Abstract
Influenza A viruses contain eight single-stranded, negative-sense RNA segments as viral genomes in the form of viral ribonucleoproteins (vRNPs). During genome replication in the nucleus, positive-sense complementary RNPs (cRNPs) are produced as replicative intermediates, which are not incorporated into progeny virions. To analyze the mechanism of selective vRNP incorporation into progeny virions, we quantified vRNPs and cRNPs in the nuclear and cytosolic fractions of infected cells, using a strand-specific qRT-PCR. Unexpectedly, we found that cRNPs were also exported to the cytoplasm. This export was chromosome region maintenance 1 (CRM1)-independent unlike that of vRNPs. Although both vRNPs and cRNPs were present in the cytosol, viral matrix (M1) protein, a key regulator for viral assembly, preferentially bound vRNPs over cRNPs. These results indicate that influenza A viruses selectively uptake cytosolic vRNPs through a specific interaction with M1 during viral assembly.
Keywords: influenza, vRNP, cRNP, nuclear export, CRM1, M1, trafficking, assembly
1. Introduction
Influenza A virus, a major respiratory pathogen that belongs to the family of Orthomyxoviridae, frequently causes seasonal epidemics and periodic pandemic outbreaks. Genetic reassortment between human and animal viruses caused emergence of many pandemic viruses (de Silva et al., 2012; Neumann et al., 2009; Y. Watanabe et al., 2012). Therefore, a thorough understanding of the molecular mechanisms underlying influenza A virus replication, genome packaging and viral assembly is needed to clarify the reassortment process, which will also allow identifying potential drug targets.
Influenza A virus contains eight segments of single-stranded, negative-sense viral RNA (vRNA) (Knipe and Howley, 2013). The viral genomes are encapsidated with nucleoproteins (NP) and associated with the trimeric polymerase complexes to form helical-shaped viral ribonucleoproteins (vRNPs) (Arranz et al., 2012; Moeller et al., 2012; Noda and Kawaoka, 2010; Zheng and Tao, 2013). Unlike other RNA viruses, influenza A virus transcribes and replicates its genome in the nucleus of the host cell (de Silva et al., 2012; Knipe and Howley, 2013; Neumann et al., 2009; Y. Watanabe et al., 2012). Within the nucleus of infected cells, positive-sense full-length complementary RNAs (cRNAs) are synthesized. The cRNPs are replicative intermediates used as templates to generate nascent vRNPs. Progeny vRNPs are exported to the cytoplasm via chromosome region maintenance 1 (CRM1) nuclear exporter. Substantial evidence indicates an important role of viral nuclear export protein (NEP) (S. Huang et al., 2013; Iwatsuki-Horimoto et al., 2004; Knipe and Howley, 2013; Ma et al., 2001; Neumann et al., 2000; O’Neill et al., 1998) and matrix (M1) protein (Arranz et al., 2012; Bui et al., 2000; Cao et al., 2012; Ma et al., 2001; Martin and Helenius, 1991; Moeller et al., 2012; Noda and Kawaoka, 2010; Zheng and Tao, 2013) in vRNP nuclear export via a CRM1-dependent pathway. The current model proposes that vRNP forms a daisy chain complex with M1 and NEP. This vRNP-M1-NEP complex interacts with CRM1 in association with RanGTP, a small GTPase, resulting in the translocation of vRNP into the cytoplasm (Akarsu et al., 2003; Baudin et al., 2001; Ma et al., 2001; Neumann et al., 2000; Shimizu et al., 2011). However, other studies suggested that NP can directly interact with CRM1, which also regulates vRNP nuclear export (Chutiwitoonchai and Aida, 2016; Elton et al., 2001). After nuclear export, vRNPs are transported to the site of assembly at the apical plasma membrane through Rab11a-regulated recycling endosome (Amorim et al., 2011; Chou et al., 2013; Eisfeld et al., 2011). At the plasma membrane, M1 protein together with viral envelope proteins are considered to play a major role in incorporation of viral genome into budding virions during viral assembly (Ali et al., 2000; Nayak et al., 2009; Noton et al., 2007; Rossman and Lamb, 2011; Wang et al., 2010; Wu et al., 2011)
Incorporation of influenza genome is a sophisticated process, as eight segments of negative-sense vRNPs are required to make an infectious particle. Electron tomography and fluorescent in situ hybridization (FISH) studies demonstrate that the majority of progeny virions incorporate a single copy of all eight vRNPs (Chou et al., 2012; Lakdawala et al., 2014; Noda and Kawaoka, 2012; Noda et al., 2006). However, how influenza virus selectively uptakes vRNPs, while excluding the replicative intermediates, cRNPs, is not known. cRNPs are produced in the nucleus, but whether they remain in the nucleus throughout infection or are exported to the cytoplasm is also not clear. In fact, a very limited analysis has been done regarding localization of cRNPs in infected cells. In this study, we investigated two possible mechanisms that facilitate selective incorporation of vRNPs over cRNPs into influenza infectious virions: 1) cRNPs remain in the nucleus throughout the virus life cycle and therefore are not incorporated into progeny virions, or 2) both vRNPs and cRNPs are exported from the nucleus, but selective uptake of vRNPs occurs during trafficking to or assembly at the plasma membrane. We used a specific and sensitive qRT-PCR approach to study production and distribution of vRNAs and cRNAs at various time points after infection. Our data indicate that both vRNPs and cRNPs are exported from the nucleus to the cytoplasm, but their export mechanism seems to be different. We also showed that influenza M1 protein preferentially interacts with vRNPs over cRNPs, suggesting that influenza A viruses selectively uptake cytosolic vRNPs through a specific interaction with M1 during viral assembly.
2. Results
2.1. Influenza cRNPs are exported from the nucleus of an infected cell
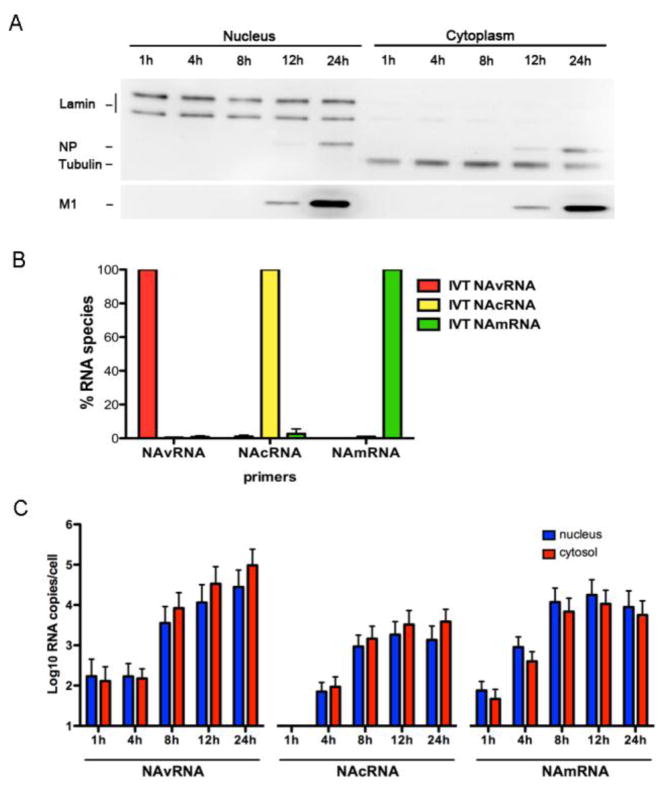
To determine the translocation of cRNPs during virus replication, we first quantified the amount of cRNAs in the nuclear and cytoplasmic fractions at various times after infection and compared them with those of vRNAs and mRNAs. We prepared nuclear and cytoplasmic fractions from MDCK cells infected with A/WSN/1933 (H1N1) at 1, 4, 8, 12, and 24 hours post infection (hpi). Extracted RNAs were applied for strand-specific qRT-PCR to quantify the amount of vRNAs, cRNAs and mRNAs of the neuraminidase (NA) RNA segment (Kawakami et al., 2011). First, we confirmed the quality of nuclear/cytosolic fractionations by Western blot analysis using lamin A/C as a nuclear marker and tubulin as a cytosolic marker. The presence of lamin A/C (nuclear marker) in the cytosolic fraction and tubulin (cytosolic marker) in the nuclear fraction was less than 2.8% and 0.9%, respectively, showing efficient separation of the nuclear and cytosolic samples (Fig. 1A). We also confirmed the specificity of strand-specific qRT-PCR primers using in vitro-transcribed RNA template (see Materials and Methods). The vRNA primer set minimally detected cRNA and mRNA (0.48% and 0.8%, respectively). Similarly, the cRNA primer set detected only 0.99% and 2.67% of vRNA and mRNA, respectively. The mRNA primer set also minimally detected vRNA and cRNA (0.08% and 0.93%, respectively) (Fig. 1B). These data indicate that the strand-specific qRT-PCR primers were highly specific to distinguish viral vRNA, cRNA, and mRNA as previously reported (Kawakami et al., 2011).
Figure 1. Influenza cRNPs are exported from the nucleus.
(A) MDCK cells infected with WSN at a MOI of 2 were fractionated at indicated time points after infection. Markers for nucleus (lamin A/C) and cytosol (tubulin), together with viral NP and M1 proteins were detected by Western blotting using specific antibodies. (B) The specificity of qRT-PCR primer sets was determined using WSN NA vRNA (red), cRNA (yellow), and mRNA (green) templates prepared in vitro. The specificity of primers for WSN NAvRNA, NAcRNA, and NAmRNA is presented in respect to the percentage of its corresponding RNA template. (C) MDCK cells infected with WSN were fractionated, and total RNAs were extracted and applied for strand-specific real-time qRT-PCR. Quantities of vRNA, cRNA and mRNA of the NA segment of WSN influenza A virus in the nuclear (blue) and cytosolic (red) fractions are shown as averages with standard deviations from six independent experiments.
Following subcellular fractionation of infected cells, we isolated RNAs, which were subjected to qRT-PCR using the specific primer sets. At 1 hpi, we detected a small amount of vRNAs (1.7 × 102 RNA copies/cell), which originated from inoculated viruses. The cRNAs were undetectable at this time-point. At 4 hpi, vRNAs remained constant, while the copy number of cRNAs began to rise, indicating initial genome replication from inoculated vRNA templates. Small quantities of viral mRNAs were detected at 1 hpi and began to increase at 4 hpi, indicating that primary transcription from vRNA templates occurs prior to genome replication. Viral mRNAs were present in both the nuclei and cytoplasm of infected cells, indicating nuclear export of viral mRNAs. Copy numbers of vRNAs and cRNAs increased over time from 8 hpi onwards. A significant quantity (> 103 RNA copies/cell) of vRNAs were present in both the nuclei and cytoplasm of infected cells at late time points (8, 12, and 24 hpi), indicating nuclear export of newly synthesized vRNPs to the cytoplasm as anticipated (Fig. 1C). Intriguingly, we also detected cRNAs in the cytoplasm at a similar ratio with vRNAs. Although the difference between cytoplasmic and nuclear cRNA was not statistically significant, we did tend to observe more cRNA in the cytoplasm than in the nucleus (Fig. 1C). These data indicate that both influenza vRNPs and cRNPs are exported from the nucleus of infected cells.
2.2. cRNPs are exported from the nucleus via a CRM1-independent pathway
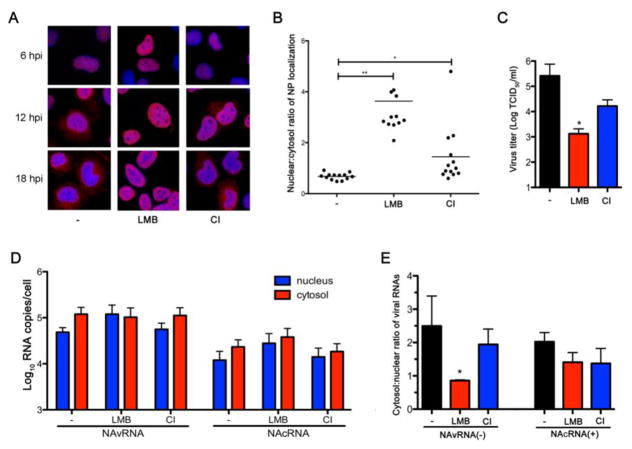
A current model of vRNP nuclear export proposes that vRNP forms a daisy chain complex with M1 and NEP, in which the N-terminal domain in NEP interacts with CRM1 to navigate vRNP nuclear export (Akarsu et al., 2003; Baudin et al., 2001; Iwatsuki-Horimoto et al., 2004; Neumann et al., 2000). In contrast, recent studies showed that vRNPs passively traverse across the caspase-induced enlarged nuclear pores caused by influenza infection (Mühlbauer et al., 2015; Wurzer et al., 2003). To test if cRNPs utilize CRM1 for their trafficking across nuclear pore complexes, we treated MDCK cells with leptomycin B (LMB), a potent CRM1 inhibitor immediately after infection and determined NP localization by immunofluorescent (IF) assay. Previous studies demonstrated nuclear retention of influenza NP as a surrogate for vRNP, when infected cells were treated with LMB (Elton et al., 2001; Ma et al., 2001; K. Watanabe et al., 2001). Consistent with previous reports, LMB treatment caused nuclear accumulation of NP at late time points (12 and 18 hpi) (Fig. 2A). Quantification of NP signals in the nucleus and cytoplasm of the 18 hpi-sample indicates a significant increase of nuclear/cytosol ratios of NP in the LMB-treated cells (P < 0.001) (Fig. 2B). We also investigated the effect of LMB on production of infectious virions and found that LMB treatment caused over a hundred-fold reduction in infectious virion production (P < 0.05) (Fig. 2C).
Figure 2. Influenza cRNPs are exported from the nucleus via a CRM-1 independent pathway.
(A) MDCK cells were infected with WSN at a MOI of 2 for 1 h and cultured in the presence or absence of either 20 nM LMB or 40 μM CI. Cells were fixed and permeabilized at 6, 12, and 18 hpi and processed for IFA using mouse anti-NP mAb and Alexa Flour 594 anti-mouse antibody, and counterstained with DAPI. (B) Intracellular NP signal was quantified from fluorescence intensity at the Alexa Flour 594 channel within the nucleus and the cytoplasm using ImageJ software. Data from cells processed at 18 hpi are represented as the nuclear/cytosol ratio of NP localization (n = 13, mean ± s.d. * P < 0.05, ** P < 0.001). (C) Infected MDCK cells were treated with inhibitors as described in A. Culture supernatant was collected at 18 hpi to perform TCID50 assay for virus titer calculation (n = 3 independent experiments, mean ± s.d. * P <0.05). (D) Infected MDCK cells were treated with inhibitors as described in A. At 18 hpi, cells were fractionated and total RNA from each fraction was extracted and used for the strand-specific real-time qRT-PCR. Quantities of vRNA and cRNA of the NA segment in the nuclear (blue) and cytosolic (red) fractions are shown as averages with standard deviations from three independent experiments. (E) Results obtained from D are represented as the nuclear/cytosol ratio of viral RNAs (n = 3 independent experiments, mean ± s.d. * P < 0.05).
To determine if cRNP nuclear export is CRM1-dependent, we quantified the amount of both vRNAs and cRNAs in the nuclear and cytosolic fractions of cells treated with LMB for 18 h. LMB treatment increased the quantity of vRNAs in the nucleus 2.5-fold, while slightly decreasing their amount in the cytosol, compared to the untreated condition (Fig. 2D). LMB reduced the cytosol/nuclear ratio of vRNAs from 2.49 to 0.86 (P < 0.05) (Fig. 2E), indicating an inhibition of vRNP nuclear export in the presence of a CRM1 inhibitor consistent with previous findings (Chase et al., 2011; Chou et al., 2013; K. Watanabe et al., 2001). In contrast, LMB treatment did not reduce nuclear export of cRNP. More cRNA copies were detected in the cytosol than nucleus as observed in untreated cells (Fig. 2D). There was no significant difference in the cRNA cytosol/nuclear ratio between treated and untreated cells (Fig. 2E). There results suggest that cRNP nuclear export is CRM1-independent.
To test if cRNPs are exported from the nucleus via a caspase-dependent passive transportation, we treated infected cells with a caspase 3/7 inhibitor (CI) at the concentration reported to block caspase activation (Mühlbauer et al., 2015) immediately after virus infection. CI treatment slightly increased nuclear accumulation of NP (Fig. 2A and B) and caused a 10-fold reduction in infectious virion production (Fig. 2C). However, we detected no significant difference in either vRNA or cRNA cytosol/nuclear ratio between CI-treated and untreated cells (Fig. 2D and E). Overall, these results suggest that cRNP nuclear export is CRM1- and caspase- independent.
2.3. Selective incorporation of vRNPs into progeny virions
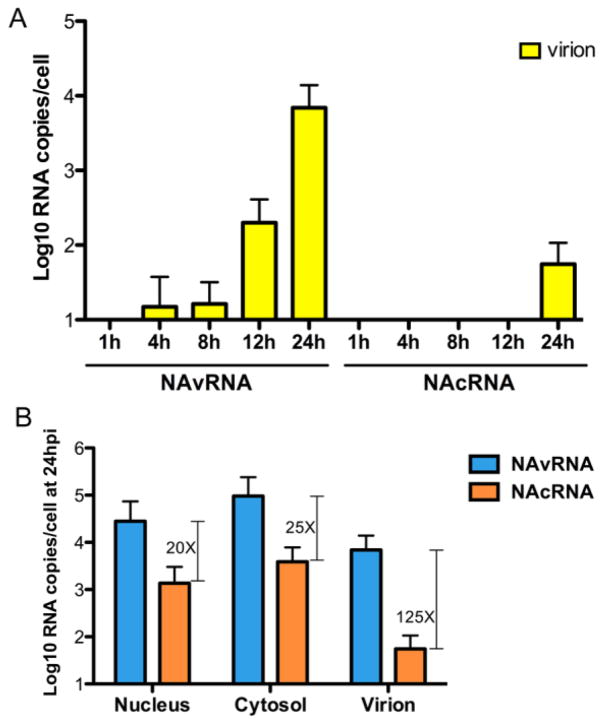
Our data indicate that cRNAs are exported from the nucleus and are present in the cytoplasm. Therefore, we next compared the vRNA/cRNA ratio in the cytoplasm and released virions at various time points after infection to determine the specificity of vRNP incorporation into progeny virions. As expected, we detected release of progeny virions containing vRNAs at late time points (1.99 × 102 RNA copies at 12 hpi and 6.93 × 103 RNA copies at 24 hpi). In contrast, cRNAs were undetectable, except for a small amount (5.5 × 101 RNA copies) at 24 hpi, which was 125-fold less than vRNA (Fig. 3A). We also quantified the vRNAs and cRNAs in the nuclei and cytosol of the same infected cells, and compared the vRNA/cRNA ratio. At 24 hpi, the ratios in the nuclear fraction, cytosolic fraction, and culture supernatant were 20, 25, and 125, respectively (Fig. 3B). The dramatically increased vRNA/cRNA ratio in the released virions indicates that progeny virions selectively incorporate vRNPs.
Figure 3. Selective incorporation of vRNPs into the virion.
(A) MDCK cells were infected as described in Fig. 1C. Virions in the culture supernatant were purified by ultracentrifugation through a 20% sucrose cushion. Extracted viral RNAs were used for strand-specific real-time qRT-PCR. Data are shown as averages with standard deviations from six independent experiments. (B) Quantities of vRNA (light blue) and cRNA (orange) in each fraction of the cells and in released virions at 24 hpi are shown as averages with standard deviations from six independent experiments. Capped lines represent the fold difference of vRNA over cRNA.
2.4. Influenza M1 preferentially interacts with vRNPs over cRNPs
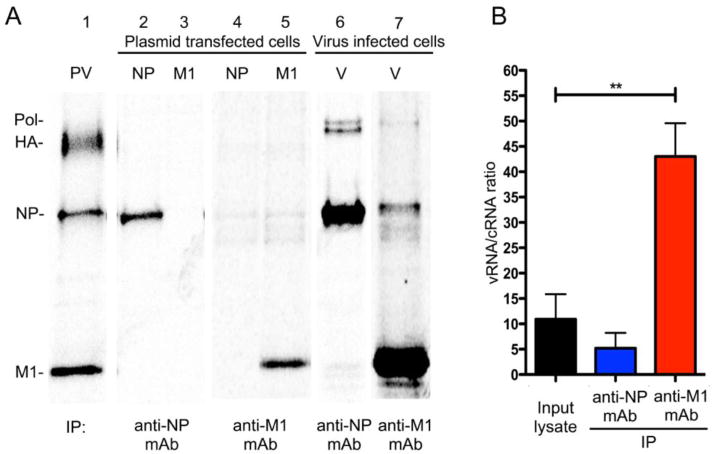
Our data suggest that there is a mechanism regulating selective incorporation of viral genome in the cytoplasm (Fig. 3). Influenza M1 protein is known to be indispensable for assembly and budding of virus particles (Ali et al., 2000; Nayak et al., 2009; Noton et al., 2007; Wang et al., 2010; Wu et al., 2011). Previous studies suggest a specific interaction between M1 and vRNP during virus assembly (Gómez-Puertas et al., 2000; Noton et al., 2007; Wu et al., 2011). Our lab also showed that M1-vRNP interaction affects morphology of progeny virions (Bialas et al., 2014). Therefore, we determined if M1 specifically recognizes and interacts with vRNPs to promote specific vRNP incorporation into infectious virions. To test the specificity of M1 interaction, we co-immunoprecipitated the RNPs associated with M1 from virus-infected cells using anti-M1 mAb, and quantified the amount of vRNAs and cRNAs associated with M1 protein. First, we tested the specificity of anti-M1 mAb to be used for immunoprecipitation. Anti-M1 mAb or a cocktail of anti-NP mAbs were used to react with radiolabeled cell lysate transfected with M1 or NP expressing plasmids. Both mAbs were highly specific for their respective proteins and displayed no cross-reactivity (Fig. 4A, lanes 2–5). When anti-NP mAbs reacted with the virus-infected cell lysate, the Abs precipitated NP and associated polymerase proteins as expected (Fig. 4A, lane 6). Importantly, anti-M1 mAb precipitated not only M1, but also NP and polymerase proteins, indicating that M1 interacts with RNPs (Fig. 4A lane 7).
Figure 4. Influenza M1 protein preferentially interacts with vRNPs over cRNPs.
(A) 293T cells transfected with NP or M1 expressing plasmids or infected with WSN (V) at a MOI of 3 were radiolabeled for 16 h and used for immunoprecipitation with either anti-NP or anti-M1 mAb. Purified virion (PV) was used as a size marker for viral proteins. (B) Cells infected with WSN at a MOI of 3 were cultured for 14 h at 37°C, and cell lysates were used for immunoprecipitation with anti-NP mAb or anti-M1 mAb. Total RNAs in immunoprecipitated materials and the input lysate were isolated and used for qRT-PCR reaction using the NA gene strand-specific primers to quantify vRNAs and cRNAs. Results are shown as averages with standard deviations of vRNA/cRNA ratios (n = 4 independent experiments, mean ± s.d. ** P < 0.001).
Using these specific mAbs, we immunoprecipitated viral nucleocapsids and isolated RNAs. Quantities of vRNAs and cRNAs of NA gene in the immunoprecipitated materials were determined by strand-specific qRT-PCR as described above. Analysis on the input lysate demonstrated 10.9-fold more vRNAs than cRNAs (Fig. 4B). Viral nucleocapsids immunoprecipitated with anti-NP mAbs showed 5.2-fold more vRNAs than cRNAs. However, viral nucleocapsids co-immunoprecipitated with anti-M1 mAb included 43-fold more vRNAs than cRNAs, indicating that M1 preferentially binds to vRNPs over cRNPs (Fig. 4B). These data suggest that influenza A virus selectively recognizes and uptakes cytosolic vRNPs through specific interaction with M1 during viral assembly.
2.5. M1-NP interaction in infected cells
M1 protein interacts with vRNP both in the nucleus to navigate vRNP nuclear export (Bui et al., 2000; Cao et al., 2012; Chase et al., 2011; X. Huang et al., 2001; Martin and Helenius, 1991; Wu et al., 2011) and at the plasma membrane together with other viral glycoproteins to facilitate viral assembly and budding (Ali et al., 2000; Baudin et al., 2001; M. Enami and K. Enami, 1996; Gómez-Puertas et al., 2000; Noton et al., 2007; Wu et al., 2011; Ye et al., 1999). Studies on cytoplasmic trafficking of vRNP indicate that Rab11-regulated recycling endosomes translocate vRNPs to the apical plasma membrane (Amorim et al., 2011; Bruce et al., 2010; Eisfeld et al., 2011). It was suggested that Rab11-containing endosome also functions as a platform to co-localize different vRNP segments along their route to the site of assembly (Chou et al., 2013; Lakdawala et al., 2014). However, it is not known if M1 remains associated with vRNP throughout its translocation from the nucleus to the plasma membrane assembly sites.
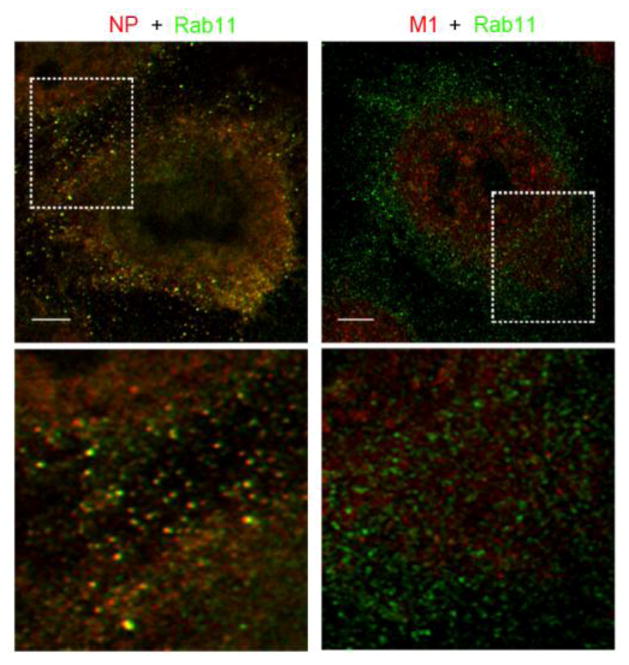
To investigate M1 interaction with vRNP during Rab11-regulated vRNP cytoplasmic trafficking, we determined co-localization of Rab11 with either NP or M1 using a super resolution microscope. A549 cells infected with the virus for 15 h were processed for dual staining of NP or M1 (red channel) with Rab11 (green channel) using specific antibodies. NP showed a distinct co-localization and/or in close association with Rab11 protein (Fig. 5, left column) as reported (Amorim et al., 2011; Bruce et al., 2010; Eisfeld et al., 2011). In sharp contrast, we did not detect obvious co-localization between M1 and Rab11 (Fig. 5, right column). This result may indicate that M1 dissociates from vRNP export complex after nuclear export.
Figure 5. Co-localization of Rab11 with cytoplasmic NP and M1.
Cells infected with WSN for 15 h were processed for IFA with dual staining of NP or M1 (red) with Rab11 (green). Images were obtained using super resolution microscopy with a 100x oil immersion objective. Areas within the white boxes are magnified and shown in the lower panels. Scale bars are 4 μm.
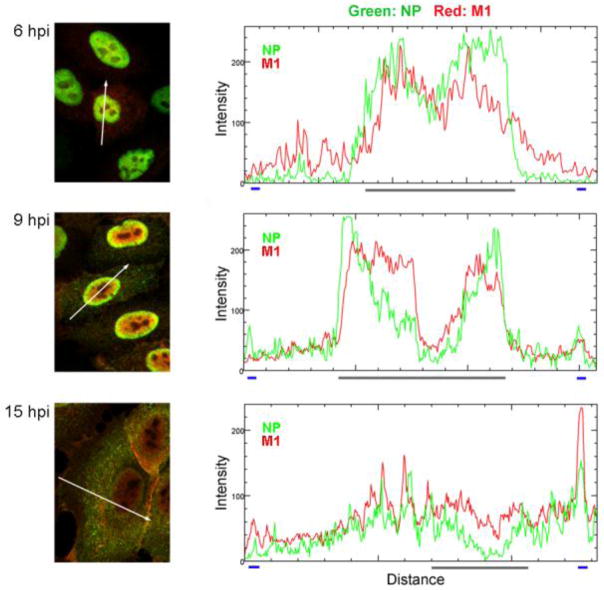
To further analyze the spatiotemporal relationship between influenza vRNP and M1, infected cells were processed for immunostaining with anti-NP and anti-M1 antibodies at 6, 9 and 15 hpi. Co-localization of NP and M1 was visualized using a confocal microscope and analyzed by ImageJ software (Fig. 6). At 6 hpi, punctate staining of influenza NP (green channel) and M1 proteins (red channel) were both distributed in the central area of cell nucleus with a high degree of co-localization (yellow). At 9 hpi, M1 proteins were dispersed throughout the cell nucleus, while NP proteins were consolidated at the internal rim of the nucleus where they also co-localized with M1 proteins (Fig. 6, middle panel). At 15 hpi, we observed that NP and M1 were distributed throughout the cytoplasm of infected cells and were strikingly co-localized around the plasma membrane area (Fig. 6, lower panel). Collectively, these data suggest that M1 specifically interacts with vRNPs in the nucleus for vRNP nuclear export, but is released from the export complex during Rab11-regulated vRNP translocation, and is then re-associated with vRNPs at the plasma membrane, which determines specific incorporation of vRNPs into progeny virion.
Figure 6. Co-localization of NP and M1 at various times after infection.
Infected cells were fixed and permeabilized at indicated times after infection and processed for IFA with anti-NP (green channel) and anti-M1 (red channel) mAbs. Images were obtained using an Olympus FV1000 confocal microscope with a 60× oil immersion objective. Histograms indicate the fluorescence intensities of NP and M1 in the area represented by the white arrow in the image. The X-axis demonstrates the arbitrary unit of distance of the marked white arrow. The blue lines indicate the plasma membrane margin, and the grey line indicates the nuclear margin.
3. Discussion
Replication and assembly processes of influenza A virus are unique and the most complicated among negative strand RNA viruses. The virus replicates its genome in the cell nucleus in which the cRNP replicative intermediates are synthesized to be used as templates to generate nascent vRNPs (Jackson et al., 1982; Krug et al., 1987; Shapiro et al., 1987). Then, progeny vRNA-containing nucleocapsids are exported to the cytoplasm and transported to the plasma membrane assembly sites whereby an entire set of eight vRNA segments is incorporated into budding virions to complete infectious virus formation. In contrast to the extensive studies on vRNP trafficking, little attention has been paid on intracellular localization of cRNP and the mechanism of selective vRNP incorporation into progeny virions. In this study, we found that cRNPs were actually exported to the cytoplasm during virus infection through a CRM1-independent manner (Figs. 1 and 2). The cytosolic cRNPs, however, were not incorporated into progeny virion (Fig. 3). Viral M1 proteins can distinguish vRNP and cRNP and preferentially interact with vRNP (Fig. 4). Our data also suggest that this specific M1-vRNP interaction takes place at the plasma membrane, but not during vRNP translocation through Rab11-mediated recycling endosomes (Figs. 5 and 6).
An early study suggested that influenza cRNAs were synthesized only at early time points (1.5–2.5 hpi) and remained in the nucleus (Shapiro et al., 1987). However, a more recent study using subcellular fractionation and a primer extension assay suggests the presence of cRNAs in the cytoplasm (Chase et al., 2011). In our study, we used highly sensitive and specific strand-specific qRT-PCR to quantify vRNAs and cRNAs for an extended period of time after infection (Fig. 1). Our findings indicate that nuclear cRNA accumulation increases over time until achieving equilibrium at 12 hpi. Interestingly, we also detected a significant amount of cRNAs in the cytosolic fraction, suggesting that, like vRNPs, cRNPs are also exported from the nucleus of infected cells (Fig. 1C). However, the mechanism of cRNP nuclear export seems to be different from that of vRNP, which usurps the cellular CRM1 nuclear exporter (Elton et al., 2001; Ma et al., 2001; K. Watanabe et al., 2001). Using a potent CRM1 inhibitor, LMB, we observed a significant inhibition of vRNP nuclear export, as determined by qRT-PCR (Fig. 2E) along with a significant reduction of infectious virion production (Fig. 2C), which are consistent with previous reports (Chase et al., 2011; Larsen et al., 2014; Mühlbauer et al., 2015). However, LMB treatment did not reduce nuclear export of cRNPs, suggesting that cRNPs are likely to be exported via a CRM1-independent pathway (Fig. 2B). These findings are in agreement with a previous study using a primer extension assay, which reported no significant differences in cRNA accumulation after treatment with LMB (Chase et al., 2011).
A recent study proposed a possibility of passive translocation of RNPs across the enlarged nuclear pores at a later stage of infection, indicating an alternative CRM1-independent nuclear export process. The study reported that influenza virus infection induced caspase activation. Activation of caspase cascades degraded nucleoporin, an essential element of nuclear pore complexes, allowing passive diffusion of macromolecules across the enlarged nuclear pores (Mühlbauer et al., 2015). We used a caspase inhibitor at the same dose reported in this study, but did not observe an inhibitory effect on cRNP nuclear export, which may suggest that the presence of cRNP in the cytosol is not due to a caspase-induced passive diffusion (Fig. 2D and E). Since cRNP nuclear export was CRM1-independent (Fig. 2) and M1 did not interact efficiently with cRNPs (Fig. 4), it is unclear whether cRNPs form a daisy chain with M1 and NEP to navigate their translocation across the nuclear pores. Importantly, the fact that M1 preferentially binds to vRNP, but not cRNP, may suggest some degree of structural difference, especially the surface-exposed and hidden NP residues along the RNA helix, between vRNP and cRNP. Thus, it is possible that cRNPs are exported from the nucleus due to nuclear export signals of NP, which are uniquely exposed on the surface of cRNP. Alternatively, a recent study suggested that influenza polymerase can adopt an alternative configuration depending on which kind of viral RNA (vRNA or cRNA) is bound (Thierry et al., 2016). In addition, NEP was also found to directly interact with the viral polymerase associated with vRNPs, in an M1-dependent manner (Brunotte et al., 2014), leaving open the possibility of a direct interaction between NEP and the viral polymerase associated with cRNPs in navigating cRNP nuclear export. Further analysis will be required to fully understand the cRNP nuclear export process, as well as its biological functions in the cytosol.
It is a striking finding that M1 specifically recognizes and interacts with vRNP, but not cRNP (Fig. 4). M1 interaction with vRNP complexes (Elster et al., 1997; Noton et al., 2007; Ye et al., 1999), as well as viral envelope proteins (Ali et al., 2000; Chen et al., 2008; M. Enami and K. Enami, 1996) facilitates virion formation. Cryogenic electron microscopy studies of RNP revealed its double-helical conformation that is comprised of two NP strands of opposite polarity, by which both strands are connected by a short loop at one end and an association with the viral RNA-dependent RNA polymerase at the other end (Arranz et al., 2012; Moeller et al., 2012). Isolation of influenza positive-sense replicative intermediate, followed by electron microscopy study, revealed that cRNP also exhibits a similar filamentous double-helical structure (York et al., 2013). Even though the viral protein components that constitute vRNPs and cRNPs are likely to be the same, we speculate that vRNP and cRNP might form a slightly different conformation in terms of surface-exposed and hidden NP residues along the RNA helix, which might determine the preferential recognition by M1. In line with this speculation, our previous study indicated that NP residues affect the morphology of progeny virions. Mutations in NP residues 214, 217, and 253, which are located and exposed at the minor groove of the helical vRNP, changed the filamentous virions to spherical, most likely through their interaction with M1 at the assembly sites (Bialas et al., 2014). However, we cannot exclude the possibility that the conformational differences in the viral polymerase associated with vRNPs and cRNPs might determine the preferential interaction of M1 with vRNPs (Thierry et al., 2016). Further investigations on vRNP and cRNP structures and their interactions with viral and cellular partners are required to elucidate the mechanism by which M1 preferentially interacts with vRNP.
As discussed above, specific interaction between M1 and vRNPs is likely to be a key determinant of selective incorporation of vRNPs into virions. However, this interaction seems to take place only during the nuclear export of vRNPs and virion assembly at the plasma membrane, but not during vRNP translocation in the cytosol. Using a super resolution microscope, we observed co-localization of Rab11 and NP, but not M1, in the cytosol (Fig. 5). In fact, associations between M1 and cytoplasmic vRNPs are not well established (Eisfeld et al., 2015). Existing data indicate that vRNPs are not exported from the nucleus as a complete set of eight vRNA segments (Chou et al., 2013; Lakdawala et al., 2014) nor as an individual segment (Lakdawala et al., 2014). During translocation through the Rab11-regulated recycling endosome, vRNPs have been shown to interact with each other and form subcomplexes containing multiple distinct vRNA segments, which are later fused and yield a complete set of eight vRNA segments to be incorporated into progeny virion at the plasma membrane (Lakdawala et al., 2014). Therefore, it is possible that M1 associated with vRNP at the perinuclear region is released from the complex, and binds again to a set of vRNP segments at the plasma membrane during virion assembly. In this case, M1 at the plasma membrane may form a different conformation through its interaction with envelope glycoproteins that allows specific recognition and interaction with a set of vRNPs at the plasma membrane for virion assembly.
Overall, our study indicates that influenza replicative intermediates, cRNPs, are exported from the nucleus to the cytoplasm, but the virus selectively uptake cytosolic vRNPs through a specific interaction with M1 during viral assembly. These data also suggest a possible structural difference between vRNP and cRNP, and temporal M1 interactions with vRNPs while being translocated from the site of synthesis in the nucleus to the site of virion formation at the plasma membrane.
4. Materials and Methods
4.1. Cells, viruses and plasmids
Madin-Darby canine kidney (MDCK), human embryonic kidney 293T, and human alveolar A549 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Corning) supplemented with 8% fetal bovine serum (FBS, Life Technologies or Seradigm) and GlutaMAX (Life Technologies). Influenza A/WSN/1933 (H1N1) was rescued using the 12-plasmid system obtained from Y. Kawaoka (University of Wisconsin, Madison, WI) (Neumann et al., 1999). Virus titers were determined by immunofluorescence assay detecting viral NP or TCID50 assay using MDCK cells. pCAGGS-WSNNP, and pCAGGS-WSNM1 were previously constructed in our lab (Bialas et al., 2014).
4.2. Subcellular fractionation
MDCK cells were seeded on a 6-well plate overnight prior to infection with WSN at a multiplicity of infection (MOI) of 2. Infected cells were maintained at 37°C, 5% CO2 in an incubator. At indicated time points, culture supernatant was collected for virion purification. Infected cells were washed with cold phosphate buffer saline (PBS), scraped and centrifuged at 3,000 rpm for 5 min at 4°C to pellet the cells. Fractionation was performed using the Nuclear/Cytosol Fractionation kit (BioVision) according to the manufacturer’s protocol.
4.3. Western blot analysis
Viral and cellular proteins were applied to a 12% SDS-PAGE gel and transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore). The blot was blocked with 2% dry milk in Tris-buffered saline with Tween 20 (TBST) and then incubated with primary antibodies as follows: anti-lamin A/C mouse mAb, 4C11 (1:1,000; Cell Signaling), anti-α/β tubulin rabbit Ab (1:1,000; Cell Signaling), anti-influenza A NP mouse mAb, NR4282 (1:1,000; BEI Resources), and anti-influenza A M1 mouse mAb GA2B (1:1,000; Abcam). The membrane was incubated with goat anti-mouse IgG conjugated with horseradish peroxidase (HRP) (1:10,000; Bio-Rad) and HRP-conjugated goat anti-rabbit IgG (1:5,000; Bio-Rad). Target proteins were visualized using SuperSignal West Femto maximum sensitivity substrate (Thermo Scientific). Images were captured using the ChemDoc XRS system (Bio-Rad) and analyzed using Quantity One 1-D analysis software (Bio-Rad).
4.4. Strand-specific qRT-PCR
We used a strand-specific real-time qRT-PCR, which has been established for the specific detection of vRNA, cRNA, and mRNA of the WSN NA segment (Kawakami et al., 2011). Virions in the culture supernatants were purified by ultracentrifugation through a 20% sucrose cushion. Total RNAs were extracted from fractionated cells or virions using illustra RNAspin Mini (GE Healthcare). cDNAs complementary to the influenza vRNA and cRNA were synthesized with the hot-start modification of the SuperScript III first-strand synthesis system (Life Technologies) using saturated trehalose (Kawakami et al., 2011). Real-time PCR (qPCR) was performed with SYBR Green qPCR. The cycle conditions of qPCR were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min (Kawakami et al., 2011). Ten-fold serial dilutions (109, 108, 107, 106, 105, 104, 103 copies/μl) of synthetic viral RNA standards were used to generate a standard curve. The quantity of vRNA and cRNA was calculated in Log10 copies/cells. The specificity of strand-specific qRT-PCR primers was tested as follows: WSN NA gene was amplified by PCR using pPolI-WSNNA plasmid. Primers were designed to include T7 promoter sequences at the right direction to produce template vRNA, cRNA and mRNA as follows: AGCGAAAGCAGGAGTTTAAATGAA (forward primer for vRNA), GGATCCTAATACGACTCACTATAGGGAGTAGAAACAAGGAGTTTTTTGAA (reverse primer for vRNA), GGATCCTAATACGACTCACTATAGGGAGCGAAAGCAGGAGTTTAAATGAA (forward primer for cRNA and mRNA), AGTAGAAACAAGGAGTTTTTTGAA (reverse primer for cRNA), and TTTTTTTTTTTTTTTTGAACAAAC (reverse primer for mRNA). PCR products were purified by phenol/chloroform extraction and ethanol precipitation. Template vRNAs, cRNAs and mRNAs were prepared from purified PCR products by in vitro transcription using MAXIscript® T7 Transcription Kit (Ambion, AM1312). Reverse transcription was performed with 109 copies of each template RNAs (vRNA, cRNA and mRNA) using strand-specific primers for WSN NAvRNA, NAcRNA, and NAmRNA (Kawakami et al., 2011). The cDNA products were quantified by qPCR reactions using the specific primer sets.
4.5. Immunofluorescence assay (IFA)
To investigate intracellular localization of NP in the presence or absence of LMB or CI, MDCK cells were infected with WSN at a MOI of 2 and treated with 20 nM LMB (Cell Signaling) or 40 uM CI (R&D Systems) at 1 hpi. At indicated time points (6, 12, and 18 hpi), infected cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.5% Triton X-100 for 10 min at room temperature. Viral NP was detected using mouse anti-NP mAb (1:1,000; BEI Resource, NR19868), followed by donkey anti-mouse IgG Alexa Flour 594 (1:1,000; Thermo Fisher Scientific) and counterstained with DAPI (1:10,000). Images were obtained using an Olympus IX50 inverted fluorescence microscope with a 60× oil immersion objective. Fluorescence intensity at the Alexa Flour 594 channel within nuclei and whole cell was quantified using ImageJ software.
For the analysis of Rab11 co-localization with NP or M1, A549 cells on a round glass cover slip were infected with WSN at a MOI of 2 for 15 h. Infected cells were fixed and permeabilized as described above and were processed for dual staining with NP or M1 with Rab11 using mouse anti-NP mAb (1:100, BEI Resources, NR4282) or anti-M1 mAb GA2B (1:1,000, Abcam) together with rabbit anti-Rab11 pAb (1:1,000, Invitrogen), followed by goat anti-mouse IgG Alexa Fluor 594 (1:1,000, Abcam) and goat anti-rabbit IgG FITC (1:1,000, Invitrogen). Images were obtained using Leica TCS SP8 STED 3X super resolution microscope with a 100×/1.4 oil immersion objective. A sequential scanning program was established based on the manufacturer’s excitation and emission spectra for each fluorophore.
Co-localization of NP and M1 in infected cells was determined as follows. After infection of A549 cells with WSN at MOI of 2, cells were fixed and permeabilized as described above at indicated time points (6, 9, and 15 hpi). Cells were processed for reaction with mouse anti-M1 mAb GA2B (1:1,000; Abcam) followed by goat anti-mouse IgG Alexa Fluor 647 (1:1000; Abcam). Then, cells were incubated with mouse anti-NP mAb directly conjugated with FITC using ReadiLinkTM KLH Conjugation Kit (AAT Bioquest). Images were obtained using an Olympus FV1000 confocal microscope with a 60× oil immersion objective. Co-localization of NP and M1 at various time points after infection was analyzed by ImageJ software.
4.6. Radioimmunoprecipitation (RIP)
293T cells infected with WSN at a MOI of 3 or transfected with pCAGGS vectors expressing NP or M1 were radiolabeled with [35S]Met/Cys (Perkin Elmer) for 16 h at 37°C. Total cell lysates were used for immunoprecipitation using Dynabeads reacted with anti-NP mAb cocktail (BEI Resources, NR4282) or anti-M1 mAb GA2B (Abcam). Eluted samples were analyzed by SDS-PAGE. For RNA analysis, lysates from cells infected with WSN as above were reacted with anti-NP or M1 mAbs. Total RNAs in immunoprecipitated materials and the crude lysates (input control) were isolated using RNAspin Mini kit (GE Healthcare) and applied to qRT-PCR reaction using WSN NA gene strand-specific primers to quantify vRNAs and cRNAs.
4.7. Statistical analysis
Statistical analysis was performed using one-way ANOVA followed by Tukey’s multiple comparison test (JMP Pro 12). P-value of < 0.05 was considered to be statistically significant.
Highlight.
Influenza cRNPs are exported from the nucleus of an infected cell via a CRM1-independent pathway.
Influenza A viruses selectively incorporate cytosolic vRNPs through a specific interaction with M1 during viral assembly
M1 dissociates from vRNP export complex after nuclear export, and is re-associated with vRNPs at the plasma membrane.
Acknowledgments
This work was supported by National Institutes of Health Grant R01AI081779 and R21-A1078130. We would like to thank Linda Callahan for the use of the URMC Confocal and Conventional Microscopy Core.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akarsu H, Burmeister WP, Petosa C, Petit I, Müller CW, Ruigrok RWH, Baudin F. Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2).\ EMBO J. 2003;22:4646–4655. doi: 10.1093/emboj/cdg449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Avalos RT, Ponimaskin E, Nayak DP. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein.\ J Virol. 2000;74:8709–8719. doi: 10.1128/jvi.74.18.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim MJ, Bruce EA, Read EKC, Foeglein A, Mahen R, Stuart AD, Digard P. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA.\ J Virol. 2011;85:4143–4156. doi: 10.1128/JVI.02606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arranz R, Coloma R, Chichón FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortín J, Martín-Benito J. The structure of native influenza virion ribonucleoproteins. Science. 2012;338:1634–1637. doi: 10.1126/science.1228172. [DOI] [PubMed] [Google Scholar]
- Baudin F, Petit I, Weissenhorn W, Ruigrok RW. In vitro dissection of the membrane and RNP binding activities of influenza virus M1 protein. Virology. 2001;281:102–108. doi: 10.1006/viro.2000.0804. [DOI] [PubMed] [Google Scholar]
- Bialas KM, Bussey KA, Stone RL, Takimoto T. Specific nucleoprotein residues affect influenza virus morphology.\ J Virol. 2014;88:2227–2234. doi: 10.1128/JVI.03354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce EA, Digard P, Stuart AD. The Rab11 pathway is required for influenza A virus budding and filament formation.\ J Virol. 2010;84:5848–5859. doi: 10.1128/JVI.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunotte L, Flies J, Bolte H, Reuther P, Vreede F, Schwemmle M. The nuclear export protein of H5N1 influenza A viruses recruits Matrix 1 (M1) protein to the viral ribonucleoprotein to mediate nuclear export.\ J Biol Chem. 2014;289:20067–20077. doi: 10.1074/jbc.M114.569178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui M, Wills EG, Helenius A, Whittaker GR. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins.\ J Virol. 2000;74:1781–1786. doi: 10.1128/jvi.74.4.1781-1786.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Liu X, Yu M, Li J, Jia X, Bi Y, Sun L, Gao GF, Liu W. A nuclear export signal in the matrix protein of Influenza A virus is required for efficient virus replication.\ J Virol. 2012;86:4883–4891. doi: 10.1128/JVI.06586-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase GP, Rameix-Welti M-A, Zvirbliene A, Zvirblis G, Götz V, Wolff T, Naffakh N, Schwemmle M. Influenza virus ribonucleoprotein complexes gain preferential access to cellular export machinery through chromatin targeting.\ PLoS Pathog. 2011;7:e1002187. doi: 10.1371/journal.ppat.1002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Leser GP, Jackson D, Lamb RA. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding.\ J Virol. 2008;82:10059–10070. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y-Y, Heaton NS, Gao Q, Palese P, Singer RH, Singer R, Lionnet T. Colocalization of different influenza viral RNA segments in the cytoplasm before viral budding as shown by single-molecule sensitivity FISH analysis.\ PLoS Pathog. 2013;9:e1003358. doi: 10.1371/journal.ppat.1003358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y-Y, Vafabakhsh R, Doğanay S, Gao Q, Ha T, Palese P. One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis.\ Proc Natl Acad Sci USA. 2012;109:9101–9106. doi: 10.1073/pnas.1206069109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutiwitoonchai N, Aida Y. NXT1, a Novel Influenza A NP Binding Protein, Promotes the Nuclear Export of NP via a CRM1-Dependent Pathway.\ Viruses. 2016;8 doi: 10.3390/v8080209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva UC, Tanaka H, Nakamura S, Goto N, Yasunaga T. A comprehensive analysis of reassortment in influenza A virus. Biol Open. 2012;1:385–390. doi: 10.1242/bio.2012281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisfeld AJ, Kawakami E, Watanabe T, Neumann G, Kawaoka Y. RAB11A is essential for transport of the influenza virus genome to the plasma membrane.\ J Virol. 2011;85:6117–6126. doi: 10.1128/JVI.00378-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisfeld AJ, Neumann G, Kawaoka Y. At the centre: influenza A virus ribonucleoproteins.\ Nat Rev Microbiol. 2015;13:28–41. doi: 10.1038/nrmicro3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elster C, Larsen K, Gagnon J, Ruigrok RW, Baudin F. Influenza virus M1 protein binds to RNA through its nuclear localization signal.\ J Gen Virol. 1997;78(Pt 7):1589–1596. doi: 10.1099/0022-1317-78-7-1589. [DOI] [PubMed] [Google Scholar]
- Elton D, Simpson-Holley M, Archer K, Medcalf L, Hallam R, McCauley J, Digard P. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway.\ J Virol. 2001;75:408–419. doi: 10.1128/JVI.75.1.408-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enami M, Enami K. Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein.\ J Virol. 1996;70:6653–6657. doi: 10.1128/jvi.70.10.6653-6657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Puertas P, Albo C, Pérez-Pastrana E, Vivo A, Portela A. Influenza virus matrix protein is the major driving force in virus budding.\ J Virol. 2000;74:11538–11547. doi: 10.1128/jvi.74.24.11538-11547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Chen J, Chen Q, Wang H, Yao Y, Chen J, Chen Z. A second CRM1-dependent nuclear export signal in the influenza A virus NS2 protein contributes to the nuclear export of viral ribonucleoproteins.\ J Virol. 2013;87:767–778. doi: 10.1128/JVI.06519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Liu T, Muller J, Levandowski RA, Ye Z. Effect of influenza virus matrix protein and viral RNA on ribonucleoprotein formation and nuclear export. Virology. 2001;287:405–416. doi: 10.1006/viro.2001.1067. [DOI] [PubMed] [Google Scholar]
- Iwatsuki-Horimoto K, Horimoto T, Fujii Y, Kawaoka Y. Generation of influenza A virus NS2 (NEP) mutants with an altered nuclear export signal sequence.\ J Virol. 2004;78:10149–10155. doi: 10.1128/JVI.78.18.10149-10155.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Caton AJ, McCready SJ, Cook PR. Influenza virus RNA is synthesized at fixed sites in the nucleus. Nature. 1982;296:366–368. doi: 10.1038/296366a0. [DOI] [PubMed] [Google Scholar]
- Kawakami E, Watanabe T, Fujii K, Goto H, Watanabe S, Noda T, Kawaoka Y. Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J Virol Methods. 2011;173:1–6. doi: 10.1016/j.jviromet.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe DM, Howley P. Fields Virology. 2013:2664. [Google Scholar]
- Krug RM, St Angelo C, Broni B, Shapiro G. Transcription and replication of influenza virion RNA in the nucleus of infected cells.\ Cold Spring Harb Symp Quant Biol. 1987;52:353–358. doi: 10.1101/sqb.1987.052.01.040. [DOI] [PubMed] [Google Scholar]
- Lakdawala SS, Wu Y, Wawrzusin P, Kabat J, Broadbent AJ, Lamirande EW, Fodor E, Altan-Bonnet N, Shroff H, Subbarao K. Influenza a virus assembly intermediates fuse in the cytoplasm.\ PLoS Pathog. 2014;10:e1003971. doi: 10.1371/journal.ppat.1003971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen S, Bui S, Perez V, Mohammad A, Medina-Ramirez H, Newcomb LL. Influenza polymerase encoding mRNAs utilize atypical mRNA nuclear export. Virology Journal. 2014;11:154. doi: 10.1186/1743-422X-11-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Roy AM, Whittaker GR. Nuclear export of influenza virus ribonucleoproteins: identification of an export intermediate at the nuclear periphery. Virology. 2001;282:215–220. doi: 10.1006/viro.2001.0833. [DOI] [PubMed] [Google Scholar]
- Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. Organization of the influenza virus replication machinery. Science. 2012;338:1631–1634. doi: 10.1126/science.1227270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlbauer D, Dzieciolowski J, Hardt M, Hocke A, Schierhorn KL, Mostafa A, Müller C, Wisskirchen C, Herold S, Wolff T, Ziebuhr J, Pleschka S. Influenza virus-induced caspase-dependent enlargement of nuclear pores promotes nuclear export of viral ribonucleoprotein complexes.\ J Virol. 2015;89:6009–6021. doi: 10.1128/JVI.03531-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak DP, Balogun RA, Yamada H, Zhou ZH, Barman S. Influenza virus morphogenesis and budding.\ Virus Res. 2009;143:147–161. doi: 10.1016/j.virusres.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Hughes MT, Kawaoka Y. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1.\ EMBO J. 2000;19:6751–6758. doi: 10.1093/emboj/19.24.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Generation of influenza A viruses entirely from cloned cDNAs.\ Proc Natl Acad Sci USA. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Kawaoka Y. Structure of influenza virus ribonucleoprotein complexes and their packaging into virions.\ Rev Med Virol. 2010;20:380–391. doi: 10.1002/rmv.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Kawaoka Y. Packaging of influenza virus genome: robustness of selection.\ Proc Natl Acad Sci USA. 2012;109:8797–8798. doi: 10.1073/pnas.1206736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Sagara H, Yen A, Takada A, Kida H, Cheng RH, Kawaoka Y. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature. 2006;439:490–492. doi: 10.1038/nature04378. [DOI] [PubMed] [Google Scholar]
- Noton SL, Medcalf E, Fisher D, Mullin AE, Elton D, Digard P. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions.\ J Gen Virol. 2007;88:2280–2290. doi: 10.1099/vir.0.82809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill RE, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins.\ EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro GI, Gurney T, Krug RM. Influenza virus gene expression: control mechanisms at early and late times of infection and nuclear-cytoplasmic transport of virus-specific RNAs.\ J Virol. 1987;61:764–773. doi: 10.1128/jvi.61.3.764-773.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Takizawa N, Watanabe K, Nagata K, Kobayashi N. Crucial role of the influenza virus NS2 (NEP) C-terminal domain in M1 binding and nuclear export of vRNP.\ FEBS Lett. 2011;585:41–46. doi: 10.1016/j.febslet.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Thierry E, Guilligay D, Kosinski J, Bock T, Gaudon S, Round A, Pflug A, Hengrung N, El Omari K, Baudin F, Hart DJ, Beck M, Cusack S. Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of PB2 Domains. Mol Cell. 2016;61:125–137. doi: 10.1016/j.molcel.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Harmon A, Jin J, Francis DH, Christopher-Hennings J, Nelson E, Montelaro RC, Li F. The lack of an inherent membrane targeting signal is responsible for the failure of the matrix (M1) protein of influenza A virus to bud into virus-like particles.\ J Virol. 2010;84:4673–4681. doi: 10.1128/JVI.02306-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Takizawa N, Katoh M, Hoshida K, Kobayashi N, Nagata K. Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B.\ Virus Res. 2001;77:31–42. doi: 10.1016/s0168-1702(01)00263-5. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Ibrahim MS, Suzuki Y, Ikuta K. The changing nature of avian influenza A virus (H5N1).\ Trends Microbiol. 2012;20:11–20. doi: 10.1016/j.tim.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Wu C-Y, Jeng K-S, Lai MM-C. The SUMOylation of matrix protein M1 modulates the assembly and morphogenesis of influenza A virus.\ J Virol. 2011;85:6618–6628. doi: 10.1128/JVI.02401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzer WJ, Planz O, Ehrhardt C, Giner M, Silberzahn T, Pleschka S, Ludwig S. Caspase 3 activation is essential for efficient influenza virus propagation.\ EMBO J. 2003;22:2717–2728. doi: 10.1093/emboj/cdg279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Liu T, Offringa DP, McInnis J, Levandowski RA. Association of influenza virus matrix protein with ribonucleoproteins.\ J Virol. 1999;73:7467–7473. doi: 10.1128/jvi.73.9.7467-7473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York A, Hengrung N, Vreede FT, Huiskonen JT, Fodor E. Isolation and characterization of the positive-sense replicative intermediate of a negative-strand RNA virus.\ Proc Natl Acad Sci USA. 2013;110:E4238–45. doi: 10.1073/pnas.1315068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Tao YJ. Structure and assembly of the influenza A virus ribonucleoprotein complex.\ FEBS Lett. 2013;587:1206–1214. doi: 10.1016/j.febslet.2013.02.048. [DOI] [PubMed] [Google Scholar]