Abstract
DNA double strand breaks (DSBs) are dangerous sources of genome instability and must be repaired by the cell. Nonhomologous end joining (NHEJ) is an evolutionarily conserved pathway to repair DSBs by direct ligation of the ends, with no requirement for a homologous template. While NHEJ is the primary DSB repair pathway in mammalian cells, conservation of the core NHEJ factors throughout eukaryotes make the pathway attractive for study in model organisms. The budding yeast, Saccharomyces cerevisiae, has been used extensively to develop a functional picture of NHEJ. In this review, we will discuss the current understanding of NHEJ in S. cerevisiae. Topics include canonical end-joining, alternative end-joining, and pathway regulation. Particular attention will be paid to the NHEJ mechanism involving core factors, including Yku70/80, Dnl4, Lif1, and Nej1, as well as the various factors implicated in the processing of the broken ends. The relevance of chromatin dynamics to NHEJ will also be discussed. This review illustrates the use of S. cerevisiae as a powerful system to understand the principles of NHEJ, as well as in pioneering the direction of the field.
Keywords: nonhomologous end-joining, Ku heterodimer, DNA ligase IV, double strand break repair, end-processing
DNA double strand breaks (DSBs) are extremely hazardous cellular lesions. Failure to repair even a single DSB can lead to cell death, whereas misrepair can introduce catastrophic chromosomal translocations (Malkova and Haber 2012). Defects in DSB repair pathways are implicated in cancer and a number of congenital disorders, including primary immunodeficiency diseases (McKinnon and Caldecott 2007; Pierce et al. 2001; Prochazkova and Loizou 2015; Rulten and Caldecott 2013).
DSB repair can occur via one of several pathways. A subset of these pathways relies on processing of 5′ DSB ends to reveal tracts of homology. These resection based pathways are referred to as homology directed repair (HDR) and have been reviewed recently elsewhere (Kowalczykowski 2015). The second major pathway of DSB repair is nonhomologous end-joining (NHEJ), in which DSB ends are ligated directly. The protein machinery of NHEJ is well-conserved throughout eukaryotes, and an analogous process is found in prokaryotes. This review focuses on NHEJ in the yeast Saccharomyces cerevisiae, a major model organism of NHEJ research. Although NHEJ is less efficient than HDR in S. cerevisiae, it is relatively easy to enrich for and study NHEJ by deletion of HDR genes (Kramer et al. 1994) or deletion of homologous templates (Schiestl et al. 1993). Thus, S. cerevisiae is a tractable model to derive insight into the general principles of NHEJ. Indeed, several NHEJ factors and processes were first characterized in S. cerevisiae, underscoring the organism’s significance in the field of DSB repair. Large scale screens in yeast continue to identify new genes that influence NHEJ (Jessulat et al. 2015; McKinney et al. 2013). Additionally, because aberrant NHEJ is a major source of chromosomal disruption in budding yeast (Yu and Gabriel 2004), like mammalian cells (Lieber et al. 2010), S. cerevisiae provides an appealing model to study the implications of NHEJ defects in human genome instability. This review will use S. cerevisiae as the context for discussion of NHEJ substrates, regulation, factors, and mechanism.
Causes of DSBs
Programmed DSBs are induced during meiosis, mating type switching in yeast, and V(D)J recombination and IgH class switch recombination in lymphoid cells (Haber 2012; Lam and Keeney 2015; Prochazkova and Loizou 2015). Outside of these contexts, causes of DNA DSBs can be classified into two broad categories: 1) toxic exposures and 2) failure of endogenous processes. Toxic exposures that generate DSBs include ionizing radiation, reactive oxygen species, and chemicals like bleomycin and camptothecin. Defects in endogenous processes, such DNA replication and mitosis can also result in DSBs. Replication fork collapse can result in the generation of single-ended DSBs. The physical stress of mitosis can pull apart chromosomes, producing DSBs (Mathieu et al. 2004). In the case of telomeres, loss of the protective telomere cap causes the natural chromosome end to be sensed as a DSB and acted upon as such. Additionally, inappropriate function of DNA enzymes, including endonucleases and topoisomerases, may be a source of endogenous breaks.
Sources of DSBs are important to consider because they influence the proficiency and error-rate of NHEJ. For example, while restriction endonucleases generate “clean” compatible ends that may be directly ligated by NHEJ, ionizing radiation creates complex lesions, or “dirty” ends, which require processing prior to end-joining (Hill 1999). In S. cerevisiae, deprotected telomeres may either be engaged in HDR pathways (Le et al. 1999) or subjected to NHEJ-mediated chromosome fusions (Marcand et al. 2008). In contrast, single-ended DSBs generated by collapsed replication forks make poor substrates for NHEJ (Michel et al. 2004) and are instead repaired by limited use of error-prone HDR prior to engagement by a converging replication fork (Mayle et al. 2015).
Repair Products and Errors
As mentioned above, restriction enzymes generate compatible ends which require no or minimal processing before repair. Ligation of these ends together to accurately restore the original genetic sequence is called precise repair (Fig. 1). In contrast, imprecise repair occurs when bases are lost or inserted at the repair junction. Subsets of imprecise repair pathways are apparent in yeast, as shown by the occurrence of only inserted or deleted base pairs (bp) at the repair scar when specific factors, such as the MRX complex and end-processing polymerases Pol4 and Pol2, are absent (Moore and Haber 1996; Tseng et al. 2008).
Figure 1.
Subtypes of end-joining repair depicted by repair of an HO endonuclease-induced DSB, a commonly used system in yeast genetics. Cleavage by HO results in a 4 nucleotide 3′ overhang at the DSB site. The c-NHEJ repair pathway requires both Yku and Dnl4, and can result in either precise or imprecise repair events. Precise repair results in rejoining of the complementary ends without sequence loss or gain. Imprecise repair results in the rejoining of ends with small deletions or insertions (e.g., bolded residues). Microhomology mediated end-joining (MMEJ) does not require c-NHEJ factors and results in larger deletions (e.g., bolded residues) that expose microhomologies for strand annealing and repair.
Traditionally, NHEJ has been considered an error-prone process, especially compared to the HDR pathways. However, while HDR’s fidelity has been called into question (Malkova and Haber 2012; Rodgers and McVey 2016), the error-prone nature of classical NHEJ repair is also being revisited (Betermier et al. 2014). Chromosomal break and plasmid repair assays both demonstrate that imprecise NHEJ repair of simple breaks is rare in yeast (1 in 103 – 104), and that precise repair is by far the most common occurrence (Bahmed et al. 2010; Daley and Wilson 2005; Moore and Haber 1996). A principle is emerging in which NHEJ is not inherently error-prone, but that the condition of DSB ends before repair is the determining factor in repair precision (Betermier et al. 2014).
Alt-NHEJ and MMEJ
While the focus of this review is classical (or canonical) NHEJ, abbreviated c-NHEJ, there has been considerable interest in alternative end-joining pathways (alt-NHEJ). C-NHEJ is defined by a dependence on core factors, Dnl4 and the Ku heterodimer (Yku), whereas alt-NHEJ is loosely defined as the homology-independent repair events observed in the absence of these factors (Ma et al. 2003) (Fig. 1). Repair products of c-NHEJ, excepting precise repair, exhibit insertions or deletions of 5 or less bp, while the products of microhomology mediated end-joining (MMEJ), a subset of alt-NHEJ, are distinguished by deletions of 5–20 bp (Lee and Lee 2007; Ma et al. 2003).
It is unclear whether alt-NHEJ in S. cerevisiae is mediated by a dedicated pathway, or whether it is simply an extension of c-NHEJ where some factors substitute for the loss of others, completing repair at the cost of larger deletions or other errors at the repair site. In human cells, a strong argument has been made for a dedicated alt-NHEJ pathway (Bennardo et al. 2008); however, in yeast, the issue is less clear (Chiruvella et al. 2013b). Several mechanisms of alt-NHEJ likely exist in yeast. Although there is agreement that alt-NHEJ requires the MRX complex (Lee and Lee 2007; Ma et al. 2003; Paull and Gellert 2000), there are contradictory findings concerning the dependency of MMEJ on factors such as Dnl4 and Pol4 (Galli et al. 2015; Lee and Lee 2007; Ma et al. 2003; Meyer et al. 2015). Additionally, some alt-NHEJ mechanisms are likely disguised by c-NHEJ activity, if the repair products are indistinguishable from those of c-NHEJ. For example, creation of a catalytically dead Dnl4 mutant revealed an imprecise repair pathway that did not fit neatly into either c-NHEJ or MMEJ categories (Chiruvella et al. 2013a). Future work to understand these pathways will be necessary, as it is now accepted that MMEJ repairs ionizing radiation damage in both yeast and mammalian cells (Scuric et al. 2009).
NHEJ Regulation
Repair pathway choice in S. cerevisiae is regulated by ploidy and cell cycle phase. Nej1 is a core NHEJ factor with haploid specific expression (Frank-Vaillant and Marcand 2001; Kegel et al. 2001; Valencia et al. 2001). In the absence of targeted modification, haploid yeast are either MATa or MATα, while diploids are MATa/MATα (Haber 2012). In diploids, the a1-α2 repressor, with its subunits encoded by MATa and MATα, respectively, inhibits expression of a set of haploid-specific proteins, including Nej1. As Nej1 is required for NHEJ, this results in suppression of NHEJ activity. This haploid-specific regulation is unique to yeast and is not found in mammals, where NHEJ predominates throughout the cell cycle except in mid S phase (Karanam et al. 2012). While haploid-specific expression of Nej1 promotes HDR in the diploid yeast where a homologous donor chromosome is present, HDR is also tightly regulated by cell cycle phase.
In haploids, pathway choice is primarily determined by resection activity, as 5′ to 3′ processing of the DSB end inhibits NHEJ and directs the DSB to HDR. Resection is the regulatory decision point in both yeast and mammalian cells, and more comprehensive reviews are recommended for this topic (Daley et al. 2015; Symington 2014; Symington and Gautier 2011). In brief, resection is inhibited in G1 by inactivation of Cdk1, also called Cdc28 (Aylon and Kupiec 2005; Ira et al. 2004). The magnitude of Cdk1’s regulatory importance is illustrated by the finding that loss of Cdk1 activity in G2 arrested cells extinguished resection (Ira et al. 2004). The role of Cdk1 in initiating resection is not completely understood, although roles for the kinase in promoting late stage resection have been revealed (Chen et al. 2011). MRX nuclease activity is critical to the early stages of short range resection, downstream of Cdk1 (Mimitou and Symington 2008). Interestingly, the MRX complex promotes both NHEJ and resection activity, a curiosity which will be discussed in detail below. MRX activity commits repair substrates to repair by HDR pathways and recruits the long range resection factors Dna2, Exo1, and Sgs1 (Shim et al. 2010; Zhu et al. 2008).
To summarize, NHEJ is promoted in G1 haploid cells by the suppression of HDR pathways via cell-cycle dependent inhibition of resection. Thus, resection and HDR are restricted to S/G2 phases. Resection degrades double stranded DNA at break ends, committing the processed single strand to a homology search and recombination-dependent repair. Yku also promotes NHEJ by inhibiting resection activity (Mimitou and Symington 2010), a role which will be discussed shortly. A model of pathway determination by regulation of resection, rather than direct inhibition of NHEJ machinery, is also supported by the observation of NHEJ activity in S/G2, where NHEJ is permitted if the repair activity is completed before resection initiation (Frank-Vaillant and Marcand 2002).
Yeast Core NHEJ Proteins
The core protein activity and overall mechanism of NHEJ show considerable conservation across eukaryotes. The core NHEJ factors in yeast are Yku70-Yku80, Dnl4-Lif1, Nej1, and the Mre11-Rad50-Xrs2 complex. The mammalian homologs of these are Ku70-Ku86, LIG4-XRCC4, Cernunnos (XLF), and Mre11-Rad50-Nbs1, respectively. Whereas the Mre11-Rad50-Nbs1 complex is not a core NHEJ factor in vertebrates, DNA-PKcs, a catalytic subunit which binds DNA-bound Ku to form the DNA-dependent protein kinase (DNA-PK), which yeast lack, is (Ceccaldi et al. 2016). The absence of DNA-PKcs in species such as yeast suggests that other factors are able to compensate for the function(s) of DNA-PKcs, such as end bridging by the MRX complex discussed below.
Yku
The Ku protein is a heterodimer encoded by copies of an ancestral gene found in prokaryotes (Doherty et al. 2001). In S. cerevisiae, the protein is composed of the Yku70 and Yku80 subunits and is often referred to as Yku. The Ku subunits combine to form a conserved β-barrel ring structure that allows the complex to avidly bind DNA ends in a sequence independent manner (Walker et al. 2001). Double stranded DNA ends are the preferred binding substrate of Ku, but Ku can also engage single stranded DNA in a limited capacity (Krasner et al. 2015). Ku functions in multiple cellular roles beside DSB repair, including telomere maintenance and nuclear organization (Fell and Schild-Poulter 2015). Whereas Yku is a nonessential protein in yeast, loss of human Ku is lethal to mammalian cells because of telomere function deficits (Wang et al. 2009).
Ku’s NHEJ activity is genetically separable from its roles at telomeres, where it also engages DNA ends (Bertuch and Lundblad 2003; Driller et al. 2000; Lopez et al. 2011; Ribes-Zamora et al. 2007; Roy et al. 2004; Stellwagen et al. 2003), The localization of the various separation-of-function mutations that have been identified combined with the polarity with which the quasi-symmetrical Ku heterodimer loads onto DNA ends have led to the proposal of a “two-face” model of Yku function wherein the Yku80 loading face mediates telomere functions, while the Yku70 surface, which is oriented toward the DSB, mediates NHEJ activity (Ribes-Zamora et al. 2007). In support of this model, mutations of the Yku80 α-helix result in telomeric silencing defects, while mutations of the related Yku70 α-helix result in NHEJ defects (Ribes-Zamora et al. 2007). Notably, mutations of the Yku80 C-terminus specifically impair NHEJ (Palmbos et al. 2005) and an intact Yku70 C-terminus is required for telomeric functions (Driller et al. 2000). To date, a crystal structure is only available for human Ku and does not account for the C-termini (Walker et al. 2001). It seems likely that the C-termini of Yku70 and Yku80 wrap around the heterodimer structure, so that the Yku80 C-terminus is oriented towards the DSB end and the Yku70 C-terminus faces inward on the DNA strand.
Inhibition of resection by Ku is the first step in a successful NHEJ reaction. Upon formation of a DSB, Yku rapidly associates with the break, presumably by loading onto the broken DNA end, blocking resection and preventing formation of the long tracts of single-stranded DNA necessary for HDR (Clerici et al. 2008; Mimitou and Symington 2010). Because of the importance of Ku’s initial binding activity, much attention has been paid to the interplay between Ku and the MRX complex at the nascent DSB. Both complexes act as “first responders,” by rapidly and independently associating with the DSB site (Wu et al. 2008). Evidence suggests that Ku and MRX act antagonistically at the end (Clerici et al. 2008; Wu et al. 2008). Loss of Yku results in an MRX dependent increase of 5′ end degradation in G1 and overexpression of Yku in G2 results in decreased MRX loading at the site of a DSB (Clerici et al. 2008). Furthermore, the MRX complex appears to play a role in removing Yku from unrepaired DSB ends (Balestrini et al. 2013; Wasko et al. 2009; Wu et al. 2008), resulting in transition to HDR. In addition to protecting the DSB end from degradation, Ku recruits the other core NHEJ proteins, Dnl4-Lif1 and Nej1, to the DSB site.
Dnl4-Lif1
DNA ligase IV, Dnl4 in S. cerevisiae, is an ATP-dependent DNA ligase strictly required for NHEJ (Wilson et al. 1997). Although previously thought to not function outside of NHEJ, Dnl4 has more recently been implicated in alternative end-joining repair as well as ribosomal DNA maintenance (Chiruvella et al. 2013a; Fritsch et al. 2010). Dnl4 is one of three DNA ligases. The catalytic activity of these DNA ligases can be summarized in three steps (Ellenberger and Tomkinson 2008). In the first step, DNA ligase displaces a pyrophosphate from ATP, leading to covalent auto-adenylation of the active site lysine (K282 in Dnl4) and the creation of a long-lived intermediate. In step two, activated AMP is transferred to the available 5′ phosphate on DNA via a 5′- 5′ phosphoanhydride bond. Ligation occurs in step 3, when an adjacent 3′ hydroxyl group on DNA attacks the phosphoanhydride bond from step two. The bond is cleaved by the attack, resulting in DNA ligation and release of AMP.
Dnl4 is strongly associated with the NHEJ factor Lif1, so much so that Dnl4 is unstable in the absence of Lif1 (Herrmann et al. 1998). This interaction was the basis for identification of Lif1, which was anticipated based on the interaction of XRCC4 with LIG4 in humans (Herrmann et al. 1998). Lif1 homodimerizes with a globular head and central coiled coil region (Deshpande and Wilson 2007; Dore et al. 2006; Herrmann et al. 1998). Dnl4-Lif1 interaction, which has a 1:2 stoichiometry, is mediated by the central coiled coil region in Lif1 and the second of two tandem BRCT domains (BRCT2) and the linker region that separates them in Dnl4, analogous to the interaction of these regions in XRCC4 and LIG4 (Chiruvella et al. 2014). Unexpectedly, a separation-of-function mutation in the BRCT1 domain, which retained Lif1 interaction, resulted in a marked reduction in the association of Dnl4 at a DSB and a large effect on NHEJ efficiency, suggesting the BRCT1 domain supplies a function to promote Dnl4 DSB association and NHEJ that is independent of Lif1.
Both in vitro and in vivo studies have shown that Dnl4-Lif1 association with DNA is strongly dependent on Yku (Wu et al. 2008; Zhang et al. 2007). More specifically, interaction between the Yku80 C-terminus and Dnl4 is critical to NHEJ function (Palmbos et al. 2008). The MRX complex also appears to influence Dnl4-Lif1 association with the DSB. Deletion of RAD50 delays the association of the Dnl4-Lif1 proteins with a chromosomal DSB (Wu et al. 2008). This may be mediated via Lif1’s C-terminus interactions with the N-terminal FHA domain of Xrs2 (Matsuzaki et al. 2008; Palmbos et al. 2005). This interaction, along with the Yku80-Dnl4 interaction, is required for stable association of Dnl4-Lif1 with a DSB (Palmbos et al. 2008). Conversely, Dnl4-Lif1 association with the DSB stabilizes Ku’s binding to the DNA end (Chen and Tomkinson 2011; Zhang et al. 2007). Stabilization of Ku by Dnl4-Lif1 attenuates the execution of HDR pathways by supporting Ku’s role in inhibiting resection activity (Zhang et al. 2007). Dnl4-Lif1 binding also promotes pair-wise interactions which are necessary to stabilize the greater repair complex formed by Yku70/80, Dnl4-Lif1, and Nej1 (Chen and Tomkinson 2011). Thus, the role of Dnl4 extends beyond its enzymatic activity of ligation.
Nej1
Deletion of NEJ1 results in NHEJ defects that are epistatic with dnl4Δ (Frank-Vaillant and Marcand 2001; Kegel et al. 2001; Valencia et al. 2001), underscoring the importance of this factor in the core NHEJ mechanism. Nej1, the most recently identified core NHEJ factor in yeast, is a haploid-specific protein whose existence was predicted by observations that diploid cells expressing the a1-α2 repressor were defective in NHEJ (Astrom et al. 1999; Lee et al. 1999). The haploid specific expression of Nej1 is the result of the binding of the a1-α2 repressor to its consensus binding site found within the NEJ1 promoter gene (Kegel et al. 2001; Valencia et al. 2001).
Nej1 interacts with several core NHEJ factors, including Lif1. Nej1 and Lif1 show robust physical interaction, as determined by yeast two hybrid and pull-down assays (Deshpande and Wilson 2007; Ooi et al. 2001). Interestingly, the LIF1 and NEJ1 genes are related and, like Lif1, Nej1 is thought to predominantly exist as a coiled-coil homodimer in vivo. However, despite their relatedness, Nej1 and Lif1 are not predicted to form stable heterodimers (Deshpande and Wilson 2007). Nej1-Lif1 interaction is mediated by the C-terminal domain of Nej1 and the N-terminal globular head of Lif1 (Deshpande and Wilson 2007; Mahaney et al. 2014; Ooi et al. 2001; Sulek et al. 2007). Notably, Nej1 is not necessary for Dnl4-Lif1 complex formation and Nej1-Lif1 interaction is independent of Dnl4 (Ooi et al. 2001). The C-terminus of Nej1 has been characterized in depth with regard to its NHEJ functions. Specifically, the C-terminal K331-K334 region is critical for nuclear targeting/recruitment and NHEJ repair, while F335-V338 residues are important for Nej1’s interaction with Lif1, but not its recruitment to a chromosomal DSB (Mahaney et al. 2014).
Interestingly, Nej1 associates with HO endonuclease-induced chromosomal DSBs independently of Dnl4-Lif1; instead, it associates in a manner strongly dependent on Yku (Chen and Tomkinson 2011; Wu et al. 2008). Chromatin immunoprecipitation experiments demonstrated that Nej1 is required for stable binding of Yku at a DSB (Chen and Tomkinson 2011), presumably to assist Yku’s role in inhibiting resection, much like Dnl4-Lif1 (Zhang et al. 2007). Nej1 interacts with Yku70 directly via its C-terminal region, the same region that interacts with Lif1 (Chen and Tomkinson 2011). In addition to supporting stable formation of the NHEJ super-complex at DSB ends, Nej1 promotes reactivation of Dnl4 primarily by enhancing deadenylation of the enzyme’s active site lysine K282, which prevents NHEJ repair from being limited by the number of available Dnl4-Lif1 molecules (Chen and Tomkinson 2011; Ellenberger and Tomkinson 2008). Finally, Nej1 also plays a role in recruiting end-processing factors to the DSB.
MRX complex
The contribution of MRX to the NHEJ pathway is less understood, due in part to its significance in several other cellular processes (D’Amours and Jackson 2002), including telomere maintenance, meiotic recombination, and the competing HDR pathways of DSB repair in mitotic cells. The complex is composed of Mre11, Rad50, and Xrs2 subunits, in a 2:2:1 stoichiometric ratio (Chen et al. 2001). Mre11 possesses 3′ exonuclease, endonuclease, and DNA unwinding activities (Ghodke and Muniyappa 2013; Trujillo and Sung 2001). Rad50 directly drives the endonuclease and DNA unwinding functions of Mre11 by conformational changes of its globular ATPase domain, which is adjacent to the Mre11-Rad50 contact site (Hopfner et al. 2001; Hopfner et al. 2000; Trujillo and Sung 2001). The ATPase domain of Rad50 is split, such that it only becomes functional upon Zn2+-dependent dimerization of two Rad50 molecules, mediated by a Zinc-hook structure (Hopfner et al. 2000). Xrs2 assists in targeting the complex to DSB ends by direct DNA binding, although all three MRX subunits are capable of binding DNA independently (Raymond and Kleckner 1993; Trujillo et al. 2003; Trujillo and Sung 2001). The association of MRX with DSBs is also enhanced by Yku-Dnl4 (Wu et al. 2008; Zhang et al. 2007), raising the possibility that the mechanism by which MRX engages a DSB may dictate pathway choice.
As discussed above, MRX acts antagonistically to Yku functions at DSB ends; however, understanding MRX activity solely as an opposition to NHEJ does not capture the complexity of MRX’s role in end-joining. Deletion of MRX subunits has been shown repeatedly to result in a dramatic decrease in NHEJ efficiency in plasmid repair assays for both compatible and incompatible ends, as well as in a chromosomal NHEJ assays (Moore and Haber 1996; Schiestl et al. 1993; Zhang and Paull 2005). Interestingly, MRX’s supporting role in NHEJ cannot be attributed to Mre11 nucleolytic activity, as demonstrated by experiments utilizing several nuclease dead alleles of Mre11 (Lewis et al. 2004; Moreau et al. 1999; Zhang and Paull 2005). However, disruption of either the ATPase or coiled coil domains of Rad50 does result in a loss of NHEJ functionality (Chen et al. 2005; Hohl et al. 2011), providing clues to MRX’s role in end-joining.
The predominant model to describe MRX function in NHEJ is one in which the complex bridges DSB ends and acts as a flexible tether to assist in ligation. The linchpin in the tethering model is the Zinc-hook structure formed by Rad50 homodimerization (Hopfner et al. 2002). Long flexible coiled coils extending outward from the Zinc hook are predicted to bind two distinct Mre11 dimers, which themselves bind DNA (Chen et al. 2001; Chen et al. 2005). This model is supported by in vitro observations that the MRX complex strongly stimulated DNA ligation by Dnl4-Lif1, as well as by atomic force microscopy visualizations of MRX-mediated oligomerization of linear DNA fragments (Chen et al. 2001). End-bridging is still possible in the absence of the MRX complex, as evidenced by electron microscopic detection of Dnl4-Lif1 forming end bridging complexes via interaction with Yku (Grob et al. 2012). However, in a chromosomal DSB assay selective for imprecise repair events, absence of MRX subunits resulted in a more than 70-fold decrease in NHEJ and a significant increase in small deletions at repair junctions, indicating that end-bridging is less efficient and may be accompanied by deletion-prone end processing in the absence of the MRX complex (Moore and Haber 1996).
Despite the appeal of the end-bridging model of MRX function, several questions remain regarding the role of the complex in NHEJ. Because the complex acts in both HDR and NHEJ pathways, it is likely to be involved in the regulation of pathway choice. However, the question of how MRX promotes one pathway over the other has yet to be totally explained. While MRX can associate with a DSB independently, its association is also enhanced by Yku-Dnl4 (Wu et al. 2008; Zhang et al. 2007), raising the possibility that the mechanism by which MRX engages a DSB may dictate pathway choice. Additionally, although Mre11 nuclease activity is not required for NHEJ, it is still plausible that this activity may contribute to one of the redundant pathways of end-processing, a role which is often speculated but has yet to be identified.
End-Processing Factors
DSBs usually require processing to achieve the complementary ends needed for successful ligation by Dnl4. Several polymerases and nucleases in S. cerevisiae have been identified as acting in the NHEJ pathway to correct damaged or mismatched DNA ends (Table 1). Nonetheless, end-processing is the least understood step in the NHEJ mechanism, in part because of the difficulty in differentiating redundant and overlapping pathways.
Table 1.
End-processing factors reported in S. cerevisiae.
| Gene in S. cerevisiae (Human Homolog) | Reported Functions in NHEJ | References |
|---|---|---|
| Pol4 (Pol β, λ, μ) | 3′ gap-filling, stabilizes 3′ overhangs, supports Dnl4 ligation activity | Bebenek et al., 2005; Daley et al., 2005; Daley and Wilson, 2008; Tseng and Tomkinson, 2002; Wilson and Lieber, 1999 |
| Rad27 (FEN1) | 5′ flap endonuclease | Daley and Wilson, 2008; Tseng and Tomkinson, 2004; Wu et al., 1999; Yang et al., 2015 |
| Pol2 (Pol ε) | 3′ flap endonuclease | Tseng et al., 2008 |
| Pol3 (Pol δ) | 3′ gap-filling, redundant with Pol4 function | Chan et al., 2008; Galli et al., 2015 |
| Exo1 (EXO1) | 5′ exonuclease, exposes microhomology | Bahmed et al., 2011 |
| Tdp1 (TDP1) | Promotes accurate repair at 5′ overhangs | Bahmed et al., 2010 |
Pol4
Of the end-processing factors, Pol4 is the best understood to date. Pol4 is a Pol X family polymerase (related to mammalian polymerases β, λ and μ) and is most closely associated with gap-filling in NHEJ (Bebenek et al. 2005; Tseng and Tomkinson 2002; Wilson and Lieber 1999). Pol4 shares a domain structure with Pol λ and Pol μ, composed of an N-terminal BRCT domain, a central lyase domain, and a C-terminal nucleotidyltransferase domain (Tseng and Tomkinson 2002). Strains lacking Pol4 show no defect in growth or meiosis, but exhibit slight sensitivity to the DNA alkylating agent MMS and hyper-recombination during meiosis (Leem et al. 1994), providing indication of a nonessential role in DSB repair. Notably, Pol4 has low processivity and is highly error-prone, which accounts for the polymerase’s ability to fill small gaps by extending termini with limited sequence homology (Bebenek et al. 2005).
Pol4 is particularly important for gap-filling at unstable 3′ overhangs, an ability which is attributed to a reduced dependence on stable primer-template pairing (Daley et al. 2005; Pardo et al. 2006; Ruiz et al. 2013). This model of Pol4 function is supported by the finding that stabilizing 3′ overhangs, either by extending the overhang, changing overhang polarity, or adding a 5′ nucleotide flap, results in reduced dependence on Pol4 for repair (Daley et al. 2005; Daley and Wilson 2008). Pol4 synthesis activity is stimulated by interaction of the Pol4 BRCT domain with the Dnl4-Lif1 complex, and this interaction also weakly stimulates ligation efficiency (Tseng and Tomkinson 2002). Interactions with other core NHEJ factors also help to recruit Pol4 to the DSB, as illustrated by the finding that independent and additive interactions of Pol4 with Dnl4 and Nej1 recruit Pol4 and stimulate its activity at a DSB (Yang et al. 2015). Finally, Pol4 utilization in NHEJ may be regulated by Tel1-dependent phosphorylation of the Pol4-Thr540 residue (Ruiz et al. 2013).
Rad27
Rad27 is a 5′ nuclease that was implicated early as an end-processing factor in NHEJ, but reports have been inconsistent regarding its importance. In a plasmid repair assay using DNA substrates predicted to form 5′ flap intermediates, loss of Rad27 resulted in a more than 4-fold decrease in successful NHEJ activity (Wu et al. 1999). However, later work found Rad27 to be dispensable for NHEJ events requiring processing of flaps, gaps, or complex overhangs (Daley and Wilson 2008). Despite this discrepancy, Rad27 interacts physically with both Pol4 and Dnl4-Lif1 (Tseng and Tomkinson 2004) and is recruited to the site of a chromosomal DSB by interaction with Nej1 (Yang et al. 2015). Interestingly, rad27Δ is synthetically lethal with mre11Δ (Debrauwere et al. 2001). Lethality is suppressed by yku70Δ (Foster et al. 2011) or overexpression of Exo1 (Moreau et al. 2001), corresponding to Rad27’s better defined role in DNA replication associated damage, rather than NHEJ. Given the absence of evidence for a clear and consistent role for Rad27 in 5′ end-processing, it seems apparent that other nucleases remain to be identified in NHEJ function.
Other end-processing factors
Several other factors have been reported to assist in end-processing at DNA breaks in S. cerevisiae, including Pol2, Pol3, Exo1 and Tdp1. Pol2 (ortholog of mammalian Pol ε) is necessary for the removal of 3′ flaps in a chromosomal break assay specific for imprecise end-joining (Tseng et al. 2008). This role for Pol2 remains to be explored in other contexts. Pol3 (mammalian Pol δ) functions similarly to Pol4 in gap-filling repair at 3′ overhangs in plasmid repair assays, suggesting redundant processing pathways (Chan et al. 2008; Galli et al. 2015). Interestingly, there are indications that Pol3, like Pol4, may help facilitate DSB repair in a capacity beyond gap-filling. Deletion of both factors results in decreased NHEJ repair of compatible and blunt ends (Chan et al. 2008), suggesting that Pol3, similar to Pol4, can support the ligation activity of Dnl4-Lif1. However, Pol3 does not appear to be as important to NHEJ as Pol4, since its effects are most apparent in the absence of Pol4 (Chan et al. 2008; Galli et al. 2015). Exo1, a 5′ exonuclease, has been suggested to support strand annealing in NHEJ by exposing small microhomologies and reversing polymerase activity (Bahmed et al. 2011), despite its well-established role in long-range resection and HDR (Zhu et al. 2008), raising questions about cross-talk between NHEJ and HDR.
Tyrosyl-DNA phosphodiesterase (Tdp1), a factor primarily associated with repair of topoisomerase 1-mediated damage (Liu et al. 2004; Nitiss et al. 2006), regulates end-processing in a manner different than the previously discussed polymerases and nucleases. Tdp1 has been shown to act specifically at 5′ DSBs, where it cleaves the recessed 3′ terminal nucleoside to generate a 3′ phosphate and block Pol4 filling (Bahmed et al. 2010). Consequently, absence of Tdp1 results in a high frequency of imprecise repair of breaks with 5′ overhangs, but does not affect the accuracy of repair of breaks with 3′ overhangs or blunt ends. Imprecise repair events in tdp1Δ strains are primarily characterized by small insertions, which were dependent on Pol4. Additionally, although human TDP1 is observed to directly bind and support the function of the human Nej1 homolog, XLF (Heo et al. 2015), no such interaction has been characterized with yeast Tdp1 and Nej1. Together, these data suggest that Tdp1’s role in yeast is to promote repair fidelity by restricting access of enzymes such as Pol4 to DSB ends. It will be interesting to see how this role for Tdp1 is reconciled with Pol4’s reported contributions to repair accuracy and Dnl4 activity (Chan et al. 2008; Tseng and Tomkinson 2002, 2004).
The NHEJ Mechanism
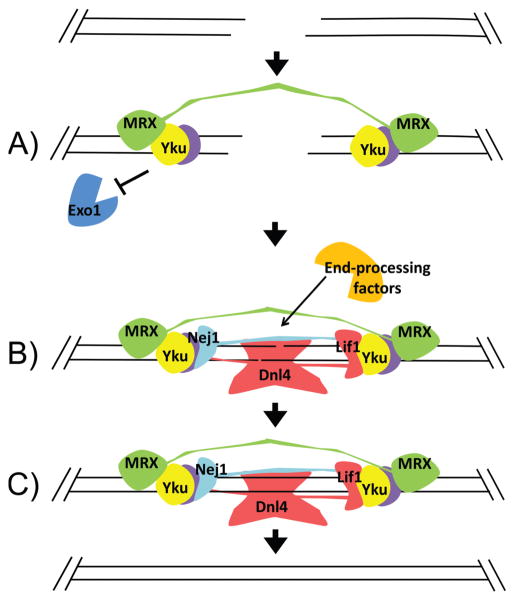
While existing data allow us to describe a model of NHEJ mechanism, it is worth noting that NHEJ is unlikely to be a strictly ordered process. Processing of the DSB is probably iterative (Ma et al. 2005), with variation in factor recruitment, strand annealing, and end-processing occurring in a manner dependent on the damage incurred. Additionally, while many interactions between different NHEJ factors have been identified, it is still unknown how each interaction contributes to NHEJ function or how stable these interactions might be. We present a model with three steps: 1) End protection and tethering, 2) Strand annealing, complex assembly and end-processing, and 3) Ligation and complex disassembly.
End protection and tethering
Upon formation of a DSB, Yku and MRX bind the site rapidly and independently (Wu et al. 2008) (Fig. 2A). It is unknown which factor binds first in yeast, but in a mammalian model, Ku associates with a laser-induced break faster than can be detected, while the MRN complex is recruited 15 to 30 seconds after damage (Hartlerode et al. 2015). If MRX is similarly delayed in yeast, it may be indicative of a signaling pathway required to regulate the seemingly contradictory activities of MRX in both NHEJ and HDR. Once associated, Yku protects ends from exonucleolytic degradation (Clerici et al. 2008; Mimitou and Symington 2010). As discussed previously, MRX is predicted to act as a flexible tether, utilizing the long coiled coils and Zn hook structure in the Rad50 subunit (Chen et al. 2001; Chen et al. 2005). Tethering and protection of the ends allows them to be brought together and stabilized by strand annealing.
Figure 2.
Model of the NHEJ mechanism. A) Upon formation of a DSB, Yku70/80 and MRX rapidly and independently bind DNA ends. Yku protects ends from resection by Exo1 and other factors. MRX acts as a flexible tether to bring ends into proximity. B) Coordinated actions between the core repair complex and end-processing factors facilitate strand annealing. Yku binds and retains core factors Dnl4-Lif1 and Nej1. Pairwise interactions stabilize the assembled repair complex. Whether this is achieved by filament formation as observed in vertebrates is not known. End-processing factors act at the junction, creating compatible ends. C) Dnl4 ligates the compatible ends. The repair complex then disassembles from the repaired site.
Strand annealing, complex assembly and end-processing
A critical requirement of the NHEJ pathway is to overcome the entropy of the disassociated DSB ends. This is facilitated by strand annealing and by the formation of the repair super-complex to stabilize the damaged site (Fig. 2B). Small microhomologies increase rejoining efficiency, especially in comparison to the joining efficiency of blunt ends (Boulton and Jackson 1996; Westmoreland et al. 2010). Extending overhang length increases NHEJ efficiency, although very long overhangs become more prone to repair by HDR pathways (Daley and Wilson 2005).
Repair complex formation, like strand annealing, stabilizes the repair joint. Yku, which serves as a scaffold, recruits and is stabilized by Dnl4-Lif1 and Nej1 (Chen and Tomkinson 2011; Zhang et al. 2007) (Fig. 2B). DNA ligase IV-XRCC4 and XLF form filaments adjacent to DSBs that bridge and stabilize DNA ends in vitro (Hammel et al. 2011; Hammel et al. 2010; Mahaney et al. 2013; Ropars et al. 2011). Whether Dnl4-Lif1 and Nej1 similarly form filaments remains to be determined. MRX also promotes formation of the core complex by interactions with Dnl4-Lif1 (Palmbos et al. 2008; Wu et al. 2008). The core complex is flexible enough to shift around the break site to accommodate the end-processing required at varying positions, due in part to Yku’s demonstrated ability to translocate along DNA (Kysela et al. 2003) and by the coiled coils in Lif1 and Nej1 (Deshpande and Wilson 2007). End-processing factors Pol4 and Rad27 act in parallel with complex formation, stabilizing the repair joint and promoting ligation (Daley and Wilson 2008; Tseng and Tomkinson 2002, 2004). This finding leaves open the possibility that other end-processing factors, discussed above, contribute in several capacities to the NHEJ mechanism. In any case, end-processing likely occurs in an iterative manner, with factors being recruited to the break depending on the damage and the position of the repair complex.
Ligation and complex disassembly
Once DSB ends are processed for compatibility, Dnl4 ligates the nicks in the annealed DNA (Wilson et al. 1997) (Fig. 2C). Little is known about how the complex disassembles after successful ligation in budding yeast. An unrepaired chromosomal DSB requires MRX activity to remove the core repair complex, but this dissociation event precedes a transition to HDR (Wu et al. 2008) and likely does not represent the events after successful ligation. Particular interest is focused on how Yku is removed from the repair site, since the ring structure of the heterodimer does not lend itself to easy dissociation from a continuous length of DNA. In mammalian systems, Ku80 is removed from DNA by ubiquitin dependent signaling and degradation (Feng and Chen 2012; Postow and Funabiki 2013; Postow et al. 2008), but no such mechanism has been described in yeast.
NHEJ and Chromatin
DNA is packaged into chromatin, comprised of orderly arrangement of DNA into nucleosomes. In the context of DSB repair, chromatin modification allows protein access to DNA and facilitates processing of DNA for repair. Extensive chromatin modification is associated with HDR, which requires processing and manipulation of a long tract of single-stranded DNA through a homology search and recombination (Bennett et al. 2013; Tsukuda et al. 2005; van Attikum et al. 2004). The initial step of NHEJ is inhibitory to chromatin modification, as most chromatin remodelling complexes are inhibited from recruitment to DSBs in G1 by Ku (Bennett et al. 2013; Chai et al. 2005; Chen et al. 2012; Tsukuda et al. 2005; van Attikum et al. 2004; Vincent et al. 2008). These findings are consistent with the understanding that NHEJ proteins need only to access the ends of the DSB for end joining repair, with minimal need for remodeling.
While most chromatin remodeling complexes have been shown to have negligible roles in NHEJ, there are some, albeit conflicting, reports of RSC influencing NHEJ (Chai et al. 2005; Liang et al. 2007). RSC, a member of the SWI/SNF family, is the most abundant ATP-dependent chromatin remodeling complex in yeast (Cairns et al. 1996). Deletion of RSC subunits results in diminished precise and imprecise NHEJ frequency at the site of an HO induced chromosomal DSB (Shim et al. 2005). Plasmid repair assays also revealed NHEJ defects upon deletion of RSC subunits (Florio et al. 2007; Moscariello et al. 2010; Shim et al. 2005), although one group detected increased plasmid repair in rsc1Δ and rsc2Δ strains (Chai et al. 2005). The RSC subunit Sth1 is detected at an HO endonuclease-induced DSB by ChIP as early as 10 minutes after break induction (Shim et al. 2007; Shim et al. 2005). At these DSB ends, RSC facilitates nucleosome rearrangement and promotes recruitment of Yku and Mre11 (Kent et al. 2007; Shim et al. 2007; Tsukuda et al. 2005). A positive feedback mechanism has been proposed for nucleosome movement and protein recruitment, given that yku70Δ results in delayed recruitment of Sth1 to a DSB (Chambers and Downs 2012; Shim et al. 2005). However, another group found that recruitment of RSC subunit Sth1 to a chromosomal DSB was inhibited by Yku70 in G1 (Bennett et al. 2013), suggesting that RSC roles may be heavily influenced by cell cycle. Thus, the premise mechanism by which RSC impacts NHEJ efficiency remains unclear.
Another complex which has been implicated in NHEJ is SWR1 (SWR-C), a member of the INO80 family of chromatin remodelers (van Attikum et al. 2007). Loss of SWR1 results in decreased binding of Yku80 to a DSB site, but does not affect Mre11 binding (van Attikum et al. 2007). Interestingly, deletion of SWR1 selectively impairs error-free NHEJ, indicating a role for the complex in the regulating NHEJ fidelity (van Attikum et al. 2007). However, like RSC, SWR1 recruitment to a DSB is inhibited by Yku70 in G1- arrested cells, suggesting cell cycle specific activity of this complex as well (Bennett et al. 2013).
In addition to chromatin remodeling complexes, histone modifications have the potential to influence NHEJ. One of the earliest events in response to DSB formation is the activation of Mec1 and Tel1 (ATR and ATM in mammals, respectively), which amplifies a signal cascade that includes phosphorylation of the yeast histone H2A on S129, forming the DNA damage marker γ-H2A (Downs et al. 2000; Gobbini et al. 2013). However, the role of this key histone modification in NHEJ signaling and repair remains unclear. In G1-arrested cells, γ-H2A formation spreads out from an induced DSB approximately 40 kilobases in either direction, but enrichment of the histone variant is relatively low in the 1 to 2 kilobases directly adjacent to the break (Bennett et al. 2013; Shroff et al. 2004). Interestingly, γ-H2A formation is more robust in G1-arrested cells, compared to G2/M synchronized cells, yet the modification does not appear to promote recruitment of NHEJ repair factors or chromatin modifiers (Bennett et al. 2013; Shroff et al. 2004).
Set1-mediated H3K4 methylation has also been implicated in NHEJ (Faucher and Wellinger 2010). Strains lacking Set1 exhibit modest decreases in both precise and imprecise NHEJ of HO-induced DSBs. Consistent with this, Ku recruitment to DSBs is reduced is set1Δ strains. Notably, Set1 recruitment to DSBs is dependent on RSC, suggesting effects of RSC on NHEJ may be due to downstream Set1-dependent histone modification.
Summary and Perspectives
NHEJ is a conserved pathway to resolve the trauma of DNA DSBs and dysfunction in DSB repair can result in genome instability and human disease (Malkova and Haber 2012; McKinnon and Caldecott 2007; Pierce et al. 2001; Prochazkova and Loizou 2015; Rulten and Caldecott 2013). In S. cerevisiae, c-NHEJ is a haploid specific process that occurs predominantly in G1. While the core NHEJ factors, Yku70/80, Dnl4-Lif1, and Nej1 are well characterized, the role of the MRX in the c-NHEJ reaction in yeast is less well understood, partly due to its additional roles in promoting HDR. Further work regarding MRX’s role in DSB repair pathway choice is needed, particularly in the context of DNA damage signaling and chromatin. End-processing factors in NHEJ are also an ongoing focus of characterization. Identified end-processing enzymes have been implicated in repair fidelity, as well as supporting the ligation activity of core NHEJ machinery. These factors have proven difficult to study due to their function in overlapping and redundant processing mechanisms, as well as playing roles in other repair pathways. It is likely that additional end-processing factors exist in addition to those already characterized.
Of the three major steps of the NHEJ mechanism the least understood is how end-processing is coordinated with strand annealing and complex assembly. So far, Pol4 is the best characterized end-processing factor with respect to how its action is integrated into the greater NHEJ mechanism (Tseng and Tomkinson 2002, 2004). Additionally, how the NHEJ repair complex is disassembled is not yet understood in yeast, though some information may be discerned from mammalian studies (Feng and Chen 2012; Postow et al. 2008).
Chromatin dynamics in NHEJ will likely be a growing focus of research, given how little is currently understood. It will be particularly exciting to see how developing imaging technologies are utilized in the field. Recent imaging studies have confirmed that HO-induced DSB ends are isolated before commitment to HDR repair (Saad et al. 2014), but the mechanism of isolation is still unknown. Yet to be identified roles for chromatin remodeling complexes will have implications for NHEJ. How H3K4 methylation exerts its effect on Yku recruitment remains an open question and other histone modifications that influence NHEJ likely remain to be identified. Additionally, the field of chromatin dynamics currently seems to be separated from the field of classical DNA repair. Integration of these major fields to come to a cohesive understanding of NHEJ mechanism in the context of chromatin will be a major achievement.
The budding yeast S. cerevisiae has been used extensively to characterize the general principles of NHEJ. Numerous and variable systems to study NHEJ in a well-characterized and tractable model organism make yeast highly attractive for pioneering avenues of research. For these reasons, yeast will continue to be a relevant model organism, even as methodologies develop for other model systems.
References
- Astrom SU, Okamura SM, Rine J. Yeast cell-type regulation of DNA repair. Nature. 1999;397(6717):310. doi: 10.1038/16833. [DOI] [PubMed] [Google Scholar]
- Aylon Y, Kupiec M. Cell cycle-dependent regulation of double-strand break repair: a role for the CDK. Cell Cycle. 2005;4(2):259–261. [PubMed] [Google Scholar]
- Bahmed K, Nitiss KC, Nitiss JL. Yeast Tdp1 regulates the fidelity of nonhomologous end joining. Proc Natl Acad Sci U S A. 2010;107(9):4057–4062. doi: 10.1073/pnas.0909917107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahmed K, Seth A, Nitiss KC, Nitiss JL. End-processing during non-homologous end-joining: a role for exonuclease 1. Nucleic Acids Res. 2011;39(3):970–978. doi: 10.1093/nar/gkq886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini A, Ristic D, Dionne I, Liu XZ, Wyman C, Wellinger RJ, Petrini JH. The Ku heterodimer and the metabolism of single-ended DNA double-strand breaks. Cell Rep. 2013;3(6):2033–2045. doi: 10.1016/j.celrep.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebenek K, Garcia-Diaz M, Patishall SR, Kunkel TA. Biochemical properties of Saccharomyces cerevisiae DNA polymerase IV. J Biol Chem. 2005;280(20):20051–20058. doi: 10.1074/jbc.M501981200. [DOI] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4(6):e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett G, Papamichos-Chronakis M, Peterson CL. DNA repair choice defines a common pathway for recruitment of chromatin regulators. Nat Commun. 2013;4:2084. doi: 10.1038/ncomms3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuch AA, Lundblad V. The Ku heterodimer performs separable activities at double-strand breaks and chromosome termini. Mol Cell Biol. 2003;23(22):8202–8215. doi: 10.1128/MCB.23.22.8202-8215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betermier M, Bertrand P, Lopez BS. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 2014;10(1):e1004086. doi: 10.1371/journal.pgen.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15(18):5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87(7):1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Ceccaldi R, Rondinelli B, D’Andrea AD. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016;26(1):52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19(14):1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers AL, Downs JA. The RSC and INO80 chromatin-remodeling complexes in DNA double-strand break repair. Prog Mol Biol Transl Sci. 2012;110:229–261. doi: 10.1016/B978-0-12-387665-2.00009-2. [DOI] [PubMed] [Google Scholar]
- Chan CY, Galli A, Schiestl RH. Pol3 is involved in nonhomologous end-joining in Saccharomyces cerevisiae. DNA Repair (Amst) 2008;7(9):1531–1541. doi: 10.1016/j.dnarep.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8(5):1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- Chen L, Trujillo KM, Van Komen S, Roh DH, Krejci L, Lewis LK, Resnick MA, Sung P, Tomkinson AE. Effect of amino acid substitutions in the rad50 ATP binding domain on DNA double strand break repair in yeast. J Biol Chem. 2005;280(4):2620–2627. doi: 10.1074/jbc.M410192200. [DOI] [PubMed] [Google Scholar]
- Chen X, Cui D, Papusha A, Zhang X, Chu CD, Tang J, Chen K, Pan X, Ira G. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature. 2012;489(7417):576–580. doi: 10.1038/nature11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Niu H, Chung WH, Zhu Z, Papusha A, Shim EY, Lee SE, Sung P, Ira G. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat Struct Mol Biol. 2011;18(9):1015–1019. doi: 10.1038/nsmb.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Tomkinson AE. Yeast Nej1 is a key participant in the initial end binding and final ligation steps of nonhomologous end joining. J Biol Chem. 2011;286(6):4931–4940. doi: 10.1074/jbc.M110.195024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiruvella KK, Liang Z, Birkeland SR, Basrur V, Wilson TE. Saccharomyces cerevisiae DNA ligase IV supports imprecise end joining independently of its catalytic activity. PLoS Genet. 2013a;9(6):e1003599. doi: 10.1371/journal.pgen.1003599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiruvella KK, Liang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol. 2013b;5(5):a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiruvella KK, Renard BM, Birkeland SR, Sunder S, Liang Z, Wilson TE. Yeast DNA ligase IV mutations reveal a nonhomologous end joining function of BRCT1 distinct from XRCC4/Lif1 binding. DNA Repair (Amst) 2014;24:37–45. doi: 10.1016/j.dnarep.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP. The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep. 2008;9(8):810–818. doi: 10.1038/embor.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3(5):317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- Daley JM, Laan RL, Suresh A, Wilson TE. DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J Biol Chem. 2005;280(32):29030–29037. doi: 10.1074/jbc.M505277200. [DOI] [PubMed] [Google Scholar]
- Daley JM, Niu H, Miller AS, Sung P. Biochemical mechanism of DSB end resection and its regulation. DNA Repair (Amst) 2015;32:66–74. doi: 10.1016/j.dnarep.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Wilson TE. Rejoining of DNA double-strand breaks as a function of overhang length. Mol Cell Biol. 2005;25(3):896–906. doi: 10.1128/MCB.25.3.896-906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Wilson TE. Evidence that base stacking potential in annealed 3′ overhangs determines polymerase utilization in yeast nonhomologous end joining. DNA Repair (Amst) 2008;7(1):67–76. doi: 10.1016/j.dnarep.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debrauwere H, Loeillet S, Lin W, Lopes J, Nicolas A. Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc Natl Acad Sci U S A. 2001;98(15):8263–8269. doi: 10.1073/pnas.121075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande RA, Wilson TE. Modes of interaction among yeast Nej1, Lif1 and Dnl4 proteins and comparison to human XLF, XRCC4 and Lig4. DNA Repair (Amst) 2007;6(10):1507–1516. doi: 10.1016/j.dnarep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty AJ, Jackson SP, Weller GR. Identification of bacterial homologues of the Ku DNA repair proteins. FEBS Lett. 2001;500(3):186–188. doi: 10.1016/s0014-5793(01)02589-3. [DOI] [PubMed] [Google Scholar]
- Dore AS, Furnham N, Davies OR, Sibanda BL, Chirgadze DY, Jackson SP, Pellegrini L, Blundell TL. Structure of an Xrcc4-DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode. DNA Repair (Amst) 2006;5(3):362–368. doi: 10.1016/j.dnarep.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408(6815):1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- Driller L, Wellinger RJ, Larrivee M, Kremmer E, Jaklin S, Feldmann HM. A short C-terminal domain of Yku70p is essential for telomere maintenance. J Biol Chem. 2000;275(32):24921–24927. doi: 10.1074/jbc.M002588200. [DOI] [PubMed] [Google Scholar]
- Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annu Rev Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher D, Wellinger RJ. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 2010;6(8) doi: 10.1371/journal.pgen.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell VL, Schild-Poulter C. The Ku heterodimer: function in DNA repair and beyond. Mutat Res Rev Mutat Res. 2015;763:15–29. doi: 10.1016/j.mrrev.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol. 2012;19(2):201–206. doi: 10.1038/nsmb.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio C, Moscariello M, Ederle S, Fasano R, Lanzuolo C, Pulitzer JF. A study of biochemical and functional interactions of Htl1p, a putative component of the Saccharomyces cerevisiae, Rsc chromatin-remodeling complex. Gene. 2007;395(1–2):72–85. doi: 10.1016/j.gene.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Foster SS, Balestrini A, Petrini JH. Functional interplay of the Mre11 nuclease and Ku in the response to replication-associated DNA damage. Mol Cell Biol. 2011;31(21):4379–4389. doi: 10.1128/MCB.05854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Vaillant M, Marcand S. NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev. 2001;15(22):3005–3012. doi: 10.1101/gad.206801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Vaillant M, Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol Cell. 2002;10(5):1189–1199. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- Fritsch O, Burkhalter MD, Kais S, Sogo JM, Schar P. DNA ligase 4 stabilizes the ribosomal DNA array upon fork collapse at the replication fork barrier. DNA Repair (Amst) 2010;9(8):879–888. doi: 10.1016/j.dnarep.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Galli A, Chan CY, Parfenova L, Cervelli T, Schiestl RH. Requirement of POL3 and POL4 on non-homologous and microhomology-mediated end joining in rad50/xrs2 mutants of Saccharomyces cerevisiae. Mutagenesis. 2015 doi: 10.1093/mutage/gev046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodke I, Muniyappa K. Processing of DNA double-stranded breaks and intermediates of recombination and repair by Saccharomyces cerevisiae Mre11 and its stimulation by Rad50, Xrs2, and Sae2 proteins. J Biol Chem. 2013;288(16):11273–11286. doi: 10.1074/jbc.M112.439315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbini E, Cesena D, Galbiati A, Lockhart A, Longhese MP. Interplays between ATM/Tel1 and ATR/Mec1 in sensing and signaling DNA double-strand breaks. DNA Repair (Amst) 2013;12(10):791–799. doi: 10.1016/j.dnarep.2013.07.009. [DOI] [PubMed] [Google Scholar]
- Grob P, Zhang TT, Hannah R, Yang H, Hefferin ML, Tomkinson AE, Nogales E. Electron microscopy visualization of DNA-protein complexes formed by Ku and DNA ligase IV. DNA Repair (Amst) 2012;11(1):74–81. doi: 10.1016/j.dnarep.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 2012;191(1):33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Rey M, Yu Y, Mani RS, Classen S, Liu M, Pique ME, Fang S, Mahaney BL, Weinfeld M, et al. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J Biol Chem. 2011;286(37):32638–32650. doi: 10.1074/jbc.M111.272641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Yu Y, Fang S, Lees-Miller SP, Tainer JA. XLF regulates filament architecture of the XRCC4.ligase IV complex. Structure. 2010;18(11):1431–1442. doi: 10.1016/j.str.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlerode AJ, Morgan MJ, Wu Y, Buis J, Ferguson DO. Recruitment and activation of the ATM kinase in the absence of DNA-damage sensors. Nat Struct Mol Biol. 2015;22(9):736–743. doi: 10.1038/nsmb.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J, Li J, Summerlin M, Hays A, Katyal S, McKinnon PJ, Nitiss KC, Nitiss JL, Hanakahi LA. TDP1 promotes assembly of non-homologous end joining protein complexes on DNA. DNA Repair (Amst) 2015;30:28–37. doi: 10.1016/j.dnarep.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann G, Lindahl T, Schar P. Saccharomyces cerevisiae LIF1: a function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J. 1998;17(14):4188–4198. doi: 10.1093/emboj/17.14.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MA. Radiation damage to DNA: the importance of track structure. Radiat Meas. 1999;31(1–6):15–23. doi: 10.1016/s1350-4487(99)00090-6. [DOI] [PubMed] [Google Scholar]
- Hohl M, Kwon Y, Galvan SM, Xue X, Tous C, Aguilera A, Sung P, Petrini JH. The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat Struct Mol Biol. 2011;18(10):1124–1131. doi: 10.1038/nsmb.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418(6897):562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105(4):473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin D, Fairley C, Tainer JA, Carney JP. Mre11 and Rad50 from Pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J Bacteriol. 2000;182(21):6036–6041. doi: 10.1128/jb.182.21.6036-6041.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431(7011):1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessulat M, Malty RH, Nguyen-Tran DH, Deineko V, Aoki H, Vlasblom J, Omidi K, Jin K, Minic Z, Hooshyar M, et al. Spindle Checkpoint Factors Bub1 and Bub2 Promote DNA Double-Strand Break Repair by Nonhomologous End Joining. Mol Cell Biol. 2015;35(14):2448–2463. doi: 10.1128/MCB.00007-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanam K, Kafri R, Loewer A, Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell. 2012;47(2):320–329. doi: 10.1016/j.molcel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel A, Sjostrand JO, Astrom SU. Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast. Curr Biol. 2001;11(20):1611–1617. doi: 10.1016/s0960-9822(01)00488-2. [DOI] [PubMed] [Google Scholar]
- Kent NA, Chambers AL, Downs JA. Dual chromatin remodeling roles for RSC during DNA double strand break induction and repair at the yeast MAT locus. J Biol Chem. 2007;282(38):27693–27701. doi: 10.1074/jbc.M704707200. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC. An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb Perspect Biol. 2015;7(11) doi: 10.1101/cshperspect.a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer KM, Brock JA, Bloom K, Moore JK, Haber JE. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol Cell Biol. 1994;14(2):1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasner DS, Daley JM, Sung P, Niu H. Interplay between Ku and Replication Protein A in the Restriction of Exo1-mediated DNA Break End Resection. J Biol Chem. 2015;290(30):18806–18816. doi: 10.1074/jbc.M115.660191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kysela B, Doherty AJ, Chovanec M, Stiff T, Ameer-Beg SM, Vojnovic B, Girard PM, Jeggo PA. Ku stimulation of DNA ligase IV-dependent ligation requires inward movement along the DNA molecule. J Biol Chem. 2003;278(25):22466–22474. doi: 10.1074/jbc.M303273200. [DOI] [PubMed] [Google Scholar]
- Lam I, Keeney S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol. 2015;7(1):a016634. doi: 10.1101/cshperspect.a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S, Moore JK, Haber JE, Greider CW. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152(1):143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176(4):2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Paques F, Sylvan J, Haber JE. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr Biol. 1999;9(14):767–770. doi: 10.1016/s0960-9822(99)80339-x. [DOI] [PubMed] [Google Scholar]
- Leem SH, Ropp PA, Sugino A. The yeast Saccharomyces cerevisiae DNA polymerase IV: possible involvement in double strand break DNA repair. Nucleic Acids Res. 1994;22(15):3011–3017. doi: 10.1093/nar/22.15.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LK, Storici F, Van Komen S, Calero S, Sung P, Resnick MA. Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics. 2004;166(4):1701–1713. doi: 10.1534/genetics.166.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Qiu J, Ratnakumar K, Laurent BC. RSC functions as an early double-strand-break sensor in the cell’s response to DNA damage. Curr Biol. 2007;17(16):1432–1437. doi: 10.1016/j.cub.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Gu J, Lu H, Shimazaki N, Tsai AG. Nonhomologous DNA end joining (NHEJ) and chromosomal translocations in humans. Subcell Biochem. 2010;50:279–296. doi: 10.1007/978-90-481-3471-7_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Pouliot JJ, Nash HA. The role of TDP1 from budding yeast in the repair of DNA damage. DNA Repair (Amst) 2004;3(6):593–601. doi: 10.1016/j.dnarep.2004.03.030. [DOI] [PubMed] [Google Scholar]
- Lopez CR, Ribes-Zamora A, Indiviglio SM, Williams CL, Haricharan S, Bertuch AA. Ku must load directly onto the chromosome end in order to mediate its telomeric functions. PLoS Genet. 2011;7(8):e1002233. doi: 10.1371/journal.pgen.1002233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol. 2003;23(23):8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Lu H, Schwarz K, Lieber MR. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005;4(9):1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- Mahaney BL, Hammel M, Meek K, Tainer JA, Lees-Miller SP. XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair. Biochem Cell Biol. 2013;91(1):31–41. doi: 10.1139/bcb-2012-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaney BL, Lees-Miller SP, Cobb JA. The C-terminus of Nej1 is critical for nuclear localization and non-homologous end-joining. DNA Repair (Amst) 2014;14:9–16. doi: 10.1016/j.dnarep.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Haber JE. Mutations arising during repair of chromosome breaks. Annu Rev Genet. 2012;46:455–473. doi: 10.1146/annurev-genet-110711-155547. [DOI] [PubMed] [Google Scholar]
- Marcand S, Pardo B, Gratias A, Cahun S, Callebaut I. Multiple pathways inhibit NHEJ at telomeres. Genes Dev. 2008;22(9):1153–1158. doi: 10.1101/gad.455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu N, Pirzio L, Freulet-Marriere MA, Desmaze C, Sabatier L. Telomeres and chromosomal instability. Cell Mol Life Sci. 2004;61(6):641–656. doi: 10.1007/s00018-003-3296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki K, Shinohara A, Shinohara M. Forkhead-associated domain of yeast Xrs2, a homolog of human Nbs1, promotes nonhomologous end joining through interaction with a ligase IV partner protein, Lif1. Genetics. 2008;179(1):213–225. doi: 10.1534/genetics.107.079236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayle R, Campbell IM, Beck CR, Yu Y, Wilson M, Shaw CA, Bjergbaek L, Lupski JR, Ira G. DNA REPAIR. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science. 2015;349(6249):742–747. doi: 10.1126/science.aaa8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney JS, Sethi S, Tripp JD, Nguyen TN, Sanderson BA, Westmoreland JW, Resnick MA, Lewis LK. A multistep genomic screen identifies new genes required for repair of DNA double-strand breaks in Saccharomyces cerevisiae. BMC Genomics. 2013;14:251. doi: 10.1186/1471-2164-14-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- Meyer D, Fu BX, Heyer WD. DNA polymerases delta and lambda cooperate in repairing double-strand breaks by microhomology-mediated end-joining in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1507833112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B, Grompone G, Flores MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proc Natl Acad Sci U S A. 2004;101(35):12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455(7214):770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29(19):3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16(5):2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19(1):556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Morgan EA, Symington LS. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159(4):1423–1433. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscariello M, Florio C, Pulitzer JF. Accurate repair of non-cohesive, double strand breaks in Saccharomyces cerevisiae: enhancement by homology-assisted end-joining. Yeast. 2010;27(10):837–848. doi: 10.1002/yea.1789. [DOI] [PubMed] [Google Scholar]
- Nitiss KC, Malik M, He X, White SW, Nitiss JL. Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage. Proc Natl Acad Sci U S A. 2006;103(24):8953–8958. doi: 10.1073/pnas.0603455103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SL, Shoemaker DD, Boeke JD. A DNA microarray-based genetic screen for nonhomologous end-joining mutants in Saccharomyces cerevisiae. Science. 2001;294(5551):2552–2556. doi: 10.1126/science.1065672. [DOI] [PubMed] [Google Scholar]
- Palmbos PL, Daley JM, Wilson TE. Mutations of the Yku80 C terminus and Xrs2 FHA domain specifically block yeast nonhomologous end joining. Mol Cell Biol. 2005;25(24):10782–10790. doi: 10.1128/MCB.25.24.10782-10790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmbos PL, Wu D, Daley JM, Wilson TE. Recruitment of Saccharomyces cerevisiae Dnl4-Lif1 complex to a double-strand break requires interactions with Yku80 and the Xrs2 FHA domain. Genetics. 2008;180(4):1809–1819. doi: 10.1534/genetics.108.095539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B, Ma E, Marcand S. Mismatch tolerance by DNA polymerase Pol4 in the course of nonhomologous end joining in Saccharomyces cerevisiae. Genetics. 2006;172(4):2689–2694. doi: 10.1534/genetics.105.053512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc Natl Acad Sci U S A. 2000;97(12):6409–6414. doi: 10.1073/pnas.110144297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Stark JM, Araujo FD, Moynahan ME, Berwick M, Jasin M. Double-strand breaks and tumorigenesis. Trends Cell Biol. 2001;11(11):S52–59. doi: 10.1016/s0962-8924(01)02149-3. [DOI] [PubMed] [Google Scholar]
- Postow L, Funabiki H. An SCF complex containing Fbxl12 mediates DNA damage-induced Ku80 ubiquitylation. Cell Cycle. 2013;12(4):587–595. doi: 10.4161/cc.23408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Ghenoiu C, Woo EM, Krutchinsky AN, Chait BT, Funabiki H. Ku80 removal from DNA through double strand break-induced ubiquitylation. J Cell Biol. 2008;182(3):467–479. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochazkova J, Loizou JI. Programmed DNA breaks in lymphoid cells: Repair mechanisms and consequences in human disease. Immunology. 2015 doi: 10.1111/imm.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond WE, Kleckner N. RAD50 protein of S.cerevisiae exhibits ATP-dependent DNA binding. Nucleic Acids Res. 1993;21(16):3851–3856. doi: 10.1093/nar/21.16.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes-Zamora A, Mihalek I, Lichtarge O, Bertuch AA. Distinct faces of the Ku heterodimer mediate DNA repair and telomeric functions. Nat Struct Mol Biol. 2007;14(4):301–307. doi: 10.1038/nsmb1214. [DOI] [PubMed] [Google Scholar]
- Rodgers K, McVey M. Error-Prone Repair of DNA Double-Strand Breaks. J Cell Physiol. 2016;231(1):15–24. doi: 10.1002/jcp.25053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropars V, Drevet P, Legrand P, Baconnais S, Amram J, Faure G, Marquez JA, Pietrement O, Guerois R, Callebaut I, et al. Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proc Natl Acad Sci U S A. 2011;108(31):12663–12668. doi: 10.1073/pnas.1100758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Meier B, McAinsh AD, Feldmann HM, Jackson SP. Separation-of-function mutants of yeast Ku80 reveal a Yku80p-Sir4p interaction involved in telomeric silencing. J Biol Chem. 2004;279(1):86–94. doi: 10.1074/jbc.M306841200. [DOI] [PubMed] [Google Scholar]
- Ruiz JF, Pardo B, Sastre-Moreno G, Aguilera A, Blanco L. Yeast pol4 promotes tel1-regulated chromosomal translocations. PLoS Genet. 2013;9(7):e1003656. doi: 10.1371/journal.pgen.1003656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten SL, Caldecott KW. DNA strand break repair and neurodegeneration. DNA Repair (Amst) 2013;12(8):558–567. doi: 10.1016/j.dnarep.2013.04.008. [DOI] [PubMed] [Google Scholar]
- Saad H, Gallardo F, Dalvai M, Tanguy-le-Gac N, Lane D, Bystricky K. DNA dynamics during early double-strand break processing revealed by non-intrusive imaging of living cells. PLoS Genet. 2014;10(3):e1004187. doi: 10.1371/journal.pgen.1004187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Dominska M, Petes TD. Transformation of Saccharomyces cerevisiae with nonhomologous DNA: illegitimate integration of transforming DNA into yeast chromosomes and in vivo ligation of transforming DNA to mitochondrial DNA sequences. Mol Cell Biol. 1993;13(5):2697–2705. doi: 10.1128/mcb.13.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuric Z, Chan CY, Hafer K, Schiestl RH. Ionizing radiation induces microhomology-mediated end joining in trans in yeast and mammalian cells. Radiat Res. 2009;171(4):454–463. doi: 10.1667/RR1329.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010;29(19):3370–3380. doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, Lee SE. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol. 2007;27(5):1602–1613. doi: 10.1128/MCB.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Ma JL, Oum JH, Yanez Y, Lee SE. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol Cell Biol. 2005;25(10):3934–3944. doi: 10.1128/MCB.25.10.3934-3944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14(19):1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003;17(19):2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulek M, Yarrington R, McGibbon G, Boeke JD, Junop M. A critical role for the C-terminus of Nej1 protein in Lif1p association, DNA binding and non-homologous end-joining. DNA Repair (Amst) 2007;6(12):1805–1818. doi: 10.1016/j.dnarep.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Symington LS. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb Perspect Biol. 2014;6(8) doi: 10.1101/cshperspect.a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Roh DH, Chen L, Van Komen S, Tomkinson A, Sung P. Yeast xrs2 binds DNA and helps target rad50 and mre11 to DNA ends. J Biol Chem. 2003;278(49):48957–48964. doi: 10.1074/jbc.M309877200. [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem. 2001;276(38):35458–35464. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- Tseng HM, Tomkinson AE. A physical and functional interaction between yeast Pol4 and Dnl4-Lif1 links DNA synthesis and ligation in nonhomologous end joining. J Biol Chem. 2002;277(47):45630–45637. doi: 10.1074/jbc.M206861200. [DOI] [PubMed] [Google Scholar]
- Tseng HM, Tomkinson AE. Processing and joining of DNA ends coordinated by interactions among Dnl4/Lif1, Pol4, and FEN-1. J Biol Chem. 2004;279(46):47580–47588. doi: 10.1074/jbc.M404492200. [DOI] [PubMed] [Google Scholar]
- Tseng SF, Gabriel A, Teng SC. Proofreading activity of DNA polymerase Pol2 mediates 3′-end processing during nonhomologous end joining in yeast. PLoS Genet. 2008;4(4):e1000060. doi: 10.1371/journal.pgen.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438(7066):379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia M, Bentele M, Vaze MB, Herrmann G, Kraus E, Lee SE, Schar P, Haber JE. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature. 2001;414(6864):666–669. doi: 10.1038/414666a. [DOI] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 2007;26(18):4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119(6):777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- Vincent JA, Kwong TJ, Tsukiyama T. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat Struct Mol Biol. 2008;15(5):477–484. doi: 10.1038/nsmb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412(6847):607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci U S A. 2009;106(30):12430–12435. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasko BM, Holland CL, Resnick MA, Lewis LK. Inhibition of DNA double-strand break repair by the Ku heterodimer in mrx mutants of Saccharomyces cerevisiae. DNA Repair (Amst) 2009;8(2):162–169. doi: 10.1016/j.dnarep.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland JW, Summers JA, Holland CL, Resnick MA, Lewis LK. Blunt-ended DNA double-strand breaks induced by endonucleases PvuII and EcoRV are poor substrates for repair in Saccharomyces cerevisiae. DNA Repair (Amst) 2010;9(6):617–626. doi: 10.1016/j.dnarep.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TE, Grawunder U, Lieber MR. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388(6641):495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- Wilson TE, Lieber MR. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J Biol Chem. 1999;274(33):23599–23609. doi: 10.1074/jbc.274.33.23599. [DOI] [PubMed] [Google Scholar]
- Wu D, Topper LM, Wilson TE. Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics. 2008;178(3):1237–1249. doi: 10.1534/genetics.107.083535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wilson TE, Lieber MR. A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc Natl Acad Sci U S A. 1999;96(4):1303–1308. doi: 10.1073/pnas.96.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Matsumoto Y, Trujillo KM, Lees-Miller SP, Osley MA, Tomkinson AE. Role of the yeast DNA repair protein Nej1 in end processing during the repair of DNA double strand breaks by non-homologous end joining. DNA Repair (Amst) 2015;31:1–10. doi: 10.1016/j.dnarep.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Gabriel A. Reciprocal translocations in Saccharomyces cerevisiae formed by nonhomologous end joining. Genetics. 2004;166(2):741–751. doi: 10.1534/genetics.166.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Paull TT. The Mre11/Rad50/Xrs2 complex and non-homologous end-joining of incompatible ends in S. cerevisiae. DNA Repair (Amst) 2005;4(11):1281–1294. doi: 10.1016/j.dnarep.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, Kwon Y, Sung P, Lee SE, Tomkinson AE. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol. 2007;14(7):639–646. doi: 10.1038/nsmb1261. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134(6):981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]