Abstract
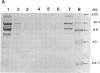
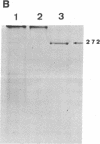
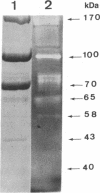
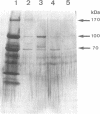
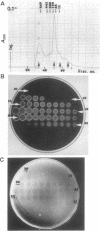
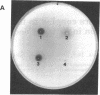
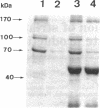
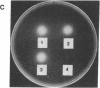
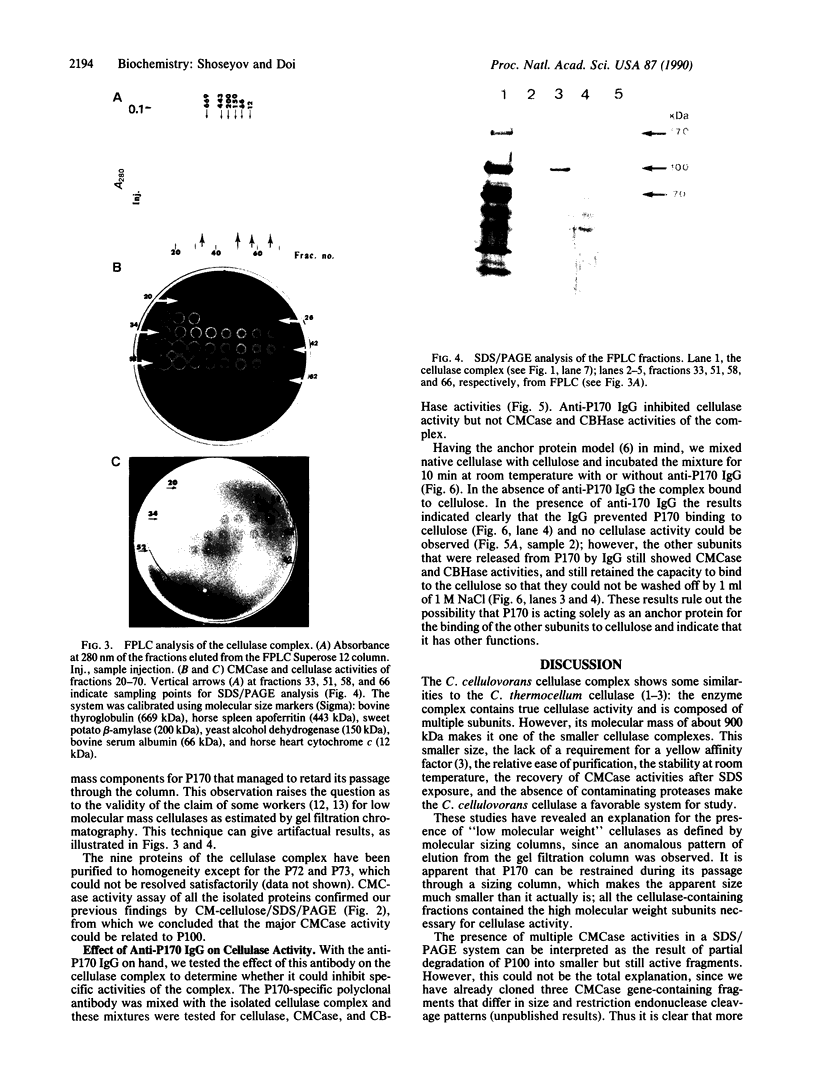
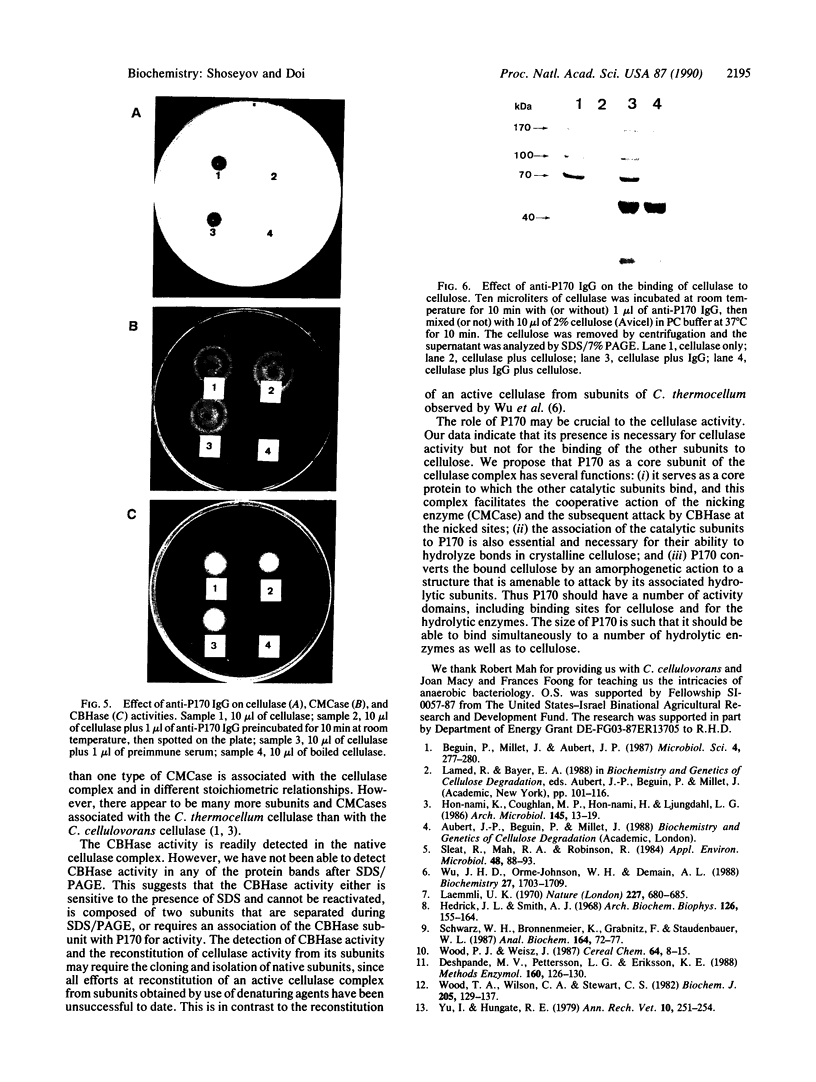
The cellulase complex from Clostridium cellulovorans has been purified and its subunit composition determined. The complex exhibits cellulase activity against crystalline cellulose as well as carboxymethylcellulase (CMCase) and cellobiohydrolase activities. Three major subunits are present with molecular masses of 170, 100, and 70 kDa. The 100-kDa subunit is the major CMCase, although at least four other, minor subunits show CMCase activity. The 170-kDa subunit has the highest affinity for cellulose, does not have detectable enzymatic activity, but is necessary for cellulase activity. Immunological studies indicate that the 170-kDa subunit is not required for binding of the catalytic subunits to cellulose and therefore does not function solely as an anchor protein. Thus this core subunit must have multiple functions. We propose a working hypothesis that the binding of the 170-kDa subunit converts the crystalline cellulose to a form that is capable of being hydrolyzed in a cooperative fashion by the associated catalytic subunits.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Béguin P., Millet J., Aubert J. P. The cloned cel (cellulose degradation) genes of Clostridium thermocellum and their products. Microbiol Sci. 1987 Sep;4(9):277–280. [PubMed] [Google Scholar]
- Hedrick J. L., Smith A. J. Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch Biochem Biophys. 1968 Jul;126(1):155–164. doi: 10.1016/0003-9861(68)90569-9. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Schwarz W. H., Bronnenmeier K., Gräbnitz F., Staudenbauer W. L. Activity staining of cellulases in polyacrylamide gels containing mixed linkage beta-glucans. Anal Biochem. 1987 Jul;164(1):72–77. doi: 10.1016/0003-2697(87)90369-1. [DOI] [PubMed] [Google Scholar]
- Sleat R., Mah R. A., Robinson R. Isolation and Characterization of an Anaerobic, Cellulolytic Bacterium, Clostridium cellulovorans sp. nov. Appl Environ Microbiol. 1984 Jul;48(1):88–93. doi: 10.1128/aem.48.1.88-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T. M., Wilson C. A., Stewart C. S. Preparation of the cellulase from the cellulolytic anaerobic rumen bacterium Ruminococcus albus and its release from the bacterial cell wall. Biochem J. 1982 Jul 1;205(1):129–137. doi: 10.1042/bj2050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I., Hungate R. E. The extracellular cellulases of Ruminococcus albus. Ann Rech Vet. 1979;10(2-3):251–254. [PubMed] [Google Scholar]