Abstract
Pseudomonas chlororaphis subsp. aurantiaca StFRB508 regulates phenazine production through N-acyl-l-homoserine lactone (AHL)-mediated quorum sensing. Two sets of AHL-synthase and AHL-receptor genes, phzI/phzR and aurI/aurR, have been identified from the incomplete draft genome of StFRB508. In the present study, the complete genome of StFRB508, comprising a single chromosome of 6,997,933 bp, was sequenced. The complete genome sequence revealed the presence of a third quorum-sensing gene set, designated as csaI/csaR. An LC-MS/MS analysis revealed that StFRB508 produced six types of AHLs, with the most important AHL being N-(3-hydroxyhexanoyl)-l-homoserine lactone (3-OH-C6-HSL). PhzI mainly catalyzed the biosynthesis of 3-OH-C6-HSL, while AurI and CsaI catalyzed that of N-hexanoyl-l-homoserine lactone and N-(3-oxohexanoyl)-l-homoserine lactone, respectively. A mutation in phzI decreased phenazine production, whereas that in aurI or csaI did not. A phzI aurI csaI triple mutant (508ΔPACI) did not produce phenazine. Phenazine production by 508ΔPACI was stimulated by exogenous AHLs and 3-OH-C6-HSL exerted the strongest effects on phenazine production at the lowest concentration tested (0.1 μM). The plant protection efficacy of 508ΔPACI against an oomycete pathogen was lower than that of wild-type StFRB508. These results demonstrate that the triplicate quorum-sensing system plays an important role in phenazine production by and the biocontrol activity of StFRB508.
Keywords: Pseudomonas chlororaphis, quorum sensing, acylhomoserine lactone, phenazine, biocontrol
Phenazine and its derivatives are orange-pigmented heterocyclic products of bacterial secondary metabolism, and are known for their broad-spectrum antifungal activity (12, 18). A wide variety of phenazine derivatives, such as phenazine-1-carboxylic acid (PCA) and phenazine-1-carboxamide (PCN), are produced by various Gram-negative and Gram-positive genera, including Brevibacterium, Burkholderia, Pseudomonas, and Streptomyces (18). The core phenazine biosynthetic gene cluster contains seven genes (phzABCDEFG) and is present in several Pseudomonas species such as Pseudomonas chlororaphis, P. aeruginosa, and P. fluorescens (12). Phenazine-producing Pseudomonas species have been studied as biocontrol agents against many plant diseases (27). P. chlororaphis PCL1391 produces PCN and controls tomato foot and root rot caused by Fusarium oxysporum (3). PCN and PCA produced by P. aeruginosa PNA1 are involved in the biocontrol of Pythium myriotylum on cocoyam (24).
Quorum sensing is a bacterial cell-cell communication process and modulates cell density-dependent gene expression (16). N-Acyl-l-homoserine lactone (AHL) has been identified as a signaling compound involved in quorum-sensing in many Gram-negative bacteria (16). The LuxI/LuxR system is used by many Gram-negative bacteria to control the expression of genes regulated by quorum sensing (16). Members of the LuxI protein family catalyze AHL biosynthesis. When AHL concentrations reach a threshold due to increases in bacterial cell density, members of the LuxR protein family, comprising AHL receptors, bind to AHL, thereby regulating the expression of many genes. Previous studies reported that the production of some antimicrobial compounds, such as carbapenem, prodigiosin, and pyrrolnitrin, is regulated by AHL-mediated quorum sensing (21, 25). The expression of the phenazine biosynthesis gene cluster in fluorescent pseudomonads is activated in the stationary growth phase in a manner that depends on the production of AHLs (17). The quorum sensing-mediated regulation of the phenazine biosynthesis gene cluster in some P. chlororaphis strains has been investigated in detail (3, 7, 27). AHLs are mainly synthesized by PhzI in phenazine-producing P. chlororaphis (27). The AHL receptor protein, PhzR, binds to AHLs and activates the expression of the phenazine biosynthetic gene cluster. Another quorum-sensing system, csaI/csaR, was detected in P. chlororaphis subsp. aureofaciens 30–84, but was not involved in the regulation of the phenazine biosynthetic gene cluster (27). We previously isolated P. chlororaphis subsp. aurantiaca StFRB508 from potato roots and identified two functional quorum-sensing systems, phzI/phzR and aurI/aurR, in the incomplete draft genome sequence of StFRB508 (13). The findings of a thin-layer chromatography (TLC) analysis revealed that PhzI catalyzes the biosynthesis of N-(3-hydroxyhexyanoyl)-l-homoserine lactone (3-OH-C6-HSL), while AurI catalyzes that of N-butanoyl-l-homoserine lactone (C4-HSL) and N-hexanoyl-l-homoserine lactone (C6-HSL). Phenazine production in AHL-negative mutants was stimulated by exogenous AHLs and the most active AHL was 3-OH-C6-HSL. In an attempt to elucidate the quorum-sensing system involved in the regulation of phenazine production in the present study, the complete genome of StFRB508 was sequenced and a third quorum-sensing system was identified. In addition, the concentrations and structures of AHLs produced by StFRB508 were accurately measured and elucidated using LC-MS/MS. We also investigated the biocontrol activity of StFRB508 and its quorum-sensing-deficient mutant.
Materials and Methods
Bacterial strains, compounds, and growth conditions
The bacterial strains used in this study are listed in Table S1. All bacterial strains were grown in Luria-Bertani (LB) medium (10 g L−1 peptone, 5 g L−1 yeast extract, and 5 g L−1 NaCl). Solid bacterial media were prepared by the addition of agar at a final concentration of 1.5%. Antibiotics were added as required at final concentrations of 100 μg mL−1 carbenicillin and 50 μg mL−1 kanamycin. The AHLs used in this study, C6-HSL, N-(3-oxohexanoyl)-l-homoserine lactone (3-oxo-C6-HSL), N-(3-oxooctanoyl)-l-homoserine lactone (3-oxo-C8-HSL), N-(3-oxodecanoyl)-l-homoserine lactone (3-oxo-C10-HSL), and N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12-HSL), were synthesized using a previously described method (2). 3-OH-C6-HSL, N-(3-hydroxyoctanoyl)-l-homoserine lactone (3-OH-C8-HSL), N-(3-hydroxydecanoyl)-l-homoserine lactone (3-OH-C10-HSL), and N-(3-hydroxydodecanoyl)-l-homoserine lactone (3-OH-C12-HSL) were prepared from 3-oxo-C6-HSL, 3-oxo-C8-HSL, 3-oxo-C10-HSL, and 3-oxo-C12-HSL, respectively, via the reduction of the ketone functional group with NaBH4 by a previously described method (19).
Genome sequencing of StFRB508
The single- and paired-end whole-genome shotgun sequencing of StFRB508 was performed using a Roche Genome Sequencer FLX Titanium pyrosequencer (Eurofins Genomics, Tokyo, Japan) as described in a previous study (13). Gap closure was attempted using gap-spanning clones and PCR products. The prediction of putative coding sequences and gene annotation were performed using the Microbial Genome Annotation Pipeline (http://www.migap.org/). Briefly, protein-coding sequences (CDSs) were predicted by the combined use of MetaGeneAnnotator (14), RNAmmer (8), tRNAScan (11), and BLAST (1).
Cloning and disruption of quorum-sensing genes of StFRB508
The phzIR, aurIR, and csaIR coding regions on the StFRB508 genome were amplified with Blend Taq plus DNA polymerase (Toyobo, Osaka, Japan) and specific primers (Table S2). PCR reaction conditions were as follows: 27 cycles at 94°C for 30 s, 60°C for 30 s, and 74°C for 2 min. PCR products were cloned into the pGEM-T easy cloning vector (Promega, Tokyo, Japan). In order to remove the internal sequence of the target genes, sequences upstream and downstream of the target gene were amplified using pGEM-T easy containing constructed plasmids as the template with specific primers for gene deletion (Table S2). The amplified PCR fragments were excised by HindIII digestion and self-ligated. The gene-coding region with a deletion in an internal sequence was excised by EcoRI and inserted into the EcoRI site of the suicide vector, pK18mobsacB (20), in order to create the plasmids for gene deletion. Gene-deletion plasmids were conjugated from Escherichia coli S17-1 λpir (22) using StFRB508 mutants as recipients. The StFRB508 recombinants corresponding to single-crossover events were selected on LB agar containing kanamycin and carbenicillin. Deletion mutants of the quorum-sensing genes in StFRB508 were generated by homologous recombination with sacB selection. The single-crossover mutants of StFRB508 were streaked onto an LB agar plate with 10% (w/v) sucrose in order to select homologous recombinants. The presence of the expected internal gene deletion in StFRB508 was confirmed by PCR.
Assessment of phenazine production
In the agar plate assay, StFRB508 and its mutants were grown on LB agar plates with or without 1% (w/v) monosaccharides such as d-glucose, d-fructose, d-galactose, l-arabinose, and d-xylose (Kanto Chemical, Tokyo, Japan). A sterile toothpick was dipped into a bacterial suspension and inoculated onto new LB agar prepared in 24-well plates. After an incubation at 30°C for 2 d, phenazine production was identified by the presence of orange pigments. Phenazine production was categorized into three levels: (i) high production (deep orange with diffusion), (ii) low production (moderate orange without diffusion), and (iii) no production (white or very pale yellow).
Phenazine production was also evaluated by the broth assay as described previously with modifications (13). StFRB508 and its mutants were grown for 15 h, inoculated into fresh LB medium (1% inoculum) with or without AHLs, and further incubated for 18 h. One milliliter of each culture was placed in a 1.5-mL microtube. After centrifugation for 5 min, the concentration of phenazine in the culture supernatant was evaluated by measuring absorbance at 365 nm (A365). The turbidity of the culture suspension was assessed by measuring optical density at 600 nm (OD600). Phenazine production was evaluated as relative production (A365/OD600), the maximum value of which equaled 100%.
Extraction and identification of AHLs
StFRB508 and its mutants were inoculated into 4 mL of LB medium containing 1% (w/v) glucose and incubated at 30°C for 18 h under shaking conditions at 150 rpm. Full-grown cultures (500 μL) were transferred into 50 mL fresh medium and incubated at 30°C for 24 h. Cells were removed by centrifugation at 10,000×g for 5 min. Culture supernatants were concentrated by evaporation using a rotary evaporator. They were then extracted with a 3-fold volume of ethyl acetate in a separation funnel. The extract was evaporated to dryness using a rotary evaporator and then dissolved in 500 μL dimethylsulfoxide. The chemical structures of extracted AHLs were analyzed by Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) as described previously (15).
Quantification of AHLs by LC-MS/MS
The quantification of AHLs was performed using a triple quadrupole/linear ion trap instrument (LIT) (QTRAP5500; AB SCIEX, Tokyo, Japan) with an electrospray ionization (ESI) source coupled to a UHPLC system (Nexera X2; Shimadzu, Kyoto, Japan) in the multiple reaction monitoring (MRM) analysis mode. Chromatographic separation and MS/MS spectra were recorded as reported previously (15). An MRM analysis was performed at 15 V collision energies for the transitions of m/z 200→102 for C6-HSL, 214→102 for 3-oxo-C6-HSL, 216→102 for 3-OH-C6-HSL, 244→102 for 3-OH-C8-HSL, 272→102 for 3-OH-C10-HSL, and 300→102 for 3-OH-C12-HSL. Data acquisition and analyses were performed with Multi Quant software ver. 3.0.1 (AB SCIEX). The quantification of AHLs was conducted using synthetic standards. Exudate samples were dissolved in 50% aqueous acetonitrile and filtered through a spin column (Ultrafree-MC 0.45 μm filter unit; Millipore, Bedford, MA). An aliquot of filtered 50% aqueous acetonitrile sample solutions was diluted with a volume of either pure 50% acetonitrile or 50% acetonitrile containing known amounts of the AHL standards. The increase in the peak area on the chromatogram corresponded to the amounts of the AHL standards added, thereby enabling the amounts of AHLs in the samples to be estimated. Results were reproduced in three repeated experiments.
Plant disease suppression assays
The biocontrol activity of each strain was evaluated as described previously (23), with slight modifications. Flasks containing 20 g of a 2:1 mixture of vermiculite and peat moss were planted with three cucumber seedlings each and treated with Pythium ultimum MAFF425494. StFRB508 mutants were added to soil as a suspension (4 mL in each flask) of cells washed twice in sterile distilled water to give 2×107 CFU g−1 soil. Control flasks received the same amount of sterile water. Seedlings were covered with 5 g of untreated soil and flasks were sealed with aerated silicon caps. Microcosms were incubated in a growth chamber at 60% relative humidity and 26°C with light for 16 h, followed by an 8-h dark period. No watering was necessary. After a 7-d incubation, biocontrol activity was assessed by counting the surviving plants in each flask. Data in Table 1 represent means from two individual repetitions of the same experiment. Data from both experiments were analyzed for a trial by a treatment interaction using an analysis of variance, which indicated that data from the two independent trials may be pooled. Means were separated using Tukey’s honest significant difference test (at P≤0.05). Statistical analyses were performed using R version 3.0.1 (http://www.r-project.org/).
Table 1.
Suppression of Pythium-induced damping-off and root rot in cucumber by Pseudomonas chlororaphis subsp. aurantiaca StFRB508 and 508ΔPACI.
| Bacterial strain addeda | Pythium addeda | Surviving plants per flask (%)b |
|---|---|---|
| None | − | 100 a |
| StFRB508 | − | 100 a |
| 508ΔPACI | − | 100 a |
| None | + | 21 c |
| StFRB508 | + | 63 b |
| 508ΔPACI | + | 25 c |
The Pseudomonas strain was added at 2×107 CFU g−1 soil to100-mL flasks (50 mL of soil per flask), after planting three 48-h-old, sterile-grown cucumber seedlings per flask. P. ultimum was added as a millet-seed inoculum at 2.5 g kg−1 of vermiculite before planting. Plants were harvested after 7 d.
Data represent the average of 10 replicates (flasks containing three cucumber plants) per treatment without P. ultimum and 16 replicates per treatment with P. ultimum. Means within the same column followed by different letters (a–c) are significantly different (P<0.05) according to Tukey’s HSD test.
Nucleotide sequence accession number
The complete genome sequence of P. chlororaphis subsp. aurantiaca StFRB508 has been deposited in the DDBJ/ENA/GenBank databases under accession no. AP014623.
Results and Discussion
Complete genome sequencing of StFRB508
We previously reported the incomplete draft genome sequence of StFRB508 (13). Two pairs of luxI and luxR homologues, designated as phzIR and aurIR, were detected in the draft genome of StFRB508. However, the draft genome sequence of StFRB508 still contains numerous long sequencing gaps. In order to obtain the complete genome sequence of StFRB508, 896,121 paired-end reads with an average read length of 154 bases were re-assembled by applying Eurofins in silico Gap Closure pipeline with manual inspection. Consequently, reads were assembled into 34 large contigs (>2,000 bp). The results of PCR for closing assembly gaps revealed that the complete genome of StFRB508 comprised a single circular chromosome of 6,997,933 bp with an average G+C content of 62.82% (Fig. S1). The genome contained 6,309 protein-coding, 16 rRNA, and 67 tRNA genes. In order to elucidate complete quorum-sensing gene networks, we searched for additional AHL-synthase gene homologues in the complete genome. One predicted an ORF (PCAU_2446), which encoded 219 amino acids, showing high identity (more than 94%) to the AHL synthase CsaI from P. chlororaphis subsp. aureofaciens 30–84 (27). A putative luxR homologous gene, designated as csaR (PCAU_2447), was mapped upstream of csaI as well as 30–84. Furthermore, a previous study reported that HdtS from Pseudomonas fluorescens F113 is a third protein family capable of AHL biosynthesis (9). The hdtS homologue was also detected in the complete genome of StFRB508 (PCAU_6164). The complete genome sequence of another P. chlororaphis subsp. aurantiaca strain JD37 has also been described (6). Therefore, we searched quorum sensing-related genes in the complete genome of JD37. Based on the results obtained, phzIR, aurIR, csaIR, and hdtS genes were found to be shared between StFRB508 and JD37. These results demonstrate that these quorum-sensing genes are conserved in P. chlororaphis subsp. aurantiaca.
Quorum sensing-regulated phenazine biosynthesis in StFRB508
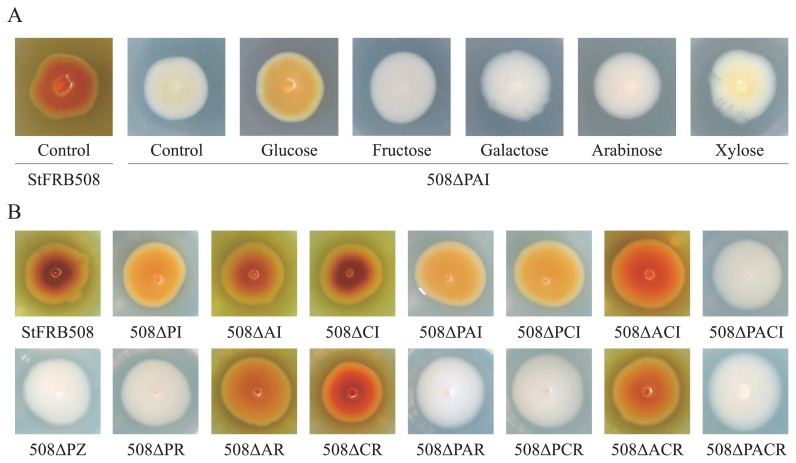
In order to elucidate the relationship between quorum sensing-related genes and phenazine production, we constructed multiple gene deletion mutants using a non-marker mutagenesis technique. Our previous findings revealed that phenazine production was not detected in the culture supernatant of the phzI aurI double mutant (13). In the present study, we examined phenazine production using the non-marker phzI aurI double mutant (508ΔPAI) on LB agar. Similar to our previous findings, no significant amount of phenazine production was observed in 508ΔPAI (Fig. 1A). With respect to growth conditions that favor phenazine production, Yuan et al. demonstrated that glucose supplementation enhanced phenazine production in Pseudomonas sp. M-18Q (26). In order to investigate the effects of monosaccharides on phenazine production by 508ΔPAI, glucose, fructose, galactose, arabinose, and xylose were added to LB agar at a concentration of 1% (w/v). 508ΔPAI showed low-level phenazine production on LB agar containing glucose, suggesting that the double mutation in phzI and aurI was not sufficient to abolish phenazine production. This effect was not observed with the addition of other monosaccharides (Fig. 1A).
Fig. 1.
Phenazine production in StFRB508 and its mutants when grown on LB agar plates. (A) The 508ΔPAI mutant strain was inoculated on LB agar containing 1% (w/v) glucose, fructose, galactose, arabinose, and xylose. StFRB508 and 508ΔPAI were inoculated on LB agar without monosaccharides as a control. (B) A series of mutants of StFRB508 were inoculated on LB agar containing 1% (w/v) glucose. After an incubation at 30°C for 2 d, phenazine production was identified by the presence of orange pigments.
We then examined phenazine production by all constructed mutants on LB agar containing glucose (Fig. 1B). In cases of AHL-synthase gene mutants, mutations in aurI and csaI did not reduce phenazine production. The mutation in phzI induced a slight reduction in phenazine production. The triple mutant of phzI, aurI, and csaI (508ΔPACI) did not produce phenazine, which was similar to 508ΔPZ as a negative control. The AHLs produced by PhzI were assumed to strongly stimulate phenazine production, whereas those produced by AurI and CsaI only yielded a slight stimulation. Although Pseudomonas sp. CMR12a and P. chlororaphis subsp. aureofaciens 30–84 have a second quorum-sensing system in addition to the phzI/phzR system, phenazine production was only regulated by the AHLs produced by PhzI (5, 27). This is the first study to show the cross-regulation of phenazine production by AHLs produced by the three independent AHL synthases. In the case of AHL-receptor gene mutants, mutations in aurR and csaR did not affect phenazine production. However, a single mutation in phzR completely diminished phenazine production by StFRB508. These results suggest that the expression of the phenazine biosynthesis gene cluster is regulated by the PhzR receptor alone. A previous study reported that the csaI/csaR quorum-sensing system regulates the cell surface properties of P. chlororaphis subsp. aureofaciens 30–84 (27). However, visual differences in colony morphologies were not confirmed between StFRB508 and mutants of the aurI/aurR or csaI/csaR quorum-sensing system.
Identification of the structures of AHLs produced by StFRB508 and its mutants
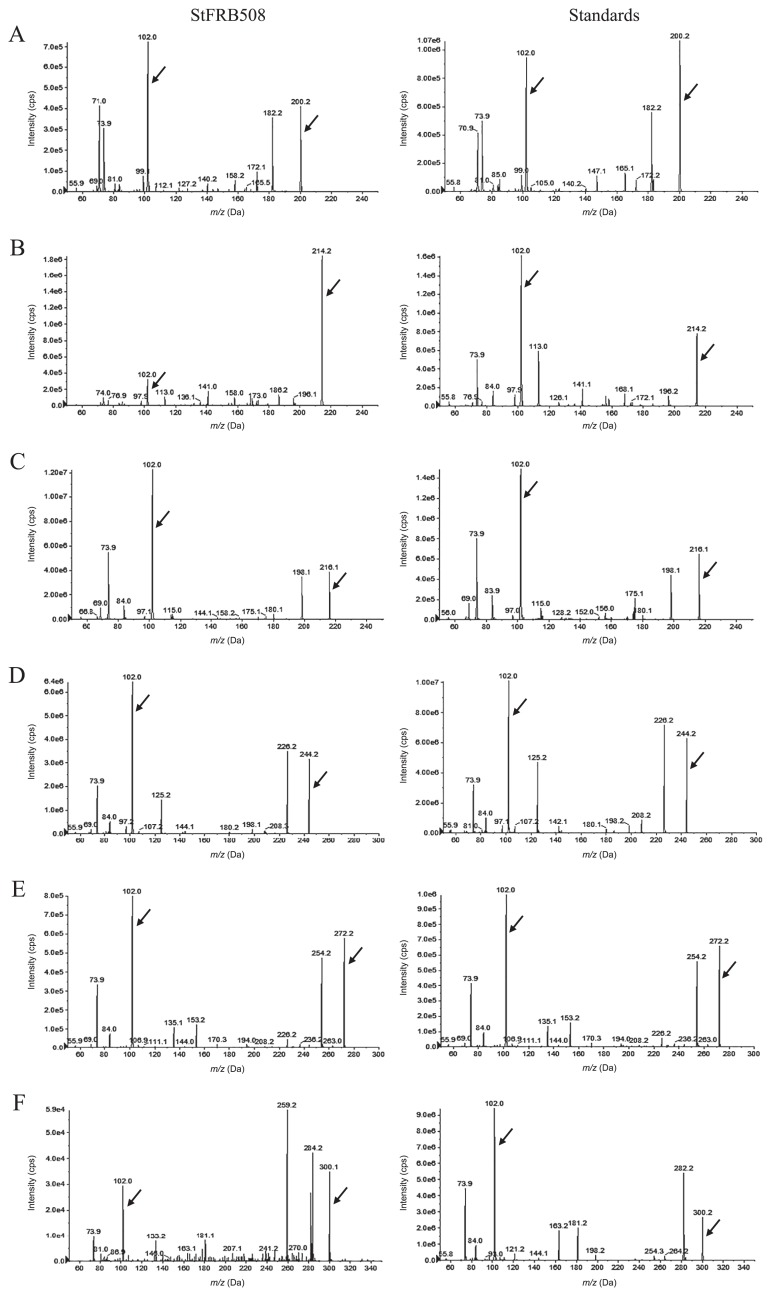
We previously identified the structures of AHLs produced by PhzI and AurI using a TLC analysis combined with those produced by AHL-reporter strains (13). Our previous findings demonstrated that PhzI catalyzed the biosynthesis of 3-OH-C6-HSL, while AurI catalyzed that of C4-HSL and C6-HSL. However, the use of a TLC analysis to detect the structure of AHL is too simple, and difficulties are associated with elucidating specific structures or small amounts of AHLs. Therefore, in the present study, the chemical structures of AHLs extracted from StFRB508 and its mutants were analyzed by LC-MS/MS. The mass spectra of AHLs extracted from StFRB508 culture supernatants and AHL standards are shown in Fig. 2. Six AHLs—C6-HSL, 3-oxo-C6-HSL, 3-OH-C6-HSL, 3-OH-C8-HSL, 3-OH-C10-HSL, and 3-OH-C12-HSL—were detected in the StFRB508 culture supernatant. In order to elucidate the AHL structures produced by three AHL-synthase mutants, the culture supernatants of 508ΔACI (including the phzI gene only), 508ΔPCI (including the aurI gene only), and 508ΔPAI (including the csaI gene only) were prepared and assayed using LC-MS/MS. The culture extract of 508ΔACI contained four AHLs, 3-OH-C6-HSL, 3-OH-C8-HSL, 3-OH-C10-HSL, and 3-OHC12-HSL (Fig. S2). The culture supernatants of 508ΔPCI and 508ΔPAI contained two AHLs, C6-HSL and 3-oxo-C6-HSL (Fig. S3 and 4). We previously demonstrated using a TLC analysis that AurI produced C4-HSL instead of 3-oxo-C6-HSL (13). C4-HSL has also been detected in the culture supernatants of other P. chlororaphis strains using a TLC analysis (4, 10). Since the Rf values of C4-HSL and 3-oxo-C6-HSL were similar, we assumed that C4-HSL and 3-oxo-C6-HSL were not separated clearly by TLC. The csaI/csaR quorum-sensing system was reported for the first time from P. chlororaphis subsp. aureofaciens 30–84 (27). However, the structures of AHLs produced by CsaI were not elucidated in that study. As described above, CsaI from StFRB508 showed greater similarity to that from 30–84. Therefore, CsaI from Pseudomonas chlororaphis strains was assumed to commonly produce C6-HSL and 3-oxo-C6-HSL.
Fig. 2.
Mass spectra of AHLs extracted from the cell-free supernatant of StFRB508 (left panel) and AHL standards (right panel). After fractionation by reverse-phase HPLC, the ESI-MS/MS fragment peaks of AHLs were analyzed. All corresponding peaks for respective C6-HSL (A; m/z 200), 3-oxo-C6-HSL (B; m/z 214), 3-OH-C6-HSL (C; m/z 216), 3-OH-C8-HSL (D; m/z 244), 3-OH-C10-HSL (E; m/z 272), and 3-OH-C12-HSL (F; m/z 300) along with the product ion peaks (m/z 102) are marked by arrows.
Effects of AHL structures on phenazine production in AHL-deficient mutants
AHLs produced by wild-type StFRB508 were extracted after a 24-h cultivation and quantified by the LC-MS/MS system (Table 2). StFRB508 mainly produced 3-hydroxy-substituted AHLs, and 3-OH-C6-HSL showed the highest concentration (approximately 6 μM) among the six AHLs produced. The concentration and composition ratio of 3-hydroxy-substituted AHLs in 508ΔACI (for PhzI) were similar to those of wild-type StFRB508. In contrast, 508ΔPCI (for AurI) and 508ΔPAI (for CsaI) produced higher concentrations of C6-HSL and 3-oxo-C6-HSL than StFRB508. Since AHLs are produced by AHL synthase from S-adenosyl-l-methionine (SAM) and an acylated acyl carrier protein (16), the inactivation of PhzI caused an excessive supply of SAM toward AurI or CsaI in 508ΔPCI and 508ΔPAI. Although AurI and CsaI both produce C6-HSL and 3-oxo-C6-HSL, the composition ratio of the two AHLs is opposite in the products of AurI and CsaI.
Table 2.
Quantification of AHLs produced by StFRB508 and its mutants.
| Strains | StFRB508 | 508ΔACI | 508ΔPCI | 508ΔPAI | 508ΔPACI |
|---|---|---|---|---|---|
|
| |||||
| Functional AHL synthase | PhzI, AurI, CsaI | PhzI | AurI | CsaI | None |
| C6-HSL | 218±43a | N.D.b | 371±86 | 4177±28 | N.D. |
| 3-oxo-C6-HSL | 159±67 | N.D. | 3205±692 | 426±235 | N.D. |
| 3-OH-C6-HSL | 6006±713 | 6653±2506 | N.D. | N.D. | N.D. |
| 3-OH-C8-HSL | 1282±412 | 1946±874 | N.D. | N.D. | N.D. |
| 3-OH-C10-HSL | 768±210 | 809±413 | N.D. | N.D. | 8±12 |
| 3-OH-C12-HSL | 2±0 | 3±1 | N.D. | N.D. | N.D. |
AHL concentrations are expressed in nM.
N.D., not detected.
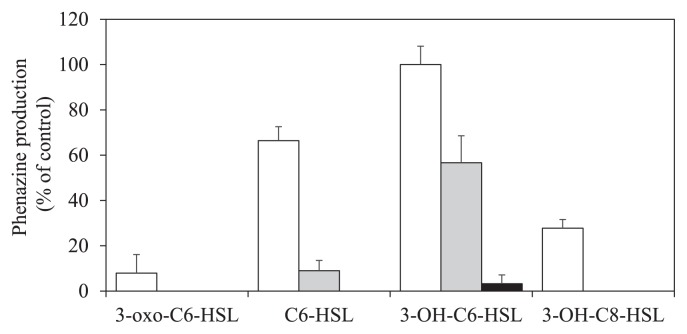
In order to evaluate the effects of AHLs produced by StFRB508 on phenazine production, 508ΔPACI was inoculated into LB medium containing AHLs at a final concentration of 0.1, 1, or 10 μM. The results of the assay for phenazine production are shown in Fig. 3. 3-OH-C6-HSL exerted the strongest effects on phenazine production and also stimulated it at the lowest concentration tested (0.1 μM). Although C6-HSL induced a certain amount of phenazine production at a concentration of 1 μM, this induction level was 6-fold lower than that of 3-OH-C6-HSL. 3-oxo-C6-HSL and 3-OH-C8-HSL slightly induced phenazine production at a concentration of 10 μM. Neither 3-OH-C12-HSL nor 3-OH-C10-HSL stimulated phenazine production at the concentrations tested (data not shown). The null mutant of three AHL synthase genes, 508ΔPACI, produced a small amount of 3-OH-C10-HSL (Table 1). A complete genome analysis of StFRB508 revealed the presence of the hdtS gene homologue, which encodes a third class of AHL synthase. E. coli harboring hdtS from P. fluorescens F113 is capable of synthesizing three kinds of AHLs (9). The small amount of 3-OH-C10-HSL detected in the AHL extracts of 508ΔPACI was assumed to be produced by HdtS. However, since phenazine production was not stimulated in the presence of 3-OH-C10-HSL, the expression of the phenazine biosynthesis gene cluster may not be practically affected by 3-OH-C10-HSL.
Fig. 3.
Phenazine production by 508ΔPACI in LB medium containing AHLs at concentrations of 0.1 (black bars), 1 (gray bars), and 10 μM (white bars). Relative phenazine production was evaluated using the value of A365/OD600; the maximum value obtained for 10 μM 3-OH-C6-HSL was set as 100%. Results were reproduced in three repeated experiments, and error bars indicate standard deviations.
A quorum-sensing mutant exhibited decreased biocontrol efficacy
In our previous study, growth inhibitory activity against the phytopathogenic fungus, F. oxysporum, was attenuated in a phenazine-deficient mutant of StFRB508 in in vitro antagonism tests (13). In order to evaluate the contribution of AHL production to the in vivo biocontrol capacity of StFRB508, we used a cucumber-P. ultimum pathosystem. The addition of strain StFRB508 to pathogen-infested soil increased plant survival by approximately 3-fold, which confirmed this strain as an effective biocontrol agent (Table 2). In contrast, the AHL-deficient mutant, 508ΔPACI, had a significantly impaired capacity to protect cucumber from disease symptoms. In the case of Pseudomonas sp. CMR12a, the quorum-sensing mutant deficient in phenazine production had a significantly lower biocontrol capacity against Rhizoctonia root rot on bean plants than the wild-type (5). Our results also demonstrated that biocontrol activity through phenazine production was regulated by AHL-mediated quorum sensing in StFRB508. Although phenazine production is considered to be a major factor endowing this strain with plant protection efficacy, its relevance to other phenotypes such as bacterial motility and colonization needs to be established in future studies.
Concluding Remarks
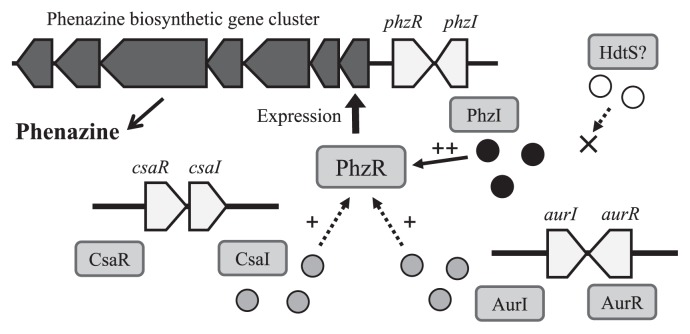
Based on the present results, we propose a mechanism for the regulation of phenazine production by AHL-mediated quorum sensing in StFRB508 (Fig. 4). The results of the AHL quantification demonstrated that of all AHLs, 3-OH-C6-HSL, which exhibited the strongest activity for activating the phenazine biosynthesis gene cluster, was produced by PhzI at the highest concentration in StFRB508. In addition, 3-oxo-C6-HSL and C6-HSL, which were produced by AurI and CsaI, induced only the slight activation of the gene cluster in parallel with phzI/phzR system-mediated regulation. Although AurR and CsaR do not function as response regulators of the phenazine biosynthetic gene cluster, AHLs produced by AurI and CsaI exerted heterologous effects on the unrelated phzI/phzR quorum-sensing system. The true target genes of the aurI/aurR and csaI/csaR quorum-sensing systems have not yet been elucidated. Regarding phenazine production, unexpected spontaneous mutations in up to two AHL-synthase genes may not affect phenazine production. AHLs produced by AurI and CsaI may function as backup signals in the phzI/phzR quorum-sensing system in StFRB508.
Fig. 4.
Schematic representation of the phenazine biosynthetic gene cluster by quorum sensing in StFRB508. Pentagons show the orientation and size of the genes. AHLs produced by PhzI (black circles) or AurI/CsaI (gray cycles), strongly (++) or weakly (+) bind to the PhzR receptor protein, respectively. The PhzR-AHL complex binds to the promoter and induces the expression of the phenazine biosynthetic gene. The putative AHL products by HdtS (open cycles) do not bind to the PhzR receptor.
Supplementary Information
Acknowledgements
We gratefully thank Dr. Satoshi Ito and Mr. Muneki Ichikawa from Utsunomiya University for synthesizing AHL standards. We thank Ms. Nanase Ichimura from Utsunomiya University for her technical assistance. This work was supported by JSPS KAKENHI Grant Number JP16K07656.
References
- 1.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Chhabra S.R., Harty C., Hooi D.S., Daykin M., Williams P., Telford G., Pritchard D.I., Bycroft B.W. Synthetic analogues of the bacterial signal (quorum sensing) molecule N-(3-oxododecanoyl)-l-homoserine lactone as immune modulators. J Med Chem. 2003;46:97–104. doi: 10.1021/jm020909n. [DOI] [PubMed] [Google Scholar]
- 3.Chin-A-Woeng T.F., Bloemberg G.V., van der Bij A.J., van der Drift K.M., Schripsema J., Kroon B., Scheffer R.J., Keel C., Bakker P.A., Tichy H.-V. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol Plant-Microbe Interact. 1998;11:1069–1077. [Google Scholar]
- 4.Chin-A-Woeng T.F., van den Broek D., de Voer G., van der Drift K.M., Tuinman S., Thomas-Oates J.E., Lugtenberg B.J., Bloemberg G.V. Phenazine-1-carboxamide production in the biocontrol strain Pseudomonas chlororaphis PCL1391 is regulated by multiple factors secreted into the growth medium. Mol Plant-Microbe Interact. 2001;14:969–979. doi: 10.1094/MPMI.2001.14.8.969. [DOI] [PubMed] [Google Scholar]
- 5.De Maeyer K., D’Aes J., Hua G.K., Perneel M., Vanhaecke L., Noppe H., Hofte M. N-Acylhomoserine lactone quorum-sensing signalling in antagonistic phenazine-producing Pseudomonas isolates from the red cocoyam rhizosphere. Microbiology. 2011;157:459–472. doi: 10.1099/mic.0.043125-0. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Q., Xiao J., Zhou C., Mu Y., Xu B., He Q., Xiao M. Complete genome sequence of the plant growth-promoting rhizobacterium Pseudomonas aurantiaca strain JD37. J Biotechnol. 2014;192:85–86. doi: 10.1016/j.jbiotec.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 7.Khan S.R., Herman J., Krank J., Serkova N.J., Churchill M.E., Suga H., Farrand S.K. N-(3-hydroxyhexanoyl)-L-homoserine lactone is the biologically relevant quormone that regulates the phz operon of Pseudomonas chlororaphis strain 30–84. Appl Environ Microbiol. 2007;73:7443–7455. doi: 10.1128/AEM.01354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagesen K., Hallin P., Rodland E.A., Staerfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laue B.E., Jiang Y., Chhabra S.R., Jacob S., Stewart G.S., Hardman A., Downie J.A., O’Gara F., Williams P. The biocontrol strain Pseudomonas fluorescens F113 produces the Rhizobium small bacteriocin, N-(3-hydroxy-7-cis-tetradecenoyl)homoserine lactone, via HdtS, a putative novel N-acylhomoserine lactone synthase. Microbiology. 2000;146:2469–2480. doi: 10.1099/00221287-146-10-2469. [DOI] [PubMed] [Google Scholar]
- 10.Liu H., He Y., Jiang H., Peng H., Huang X., Zhang X., Thomashow L.S., Xu Y. Characterization of a phenazine-producing strain Pseudomonas chlororaphis GP72 with broad-spectrum antifungal activity from green pepper rhizosphere. Curr Microbiol. 2007;54:302–306. doi: 10.1007/s00284-006-0444-4. [DOI] [PubMed] [Google Scholar]
- 11.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mavrodi D.V., Blankenfeldt W., Thomashow L.S. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol. 2006;44:417–445. doi: 10.1146/annurev.phyto.44.013106.145710. [DOI] [PubMed] [Google Scholar]
- 13.Morohoshi T., Wang W.Z., Suto T., Saito Y., Ito S., Someya N., Ikeda T. Phenazine antibiotic production and antifungal activity are regulated by multiple quorum-sensing systems in Pseudomonas chlororaphis subsp. aurantiaca StFRB508. J Biosci Bioeng. 2013;116:580–584. doi: 10.1016/j.jbiosc.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 14.Noguchi H., Taniguchi T., Itoh T. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 2008;15:387–396. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okutsu N., Morohoshi T., Xie X., Kato N., Ikeda T. Characterization of N-acylhomoserine lactones produced by bacteria isolated from industrial cooling water systems. Sensors. 2015;16:44. doi: 10.3390/s16010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsek M.R., Greenberg E.P. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc Natl Acad Sci USA. 2000;97:8789–8793. doi: 10.1073/pnas.97.16.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierson L.S., III, Wood D.W., Pierson E.A. Homoserine lactone-mediated gene regulation in plant-associated bacteria. Annu Rev Phytopathol. 1998;36:207–225. doi: 10.1146/annurev.phyto.36.1.207. [DOI] [PubMed] [Google Scholar]
- 18.Pierson L.S., III, Pierson E.A. Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotechnol. 2010;86:1659–1670. doi: 10.1007/s00253-010-2509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pomini A.M., Cruz P.L., Gai C., Araujo W.L., Marsaioli A.J. Long-chain acyl-homoserine lactones from Methylobacterium mesophilicum: synthesis and absolute configuration. J Nat Prod. 2009;72:2125–2129. doi: 10.1021/np900043j. [DOI] [PubMed] [Google Scholar]
- 20.Schafer A., Tauch A., Jager W., Kalinowski J., Thierbach G., Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt S., Blom J.F., Pernthaler J., Berg G., Baldwin A., Mahenthiralingam E., Eberl L. Production of the antifungal compound pyrrolnitrin is quorum sensing-regulated in members of the Burkholderia cepacia complex. Environ Microbiol. 2009;11:1422–1437. doi: 10.1111/j.1462-2920.2009.01870.x. [DOI] [PubMed] [Google Scholar]
- 22.Simon R., Priefer U., Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol. 1983;1:784–791. [Google Scholar]
- 23.Takeuchi K., Yamada K., Haas D. ppGpp controlled by the Gac/Rsm regulatory pathway sustains biocontrol activity in Pseudomonas fluorescens CHA0. Mol Plant-Microbe Interact. 2012;25:1440–1449. doi: 10.1094/MPMI-02-12-0034-R. [DOI] [PubMed] [Google Scholar]
- 24.Tambong J.T., Höfte M. Phenazines are involved in biocontrol of Pythium myriotylum on cocoyam by Pseudomonas aeruginosa PNA1. Eur J Plant Pathol. 2001;107:511–521. [Google Scholar]
- 25.Thomson N., Crow M., McGowan S., Cox A., Salmond G. Biosynthesis of carbapenem antibiotic and prodigiosin pigment in Serratia is under quorum sensing control. Mol Microbiol. 2000;36:539–556. doi: 10.1046/j.1365-2958.2000.01872.x. [DOI] [PubMed] [Google Scholar]
- 26.Yuan L.-L., Li Y.-Q., Wang Y., Zhang X.-H., Xu Y.-Q. Optimization of critical medium components using response surface methodology for phenazine-1-carboxylic acid production by Pseudomonas sp. M-18Q. J Biosci Bioeng. 2008;105:232–237. doi: 10.1263/jbb.105.232. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z., Pierson L.S., III A second quorum-sensing system regulates cell surface properties but not phenazine antibiotic production in Pseudomonas aureofaciens. Appl Environ Microbiol. 2001;67:4305–4315. doi: 10.1128/AEM.67.9.4305-4315.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.