Abstract
Undoubtedly, the discovery of penicillin is one of the greatest milestones in modern medicine. 2016 marks the 75th anniversary of the first systemic administration of penicillin in humans, and is therefore an occasion to reflect upon the extraordinary impact that penicillin has had on the lives of millions of people since. This perspective presents a historical account of the discovery of the wonder drug, describes the biological nature of penicillin, and considers lessons that can be learned from the golden era of antibiotic research, which took place between the 1940s and 1960s. Looking back at the history of penicillin might help us to relive this journey to find new treatments and antimicrobial agents. This is particularly relevant today as the emergence of multiple drug resistant bacteria poses a global threat, and joint efforts are needed to combat the rise and spread of resistance.
Keywords: Penicillin, antimicrobial resistance, Howard Florey
Fleming’s discovery and Oxford’s breakthrough 1928 to 1941
The discovery of penicillin was much more than just a lucky accident, although it did start with a mistake. In 1928, after returning from holiday, Alexander Fleming, a bacteriologist working at St. Mary’s hospital in London, noticed that one of his petri dishes containing staphylococci that he left on a bench was contaminated [1]. He observed that a fungal contaminant was affecting the growth of the nearby bacteria. The fungus turned out to be Penicillium notatum, and the antibacterial molecule that it produced was named penicillin. Fleming recorded his observations in the article in The British Journal of Experimental Pathology in 1929 [2], where he showed that penicillin is able to inhibit bacterial growth in vitro. Fleming thought that penicillin could be useful as a local antiseptic, but did not manage to purify penicillin or characterize its activity [1,3].
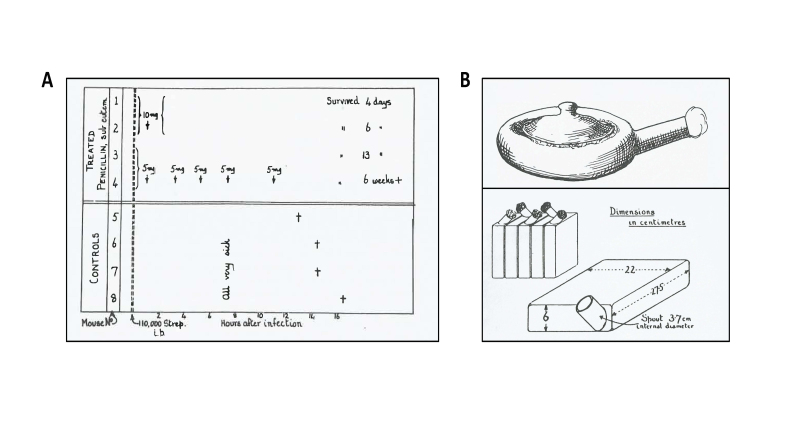
Fleming’s article on penicillin served as a basis for scientists at Oxford to begin research on new antimicrobials in 1939. At that time, Howard Florey was working on lysozyme and its ability to kill bacteria. Together with Ernst Chain, a chemist in the laboratory, Florey took interest in Fleming’s observation of the antimicrobial capacity of Penicillium. Chain and Florey decided to design a method to culture the fungus and aimed to produce it in sufficient quantities to allow further testing of its antimicrobial roles. Norman Heatley, a young chemist in Florey’s lab, played a key role in the process of penicillin purification. Heatley and Chain designed early methods for extracting penicillin, which was needed to obtain enough material to perform the first trials with the drug. By the mid-1940s, enough penicillin was available at the Sir William Dunn School of Pathology in Oxford to set up trials of its efficacy in mice. It was this experiment performed by Heatley and colleagues that provided key data to demonstrate the effect of penicillin in vivo [4,5]. Eight mice were injected with a fatal dose of Group A streptococcus. After one hour, two mice were given a single dose of penicillin (10 mg) and two were given 5 mg of penicillin, plus three additional doses of 5 mg at 3, 5, 7, and 11 hours after infection. Four mice served as controls and received no penicillin. Seventeen hours after the initial infection, all mice in the control group had died, while all the mice that had received a penicillin dose survived (Figure 1A). This remarkable observation provided the key evidence that penicillin had potential as a life-saving drug.
Figure 1.
The first mouse experiment and ceramic vessels which were used to grow the fungus. A. Results from the first penicillin trial experiment involving mice which was performed by the scientists at Oxford in 1940 (left panel). B. Old-fashioned bedpan, acquired from Radcliffe Infirmary hospital in Oxford to grow the first batch of penicillin. Ceramic culture vessel designed by Florey and colleagues to increase the yield of penicillin production (right panel). (Figures from [4]).
Penicillin Production 1941 to 1943
The main challenge faced by Florey and his team was to produce enough penicillin for further experimentation on mice, while human trials required much larger doses. The yield and production had to be increased, but during wartime no help was available from commercial firms because resources were so scarce. Production was therefore set up in Florey’s department. Originally, old-fashioned bedpans were used to grow Penicillium, but these did not generate sufficient yield [4]. With limited equipment, Florey and colleagues designed their own ceramic culture vessel. Heatley and Florey then arranged the vessel to be mass produced by a pottery firm about 100 meters away from Oxford, which was suggested to him by his acquaintance in potteries (Figure 1B). In the end of 1940, Florey and colleagues received the vessels, and Heatley inoculated them with the fungus on December 25th. By the start of February 1941, the Oxford team had purified sufficient material for clinical trials in humans.
The First Trials in Humans
The first patient to receive the penicillin as part of the toxicity test was a woman with terminal cancer. Shortly after she was given penicillin by injection, she developed fever and rigor, which were caused by impurities of pyrogenic origin that were contained in the penicillin mix [5].
Edward Abraham was a biochemist in Florey’s lab, and together with Chain he suggested that the penicillin could be further purified to remove any residual pyrogens before it was given to patients. The second patient to receive a purified dose of penicillin was a policeman at the Radcliffe Infirmary who had severe staphylococcal and streptococcal infection [5]. Repeated intravenous injections of penicillin over 5 days had a profound effect on his recovery. Due to very low supplies of penicillin, the patient’s urine had to be collected and recycled to re-extract more penicillin for further injections. Eventually, the overall shortage of penicillin forced the treatment to be terminated, and the patient relapsed and died shortly after [6].
Florey, Heatley, and Chain conducted a series of further clinical trials between 1941 to 1942 which involved 170 patients. The results demonstrated a remarkable effect of penicillin in combating bacterial infections without any toxic side effects [7].
The Oxford Team immediately recognized the potential of penicillin for treatment of injured soldiers and wounded civilians during the war. However, the funding and the capacity for mass production of penicillin were not available in the United Kingdom, so in 1941 Florey and Heatley went to the United States, which was not yet at war, to seek support. Florey and Heatley, together with American colleagues, helped to establish what later became known as “The Penicillin Project”. The work of the project had three main streams: the first was focused on improving the purification of penicillin; the second aimed to find more potent strains of Penicillium, and Heatley worked closely with the US Department of Agriculture to characterize these strains; the last stream, headed by Florey and his American partners, was focused on finding pharmaceutical companies which would take on mass production of penicillin. Eventually, 15 drug companies in both the U.K. and the U.S. worked on penicillin production, and clinical trials took place across the U.S. to further prove the effectiveness of the drug. The first trials of penicillin in the war setting were conducted by Florey in the military hospitals in north Africa in 1942, and showed that penicillin was effective when used on both fresh and infected wounds [8]. It was evident that penicillin would be instrumental in the war effort to save the lives of many soldiers. Once fatal bacterial infections, which took thousands of servicemen’s lives during WWI, became curable thanks to penicillin. Soon, collaborative efforts between the government, industry, and British and American scientists led to sufficient supplies of penicillin being manufactured by D-Day in 1944, when Allied troops landed in France [8,9]. After the war, by 1946, penicillin was widely available for prescription.
In 1945, Fleming, Florey and Chain were awarded the Nobel Prize for “the discovery of penicillin and its curative effect in various infectious diseases” [10]. Chain and Abraham had continued to work on the structure of penicillin until 1943, when Abraham first proposed the beta-lactam structure. This was confirmed by Dorothy Hodgkin and Barbara Low using X-ray crystallography [11,12].
The discovery of penicillin changed the course of history. Penicillin saved thousands of wounded soldiers and civilians during the biggest of the wars, and its discovery laid the foundations of the antibiotic era and subsequent development of other more potent antibiotics.
Mechanisms of Penicillin, a Revolutionary and Inspirational Therapeutic of Modern Medicine
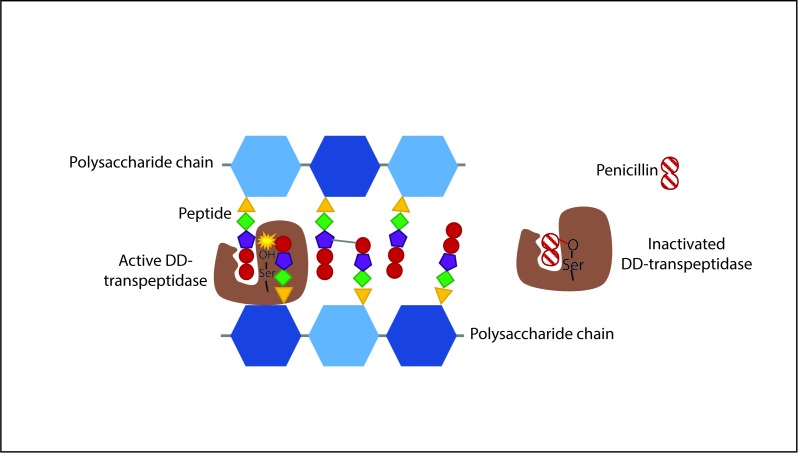
An essential structural element for most bacteria is the cell wall, a protective layer of peptidoglycan (PGN) whose main function is to preserve cell integrity and shape and prevent macromolecules from penetrating into the cell [13]. PGN is located just outside the cytoplasmic membrane, and is composed of chains of alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) residues, which are covalently crosslinked via short peptides (Figure 2). During growth and division, PGN is continuously synthesized and remodeled. Therefore, it is essential for bacteria to be able to synthesize the components of PGN and assemble them into a single macromolecule. The characteristic strength of PGN resides in its net-like conformation that is mainly derived from peptide cross-linkages [13]. These linkages are formed by the activity of specific enzymes called transpeptidases or Penicillin-Binding Proteins (PBPs). Penicillin, like other components of the beta-lactam antibiotics, contains a four-membered beta-lactam ring (Figure 3), which is responsible for the inhibition of transpeptidase [14]. By mimicking the last two D-alanine residues of the peptide, penicillin is able to bind irreversibly the active site of the transpeptidase, preventing the enzyme from cross-linking the peptidoglycan strands. Therefore, by blocking the formation of peptide bridges, penicillin prevents new PGN formation and the cell is susceptible to lysis, as the PGN is no longer able to provide resistance against osmotic stress. Furthermore, penicillin specifically targets bacteria, as eukaryotic cells lack both PGN and the enzymes responsible for PGN synthesis.
Figure 2.
Schematic representation of the mechanism of penicillin action. PGN is composed of polysaccharide chains made of GlcNAc and MurNAc units (shown in different shades of blue) which in turn have small peptides attached to them. The transpeptidase enzyme (PBP) (in brown) catalyzes the formation of cross-linkages between these peptides, by specifically binding the last two D-alanine residues of one peptide (red circles). Penicillin mimics the structure of these residues and inactivates the PBP by forming an irreversible covalent bond to the catalytic serine residue of the enzyme [69].
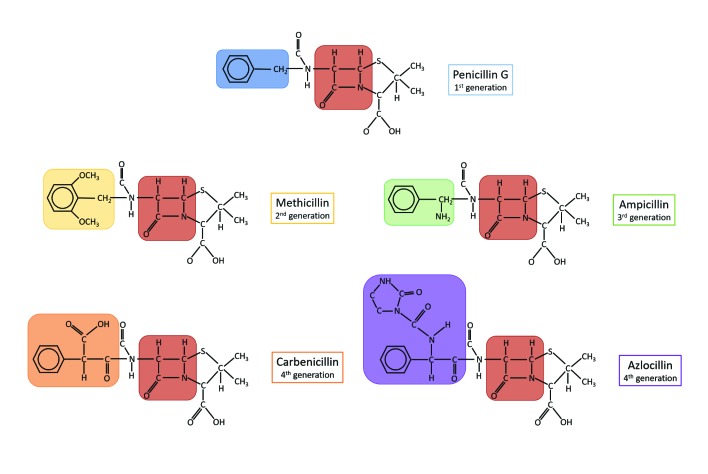
Figure 3.
Chemical structures of different classes of penicillins. Examples of different generations of penicillins are shown. Beta-lactam ring, the common feature of all classes, is highlighted in brown and the corresponding chemical substitute on the side chain is color-coded: blue, penicillin G (benzylpenicillin class, 1st generation); yellow, methicillin (2nd generation); green, ampicillin (aminopenicillin class, 3rd generation); orange, carbenicillin (carboxypenicillin class, 4th generation); purple, azlocillin (ureidopenicillin class, 4th generation).
In current times, the name “penicillin” is generically used to refer to different molecules that a have beta lactam-based structure and the same antibacterial activity as benzylpenicillin (penicillin G) – the original molecule extracted from P. notatum. The classification of penicillins relies on chemical substitutions on the residue attached to the beta-lactam ring, which confer different activities. Benzylpenicillins, for example, are more active against Gram-positive bacteria in particular cocci, such as staphylococci, pneumococci, and other streptococci, and bacilli, including Bacillus anthracis, Clostridium perfringens, and Corynebacterium diphtheriae, but less efficacious against Gram-negative bacteria. The inability to act against Gram-negative bacteria is observed not only among benzylpenicillins but also across many different antibiotics. This is largely due to two factors: firstly, unlike Gram positive bacteria, Gram negative species contain the outer membrane, which acts as a selective barrier, blocking the penetration of penicillin [15]; secondly, some Gram-negative bacteria have acquired specific genes which encode for penicillinases (also known as beta-lactamases), a class of enzymes that inactivate penicillin by hydrolysis of the beta-lactam ring [16].
Following a wide use of the “natural penicillins,” penicillinase-producing strains also emerged among Gram-positive species. This shifted pharmacological research towards the development of semisynthetic, beta-lactamase-resistant penicillins (i.e. second generation penicillins): oxacillin, methicillin, and dicloxacillin, also defined as anti-staphylococcal penicillins, due to their ability to resist penicillinases present in staphylococci. The relatively narrow spectrum of activity of these antibiotics and the need for broader coverage against Gram-negative organisms, served as an incentive to expand the second generation penicillins. In the 1960s, the third generation and broad-spectrum penicillins also known as aminopenicillins, were introduced. Amoxicillin and ampicillin are the main examples of this group and unlike their predecessors, third generation penicillins proved to be more effective against a wider group of Gram-negative bacteria (including Haemophilus influenzae, Escherichia coli, Salmonella spp., and Shigella spp.), thanks to their higher stability to penicillinases [16]. The last generation of penicillins which includes carboxypenicillins and ureidopenicillins further broadened the spectrum of penicillin coverage against Gram-negative bacteria and displayed potent activity against Pseudomonas aeruginosa [17].
In addition to penicillins, other classes of beta-lactam compounds were discovered and have been introduced for clinical use. In 1945, the first component of the cephalosporin family was isolated from the fungus Cephalosporium acremonium [18]. Several generations of this novel antibiotic have been developed through different chemical modifications of the natural compound originally isolated, increasing the spectrum of their activity. Since the late 1970s, both new discoveries and advances in chemical alterations of the basic beta-lactam structure allowed production of more beta-lactam antibiotics, including the penems, carbapenems, and monobactams [19].
Penicillin Resistance: First Signs, Progression and the Global Problem
The first sign of antibiotic resistance became apparent soon after the discovery of penicillin. In 1940, Abraham and Chain reported that an E. coli strain was able to inactivate penicillin by producing penicillinase [20].
The spread of penicillin resistance was already documented by 1942, when four Staphylococcus aureus strains were found to resist the action of penicillin in hospitalized patients [21]. During the next few years, the proportion of infections caused by penicillin-resistant S. aureus rapidly rose, spreading quickly from hospitals to communities. By the late 1960s, more than 80 percent of both community and hospital-acquired strains of S. aureus were penicillin-resistant [22]. The rapid spread of penicillin resistance temporarily came to a halt after the introduction of the second-generation, semisynthetic methicillin in the 1960s. However, methicillin-resistant strains soon emerged, and only in 1981 was this mechanism of resistance unraveled [23]: these strains harbored an altered PBP, designated PBP-2a, which showed a reduced affinity for penicillin, thereby conferring resistance to penicillin. PBP-2a is encoded by mecA, a gene located on the S. aureus chromosome [24], which resides within the mobile genomic island SCCmec (staphylococcal cassette chromosome mec) [25]. In approximately 20 years, methicillin resistance became endemic in the U.S., reaching 29 percent of hospitalized S. aureus-infected patients [26,27].
In 1967, strains of S. pneumoniae also became resistant to penicillin [28]. By 1999, the percentage of cases associated with antibiotic-resistant pneumococcus had tripled compared to 1979, reaching 14.4 percent in South Africa [29]. In 1976, beta lactamase-producing gonococci were isolated in England and the U.S. [30-32]. Rapid spread of gonococcus resistance followed [33] and in the 10-year period after the first introduction of penicillin to treat gonorrhea, the prevalence of gonococcal penicillin-resistant strains reached its peak, particularly in Asia [30]. Furthermore, in 1983, a large outbreak of resistant non-beta-lactamase producing gonococcus affected Durham city in North Carolina (U.S.) [34]. Resistance of these strains was chromosomally-mediated, due to the emergence of mutations that modified the penicillin target PBP2 and expression of drug efflux pumps systems [35,36]. Together, these events led to the prohibition of penicillin use as the first-line drug for gonococcus treatment in most parts of the world [37].
Another group of bacteria with high rates of penicillin resistance is the Enterobacteriaceae, of which several strains are intrinsically aminopenicillin-resistant, particularly among E. coli species [38-40]. Between 1950 and 2001, approximately two-thirds of E. coli causing human diseases were ampicillin-resistant in the U.S. [40], and the rate of aminopenicillin resistance is still on the rise [41].
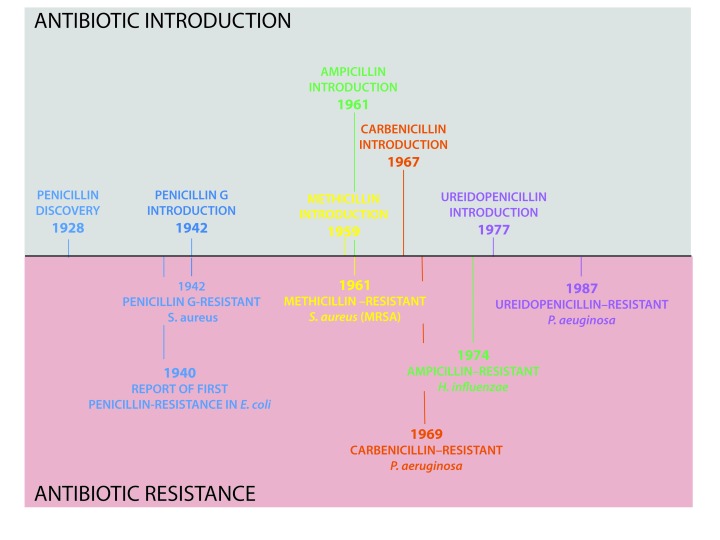
The development of resistance went hand in hand with the introduction of new generations of penicillin into clinical practice (Figure 4). More than 150 antibiotics have been found since the discovery of penicillin, and for the majority of antibiotics available, resistance has emerged. Moreover, the recent rise of multi/pan-drug resistant strains has correlated with enhanced morbidity and mortality. Overall, ineffectiveness of the antibiotic treatments to “superbug” infections has resulted in persistence and spread of multi-resistant species [42] across the globe. This represents a serious worldwide threat to public health [41].
Figure 4.
The journey of antibiotic development and resistance. Key dates for introduction of different penicillin classes and corresponding emergence of resistance are shown (top and bottom, respectively). Examples of different generations of penicillin are indicated and color-coded as in Figure 3.
Post-antibiotic Era
In early 1945, Fleming predicted that the high public demand of antibiotics would determine an “era of abuse”; this eventually became a reality [43-45]. No sooner had the miraculous effects of penicillin become apparent to the general public, then the antibiotic started to be overused. This triggered selective pressure for the emergence of penicillin-resistant strains, which over a few years spread across different countries. The discovery of each new generation of antibiotic quickly followed the same trend. Several studies have demonstrated the association of antibiotic use with the emergence of resistance. In 2001, the European Surveillance of Antimicrobial Consumption (ESAC) documented in a project variations of antimicrobial resistance (AMR) in selected bacteria, and found a clear link between resistance and antimicrobial use in European countries [46]. This was particularly evident for S. pneumoniae, for which higher rates of antibiotic resistance were found in countries of southern and eastern Europe, such as France, Spain, Portugal, and Slovenia where antibiotics are consumed in higher amounts compared to northern Europe. [46].
There is a hypothesis that the excessive use of antibiotics is correlated with inappropriate prescription and administration of antibiotic therapy [45]. A recent study conducted in the U.S. analyzed the data from antibiotic prescriptions written between 2010 to 2011, and showed that approximately 30 percent of prescriptions of orally-administrated antibiotic were unnecessary [47]. Moreover, as stated by the World Health Organization in 2014, antibiotics can be bought legally over the counter in 19 European countries, and in five countries they can be bought on the Internet without prescription. Therefore, easy access to antibiotics is likely to have promoted overuse of these drugs and contributed to the current problem of antibiotic resistance.
Another factor that is thought to contribute to antibiotic resistance is the extensive agricultural use of antibiotics, primarily as growth promoters and to prevent infection in livestock [45]. The majority of antibiotic consumption is for agricultural purposes, accounting for 63,000 to over 240,000 tons of annual global antibiotic use [48-50]. This large use of antibiotics had a positive influence on the emergence of resistant strains that could be directly transferred from animals to humans through the food chain. A WHO report in 2013 showed that levels of Campylobacter resistance to several antibiotics is likely to be linked to consumption of infected poultry in many parts of the world [61]. The overuse of antibiotics is also thought to damage the environmental microbiome. Antibiotics can reach the environment not only by the direct use of preservatives for plants, but also through urine and stools excreted by antibiotic-treated animals. In this way, environmental non-pathogenic or opportunistic species could be exposed to antibiotics and act as a reservoir of resistance genes [43,52].
As stated in the first major WHO report on AMR, appropriate global data on AMR is missing [53]. Collecting data globally to monitor AMR is necessary to establish the link between antibiotic use and resistance across different countries and to design intervention strategies to target AMR on a global scale. However, there are challenges in the methods used for global collection of data. AMR is constantly evolving, and so it is clear that resistance data collected from several countries provides us with only a snapshot of a highly dynamic situation. Better systems to monitor AMR have to be designed to take into account people exposed to antimicrobials, the density of the human population, as well as the effect of individual antibiotic classes on specific bacteria [54].
Recently, the U.K. Government commissioned a Review on Antimicrobial Resistance [55]. This has highlighted one crucial point: the AMR epidemic that we are facing now is a global problem, which involves not just the medical and scientific communities, but also society as a whole. The authors examined the financial burden of AMR as well as the human cost, and outlined the steps the global community should take to target AMR and new approaches for antimicrobial therapies. A ten-point plan proposed in this report (Table 1) aims to present key strategies that have to be implemented to address the problem of AMR. At the heart of the plan lies a call for international cooperation, which is essential to stop inappropriate antibiotic administration, to increase surveillance, and to develop new therapies. Significant progress has been made by creating several global funds including the Global Innovation Fund that combines the action of several governments with existing funding bodies [56]. Together, this fund aims to create better diagnostics for AMR and invest in research for antibiotic development. In addition to the U.K. Review, other countries recognize the importance of coordinated action to combat AMR. In 2014, the U.S. government designed the National Action plan for combating AMR bacteria, which aims to reduce and prevent the emergence of diseases caused by drug-resistant bacteria [57]. The plan also emphasizes the importance of improving the infrastructure for public health surveillance and better design of lab diagnostics to identify resistant bacteria. Similarly, the European Union Joint Programming Initiative on AMR (JPIAMR) was created to establish the international collaborative platform to reduce AMR. Between 2014-2016, JPIAMR has guaranteed 55 million euro funding for key AMR projects and has successfully created a map of all AMR research and associated investment in its member countries, which include 19 European countries as well partners outside Europe, such as Japan, Canada, and Argentina. Interestingly, in February 2017 the European Commission for Research and Innovation awarded the 1 million euro Horizon Prize for “better use of antibiotics” to the company Minicare HNL [58]. The machine designed by Minicare HNL can perform a finger prick test to determine in a few minutes whether the patient has a bacterial or viral infection, thus providing accurate diagnosis for appropriate administration of antibiotics. This affordable and easy-to-use machine is expected to be available for physicians in 2018. On a global level, AMR prevention and intervention strategies have also been endorsed by WHO. As one of these initiatives, all WHO member states are expected to develop national action plans to combat AMR by 2017 [59]. This will undoubtedly make the first key step in implementing all main action plans currently available, which share the same goals: stop and prevent the spread of multi-drug resistant bacterial strains.
Table 1. Tackling AMR. ten-point plan proposed by Jim O’Neill and colleagues to the UK Government as part of the Review on the Antimicrobial Resistance [55].
| 1 Public Awareness |
| •Public health programs across the countries |
| 2 Prevent the spread of infection |
| •Expansion of the access to clean water and appropriate sanitation |
| •Reduction of infection in hospitals and care settings |
| 3 Reduction of antibiotic use in agriculture |
| •Restriction on the use of highly critical antibiotics in farming |
| •Prevention of antibiotic dissemination into environment |
| 4 Global surveillance |
| •Major surveillance programs including USA Global Health Security Agenda, UK Fleming Fund, WHO Global AMR Surveillance System |
| •Easy data accessibility around the world |
| 5 Rapid new diagnostics |
| •Support research and innovation in this area |
| 6 Use of alternative antimicrobials |
| •Vaccines |
| •Bacteriophage therapy, engineered bacteria, antimicrobial peptides |
| 7 Recognition of researchers in infectious disease |
| •Clear career paths and rewards for scientists in the field |
| 8 Global Innovation Fund |
| •Link and expand major initiatives |
| •Fund different projects (e.g. R&D that might lack commercial imperative) |
| 9 Better investment for new drugs |
| •Governments should find new ways to reward innovation |
| •Link between profit and volume of sales should be reduced |
| 10 Global Coalition for real action |
| •Joint efforts from G20 and UN are needed |
| •Step change plan to fight AMR has to be redesigned |
Alternative Approaches to Treat Infectious Diseases
The route to find new antibiotics and develop them into drugs is long and expensive. It costs 800 million to 1 billion dollars to bring a new drug to market, and it takes on average over 10 years for it to enter the clinic. Due to the time pressure that we are currently facing in the battle against AMR, a different approach is to explore alternatives to antibiotic therapy.
It is well-known that some metals have antimicrobial properties; therefore, exploring metal nanoparticles as a new antimicrobial therapy could eliminate antibiotic-resistant bacteria. There are several ways by which metal nanoparticles can affect bacterial survival. Silver-containing antimicrobials can for instance exert a physical stress on bacterial cells. Other evidence suggests that gallium can be effective in interfering with bacterial metabolic pathways by interrupting bacterial metal ion uptake [60], which in turn affects biofilm-forming P. aeruginosa in vitro. Gallium is now entering clinical trials as an antimicrobial treatment for patients with cystic fibrosis; however, the toxicity and narrow spectrum activity of metal nanoparticles overall remains a challenge [60,61].
Some studies have suggested that genetically engineered bacteria might serve as a tool to eliminate pathogenic bacteria. Hwang and colleagues have shown that laboratory-engineered E. coli is able to secrete antimicrobial peptides in response to quorum-sensing molecules released by P. aeruginosa [62]. These antimicrobial peptides were able to degrade biofilms formed by P. aeruginosa, which suggests that “predator” bacteria can be specifically engineered to target important pathogens.
Using antimicrobial peptides on their own is another approach. For instance, pexiganan, a natural peptide identified over ten years ago in the skin of the African clawed frog, was shown to be effective in killing both Gram-positive and Gram-negative bacteria [63]. This drug has now entered a Phase III clinical trial as a treatment against diabetic ulcers [64,65].
Phage therapy is emerging as an alternative to target antimicrobial resistant bacteria. In some countries, phage therapy has been used for a number of years and there are designated centers for phage therapy, such as those in Georgia. European countries including Switzerland, Belgium, and France have begun to explore phage therapy by creating the Phagoburn project, which focuses on using a combination of phage therapies to treat bacteria-infected burns. This treatment is now in Phase I-II clinical trials [66]. The scientific community is still looking for ways to overcome the hurdles of phage therapy. For example, it remains difficult to validate production, as combinations of phages or phage cocktails remain highly variable. Furthermore, stability of the phages and their antibacterial activity has to be validated.
Preventing infections in the first place is another strategy in tackling AMR. Access to clean water supply and effective healthcare systems will substantially reduce the burden of AMR by limiting the spread of infections and decreasing the overall number of infected individuals. Furthermore, improving the hygiene and sanitation in hospitals can lower the number of cases associated with multiple-drug resistant bacteria [55].
Unlike antimicrobial therapies, vaccines have several advantages. Firstly, they can prevent infections by both antibiotic-resistant and antibiotic-sensitive bacteria. Secondly, vaccines are able to provide herd immunity, offering protection to unvaccinated individuals by reducing the transmission of pathogens. Furthermore, vaccination may affect bacterial colonization, thus reducing the overall population of bacteria and possibly the spread of antimicrobial genes to the commensal microbiota [67]. Thirdly, antibiotics are often given to individuals with viral infection to prevent any potential secondary implications caused by bacterial infection. Vaccination programs have the capacity to prevent viral infections which would subsequently lower antibiotic administration and combat the rising AMR. Similar to human immunization, vaccines have a potential to be exploited in agriculture to reduce the antibiotic usage in the farming sector.
An example to show the successful impact of vaccines on AMR is the introduction of pneumococcal conjugate vaccine (PCV). The vaccine was introduced in the U.S. in children during 2001 and led to a striking decrease in pneumococcal diseases, including antibiotic resistant infections. Penicillin-resistant cases dropped by 81 percent, and a general downturn of resistant pneumococcal infections was observed among older children as well as adults who did not receive the vaccine. This demonstrated herd immunity, leading to an estimated 50 percent reduction of the total number of penicillin resistant cases [68].
Similar results were obtained following the introduction of Haemophilus influenzae type b (Hib) conjugate vaccine in 1990, which slowed down the incessant evolution of resistant strains [67].
For vaccines to be effective as an antimicrobial strategy, several challenges have to be overcome. For instance, the pneumococcal vaccine introduced in 2001 was able to protect individuals against seven serotypes defined by their capsular polysaccharide. Over time, there was an increased incidence of disease caused by non-vaccine pneumococcal serotypes. This has led to reintroduction of a modified PCV, which covers six additional serotypes and offers broader protection. Therefore, the impact of vaccines has to be monitored so that the vaccines can be updated to cover emerging strains.
The alternative therapies demand rapid diagnostic tools to identify bacterial infection quickly and efficiently. This will allow more targeted approaches to therapy by determining the bacteria species causing disease. Effective diagnostic techniques and rapid screens will also reduce empirical administration of the existing antibiotics, hopefully lowering the selective benefit for antibiotic-resistant strains.
Understanding the mechanisms that underlie resistance remains a key priority, as the acquisition and development of resistance varies depending on the species as well as on the stage of the infection. Subverting resistance mechanisms themselves, for instance through the development of beta lactamase inhibitors, has proven highly successful in the past, and might prolong the effective lifespan of our current stock of antibiotics.
Conclusions
The discovery and development of penicillin by Alexander Fleming, Howard Florey, Ernst Chain, and Norman Heatley, opened a new chapter in modern medicine. The purification and characterization of penicillin resulted in identification of next generation penicillins and has led to the discovery of different classes of antibiotics, which has had a profound impact, saving many lives throughout the last 75 years.
Unfortunately, the influence of antibiotics is now fading due to the progressive rise of resistance, and this phenomenon is observed among all antimicrobial drugs. Increasingly, there are reports of bacterial species which are resistant to all known antibiotics, leaving us very vulnerable to common infections. We now must respond to this challenge, keeping in mind the lessons we learned from the discovery of penicillin and the subsequent events. To begin to tackle AMR, countries across the world must unite to implement strict regulations for antibiotic administration in the medical and agricultural sectors. Secondly, alternative strategies such as phage therapy, antimicrobial peptides, and vaccines should be explored, as they offer another route to halt and prevent AMR. Sufficient funding and joint efforts from pharmaceutical companies, governments, and academia are necessary to turn promising therapies into valuable drugs. Thirdly, new techniques for rapid diagnostics of bacterial infection and better AMR surveillance schemes should be designed. These in fact are crucial for early identification of resistance and for the implementation of appropriate interventions to combat the spread of AMR. The solution to stop AMR will undoubtedly require global efforts from researchers across all disciplines, doctors, politicians, policymakers, and the public.
Glossary
- PGN
peptidoglycan
- GlcNAc
N-acetylglucosamine
- MurNAc
N-acetylmuramic acid
- PBPs
Penicillin-Binding Proteins
- SCCmec
staphylococcal cassette chromosome mec
- ESAC
European Surveillance of Antimicrobial Consumption
- AMR
antimicrobial resistance
- JPIAMR
European Union Joint Programming Initiative on AMR
- PCV
pneumococcal conjugate vaccine
- Hib
Haemophilus influenzae type b
Author Contributions
ML contributed to the following sections: “Fleming’s discovery and Oxford’s breakthrough 1928 to 1941,” “Penicillin production 1941 to 1943,” and “The First Trials in Human.” GP contributed to the following sections: “Mechanisms of penicillin, a revolutionary and inspirational therapeutic of modern medicine,” “Penicillin resistance: first signs, progression and the global problem.” Both GP and ML contributed equally to the following sections “Post-antibiotic era,” and “Alternative approaches to treat infectious diseases.”
References
- Swann JP. The Search for Synthetic Penicillin during World War II. Br J Hist Sci. 1983;16(2):154–190. doi: 10.1017/s0007087400026789. [DOI] [PubMed] [Google Scholar]
- Fleming A. On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to their Use in the Isolation of B. influenzæ. Br J Exp Pathol. 1929;10(3):226–236. [Google Scholar]
- Aminov RI. A Brief History of the Antibiotic Era: Lessons Learned and Challenges for the Future. Front Microbiol. 2010;1:134. doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heatley N. Penicillin and Luck: Good Fortune in the Development of the 'miracle Drug. RCJT Books; 2004. [Google Scholar]
- Cranston D, Sidebottom E. Penicillin and the legacy of Norman Heatley. Wordsbydesign. 2016 [Google Scholar]
- Fletcher C. First clinical use of penicillin. BMJ. 1984;289(6462):1721–1723. doi: 10.1136/bmj.289.6460.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon BL. Penicillin: its discovery and early development. Semin Pediatr Infect Dis. 2004;15(1):52–57. doi: 10.1053/j.spid.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Fraser I. Penicillin: early trials in war casualties. Br Med J (Clin Res Ed) 1984;289(6460):1723–1725. doi: 10.1136/bmj.289.6460.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bud R. Penicillin: Triumph and Tragedy. Oxford: Oxford University Press; 2007. [Google Scholar]
- The Nobel Prize in Physiology or Medicine 1945. Nobelprize.org. Available from http://www.nobelprize.org/nobel_prizes/medicine/laureates/1945/
- Hodgkin DC. The X-ray analysis of the structure of penicillin. Adv Sci. 1946;6(22):85–89. [PubMed] [Google Scholar]
- Glusker JP. Dorothy Crowfoot Hodgkin (1910-1994). Protein Sci. 1994;3(12):2465–2469. doi: 10.1002/pro.5560031233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32(2):149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- Tipper DJ, Strominger JL. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc Natl Acad Sci U S A. 1965;54(4):1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MG. The role of the outer membrane of Gram-negative bacteria in antibiotic resistance: Ajax' shield or Achilles' heel? Handb Exp Pharmacol. 2012;211:67–86. doi: 10.1007/978-3-642-28951-4_5. [DOI] [PubMed] [Google Scholar]
- Sutherland R. The nature of the insensitivity of gram-negative bacteria towards penicillins. J Gen Microbiol. 1964;34:85–98. doi: 10.1099/00221287-34-1-85. [DOI] [PubMed] [Google Scholar]
- Preston SL, Drusano GL. Penicillins Antimicrobe. E-Sun Technologies, Inc. 2016. [[cited 8-12-2016]]. Available from http://www.antimicrobe.org/d24.asp .
- Bo G. Giuseppe Brotzu and the discovery of cephalosporins. Clin Microbiol Infect. 2000;6(Suppl 3):6–9. doi: 10.1111/j.1469-0691.2000.tb02032.x. [DOI] [PubMed] [Google Scholar]
- Dalhoff A, Janjic N, Echols R. Redefining penems. Biochem Pharmacol. 2006;71(7):1085–1095. doi: 10.1016/j.bcp.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Abraham EP, Chain E. An enzyme from bacteria able to destroy penicillin. 1940. Rev Infect Dis. 1988;10(4):677–678. [PubMed] [Google Scholar]
- Rammelkamp T. Resistance of Staphylococcus aureus to the action of penicillin. Exp Biol Med. 1942;51:386–389. [Google Scholar]
- Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest. 2003;111(9):1265–1273. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman B, Tomasz A. Altered penicillin-binding proteins in methicillin-resistant strains of Staphylococcus aureus. Antimicrob Agents Chemother. 1981;19(5):726–735. doi: 10.1128/aac.19.5.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuhashi M, Song MD, Ishino F. et al. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to beta-lactam antibiotics in Staphylococcus aureus. J Bacteriol. 1986;167(3):975–980. doi: 10.1128/jb.167.3.975-980.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44(6):1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panlilio AL, Culver DH, Gaynes RP. et al. Methicillin-resistant Staphylococcus aureus in U.S. hospitals, 1975-1991. Infect Control Hosp Epidemiol. 1992;13(10):582–586. doi: 10.1086/646432. [DOI] [PubMed] [Google Scholar]
- Zaffiri L, Gardner J, Toledo-Pereyra LH. History of antibiotics: from fluoroquinolones to daptomycin (Part 2). J Invest Surg. 2013;26(4):167–179. doi: 10.3109/08941939.2013.808461. [DOI] [PubMed] [Google Scholar]
- Hansman D, Devitt L, Miles H. et al. Pneumococci relatively insensitive to penicillin in Australia and New Guinea. Med J Aust. 1974;2(10):353–356. doi: 10.5694/j.1326-5377.1974.tb70836.x. [DOI] [PubMed] [Google Scholar]
- Koornhof HJ, Wasas A, Klugman K. Antimicrobial resistance in Streptococcus pneumoniae: a South African perspective. Clin Infect Dis. 1992;15(1):84–94. doi: 10.1093/clinids/15.1.84. [DOI] [PubMed] [Google Scholar]
- Lind I. Antimicrobial resistance in Neisseria gonorrhoeae. Clin Infect Dis. 1997;24(Suppl 1):S93–S97. doi: 10.1093/clinids/24.supplement_1.s93. [DOI] [PubMed] [Google Scholar]
- Phillips I. Beta-lactamase-producing, penicillin-resistant gonococcus. Lancet. 1976;2(7987):656–657. doi: 10.1016/s0140-6736(76)92466-1. [DOI] [PubMed] [Google Scholar]
- Ashford WA, Golash RG, Hemming VG. Penicillinase-producing Neisseria gonorrhoeae. Lancet. 1976;2(7987):657–658. doi: 10.1016/s0140-6736(76)92467-3. [DOI] [PubMed] [Google Scholar]
- Unemo M, Shafer WM. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev. 2014;27(3):587–613. doi: 10.1128/CMR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruki H, Kohmescher RN, McKinney WP. et al. A community-based outbreak of infection with penicillin-resistant Neisseria gonorrhoeae not producing penicillinase (chromosomally mediated resistance). N Engl J Med. 1985;313(10):607–611. doi: 10.1056/NEJM198509053131004. [DOI] [PubMed] [Google Scholar]
- Faruki H, Sparling PF. Genetics of resistance in a non-beta-lactamase-producing gonococcus with relatively high-level penicillin resistance. Antimicrob Agents Chemother. 1986;30(6):856–860. doi: 10.1128/aac.30.6.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman KE, Pan W, Spratt BG. et al. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995;141(Pt 3):611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- Patel AL, Chaudhry U, Sachdev D. et al. An insight into the drug resistance profile & mechanism of drug resistance in Neisseria gonorrhoeae. Indian J Med Res. 2011;134:419–431. [PMC free article] [PubMed] [Google Scholar]
- Nordmann P. Trends in beta-lactam resistance among Enterobacteriaceae. Clin Infect Dis. 1998;27(Suppl 1):S100–S106. doi: 10.1086/514905. [DOI] [PubMed] [Google Scholar]
- Paterson DL. Resistance in gram-negative bacteria: Enterobacteriaceae. Am J Infect Control. 2006;34(5 Suppl 1):S20–S28. doi: 10.1016/j.ajic.2006.05.238. [DOI] [PubMed] [Google Scholar]
- Bouza E, Cercenado E. Klebsiella and enterobacter: antibiotic resistance and treatment implications. Semin Respir Infect. 2002;17(3):215–230. doi: 10.1053/srin.2002.34693. [DOI] [PubMed] [Google Scholar]
- Bergman M, Nyberg ST, Huovinen P. et al. Resistance FSGfA. Association between antimicrobial consumption and resistance in Escherichia coli. Antimicrob Agents Chemother. 2009;53(3):912–917. doi: 10.1128/AAC.00856-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanwar J, Das S, Fatima Z. et al. Multidrug resistance: an emerging crisis. Interdiscip Perspect Infect Dis. 2014;2014:541340. doi: 10.1155/2014/541340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett JG, Gilbert DN, Spellberg B. Seven ways to preserve the miracle of antibiotics. Clin Infect Dis. 2013;56(10):1445–1450. doi: 10.1093/cid/cit070. [DOI] [PubMed] [Google Scholar]
- Spellberg B, Gilbert DN. The future of antibiotics and resistance: a tribute to a career of leadership by John Bartlett. Clin Infect Dis. 2014;59(Suppl 2):S71–S75. doi: 10.1093/cid/ciu392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40(4):277–283. [PMC free article] [PubMed] [Google Scholar]
- Goossens H, Ferech M, Vander Stichele R. et al. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet. 2005;365(9459):579–587. doi: 10.1016/S0140-6736(05)17907-0. [DOI] [PubMed] [Google Scholar]
- Fleming-Dutra KE, Hersh AL, Shapiro DJ. et al. Prevalence of Inappropriate Antibiotic Prescriptions Among US Ambulatory Care Visits, 2010-2011. JAMA. 2016;315(17):1864–1873. doi: 10.1001/jama.2016.4151. [DOI] [PubMed] [Google Scholar]
- Landers TF, Cohen B, Wittum TE. et al. A review of antibiotic use in food animals: perspective, policy, and potential. Public Health Rep. 2012;127(1):4–22. doi: 10.1177/003335491212700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Boeckel TP, Brower C, Gilbert M. et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci U S A. 2015;112(18):5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace D. Review of Evidence on Antimicrobial Resistance and Animal Agriculture in Developing Countries, Evidence on Demand. International Livestock Research Institute. 2015. Available from: https://www.gov.uk/dfid-research-outputs/review-of-evidence-on-antimicrobial-resistance-and-animal-agriculture-in-developing-countries-201309 .
- WHO. The Global View of Campylobacteriosis. Report of expert consultation. 2012. Available from: http://www.who.int/foodsafety/publications/campylobacteriosis/en/
- Wright GD. Antibiotic resistance in the environment: a link to the clinic? Curr Opin Microbiol. 2010;13(5):589–594. doi: 10.1016/j.mib.2010.08.005. [DOI] [PubMed] [Google Scholar]
- WHO. Antimicrobial resistance: global report on surveillance 2014. 2014. [[cited 2016]]. Available from: http://www.who.int/drugresistance/documents/surveillancereport/en/
- Turnidge J. Antibiotic use and resistance;proving the obvious. The Lancet. 2014;365(9459):548–549. doi: 10.1016/S0140-6736(05)17920-3. [DOI] [PubMed] [Google Scholar]
- Review on Antimicrobial Resistance. Tackling drug-resitant infections globally: final report and recommendations. 2016 Available from: https://amr-review.org/home.html . [Google Scholar]
- Global Innovation Fund 2016. Available from: http://www.globalinnovation.fund/
- CDC. U.S. Activities to Combat AR. Available from: https://www.cdc.gov/drugresistance/us-activities.html .
- Prizes H. Better use of antibiotics - €1 million. Available from: https://ec.europa.eu/research/horizonprize/index.cfm?prize=better-use-antibiotics .
- WHO. Global action plan on antimicrobial resistance. Available from: http://www.who.int/antimicrobial-resistance/global-action-plan/en/
- Minandri F, Bonchi C, Frangipani E. et al. Promises and failures of gallium as an antibacterial agent. Future Microbiol. 2014;9(3):379–397. doi: 10.2217/fmb.14.3. [DOI] [PubMed] [Google Scholar]
- Lemire JA, Harrison JJ, Turner RJ. Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat Rev Microbiol. 2013;11(6):371–384. doi: 10.1038/nrmicro3028. [DOI] [PubMed] [Google Scholar]
- Hwang IY, Tan MH, Koh E. et al. Reprogramming microbes to be pathogen-seeking killers. ACS Synth Biol. 2014;3(4):228–237. doi: 10.1021/sb400077j. [DOI] [PubMed] [Google Scholar]
- Flamm RK, Rhomberg PR, Simpson KM. et al. In vitro spectrum of pexiganan activity when tested against pathogens from diabetic foot infections and with selected resistance mechanisms. Antimicrob Agents Chemother. 2015;59(3):1751–1754. doi: 10.1128/AAC.04773-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A Randomized, Double-Blind, Multicenter, Superiority, Placebo-Controlled Phase 3 Study of Pexiganan Cream 0.8% Applied Twice Daily for 14 Days in the Treatment of Adults With Mild Infections of Diabetic Foot Ulcers. 2016. Available from: http://adisinsight.springer.com/trials/700217169 .
- Reardon S. Antibiotic alternatives rev up bacterial arms race. Nature. 2015;521:402–403. doi: 10.1038/521402a. [DOI] [PubMed] [Google Scholar]
- Gabard J. Phage Therapy - Back to the Future! AMR Control. 2015;1:106–109. [Google Scholar]
- Lipsitch M, Siber GR. How Can Vaccines Contribute to Solving the Antimicrobial Resistance Problem? MBio. 2016;7(3) doi: 10.1128/mBio.00428-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyaw MH, Lynfield R, Schaffner W. et al. Effect of introduction of the pneumococcal conjugate vaccine on drug-resistant Streptococcus pneumoniae. N Engl J Med. 2006;354(14):1455–1463. doi: 10.1056/NEJMoa051642. [DOI] [PubMed] [Google Scholar]
- Yocum RR, Waxman DJ, Rasmussen JR. et al. Mechanism of penicillin action: penicillin and substrate bind covalently to the same active site serine in two bacterial D-alanine carboxypeptidases. Proc Natl Acad Sci U S A. 1979;76(6):2730–2734. doi: 10.1073/pnas.76.6.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]