Abstract
The transport of drug molecules is mainly determined by the distribution of influx and efflux transporters for which they are substrates. To enable tissue targeting, we sought to develop the idea that we might affect the transporter-mediated disposition of small-molecule drugs via the addition of a second small molecule that of itself had no inhibitory pharmacological effect but that influenced the expression of transporters for the primary drug. We refer to this as a “binary weapon” strategy. The experimental system tested the ability of a molecule that on its own had no cytotoxic effect to increase the toxicity of the nucleoside analog gemcitabine to Panc1 pancreatic cancer cells. An initial phenotypic screen of a 500-member polar drug (fragment) library yielded three “hits.” The structures of 20 of the other 2,000 members of this library suite had a Tanimoto similarity greater than 0.7 to those of the initial hits, and each was itself a hit (the cheminformatics thus providing for a massive enrichment). We chose the top six representatives for further study. They fell into three clusters whose members bore reasonable structural similarities to each other (two were in fact isomers), lending strength to the self-consistency of both our conceptual and experimental strategies. Existing literature had suggested that indole-3-carbinol might play a similar role to that of our fragments, but in our hands it was without effect; nor was it structurally similar to any of our hits. As there was no evidence that the fragments could affect toxicity directly, we looked for effects on transporter transcript levels. In our hands, only the ENT1-3 uptake and ABCC2,3,4,5, and 10 efflux transporters displayed measurable transcripts in Panc1 cultures, along with a ribonucleoside reductase RRM1 known to affect gemcitabine toxicity. Very strikingly, the addition of gemcitabine alone increased the expression of the transcript for ABCC2 (MRP2) by more than 12-fold, and that of RRM1 by more than fourfold, and each of the fragment “hits” served to reverse this. However, an inhibitor of ABCC2 was without significant effect, implying that RRM1 was possibly the more significant player. These effects were somewhat selective for Panc cells. It seems, therefore, that while the effects we measured were here mediated more by efflux than influx transporters, and potentially by other means, the binary weapon idea is hereby fully confirmed: it is indeed possible to find molecules that manipulate the expression of transporters that are involved in the bioactivity of a pharmaceutical drug. This opens up an entirely new area, that of chemical genomics-based drug targeting.
Keywords: binary weapon, cheminformatics, gemcitabine, anticancer drugs, pancreatic cancer, drug transporters, phenotypic screening
Introduction
In a typical small molecule drug discovery programme pipeline, candidate (“hit”) compounds for treating a particular disease are selected from a large chemical library, and after various modifications (to form “leads” and variants thereof) enter “phase 1,” a testing for safety at low doses in healthy volunteers. “Attrition” is a term used to describe the failure of such molecules to progress further to market, via phases 2 and 3 (small and larger clinical trials) (Kola and Landis, 2004; Empfield and Leeson, 2010; Leeson and Empfield, 2010; Leeson, 2016). Nowadays attrition occurs largely for reasons of toxicity or lack of efficacy (Kola and Landis, 2004; Arrowsmith and Miller, 2013), and runs in excess of 90% (e.g., Kola and Landis, 2004; Kell, 2013, and see for full details http://csdd.tufts.edu/files/uploads/Tufts_CSDD_briefing_on_RD_cost_study_-_Nov_18,_2014.pdf), with gross pharmacokinetics and pharmacodynamics (as assessed at the whole organ level) being seen as less of an issue than it once was (Kola and Landis, 2004). The simple consequence of this level of attrition is that it costs ~10 times more than it might, per molecule, now as much as $2.5 Bn, to bring a drug successfully to market.
Role of transporters in cellular drug uptake
We have argued that a lack of understanding of human metabolism and of the transporters necessary to get orally active drugs across intestinal epithelia and into target cells is one of the chief causes of attrition. By now, following a similar programme in yeast (Herrgård et al., 2008), we do have a reasonable model of the human metabolic network (Swainston et al., 2013, 2016; Thiele et al., 2013), with fully one third of the steps involving some kind of transport(er), and with uptake transporters of the SoLute Carrier families (SLCs) (Hediger et al., 2004, 2013) being woefully understudied (César-Razquin et al., 2015). In particular, although it remains underappreciated, we have rehearsed on multiple occasions the abundant evidence that the non-transporter- (i.e., bilayer-) mediated uptake of drugs through intact cell membranes is normally negligible (e.g., Dobson and Kell, 2008; Dobson et al., 2009a,b; Kell et al., 2011, 2013, 2015; Lanthaler et al., 2011; Kell, 2013, 2015a,b, 2016a,b; Kell and Goodacre, 2014; Kell and Oliver, 2014; Mendes et al., 2015; O'Hagan and Kell, 2015a; Kell, 2015a,b), a striking recent example being that of Superti-Furga and colleagues (Winter et al., 2014). This shifts the agenda to one of molecular enzymology and systems biology, in which we need to discover (i) which transporters transport which drugs (Giacomini et al., 2010; Sugiyama and Steffansen, 2013), (ii) their expression profiles in different membranes and tissues, and (iii) their kinetic properties. In other words it leads us to recognize that this is fundamentally a problem of systems pharmacology (e.g., van der Greef and Mcburney, 2005; Berger and Iyengar, 2009; van der Graaf and Benson, 2011; Antman et al., 2012; Rostami-Hodjegan, 2012; Waldman and Terzic, 2012; Zhao and Iyengar, 2012; Kell and Goodacre, 2014; Westerhoff et al., 2015; Kell, 2015a).
Transporter-mediated drug targeting
A particularly nice example of the overwhelming use of transporters for drug uptake comes from the study of Superti-Furga and colleagues (Winter et al., 2014) using haploid cells and determining that very much less than 1% of sepantronium uptake could have occurred other than via a specific SLC called SLC35F2. In a similar and complementary vein, the expression profile of specific transporters allows one to target drug substrates to the particular tissues in which the relevant transporters are most highly expressed. This has been illustrated beautifully by Pfefferkorn and colleagues for both a glucokinase activator (Pfefferkorn et al., 2012; Pfefferkorn, 2013; Sharma et al., 2015) and a “statin”-type drug (Pfefferkorn et al., 2011) that are both targeted to the liver via proteins of the Organic Anion Transport Protein (OATP/SLCO/SLC21) (Hagenbuch and Stieger, 2013) family. In this case substantial concentration ratios of e.g., hepatocyte: pancreas of 50:1 (Pfefferkorn et al., 2012) and hepatocyte:myocyte of 250,000:1 (Pfefferkorn et al., 2011) could be achieved (a finding hard to explain on the basis of any significant bilayer permeability!). Other examples of tissue-selective drug targeting include a liver-targeted stearoyl desaturase inhibitor (Oballa et al., 2011; Ramtohul et al., 2011; Liu, 2013), various other liver-targeted drugs based on OATPs (Buxhofer-Ausch et al., 2013; Tu et al., 2013), and a prostate-specific targeting of an iodide transporter for radio-iodine-mediated cell killing (Kakinuma et al., 2003).
These examples show what can be achieved in terms of drug targeting if the transporter distribution happens to work to one's advantage “naturally,” but cannot be exploited directly when it does not.
The glucokinase activator case is important, since if such drug molecules were allowed to enter all tissues they proved toxic (Pfefferkorn et al., 2012; Pfefferkorn, 2013). A similar and particular case of interest is that of broadly cytotoxic anticancer drugs, where we evidently need mechanisms to target them solely to the tissue of interest, and where we might then greatly improve their therapeutic index. Since the tissue-dependent expression of such transporter molecules is highly heterogeneous (see e.g., almost any dataset in the human protein atlas http://proteinatlas.org/ Uhlén et al., 2015, including those for SLC28 http://www.proteinatlas.org/search/slc28 and SLC29 http://www.proteinatlas.org/search/slc29), it must be subject to regulation (e.g., Pennycooke et al., 2001; Del Santo et al., 2001; Fernández-Veledo et al., 2004, 2007; Plant, 2016). Thus, just as with the small-molecule-driven induction of pluripotent stem cells (Okita et al., 2007; Feng et al., 2009; Desponts and Ding, 2010; Li and Ding, 2010; Zhang, 2010; Grskovic et al., 2011; Li et al., 2012, 2014; Li X. et al., 2015; Jung et al., 2014; Kang et al., 2014), that regulation can similarly be affected by pharmacological intervention with small molecule effectors. Thus, our aim was to seek small molecules that were themselves without cytotoxic effects but that could increase the response of different target cells to anti-cancer drugs that are otherwise present at only a barely cytotoxic level, in particular by modulating the level of activities of specific uptake transporters. It differs from the use of pairs of existing drugs of known activities (e.g., Borisy et al., 2003; Lehár et al., 2007, 2008, 2009a,b; Zimmermann et al., 2007; Wright, 2016), but, interestingly, bears a clear resemblance to the overall strategy used in traditional Chinese medicine where a “shi” (“courier”) herb is used to assist the delivery of the main ingredient (“Jun” or “Emperor” herb) to its site of action (Zhao et al., 2015). We refer to this combination as a “binary weapon.”
Gemcitabine and pancreatic cancer
The nucleoside analog gemcitabine (2',2'-difluorodeoxycytidine, Gemzar®) (Alvarellos et al., 2014) is one of the most commonly used chemotherapeutic agents in pancreatic adenocarcinoma, the carcinoma with arguably the least favorable prognosis (5-year survival time) of any (Bhattacharjee et al., 2014; Waddell et al., 2015). Like all nucleoside inhibitors of this type, it must first be transported into the cell and then be metabolized (phosphorylated) to exert its clinical action (thereby lowering its ability to act as a substrate for efflux pumps Fukuda and Schuetz, 2012, though see below). Gemcitabine has multiple intracellular targets, and up-regulation of these targets or nucleoside-metabolizing enzymes such as ribonucleotide reductase (RRM1) may confer resistance to this drug (Bergman et al., 2002, 2005; Nakano et al., 2007; Minami et al., 2015). The main uptake transporters are considered to be ENT1 (SLC29A1) and CNT1/3 (SLC28A1/3) of the SLC28/29 families (Kong et al., 2004; Podgorska et al., 2005; Veltkamp et al., 2008; Young et al., 2008, 2013; Molina-Arcas et al., 2009; Cano-Soldado and Pastor-Anglada, 2012; Molina-Arcas and Pastor-Anglada, 2013) (Table 1). SLC28 transporters are sodium-dependent concentrative nucleoside transporters (Smith et al., 2007), while SLC29 are equilibrative. Notably, there is considerable evidence that the potency (cytotoxicity) of gemcitabine is strongly related to the expression level(s) of these transporters (e.g., Burke et al., 1998; Mackey et al., 1998a,b; Baldwin et al., 1999; Rauchwerger et al., 2000; Cass, 2001; Achiwa et al., 2004; Spratlin et al., 2004; Giovannetti et al., 2005, 2006, 2007; King et al., 2006; Marcé et al., 2006; Mey et al., 2006; Mini et al., 2006; Leung and Tse, 2007; Mori et al., 2007; Oguri et al., 2007; Zhang et al., 2007; Cano-Soldado et al., 2008; Molina-Arcas et al., 2008; Pérez-Torras et al., 2008; Veltkamp et al., 2008; Andersson et al., 2009; Damaraju et al., 2009; Farrell et al., 2009, 2016; Köse and Schiedel, 2009; Maréchal et al., 2009, 2012; Wong et al., 2009; Hagmann et al., 2010; Lane et al., 2010; Molina-Arcas and Pastor-Anglada, 2010; Okazaki et al., 2010; Paproski et al., 2010, 2013; Santini et al., 2010, 2011; Spratlin and Mackey, 2010; Tanaka et al., 2010; Bhutia et al., 2011; De Pas et al., 2011; Gusella et al., 2011; Komori et al., 2011; Matsumura et al., 2011; Borbath et al., 2012; Choi, 2012; Gesto et al., 2012; Kobayashi et al., 2012; Koczor et al., 2012; Morinaga et al., 2012; Murata et al., 2012; Eto et al., 2013; Nakagawa et al., 2013; Skrypek et al., 2013; Xiao et al., 2013; Chan et al., 2014; Deng et al., 2014; Greenhalf et al., 2014; Khatri et al., 2014; Koay et al., 2014; Lee et al., 2014; Lemstrová et al., 2014; Liu et al., 2014; Nordh et al., 2014; Tavano et al., 2014; Wu et al., 2014; de Sousa Cavalcante and Monteiro, 2014; Hung et al., 2015; Pastor-Anglada and Pérez-Torras, 2015; Yamada et al., 2016).
Table 1.
The main human nucleoside transporters and some of their properties.
| Name | Notes | Nucleosides transported | |||
|---|---|---|---|---|---|
| Adenosine | Thymidine | Cytidine | Guanosine | ||
| ENT1/SLC29A1 | Transports adenosine, guanosine, inosine, uridine, cytidine, thymidine with Km-values ranging from 50 to 580 μM (You and Morris, 2014) | High affinity (Aran and Plagemann, 1992; Mackey et al., 1998b; Ward et al., 2000; Vickers et al., 2002; Spratlin et al., 2004) | High affinity (Aran and Plagemann, 1992; Mackey et al., 1998b; Ward et al., 2000; Vickers et al., 2002; Spratlin et al., 2004) | High affinity (Aran and Plagemann, 1992; Mackey et al., 1998b; Ward et al., 2000; Vickers et al., 2002; Spratlin et al., 2004) | High affinity (Aran and Plagemann, 1992; Mackey et al., 1998b; Ward et al., 2000; Spratlin et al., 2004) |
| ENT2/SLC29A2 | To date, hENT2 (and intracellular hENT3) are the first discovered, and so far only identified, transporter proteins for nucleobases inside human cells and tissues (Yao et al., 2011) | hENT2 is a generally low affinity nucleoside transporter with 2.6-, 2.8-, 7.7-, and 19.3-fold lower affinity than hENT1 for thymidine, adenosine, cytidine, and guanosine, respectively (Aran and Plagemann, 1992; Mackey et al., 1998b; Ward et al., 2000; Spratlin et al., 2004; You and Morris, 2014). High affinity toward adenosine metabolites (inosine and hypoxanthine) (Ward et al., 2000). 4-fold higher toward inosine (Aran and Plagemann, 1992; Ward et al., 2000; Vickers et al., 2002; Spratlin et al., 2004; You and Morris, 2014) | |||
| ENT3/SLC29A3 | Also carries purines and pyrimidines nucleosides but functions predominantly in intracellular membranes (Baldwin et al., 2005; Yao et al., 2011) | Low affinity (Baldwin et al., 2005). Transport exhibits a pH optimum (Baldwin et al., 2005; You and Morris, 2014) | Also transports adenine (Yao et al., 2011; You and Morris, 2014) | ||
| CNT1/SLC28A1 | Pyrimidine selective (Ward et al., 2000; Ritzel et al., 2001; You and Morris, 2014) | Also transports adenosine with high affinity (Ritzel et al., 2001) | |||
| CNT2/SLC28A2 | Purine selective (Loewen et al., 1999; Wang and Giacomini, 1999; Ward et al., 2000; Saunders et al., 2011) | High affinity for purines (Km < 10 μM) (Cansev, 2006; Huber-Ruano et al., 2010; You and Morris, 2014) | Low affinity for pyrimidines? (Cansev, 2006) | ||
| CNT3/SLC28A3 | Non-selective for purine and pyrimidine (Ward et al., 2000; You and Morris, 2014) | ||||
SLC29 family and concentrative and Na+-coupled, while SLC28 family is equilibrative. ENT4 also exists but seems to play little or any role.
ABC-type efflux transporters are heavily involved in drug resistance in both mammals (e.g., Liu et al., 2005; Fukuda and Schuetz, 2012; Rosenberg et al., 2015; Silva et al., 2015) and microbes (e.g., Putman et al., 2000; Du et al., 2014; Prasad and Rawal, 2014; Li X.-Z. et al., 2015), and may be of value in industrial biotechnology (Kell et al., 2015). Gemcitabine may also be a substrate for certain efflux transporters such as ABCG2/BRCP (König et al., 2005; Keppler, 2011; Chen et al., 2012; Lemstrová et al., 2014), although “knockdown of ABCC3, ABCC5 or ABCC10 individually did not significantly increase gemcitabine sensitivity” (Rudin et al., 2011). Finally, gemcitabine may also be deaminated in plasma, leading to its clearance (Hodge et al., 2011).
Other small molecules known to affect the response of pancreatic cancer cells to gemcitabine include nicotine (Banerjee et al., 2013, 2014), while molecules that affect nucleoside transporter expression include bile acids (Klein et al., 2009). Finally, erlotinib, gefitinib, and vandetanib inhibit human nucleoside transporters and thereby protect cancer cells from gemcitabine cytotoxicity (Damaraju et al., 2014), while a variety of kinase inhibitors (Huang et al., 2002, 2003, 2004) and dihydropyridine-type calcium channel antagonists (Li et al., 2007) may also affect nucleoside transport.
In particular, however, and not least since the small molecule indole-3-carbinol (which is probably converted to 3,3′-diindolylmethane Banerjee et al., 2009) had been stated to increase both ENT1 expression and the sensitivity of pancreatic carcinoma cells to gemcitabine (Wang et al., 2011), possibly acting via miRNA-21 (Giovannetti et al., 2010; Hwang et al., 2010; Melkamu et al., 2010; Paik et al., 2013), as too did the molecule “S-1” (Nakahira et al., 2008; Jordheim and Dumontet, 2013), the gemcitabine/nucleoside transporter system seemed ideal for the test of our “binary weapon” strategy. The present paper reports the results of this approach.
Materials and methods
Cells and reagents
The human pancreatic duct epithelioid carcinoma cell line, Panc1 (see Gou et al., 2007), and the human embryonic kidney cell line, HEK293 were grown in Dulbecco's modified Eagle's medium (DMEM) (Sigma). The human bone marrow neuroblastoma cell line, SH-SY5Y was grown in a 1:1 mixture of Eagle's Minimum Essential Medium (Sigma) and F12 Medium (Sigma). All cell culture media were supplemented with 10% heat-inactivated fetal bovine serum (FBS), 200 mM L-glutamine, and a 5 mL solution containing 10,000 units.mL-1 penicillin and 10 mg.mL-1 streptomycin. The immortal human pancreatic duct epithelial cell line, hPDE was grown in Keratinocyte-SFM (1X) medium (ThermoFisher), supplemented with 10 mg.mL-1 streptomycin. All four cell lines were obtained and karyotyped locally. Cells were routinely maintained at 37°C in a humidified 5% CO2 atmosphere, in continuous exponential growth at a cell density ranging between 1 × 105 and 1 × 106 cells.mL-1, by passaging every 3 or 4 days. Cell line authenticity was confirmed through karyotype testing (University of Manchester, UK).
Cell growth/viability assay
Cells were seeded in a 96-well plate at a density of 5,000 cells/well, in triplicate, and left to attach. Gemcitabine, present at different concentrations, was added directly to the cells, and left to incubate for an additional 96 h. Cells were then subjected to the MTT Cell Proliferation Assay as per the manufacturer's instructions (Sigma). Absorbance at 570 nm was measured 3 h after the addition of 10 μL of MTT salt reagent/well.
Maybridge fragment screening
Maybridge fragments (MBFs) obeying the “rule of three” (Congreve et al., 2003) were supplied at 100 mM in DMSO and were deployed into the assay plates using an ECHO contactless liquid handler (Labcyte, Inc). For screening purposes, the first 500 MBFs (Library 1) were pooled, i.e., each well in a 96-well plate had a pool of six MBFs. Cells were seeded in a 96-well plate at a density of 5,000 cells/well, in triplicate, and left to attach overnight. Following incubation, the growth medium was replaced with fresh medium containing the pooled MBFs, each fragment present at 10 μM, followed by an additional 24 h incubation. The cells were further incubated with the fragments in the presence of gemcitabine at 20 nM for 96 h. Cells were then subjected to the MTT Cell Proliferation Assay as described above.
To study the effect of each MBF on its own rather than in a pool, the candidate pooled fragments (i.e., showing activity) were de-convolved, i.e., one MBF/well, and cells were plated and treated as described above.
Specificity experiments
SH-SY5Y cells were seeded at a density of 12,500 cells/well, HEK293 and hPDE cells at a density of 10,000 cells/well, in triplicate in a 96-well plate, and left to attach overnight. Following incubation, the medium was replaced with fresh medium containing the MBF hits (i.e., MBF D1, B1, 10, 11, 12, and 20) at 10 μM, followed by an additional 24 h incubation period. Cells were further incubated with the fragments in the presence of gemcitabine at 100 nM for 72 h (SH-SY5Y cells) and 96 h (HEK293 and hPDE cells). Cell viability was then assessed using the MTT Cell Proliferation Assay.
Cheminformatic analyses
These were all performed as in our previous work of this type (O'Hagan and Kell, 2015a,b,c; O'Hagan et al., 2015; O'Hagan and Kell, 2016), using the KNIME workflow system (see e.g., Berthold et al., 2008; Mazanetz et al., 2012; Meinl et al., 2012; Warr, 2012; O'Hagan and Kell, 2015b and http://knime.org/).
Maybridge fragment titration experiments
Cells were seeded in a 96-well plate at a density of 5,000 cells/well, in triplicate, and left to attach overnight. Following incubation, the medium was replaced with fresh medium containing MBFs at different concentrations (3, 10, 30, 100, and 300 μM) followed by an additional 24 h incubation. Cells were further incubated with the fragments in the presence of gemcitabine at 100 nM for 96 h. Cells were then assessed using the MTT Cell Proliferation Assay as described earlier.
Cell culture treatments for gene dysregulation studies
To examine the effect of gemcitabine and MBFs, alone or in combination; on expression of the influx and efflux transporter genes and of the RRM1 gene, cells were seeded in a 6-well plate at a density of 30,000 cells/well, in duplicate, and left to attach overnight. Following incubation, in studies where the effects of the MBFs alone were studied, the medium was replaced with fresh medium containing MBFs at 10 μM, followed by further incubation for 24 h. For studies where the effects of gemcitabine alone were studied, the medium was replaced with fresh medium containing gemcitabine at 100 nM, followed by further incubation for 96 h. For studies in which cells were treated with gemcitabine in combination with the fragments, the cells were first pre-treated with MBFs at 10 μM for 24 h, followed by further incubation with the fragments at 10 μM in the presence of gemcitabine at 100 nM for 96 h. Cells were harvested using TRIzol® reagent (Life Technologies) and stored in −80°C until use.
Total RNA isolation and quantitative real-time reverse transcription polymerase chain reaction (RT-qPCR)
Following treatment as described above, total cellular RNA was isolated from the cells using the RNeasy isolation kit (Qiagen) according to the manufacturer's instructions. RNA concentration was determined using a NanoDrop® Spectrophotometer (NanoDrop ND-1000, NanoDrop Technologies, Wilmington, USA). The OD_260/280 nm ratios of all RNA samples were determined to be between 1.9 and 2.0, suggesting that all RNA samples were highly pure. RNA integrity was verified by the Agilent RNA 6000 Nano assay kit (Agilent Bioanalyser 2100, Agilent Technologies, Cheadle, UK) as described by the manufacturer. Single-strand cDNA used for RT-qPCR analyses was synthesized from purified total RNA using SuperScript® III Reverse Transcriptase (Life Technologies, Paisley, UK). RT-qPCR were performed using 384-well plates, with a final volume of 10 μL in each well, consisting of 4 μL of cDNA, 5 μL of 2x SYBR Green LightCycler 480_TM PCR master mix (Roche Life Sciences), 0.8 μL of sterile distilled water, 0.1 μL each of 20 μM reverse and forward primers. Samples were performed in triplicates. In the no template controls (negative controls) 4 μL of H2O were added, instead of the cDNA samples. RT-qPCR reactions were carried-out using the Roche LightCycler LC_480-qPCR platform, where fluorescence signals were measured in real-time. The protocol, set-up with thermal cycling conditions, consisted of one cycle at 95°C for 10 min, followed by 45 cycles of amplification at 95°C for 10 s, and 60°C for 30 s. Roche LightCycler Data Analysis Software was used to determine the melt curve data as well as the quantification cycle values (Cq values). The changes in expression levels were normalized against two reference gene as determined via GeNorm (REF), and the relative mRNA levels of genes following treatment were calculated using “The Comparative CT Method” (ΔΔCT Method).
Design of primers for RT-qPCR
The National Centre for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/) was used to identify and obtain mRNA sequences. Exon boundaries were determined from the “European Molecular Biology Laboratories” website (http://www.ensembl.org). This procedure was performed until sets of primers were selected for each target gene. The final step involved checking the primers for similarity using NCBI BLAST (Basic Local Alignment Search Tool) (http://www.ncbi.nlm.nih.gov/BLAST), reducing the chance of primers binding non-specifically.
Identification of reference genes for RT-qPCR analysis
Samples were analyzed for the expression of each of eight candidate reference genes, namely: ACTB (Beta-Actin), B2M (Beta-2-microglobulin), GAPDH (glyceraldehyde-3-phosphate dehydrogenase), HMBS (hydroxymethyl-bilane synthase), HPRT1 (hypoxanthine phosphoribosyl transferase 1), RPL13A (ribosomal protein L13a), RPL32 (ribosomal protein L32), SDHA (succinate dehydrogenase complex, subunit A) as recommended by Vandesompele et al. (2002). RT-qPCR was performed as described previously, using the primers specific for each candidate reference gene. The GeNorm algorithm software package was used to determine the two most stable reference genes from the set of tested candidate genes by calculating a gene normalization factor, eliminating the least stable genes until a stability value (M) of 0.4 or less was reached (Vandesompele et al., 2002).
Results
Effects of gemcitabine and drug fragments on the viability of Panc-1 cells
A standard strategy is to choose a series of molecules that cover chemical space effectively, and for this we chose initially the main Maybridge drug fragment library. It consists of 500 rule-of-three-compliant (Congreve et al., 2003) polar molecules that cover chemical space widely, and where the molecular properties include molecular weight <300, number of hydrogen bond donors ≤3, number of hydrogen bond acceptors ≤3, ClogP ≤3, and in addition, the number of rotatable bonds ≤3 and the polar surface area ≤60Å2. While the use of fragments is commonplace in target-based assays, especially where structures are known (e.g., Erlanson and Hansen, 2004; Erlanson et al., 2004; Rees et al., 2004; Carr et al., 2005; Alex and Flocco, 2007; Ciulli and Abell, 2007; Jhoti, 2007; Jhoti et al., 2007; Hubbard, 2008; Fischer and Hubbard, 2009; Schulz and Hubbard, 2009; Whittaker et al., 2010; Leach and Hann, 2011; Erlanson, 2012; Caliandro et al., 2013), we here prefer the use of the rather more successful phenotypic screens (Swinney and Anthony, 2011; Swinney, 2013). Although it is hard to find published examples of phenotypic screens that used fragment-based libraries, we merely point out that 25% of successful (marketed) drugs are no larger than fragments (i.e., <300 Da) (O'Hagan and Kell, 2015c). The fragment-based approach also has the advantage of avoiding the increasing “molecular obesity” (Hann, 2011; Meanwell, 2011) that is seen in some cases as inimical to the finding of successful drugs (Leeson and Springthorpe, 2007; Leeson and Empfield, 2010).
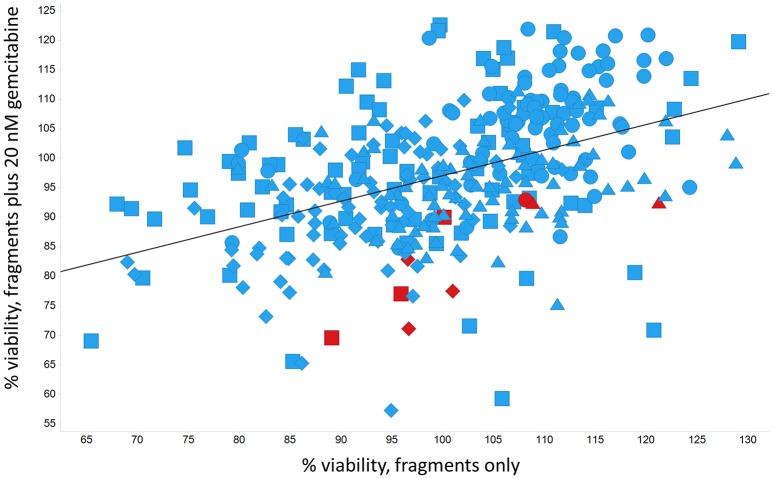
Panc1 cells are a pancreatic cancer cell line (e.g., Gradiz et al., 2016). Figure 1 shows four separate experiments in which the effect of the pools of the Maybridge fragments (6 at a time) on the viability of cells was assessed in the presence and absence of 20 nM gemcitabine, pointing up three pools containing “hits” (which occurred in at least 3 experiments; there are a total of 336 experiments here). Figure 2 shows the % viability of one set of Panc1 cells as a function of the gemcitabine concentration, as a result of which we later chose 100 nM gemcitabine to assess the efficacy of the individual fragments in increasing its toxicity. Figure 3 shows a titration curve for three repeats with one of the “hits,” the plot also serving to illustrate the variability of the toxicity of gemcitabine alone on different days. Figures 4, 5 show the distribution in chemical space of all 500 fragments in the first Maybridge library and three “hits” at 10 μM that lowered the viability of cells by at least 10% in the presence, but not the absence, of 100 nM gemcitabine. These were retested singly, then together pairwise, resulting in three hits, viz B1, D1, and B12. B12 seemed to interfere with the other two fragments by binding to them directly (UV evidence) and was not used further. Note that a significant issue is that although for a given batch of Panc1 cells the titration curves were reasonably reproducible, they were considerably less so between batches (for reasons that will become apparent below). This meant that each culture had to be used as its own control, as we did e.g., in Figure 2. Another interesting feature was that quite a significant fraction of the fragments (as in Figure 1, and see below) were even somewhat stimulatory to cell growth in the absence of gemcitabine.
Figure 1.
Effect of 500 Maybridge fragments on the viability of Panc1 cells in the absence and presence of 20 nM gemcitabine. Experiment number is encoded by shape. Fragments were added in pools of 6. Pools in which there was a hit relative to the same control are marked in red. The line is a line of best fit.
Figure 2.
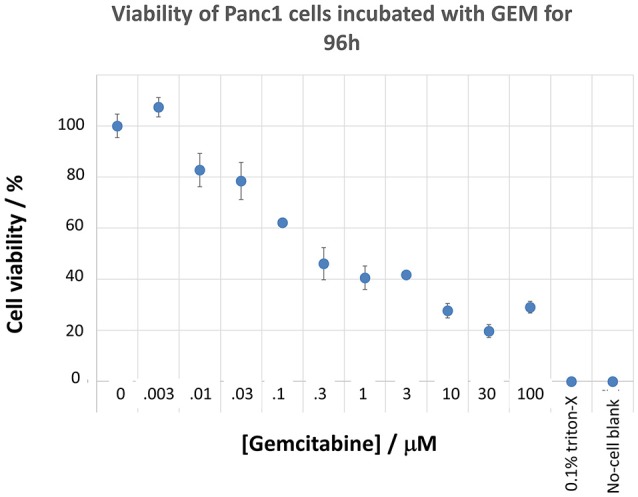
Effect of gemcitabine concentration on the viability of Panc1 cells. Cells were grown and pre-incubated with the stated concentration of gemcitabine, and their viability was assessed, as described in the Methods section.
Figure 3.
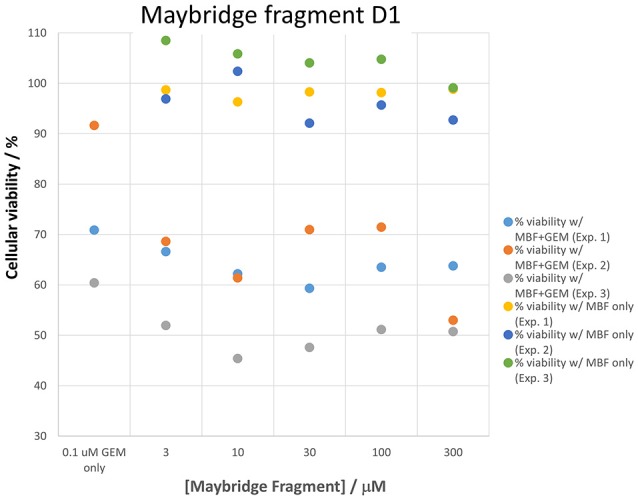
Variability in gemcitabine sensitivity and the effect of a “hit” (fragment D1) on cellular viability when measured on three sets of cells in cultures grown on different days. The differences between gemcitabine and gemcitabine plus all “hit” fragments such as D1 is statistically significant at the P < 0.05 level (n = 3).
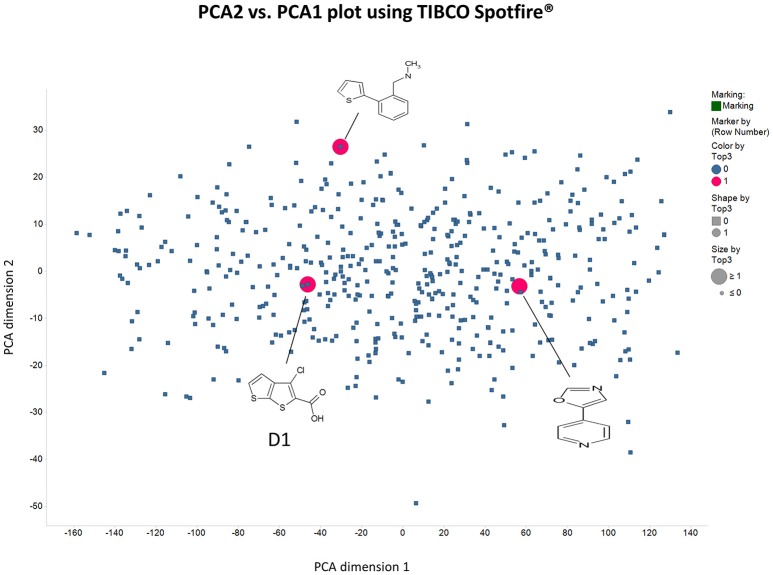
Figure 4.
Distribution in chemical space of the first 500 Maybridge fragments as judged using the principal components of the variance in a set of their biophysical properties (see Methods) as produced using RDKit in KNIME.
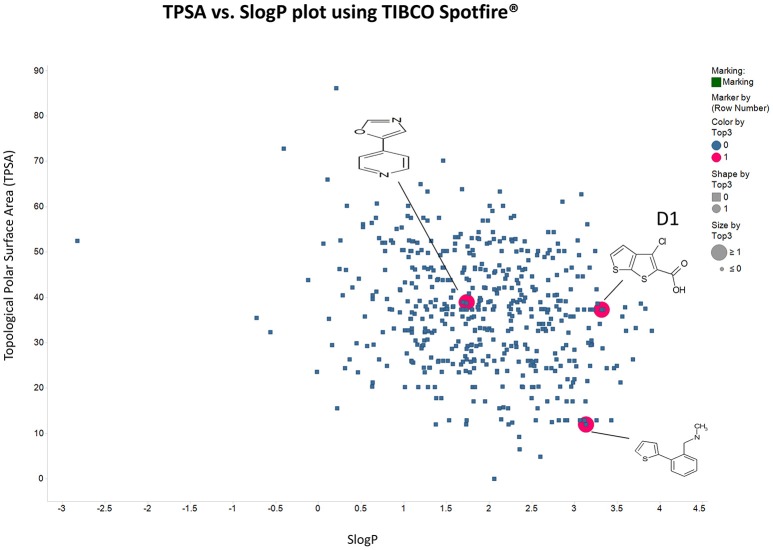
Figure 5.
As in Figure 4 save that the axes are Total Polar Surface area and S log P as calculated using RDKit.
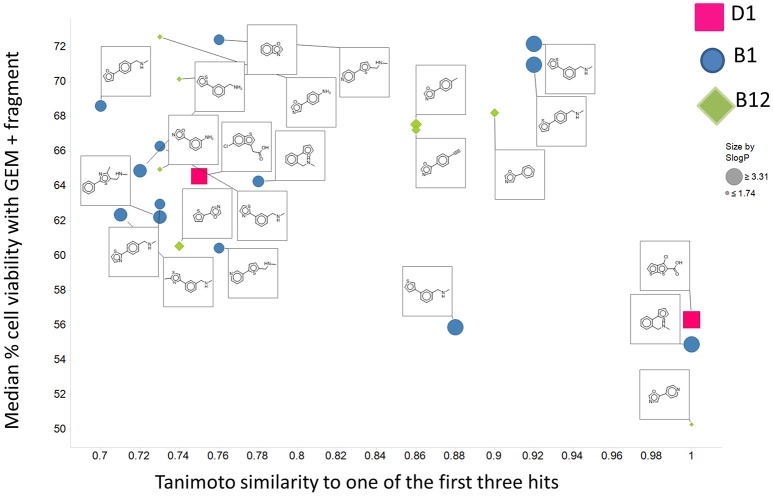
There are four other Maybridge fragment libraries of 500 molecules each, covering broadly the same chemical space but in more detail (O'Hagan and Kell, 2015c), and we performed a cheminformatics analysis (MACCS encoding, Tanimoto similarity) to establish which other molecules might be similar, exactly as per the analyses in (O'Hagan et al., 2015). Some 20 molecules had a Tanimoto similarity within 0.7 of one of the three remaining hits and were tested. In this case, the starting % viability was much higher than those in Figure 2. All 20 of these fragments are in fact active, which shows that these molecules (Figure 6) exhibit a very considerable enrichment over the whole library, and illustrates the utility of the principle of molecular similarity (Gasteiger, 2003; Bender and Glen, 2004; Stumpfe and Bajorath, 2011; Maggiora et al., 2014). The figure also illustrates which of the original three hits the new hits are closest to, and encodes their S log P-values as the size of the marker. This enormous cheminformatics-based enrichment also gives considerable confidence in our strategy, despite the variability in sensitivity of the Panc1 cells to gemcitabine alone, since such a huge enrichment could not conceivable occur for molecules that were not active. Although none was quite as active as the original hits, all exhibited some kind of activity (Figure 2) (the starting viabilities for two different experiments in the presence of gemcitabine only were 78 and 84%). Of all of these, the seven most potent molecules exhibited activity at 3 μM. One was rather expensive and was again excluded. Thus, we had a total of 6 hits to consider [two from library 1 (B1 and D1), and a total of four from the other four libraries, referred to as fragments 10, 11, 12, and 20]. Table 2 gives their names, SMILES encodings and 2D structures, along with that of indole-3-carboxylic acid (see later).
Figure 6.
Tanimoto similarity to the set of three hits in the first 500 Maybridge fragments of 20 molecules selected from the other four libraries. The average % viability of the cells in the presence of gemcitabine but the absence of Maybridge fragments in this experiment was 81. The starting fragment to which the molecule was most similar is encoded by shape and color, while the S log P-value is encoded by size.
Table 2.
Six hits in the “binary weapon” assay given in three formats, plus indole-3-carbinol.
| MBF | SMILES | Name | |
|---|---|---|---|
| D1 | OC(= O)c1sc2sccc2c1Cl | 3-chlorothieno[2,3-b]thiophene-2-carboxylic acid | 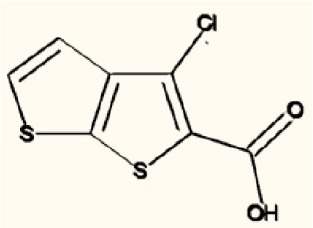 |
| B1 | CNCc1ccccc1c2cccs2 | N-methyl-N-(2-thien-2-ylbenzyl)amine | 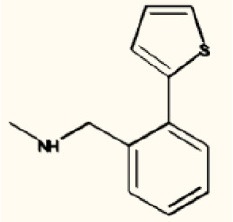 |
| 10 | S1C(= CC = C1CNC)c1cccnc1 | N-methyl-(5-pyrid-3-ylthien-2-yl)methylamine | 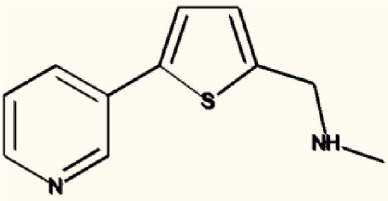 |
| 11 | S1C(= CC = C1CNC)c1ccncc1 | N-methyl-(5-pyrid-4-ylthien-2-yl)methylamine | 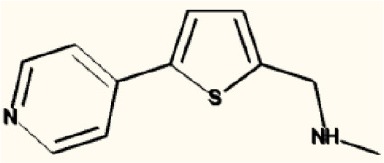 |
| 12 | S1C = C(c2c1ccc(c2)Cl)CC(= O)O | 2-(5-Chlorobenzo[b]thiophen-3-yl)acetic acid | 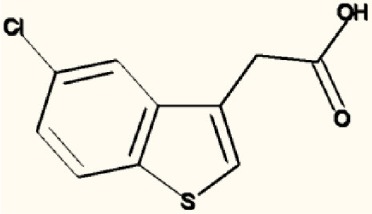 |
| 20 | N1 = COC(= C1)c1ccc(cc1)N | 4-(1, 3-Oxazol-5-yl)aniline | 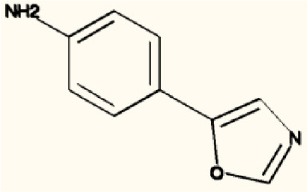 |
| I3C | C1 = CC = C2C(= C1)C(= CN2)CO | Indole-3-carbinol |  |
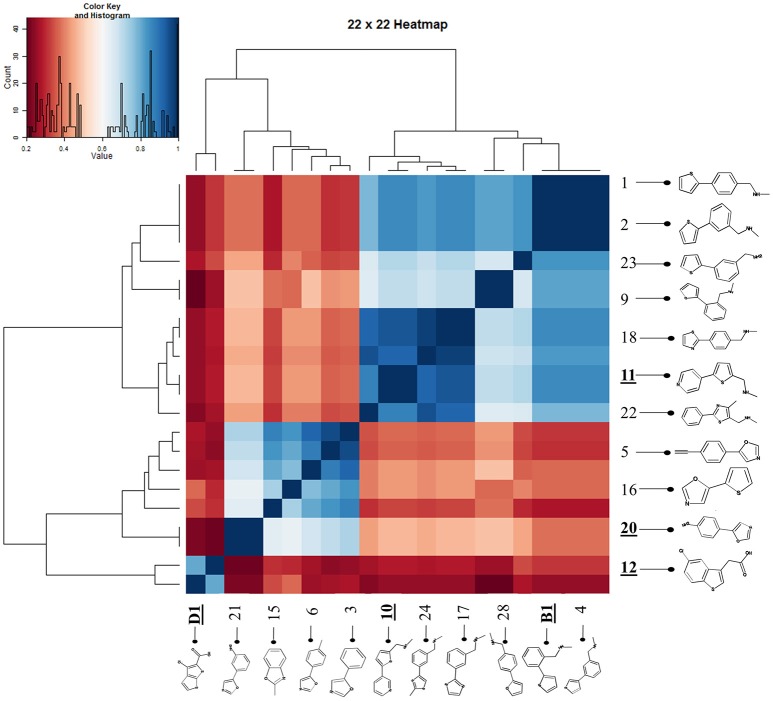
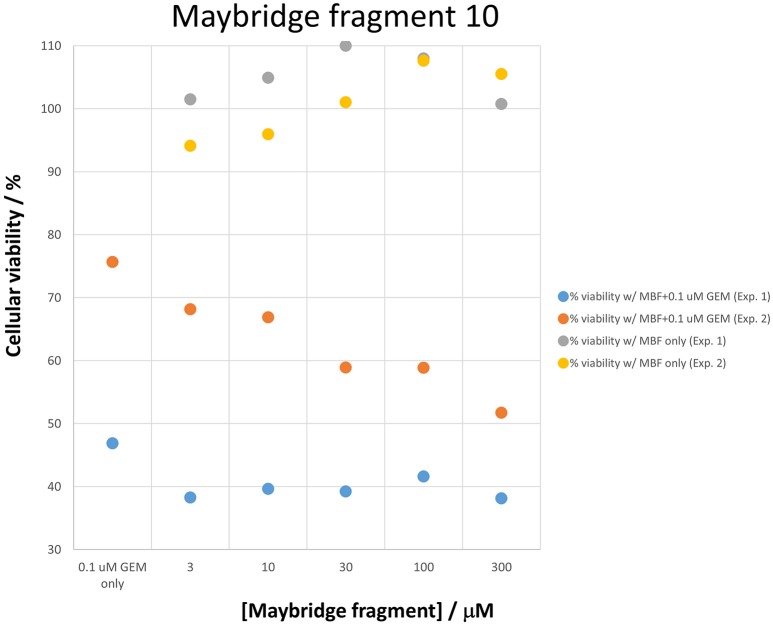
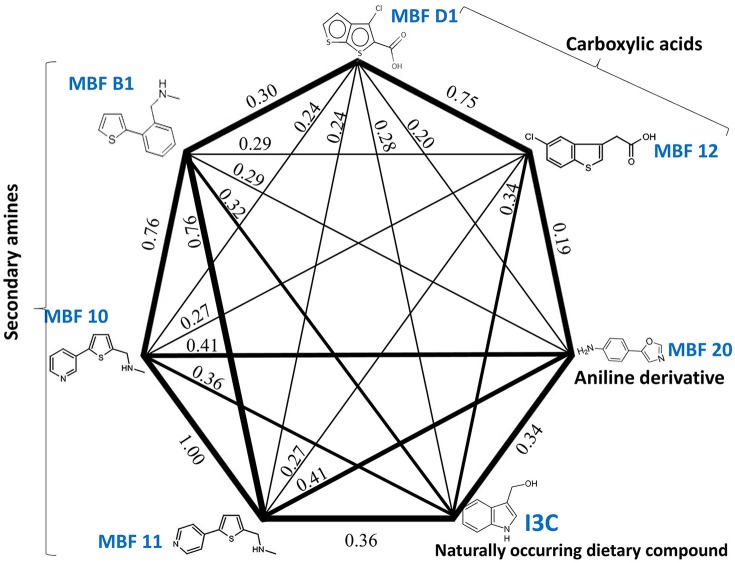
Figure 7 shows a (symmetrical) heatmap (MACCS encoding) of the Tanimoto similarities of the 22 most potent molecules, where it can again be seen that the hits are in three clusters. These are B1, 10, and 11 (all are amines), D1 and 12 (carboxylic acids), and 20 (an aniline derivative—possibly to be avoided Benigni and Passerini, 2002; Benigni et al., 2009; Franke et al., 2010). One implication is that they each have different targets (probably plural) but attempts even to show additivity, let alone synergy, met with failure, possibly because the molecules were indeed rather similar to each other in terms of the larger chemical space. Figure 8—equivalent to Figure 3—shows data for two experiments with fragment 10, again illustrating the stimulation of growth by the fragment alone, and its inhibition in the presence of a relatively weakly inhibiting concentration of gemcitabine. Finally, Figure 9 shows the Tanimoto similarities (TS, based on the MACCS encoding) between the six hits plus Indole-3-carbinol (I3C, see below). Fragments within a group showed a Tanimoto similarity of 0.75 or greater, while those between groups were less than 0.5. I3C was not really similar to any of the hits; its highest TS to any of the hits was 0.36. It is especially gratifying to note that MBF10 and MBF11 were both selected and had a TS to each other of 1, as they are in fact structural isomers. Along with the other clusterings, this adds considerable weight to the validity of our assays.
Figure 7.
Chemical similarities of the various hits to each other, and effect of Maybridge fragment 10 on cell viability. A heatmap showing the three clusters of molecules that could be observed.
Figure 8.
Two experiments illustrating the effect of Maybridge fragment 10 on cell viability in the absence and presence of gemcitabine, again showing the stimulation in the absence of gemcitabine. The differences between gemcitabine and gemcitabine plus fragment 10 is statistically significant at the P < 0.05 level (n = 3).
Figure 9.
Tanimoto similarities of the main hits in the three clusters of Figure 7 (plus I3C).
Effect of indole-3-carbinol on gemcitabine toxicity
Cruciferous vegetables such as Brassica spp. are considered to have certain anticancer properties (Higdon et al., 2007; Juge et al., 2007; Fujioka et al., 2016b), and small molecules derived from the hydrolysis of glucosinolates, such as sulforaphane and indole-3-carbinol (I3C), have been implicated in a variety of anticarcinogenic mechanisms (e.g., Chen et al., 2014; Fujioka et al., 2016a). I3C is a small molecule (MW 147.17, well within the range of “fragments”), and Lyn-Cook and colleagues (Lyn-Cook et al., 2010; Wang et al., 2011; Paik et al., 2013) have published that I3C can enhance the sensitivity of pancreatic cancer cells to gemcitabine, possibly via upregulation of ENT1 expression (Wang et al., 2011). It was thus of interest to compare I3C with the hits that we found. In our hands, however, I3C had no measurable effect on either the cell viability in the presence or absence of gemcitabine (nor on the expression profiles discussed below). This is entirely consistent with its low structural similarity to the other hits as indicated above.
Effect of fragments on the growth of Panc1 cells
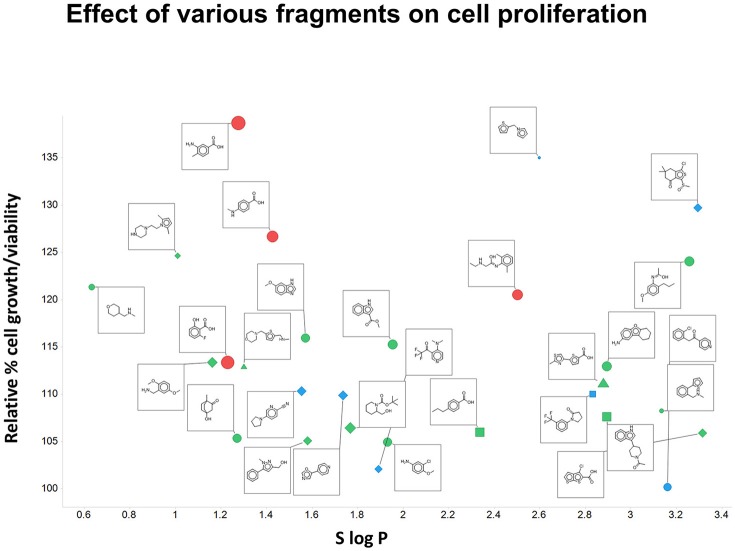
Although this was not the main focus of the present paper, we did note (as mentioned above) that the fragments themselves could stimulate the growth of Panc1 cells relative to that of controls (as measured by OD). This is illustrated in Figure 10 for 28 of the fragments on which we focussed. Also encoded with the structures are the number of H-bond donors and acceptors, the total polar surface area of the fragments, and (on the abscissa) the S log P-values. It is clear (i) that virtually every fragment could stimulate the growth of the cells, and (ii) that there was no particularly obvious relationship of the extent of such stimulation with any of the descriptors stated.
Figure 10.
Effect of various fragments on cell growth/viability relative to untreated controls. Also plotted are the number of H-bond acceptors (by shape; square 1, circle 2, diamond 3, triangle 4), H-bond donors (by color, blue 0, green 1, red 2, yellow 3), total polar surface area (by size of symbol, up to 63 Å2) and S log P (on the abscissa).
Effect of gemcitabine and fragments on the expression of selected transcripts in Panc 1 cells
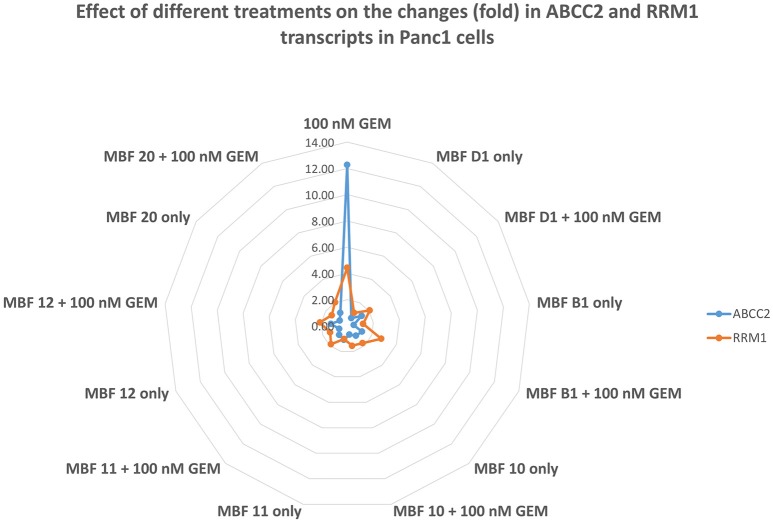
Given that there was evidence that the fragments did not affect gemcitabine uptake directly, we assumed that they must be working by influencing the activity or expression of appropriate targets (and certainly small molecules can affect transporter expression, (e.g., Mrozikiewicz et al., 2014). To this end, we designed primers to enable PCR of transcripts relevant to gemcitabine transport and metabolism. Table 3 shows each of those that were detectable within 35 PCR cycles when treated (i) with gemcitabine alone, (ii) with Maybridge fragment D1 alone, and (iii) with both gemcitabine and D1. Strikingly, gemcitabine increases the expression of the ABCC2 efflux transporter (MRP2) more than 12-fold, and that of RRM1 more than fourfold, while the addition of D1 largely reverses both of these effects. It would seem that these are by far the largest contributors to the efficacy of fragment D1 in enhancing the cytotoxicity of gemcitabine, and the same is true for each of the other fragments (Table 4 and Figure 11). However, the ABCC2 inhibitor MK-571 (e.g., Weiss et al., 2007; Noma et al., 2008) at 20 μM had no effect on the viability of Panc1 cells treated with Gemcitabine alone (data not shown), possibly implying that RRM1 was the more significant contributor to the phenotypic changes in resistance.
Table 3.
Changes in the transcript level of relevant transporters and other genes when treated with gemcitabine and/or fragment D1.
| Gene | Fold changes | ||
|---|---|---|---|
| Treatment with 100 nM GEM | Treatment with MBF D1 only | Treatment with MBF D1 and 100 nM GEM | |
| ENT1 | 0.87 ± 0.13 | 0.79 ± 0.12 | 1.08 ± 0.17 |
| ENT2 | 0.57 ± 0.13 | 0.98 ± 0.27 | 0.59 ± 0.17 |
| ENT3 | 2.58 ± 0.11 | 1.18 ± 0.64* | 0.89 ± 0.20*** |
| ABCC2 | 12.27 ± 0.34 | 0.66 ± 0.14*** | 1.33 ± 0.33*** |
| ABCC3 | 0.16 ± 0.48 | 2.10 ± 0.09** | 0.54 ± 0.18 |
| ABCC4 | 0.53 ± 0.10 | 0.90 ± 0.23 | 0.36 ± 0.14 |
| ABCC5 | 0.50 ± 0.11 | 1.18 ± 0.32* | 1.21 ± 0.15** |
| ABCC10 | 1.61 ± 0.48 | 0.53 ± 0.08* | 0.48 ± 0.16* |
| RRM1 | 4.43 ± 0.13 | 1.11 ± 0.17*** | 2.07 ± 0.16*** |
Only those transcripts detectable within 35 PCR cycles are shown. Data are given as mean ± standard deviation. A 2-sided T-test was performed to assess statistical significance against GEM alone, P-values being encoded as
< 0.05,
< 0.01,
< 0.001.
Table 4.
Changes in the transcript level of ABCC2 and RRM1 when treated with gemcitabine and/or the other fragment hits.
| Treatment | Gene fold changes | ||||
|---|---|---|---|---|---|
| ABCC2 | STDEV | RRM1 | STDEV | ||
| GEM | 100 nM GEM | 12.27 | ±0.34 | 4.43 | ±0.13 |
| MBF D1 | MBF D1 only | 0.66*** | ±0.14 | 1.11*** | ±0.17 |
| MBF D1 + 100 nM GEM | 1.33*** | ±0.33 | 2.07*** | ±0.16 | |
| MBF B1 | MBF B1 only | 0.49*** | ±0.08 | 1.22*** | ±0.12 |
| MBF B1 + 100 nM GEM | 1.21*** | ±0.53 | 2.77*** | ±0.11 | |
| MBF 10 | MBF 10 only | 1.00*** | ±0.23 | 1.76*** | ±0.14 |
| MBF 10 + 100 nM GEM | 0.68*** | ±0.05 | 1.56*** | ±0.08 | |
| MBF 11 | MBF 11 only | 1.09*** | ±0.09 | 1.04*** | ±0.11 |
| MBF 11 + 100 nM GEM | 0.93*** | ±0.15 | 1.88*** | ±0.20 | |
| MBF 12 | MBF 12 only | 0.65*** | ±0.13 | 1.39*** | ±0.09 |
| MBF 12 + 100 nM GEM | 1.25*** | ±0.19 | 2.11*** | ±0.15 | |
| MBF 20 | MBF 20 only | 0.7*** | ±0.06 | 1.43*** | ±0.11 |
| MBF 20 + 100 nM GEM | 1.13*** | ±0.05 | 2.02*** | ±0.12 | |
A 2-sided T-test was performed to assess statistical significance vs. GEM alone, P-values being encoded as * < 0.05, ** < 0.01,
< 0.001.
Figure 11.
Effect of gemcitabine ± various Maybridge fragments on the expression of transcripts for ABCC2 and RRM1. Each experiment was performed three times, as described in Materials and Methods, and the mean is shown. For clarity, SD and statistical significance data are given only in the legends to Tables 3, 4.
Selectivity of fragments for increasing transporter expression
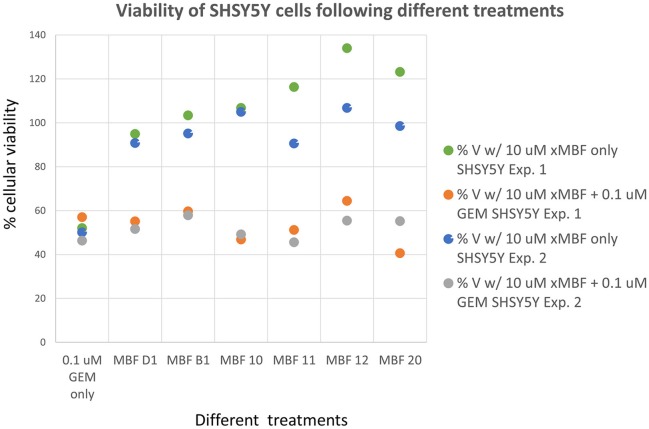
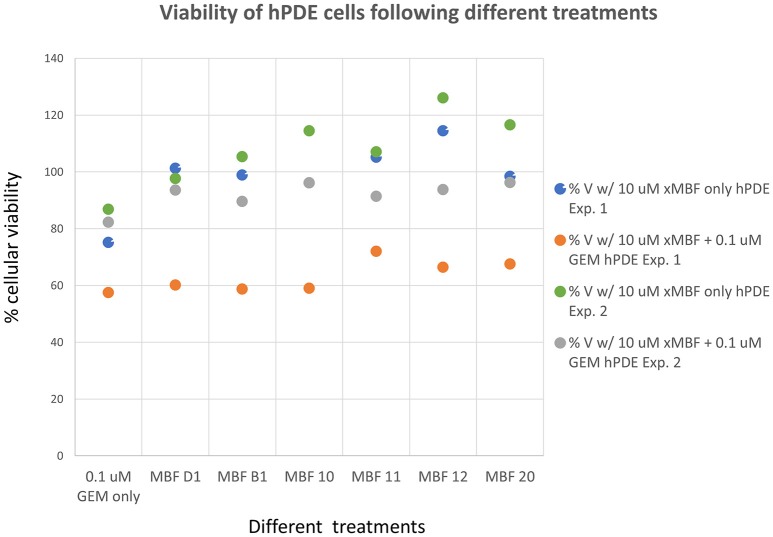
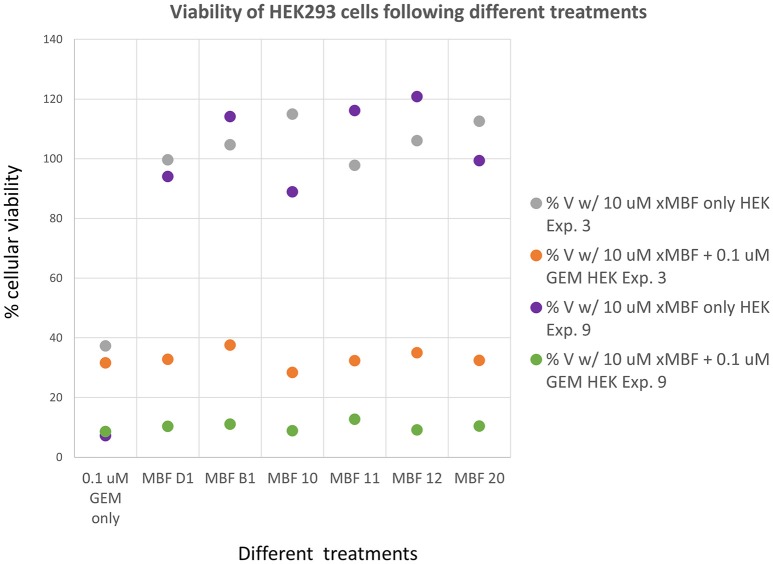
Having seen that various of the fragments could increase the toxicity of gemcitabine to Panc1 cells, it was of interest to see whether this was a cell-selective phenomenon. Although time did not permit an exhaustive study, we noted that fragments 10 and 20 also had these toxicity-enhancing effect for the neuroblastoma SH-SY5Y cell line while B1, D1, 11, and 12 did not (Figure 12). No fragments seemed to have any such effects on the non-cancerous pancreatic cell line hPDE (Figure 13) and HEK293 cells (Figure 14), implying that there is or can be at least some degree of specificity in our “binary weapon” approach. Clearly a larger-scale study (including both larger libraries and more cell lines) would be able to discover molecules with both potency and selectivity.
Figure 12.
Effects of gemcitabine and gemcitabine plus fragments on the viability of SH-SY5Y cells. Apart from fragments 10 and 20, the effects of the fragments were not statistically significant at the P < 0.05 level, n = 3 per experiment.
Figure 13.
Effects of gemcitabine and gemcitabine plus fragments on the viability of hPDE cells. The effects of the fragments were not statistically significant at the P < 0.05 level, n = 3 per experiment.
Figure 14.
Effects of gemcitabine and gemcitabine plus fragments on the viability of HEK293 cells. Experiments were performed as described, and as per the legend to Figures 3, 8. The effects of the fragments were not statistically significant at the P < 0.05 level, n = 3 per experiment.
Discussion
In the present work, we sought to develop the idea that we might affect the transporter-mediated disposition of small-molecule drugs via the addition of a second small molecule that of itself had no inhibitory pharmacological effect but that influenced the expression of transporters for the primary drug (Figure 15). We refer to this as a “binary weapon” strategy. The specific phenotypic effect we sought was for a molecule that on its own had no such effect to increase the toxicity of the nucleoside analog gemcitabine to Panc1 pancreatic cancer cells (Figures 1–3).
Figure 15.
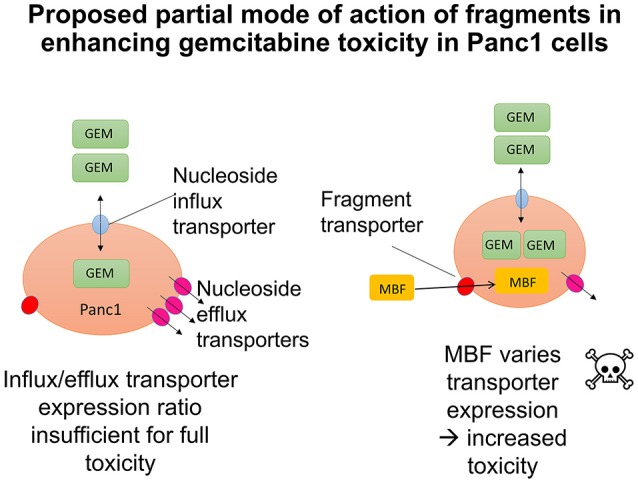
Cartoons illustrating the potential modes of action of fragments in enhancing transporter-mediated gemcitabine toxicity in Panc1 cells. (The smaller effects on RRM1 are ignored for clarity). Left: Original hypothesis that fragments would stimulate the activity of uptake transporters. Right: Actual mechanism based on PCR data.
Given the recognition (O'Hagan and Kell, 2015c) that more some 25% of marketed drugs are in fact no larger than the polar “rule-of-three”-compliant (Congreve et al., 2003) molecules used in fragment-based drug discovery, we used an initial screen of a 500-member polar drug fragment library. This yielded three “hits” (Figures 4, 5). The structures of 20 of the other 2000 members of this library had a Tanimoto similarity greater than 0.7 to those of the initial hits, and each was itself a hit (Figure 6) (with the cheminformatics thus providing for a massive enrichment in the fraction of successful experiments). We chose the top six representatives for further study. They each bore reasonable structural similarities to each other (two were in fact isomers), lending strength to the self-consistency of both our conceptual and experimental strategies (Figures 7, 8).
Existing literature had suggested that indole-3-carbinol might play a similar role to that of our fragments, but in our hands it was without effect, and nor was it structurally similar to any of our hits (Figure 9). We therefore discounted it.
There is an interesting issue when the phenotypic activity being measured is in fact cell death, as it is then impossible legitimately to compare bulk measurements of biochemical changes with individual-cell viabilities. This is because with bulk or ensemble measurements one does not know if say a lowering of a biochemical parameter by 50% means that all of the cells have lost half of it or half of the cells have lost all of it (or anything in between) (Kell et al., 1991, 1998; Davey and Kell, 1996). In the event, the mechanism was very clear, however.
Because the fragments were themselves without negative effects on the cells in the absence of gemcitabine (interestingly, many of them actually stimulated cell growth, Figure 1, so each had to be compared to the appropriate control!), we next designed suitable primers to assess the expression levels of all the candidate transporters plus ribonucleotide reductase. In our hands, only the ENT1-3 uptake and ABCC2,3,4,5, and 10 efflux transporters displayed measurable transcripts, along with RRM1. Very strikingly, the addition of gemcitabine alone increased the expression of the transcript for ABCC2 (MRP2) by more than 12-fold, and that of RRM1 by more than fourfold, and each of the fragment “hits” served to reverse this, at least in part (Figure 11). The effects on ABCC2 are thus consistent with the finding (Horiguchi et al., 2013) that it may be a major efflux pump for gemcitabine.
It seems, therefore, that while the effect was here mediated more by efflux than influx transporters, the binary weapon idea is hereby fully confirmed: our results show that it is possible to find molecules that manipulate the expression of transporters that are involved in the bioactivity of a pharmaceutical drug, and that there is a certain degree of specificity in this for pancreatic cancer cells (Figures 12–14). This could explain, at least in part, the basis for the selective toxicity of a drug that is otherwise cytotoxic generally (Figure 15). The next steps will involve determining much more extensively how much any such activity differs, or can be made to differ (as do most transcript levels), between different cells.
Author contributions
DK and PD designed the study. All the experimental work was performed by JG, who was supervised by PD and DK. Some of the cheminformatics analyses were performed by SO. All authors contributed to and approved the writing of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Biotechnology and Biological Sciences Research Council for supporting this work (BBSRC grants BB/K019783/1 and BB/M017702/1). This is a contribution from the Manchester Centre for Synthetic Biology of Fine and Speciality Chemicals (SYNBIOCHEM).
References
- Achiwa H., Oguri T., Sato S., Maeda H., Niimi T., Ueda R. (2004). Determinants of sensitivity and resistance to gemcitabine: the roles of human equilibrative nucleoside transporter 1 and deoxycytidine kinase in non-small cell lung cancer. Cancer Sci. 95, 753–757. 10.1111/j.1349-7006.2004.tb03257.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alex A. A., Flocco M. M. (2007). Fragment-based drug discovery: what has it achieved so far? Curr. Top. Med. Chem. 7, 1544–1567. 10.2174/156802607782341082 [DOI] [PubMed] [Google Scholar]
- Alvarellos M. L., Lamba J., Sangkuhl K., Thorn C. F., Wang L., Klein D. J., et al. (2014). PharmGKB summary: gemcitabine pathway. Pharmacogenet. Genomics 24, 564–574. 10.1097/FPC.0000000000000086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R., Aho U., Nilsson B. I., Peters G. J., Pastor-Anglada M., Rasch W., et al. (2009). Gemcitabine chemoresistance in pancreatic cancer: molecular mechanisms and potential solutions. Scand. J. Gastroenterol. 44, 782–786. 10.1080/00365520902745039 [DOI] [PubMed] [Google Scholar]
- Antman E., Weiss S., Loscalzo J. (2012). Systems pharmacology, pharmacogenetics, and clinical trial design in network medicine. Wiley interdisciplinary reviews. Syst. Biol. Med. 4, 367–383. 10.1002/wsbm.1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran J. M., Plagemann P. G. W. (1992). Nucleoside transport-deficient mutants of PK-15 pig kidney cell line. Biochim. Biophys. Acta 1110, 51–58. 10.1016/0005-2736(92)90293-U [DOI] [PubMed] [Google Scholar]
- Arrowsmith J., Miller P. (2013). Trial watch: phase II and phase III attrition rates 2011-2012. Nat. Rev. Drug Discov. 12, 569. 10.1038/nrd4090 [DOI] [PubMed] [Google Scholar]
- Baldwin S. A., Mackey J. R., Cass C. E., Young J. D. (1999). Nucleoside transporters: molecular biology and implications for therapeutic development. Mol. Med. Today 5, 216–224. 10.1016/S1357-4310(99)01459-8 [DOI] [PubMed] [Google Scholar]
- Baldwin S. A., Yao S. Y. M., Hyde R. J., Ng A. M. L., Foppolo S., Barnes K., et al. (2005). Functional characterization of novel human and mouse equilibrative nucleoside transporters (hENT3 and mENT3) located in intracellular membranes. J. Biol. Chem. 280, 15880–15887. 10.1074/jbc.M414337200 [DOI] [PubMed] [Google Scholar]
- Banerjee J., Al-Wadei H. A., Al-Wadei M. H., Dagnon K., Schuller H. M. (2014). Differential modulation of nicotine-induced gemcitabine resistance by GABA receptor agonists in pancreatic cancer cell xenografts and in vitro. BMC Cancer 14:725. 10.1186/1471-2407-14-725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee J., Al-Wadei H. A., Schuller H. M. (2013). Chronic nicotine inhibits the therapeutic effects of gemcitabine on pancreatic cancer in vitro and in mouse xenografts. Eur. J. Cancer 49, 1152–1158. 10.1016/j.ejca.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Wang Z., Kong D., Sarkar F. H. (2009). 3,3′-Diindolylmethane enhances chemosensitivity of multiple chemotherapeutic agents in pancreatic cancer. Cancer Res. 69, 5592–5600. 10.1158/0008-5472.CAN-09-0838 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bender A., Glen R. C. (2004). Molecular similarity: a key technique in molecular informatics. Org. Biomol. Chem. 2, 3204–3218. 10.1039/b409813g [DOI] [PubMed] [Google Scholar]
- Benigni R., Passerini L. (2002). Carcinogenicity of the aromatic amines: from structure-activity relationships to mechanisms of action and risk assessment. Mutat. Res. 511, 191–206. 10.1016/S1383-5742(02)00008-X [DOI] [PubMed] [Google Scholar]
- Benigni R., Worth A., Netzeva T., Jeliazkova N., Bossa C., Gruska A., et al. (2009). Structural motifs modulating the carcinogenic risk of aromatic amines. Environ. Mol. Mutagen. 50, 152–161. 10.1002/em.20461 [DOI] [PubMed] [Google Scholar]
- Berger S. I., Iyengar R. (2009). Network analyses in systems pharmacology. Bioinformatics 25, 2466–2472. 10.1093/bioinformatics/btp465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman A. M., Eijk P. P., Ruiz Van Haperen V. W., Smid K., Veerman G., Hubeek I., et al. (2005). In vivo induction of resistance to gemcitabine results in increased expression of ribonucleotide reductase subunit M1 as the major determinant. Cancer Res. 65, 9510–9516. 10.1158/0008-5472.CAN-05-0989 [DOI] [PubMed] [Google Scholar]
- Bergman A. M., Pinedo H. M., Peters G. J. (2002). Determinants of resistance to 2′,2′-difluorodeoxycytidine (gemcitabine). Drug Res. Updat. 5, 19–33. 10.1016/S1368-7646(02)00002-X [DOI] [PubMed] [Google Scholar]
- Berthold M. R., Cebron N., Dill F., Gabriel T. R., Kötter T., Meinl T., et al. (2008). KNIME: the Konstanz Information Miner, in Data Analysis, Machine Learning and Applications, eds Preisach C., Burkhardt H., Schmidt-Thieme L., Decker R. (Berlin: Springer; ), 319–326. [Google Scholar]
- Bhattacharjee V., Zhou Y., Yen T. (2014). A synthetic lethal screen identifies the Vitamin D receptor as a novel gemcitabine sensitizer in pancreatic cancer cells. Cell Cycle 13, 3839–3856. 10.4161/15384101.2014.967070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutia Y. D., Hung S. W., Patel B., Lovin D., Govindarajan R. (2011). CNT1 expression influences proliferation and chemosensitivity in drug-resistant pancreatic cancer cells. Cancer Res. 71, 1825–1835. 10.1158/0008-5472.CAN-10-2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbath I., Verbrugghe L., Lai R., Gigot J. F., Humblet Y., Piessevaux H., et al. (2012). Human equilibrative nucleoside transporter 1 (hENT1) expression is a potential predictive tool for response to gemcitabine in patients with advanced cholangiocarcinoma. Eur. J. Cancer 48, 990–996. 10.1016/j.ejca.2011.11.006 [DOI] [PubMed] [Google Scholar]
- Borisy A. A., Elliott P. J., Hurst N. W., Lee M. S., Lehar J., Price E. R., et al. (2003). Systematic discovery of multicomponent therapeutics. Proc. Natl. Acad. Sci. U.S.A. 100, 7977–7982. 10.1073/pnas.1337088100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke T., Lee S., Ferguson P. J., Hammond J. R. (1998). Interaction of 2′,2′-difluorodeoxycytidine (gemcitabine) and formycin B with the Na+-dependent and -independent nucleoside transporters of Ehrlich ascites tumor cells. J. Pharmacol. Exp. Ther. 286, 1333–1340. [PubMed] [Google Scholar]
- Buxhofer-Ausch V., Secky L., Wlcek K., Svoboda M., Kounnis V., Briasoulis E., et al. (2013). Tumor-specific expression of organic anion-transporting polypeptides: transporters as novel targets for cancer therapy. J. Drug Deliv. 2013:863539. 10.1155/2013/863539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliandro R., Belviso D. B., Aresta B. M., De Candia M., Altomare C. D. (2013). Protein crystallography and fragment-based drug design. Future Med. Chem. 5, 1121–1140. 10.4155/fmc.13.84 [DOI] [PubMed] [Google Scholar]
- Cano-Soldado P., Molina-Arcas M., Algueró B., Larráyoz I., Lostao M. P., Grandas A., et al. (2008). Compensatory effects of the human nucleoside transporters on the response to nucleoside-derived drugs in breast cancer MCF7 cells. Biochem. Pharmacol. 75, 639–648. 10.1016/j.bcp.2007.10.005 [DOI] [PubMed] [Google Scholar]
- Cano-Soldado P., Pastor-Anglada M. (2012). Transporters that translocate nucleosides and structural similar drugs: structural requirements for substrate recognition. Med. Res. Rev. 32, 428–457. 10.1002/med.20221 [DOI] [PubMed] [Google Scholar]
- Cansev M. (2006). Uridine and cytidine in the brain: their transport and utilization. Brain Res. Rev. 52, 389–397. 10.1016/j.brainresrev.2006.05.001 [DOI] [PubMed] [Google Scholar]
- Carr R. A., Congreve M., Murray C. W., Rees D. C. (2005). Fragment-based lead discovery: leads by design. Drug Discov. Today 10, 987–992. 10.1016/S1359-6446(05)03511-7 [DOI] [PubMed] [Google Scholar]
- Cass C. E. (2001). Nucleoside transporter proteins: from Membrane Biology to Therapeutic Applications, in The Jeanne Manery Fisher Memorial Lecture, (Alliston, ON: CSBMCB/SCBBMC Bulletin; ), 52–59. [Google Scholar]
- César-Razquin A., Snijder B., Frappier-Brinton T., Isserlin R., Gyimesi G., Bai X., et al. (2015). A call for systematic research on solute carriers. Cell 162, 478–487. 10.1016/j.cell.2015.07.022 [DOI] [PubMed] [Google Scholar]
- Chan S. L., Chan S. T., Chan E. H., He Z. X. (2014). Systemic treatment for inoperable pancreatic adenocarcinoma: review and update. Chin. J. Cancer 33, 267–276. 10.5732/cjc.013.10134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Cheng P. H., Rao X. M., Mcmasters K. M., Zhou H. S. (2014). Indole-3-carbinol (I3C) increases apoptosis, represses growth of cancer cells, and enhances adenovirus-mediated oncolysis. Cancer Biol. Ther. 15, 1256–1267. 10.4161/cbt.29690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Xue X., Wang F., An Y., Tang D., Xu Y., et al. (2012). Expression and promoter methylation analysis of ATP-binding cassette genes in pancreatic cancer. Oncol. Rep. 27, 265–269. 10.3892/or.2011.1475 [DOI] [PubMed] [Google Scholar]
- Choi M. K. (2012). Variability of gemcitabine accumulation and its relationship to expression of nucleoside transporters in peripheral blood mononuclear cells. Arch. Pharm. Res. 35, 921–927. 10.1007/s12272-012-0518-8 [DOI] [PubMed] [Google Scholar]
- Ciulli A., Abell C. (2007). Fragment-based approaches to enzyme inhibition. Curr. Opin. Biotechnol. 18, 489–496. 10.1016/j.copbio.2007.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congreve M., Carr R., Murray C., Jhoti H. (2003). A rule of three for fragment-based lead discovery? Drug Discov. Today 8, 876–877. 10.1016/S1359-6446(03)02831-9 [DOI] [PubMed] [Google Scholar]
- Damaraju V. L., Sawyer M. B., Mackey J. R., Young J. D., Cass C. E. (2009). Human nucleoside transporters: biomarkers for response to nucleoside drugs. Nucleosides Nucleotides Nucleic Acids 28, 450–463. 10.1080/15257770903044499 [DOI] [PubMed] [Google Scholar]
- Damaraju V. L., Scriver T., Mowles D., Kuzma M., Ryan A. J., Cass C. E., et al. (2014). Erlotinib, gefitinib, and vandetanib inhibit human nucleoside transporters and protect cancer cells from gemcitabine cytotoxicity. Clin. Cancer Res. 20, 176–186. 10.1158/1078-0432.CCR-13-2293 [DOI] [PubMed] [Google Scholar]
- Davey H. M., Kell D. B. (1996). Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analysis. Microbiol. Rev. 60, 641–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Santo B., Tarafa G., Felipe A., Casado F. J., Pastor-Anglada M. (2001). Developmental regulation of the concentrative nucleoside transporters CNT1 and CNT2 in rat liver. J. Hepatol. 34, 873–880. 10.1016/S0168-8278(01)00036-8 [DOI] [PubMed] [Google Scholar]
- Deng T., Pan H., Han R., Huang D., Li H., Zhou L., et al. (2014). Gemcitabine sensitivity factors, hENT1 and RRM1 as potential prognostic biomarker for advanced biliary tract cancer. Int. J. Clin. Exp. Med. 7, 5041–5049. [PMC free article] [PubMed] [Google Scholar]
- De Pas T. M., Toffalorio F., Giovannetti E., Radice D., Russo F., Angeli I., et al. (2011). Optimizing pemetrexed-gemcitabine combination in patients with advanced non-small cell lung cancer: a pharmacogenetic approach. J. Thorac. Oncol. 6, 768–773. 10.1097/JTO.0b013e31820d7818 [DOI] [PubMed] [Google Scholar]
- de Sousa Cavalcante L., Monteiro G. (2014). Gemcitabine: metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. Eur. J. Pharmacol. 741C, 8–16. 10.1016/j.ejphar.2014.07.041 [DOI] [PubMed] [Google Scholar]
- Desponts C., Ding S. (2010). Using small molecules to improve generation of induced pluripotent stem cells from somatic cells. Methods Mol. Biol. 636, 207–218. 10.1007/978-1-60761-691-7_13 [DOI] [PubMed] [Google Scholar]
- Dobson P. D., Kell D. B. (2008). Carrier-mediated cellular uptake of pharmaceutical drugs: an exception or the rule? Nat. Rev. Drug Discov. 7, 205–220. 10.1038/nrd2438 [DOI] [PubMed] [Google Scholar]
- Dobson P. D., Patel Y., Kell D. B. (2009b). “Metabolite-likeness” as a criterion in the design and selection of pharmaceutical drug libraries. Drug Discov. Today 14, 31–40. 10.1016/j.drudis.2008.10.011 [DOI] [PubMed] [Google Scholar]
- Dobson P. D., Lanthaler K., Oliver S. G., Kell D. B. (2009a). Implications of the dominant role of cellular transporters in drug uptake. Curr. Top. Med. Chem. 9, 163–184. 10.2174/156802609787521616 [DOI] [PubMed] [Google Scholar]
- Du D., Wang Z., James N. R., Voss J. E., Klimont E., Ohene-Agyei T., et al. (2014). Structure of the AcrAB-TolC multidrug efflux pump. Nature 509, 512–515. 10.1038/nature13205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Empfield J. R., Leeson P. D. (2010). Lessons learned from candidate drug attrition. IDrugs 13, 869–873. [PubMed] [Google Scholar]
- Erlanson D. A. (2012). Introduction to fragment-based drug discovery. Top. Curr. Chem. 317, 1–32. 10.1007/128_2011_180 [DOI] [PubMed] [Google Scholar]
- Erlanson D. A., Hansen S. K. (2004). Making drugs on proteins: site-directed ligand discovery for fragment-based lead assembly. Curr. Opin. Chem. Biol. 8, 399–406. 10.1016/j.cbpa.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Erlanson D. A., Mcdowell R. S., O'Brien T. (2004). Fragment-based drug discovery. J. Med. Chem. 47, 3463–3482. 10.1021/jm040031v [DOI] [PubMed] [Google Scholar]
- Eto K., Kawakami H., Kuwatani M., Kudo T., Abe Y., Kawahata S., et al. (2013). Human equilibrative nucleoside transporter 1 and Notch3 can predict gemcitabine effects in patients with unresectable pancreatic cancer. Br. J. Cancer 108, 1488–1494. 10.1038/bjc.2013.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell J. J., Elsaleh H., Garcia M., Lai R., Ammar A., Regine W. F., et al. (2009). Human equilibrative nucleoside transporter 1 levels predict response to gemcitabine in patients with pancreatic cancer. Gastroenterology 136, 187–195. 10.1053/j.gastro.2008.09.067 [DOI] [PubMed] [Google Scholar]
- Farrell J. J., Moughan J., Wong J. L., Regine W. F., Schaefer P., Benson A. B., III, et al. (2016). Precision medicine and pancreatic cancer: a gemcitabine pathway approach. Pancreas 45, 1485–1493. 10.1097/MPA.0000000000000710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B., Ng J. H., Heng J. C., Ng H. H. (2009). Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell 4, 301–312. 10.1016/j.stem.2009.03.005 [DOI] [PubMed] [Google Scholar]
- Fernández-Veledo S., Jover R., Casado F. J., Gómez-Lechón M. J., Pastor-Anglada M. (2007). Transcription factors involved in the expression of SLC28 genes in human liver parenchymal cells. Biochem. Biophys. Res. Commun. 353, 381–388. 10.1016/j.bbrc.2006.12.021 [DOI] [PubMed] [Google Scholar]
- Fernández-Veledo S., Valdés R., Wallenius V., Casado F. J., Pastor-Anglada M. (2004). Up-regulation of the high-affinity pyrimidine-preferring nucleoside transporter concentrative nucleoside transporter 1 by tumor necrosis factor-alpha and interleukin-6 in liver parenchymal cells. J. Hepatol. 41, 538–544. 10.1016/j.jhep.2004.06.008 [DOI] [PubMed] [Google Scholar]
- Fischer M., Hubbard R. E. (2009). Fragment-based ligand discovery. Mol. Interv. 9, 22–30. 10.1124/mi.9.1.7 [DOI] [PubMed] [Google Scholar]
- Franke R., Gruska A., Bossa C., Benigni R. (2010). QSARs of aromatic amines: identification of potent carcinogens. Mutat. Res. 691, 27–40. 10.1016/j.mrfmmm.2010.06.009 [DOI] [PubMed] [Google Scholar]
- Fujioka N., Fritz V., Upadhyaya P., Kassie F., Hecht S. S. (2016a). Research on cruciferous vegetables, indole-3-carbinol, and cancer prevention: a tribute to Lee W. Wattenberg. Mol. Nutr. Food Res. 60, 1228–1238. 10.1002/mnfr.201500889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka N., Ransom B. W., Carmella S. G., Upadhyaya P., Lindgren B. R., Roper-Batker A., et al. (2016b). Harnessing the power of cruciferous vegetables: developing a biomarker for Brassica vegetable consumption using urinary 3,3′-diindolylmethane. Cancer Prev. Res. (Phila). 9, 788–793. 10.1158/1940-6207.CAPR-16-0136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y., Schuetz J. D. (2012). ABC transporters and their role in nucleoside and nucleotide drug resistance. Biochem. Pharmacol. 83, 1073–1083. 10.1016/j.bcp.2011.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger J. (ed.). (2003). Handbook of Chemoinformatics: From Data to Knowledge. Weinheim: Wiley/VCH. [Google Scholar]
- Gesto D. S., Cerqueira N. M., Fernandes P. A., Ramos M. J. (2012). Gemcitabine: a critical nucleoside for cancer therapy. Curr. Med. Chem. 19, 1076–1087. 10.2174/092986712799320682 [DOI] [PubMed] [Google Scholar]
- Giacomini K. M., Huang S. M., Tweedie D. J., Benet L. Z., Brouwer K. L., Chu X., et al. (2010). Membrane transporters in drug development. Nat. Rev. Drug Discov. 9, 215–236. 10.1038/nrd3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti E., Del Tacca M., Mey V., Funel N., Nannizzi S., Ricci S., et al. (2006). Transcription analysis of human equilibrative nucleoside transporter-1 predicts survival in pancreas cancer patients treated with gemcitabine. Cancer Res. 66, 3928–3935. 10.1158/0008-5472.CAN-05-4203 [DOI] [PubMed] [Google Scholar]
- Giovannetti E., Funel N., Peters G. J., Del Chiaro M., Erozenci L. A., Vasile E., et al. (2010). MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 70, 4528–4538. 10.1158/0008-5472.CAN-09-4467 [DOI] [PubMed] [Google Scholar]
- Giovannetti E., Mey V., Loni L., Nannizzi S., Barsanti G., Savarino G., et al. (2007). Cytotoxic activity of gemcitabine and correlation with expression profile of drug-related genes in human lymphoid cells. Pharmacol. Res. 55, 343–349. 10.1016/j.phrs.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Giovannetti E., Mey V., Nannizzi S., Pasqualetti G., Marini L., Del Tacca M., et al. (2005). Cellular and pharmacogenetics foundation of synergistic interaction of pemetrexed and gemcitabine in human non-small-cell lung cancer cells. Mol. Pharmacol. 68, 110–118. 10.1124/mol.104.009373 [DOI] [PubMed] [Google Scholar]
- Gou S., Liu T., Wang C., Yin T., Li K., Yang M., et al. (2007). Establishment of clonal colony-forming assay for propagation of pancreatic cancer cells with stem cell properties. Pancreas 34, 429–435. 10.1097/MPA.0b013e318033f9f4 [DOI] [PubMed] [Google Scholar]
- Gradiz R., Silva H. C., Carvalho L., Botelho M. F., Mota-Pinto A. (2016). MIA PaCa-2 and PANC-1 - pancreas ductal adenocarcinoma cell lines with neuroendocrine differentiation and somatostatin receptors. Sci. Rep. 6:21648. 10.1038/srep21648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalf W., Ghaneh P., Neoptolemos J. P., Palmer D. H., Cox T. F., Lamb R. F., et al. (2014). Pancreatic cancer hENT1 expression and survival from gemcitabine in patients from the ESPAC-3 trial. J. Natl. Cancer Inst. 106:djt347. 10.1093/jnci/djt347 [DOI] [PubMed] [Google Scholar]
- Grskovic M., Javaherian A., Strulovici B., Daley G. Q. (2011). Induced pluripotent stem cells–opportunities for disease modelling and drug discovery. Nat. Rev. Drug Discov. 10, 915–929. 10.1038/nrd3577 [DOI] [PubMed] [Google Scholar]
- Gusella M., Pasini F., Bolzonella C., Meneghetti S., Barile C., Bononi A., et al. (2011). Equilibrative nucleoside transporter 1 genotype, cytidine deaminase activity and age predict gemcitabine plasma clearance in patients with solid tumours. Br. J. Clin. Pharmacol. 71, 437–444. 10.1111/j.1365-2125.2010.03838.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuch B., Stieger B. (2013). The SLCO (former SLC21) superfamily of transporters. Mol. Aspects Med. 34, 396–412. 10.1016/j.mam.2012.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann W., Jesnowski R., Lohr J. M. (2010). Interdependence of gemcitabine treatment, transporter expression, and resistance in human pancreatic carcinoma cells. Neoplasia 12, 740–747. 10.1593/neo.10576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann M. M. (2011). Molecular obesity, potency and other addictions in drug discovery. MedChemComm 2, 349–355. 10.1039/c1md00017a [DOI] [Google Scholar]
- Hediger M. A., Clemencon B., Burrier R. E., Bruford E. A. (2013). The ABCs of membrane transporters in health and disease (SLC series): introduction. Mol. Aspects Med. 34, 95–107. 10.1016/j.mam.2012.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger M. A., Romero M. F., Peng J. B., Rolfs A., Takanaga H., Bruford E. A. (2004). The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteinsintroduction. Pflügers Arch. 447, 465–468. 10.1007/s00424-003-1192-y [DOI] [PubMed] [Google Scholar]
- Herrgård M. J., Swainston N., Dobson P., Dunn W. B., Arga K. Y., Arvas M., et al. (2008). A consensus yeast metabolic network obtained from a community approach to systems biology. Nat. Biotechnol. 26, 1155–1160. 10.1038/nbt1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higdon J. V., Delage B., Williams D. E., Dashwood R. H. (2007). Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacol. Res. 55, 224–236. 10.1016/j.phrs.2007.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge L. S., Taub M. E., Tracy T. S. (2011). Effect of its deaminated metabolite, 2′,2′-difluorodeoxyuridine, on the transport and toxicity of gemcitabine in HeLa cells. Biochem. Pharmacol. 81, 950–956. 10.1016/j.bcp.2011.01.016 [DOI] [PubMed] [Google Scholar]
- Horiguchi S., Shiraha H., Nagahara T., Kataoka J., Iwamuro M., Matsubara M., et al. (2013). Loss of runt-related transcription factor 3 induces gemcitabine resistance in pancreatic cancer. Mol. Oncol. 7, 840–849. 10.1016/j.molonc.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Wang Y., Cogut S. B., Mitchell B. S., Graves L. M. (2003). Inhibition of nucleoside transport by protein kinase inhibitors. J. Pharmacol. Exp. Ther. 304, 753–760. 10.1124/jpet.102.044214 [DOI] [PubMed] [Google Scholar]
- Huang M., Wang Y., Collins M., Gu J. J., Mitchell B. S., Graves L. M. (2002). Inhibition of nucleoside transport by p38 MAPK inhibitors. J. Biol. Chem. 277, 28364–28367. 10.1074/jbc.C200321200 [DOI] [PubMed] [Google Scholar]
- Huang M., Wang Y., Mitchell B. S., Graves L. M. (2004). Regulation of equilibrative nucleoside uptake by protein kinase inhibitors. Nucleosides Nucleotides Nucleic Acids 23, 1445–1450. 10.1081/NCN-200027667 [DOI] [PubMed] [Google Scholar]
- Hubbard R. E. (2008). Fragment approaches in structure-based drug discovery. J. Synchrotron Radiat. 15, 227–230. 10.1107/S090904950705666X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber-Ruano I., Pinilla-Macua I., Torres G., Casado F. J., Pastor-Anglada M. (2010). Link between high-affinity adenosine concentrative nucleoside transporter-2 (CNT2) and energy metabolism in intestinal and liver parenchymal cells. J. Cell. Physiol. 225, 620–630. 10.1002/jcp.22254 [DOI] [PubMed] [Google Scholar]
- Hung S. W., Marrache S., Cummins S., Bhutia Y. D., Mody H., Hooks S. B., et al. (2015). Defective hCNT1 transport contributes to gemcitabine chemoresistance in ovarian cancer subtypes: overcoming transport defects using a nanoparticle approach. Cancer Lett. 359, 233–240. 10.1016/j.canlet.2015.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J. H., Voortman J., Giovannetti E., Steinberg S. M., Leon L. G., Kim Y. T., et al. (2010). Identification of MicroRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS ONE 5:e10630. 10.1371/journal.pone.0010630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhoti H. (2007). Fragment-based drug discovery using rational design. Ernst Schering Found. Symp. Proc. 2007, 169–185. 10.1007/2789_2007_064 [DOI] [PubMed] [Google Scholar]
- Jhoti H., Cleasby A., Verdonk M., Williams G. (2007). Fragment-based screening using X-ray crystallography and NMR spectroscopy. Curr. Opin. Chem. Biol. 11, 485–493. 10.1016/j.cbpa.2007.07.010 [DOI] [PubMed] [Google Scholar]
- Jordheim L. P., Dumontet C. (2013). Do hENT1 and RRM1 predict the clinical benefit of gemcitabine in pancreatic cancer? Biomark. Med. 7, 663–671. 10.2217/bmm.13.48 [DOI] [PubMed] [Google Scholar]
- Juge N., Mithen R. F., Traka M. (2007). Molecular basis for chemoprevention by sulforaphane: a comprehensive review. Cell. Mol. Life Sci. 64, 1105–1127. 10.1007/s00018-007-6484-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D. W., Kim W. H., Williams D. R. (2014). Reprogram or reboot: small molecule approaches for the production of induced pluripotent stem cells and direct cell reprogramming. ACS Chem. Biol. 9, 80–95. 10.1021/cb400754f [DOI] [PubMed] [Google Scholar]
- Kakinuma H., Bergert E. R., Spitzweg C., Cheville J. C., Lieber M. M., Morris J. C. (2003). Probasin promoter (ARR(2)PB)-driven, prostate-specific expression of the human sodium iodide symporter (h-NIS) for targeted radioiodine therapy of prostate cancer. Cancer Res. 63, 7840–7844. [PubMed] [Google Scholar]
- Kang P. J., Moon J. H., Yoon B. S., Hyeon S., Jun E. K., Park G., et al. (2014). Reprogramming of mouse somatic cells into pluripotent stem-like cells using a combination of small molecules. Biomaterials 35, 7336–7345. 10.1016/j.biomaterials.2014.05.015 [DOI] [PubMed] [Google Scholar]
- Kell D. B. (2013). Finding novel pharmaceuticals in the systems biology era using multiple effective drug targets, phenotypic screening, and knowledge of transporters: where drug discovery went wrong and how to fix it. FEBS J. 280, 5957–5980. 10.1111/febs.12268 [DOI] [PubMed] [Google Scholar]
- Kell D. B. (2015a). The transporter-mediated cellular uptake of pharmaceutical drugs is based on their metabolite-likeness and not on their bulk biophysical properties: towards a systems pharmacology. Perspect. Sci. 6, 66–83. 10.1016/j.pisc.2015.06.004 [DOI] [Google Scholar]
- Kell D. B. (2015b). What would be the observable consequences if phospholipid bilayer diffusion of drugs into cells is negligible?. Trends Pharmacol. Sci. 36, 15–21. 10.1016/j.tips.2014.10.005 [DOI] [PubMed] [Google Scholar]
- Kell D. B. (2016a). How drugs pass through biological cell membranes – a paradigm shift in our understanding? Beilstein Magazine 2:5 10.3762/bmag.5 [DOI] [Google Scholar]
- Kell D. B. (2016b). Implications of endogenous roles of transporters for drug discovery: hitchhiking and metabolite-likeness. Nat. Rev. Drug Discov. 15, 143–144. 10.1038/nrd.2015.44 [DOI] [PubMed] [Google Scholar]
- Kell D. B., Dobson P. D., Bilsland E., Oliver S. G. (2013). The promiscuous binding of pharmaceutical drugs and their transporter-mediated uptake into cells: what we (need to) know and how we can do so. Drug Discov. Today 18, 218–239. 10.1016/j.drudis.2012.11.008 [DOI] [PubMed] [Google Scholar]
- Kell D. B., Dobson P. D., Oliver S. G. (2011). Pharmaceutical drug transport: the issues and the implications that it is essentially carrier-mediated only. Drug Discov. Today 16, 704–714. 10.1016/j.drudis.2011.05.010 [DOI] [PubMed] [Google Scholar]
- Kell D. B., Goodacre R. (2014). Metabolomics and systems pharmacology: why and how to model the human metabolic network for drug discovery. Drug Discov. Today 19, 171–182. 10.1016/j.drudis.2013.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell D. B., Kaprelyants A. S., Weichart D. H., Harwood C. L., Barer M. R. (1998). Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie van Leeuwenhoek 73, 169–187. 10.1023/A:1000664013047 [DOI] [PubMed] [Google Scholar]
- Kell D. B., Oliver S. G. (2014). How drugs get into cells: tested and testable predictions to help discriminate between transporter-mediated uptake and lipoidal bilayer diffusion. Front. Pharmacol. 5:231. 10.3389/fphar.2014.00231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell D. B., Ryder H. M., Kaprelyants A. S., Westerhoff H. V. (1991). Quantifying heterogeneity: flow cytometry of bacterial cultures. Antonie van Leeuwenhoek 60, 145–158. 10.1007/BF00430362 [DOI] [PubMed] [Google Scholar]
- Kell D. B., Swainston N., Pir P., Oliver S. G. (2015). Membrane transporter engineering in industrial biotechnology and whole-cell biocatalysis. Trends Biotechnol. 33, 237–246. 10.1016/j.tibtech.2015.02.001 [DOI] [PubMed] [Google Scholar]
- Keppler D. (2011). Multidrug resistance proteins (MRPs, ABCCs): importance for pathophysiology and drug therapy. Handb. Exp. Pharmacol. 201, 299–323. 10.1007/978-3-642-14541-4_8 [DOI] [PubMed] [Google Scholar]
- Khatri A., Williams B. W., Fisher J., Brundage R. C., Gurvich V. J., Lis L. G., et al. (2014). SLC28A3 genotype and gemcitabine rate of infusion affect dFdCTP metabolite disposition in patients with solid tumours. Br. J. Cancer 110, 304–312. 10.1038/bjc.2013.738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. E., Ackley M. A., Cass C. E., Young J. D., Baldwin S. A. (2006). Nucleoside transporters: from scavengers to novel therapeutic targets. Trends Pharmacol. Sci. 27, 416–425. 10.1016/j.tips.2006.06.004 [DOI] [PubMed] [Google Scholar]
- Klein K., Kullak-Ublick G. A., Wagner M., Trauner M., Eloranta J. J. (2009). Hepatocyte nuclear factor-4α and bile acids regulate human concentrative nucleoside transporter-1 gene expression. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G936–G947. 10.1152/ajpgi.90678.2008 [DOI] [PubMed] [Google Scholar]
- Koay E. J., Truty M. J., Cristini V., Thomas R. M., Chen R., Chatterjee D., et al. (2014). Transport properties of pancreatic cancer describe gemcitabine delivery and response. J. Clin. Invest. 124, 1525–1536. 10.1172/JCI73455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H., Murakami Y., Uemura K., Sudo T., Hashimoto Y., Kondo N., et al. (2012). Human equilibrative nucleoside transporter 1 expression predicts survival of advanced cholangiocarcinoma patients treated with gemcitabine-based adjuvant chemotherapy after surgical resection. Ann. Surg. 256, 288–296. 10.1097/SLA.0b013e3182536a42 [DOI] [PubMed] [Google Scholar]
- Koczor C. A., Torres R. A., Lewis W. (2012). The role of transporters in the toxicity of nucleoside and nucleotide analogs. Expert Opin. Drug Metab. Toxicol. 8, 665–676. 10.1517/17425255.2012.680885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kola I., Landis J. (2004). Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 3, 711–715. 10.1038/nrd1470 [DOI] [PubMed] [Google Scholar]
- Komori S., Osada S., Yoshida K. (2011). Novel strategy with gemcitabine for advanced pancreatic cancer. ISRN Oncol. 2011:936893. 10.5402/2011/936893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W., Engel K., Wang J. (2004). Mammalian nucleoside transporters. Curr. Drug Metab. 5, 63–84. 10.2174/1389200043489162 [DOI] [PubMed] [Google Scholar]
- König J., Hartel M., Nies A. T., Martignoni M. E., Guo J., Büchler M. W., et al. (2005). Expression and localization of human multidrug resistance protein (ABCC) family members in pancreatic carcinoma. Int. J. Cancer 115, 359–367. 10.1002/ijc.20831 [DOI] [PubMed] [Google Scholar]
- Köse M., Schiedel A. C. (2009). Nucleoside/nucleobase transporters: drug targets of the future? Future Med. Chem. 1, 303–326. 10.4155/fmc.09.29 [DOI] [PubMed] [Google Scholar]
- Lane J., Martin T. A., Mcguigan C., Mason M. D., Jiang W. G. (2010). The differential expression of hCNT1 and hENT1 in breast cancer and the possible impact on breast cancer therapy. J. Exp. Ther. Oncol. 8, 203–210. [PubMed] [Google Scholar]
- Lanthaler K., Bilsland E., Dobson P., Moss H. J., Pir P., Kell D. B., et al. (2011). Genome-wide assessment of the carriers involved in the cellular uptake of drugs: a model system in yeast. BMC Biol. 9:70. 10.1186/1741-7007-9-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach A. R., Hann M. M. (2011). Molecular complexity and fragment-based drug discovery: ten years on. Curr. Opin. Chem. Biol. 15, 489–496. 10.1016/j.cbpa.2011.05.008 [DOI] [PubMed] [Google Scholar]
- Lee S. Y., Im S. A., Park Y. H., Woo S. Y., Kim S., Choi M. K., et al. (2014). Genetic polymorphisms of SLC28A3, SLC29A1 and RRM1 predict clinical outcome in patients with metastatic breast cancer receiving gemcitabine plus paclitaxel chemotherapy. Eur. J. Cancer 50, 698–705. 10.1016/j.ejca.2013.11.028 [DOI] [PubMed] [Google Scholar]
- Leeson P. D. (2016). Molecular inflation, attrition and the rule of five. Adv. Drug Deliv. Rev. 101, 22–33. 10.1016/j.addr.2016.01.018 [DOI] [PubMed] [Google Scholar]
- Leeson P. D., Empfield J. R. (2010). Reducing the risk of drug attrition associated with physicochemical properties. Annu. Rep. Med. Chem. 45, 393–407. 10.1016/S0065-7743(10)45024-1 [DOI] [Google Scholar]
- Leeson P. D., Springthorpe B. (2007). The influence of drug-like concepts on decision-making in medicinal chemistry. Nat. Rev. Drug Discov. 6, 881–890. 10.1038/nrd2445 [DOI] [PubMed] [Google Scholar]
- Lehár J., Krueger A. S., Avery W., Heilbut A. M., Johansen L. M., Price E. R., et al. (2009a). Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat. Biotechnol. 27, 659–666. 10.1038/nbt.1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehár J., Krueger A. S., Zimmermann G. R., Borisy A. A. (2009b). Therapeutic selectivity and the multi-node drug target. Discov. Med. 8, 185–190. [PubMed] [Google Scholar]
- Lehár J., Krueger A., Zimmermann G., Borisy A. (2008). High-order combination effects and biological robustness. Mol. Syst. Biol. 4, 215. 10.1038/msb.2008.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehár J., Zimmermann G. R., Krueger A. S., Molnar R. A., Ledell J. T., Heilbut A. M., et al. (2007). Chemical combination effects predict connectivity in biological systems. Mol. Syst. Biol. 3, 80. 10.1038/msb4100116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemstrová R., Souček P., Melichar B., Mohelnikova-Duchonova B. (2014). Role of solute carrier transporters in pancreatic cancer: a review. Pharmacogenomics 15, 1133–1145. 10.2217/pgs.14.80 [DOI] [PubMed] [Google Scholar]
- Leung G. P. H., Tse C. M. (2007). The role of mitochondrial and plasma membrane nucleoside transporters in drug toxicity. Expert Opin. Drug Metab. Toxicol. 3, 705–718. 10.1517/17425255.3.5.705 [DOI] [PubMed] [Google Scholar]
- Li K., Zhu S., Russ H. A., Xu S., Xu T., Zhang Y., et al. (2014). Small molecules facilitate the reprogramming of mouse fibroblasts into pancreatic lineages. Cell Stem Cell 14, 228–236. 10.1016/j.stem.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. W. S., Tse C. M., Man R. Y., Vanhoutte P. M., Leung G. P. H. (2007). Inhibition of human equilibrative nucleoside transporters by dihydropyridine-type calcium channel antagonists. Eur. J. Pharmacol. 568, 75–82. 10.1016/j.ejphar.2007.04.033 [DOI] [PubMed] [Google Scholar]
- Li W., Ding S. (2010). Small molecules that modulate embryonic stem cell fate and somatic cell reprogramming. Trends Pharmacol. Sci. 31, 36–45. 10.1016/j.tips.2009.10.002 [DOI] [PubMed] [Google Scholar]
- Li W., Jiang K., Ding S. (2012). Concise review: a chemical approach to control cell fate and function. Stem Cells 30, 61–68. 10.1002/stem.768 [DOI] [PubMed] [Google Scholar]
- Li X.-Z., Plesiat P., Nikaido H. (2015). The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. 10.1128/CMR.00117-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zuo X., Jing J., Ma Y., Wang J., Liu D., et al. (2015). Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell 17, 195–203. 10.1016/j.stem.2015.06.003 [DOI] [PubMed] [Google Scholar]
- Liu G. (2013). Stearoyl-CoA Desaturase 1 (SCD1) inhibitors: bench to bedside must only go through liver. RSC Drug Discov. 27, 249–269. 10.1039/9781849735322-00249 [DOI] [Google Scholar]
- Liu Y., Peng H., Zhang J. T. (2005). Expression profiling of ABC transporters in a drug-resistant breast cancer cell line using AmpArray. Mol. Pharmacol. 68, 430–438. 10.1124/mol.105.011015 [DOI] [PubMed] [Google Scholar]
- Liu Z. Q., Han Y. C., Zhang X., Chu L., Fang J. M., Zhao H. X., et al. (2014). Prognostic value of human equilibrative nucleoside transporter1 in pancreatic cancer receiving gemcitabin-based chemotherapy: a meta-analysis. PLoS ONE 9:e87103. 10.1371/journal.pone.0087103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen S. K., Ng A. M., Yao S. Y., Cass C. E., Baldwin S. A., Young J. D. (1999). Identification of amino acid residues responsible for the pyrimidine and purine nucleoside specificities of human concentrative Na+ nucleoside cotransporters hCNT1 and hCNT2. J. Biol. Chem. 274, 24475–24484. 10.1074/jbc.274.35.24475 [DOI] [PubMed] [Google Scholar]
- Lyn-Cook B. D., Mohammed S. I., Davis C., Word B., Haefele A., Wang H., et al. (2010). Gender differences in gemcitabine (Gemzar) efficacy in cancer cells: effect of indole-3-carbinol. Anticancer Res. 30, 4907–4913. [PubMed] [Google Scholar]
- Mackey J. R., Baldwin S. A., Young J. D., Cass C. E. (1998a). Nucleoside transport and its significance for anticancer drug resistance. Drug Resist. Updat. 1, 310–324. 10.1016/S1368-7646(98)80047-2 [DOI] [PubMed] [Google Scholar]
- Mackey J. R., Mani R. S., Selner M., Mowles D., Young J. D., Belt J. A., et al. (1998b). Functional nucleoside transporters are required for gemcitabine influx and manifestation of toxicity in cancer cell lines. Cancer Res. 58, 4349–4357. [PubMed] [Google Scholar]
- Maggiora G., Vogt M., Stumpfe D., Bajorath J. (2014). Molecular similarity in medicinal chemistry. J. Med. Chem. 57, 3186–3204. 10.1021/jm401411z [DOI] [PubMed] [Google Scholar]
- Marcé S., Molina-Arcas M., Villamor N., Casado F. J., Campo E., Pastor-Anglada M., et al. (2006). Expression of human equilibrative nucleoside transporter 1 (hENT1) and its correlation with gemcitabine uptake and cytotoxicity in mantle cell lymphoma. Haematologica 91, 895–902. [PubMed] [Google Scholar]
- Maréchal R., Bachet J. B., Mackey J. R., Dalban C., Demetter P., Graham K., et al. (2012). Levels of gemcitabine transport and metabolism proteins predict survival times of patients treated with gemcitabine for pancreatic adenocarcinoma. Gastroenterology, 143, 664–674.e661–e666. 10.1053/j.gastro.2012.06.006 [DOI] [PubMed] [Google Scholar]
- Maréchal R., Mackey J. R., Lai R., Demetter P., Peeters M., Polus M., et al. (2009). Human equilibrative nucleoside transporter 1 and human concentrative nucleoside transporter 3 predict survival after adjuvant gemcitabine therapy in resected pancreatic adenocarcinoma. Clin. Cancer Res. 15, 2913–2919. 10.1158/1078-0432.CCR-08-2080 [DOI] [PubMed] [Google Scholar]
- Matsumura N., Nakamura Y., Kohjimoto Y., Inagaki T., Nanpo Y., Yasuoka H., et al. (2011). The prognostic significance of human equilibrative nucleoside transporter 1 expression in patients with metastatic bladder cancer treated with gemcitabine-cisplatin-based combination chemotherapy. BJU Int. 108, E110–E116. 10.1111/j.1464-410X.2010.09932.x [DOI] [PubMed] [Google Scholar]
- Mazanetz M. P., Marmon R. J., Reisser C. B. T., Morao I. (2012). Drug discovery applications for KNIME: an open source data mining platform. Curr. Top. Med. Chem. 12, 1965–1979. 10.2174/156802612804910331 [DOI] [PubMed] [Google Scholar]
- Meanwell N. A. (2011). Improving drug candidates by design: a focus on physicochemical properties as a means of improving compound disposition and safety. Chem. Res. Toxicol. 24, 1420–1456. 10.1021/tx200211v [DOI] [PubMed] [Google Scholar]
- Meinl T., Jagla B., Berthold M. R. (2012). Integrated data analysis with KNIME, in Open Source Software in Life Science Research, eds Harland L., Forster M. (Sawston: Woodhead Publishing; ), 151–171. [Google Scholar]
- Melkamu T., Zhang X. X., Tan J. K., Zeng Y., Kassie F. (2010). Alteration of microRNA expression in vinyl carbamate-induced mouse lung tumors and modulation by the chemopreventive agent indole-3-carbinol. Carcinogenesis 31, 252–258. 10.1093/carcin/bgp208 [DOI] [PubMed] [Google Scholar]
- Mendes P., Oliver S. G., Kell D. B. (2015). Fitting transporter activities to cellular drug concentrations and fluxes: why the bumblebee can fly. Trends Pharmacol. Sci. 36, 710–723. 10.1016/j.tips.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey V., Giovannetti E., De Braud F., Nannizzi S., Curigliano G., Verweij F., et al. (2006). In vitro synergistic cytotoxicity of gemcitabine and pemetrexed and pharmacogenetic evaluation of response to gemcitabine in bladder cancer patients. Br. J. Cancer 95, 289–297. 10.1038/sj.bjc.6603242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami K., Shinsato Y., Yamamoto M., Takahashi H., Zhang S., Nishizawa Y., et al. (2015). Ribonucleotide reductase is an effective target to overcome gemcitabine resistance in gemcitabine-resistant pancreatic cancer cells with dual resistant factors. J. Pharmacol. Sci. 127, 319–325. 10.1016/j.jphs.2015.01.006 [DOI] [PubMed] [Google Scholar]
- Mini E., Nobili S., Caciagli B., Landini I., Mazzei T. (2006). Cellular pharmacology of gemcitabine. Ann. Oncol. 17(Suppl. 5), v7–v12. 10.1093/annonc/mdj941 [DOI] [PubMed] [Google Scholar]
- Molina-Arcas M., Casado F. J., Pastor-Anglada M. (2009). Nucleoside transporter proteins. Curr. Vasc. Pharmacol. 7, 426–434. 10.2174/157016109789043892 [DOI] [PubMed] [Google Scholar]
- Molina-Arcas M., Pastor-Anglada M. (2010). Role of nucleoside transporters in nucleoside-derived drug sensitivity. Nucleosides Nucleotides Nucleic Acids 29, 335–346. 10.1080/15257771003729823 [DOI] [PubMed] [Google Scholar]
- Molina-Arcas M., Pastor-Anglada M. (2013). Nucleoside transporters (SLC28 and SLC29) family, in Pharmacogenomics of Human Drug Transporters: Clinical Impacts, eds Ishikawa T., Kim R. B., König J. (New York, NY: Wiley; ), 243–270. [Google Scholar]
- Molina-Arcas M., Trigueros-Motos L., Casado F. J., Pastor-Anglada M. (2008). Physiological and pharmacological roles of nucleoside transporter proteins. Nucleosides Nucleotides Nucleic Acids 27, 769–778. 10.1080/15257770802145819 [DOI] [PubMed] [Google Scholar]
- Mori R., Ishikawa T., Ichikawa Y., Taniguchi K., Matsuyama R., Ueda M., et al. (2007). Human equilibrative nucleoside transporter 1 is associated with the chemosensitivity of gemcitabine in human pancreatic adenocarcinoma and biliary tract carcinoma cells. Oncol. Rep. 17, 1201–1205. 10.3892/or.17.5.1201 [DOI] [PubMed] [Google Scholar]
- Morinaga S., Nakamura Y., Watanabe T., Mikayama H., Tamagawa H., Yamamoto N., et al. (2012). Immunohistochemical Analysis of Human Equilibrative Nucleoside Transporter-1 (hENT1) predicts survival in resected pancreatic cancer patients treated with adjuvant gemcitabine monotherapy. Ann. Surg. Oncol. 19(Suppl. 3), 558–564. 10.1245/s10434-011-2054-z [DOI] [PubMed] [Google Scholar]
- Mrozikiewicz P. M., Bogacz A., Bartkowiak-Wieczorek J., Kujawski R., Mikolajczak P. L., Ozarowski M., et al. (2014). Screening for impact of popular herbs improving mental abilities on the transcriptional level of brain transporters. Acta Pharm. 64, 223–232. 10.2478/acph-2014-0020 [DOI] [PubMed] [Google Scholar]
- Murata Y., Hamada T., Kishiwada M., Ohsawa I., Mizuno S., Usui M., et al. (2012). Human equilibrative nucleoside transporter 1 expression is a strong independent prognostic factor in UICC T3-T4 pancreatic cancer patients treated with preoperative gemcitabine-based chemoradiotherapy. J. Hepatobiliary Pancreat. Sci. 19, 413–425. 10.1007/s00534-011-0440-3 [DOI] [PubMed] [Google Scholar]
- Nakagawa N., Murakami Y., Uemura K., Sudo T., Hashimoto Y., Kondo N., et al. (2013). Combined analysis of intratumoral human equilibrative nucleoside transporter 1 (hENT1) and ribonucleotide reductase regulatory subunit M1 (RRM1) expression is a powerful predictor of survival in patients with pancreatic carcinoma treated with adjuvant gemcitabine-based chemotherapy after operative resection. Surgery 153, 565–575. 10.1016/j.surg.2012.10.010 [DOI] [PubMed] [Google Scholar]
- Nakahira S., Nakamori S., Tsujie M., Takeda S., Sugimoto K., Takahashi Y., et al. (2008). Pretreatment with S-1, an oral derivative of 5-fluorouracil, enhances gemcitabine effects in pancreatic cancer xenografts. Anticancer Res. 28, 179–186. [PubMed] [Google Scholar]
- Nakano Y., Tanno S., Koizumi K., Nishikawa T., Nakamura K., Minoguchi M., et al. (2007). Gemcitabine chemoresistance and molecular markers associated with gemcitabine transport and metabolism in human pancreatic cancer cells. Br. J. Cancer 96, 457–463. 10.1038/sj.bjc.6603559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma B., Sasaki T., Fujimoto Y., Serikawa M., Kobayashi K., Inoue M., et al. (2008). Expression of multidrug resistance-associated protein 2 is involved in chemotherapy resistance in human pancreatic cancer. Int. J. Oncol. 33, 1187–1194. 10.3892/ijo_00000108 [DOI] [PubMed] [Google Scholar]
- Nordh S., Ansari D., Andersson R. (2014). hENT1 expression is predictive of gemcitabine outcome in pancreatic cancer: a systematic review. World J. Gastroenterol. 20, 8482–8490. 10.3748/wjg.v20.i26.8482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oballa R. M., Belair L., Black W. C., Bleasby K., Chan C. C., Desroches C., et al. (2011). Development of a liver-targeted stearoyl-CoA desaturase (SCD) inhibitor (MK-8245) to establish a therapeutic window for the treatment of diabetes and dyslipidemia. J. Med. Chem. 54, 5082–5096. 10.1021/jm200319u [DOI] [PubMed] [Google Scholar]
- Oguri T., Achiwa H., Muramatsu H., Ozasa H., Sato S., Shimizu S., et al. (2007). The absence of human equilibrative nucleoside transporter 1 expression predicts nonresponse to gemcitabine-containing chemotherapy in non-small cell lung cancer. Cancer Lett. 256, 112–119. 10.1016/j.canlet.2007.06.012 [DOI] [PubMed] [Google Scholar]
- O'Hagan S., Kell D. B. (2015a). The apparent permeabilities of Caco-2 cells to marketed drugs: magnitude, and independence from both biophysical properties and endogenite similarities PeerJ 3:E1405. 10.7717/peerj.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan S., Kell D. B. (2015b). Software review: the KNIME workflow environment and its applications in Genetic Programming and machine learning. Genetic Progr. Evol. Mach. 16, 387–391. 10.1007/s10710-015-9247-3 [DOI] [Google Scholar]
- O'Hagan S., Kell D. B. (2015c). Understanding the foundations of the structural similarities between marketed drugs and endogenous human metabolites. Front. Pharmacol. 6:105. 10.3389/fphar.2015.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan S., Kell D. B. (2016). MetMaxStruct: a Tversky-similarity-based strategy for analysing the (sub)structural similarities of drugs and endogenous metabolites. Front. Pharmacol. 7:266. 10.3389/fphar.2016.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan S., Swainston N., Handl J., Kell D. B. (2015). A ‘rule of 0.5’ for the metabolite-likeness of approved pharmaceutical drugs. Metabolomics 11, 323–339. 10.1007/s11306-014-0733-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki T., Javle M., Tanaka M., Abbruzzese J. L., Li D. (2010). Single nucleotide polymorphisms of gemcitabine metabolic genes and pancreatic cancer survival and drug toxicity. Clin. Cancer Res. 16, 320–329. 10.1158/1078-0432.CCR-09-1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. (2007). Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317. 10.1038/nature05934 [DOI] [PubMed] [Google Scholar]
- Paik W. H., Kim H. R., Park J. K., Song B. J., Lee S. H., Hwang J. H. (2013). Chemosensitivity induced by down-regulation of microRNA-21 in gemcitabine-resistant pancreatic cancer cells by indole-3-carbinol. Anticancer Res. 33, 1473–1481. 10.1371/journal.pone.0056423 [DOI] [PubMed] [Google Scholar]
- Paproski R. J., Yao S. Y. M., Favis N., Evans D., Young J. D., Cass C. E., et al. (2013). Human concentrative nucleoside transporter 3 transfection with ultrasound and microbubbles in nucleoside transport deficient HEK293 cells greatly increases gemcitabine uptake. PLoS ONE 8:e56423. 10.1371/journal.pone.0056423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paproski R. J., Young J. D., Cass C. E. (2010). Predicting gemcitabine transport and toxicity in human pancreatic cancer cell lines with the positron emission tomography tracer 3′-deoxy-3′-fluorothymidine. Biochem. Pharmacol. 79, 587–595. 10.1016/j.bcp.2009.09.025 [DOI] [PubMed] [Google Scholar]
- Pastor-Anglada M., Pérez-Torras S. (2015). Nucleoside transporter proteins as biomarkers of drug responsiveness and drug targets. Front. Pharmacol. 6:13. 10.3389/fphar.2015.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennycooke M., Chaudary N., Shuralyova I., Zhang Y., Coe I. R. (2001). Differential expression of human nucleoside transporters in normal and tumor tissue. Biochem. Biophys. Res. Commun. 280, 951–959. 10.1006/bbrc.2000.4205 [DOI] [PubMed] [Google Scholar]
- Pérez-Torras S., García-Manteiga J., Mercadé E., Casado F. J., Carbó N., Pastor-Anglada M., et al. (2008). Adenoviral-mediated overexpression of human equilibrative nucleoside transporter 1 (hENT1) enhances gemcitabine response in human pancreatic cancer. Biochem. Pharmacol. 76, 322–329. 10.1016/j.bcp.2008.05.011 [DOI] [PubMed] [Google Scholar]
- Pfefferkorn J. A. (2013). Strategies for the design of hepatoselective glucokinase activators to treat type 2 diabetes. Expert Opin. Drug Discov. 8, 319–330. 10.1517/17460441.2013.748744 [DOI] [PubMed] [Google Scholar]
- Pfefferkorn J. A., Guzman-Perez A., Litchfield J., Aiello R., Treadway J. L., Pettersen J., et al. (2012). Discovery of (S)-6-(3-cyclopentyl-2-(4-(trifluoromethyl)-1H-imidazol-1-yl)propanamido)nicotini c acid as a hepatoselective glucokinase activator clinical candidate for treating type 2 diabetes mellitus. J. Med. Chem. 55, 1318–1333. 10.1021/jm2014887 [DOI] [PubMed] [Google Scholar]
- Pfefferkorn J. A., Litchfield J., Hutchings R., Cheng X. M., Larsen S. D., Auerbach B., et al. (2011). Discovery of novel hepatoselective HMG-CoA reductase inhibitors for treating hypercholesterolemia: a bench-to-bedside case study on tissue selective drug distribution. Bioorg. Med. Chem. Lett. 21, 2725–2731. 10.1016/j.bmcl.2010.11.103 [DOI] [PubMed] [Google Scholar]
- Plant N. (2016). Enabling dynamic response to chemical challenge: nuclear receptor-mediated control of transporter expression, in Drug Transporters, Vol. 2, Recent Advances and Emerging Technologies, eds Nicholls G., Youdim K. (London: RSC; ), 19–43. [Google Scholar]
- Podgorska M., Kocbuch K., Pawelczyk T. (2005). Recent advances in studies on biochemical and structural properties of equilibrative and concentrative nucleoside transporters. Acta Biochim. Pol. 52, 749–758. [PubMed] [Google Scholar]
- Prasad R., Rawal M. K. (2014). Efflux pump proteins in antifungal resistance. Front. Pharmacol. 5:202. 10.3389/fphar.2014.00202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putman M., Van Veen H. W., Konings W. N. (2000). Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64, 672–693. 10.1128/MMBR.64.4.672-693.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramtohul Y. K., Powell D., Leclerc J. P., Leger S., Oballa R., Black C., et al. (2011). Bicyclic heteroaryl inhibitors of stearoyl-CoA desaturase: from systemic to liver-targeting inhibitors. Bioorg. Med. Chem. Lett. 21, 5692–5696. 10.1016/j.bmcl.2011.08.037 [DOI] [PubMed] [Google Scholar]
- Rauchwerger D. R., Firby P. S., Hedley D. W., Moore M. J. (2000). Equilibrative-sensitive nucleoside transporter and its role in gemcitabine sensitivity. Cancer Res. 60, 6075–6079. [PubMed] [Google Scholar]
- Rees D. C., Congreve M., Murray C. W., Carr R. (2004). Fragment-based lead discovery. Nat. Rev. Drug Discov. 3, 660–672. 10.1038/nrd1467 [DOI] [PubMed] [Google Scholar]
- Ritzel M. W. L., Ng A. M. L., Yao S. Y. M., Graham K., Loewen S. K., Smith K. M., et al. (2001). Molecular identification and characterization of novel human and mouse concentrative Na+-nucleoside cotransporter proteins (hCNT3 and mCNT3) broadly selective for purine and pyrimidine nucleosides (system cib). J. Biol. Chem. 276, 2914–2927. 10.1074/jbc.M007746200 [DOI] [PubMed] [Google Scholar]
- Rosenberg M. F., Bikadi Z., Hazai E., Starborg T., Kelley L., Chayen N. E., et al. (2015). Three-dimensional structure of the human breast cancer resistance protein (BCRP/ABCG2) in an inward-facing conformation. Acta Crystallogr. D Biol. Crystallogr. 71, 1725–1735. 10.1107/S1399004715010676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostami-Hodjegan A. (2012). Physiologically based pharmacokinetics joined with in vitro-in vivo extrapolation of ADME: a marriage under the arch of systems pharmacology. Clin. Pharmacol. Ther. 92, 50–61. 10.1038/clpt.2012.65 [DOI] [PubMed] [Google Scholar]
- Rudin D., Li L., Niu N., Kalari K. R., Gilbert J. A., Ames M. M., et al. (2011). Gemcitabine cytotoxicity: interaction of efflux and deamination. J. Drug Metab. Toxicol. 2, 1–10. 10.4172/2157-7609.1000107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini D., Schiavon G., Vincenzi B., Cass C. E., Vasile E., Manazza A. D., et al. (2011). Human equilibrative nucleoside transporter 1 (hENT1) levels predict response to gemcitabine in patients with biliary tract cancer (BTC). Curr. Cancer Drug Targets 11, 123–129. 10.2174/156800911793743600 [DOI] [PubMed] [Google Scholar]
- Santini D., Vincenzi B., Fratto M. E., Perrone G., Lai R., Catalano V., et al. (2010). Prognostic role of human equilibrative transporter 1 (hENT1) in patients with resected gastric cancer. J. Cell. Physiol. 223, 384–388. 10.1002/jcp.22045 [DOI] [PubMed] [Google Scholar]
- Saunders N. R., Ek C. J., Habgood M. D., Johansson P., Liddelow S., Dziegielewska K. M. (2011). Assessing blood-cerebrospinal fluid barrier permeability in the rat embryo. Methods Mol. Biol. 686, 247–265. 10.1007/978-1-60761-938-3_11 [DOI] [PubMed] [Google Scholar]
- Schulz M. N., Hubbard R. E. (2009). Recent progress in fragment-based lead discovery. Curr. Opin. Pharmacol. 9, 615–621. 10.1016/j.coph.2009.04.009 [DOI] [PubMed] [Google Scholar]
- Sharma R., Litchfield J., Bergman A., Atkinson K., Kazierad D., Gustavson S. M., et al. (2015). Comparison of the circulating metabolite profile of PF-04991532, a hepatoselective glucokinase activator, across preclinical species and humans: potential implications in metabolites in safety testing assessment. Drug Metab. Dispos. 43, 190–198. 10.1124/dmd.114.061218 [DOI] [PubMed] [Google Scholar]
- Silva R., Vilas-Boas V., Carmo H., Dinis-Oliveira R. J., Carvalho F., De Lourdes Bastos M., et al. (2015). Modulation of P-glycoprotein efflux pump: induction and activation as a therapeutic strategy. Pharmacol. Ther. 149, 1–123. 10.1016/j.pharmthera.2014.11.013 [DOI] [PubMed] [Google Scholar]
- Skrypek N., Duchêne B., Hebbar M., Leteurtre E., Van Seuningen I., Jonckheere N. (2013). The MUC4 mucin mediates gemcitabine resistance of human pancreatic cancer cells via the concentrative nucleoside transporter family. Oncogene 32, 1714–1723. 10.1038/onc.2012.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. M., Slugoski M. D., Cass C. E., Baldwin S. A., Karpinski E., Young J. D. (2007). Cation coupling properties of human concentrative nucleoside transporters hCNT1, hCNT2 and hCNT3. Mol. Membr. Biol. 24, 53–64. 10.1080/09687860600942534 [DOI] [PubMed] [Google Scholar]
- Spratlin J. L., Mackey J. R. (2010). Human Equilibrative Nucleoside Transporter 1 (hENT1) in pancreatic adenocarcinoma: towards individualized treatment decisions. Cancers 2, 2044–2054. 10.3390/cancers2042044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratlin J., Sangha R., Glubrecht D., Dabbagh L., Young J. D., Dumontet C., et al. (2004). The absence of human equilibrative nucleoside transporter 1 is associated with reduced survival in patients with gemcitabine-treated pancreas adenocarcinoma. Clin. Cancer Res. 10, 6956–6961. 10.1158/1078-0432.CCR-04-0224 [DOI] [PubMed] [Google Scholar]
- Stumpfe D., Bajorath J. (2011). Similarity searching. Wires Comput. Mol. Sci. 1, 260–282. 10.1002/wcms.23 [DOI] [Google Scholar]
- Sugiyama Y., Steffansen B. (eds.). (2013). Transporters in Drug Development: Discovery, Optimization, Clinical Study and Regulation. New York, NY: AAPS/Springer. [Google Scholar]
- Swainston N., Mendes P., Kell D. B. (2013). An analysis of a ‘community-driven’ reconstruction of the human metabolic network. Metabolomics 9, 757–764. 10.1007/s11306-013-0564-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swainston N., Smallbone K., Hefzi H., Dobson P. D., Brewer J., Hanscho M., et al. (2016). Recon 2.2: from reconstruction to model of human metabolism. Metabolomics 12, 109. 10.1007/s11306-016-1051-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinney D. C. (2013). Phenotypic vs. target-based drug discovery for first-in-class medicines. Clin. Pharmacol. Ther. 93, 299–301. 10.1038/clpt.2012.236 [DOI] [PubMed] [Google Scholar]
- Swinney D. C., Anthony J. (2011). How were new medicines discovered? Nat. Rev. Drug Discov. 10, 507–519. 10.1038/nrd3480 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Javle M., Dong X., Eng C., Abbruzzese J. L., Li D. (2010). Gemcitabine metabolic and transporter gene polymorphisms are associated with drug toxicity and efficacy in patients with locally advanced pancreatic cancer. Cancer 116, 5325–5335. 10.1002/cncr.25282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavano F., Fontana A., Pellegrini F., Burbaci F., Rappa F., Cappello F., et al. (2014). Modeling interactions between human equilibrative nucleoside transporter-1 and other factors involved in the response to gemcitabine treatment to predict clinical outcomes in pancreatic ductal adenocarcinoma patients. J. Transl. Med. 12, 248. 10.1186/s12967-014-0248-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I., Swainston N., Fleming R. M. T., Hoppe A., Sahoo S., Aurich M. K., et al. (2013). A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 31, 419–425. 10.1038/nbt.2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu M. H., Mathiowetz A. M., Pfefferkorn J. A., Cameron K. O., Dow R. L., Litchfield J., et al. (2013). Medicinal chemistry design principles for liver targeting through OATP transporters. Curr. Top. Med. Chem. 13, 857–866. 10.2174/1568026611313070008 [DOI] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallstrom B. M., Lindskog C., Oksvold P., Mardinoglu A., et al. (2015). Tissue-based map of the human proteome. Science 347:1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- van der Graaf P. H., Benson N. (2011). Systems pharmacology: bridging systems biology and pharmacokinetics-pharmacodynamics (PKPD) in drug discovery and development. Pharm. Res. 28, 1460–1464. 10.1007/s11095-011-0467-9 [DOI] [PubMed] [Google Scholar]
- van der Greef J., Mcburney R. N. (2005). Rescuing drug discovery: in vivo systems pathology and systems pharmacology. Nat. Rev. Drug Discov. 4, 961–967. 10.1038/nrd1904 [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034. 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltkamp S. A., Pluim D., Van Eijndhoven M. A., Bolijn M. J., Ong F. H., Govindarajan R., et al. (2008). New insights into the pharmacology and cytotoxicity of gemcitabine and 2′,2′-difluorodeoxyuridine. Mol. Cancer Ther. 7, 2415–2425. 10.1158/1535-7163.MCT-08-0137 [DOI] [PubMed] [Google Scholar]
- Vickers M. F., Kumar R., Visser F., Zhang J., Charania J., Raborn R. T., et al. (2002). Comparison of the interaction of uridine, cytidine, and other pyrimidine nucleoside analogues with recombinant human equilibrative nucleoside transporter 2 (hENT2) produced in Saccharomyces cerevisiae. Biochem. Cell Biol. 80, 639–644. 10.1139/o02-148 [DOI] [PubMed] [Google Scholar]
- Waddell N., Pajic M., Patch A. M., Chang D. K., Kassahn K. S., Bailey P., et al. (2015). Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501. 10.1038/nature14169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman S. A., Terzic A. (2012). Advancing pharmacometrics and systems pharmacology. Clin. Pharmacol. Ther. 92, 535–537. 10.1038/clpt.2012.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Word B. R., Lyn-Cook B. D. (2011). Enhanced efficacy of gemcitabine by indole-3-carbinol in pancreatic cell lines: the role of human equilibrative nucleoside transporter 1. Anticancer Res. 31, 3171–3180. [PubMed] [Google Scholar]
- Wang J., Giacomini K. M. (1999). Serine 318 is essential for the pyrimidine selectivity of the N2 Na+-nucleoside transporter. J. Biol. Chem. 274, 2298–2302. 10.1074/jbc.274.4.2298 [DOI] [PubMed] [Google Scholar]
- Ward J. L., Sherali A., Mo Z. P., Tse C. M. (2000). Kinetic and pharmacological properties of cloned human equilibrative nucleoside transporters, ENT1 and ENT2, stably expressed in nucleoside transporter-deficient PK15 cells. Ent2 exhibits a low affinity for guanosine and cytidine but a high affinity for inosine. J. Biol. Chem. 275, 8375–8381. 10.1074/jbc.275.12.8375 [DOI] [PubMed] [Google Scholar]
- Warr W. A. (2012). Scientific workflow systems: pipeline pilot and KNIME. J. Comput. Aided Mol. Des. 26, 801–804. 10.1007/s10822-012-9577-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J., Theile D., Ketabi-Kiyanvash N., Lindenmaier H., Haefeli W. E. (2007). Inhibition of MRP1/ABCC1, MRP2/ABCC2, and MRP3/ABCC3 by nucleoside, nucleotide, and non-nucleoside reverse transcriptase inhibitors. Drug Metab. Dispos. 35, 340–344. 10.1124/dmd.106.012765 [DOI] [PubMed] [Google Scholar]
- Westerhoff H. V., Nakayama S., Mondeel T. D. G. A., Barberis M. (2015). Systems pharmacology: an opinion on how to turn the impossible into grand challenges. Drug Discov. Today 15, 23–31. 10.1016/j.ddtec.2015.06.006 [DOI] [PubMed] [Google Scholar]
- Whittaker M., Law R. J., Ichihara O., Hesterkamp T., Hallett D. (2010). Fragments: past, present and future. Drug Discov. Today 7, e163–e171. 10.1016/j.ddtec.2010.11.007 [DOI] [Google Scholar]
- Winter G. E., Radic B., Mayor-Ruiz C., Blomen V. A., Trefzer C., Kandasamy R. K., et al. (2014). The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat. Chem. Biol. 10, 768–773. 10.1038/nchembio.1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A., Soo R. A., Yong W. P., Innocenti F. (2009). Clinical pharmacology and pharmacogenetics of gemcitabine. Drug Metab. Rev. 41, 77–88. 10.1080/03602530902741828 [DOI] [PubMed] [Google Scholar]
- Wright G. D. (2016). Antibiotic adjuvants: rescuing antibiotics from resistance. Trends Microbiol. 24, 862–871. 10.1016/j.tim.2016.06.009 [DOI] [PubMed] [Google Scholar]
- Wu F., Zhang J., Hu N., Wang H., Xu T., Liu Y., et al. (2014). Effect of hENT1 polymorphism G-706C on clinical outcomes of gemcitabine-containing chemotherapy for Chinese non-small-cell lung cancer patients. Cancer Epidemiol. 38, 728–762. 10.1016/j.canep.2014.08.008 [DOI] [PubMed] [Google Scholar]
- Xiao J. C., Zhang T. P., Zhao Y. P. (2013). Human Equilibrative Nucleoside Transporter 1 (hENT1) predicts the asian patient response to gemcitabine-based chemotherapy in pancreatic cancer. Hepato-Gastroenterology 60, 258–262. 10.5754/hge12687 [DOI] [PubMed] [Google Scholar]
- Yamada R., Mizuno S., Uchida K., Yoneda M., Kanayama K., Inoue H., et al. (2016). Human equilibrative nucleoside transporter 1 expression in endoscopic ultrasonography-guided fine-needle aspiration biopsy samples is a strong predictor of clinical response and survival in the patients with pancreatic ductal adenocarcinoma undergoing gemcitabine-based chemoradiotherapy. Pancreas 45, 761–771. 10.1097/MPA.0000000000000597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S. Y. M., Ng A. M. L., Cass C. E., Baldwin S. A., Young J. D. (2011). Nucleobase transport by human equilibrative nucleoside transporter 1 (hENT1). J. Biol. Chem. 286, 32552–32562. 10.1074/jbc.M111.236117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You G., Morris M. E. (eds.). (2014). Drug Transporters: Molecular Characterization and Role in Drug Disposition. New York, NY: Wiley. [Google Scholar]
- Young J. D., Yao S. Y. M., Baldwin J. M., Cass C. E., Baldwin S. A. (2013). The human concentrative and equilibrative nucleoside transporter families, SLC28 and SLC29. Mol. Aspects Med. 34, 529–547. 10.1016/j.mam.2012.05.007 [DOI] [PubMed] [Google Scholar]
- Young J. D., Yao S. Y., Sun L., Cass C. E., Baldwin S. A. (2008). Human equilibrative nucleoside transporter (ENT) family of nucleoside and nucleobase transporter proteins. Xenobiotica 38, 995–1021. 10.1080/00498250801927427 [DOI] [PubMed] [Google Scholar]
- Zhang J., Visser F., King K. M., Baldwin S. A., Young J. D., Cass C. E. (2007). The role of nucleoside transporters in cancer chemotherapy with nucleoside drugs. Cancer Metastasis Rev. 26, 85–110. 10.1007/s10555-007-9044-4 [DOI] [PubMed] [Google Scholar]
- Zhang X. Z. (2010). Modulation of embryonic stem cell fate and somatic cell reprogramming by small molecules. Reprod. Biomed. Online 21, 26–36. 10.1016/j.rbmo.2010.03.021 [DOI] [PubMed] [Google Scholar]
- Zhao S., Iyengar R. (2012). Systems pharmacology: network analysis to identify multiscale mechanisms of drug action. Annu. Rev. Pharmacol. Toxicol. 52, 505–521. 10.1146/annurev-pharmtox-010611-134520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Zheng X., Fan T.-P., Li Z., Zhang Y., Zhang J. (2015). A novel drug discovery strategy inspired by traditional medicine philosophies. Science 347, S38–S40. [Google Scholar]
- Zimmermann G. R., Lehár J., Keith C. T. (2007). Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov. Today 12, 34–42. 10.1016/j.drudis.2006.11.008 [DOI] [PubMed] [Google Scholar]