Abstract
Prostate cancer (PC) is the most common and the second leading cause of cancer-related death among American men. Early diagnosis is a prerequisite to improving therapeutic benefits. However, the current clinical biomarkers for PC do not reliably decipher indolent PC from other urogenital disorders. Thus, effective clinical intervention necessitates development of new biomarkers for early detection of PC. The present study aimed to identify the miRNA signature in organ-confined (Gleason Score 6) prostate tumors. MicroRNA (miRNA/miR) array analysis identified 118 upregulated and 73 downregulated miRNAs in microdissected tumors in comparison to matched neighboring normal prostate epithelium. The miRs-Plus-A1083, -92b-5p, -18a-3p, -19a-3p, -639, -3622b-3p, -3189-3p, -155-3p, -410, -1179, 548b-5p, and -4469 are predominantly expressed (7–11-fold), whereas miRs-595, 4490, -3120-5p, -1299, -21-5p, -3677-3, -let-7b-5p, -5189, 3-121-5p, -4518, -200a-5p, -3682-5p, -3689d, -3149 represent the most downregulated (12–113-fold) miRNAs in microdissected prostate tumors. The array expression profile of selected miRNA signature and their potential mRNA targets was validated by qRT-PCR analysis in PC cell lines. Integrated in silico and computational prediction analyses demonstrated that the dysregulated miRNA signature map to key regulatory factors involved in tumorigenesis, including cell cycle, apoptosis, and p53 pathways. The newly identified miRNA signature has potential clinical utility as biomarkers, prognostic indicators, and therapeutic targets for early detection of PC. Further studies are needed to assess the functional significance and clinical usefulness of the identified miRNAs.
Impact Statement
To our knowledge his is the first study of identifying miRNA signatures in microdissected indolent (Gleason score 6) prostate cancer in comparison to matched normal prostate epithelium. By employing in silico and computational prediction analysis, the study provides a landscape of potential miRNA targets and key cellular pathways involved in prostate tumorigenesis. Identification if miRNAs and their relevant targets and pathways pave the way for underpinning their mechanistic role of miRNAs in human prostate tumorigenesis, and possibly other human cancers. Importantly, the outcome of the study has important clinical implications for the management of prostate cancer, including the use of miRNA(s) as biomarkers for early detection of prostate cancer.
Keywords: Prostate Cancer, microdissection, microRNA signature, predicted pathways
Introduction
Despite decreasing death rates since the early 1990s, prostate cancer (PC) remains the most predominant form and the second leading cause of cancer-related death among American men.1 In the USA, an estimated 220,800 men will be diagnosed with PC, with an estimated 27,540 deaths in 2015.1 For unknown reasons, both incidence and mortality rates remain higher among African Americans than non-Hispanic Caucasian Americans. Although most patients are highly responsive to androgen-deprivation therapy, the disease becomes unresponsive and progresses to castration-resistant phenotype.2 The morbidity and mortality of PC is attributed to its propensity to metastasize to predilection sites, such as bone and other organs.3,4
Treatment options for clinically localized PC mostly curb the ability of the disease to progress to advanced stages. In contrast, although several treatment options are available for patients diagnosed with high-risk PC including radical prostatectomy, androgen deprivation therapy, and/or radiotherapy, the relapse inevitably occurs regardless of treatment type. This necessitates development of biomarkers for early detection of PC. Prostate-specific antigen is considered the gold standard biomarker for PC screening. However, this screening tool has limitations due to its poor diagnostic ability, especially in patients with indolent PC.5 Despite some success, other biomarkers are still in the validation process, including serum early PC antigen and prostatic acid phosphatase, α-methylacyl coenzyme A racemase methylated glutathione-S-transferase II in PC tissue, and urine differential display code 3 (DD3PCA3/UPM-3).6 Moreover, it remains to be seen whether multiple biomarkers, rather than a single biomarker, are more specific to detect early onset and discern PC from other genitourinary diseases. Thus, there is a need to establish a novel and reliable prognostic and/or diagnostic tool that could also serve as a new therapeutic target for disease management in a clinical setting.
MicroRNAs (miRNAs/miRs) are a class of endogenous, non-coding, short (20–22nt) RNA molecules that regulate their target gene either by mRNA degradation or translational suppression.7 They are transcribed in the nucleus by RNA polymerase into primary transcripts, which in turn are further processed by Drosha–Pasha enzyme to mature miRNAs. Exportin 5 incorporates to mature miRNA and facilitates its transportation to the cytosol where it binds to RNA-induced silencing complex (RISC). The miRNAs–RISC complex mediates downregulation of target gene by mRNA degradation or translational repression.7 Emerging evidence suggests that miRNAs play a crucial role in the pathogenesis of human cancers. Aberrant expression of miRNAs may be associated with many human cancers.8,9 Notably, among the most dysregulated are the onco-miRs and the tumor-suppressive miRs.10 For instance, it was shown that the tumor-suppressor let-7 miRNA family targets RAS11 and MYC,12 two key regulatory factors involved in tumorigenesis. In contrast, miR-125 is upregulated and acts as oncogene in many human cancers, including PC, by targeting proapoptotic genes.13 Furthermore, in glioblastoma cells miR-21 exerts antiapoptotic effect by targeting phosphatase and tensin homolog (PTEN).14
The high prevalence of PC heterogeneity represents a clinical challenge in the management of newly diagnosed patients. Therefore, functional genomics studies may help unravel the molecular mechanisms potentially involved in the stepwise transition into prostate adenocarcinoma. Identification of PC-related genomic signature may not only serve as early diagnostic biomarkers, but may also furnish a platform of novel therapeutic targets for more informed decision making in clinical settings. In the present study, an attempt was made to identify PC-related miRNA signatures in microdissected specimens and further define their functional targets and potential clinical utility in PC.
Material and methods
Clinical PC specimens and microdissection
Human PC specimens of organ-confined disease (Gleason score 6) were obtained from the Biospecimen Core of the Louisiana Cancer Research Consortium (http://www.louisianacancercenter.org/research/shared-resources/biospecimen-core) in New Orleans, Louisiana. The specimens were procured from PC patients undergoing radical prostatectomies at Tulane University Hospital and were de-identified based on a written informed consent and an approved protocol from the Tulane University Institutional Review Board. Using hematoxylin and eosin-stained sections, the disease stage, Gleason score, and cancerous and normal prostate glands were determined by a pathologist (KM). For laser capture microdissection (LCM), replica sections mounted onto uncoated glass slides were cleared in xylene two times followed by hydration in descending series of alcohol (absolute, 95 and 75%) and then washed in nuclease-free water for 30 s. Samples were then stained using Arcturus® Paradise® PLUS Reagent System (Applied Biosystems, Grand Island, NY) for 45 s and then dehydrated in ascending series of alcohol (75, 95%, and absolute). The slides were held in xylene until ready for microdissection. Target cells were microdissected using Arcturus Pixcell II system (Arcturus Engineering, CA) as we described.15 Briefly, the stained prostate tumors and matched normal prostate epithelium were harvested using 2000–3000 pulses, spot diameter of 15 µm, and 25–35 mW laser power. The captured cells were collected in CapSure® HS LCM caps (Applied Biosystems) and incubated with proteinase K extraction solution and Arcturus® Paradise® extraction and isolation kit (Applied Biosystems) for 16 h at 37℃ in an air incubator (AFAB Lab Resources, Buckeystown, MD). After incubation, samples were centrifuged and the supernatants were frozen at −80℃ until used.
RNA labeling and array analysis
Total RNA was isolated from supernatants of pooled captured cells and stored according to the manufacturer’s instructions provided by Arcturus® Paradise® extraction and isolation kit (Applied Biosystems). Total RNA was quantified using Nanodrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, DE). About 140 ng of total RNA from prostate tumors and matched normal glands was labeled by CY3 and CY5, using Label IT® miRNA labeling kit in accordance with the manufacturer’s protocol (Mirus Bio LLC, Madison, WI), in the presence of the miRCURY LNA array spike-in miRNAs (Exiqon Inc., MA). Relative miRNA expression profile analysis was performed using miRCURY LNA™ microRNA array kit (Cat#208502) (Exiqon Inc., MA), encompassing 3100 capture probes, complementary to most of human, mouse, rat, and their related viral sequences from the v.19.0 release of miRBase. The Cy3- and Cy5-labeled probes were mixed (1:1 ratio) and loaded onto the array slides and allowed to hybridize at 56℃ for 16 h using GeneTAC™ hybridization station (Genomic Solution, Ann Arbor, MI). The slides were washed and scanned using the dual channel scanner GeneTAC™ UC-4 (Genomic Solution, Ann Arbor, MI) and GeneTAC™ GT scanning software. The background-subtracted signal intensities were normalized and Cy5/Cy3 signal ratios were used to compare the relative expression of miRNAs as we described.15 The intensity of hybridization signals was evaluated by GenTAC integrator software (GeneTAC) and normalized to the average signals of the housekeeping genes. Raw microarray measurements were then log transformed to achieve normality and make the datasets from different hybridizations comparable. Genes with significant differential expression in prostate tumor cells were reported as fold change relative to matched control values. Heat maps were constructed using the R-software (The R Foundation for Statistical Computing) with a cut-off value between 2- and 2.5-fold.
Target and pathway prediction of miRNAs
DIANA-micro T-CDS (DIANA-microT web server v5.0) was used to determine target prediction of both downregulated and upregulated mRNAs in prostate tumor cells as described.16 Other databases like microRNA.org-Targets and Expression17 and TargetScan databases18 were also used to confirm our findings. In addition, DIANA-miRPath (DIANA miRPath v.2.0)19 was employed to predict the pathways potentially modulated by the differentially expressed miRNAs in prostate tumor cells.
Validation of expression of dysregulated miRNAs
The differentially expressed miRNAs detected by array analysis were validated by qRT-PCR in PC cells. The normal prostate epithelial cell line RWPE-1 was obtained from American Type Culture Collection (Manassas, VA). The bone metastatic PC cell line (C4-2B) was a generous gift from Dr L.W. Chung, Emory University, Atlanta, GA. RWPE-1 cells were cultured in keratinocyte serum-free medium supplemented with 5 ng/mL human recombinant epidermal growth factor and 0.05 mg/mL bovine pituitary extract (Invitrogen Life Technologies, MD). C4-2B cells were cultured in RPMI-1640 medium (ATCC) supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin/streptomycin (Invitrogen Life Technologies, MD). Cells were maintained at 37℃ in an air incubator with 5% CO2.
Total RNA was isolated from normal prostate epithelial cells (RWPE-1) and prostate malignant C4-2B cells using miRCURY™ RNA Isolation Kits (Exiqon Inc., MA) according to manufacturer’s instructions. Total RNA was quantified using Nanodrop ND-1000 Spectrophotometer (ThermoFisher Scientific, DE). Validation of miRNA array expression profiles was performed by selection of 11 microRNAs with potential targets in cancer pathways. The validation was performed using qRT-PCR. Reverse transcription was performed using miScript II RT Kit (Qiagen GMbH, Hilden, Germany). First-strand cDNA synthesis was performed at 37℃ for 60 min followed by inactivation of the enzyme by incubation at 95℃ for 5 min. The PCR reaction was carried out using miScript SYBR Green PCR kit (Bio-Rad, Hercules, CA) in presence of primer sets specific for miRNA targets and the housekeeping miRNA RNU6, designed by Quanta BioSciences (Beverly, MA) and purchased from Integrated DNA Technologies (IDT) Inc. (Coralville, Iowa). PCR primer sequences are listed in Supplementary Table 1. The amplicon levels were detected using the CFX96 Real-time System (Bio-Rad). The results were normalized using RNU6 as a reference housekeeping gene. All experiments were performed in triplicates by three independent experiments. Data were represented as the mean ± standard deviation.
Validation of dysregulated miRNA targets
Total RNA was isolated from RWPE-1 and C4-2B using RNeasy plus mini kit (Qiagen). Total RNA (1 µg) was employed for first-strand cDNA synthesis using iScript™ cDNA Synthesis Kit according to the manufacturer’s protocol (Bio-Rad). Conventional PCR was performed using IQ SYBR Green Supermix according to manufacturer’s protocol (Bio-Rad). Transcript-specific primers for protein kinase B (also known as Akt), H-ras, K-ras, c-fos, c-jun, B-cell lymphoma 2 (Bcl2), ataxia telangiectasia mutated (ATM), and the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase were purchased from IDT. The primer sequences are listed in Supplementary Table 2.
Statistical analysis
The gene expression levels are measured by the fluorescence intensities of the two dyes (green Cy3 and red Cy5 dye). The intensities were corrected by subtracting the background intensities. These adjusted intensities were then used for comparing the distributions of log intensities or log ratios of genes. T-test statistic criterion with multiple-testing adjustments was performed for comparison of differential transcriptions and selecting genes. All analyses, summaries, and listings were performed using R-statistical computing software (http://www.R-project.org).
Results
Prostate tumor miRNA signature
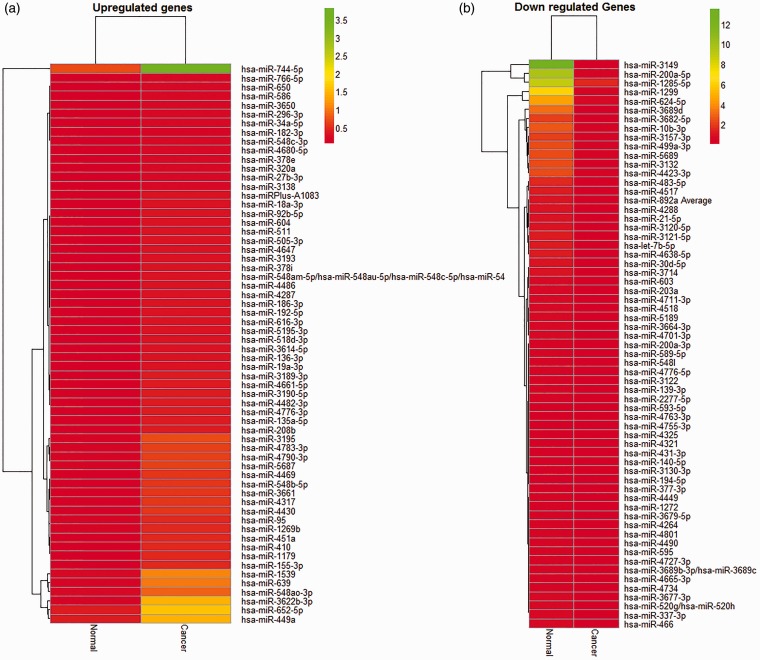
Tumor cells and matched normal epithelial cells were harvested by LCM from PC patients’ section slides according to a standard protocol (Supplementary Figure 1). Microarray profiling of miRNA expression was then performed to assess miRNA signature in microdissected prostate tumor cells compared to adjacent normal tissues. We characterized a unique set of differentially expressed miRNAs in PC tissue (Figure 1). The results depicted in heat maps and tables demonstrate selective upregulation of 118 miRNAs with a cut-off value of twofold (Figure 1(a); Supplementary Table 3) and 73 downregulated miRNAs with a cut-off value of 2.5-fold (Figure 1(b); Supplementary Table 4). In comparison to matched neighboring normal prostate epithelium, the most highly upregulated (7–11-fold) miRNAs are miRs-Plus-A1083, -92b-5p, -18a-3p, -19a-3p, -639, -3622b-3p, -3189-3p, -155-3p, -410, -1179, 548b-5p, and -4469 (Supplementary Table 3). The miRs-595, 4490, -3120-5p, -1299, -21-5p, -3677-3, -let-7b-5p, -5189, 3-121-5p, -4518, -200a-5p, -3682-5p, -3689d, -3149 represent the most downregulated (12–113-fold) miRNAs in microdisscted prostate tumors in comparison to matched neighboring normal prostate epithelium (Supplementary Table 4).
Figure 1.
Heat Map of dysregulated microRNAs in microdissected prostate tumor cells. Heat map of upregulated (a) and downregulated (b) miRNA expression is presented in hierarchical clustering of miRNA expression in microdissected prostate tumor cells normalized to adjacent non-tumorous tissues. The palette of red and green colors depicts fold change in up- and downregulated miRNAs in both groups. (A color version of this figure is available in the online journal.)
Validation of expression of dysregulated miRNAs in PC cell lines
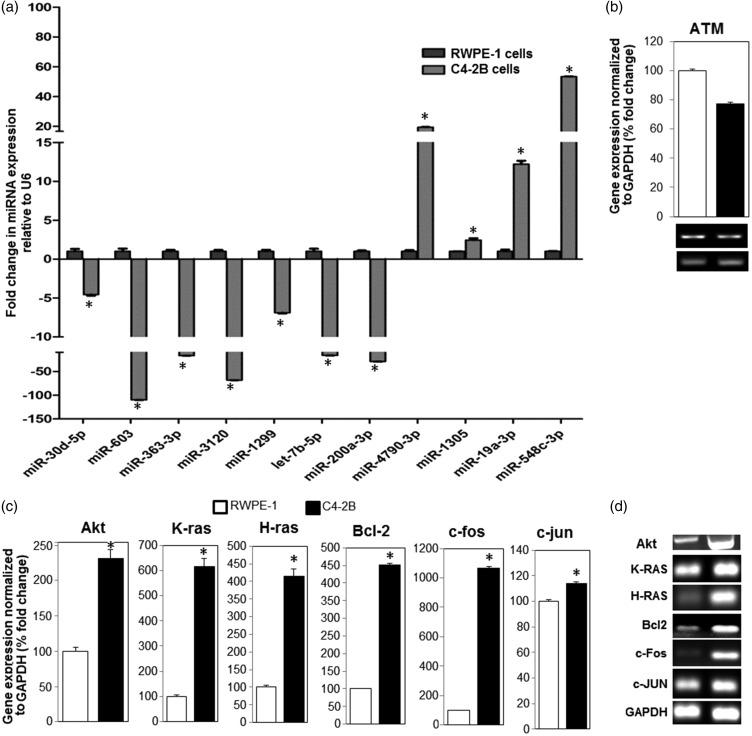
The differentially expressed miRNAs initially identified in prostate tumors by miRNA array analysis were validated by qRT-PCR. This was achieved by comparing miRNA target expression levels in the normal prostate epithelial (RWPE-1) and PC (C4-2B) cells. A total of 11 miRNAs with potential targets in PC progression and cancer pathways were selected. The endogenous levels of miR-30d-5p, 603, 363-3p, 3120, 1299, let-7b-5p, and 200a-3p were found to be downregulated, whereas those of miR-4790-3p, 1305, 19a-3p, and 548 were found to be upregulated in C4-2B cells compared to RWPE-1 cells (Figure 2(a)). The fold changes in the expression levels of the selected miRNAs in C4-2B cells compared to RWPE1 cells are shown in Table 1. Taken together, the qRT-PCR analysis further corroborated our microarray findings.
Figure 2.
Validation of differentially expressed miRNAs and their potential targets in RWPE-1 and C4-2B cell lines. (a) Quantitative RT-PCR (qRT-PCR) validation analysis depicting overexpression and downregulation expression levels of selected cancer-related miRNAs in C4-2B cells in comparison to RWPE-1 cells. (b) Expression of ataxia telangiectasia mutated (ATM) mRNA transcripts, a presumable target for upregulated miR-548c-3p in C4-2B cells compared to RWPE1 cells. The corresponding qRT-PCR products of ATM in both cells are shown in ethidium bromide-stained gels (lower panel). (c) Comparative qRT-PCR analysis of constitutive mRNA expression levels of Akt, K-ras, H-ras, Bcl-2, c-fos, and c-jun in C4-2B and RWPE1 cells. These are putative targets of the downregulated miR-3120, miR-let7b-5p, miR-603, and miR-1299 in C4-2B compared to RWPE1 cells. (d) The corresponding qRT-PCR products of the potential miRNA targets depicted in Figure 3(c) are shown as ethidium bromide-stained gels. The mRNA levels are normalized to GAPDH. * denotes significance at P < 0.05. GAPDH: glyceraldehyde 3-phosphate dehydrogenase; RWPE1: a control prostate epithelial cell line.
Table 1.
Relative fold change of expression and potential targets of a selected subset of miRNAs detected in microdissected prostate tumors by miRNA array and validated by qRT-PCR analysis in PC cell lines
| Expression | miRNA | Array fold change | PCR fold change | Predicted targets |
|---|---|---|---|---|
| Downregulated miRNAs | miR-30d-5p | −10.7 | −4.54 | CCNE1, HRAS, EGF, and SOS1 |
| miR-603 | −4.9 | −109.89 | EGFR, RB2, CDK4, CDK2, JUN, GRB2 and STAT5A | |
| miR-363-3p | −4.5 | −16.50 | PIK3R5 | |
| miR-3120 | −12.4 | −68.11 | MDM4, AKT3 and EGFR | |
| miR-1299 | −13.0 | −6.89 | CDK4, CDK2, JUN, PRKCA, and PIK3R5 | |
| Let-7b-5p | −14.6 | −16.22 | RAS, CCND1, MDM4, MYB, BCL2L1, and NGF | |
| miR-200a-3p | −20.0 | −29.04 | CCND1, CDK4, CDK2, CCNE1, PRKCA, and PRKACA | |
| Upregulated miRNAs | miR-4790-3p | 4.4 | 19.45 | RB1 |
| miR-1305 | 2.0 | 2.44 | APAF1, CYCS, BBC3, P53/TP53, SESN3, and PTEN | |
| miR-19a-3p | 3.1 | 12.21 | CSF2RB, IL1R1, PTEN, and SESN3 | |
| miR548c-3p | 4.4 | 53.40 | RB1, FOXO1, PTEN, SESNS, SERPINE1, CASP9, APAF1, CYCS, BBC3, and ATM |
ATM: ataxia telangiectasia mutated; EGF: epidermal growth factor; AKT3: RAC-gamma serine/threonine-protein kinase; APAF1: apoptotic protease-activating factor 1; BBC3: Bcl-2-binding component 3; BCL2L1: Bcl-2-like protein 1; CASP9: caspase 9; CCND1: G1/S-specific cyclin-D1; CCNE: G1/S-specific cyclin-E1; CDK2: cyclin-dependent kinase 2; CDK4: cyclin-dependent kinase 4; CSF2RB: Colony Stimulating Factor 2 Receptor Beta Common Subunit; CYCS: cytochrome c; EGFR: EGF receptor; FOXO1: forkhead box protein O1; GRB2: growth factor receptor-bound protein 2; IL1R1: interleukin-1 receptor type 1; JUN: proto-oncogene c-Jun; MDM4: double minute 4 protein; MYB: proto-oncogene c-Myb; NGF: nerve growth factor; P53/TP53: tumor suppressor p53; PIK3R5: Phosphoinositide 3-kinase regulatory subunit 5; PRKACA: cAMP-dependent protein kinase catalytic subunit alpha; PRKCA: protein kinase C alpha type; PTEN: phosphatase and tensin homolog; RAS: a https://en.wikipedia.org/wiki/Protein_superfamily protein superfamily of https://en.wikipedia.org/wiki/Small_GTPase small GTPases; RB1: Retinoblastoma-associated protein; RB2: Retinoblastoma-like protein 2; SERPINE1: plasminogen activator inhibitor 1; SESN3: sestrin-3; SOS1: Son Of Sevenless Homolog 1; STAT5A: Signal transducer and activator of transcription 5.
Modulation of predicted targets by dysregulated miRNAs in PC cells
A list of qRT-PCR-validated miRNAs and their predicted cellular targets are listed in Table 1. Initially, we examined one of the known cellular targets for miR-548c-3p, which is overexpressed in prostate tumors (array analysis) and validated in PC cells compared to normal adjacent gland and normal RWPE-1 cells (Figure 2(a)). One known target of miR-548c-3p is ATM mRNA, a serine/threonine protein kinase required for p53 phosphorylation and blockade of Cdc25 family of dual-specificity phosphatases. PCR analysis demonstrated low levels of ATM transcripts in C4-2B cells compared to RWPE-1 cells (Figure 2(b)), due to high endogenous level of miR-548c-3p.
Next, we examined by PCR the correlation of the endogenous gene expression levels of predicted targets with the expression levels of the downregulated miRNAs in PC cells. According to our target prediction analysis, one of the targets of miR-3120 is Akt. The transcript levels of Akt were found to be upregulated in C4-2B cells compared to RWPE-1 cells. Moreover, RAS oncogene and antiapoptotic Bcl-2 were predicted as two potential targets of the tumor-suppressor miRNA let-7b-5p. The downregulation of miRNA let-7b-5p transcripts was associated with an increase in the transcriptional upregulation of K-ras, H-ras, and Bcl-2 in C4-2B cells, as opposed to RWPE-1 cells (Figure 2(c)). Additionally, the early response proto-oncogenes c-fos and c-jun are predicted targets of the downregulated miR-603 and miR-1299, respectively. In agreement, the mRNAs of both putative targets (c-fos and c-jun) were found to be elevated in C4-2B cells compared to RWPE-1 cells (Figure 2(c)). The corresponding qRT-PCR products of the miRNA targets are depicted on ethidium bromide-stained gels (Figure 2(d)).
Target and pathway prediction
We employed a bioinformatics workflow to identify putative target genes associated with our selected miRNAs. We used DIANA-micro T-CDS “DIANA-microT web server v5.0” (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microtv4/index) to predict the possible targets of both downregulated and upregulated miRNA. The output of potential targets was validated by the other two algorithms: TargetScan (http://www.targetscan.org/) and microRNA.org-Targets and Expression, which represent some of the most used target prediction tools employing a conservation filter. For the final analysis, we considered predicted genes by all prediction programs. A total of 35 and 38 genes were identified as targets for all (>2-fold) upregulated (Supplementary Table 5) and downregulated (Supplementary Table 6) miRNAs detected in microdissected prostate tumors, respectively. Moreover, validated targets for 17 upregulated and downregulated prostate tumor miRNAs are listed in Table 2.
Table 2.
A list of validated targets for dysregulated prostate tumor miRNAs
| Expression | MicroRNA | Validated target | References |
|---|---|---|---|
| Upregulated miRs | has-miR-410 | Four and a half LIM domain 1 (FHL1) tumor suppressor | Wang et al.20 |
| has-miR-18a-3p | Serine/threonine kinase 4 (STK4) | Hsu et al.21 | |
| has-miR-650 | Cyclin-dependent kinase 1 (CDK1), inhibitor of growth 4 (ING4), and early B-cell factor 3 (EBF3) | Mraz et al.22 | |
| hsa-miR-744 | Rho GTPase activating protein 5 (ARHGAP5) | Fang et al.23 | |
| hsa-miR-27b | Suppression of tumorigenicity 14 (ST14) | Wang et al.24 | |
| hsa-miR-182 | FOXO1 | Myatt et al.25 | |
| hsa-miR-221 | Proapoptotic gene (PUMA), p27, and p57 | le Sage et al.26 | |
| hsa-miR-616 | Tissue factor pathway inhibitor 2 (TFPI-2) tumor suppressor | Ma et al.27 | |
| hsa-miR-490 | Endoplasmic reticulum-Golgi intermediate compartment protein3 (ERGIC3) | Zhang et al.28 | |
| Downregulated miRs | has-miR-21 | Tropomyosin 1 (alpha) (TPM1), programmed cell death 4 (PDCD4) | Zhang et al.29 |
| hsa-miR-595 | Jun proto-oncogene (JUN) | Guled et al.30 | |
| hsa-miR-139-3p | Nuclear receptor super family 5 (NR5A2) | Liu et al.31 | |
| hsa-miR-337-3p | Signal transducer and activator of transcription 3 (STAT3) and member of RAS oncogene family (RAP1A) | Du et al.32 | |
| hsa-miR-140-5p | SMAD family member 2 (Smad2) | Zhai et al.33 | |
| hsa-miR-203 | Snail family zinc finger 2 (SNAI2) | Zhang et al.34 | |
| hsa-miR-431-3p | Zinc finger E-box binding homeobox 1 (ZEB1) | Sun et al.35 | |
| hsa-miR-194 | Polycomb ring finger oncogene (BMI-1) | Dong et al.36 |
FOXO1: forkhead box protein O1; PDCD4: programmed cell death protein 4
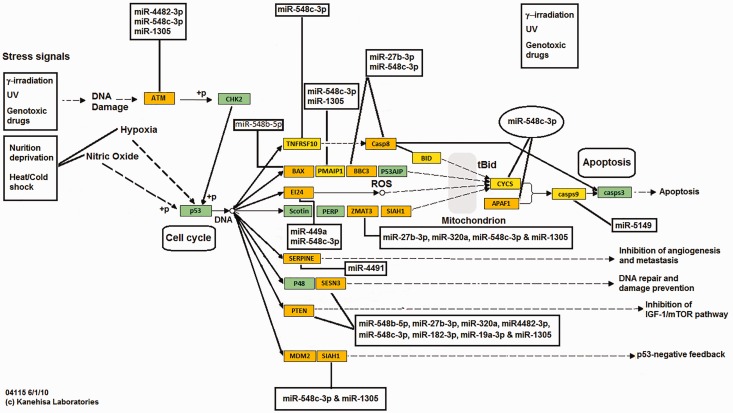
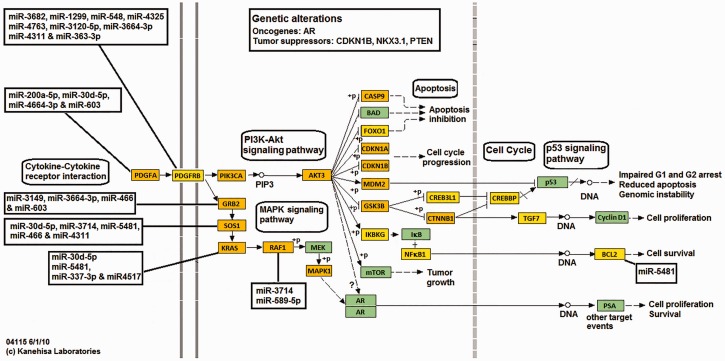
An integrated in silico approach and computational prediction analysis (DIANA-miRPath “miRPath v2.0”; http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=mirpath/index) was performed to identify pathways potentially regulated by a selected subset of highly upregulated and downregulated (>2-fold) miRNAs in microdissected prostate tumors. The results demonstrated that the upregulated miRNAs in prostate tumors target genes associated with p53 pathway (Figure 3), apoptosis (Supplementary Figure 2), and cell cycle (Supplementary Figure 3). In contrast, genes involved in prostate tumorigenesis and survival (Figure 4) and cell cycle (Supplementary Figure 4) were found to be linked with downregulated miRNAs in prostate tumors.
Figure 3.
Diagrammatic scheme depicting targeted p53 pathway-related genes by upregulated miRNAs in prostate tumor cells. An integrated in silico and computational prediction databases (DIANA-micro T-CDS, miRBase, and TargetScan) were employed as described in “Materials and methods” section to predict potential targets of a subset of highly upregulated miRNAs (>2-fold) in prostate tumors. DIANA-miRPath “miRPath v2.0” was used to construct potential miRNA interactions in modulating p53 pathway-related genes. (A color version of this figure is available in the online journal.)
Figure 4.
Diagrammatic scheme depicting potential survival pathway-related gene targets of the downregulated microRNAs and their signaling pathways in prostate cancer cells. An integrated in silico and computational prediction databases (DIANA-micro T-CDS, miRBase, and TargetScan) were employed to predict potential targets of a subset of highly downregulated miRNAs (>2-fold) in prostate tumors as described in “Material and methods” section. DIANA-miRPath “miRPath v2.0” was used to construct potential key targets in prostate cancer survival pathways. (A color version of this figure is available in the online journal.)
Differentially expressed miRNAs potentially target proapoptotic, cell cycle inhibitors, and tumor suppressor genes in prostate tumor cells
The upregulated miRNAs target at least three apoptotic pathways in prostate tumor cells (Supplementary Figure 2). The miRNAs 19a-3p, 548c-3p, 766-5p, and 3190-5p potentially inhibit caspase-8-mediated apoptosis through degradation of interleukin 1 receptor, type 1. Additionally, miRNAs 548au-5p, 548b-5p, 548c-5p, 766-5p, and 4647 inhibit Bax (as known as bcl-2-like protein 4) mediated apoptotic signaling (Supplementary Figure 2). Others potentially inhibit apoptosis by targeting caspases; miR-5194 targets caspase 9 whereas miR-27b-3p and 548c-3p degrade caspase 8 and Bcl2-binding component 3 (BBC3) transcripts, a p53 upregulated modulator of apoptosis (Figure 2 and Supplementary Figure 2). P53-mediated activation of caspases can also be inhibited through targeted degradation of Bax mRNA(miR-548b-5p) and Zinc Finger Matrin-Type 3 (miR-27b, miR-320a, miR-548c-3p, and miR-1305) (Supplementary Figure 2). In contrast, a miR-451a inhibits p15 and upregulated miRNAs 548c-3p, 4287, and 4482-3p potentiate cyclin-cdk and cell cycle progression through targeted degradation of p27, a cdk inhibitor, ATM (Figure 3 and Supplementary Figure 3). Other miRNAs (548c-3p, 4790-3p) prime cell cycle progression through targeted degradation of retinoblastoma (Rb) transcripts (Figure 3). We also identified a number of miRNAs (19a-3p, 320a, 387e, 410, 448c-3p, 548c-3p, 616-3p, 4482, and 4776) that may preferentially target PTEN mRNA, a tumor suppressor that inhibits AKT-mediated survival pathways in prostate tumor cells (Figure 3).
The miRNAs 378e and 378i can potentially target homeobox NKX-3.1 mRNA and inhibit its selective prostate tumor suppressor activity. The inhibition of the proapoptotic activity of forkhead box protein O1, also known as forkhead in rhabdomyosarcoma and a downstream target of Akt phosphorylation, can be suppressed by several miRNAs, including 548c-3p, 578, 1185-1-3p and 4445-5p, and 4719 (Supplementary Figure 3). A list of upregulated miRNAs in microdissected prostate tumors, and their predicted targets and cellular pathways are included in Supplementary Table 5. Collectively, our computational prediction analysis demonstrated that the vast majority of the upregulated miRNAs target key pathways involved in prostate tumorigenesis.
Downregulated miRNAs block tumor cell survival
Based on pathway prediction analysis of downregulated miRNA signature, the Akt signaling pathway triggered by platelet-derived growth factor is potentially blocked in prostate tumors cells through miRNAs 20d-5p, 200a-5p, 363-3p, and 548 (Figure 4). Thus, downregulation of these transcripts augments platelet-derived growth factor-mediated survival and antiapoptotic signaling through inhibition of Akt gene expression in prostate tumor cells. Other miRNAs, such as 30d-5p, 466, and 603 can block cell survival mediated by receptor tyrosine kinase pathways through targeted degradation of adaptor protein-encoding genes Grb2 and SOS in prostate tumor cells (Figure 4). Additionally, mRNAs 30d-5p and 589-5p may potentially target K-ras and Raf1 transcripts and inhibit their downstream survival pathways (Figure 4). The downregulation of miR-548l may also augment antiapoptotic and survival pathways by facilitating expression of MEM2 and Bcl2 genes in prostate tumor cells (Figure 4). Thus, dysregulation of these putative tumor suppressor miRNAs may augment and provide a survival advantage for prostate tumor cells.
A number of cell cycle-targeting miRNAs were found to be downregulated in prostate tumor cells compared to matched normal epithelium (Supplementary Figure 4). For instance, CCND1 transcript, which encodes cyclin D1, is a putative target of let-7b-5p, 200a-3p, and 593-5p miRNAs in prostate tumors. In addition, miRNAs 200a-3p, 548l, 603, and 621 target the cyclic-dependent kinase 6 mRNA. Others, such as miR-548e, can potentially inhibit the cell cycle by targeting CDK1 mRNA (Supplementary Figure 4). A list of downregulated miRNAs in microdissected prostate tumors, and their potential targets and cellular pathways are included in Supplementary Table 6.
Taken together, a number of newly identified tumor suppressor miRNAs, which presumably inhibit growth of normal prostate epithelial cells by targeting key molecules involved in the cell cycle and survival pathways, were found to be downregulated in microdissected prostate tumor cells.
Discussion
MicroRNAs are small noncoding oligonucleotides that regulate gene expression either by mRNA degradation or translational repression. The number of encoded miRNAs varies with the organism and may reach 1500 miRNAs in humans.37 It is well established that miRNAs have multiple mRNA targets resulting in regulation of more than half of the mammalian cellular proteins.38 Emerging evidence suggests dysregulation of miRNAs is associated with human diseases, including cancer.39 In this study, we implemented, for the first time, a miRNA array analysis to identify the differential expression profile of upregulated and downregulated miRNAs in microdissected prostate tumor cells compared to normal adjacent cells procured from patients with Gleason score 6 indolent disease. We have found that 118 miRNAs were upregulated and 73 miRNAs were downregulated in PC tissue compared to adjacent normal tissues.
Among the most highly expressed miRNAs in prostate tumors are miR-4469, miR-548b-5p, miR-410, miR-1179, miR-155-3p, miR-3189-3p, miR-3622b-3p, miR-221, miR-182, miR-10a, miR-31, miR-125b, miR-191, and miR-34a. The validation of expression of this subset of mRNAs by qRT-PCR analysis further corroborated their selective expression in microdissected tumors procured from PC patients. We recently reported that trafficking of miR-125b and miR-155 by PC-associated exosomes contributes to neoplastic reprogramming of patient-derived mesenchymal stem cells.40 We demonstrated that miR-125b and miR-155 potentially prime oncogenic reprogramming by targeting large tumor suppressor homolog2 (Lats2) and the programmed cell death protein 4, a neoplastic transformation inhibitor, in the recipient cells.40 Consistent with our finding is that miR-155, miR-191, and miR-125 were shown to be upregulated in breast cancer.41 The dysregulation of miR-125b and miR-155 may be crucial to underlying mechanisms involved in early prostate tumorigenesis and possibly in other types of human cancers. Additionally, upregulation of miR-221 has been reported in cervical cancers.42 Consistent with our findings, miR-4469 has been identified as a potential biomarker for colorectal cancer.43 In another study, the upregulation of miR-182 in PC and its potential clinical utility as a biomarker for early diagnosis of PC has been reported.44 Likewise, miR-10a and miR-34a have been shown to be highly expressed in bladder and cervical cancers.45 Taken together, our data attest to the potential clinical utility of the upregulated miRNAs as early diagnostic biomarkers of PC cancer. However, further studies are required to validate these findings in a large cohort of PC patients.
In the present study, the most downregulated miRNAs in prostate tumor cells, compared to its adjacent normal glands, were found to be miR-3149, miR3689d, miR3682-5p, miR-200a-5p, miR4518, miRNA3121-5p, miR5189, miR-200a, miR-1285, miR-203, and miR-145. In hepatocellular carcinoma, miR-200a, which targets histone deacetylase 4 (HDAC4) mRNA, was reported to be downregulated.46 The miR-145 was found to be downregulated in bladder cancer47 and in breast cancer.48 The downregulation of miR-203 and its regulatory role in PC progression and metastasis has been documented.49 Collectively, while some of the downregulated miRNAs have been shown to be associated with tumorigenesis, additional studies are needed to establish the expression, the functional significance in PC development, and the potential clinical utilities of the remaining miRNAs.
A number of mRNA targets associated with the dysregulated miRNAs and their predicted pathways in prostate tumor cells were delineated. For instance, we identified PTEN and P53/TP53 pathways as two potential targets of miR-19a-3p and miR-1305, respectively. Our findings come in agreement with previous reports that miR-19 and miR-1305 gain their oncogenic activities by targeting PTEN50 and P5351 transcripts, respectively. Among the downregulated miRNAs we found that cyclin D1 mRNA is a possible target of miR-200a and let-7b. Higher apoptotic rate was observed in B16F10 melanoma cells in which let-7b was reconstituted, and this occurred by downregulation of cyclin D1 mRNA expression.52 In contrast, increased expression of cyclin D1 and beta-catenin transcripts was observed following miR-200a downregulation.53 Taken together, our combined array-prediction approach proved to be successful not only in identifying novel miRNAs, but also in deciphering their functional significance by defining their targets and pathways in prostate tumor cells.
The present study unravels a novel subset of dysregulated miRNA signature and their potential clinical implications in organ-confined PC. Identification of mRNA targets and key regulatory pathways that can be modulated by these miRNA further enhances our understanding of the underlying key regulatory molecular mechanisms potentially involved in early prostate tumorigenesis. The aberrant expression of miRNAs paves the way for assessment of their mechanistic roles and clinical utilities as biomarkers, prognostic indicators, and/or therapeutic targets in early prostate tumorigenesis.
Supplementary Material
Acknowledgements
This work was supported by a research fund from the Cultural Affairs Section and Missions, Ministry of Higher Education, Cairo, Egypt, to A.A.M. We thank Ms. Jessica A. Daigle for editing the manuscript.
Authors’ contributions
Conceived and designed the experiments: ABAM, AAM, MZ, AE
Performed experiments: AAM, MZ, AE, AD, HK
Analyzed data: AAM, MZ, KM, SS, ABAM
Provided reagents, materials and tools: RT, KM, SS, JLS, KM
Writing of the manuscript: AAM, MZ, FEHZ, OHEH, ABAM
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin 2015; 65: 5–29. [DOI] [PubMed] [Google Scholar]
- 2.Masson S, Bahl A. Metastatic castrate-resistant prostate cancer: dawn of a new age of management. BJU Int 2012; 110: 1110–14. [DOI] [PubMed] [Google Scholar]
- 3.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 2005; 120: 21–4. [DOI] [PubMed] [Google Scholar]
- 4.Logothetis CJ, Lin SH. Osteoblasts in prostate cancer metastasis to bone. Nat Rev Cancer 2005; 5: 21–28. [DOI] [PubMed] [Google Scholar]
- 5.Draisma G, Etzioni R, Tsodikov A, Mariotto A, Wever E, Gulati R, Feuer E, de Koning H. Lead time and overdiagnosis in prostate-specific antigen screening: importance of methods and context. J Natl Cancer Inst 2009; 101: 374–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarov DV, Loeb S, Getzenberg RH, Partin AW. Biomarkers for prostate cancer. Annu Rev Med 2009; 60: 139–51. [DOI] [PubMed] [Google Scholar]
- 7.Lan H, Lu H, Wang X, Jin H. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed Res Int 2015; 2015: 125094–125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esquela-Kerscher A, Slack FJ. Oncomirs – micrornas with a role in cancer. Nat Rev Cancer 2006; 6: 259–69. [DOI] [PubMed] [Google Scholar]
- 9.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death and tumorigenesis. Br J Cancer 2006; 94: 776–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koturbash I, Zemp FJ, Pogribny I, Kovalchuk O. Small molecules with big effects: the role of the microRNAome in cancer and carcinogenesis. Mutat Res 2011; 722: 94–105. [DOI] [PubMed] [Google Scholar]
- 11.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. Ras is regulated by the let-7 microRNA family. Cell 2005; 120: 635–47. [DOI] [PubMed] [Google Scholar]
- 12.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates myc and reverts myc-induced growth in Burkitt lymphoma cells. Cancer Res 2007; 67: 9762–70. [DOI] [PubMed] [Google Scholar]
- 13.Shi XB, Xue L, Ma AH, Tepper CG, Kung HJ, White RW. Mir-125b promotes growth of prostate cancer xenograft tumor through targeting pro-apoptotic genes. Prostate 2011; 71: 538–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan JA, Krichevsky AM, Kosik KS. Microrna-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 2005; 65: 6029–33. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Jia D, Kim H, Abd Elmageed ZY, Datta A, Davis R, Srivastav S, Moroz K, Crawford BE, Moparty K, Thomas R, Hudson RS, Ambs S, Abdel-Mageed AB. Dysregulation of microRNA-212 promotes prostate tumorigenesis and castration resistance through hnRNPH1-mediated regulation of AR and its splice variant AR-V7: implications for racial disparity of prostate cancer progression. Clin Cancer Res 2016; 22: 1744–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maragkakis M, Vergoulis T, Alexiou P, Reczko M, Plomaritou K, Gousis M, Kourtis K, Koziris N, Dalamagas T, Hatzigeorgiou AG. Diana-microt web server upgrade supports fly and worm mirna target prediction and bibliographic mirna to disease association. Nucleic Acids Res 2011; 39: W145–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.Org resource: targets and expression. Nucleic Acids Res 2008; 36: D149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witkos TM, Koscianska E, Krzyzosiak WJ. Practical aspects of microRNA target prediction. Curr Mol Med 2001; 11: 93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vlachos IS, Kostoulas N, Vergoulis T, Georgakilas G, Reczko M, Maragkakis M, Paraskevopoulou MD, Prionidis K, Dalamagas T, Hatzigeorgiou AG. Diana mirpath v.2.0: Investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res 2012; 40: W498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Fu J, Jiang M, Zhang X, Cheng L, Xu X, Fan Z, Zhang J, Ye Q, Song H. Mir-410 is overexpressed in liver and colorectal tumors and enhances tumor cell growth by silencing fhl1 via a direct/indirect mechanism. PLoS One 2014; 9: e108708–e108708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu TI, Hsu CH, Lee KH, Lin JT, Chen CS, Chang KC, Su CY, Hsiao M, Lu PJ. Microrna-18a is elevated in prostate cancer and promotes tumorigenesis through suppressing stk4 in vitro and in vivo. Oncogenesis 2014; 3: e99–e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mraz M, Dolezalova D, Plevova K, Stano Kozubik K, Mayerova V, Cerna K, Musilova K, Tichy B, Pavlova S, Borsky M, Verner J, Doubek M, Brychtova Y, Trbusek M, Hampl A, Mayer J, Pospisilova S. Microrna-650 expression is influenced by immunoglobulin gene rearrangement and affects the biology of chronic lymphocytic leukemia. Blood 2012; 119: 2110–13. [DOI] [PubMed] [Google Scholar]
- 23.Fang Y, Zhu X, Wang J, Li N, Li D, Sakib N, Sha Z, Song W. Mir-744 functions as a proto-oncogene in nasopharyngeal carcinoma progression and metastasis via transcriptional control of arhgap5. Oncotarget 2015; 6: 13164–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Rathinam R, Walch A, Alahari SK. St14 (suppression of tumorigenicity 14) gene is a target for mir-27b, and the inhibitory effect of st14 on cell growth is independent of mir-27b regulation. J Biol Chem 2009; 284: 23094–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myatt SS, Wang J, Monteiro LJ, Christian M, Ho KK, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S, Lam EW. Definition of micrornas that repress expression of the tumor suppressor gene foxo1 in endometrial cancer. Cancer Res 2010; 70: 367–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafre SA, Farace MG, Agami R. Regulation of the p27(kip1) tumor suppressor by mir-221 and mir-222 promotes cancer cell proliferation. EMBO J 2007; 26: 3699–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma S, Chan YP, Kwan PS, Lee TK, Yan M, Tang KH, Ling MT, Vielkind JR, Guan XY, Chan KW. Microrna-616 induces androgen-independent growth of prostate cancer cells by suppressing expression of tissue factor pathway inhibitor tfpi-2. Cancer Res 2011; 71: 583–92. [DOI] [PubMed] [Google Scholar]
- 28.Zhang LY, Liu M, Li X, Tang H. Mir-490-3p modulates cell growth and epithelial to mesenchymal transition of hepatocellular carcinoma cells by targeting endoplasmic reticulum-golgi intermediate compartment protein 3 (ergic3). J Biol Chem 2013; 288: 4035–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Gee H, Rose B, Lee CS, Clark J, Elliott M, Gamble JR, Cairns MJ, Harris A, Khoury S, Tran N. Regulation of the tumour suppressor pdcd4 by mir-499 and mir-21 in oropharyngeal cancers. BMC Cancer 2015; 16: 86–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guled M, Lahti L, Lindholm PM, Salmenkivi K, Bagwan I, Nicholson AG, Knuutila S. Cdkn2a, nf2, and jun are dysregulated among other genes by mirnas in malignant mesothelioma -a mirna microarray analysis. Genes Chromosomes Cancer 2009; 48: 615–23. [DOI] [PubMed] [Google Scholar]
- 31.Liu R, Yang M, Meng Y, Liao J, Sheng J, Pu Y, Yin L, Kim SJ. Tumor-suppressive function of mir-139-5p in esophageal squamous cell carcinoma. PLoS One 2013; 8: e77068– e77068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du L, Subauste MC, DeSevo C, Zhao Z, Baker M, Borkowski R, Schageman JJ, Greer R, Yang CR, Suraokar M, Wistuba II, Gazdar AF, Minna JD, Pertsemlidis A. Mir-337-3p and its targets stat3 and rap1a modulate taxane sensitivity in non-small cell lung cancers. PLoS One 2012; 7: e39167–e39167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhai H, Fesler A, Ba Y, Wu S, Ju J. Inhibition of colorectal cancer stem cell survival and invasive potential by hsa-mir-140-5p mediated suppression of smad2 and autophagy. Oncotarget 2015; 6: 19735–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, Zhang B, Li W, Fu L, Zhu Z, Dong JT. Epigenetic silencing of mir-203 upregulates snai2 and contributes to the invasiveness of malignant breast cancer cells. Genes Cancer 2011; 2: 782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun K, Zeng T, Huang D, Liu Z, Huang S, Liu J, Qu Z. Microrna-431 inhibits migration and invasion of hepatocellular carcinoma cells by targeting the zeb1-mediated epithelial-mensenchymal transition. FEBS Open Bio 2015; 5: 900–7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Dong P, Kaneuchi M, Watari H, Hamada J, Sudo S, Ju J, Sakuragi N. MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene bmi-1. Mol Cancer 2011; 10: 99–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ameres SL, Zamore PD. Diversifying microrna sequence and function. Nat Rev Mol Cell Biol 2013; 14: 475–88. [DOI] [PubMed] [Google Scholar]
- 38.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mrnas are conserved targets of microRNAs. Genome Res 2009; 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hesse M, Arenz C. MicroRNA maturation and human disease. Methods Mol Biol 2014; 1095: 11–25. [DOI] [PubMed] [Google Scholar]
- 40.Abd Elmageed ZY, Yang Y, Thomas R, Ranjan M, Mondal D, Moroz K, Fang Z, Rezk BM, Moparty K, Sikka SC, Sartor O, Abdel-Mageed AB. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells 2014; 32: 983–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hui AB, Shi W, Boutros PC, Miller N, Pintilie M, Fyles T, McCready D, Wong D, Gerster K, Waldron L, Jurisica I, Penn LZ, Liu FF. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest 2009; 89: 597–606. [DOI] [PubMed] [Google Scholar]
- 42.Hu X, Schwarz JK, Lewis JS, Jr, Huettner PC, Rader JS, Deasy JO, Grigsby PW, Wang X. A microRNA expression signature for cervical cancer prognosis. Cancer Res 2010; 70: 1441–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu X, Xu X, Li S, Wu S, Chen R, Jiang Q, Liu H, Sun Y, Li Y, Xu Y. Identification and validation of potential biomarkers for the detection of dysregulated microRNA by qPCR in patients with colorectal adenocarcinoma. PLoS One 2015; 10: e0120024–e0120024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casanova-Salas I, Rubio-Briones J, Calatrava A, Mancarella C, Masia E, Casanova J, Fernandez-Serra A, Rubio L, Ramirez-Backhaus M, Arminan A, Domínguez-Escrig J, Martínez F, García-Casado Z, Scotlandi K, Vicent MJ, López-Guerrero JA. Identification of mir-187 and mir-182 as biomarkers of early diagnosis and prognosis in patients with prostate cancer treated with radical prostatectomy. J Urol 2014; 192: 252–59. [DOI] [PubMed] [Google Scholar]
- 45.Segersten U, Spector Y, Goren Y, Tabak S, Malmstrom PU. The role of microRNA profiling in prognosticating progression in ta and t1 urinary bladder cancer. Urol Oncol 2014; 32: 613–18. [DOI] [PubMed] [Google Scholar]
- 46.Yuan JH, Yang F, Chen BF, Lu Z, Huo XS, Zhou WP, Wang F, Sun SH. The histone deacetylase 4/sp1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology 2011; 54: 2025–35. [DOI] [PubMed] [Google Scholar]
- 47.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K, Tsujimoto G, Nakagawa M, Seki N. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer 2009; 125: 345–52. [DOI] [PubMed] [Google Scholar]
- 48.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN. Altered microRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res 2007; 67: 11612–20. [DOI] [PubMed] [Google Scholar]
- 49.Saini S, Majid S, Yamamura S, Tabatabai L, Suh SO, Shahryari V, Chen Y, Deng G, Tanaka Y, Dahiya R. Regulatory role of mir-203 in prostate cancer progression and metastasis. Clin Cancer Res 2011; 17: 5287–98. [DOI] [PubMed] [Google Scholar]
- 50.Olive V, Jiang I, He L. Mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol 2010; 42: 1348–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slattery ML, Wolff E, Hoffman MD, Pellatt DF, Milash B, Wolff RK. MicroRNAs and colon and rectal cancer: differential expression by tumor location and subtype. Genes Chromosomes Cancer 2011; 50: 196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu D, Tan J, Zhou M, Jiang B, Xie H, Nie X, Xia K, Zhou J. Let-7b and microRNA-199a inhibit the proliferation of b16f10 melanoma cells. Oncol Lett 2012; 4: 941–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saydam O, Shen Y, Wurdinger T, Senol O, Boke E, James MF, Tannous BA, Stemmer-Rachamimov AO, Yi M, Stephens RM, Fraefel C, Gusella JF, Krichevsky AM, Breakefield XO. Downregulated microrna-200a in meningiomas promotes tumor growth by reducing e-cadherin and activating the wnt/beta-catenin signaling pathway. Mol Cell Biol 2009; 29: 5923–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.