Abstract
This study aimed to investigate the associations of XPC c.2815A>C, XPD c.934G>A and c.2251A>C, XPF c.2505T>C and ERCC1 c.354C>T single nucleotide polymorphisms (SNPs) of nucleotide excision repair pathway in outcome of head and neck squamous cell carcinoma (HNSCC) patients treated with cisplatin (CDDP) chemoradiation. Patients with XPC c.2815AC or CC and XPD c.934GA or AA genotypes had 0.20 and 0.38 less chances of presenting moderate/severe ototoxicity and nausea, respectively. Patients with XPD c.934AA and c.2251AC or CC genotypes had 8.64, 12.29 and 3.55 more chances of achieving complete response (CR), consistent ototoxicity and nephrotoxicity, respectively. AA haplotype of XPD and ACT haplotype of XPD and ERCC1 SNPs were associated with 9.30 and 3.41 more chances of achieving CR and consistent nephrotoxicity, respectively. At 24 months of follow-up, patients with XPD c.934AA genotype presented lower progression-free survival and overall survival in Kaplan-Meier estimates, and differences between groups remained the same in univariate Cox analysis. Patients with XPD c.934AA genotype had 2.13 and 2.04 more risks of presenting tumor progression and death than others in multivariate Cox analysis. Our data present preliminary evidence that XPC c.2815A>C, XPD c.934G>A and c.2251A>C, and ERCC1 c.354C>T SNPs alter outcome of HNSCC patients treated with CDDP chemoradiation.
Keywords: head and neck squamous cell carcinoma, cisplatin, nucleotide excision repair pathway, single nucleotide polymorphisms, outcome
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common human cancer, with a worldwide incidence of 600,000 new cases and approximately 350,000 deaths are attributed to tumor each year [1].
About two-thirds of HNSCC patients exhibit advanced stage disease at diagnosis [2], and cisplatin (CDDP) associated with radiotherapy (RT) has been used in their treatment [3]. RT induces DNA damage directly by action of photons and indirectly by liberation of free radicals [4]. CDDP develops adducts with cellular DNA and also releases free radicals [5]. In both cases, damaged cells are induced to apoptosis when not adequately repaired, particularly by nucleotide excision repair (NER) pathway [6].
The xeroderma pigmentosum (XP) genes, including complementation group C (XPC), D (XPD), F (XPF) and excision repair cross-complementation group1 (ERCC1), operate in NER pathway, and participate of recognition, demarcation and removal of DNA damage induced by CDDP and RT [7].
Variations in tumor sensitivity to CDDP [8–21], RT [22, 23] and CDDP associated with RT [24–26], as well as in side effects of therapeutic modalities [10, 14, 27, 28], have been attributed to distinct activities of proteins encoded by single nucleotide polymorphisms (SNPs) in genes involved in DNA repair through NER pathway.
The variant alleles of XPC c.2815A>C (p.Lys939Gln) (rs2228001), XPD c.934G>A (p.Asp312Asn) (rs1799793) and XPD c.2251A>C (p.Lys751Gln) (rs13181) SNPs determine activity of protein reduction, with consequent lower function in DNA repair capacity (DRC) [29, 30]. The variant alleles of XPF c.2505T>C (p.Ser835Ser) (rs1799801) and ERCC1 c.354C>T (p.Asn118Asn) (rs11615) SNPs can be associated with a reduction of mRNA stability or processing, and lower DRC [31–33].
The XPC c.2815A>C [14–17], XPD c.934G>A and c.2251A>C [8–10, 13, 14, 19, 20, 24, 26, 28], XPF c.2505T>C [26], and ERCC1 c.354C>T [10–12, 15, 16, 18, 21, 25, 26, 28] SNPs were associated with variable response rate (RR), toxicity, progression-free survival (PFS) and overall survival (OS) in patients with different tumors treated with CDDP-based chemotherapy with or without RT; however only few studies were conducted in HNSCC patients [24, 25].
In the present study, we investigated whether the above-mentioned SNPs alter the outcome of HNSCC patients treated with CDDP and RT.
RESULTS
Study population
Most of 90 patients enrolled in study were male and with a history of tobacco and alcohol consumption. About two-thirds of cases had tumor in pharynx and most of patients presented well or moderately differentiated tumor and tumor in advanced stages. Human papillomavirus (HPV) type 16 was negative in all analyzed cases (Table 1).
Table 1. Clinical characteristics and tumor aspects of head and neck squamous cell carcinoma patients.
| Variable | Median (range) or N (%) |
|---|---|
| Age (years) | 56 (27-74) |
| Gender | |
| Male | 83 (92.2) |
| Female | 7 (7.8) |
| Body mass index (kg/m2) | 19 (13-31) |
| Tobacco consumption | |
| Smokers | 88 (97.8) |
| Non-smokers | 2 (2.2) |
| Alcohol consumption | |
| Drinkers | 83 (92.2) |
| Abstainers | 7 (7.8) |
| Tumor location | |
| Oral cavity | 12 (13.3) |
| Pharynx | 55 (61.1) |
| Larynx | 23 (25.6) |
| Histological grade* | |
| Well + moderately | 60 (82.2) |
| Poorly + undifferentiated | 13 (17.8) |
| Tumor stage | |
| I + II | 6 (6.7) |
| III + IV | 84 (93.3) |
| Human papillomavirus type 16* | |
| Positive | 0 (0.0) |
| Negative | 57 (100.0) |
(N) number of patients. *The number of patients differed from the total quoted in the study (n= 90), because it was not possible to obtain consistent information about histological grade and human papillomavirus type 16 status in some cases.
All patients received RT with a total dose of 70 Gy and CDDP at initial dose of 80-100 mg/m2. Thirteen patients with consistent side effects after the first infusion of CDDP, received lower dose (50-75 mg/m2) of agent in following administrations. Sixty-eight patients (75.5%) received three infusions of CDDP and 22 patients (24.5%) received only two CDDP infusions due to renal or hematologic toxicities; the median cumulative dose of CDDP in patients was 265 mg (range: 100 to 616). Most of patients (97.7%) had medium or high adherence to antiemetics.
Partial response and stable disease were seen in near 80.0% of patients. About two-thirds and one-third of cases had moderate/severe nausea and vomiting, respectively, one-third to half of cases presented moderate/severe hematologic toxicities and half of cases had moderate/severe nephrotoxicity or ototoxicity (Table 2).
Table 2. Responses and toxicities to chemoradiotherapy of head and neck squamous cell carcinoma patients.
| Variable | Ideal or mild | Non-ideal, moderate or severe | ||
|---|---|---|---|---|
| Type of response or grade of toxicity | N (%) | Type of response or grade of toxicity | N (%) | |
| Response rate | CR+PR | 68 (93.2) | SD | 5 (6.8) |
| CR | 15 (20.5) | PR+SD | 58 (79.5) | |
| Gastrointestinal toxicities | ||||
| Nausea | G0+G1 | 37 (42.0) | G2+G3 | 51 (58.0) |
| Vomiting | G0+G1 | 59 (67.0) | G2+G3+G4 | 29 (33.0) |
| Hematologic toxicities | ||||
| Anemia | G0+G1 | 37 (44.0) | G2+G3+G4 | 47 (56.0) |
| Leukopenia | G0+G1 | 47 (56.0) | G2+G3+G4 | 37 (44.0) |
| Neutropenia | G0+G1+G2 | 67 (79.8) | G3+G4 | 17 (20.2) |
| Lymphopenia | G0+G1+G2 | 42 (50.0) | G3+G4 | 42 (50.0) |
| Thrombocytopenia | G0 | 54 (64.3) | G1+G2+G3+G4 | 30 (35.7) |
| Nephrotoxicity | G0+G1 | 36 (52.2) | G2+G3+G4+G5 | 33 (47.8) |
| Ototoxicity | G0+G1 | 36 (51.4) | G2+G3+G4 | 34 (48.6) |
(N) number of patients; (CR) complete response; (PR) partial response; (SD) stable disease; (G) grade of toxicity. The total number of patients differed from the total quoted in the study (n= 90), because it was not possible to obtain consistent information about response rate, nausea and vomiting, hematologic exams, glomerular filtration rate or audiometry test after chemoradiotherapy in some cases.
The mean ± standard deviation of urinary CDDP was 237.0 μg/mg ± 116.2.
The median follow-up time of 90 HNSCC patients enrolled in study was 18.6 months (range: 3.3-48.9). The estimated probabilities of 24-months PFS and OS were 37.6% and 42.4%, respectively. At the date of analysis, September 2015, 31 patients were alive, 7 of them with HNSCC and 24 without HNSCC and 59 patients died, 56 of them by the tumor effects and 3 by unrelated causes.
The linkage disequilibrium (LD) analysis revealed a LD between XPD c.934G>A and XPD c.2251A>C (D’= 64%), XPD c.934G>A and ERCC1 c.354C>T (D’= 54%), and XPD c.2251A>C and ERCC1 c.354C>T (D’= 51%) SNPs. From the theoretical eight possible XPD haplotypes for c.934G>A and c.2251A>C SNPs, four were found to be common (frequency > 1%: GA, GC, AA, AC). Only seven out of eighteen possible XPD and ERCC1 haplotypes for c.934G>A, c.2251A>C and c.354C>T SNPs were found to be common (frequency > 1%: GAC, GCC, AAC, ACC, GAT, GCT, ACT). The common haplotypes of referred SNPs were included in further analysis.
Polymorphisms, response rate and toxicity
The frequencies of referred genotypes and haplotypes of HNSCC patients stratified by RR and toxicity to chemoradiotherapy are presented in Table 3. The XPC c.2815AC or CC genotypes were less common than AA genotype in patients with moderate/severe ototoxicity (40.4% versus 65.2%). Patients with AC or CC genotypes had 0.20 less chance of moderate/severe ototoxicity than others. The XPD c.934AA variant genotype was more frequent than GG or GA genotypes in patients with complete response (CR) after chemoradiotherapy (42.9% versus 18.2%). Carriers of variant genotype AA had 8.64 more chances of achieving CR than others. The maximum changes from baseline in the sum of reference diameters of target lesions in HNSCC patients with XPD c.934G>A SNP genotypes are presented in Figure 1A and 1B; patients with variant genotype had more median change than those with wild-type or heterozygous genotypes of XPD c.934G>A SNP (-63.0% versus -52.5%) of presenting response to chemoradiotherapy. The XPD c.934AA genotype was also more frequent than the GG or GA genotypes in patients with moderate/severe ototoxicity (85.7% versus 44.4%). Patients with AA genotype had 12.29 more chances of consistent ototoxicity than others. In contrast, the XPD c.934GA or AA genotypes were less common than the GG genotype in patients with moderate/severe nausea (48.8% versus 66.0%). Carriers of variant A allele had 0.38 less chance of moderate/severe nausea than those with the wild-type genotype. An excess of XPD c.2251AC or CC genotypes compared to the AA genotype were seen in patients with moderate/severe nephrotoxicity (62.2% versus 31.3%). Carriers of variant C allele had 3.55 more chances of consistent nephrotoxicity than others. The AA haplotype (variant allele of XPD c.934G>A and wild-type allele of XPD c.2251A>C) was more common in patients with CR than those with other common haplotypes (44.4% versus 19.0%). Individuals with AA haplotype had 9.30 more chances of achieving CR than others. The ACT haplotype (variant alleles of XPD c.934G>A, XPD c.2251A>C and ERCC1 c.354C>T; respectively) was also more common in patients with moderate/severe nephrotoxicity than other haplotypes (70.0% versus 44.1%). Individuals with ACT haplotype had 3.41 more chances of consistent nephrotoxicity than others.
Table 3. Frequencies of XPC c.2815A>C, XPD c.934G>A, XPD c.2251A>C, XPF c.2505T>C and ERCC1 c.354C>T single nucleotide polymorphisms genotypes and haplotypes of head and neck squamous cell carcinoma patients stratified by response rate and toxicity to chemoradiotherapy.
| Variable | Response rate | Nausea | Vomiting | Nephrotoxicity | Ototoxicity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CR+PR N (%) | SD N (%) | CR N (%) | PR+SD N (%) | G0+G1 N (%) | G2+G3 N (%) | G0+G1 N (%) | G2-G4 N (%) | G0+G1 N (%) | G2-G5 N (%) | G0+G1 N (%) | G2-G4 N (%) | |
| XPC c.2815A>C | ||||||||||||
| AA+AC | 57 (93.4) | 4 (6.6) | 12 (19.7) | 49 (80.3) | 32 (42.1) | 44 (57.9) | 52 (68.4) | 24 (31.6) | 28 (48.3) | 30 (51.7) | 30 (51.7) | 28 (48.3) |
| CC | 11 (91.7) | 1 (8.3) | 3 (25.0) | 9 (75.0) | 5 (41.7) | 7 (58.3) | 7 (58.3) | 5 (41.7) | 8 (72.7) | 3 (27.3) | 6 (50.0) | 6 (50.0) |
| P-value | 0.80 | 0.34 | 0.79 | 0.46 | 0.07 | 0.65 | ||||||
| OR (95% CI) | 0.72 (0.05-8.93) | 2.20 (0.42-11.51) | 1.19 (0.31-4.51) | 1.63 (0.43-6.09) | 0.23 (0.05-1.14) | 1.36 (0.34-5.41) | ||||||
| AA | 24 (100.0) | 0 (0.0) | 7 (29.2) | 17 (70.8) | 11 (34.4) | 21 (65.6) | 20 (62.5) | 12 (37.5) | 12 (52.2) | 11 (47.8) | 8 (34.8) | 15 (65.2) |
| AC+CC | 44 (89.8) | 5 (10.2) | 8 (16.3) | 41 (83.7) | 26 (46.4) | 30 (53.6) | 39 (69.6) | 17 (30.4) | 24 (52.2) | 22 (47.8) | 28 (59.6) | 19 (40.4) |
| P-value | 0.99 | 0.32 | 0.27 | 0.65 | 0.79 | 0.01 | ||||||
| OR (95% CI) | NE | 0.52 (0.14-1.88) | 0.58 (0.22-1.52) | 0.80 (0.30-2.10) | 0.87 (0.30-2.49) | 0.20 (0.06-0.70) | ||||||
| XPD c.934G>A | ||||||||||||
| GG+GA | 61 (92.4) | 5 (7.6) | 12 (18.2) | 54 (81.8) | 32 (41.0) | 46 (59.0) | 53 (68.8) | 25 (32.1) | 33 (53.2) | 29 (46.8) | 35 (55.6) | 28 (44.4) |
| AA | 7 (100.0) | 0 (0.0) | 3 (42.9) | 4 (57.1) | 5 (50.0) | 5 (50.0) | 6 (60.0) | 4 (40.0) | 3 (42.9) | 4 (57.1) | 1 (14.3) | 6 (85.7) |
| P-value | 0.99 | 0.04 | 0.67 | 0.40 | 0.42 | 0.03 | ||||||
| OR (95% CI) | NE | 8.64 (1.04-71.76) | 0.73 (0.17-3.11) | 1.84 (0.43-7.86) | 2.00 (0.36-10.96) | 12.29 (1.19-126.04) | ||||||
| GG | 36 (94.7) | 2 (5.3) | 8 (21.1) | 30 (78.9) | 16 (34.0) | 31 (66.0) | 33 (70.2) | 14 (29.8) | 21 (58.3) | 15 (41.7) | 17 (47.2) | 19 (52.8) |
| GA+AA | 32 (91.4) | 3 (8.6) | 7 (20.0) | 28 (80.0) | 21 (51.2) | 20 (48.8) | 26 (63.4) | 15 (36.6) | 15 (45.5) | 18 (54.5) | 19 (55.9) | 15 (44.1) |
| P-value | 0.66 | 0.77 | 0.04 | 0.58 | 0.23 | 0.54 | ||||||
| OR (95% CI) | 0.65 (0.09-4.38) | 0.83 (0.25-2.79) | 0.38 (0.14-0.98) | 1.29 (0.50-3.33) | 1.83 (0.68-7.97) | 0.73 (0.26-2.01) | ||||||
| XPD c.2251A>C | ||||||||||||
| AA+AC | 60 (92.3) | 5 (7.7) | 13 (20.0) | 52 (80.0) | 35 (43.8) | 45 (56.2) | 55 (68.7) | 25 (31.3) | 33 (54.1) | 28 (45.9) | 31 (50.0) | 31 (50.0) |
| CC | 8 (100.0) | 0 (0.0) | 2 (25.0) | 6 (75.0) | 2 (25.0) | 6 (75.0) | 4 (50.0) | 4 (50.0) | 3 (37.5) | 5 (62.5) | 5 (62.5) | 3 (37.5) |
| P-value | 0.99 | 0.95 | 0.16 | 0.09 | 0.61 | 0.76 | ||||||
| OR (95% CI) | NE | 1.05 (0.14-7.63) | 3.50 (0.58-20.95) | 4.11 (0.77-21.84) | 1.51 (0.29-7.66) | 0.77 (0.14-4.26) | ||||||
| AA | 32 (91.4) | 3 (8.6) | 7 (20.0) | 28 (80.0) | 17 (39.5) | 26 (60.5) | 27 (62.8) | 16 (37.2) | 22 (68.7) | 10 (31.3) | 16 (47.1) | 18 (52.9) |
| AC+CC | 36 (94.7) | 2 (5.3) | 8 (21.1) | 30 (78.9) | 20 (44.4) | 25 (55.6) | 32 (71.1) | 13 (28.9) | 14 (37.8) | 23 (62.2) | 20 (55.6) | 16 (44.4) |
| P-value | 0.73 | 0.81 | 0.46 | 0.40 | 0.01 | 0.75 | ||||||
| OR (95% CI) | 1.39 (0.20-9.63) | 1.15 (0.34-3.87) | 0.70 (0.27-1.80) | 0.65 (0.24-1.75) | 3.55 (1.27-9.87) | 0.85 (0.31-2.34) | ||||||
| XPF c.2505T>C | ||||||||||||
| TT+TC | 60 (92.3) | 5 (7.7) | 14 (21.2) | 52 (78.8) | 33 (40.7) | 48 (59.3) | 53 (65.4) | 28 (34.6) | 33 (53.2) | 29 (46.8) | 34 (53.1) | 30 (46.9) |
| CC | 8 (100.0) | 0 (0.0) | 1 (14.3) | 6 (85.7) | 4 (57.1) | 3 (42.9) | 6 (85.7) | 1 (14.3) | 3 (42.9) | 4 (57.1) | 2 (33.3) | 4 (66.7) |
| P-value | 0.99 | 0.65 | 0.28 | 0.21 | 0.49 | 0.41 | ||||||
| OR (95% CI) | NE | 0.58 (0.05-6.46) | 0.39 (0.07-2.16) | 0.23 (0.02-2.28) | 1.77 (0.33-9.26) | 2.17 (0.33-14.07) | ||||||
| TT | 25 (86.2) | 4 (13.8) | 7 (24.1) | 22 (75.9) | 16 (40.0) | 24 (60.0) | 28 (70.0) | 12 (30.0) | 20 (66.7) | 10 (33.3) | 13 (44.8) | 16 (55.2) |
| TC+CC | 43 (97.7) | 1 (2.3) | 8 (18.2) | 36 (81.8) | 21 (43.8) | 27 (56.2) | 31 (64.6) | 17 (35.4) | 16 (41.0) | 23 (59.0) | 23 (56.1) | 18 (43.9) |
| P-value | 0.11 | 0.53 | 0.69 | 0.56 | 0.07 | 0.30 | ||||||
| OR (95% CI) | 7.17 (0.64-80.32) | 0.67 (0.19-2.34) | 0.83 (0.33-2.05) | 1.32 (0.51-3.40) | 2.56 (0.92-7.11) | 0.58 (0.20-1.64) | ||||||
| ERCC1 c.354C>T | ||||||||||||
| CC+CT | 57 (93.4) | 4 (6.6) | 12 (19.7) | 49 (80.3) | 33 (45.2) | 40 (54.8) | 48 (65.8) | 25 (34.2) | 33 (57.9) | 24 (42.1) | 31 (52.5) | 28 (47.5) |
| TT | 11 (91.7) | 1 (8.3) | 3 (25.0) | 9 (75.0) | 4 (26.7) | 11 (73.3) | 11 (73.3) | 4 (26.7) | 3 (25.0) | 9 (75.0) | 5 (45.5) | 6 (54.5) |
| P-value | 0.67 | 0.79 | 0.15 | 0.61 | 0.06 | 0.47 | ||||||
| OR (95% CI) | 0.58 (0.05-6.90) | 1.23 (0.26-5.67) | 2.50 (0.69-9.03) | 0.71 (0.19-2.58) | 4.00 (0.95-16.69) | 1.63 (0.42-6.38) | ||||||
| CC | 20 (95.2) | 1 (4.8) | 6 (28.6) | 15 (71.4) | 11 (44.0) | 14 (56.0) | 16 (64.0) | 9 (36.0) | 12 (63.2) | 7 (36.8) | 12 (60.0) | 8 (40.0) |
| CT+TT | 48 (92.3) | 4 (7.7) | 9 (17.3) | 43 (82.7) | 26 (41.3) | 37 (58.7) | 43 (68.3) | 20 (31.7) | 24 (48.0) | 26 (52.0) | 24 (48.0) | 26 (52.0) |
| P-value | 0.87 | 0.54 | 0.88 | 0.47 | 0.13 | 0.68 | ||||||
| OR (95% CI) | 0.82 (0.07-8.58) | 0.66 (0.18-2.44) | 1.07 (0.39-2.95) | 0.68 (0.24-1.94) | 2.43 (0.75-7.85) | 1.27 (0.39-4.11) | ||||||
| XPD+XPD | ||||||||||||
| AA | 8 (88.9) | 1 (11.1) | 4 (44.4) | 5 (55.6) | 7 (58.3) | 5 (41.7) | 6 (50.0) | 6 (50.0) | 5 (71.4) | 2 (28.6) | 4 (44.4) | 5 (55.6) |
| Other haplotypes | 128 (93.4) | 9 (6.6) | 26 (19.0) | 111 (81.0) | 67 (40.9) | 97 (59.1) | 112 (68.3) | 52 (31.7) | 67 (51.1) | 64 (48.9) | 68 (51.9) | 63 (48.1) |
| P-value | 0.87 | 0.01 | 0.13 | 0.24 | 0.49 | 0.63 | ||||||
| OR (95% CI) | 0.83 (0.08-7.98) | 9.30 (1.67-51.77) | 0.36 (0.09-1.38) | 2.05 (0.60-7.00) | 0.55 (0.10-3.03) | 1.42 (0.33-6.11) | ||||||
| XPD+XPD+ERCC1 | ||||||||||||
| ACT | 18 (94.7) | 1 (5.3) | 3 (15.8) | 16 (84.2) | 11 (47.8) | 12 (52.2) | 15 (65.2) | 8 (34.8) | 6 (30.0) | 14 (70.0) | 9 (50.0) | 9 (50.0) |
| Other haplotypes | 118 (92.9) | 9 (7.1) | 27 (21.3) | 100 (78.7) | 63 (41.2) | 90 (58.8) | 103 (67.3) | 50 (32.7) | 66 (55.9) | 52 (44.1) | 63 (51.6) | 59 (48.4) |
| P-value | 0.70 | 0.56 | 0.36 | 0.97 | 0.02 | 0.79 | ||||||
| OR (95% CI) | 1.54 (0.16-14.26) | 0.66 (0.16-2.63) | 0.64 (0.24-1.66) | 1.01 (0.38-2.70) | 3.41 (1.18-9.87) | 1.15 (0.39-3.42) | ||||||
Figure 1. Characteristics of response to cisplatin-chemoradiotherapy and probability of progression-free and overall survival of head and neck squamous cell carcinoma (HNSCC) patients.
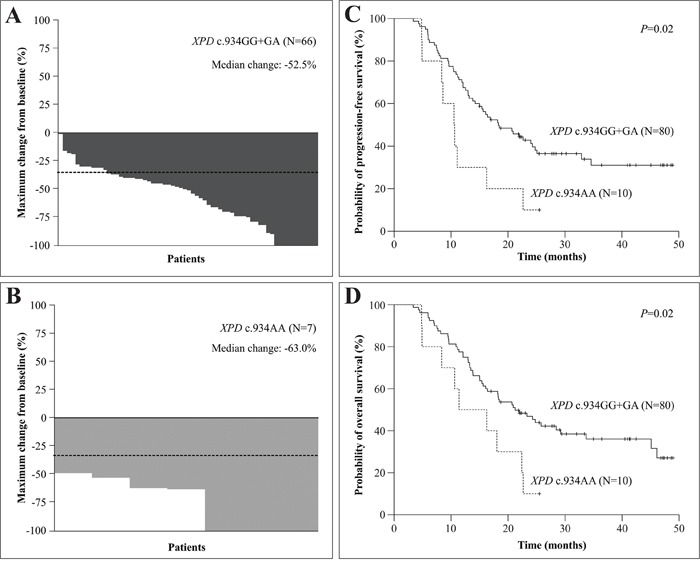
Panels A and B. show the waterfall plots indicate the maximum change from baseline in the sum of reference diameters of target lesion in with XPD c.934G>A genotypes. The dashed lines indicate a 30% reduction in the tumor burden in the target lesion, as defined by Response Evaluation Criteria in Solid Tumors version 1.1. Panels C and D. show Kaplan-Meier curve for progression-free and overall survival among HNSCC patients with XPD c.934G>A genotypes.
Similar frequencies of genotypes, alone or combined, and haplotypes of corresponding SNPs were seen in patients stratified by hematologic toxicities and concentration of CDDP in urine (data not shown).
Polymorphisms and survival analysis
At 24 months of follow-up, shorter PFS was observed in patients with advanced tumor stage (34.4% versus 83.3%, P= 0.03) and XPD c.934AA genotype (10.0% versus 41.2%, P= 0.02) (Figure 1C); a shorter OS was also observed in patients with advanced tumor stage (38.4% versus 100.0%, P= 0.02) and XPD c.934AA genotype (10.0% versus 46.9%, P= 0.02) (Figure 1D) at this time (Kaplan-Meier estimates).
Associations of clinical and tumor characteristics and referred genotypes with survival of HNSCC patients in Cox analyses are presented in Table 4. In univariate Cox analysis, a tendency for shorter PFS and shorter PFS were seen in patients with advanced tumor stage and in those with XPD c.934AA genotype, respectively. Only the XPD c.934AA genotype was associated with shorter PFS in multivariate Cox analysis; individuals with XPD c.934AA genotype had 2.13 more risks to present tumor progression than those with the remaining genotypes. In univariate Cox analysis, a shorter OS was seen in patients with advanced tumor stage and in those with XPD c.934AA genotype. Only the XPD c.934AA genotype was associated with shorter OS in multivariate Cox analysis; individuals with XPD c.934AA genotype had 2.04 more risks of death than those with the remaining genotypes.
Table 4. Association of clinical and tumor characteristics, XPC c.2815A>C, XPD c.934G>A, XPD c.2251A>C, XPF c.2505T>C and ERCC1 c.354C>T single nucleotide polymorphisms genotypes with survival of head and neck squamous cell carcinoma patients treated with chemoradiotherapy in univariate Cox analysis.
| Variables | Progression-free survival | Overall survival | ||||
|---|---|---|---|---|---|---|
| N with event/N total | P value | HR (95% CI) | N with event/N total | P value | HR (95% CI) | |
| Age (years) | ||||||
| ≤ 56 | 32/46 | 0.40 | 1.24 (0.74-2.06) | 29/46 | 0.79 | 1.08 (0.57-2.05) |
| > 56 | 28/44 | 30/44 | ||||
| Gender | ||||||
| Male | 54/83 | 0.22 | 1.68 (0.72-3.93) | 55/83 | 0.86 | 1.10 (0.34-3.60) |
| Female | 6/7 | 4/7 | ||||
| Tobacco consumption | ||||||
| Smokers | 58/88 | 0.12 | 3.05 (0.73-12.67) | 58/88 | 0.72 | 1.43 (0.19-10.44) |
| Non-smokers | 2/2 | 1/2 | ||||
| Alcohol consumption | ||||||
| Drinkers | 57/83 | 0.30 | 1.83 (0.57-5.89) | 57/83 | 0.21 | 3.52 (0.48-25.71) |
| Abstainers | 3/7 | 2/7 | ||||
| Tumor location | ||||||
| Oral cavity/oropharynx | 36/51 | 0.47 | 1.20 (0.71-2.03) | 36/51 | 0.24 | 1.48 (0.76-2.91) |
| Hypopharynx/larynx | 24/39 | 23/39 | ||||
| Histological grade | ||||||
| Well/moderately | 39/60 | 0.28 | 1.48 (0.71-3.08) | 38/60 | 0.44 | 1.32 (0.64-2.75) |
| Poorly/undifferentiated | 9/13 | 9/13 | ||||
| Tumor stage | ||||||
| I + II | 1/6 | 0.06 | 6.64 (0.91-48.05)* | 1/6 | 0.05 | 7.08 (0.97-51.28)* |
| III + IV | 59/84 | 58/84 | ||||
| XPC c.2815A>C | ||||||
| AA+AC | 51/77 | 0.90 | 1.04 (0.51-2.12) | 51/77 | 0.47 | 1.31 (0.62-2.77) |
| CC | 9/13 | 8/13 | ||||
| AA | 21/32 | 0.82 | 1.06 (0.62-1.80) | 22/32 | 0.58 | 1.15 (0.68-1.96) |
| AC+CC | 39/58 | 37/58 | ||||
| XPD c.934G>A | ||||||
| GG+GA | 51/80 | 0.02 | 2.33 (1.13-4.77)*** | 50/80 | 0.02 | 2.24 (1.09-4.61)**** |
| AA | 9/10 | 9/10 | ||||
| GG | 30 / 48 | 0.38 | 1.25 (0.75-2.07) | 31/48 | 0.75 | 1.08 (0.65-1.81) |
| GA+AA | 30/42 | 28/42 | ||||
| XPD c.2251A>C | ||||||
| AA+AC | 56/82 | 0.46 | 1.45 (0.52-4.02) | 55/82 | 0.51 | 1.40 (0.50-3.88) |
| CC | 4/8 | 4/8 | ||||
| AA | 26/44 | 0.19 | 1.40 (0.84-2.33) | 28/44 | 0.71 | 1.10 (0.66-1.83) |
| AC+CC | 34/46 | 31/46 | ||||
| XPF c.2505T>C | ||||||
| TT+TC | 56/83 | 0.75 | 1.17 (0.42-3.24) | 55/83 | 0.85 | 1.10 (0.39-3.04) |
| CC | 4/7 | 4/7 | ||||
| TT | 26/41 | 0.60 | 1.14 (0.68-1.90) | 25/41 | 0.40 | 1.24 (0.74-2.10) |
| TC+CC | 34/49 | 34/49 | ||||
| ERCC1 c.354C>T | ||||||
| CC+CT | 52/74 | 0.11 | 1.92 (0.91-4.06) | 51/74 | 0.15 | 1.72 (0.81-3.65) |
| TT | 8/16 | 8/16 | ||||
| CC | 19/25 | 0.35 | 1.29 (0.74-2.22) | 18/25 | 0.64 | 1.13 (0.65-1.98) |
| CT+TT | 41/65 | 41/65 | ||||
(N) number of patients; (HR) hazard ratio; (CI) confidence interval. Significant differences between groups are presented in bold letters. In multivariate Cox analysis (adjusted by tumor stage and XPD c.934G>A polymorphism): *P= 0.07, HR: 6.15, 95% CI: 0.84-44.68; *P= 0.06, HR: 6.59, 95% CI: 0.90-47.90; ***P= 0.03, HR: 2.13, 95% CI: 1.04-4.38; ****P= 0.05, HR: 2.04, 95% CI: 1.00-4.20.
DISCUSSION
We initially found that clinical and tumor aspects [3, 22, 24, 34, 35], RR, toxicity to chemoradiation and short survival in advanced tumor stages [3, 34–36] in our sample were similar to those previously described in other parts of world. Therefore, they were adequate for analysis of new prognostic factors in disease. Low prevalence of HPV infection was seen in our cases, as previously reported [37, 38], suggesting that the major factors enrolled in tumor development were tobacco and alcohol consumption.
Secondly, we found that XPC c.2815AC or CC genotype was associated with reduced ototoxicity. The AA genotype was related to less hearing impairment in osteosarcoma patients treated with CDDP [14]. Our finding was not expected, since the wild-type A allele of XPC c.2815A>C SNP was previously associated with higher DRC [30], and possibly with protection against hearing loss. However, variant C and wild-type A alleles of XPC c.2815A>C were also associated with similar DRC [39]. Thus, additional studies are required to evaluate the bind of XPC c.2815A>C with ototoxicity in HNSCC patients treated with CDDP chemoradiation.
Third, as previously reported, we found that AA genotype of XPD c.934G>A SNP was associated with CR in HNSCC patients [24]. The XPD c.934GA or AA genotypes and AA genotype were associated with reduced manifestation of nausea and moderate/severe ototoxicity in our cases, respectively. Nephrotoxicity was also more common in our patients with the XPD c.2251AC or CC genotypes. A possible explanation for these associations is that variant A and C alleles of XPD c.934G>A and c.2251A>C SNPs determine lower DRC [29], which could induced more apoptosis in tumor cells and normal outer hairs and renal tubular cells in response to CDDP chemoradiation. The reduced DRC in patients with GA or AA genotypes of XPD c.934G>A SNP may also induce more apoptosis in epithelial enterochromaffin cells of intestine of treated patients, resulting in absence of serotonin release, and consequent lack of stimuli in chemoreceptor trigger zone and vomiting center. No associations of XPD c.934G>A and c.2251A>C SNPs with RR and toxicities were seen in non-small cell lung cancer [8, 10, 12, 15, 19, 20, 26], osteosarcoma [14] and ovarian cancer [28] patients treated with CDDP with or without RT. The divergent results may be caused by differences in sample sizes, tumor types, antiemetic therapies, hydration conditions, and doses of CDDP in our and previously reported studies.
Fourth, the AA haplotype of XPD c.934G>A and c.2251A>C SNPs was associated with increased chance of obtaining CR, and nephrotoxicity was predominately seen in patients with ACT haplotype of XPD c.934G>A, c.2251A>C and ERCC1 c.354C>T SNPs, indicating that the SNPs in XPD and ERCC1 genes may act together in DRC, with effects on clinical manifestation in those patients.
Finally, we found shorter PFS and OS in patients with XPD c.934AA variant genotype. Corroborating our findings, this genotype was previously reported with shorter OS in non-small cell lung cancer patients treated with platinum [9, 19]. In contrast, XPD c.934AA variant genotype was related with longer PFS and/or OS in HNSCC [24] and esophageal cancer [13] patients, and did not influence survival in non-small cell lung cancer [8, 10, 12, 15, 20, 26] and ovarian cancer [28] patients treated with platinum or CDDP with or without RT. The variant allele of XPD c.934G>A SNP determines lower function in DRC [29], which may induced high apoptosis in response to CDDP chemoradiation in tumor cells and high RR in HNSCC. It is well known that the TP53 gene has a crucial role in induction of apoptosis [40]; however, the tobacco consumption was associated with increased risk of TP53 mutations in previous analyzed HNSCC patients [41], and might have produced the same effect in our cases. Facing these descriptions, we hypothesized that HNSCC patients with XPD c.934AA variant genotype and non-functional TP53 protein could lead to decrease DRC induced by CDDP chemoradiation and consequent decreased apoptosis of tumor cells. This could constitute a possible reason for the initial sensitivity to chemoradiotherapy and further poor prognosis (PFS and OS) seen in our cases, as previously reported in small cell lung cancer patients [42]. The divergent results seen in previous studies and our study may be attributed to different sample sizes, treatment types, antiemetic therapies, hydration conditions and follow-up times.
In conclusion, our findings presented preliminary evidence that XPC c.2815A>C, XPD c.934G>A, XPD c.2251A>C and ERCC1 c.354C>T SNPs alter clinical outcome of HNSCC patients treated with CDDP chemoradiation. We believe that in the near future, pharmacogenetic studies in HNSCC can contribute to personalize treatment and optimize patient outcome.
PATIENTS AND METHODS
Patients, treatment and clinical variables
This prospective study comprised HNSCC patients seen at diagnosis at the Clinical Oncology Service of General Hospital of University of Campinas between June 2011 and February 2014. All patients were selected to CDDP chemoradiation as definitive treatment due to locoregional unresectable tumor, refusal of surgery facing expected functional or anatomic sequels, or an organ preservation protocol. Exclusion criteria were refusing to participate in study, low Karnofsky performance scale score and renal dysfunction. The study was conducted according to the Declaration of Helsinki and was approved by the institutional review board guidelines (n° 274/2011).
The data relating to age, gender, body mass index, tobacco and alcohol consumption, hematologic and biochemistry exams, tumor location, histological grade and stage were obtained from patient charts. Subjects were classified as smokers or non-smokers and drinkers or abstainers as previously reported [43]. The tumor was diagnosed by standard criteria [44] and staged by the criteria of American Joint Committee of Cancer [45].
HPV testing consisted of P16 immunohistochemistry in tumor fragments embedded in paraffin. Staining was regarded as positive if it was strong and diffuse (> 80% of tumor cells) and it was regarded as negative if absent or focal [46]. Wide spectrum HPV in situ hybridization was reserved for P16-positive cases. Punctate hybridization signals localized to the tumor cell nuclei in either analysis defined an HPV-positive tumor [47].
Concurrent single daily fractionated radiation (2 Gy/day) during 35 days and intravenous CDDP at initial dose of 80-100 mg/m2 on days 1, 22 and 43 was administered to patients; patients with consistent side effects during treatment received CDDP at lower dose [3, 48]. RR to chemoradiotherapy was assessed using Response Evaluation Criteria in Solid Tumors (RECIST) guidelines version 1.1 [49].
As hydration and antiemetic protocols, the patients received intravenous 3,000 ml of saline 0.9%, 125 ml of 20% mannitol, ondansetron 24 mg and dexamethasone 20 mg before CDDP infusion, as well as intravenous 2,000 ml of saline 0.9% and oral dexamethasone 8 mg (every 12 hours) and metoclopramida 10 mg (every 6 hours) during three days after each CDDP infusion [50, 51]. The antiemetics adherence was classified as high or medium adherence or non-adherence [52].
Nausea, vomiting, hematologic toxicities, nephrotoxicity and ototoxicity were assessed using information of adverse effects, hematologic exams, 51Cr-EDTA glomerular filtration rate and audiometric tests performed before and after chemoradiotherapy. The toxicities were evaluated according to the National Cancer Institute (NCI) criteria version 4.0 [53], and the worst grade for each toxicity in each patient was included in analysis.
CDDP in urine of patients collected 0 to 48 hours after each dose of CDDP was measured by high-performance liquid chromatographic [54]. The final concentration of urinary CDDP was considered as the sum of all measurements obtained after each administration of agent.
Surgical tumor resection was offered to patients with good clinical condition and partial response or tumor relapse. Patients not amenable to resection and with progressive disease or relapse received intravenous methotrexate at dose of 40 mg/m2 once a week until best response, limiting toxicity or progression of disease [55]. The follow-up of patients was performed at 3-month intervals. The end of follow-up period was September 2015.
DNA extraction and genotyping
The genotyping procedure was performed using genomic DNA obtained from peripheral blood of patients and involved a polymerase chain reaction followed by the enzymatic digestion, as reported for XPC c.2815A>C [56], XPD c.934G>A and XPD c.2251A>C [57], XPF c.2505T>C [33] and ERCC1 c.354C>T [58] SNPs. Positive and negative controls were used in all genotyping reactions. The amount of 15% of genotype determinations was carried out twice in independent experiments with 100% of concordance.
Statistical analysis
The pairwise LD was performed using the Haploview 4.2 software to ensure that the markers were appropriate for inclusion in the XPD and ERCC1 haplotype estimates. The LD was measured by the disequilibrium coefficient (D’). The D’ values ≤ 1 indicate LD.
The differences between groups were analyzed by chi-square (χ2) or Fisher’s exact test. Logistic regression model served to obtain odds ratios values, adjusted for clinicopathological aspects with P-values ≤ 0.10, with 95% confidence intervals (95% CI), to assess associations between SNPs genotypes, RR, nausea, vomiting, hematologic toxicities, nephrotoxicity and ototoxicity. ANOVA served to obtain values, adjusted for clinicopathological aspects with P-values ≤ 0.10, in assessment of associations between SNPs genotypes and urinary CDDP. This variable was transformed into ranks to perform the comparative analysis, since it was not normally distributed.
PFS and OS were defined as time interval between the date of diagnosis and the date of progression or relapse of disease, and the date of death by any causes or last follow-up, respectively. Kaplan-Meier method was used to plot PFS and OS curves, and log-rank test was applied to compare the distribution between groups. Multivariate Cox regression served to estimate hazard ratios values, adjusted for variables with P-values ≤ 0.10, with 95% CI, with the purpose of to assess the associations between SNPs genotypes, PFS and OS.
For statistical tests, significance was two-sided and achieved when P-values were ≤ 0.05. All tests were done using the SPSS 21.0 software.
Acknowledgments
This work was supported by São Paulo Research Foundation (FAPESP) (grant numbers 2011/15089-1 and 2012/01807-2).
Footnotes
CONFLICTS OF INTEREST
The authors declare no competing financial interests.
REFERENCES
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. The Lancet. 2008;371:1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adelstein DJ, Li Y, Adams GL, Wagner HJ, Kish JA, Ensley JF, Schuller DE, Forastiere AA. An intergroup phase III comparison of standard radiation therapy and two schedules of concurrent chemoradiotherapy in patients with unresectable squamous cell head and neck cancer. J Clin Oncol. 2003;21:92–98. doi: 10.1200/JCO.2003.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Lomax ME, Folkes LK, O'Neill P. Biological consequences of radiation-induced DNA damage: relevance to radiotherapy. Clin Oncol (R Coll Radiol) 2013;25:578–585. doi: 10.1016/j.clon.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Deavall DG, Martin EA, Horner JM, Roberts R. Drug-induced oxidative stress and toxicity. J Toxicol. 2012;2012:645460. doi: 10.1155/2012/645460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shuck SC, Short EA, Turchi JJ. Eukaryotic nucleotide excision repair: From understanding mechanisms to influencing biology. Cell Res. 2008;18:64–72. doi: 10.1038/cr.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rouillon C, White MF. The evolution and mechanisms of nucleotide excision repair proteins. Res Microbiol. 2011;162:19–26. doi: 10.1016/j.resmic.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Camps C, Sarries C, Roig B, Sanchez JJ, Queralt C, Sancho E, Martinez N, Tarón M, Rosell R. Assessment of nucleotide excision repair XPD polymorphisms in the peripheral blood of gemcitabine/cisplatin-treated advanced non-small-cell lung cancer patients. Clin Lung Cancer. 2003;4:237–241. doi: 10.3816/clc.2003.n.004. [DOI] [PubMed] [Google Scholar]
- 9.Gurubhagavatula S, Liu G, Park S, Zhou W, Su L, Wain JC, Lynch TJ, Neuberg DS, Christiani DC. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J Clin Oncol. 2004;22:2594–2601. doi: 10.1200/JCO.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 10.Isla D, Sarries C, Rosell R, Alonso G, Domine M, Taron M, Lopez-Vivanco G, Camps C, Botia M, Nuñez L, Sanchez-Ronco M, Sanchez JJ, Lopez-Brea M, et al. Single nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated advanced non-small-cell lung cancer. Ann Oncol. 2004;15:1194–1203. doi: 10.1093/annonc/mdh319. [DOI] [PubMed] [Google Scholar]
- 11.Kamikozuru H, Kuramochi H, Hayashi K, Nakajima G, Yamamoto M. ERCC1 codon 118 polymorphism is a useful prognostic marker in patients with pancreatic cancer treated with platinum-based chemotherapy. Int J Oncol. 2008;32:1091–1096. [PubMed] [Google Scholar]
- 12.Tibaldi C, Giovannetti E, Vasile E, Mey V, Laan AC, Nannizzi S, Di Marsico R, Antonuzzo A, Orlandini C, Ricciardi S, M Del Tacca, Peters GJ, Falcone A, et al. Correlation of CDA, ERCC1, and XPD polymorphisms with response and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2008;14:1797–1803. doi: 10.1158/1078-0432.CCR-07-1364. [DOI] [PubMed] [Google Scholar]
- 13.Bradbury PA, Kulke MH, Heist RS, Zhou W, Ma C, Xu W, Marshall AL, Zhai R, Hooshmand SM, Asomaning K, Su L, Shepherd FA, Lynch TJ, et al. Cisplatin pharmacogenetics, DNA repair polymorphisms, and esophageal cancer outcomes. Pharmacogenet Genomics. 2009;19:613–625. doi: 10.1097/FPC.0b013e32832f3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caronia D, Patiño-García A, Milne RL, Zalacain-Díez M, Pita G, Alonso MR, Moreno LT, Sierrasesumaga-Ariznabarreta L, Benítez J, González-Neira A. Common variations in ERCC2 are associated with response to cisplatin chemotherapy and clinical outcome in osteosarcoma patients. Pharmacogenomics J. 2009;9:347–353. doi: 10.1038/tpj.2009.19. [DOI] [PubMed] [Google Scholar]
- 15.Kalikaki A, Kanaki M, Vassalou H, Souglakos J, Voutsina A, Georgoulias V, Mavroudis D. DNA repair gene polymorphisms predict favorable clinical outcome in advanced non-small-cell lung cancer. Clin Lung Cancer. 2009;10:118–123. doi: 10.3816/CLC.2009.n.015. [DOI] [PubMed] [Google Scholar]
- 16.Zhou C, Ren S, Zhou S, Zhang L, Su C, Zhang Z, Deng Q, Zhang J. Predictive effects of ERCC1 and XRCC3 SNP on efficacy of platinum-based chemotherapy in advanced NSCLC patients. Jpn J Clin Oncol. 2010;40:954–960. doi: 10.1093/jjco/hyq071. [DOI] [PubMed] [Google Scholar]
- 17.Zhu XL, Sun XC, Chen BA, Sun N, Cheng HY, Li F, Zhang HM, Feng JF, Qin SK, Cheng L, Lu ZH. XPC Lys939Gln polymorphism is associated with the decreased response to platinum based chemotherapy in advanced non-small-cell lung cancer. Chin Med J (Engl) 2010;123:3427–3432. [PubMed] [Google Scholar]
- 18.Wei SZ, Zhan P, Shi MQ, Shi Y, Qian Q, Yu LK, Song Y. Predictive value of ERCC1 and XPD polymorphism in patients with advanced non-small cell lung cancer receiving platinum-based chemotherapy: a systematic review and meta-analysis. Med Oncol. 2011;28:315–321. doi: 10.1007/s12032-010-9443-1. [DOI] [PubMed] [Google Scholar]
- 19.Wu W, Li H, Wang H, Zhao X, Gao Z, Qiao R, Zhang W, Qian J, Wang J, Chen H, Wei Q, Han B, Lu D. Effect of polymorphisms in XPD on clinical outcomes of platinum-based chemotherapy for Chinese non-small cell lung cancer patients. PLoS One. 2012;7:e33200. doi: 10.1371/journal.pone.0033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P, Wang YD, Cheng J, Chen JC, Ha MW. Association between polymorphisms of BAG-1 and XPD and chemotherapy sensitivity in advanced non-small-cell lung cancer patients treated with vinorelbine combined cisplatin regimen. Tumour Biol. 2015 doi: 10.1007/s13277-015-3672-z. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Shi ZH, Shi GY, Liu LG. Polymorphisms in ERCC1 and XPF gene and response to chemotherapy and overall survival of non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:3132–3137. [PMC free article] [PubMed] [Google Scholar]
- 22.Carles J, Monzo M, Amat M, Jansa S, Artells R, Navarro A, Foro P, Alameda F, Gayete A, Gel B, Miguel M, Albanell J, Fabregat X. Single-nucleotide polymorphisms in base excision repair, nucleotide excision repair, and double strand break genes as markers for response to radiotherapy in patients with stage I to II head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2006;66:1022–1030. doi: 10.1016/j.ijrobp.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 23.Mahimkar MB, Samant TA, Kannan S, Tulsulkar J, Pai PS, Anantharaman D. Polymorphisms in GSTM1 and XPD genes predict clinical outcome in advanced oral cancer patients treated with postoperative radiotherapy. Mol Carcinog. 2012;51:E94–103. doi: 10.1002/mc.21868. [DOI] [PubMed] [Google Scholar]
- 24.Quintela-Fandino M, Hitt R, Medina PP, Gamarra S, Manso L, Cortes-Funes H, Sanchez-Cespedes M. DNA-repair gene polymorphisms predict favorable clinical outcome among patients with advanced squamous cell carcinoma of the head and neck treated with cisplatin-based induction chemotherapy. J Clin Oncol. 2006;24:4333–4339. doi: 10.1200/JCO.2006.05.8768. [DOI] [PubMed] [Google Scholar]
- 25.De Castro G, Jr, Pasini FS, Siqueira SA, Ferraz AR, Villar RC, Snitcovsky IM, Federico MH. ERCC1 protein, mRNA expression and T19007C polymorphism as prognostic markers in head and neck squamous cell carcinoma patients treated with surgery and adjuvant cisplatin-based chemoradiation. Oncol Rep. 2011;25:693–699. doi: 10.3892/or.2011.1133. [DOI] [PubMed] [Google Scholar]
- 26.Sullivan I, Salazar J, Majem M, Pallarés C, E Del Río, Páez D, Baiget M, Barnadas A. Pharmacogenetics of the DNA repair pathways in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer Lett. 2014;353:160–166. doi: 10.1016/j.canlet.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 27.Chen S, Huo X, Lin Y, Ban H, Lin Y, Li W, Zhang B, Au WW, Xu X. Association of MDR1 and ERCC1 polymorphisms with response and toxicity to cisplatin-based chemotherapy in non-small-cell lung cancer patients. Int J Hyg Environ Health. 2010;213:140–145. doi: 10.1016/j.ijheh.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 28.Khrunin AV, Moisseev A, Gorbunova V, Limborska S. Genetic polymorphisms and the efficacy and toxicity of cisplatin-based chemotherapy in ovarian cancer patients. Pharmacogenomics J. 2010;10:54–61. doi: 10.1038/tpj.2009.45. [DOI] [PubMed] [Google Scholar]
- 29.Qiao Y, Spitz MR, Shen H, Guo Z, Shete S, Hedayati M, Grossman L, Mohrenweiser H, Wei Q. Modulation of repair of ultraviolet damage in the host-cell reactivation assay by polymorphic XPC and XPD/ERCC2 genotypes. Carcinogenesis. 2002;23:295–299. doi: 10.1093/carcin/23.2.295. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y, Yang H, Chen Q, Lin J, Grossman HB, Dinney CP, Wu X, Gu J. Modulation of DNA damage/DNA repair capacity by XPC polymorphisms. DNA Repair (Amst) 2008;7:141–148. doi: 10.1016/j.dnarep.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winsey SL, Haldar NA, Marsh HP, Bunce M, Marshall SE, Harris AL, Wojnarowka F, Welsh KI. A variant within the DNA repair gene XRCC3 is associated with the development of melanoma skin cancer. Cancer Res. 2000;60:5612–5616. [PubMed] [Google Scholar]
- 32.Yu JJ, Lee KB, Mu C, Li Q, Abernathy TV, Bostick-Bruton F, Reed E. Comparison of two human ovarian carcinoma cell lines (A2780/CP70 and MCAS) that are equally resistant to platinum, but differ at codon 118 of the ERCC1 gene. Int J Oncol. 2000;16:555–560. doi: 10.3892/ijo.16.3.555. [DOI] [PubMed] [Google Scholar]
- 33.Povey JE, Darakhshan F, Robertson K, Bisset Y, Mekkey M, Ress J, Doherty V, Kavanag G, Anderson N, Campbell H, Mackie RM, Melton DW. DNA repair gene polymorphisms and genetic predisposition to cutaneous melanoma. Carcinogenesis. 2007;28:1087–1093. doi: 10.1093/carcin/bgl257. [DOI] [PubMed] [Google Scholar]
- 34.De Castro G, Jr, Snitcovsky IM, Gebrim EM, Leitão GM, Nadalin W, Ferraz AR, Federico MH. High-dose cisplatin concurrent to conventionally delivered radiotherapy is associated with unacceptable toxicity in unresectable, non-metastatic stage IV head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2007;264:1475–1482. doi: 10.1007/s00405-007-0395-9. [DOI] [PubMed] [Google Scholar]
- 35.Martins RG, Parvathaneni U, Bauman JE, Sharma AK, Raez LE, Papagikos MA, Yunus F, Kurland BF, Eaton KD, Liao JJ, Mendez E, Futran N, Wang DX, et al. Cisplatin and radiotherapy with or without erlotinib in locally advanced squamous cell carcinoma of the head and neck: a randomized phase II trial. J Clin Oncol. 2013;31:1415–1421. doi: 10.1200/JCO.2012.46.3299. [DOI] [PubMed] [Google Scholar]
- 36.Dell'Aringa AH, Isaac ML, Arruda GV, Esteves MC, Dell'aringa AR, Júnior JL, Rodrigues AF. Audiological findings in patients treated with radio- and concomitant chemotherapy for head and neck tumors. Radiat Oncol. 2009;4:53. doi: 10.1186/1748-717X-4-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ribeiro KB, Levi JE, Pawlita M, Koifman S, Matos E, Eluf-Neto J, Wunsch-Filho V, Curado MP, Shangina O, Zaridze D, Szeszenia-Dabrowska N, Lissowska J, Daudt A, et al. Low human papillomavirus prevalence in head and neck cancer: results from two large case-control studies in high-incidence regions. Int J Epidemiol. 2011;40:489–502. doi: 10.1093/ije/dyq249. [DOI] [PubMed] [Google Scholar]
- 38.López RV, Levi JE, Eluf-Neto J, Koifman RJ, Koifman S, Curado MP, Michaluart-Junior P, Figueiredo DL, Saggioro FP, de Carvalho MB, Kowalski LP, Abrahão M, de Góis-Filho F, et al. Human papillomavirus (HPV) 16 and the prognosis of head and neck cancer in a geographical region with a low prevalence of HPV infection. Cancer Causes Control. 2014;25:461–471. doi: 10.1007/s10552-014-0348-8. [DOI] [PubMed] [Google Scholar]
- 39.Khan SG, Metter EJ, Tarone RE, Bohr VA, Grossman L, Hedayati M, Bale SJ, Emmert S, Kraemer KH. A new xeroderma pigmentosum group C poly(AT) insertion/deletion polymorphism. Carcinogenesis. 2000;21:1821–1825. doi: 10.1093/carcin/21.10.1821. [DOI] [PubMed] [Google Scholar]
- 40.Zuckerman V, Wolyniec K, Sionov RV, Haupt S, Haupt Y. Tumour suppression by p53: the importance of apoptosis and cellular senescence. J Pathol. 2009;219:3–15. doi: 10.1002/path.2584. [DOI] [PubMed] [Google Scholar]
- 41.Urashima M, Hama T, Suda T, Suzuki Y, Ikegami M, Sakanashi C, Akutsu T, Amagaya S, Horiuchi K, Imai Y, Mezawa H, Noya M, Nakashima A, et al. Distinct effects of alcohol consumption and smoking on genetic alterations in head and neck carcinoma. PLoS One. 2013;8:e80828. doi: 10.1371/journal.pone.0080828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodin SN, Rodin AS. Origins and selection of p53 mutations in lung carcinogenesis. Semin Cancer Biol. 2005;15:103–112. doi: 10.1016/j.semcancer.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Hayes RB, Bravo-Otero E, Kleinman DV, Brown LM, Fraumeni JF, Jr, Harty LC, Winn DM. Tobacco and alcohol use and oral cancer in Puerto Rico. Cancer Causes Control. 1999;10:27–33. doi: 10.1023/a:1008876115797. [DOI] [PubMed] [Google Scholar]
- 44.Mori S, Nose M, Morikawa H, Sato A, Saito T, Song ST, Tanda N, Teshima T. A novel evaluation system of metastatic potential of oral squamous cell carcinoma according to the histopathological and histochemical grading. Oral Oncol. 1998;34:549–557. doi: 10.1016/s1368-8375(98)00048-7. [DOI] [PubMed] [Google Scholar]
- 45.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. American Joint Committee on Cancer: Cancer Staging Manual. 7. New York: Springer; 2010. pp. 21–97. [Google Scholar]
- 46.Begum S, Gillison ML, Ansari-Lari MA, Shah K, Westra WH. Detection of human papillomavirus in cervical lymph nodes: a highly effective strategy for localizing site of tumor origin. Clin Cancer Res. 2003;9:6469–6475. [PubMed] [Google Scholar]
- 47.Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer. 2010;116:2166–2173. doi: 10.1002/cncr.25033. [DOI] [PubMed] [Google Scholar]
- 48.Nogueira GAS, Lourenço GJ, Oliveira CBM, Marsom FAL, Lopes-Aguiar L, Costa EFD, Lima TRP, Liutti VT, Leal F, Santos VCA, Rinck-Junior JA, Lima CSP. Association between genetic polymorphisms in DNA mismatch repair-related genes with risk and prognosis of head and neck squamous cell carcinoma. Int J Cancer. 2015;137:810–818. doi: 10.1002/ijc.29435. [DOI] [PubMed] [Google Scholar]
- 49.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 50.Cunningham D, Dicato M, Verweij J, Crombez R, de Mulder P, A du Bois, Stewart A, Smyth J, Selby P, van Straelen D, Parideans R, McQuade B, McRae J. Optimum anti-emetic therapy for cisplatin induced emesis over repeat courses: ondansetron plus dexamethasone compared with metoclopramide, dexamethasone plus lorazepam. Ann Oncol. 1996;7:277–282. doi: 10.1093/oxfordjournals.annonc.a010572. [DOI] [PubMed] [Google Scholar]
- 51.Kris MG, Tonato M, Bria E, Ballatori E, Espersen B, Herrstedt J, Rittenberg C, Einhorn LH, Grunberg S, Saito M, Morrow G, Hesketh P. Consensus recommendations for the prevention of vomiting and nausea following high-emetic-risk chemotherapy. Support Care Cancer. 2011;19:S25–32. doi: 10.1007/s00520-010-0976-9. [DOI] [PubMed] [Google Scholar]
- 52.Morisky DE, Green LW, Levine DM. Concurrent and predictive validity of a self-reported measure of medication adherence. Med Care. 1986;24:67–74. doi: 10.1097/00005650-198601000-00007. [DOI] [PubMed] [Google Scholar]
- 53.National Cancer Institute Cancer therapy evolution program. Common terminology criteria for adverse events (CTCAE) v 4.0, 2009. Available at: http://ctep.cancer.gov Accessed July 15, 2015.
- 54.Lopez-Flores A, Jurado R, Garcia-Lopez P. A high-performance liquid chromatographic assay for determination of cisplatin in plasma, cancer cell, and tumor samples. J Pharmacol Toxicol Methods. 2005;52:366–372. doi: 10.1016/j.vascn.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 55.Forastiere AA, Metch B, Schuller DE, Ensley JF, Hutchins LF, Triozzi P, Kish JA, McClure S, VonFeldt E, Williamson SK, Hoff DDV. Randomized comparison of cisplatin plus fluorouracil and carboplatin plus fluorouracil versus methotrexate in advanced squamous cell carcinoma of the head and neck: a Southwest Oncology Group study. J Clin Oncol. 1992;10:1245–1251. doi: 10.1200/JCO.1992.10.8.1245. [DOI] [PubMed] [Google Scholar]
- 56.Liang J, Gu A, Xia Y, Wu B, Lu N, Wang W, Lu C, Zheng Q, Wang S, Wang X. XPC gene polymorphisms and risk of idiopathic azoospermia or oligozoospermia in a Chinese population. Int J Androl. 2009;32:235–241. doi: 10.1111/j.1365-2605.2007.00842.x. [DOI] [PubMed] [Google Scholar]
- 57.Spitz MR, Wu X, Wang Y, Wang LE, Shete S, Amos CI, Guo Z, Lei L, Mohrenweiser H, Wei Q. Modulation of nucleotide excision repair capacity by XPD polymorphisms in lung cancer patients. Cancer Res. 2001;61:1354–1357. [PubMed] [Google Scholar]
- 58.Wang SL, Zhao H, Zhou B, Chen YL, Zou Y, Zhu XF, Li QS, Han MZ, Yang RC, Han ZC. Polymorphisms in ERCC1 and susceptibility to childhood acute lymphoblastic leukemia in a Chinese population. Leuk Res. 2006;30:1341–1345. doi: 10.1016/j.leukres.2006.03.027. [DOI] [PubMed] [Google Scholar]


