Abstract
Objective
Poly(ADP-ribose) polymerase (PARP) inhibitors have yielded encouraging responses in high-grade serous ovarian carcinomas (HGSOCs), but the optimal treatment setting remains unknown. We assessed the effect of niraparib on HGSOC patient-derived xenograft (PDX) models as well as the relationship between certain markers of homologous recombination (HR) status, including BRCA1/2 mutations and formation of RAD51 foci after DNA damage, and response of these PDXs to niraparib in vivo.
Methods
Massively parallel sequencing was performed on HGSOCs to identify mutations contributing to HR deficiency. HR pathway integrity was assessed using fluorescence microscopy-based RAD51 focus formation assays. Effects of niraparib (MK-4827) on treatment-naïve PDX tumor growth as monotherapy, in combination with carboplatin/paclitaxel, and as maintenance therapy were assessed by transabdominal ultrasound. Niraparib responses were correlated with changes in levels of poly(ADP-ribose), PARP1, and repair proteins by western blotting.
Results
Five PDX models were evaluated in vivo. Tumor regressions were induced by single-agent niraparib in one of two PDX models with deleterious BRCA2 mutations and in a PDX with RAD51C promoter methylation. Diminished formation of RAD51 foci failed to predict response, but Artemis loss was associated with resistance. Niraparib generally failed to enhance responses to carboplatin/paclitaxel chemotherapy, but maintenance niraparib therapy delayed progression in a BRCA2-deficient PDX.
Conclusions
Mutations in HR genes are neither necessary nor sufficient to predict response to niraparib. Assessment of repair status through multiple complementary assays is needed to guide PARP inhibitor therapy, design future clinical trials and identify ovarian cancer patients most likely to benefit from PARP inhibition.
Keywords: Ovarian cancer, Xenografts, PARP inhibitors, Niraparib, BRCA, Homologous recombination, DNA repair
1. Introduction
High-grade serous ovarian carcinoma (HGSOC)1, the most common and most lethal subtype, is often characterized by genomic scarring as well as alterations in genes encoding components of the homologous recombination (HR) DNA repair pathway [1,2]. In particular, BRCA1 or BRCA2 (BRCA1/2) mutations occur in up to 35% of HGSOCs (15–25% germline and 6–10% somatic), while an additional 6–10% of cases harbor germline or somatic mutations in at least 10 other genes in the HR pathway [1,3–5].
Because HGSOC has few driving oncogenes, targeted therapy remains a challenge [6]. However, the frequent impairment of HR in HGSOC provides a potential explanation for the initial response to DNA damaging agents such as platinum compounds [3,7–10] as well as a potential therapeutic opportunity because inhibition of the repair enzyme poly(ADP-ribose) polymerase 1 (PARP1) selectively kills cells with inactivation or silencing of BRCA1, BRCA2, or several other HR genes, such as RAD51, ATR, ATM, and CHK1 [11–13]. Importantly, cells are sensitized to PARP inhibitors even by mutations in certain genes that are not directly involved in DNA repair. For example, mutational inactivation of CDK12 [1], which encodes a kinase that facilitates expression of BRCA1 and several other repair proteins [14], also sensitizes ovarian cancer cells to PARP inhibitors [15,16].
Although the mechanistic basis for the synthetic lethality of PARP inhibition in HR-deficient cells remains incompletely understood [17], these observations have led to clinical development and regulatory approval of PARP inhibitors for HGSOC [17,18]. The PARP inhibitor olaparib has monotherapy response rates as high as 46% in BRCA1/2-mutant platinum-sensitive relapsed HGSOC [19–21] and maintenance therapy after platinum-based chemotherapy prolongs progression-free survival [22], leading to approval for maintenance in Europe. Other PARP inhibitors are in various stages of clinical development [17,18]. Niraparib (MK-4827), a potent and selective PARP inhibitor (IC50 = 3.8 nmol/l for PARP1 and 2.1 nmol/l for PARP2) [23], is safe and effective in both hereditary and sporadic HGSOCs [24].
Although olaparib is approved for the treatment of BRCA1/2-mutant HGSOCs, 40–70% of BRCA1/2-mutant ovarian cancers fail to respond [19–21,24] and clinical activity has been observed in patients with wildtype (wt) BRCA1/2 [20,24]. Accurately identifying ovarian cancers that will respond to PARP inhibitors is an area of intense interest [17]. The ability of RAD51, a downstream component of the HR pathway, to form foci after DNA damage has been proposed as a potential predictor of response to the PARP inhibitor rucaparib ex vivo [25] and reported to correlate with response of conventional and ovarian cancer patient-derived xenografts (PDXs) to the PARP inhibitor veliparib and carboplatin [26].
We have developed a large bank of ovarian PDXs that recapitulate many aspects of the source tumors in patients, including histological subtype, gene copy number variation, degree of stromal involvement, absence or presence of ascites, homing to the bowel wall, and response to platinum/taxane therapy [27]. In the present study, we have used several of these models to assess the effect of niraparib, alone and in conjunction with platinum/taxane therapy, on HGSOC in vivo. In addition, we have assessed the relationship between certain markers of HR status, including BRCA1/2 mutations and formation of RAD51 foci after DNA damage, and PDX response to niraparib.
2. Materials and methods
2.1. Reagents and antibodies
Niraparib was provided by Merck and TesaroBio. Antibodies to poly(ADP-ribose) polymer (pADPr, rabbit polyclonal 96–10) and PARP1 (mouse monoclonal C-2-10) were from G. Poirier (Universite Laval, Quebec, Canada). Goat anti-β-actin and rabbit anti-RAD51C were from Santa Cruz Biotechnology. Other antibodies were obtained as described [28].
2.2. RAD51 focus assay
Single cell PDX suspensions were isolated from fresh tissue immediately following resection using a gentleMACS™ dissociator (Miltenyl Biotec, Auburn, CA) and enzymatic digestion per the manufacturer's instructions. PEO1 and PEO4 cells (kind gifts from Fergus Couch, Mayo Clinic) were authenticated by short tandem repeat profiling, confirmed to have BRCA2 mutations by Sanger sequencing, and cultured in DMEM containing 10% (vol/vol) heat-inactivated fetal bovine serum (FBS), 100 μM nonessential amino acids and 10 μg/ml insulin (medium A). Dissociated PDX cells and cell lines in medium A were plated on sterile coverslips and allowed to adhere overnight.
Cells were irradiated (10 Gy) or supplemented with niraparib (8 μM, two times the IC90 in 48-h clonogenic assays in PEO1 cells) and incubated for 6 h. Working at 4 °C, cells were washed twice with PBS, fixed in 4% (wt/vol) paraformaldehyde for 15 min, permeabilized with 0.25% (wt/ vol) Triton X-100 in PBS, and blocked overnight with 5% (v/v) goat serum in PBS. Coverslips were incubated with rabbit monoclonal antibody to RAD51 (Abcam 133,534, 1:500) and mouse monoclonal antibody to geminin (Abcam 1,043,306, 1:25) in blocking buffer overnight at 4 °C, washed 6 times over 20 min with wash buffer (PBS containing 0.1% Triton X-100 and 0.1% bovine serum albumin), incubated with secondary antibody (Alexa fluor 488 goat anti-rabbit IgG and Alexa fluor 568 goat anti-mouse IgG, each at 1:1000) in blocking buffer for 1 h at 21 °C in the dark, washed, and counterstained with 1 μg/ml Hoechst 33258 in PBS. Samples were examined by immunofluorescence microscopy on a Zeiss Axiovert with a N.A. 1.40 63× lens and photographed on a Zeiss Axiocam MRm CCD camera using Zeiss Zen software. RAD51 foci were quantified manually in a blinded fashion in at least 100 cells per slide and considered positive if ≥10 RAD51 foci were visible.
2.3. PDX passaging and treatment
Five individual low-passage HGSOC models (labeled PH#) were chosen based on previous experience and ease of engraftment and established intraperitoneally in up to 35 female SCID mice as described [27]. Tumor diameter and cross-sectional area were followed by transabdominal ultrasound using a Fujifilm SonoSite S-Series with onboard measurement tools and ImageJ software after calibration as described [27].
When tumor area reached 0.5–1 cm2, mice were randomized to treatment arms. Niraparib was given by daily oral gavage in 15% methylcellulose (100 mg/kg as monotherapy or 50 mg/kg with chemotherapy). Intraperitoneal (IP) carboplatin (Novaplus #61703-360-18) and paclitaxel (Novaplus #55390-304-05) at 50 mg/kg and 15 mg/kg, respectively, were administered on days 1, 8, 15, and 22 with or without niraparib. Largest tumor cross-sectional area was measured twice weekly through day 28. Mice were euthanized individually when moribund or as a cohort at day 28. The primary endpoint was tumor area by ultrasound, normalized to the day 1 area of the same tumor and plotted as a ratio vs. time.
2.4. BROCA sequencing
DNA was harvested from PDXs and assayed at the University of Washington by massively parallel sequencing [4] for mutations in a panel of genes involved in various aspects of DNA repair or its regulation, including ATM, ATR, BARD1, BRCA1, BRCA2, CDK12, CHEK1, PALB2, RAD51C, TP53, and 43 others (Table S1). All deleterious mutations were confirmed by Sanger sequencing. Only clear loss of function mutations and missense mutations with experimental evidence of functional consequences were considered deleterious. Germline mutations were assayed in peripheral leukocytes of matched patients when available, or non-neoplastic tissue when not, because computational methods to assess germline mutations in tumors are less reliable in the setting of high neoplastic cellularity, such as PDX tumors.
2.5. Immunoblotting
PEO1 cells and PDX tissue (cut to 1 mm sections) were washed twice with calcium- and magnesium-free Dulbecco's phosphate buffered saline (PBS), solubilized in buffered 6 M guanidine hydrochloride under reducing conditions, and prepared for electrophoresis as described [29]. Aliquots containing 50 μg of protein were separated on SDS-polyacrylamide gels containing 8% (wt/vol) acrylamide, transferred to nitro-cellulose and probed with antibodies [30].
2.6. Artemis knockout
The oligonucleotides (5'-TCTCCATAGACCGCTTCGAT-3') guiding to Artemis 463–485 (accession number: NM_001033855) were synthesized, annealed, and cloned into BsmBI site of lentiCRISPRv2 (Addgene) plasmid. Viruses were packaged in HEK293T cells by transfecting with the packaging vector psPAX2, envelope vector pMD2.G, and lentiCRISPRv2-artemis 463–485 using Lipofectamine 2000. Two days after transduction with virus, PEO1 cells were selected with 3 μg/ml puromycin and cloned by limiting dilution. Artemis knockout was verified by genomic DNA sequencing and immunoblotting.
2.7. DNA methylation
250 ng of DNA was bisulfite converted with an EZ Methylation Direct kit (Zymo Research, Irvine, CA) and evaluated with methylation sensitive PCR for BRCA1 as previously described [31]. Methylation sensitive PCR for RAD51C was performed with primers for the methylated (M) reaction (sense-5'-TGtAAGGttCGGAGtttCGTGC-3' and antisense-5'-TCGCTaaaaCGTaCGaCGTaACG-3', 85 nt) and unmethylated (UM) reaction (sense-5'-GtGtAA-AGtTGtAAGGtttGGAGttttGTGtG-3' and antisense-5'-CaCACaCCCTCaCTaaaaCaTaCaaCa-TaACa-3', 103 nt). Methylated and unmethylated DNA (#D5014) was used as positive M control (M ctrl) and UM control (UM ctrl) for bisulfite conversion and validation of amplicon size. Water (H2O) was substituted for DNA to rule out cross-contamination of samples.
2.8. Quantitative polymerase chain reaction
Quantitative PCR was performed using a LightCycler 480 II (Roche Molecular Biochemicals, Mannheim, Germany) with software v1.5.1.62 to calculate ΔΔCp using the second derivative maximum method against RPLP0. Primers were purchased from Integrated DNA Technology (Coralville, IA) for BRCA1 (FWD-5'-CTGAAGACTG CTCAGGGCTATC-3', REV-5'-AGGGTAGCTGTTAGAAGGCTGG-3'), RAD51C (FWD-5'-TTTGGTGAGTTTCCCGCTGTC-3', REV-5'-CTCAGCA GTCTGGAACCCC-3'), and RPLP0 (FWD-5'-TGCTGATGGGC-AAGAA CAC-3'; REV-5'-GAACACAAAGCCCACATTCC-3').
2.9. Statistical analysis
Linear mixed effects modeling, performed in the SAS PROC MIXED procedure [32], was used to assess differences between groups. Change in tumor area from baseline on the natural log scale was compared between treatments using a two-parameter growth model framework. Due to zeroes in the data, a small constant (0.02 for PH039, PH095, PH080, PH087, and 0.3 for PH077) was added prior to log transformation to linearize growth trajectories where possible. Models were fit separately for each PDX, and the time variable was centered for hypothesis testing. Akaike information criterion (AIC) and Bayesian information criterion (BIC) were used to choose the random effects and structure of the correlation between repeated measurements [33]. A RANDOM statement was used to specify the intercept and slope as random effects with unstructured correlation, allowing per-mouse regression lines. An autoregressive correlation structure, which assumes any two observations from the same mouse are correlated and this correlation decreases exponentially with time between the observations [33], was specified in the REPEATED statement. Residual plots were used to choose between linear (PH077, PH080, PH087) versus quadratic (PH039, PH095, regrowth experiments) time functions. If the test of differing slopes (growth trajectories) was non-significant, the test of mean area differences was reported. For visualization, both model estimates with standard errors and observed averages are included for visual demonstration that the models fit the data well. For other studies, two group or multiple group comparisons were performed using student t-test or ANOVA with Dunnett post-test, where appropriate. For these studies, p < 0.05 was considered significant.
3. Results
3.1. Genomic PDX characterization
Using massively parallel DNA sequencing (BROCA), deleterious mutations in HR or related genes were detected in 11 out of 48 (23%) founder PDXs (Table S2). One model had mutations in three genes (ATM, FAM175A, PTEN), one model had mutations in two (BRCA2, CHEK2), and nine models had mutations in one gene (BRCA1, BRCA2, CHEK2, XRCC2, or CDK12). BRCA2 was mutated in four models (8.3%), followed by BRCA1 and CHEK2 in two models (4.2%) each.
Five PDX models (PH039, PH077, PH080, PH087, and PH095) selected for niraparib treatment all demonstrated TP53 mutations accompanied by loss of the wildtype allele (Table 1). PH077 contained a BRCA2 mutation (c.5042_5043delTG) in the primary carcinoma specimen and PDX but not in the patient's germline DNA (Fig. S1A). PH095 contained a germline BRCA2 nonsense mutation (p.L771X, Fig. S1B). Both of these BRCA2 mutations were accompanied by loss of the wildtype allele and were present in 100% of sequence reads. PH080 harbored a frameshift mutation in the second exon of CDK12 with loss of the wildtype allele (c.1479delA) (Fig. S1C). Notably, all mutations were preserved across PDX passages tested; and no reversion mutations were detected. From this sequence analysis, BRCA2-mutant PH077 and PH095 as well as CDK12-mutant PH080 were predicted to be HR deficient, while PDXs without mutations in HR-related genes (PH039 and PH087) were tentatively predicted to be HR-proficient.
Table 1.
Characterization of source tumors and responses of corresponding PDXs.
| PDX model | Grade | Stage | Histology | Platinum-free interval (months) | Source tumor platinum responsea | Non TP53 mutations by BROCAb | TP53 gene mutation | Rad51focic | Rad51C and BRCA1 methylationd | PDX niraparib responsee | PDX carboplatin/paclitaxel responsee | Enhanced carboplatin/paclitaxel effect by niraparibf |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PH077 | 3 | IV | Serous | 4.4 | Sensitive* | BRCA2 c.5040delTG (somatic) | c.672+1G>A (splice) | − | − | Regression | Regression | − |
| PH095 | 3 | IB | Serous | 3.1 | Sensitive** | BRCA2 p.L771X (germline) | p.S215I | − | − | Refractory | Regression | − |
| PH080 | 3 | IC | Serous | No recurrence to date | Sensitive | CDK12 c.1476delA | p.R249S | + | − | Attenuation | Regression | − |
| PH087 | 3 | IIIC | Serous | 22.8 | Sensitive | None | p.V173M | ++ | − | Attenuation | Regression | − |
| PH039 | 3 | IIIC | Serous | 11.7 | Sensitive | None | p.N239T | +++ | + | Regression | Regression | + |
Source patients for PH077, PH087, PH080, and PH039 were clinically sensitive to platinum based therapy as defined by the absence of recurrence >6 months after treatment, complete response to repeat platinum (*), or a combination of CA-125 and clinical factors (**).
Patient developed postoperative complications, which made chemotherapy sub-optimal. At recurrence, she was retreated with a platinum doublet and achieved a complete radiographic and CA-125 response, consistent with platinum sensitive disease.
Patient’s CA-125 normalized while on chemotherapy (1840 pre-operative to 17 after chemotherapy), recurrent pleural effusions resolved, and there was no radiographic evidence of disease, consistent with platinum sensitive ovarian cancer. The patient opted not to receive more than 4 cycles. Due to failure to thrive, she died 3.1 months after adjuvant therapy and to the authors’ knowledge, she did not have recurrent cancer.
BROCA sequencing for genes implicated in homologous recombination shows BRCA2 mutations in PH077 (somatic) and PH095 (germline) in addition to CDK12 in PH080 (verified by Sanger sequencing).
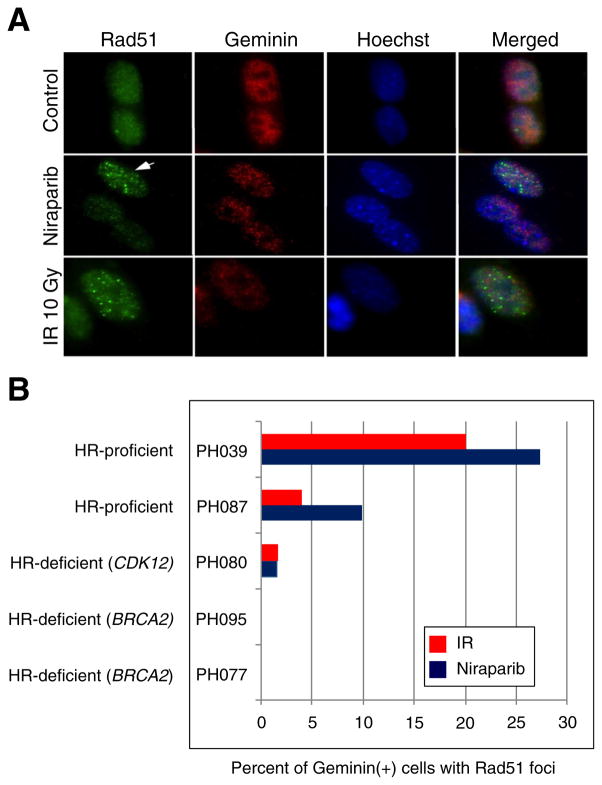
RAD51 foci from Figure 1 with absent (−), low (+), moderate (++), or high (+++) foci formation.
RAD51C methylation and expression data from Figure 5 and associated experiments described in the text.
Response of PDXs is defined as Regression (mean tumor area on day 28 is below baseline), Attenuation (mean tumor area on day 28 is above baseline but below diluent control), or Refractory (no difference from diluent-treated control).
(−) indicates no enhancement of carboplatin/paclitaxel effect by addition of niraparib 50 mg/kg daily, whereas (+) indicates enhanced tumor shrinkage relative to carboplatin/pac-litaxel doublet without niraparib. See Figure 4 for additional details.
3.2. RAD51 staining
To complement this sequence analysis, we assessed formation of RAD51 foci after DNA damage. Cells isolated from the xenografts were transiently cultured ex vivo; exposed to ionizing radiation, diluent or niraparib; and examined by immunofluorescence after staining with monoclonal antibodies specific for human RAD51 and, to mark cells in S and G2 phases of the cell cycle where HR is active, human geminin [34]. PEO1 cells (BRCA2-mutant) and PEO4 cells (BRCA2 revertant [35]) served as negative and positive controls, respectively. Fig. 1A shows an example of these assays in PH039 cells; and Fig. 1B and Table 1 summarize the results. PH087 and PH039, which lacked deleterious HR mutations, formed RAD51 foci after DNA damage (Fig. 1B). In contrast, PH095, PH080 and PH077, which harbored deleterious mutations in HR genes, formed fewer RAD51 foci. Interestingly, the two PDXs with BRCA2 mutations had no nuclei with >10 RAD51 foci, while the PDX with a CDK12 mutation (PH080), which would be predicted to down-regulate BRCA1 [14], showed reduced but detectable RAD51 foci after DNA damage. In short, the pre-treatment BROCA sequencing of HR genes and RAD51 focus assays in the five tested PDXs were concordant.
Fig. 1. RAD51 focus assay on PDX tumor cells.
A. Immunofluorescence microscopy of niraparib-treated PDX cells (PH039) and irradiated cells showing RAD51 foci (arrow) within geminin positive cells and lack of RAD51 foci formation in control cells. B. Percentage of geminin positive cells that were RAD51 foci positive after treatment with ionizing irradiation (IR) or niraparib. Similar results were observed using multiple independent tumorgrafts from each model, including 5 separate PH095 xenografts.
3.3. Single agent niraparib therapy
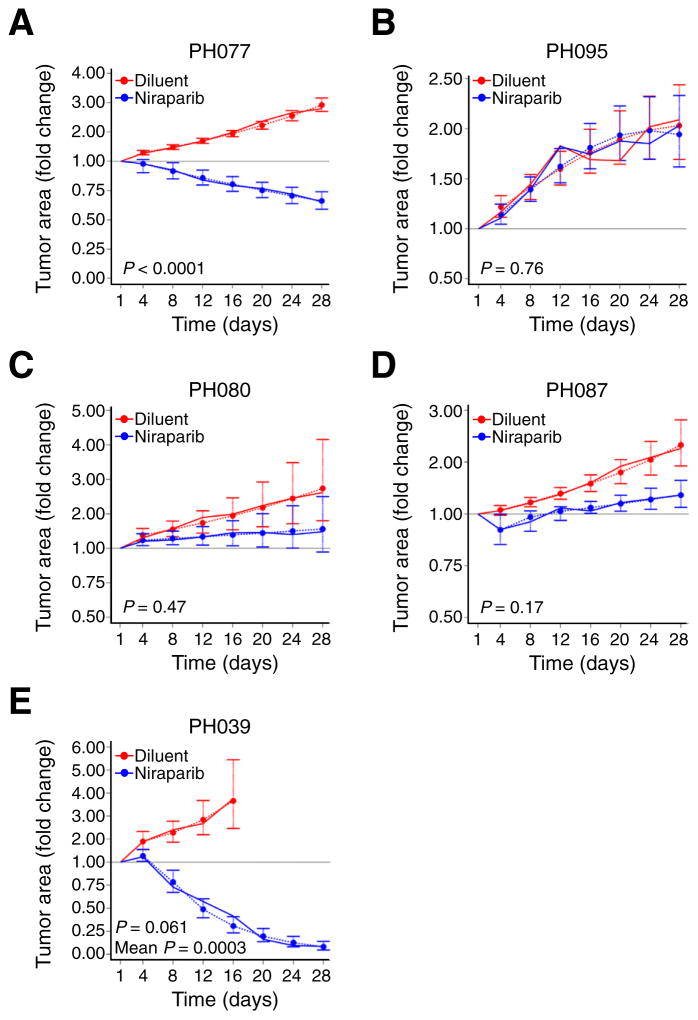
Niraparib monotherapy was well tolerated with no weight loss (Fig. S2). The two BRCA2-mutant models, PH077 and PH095, exhibited contrasting responses to treatment: Whereas PH077 tumor size declined below baseline (regression) (trajectory p < 0.0001) (Fig. 2A and Table 1), PH095 growth was similar to controls (refractory) (Fig. 2B). LIkewise, CDK12-mutant PH080 showed only a trend toward slowed growth relative to controls but no regression below baseline (attenuation) (Fig. 2C).
Fig. 2. Response of PDXs to single-agent niraparib.
Change in mean tumor area (solid lines) in niraparib treated (blue line) and control (red line) arms. Mean tumor area has been normalized to day 1 mean area as described in the statistical methods. p-Values for differences in growth trajectories in each PDX model are presented. Estimated (predicted) mean tumor areas together with standard error bars from the regression model are included for visual demonstration that the model fits the data well (dashed lines). “P” indicates test for different growth rates (slope), while “Mean P” indicates test for different average area (centered intercept).
PDX models PH087 and PH039 lacked HR gene mutations. Although niraparib-treated PH087 tumors initially regressed, resulting in attenuated average tumor area relative to controls (mean area p = 0.03), tumors were on average 1.4-fold their baseline area by day 28 (Fig. 2D). On the other hand, PH039 demonstrated the most profound tumor regression (Fig. 2E) to an average of 8% of baseline area in response to niraparib (p = 0.0003).
3.4. Niraparib combined with platinum-based chemotherapy and as maintenance
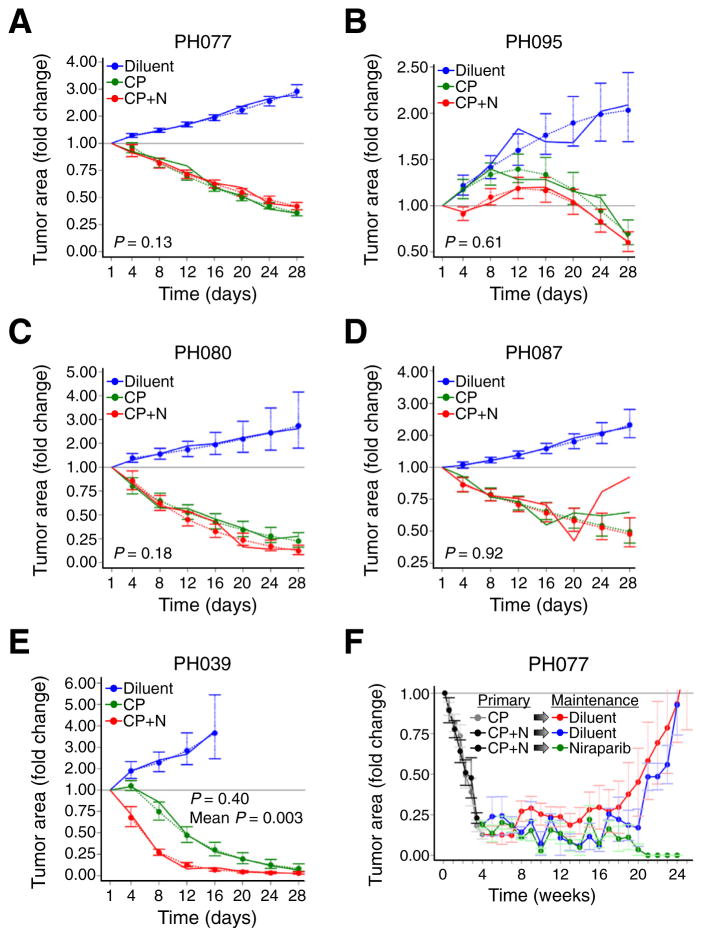
We also assessed whether niraparib would augment response to platinum/taxane therapy in vivo. Carboplatin/paclitaxel by itself induced marked attenuation of growth relative to control in all PDX models (Fig. 3A–E, p values for trajectory vs. control animals ranging from <0.0001 to 0.0016). The PDX with the greatest niraparib sensitivity (PH039) was also most sensitive to carboplatin/paclitaxel therapy, with tumor areas shrinking to 8% of baseline (Fig. 3E). PH077, PH080, PH087, and PH095 also regressed, decreasing on average to 36%, 23%, 50%, and 87% of baseline, respectively. These results are consistent with the clinical response of the source tumors, which were all platinum sensitive (Table 1).
Fig. 3. Response of PDXs to paclitaxel/carboplatin with and without niraparib.
A–E. Tumor area of carboplatin/paclitaxel-treated models relative to control. Although tumors regressed below baseline for PDXs PH077, PH080, PH087 and PH095, the addition of niraparib (CP + N, red line) did not result in statistically significant differences from carboplatin/paclitaxel without niraparib (CP, green line). Solid lines indicate change in mean tumor area normalized to day 1 mean area as described in the statistical methods, and dashed lines indicate estimated or predicted mean tumor areas together with standard error bars from the regression model. p-Values shown on graphs are for differences in trajectory between PDXs treated with carboplatin/paclitaxel and carboplatin/paclitaxel/niraparib. p-Values for differences in mean area over time also were >0.05. In PH039, the growth trajectories were not statistically significantly different (p = 0.40), but the average change from baseline over time was significantly larger with addition of niraparib (p = 0.003). F. Effect of maintenance niraparib therapy in PH077. Initial treatments (weeks 0 to 3) are denoted as “primary” and consisted of carboplatin/paclitaxel (CP, gray line) or carboplatin/paclitaxel/niraparib (CP + N, black line). Niraparib “maintenance” was initiated following primary carboplatin/paclitaxel/niraparib (Niraparib, green line) and diluent controls included for both carboplatin/paclitaxel CP (Diluent, red line) and CP + N (Diluent, blue line).
Addition of niraparib failed to augment the response of PH077, PH080, PH087 or PH095 to carboplatin/paclitaxel treatment (Fig. 3A–D, p = 0.13–0.95 for trajectory of carboplatin/paclitaxel vs. triple therapy and p = 0.44–0.87 for tumor area). In contrast, PH039 xenografts treated with this trio began to regress immediately, whereas those treated with carboplatin/paclitaxel grew to day 4 before shrinking, resulting in statistically smaller tumor areas over the time-course (p = 0.003) and a trend toward smaller xenografts in the combination arm at day 28 (2% vs. 8% of baseline, p = 0.051). These results suggest some potential benefit from adding niraparib to platinum/taxane therapy in this PDX.
In light of the phase 2 clinical trial by Ledermann et al. showing efficacy of maintenance olaparib in platinum sensitive recurrent BRCA1/2-mutant HGSOC [17,22], we investigated the role of niraparib maintenance therapy after treatment with combination niraparib/ carboplatin/paclitaxel. Five mice bearing PH077 (niraparib-sensitive BRCA2 mutant tumor, Fig. 2A) were randomized at day 28 after the start of triple therapy to maintenance with niraparib 50 mg/kg/day, a dose previously shown to be efficacious [36], and four were randomized to observation. Ten additional mice from the carboplatin/paclitaxel arm entered observation. One mouse receiving niraparib maintenance died unexpectedly at week, but carcinomas in the remaining four mice on niraparib maintenance therapy became undetectable over 20 weeks, whereas tumors in mice randomized to observation grew to their baseline size (Fig. 3F).
3.5. Effect of niraparib on pADPr formation
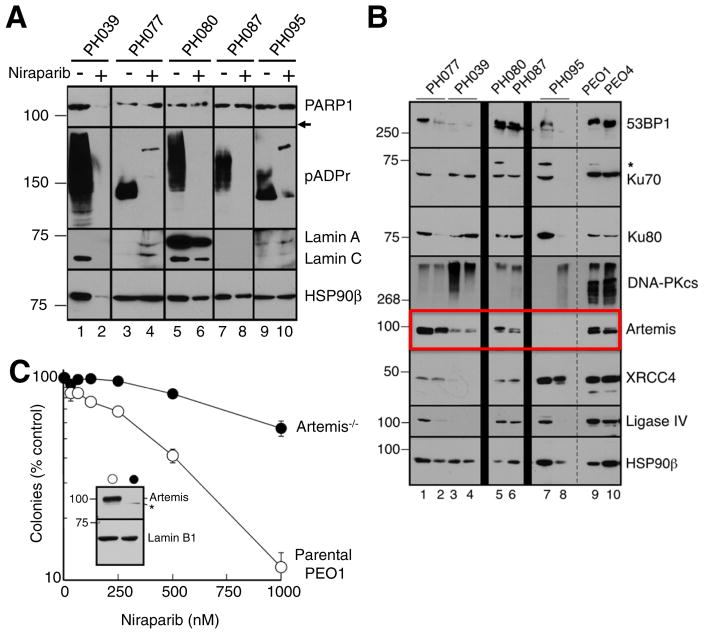
Further studies were performed to understand the disparity between HR status and response (Table 1). Inhibition of PARP activity was assessed by comparing pADPr, the product of the polymerization reaction, in PDX tissue harvested from diluent vs. nirparib-treated PDXs on day 28. Suppression of pADPr, suggesting effective drug delivery, was observed in all niraparib-treated PDXs regardless of response (Fig. 4A). Interestingly, niraparib treatment was associated with loss of PARP1 protein expression in PH039, which exhibited the greatest tumor regression in response to single agent therapy (Fig. 4A). The signature 89 kDa caspase-generated PARP1 fragment was not detectable, suggesting loss of PARP1 in PH039 is distinct from apoptosis-associated cleavage. Blotting for lamins A and C, heat shock protein 90β and topoisomerase I (Fig. 4A and additional experiments not shown) also demonstrated diminished signal in niraparib-treated PH039, suggesting a paucity of viable epithelial cells in this model after PARP inhibitor treatment as a consequence of its profound PARP inhibitor sensitivity.
Fig. 4. PARP is inhibited in all PDX models.
A. Three in-vivo tumor replicates of treated models and controls were harvested at the end of treatment (day 28), pooled, and subjected immunoblotting of the indicated antigens. Formation of pADPr is inhibited in niraparib treated tumors. Absence of PARP1 protein is seen in model PH039 treated with niraparib. Arrow indicates expected location of 89 kDa caspase-generated PARP1 fragment if it were present. B. Lysates containing 50 μg of protein from one or two separate aliquots of the indicated PDX were subjected to SDS-PAGE and blotted with antibodies to the indicated antigen. PE01 and PE04 cells, which are known to express all of these proteins, were included as positive controls. HSP90β served as a loading control. * indicates nonspecific band. C. Parental PEO1 cells (open circles) or Artemis−/− PEO1 cells derived as described in the Materials and methods (closed circles) were plated (750 cells/plate) and treated beginning 18 h later with diluent (0.1% DMSO) or the indicated concentration of niraparib. Error bars, ±SEM from triplicate aliquots.
3.6. Deficiencies in Artemis and RAD51C predict niraparib response
Because downregulation of proteins involved in nonhomologous endjoining (NHEJ), including Ku80, DNA-PKcs and Artemis, has previously been reported to render BRCA2-deficient cells resistant to PARP inhibitors [28,37,38], we examined expression of these proteins in PH095 xenografts to assess whether their loss might contribute to PARP inhibitor resistance. While the majority of NHEJ pathway proteins, including 53BP1, Ku70, Ku80, DNA-PKcs, XRCC4 and DNA ligase IV were readily detected in all five xenograft models, Artemis was undetectable in PH095 (Fig. 4B, lanes 7 and 8). In further experiments, Artemis gene knockout was observed to render PE01 cells niraparib resistant (Fig. 4C), suggesting intact NHEJ is needed for niraparib activity and providing evidence that the Artemis deficiency observed in PH095 is sufficient to render BRCA2-mutant ovarian cancer cells PARP inhibitor resistant.
3.7. RAD51C promoter methylation is associated with niraparib response
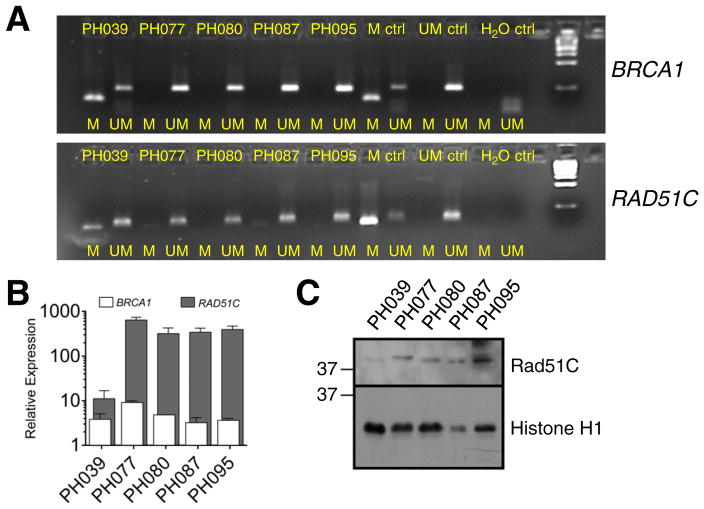
During the course of these experiments, we also observed that RAD51C message was particularly low in PH039. Given previous data showing that promoter methylation correlates with RAD51C loss in HGSOC [2], further analysis revealed that the RAD51C and BRCA1 promoters were hypermethylated in PH039 but not the other four PDXs (Fig. 5A). At the mRNA level, PH039 had lower expression of RAD51C, but not BRCA1, compared to the other four models (Fig. 5B). Consistent with these results, immunoblotting also demonstrated diminished RAD51C in PH039 (Fig. 5C).
Fig. 5. Gene expression and RAD51C methylation analysis in PDXs.
A. Using bisulfite sequencing and methylation specific PCR, methylated (M) and unmethylated (UM) RAD51C and BRCA1 in PDX models is shown with a control (ctrl) for M and UM. Water (H2O) was used as a blank control. B. Relative RAD51C and BRCA1 mRNA levels across the 5 PDX models shows lower expression of RAD51C in niraparib-responsive PDX PH039. C. Immunoblot for RAD51C in xenograft samples.
4. Discussion
In this study, we assessed the effectiveness of niraparib in a series of human PDX models. To our knowledge this was the first attempt to assess the ability of HR gene mutations and formation of RAD51 foci, two frequently discussed biomarkers, for their ability to predict PARP inhibitor response. Importantly, we observed that responses of HGS ovarian carcinoma PDXs to niraparib monotherapy do not necessarily track with these markers. In particular, responses to niraparib were seen in a BRCA2-mutant and a BRCA1/2-wt model, while another PDX with an underlying BRCA2 mutation failed to respond. These results are consistent with data from previous clinical trials showing responses to PARP inhibitors in some ovarian cancer patients with germline BRCA1/2 mutations as well as some with sporadic cancers but lack of response in up to half of cases with germline BRCA1 or BRCA2 mutations [17,19,20].
DNA damage-induced formation of RAD51 foci can purportedly predict response of ovarian cancer to PARP inhibitors and platinum [25,26]. In our study, ability to form RAD51 foci tracked with mutation status of HR genes. In particular, PDXs with BRCA2 or CDK12 mutations formed few if any RAD51 foci after ionizing radiation or niraparib treatment (Fig. 1). However, the ability to form RAD51 foci was not a good predictor of niraparib response in vivo (Table 1). The differences between our findings and previously published data [26] might reflect our larger PDX sample size, assessment of RAD51 foci in G2-S phase with geminin staining, or different RAD51 monoclonal antibody specificity.
Results observed with the PH039 model are particularly informative. Both the BRCA1 and RAD51C promoters were hypermethylated, but only RAD51C was diminished at the mRNA and protein levels (Fig. 5). These observations suggest RAD51C promoter methylation as a likely explanation for the response of PH039 to niraparib in the absence of identifiable HR gene mutations. Despite absence of this critical HR protein, DNA damage induced RAD51 foci in PH039, consistent with previous results in other cells [39] and with studies showing that RAD51C acts downstream of RAD51 during HR. Even though RAD51 foci are formed in this model in response to DNA damage (Fig. 1), this model was exquisitely sensitive to niraparib and platinum/taxane (Fig. 2E and 3E). To our knowledge, this is the first documentation of PARP inhibitor sensitivity correlating with RAD51C promoter hypermethylation in vivo, suggesting this could be an infrequent but important molecular alteration leading to PARP inhibitor sensitivity in HGSOC. These data are also consistent with low RAD51C expression and sensitivity to olaparib in gastric cancer cell lines [40].
Results obtained with the PH095 model are also informative. Previous studies in tissue culture have shown that killing by PARP inhibitors involves activation of the error-prone NHEJ pathway when HR is disabled [28,37,38]. Conversely, downregulation of any of a number of NHEJ proteins, including DNA-PKcs, Ku80, or Artemis impairs PARP inhibitor sensitivity in vitro. Here we have shown that the BRCA2-mutant xenograft PH095, which fails to respond to niraparib in vivo, lacks detectable Artemis (Fig. 4B). Moreover, we have demonstrated that Artemis deficiency is sufficient to confer niraparib resistance (Fig. 4C), providing a potential explanation for resistance of this BRCA2-mutant model in vivo.
Our observations in PDX PH080 (CDK12 mutation) are also intriguing. CDK12 regulates transcription of several genes involved in the Fanconi anemia/HR repair pathway, including BRCA1, ATR, FANC1 and FANCD2 [14]. Previous studies suggest that CDK12 inactivation sensitizes HGSOC cells to PARP inhibitors [15,16]. PDX PH080, on the other hand, was not as sensitive to niraparib as BRCA2-mutated PH077 (Fig. 2C), but instead demonstrated an intermediate phenotype with partial diminution of damage-induced RAD51 foci (Fig. 1B) and diminished growth rather than regression below baseline after PARP inhibitor monotherapy (Fig. 2C and Table 1). This intermediate response may be consistent with the partial but incomplete suppression of HR reported after CDK12 silencing [15].
We have shown that platinum response in ovarian cancer PDXs correlates strongly with platinum response of corresponding patients [27]. For the five PDX models derived from platinum-sensitive HGSOCs, the relationship between platinum and PARP inhibitor response is complicated. Although all five models regressed on platinum below baseline, only two models regressed with niraparib. These results parallel the 40–60% clinical response of platinum sensitive BRCA1/2-mutant HGS ovarian cancers to PARP inhibitors [17] and highlight the need for better understanding of disparities between platinum and PARP inhibitor response.
One of the most notable findings in the present study is that therapeutic response to platinum/taxane was not generally augmented by the addition of niraparib in either HR-deficient or HR-proficient PDXs. However, as illustrated by PH039, extreme sensitivity to niraparib monotherapy may predict an augmented response to the carboplatin/ paclitaxel/niraparib combination. On the other hand, we readily observed prolonged time to recurrence in a PDX undergoing maintenance niraparib. Results of two ongoing phase III trials, NCT01847274/NOVA with maintenance niraparib in recurrent HGSOCs and NCT02470585/ GOG-3005 with up-front and maintenance ABT-888, will be essential in corroborating the present preclinical findings.
The assessment of tumor genotype through gene sequencing is anticipated to become a routine component of HGSOC evaluation. Although the combination of mutational analysis and validated functional assays may allow patient stratification into distinct subgroups, the present results suggest that these analyses might not be adequate for predicting the outcome of PARP inhibitor therapy. Alternative mechanisms, such as RAD51C promoter hypermethylation or loss of Artemis, may also contribute to PARP inhibitor response. The present observations highlight the need for further study of factors that affect response to PARP inhibitors and raise the possibility that multimodality assessments of DNA repair status, such as targeted gene sequencing and DNA methylation studies of fresh patient biopsies, might be required to more effectively predict PARP inhibitor response.
Supplementary Material
HIGHLIGHTS.
Niraparib is a PARP inhibitor under clinical development.
Predictive biomarkers of PARP inhibitor response may help with patient selection.
Gene mutations are neither necessary nor sufficient for in vivo niraparib response.
Diminished RAD51 foci failed to predict response.
Assessment of repair status through multiple complementary assays is needed.
Abbreviations
- BRCA1/2
BRCA1 or BRCA2
- CDK
cyclin-dependent kinase
- FBS
heat-inactivated fetal bovine serum
- HGSOC
high-grade serous ovarian carcinoma
- HR
homologous recombination
- IP
intraperitoneal
- pADPr
poly(ADP-ribose) polymer
- PARP
poly(ADP-ribose) polymerase
- PBS
Dulbecco's calcium- and magnesium-free phosphate buffered saline
- PDX
patient-derived xenograft
- SCID
severe combined immunodeficiency
- TP53
tumor protein p53
Footnotes
Supported by the Mayo Clinic Ovarian SPORE P50 CA136393, Ovarian Cancer Research Fund (to ES, PH and SK), the Pacific Ovarian Cancer Research Consortium SPORE P50 CA0836 (ES), the Wendy Feuer Ovarian Cancer Research Fund (ES), the Cori and Tony Bates Novel Technologies for Cancer Prevention Fund (ES), the Fred C. and Katherine B. Andersen Foundation (PH), T32 DK07352 (MAB) and T32 GM65841 (RMH).
Authors' disclosure of potential conflicts of interest: PH was an unpaid consultant for Merck and received research funding from Merck and Tesaro.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ygyno.2016.08.328.
Conflict of interest statement
Dr. Becker reports personal fees from Ovarian Cancer Tumorgrafts - Avatar System, outside the submitted work; Dr. Haluska reports non-financial support from Tesaro, during the conduct of the study through the supply of niraparib; and intellectual property (non-patent) involving the use of the PDX models employed in this work, which includes the awarding of royalties. Dr. Kaufmann reports grants from National Cancer Institute and Ovarian Cancer Research Fund during the conduct of the study. In addition, Mayo Clinic has a patent related to methods for assessing responsiveness to PARP inhibitors on which Dr. Kaufmann is a co-inventor. Dr. Weroha reports grants from Tesaro, during the conduct of the study.
References
- 1.Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abkevich V, Timms KM, Hennessy BT, Potter J, Carey MS, Meyer LA, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107:1776–1782. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pennington KP, Walsh T, Harrell MI, Lee MK, Pennil CC, Rendi MH, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20:764–775. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wickramanyake A, Bernier G, Pennil C, Casadei S, Agnew KJ, Stray SM, et al. Loss of function germline mutations in RAD51D in women with ovarian carcinoma. Gynecol Oncol. 2012;127:552–555. doi: 10.1016/j.ygyno.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bast RC, Jr, Mills GB. Personalizing therapy for ovarian cancer: BRCAness and beyond. J Clin Oncol. 2010;28:3545–3548. doi: 10.1200/JCO.2010.28.5791. [DOI] [PubMed] [Google Scholar]
- 7.Cass I, Baldwin RL, Varkey T, Moslehi R, Narod SA, Karlan BY. Improved survival in women with BRCA-associated ovarian carcinoma. Cancer. 2003;97:2187–2195. doi: 10.1002/cncr.11310. [DOI] [PubMed] [Google Scholar]
- 8.Ben David Y, Chetrit A, Hirsh-Yechezkel G, Friedman E, Beck BD, Beller U, et al. Effect of BRCA mutations on the length of survival in epithelial ovarian tumors. J Clin Oncol. 2002;20:463–466. doi: 10.1200/JCO.2002.20.2.463. [DOI] [PubMed] [Google Scholar]
- 9.Gallagher DJ, Konner JA, Bell-McGuinn KM, Bhatia J, Sabbatini P, Aghajanian CA, et al. Survival in epithelial ovarian cancer: a multivariate analysis incorporating BRCA mutation status and platinum sensitivity. Ann Oncol. 2011;22:1127–1132. doi: 10.1093/annonc/mdq577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukhopadhyay A, Plummer ER, Elattar A, Soohoo S, Uzir B, Quinn JE, et al. Clin-icopathological features of homologous recombination-deficient epithelial ovarian cancers: sensitivity to PARP inhibitors, platinum, and survival. Cancer Res. 2012;72:5675–5682. doi: 10.1158/0008-5472.CAN-12-0324. [DOI] [PubMed] [Google Scholar]
- 11.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 12.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 13.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 14.Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z, et al. The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011;25:2158–2172. doi: 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bajrami I, Frankum JR, Konde A, Miller RE, Rehman FL, Brough R, et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 2014;74:287–297. doi: 10.1158/0008-5472.CAN-13-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joshi PM, Sutor SL, Huntoon CJ, Karnitz LM. Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. J Biol Chem. 2014;289:9247–9253. doi: 10.1074/jbc.M114.551143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott CL, Swisher EM, Kaufmann SH. Poly (ADP-ribose) polymerase inhibitors: recent advances and future development. J Clin Oncol. 2015;33:1397–1406. doi: 10.1200/JCO.2014.58.8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JM, Ledermann JA, Kohn EC. PARP inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Ann Oncol. 2014;25:32–40. doi: 10.1093/annonc/mdt384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 20.Gelmon KA, Tischkowitz M, Mackay H, Swenerton K, Robidoux A, Tonkin K, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–861. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- 21.Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010;28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 22.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15:852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 23.Jones P, Altamura S, Boueres J, Ferrigno F, Fonsi M, Giomini C, et al. Discovery of 2-{4-[(3S)-piperidin-3-yl]phenyl}-2H-indazole-7-carboxamide (MK-4827): a novel oral poly(ADP-ribose)polymerase (PARP) inhibitor efficacious in BRCA-1 and -2 mutant tumors. J Med Chem. 2009;52:7170–7185. doi: 10.1021/jm901188v. [DOI] [PubMed] [Google Scholar]
- 24.Sandhu SK, Schelman WR, Wilding G, Moreno V, Baird RD, Miranda S, et al. The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet Oncol. 2013;14:882–892. doi: 10.1016/S1470-2045(13)70240-7. [DOI] [PubMed] [Google Scholar]
- 25.Mukhopadhyay A, Elattar A, Cerbinskaite A, Wilkinson SJ, Drew Y, Kyle S, et al. Development of a functional assay for homologous recombination status in primary cultures of epithelial ovarian tumor and correlation with sensitivity to poly(ADP-ribose) polymerase inhibitors. Clin Cancer Res. 2010;16:2344–2351. doi: 10.1158/1078-0432.CCR-09-2758. [DOI] [PubMed] [Google Scholar]
- 26.Shah MM, Dobbin ZC, Nowsheen S, Wielgos M, Katre AA, Alvarez RD, et al. An ex vivo assay of XRT-induced Rad51 foci formation predicts response to PARP-inhibition in ovarian cancer. Gynecol Oncol. 2014;134:331–337. doi: 10.1016/j.ygyno.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weroha SJ, Becker MA, Enderica-Gonzalez S, HSC, OAL, MMJ, et al. Tumorgrafts as in vivo surrogates for women with ovarian cancer. Clin Cancer Res. 2014 doi: 10.1158/1078-0432.CCR-13-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel A, Sarkaria J, Kaufmann SH. Nonhomologous end-joining drives PARP inhibitor synthetic lethality in homologous recombination-deficient cells. Proc Natl Acad Sci U S A. 2011;108:3406–3411. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaufmann SH, Svingen PA, Gore SD, Armstrong DK, Cheng YC, Rowinsky EK. Altered formation of topotecan-stabilized topoisomerase I-DNA adducts in human leukemia cells. Blood. 1997;89:2098–2104. [PubMed] [Google Scholar]
- 30.Kaufmann SH. Reutilization of immunoblots after chemiluminescent detection. Anal Biochem. 2001;296:283–286. doi: 10.1006/abio.2001.5313. [DOI] [PubMed] [Google Scholar]
- 31.Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 32.SAS Institute I, SAS Institute I. SAS/STAT User Guide 9.0. SAS Institute Inc; Carey, NC: 2005. [Google Scholar]
- 33.Littell RC, Pendergast J, Natarajan R. Modelling covariance structure in the analysis of repeated measures data. Stat Med. 2000;19:1793–1819. doi: 10.1002/1097-0258(20000715)19:13<1793::aid-sim482>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 34.Williams GH, Stoeber K. Cell cycle markers in clinical oncology. Curr Opin Cell Biol. 2007;19:672–679. doi: 10.1016/j.ceb.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Sakai W, Swisher EM, Jacquemont C, Chandramohan KV, Couch FJ, Langdon SP, et al. Functional restoration of BRCA2 protein by secondary BRCA2 mutations in BRCA2-mutated ovarian carcinoma. Cancer Res. 2009;69:6381–6386. doi: 10.1158/0008-5472.CAN-09-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilcoxen Keith M, Brooks David G, Tiruchinapalli Dhanrajan, Anderson Nathan, Donaldson Rodney, Nivens McIver, Cook Charlie, Khor Tin, Lu Bin, De Oliveira Elizabeth, Hawley Erin, Bruckheimer Elizabeth. The PARP inhibitor niraparib demonstrates synergy with chemotherapy in treatment of patient derived Ewing's sarcoma tumorGraft models. Mol Cancer Ther; Proceedings of the AACR-NCI-EORTC International Conference: Molecular Targets and Cancer Therapeutics; Oct 19–23, 2013; Boston, MA. Philadelphia (PA): AACR; 2013. Abstract nr A258. [Google Scholar]
- 37.Williamson CT, Kubota E, Hamill JD, Klimowicz A, Ye R, Muzik H, et al. Enhanced cytotoxicity of PARP inhibition in mantle cell lymphoma harbouring mutations in both ATM and p53. EMBO Mol Med. 2012;4:515–527. doi: 10.1002/emmm.201200229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Aroumougame A, Lobrich M, Li Y, Chen D, Chen J, et al. PTIP associates with Artemis to dictate DNA repair pathway choice. Genes Dev. 2014;28:2693–2698. doi: 10.1101/gad.252478.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chun J, Buechelmaier ES, Powell SN. Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol Cell Biol. 2013;33:387–395. doi: 10.1128/MCB.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Min A, Im SA, Yoon YK, Song SH, Nam HJ, Hur HS, et al. RAD51C-deficient cancer cells are highly sensitive to the PARP inhibitor olaparib. Mol Cancer Ther. 2013;12:865–877. doi: 10.1158/1535-7163.MCT-12-0950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.