing step of transjugation. Among candidates encoded by T. thermophilus HB27, the TdtA protein was found to be required for DNA pushing but not for DNA pulling during transjugation, without affecting other cellular processes. Purified TdtA shows ATPase activity and oligomerizes as hexamers with a central opening that can accommodate double-stranded DNA. The tdtA gene was found to belong to a mobile 14 kbp-long DNA element inserted within the 3′ end of a tRNA gene, flanked by 47 bp direct repeats. The insertion also encoded a homolog of bacteriophage site-specific recombinases and actively self-excised from the chromosome at high frequency to form an apparently non-replicative circular form. The insertion also encoded a type II restriction endonuclease and a NurA-like nuclease, whose activities were required for efficient transjugation. All these data support that TdtA belongs to a new type of Integrative and Conjugative Element which promotes the generalized and efficient transfer of genetic traits that could facilitate its co-selection among bacterial populations.
Author summary
Transjugation is a new type of horizontal gene transfer process in which a donor cell pushes out genomic DNA upon cell contact and a recipient cell pulls this DNA inside by natural transformation. Here we describe TdtA, a DNA translocase of the pushing system of T. thermophilus, which is encoded within ICEth1, a new class of Integrative and Conjugative Element whose presence leads to generalized cell-to-cell transfer of any gene marker, circumventing the Argonaute surveillance system that controls access of extracellular DNA acquired by transformation.
Introduction
Conjugation is the most widely distributed mechanism for horizontal sharing of genetic information in bacteria, favoring the dissemination of antibiotic resistance and spread of adaptive traits, providing an extraordinary source of intra- and interspecies genetic plasticity [1–4]. In well-known conjugation models, DNA transfer starts at a cis-acting site (oriT) and proceeds unidirectionally from the donor to a recipient cell that is in intimate contact. The process requires highly specialized machinery in the donor cell, including a complex type IV secretion system (T4SS) mobilization apparatus and a “relaxosome complex”. Relaxase is an enzyme that identifies a given oriT sequence, cuts its specifically, and remains covalently bound to the generated 5′ end. In the conjugation process, a tip protein of type 4 pilus binds to a compatible recipient cell, while further pilus retraction forms direct contacts through which the T4SS of the donor manages to generate a bridge to the cytoplasm of the recipient, allowing the recruitment and head-first translocation of the relaxase-ssDNA (single-stranded DNA) complex. In this classical conjugation model, the recipient cell remains passive, awaiting the arrival of DNA (reviewed by Lanka et al [5] and Zechner et al [6]).
Although also encoded by plasmids, the machinery for classical conjugation is frequently found within Integrative and Conjugative Elements (ICEs), mobile DNA elements that encode a phage-like site-specific integrase, which recognizes attachment sites (att) in the ICE and the genome, catalyzing its integration. Most frequently, the specific att site is located within the 3′ end of a tRNA gene, but att sites have also been observed in other genes. The att site (as much as 60 bp in length) duplicates as a consequence of insertion, its direct repeats allowing the identification of the ICE boundaries. Different stress conditions increase the frequency by which ICEs excise from the host genome, promoting self-encoded conjugative transfer to recipient cells. Some defective ICEs are also known to rely on conjugative plasmids, or even on other ICEs, to be mobilized. Also, defective plasmids can be mobilized by complementation with ICE-encoded proteins [7,8].
Alternative models for direct cell-to-cell DNA transfer have been reported in bacterial genera such as Mycobacterium and Streptomyces [9–11]. In Mycobacterium, a chromosome-encoded Distributive Conjugal Transfer (DCT) model [11,12] has been described, in which extensive fragments of non-contiguous chromosomal DNA are transferred in a unidirectional fashion from the donor to recipient cells, in the absence of known plasmids or ICEs [13]. Indeed, cis-acting sites show similar transfer efficiencies regardless of their chromosomal location, leading to extensive genomic complexity in the progeny, which have a mosaic-like architecture derived from both parental strains [11]. Some of these transconjugants can become donors in subsequent DCT [13].
In Streptomyces spp., conjugative plasmids encode a single DNA translocase (TraB), which is the only protein required for primary transfer of the plasmid to a recipient cell [14]. TraB is a large DNA translocase of the FtsK family that forms hexameric structures and catalyzes ATP-dependent double-stranded DNA (dsDNA) transfer to the recipient cell. The TraB protein also has a DNA recognition motif at its C-terminus which is responsible for identification of specific cis-acting locus of transfer (clt) sequences in the plasmid and, with lower frequency, in the chromosome. The conjugative plasmid also encodes a few Spd proteins which are required along with TraB for further intramycelial transfer across the septa, leading to rapid colonization of the whole mycelium [15].
In Thermus thermophilus, an unconventional conjugation-like mechanism has been described too. In this ancient phylum, cell contact-dependent DNA transfer occurs between completely isogenic strains, with no apparent order in the transfer and without the requirement for homologs of T4SS or relaxases. In T. thermophilus HB27, the process is even more efficient than transformation, with a 10-fold preference for transfer of genes encoded in the pTT27 megaplasmid compared to chromosomal genes. In this DNA transfer process, the natural competence machinery of the recipient cell plays an active role that is essential for the transfer, whereas competence is not required at all in the donor. For this reason, we proposed a two-step “push-pull” DNA model [16], where both mates play an active role. In this model, termed “transjugation” (from transformation-dependent conjugation), the donor cell actively pumps DNA via a hypothetical DNA translocase that is independent from the transformation machinery. In the second step, the competence apparatus actively pulls the DNA into the recipient cell.
In this work, we identified components of the pushing machinery by a combination of in silico screening and further genetic analysis. Through bioinformatics we identified putative orphan DNA translocases as targets for mutagenesis. Among the screened proteins, we identified TdtA as a major component required for DNA donation in transjugation. We determined that TdtA is a hexameric ATPase that binds to the membrane. We also found that the tdtA gene belongs to an active mobile genetic island integrated within a tRNA gene in what is the first evidence of the existence of an active new class of ICE-like elements in these phylogenetically ancient thermophiles.
Results
T. thermophilus HB27 encodes four putative DNA translocases
Because the hypothesized pushing step in transjugation requires DNA pumping from the donor cell, we performed a bioinformatics search across the genome of T. thermophilus HB27, looking for putative proteins harboring ATPase motifs (Walker A and B) and HAS-barrel motifs found in proteins of the FtsK-HerA family of ATPases, which are frequently involved in DNA translocation [17]. Four putative members of this protein family were found within the genome of T. thermophilus HB27, encoded by genes TTC0474 (FtsK, AAS80822, 867 aa), TTC0147 (AAS80495, 576 aa), TTC1430 (AAS81772, 611 aa), and TTC1879 (AAS82221, 568 aa). The putative amino acid sequence of the TTC0147 product shares 31% identity with that of TTC1879, whereas no significant sequence similarity was detected between the product of TTC1430 and that of TTC0147 or TTC1879.
To determine the roles of the putative proteins in DNA repair or as DNA translocases in segregation/conjugation, we attempted to generate knockout insertion mutants (Δgene::kat). For TTC0474, the gene encoding the likely homolog to FtsK, mutant recovery was unsuccessful despite the use of multiple different mutational strategies, suggesting its requirement for cell survival. In contrast, knockouts of the other three hypothetical genes were viable. As transjugation was not expected to affect viability or any other physiologically relevant cellular process, these mutants were subjected to UV-light and heat-shock, which increases mutation rates [18] and stress to discard those involved in cell survival. The results shown in Table 1 demonstrate significant (10 to 100-fold) differences between the TTC0147 and TTC1430 mutants compared to the wild type both for UV and heat shock survival assays (t-test; p-value< 0.001), whereas no difference was observed between ΔTTC1879::kat and the wild type in heat shock studies (t-tests; p-value: 0.678), and only marginal differences were found by the UV tests (t-tests; p-value: 0.038) (n = 8). On the other hand, transformation assays revealed a small decrease in efficiency with respect to the wild type for the mutants in TTC0147 and TTC1430, with the TTC1879::kat mutant not significantly affected.
Table 1. Phenotypic effects of deletion of FtsK-HerA homologs encoded by T. thermophilus HB27.
| Strain | Survival to UV treatment | Survival to heat shock | Transformation efficiency |
|---|---|---|---|
| Wild type | 1.07 ± 0.23 | 1.02 ± 0.86 | 18.51 ± 6.02 E-5 |
| ΔTTC0147::kat | 0.45 ± 0.03 | 0.02 ± 0.01 | 6.71 ± 2.47 E-5 |
| ΔTTC1430::kat | 0.02 ± 0.01 | 0.13 ± 0.04 | 9.02 ± 0.76 E-5 |
| ΔTTC1879::kat | 0.75 ± 0.04 | 0.87 ± 0.51 | 12.47 ± 4.02 E-5 |
Therefore, TTC0147 and TTC1430 seem to play some relevant role in cell physiology, likely in DNA repair after stress, whereas TTC1879 is dispensable, as could be expected for a transjugation-associated gene.
TTC1879 is required for transjugation
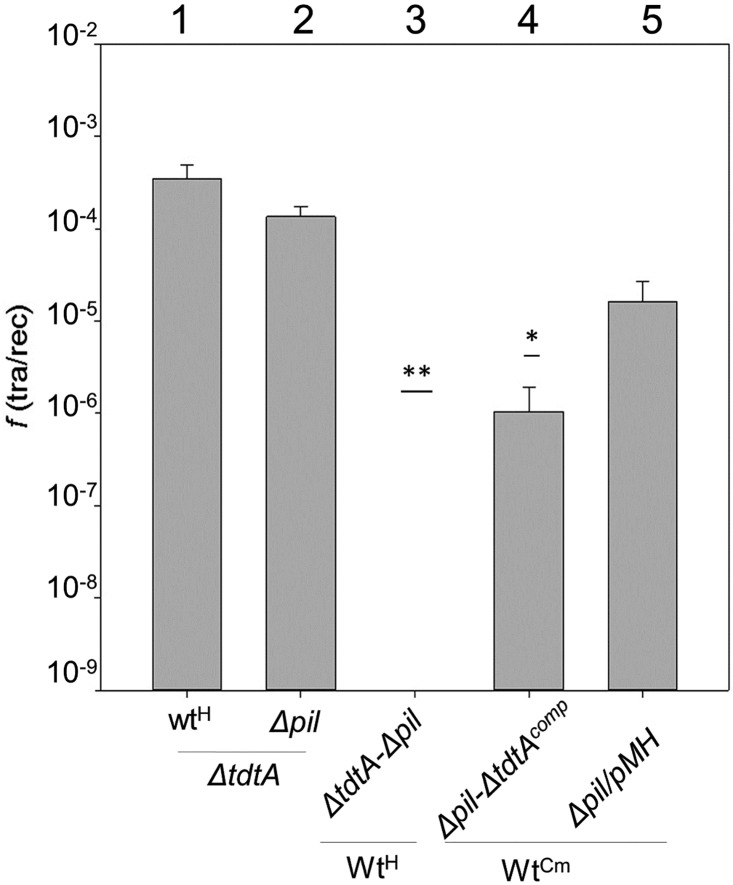
To analyze the role of TTC1879 in DNA donation (pushing step) we introduced the ΔTTC1879::kat mutation into a ΔpilA4 background, which is unable to serve as a recipient in transjugation assays [16], and mated it with a wild type strain labeled with hygromycin (Hyg) resistance. No transjugants were generated in this mating (Fig 1, bar n° 3), implying that the ΔTTC1879::kat mutant cannot donate DNA in transjugation. As expected from the bidirectionality of the transjugation process, mating assays between the single ΔTTC1879::kat mutant and the wild type produced transjugants resistant to kanamycin (Km) and Hyg at high frequencies (Fig 1, bar n° 1), illustrating that the ΔTTC1879::kat mutant can act as a recipient in transjugation. On the other hand, transjugation between a chloramphenicol (Cm)-resistant wild type strain and a ΔTTC1879::kat-ΔpilA4 double mutant carrying a multicopy plasmid (pMHTTC1879, Hygr) ectopically expressing the product of TTC1879, complemented the transjugation capability(Fig 1, bar n° 4). A control pilA4 mutant (wild type for TTC1879) carrying an empty Hygr plasmid had transjugation frequencies about one order of magnitude higher (Fig 1, bar n° 5). On the other hand efficient transjugation was detected between ΔTTC1879::kat and ΔpilA::hyg single mutants (Fig 1, bar n° 2). As two strains defective in competence cannot mate [16,19], this result confirmed that the product of TTC1879 was not involved in natural competence, and provides evidence that TTC1879 is required for DNA donation in transjugation, and not for DNA acquisition. The product of TTC1879 was thus designated the Transjugation Donor Translocase A (TdtA).
Fig 1. The product of TTC1879 (TdtA) is required for transjugation.
Transfer frequencies are expressed as the ratio of transjugant: wild type CFU. Bars 1 and 2 correspond to matings between a tdtA mutant (ΔtdtA) and a Hygr wild type strain (wtH) (1) or a ΔpilA4 competence mutant (Δpil) (2). Mating between the wtH strain and a double tdtA and ΔpilA4 mutant (ΔtdtA-ΔpilA) rendered no transjugants (3). The expression of TdtA from a plasmid in this double mutant (Δpil-ΔtdtAcomp) allowed for the generation of transjugants in matings with a Cmr wild type (wtCm) (4). Control matings between the same wtCm strain and a single ΔpilA4 mutant carrying the empty plasmid (Δpil/pMH) showed a 10-fold higher transjugation frequency (5). ANOVA tests showed significant differences among frequencies of transfer of all the derivatives plotted (p-value< 0.001) and post-hoc Holm Sidak tests proved that absence of tdtA has an effect on transjugation (n = 8). Asterisks indicate significant statistical differences compared to the wild type (*: p-value>0.05;**p-value<0.001).
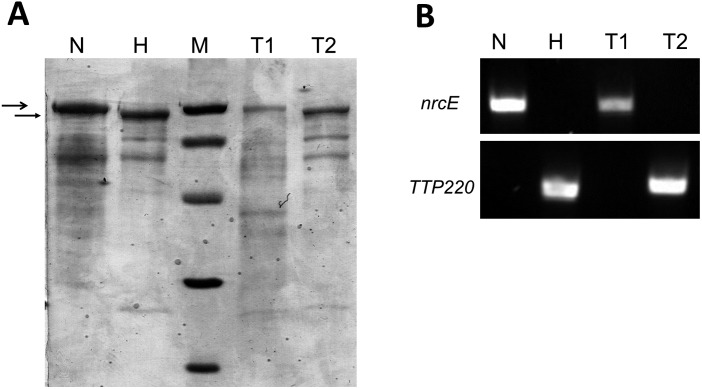
To further confirm that TdtA was required only for DNA donation in transjugation, PCR was used to screen different strains of T. thermophilus, finding a homolog with 100% sequence identity to TdtA in the nitrate-respiring NAR1 strain. This strain can be distinguished from HB27 both by PCR and by differences in the pattern of their membrane proteins, specifically size differences of S-layer proteins (SlpA) [20], which enables inference of the directionality of DNA transfer by parenthood analysis. The NAR1 and HB27 wild type strains transjugate efficiently in both directions [16]. To analyze the role of TdtA, a ΔtdtA::kat mutant was derived from NAR1 and used in transjugation assays with a Hygr derivative of the HB27 strain. The whole pool of transjugants from this mating experiment was harvested from the plates and analyzed to identify the parenthood. The membrane protein pattern of the transjugants was similar to that of NAR1, characterized by a 100 kDa SlpA protein (Fig 2A; lane N vs. lane T1), and PCR assays revealed the presence of the NAR1-specific nrcE gene but the absence of the HB27-specific TTP220 gene (Fig 2B; lane N vs. lane T1). These results show that the NAR1 ΔtdtA::kat mutant serves as a recipient and not as a donor in transjugation. Conversely, transjugants from a mating between an HB27-derived ΔtdtA::kat mutant and a Hygr NAR1 wild type strain possessed the smaller SlpA protein of HB27 (Fig 2A; lane H vs. lane T2), the HB27-specific TTP220 gene, and lacked nrcE (Fig 2B; lane H vs. lane T2), indicating that the HB27 ΔtdtA::kat mutant can function only as recipient in transjugation and not as donor. These experiments provide solid evidence that the absence of TdtA impairs the ability of the cell to act as a donor in transjugation, but does not affect its ability to serve as recipient.
Fig 2. TdtA is required in the donor strain for transjugation.
(A) SDS-PAGE electrophoresis showing membrane protein profiles. Patterns corresponding to the ΔtdtA::kat mutants derived from NAR1 (N) and HB27 (H) and the corresponding patterns after transjugation between NAR1-ΔtdtA and HB27-Wt (T1) and between HB27-ΔtdtA and NAR1-Wt (T2). Lane M shows protein molecular standards at 97.4, 66.2, 45, 31, and 21.5 kDa. Large and small arrowheads signal the S layer proteins of 100 kDa and 97 kDa corresponding to the NAR1 and HB27 strains, respectively, used as main strain identification marker. Note the similarities between lanes N and T1, and between lanes H and T2, supporting that transjugants derive from the respective tdtA mutant in the matings. (B) Agarose gel electrophoresis showing PCR amplicons of nrcE (upper panel), specific to the NAR1 strain, and ttp0220 (lower panel), specific to the HB27 strain using DNA extracted from ΔtdtA::kat mutants derived from NAR1 (N) and HB27 (H), the transjugants pool from mating experiments between NAR1-ΔtdtA and HB27-Wt (T1), and the transjugants pool of the reciprocal mating between HB27-ΔtdtA and NAR1-Wt (T2). Oligonucleotide sequences are shown in S2 Table.
TdtA is expressed from a 4-gene operon, and localizes to the cytoplasmic membrane
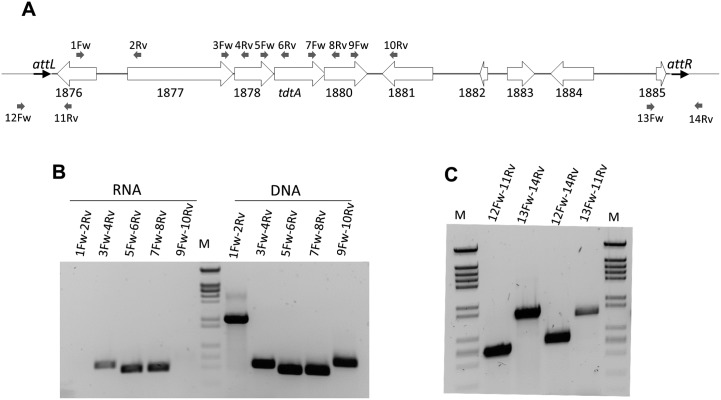
Analysis of tdtA’s position within the genome of T. thermophilus HB27 is presented in Fig 3A. Upstream of tdtA is a gene that encodes a putative nuclease of the NurA family (TTC1878; 377 aa), which are frequently found immediately upstream of proteins from the FtsK-HerA superfamily. Further upstream, TTC1877 encodes a large protein (1106 aa) that is identical to Tth111II, a type IIG restriction endonuclease previously characterized from another strain of T. thermophilus [21]. Downstream from tdtA, TTC1880 encodes a hypothetical 410 aa protein of the N4-N6 methylase family (pfam 01555).
Fig 3. TdtA is encoded within an active ICE-like element.
(A) Scheme showing the context of tdtA in the HB27 chromosome. White arrows represent ORFs encoded by the genes indicated underneath and are scaled proportionally to their size. The thick grey line is also proportional to the length of the whole ICEth1. Small grey arrows represent the relative position of primers employed in RT-PCR assays shown in panel B. (B) RT-PCR assays with the indicated primer pairs were conducted to amplify the intergenic regions between the genes TTC1877-1878-tdtA-1880. Parallel control PCRs performed on genomic DNA are shown at the right. (C) Agarose gel showing PCR amplification from genomic DNA of exponential cultures of T. thermophilus HB27 with the indicated primers to detect the ends of the ICEth1 in its integrated form (12Fw-11Rv and 13Fw-14Rv), the DNA scar produced by it excision (12Fw-14Rv), and the excised circular form (13Fw-11Rv).
These four genes are separated by 4 (TTC1877-1878), 2 (TTC1878-tdtA), and -4 (tdtA-TTC1880, overlapping) bp, and all other nearby genes are encoded in the opposite orientation. Therefore, these four genes are likely co-transcribed from a promoter upstream of TTC1877. In fact, RT-PCRs spanning each intergenic region between TTC1877 and TTC1880 produced successful amplification from total mRNA of T. thermophilus HB27, whereas amplification failed using primers bridging the TTC1876-1877 and TTC1880-1881 intergenic regions (Fig 3B), thus confirming co-transcription of a four-gene operon.
To analyze the expression of the operon throughout the growth cycle, the TdtA protein was tagged with a thermostable fluorescent protein (TdtA-sYFP), allowing its detection by western blot. TdtA was detected at similar levels from the exponential through the stationary phase of growth (Fig 4A).
Fig 4. Expression and subcellular localization of TdtA.
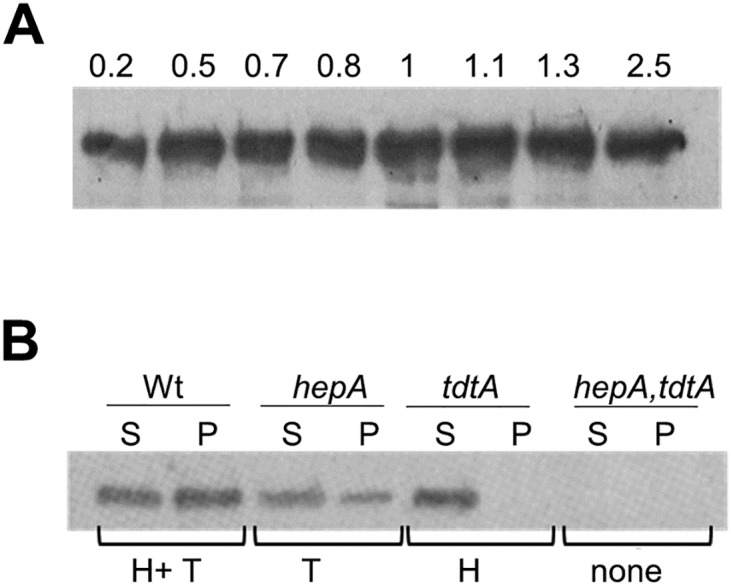
(A) The expression of the TdtA-YFP fusion from its native promoter in the chromosome was followed by western blot with anti-GFP antiserum throughout growth at 60°C. Identical cell mass was analyzed at the indicated optical densities at 550 nm (B) Western blot with an antiserum that cross-reacts with both TdtA and a HerA-like protein (product of TTC0147 baptized as HepA) was used to localize the proteins in soluble (S) and non-soluble (P) fractions from the following strains of T. thermophilus HB27: wild type (Wt), ΔhepA::kat (hepA), ΔtdtA::kat (tdtA) and ΔtdtA::kat, hepA::hyg (hepA,tdtA). The proteins detected in each case are indicated underneath: TdtA (T) and HepA (H).
The role of TdtA in pushing DNA out of the cells could imply a connection to the membrane. To test this, a rabbit antiserum that cross-reacts with both TdtA and the product of TTC0147 (a putative HerA-like helicase) was employed. Proteins of the expected size (approximately 64 kDa for both proteins) were detected in both the soluble (S) and the cell envelope insoluble (P) fractions of the wild type strain, whereas no signal was detected in a ΔtdtA-TTC0147 double-deletion mutant (Fig 4B). In the TTC0147 single-deletion mutant, TdtA was detected in both the S and P fractions. In contrast, in a ΔtdtA context the TTC0147 product was detected only in the S fraction. Keeping in mind that part of the TdtA protein is found in the soluble fraction, and that its primary sequence does not contain a putative secretion signal, the most likely location for the TdtA protein is the cytoplasmic membrane.
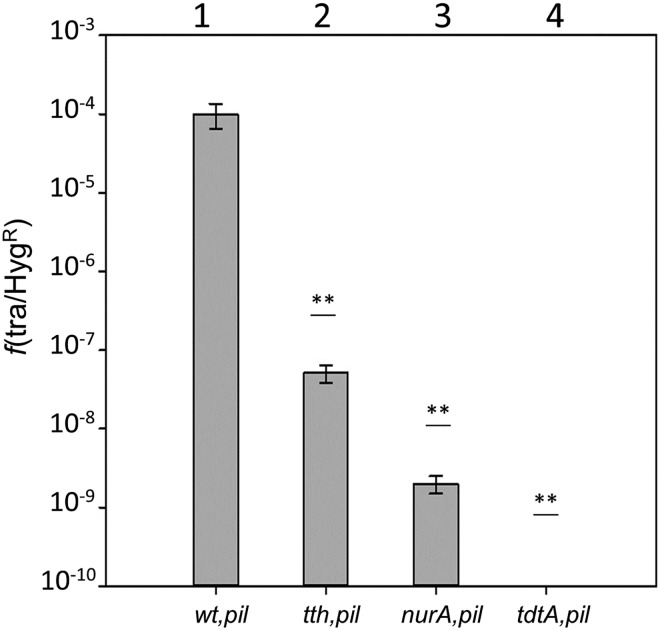
The NurA-homolog and the Tth111II restriction enzyme encoded by the tdtA operon are required for efficient transjugation
The putative role in transjugation of the NurA-like protein and Tth111II restriction enzyme was analyzed using the same method as done previously for TdtA. In essence, ΔpilA4 -TTC1877::kat and ΔpilA4 -TTC1878::kat double mutants were used in mating experiments with a Hygr wild type. The results show that the strain lacking TTC1878 (nurA-like) suffered a decrease in transjugation efficiency of 4–5 orders of magnitude with respect to a ΔpilA4 control strain labeled at another position in the chromosome (Fig 5, bars 1 vs. 2), whereas the mutant without the Tth111II restriction enzyme showed a decrease of around 3 orders of magnitude (Fig 5, bars 1 vs. 3). As expected, a ΔpilA4-tdtA::kat control rendered no detectable transjugants in mates with the wild type (Fig 5, bar 4). These data support that both genes somehow cooperate with TdtA to enhance the transjugation process.
Fig 5. Effects of the NurA-homolog and Tth111II on transjugation.
The frequencies of transjugation assays between pilA mutants labeled with kanamycin at the gdh (wt,pil) locus (1), or at the genes encoding Tth111II (tth,pil) (2), NurA-like (nurA,pil) (3) or TdtA (tdtA,pilA) (4) and a wild type strain labeled with Hygr are shown. Asterisks indicate significant statistical differences compared to the wild type (p-value<0.001) (n = 6).
TdtA is encoded within a new ICE
Upstream of the tdtA operon, the TTC1876 gene encodes a homolog to phage and ICE integrases of the XerC family (392 aa), while a putative DDE transposase (TTC1881, 542 aa) was found downstream. With the exception of this transposase (of which two more chromosomal copies exist), the whole region encompassing tdtA has a lower G+C content (58%) than the average of its genome (68%). Actually, the limits of this low G+C content region were defined by two 47-bp direct repeats (labeled attL and attR in Fig 3A) that are identical in sequence to the 3′ end of an isoleucine tRNA. The whole 14,857 bp region is absent from the closely related strain T. thermophilus HB8.
As these properties are common to ICEs, the region was designated ICEth1. To determine whether ICEth1 was able to excise from the chromosome, PCR assays were used (Fig 3C); primer pairs 12Fw/11Rv and 13Fw/14Rv allowed amplification of the ends of ICEth1 in its integrated form (amplicons of 566 and 1,097 bp for the left and right sides, respectively), whereas primers 12Fw/14Rv produce a 646 bp amplicon only from genomes in which the ICEth1 had been excised, leaving the corresponding scar behind. Both the left and right arms of the integrated form and the scar were detected by PCR, showing that the ICEth1 had excised from the chromosome in a fraction of the cells in the culture. In addition, primers 13Fw/11Rv detected a 1,017 bp amplicon corresponding to the circularized form of ICEth1, albeit present at lower levels compared to the attB scar produced by its excision.
To quantify the frequency of this excision during growth in rich medium, the percentage loss of the Kmr phenotype was compared between the ΔtdtA::kat mutant and a non-excisable Kmr chromosomal marker (Δgdh::kat). Up to 8% of the ΔtdtA::kat colonies from exponential cultures (OD600 = 0.6) had lost the Kmr marker, whereas no loss was detected in the Δgdh::kat mutant under the same experimental conditions (p-value < 0.001). These data demonstrate that ICEth1 excises from the chromosome at high frequencies during exponential growth, in a TdtA-independent manner.
The presence of the ICEth1 enhances transjugation dramatically
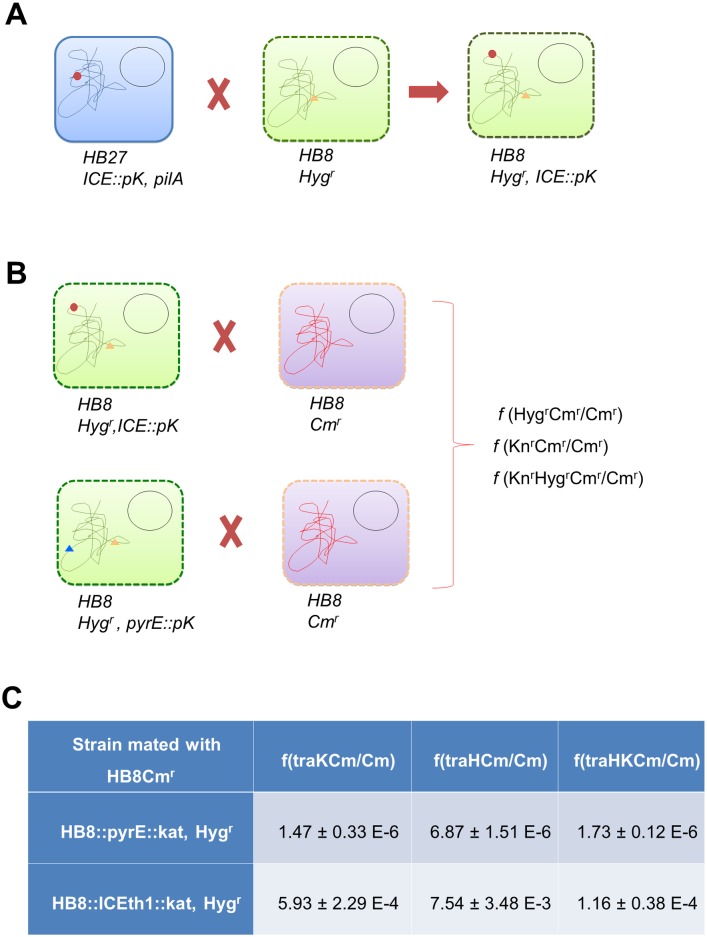
To establish a clear relation between the presence of ICEth1 and transjugation capability in another genetic background, the experimental scheme shown in Fig 6 was followed. First, the ICEth1 was labeled with Kmr in a non-coding region, downstream of the tdtA operon in a HB27 ΔpilA4 background to prevent this strain from serving as a recipient in transjugation. Then, mating experiments were carried out between this strain and a Hygr derivative of the HB8 strain, which does not contain the ICEth1, but does contain its attB site (Fig 6A). The resultant transjugants (HB8::ICEth1::kat, Hygr) were confirmed by PCR. Finally, seven different HB8::ICEth1::kat, Hygr clones from the above mating were assayed in transjugation experiments with a Cmr derivative of HB8 serving as recipient (Fig 6B). As a donor control, we used an HB8 strain (lacking ICEth1) with the same Hygr marker and also a Kmr marker, located at another gene in the chromosome (pyrE). Transjugation efficiencies were measured for the Kmr and Hygr markers with respect to the number of Cmr recipients. In the absence of ICEth1, cells with a combination of resistances from both mates (Hyg or Km and Cm) were detected at frequencies ranging from 1.5–6.9 x 10−6 (Fig 6C), supporting that alternative horizontal gene transfer (HGT) mechanisms are acting on this strain. Such an alternative HGT mechanism has been already described, involving DNAse-resistant vesicle-protected DNA [22], and may be the most likely explanation for the observed transfer frequencies in the absence of ICEth1. In any case, presence of ICETh1 in the HB8 strain produced a 2.5 order of magnitude increase in the transfer of the Kmr marker (ICEth1) compared to the control strain (pyrE::kat) lacking the ICEth1. Moreover, the presence of the ICEth1 produced an even more dramatic increase (3 orders of magnitude) in the transfer efficiency of the chromosomal Hygr marker relative to the ICEth1-deficient control strain.
Fig 6. Transjugation is associated with presence of ICEth1.
Scheme of the experimental design followed to unequivocally associate ICEth1 and transjugation efficiency. (A) ICEth1 (red dot) was labeled with Kmr (ICEth1::pK) in a HB27ΔpilA4 background and transjugated into an HB8 strain labeled with Hygr (orange triangle), which naturally lacks this element. (B) The HB8 containing ICEth1 (Hygr, Kmr) was mated with a Cmr derivative of HB8, using a Hygr, pyrE::pK (blue triangle) strain as a control. (C) Transfer frequencies detected for the matings described in B above. Frequencies of the ICEth1-containing strain are the average value from 7 donor clones in three independent experiments. A similar number of assays were carried out for the strain lacking ICEth1.
TdtA assembles into ring-shaped hexamers with ATPase activity
Sequence analysis revealed the presence of putative Walker A and B ATPase motifs in TdtA (S1 Fig). To test whether the protein actually had ATPase activity, we overexpressed and purified a His-tagged TdtA protein in E. coli (S2 Fig). Subsequently, ATPase assays were carried out at 65°C with the purified protein, revealing efficient ATP consumption that correlated with increasing TdtA concentration (S2 Fig). Presence of protein contaminants with ATPase activity or DNA from the E. coli overproducer cells was unlikely due to the harsh conditions used for purification including urea denaturation and IMAC-bound renaturation. Moreover as the ATPase assays were carried out at 65°C the putative activity of any E. coli enzyme is unlikely. As the addition of genomic dsDNA from T. thermophilus (its likely natural substrate) to these assays produced only a moderate increase in the activity (S2 Fig), the putative dependence on nucleic acids of the ATPase activity of TdtA constitutes a possibility that will require more detailed experimentation.
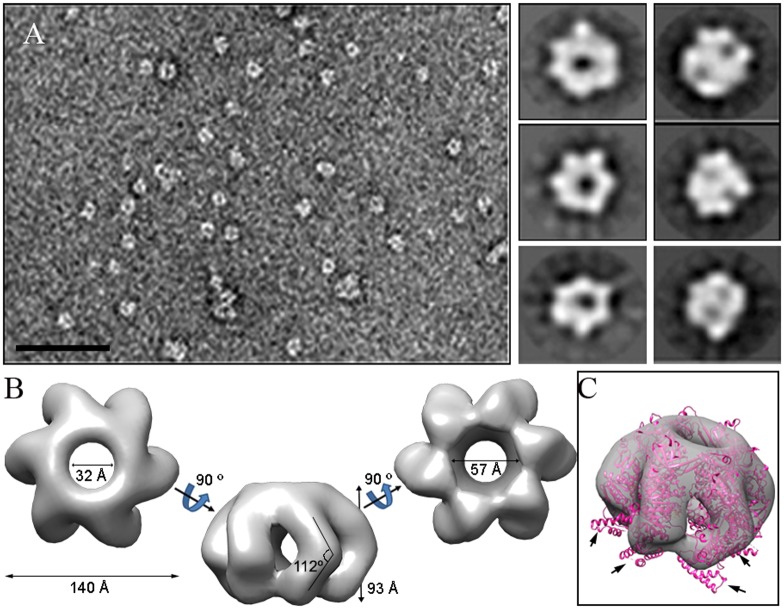
Regular ring-shaped TdtA assemblies were detected by negative stain electron microscopy after incubation at 65°C with ATP (Fig 7A, left). In the absence of ATP or without incubation at 65°C, no such complexes were detected. Two-dimensional averaged images exhibited six-fold symmetry (Fig 7A, right), and 103,550 images were used to generate a three-dimensional reconstruction of TdtA, with a final resolution for the model estimated to be 16 Å. Oligomeric TdtA was observed to be formed by six sausage-like structures connected at their ends, with each elongated structure slightly bent at the equatorial plane (Fig 7B). The top and bottom views of this hexameric ring show a star-shaped morphology with a maximum external diameter of 140 Å and a channel with a diameter of 32–57 Å at its ends, wide enough to allow dsDNA passage.
Fig 7. TdtA single-particle electron microscopy reconstruction.
(A) Representative electron micrograph of a negatively stained TdtA sample; bar = 50 nm. Six two-dimensional averaged classes of the oligomeric TdtA are shown (right). (B) Three-dimensional reconstruction of the hexameric TdtA. (C) Semitransparent model of the hexameric TdtA with the fitted atomic model of the hexameric HerA from S. solfataricus (pink). Arrows indicate the HerA region (residues 216–289), which remains outside the TdtA ring.
We compared the TdtA model with structural homologs such as the hexameric HerA map from Sulfolobus solfataricus [23], which has similar protein monomer sizes. The HerA (Protein Data Bank (PDB) ID 4D2I) model fits quite well within the TdtA model except for a four-helix bundle (HerA residues 216–289) that remained outside (Fig 7C, arrows). These results suggest that a region of TdtA is not visible, due either to lack of contrast agent or to structural disorganization.
In conclusion, our data show TdtA to be a hexameric ATPase with an internal channel whose diameter is sufficient to accommodate dsDNA [23].
Discussion
Cell-to-cell contact-dependent DNA transfer in T. thermophilus relies not only on the competence machinery of recipient cells to take up DNA [16], but also on a transformation-independent process for donor cells to push DNA out DNA. This “push-pull” model is termed “transjugation”, an intermediate process between classical conjugation and transformation. An in silico search for putative DNA translocases involved in DNA donation pointed to orphan members of the FtsK-HerA protein family, of which four were identified in the genome of T. thermophilus HB27. One of these (TTC0474; UniProtKB-Q72K53) was essential, consistent with a role as the actual FtsK division protein, whereas the proteins encoded by TTC0147 and TTC1430 seemed to be somehow involved in DNA repair, as corresponding knockouts were much more sensitive to UV light and/or heat shock treatments and also affecting transformation. Actually, the TTC0147 product has greater similarity to the HerA helicase of Sulfolobus solfataricus, described as a component of the NurA-HerA DNA-repair system [24], and also to the Deinococcus radiodurans HerA protein [25], which is in the same phylum as Thermus spp. In contrast, mutants in TTC1879 showed similar sensitivity to stress and transformation capability compared to the wild type, but a complete inability to act as a donor in transjugation experiments (Fig 1), leading us to rename its product as TdtA. In the following subsections we discuss these results.
The role of TdtA
Mutants lacking TdtA (push-less) that were also defective in the transformation apparatus (pull-less) could not participate in transjugation with wild type strains (Fig 1), as predicted by the transjugation model [16]. Further characterization of recombinant TdtA demonstrated its ATPase activity as well as the hexameric structure common to FtsK-like DNA translocases and coupling conjugation proteins. In this context, HerA helicases involved in DNA repair have ATPase activity only in the presence of DNA [26,27]. In contrast, TdtA purified from inclusion bodies and further solubilized in urea and renatured after binding to an IMAC support (thus much likely without DNA) showed a strong in vitro ATPase activity in the absence of DNA, with, addition of dsDNA producing only a slight increase in the activity (S2 Fig). However, the risk of cell damage from uncontrolled ATPases such as TdtA makes more likely the existence of some class of regulation, either by timed synthesis or through posttranslational means. Considering that TdtA is present along all the culture phases some class of posttranslational control of the activity seems more likely.
The role of TdtA in transjugation could parallel that described for TraB, the hexameric ATPase required for plasmid-encoded conjugation in Streptomyces spp. and other Actinomycetes [9,14] or that of the TcpA protein involved in conjugal transfer of the pCW3 plasmid in Clostridium spp. [28]. However, whereas conjugation in Clostridium depends on a T4SS-like system with TcpA serving as a coupling protein, TraB functions independently, allowing for transfer of traB-containing plasmids as small as 10 kbp [15], and also of chromosomal genes, although with lower efficiency. Interestingly, TraB transfers dsDNA [29], in contrast to the ssDNA-transfer mediated by classical T4SS-based conjugation systems. To accommodate dsDNA, the TraB hexamer forms a 31 Å central pore [9], similar to the 32 Å central pore predicted by our electron microscopy reconstruction of the TdtA hexamer (Fig 7). Moreover, recent X-ray analysis revealed that contrary to previous reports [30], the HerA protein from Sulfolobus solfataricus used for the TdtA reconstruction in Fig 7 can also accommodate and push dsDNA [23]. Therefore, TdtA of T. thermophilus most likely pumps dsDNA. However, the fact that the NurA-like ssDNA-nuclease encoded upstream of tdtA clearly plays a role in transjugation (Fig 5) suggests that nucleolytic degradation of dsDNA is likely required in the process. As transformation in T. thermophilus is highly efficient regardless of the origin of the dsDNA [31], and transjugation depends on the competence apparatus of the recipient cell, it is likely that the actual extruded substrate through the TdtA protein is dsDNA. In this scenario, NurA could be involved either in trimming of the dsDNA substrate as proposed for the NurA protein of Sulfolobus spp. [23] or in repairing the targeted DNA in the chromosome of the donor.
A model for transjugation
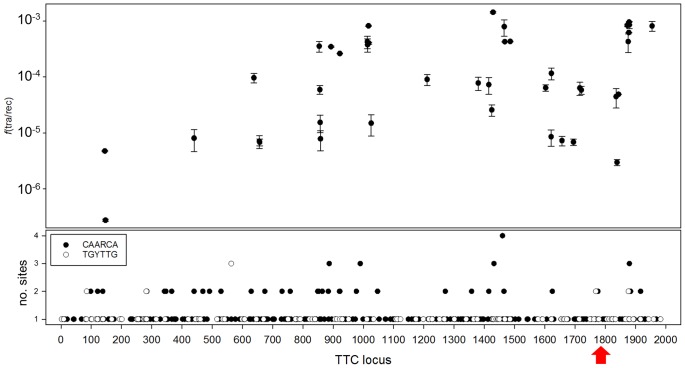
The TraB protein of Streptomyces spp. recognizes 8-bp repeats through its C-terminal HTH motif [14] at the clt, and starts dsDNA transfer to the recipient strains. Compared to TraB, TdtA is smaller with no putative DNA binding domains detected by sequence analysis, thus the recognition of any hypothetical oriT would likely depend on other proteins. The best candidate for this role is the endonuclease Tth111II [21], whose deletion produced a 1,000-fold decrease in transjugation efficiency. This type IIG restriction enzyme has a rather low specific activity and preferentially nicks the top strand at position N11 downstream from its CAARCA recognition sequence. Interestingly, homologs of the same family of restriction endonucleases are also found in the tdtA-like gene clusters of T. scotoductus DSM8553 (WP_020700619.1, 50% identity), and Chloroacidobacterium thermophilus B (WP_014099240.1, 66% identity). Thus, it is tempting to speculate about the role of these restriction enzymes in the nicking of DNA to signal the origin of transfer sites during transjugation in Thermus spp or in conjugation-like processes in other bacteria. In this context, it is of note the existence of at least a conjugative relaxase described that contains a C-terminal domain with structural similarity to restriction endonucleases [32], suggesting some connection of these two classes of enzymes. As the transjugation process in T. thermophilus seems to start simultaneously at several points in the chromosome [16], and having in mind the ancestral character of the genus Thermus spp, it is tempting to speculate on primitive restriction-like enzymes acting in HGT processes. Actually, a map of transfer frequencies for 42 genes against their chromosome locations shows greater transfer of genes located in regions with more Tth111II recognition sites, whereas less-frequently transferred genes are found in regions with fewer recognition sites (Fig 8). Therefore, a tentative but preliminary hypothesis involves Tth111II as the enzyme that recognizes multiple origin of transfer for the concerted action of a likely NurA-TdtA complex.
Fig 8. Distribution of Tth111II recognition sites and transjugation efficiencies in the HB27 chromosome.
The bottom panel shows the gene locus linear map representation of the HB27 chromosome (TTC locus 1 to 1988) and the number of Tth111II recognition sites per gene found on the top (black circles) or bottom strand (empty circles). The upper panel shows the transjugation frequencies for 42 chromosomal genes (TTC::kat mutants described in Table 2) represented at their corresponding position in the linear map of the chromosome (n = 3). Note how the transjugation frequencies are higher in regions with greater presence of Tth111II recognition sites. The red arrow indicates the position of ICEth1.
TdtA is encoded by a new class of ICE
In a bioinformatics study searching for mobility of R-M systems, Furuta and co-workers identified the TTC1877 (encoding Tth111II) and TTC1880 (encoding a putative methylase) genes of T. thermophilus HB27 as part of an integrated element that was absent in the HB8 strain [33]. Our data support that the 14,857 bp region located between 47-bp direct repeats (i.e., attL and attR) actually constitutes the first active ICE described for T. thermophilus (ICEth1). However, our studies on ICEth1 excision by PCR and frequency of loss during exponential growth (up to 8%) suggested that the ICEth1 does not replicate after excision from the chromosome like other ICEs do [34]. Therefore, for the ICEth1 to be maintained in the population, a continuous mobilization and invasion process must take place in order to spread to those cells that had lost the element during exponential growth.
The ability of ICEth1 to spread to ICE-less strains was shown using the HB8 as recipient. In our experiments, the new ICEth1+ HB8 behaves as a donor of the inherited ICEth1, as expected for any ICE. However, in contrast to most ICEs, the ICEth1 promotes the efficient transfer of genes from other locations in the genome, in what seems to be a sort of generalized transfer that mimics the well-known generalized transduction in classical bacterial HGT processes. Thus, in the absence of selectable properties encoded by ICEth1, it is tempting to speculate that the selection of favorable phenotypic traits (i.e., anaerobic respiration, heavy metal resistance, UV resistance, etc.) under given environmental conditions would also favor the co-selection and spreading of ICEth1 itself throughout a population of recipient strains. In this context, the noteworthy ability of ICEth1-mediated transjugation to escape the Argonaute surveillance system, which is an effective barrier against environmental [16] and vesicle protected DNA [22], enhances its relevance in intraspecies HGT and facilitates its own spreading.
Methods
Bacterial strains and growth conditions
Table 2 summarizes the bacterial strains employed in this study. Escherichia coli strains DH5α (for plasmid construction) and BL21 (DE3) (for protein overexpression and purification) were grown at 37°C in LB (Luria-Bertani) selective media. Aerobic growth of T. thermophilus strains was carried out with rotational shaking (150 rpm) at 60°C in TB (Thermus broth) liquid or solid media, unless otherwise indicated. Selection with Km (30 μg/ml), ampicillin (Am, 100 μg/ml), Cm (20 μg/ml) and/or Hyg (100 μg/ml) was employed as required.
Table 2. Bacterial strains used in this work.
| Strain | Genotype | Phenotype/use | Source |
|---|---|---|---|
| E. coli DH5α | supE44 ΔlacU169 (Φ80 lacZΔM15) hsdR17, recA1, endA1, gyrA96, thi-1 relA1 | Ordinary cloning | [35] |
| E. coli BL21 (DE3) | F- ompT gal dcm lon HsdSB (rB-mB-) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | Protein overexpression and purification | [36] |
| T. thermophilus strains | |||
| HB27wt | ATCC BAA-163/DSM7039 | Wt | Y. Koyama |
| NAR1 | [pTT27::nar] | wt. NCE, partial denitrifer | [37] |
| HB8 | ATCC 27634 | ICEth1-less | Y. Koyama |
| HB27EC | HB27 ago::agoISTth7 | enhanced competence | This work |
| WtCm | HB27 Cmr | This work | |
| Δago | HB27 Δago | Argonaute less | [38] |
| ΔpilA4 | HB27EC ΔpilA4 | Non competent | [16] |
| ΔpilA4::hyg | HB27EC ΔTTC0858::hyg | Non competent | [16] |
| ΔtdtA | HB27EC ΔTTC1879 | Deficient in DNA donation | This work |
| tdtA::pk | HB27ECTTC1879::pK18 | Kmr Deficient in DNA donation. | This work |
| ΔtdtA::kat | HB27EC ΔTTC1879::kat | Kmr. Deficient in DNA donation. | This work |
| ΔtdtA::hyg | HB27EC ΔTTC1879::hyg, | Hygr. Impaired in DNA donation. | This work |
| tdtA::kat pilA4 | HB27EC ΔTTC1879::kat, ΔpilA4 | Kmr. Deficient in DNA donation and transformation. | This work |
| WtH | HB27ECΔTTC0313::hyg | Hygr | [16] |
| Nar tdtA::kat | NAR1 [ΔTTC1879::kat] | Nitrate respiring. Kmr | This work |
| Nar TTC::hyg | NAR1 [ΔTTC0313::hyg] | Nitrate respiring. Hygr | [16] |
| gdh::kat | HB27 [ΔTTC1211::kat] | Kmr | [37] |
| HB8 hyg | HB8 [ΔTTC0313::hyg] | ICEth1-less. Hygr | This work |
| HB8 Cmr | HB8 spontaneous chloramphenicol resistant | ICEth1-less. Cmr | This work |
| HB8::ICEth1::kat, Hygr | HB8 [ΔTTC0313::hyg], ICEth1::pK18 | ICEth1, Kmr, Hygr | This work |
| tdtAYFPph | HB27EC [TTC1879-sYFP:pH118] | Hygr. Single copy expression of TdtA fused to sYFP | This work |
| ΔtdtA, ΔpilA4 PMH | HB27EC ΔTTC1879::kat, ΔpilA4 [pMH::TTC1879] | Complementation of TdtA | This work |
| ΔtdtA, ΔpilA4 PMHY | HB27EC ΔTTC1879::kat, ΔpilA4 [pMH::TTC1879::sYFP] | Complementation of TdtA with TdtA-sYFP | This work |
| ICEth13’kat | HB27 TTC1880-TTC1881::pK18 | Intergenic tag (TTC1880-TTC1881) of kanamycin marker. | This work |
| ΔTh111II::kat, ΔpilA4 | HB27EC ΔTTC1877::kat | Kmr. Impaired in DNA donation and transformation | This work |
| ΔnurA::kat, ΔpilA4 | HB27EC ΔTTC1878::kat | Kmr. Impaired in DNA donation and transformation | This work |
| ΔTTC0147::kat | HB27EC ΔTTC0147::kat | Kmr. Affected in stress survival | This work |
| ΔTTC1430::kat | HB27EC ΔTTC1430::kat | Kmr. Affected in stress survival | This work |
| ΔTTC1844::kat | HB27EC ΔTTC1844::kat | Kmr. Affected in stress survival and transformation | [16] |
| Δgdh::kat | HB27 ΔTTC1211::kat | Kmr | [16] |
| ΔpilA4::kat | HB27 ΔTTC0858::kat | Kmr. Impaired in transformation | [16] |
| TTC1017::kat | HB27 ΔTTC1017::kat | Kmr. Impaired in transformation | [16] |
| TTC1622:kat | HB27 ΔTTC1622::kat | Kmr. Impaired in transformation | [39] |
| TTC854::kat | HB27 ΔTTC0854::kat | Kmr | [40] |
| TTC1621::kat | HB27 ΔTTC1621::kat | Kmr | [39] |
| TTC857::kat | HB27 ΔTTC0857::kat | Kmr | [40] |
| TTC0856::kat | HB27 ΔTTC0856::kat | Kmr | [40] |
| TTC1716::kat | HB27 ΔTTC1716::kat | Kmr | [40] |
| TTTC1603::kat | HB27 ΔTTC1603::kat | Kmr | [39] |
| TTC1415::kat | HB27 ΔTTC1415::kat | Kmr | [16] |
| TTC0638::kat | HB27 ΔTTC0638::kat | Kmr | [16] |
| TTC0893::kat | HB27 ΔTTC0893::kat | Kmr | [16] |
| TTC1425::kat | HB27 ΔTTC1425::kat | Kmr | This work |
| TTC1429::kat | HB27 ΔTTC1429::kat | Kmr | This work |
| TTC0474::kat | HB27 ΔTTC0474::kat | Kmr | This work |
| TTC0656::kat | HB27 ΔTTC0656::kat | Kmr | This work |
| TTC1026::kat | HB27 ΔTTC1026::kat | Kmr | This work |
| TTC1878::kat | HB27 ΔTTC1878::kat | Kmr | This work |
| TTC1877::kat | HB27 ΔTTC1877::kat | Kmr | This work |
| TTC1380::kat | HB27 ΔTTC1380::kat | Kmr | This work |
| TTC1013::kat | HB27 ΔTTC1013::kat | Kmr | [40] |
| TTC1873::kat | HB27 ΔTTC1873::kat | Kmr | This work |
| TTC440::kat | HB27 ΔTTC0440::kat | Kmr | [40] |
| TTC1695::kat | HB27 ΔTTC1695::kat | Kmr | This work |
| TTC1468::kat | HB27 ΔTTC1468::kat | Kmr | This work |
| TTC1839::kat | HB27 ΔTTC1839::kat | Kmr | This work |
| TTC1836::kat | HB27 ΔTTC1836::kat | Kmr | This work |
| TTC1876::kat | HB27 ΔTTC1876::kat | Kmr | This work |
| TTC1466::kat | HB27 ΔTTC1466::kat | Kmr | This work |
| TTC922::kat | HB27 ΔTTC0922::kat | Kmr | [16] |
| TTC1955::kat | HB27 ΔTTC1955::kat | Kmr | This work |
| TTC1721::kat | HB27 ΔTTC1721::kat | Kmr | This work |
| TTC1486::kat | HB27 ΔTTC1486::kat | Kmr | This work |
| TTC1657::kat | HB27 ΔTTC1657::kat | Kmr | This work |
| TTC657::kat | HB27 ΔTTC0657::kat | Kmr | This work |
| TTC1880::kat | HB27 ΔTTC1880::kat | Kmr | This work |
| TTC145::kat | HB27 ΔTTC0145::kat | Kmr | This work |
| TTC1014::kat | HB27 ΔTTC1014::kat | Kmr | [40] |
| TTC1018::kat | HB27 ΔTTC1018::kat | Kmr | [40] |
| TTC1844::kat | HB27 ΔTTC1844::kat | Kmr | [16] |
Generation of bacterial mutants
The plasmids employed in this work are listed in S1 Table. For initial screening of gene functionality, single-insertion mutants were obtained by recombination with central regions of the target gene cloned in suicide vectors pK18 or pH118, encoding thermostable resistance to Km or Hyg, respectively. For further genetic analysis, deletion derivatives were generated by double recombination between specific upstream and downstream target arms separated by the kat or the hyg genes (encoding resistance to Km and Hyg, respectively). For marker-less mutants, these Δgene::cassette mutants were transformed again with a similar upstream-downstream construct and cloned into a suicide vector carrying a different thermostable resistance gene. After isolation of single recombinants with both antibiotic resistances, spontaneous loss of both markers by back recombination allowed the isolation of marker-less Δgene mutants. All mutants were confirmed by PCR and sequencing. Spontaneous Cmr strains were obtained when necessary. The isolation of mutants in non-competent ΔpilA4 cells required electroporation.
Isolation of strains expressing fusions to thermostable fluorescent proteins was performed by fusing either the whole gene or its C-terminal region to superfolder yellow fluorescent protein (sYFP, [41]). Fusions of C-terminal regions to sYFP were cloned into suicide plasmid pK18, and subsequently transformed into T. thermophilus. Transformants were checked by fluorescence microscopy, PCR and sequencing.
Complementation of mutants was carried out using derivatives of plasmid pMH184 (Hygr), which constitutively expressed the corresponding gene at moderate levels.
Transformation and transjugation assays
Quantitative transformation assays were performed as described [16,42] using exponential cultures grown at 65°C and 10 to 200 ng of genomic or plasmid DNA. Transformation frequencies were estimated by the number of CFU on selective plates per viable cell.
Mating experiments were conducted as described by Blesa et al [16]. Briefly, 100 μl of saturated cultures of Hygr and Kmr strains were mixed in the presence of DNase I (5 units; Roche), washed in 1 volume of TB, resuspended in 10 μl of TB containing DNase I (5 units; Roche) and plated onto sterile 0.22 μm nitrocellulose filters (GSWP, Millipore) on TB agar plates prior to incubation for 4 h at 60°C. Cells on the filters were then resuspended in TB and appropriate serial dilutions were plated onto selective agar plates. Transjugation frequencies were expressed as the number of CFU grown in selective media (Hyg plus Km, and in some experiments, Cm) per CFU grown on agar plates with Hyg or Cm.
Statistical analysis of the transfer frequencies was performed using SPSS ®Statistics v.21.0 (SPSS Inc., Chicago, USA; 2008), considered statistically significant when p–value < 0.05. Inferential and comparative assays were performed when necessary and included Student’s t-tests, Kruskal-Wallis one-way analysis, Wilcoxon tests and one-way ANOVA tests.
The parenthood analysis of transjugants in Fig 2 was carried out by collecting in a single pool all the colonies grown on the selection plates of the assays described above (>103 transjugants). Aliquots of these pools were used for genomic DNA isolation and for membrane protein profile analysis. PCR assays specific for the NAR1 and HB27 strains were carried out using primer sets nrcE_fw/nrcE_rev and TTP0220_fw/TTP0220_rev, respectively (S2 Table). Membrane isolation was done after breaking the cells by sonication (4°C, 3 pulses of 1 min with 0.5 s cycles, 0.9 intensity, Labsonic, Braun), elimination of unbroken cells by low speed centrifugation (5,000 x g 5 min) and further recovering of membrane insoluble fraction by higher speed centrifugation (15,000 x g for 20 min). Membrane proteins were analyzed by PAGE and gene staining as described below.
Stress assays
Resistance to UV radiation and heat shock were measured as the ratio of viable cells after 60 min of UV light exposure (Osram Sylvania G8T5, λ = 254 nm) and 60 min incubation at 90°C, and expressed as the ratio with respect to untreated controls. The strain used as wild type in these analyses was the Δgdh::kat mutant, to control for any role of Km resistance in the assays. Statistical significance of differences between mutants and wild type both for UV and “heat shock” assays was checked as above.
Recombinant protein expression and purification
The tdtA gene was cloned into pET28-b and electroporated into BL21(DE3) E. coli cells. Cells were grown in LB with Km at 30°C to an OD600 = 0.6, at which point 0.4 mM IPTG was added and incubated for 12 h at 24°C. Cells were harvested by centrifugation (23,700 x g for 20 min at 4°C) and 0.4 g of wet weight cells were resuspended in 30 ml of buffer A (25 mM sodium phosphate buffer pH 8, 100 mM NaCl and 0.1% Tween 20). Lysis was accomplished by sonication (3 pulses for 1 min, 0.8 power, 0.9 intensity, 0.5 s of cycle frequency, in a Labsonic U, B. Braun device), and cell debris was removed by centrifugation (23,700 x g for 20 min at 4°C). The insoluble fraction, containing inclusion bodies of the His-tagged TdtA protein, was washed in 7 ml of buffer A and further incubated for 30 min at 30°C in 4 ml of the same buffer with Triton 0.2% (w/v). After detergent removal by centrifugation (23,700 x g for 15 min at 4°C) the pellet was washed again with 1.5 ml buffer A and centrifuged as before. In order to solubilize aggregated proteins, the pellet was resuspended in buffer A containing 6 M urea (Merck) and incubated for 12 h at 4°C. After centrifugation to remove contaminants, the supernatant was loaded onto a pre-equilibrated Cobalt TALON resin (ClonTech Laboratories, Inc.). Renaturation by step-down urea gradient was performed and the final 2 M urea fraction eluted from the column with imidazole was dialyzed against 25 mM phosphate buffer pH 8.0 with 100 mM NaCl and loaded again onto pre-equilibrated Cobalt TALON resin. Proteins were eluted with 200 mM imidazole, dialyzed against the above buffer and stored at -20 and 4°C with 50% and 10% glycerol, respectively. Glycerol-free purified protein fractions were used directly in electron microscopy and ATPase assays.
Single-particle electron microscopy and image processing
Samples of purified TdtA (2–5 μl at 1.5 mg/ml in 25 mM Tris- HCl pH 7.5), previously incubated with 1–10 mM ATP (30 min at 65°C with shaking), were applied to glow-discharged carbon-coated grids for 2 min. Grids were washed twice with water and negatively stained with 2% (w/v) aqueous uranyl acetate. Electron microscopy images were recorded with a CCD camera (4k x 4k TEMCam-F416, TVIPS) on a JEOL 1010 JEM electron microscope operating at 80 kV. Images were recorded at a sampling rate of 2.44 Å/pixel, with an under focus ranging from 0.7 to 1.5 μm.
General image processing operations were performed using Xmipp software [43], and graphics were produced by UCSF Chimera [44]. The contrast transfer function (CTF) was corrected with Ctffind3 [45] and images were down-sampled to a factor of 2 (final sampling ratio 4.88 Å/pixel). The Xmipp automatic picking routine was used to select 103,745 TdtA particles which were classified using a reference-free clustering approach with the CL2D program [46] to select a homogeneous population of 103,550 particles. An artificial noise model was used as a starting reference for iterative angular refinement using the EMAN program [47]. The resulting model was selected and refined using the Xmipp iterative projection matching routine [48]. After independent refinement processes, 90% of particles were included in the final three-dimensional reconstruction and the resolution of the model was determined by the Fourier shell correlation (FSC) criterion between independent half-dataset maps applying a correlation limit of 0.5.
The CHIMERA fitting routine was used to dock the HerA atomic model in the three-dimensional map of TdtA after initial manual placement. Each protomer of the crystallographic hexamer was fitted in the cryo-EM map as an independent rigid body.
Protein detection
For cell fractionation, cell samples were recovered and washed by centrifugation (5000 x g 5 min), resuspended in 50 mM phosphate buffer, pH 7, and broken by sonication in the same buffer (4°C, 3 pulses of 1 min with 0.5 s cycles, 0.9 intensity, Labsonic, Braun). Residual unbroken cells and large cell fragments were eliminated by centrifugation (5,000 x g 5 min) and the soluble fractions was obtained after two consecutive 15,000 x g for 20 min centrifugation steps to avoid contamination with membranes. The insoluble fraction was also washed twice by centrifugation in the same buffer and conditions before SDS-PAGE and western blot analysis. PAGE protein analysis was carried out by standard procedures in 12% acrylamide/bis-acrylamide gels stained with Coomassie-Blue. Inmunodetection of TdtA and HepA proteins in the soluble and insoluble cell fractions of T. thermophilus HB27 was carried out by western blot using rabbit antiserum raised against the HepA protein from T. thermophilus, which also cross-reacts with its TdtA homolog, allowing the detection of both proteins simultaneously. Inmunodetection of protein fusions to sYFP was carried out with commercial antiserum. Goat anti-rabbit secondary antibodies conjugated to horseradish peroxidase were used for specific chemiluminescent detection of the protein (ECL, Amersham International).
Measurement of ATPase activity
ATP hydrolysis was estimated using the luciferin-luciferase ATP Bioluminescence Assay Kit CLS II (Roche), following manufacturer’s instructions. Different amounts of the analyzed protein were incubated for 1 h at 65°C in a buffer containing 5 mM MgSO4, 50 mM NaCl, 25 mM Tris-HCl pH 7.5 and 0.1 mM of ATP (Sigma-Aldrich). End-point kinetic ATPase assays were run in triplicate and results were expressed as percentage of ATP consumed against protein concentration (nM). No-protein controls, both with and without substrate, were included in every assay.
Reverse transcription PCR
RNA was isolated from T. thermophilus HB27 using an RNeasy Mini Kit (QIAGEN), and after DNAse I treatment (RQ1, Promega), all samples were tested by conventional PCR to verify absence of DNA contamination. Reverse transcription was performed using the SuperScript III first strand synthesis kit (Invitrogen) according to the manufacturer’s instructions. Subsequent PCRs to amplify cDNA were performed using Pfu Ultra II Fusion HS DNA Polymerase (Agilent Technologies) with the primers indicated in S2 Table.
Supporting information
BLASTp results for TdtA with its best homologs ordered by highest TdtA sequence similarity: Chloracidobac (Chloracidobacterium thermophilum), T. scotoductus (Thermus scotoductus SA01), T. antranikianii (Thermus antranikianii) and Thermotoga (Thermotoga napholitana). Common amino acids among the five sequences are represented in red, with conserved ATPase (Walker A, Walker B) and HAS-barrel motifs shown within red boxes.
(TIF)
SDS-PAGE gels showing: (A) total protein content of E. coli BL21 cells carrying plasmid pAB201 before (ni) and after induction (i). (B) proteins of the soluble (s) and insoluble (p) cell fractions. (C) IMAC affinity-purified TdtA protein (pp). Lane M corresponds to protein size markers of: 97.4, 66.2, 45, 31, 21.5 and 14.4 kDa. (D) % of ATP consumed after incubation for 1 h at 65°C with the indicated concentrations of TdtA in the absence of DNA (diamonds) or in the presence of 1 mg (1.2 nM) of genomic dsDNA from T. thermophilus (squares). Initial concentration of ATP was 10−4 M.
(TIF)
(DOCX)
(DOCX)
Acknowledgments
Critical reading of this manuscript by J. Casadesús, J. Penadés and M. Mencía is acknowledged. Technical assistance of E. Sánchez is also acknowledged.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the Spanish Ministry of Economy and Competitiveness (BIO2013-44963-R and BIO2016-77031-R to JB and BFU2014-55475-R to JRC), the FP7-PEOPLE-2012-IAPP from the European Union (Grant number 324439 to JB), and the Comunidad Autónoma de Madrid (S2013/MIT-2807 to JRC). An institutional grant from Fundación Ramón Areces to CBMSO is also acknowledged. AB and IB hold FPI fellowships from the Spanish Ministry of Education. CPM is a PhD fellow in the La Caixa Foundation International Fellowship Program (La Caixa/CNB). Hiring of NGQ through project BIO2013-50068-EXP is also acknowledged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ochman H, Lawrence JG, Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405: 299–304. 10.1038/35012500 [DOI] [PubMed] [Google Scholar]
- 2.Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, de la Cruz F (2010) Mobility of plasmids. Microbiol Mol Biol Rev 74: 434–452. 10.1128/MMBR.00020-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas CM, Nielsen KM (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3: 711–721. 10.1038/nrmicro1234 [DOI] [PubMed] [Google Scholar]
- 4.Nelson K, Clayton R, Gill S, Gwinn M, Dodson R, et al. (1999) Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399: 323–329. 10.1038/20601 [DOI] [PubMed] [Google Scholar]
- 5.Lanka E, Wilkins BM (1995) DNA Processing Reactions in Bacterial Conjugation. Annual Review of Biochemistry 64: 141–169. 10.1146/annurev.bi.64.070195.001041 [DOI] [PubMed] [Google Scholar]
- 6.Zechner E, De La Cruz F, Eisenbrandt R, Grahn A, Koraimann G, et al. (2000) Conjugative-DNA transfer processes The horizontal gene pool: bacterial plasmids and gene spread: 87–174. [Google Scholar]
- 7.Guglielmini J, Quintais L, Garcillán-Barcia MP, De La Cruz F, Rocha EP (2011) The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS genetics 7: e1002222 10.1371/journal.pgen.1002222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wozniak RA, Waldor MK (2010) Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8: 552–563. 10.1038/nrmicro2382 [DOI] [PubMed] [Google Scholar]
- 9.Vogelmann J, Ammelburg M, Finger C, Guezguez J, Linke D, et al. (2011) Conjugal plasmid transfer in Streptomyces resembles bacterial chromosome segregation by FtsK/SpoIIIE. The EMBO journal 30: 2246–2254. 10.1038/emboj.2011.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo Y, Wasserfallen A (2001) Gene Transfer Systems and Their Applications in Archaea. Systematic and Applied Microbiology 24: 15–25. 10.1078/0723-2020-00005 [DOI] [PubMed] [Google Scholar]
- 11.Derbyshire KM, Gray TA (2014) Distributive Conjugal Transfer: New Insights into Horizontal Gene Transfer and Genetic Exchange in Mycobacteria. Microbiology spectrum 2: 04 10.1128/microbiolspec.MGM2-0022-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mortimer TD, Pepperell CS (2014) Genomic Signatures of Distributive Conjugal Transfer among Mycobacteria. Genome Biology and Evolution 6: 2489–2500. 10.1093/gbe/evu175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Karnati PK, Takacs CM, Kowalski JC, Derbyshire KM (2005) Chromosomal DNA transfer in Mycobacterium smegmatis is mechanistically different from classical Hfr chromosomal DNA transfer. Molecular Microbiology 58: 280–288. 10.1111/j.1365-2958.2005.04824.x [DOI] [PubMed] [Google Scholar]
- 14.Thoma L, Muth G (2012) Conjugative DNA transfer in Streptomyces by TraB: is one protein enough? FEMS Microbiol Lett 337: 81–88. 10.1111/1574-6968.12031 [DOI] [PubMed] [Google Scholar]
- 15.Thoma L, Muth G (2015) The conjugative DNA-transfer apparatus of Streptomyces. International Journal of Medical Microbiology 305: 224–229. 10.1016/j.ijmm.2014.12.020 [DOI] [PubMed] [Google Scholar]
- 16.Blesa A, César CE, Averhoff B, Berenguer J (2014) Non canonical cell-to-cell DNA transfer in Thermus spp. is insensitive to Argonaute-mediated interference. Journal of Bacteriology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaimer C, Graumann PL (2011) Players between the worlds: multifunctional DNA translocases. Curr Opin Microbiol 14: 719–725. 10.1016/j.mib.2011.10.004 [DOI] [PubMed] [Google Scholar]
- 18.Castan P, Casares L, Barbe J, Berenguer J (2003) Temperature-dependent hypermutational phenotype in recA mutants of Thermus thermophilus HB27. J Bacteriol 185: 4901–4907. 10.1128/JB.185.16.4901-4907.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.César CE, Álvarez L, Bricio C, van Heerden E, Littauer D, et al. (2012) Unconventional lateral gene transfer in extreme thermophilic bacteria. International Microbiology 14: 187–199. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Herrero LA, Olabarria G, Caston JR, Lasa I, Berenguer J (1995) Horizontal transference of S-layer genes within Thermus thermophilus. J Bacteriol 177: 5460–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Z, Guan S, Robinson D, El Fezzazi H, Quimby A, et al. (2014) Characterization of cleavage intermediate and star sites of RM. Tth111II. Scientific reports 4: 3838 10.1038/srep03838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blesa A, Berenguer J (2015) Contribution of vesicle-protected extracellular DNA to horizontal gene transfer in Thermus spp. Int Microbiol 18: 177–187. 10.2436/20.1501.01.248 [DOI] [PubMed] [Google Scholar]
- 23.Rzechorzek NJ, Blackwood JK, Bray SM, Maman JD, Pellegrini L, et al. (2014) Structure of the hexameric HerA ATPase reveals a mechanism of translocation-coupled DNA-end processing in archaea. Nat Commun 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rolfsmeier ML, Laughery MF, Haseltine CA (2010) Repair of DNA double-strand breaks following UV damage in three Sulfolobus solfataricus strains. Journal of bacteriology 192: 4954–4962. 10.1128/JB.00667-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng K, Chen X, Xu G, Wang L, Xu H, et al. (2015) Biochemical and Functional Characterization of the NurA-HerA Complex from Deinococcus radiodurans. Journal of Bacteriology 197: 2048–2061. 10.1128/JB.00018-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Constantinesco F, Forterre P, Koonin EV, Aravind L, Elie C (2004) A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea. Nucleic Acids Research 32: 1439–1447. 10.1093/nar/gkh283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer LM, Makarova KS, Koonin EV, Aravind L (2004) Comparative genomics of the FtsK—HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Research 32: 5260–5279. 10.1093/nar/gkh828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parsons JA, Bannam TL, Devenish RJ, Rood JI (2007) TcpA, an FtsK/SpoIIIE homolog, is essential for transfer of the conjugative plasmid pCW3 in Clostridium perfringens. J Bacteriol 189: 7782–7790. 10.1128/JB.00783-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Possoz C, Ribard C, Gagnat J, Pernodet JL, Guérineau M (2001) The integrative element pSAM2 from Streptomyces: kinetics and mode of conjugal transfer. Molecular microbiology 42: 159–166. [DOI] [PubMed] [Google Scholar]
- 30.Blackwood JK, Rzechorzek NJ, Abrams AS, Maman JD, Pellegrini L, et al. (2012) Structural and functional insights into DNA-end processing by the archaeal HerA helicase-NurA nuclease complex. Nucleic Acids Res 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarzenlander C, Averhoff B (2006) Characterization of DNA transport in the thermophilic bacterium Thermus thermophilus HB27. FEBS J 273: 4210–4218. 10.1111/j.1742-4658.2006.05416.x [DOI] [PubMed] [Google Scholar]
- 32.Francia MV, Clewell DB, de la Cruz F, Moncalian G (2013) Catalytic domain of plasmid pAD1 relaxase TraX defines a group of relaxases related to restriction endonucleases. Proc Natl Acad Sci U S A 110: 13606–13611. 10.1073/pnas.1310037110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furuta Y, Abe K, Kobayashi I (2010) Genome comparison and context analysis reveals putative mobile forms of restriction—modification systems and related rearrangements. Nucleic acids research: gkp1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carraro N, Poulin D, Burrus V (2015) Replication and Active Partition of Integrative and Conjugative Elements (ICEs) of the SXT/R391 Family: The Line between ICEs and Conjugative Plasmids Is Getting Thinner. PLoS Genet 11: e1005298 10.1371/journal.pgen.1005298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166: 557–580. [DOI] [PubMed] [Google Scholar]
- 36.Rosenberg AH, Lade BN, Dao-shan C, Lin S-W, Dunn JJ, et al. (1987) Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene 56: 125–135. [DOI] [PubMed] [Google Scholar]
- 37.Cava F, Zafra O, Magalon A, Blasco F, Berenguer J (2004) A new type of NADH dehydrogenase specific for nitrate respiration in the extreme thermophile Thermus thermophilus. Journal of Biological Chemistry 279: 45369–45378. 10.1074/jbc.M404785200 [DOI] [PubMed] [Google Scholar]
- 38.Swarts DCJ, Matthijs M.; Westra Edze R.; Zhu Yifan; Janssen Jorijn H.; Snijders Ambrosius P.; Wang Yanli; Patel Dinshaw J.; Berenguer José; Brouns Stan J.; van der Oost John (2014) DNA- guided DNA interference by a prokaryotic Arognaute. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedrich A, Hartsch T, Averhoff B (2001) Natural transformation in mesophilic and thermophilic bacteria: identification and characterization of novel, closely related competence genes in Acinetobacter sp. strain BD413 and Thermus thermophilus HB27. Appl Environ Microbiol 67: 3140–3148. 10.1128/AEM.67.7.3140-3148.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedrich A, Rumszauer J, Henne A, Averhoff B (2003) Pilin-like proteins in the extremely thermophilic bacterium Thermus thermophilus HB27: implication in competence for natural transformation and links to type IV pilus biogenesis. Appl Environ Microbiol 69: 3695–3700. 10.1128/AEM.69.7.3695-3700.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-ramahi Y (2013) Ingeniería de proteínas fluorescentes y aplicaciones de localización celular en microorganismos termófilos. Madrid: Universidad Autónoma de Madrid; 141 p. [Google Scholar]
- 42.Koyama Y, Hoshino T, Tomizuka N, Furukawa K (1986) Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J Bacteriol 166: 338–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marabini R, Masegosa IM, San Martin MC, Marco S, Fernandez JJ, et al. (1996) Xmipp: An Image Processing Package for Electron Microscopy. J Struct Biol 116: 237–240. [DOI] [PubMed] [Google Scholar]
- 44.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 45.Mindell JA, Grigorieff N (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142: 334–347. [DOI] [PubMed] [Google Scholar]
- 46.Sorzano CO, Bilbao-Castro JR, Shkolnisky Y, Alcorlo M, Melero R, et al. (2010) A clustering approach to multireference alignment of single-particle projections in electron microscopy. J Struct Biol 171: 197–206. 10.1016/j.jsb.2010.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97. 10.1006/jsbi.1999.4174 [DOI] [PubMed] [Google Scholar]
- 48.Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, Marabini R (2008) Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc 3: 977–990. 10.1038/nprot.2008.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BLASTp results for TdtA with its best homologs ordered by highest TdtA sequence similarity: Chloracidobac (Chloracidobacterium thermophilum), T. scotoductus (Thermus scotoductus SA01), T. antranikianii (Thermus antranikianii) and Thermotoga (Thermotoga napholitana). Common amino acids among the five sequences are represented in red, with conserved ATPase (Walker A, Walker B) and HAS-barrel motifs shown within red boxes.
(TIF)
SDS-PAGE gels showing: (A) total protein content of E. coli BL21 cells carrying plasmid pAB201 before (ni) and after induction (i). (B) proteins of the soluble (s) and insoluble (p) cell fractions. (C) IMAC affinity-purified TdtA protein (pp). Lane M corresponds to protein size markers of: 97.4, 66.2, 45, 31, 21.5 and 14.4 kDa. (D) % of ATP consumed after incubation for 1 h at 65°C with the indicated concentrations of TdtA in the absence of DNA (diamonds) or in the presence of 1 mg (1.2 nM) of genomic dsDNA from T. thermophilus (squares). Initial concentration of ATP was 10−4 M.
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.