ABSTRACT
Histone proteins are main structural components of the chromatin and major determinants of gene regulation. Expression of canonical histone genes is strictly controlled during the cell cycle in order to couple DNA replication with histone deposition. Indeed, reductions in the levels of canonical histones or defects in chromatin assembly cause genetic instability. Early data from yeast demonstrated that severe histone depletion also causes strong gene expression changes. We have recently reported that a moderated depletion of canonical histones in human cells leads to an open chromatin configuration, which in turn increases RNA polymerase II elongation rates and causes pre-mRNA splicing defects. Interestingly, some of the observed defects accompany the scheduled histone depletion that is associated with several senescence and aging processes. Thus, our comparison of induced and naturally-occurring histone depletion processes suggests that a programmed reduction of the level of canonical histones might be a strategy to control gene expression during specific physiological processes.
keywords: Aging, DNA damage, histones, RNA splicing, transcription
The dynamics of DNA during the cell cycle have long been known to be tightly linked to its packaging into chromatin. This association is manifested in two 2 ways: first, the processes that deal with DNA, such as transcription, RNA processing, replication, DNA repair, and chromosome segregation, have to overcome the physical restrains that histones pose for proteins to bind DNA; and second, chromatin dynamics per se provide an additional level of regulation for those processes. Chromatin organization occurs first during DNA replication and involves genetic and physical interactions between chromatin assembly factors and replisome components, to ensure a rapid and correct deposition of histones behind the replication fork.1 Accordingly, canonical histone synthesis is coupled to DNA replication and cell cycle.2 Notably, an evolutionarily conserved feature from yeast to humans is the synthesis of huge amounts of histones, which avoids a deficit in the histone supply during chromosome duplication. Thirty years ago, Michael Grunstein and colleagues provided the first explanation for this observation, showing with an elegant genetic approach in yeast that histone depletion causes transcriptional defects and cell cycle arrest.3,4 Since then, many different studies have highlighted how deleterious a deficit in histone supply can be for cell fitness. Interestingly, scheduled histone depletion mechanisms seem to have also evolved to regulate replicative senescence and aging, providing further support for the importance of a strict regulation of histone synthesis. Here we briefly discuss recent data that suggest that histone availability is a regulatory variable in the intersection between DNA replication, transcription, pre-mRNA processing, and genome integrity.
Histone depletion and genome integrity
Cell cycle arrest by histone depletion in yeast occurs in mitosis and is associated with alterations in the structure of the centromeric chromatin.5 Recently, this arrest was shown to be mediated by activation of the Aurora kinase–dependent branch of the spindle assembly checkpoint (SAC), which detects problems of tension at the kinetochore. Indeed, histone depletion also impairs chromatid decatenation. Interestingly, both phenotypes can be suppressed by a reduction in the activity of topoisomerase II, condensin, or cohesin. These results establish a not yet understood connection between histone deposition and chromosome architecture.6
In vertebrates, studying the effects of histone depletion was initially hampered by having to deal with the large number of genes that encode for each histone subunit. This has now been resolved by knocking down key regulators of histone mRNA expression, such as the histone-specific mRNA 3′-end processing factors SLBP and FLASH or the transcription factor HINFP. These studies revealed mitotic aberrations, including aneuploidy and multipolar spindles, in response to histone depletion.7 However, the major cell cycle arrest following these knockdowns occurs in S phase,8-11 a phenotype also observed in human cells defective for the chromatin assembly factor CAF1.12 This S-phase delay is a consequence of the direct effects that defective chromatin integrity—caused either by histone depletion or the lack of chromatin assembly factors—have on the stability of the advancing replication forks, with a consequent accumulation of DNA damage, hyper-recombination, and checkpoint activation.7,13-16 This genetic instability is observed from yeast to humans and underscores the evolutionarily relevance of a rapid and correct deposition of nucleosomes at the newly synthesized DNA for genome integrity. Indeed, accumulation of DNA damage in human cells depleted for the chromatin assembly factor ASF1 induces the alternative lengthening of telomeres (ALT) pathway, which accounts for telomere maintenance in approximately 10% of human cancers.17 Moreover, certain types of malignant cancers can result from uncoupling DNA synthesis and histone deposition. For example, histone reduction is induced by the human T-cell leukemia/lymphotropic virus type 1 (HTLV-1) Tax protein in adult T-cell leukemia/lymphoma (ATLL).18
Histone depletion, transcription, and pre-mRNA processing
Packaging DNA into nucleosomes hinders its accessibility for the RNA polymerase II (RNAPII) at promoters and regulatory elements,19,20 and nucleosomes act as barriers for elongating RNAPII at gene bodies.21-25 Consistently, histone depletion studies in yeast result in a genome-wide transcriptional de-repression of hundreds of genes.26 More sensitive RNA-seq analyses have recently reinforced the notion that nucleosomes are major transcription repressors. Interestingly, mapping nucleosome positioning in histone H3–depleted cells showed that not all nucleosomes present similar reductions of occupancy, with the most labile nucleosomes associated with promoters.27 Nevertheless, nucleosomes should not only be seen as passive repressors, as they also play active roles in transcription activation, as reflected by the huge number of genes that are repressed in response to histone depletion.26,27 Nucleosomes act as dynamic scaffolds that determine the timing and order of entry for structural and regulatory transcription factors.28 One way in which nucleosomes perform this function is by their position relative to the DNA sequence, which can determine the ability of activators to synergise;29 consistently, histone depletion causes the loss of nucleosome positioning and transcriptional synergism.30 It has been also suggested that nucleosome-mediated RNAPII pausing and/or recruitment of splicing factors can influence splicing, based in the fact that exons display higher nucleosome occupancy than surrounding introns.31-35
Despite the huge number of studies that have addressed the importance of chromatin integrity in different organisms, a genome-wide analysis of the effects of histone depletion on gene expression in human cells was missing until now. We have recently addressed this by generating a stable cell line that expresses an inducible shRNA against SLBP.36 Histone genes in vertebrates are the only genes whose mRNAs are not polyadenylated; instead, their mRNAs form a loop at the 3′-end that binds SLBP, to promote proper RNA termination, stability, and translation.2 While other histone-depletion approaches relied on a dramatic block in DNA synthesis and cell-cycle progression,7,10,13 we performed a moderated silencing of SLBP that led to a subtle reduction in the pool of available histones, so that the cell cycle was only mildly affected. This was particularly important for avoiding cell cycle– and DNA damage–mediated transcriptional changes to the greatest extent possible. Depletion of canonical histones results in a more open chromatin conformation, characterized by a reduced histone:DNA ratio and an increased level of the SLBP-independent variant histone H3.3. This increase is probably due to the fact that H3.3 can partially replace the canonical H3.1 and H3.2 histones. H3.3 is normally enriched in transcriptionally active genes and impairs higher-ordered chromatin folding, contributing to an open chromatin configuration.37,38 Under these chromatin conditions, we found that RNAPII transcription elongation rate increases, consistent with the idea that histone depletion lowers the nucleosomal barrier effect. We also observed that an increased RNAPII elongation rate does not increase the kinetics of co-transcriptional splicing, suggesting that splicing is a rate-limiting step for gene expression. A similar conclusion was recently reported from a study using a single-molecule nascent RNA-seq strategy.39
Consistent with the subtle alterations in chromatin structure induced in our SLBP-depleted cell line, genome-wide analysis showed a low number of misregulated transcripts; however, widespread alterations in pre-mRNA splicing were observed. The kinetic model for co-transcriptional splicing proposes that a slow elongation rate facilitates the recognition of weak splice sites, promoting the inclusion of alternative exons. In contrast, a fast elongation rate would promote skipping of variable exons.40,41 In line with this, we observed that an increased elongation rate is associated with skipping of the variable exons of the CD44 gene. Importantly, a slow elongation mutant of RNAPII is able to rescue this defect, indicating that the splicing defect induced by histone depletion in CD44 is a direct consequence of the increased elongation rate. Higher inclusion of some cassette exons were also found under conditions of histone depletion, in agreement with the recent data suggesting that the classical kinetic model can explain only part of the elongation-dependent splicing defects.41,42 We also observed that highly expressed genes display increased intron retention under conditions of defective histone supply. We hypothesize that the chromatin structure of highly expressed genes may be more severely altered by histone depletion than that of lowly expressed genes, due to a higher rate of histone exchange. As a consequence, the RNAPII elongation rate might be further accelerated, impairing recognition of splicing sites and hence promoting intron retention.
Strikingly, intron retention was often observed for genes encoding RNA processing and RNA-binding factors (36 and our unpublished observations). Indeed, the expression of a significant number of RNA processing genes was decreased in this case, probably due to mRNA degradation by the nonsense-mediated decay (NMD) pathway induced by the retained intron.36 Intriguingly, work from the group of Benjamin J. Blencowe has shown that RNA processing and RNA-binding factors are often regulated by alternative splicing through NMD.43,44 Taken together, these results suggest the existence of a fine-tuned regulatory mechanism that impairs expression of the RNA processing machinery under conditions of histone stress.
Interestingly, inclusion of non-coding second exons that contain a polyadenylation site in some histone mRNAs was observed upon SLBP depletion, suggesting that cells contain a set of histone genes that can respond to SLBP depletion to maintain a certain level of histones.11,36
Histone depletion, senescence, and aging
Despite the wide range of deleterious consequences of a defective histone supply, scheduled histone depletion can also be a regulatory mechanism to control processes such as DNA repair, transcription, senescence, and aging. For instance, eukaryotic cells have evolved multiple mechanisms to reduce the accumulation of free histones that result from fork stalling by replicative stress, which could interfere with DNA metabolism.45 Likewise, histone depletion is actively promoted in response to DNA breaks in human fibroblasts.46 The biological meaning of this reduction is unknown; however, physical interactions between the recombination proteins Rad52 and Rad51 with histones seem to modulate their accessibility to DNA double-strand breaks in yeast, suggesting that DNA repair by homologous recombination might be regulated by the histone levels.47
Histone depletion is associated with the process of cellular senescence that is triggered in response to telomere shortening. In yeast, the study of this mechanism has revealed for the first time a transcriptional regulatory circuit triggered by histone depletion.48 Telomere shortening causes the loss of the telomere binding sites and the subsequent relocation of the protein Rap1 (repressor activator protein 1) from telomeres to the promoters of hundreds of senescence-specific genes, which contributes to their activation. Notably, Rap1 also binds to and represses histone genes, and this facilitates the activation of the senescence program by reducing nucleosome occupancy at the Rap1-targeted promoters. Consistently, overexpression of core histones delays senescence. More importantly, this process depends on the checkpoint protein Mec1, demonstrating that histone depletion is a regulated mechanism to control the transcriptional program of senescence.48 A decreased level of canonical histones has also been observed during replicative senescence46 and oncogene-induced senescence in mammals.49 In late passage human fibroblasts, telomere shortening leads to reduced SLBP levels, a phenotype that might explain the reduction in the amount of canonical histones. Interestingly, genes encoding the replication-associated chromatin assembly factors CAF1 and ASF1 are also downregulated.46 Depletion of histones and chromatin assembly factors lead to a loss of nucleosome occupancy and a redistribution of epigenetic marks at the telomeric chromatin. These structural changes accompany and seem to reinforce the activation of the DNA damage response that eventually causes senescence. Importantly, expression of the wild-type telomerase in late passage fibroblasts restores not only telomere lengthening but also the levels of SLBP, CAF1, ASF1, and histones.46
Consistent with a connection between senescence and histone depletion, senescent cells display some of the transcriptional features observed after inducing histone depletion in our SLBP-depleted cells.36 Thus, a characteristic of senescent chromatin is the accumulation of the histone variant H3.3 by the deposition activity of the replication-independent chaperone HIRA.50,51 In addition, replicative and oncogene-induced senescent fibroblasts present increased inclusion of cryptic noncoding second exons with polyadenylation signals in some mature histone mRNAs.50 Although its function is not yet clear, this mechanism might contribute to maintaining a certain level of expression of canonical histones in senescent cells.
Histone reduction is also a hallmark of replicative aging in yeast. This type of aging, which reproduces some of the aging features in mammals, is independent of telomere shortening and determines the number of times a mother cell can divide before dying.52 Replicative aging in yeast is associated with a significant decrease in the amount of histones, and importantly, life span can be shortened and extended by decreasing or increasing, respectively, histone expression.53 In addition, nucleosome loss during yeast aging causes transcriptional upregulation and genetic instability,54 as observed after inducing histone depletion in growing cells.14,26 A relationship between histones levels and aging has been also observed in mammals. In quiescent stem cells, chronological aging can be studied in the absence of replicative aging due to the extremely low turnover of these cells. Interestingly, quiescent skeletal muscle stem cells isolated from old mice also show low histone levels as compared to those isolated from younger animals.55
Recent studies have shown strong alterations in alternative splicing linked to aging56,57 and age-related disorders, such as Alzheimer's.58 Notably, these aging-related splicing defects affect a group of genes that are significantly enriched in RNA processing functions,56,57 as we have also observed after inducing histone depletion.36 Moreover, large-scale, human population-based studies have reported a robust negative correlation between aging and levels of expression of mRNA processing factors.59,60 Finally, accumulation of intron retention in response to histone depletion in human cell lines36 phenocopies the most abundant age-associated splicing changes detected in human brain.57 Therefore, our data suggest that cells depleted of histones by SLBP knockdown recapitulate, at least partially, the splicing defects observed in aged cells and uncover an additional connection between histone depletion, RNA processing, and aging.
Concluding remarks and future perspectives
Altogether, these studies suggest that replicative and chronologic aging, as well as senescence processes in eukaryotes, are associated with a reduction in the supply of histones, which likely causes a deterioration of chromatin structures and signaling that in turn leads to transcriptional changes and DNA damage accumulation.61 Our data indicate that human cells depleted of canonical histones show transcription and splicing changes that are often observed in aged tissues. Therefore, our results suggest that changes in gene expression associated to aging and senescence might be caused by histone depletion. It is tempting to speculate that histone availability can be also used in other cellular physiological or pathological processes as a regulatory mechanism (Fig. 1). For example, it was reported that histone mRNA levels increase gradually through early zygotic cell divisions,62 and in differentiating embryonic stem cells63 and decrease in certain type of cancer.18 Whether control of the general level of histones is a regulatory system for regulating gene expression during development, differentiation or dedifferentiation processes is an open question that requires further investigation.
Figure 1.
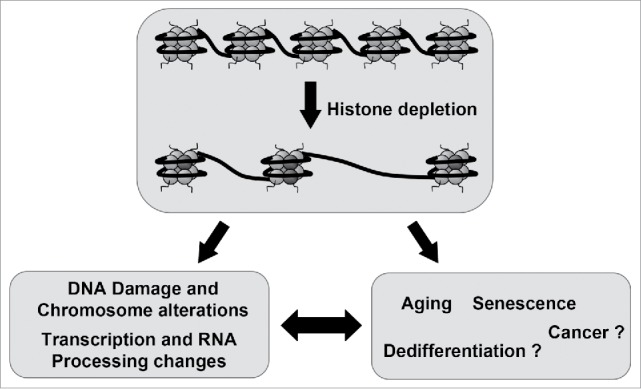
Histone availability as a strategy to control gene expression. Induced depletion of canonical histones leads to DNA damage and chromosomal alterations and also to changes in transcription and RNA processing. Many of these changes recapitulate those observed in the senescence and aging process, which are associated to histone depletion. This observation leads to the hypothesis that senescence and aging and maybe other processes such as dedifferentiation and cancer are, to some extend, a consequence of a programmed reduction in the levels of canonical histones.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was funded by the Spanish Ministry of Economy and Competitiveness (MINECO) [BFU-2011-23442 and BFU2014-53543-P to J.C.R.] and the Andalusian Government [P12CT52270 to F.P.].
References
- 1.Corpet A, Almouzni G. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol 2009; 19:29-41; PMID:19027300; http://dx.doi.org/ 10.1016/j.tcb.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 2.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 2008; 9:843-54; PMID:18927579; http://dx.doi.org/ 10.1038/nrg2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han M, Chang M, Kim UJ, Grunstein M. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell 1987; 48:589-97; PMID:3815518; http://dx.doi.org/ 10.1016/0092-8674(87)90237-6 [DOI] [PubMed] [Google Scholar]
- 4.Kim UJ, Han M, Kayne P, Grunstein M. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. Embo J 1988; 7:2211-9; PMID:3046933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saunders MJ, Yeh E, Grunstein M, Bloom K. Nucleosome depletion alters the chromatin structure of Saccharomyces cerevisiae centromeres. Mol Cell Biol 1990; 10:5721-7; PMID:2233714; http://dx.doi.org/ 10.1128/MCB.10.11.5721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murillo-Pineda M, Cabello-Lobato MJ, Clemente-Ruiz M, Monje-Casas F, Prado F. Defective histone supply causes condensin-dependent chromatin alterations, SAC activation and chromosome decatenation impairment. Nucleic Acids Res 2014; 42:12469-82; PMID:25300489; http://dx.doi.org/ 10.1093/nar/gku927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghule PN, Xie RL, Medina R, Colby JL, Jones SN, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Fidelity of histone gene regulation is obligatory for genome replication and stability. Mol Cell Biol 2014; 34:2650-9; PMID:24797072; http://dx.doi.org/ 10.1128/MCB.01567-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao X, McKillop-Smith S, Muller B. The human histone gene expression regulator HBP/SLBP is required for histone and DNA synthesis, cell cycle progression and cell proliferation in mitotic cells. J Cell Sci 2004; 117:6043-51; PMID:15546920; http://dx.doi.org/ 10.1242/jcs.01523 [DOI] [PubMed] [Google Scholar]
- 9.Wagner EJ, Berkow A, Marzluff WF. Expression of an RNAi-resistant SLBP restores proper S-phase progression. Biochem Soc Trans 2005; 33:471-3; PMID:15916543; http://dx.doi.org/ 10.1042/BST0330471 [DOI] [PubMed] [Google Scholar]
- 10.Barcaroli D, Bongiorno-Borbone L, Terrinoni A, Hofmann TG, Rossi M, Knight RA, Matera AG, Melino G, De Laurenzi V. FLASH is required for histone transcription and S-phase progression. Proc Natl Acad Sci U S A 2006; 103:14808-12; PMID:17003125; http://dx.doi.org/ 10.1073/pnas.0604227103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan KD, Mullen TE, Marzluff WF, Wagner EJ. Knockdown of SLBP results in nuclear retention of histone mRNA RNA New York, NY 2009; 15:459-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol Cell 2003; 11:341-51; PMID:12620223; http://dx.doi.org/ 10.1016/S1097-2765(03)00037-6 [DOI] [PubMed] [Google Scholar]
- 13.Mejlvang J, Feng Y, Alabert C, Neelsen KJ, Jasencakova Z, Zhao X, Lees M, Sandelin A, Pasero P, Lopes M, Groth A. New histone supply regulates replication fork speed and PCNA unloading. J Cell Biol 2014; 204:29-43; PMID:24379417; http://dx.doi.org/ 10.1083/jcb.201305017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clemente-Ruiz M, Prado F. Chromatin assembly controls replication fork stability. EMBO Rep 2009; 10:790-6; PMID:19465889; http://dx.doi.org/ 10.1038/embor.2009.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clemente-Ruiz M, Gonzalez-Prieto R, Prado F. Histone H3K56 acetylation, CAF1, and Rtt106 coordinate nucleosome assembly and stability of advancing replication forks. PLoS Genet 2011; 7:e1002376; PMID:22102830; http://dx.doi.org/15684401 10.1371/journal.pgen.1002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prado F, Aguilera A. Partial depletion of histone H4 increases homologous recombination-mediated genetic instability. Mol Cell Biol 2005; 25:1526-36; PMID:15684401; http://dx.doi.org/ 10.1128/MCB.25.4.1526-1536.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Sullivan RJ, Arnoult N, Lackner DH, Oganesian L, Haggblom C, Corpet A, Almouzni G, Karlseder J. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat Struct Mol Biol 2014; 21:167-74; PMID:24413054; http://dx.doi.org/ 10.1038/nsmb.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogenberger JM, Laybourn PJ. Human T Lymphotropic Virus Type 1 protein Tax reduces histone levels. Retrovirology 2008; 5:9; PMID:18237376; http://dx.doi.org/ 10.1186/1742-4690-5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwak H, Lis JT. Control of Transcriptional Elongation. Annu Rev Genet 2013; 47:501-26; PMID:24050178; http://dx.doi.org/25650798 10.1146/annurev-genet-110711-155440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol 2015; 16:178-89; PMID:25650798; http://dx.doi.org/ 10.1038/nrm3941 [DOI] [PubMed] [Google Scholar]
- 21.Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell 2005; 18:97-108; PMID:15808512; http://dx.doi.org/ 10.1016/j.molcel.2005.02.027 [DOI] [PubMed] [Google Scholar]
- 22.Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev 1991; 5:683-96; PMID:2010092; http://dx.doi.org/ 10.1101/gad.5.4.683 [DOI] [PubMed] [Google Scholar]
- 23.Subtil-Rodriguez A, Reyes JC. BRG1 helps RNA polymerase II to overcome a nucleosomal barrier during elongation, in vivo. EMBO Rep 2010; 11:751-7; PMID:20829883; http://dx.doi.org/ 10.1038/embor.2010.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 2009; 325:626-8; PMID:19644123; http://dx.doi.org/ 10.1126/science.1172926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jimeno-Gonzalez S, Ceballos-Chavez M, Reyes JC. A positioned +1 nucleosome enhances promoter-proximal pausing. Nucleic Acids Res 2015; 43:3068-78; PMID:25735750; http://dx.doi.org/ 10.1093/nar/gkv149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 1999; 402:418-21; PMID:10586882; http://dx.doi.org/ 10.1038/46567 [DOI] [PubMed] [Google Scholar]
- 27.Gossett AJ, Lieb JD. In vivo effects of histone H3 depletion on nucleosome occupancy and position in Saccharomyces cerevisiae. PLoS Genet 2012; 8:e1002771; PMID:22737086; http://dx.doi.org/ 10.1371/journal.pgen.1002771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell 2007; 128:707-19; PMID:17320508; http://dx.doi.org/ 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- 29.Prado F, Koop R, Beato M. Accurate chromatin organization of the mouse mammary tumor virus promoter determines the nature of the synergism between transcription factors. J Biol Chem 2002; 277:4911-7; PMID:11733499; http://dx.doi.org/ 10.1074/jbc.M110094200 [DOI] [PubMed] [Google Scholar]
- 30.Chavez S, Beato M. Nucleosome-mediated synergism between transcription factors on the mouse mammary tumor virus promoter. Proc Natl Acad Sci U S A 1997; 94:2885-90; PMID:9096316; http://dx.doi.org/ 10.1073/pnas.94.7.2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell 2009; 36:245-54; PMID:19854133; http://dx.doi.org/ 10.1016/j.molcel.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res 2009; 19:1732-41; PMID:19687145; http://dx.doi.org/ 10.1101/gr.092353.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol 2009; 16:996-1001; PMID:19684599; http://dx.doi.org/ 10.1038/nsmb.1658 [DOI] [PubMed] [Google Scholar]
- 34.Nahkuri S, Taft RJ, Mattick JS. Nucleosomes are preferentially positioned at exons in somatic and sperm cells Cell Cycle Georgetown, Tex 2009; 8:3420-4 [DOI] [PubMed] [Google Scholar]
- 35.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol 2009; 16:990-5; PMID:19684600; http://dx.doi.org/ 10.1038/nsmb.1659 [DOI] [PubMed] [Google Scholar]
- 36.Jimeno-Gonzalez S, Payan-Bravo L, Munoz-Cabello AM, Guijo M, Gutierrez G, Prado F, Reyes JC. Defective histone supply causes changes in RNA polymerase II elongation rate and cotranscriptional pre-mRNA splicing. Proc Natl Acad Sci U S A 2015; 112:14840-5; PMID:26578803; http://dx.doi.org/ 10.1073/pnas.1506760112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci U S A 2004; 101:1525-30; PMID:14732680; http://dx.doi.org/ 10.1073/pnas.0308092100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen P, Zhao J, Wang Y, Wang M, Long H, Liang D, Huang L, Wen Z, Li W, Li X et al., H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev 2013; 27:2109-24; PMID:24065740; http://dx.doi.org/ 10.1101/gad.222174.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrillo Oesterreich F, Herzel L, Straube K, Hujer K, Howard J, Neugebauer KM. Splicing of Nascent RNA Coincides with Intron Exit from RNA Polymerase II. Cell 2016; 165:372-381; PMID:27020755; http://dx.doi.org/14536091 10.1016/j.cell.2016.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 2003; 12:525-32; PMID:14536091; http://dx.doi.org/ 10.1016/j.molcel.2003.08.001 [DOI] [PubMed] [Google Scholar]
- 41.Dujardin G, Lafaille C, de la Mata M, Marasco LE, Munoz MJ, Le Jossic-Corcos C, Corcos L, Kornblihtt AR. How slow RNA polymerase II elongation favors alternative exon skipping. Mol Cell 2014; 54:683-90; PMID:24793692; http://dx.doi.org/ 10.1016/j.molcel.2014.03.044 [DOI] [PubMed] [Google Scholar]
- 42.Fong N, Kim H, Zhou Y, Ji X, Qiu J, Saldi T, Diener K, Jones K, Fu XD, Bentley DL. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev 2014; 28:2663-76; PMID:25452276; http://dx.doi.org/ 10.1101/gad.252106.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol 2008; 28:4320-30; PMID:18443041; http://dx.doi.org/ 10.1128/MCB.00361-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saltzman AL, Pan Q, Blencowe BJ. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev 2011; 25:373-84; PMID:21325135; http://dx.doi.org/ 10.1101/gad.2004811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gunjan A, Paik J, Verreault A. The emergence of regulated histone proteolysis. Curr Opin Genet Dev 2006; 16:112-8; PMID:16510276; http://dx.doi.org/ 10.1016/j.gde.2006.02.010 [DOI] [PubMed] [Google Scholar]
- 46.O'Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol 2010; 17:1218-25; PMID:20890289; http://dx.doi.org/ 10.1038/nsmb.1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang D, Burkhart SL, Singh RK, Kabbaj MH, Gunjan A. Histone dosage regulates DNA damage sensitivity in a checkpoint-independent manner by the homologous recombination pathway. Nucleic Acids Res 2012; 40:9604-20; PMID:22850743; http://dx.doi.org/ 10.1093/nar/gks722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Platt JM, Ryvkin P, Wanat JJ, Donahue G, Ricketts MD, Barrett SP, Waters HJ, Song S, Chavez A, Abdallah KO, Master SR, Wang LS, Johnson FB. Rap1 relocalization contributes to the chromatin-mediated gene expression profile and pace of cell senescence. Genes Dev 2013; 27:1406-20; PMID:23756653; http://dx.doi.org/ 10.1101/gad.218776.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, Shah PP, Hewitt G, Korolchuk VI, Passos JF, Wu H, Berger SL, Adams PD. Lysosome-mediated processing of chromatin in senescence. J Cell Biol 2013; 202:129-43; PMID:23816621; http://dx.doi.org/ 10.1083/jcb.201212110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rai TS, Cole JJ, Nelson DM, Dikovskaya D, Faller WJ, Vizioli MG, Hewitt RN, Anannya O, McBryan T, Manoharan I, et al.. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev 2014; 28:2712-25; PMID:25512559; http://dx.doi.org/ 10.1101/gad.247528.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogakou EP, Sekeri-Pataryas KE. Histone variants of H2A and H3 families are regulated during in vitro aging in the same manner as during differentiation. Exp Gerontol 1999; 34:741-54; PMID:10579635; http://dx.doi.org/ 10.1016/S0531-5565(99)00046-7 [DOI] [PubMed] [Google Scholar]
- 52.Tissenbaum HA, Guarente L. Model organisms as a guide to mammalian aging. Dev Cell 2002; 2:9-19; PMID:11782310; http://dx.doi.org/ 10.1016/S1534-5807(01)00098-3 [DOI] [PubMed] [Google Scholar]
- 53.Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. Elevated histone expression promotes life span extension. Mol Cell 2010; 39:724-35; PMID:20832724; http://dx.doi.org/ 10.1016/j.molcel.2010.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu Z, Chen K, Xia Z, Chavez M, Pal S, Seol JH, Chen CC, Li W, Tyler JK. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev 2014; 28:396-408; PMID:24532716; http://dx.doi.org/ 10.1101/gad.233221.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu L, Cheung TH, Charville GW, Hurgo BM, Leavitt T, Shih J, Brunet A, Rando TA. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell reports 2013; 4:189-204; PMID:23810552; http://dx.doi.org/ 10.1016/j.celrep.2013.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez SA, Grochova D, McKenna T, Borate B, Trivedi NS, Erdos MR, Eriksson M. Global genome splicing analysis reveals an increased number of alternatively spliced genes with aging. Aging cell 2016; 15:267-78; PMID:26685868; http://dx.doi.org/ 10.1111/acel.12433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mazin P, Xiong J, Liu X, Yan Z, Zhang X, Li M, He L, Somel M, Yuan Y, Phoebe Chen YP, et al.. Widespread splicing changes in human brain development and aging. Mol System Biol 2013; 9:633; PMID:23340839; http://dx.doi.org/21846794 10.1038/msb.2012.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tollervey JR, Wang Z, Hortobagyi T, Witten JT, Zarnack K, Kayikci M, Clark TA, Schweitzer AC, Rot G, Curk T, et al.. Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res 2011; 21:1572-82; PMID:21846794; http://dx.doi.org/ 10.1101/gr.122226.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harries LW, Hernandez D, Henley W, Wood AR, Holly AC, Bradley-Smith RM, Yaghootkar H, Dutta A, Murray A, Frayling TM, et al.. Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging Cell 2011; 10:868-78; PMID:21668623; http://dx.doi.org/ 10.1111/j.1474-9726.2011.00726.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holly AC, Melzer D, Pilling LC, Fellows AC, Tanaka T, Ferrucci L, Harries LW. Changes in splicing factor expression are associated with advancing age in man. Mechan Age Dev 2013; 134:356-66; PMID:23747814; http://dx.doi.org/22959736 10.1016/j.mad.2013.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Sullivan RJ, Karlseder J. The great unravelling: chromatin as a modulator of the aging process. Trends Biochem Sci 2012; 37:466-76; PMID:22959736; http://dx.doi.org/ 10.1016/j.tibs.2012.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Graves RA, Marzluff WF, Giebelhaus DH, Schultz GA. Quantitative and qualitative changes in histone gene expression during early mouse embryo development. Proc Natl Acad Sci U S A 1985; 82:5685-9; PMID:3862090; http://dx.doi.org/ 10.1073/pnas.82.17.5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karnavas T, Pintonello L, Agresti A, Bianchi ME. Histone content increases in differentiating embryonic stem cells. Front Physiol 2014; 5:330; PMID:25221520; http://dx.doi.org/ 10.3389/fphys.2014.00330 [DOI] [PMC free article] [PubMed] [Google Scholar]


