Abstract
Intrinsic cell chirality has been implicated in the left–right (LR) asymmetry of embryonic development. Impaired cell chirality could lead to severe birth defects in laterality. Previously, we detected cell chirality with an in vitro micropatterning system. Here, we demonstrate for the first time that chirality can be quantified as the coordination of multiaxial polarization of individual cells and nuclei. Using an object labeling, connected component based method, we characterized cell chirality based on cell and nuclear shape polarization and nuclear positioning of each cell in multicellular patterns of epithelial cells. We found that the cells adopted a LR bias the boundaries by positioning the sharp end towards the leading edge and leaving the nucleus at the rear. This behavior is consistent with the directional migration observed previously on the boundary of micropatterns. Although the nucleus is chirally aligned, it is not strongly biased towards or away from the boundary. As the result of the rear positioning of nuclei, the nuclear positioning has an opposite chirality to that of cell alignment. Overall, our results have revealed deep insights of chiral morphogenesis as the coordination of multiaxial polarization at the cellular and subcellular levels.
Electronic supplementary material
The online version of this article (doi:10.1007/s12195-016-0467-2) contains supplementary material, which is available to authorized users.
Keywords: Cell chirality, Cell polarity, Cell morphology, Nuclear morphology
Introduction
Cell chirality is an intrinsic property of the cell, and has recently been observed undisputedly as a consistent left–right bias in multicellular structures5,47 as well as in individual cells.52,53 This LR asymmetry can be seen in vivo in the tissue level with the alignment of epithelial cells in the hindgut of Drosophila.12,36 On an organism level, the asymmetric morphology and positioning of internal organs in the thorax of animals and morphological asymmetry of the central nervous system (e.g., differences between the left and right frontal lobes) of higher mammals have long been appreciated.22,26 Changes in cell chirality have been associated with cell phenotype and oxidative stress.35,47 Cancer, diabetes, and other disorders can also cause birth defects in LR asymmetry.2,22,32 An understanding of biophysical mechanisms underlying multicellular chiral morphogenesis will facilitate the elucidation of etiology of birth defects in laterality.
The determination of LR axis requires the pre-establishment of the other two axes, anterior-posterior and dorsal–ventral axes, in embryonic development. Coordination among these axes is critical as any misinterpretation could lead to severe birth defects.4,22,33 On a 2D substrate, cell migration direction is directly linked to the back-front (BF) polarity through the polarized shape that migrating cells take on.14,21,30,41,44,49 2D cell chirality is a handedness, and can be considered as the relation between the LR and BF polarity,38,47,51,52 or towards which direction, left or right, the cell will be biased when the BF axis is defined. Therefore, the coordination of multiaxial polarity is crucial to the determination of chiral biases in cells and important to the understanding of the biophysical mechanisms of LR asymmetry.
Previously, chirality was measured through the bias in alignment angle as found in cell edges,12,36 the minimum circumscribed rectangle method,5 and using intensity gradients of phase contrast images.16,35,47,51 None of these analyses, however, were based on individual cells, and the physical nature behind cell chirality (i.e., the coordination of multiaxial polarity) was largely ignored. Here, we utilized a recently developed Python-based algorithm to evaluate the polarizations of individual cells, nuclei and their relative positioning within a geometrically controlled cellular monolayer.29 Such an approach enables accurate and robust analyses of epithelial chiral morphogenesis.
With this approach, we demonstrated that the cells are polarized by positioning their sharp ends towards the boundary and biasedly along the direction of cell migration. Cell chirality is the result of interactions between three axes of polarity while nuclei simply take the same orientation of the cell without a directional bias towards the leading edge or the rear. We believe that this new analytical approach based on polarity analysis is potentially a powerful tool to unveil the physical mechanisms of cell chirality and provide deep insights into the nature of multicellular chiral morphogenesis.
Materials and Methods
Cell Culture and Immunostaining
MDCK cells were maintained in flasks with media composed of Dulbecco’s Modified Eagle Medium (DMEM) with High Glucose (Life Technologies), 10% fetal bovine serum, 1 mM sodium pyruvate, and 1% penicillin–streptomycin. Microcontact printing was performed using polydimethylsiloxane (PDMS) stamps and self-assembled monolayers (SAMs) as previously described.45,47 All micropatterns were in the shape of rings with an inner diameter of 90 μm and a width of 235 µm.
Cells were trypsinized and seeded onto the patterned surfaces. Once attached, the excess cells were washed off using phosphate buffered serum (PBS). Micropatterned cells were then cultured for 24–36 h until confluency. Cells were fixed with 4% paraformaldehyde and stained for nuclei and tight junction with DAPI and ZO-1 antibody, respectively. Samples were subsequently imaged for the cells with phase contrast imaging, and nuclei and ZO-1 with fluorescence imaging.
Image Processing
We used a Python-based automated object-labeling connected-component method to analyze individual cells and their nuclei, as reported before.29 Briefly, ZO-1 images were adjusted for brightness, and the background was subtracted to highlight the cell edges and to minimize artifacts in ImageJ. Edges were further corrected for discontinuities using the brush tool. Nuclei images were adjusted for brightness and thresholded. Pre-processed ZO-1 images were doubled in size, thresholded, run through a watershed algorithm, and skeletonized. Images were then converted into a binary array and run through a nearest neighbor protocol. Nearest neighbor analyzed all the elements in the array and identified all the “on” pixels which had a value of 1. Pixels with a 0 value were designated as “off” pixels. “On” pixels were then analyzed to identify nodes, which are pixels with three or more neighbors. Nodes were paired to create edges through the pixels connected between them. These were simplified to polygonized images. Individual cells and their edges were identified in the polygonized images through a connected component protocol. Pre-processed nuclei were run through a connected component protocol and paired with cells. Any cells with multiple or no nuclei were removed from analysis. Cells and nuclei were also analyzed for morphological features, alignment angles, and polarity-based features. This algorithm’s pipeline is highlighted in Fig. S1.
Determination of Alignment and Polarization
Alignment angles (θ) are the orientation angle of the major axis between −90° and 90° and defined as either clockwise (CW) ranging from −90° to −0° and counterclockwise (CCW) from 0° to 90°. Shape polarization angles (φ) are also along the major axis, but defined to point towards the sharp end. Therefore φ is between −180° and 180°. Nuclear positioning polarization angle (β) is between −180° and 180° and is the direction from the centroid of the cell to the centroid of the nuclei. Nuclear positioning polarization distance, or more simply polarization distance, is defined as the distance between the centroids of the cell and its nucleus.
Statistical Analysis
All data is presented as mean ± standard error of mean. Alignment bias was determined using a binomial test. The variation of cellular and nuclear percentages and morphological parameters with their polarization directions and radial positions was assessed using a two-way analysis of variance (ANOVA). Tukey’s HSD post hoc tests were performed at a 95% confidence interval to determine statistical significance. The p values were reported for post hoc tests, unless noted otherwise. Error bars in all graphs are displayed as standard error.
Results
Cells and Nuclei Exhibit Chiral Alignment
We defined cell and nuclear polarization as vectors starting from the centroid and pointing towards the sharp end as shown in Fig. 1a. This definition is based on our observation that on confluent substrates the cell moves through the monolayer with the sharp ends at the leading edge (see Supplementary Video S1).47,51 For each of these vectors, an angle was assigned as defined in Fig. 1b.
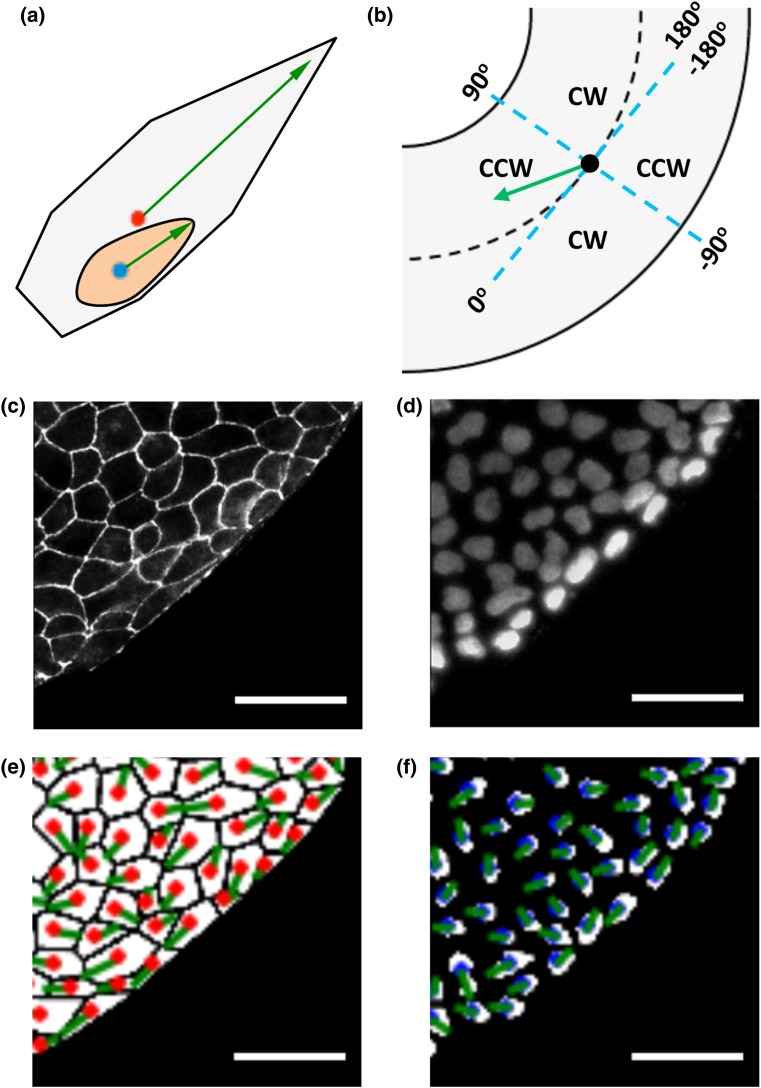
Figure 1.
Analysis of polarization biases and chirality in cell and nuclear shape. Scale = 50 μm. (a) The front of cells and nuclei is defined as the sharp end, as indicated by the green arrows (red dot for cell centroid and blue dot for nuclear centroid). (b) Definition of alignment angles in a polarization axis (blue dashed lines representing cylindrical coordinates). (c) A region of interest of ZO-1 (tight junctions) fluorescence image. (d) Nuclei with DAPI staining. (e) ZO-1 images are processed and cell shape polarization angles are calculated, with red dots for cell centroids and green lines for cell polarization direction. (f) Nuclear shape polarization was determined and shown with green lines. Blue dots represent nuclear centroids.
Figures 1c and 1d show typical images of the ZO-1 (tight junctions) and nuclei staining from a region on the micropatterned ring. After image processing, cell boundary and nuclei are identified as shown Fig. 1e and 1f, and the cell and nucleus shape polarization angles were highlighted with green lines, and evaluated based on Fig. 1b. Among four quadrants defined by circumferential and radial axes, two (−180° to −90° and 0°−90°) were defined as counterclockwise (CCW) alignment and two (−90° to 0° and 90°−180°) as clockwise (CW) alignment, following the convention outlined before.47 Consistently with previous analyses,29 both cell and nucleus based analyses had a significant bias towards counterclockwise (CCW) alignment among chiral rings.
Polarization Switches Directions on the Micropattern Borders
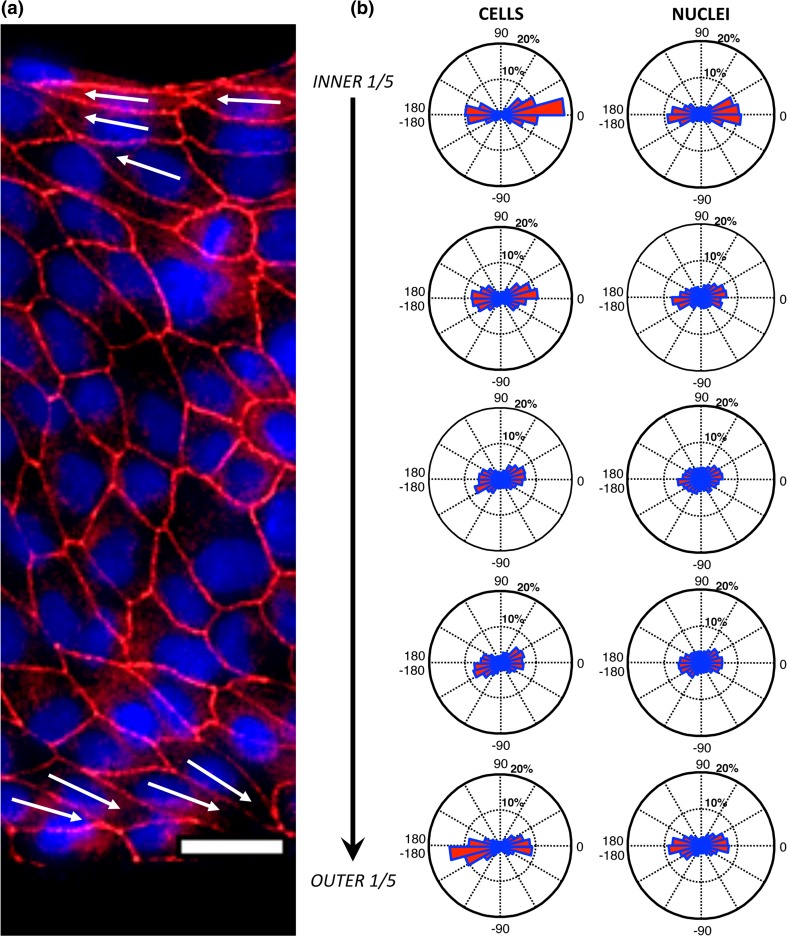
We next sought to determine how shape polarization direction changes between borders. Figure 2a shows a region of a typical ring in which the cells show a CCW alignment and there is a clear switch of cell shape polarization between the inner and outer ring, as shown by the arrows. In order to quantify this change, we analyzed the distributions of cell shape polarizations at five equally sized ranges of radial distance as shown in Fig. 2b. We found that the cells switched from a polarization that is pointed towards 0° and tilted towards a positive value on the inner boundary, to a direction that is pointed towards ±180°, and biased to negative values on the outer boundary. Similar but less significant trend was found with nuclear shape polarizations. Although the chiral alignment biases were weak as indicated by the small deviation of alignment direction from the circumferential direction, such polarizations on boundaries would be both characterized as the CCW alignment based on the definition of Fig. 1b. In contrast, the direction of polarization in the circumferential direction was exactly opposite on the two boundaries. This highlights the necessity of considering the direction of polarization, instead of simply its orientation.47
Figure 2.
Variation of cell and nuclear shape polarization between the inner and outer boundary. Scale bar = 25 µm. (a) A region of a CCW ring stained for ZO-1 (red) and nuclei (blue). White arrows indicate cell shape polarization direction of representative cells at the boundaries. (b) Circular histograms show the distribution of shape polarization angles of cells (left) and nuclei (right) in 5 regions of the 38 rings that are equally divided in the radial distance from the inner to the outer ring.
Cell Chirality Demonstrate Polarity Bias and Nuclei Are Chirally Biased
As the largest directional bias has been observed on two boundaries (Fig. 2), we therefore focused on the inner and outer one fifth (1/5) of the rings. If we define that the cells face towards the boundary (Fig. 3a), the opposite polarization of the cells observed on inner and outer boundaries (Fig. 2a) is actually consistent and both have a leftward bias.47 To compare cell behavior on inner and outer boundaries, we adopt local coordinates with the BF (from back to front) and LR (from left to right) axes defined separately on two boundaries. This effectively divides the polarization into four quadrants: front-right (FR), back-left (BL), front-left (FL), and back-right (BR). We first performed morphological analyses (Fig. S2). We found that the cells with BL polarization had a slightly smaller area and a lower aspect ratio, while the nuclear morphology didn’t show a significant variation between the four quadrants.
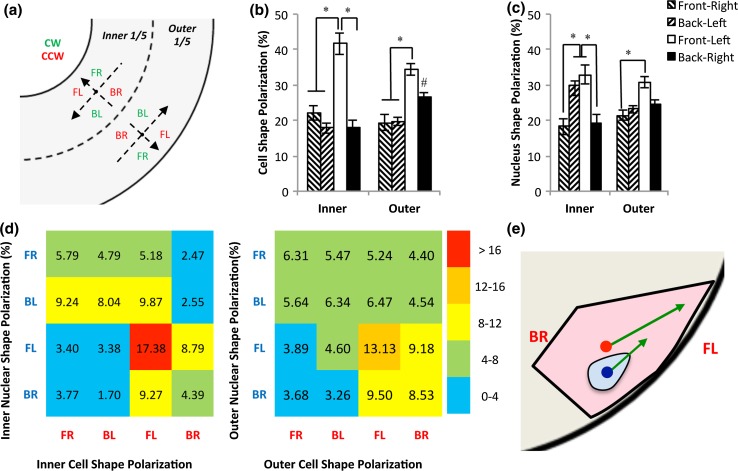
Figure 3.
Analysis of cell and nuclear shape polarization at boundaries. (a) Each boundary region adopts local coordinates with the cells facing the closest boundary, dividing the entire plane into four quadrants (FR: front-right, BL: back-left, FL: front-left, and BR: back-right). Among them, BR and FL (red) represent a CCW bias and FR and BL (green) represent a CW bias. (b) Analysis of cell shape polarization reveals a bias towards front left (FL) polarization at the inner ring, and FL and BR polarizations at the outer. * significantly different between quadrants, and # significantly different from the inner ring. (c) Nuclear shape polarization analysis reveals a strong bias towards FL and BL polarizations at boundaries. * significantly different between quadrants. (d) Heat maps between cell shape polarization and nuclear shape polarization at the inner (left) and outer (right) rings. (e) Cells tend to polarize themselves towards FL in regions near the micropatterned boundaries, while nuclei are biased to the left with no significant bias towards front or back (arrows: cellular or nuclear shape polarization, blue dot: nuclear centroid, and red dot: cell centroid).
When comparing the percentage of the cells in four quadrants, we found that the quadrant location had a significant effect on the percentage of cells present (p < 0.001, ANOVA; Fig. 3b), but effects of physical location (inner ring and outer ring) were not significant (p > 0.05, ANOVA). There was a strong bias towards a front-left (FL) polarization of cell shape (p < 0.05), with a larger bias on the inner boundary than on the outer (FL fraction: 41.7 ± 3.1% vs. 34.3 ± 1.6%; p < 0.05). On the outer ring, the back-right (BR) portion was elevated (26.7 ± 1.2% on the outer vs. 18.2 ± 1.9% on the inner; p < 0.05).
Analysis of nuclear shape polarization also revealed a strong dependence on quadrant location (p < 0.001, ANOVA), but not between inner and outer ring (p > 0.05, ANOVA; Fig. 3c). Differently from cellular strong polarization towards FL, nuclei strongly polarized towards FL and BL on the inner ring (p < 0.05), and towards FL on the outer boundary.
To further understand the nature of cellular and nuclear polarizations, we performed additional statistical tests, as shown in Table 1. For the cells, on both the inner and outer boundaries there was a significant bias towards the front (i.e., towards the boundaries; F vs. B in Table 1), while the nuclei did not show a significant bias. Both the cells and nuclei demonstrated a significant leftward bias (L vs. R) and towards CCW (CW vs. CCW) at both boundaries. The cells or the nuclei that were polarized towards the front tended to bias towards the left side (FL vs. FR). Whereas the cells polarizing towards back were biased to the right side (BR) on the outer ring (BL vs. BR), the nuclei were biased towards to BL on the inner ring (BL vs. BR). The data suggest that while the leftward bias of the cells is a chiral behavior of the cell based on its front polarization, the nuclei seem to bias towards the left side in general, especially on the inner ring.
Table 1.
Analysis of polarity and chirality of cell and nuclear shape at the micropatterned boundaries.
| Polarization Type | Cell | Nucleus | |||
|---|---|---|---|---|---|
| Location | Measures | Bias | p value | Bias | p value |
| Inner | F vs. B | F*** | 1.4E − 06 | F | 3.1E − 01 |
| L vs. R | L*** | 1.2E − 04 | L*** | 3.8E − 04 | |
| CW vs. CCW | CCW** | 2.5E − 03 | CCW* | 2.2E − 01 | |
| FL vs. FR | FL*** | 8.1E − 05 | FL*** | 6.8E − 04 | |
| BR vs. BL | BR | 4.7E − 01 | BL* | 1.2E − 02 | |
| Outer | F vs. B | F*** | 7.0E − 04 | F | 5.6E − 02 |
| L vs. R | L** | 2.8E − 03 | L* | 1.6E − 02 | |
| CW vs. CCW | CCW*** | 4.2E − 06 | CCW*** | 5.1E − 04 | |
| FL vs. FR | FL*** | 5.1E − 06 | FL*** | 7.0E − 04 | |
| BR vs. BL | BR*** | 7.4E − 04 | BR | 2.1E − 01 | |
| Combined | F vs. B | F*** | 5.3E − 08 | F | 1.2E − 01 |
| L vs. R | L*** | 5.7E − 06 | L*** | 6.2E − 04 | |
| CW vs. CCW | CCW*** | 2.0E − 05 | CCW* | 1.6E − 02 | |
| FL vs. FR | FL*** | 6.7E − 07 | FL*** | 1.6E − 04 | |
| BR vs. BL | BR*** | 9.0E − 08 | BL* | 1.8E − 02 | |
Cell and nuclear shape polarization angles for the 38 rings were analyzed for biases at the inner, outer, and combined inner-outer fifths of the ring
CW, clockwise; CCW, counter clockwise; NC, non-chiral; L, left; R, right; FL, front-left polarization; FR, front-back polarization; BL, back-left polarization; BR, back-right polarization
* Significant at p < 0.05, ** significant at p < 0.01, and *** significant at p < 0.001
To determine the relationship between cell and nuclear shape polarizations, heat maps were generated for the inner and outer boundaries (Fig. 3d). The heat maps demonstrated that there was a simultaneous bias towards FL polarization for both cells and nuclei at both boundaries (17.4 and 13.1%). Further, among the cells that were chirally aligned (i.e., FL or BR polarization), a significant number of nuclei demonstrated a leftward bias (i.e., FL and BL polarizations) on both the inner (64.4%) and outer (54.6%) fifths of the ring. Overall, the cells have a strong FL polarization, while the nucleus has a bias toward the left side (Fig. 3e).
Chiral Measures for Cell and Nucleus Polarization
Based on these analyses, we proposed four possible chiral measures: L vs. R, CCW vs. CW (i.e., FL + BR vs. FR + BL), FL vs. FR, and BL vs. BR as seen in Table 2. We found that the first three measures provided similar results for determining cell chirality, while the measure BL vs. BR could not do as well. The measure FL vs. FR gave the least number of the opposite chirality (CW), indicating that this measure might be most robust for detecting a chiral bias. The nuclei were always biased to the left side no matter which direction of the cells take on in the BF axis. In particular, on the inner ring, the nuclei are biased towards BL instead of BR (BL vs. BR). Therefore, L vs. R or FL vs. FR will be a good measure of nuclear chirality, but CW vs. CCW (FL+BR vs. FR+BL) will not be able to capture the biased nature behind the chiral alignment of the nuclei.
Table 2.
Chirality analysis of patterned epithelial cells with four chiral measures.
| A. Cells and Nuclei Polarized Left vs. Right | |||||||
|---|---|---|---|---|---|---|---|
| Method | Total | Cells | Nuclei | ||||
| CW (R) | CCW (L) | NC | CW (R) | CCW (L) | NC | ||
| Inner ring L vs. R | 38 | 7 | 27 | 4 | 8 | 27 | 3 |
| Outer ring L vs. R | 38 | 7 | 21 | 10 | 8 | 21 | 9 |
| Combined ring L vs. R | 38 | 5 | 28 | 5 | 5 | 23 | 11 |
| B. Cell and Nuclei Alignment | |||||||
|---|---|---|---|---|---|---|---|
| Method | Total | Cells | Nuclei | ||||
| CW | CCW | NC | CW | CCW | NC | ||
| Inner ring alignment | 38 | 6 | 27 | 5 | 12 | 18 | 8 |
| Outer ring alignment | 38 | 4 | 25 | 9 | 5 | 25 | 8 |
| Combined ring alignment | 38 | 5 | 27 | 6 | 8 | 20 | 10 |
| C. Cells and Nuclei Polarized Towards Front Axis | |||||||
|---|---|---|---|---|---|---|---|
| Method | Total | Cells | Nuclei | ||||
| CW (FR) | CCW (FL) | NC | CW (FR) | CCW (FL) | NC | ||
| Inner ring FL vs. FR | 38 | 4 | 25 | 9 | 8 | 23 | 7 |
| Outer ring FL vs. FR | 38 | 2 | 25 | 11 | 6 | 23 | 9 |
| Combined ring FL vs. FR | 38 | 3 | 27 | 8 | 9 | 23 | 6 |
| D. Cells and Nuclei Polarized Towards Back Axis | |||||||
|---|---|---|---|---|---|---|---|
| Method | Total | Cells | Nuclei | ||||
| CW (BL) | CCW (BR) | NC | CW (BL) | CCW (BR) | NC | ||
| Inner ring BR vs. BL | 38 | 13 | 15 | 10 | 24 | 11 | 3 |
| Outer ring BR vs. BL | 38 | 4 | 18 | 14 | 9 | 11 | 18 |
| Combined ring BR vs. BL | 38 | 8 | 17 | 13 | 19 | 10 | 9 |
Bold values indicate statistical significance (p < 0.05)
Cell and nuclear shape polarization over individual rings was determined for the various types of chiral measures as listed below. The number is highlighted in bold to indicate the direction that a significant bias is detected among chiral rings with a binomial test
CW, clockwise; CCW, counter clockwise; NC, non-chiral; L, left; R, right; FL, front-left polarization; FR, front-back polarization; BL, back-left polarization; BR, back-right polarization
Polarity and Chirality of Nucleus Positioning in the Cell
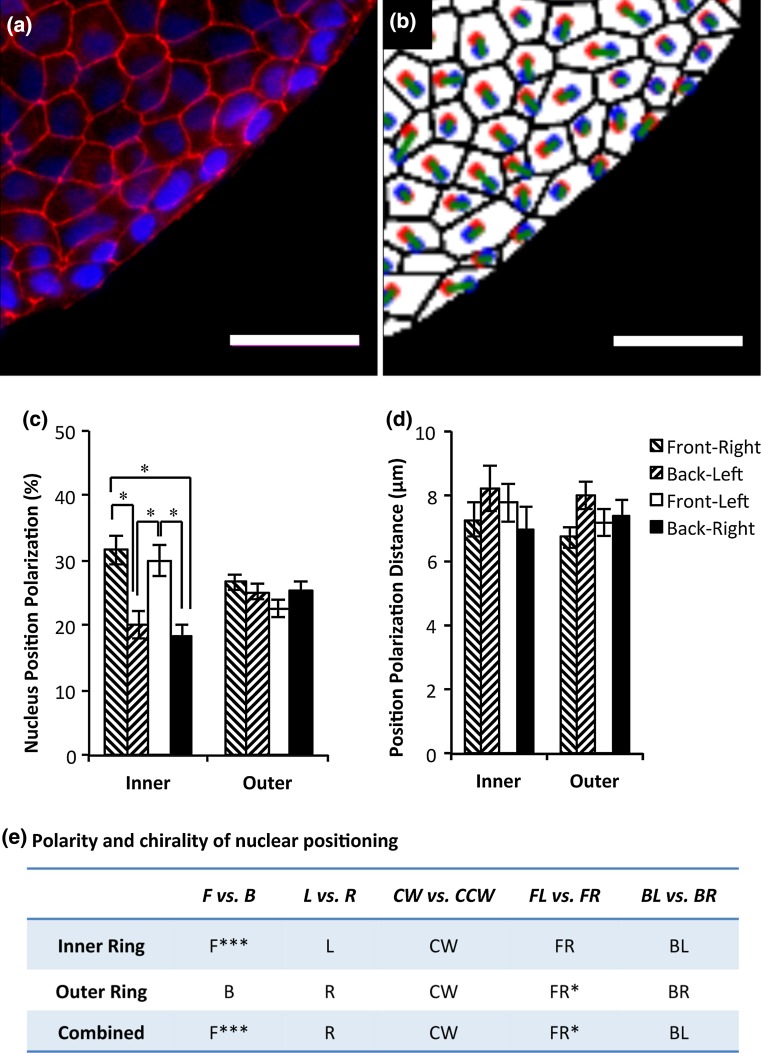
We next investigated the directional bias in nuclear positing within a cell. We first determined the centroids of cells and their nuclei as shown in Figs. 4a and 4b. The nuclear positioning was defined as a vector from cell centroid to its nuclear centroid, and the angle of this vector was determined as the biased angle for nuclear positioning and characterized similarly as in Figs. 1b and 3a. 61.6% of nuclei oriented themselves towards boundaries (FR+FL) at the inner ring, while at the outer ring the nuclei were more randomly polarized (relative to cell centroid) with 49.3% towards boundaries (FR+FL) (Fig. 4c). Cell nuclei taking on a BL position had the largest polarization distances at both the inner and outer border (7.79 ± 0.70 µm and 8.02 ± 0.45 µm, respectively; Fig. 4d), although no significant difference between any two quadrants was detected. We also generated heat maps to visualize the relationships between nuclear positioning polarization and cell and nucleus shape polarization angles with heat maps (Fig. S3). Further analysis showed that there was a bias, though not significant, in nuclear positioning towards CW alignments (Fig. 4e). A trend of rightward bias in polarization was also observed. Interestingly, there was also a bias towards FR, significantly at the outer ring (p < 0.05). When the biases were calculated for individual rings in Table S1, to determine chiral biases of nuclear positioning, among the four chiral measures, only the measure FL vs. FR was able to detect a significant chiral bias (towards FR) of patterned cellular rings.
Figure 4.
Nuclear positioning polarization. Scale bars: 50 μm. (a) ZO-1 (red) and nuclei (blue) fluorescence images. (b) Nuclear positioning polarization, defined as a vector from cell centroid to nucleus centroid, was determined from image analysis. (red: cell centroids, blue: nucleus centroids, green: nucleus positioning vector connecting two centroids with a cell). (c) Analysis of nuclear positioning polarization angles. (d) Analysis of nuclear positioning distances. (e) Determining polarity and chirality of nuclear positioning inside the cells at the patterned boundaries. * Significantly different at p < 0.05 and ** Significantly different at p < 0.01.
Discussion
The objective of this study was to investigate the possibility of using cell and nuclear polarity in multiple axes to determine the chirality of individual cells and nuclei in a geometrically defined multicellular structure. Using a combined approach of cell patterning, fluorescence straining, and advanced image processing, we showed that cell shape, nuclear shape, and nuclear positioning inside the cell were chirally polarized, consistent with directional cell migration on the boundaries. While the cells could clearly sense and polarize themselves with the sharp end towards the boundary, the nuclear shape didn’t have an apparent preference. The nucleus was pushed towards the rear end of the cell, resulting in an opposite chirality in nuclear positioning. Taken together, these data suggest that epithelial chiral morphogenesis can be characterized as the coordination of multiaxial polarization at the cellular and subcellular levels.
Cell chirality is the coordination of multiaxial polarity. Chirality is known as handedness, or LR asymmetry, and recently recognized as a fundamental intrinsic property of the cell.5,12,36,47,51,52 From a physics point of view, chirality requires the coordination of multiple axes.47,52 One example is the embryonic development, which has three main axes of polarity (anterior-posterior, ventral-dorsal and left–right). Loss or disturbance of the cross-talk between these axes can lead to severe malformations.22,23,26,31,39 The other example is the Cartesian coordinates, which are typically right handed. The directionality in LR axis (say y axis) is completely defined after the establishment of the bottom-top (z axis) and back-front (x) axes. On our micropatterned ring-shaped surfaces, with cell attachment defining the z axis and patterned microscale boundaries defining the back-front (BF) x-axis, the cells on boundaries adopted a consistent dominant bias towards either CW or CCW, depending the cell phenotype.47 A loss of chiral alignment could be due to disturbed chiral machinery, but it also could be caused by a loss in polarity along the BF axis. Therefore, it is very important to study how the polarity in different axes determines cell chirality. From Fig. 3b and Table 1, we can see that the cells had a stronger bias towards FL rather than BR, although they are both considered as the CCW alignment. In addition, 68% of the cells on the inner ring and 64% on the outer adopting a front bias (i.e., FL+FR) have a significant bias towards the left side (i.e., FL), while only 50% cells on the inner ring and 57% cells on the outer ring adopting a back bias [i.e., BL+BR)] have a bias towards the right side. This suggests that the front-biased cells have a higher chance to take the correct chirality. To our knowledge, this is the first data revealing that the tight control of BF polarity has a significant influence on the chirality observed on micropatterned surfaces.
The back-front (BF) axis control is critical for chiral morphogenesis. Epithelial cells can polarize and form different structures such as planar sheets and epithelial tubes.1,3,27,28,34 Previously it was demonstrated that the cell tends to position its centrosome and Golgi apparatus closer to the boundary than its nucleus.47 In this study, we demonstrated that cell shape is also polarized with the sharp end towards the boundary. At least two mechanisms could potentially contribute to the establishment of the BF axis: 1) the difference between cell adhesive and non-adhesive regions controlling polarization on boundaries through cell active protrusion and strong cell-substrate adhesion,47 and 2) cell–cell adhesion that further propagates the BF axis to the interior region near the boundaries.8,51
As the BF axis control largely relies on the boundary, we expect that the chiral phenomena will be mostly observed in the vicinity of boundaries of the ring. Indeed, the boundary region based analysis for cells shows that 71% of rings (27 out of 38; Table 2B), rather than 58% seen in whole ring analysis (22 out of 38), have a CCW bias. For nuclei, LR analysis demonstrates that 60% have a leftward bias, which corresponds to the CCW bias, compared to the 50% seen in the whole ring alignment analysis. Therefore, boundary region based analysis may be a better choice for determining chirality in cell culture.
Cell shape polarization was demonstrated to switch directions from the inner to the outer boundaries in Fig. 2b. Previously, we also found a reversal in cell migration was also reported on two opposite boundaries.47,51 These findings further demonstrated that the boundary is critical for establishing chirality in cell culture, and suggested that cell shape polarization and migration direction may be closely related.
In this study, we define the sharp end of the cell as the leading edge, rather than the large, blunt-end used in the traditional definition for migration (Fig. 2e).21,30,46,48 This is because in multicellular culture, the cells need to wedge their way between each other in order to migrate, just like neutrophil transendothelial migration and extravasation.10,15,20,43 With this definition, shape polarization direction is consistent with the reported migration directions. The sharp end of the cell at the leading edge pulls forward the bulk of cell body, which is drawn back by cell–cell adhesion and cell matrix adhesion. Therefore, the analysis of cell shape polarization was able to predict the direction of cell migration from a still image without tedious time-lapse imaging. In particular, in the future, we will further incorporate the labeling of other organelles such as centrosomes to examine the relationship between cell polarity, cell shape and cell chirality in multicellular patterns with fluorescence live cell imaging.
Nuclear shape polarization is towards the left side in the inner ring, not specific to FL or BL (Fig. 4c). This is different from cell shape, which is strongly biased to the left side as well as the front side, creating the strong CCW alignment with a front bias (Fig. 4b). Specifically, nuclei did not have a strong bias towards the front or the back (Table 1, F vs. B), but no matter which direction the nuclei took on in the BF axis, they were always biased to the left side (Table 1, FL vs. FB & BL vs. BR). Also, as a result of the nature underlying nuclear shape polarization, the comparison between left and right side (L vs. R) will be a much better marker to assess the chiral bias, rather than FL + BR vs. FR + BL (Table 2). These findings suggest that the nuclear shape is not polarized based on the cellular front-back axis, and that the nuclear shape chirality is probably directly derived from the chiral alignment of the cell.
It is widely reported that the nucleus tends to align with the cell body. On microscale or nanoscale grooves, cell nuclei follow the groove direction but not as closely as the cells do.7,11,13,50 The nuclear deformation mediated by cell shape is through aligned actin filaments and regulated by actomyosin contractility.18,25,40 Both the dome-like actin structure on the top of nuclear membrane and the lateral tensed actin filaments surrounding the nucleus have been reported to be responsible for the nuclear alignment. The cytoskeleton such as actin filaments is mechanically linked to the nucleus through LINC complex.6,24 In this complex, the SUN-domain proteins cross the inner nuclear membrane and bind to nuclear lamina. At the other end, they are also connected to the KASH-domain containing Nesprin family of proteins that cross the outer nuclear membrane. Outside of the nuclear membrane, the Nesprin proteins can bind to actin filaments or through plectin to intermediate filaments. In this study, as the cell migrates towards the left with a sharp end as the leading edge, actin filaments aligned from the leading edge to the rear end generate the normal and lateral compression on the nucleus. As the leading edge is confined by the actin filaments, the nucleus appears smaller or elongated. Therefore, the observed nuclear alignment and polarization are possibly caused by aligned actin filaments in the chirally aligned cells.
The BF polarity of nuclear positioning, differently from cell shape polarization, is not determined relative to the boundary. At the inner ring, the nucleus tends to position closer to the boundary, relative to the centroid of the cell, while at the outer ring, it is far away from the boundary (Fig. 4c–Fig. 4e). This difference between the inner and outer boundary can be explained by curvature sensation of the cells and nuclei.42 On the ring patterns, the actin filaments tend to align in the circumferential direction, pushing the nuclei towards the center of the ring. For the concave adhesive surfaces on the inner ring, it is towards the boundary while for the convex surfaces on the outer ring, it is away from the boundary.
The chiral nuclear positioning is possibly regulated by cellular cytoskeleton. We found that the nuclei were positioned towards FR and BL, which are in the opposite circumferential direction of cellular shape polarization (towards FL and BR instead). It is reasonable since many previous studies have also implicated that the nucleus positions itself away from the leading edge in cell migration,9,19,30,41 possibly through the regulation of connections between the nucleus and the cytoskeleton.17 Therefore the cell migrating leftward will have its nucleus positioned towards the right in the cell, and vice visa. Taken together, the polarity and chirality of nuclear positioning is largely associated with cell migration and its related cytoskeleton organization.
There are a few limitations of this study. First, for simplicity, we have arbitrarily divided the ring into five zones in the radial direction and categorized the direction into the four quadrants. Although it serves the purpose of the current study, the use of more sophisticated statistical analyses without such categorization might be necessary for other studies. Second, the new measures and insights of multicellular chirality revealed by our analyses need to be further validated through fluorescence live imaging using multiple cell types under pharmacological and genetic manipulations in the future. Finally, the chirality of individual cells determined in multicellular chiral patterns should be further examined in the future with putative biomarkers associated with cell chirality such as α-actinin-1.37
In summary, the chirality of multicellular morphogenesis was characterized in great detail with the multi-axial polarity analysis of individual cells on micro-patterned surfaces. On the boundary, cells sense the boundary and adopt a LR bias, while their nuclei align with them accordingly. On a confluent cell layer, the cells move forward by positioning the sharp end at the leading edge and their nuclei at the rear, resulting an opposite chirality of nuclear positioning. The individual cell-based multiaxial polarization analysis provides insights into biophysical mechanisms of epithelial multicellular chiral morphogenesis and therefore can potentially benefit the in-depth analysis of birth defects in laterality.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank Parker Haynes for his help on Python coding. The authors would like to thank National Institutes of Health, National Science Foundation, American Heart Association, and March of Dimes for funding Support. Leo Q. Wan is a Pew Scholar in Biomedical Sciences, supported by the Pew Charitable Trusts.
Conflict of interest
All authors, Michael J. Raymond, Poulomi Ray, Gurleen Kaur, Michael Fredericks, Ajay V. Singh, and Leo Q. Wan, declare that they have no conflict of interest.
Statements of Human and Animal Rights and Informed Consent
No human or animal research was conducted in this study.
Abbreviations
- CW
Clockwise
- CCW
Counterclockwise
- NC
Non-chiral
References
- 1.Andrew DJ, Ewald AJ. Morphogenesis of epithelial tubes: insights into tube formation, elongation, and elaboration. Dev. Biol. 2010;341:34–55. doi: 10.1016/j.ydbio.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aylsworth AS. Clinical aspects of defects in the determination of laterality. Am. J. Med. Genet. 2001;101:345–355. doi: 10.1002/ajmg.1219. [DOI] [PubMed] [Google Scholar]
- 3.Balcarova-Stander J, Pfeiffer SE, Fuller SD, Simons K. Development of cell surface polarity in the epithelial Madin–Darby canine kidney (MDCK) cell line. EMBO J. 1984;3:2687–2694. doi: 10.1002/j.1460-2075.1984.tb02194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callander DC, Alcorn MR, Birsoy B, Rothman JH. Natural reversal of left-right gut/gonad asymmetry in C. elegans males is independent of embryonic chirality. Genesis. 2014;52:581–587. doi: 10.1002/dvg.22762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen TH, Hsu JJ, Zhao X, Guo C, Wong MN, Huang Y, Li Z, Garfinkel A, Ho CM, Tintut Y, Demer LL. Left-right symmetry breaking in tissue morphogenesis via cytoskeletal mechanics. Circ. Res. 2012;110:551–559. doi: 10.1161/CIRCRESAHA.111.255927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahl KN, Ribeiro AJ, Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalby MJ, Riehle MO, Yarwood SJ, Wilkinson CD, Curtis AS. Nucleus alignment and cell signaling in fibroblasts: response to a micro-grooved topography. Exp. Cell Res. 2003;284:274–282. doi: 10.1016/S0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 8.Desai RA, Gao L, Raghavan S, Liu WF, Chen CS. Cell polarity triggered by cell-cell adhesion via E-cadherin. J. Cell Sci. 2009;122:905–911. doi: 10.1242/jcs.028183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupin I, Camand E, Etienne-Manneville S. Classical cadherins control nucleus and centrosome position and cell polarity. J. Cell Biol. 2009;185:779–786. doi: 10.1083/jcb.200812034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelhardt B, Wolburg H. Mini-review: transendothelial migration of leukocytes: through the front door or around the side of the house? Eur. J. Immunol. 2004;34:2955–2963. doi: 10.1002/eji.200425327. [DOI] [PubMed] [Google Scholar]
- 11.Freytes DO, Wan LQ, Vunjak-Novakovic G. Geometry and force control of cell function. J. Cell. Biochem. 2009;108:1047–1058. doi: 10.1002/jcb.22355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatori R, Ando T, Sasamura T, Nakazawa N, Nakamura M, Taniguchi K, Hozumi S, Kikuta J, Ishii M, Matsuno K. Left-right asymmetry is formed in individual cells by intrinsic cell chirality. Mech. Dev. 2014;133:146–162. doi: 10.1016/j.mod.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Itano N, Okamoto S, Zhang D, Lipton SA, Ruoslahti E. Cell spreading controls endoplasmic and nuclear calcium: a physical gene regulation pathway from the cell surface to the nucleus. Proc. Natl. Acad. Sci. USA. 2003;100:5181–5186. doi: 10.1073/pnas.0531397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang X, Bruzewicz DA, Wong AP, Piel M, Whitesides GM. Directing cell migration with asymmetric micropatterns. Proc. Natl. Acad. Sci. USA. 2005;102:975–978. doi: 10.1073/pnas.0408954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson-Leger C, Aurrand-Lions M, Imhof BA. The parting of the endothelium: miracle, or simply a junctional affair? J. Cell Sci. 2000;113:921–933. doi: 10.1242/jcs.113.6.921. [DOI] [PubMed] [Google Scholar]
- 16.Karlon WJ, Hsu PP, Li S, Chien S, McCulloch AD, Omens JH. Measurement of orientation and distribution of cellular alignment and cytoskeletal organization. Ann. Biomed. Eng. 1999;27:712–720. doi: 10.1114/1.226. [DOI] [PubMed] [Google Scholar]
- 17.Khatau SB, Bloom RJ, Bajpai S, Razafsky D, Zang S, Giri A, Wu P-H, Marchand J, Celedon A, Hale CM. The distinct roles of the nucleus and nucleus-cytoskeleton connections in three-dimensional cell migration. Sci. Rep. 2012;2:488. doi: 10.1038/srep00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. USA. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D-H, Cho S, Wirtz D. Tight coupling between nucleus and cell migration through the perinuclear actin cap. J. Cell Sci. 2014;127:2528–2541. doi: 10.1242/jcs.144345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kvietys PR, Sandig M. Neutrophil diapedesis: paracellular or transcellular? News Physiol. Sci. 2001;16:15–19. doi: 10.1152/physiologyonline.2001.16.1.15. [DOI] [PubMed] [Google Scholar]
- 21.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/S0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 22.Levin M. Left-right asymmetry in embryonic development: a comprehensive review. Mech. Dev. 2005;122:3–25. doi: 10.1016/j.mod.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Levin M, Thorlin T, Robinson KR, Nogi T, Mercola M. Asymmetries in H +/K + -ATPase and cell membrane potentials comprise a very early step in left-right patterning. Cell. 2002;111:77–89. doi: 10.1016/S0092-8674(02)00939-X. [DOI] [PubMed] [Google Scholar]
- 24.Lovett DB, Shekhar N, Nickerson JA, Roux KJ, Lele TP. Modulation of Nuclear Shape by Substrate Rigidity. Cell. Mol. Bioeng. 2013;6:230–238. doi: 10.1007/s12195-013-0270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mercola M, Levin M. Left-right asymmetry determination in vertebrates. Annu. Rev. Cell Dev. Biol. 2001;17:779–805. doi: 10.1146/annurev.cellbio.17.1.779. [DOI] [PubMed] [Google Scholar]
- 27.Nakaya Y, Sheng G. EMT in developmental morphogenesis. Cancer Lett. 2013;341:9–15. doi: 10.1016/j.canlet.2013.02.037. [DOI] [PubMed] [Google Scholar]
- 28.Okada Y, Takeda S, Tanaka Y, Izpisua JC. Belmonte and N. Hirokawa. Mechanism of nodal flow: a conserved symmetry breaking event in left-right axis determination. Cell. 2005;121:633–644. doi: 10.1016/j.cell.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Raymond MJ, Jr, Ray P, Kaur G, Singh AV, Wan LQ. Cellular and nuclear alignment analysis for determining epithelial cell chirality. Ann. Biomed. Eng. 2015;44(5):1475–1486. doi: 10.1007/s10439-015-1431-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 31.Roychoudhuri R, Putcha V, Moller H. Cancer and laterality: a study of the five major paired organs (UK) Cancer Cause. Control. 2006;17:655–662. doi: 10.1007/s10552-005-0615-9. [DOI] [PubMed] [Google Scholar]
- 32.Sandson TA, Wen PY, LeMay M. Reversed cerebral asymmetry in women with breast cancer. Lancet. 1992;339:523–524. doi: 10.1016/0140-6736(92)90341-Y. [DOI] [PubMed] [Google Scholar]
- 33.Shibazaki Y, Shimizu M, Kuroda R. Body handedness is directed by genetically determined cytoskeletal dynamics in the early embryo. Curr. Biol. 2004;14:1462–1467. doi: 10.1016/j.cub.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 34.Simons K, Fuller SD. Cell surface polarity in epithelia. Annu. Rev. Cell Biol. 1985;1:243–288. doi: 10.1146/annurev.cb.01.110185.001331. [DOI] [PubMed] [Google Scholar]
- 35.Singh AV, Mehta KK, Worley K, Dordick JS, Kane RS, Wan LQ. Carbon nanotube-induced loss of multicellular chirality on micropatterned substrate is mediated by oxidative stress. ACS Nano. 2014;8:2196–2205. doi: 10.1021/nn405253d. [DOI] [PubMed] [Google Scholar]
- 36.Taniguchi K, Maeda R, Ando T, Okumura T, Nakazawa N, Hatori R, Nakamura M, Hozumi S, Fujiwara H, Matsuno K. Chirality in planar cell shape contributes to left-right asymmetric epithelial morphogenesis. Science. 2011;333:339–341. doi: 10.1126/science.1200940. [DOI] [PubMed] [Google Scholar]
- 37.Tee YH, Shemesh T, Thiagarajan V, Hariadi RF, Anderson KL, Page C, Volkmann N, Hanein D, Sivaramakrishnan S, Kozlov MM, Bershadsky AD. Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat. Cell Biol. 2015;17:445–457. doi: 10.1038/ncb3137. [DOI] [PubMed] [Google Scholar]
- 38.Thery M, Racine V, Piel M, Pepin A, Dimitrov A, Chen Y, Sibarita JB, Bornens M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci. USA. 2006;103:19771–19776. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandenberg LN, Levin M. A unified model for left–right asymmetry? Comparison and synthesis of molecular models of embryonic laterality. Dev. Biol. 2013;379:1–15. doi: 10.1016/j.ydbio.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Versaevel M, Grevesse T, Gabriele S. Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat. Commun. 2012;3:671. doi: 10.1038/ncomms1668. [DOI] [PubMed] [Google Scholar]
- 41.Vicente-Manzanares M, Webb DJ, Horwitz AR. Cell migration at a glance. J. Cell Sci. 2005;118:4917–4919. doi: 10.1242/jcs.02662. [DOI] [PubMed] [Google Scholar]
- 42.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 43.Wagner JG, Roth RA. Neutrophil migration mechanisms, with an emphasis on the pulmonary vasculature. Pharmacol. Rev. 2000;52:349–374. [PubMed] [Google Scholar]
- 44.Wakida NM, Botvinick EL, Lin J, Berns MW. An intact centrosome is required for the maintenance of polarization during directional cell migration. PLoS One. 2010;5:e15462. doi: 10.1371/journal.pone.0015462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wan LQ, Kang SM, Eng G, Grayson WL, Lu XL, Huo B, Gimble J, Guo XE, Mow VC, Vunjak-Novakovic G. Geometric control of human stem cell morphology and differentiation. Integr. Biol. 2010;2:346–353. doi: 10.1039/c0ib00016g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wan LQ, Ronaldson K, Guirguis M, Vunjak-Novakovic G. Micropatterning of cells reveals chiral morphogenesis. Stem Cell Res. Ther. 2013;4:24. doi: 10.1186/scrt172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan LQ, Ronaldson K, Park M, Taylor G, Zhang Y, Gimble JM, Vunjak-Novakovic G. Micropatterned mammalian cells exhibit phenotype-specific left-right asymmetry. Proc. Natl. Acad. Sci. USA. 2011;108:12295–12300. doi: 10.1073/pnas.1103834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wan LQ, Vunjak-Novakovic G. Micropatterning chiral morphogenesis. Commun. Integr. Biol. 2011;4:745–748. doi: 10.4161/cib.17649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weber GF, Bjerke MA, DeSimone DW. A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration. Dev. Cell. 2012;22:104–115. doi: 10.1016/j.devcel.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Worley K, Certo A, Wan LQ. Geometry-force control of stem cell fate. BioNanoScience. 2013;3:43–51. doi: 10.1007/s12668-012-0067-0. [DOI] [Google Scholar]
- 51.Worley KE, Shieh D, Wan LQ. Inhibition of cell-cell adhesion impairs directional epithelial migration on micropatterned surfaces. Integr. Biol. 2015;7(5):580–590. doi: 10.1039/C5IB00073D. [DOI] [PubMed] [Google Scholar]
- 52.Xu J, Van Keymeulen A, Wakida NM, Carlton P, Berns MW, Bourne HR. Polarity reveals intrinsic cell chirality. Proc. Natl. Acad. Sci. USA. 2007;104:9296–9300. doi: 10.1073/pnas.0703153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamanaka H, Kondo S. Rotating pigment cells exhibit an intrinsic chirality. Genes Cells. 2015;20:29–35. doi: 10.1111/gtc.12194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.