LETTER
Colistin is used as a last-resort antibiotic for treating infections caused by multidrug-resistant members of the family Enterobacteriaceae, in particular for carbapenem-resistant isolates. Resistance to colistin in Enterobacteriaceae species has been attributed to chromosomal mutations (1), but recently a plasmid-encoded colistin resistance gene, mcr-1, was described in isolates from different sources and in many countries worldwide (2, 3). The emergence of plasmid-encoded colistin resistance is critical, as its transfer could render carbapenem-resistant bacteria pandrug resistant, resulting in virtually untreatable pathogens.
We recently reported the first mcr-1/blaKPC-2-encoding carbapenem-resistant Escherichia coli isolate (NRZ14408) from a patient with a wound infection (4). In order to characterize the isolate, we sequenced the whole genome to completion (see the supplemental material).
E. coli NRZ14408 harbored a chromosome of 5,344,876 bp with a GC content of 50.65%, four plasmids of about 12 to 238 kbp (p14408_M, p14408_1, p14408_2, and p14408_3; see Table S1 in the supplemental material), and two phages of 41 and 19 kbp (14408_1, 14408_2; see Table S1 in the supplemental material). E. coli NRZ14408 is an O7:H6 isolate of multilocus sequence type 362 (ST362) and phylogenetic group D, which is associated with extraintestinal infections (5).
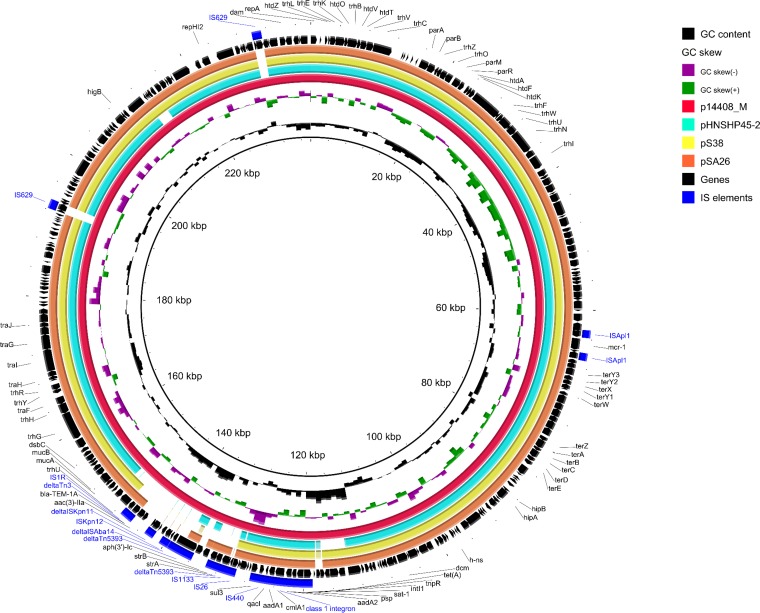
Plasmid p14408_3 harbored no antibiotic resistance genes but did have colicin R genes (cra, cri, and crl) and the mobC gene for mobilization of the plasmid. The mcr-1 gene was flanked by two ISApl1 elements and located on an IncHI2 plasmid (p14408_M) with high similarity to other mcr-1-encoding IncHI2 plasmids (pHNSHP45-2 [accession no. KU341381.1], pSA26-MCR-1 [KU743384.1], and pS38 [KX129782]). p14408_M differed from these plasmids by the presence of five additional insertion sequences (ISKpn11, ISKpn12, ISAba14, and two copies of IS629; Fig. 1) and a different set of antibiotic resistance genes (see Table S2 in the supplemental material). Plasmids p14408_1 and p14408_2 harbored the same 36,152-bp region, including nine antibiotic resistance genes (see Table S1 in the supplemental material) and a large portion of the IncN plasmid conjugation machinery (see Fig. S2 in the supplemental material). It displayed a unique blaKPC-2 cassette [IS26, aac(3)-IId, ISCfr1, blaKPC-2, blaTEM-1B, and ISKpn16], present only in the IncN plasmid pCF8698_KPC2, which was isolated during an Enterobacteriaceae blaKPC-2 outbreak in southern Hesse in Germany (6). The region was flanked by two IS26 insertion sequences in p14408_1, indicating an IS26-mediated transfer (7, 8).
FIG 1.
Whole plasmid comparison of three mcr-1-encoding IncHI2 plasmids, p14408_M, pHNSHP45-2 and pSA26-MCR-1, performed using BRIG (9). Mobile elements are marked in blue color; genes are marked with black arrows.
The genome of E. coli NRZ14408 displayed a distinct distribution of antibiotic resistance and virulence genes. Antibiotic resistance genes were located exclusively on plasmids (see Table S1 in the supplemental material), whereas virulence genes were located solely on the chromosome (see Table S3 in the supplemental material). Analysis of other E. coli ST362 isolates from 1952 to 2014 (see Table S4 in the supplemental material) harboring virulence determinants or antibiotic resistance genes provided further evidence supporting this observation (see Fig. S2 in the supplemental material). In addition, the total number of virulence genes was generally higher in ST362 isolates harboring antibiotic resistance genes than in antibiotic-susceptible isolates (see Fig. S3 in the supplemental material).
As ST362 is associated with extraintestinal infections, it may be a representative of those E. coli clones where increased antibiotic resistance together with enhanced virulence is being selected through the use of injudicious antibiotic regimens.
Accession number(s).
The closed genome and plasmid sequences obtained in this study were deposited in the European Nucleotide Archive under accession numbers LT599825 to LT599831.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christina Gerstmann, Natalia Lest, Simone Severitt, and Nicole Heyer for excellent technical assistance.
This study was supported by grants within the framework of the German Center of Infection Research (DZIF) through the German Federal Ministry of Education and Research (BMBF) to T.C. and C.I. (grant 8000 701–3 [HZI]) and to T.C. (grant TI06.001, 8032808811).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02359-16.
REFERENCES
- 1.Olaitan AO, Morand S, Rolain J-M. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y-Y, Wang Y, Walsh TR, Yi L- X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 3.Schwarz S, Johnson AP. 2016. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother 71:2066–2070. doi: 10.1093/jac/dkw274. [DOI] [PubMed] [Google Scholar]
- 4.Falgenhauer L, Waezsada S-E, Yao Y, Imirzalioglu C, Käsbohrer A, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbrock L, Chakraborty T. 2016. Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis 16:282–283. doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]
- 5.Clermont O, Bonacorsi S, Bingen E, Bonacorsi P. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao Y, Imirzalioglu C, Hain T, Kaase M, Gatermann S, Exner M, Mielke M, Hauri A, Dragneva Y, Bill R, Wendt C, Wirtz A, Domann E, Chakraborty T. 2014. Complete nucleotide sequence of a Citrobacter freundii plasmid carrying KPC-2 in a unique genetic environment. Genome Announc 2:e01157-14. doi: 10.1128/genomeA.01157-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harmer CJ, Moran RA, Hall RM. 2014. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 5:e01801-14. doi: 10.1128/mBio.01801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh H, Doijad S, Bunk B, Falgenhauer L, Yao Y, Spröer C, Gentil K, Schmiedel J, Imirzalioglu C, Overmann J, Chakraborty T. 2016. Detection of translocatable units in a blaCTX-M-15 extended-spectrum β-lactamase-producing ST131 Escherichia coli isolate using a hybrid sequencing approach. Int J Antimicrob Agents 47:245–247. doi: 10.1016/j.ijantimicag.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.