Abstract
Fungi are morphologically, ecologically, metabolically, and phylogenetically diverse. They are known to produce numerous bioactive molecules, which makes them very useful for natural products researchers in their pursuit of discovering new chemical diversity with agricultural, industrial, and pharmaceutical applications. Despite their importance in natural products chemistry, identification of fungi remains a daunting task for chemists, especially those who do not work with a trained mycologist. The purpose of this review is to update natural products researchers about the tools available for molecular identification of fungi. In particular, we discuss (1) problems of using morphology alone in the identification of fungi to the species level; (2) the three nuclear ribosomal genes most commonly used in fungal identification and the potential advantages and limitations of the ITS region, which is the official DNA barcoding marker for species-level identification of fungi; (3) how to use NCBI-BLAST search for DNA barcoding, with a cautionary note regarding its limitations; (4) the numerous curated molecular databases containing fungal sequences; (5) the various protein-coding genes used to augment or supplant ITS in species-level identification of certain fungal groups; and (6) methods used in the construction of phylogenetic trees from DNA sequences to facilitate fungal species identification. We recommend that, whenever possible, both morphology and molecular data be used for fungal identification. Our goal is that this review will provide a set of standardized procedures for the molecular identification of fungi that can be utilized by the natural products research community.
Fungi represent the second largest group of eukaryotic organisms on earth, with estimates ranging from 1.5 to 5.1 million species.1−3 Members of the fungal kingdom play significant roles in human life and have the ability to occupy a wide variety of natural and artificial niches.4 Identification of fungi to species level is paramount in both basic (ecology, taxonomy) and applied (genomics, bioprospecting) applications in scientific research. This is especially true for natural products researchers working with fungi as a source of bioactive secondary metabolites. Scientific names are crucial in communicating information about fungi, enabling researchers to identify other closely related species to better predict evolution of chemical gene clusters,5 or to prioritize taxonomically related strains, when a productive strain may attenuate production of key bioactive compounds.6 More importantly, taxonomic identification of fungi is essential if industrial, agrochemical, or pharmaceutical products are to be derived from a fungal strain.
Fungi produce a wealth of natural products; they have major industrial applications7 and are well-known for their ability to produce secondary metabolites with biological activities that can be used for drug discovery.8,9 Secondary metabolites from fungi represent a substantial fraction of our current pharmaceuticals, including the often-cited penicillin, as well as those used as cholesterol-lowering, antibiotic, or immunomodulatory agents.10−12 Some notable examples include pravastatin (∼$3.6 billion/year),13 cyclosporine (∼$1.4 billion/year), amoxicillin (∼$11.7 billion/year),8 and fingolimod (sold as Gilenya; ∼$1 billion/year).14 Thus, it is likely that new fungal-derived chemical compounds will be on the market in the coming years, and identifying those fungi would thus be a critical task.
The Journal of Natural Products has published an average of 37 fungal natural product articles every year for the past 16 years (2000–2015) (Figure 1). A survey of these studies revealed that ∼31% provided fungal identification based solely on morphology; ∼28% of them did not report any form of identification for the fungus from which secondary metabolites were isolated; 27% of the studies used molecular data only (mostly from the internal transcribed spacer (ITS) region) for fungal identification; and ∼14% used a combination of morphology and molecular data (both rRNA and protein-coding genes) to identify fungi (Figure 2A). This suggests that the taxonomic identification of fungi is a topic that could use standardization, especially as applied toward the discovery of bioactive secondary metabolites. However, if one examines the most recent six years (Figure 2B), the trend is moving in the right direction. In fact, regardless of the source of organism, be it plant, microbe, or animal in origin, taxonomic identification is a critical step to ensure reproducibility, and fungal cultures are no exception to this rule. Specific to drug discovery, accurate species identification can unlock important information regarding a species and its possible biochemical properties. More broadly, this in turn can provide insights into developing better screening programs for discovery of interesting natural products, as well as additional information regarding ecology, phylogenetic relationships, genomics, and transcriptomics among fungi.
Figure 1.
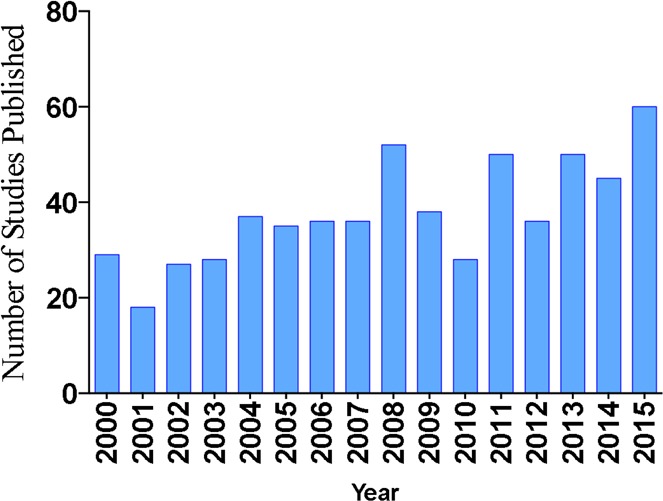
Number of studies that used fungi as a source material, as reported annually in the Journal of Natural Products from 2000 to 2015.
Figure 2.
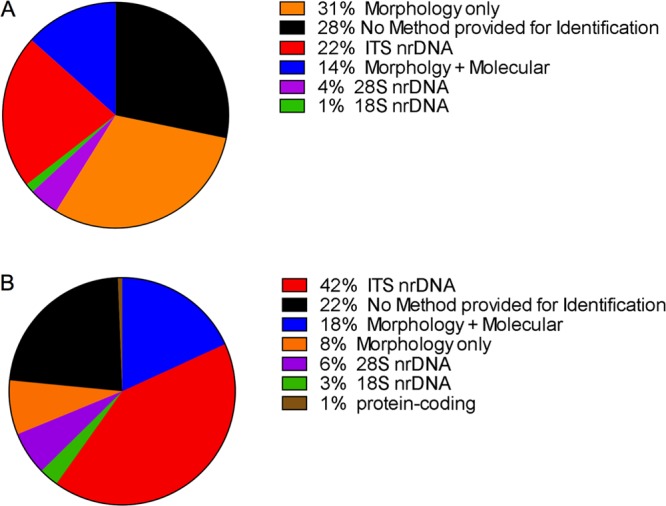
(A) Percentage of studies published in the Journal of Natural Products (2000–2015) employing various methods, or no method at all, for the taxonomic identification of fungi. (B) The same data categories, but focusing on the time period of 2010–2016. These charts illustrate how the use of molecular data in fungal identification is increasing, especially ITS data, which is the official fungal barcode marker. nr: nuclear ribosomal.
In light of the importance of fungi in natural products research and given the growing trend of studies that utilize fungi as a source material, this paper aims to address the need for a set of adoptable standardized procedures for the identification of fungi by discussing (1) problems of using morphology alone in the identification of fungi to the species level; (2) the three nuclear ribosomal genes most commonly used in fungal identification and the potential advantages and limitations of the ITS region, which is the official DNA barcoding marker for species-level identification of fungi; (3) how to use NCBI-BLAST search for DNA barcoding, with a cautionary note regarding its limitations; (4) the numerous curated molecular databases containing fungal sequences; (5) the various protein-coding genes used to augment or supplant ITS in species-level identification of certain fungal groups; and (6) methods used in the construction of phylogenetic trees from DNA sequences to facilitate fungal species identification. While the use of these methods may be weighted to a greater or lesser extent depending on the goals of the study, and while new developments, particularly with respect to DNA sequencing, may augment or even supplant some of these methods, their use and reporting in a consistent manner should add more reproducibility to the natural products research community. This is especially important, since it is likely that in the near future the identification of fungi, including description of new species, may rely heavily on DNA-based methods, including those derived solely from environmental or metabarcoding molecular sequences.15−18
1. Problems of Using Morphology Alone in the Identification of Fungi to the Species Level
Mycologists have traditionally used morphology (phenotypic characters), such as spore-producing structures formed as a result of asexual (mitosis) or sexual (meiosis) reproduction, as a sole means of identifying fungal species,19 and even today it is still adopted as a means of species identification within the mycological community. Use of morphology in fungal species identification is very important to understand the evolution of morphological characters. However, morphological approaches to fungal systematics, although routinely used in fungal taxonomic studies for classification of fungi at the ordinal or familial level,20 may not always perform well for lower-level (species) classifications21 due to the reasons outlined below. In some highly speciose lineages of fungi, morphological characters can be contentious or problematic even for trained mycologists, as they may not always provide accurate groupings within an evolutionary framework, mainly at the species level.22 Morphological characters can often be misleading due to hybridization,23,24 cryptic speciation,25−29 and convergent evolution.30 In addition, until recently it was common practice in mycology to name both asexual and sexual stages of the same fungus, most times with a different name (termed dual nomenclature), which caused confusion. This practice is no longer acceptable according to the latest International Code of Nomenclature for algae, fungi, and plants.16,31 Moreover, identifying fungi based on morphology alone can be challenging, especially when nonexperts are dealing with cultures of fungi, since there are a limited number of morphological characters that can be used for identification. For example, endosymbiotic fungal strains, such as endophytes and endolichenic fungi, which are routinely used for isolating secondary metabolites, do not always sporulate in culture, thereby providing no phenotypic characters for which to identify them based on morphology.32,33 For the fungi that do sporulate in culture, the asexual structures, such as conidia/spore shape and size, can often show highly plastic characters, making identification challenging. For sexual states (teleomorphs), ascomata are not often produced in culture, thus making it difficult to identify them based solely on morphology. As a consequence, DNA sequence-based methods have emerged for identifying species within the megadiverse fungi.15,16,34−36
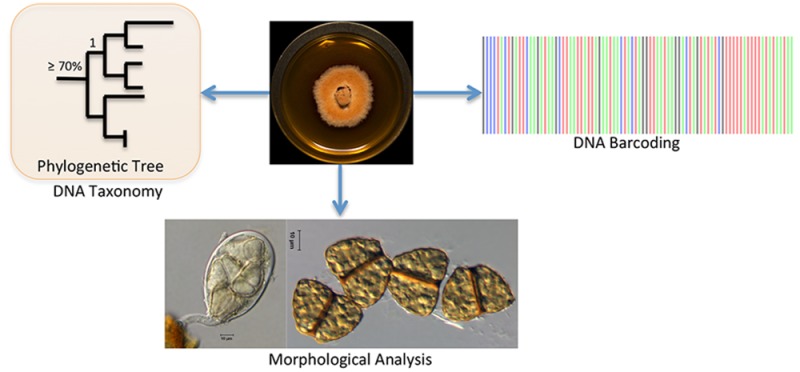
In the following sections, we discuss two methods used in mycology for sequence-based identification of fungi, namely, DNA barcoding using the ITS region and DNA taxonomy using one or multiple genes in sequence alignments and employing tree-building tools to estimate phylogenetic relationships. In DNA barcoding, the user compares an unknown sequence against a sequence database, such as either International Sequence Database (INSD: GenBank at the National Center for Biotechnology Information, GenBank, NCBI; the European Nucleotide Sequence Archive of the European Molecular Biology Laboratory, EMBL; and the DNA Data Bank of Japan, DDBJ) or UNITE (User-friendly Nordic ITS Ectomycorrhiza Database; see Table 1), and identifies species based on sequence similarity (Figure 3A). Alternatively, in DNA taxonomy the user aims to identify an unknown species by placing it into an evolutionary framework with other homologous sequences using a phylogenetic approach (Figure 3B). It is important to realize that some barcodes, due to their rapid evolution and thus high divergence at shallower (species and genus) taxonomic levels, are not useful for phylogenetic reconstruction at deeper (familial or ordinal) levels. Sections 2–5 are dedicated to the molecular identification of fungi, fungal DNA barcoding, BLAST search, curated fungal barcoding databases, and secondary barcoding markers in fungi. Section 6 then provides an overview of multiple sequence alignment and tree-building techniques to analyze phylogenetic relationships in order to identify and/or place an unknown fungal species using an evolutionary and taxonomic framework.
Table 1. List of Curated Databases for Fungal Species Identification (Adapted from Yahr et al.)75 a.
| name of the database | URL | region utilized |
|---|---|---|
| Barcode of Life Database, BOLD | http://www.boldsystems.org/index.php/IDS_OpenIdEngine | ITS |
| CBS-KNAW | http://www.cbs.knaw.nl/Collections/BioloMICSSequences.aspx | ITS |
| FUSARIUM-ID | http://isolate.fusariumdb.org | ITS, tef1, RPB1, RPB2, tub2 |
| Fungal Barcoding | http://www.fungalbarcoding.org | ITS |
| Fungal MLST database Q-Bank | http://www.q-bank.eu/Fungi/ | partial actin, tub2, RPB1. RPB2, tef1 among others |
| ISHAM, The International Society for Human and Animal Mycology | http://its.mycologylab.org | ITS |
| Naïve Bayesian Classifier | http://rdp.cme.msu.edu/classifier/classifier.jsp | 28S, ITS |
| RefSeq Target Loci (RTL) | http://www.ncbi.nlm.nih.gov/refseq/targetedloci/ | ITS, 18S, 28S |
| International Subcommision on Hypocrea and Trichoderma (ISHT) TrichoKey and TrichoBLAST (Trichoderma) | http://www.isth.info/tools/blast/ | ITS and tef1, RPB2 |
| UNITE, User-friendly Nordic ITS Ectomycorrhiza Database | https://unite.ut.ee/ | ITS |
For an exhaustive list, see Robert et al.110
Figure 3.
DNA barcoding (A) vs DNA taxonomy (B) (adapted from Vogler and Monaghan).37 For DNA barcoding (A), sequence data [ITS or other taxon-specific gene(s)] are obtained directly from a fungal fruiting body or a fungal culture, and a BLAST search is conducted in INSD, such as GenBank or other specialized databases. Identification is based on overall sequence similarity (e.g., 97–100% sequence similarity) with other reference sequences in INSD, thereby providing an identification of an unknown sequence. The example is of Lentinula edodes, which was recently published.38 In DNA taxonomy (B), after BLAST search, the most homologous sequences (usually the top 50–100 sequences) are downloaded from the INSD and incorporated into a multiple sequence alignment, and phylogenetic tree building methods, such as maximum likelihood and Bayesian inferences, are applied. Using these methods, the unknown sequence is identified within an evolutionary framework. The example is from our research on strain G77, which was published previously in this journal and identified as Aspergillus iizukae.39 Note: In some cases, Aspergillus identification can be challenging with ITS alone (see Section 5).
2. The Three Nuclear Ribosomal Genes Most Commonly Used in Fungal Identification and the Potential Advantages and Limitations of the ITS Region, Which Is the Official DNA Barcoding Marker for Species-Level Identification of Fungi
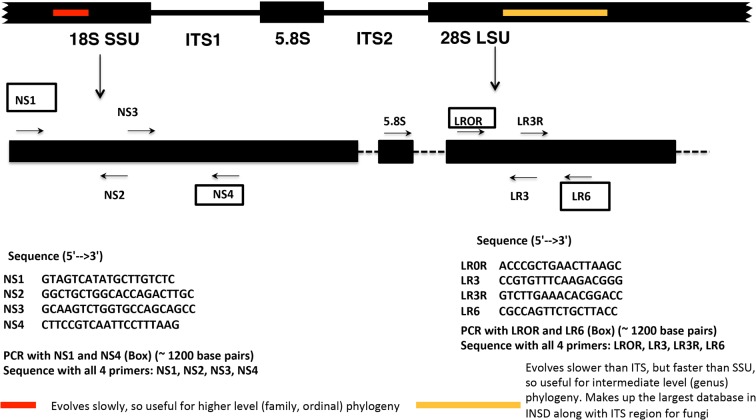
Use of molecular data for the identification of fungi began over two decades ago with the seminal paper describing fungal nuclear ribosomal operon primers by White et al.40 The fungal DNA sequences generated with these primers for the large subunit (nrLSU-26S or 28S), small subunit (nrSSU-18S), and the entire internal transcribed spacer region (ITS1, 5.8S, ITS2; ca. 0.45–0.80 kb) ushered in a new era of molecular phylogenetic sequence identification in the kingdom fungi.41,42 Different rates of evolution, resulting in varying levels of genetic variation, have been observed for these three separate regions, with SSU evolving the slowest, thus possessing the lowest amount of variation among taxa, while the ITS evolves the fastest and exhibits the highest variation.41,43 If a researcher is interested in the phylogenetic placement of a fungus at higher taxonomic levels (family, order, class, and phyla), the SSU can be amplified and sequenced using the primer combination NS1 and NS4 (Figure 4).40 Alternatively, if the identification needs to be made at the intermediate taxonomic levels (family, genera), then the user can amplify and sequence the LSU using the primer combination LROR and LR6 (Figure 4).44,45 The LSU region, which contains the D1 and D2 hypervariable domains, on its own,46 or when combined with the ITS region, can also be valuable for species identification in fungi.47−49 For species-level identification, the ITS is the most useful, as it is the fastest evolving portion of the rRNA cistron (Figure 5). Due to its ease in amplification, widespread use, and appropriately large barcode gap (i.e., the difference between interspecific and infraspecific variation), the ITS was chosen as the official barcode for fungi by a consortium of mycologists.48 Hence, for natural products research, the first two domains of the LSU gene and the entire ITS region should be sequenced due to their high prevalence in fungal taxonomy and systematics (for protocols, see Experimental Section and Table S1). While the LSU can be used in phylogenetic analyses (Section 6) to determine species relationships, the ITS can be used alone or in conjunction with other protein coding genes (Section 5) for species identification. These genes have been widely used in major landmark studies in fungal systematics, such as Assembling the Fungal Tree of Life (AFTOL).50,51 Thus, large numbers of nrDNA sequences of fungal rRNA exist in GenBank for species identification via barcoding (section 3) and phylogenetic analysis (Section 6).
Figure 4.
Primers for amplification of small-subunit (SSU) nrRNA and large-subunit (LSU) nrRNA. Adapted from (http://sites.biology.duke.edu/fungi/mycolab/primers.htm) Vilgalys Lab, Duke University (NS primers;40 LR primers44,45).
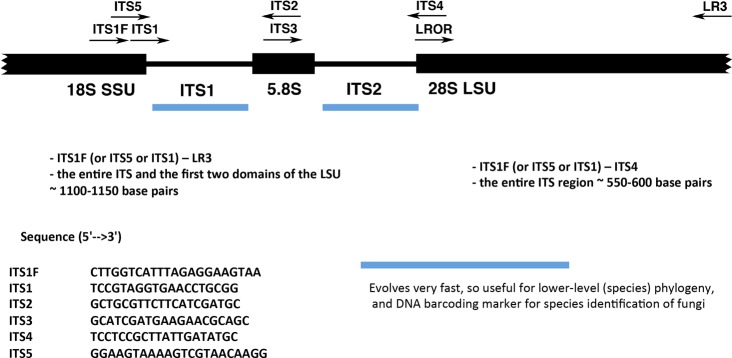
Figure 5.
Advantages and Limitations of Using the ITS Region as a DNA Barcoding Marker in Fungi
DNA barcoding was coined originally for species identification in animals.52,53 DNA barcoding systems employ a short standardized region (between 400 and 800 base pairs) to identify species.54 The premise of DNA barcoding is that interspecific variation should exceed infraspecific variation. Thus, this difference, known as the barcode gap, could be exploited for species-level identification. DNA barcoding and its application and methodology have been reviewed previously.54−57
In 2007, a group of mycologists organized a workshop in Virginia, USA, to discuss various genes that could be used for fungal barcoding.58−61 These discussions set the stage for a multinational consortium of mycologists to meet in Amsterdam in 2011 to evaluate six DNA regions (SSU, LSU, ITS, RPB1, RPB2, MCM7) and nominate the official fungal barcode, the ITS region, which was later approved by the Consortium for the Barcode of Life.48 The resulting paper (coauthored by >100 mycologists) included taxa sampled from all major fungal phyla, including the Ascomycota and Basidiomycota, the largest phyla within the kingdom fungi.51 These two phyla include the fungi from which the greatest number of secondary metabolites have been isolated.8,9 The recognition of ITS as the official DNA barcode marker for fungi represents a noteworthy advance, which has greatly benefited the research community.
Schoch et al.48 found the ITS region to be among the markers with highest probability of correct identifications for a very broad group of sampled fungi. Additional fungal studies have provided support for the ITS region as a suitable fungal barcode.63−65 Protein-coding genes are often difficult to amplify and sequence, since they occur as a single copy within the genome rather than as multiple copy tandem repeats as with the ribosomal genes. Moreover, most environmental surveys (metagenomic studies) of fungi are using the ITS region in their studies1,15,66−73 to identify fungi using modern sequencing technology, such as next-generation sequencing, which can aid in rapid identification of fungi without the need to clone the amplicons. This sequencing approach has therefore created a surge in the number of ITS sequences that are available in GenBank.59,74 According to Schoch and colleagues,48 currently about 172 000 full-length fungal ITS sequences are deposited in GenBank, of which 56% have a Latin binomial associated, representing approximately 2500 genera and 15 500 species. Sequencing the ITS region of voucher specimens deposited in fungaria provides additional authoritative data. The ITS region is a useful barcode marker because it usually can be sequenced from previously described fungi for which no sequence data are currently available by sequencing the type material (i.e., the specimen on which a species is originally described and deposited in a fungarium). Therefore, the practicality and broad kingdom-wide taxonomic applicability make ITS a useful tool for fungal barcoding for most (∼70% of all fungi tested)48,75 if not all lineages of fungi.76−79
Even though the ITS region performs well as a suitable fungal barcoding marker, it has been subject to debate.78 The ITS region does not work well in some highly speciose genera, such as Aspergillus, Cladosporium, Fusarium, Penicillium, and Trichoderma, as these taxa have narrow or no barcode gaps in their ITS regions.48,79−84 This problem is significant for natural products research since these genera yield many important secondary metabolites, most notably penicillin. In addition, intragenomic ITS variation occurs in some groups of fungi,85−87 although more recent studies suggest that it is not largely prevalent in fungi (occurring in ∼3–5% of 127 fungi belonging to Ascomycota and Basidiomycota).88 Partial solutions for identification of these important genera are discussed in Section 5. Moreover, lineage-specific cutoff values for species determination using the ITS region are still being evaluated, and no single value can be applied across all groups;76 this is especially true for morphologically similar cryptic species, which are often revealed through a DNA-based phylogenetic approach based on single or multiple protein-coding loci89,90 (see Section 6).
3. How to Use NCBI-BLAST Search for DNA Barcoding, with a Cautionary Note Regarding Its Limitations
Each ITS sequence fragment is subjected to an individual Basic Local Alignment Search Tool (BLAST)91 in GenBank to verify identity.92 BLAST search is usually employed using nucleotide collection (BLASTn), where curated RefSeq records, as well as their GenBank duplicates, are included by default in BLAST.93−95 There are several ways to do more limited BLAST searches in order to improve results such as limiting a BLAST search to RefSeq sequences only (using the BioProject numbers; see Section 4 and https://www.ncbi.nlm.nih.gov/bioproject/PRJNA177353/). This Entrez query can be added to limit the RefSeq to only the 177353 [i.e., BioProject], which is a Fungal Internal Transcribed Spacer RNA (ITS) RefSeq Targeted Loci Project. Alternatively, there is a dedicated BLAST for each of the various RefSeq projects: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&BLAST_SPEC=TargLociBlast. For further details on the NCBI taxonomy database, the user is encouraged to read a review by Federhen.96
Experts in the mycological community, including both taxonomists and ecologists, have recently proposed a set of working rules for ITS data. These rules include the submission of molecular data to public databases for the identification of fungi. Briefly, the most important rules are (1) verify that all query sequences are representative of the complete ITS region; (2) verify for orientation and chimeric sequences via BLAST; and (3) verify taxonomic annotations carefully, by using only authenticated sequences, where possible.97−99
Due to the lack of a definitive percentage sequence similarity that could precisely indicate conspecific taxa, no single cutoff value has been universally established for species identification across the kingdom fungi. In the past, mycologists have used an arbitrary cutoff value ranging from ≤3% to 5% for ITS sequence divergence to indicate conspecificity among fungi.1,100−104 For BLAST search, it is reasonable to start with ≥80% query coverage and ≥97–100% sequence similarity (i.e., up to 3% sequence divergence) for assigning a species name based on consideration of results from the GenBank BLAST search since the average weighted infraspecific ITS variability in the kingdom fungi was calculated to be 2.51% with a standard deviation of 4.57%.76 The average weighted infraspecific ITS variability for Ascomycota was calculated as 1.96% with a standard deviation of 3.73%, while the average weighted infraspecific ITS variability for Basidiomycota was calculated as 3.33% with a standard deviation of 5.62%.70 These values require further evaluation, as no single value appears to fit well with current morphological-based identification across the fungi.76,105
In addition, it is imperative that fungal sequences be deposited in GenBank and their accession numbers included in manuscripts: this is now standard practice for this journal. The user must always deposit the sequence data to GenBank and, after the manuscript in which the sequences are reported is published, inform GenBank about the pagination; this is often not accomplished by many researchers, which results in many unpublished sequences in GenBank. Failure to do so may result in your sequence remaining unpublished in GenBank, until informed by a third party. Over the long term, releasing data to GenBank would greatly improve this resource for the scientific community.
Limitations of Using GenBank BLAST Search
It has been advised that BLAST search identifications against GenBank should be made with caution, as approximately 27% of GenBank fungal ITS sequences were submitted with insufficient taxonomic identification.106 In addition, about 20% of fungal sequences in GenBank can be incorrectly annotated, but this will vary greatly according to taxonomic group.107,108 Additional concerns expressed by the mycological community include the following:
-
(1)
Third-party annotations of records are not allowed in GenBank (although records can be removed from BLAST if they are clearly misidentified).
-
(2)
Taxonomic names are not up to date due to the rapidly changing nature of fungal taxonomy.
-
(3)
Most fungal species described (about 70%) thus far have not been sequenced.109
-
(4)
Type strains are not obvious or clearly indicated (see caveats to this point below).
-
(5)
Many sequences are unnamed or only partially named.110
There are a few caveats to point 4, and the general solution to most of these would necessitate working with a mycologist. For example, in order to find type strains, one needs to find the original publications of fungal species and search within the RefSeq database. However, this challenge is compounded by the fact that not all fungal species are available in the RefSeq database. One can now insert their type data into the RefSeq database; see Federhen.111
In addition, there are many unpublished sequences in GenBank, which may decrease the authenticity of the data, because the unpublished sequence name could be incorrect or the researcher who submitted the sequence to GenBank might have erroneously interpreted the name. We strongly recommend the user to check for the presence of specimen vouchers and other collection information connected to the unpublished sequences prior to using them. If further doubt about the authenticity of a sequence persists, it can always be eliminated from the BLAST (see point 1, above).
In a recent study on the phylogeny of xylariaceous fungi isolated as endophytes, U’Ren et al.112 indicated that the user should be careful while assigning higher-level taxonomic information to ITS sequences via GenBank BLAST search alone. These authors concluded that blindly assigning taxonomy for unknown isolates based solely on BLAST search results could lead to misidentifying the fungi;112 several other studies have also highlighted concerns of simply using GenBank BLAST search data without careful analysis of the sequence data107−109,113−115 (see Section 4). We echo these concerns, but in doing so, we also stress that there are ways to mitigate these shortcomings, particularly if one is working with a trained mycologist (see Conclusions). It is also highly recommended that the users cite references to the sequences that were used from GenBank.113
4. The Numerous Curated Molecular Databases Containing Fungal Sequences
There are several curated databases that are dedicated to the identification of ITS and other ribosomal and protein-coding sequences that have been established for different groups of fungi (see Table 1 and refs (110) and (116)–119). Robert et al.110 provide an inclusive list of Web sites and online databases where pairwise alignments or polyphasic identifications are possible. Most of these databases are curated; thus identification of fungi via ITS can be accomplished against authenticated sequence data, which in a number of cases can be linked to a vouchered specimen. As mentioned earlier, GenBank may contain erroneous names associated with ITS sequences, and, despite proposals by the mycological community to allow third-party annotation,120 GenBank has limitations (Section 3). In an attempt to resolve this issue of providing a database in GenBank of properly identified taxonomic names associated with ITS sequences, a collaborative study focused on authenticated sequences from the ITS region that were obtained from type specimens111 and/or ex-type cultures (a living culture obtained from a type species is referred to as an ex-type).95 In this study, the authors reannotated and verified sequences in a curated public database referred to as the RefSeq Targeted Loci (RTL) database.94 There is a dedicated BLAST database for ITS RefSeq at its BioProject page (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA177353/). Users can also restrict any BLAST results by only comparing sequences from type material by checking the “sequences from type material” box in the general BLAST page. For further information on fungal RefSeq at NCBI-GenBank, we refer the user to a recent paper by O’Leary et al.94 Another example, Q-Bank, is a curated database used for the identification of phytopathogenic genera such as Ceratocystis, Colletotrichum, Melampsora, Monilinia, the Mycosphaerella generic complex, Phoma, Phytophthora, Puccinia, Stenocarpella, Thecaphora, and Verticillium.(121) The ISHAM (The International Society for Human and Animal Mycology) working group for DNA barcoding has recently established a database, with special focus on the majority of human and animal pathogenic fungi.118 In addition to GenBank, another popular database, termed UNITE (http://unite.ut.ee/), is often used to identify unknown ITS sequences. UNITE, which provides species hypotheses based on ITS sequence similarity,116 is curated by experts. There are direct links from GenBank to sequences at the ISHAM and UNITE databases that are listed under “LinkOut to external resources” (when available) in the individual records. FUSARIUM-ID122 and TrichoBLAST/TrichoKeys123 are curated databases for identification of Fusarium spp. and Trichoderma spp., respectively. The Naïve Bayesian Classifier matches rRNA sequences from both the LSU and ITS regions and provides taxonomic identities by comparing the sequences against a set of sequences with known taxonomy,47,124 thus providing reliable and accurate identifications. The CBS-KNAW database allows the user to select several different curated databases simultaneously and permits sequence identification across the selected databases. The BOLD database has some curated ITS data from fungi, but it is not adequately populated and thus less likely to return a fully identified species name.110 In short, given the wide variety of databases available (Table 1), the user is encouraged to examine those most appropriate for the fungi under study and, where possible, search across more than one database to compare results based on mutually supportive data.
5. The Various Protein-Coding Genes Used to Augment or Supplant ITS in Species-Level Identification of Certain Fungal Groups
Protein-coding genes are utilized for species identifications via barcoding due to the presence of intron regions, which sometimes evolve at a faster rate compared to ITS22 and are employed in phylogenetic analyses due to their better resolution at higher taxonomic levels compared to rRNA genes.125 Moreover, these genes allow for an easy recognition of homology and convergence, as they are believed to occur as a single copy in fungi, are less variable in their length since they accumulate fewer mutations in their exons, and are easier to align over rRNA genes, as they contain less ambiguity due to codon constraints.126,127 Protein-coding genes in fungal systematics have been used widely for constructing molecular phylogenies to aid in the identification and classification of taxa, largely due to the landmark studies in systematic and taxonomic mycology supported by the National Science Foundation, such as Assembling the Fungal Tree of Life.21,50,51,125,128 These efforts among fungal systematists lead the way to a more stable classification of the fungal kingdom51 and impart knowledge regarding gaps in our understanding of evolutionary relationships among fungi.129 Among protein-coding markers, the largest (RPB1) and second largest (RPB2) subunits of RNA polymerase,130−133 translation elongation factor 1-alpha (tef1),134 and beta-tubulin (tub2/BenA)135,136 have been most commonly used for inferring phylogenetic relationships among fungi.50,51,125 More recently, the mini-chromosome maintenance protein (MCM7) has shown promise as a new marker for inferring both higher- and lower-level phylogenetic relationships.137−141 The MCM7 region has been shown to be a superior gene for phylogenetics when compared to numerous, routinely utilized protein-coding markers.142 It also works well when used in conjunction with the LSU gene, as shown for members of the Ascomycota137 (for protocols of commonly used genes, see Table S1).
Secondary Barcode Marker for Fungi
Using the ITS marker alone for identification might not suffice in certain fungal clades, and it may be necessary for the user to sequence one or more single-copy protein-coding genes (Table S1) for certain fungal genera and/or lineages to obtain a more precise identification at the species level (e.g., Aspergillus, Penicillium, and Trichoderma). Due to the limitations of a single-marker barcoding system in fungi, a group of mycologists recently completed a study testing >1500 species (1931 strains or specimens) of Dikarya (Ascomycota and Basidiomycota) for different ribosomal and single-copy protein-coding markers.83 The study concluded that a novel, high-fidelity primer pair (EF1–1018F GAYTTCATCAAGAACATGAT and EF1–1620R GACGTTGAADCCRACRTTGTC) for tef-1, which is already widely used as a phylogenetic marker in mycology,134 has the best potential to serve as a secondary DNA barcode, offering superior resolution to ITS83 (for protocol, see Table S1).
The gene regions RPB1, RPB2, tub2/BenA, and partial calmodulin (CaM) are useful for species-level identification in certain lineages of fungi such as Eurotiales, which includes Aspergillus and Penicillium, two of the most prolific genera of fungi for secondary metabolites and which include numerous medicinally and industrially important species.81,143−146 For barcoding of Penicillium spp., tub2/BenA is recommended as a secondary barcoding marker.146 For phylogenetic analyses of Penicillium spp., it is recommended that either RPB2 or CaM genes be sequenced since they are easier to align than tub2/BenA, which is difficult to align across different sections of Penicillium due to the presence of numerous ambiguous regions.146 For Aspergillus, CaM is recommended as a secondary barcoding marker, while tub2/BenA and RPB2 genes are also highly preferred.81 For PCR protocols, including primer sequences of forward and reverse primers of genes used for sections in Aspergillus and Penicillium, the user is referred to Samson et al.81,146 (see Table S1).
Trichoderma spp. have huge implications in human health, the environment, agriculture, and industry;147,148 therefore, identification of Trichoderma is important for natural products research, especially when working on peptaibols.149 Unfortunately, identification of Trichoderma using the ITS region can be challenging, as it does not contain sufficient variation for differentiating among species. For this genus, the ITS region can be used for BLAST search using the TrichoKeys database, which is hosted on the ISHT webpage (Table 1), but doing so may likely provide information only on species clusters. Thus, for species-level identification, the tef-1 region has been widely used in the taxonomy and systematics of this genus.123,150−153 The tef-1 intron 4, in combination with intron 5, is proving to be very useful for species-level identification.154,155 In addition, a 1200 bp fragment of RPB2, which can be amplified and sequenced using primer pairs RPB2–5f and RPB2–7cR,156 is also proving useful for Trichoderma identification using phylogenetic methods. PCR protocols, including the appropriate primers, for the identification of Trichoderma sp. are summarized in Table S1.
6. Methods Used in the Construction of Phylogenetic Trees from DNA Sequences to Facilitate Fungal Species Identification
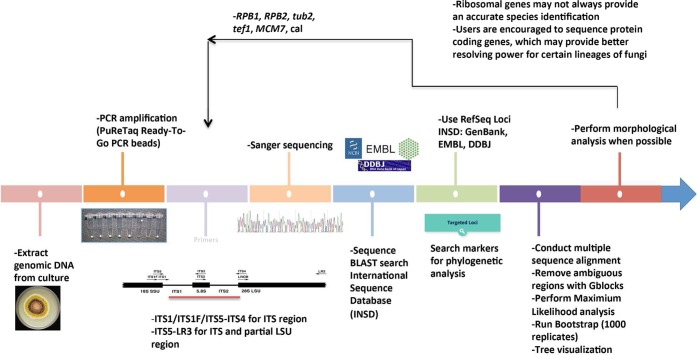
Like DNA barcoding, DNA taxonomy is based on the premise of using the genetic variation inherent among sequences of different individuals to identify taxa. However, the difference between these two methods is that in the latter an unknown is identified based on a phylogenetic hypothesis,157 thus advocating an evolutionary perspective for recognizing fungal species and utilizing an approach with predictive value.22 DNA taxonomy for a particular group of fungi may be based on one or more regions of protein coding or rDNA and can be derived from phylogenetic methods using any gene region individually or in combination.37 While DNA-based taxonomy using phylogenetic theory may not always help with identifying the exact species of a fungus, it will certainly help by placing an unknown species within a phylogenetic clade or group, thus providing a putative species identification. A synopsis of methods and software useful for carrying out analyses inferring or using phylogenetic trees was recently published by Schmitt and Barker.5 When species are identified using sequences that share a common ancestry (homologous sequences), it may be possible to predict species attributes, such as ecology and metabolism.22 Taylor et al.90 established the Genealogical Concordance Phylogenetic Species Recognition (GCPSR) concept to define the limits of sexual species based on phylogenetic concordance of multiple unlinked genetic loci using common housekeeping genes.89 The GCPSR concept is greatly beneficial in identifying fungi, because it has stronger discriminatory power than other species concepts, such as morphological species and biological species concepts.25 Although GCPSR requires two or more genetic loci to define species, according to Geiser et al.,22 a single gene marker could be utilized to provide a general species identification. A workflow of fungal identification via ITS-LSU sequencing, BLAST search, and phylogenetic analysis is shown in Figure 6. Once the ribosomal genes provide a clue to the taxonomic group, such as family or genus, nuclear protein-coding genes could be utilized via BLAST search and/or phylogenetic analyses to provide a more conclusive species-level identification.
Figure 6.
Flowchart for fungal identification using molecular phylogenetic analysis.
To assemble a phylogenetic tree, traditional approaches such as neighbor-joining algorithm (distance based) and parsimony (based on the assertion that the best phylogenetic hypothesis requires the smallest number of evolutionary steps) were used previously. However, both of these have been supplanted by faster model-based methods such as maximum likelihood and Bayesian inference. This is largely because distance-based and parsimony-based methods do not perform well with molecular sequences that are evolving at different rates.158 A summary of strengths and weaknesses of different tree reconstruction methods is provided by Yang and Rannala.159
Maximum Likelihood
The optimality criterion in maximum likelihood methods is to find the phylogenetic tree with the highest likelihood score and is based on a stochastic model of nucleotide or amino acid sequence evolution. For theory on maximum likelihood based phylogeny estimation, the user should refer to Felsenstein,160 who was one of the first to use this approach for phylogenetic estimation via DNA sequence data. For an extensive guide to phylogeny (i.e., tree) building methodology, see Harrison and Langdale,161 while Schmitt and Barker5 provide a useful outline and decision scheme including an extensive list of different software programs and packages available for phylogenetic analyses. In general, to conduct a maximum likelihood analysis for the identification of a fungus, the typical natural product researcher would likely benefit by working with a trained mycologist who utilizes phylogenetic methods for building trees or a bioinformatician.
Bayesian Inference
Bayesian inference, based on the Bayesian theorem, was introduced to molecular phylogenetics in early 2000 due to the availability of complex evolutionary models.162,163 Bayesian inference of phylogenetics is similar to maximum likelihood in that the user postulates a model of evolution and the software program searches for the best tree that is consistent with the model and the data (i.e., the alignment). However, Bayesian inference differs from maximum likelihood in that while the latter seeks the tree that maximizes the probability of the sequence data given in the tree, Bayesian inference seeks the tree that maximizes the probability of the tree given the data and model of sequence evolution. While the Bayesian method has a strong connection to maximum likelihood, it provides a faster measure of clade support and differs from maximum likelihood in that it allows for complex models of nucleotide or amino acid evolution to be implemented.158 The theory on Bayesian inference based phylogeny estimates has been reviewed.5,158,159 As with maximum likelihood, most researchers may benefit by working with a trained mycologist or a bioinformatician to implement Bayesian inference.
Conclusions
1. Whenever possible, identification of fungi should be made using a combination of micromorphological, cultural, and molecular characters. The entire ITS region alone or in combination with the first two domains of the LSU (and one or more protein-coding genes in specific cases) should always be compared with authenticated, published sequences; such information would be appropriate for the Supporting Information. For manuscripts in the chemical literature, one should strive for at least genus- and/or family-level identification. If a fungus garners enough interest for further study, then it may be possible to follow up with species-level identification in the future using methods outlined herein.
2. The ITS region works well in most cases, but should be used with caution, especially when the user is employing only the GenBank BLAST search for identification. It is recommended that the user confirm GenBank identifications against curated sequence data (Table 1), as these data can be more reliable. For cases where the ITS region is not ideal, such as (a) when there is not sufficient resolution for species identification as in Aspergillus, Fusarium, Penicillium, and Trichoderma and/or (b) when one is working with multiple, morphologically similar species, which may exhibit rampant cryptic speciation, the user should analyze other protein-coding gene regions (see Table S1).
3. One should conduct phylogenetic analyses with nuclear ribosomal genes first since more data are available for comparison in public databases. Once the user is able to narrow in on a taxonomic group or lineage, or if ribosomal genes are inconclusive, protein-coding data can be utilized in combination with ribosomal genes or separately for finer-scale species-level identification. DNA taxonomy employing phylogenetic analyses is useful for placing an unknown sequence into an existing classification in an evolutionary framework.
4. It is extremely important to use sequences that are published for comparisons and, where possible, to reference the original source of the sequences. The user must always deposit the sequence data to GenBank and, after the manuscript in which the sequences are reported is published, inform GenBank about the pagination; this is often not done by many researchers, which results in many unpublished and unreliable sequences in GenBank.
5. Both basic and applied fungal research would benefit tremendously from DNA barcoding, especially when morphological examination is not possible in sterile cultures or when morphological data are inconclusive.
6. It is important to understand that very few fungi (<1% of the approximate 1.5 million fungal species thought to exist) have been sequenced for the ITS region and made available in INSD.106 Fungi are an extremely diverse group of organisms, and as such, there is no one panacea for identification of all groups. It is possible that one may be working with an undescribed/unknown fungus or one that has previously not been sequenced. For example, it is likely that a fungal culture producing new compounds164 may belong to a new species.165 Alternatively, a fungal culture producing known compounds might also be a new species that has not been previously described (e.g., see Bills et al.166). Hence, with all of the subspecialties in natural products research, it is highly beneficial to work with a contemporary mycologist, as the taxonomy and practices of the mycological community are evolving rapidly. We specify a list of “dos and don’ts” (Table S2), which will provide natural product researchers with some important guidelines. These suggestions will ensure that common misconceptions are avoided when working on molecular identification of fungi.
7. A variety of fungal sources have been examined for secondary metabolites in this journal, and selected examples that embrace the spirit of the suggestions in this review include studies on endosymbiotic fungi (fungal endophytes and endolichenic fungi),167−169 saprobic fungi,164,170−172 and coprophilous fungi.173 This list is certainly not exhaustive, and there are numerous other examples that the user can follow that provide good methodological practice for molecular identification of fungal strains.
Experimental Section
Detailed Protocol for Sequencing the rDNA Region (SSU-ITS-LSU)
For genomic DNA extractions, approximately 5 mg of mycelial powder is used; mycelial powder is obtained by grinding a small portion of the fungal colony in liquid nitrogen using a mortar and pestle. The powder is transferred to a bashing bead tube with DNA lysis buffer provided in the Zymo Research fungal/bacterial DNA extraction kit and vortexed vigorously for 10 min. Alternatively, for Aspergillus, Penicillium, and Trichoderma spp., obtaining mycelium powder is not necessary; a small piece of agar from the leading edge of the fungal culture is aseptically transferred to DNA lysis buffer and vortexed vigorously. Subsequently, genomic DNA is extracted using procedures outlined in the Zymo fungal/bacterial DNA miniprep. The DNA is finally eluted in 30–50 μL of molecular biology grade water or storage buffer provided by the manufacturer of the kit. DNA extraction can also be accomplished using the DNAeasy plant mini kit (QIAGEN Inc., Valencia, CA, USA) following the manufacturer’s protocol. There are numerous other commercially available DNA kits on the market, which can be utilized for extracting DNA from fungal cultures.
After the genomic DNA is obtained, the entire ITS region (Figure 5) can be PCR-amplified using PuReTaq Ready-To-Go PCR beads (GE Healthcare, Buckinghamshire, UK) with primers ITS1F and ITS4.40,174 The PCR is carried out in 25 μL reactions containing 1–3 μL of template DNA and 1 μL of each 10 μM forward (ITS1F) and reverse (ITS4) primer. Addition of 2.5 μL of BSA (New England BioLabs Inc.) and/or 2.5 μL of 50% DMSO (Sigma, St Louis, MO, USA) is optional. The remaining volume consists of molecular biology grade water (Fisher Scientific). The following thermocycling parameters are typically utilized: initial denaturation at 95 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 52 °C for 30 s, and 72 °C for 1 min with a final extension step of 72 °C for 8 min.48,175 If PCR amplifications using the above primers and annealing temperatures are not successful, either ITS5 or ITS1 primer62 can be used in addition to reducing the annealing temperature from 52 °C to 48 °C (or as low as 41 °C). When possible, negative controls should be included to ensure that PCR amplicons are not contaminated. PCR products are then run on an ethidium bromide-stained 1% agarose gel (Fisher Scientific) along with a 1 kb DNA ladder (Promega) to estimate the size of the amplified band. Prior to Sanger sequencing, PCR products are purified using a Wizard SV gel and PCR clean-up system (Promega). Sanger sequencing of the purified PCR products is performed using BigDye Terminator v3.1 cycle sequencing with 3 μL of DNA template and 1 μL of 2 μM of each primer. Both strands are sequenced (i.e., bidirectionally) using a combination of the following primers: ITS1F, ITS5 or ITS1 (forward), and ITS4 (reverse). Sequences are generated on an Applied Biosystems 3730XL high-throughput capillary sequencer. Sequences are assembled with Sequencher 5.3 (Gene Codes), optimized by eye, and then corrected manually when necessary; the latter step is to ensure that the computer algorithm is assigning proper base calls. For assembly of sequence data, other programs such as Bioedit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and Geneious (http://www.geneious.com/) can also be used.
These methods are used routinely in our laboratories to obtain DNA sequences of the entire ITS region.38,175 We also obtain the entire ITS region and the D1/D2 regions of the LSU using primer combination ITS1F/ITS5 and LR3. The consensus sequence of the ITS region is submitted for a BLAST search using the NCBI GenBank database to obtain species-level information and then utilized in combination with the D1/D2 regions of the LSU for subsequent phylogenetic analyses.39,172,176
The same PCR protocol is utilized for SSU and LSU regions, except we use primers NS1 and NS4 for PCR for the SSU region40 and LROR and LR6 for the LSU region44,45 (Table S1). For sequencing these genes, a combination of primers are used: SSU = NS1, NS2, NS3, and NS4; LSU = LROR, LR3, LR3R, and LR6. Additional detailed protocols for DNA extraction, PCR amplification, and sequencing of the ITS region for fungi are outlined elsewhere.177
Maximum Likelihood
Sequences are edited using Sequencher 5.3 (Gene Codes Corp.) or other sequence-editing programs as part of Geneious software. Each sequence is subjected to an individual BLAST search to verify its identity in GenBank. The newly obtained sequences are aligned with highly similar, homologous sequences from GenBank using the multiple sequence alignment program MUSCLE,178 with default parameters. MUSCLE can be implemented in Sequencher or through the program Seaview v. 4.1.179 The final alignment is optimized by eye and manually corrected. Maximum likelihood methods are used in phylogenetic analyses for all genes. The Akaike information criterion,180 as implemented in jModeltest v. 2.1.4,181 is used to determine the best-fit model of evolution for each data set for maximum likelihood analyses. A likelihood analysis is conducted using PhyML182,183 under the following parameters: with six rate classes and invariable sites, across-site variation is fixed using parameter values obtained from jModeltest, and 1000 bootstrap replicates are performed from a BioNeighborJoining starting tree employing the best of nearest neighbor interchange and subtree pruning and regrafting branch swapping using Seaview v. 4.1. Alternatively, the user can also optimize for all parameters in the PhyML analysis conducted using Seaview. For larger data sets, or to corroborate the tree generated from PhyML, maximum likelihood analyses are performed using RAxML v. 7.0.4184,185 run on the CIPRES Portal v. 2.0186 with the default rapid hill-climbing algorithm and GTR model or the model selected by jModeltest employing 1000 fast bootstrap searches. Clades with bootstrap values ≥ 70% are considered significant and strongly supported.187
Bayesian Inference
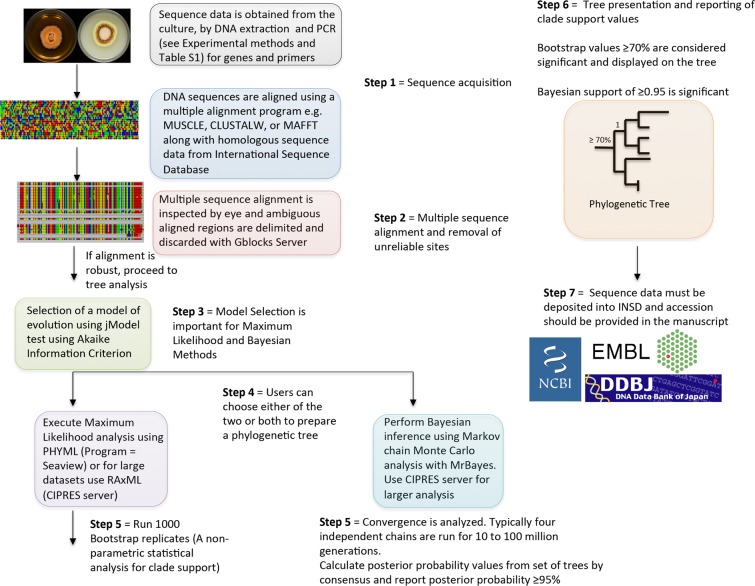
Bayesian inference employing a Markov chain Monte Carlo algorithm is performed on the data set(s) with MrBayes 3.12162,163 using the CIPRES Portal v3.3186 as an additional measure of clade support. The selected model from jModeltest is implemented, and constant characters are included. Four independent chains of Metropolis coupled Markov chain Monte Carlo are run for either 10 million or 100 million generations with trees sampled every 1000th generation, resulting in 10 000 or 100 000 total trees. The number of generations is usually determined by the amount of time it takes for the analysis to reach a stationary phase. Independent Bayesian inference analysis ensures that the same tree space is being sampled during each analysis and that the trees are not trapped in local optima. For a Bayesian analysis that is run using multiple gene markers, the data are partitioned based on the gene as well as codon position using flat priors and unlinked model parameters across partitions. The program AWTY188 is utilized to compare the split frequencies of independent runs to ensure that the stationary phase was reached. TRACER 1.5189 is used to plot the postanalyses parameters against generation time to confirm that these values have reached a stable equilibrium. To conservatively estimate that the log-likelihood values reached a stable equilibrium, the first 10 000 trees that extend beyond the burn-in phase are discarded, and the remaining 9000 or 90 000 trees can be used to calculate the posterior probabilities in each analysis. Consensus trees are generated and viewed in PAUP 4.0b10.190 Clades with a posterior probability ≥ 95% are considered significant and strongly supported.191 A workflow of these steps is outlined in Figure 7.
Figure 7.
Diagrammatic illustration of the critical steps (1–7) used for construction of phylogenetic trees for fungal species identification employing either maximum likelihood (ML) or Bayesian inference (adapted from Schmitt and Barker).5
For additional methods for tree-building protocols, the user can refer to Hall,192 while detailed information on theory and practice of phylogenetic methodology is outlined by Salemi and Mieke.193
Acknowledgments
This research was supported by program project grant P01 CA125066 from the National Cancer Institute/National Institutes of Health, Bethesda, MD, USA. We thank the reviewers and the Associate Editor for insightful suggestions that helped improve the manuscript.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jnatprod.6b01085.
Table S1 provides information on primers and PCR protocols for acquiring ribosomal genes and commonly utilized single-copy protein-coding genes for fungal identification; Table S2 specifies a list of dos and don’ts that will help the user avoid common errors made during molecular identification of fungi (PDF)
The authors declare no competing financial interest.
Dedication
Dedicated to Professor Phil Crews, of the University of California, Santa Cruz, for his pioneering work on bioactive natural products.
Supplementary Material
References
- O’Brien H. E.; Parrent J. L.; Jackson J. A.; Moncalvo J. M.; Vilgalys R. Appl. Environ. Microbiol. 2005, 71, 5544–5550. 10.1128/AEM.71.9.5544-5550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell M. Am. J. Bot. 2011, 98, 426–438. 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- Hawksworth D. L. Mycol. Res. 1991, 95, 641–655. 10.1016/S0953-7562(09)80810-1. [DOI] [Google Scholar]
- Stajich J. E.; Berbee M. L.; Blackwell M.; Hibbett D. S.; James T. Y.; Spatafora J. W.; Taylor J. W. Curr. Biol. 2009, 19, R840–R845. 10.1016/j.cub.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt I.; Barker K. Nat. Prod. Rep. 2009, 26, 1585–1602. 10.1039/b910458p. [DOI] [PubMed] [Google Scholar]
- Sudhakar T.; Dash S.; Rao R.; Srinivasan R.; Zacharia S.; Atmanand M.; Subramaniam B.; Nayak S. Curr. Sci. 2013, 104, 178. [Google Scholar]
- Hofrichter M.The Mycota: A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research: Industrial Applications, 2nd ed.; Springer-Verlag: New York, 2010. [Google Scholar]
- Smith D.; Ryan M. J.. Fungal sources for new drug discovery. In Access Science; McGraw-Hill Companies, 2009. [Google Scholar]
- Aly A. H.; Debbab A.; Proksch P. Fungal Divers. 2011, 50, 3–19. 10.1007/s13225-011-0116-y. [DOI] [Google Scholar]
- Newman D. J.; Cragg G. M. J. Nat. Prod. 2007, 70, 461–477. 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- Newman D. J.; Cragg G. M. J. Nat. Prod. 2012, 75, 311–335. 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. J.; Cragg G. M.; Snader K. M. J. Nat. Prod. 2003, 66, 1022–1037. 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- Downton C.; Clark I. Nat. Rev. Drug Discovery 2003, 2, 343–344. 10.1038/nrd1090. [DOI] [PubMed] [Google Scholar]
- Strader C. R.; Pearce C. J.; Oberlies N. H. J. Nat. Prod. 2011, 74, 900–907. 10.1021/np2000528. [DOI] [PubMed] [Google Scholar]
- Hibbett D. S.; Ohman A.; Glotzer D.; Nuhn M.; Kirk P. M.; Nilsson R. H. Fungal Biol. Rev. 2011, 25, 38–47. 10.1016/j.fbr.2011.01.001. [DOI] [Google Scholar]
- Hibbett D. S.; Taylor J. W. Nat. Rev. Microbiol. 2013, 11, 129–133. 10.1038/nrmicro2963. [DOI] [PubMed] [Google Scholar]
- Hibbett D. Science 2016, 351, 1150–1151. 10.1126/science.aae0380. [DOI] [PubMed] [Google Scholar]
- Hibbett D.; Abarenkov K.; Koljalg U.; Opik M.; Chai B.; Cole J. R.; Wang Q.; Crous P. W.; Robert V. A.; Helgason T.. Mycologia 2016, , in press [DOI] [PubMed]
- Hyde K. D.; Abd-Elsalam K.; Cai L. Mycotaxon 2010, 114, 439–451. 10.5248/114.439. [DOI] [Google Scholar]
- Wang Z.; Nilsson R. H.; James T. Y.; Dai Y.; Townsend J. P. In Biology of Microfungi; Springer, 2016; pp 25–46. [Google Scholar]
- Lutzoni F.; Kauff F.; Cox C. J.; McLaughlin D.; Celio G.; Dentinger B.; Padamsee M.; Hibbett D.; James T. Y.; Baloch E.; Grube M.; Reeb V.; Hofstetter V.; Schoch C.; Arnold A. E.; Miadlikowska J.; Spatafora J.; Johnson D.; Hambleton S.; Crockett M.; Shoemaker R.; Hambleton S.; Crockett M.; Shoemaker R.; Sung G. H.; Lucking R.; Lumbsch T.; O’Donnell K.; Binder M.; Diederich P.; Ertz D.; Gueidan C.; Hansen K.; Harris R. C.; Hosaka K.; Lim Y. W.; Matheny B.; Nishida H.; Pfister D.; Rogers J.; Rossman A.; Schmitt I.; Sipman H.; Stone J.; Sugiyama J.; Yahr R.; Vilgalys R. Am. J. Bot. 2004, 91, 1446–1480. 10.3732/ajb.91.10.1446. [DOI] [PubMed] [Google Scholar]
- Geiser D. M. In Advances in Fungal Biotechnology for Industry, Agriculture, and Medicine; Springer, 2004; pp 3–14. [Google Scholar]
- Olson Å.; Stenlid J. Microbes Infect. 2002, 4, 1353–1359. 10.1016/S1286-4579(02)00005-9. [DOI] [PubMed] [Google Scholar]
- Hughes K. W.; Petersen R. H.; Lodge D. J.; Bergemann S. E.; Baumgartner K.; Tulloss R. E.; Lickey E.; Cifuentes J. Mycologia 2013, 105, 1577–1594. 10.3852/13-041. [DOI] [PubMed] [Google Scholar]
- Harrington T. C.; Rizzo D. M. In Structure and Dynamics of Fungal Populations; Worrall J. J., Ed.; Kluwer Press: Dordrecht, The Netherlands, 1999; pp 43–71. [Google Scholar]
- Foltz M. J.; Perez K. E.; Volk T. J. Mycologia 2013, 105, 447–461. 10.3852/12-181. [DOI] [PubMed] [Google Scholar]
- Kohn L. M. Annu. Rev. Phytopathol. 2005, 43, 279–308. 10.1146/annurev.phyto.43.040204.135958. [DOI] [PubMed] [Google Scholar]
- Giraud T.; Refrégier G.; Le Gac M.; de Vienne D. M.; Hood M. E. Fungal Genet. Biol. 2008, 45, 791–802. 10.1016/j.fgb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Lücking R.; Dal-Forno M.; Sikaroodi M.; Gillevet P. M.; Bungartz F.; Moncada B.; Yánez-Ayabaca A.; Chaves J. L.; Coca L. F.; Lawrey J. D. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 11091–11096. 10.1073/pnas.1403517111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun S.; Silar P. In Evolutionary Biology – Concepts, Molecular and Morphological Evolution; Pontarotti P., Ed.; Springer: Berlin Heidelberg, 2010; pp 317–328. [Google Scholar]
- Hawksworth D. L. IMA Fungus 2012, 3, 15–24. 10.5598/imafungus.2012.03.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde K. D.; Soytong K. Fungal Divers. 2008, 33, 163–173. [Google Scholar]
- Ko T. W. K.; Stephenson S. L.; Bahkali A. H.; Hyde K. D. Fungal Divers. 2011, 50, 113–120. 10.1007/s13225-011-0130-0. [DOI] [Google Scholar]
- Hibbett D. Trans. Mycol. Soc. Japan 1992, 33, 533–556. [Google Scholar]
- Taylor J. W.; Hibbett D. S. IMA Fungus 2013, 4, 33–34. [Google Scholar]
- Bridge P.; Spooner B.; Roberts P. Adv. Bot. Res. 2005, 42, 33–67. 10.1016/S0065-2296(05)42002-9. [DOI] [Google Scholar]
- Vogler A.; Monaghan M. J. Zoolog. Syst. Evol. Res. 2007, 45, 1–10. 10.1111/j.1439-0469.2006.00384.x. [DOI] [Google Scholar]
- Raja H. A.; Baker T. R.; Little J. G.; Oberlies N. H. Food Chem. 2017, 214, 383–392. 10.1016/j.foodchem.2016.07.052. [DOI] [PubMed] [Google Scholar]
- El-Elimat T.; Raja H. A.; Graf T. N.; Faeth S. H.; Cech N. B.; Oberlies N. H. J. Nat. Prod. 2014, 77, 193–199. 10.1021/np400955q. [DOI] [PubMed] [Google Scholar]
- White T. J.; Bruns T.; Lee S. H.; Taylor J. W. PCR protocols: a guide to methods and application. San Diego 1990, 315–322. 10.1016/B978-0-12-372180-8.50042-1. [DOI] [Google Scholar]
- Bruns T. D.; White T. J.; Taylor J. W. Annu. Rev. Ecol. Syst. 1991, 22, 525–564. 10.1146/annurev.es.22.110191.002521. [DOI] [Google Scholar]
- Seifert K. A.; Wingfield B. D.; Wingfield M. J. Can. J. Bot. 1995, 73, 760–767. 10.1139/b95-320. [DOI] [Google Scholar]
- Mitchell J. I.; Zuccaro A. Mycologist 2006, 20, 62–74. 10.1016/j.mycol.2005.11.004. [DOI] [Google Scholar]
- Vilgalys R.; Hester M. J. Bacteriol. 1990, 172, 4238–4246. 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehner S. A.; Samuels G. J. Can. J. Bot. 1995, 73, S816–S823. 10.1139/b95-327. [DOI] [Google Scholar]
- Liu K.-L.; Porras-Alfaro A.; Kuske C. R.; Eichorst S. A.; Xie G. Appl. Environ. Microbiol. 2012, 78, 1523–1533. 10.1128/AEM.06826-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porras-Alfaro A.; Liu K. L.; Kuske C. R.; Xiec G. Appl. Environ. Microbiol. 2014, 80, 829–840. 10.1128/AEM.02894-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C. L.; Seifert K. A.; Huhndorf S.; Robert V.; Spouge J. L.; Levesque C. A.; Chen W. Fungal Barcoding, C.; Fungal Barcoding Consortium Author, L. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C. L.; Seifert K. A.. DNA barcoding in fungi. In Access Science; McGraw-Hill, 2011. [Google Scholar]
- James T. Y.; Kauff F.; Schoch C. L.; Matheny P. B.; Hofstetter V.; Cox C. J.; Celio G.; Gueidan C.; Fraker E.; Miadlikowska J. Nature 2006, 443, 818–822. 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Hibbett D. S.; Binder M.; Bischoff J. F.; Blackwell M.; Cannon P. F.; Eriksson O. E.; Huhndorf S.; James T.; Kirk P. M.; Lucking R.; Lumbsch H. T.; Lutzoni F.; Matheny P. B.; Mclaughlin D. J.; Powell M. J.; Redhead S.; Schoch C. L.; Spatafora J. W.; Stalpers J. A.; Vilgalys R.; Aime M. C.; Aptroot A.; Bauer R.; Begerow D.; Benny G. L.; Castlebury L. A.; Crous P. W.; Dai Y. C.; Gams W.; Geiser D. M.; Griffith G. W.; Gueidan C.; Hawksworth D. L.; Hestmark G.; Hosaka K.; Humber R. A.; Hyde K. D.; AIronside J. E.; Koljalg U.; Kurtzman C. P.; Larsson K. H.; Lichtwardt R.; Longcore J.; Miadlikowska J.; Miller A.; Moncalvo J. M.; Mozley-Standridge S.; Oberwinkler F.; Parmasto E.; Reeb V.; Rogers J. D.; Roux C.; Ryvarden L.; Sampaio J. P.; Schussler A.; Sugiyama J.; Thorn R. G.; Tibell L.; Untereiner W. A.; Walker C.; Wang Z.; Weir A.; Weiss M.; White M. M.; Winka K.; Yao Y. J.; Zhang N. Mycol. Res. 2007, 111, 509–547. 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Hebert P. D. N.; Cywinska A.; Ball S. L.; DeWaard J. R. Proc. R. Soc. London, Ser. B 2003, 270, 313–321. 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P. D. N.; Ratnasingham S.; deWaard J. R. Proc. R. Soc. London, Ser. B 2003, 270, S96–S99. 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress W. J.; Erickson D. L.. DNA Barcodes: Methods and Protocols; Springer, 2012. [DOI] [PubMed] [Google Scholar]
- Hebert P. D.; Gregory T. R. Syst. Biol. 2005, 54, 852–859. 10.1080/10635150500354886. [DOI] [PubMed] [Google Scholar]
- Hajibabaei M.; Singer G. A.; Hebert P. D.; Hickey D. A. Trends Genet. 2007, 23, 167–172. 10.1016/j.tig.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Casiraghi M.; Labra M.; Ferri E.; Galimberti A.; De Mattia F. Briefings Bioinf. 2010, 11, bbq003. 10.1093/bib/bbq003. [DOI] [PubMed] [Google Scholar]
- Eberhardt U. New Phytol. 2010, 187, 266–268. 10.1111/j.1469-8137.2010.03329.x. [DOI] [PubMed] [Google Scholar]
- Seifert K. A. Mol. Ecol. Resour. 2009, 9, 83–89. 10.1111/j.1755-0998.2009.02635.x. [DOI] [PubMed] [Google Scholar]
- Seifert K. A. Persoonia 2008, 21, 158–168. 10.3767/003158508X395706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman A. Inoculum 2007, 58, 1–5. [Google Scholar]
- Gardes M.; Bruns T. D. Mol. Ecol. 1993, 2, 113–118. 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Kelly L. J.; Hollingsworth P. M.; Coppins B. J.; Ellis C. J.; Harrold P.; Tosh J.; Yahr R. New Phytol. 2011, 191, 288–300. 10.1111/j.1469-8137.2011.03677.x. [DOI] [PubMed] [Google Scholar]
- Dentinger B. T. M.; Maryna Y. D.; Moncalvo J.-M. PLoS One 2011, 6, e25081. 10.1371/journal.pone.0025081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seena S.; Pascoal C.; Marvanovä L.; Cássio F. Fungal Divers. 2010, 44, 77–87. 10.1007/s13225-010-0056-y. [DOI] [Google Scholar]
- Bellemain E.; Carlsen T.; Brochmann C.; Coissac E.; Taberlet P.; Kauserud H.. BMC Microbiol.. 2010, 10.189. 10.1186/1471-2180-10-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Nilsson R. H.; Lopez-Giraldez F.; Zhuang W. Y.; Dai Y. C.; Johnston P. R.; Townsend J. P. PLoS One 2011, 6, e19039. 10.1371/journal.pone.0019039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K. J.; Rygiewicz P. T. BMC Microbiol. 2005, 5, 28. 10.1186/1471-2180-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosling A.; Cox F.; Cruz-Martinez K.; Ihrmark K.; Grelet G. A.; Lindahl B. D.; Menkis A.; James T. Y. Science 2011, 333, 876–879. 10.1126/science.1206958. [DOI] [PubMed] [Google Scholar]
- Amend A. S.; Seifert K. A.; Samson R.; Bruns T. D. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 13748–13753. 10.1073/pnas.1000454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrmark K.; Bödeker I. T.; Cruz-Martinez K.; Friberg H.; Kubartova A.; Schenck J.; Strid Y.; Stenlid J.; Brandström-Durling M.; Clemmensen K. E.; Lindahl B. D. FEMS Microbiol. Ecol. 2012, 82, 666–677. 10.1111/j.1574-6941.2012.01437.x. [DOI] [PubMed] [Google Scholar]
- Ovaskainen O.; Nokso-Koivista J.; Hottola J.; Rajala T.; Pennanen T.; Ali-Kovero H.; Miettinen O.; Oinonen P.; Auvinen P.; Paulin L.; Larsson K. H.; Makipaa R. Fungal Ecol. 2010, 3, 274–283. 10.1016/j.funeco.2010.01.001. [DOI] [Google Scholar]
- Nilsson R. H.; Tedersoo L.; Lindahl B. D.; Kjoller R.; Carlsen T.; Quince C.; Abarenkov K.; Pennanen T.; Stenlid J.; Bruns T.; Larsson K. H.; Koljalg U.; Kauserud H. New Phytol. 2011, 191, 314–318. 10.1111/j.1469-8137.2011.03755.x. [DOI] [PubMed] [Google Scholar]
- Begerow D.; Nilsson H.; Unterseher M.; Maier W. Appl. Microbiol. Biotechnol. 2010, 87, 99–108. 10.1007/s00253-010-2585-4. [DOI] [PubMed] [Google Scholar]
- Yahr R.; Schoch C. L.; Dentinger B. T. M.. Philos. Trans. R. Soc., B 2016, 371.20150336. 10.1098/rstb.2015.0336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R. H.; Kristiansson E.; Ryberg M.; Hallenberg N.; Larsson K. H. Evolutionary Bioinformatics Online 2008, 4, 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrålstad T. Mol. Ecol. 2011, 20, 2873–2875. 10.1111/j.1365-294X.2011.05149.x. [DOI] [PubMed] [Google Scholar]
- Kiss L. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, E1811–E1811. 10.1073/pnas.1207143109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert K. A.; Samson R. A.; DeWaard J. R.; Houbraken J.; Lévseque C. A.; Moncalvo J. M.; Louis-Seize G.; Hebert P. D. N. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 3901–3906. 10.1073/pnas.0611691104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R. A.; Pitt J. I.. Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification; CRC Press, 2000. [Google Scholar]
- Samson R. A.; Visagie C. M.; Houbraken J.; Hong S.-B.; Hubka V.; Klaassen C. H.; Perrone G.; Seifert K. A.; Susca A.; Tanney J. B. Stud. Mycol. 2014, 78, 141–173. 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hatmi A. M.; Van Den Ende A. G.; Stielow J. B.; Van Diepeningen A. D.; Seifert K. A.; McCormick W.; Assabgui R.; Gräfenhan T.; De Hoog G. S.; Levesque C. A. Fungal Biol. 2016, 120, 231–245. 10.1016/j.funbio.2015.08.006. [DOI] [PubMed] [Google Scholar]
- Stielow J.; Lévesque C.; Seifert K.; Meyer W.; Irinyi L.; Smits D.; Renfurm R.; Verkley G.; Groenewald M.; Chaduli D. Persoonia 2015, 35, 242–263. 10.3767/003158515X689135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieckfeldt E.; Seifert K. A. Stud. Mycol. 2000, 35–44. [Google Scholar]
- Lindner D. L.; Banik M. T. Mycologia 2011, 103, 731–40. 10.3852/10-331. [DOI] [PubMed] [Google Scholar]
- Chen J.; Moinard M.; Xu J.; Wang S.; Foulongne-Oriol M.; Zhao R.; Hyde K. D.; Callac P. PLoS One 2016, 11, e0156250. 10.1371/journal.pone.0156250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Jiao L.; Yao Y.-J. Mol. Phylogenet. Evol. 2013, 68, 373–379. 10.1016/j.ympev.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Lindner D. L.; Carlsen T.; Henrik Nilsson R.; Davey M.; Schumacher T.; Kauserud H. Ecol. Evol. 2013, 3, 1751–1764. 10.1002/ece3.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W.; Fisher M. C. Curr. Opin. Microbiol. 2003, 6, 351–356. 10.1016/S1369-5274(03)00088-2. [DOI] [PubMed] [Google Scholar]
- Taylor J. W.; Jacobson D. J.; Kroken S.; Kasuga T.; Geiser D. M.; Hibbett D. S.; Fisher M. C. Fungal Genet. Biol. 2000, 31, 21–32. 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- Altschul S. F.; Gish W.; Miller W.; Myers E. W.; Lipman D. J. J. Mol. Biol. 1990, 215, 403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Hennell J. R.; D’Agostino P. M.; Lee S.; Khoo C. S.; Sucher N. J., Using GenBank® for genomic authentication: a tutorial. In Plant DNA Fingerprinting and Barcoding: Methods and Protocols; Sucher N. J., Ed.; Humana Press, Springer: New York, 2012; pp 181–200. [Google Scholar]
- Pevsner J., Access to Sequence Data and Literature Information. In Bioinformatics and Functional Genomics, Second ed.; John Wiley & Sons, Inc., 2009; pp 12–45. [Google Scholar]
- O’Leary N. A.; Wright M. W.; Brister J. R.; Ciufo S.; Haddad D.; McVeigh R.; Rajput B.; Robbertse B.; Smith-White B.; Ako-Adjei D. Nucleic Acids Res. 2016, 44, gkv1189. 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C. L.; Robbertse B.; Robert V.; Vu D.; Cardinali G.; Irinyi L.; Meyer W.; Nilsson R. H.; Hughes K.; Miller A. N.; Kirk P. M.; Abarenkov K.; Aime M. C.; Ariyawansa H. A.; Bidartondo M.; Boekhout T.; Buyck B.; Cai Q.; Chen J.; Crespo A.; Crous P. W.; Damm U.; De Beer Z. W.; Dentinger B. T.; Divakar P. K.; Duenas M.; Feau N.; Fliegerova K.; Garcia M. A.; Ge Z. W.; Griffith G. W.; Groenewald J. Z.; Groenewald M.; Grube M.; Gryzenhout M.; Gueidan C.; Guo L.; Hambleton S.; Hamelin R.; Hansen K.; Hofstetter V.; Hong S. B.; Houbraken J.; Hyde K. D.; Inderbitzin P.; Johnston P. R.; Karunarathna S. C.; Koljalg U.; Kovacs G. M.; Kraichak E.; Krizsan K.; Kurtzman C. P.; Larsson K. H.; Leavitt S.; Letcher P. M.; Liimatainen K.; Liu J. K.; Lodge D. J.; Luangsa-ard J. J.; Lumbsch H. T.; Maharachchikumbura S. S.; Manamgoda D.; Martin M. P.; Minnis A. M.; Moncalvo J. M.; Mule G.; Nakasone K. K.; Niskanen T.; Olariaga I.; Papp T.; Petkovits T.; Pino-Bodas R.; Powell M. J.; Raja H. A.; Redecker D.; Sarmiento-Ramirez J. M.; Seifert K. A.; Shrestha B.; Stenroos S.; Stielow B.; Suh S. O.; Tanaka K.; Tedersoo L.; Telleria M. T.; Udayanga D.; Untereiner W. A.; Dieguez Uribeondo J.; Subbarao K. V.; Vagvolgyi C.; Visagie C.; Voigt K.; Walker D. M.; Weir B. S.; Weiss M.; Wijayawardene N. N.; Wingfield M. J.; Xu J. P.; Yang Z. L.; Zhang N.; Zhuang W. Y.; Federhen S.. Database (Oxford) 2014, 2014, bau061. 10.1093/database/bau061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federhen S. Nucleic Acids Res. 2012, 40, D136–D143. 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R. H.; Tedersoo L.; Abarenkov K.; Ryberg M.; Kristiansson E.; Hartmann M.; Schoch C. L.; Nylander J. A. A.; Bergsten J.; Porter T. M.; Jumpponen A.; Vaishampayan P.; Ovaskainen O.; Hallenberg N.; Bengtsson-Palme J.; Eriksson M. K.; Larsson K. H.; Larsson E.; Koljalg U. MycoKeys 2012, 4, 37–63. 10.3897/mycokeys.4.3606. [DOI] [Google Scholar]
- Lindahl B. D.; Nilsson R. H.; Tedersoo L.; Abarenkov K.; Carlsen T.; Kjøller R.; Kõljalg U.; Pennanen T.; Rosendahl S.; Stenlid J.; Kauserud H. New Phytol. 2013, 199, 288–299. 10.1111/nph.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde K. D.; Udayanga D.; Manamgoda D. S.; Tedersoo L.; Larsson E.; Abarenkov K.; Bertrand Y. J. K.; Oxelman B.; Hartmann M.; Kauserud H.; Ryberg M.; Kristiansson E.; Nilsson R. H. Curr. Res. Env. Appl. Mycol. 2013, 3, 1–32. [Google Scholar]
- Smith M. E.; Douhan G. W.; Rizzo D. M. New Phytol. 2007, 174, 847–863. 10.1111/j.1469-8137.2007.02040.x. [DOI] [PubMed] [Google Scholar]
- Morris M. H.; Smith M. E.; Rizzo D. M.; Rejmanek M.; Bledsoe C. S. New Phytol. 2008, 178, 167–176. 10.1111/j.1469-8137.2007.02348.x. [DOI] [PubMed] [Google Scholar]
- Izzo A.; Agbowo J.; Bruns T. D. New Phytol. 2005, 166, 619–630. 10.1111/j.1469-8137.2005.01354.x. [DOI] [PubMed] [Google Scholar]
- Ryberg M.; Nilsson R. H.; Kristiansson E.; Topel M.; Jacobsson S.; Larsson E. BMC Evol. Biol. 2008, 8, 50. 10.1186/1471-2148-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. W.; Petersen R. H.; Lickey E. B. New Phytol. 2009, 182, 795–798. 10.1111/j.1469-8137.2009.02802.x. [DOI] [PubMed] [Google Scholar]
- Blaalid R.; Kumar S.; Nilsson R. H.; Abarenkov K.; Kirk P. M.; Kauserud H. Mol. Ecol. Resour. 2013, 13, 218–24. 10.1111/1755-0998.12065. [DOI] [PubMed] [Google Scholar]
- Nilsson R. H.; Ryberg M.; Kristiansson E.; Abarenkov K.; Larsson K. H. PLoS One 2006, 1, e59. 10.1371/journal.pone.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge P. D.; Roberts P. J.; Spooner B. M.; Panchal G. New Phytol. 2003, 160, 43–48. 10.1046/j.1469-8137.2003.00861.x. [DOI] [PubMed] [Google Scholar]
- Vilgalys R. New Phytol. 2003, 160, 4–5. 10.1046/j.1469-8137.2003.00894.x. [DOI] [PubMed] [Google Scholar]
- Rossman A. Y.; Palm-Hernández M. E. Plant Dis. 2008, 92, 1376–1386. 10.1094/PDIS-92-10-1376. [DOI] [PubMed] [Google Scholar]
- Robert V.; Cardinali G.; Stielow B.; Vu T.; dos Santos F. B.; Meyer W.; Schoch C. Fungal DNA Barcoding. Molecular Biology of Food and Water Borne Mycotoxigenic and Mycotic Fungi 2015, 37–56. [Google Scholar]
- Federhen S. Nucleic Acids Res. 2015, 43, D1086. 10.1093/nar/gku1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U’Ren J. M.; Miadlikowska J.; Zimmerman N. B.; Lutzoni F.; Stajich J. E.; Arnold A. E. Mol. Phylogenet. Evol. 2016, 98, 210–232. 10.1016/j.ympev.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Stadler M.; Kuhnert E.; Peršoh D.; Fournier J. Mycology 2013, 4, 5–21. [Google Scholar]
- Gazis R.; Miadlikowska J.; Lutzoni F.; Arnold A.; Chaverri P. Mol. Phylogenet. Evol. 2012, 65, 294–304. 10.1016/j.ympev.2012.06.019. [DOI] [PubMed] [Google Scholar]
- Harris D. J. Trends Ecol. Evol. 2003, 18, 317–319. 10.1016/S0169-5347(03)00150-2. [DOI] [Google Scholar]
- Kõljalg U.; Nilsson R. H.; Abarenkov K.; Tedersoo L.; Taylor A. F.; Bahram M.; Bates S. T.; Bruns T. D.; Bengtsson-Palme J.; Callaghan T. M. Mol. Ecol. 2013, 22, 5271–5277. 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- Abarenkov K.; Henrik Nilsson R.; Larsson K. H.; Alexander I. J.; Eberhardt U.; Erland S.; Hoiland K.; Kjoller R.; Larsson E.; Pennanen T.; Sen R.; Taylor A. F.; Tedersoo L.; Ursing B. M.; Vralstad T.; Liimatainen K.; Peintner U.; Koljalg U. New Phytol. 2010, 186, 281–5. 10.1111/j.1469-8137.2009.03160.x. [DOI] [PubMed] [Google Scholar]
- Irinyi L.; Lackner M.; De Hoog G. S.; Meyer W. Fungal Biol. 2016, 120, 125–136. 10.1016/j.funbio.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Irinyi L.; Serena C.; Garcia-Hermoso D.; Arabatzis M.; Desnos-Ollivier M.; Vu D.; Cardinali G.; Arthur I.; Normand A. C.; Giraldo A.; da Cunha K. C.; Sandoval-Denis M.; Hendrickx M.; Nishikaku A. S.; de Azevedo Melo A. S.; Merseguel K. B.; Khan A.; Rocha J. A.; Sampaio P.; da Silva Briones M. R.; Carmona E. F. R.; Muniz M. M.; Castanon-Olivares L. R.; Estrada-Barcenas D.; Cassagne C.; Mary C.; Duan S. Y.; Kong F.; Sun A. Y.; Zeng X.; Zhao Z.; Gantois N.; Botterel F.; Robbertse B.; Schoch C.; AGams W.; Ellis D.; Halliday C.; Chen S.; Sorrell T. C.; Piarroux R.; Colombo A. L.; Pais C.; de Hoog S.; Zancope-Oliveira R. M.; Taylor M. L.; Toriello C.; de Almeida Soares C. M.; Delhaes L.; Stubbe D.; Dromer F.; Ranque S.; Guarro J.; Cano-Lira J. F.; Robert V.; Velegraki A.; Meyer W. Med. Mycol. 2015, 53, 313–337. 10.1093/mmy/myv008. [DOI] [PubMed] [Google Scholar]
- Bidartondo M. I.; Bruns T. D.; Blackwell M.; Edwards I.; Taylor A. F. S.; Horton T.; Zhang N.; Koljalg U.; May G.; Kuyper T. W.; Bever J. D.; Gilbert G.; Taylor J. W.; DeSantis T. Z.; Pringle A.; Borneman J.; Thorn G.; Berbee M.; Mueller G. M.; Andersen G. L.; Vellinga E. C.; Branco S.; Anderson I.; Dickie I. A.; Avis P.; Timonen S.; Kjoller R.; Lodge D. J.; Bateman R. M.; Purvis A.; Crous P. W.; Hawkes C.; Barraclough T.; Burt A.; Nilsson R. H.; Larsson K. H.; Alexander I.; Moncalvo J. M.; Berube J.; Spatafora J.; Lumbsch H. T.; Blair J. E.; Suh S. O.; Pfister D. H.; Binder M.; Boehm E. W.; Kohn L.; Mata J. L.; Dyer P.; Sung G. H.; Dentinger B.; Simmons E. G.; Baird R. E.; Volk T. J.; Perry B. A.; Kerrigan R. W.; Campbell J.; Rajesh J.; Reynolds D. R.; Geiser D.; Humber R. A.; Hausmann N.; Szaro T.; Stajich J.; Gathman A.; Peay K. G.; Henkel T.; Robinson C. H.; Pukkila P. J.; Nguyen N. H.; Villalta C.; Kennedy P.; Bergemann S.; Aime M. C.; Kauff F.; Porras-Alfaro A.; Gueidan C.; Beck A.; Andersen B.; Marek S.; Crouch J. A.; Kerrigan J.; Ristaino J. B.; Hodge K. T.; Kuldau G.; Samuels G. J.; Raja H. A.; Voglmayr H.; Gardes M.; Janos D. P.; Rogers J. D.; Cannon P.; Woolfolk S. W.; Kistler H. C.; Castellano M. A.; Maldonado-Ramirez S. L.; Kirk P. M.; Farrar J. J.; Osmundson T.; Currah R. S.; Vujanovic V.; Chen W. D.; Korf R. P.; Atallah Z. K.; Harrison K. J.; Guarro J.; Bates S. T.; Bonello P.; Bridge P.; Schell W.; Rossi W.; Stenlid J.; Frisvad J. C.; Miller R. M.; Baker S. E.; Hallen H. E.; Janso J. E.; Wilson A. W.; Conway K. E.; Egerton-Warburton L.; Wang Z.; Eastburn D.; Ho W. W. H.; Kroken S.; Stadler M.; Turgeon G.; Lichtwardt R. W.; Stewart E. L.; Wedin M.; Li D. W.; Uchida J. Y.; Jumpponen A.; Deckert R. J.; Beker H. J.; Rogers S. O.; Xu J. A. P.; Johnston P.; Shoemaker R. A.; Liu M. A.; Marques G.; Summerell B.; Sokolski S.; Thrane U.; Widden P.; Bruhn J. N.; Bianchinotti V.; Tuthill D.; Baroni T. J.; Barron G.; Hosaka K.; Jewell K.; Piepenbring M.; Sullivan R.; Griffith G. W.; Bradley S. G.; Aoki T.; Yoder W. T.; Ju Y. M.; Berch S. M.; Trappe M.; Duan W. J.; Bonito G.; Taber R. A.; Coelho G.; Bills G.; Ganley A.; Agerer R.; Nagy L.; Roy B. A.; Laessoe T.; Hallenberg N.; Tichy H. V.; Stalpers J.; Langer E.; Scholler M.; Krueger D.; Pacioni G.; Poder R.; Pennanen T.; Capelari M.; Nakasone K.; Tewari J. P.; Miller A. N.; Decock C.; Huhndorf S.; Wach M.; Vishniac H. S.; Yohalem D. S.; Smith M. E.; Glenn A. E.; Spiering M.; Lindner D. L.; Schoch C.; Redhead S. A.; Ivors K.; Jeffers S. N.; Geml J.; Okafor F.; Spiegel F. W.; ADewsbury D.; Carroll J.; Porter T. M.; Pashley C.; Carpenter S. E.; Abad G.; Voigt K.; Arenz B.; Methven A. S.; Schechter S.; Vance P.; Mahoney D.; Kang S. C.; Rheeder J. P.; Mehl J.; Greif M.; Ngala G. N.; Ammirati J.; Kawasaki M.; Gwo-Fang Y. A.; Matsumoto T.; Smith D.; Koenig G.; Luoma D.; May T.; Leonardi M.; Sigler L.; Taylor D. L.; Gibson C.; Sharpton T.; Hawksworth D. L.; Dianese J. C.; Trudell S. A.; Paulus B.; Padamsee M.; Callac P.; Lima N.; White M.; Barreau C.; Juncai M. A.; Buyck B.; Rabeler R. K.; Liles M. R.; Estes D.; Carter R.; Herr J. M.; Chandler G.; Kerekes J.; Cruse-Sanders J.; Marquez R. G.; Horak E.; Fitzsimons M.; Doring H.; Yao S.; Hynson N.; Ryberg M.; Arnold A. E.; Hughes K. Science 2008, 319, 1616. [Google Scholar]
- Crous P. W.; Hawksworth D. L.; Wingfield M. J. Annu. Rev. Phytopathol. 2015, 53, 247–267. 10.1146/annurev-phyto-080614-120245. [DOI] [PubMed] [Google Scholar]
- Geiser D.; Jimenez-Gasco M.; Kang S.; Makalowska I.; Veeraraghavan N.; Ward T.; Zhang N.; Kuldau G. Eur. J. Plant Pathol. 2004, 110, 473–479. [Google Scholar]
- Druzhinina I. S.; Kopchinskiy A. G.; Komoń M.; Bissett J.; Szakacs G.; Kubicek C. P. Fungal Genet. Biol. 2005, 42, 813–828. 10.1016/j.fgb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Deshpande V.; Wang Q.; Greenfield P.; Charleston M.; Porras-Alfaro A.; Kuske C. R.; Cole J. R.; Midgley D. J.; Tran-Dinh N. Mycologia 2016, 108, 1–5. 10.3852/14-293. [DOI] [PubMed] [Google Scholar]
- Schoch C. L.; Sung G.-H.; López-Giráldez F.; Townsend J. P.; Miadlikowska J.; Hofstetter V.; Robbertse B.; Matheny P. B.; Kauff F.; Wang Z. Syst. Biol. 2009, 58, 224. 10.1093/sysbio/syp020. [DOI] [PubMed] [Google Scholar]
- Berbee M. L.; Taylor J. W. In System Evolution; Springer, 2001; pp 229–245. [Google Scholar]
- Einax E.; Voigt K. Org. Divers. Evol. 2003, 3, 185–194. 10.1078/1439-6092-00069. [DOI] [Google Scholar]
- Blackwell M.; Hibbett D. S.; Taylor J. W.; Spatafora J. W. Mycologia 2006, 98, 829–837. 10.3852/mycologia.98.6.829. [DOI] [PubMed] [Google Scholar]
- McLaughlin D. J.; Hibbett D. S.; Lutzoni F.; Spatafora J. W.; Vilgalys R. Trends Microbiol. 2009, 17, 488–497. 10.1016/j.tim.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Liu Y. J.; Hall B. D. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 4507–4512. 10.1073/pnas.0400938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeb V.; Lutzoni F.; Roux C. Mol. Phylogenet. Evol. 2004, 32, 1036–1060. 10.1016/j.ympev.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Stiller J. W.; Hall B. D. Proc. Natl. Acad. Sci. U. S. A. 1997, 94, 4520–4525. 10.1073/pnas.94.9.4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny P. B.; Liu Y. J.; Ammirati J. F.; Hall B. D. Am. J. Bot. 2002, 89, 688–698. 10.3732/ajb.89.4.688. [DOI] [PubMed] [Google Scholar]
- Rehner S. A.; Buckley E. Mycologia 2005, 97, 84–98. 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- Glass N. L.; Donaldson G. C. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K.; Cigelnik E. Mol. Phylogenet. Evol. 1997, 7, 103–116. 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- Raja H.; Schoch C. L.; Hustad V.; Shearer C.; Miller A. MycoKeys 2011, 1, 63–94. 10.3897/mycokeys.1.1966. [DOI] [Google Scholar]
- Schmitt I.; Crespo A.; Divakar P. K.; Fankhauser J. D.; Herman-Sackett E.; Kalb K.; Nelsen M. P.; Nelson N. A.; Rivas-Plata E.; Shimp A. D.; Widhelm T.; Lumbsch H. T. Persoonia 2009, 23, 35–40. 10.3767/003158509X470602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern I.; Powlowski J.; Ishmael N.; Darmond C.; Marqueteau S.; Moisan M. C.; Quenneville G.; Tsang A. Fungal Biol. 2012, 116, 489–502. 10.1016/j.funbio.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Gillot G.; Jany J. L.; Coton M.; Le Floch G.; Debaets S.; Ropars J.; López-Villavicencio M.; Dupont J.; Branca A.; Giraud T.; Coton E.. PLoS One 2015, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustad V. P.; Miller A. N. Mycologia 2015, 107, 647–657. 10.3852/14-328. [DOI] [PubMed] [Google Scholar]
- Aguileta G.; Marthey S.; Chiapello H.; Lebrun M.-H.; Rodolphe F.; Fournier E.; Gendrault-Jacquemard A.; Giraud T. Syst. Biol. 2008, 57, 613–627. 10.1080/10635150802306527. [DOI] [PubMed] [Google Scholar]
- Houbraken J.; Frisvad J. C.; Samson R. Stud. Mycol. 2011, 70, 53–138. 10.3114/sim.2011.70.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J.; Samson R. A. Stud. Mycol. 2011, 70, 1–51. 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R.; Yilmaz N.; Houbraken J.; Spierenburg H.; Seifert K.; Peterson S.; Varga J.; Frisvad J. C. Stud. Mycol. 2011, 70, 159–183. 10.3114/sim.2011.70.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C.; Houbraken J.; Frisvad J. C.; Hong S.-B.; Klaassen C.; Perrone G.; Seifert K.; Varga J.; Yaguchi T.; Samson R. Stud. Mycol. 2014, 78, 343–371. 10.1016/j.simyco.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P. K.; Horwitz B. A.; Singh U. S.; Mukherjee M.; Schmoll M.. Trichoderma: Biology and Applications; CABI, 2013. [Google Scholar]
- Schuster A.; Schmoll M. Appl. Microbiol. Biotechnol. 2010, 87, 787–799. 10.1007/s00253-010-2632-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann N. K.; Stoppacher N.; Zeilinger S.; Degenkolb T.; Brückner H.; Schuhmacher R. Chem. Biodiversity 2015, 12, 743–751. 10.1002/cbdv.201400393. [DOI] [PubMed] [Google Scholar]
- Druzhinina I.; Kubicek C. P. J. Zhejiang Univ., Sci. 2005, 6, 100. 10.1631/jzus.2005.B0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B.; Druzhinina I. S.; Fallah P.; Chaverri P.; Gradinger C.; Kubicek C. P.; Samuels G. J. Mycologia 2004, 96, 310–342. 10.2307/3762066. [DOI] [PubMed] [Google Scholar]
- Degenkolb T.; Dieckmann R.; Nielsen K. F.; Gräfenhan T.; Theis C.; Zafari D.; Chaverri P.; Ismaiel A.; Brückner H.; Von Döhren H. Mycol. Prog. 2008, 7, 177–219. 10.1007/s11557-008-0563-3. [DOI] [Google Scholar]
- Montoya Q. V.; Meirelles L. A.; Chaverri P.; Rodrigues A. Antonie van Leeuwenhoek 2016, 109, 633–651. 10.1007/s10482-016-0666-9. [DOI] [PubMed] [Google Scholar]
- Jaklitsch W.; Voglmayr H. Stud. Mycol. 2015, 80, 1–87. 10.1016/j.simyco.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy V.; Seidl V.; Szakacs G.; Komoń-Zelazowska M.; Kubicek C. P.; Druzhinina I. S. Appl. Environ. Microbiol. 2007, 73, 7048–7058. 10.1128/AEM.00995-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. J.; Whelen S.; Hall B. D. Mol. Biol. Evol. 1999, 16, 1799–1808. 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Tautz D.; Arctander P.; Minelli A.; Thomas R. H.; Vogler A. P. Trends Ecol. Evol. 2003, 18, 70–74. 10.1016/S0169-5347(02)00041-1. [DOI] [Google Scholar]
- Holder M.; Lewis P. O. Nat. Rev. Genet. 2003, 4, 275–284. 10.1038/nrg1044. [DOI] [PubMed] [Google Scholar]
- Yang Z.; Rannala B. Nat. Rev. Genet. 2012, 13, 303–314. 10.1038/nrg3186. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. J. Mol. Evol. 1981, 17, 368–376. 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Harrison J. C.; Langdale J. A. Plant J. 2006, 45, 561–572. 10.1111/j.1365-313X.2005.02611.x. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J. P.; Ronquist F. Bioinformatics 2001, 17, 754–755. 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J. P.; Ronquist F.. Bayesian Analysis of Molecular Evolution Using Mr. Bayes; Springer, 2005; pp 186–226. [Google Scholar]
- Richter C.; Helaly S. E.; Thongbai B.; Hyde K. D.; Stadler M. J. Nat. Prod. 2016, 79, 1684–1688. 10.1021/acs.jnatprod.6b00194. [DOI] [PubMed] [Google Scholar]
- Hyde K. D.; Hongsanan S.; Jeewon R.; Bhat D. J.; McKenzie E. H. C.; Jones E. B. G.; Phookamsak R.; Ariyawansa H. A.; Boonmee S.; Zhao Q.; Abdel-Aziz F. A.; Abdel-Wahab M. A.; Banmai S.; Chomnunti P.; Cui B.-K.; Daranagama D. A.; Das K.; Dayarathne M. C.; de Silva N. I.; Dissanayake A. J.; Doilom M.; Ekanayaka A. H.; Gibertoni T. B.; Góes-Neto A.; Huang S.-K.; Jayasiri S. C.; Jayawardena R. S.; Konta S.; Lee H. B.; Li W.-J.; Lin C.-G.; Liu J.-K.; Lu Y.-Z.; Luo Z.-L.; Manawasinghe I. S.; Manimohan P.; Mapook A.; Niskanen T.; Norphanphoun C.; Papizadeh M.; Perera R. H.; Phukhamsakda C.; Richter C.; de A. Santiago A. L. C. M.; Drechsler-Santos E. R.; Senanayake I. C.; Tanaka K.; Tennakoon T. M. D. S.; Thambugala K. M.; Tian Q.; Tibpromma S.; Thongbai B.; Vizzini A.; Wanasinghe D. N.; Wijayawardene N. N.; Wu H.-X.; Yang J.; Zeng X.-Y.; Zhang H.; Zhang J.-F.; Bulgakov T. S.; Camporesi E.; Bahkali A. H.; Amoozegar M. A.; Araujo-Neta L. S.; Ammirati J. F.; Baghela A.; Bhatt R. P.; Bojantchev D.; Buyck B.; da Silva G. A.; de Lima C. L. F.; de Oliveira R. J. V.; de Souza C. A. F.; Dai Y.-C.; Dima B.; Duong T. T.; Ercole E.; Mafalda-Freire F.; Ghosh A.; Hashimoto A.; Kamolhan S.; Kang J.-C.; Karunarathna S. C.; Kirk P. M.; Kytövuori I.; Lantieri A.; Liimatainen K.; Liu Z.-Y.; Liu X.-Z.; Lücking R.; Medardi G.; Mortimer P. E.; Nguyen T. T. T.; Promputtha I.; Raj K. N. A.; Reck M. A.; Lumyong S.; Shahzadeh-Fazeli S. A.; Stadler M.; Soudi M. R.; Su H.-Y.; Takahashi T.; Tangthirasunun N.; Uniyal P.; Wang Y.; Wen T.-C.; Xu J.-C.; Zhang Z.-K.; Zhao Y.-C.; Zhou J.-L.; Zhu L. Fungal Divers. 2016, 80, 1–270. 10.1007/s13225-016-0373-x. [DOI] [Google Scholar]
- Bills G. F.; González-Menéndez V.; Martín J.; Platas G.; Fournier J.; Peršoh D.; Stadler M. PLoS One 2012, 7, e46687. 10.1371/journal.pone.0046687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne E. K.; Gunaherath G. K. B.; Chapla V. M.; Tillotson J.; De La Cruz F.; Kang M.; U'Ren J. M.; Araujo A. R.; Arnold A. E.; Chapman E. J. Nat. Prod. 2016, 79, 340–352. 10.1021/acs.jnatprod.5b00986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega H. E.; Graupner P. R.; Asai Y.; Tendyke K.; Qiu D.; Shen Y. Y.; Rios N.; Arnold A. E.; Coley P. D.; Kursar T. A.; Gerwick W. H.; Cubilla-Rios L. J. Nat. Prod. 2013, 76, 741–744. 10.1021/np300792t. [DOI] [PubMed] [Google Scholar]
- Figueroa M.; Jarmusch A. K.; Raja H. A.; El-Elimat T.; Kavanaugh J. S.; Horswill A. R.; Cooks R. G.; Cech N. B.; Oberlies N. H. J. Nat. Prod. 2014, 77, 1351–1358. 10.1021/np5000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto A.; Laub A.; Wendt L.; Porzel A.; Schmidt J.; Palfner G.; Becerra J.; Krüger D.; Stadler M.; Wessjohann L.; Westermann B.; Arnold N. J. Nat. Prod. 2016, 79, 929–938. 10.1021/acs.jnatprod.5b01018. [DOI] [PubMed] [Google Scholar]
- Mudalungu C. M.; Richter C.; Wittstein K.; Abdalla M. A.; Matasyoh J. C.; Stadler M.; Süssmuth R. D. J. Nat. Prod. 2016, 79, 894–898. 10.1021/acs.jnatprod.5b00950. [DOI] [PubMed] [Google Scholar]
- El-Elimat T.; Raja H. A.; Day C. S.; Chen W.-L.; Swanson S. M.; Oberlies N. H. J. Nat. Prod. 2014, 77, 2088–2098. 10.1021/np500497r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Yue Q.; Krausert N. M.; An Z.; Gloer J. B.; Bills G. F. J. Nat. Prod. 2016, 79, 2357. 10.1021/acs.jnatprod.6b00498. [DOI] [PubMed] [Google Scholar]
- Gardes M.; White T. J.; Fortin J. A.; Bruns T. D.; Taylor J. W. Can. J. Bot. 1991, 69, 180–190. 10.1139/b91-026. [DOI] [Google Scholar]
- Promputtha I.; Miller A. N. Mycologia 2010, 102, 574–587. 10.3852/09-051. [DOI] [PubMed] [Google Scholar]
- El-Elimat T.; Raja H. A.; Figueroa M.; Falkinham J. O. III; Oberlies N. H. Phytochemistry 2014, 104, 114–120. 10.1016/j.phytochem.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Eberhardt U. In DNA Barcodes: Methods and Protocols; Kress J. W.; Erickson L. D., Eds.; Humana Press: Totowa, NJ, 2012; pp 183–205. [Google Scholar]
- Edgar R. C. Nucleic Acids Res. 2004, 32, 1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M.; Guindon S.; Gascuel O. Mol. Biol. Evol. 2010, 27, 221–224. 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Posada D.; Buckley T. R. Syst. Biol. 2004, 53, 793–808. 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Posada D. Mol. Biol. Evol. 2008, 25, 1253–1256. 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Guindon S.; Delsuc F.; Dufayard J. F.; Gascuel O. Methods Mol. Biol. 2009, 537, 113–37. 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- Guindon S.; Dufayard J. F.; Hordijk W.; Lefort V.; Gascuel O. Infect. Genet. Evol. 2009, 9, 384–385. [Google Scholar]
- Stamatakis A. Bioinformatics 2006, 22, 2688–2690. 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A.; Hoover P.; Rougemont J. Syst. Biol. 2008, 57, 758–771. 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Miller M. A.; Pfeiffer W.; Schwartz T. Creating the CIPRES Science Gateway for Inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE) 2010, 1–8. 10.1109/GCE.2010.5676129. [DOI] [Google Scholar]
- Hillis D. M.; Bull J. J. Syst. Biol. 1993, 42, 182–192. 10.2307/2992540. [DOI] [Google Scholar]
- Nylander J. A.; Wilgenbusch J. C.; Warren D. L.; Swofford D. L. Bioinformatics 2008, 24, 581–3. 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- Rambaut A.; Drummond A. J.. Tracer v.1.5. http://tree.bio.ed.ac.uk/software/tracer.
- Swofford D. L.PAUP*: Phylogenetic Analysis Using Parsimony (* and other methods), Version 4; Sinauer Associates: Sunderland, MA, 2002.
- Alfaro M. E.; Zoller S.; Lutzoni F. Mol. Biol. Evol. 2003, 20, 255–266. 10.1093/molbev/msg028. [DOI] [PubMed] [Google Scholar]
- Hall B.Phylogenetic Trees Made Easy: A How-to Manual; Sinauer Associates, 2004. [Google Scholar]
- Salemi M.; Vandamme A.-M.. The Phylogenetic Handbook: A Practical Approach to DNA and Protein Phylogeny; Cambridge University Press, 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.