Abstract
Background
Copy number variations (CNVs) are important and widely distributed in the genome. CNV detection opens a new avenue for exploring genes associated with complex traits in humans, animals and plants. Herein, we present a genome-wide assessment of CNVs that are potentially associated with milk composition traits in dairy cattle.
Results
In this study, CNVs were detected based on whole genome re-sequencing data of eight Holstein bulls from four half- and/or full-sib families, with extremely high and low estimated breeding values (EBVs) of milk protein percentage and fat percentage. The range of coverage depth per individual was 8.2–11.9×. Using CNVnator, we identified a total of 14,821 CNVs, including 5025 duplications and 9796 deletions. Among them, 487 differential CNV regions (CNVRs) comprising ~8.23 Mb of the cattle genome were observed between the high and low groups. Annotation of these differential CNVRs were performed based on the cattle genome reference assembly (UMD3.1) and totally 235 functional genes were found within the CNVRs. By Gene Ontology and KEGG pathway analyses, we found that genes were significantly enriched for specific biological functions related to protein and lipid metabolism, insulin/IGF pathway-protein kinase B signaling cascade, prolactin signaling pathway and AMPK signaling pathways. These genes included INS, IGF2, FOXO3, TH, SCD5, GALNT18, GALNT16, ART3, SNCA and WNT7A, implying their potential association with milk protein and fat traits. In addition, 95 CNVRs were overlapped with 75 known QTLs that are associated with milk protein and fat traits of dairy cattle (Cattle QTLdb).
Conclusions
In conclusion, based on NGS of 8 Holstein bulls with extremely high and low EBVs for milk PP and FP, we identified a total of 14,821 CNVs, 487 differential CNVRs between groups, and 10 genes, which were suggested as promising candidate genes for milk protein and fat traits.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-017-3636-3) contains supplementary material, which is available to authorized users.
Keywords: Copy number variation, Chinese Holstein, Whole genome re-sequencing
Background
In dairy cattle, milk yield and composition are the most important economic traits. Compared with traditional dairy cattle breeding programs, DNA-based marker-assisted selection has obvious advantages to shorten generation interval and enhance the accuracy of selection. There are several main strategies of identifying the genes with large genetic effects on milk production traits, including marker-QTL linkage analysis (LA), candidate gene approach, genome-wide association analysis (GWAS) and next-generation sequencing (NGS). Many studies have been performed to investigate milk production traits in dairy cattle [1–15].
Copy number variations (CNVs) are DNA segments that are 1 kb or larger and present at variable copy number in comparison with a reference genome [16]. CNVs are widely distributed in the genome [17]. As a complementary genetic variant to single nucleotide polymorphisms (SNPs), CNVs have attracted increasing attention in recent years. Compared with the SNP, CNV can affect a larger portion of the genome and cause effects, like changing gene structure and dosage, altering gene regulation and exposing recessive alleles [18]. CNVs are also associated with various diseases [19, 20] and may contribute to a fraction of the missing heritability [21, 22]. Along with the development of large-scale CNV studies in human [16], substantial progress has been made in the CNV identification in domestic animals, including cattle [13, 23–34], dog [35–37], sheep [38], goat [39], chicken [40, 41] and pig [42–44]. So far, there are four mechanisms known to allow formation of CNV, i.e., non-allelic homologous recombination (NAHR), non-homologous end joining (NHEJ), fork stalling and template switching (FoSTeS) and retrotransposition [18, 45]. In addition, previous studies also suggested that segmental duplications (SDs) are one of the catalysts and hotspots for CNV formation [46, 47].
Traditionally, there were two array-based methods for CNV discovery, array comparative genomic hybridization (aCGH) and SNP arrays [48, 49]. Although they promoted the progress of CNV studies, these two array-based methods have limitations [50, 51]. They cannot detect small CNVs [52]. In addition, it is also a great challenge for microarrays to detect CNVs in the SD regions due to insufficient coverage [53] although customized chips can be designed to cover SDs.
Recently, the advent of next-generation sequencing (NGS) technology has sped up the study of CNV [23, 27, 28, 54]. Four basic strategies have been applied for detecting CNVs with NGS data in the 1000 Genomes Project pilot studies [55]. Read pair (RP or paired-end mapping) method [56, 57] analyzes discordant mapping pairs of clone reads or high-throughput sequencing fragments whose distances are different from the normal average insert size. Read-depth (RD) [58–60] analysis detects CNVs based on the read depth-of-coverage, i.e., the density of aligned reads along the chromosomes. A random distribution (Poisson distribution or corrected Poisson distribution) is assumed first in this method. Based on the depth-of-coverage, CNVs are detected with duplication regions showing high coverage, while deletion regions show low coverage. Split-read (SR) [59, 61] analysis can evaluate gapped sequence alignments for CNV detection. This method first splits one read into multiple fragments randomly. Then the first and last fragment aligned along the reference genome respectively. According to whether the fragments align or not, and the locations and directions if aligned, CNVs can be detected. The mechanism of SR is similar to RP to some extent. Sequence assembly (AS) method [62, 63] could discover all kinds of genetic variations theoretically because of its fine-scale working. The direct assembly of short reads without reference genome is called de novo assembly and the general strategy is to reconstruct DNA fragments, i.e., contigs, based on assembling overlapping reads firstly. Then by comparing the assembled fragments to the reference genome, the abnormal genomic regions with discordant copy number (CN) can be identified. Additionally, AS-based methods can also use a reference genome to improve the computational efficiency and contig quality.
RD methods applied in the 1000 Genomes Project data have been shown to predict accurate copy number values due to its capability of high-resolution CNV calls [19]. There have been several approaches based on RD, such as MAQ [52, 64], SegSeq [58], mrFAST [47] and CNVnator [65]. CNVnator can overcome some disadvantages, including unique regions of the genome [52, 58, 64], poor breakpoint resolution [47, 52, 58, 64], and detect different sizes of CNVs, from a few hundred bases to megabases in the whole genome. For CNVs accessible by RD described Abyzov et al. [65], CNVnator has high sensitivity (86 ~ 96%), low false-discovery rate (3% ~ 20%), high genotyping accuracy (93% ~ 95%), and high resolution in breakpoint discovery. In addition, they estimated that at least 11% of all CNV loci involve complex, multi-allelic events, a considerably higher estimate than reported earlier [66].
For the CNV detection in the cattle genome, there have been several studies reported using such methods, including CGH [67, 68], BovineSNP50 Beadchip [32, 69, 70], BovineHD SNP Beadchip [25, 31] and NGS [23, 27–30]. In this study, the objective was to identify candidate genes for milk protein and fat traits of dairy cattle through CNV detection based on NGS data of specific Holstein bulls that have extremely high and low estimated breeding values (EBVs) for milk protein and fat percentages.
Methods
Animals and re-sequencing
Eight proven Holstein bulls with high reliabilities (>0.90) of estimated breeding values (EBVs) for milk protein percentage (PP) and fat percentage (FP), born between 1993 and 1996, were selected from the Beijing Dairy Cattle Center (http://www.bdcc.com.cn/) according to their EBVs for PP and FP. EBVs were calculated based on a multiple trait random regression test-day model using the software RUNGE by the Dairy Data Center of China (http://www.holstein.org.cn/). The bulls were from two half sib families and two full sib families with two bulls in each family. The two bulls in each group showed extremely high and low EBV for milk PP and FP, respectively. The detailed information of the 8 bulls is present in Table 1.
Table 1.
The estimated breeding values and family information about 8 Holstein bulls
| Family | Sample | Relationship | EBV for PP | EBV for FP | Reliability |
|---|---|---|---|---|---|
| 1 | 1 | Full-sib | 0.03 | 0.1 | 0.99 |
| 2 | −0.13 | −0.31 | 0.97 | ||
| 2 | 3 | Full-sib | −0.03 | 0.27 | 0.98 |
| 4 | 0.08 | 0.56 | 0.99 | ||
| 3 | 5 | Half-sib | 0.01 | −0.26 | 0.99 |
| 6 | 0.22 | 0.09 | 0.91 | ||
| 4 | 7 | Half-sib | 0.07 | −0.14 | 0.98 |
| 8 | −0.06 | −0.26 | 0.99 |
Re-sequencing, data filter and sequence alignment
Genomic DNA of each bull was extracted from frozen sperms by a standard phenol-chloroform method [71]. DNA degradation and contamination were monitored on 1% agarose gels and the concentration and purity were assessed on NanoDrop 2000 (Thermo Scientific Inc. Waltham, DE, USA); the high-quality DNAs were then used for library construction. Two paired-end libraries were constructed for each individual, the read length was 2 × 100 bp, and whole genome sequencing was performed using Illumina Hiseq2000 instruments (Illumina Inc., San Diego, CA, USA). All processes were performed according to the standard manufacturer’s protocols. In order to get high-quality data, we removed low-quality reads and those containing primer/adaptor contamination which existing in the raw sequencing data by utilizing NGS QC Toolkit with default parameters [-l 75 -q 30] [72]. After data filtering, we used the Burrows-Wheeler Aligner (BWA) program [73] with default parameters [-A1 –B4] to perform sequence alignment based on the UMD3.1 genome assembly which was retrieved from the UCSC website (http://genome.ucsc.edu/). To save run time during the downstream analysis, we converted the SAM files to BAM files and then sorted and merged them by SAMtools [74].
Detection of CNV
CNVnator was run on merged BAM files with a bin size of 200 bp following the authors’ recommendations [65]. After calling, quality control was performed on the raw CNVs for each bull. The filtering criteria included P-value <0.01 (pval1 calculated using t-test statistics), size >1 kb, and q0 < 0.5. P-value <0.01 means that the region between two calls is not a same CNV and q0 means fraction of mapped reads with zero quality. In addition, the CNVs that overlapped with gaps or unplaced chromosomes (chrUn in UCSC) were removed.
Statistical analysis
According to the EBVs for PP and FP, the 8 Holstein bulls were divided into 2 groups, high-group and low-group, and the differential CNVs between the high and low groups were obtained as the following steps. Here, a differential CNVR describes a CNVR that was segregating within the two populations. Firstly, as for any two CNVs from any two individuals of 4 bulls within each group, they were considered to be common CNVs if they have >30% reciprocal overlap, then we obtained the common CNV regions (CNVRs) by merging the common CNVs across the four individuals in either the high or low groups, respectively. Secondly, after getting the common CNVRs in each group, differential CNVRs were identified between the high and low groups of bulls with extremely high/low PP and FP. To compare our results with previous studies, we used the UCSC liftOver tool [75] to convert the coordinates of CNVRs between UMD3.1 and Btau4.0.
Quantitative PCR validation
Quantitative PCR (qPCR) was used to validate CNVRs detected by CNVnator. A total of 11 CNVRs was randomly chosen. For each CNVR, we firstly determined the best primers after designing multiple pairs of primers because of the uncertainty of the CNVR boundaries using Primer3 webtool (http://bioinfo.ut.ee/primer3-0.4.0/primer3/). To ensure the amplification efficiencies of all pairs of primers, a serial diluted genomic DNA sample from a common cattle was used as template for creating a standard curve of each pair of primer. The Basic Transcription Factor 3 (BTF3) gene was chosen as the control with the assumption that there were two copies of DNA segment in this region [69]. With a total volume of 15 μL reagents in a 96-well plate, qPCR was conducted using SYBR green chemistry in triplicate reactions on LightCycler® 480, Roche. The condition for thermal cycle was as follows: 5 min at 95 °C followed by 45 cycles at 95 °C for 10 s, 60 °C for 10 s and 72 °C for 15 s. The 2-ΔΔCt method was used to calculate the relative copy number for each test region. First, we obtained the average Ct value of three replications of each sample and normalized against the control gene. Then we calculated the ΔCt value between the test sample and reference sample detected with normal status (i.e. two copy numbers) by CNVnator. Finally, a value around 3 or above was considered as gain and a value around 1 or below was considered as loss.
Gene contents and functional annotation
Using the BioMart Database, the genes within the detected CNVRs were retrieved based on UMD3.1 sequence assembly (http://asia.ensembl.org/biomart/martview/). Ensembl genes overlapping with CNVRs completely or partially were considered as copy number variable and selected for further analysis. To provide insight into the functional enrichment of genes picked out above, we carried out annotation analysis, including GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes), using KOBAS 2.0 [76], which annotates an input set of genes with putative pathways and disease relationships based on mapping to genes with known annotation. KOBAS 2.0 accepts ID and cross-species sequence similarity mapping and then performs statistical tests to identify statistically significantly enriched pathways and diseases. KOBAS 2.0 incorporates knowledge across 1327 species from 5 pathway databases (KEGG PATHWAY, PID, BioCyc, Reactome and Panther) and 5 human disease databases (OMIM, KEGG DISEASE, FunDO, GAD and NHGRI GWAS Catalog). All annotated Ensembl genes are used as background. In addition, we compared CNVRs with the reported cattle QTLs for milk PP and FP traits in the Animal QTL database [77].
Results
Sequencing data set statistics and CNV discovery
With Illumina paired-end sequencing technology, we obtained NGS data from the 8 Holstein bulls (Table 2). After we mapped them on the UMD3.1 bovine genome assembly and excluded potential PCR duplicates, the depth of coverage for each individual varied from 8.2× (sample 6) to 11.9× (sample 5). As shown previously, a 4x coverage is sufficient for CNV detection using a RD method [19, 23, 78]. With CNVnator, CNVs were detected for 8 individuals. After quality control, the number of duplication ranged from 687 (sample 6) to 777 (sample 4), and the number of deletion varied from 1091 (sample 1) to 1620 (sample 3) (Table 2). In order to determine how many CNVRs were detected from all 8 bulls, all the CNVs were merged if overlaps were 1 bp or greater, and a total of 6015 CNVRs were obtained. The detailed information about CNVs is shown in Additional file 1: Table S1. From Fig. 1, we can see the CNV landscapes roughly. Although chromosome 1 was the longest, the number of CNVs it contained was not the most in any individual. Chromosome X occupied the most CNVs and simultaneously the largest CNVs.
Table 2.
Summary statistics of sequencing data and CNV of 8 Holstein bulls
| No of bulls | Numbers of mapped reads | Depth | Duplication | Deletion |
|---|---|---|---|---|
| 1 | 257615849 | 9.8 | 743 | 1091 |
| 2 | 246773374 | 9.4 | 708 | 1295 |
| 3 | 237335344 | 9.0 | 705 | 1620 |
| 4 | 252933841 | 9.6 | 777 | 1210 |
| 5 | 312273373 | 11.9 | 756 | 1348 |
| 6 | 215134987 | 8.2 | 687 | 1397 |
| 7 | 249257655 | 9.5 | 706 | 1373 |
| 8 | 251909753 | 9.6 | 705 | 1410 |
Fig. 1.
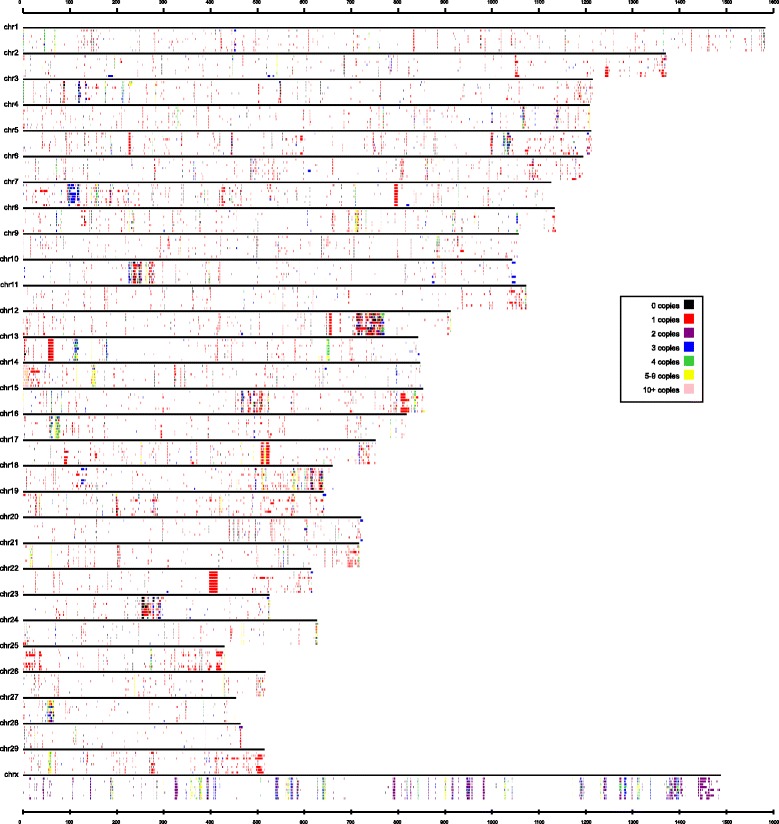
Individualized cattle CNV map. The tracks under every black bar represent the CNVs for sample 1 to sample 8 (in order from top to bottom). The colors for each bar in the animal data se tracks represent the average estimated CN for each CNV as shown in the legend
To confirm the CNVs detected by CNVnator, qPCR based on the relative comparative threshold cycle (CT) method was performed to validate 11 randomly chosen predicted CNVRs representing different types of duplication, deletion and both, on the same 8 samples for whole genome sequencing (Additional file 2: Table S2). It was shown that the validate rates of the 8 samples varied from 57.14% to 90% with an average of 73.04%.
Identification of differential CNVRs between high and low groups
According to the experimental design and filtering standards, we first screened common CNVRs shared by the high and low groups. Then these common CNVRs were excluded from whole CNVRs of high and low group respectively, and 268 and 280 CNVRs as group-specific in high and low group were remained. Finally, a total of 487 differential CNVRs were obtained after merging two group-specific CNVRs if overlaps were 1 bp or greater, covering chromosomes 1-X (Additional file 3: Table S3), which amounted to 8.23 Mb of the cattle genome (Fig. 2). The length of CNVRs varied from 1.6 kb to 275.6 kb with an average of 16.91 kb and a median of 9.4 kb (Table 3) and 31.3% of all CNVRs had sizes ranging from 5 kb to 10 kb (Fig. 3). The CNVRs were divided into 3 categories, i.e. 242 deletions, 229 duplications and 16 both events (Fig. 4). In terms of count and length, deletion and duplication CNVRs were almost similar (242 vs 229, 3.89 Mb vs 3.58 Mb).
Fig. 2.
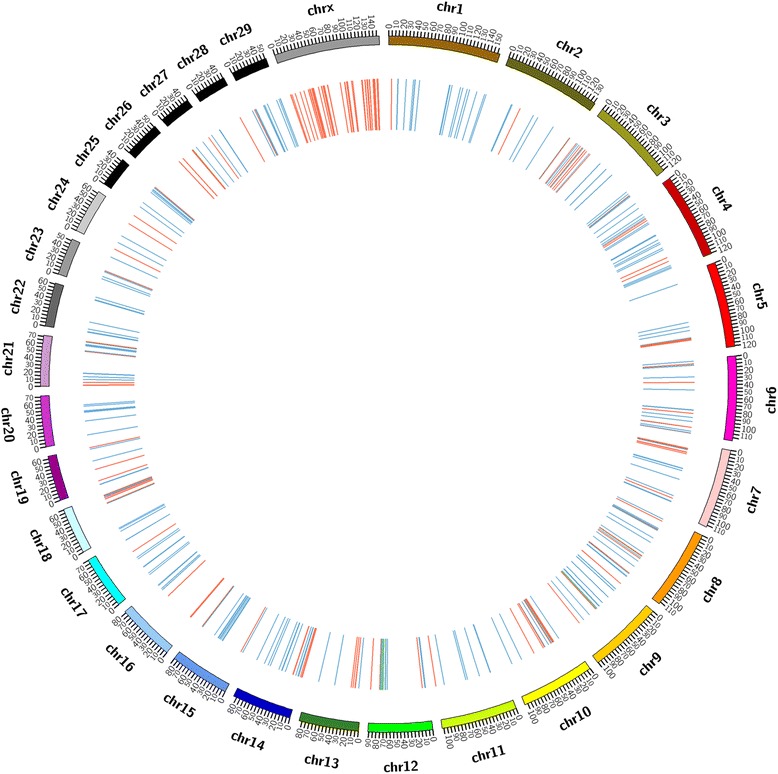
Different CNVRs between high and low group. Based on UMD3.1, 487 CNVRs derived from difference between high and low group were shown in blue (deletion), red (duplication), and green (both)
Table 3.
Characterization of cattle CNVs detected in different studies based on re-sequencing data
| Studies | Summary statistics of CNVRs | |||||
|---|---|---|---|---|---|---|
| Mean (Kb) | Median (Kb) | Min (Kb) | Max (Kb) | Std | Total (Mb) | |
| Bickhart et al. | 43.95 | 23.63 | 10.02 | 510.94 | 54.45 | 55.59 |
| Zhan et al. | 6.98 | 3.8 | 3.17 | 129.97 | 10.29 | 3.63 |
| Stothard et al. | 4.16 | 3.17 | 1.84 | 28.03 | 2.96 | 3.29 |
| This study | 12.47 | 7.2 | 1.2 | 422.8 | 19.82 | 72.02 |
Fig. 3.
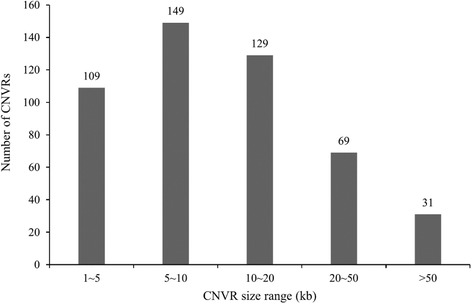
The length and frequency distribution of differential CNVRs
Fig. 4.
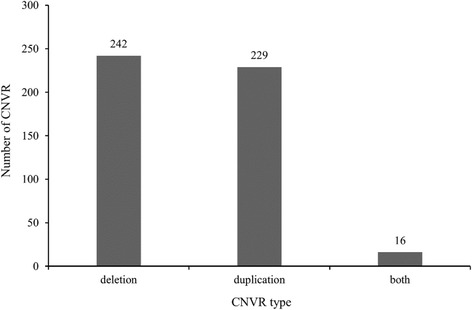
Types of differential CNVRs
Gene contents of differential and group-specific CNVRs
Differential CNVRs
Utilizing BioMart in the Ensembl database (Ensembl Genes 79), we obtained the IDs for the genes that are located within or overlapped with the detected CNVRs. As a result, a total of 235 genes were identified, including 218 protein-coding genes, 5 miRNA genes, 4 snRNA, 3 pseudogenes, 3 rRNA and 2 snoRNA (Additional file 4: Table S4) and 29.98% of the CNVRs encompass 1 or more genes. To know about the biological functions of these genes, GO and KEGG pathway analysis were performed with KOBAS. We found that there were 163 significant GO terms and 8 significant KEGG pathways. GO terms related to protein and lipid metabolism were enriched (p < 0.05), such as long-chain fatty acid binding, protein glycosylation, asymmetric protein localization, glycoprotein biosynthetic process, protein serine/threonine kinase activator activity and negative regulation of protein acetylation. Also, the enriched KEGG pathways included several well-known protein and lipid metabolisms pathways (p < 0.05) such as insulin/IGF pathway-protein kinase B signaling cascade, prolactin signaling pathway and AMPK signaling pathway (Additional file 5: Table S5).
Group-specific CNVRs
Furthermore, we obtained 106 and 139 genes based on the 268 and 280 CNVRs across 4 individuals in the high and low groups, respectively. In high group, there were 2 significant GO terms, including lipid metabolism and glucose metabolic processes, and 1 significant KEGG pathways, a well-known lipid metabolisms pathways (prolactin signaling pathway) (p < 0.05) (Additional file 6: Table S6). In low group, 3 significant GO terms, i.e., dopamine biosynthetic process and insulin receptor binding, and 1 significant KEGG pathway (olfactory transduction) were enriched (p < 0.05) (Additional file 6: Table S6).
Quantitative traits locus overlapped with differential and group-specific CNVRs
Differential CNVRS
We compared the detected differential CNVRs between high and low groups with the previously reported cattle QTL regions for milk production traits (cattle QTL database, http://www.animalgenome.org/cattle/) in order to further study the potential genetic effects of these CNVRs. Finally, 75 QTLs for protein yield, protein percentage, fat yield and fat percentage were found to be overlapped with 95 CNVRs (Additional file 7: Table S7), implying the functional genes within these CNVRs are likely candidates for milk protein and fat traits.
Group-specific CNVRs
In addition, we compared 268 CNVRs in the high group with cattle QTL regions for same traits as above. Totally, 46 QTLs were obtained which overlap with 40 CNVRs (Additional file 8: Table S8). Similarly, we compared 280 CNVRs with corresponding traits in the low group and 55 QTLs overlapped with 52 CNVRs were found (Additional file 8: Table S8).
Discussion
In this study, we detected genome-wide CNVs of 8 Holstein bulls with extremely high and low EBVs for PP and FP based on NGS using CNVnator. We obtained 1834 ~ 2326 CNVs with an average of 2066.5 per bull. Compared with the previous methods based on SNP chip and aCGH of detecting CNV, NGS has many advantages in terms of both CNV numbers and sizes because the sequencing approach overcomes the sensitivity limits in the previous methods, and can more precisely identify CNV boundaries [79]. With the ongoing developments and cost decreases in NGS, the sequencing approaches has become more and more popular for CNV detection. Due to the fact that it was not designed for CNV detection specifically and incomplete coverage of the whole genome, SNP chip was restricted in the application of CNV detection.
Based on the observation of CNV distribution, they were enriched in centromeric and the subtelomeric which is in agreement with the distribution of the cattle SD regions as reported before [80]. The number of CNVs (14,821) identified in this study was more than the reports based on NGS data by Bichkhart et al. (1265) in Angus, Holstein, Hereford and Nelore cattle [23], Stothard et al. (790) in Holstein and Black Angus [28], Zhan et al. (520) in Holstein [27], Boussaha et al. (957) in Holstein, Montbéliarde and Normande [29] and Ben et al. (823) in Holstein [30]. In addition, Jiang et al. detected CNVs based on Illumina BovineSNP50 (99) and BovineHD chips (367) data in Chinese Holstein population [24, 25], which were less than what we detected in this study. After converting 6015 CNVRs to corresponding results based on Btau4.0 using the UCSC liftOver tool with 50% of bases that must remap, 3996 CNVRs of which were successfully converted amounting to about 45.06 Mb. We found that ~80% of the 3996 CNVRs overlapped with those reported by previous investigations [23, 27–30] by 1 bp or greater (Fig. 5), and the largest overlap was ~7.92 Mb of the reported by Bickhart et al. [23]. As for the above inconsistencies, there are likely due to different detection methods and different samples. Bickhart et al. used mrFAST/mrsFAST and WSSD [23], and both Zhan et al. [27] and Stothard et al. [28] performed CNV-seq, and Boussaha et al. [29] used GATK, Pindel, and Ben et al. [30] performed control-FREE. While in this study, we used CNVnator. In addition, different cattle breed with specific genetic background may induce the inconsistencies of number of CNVs and CNVRs among various studies as well. In this study, the qPCR validation rates of the detected CNVs was 57.14% to 90%, which was similar to those reported by Bickhart et al. (82%) [23], Zhan et al. (86%) [27], Stothard et al. (100%) [28] and Yi et al. (91.7%) [41], showing the high accuracy of NGS-based CNV detection. The relatively lower validation rate in this study may be due to the following reasons: (1) false positive in CNV calling even if CNVnator has a low false-discovery rate (3% ~ 20%) [65], (2) primers in qPCR experiment were not the best although we tried multiple primers. As we know, there may be potential SNPs and small INDELs in the genome, and the negative impact of these potential variants could result in the reduced primer efficiency.
Fig. 5.
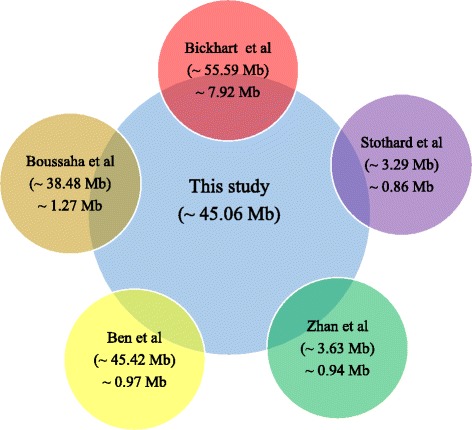
Comparison between 3996 CNVRs in this study and the other cattle CNVR datasets on Btau4.0
Genome wide CNV detection is also a strategy to identify the potential key genes for the traits of interest by mining the genes within the CNVRs in a specific experimental design. Hence, the different CNVRs between the high and low groups of Holstein bulls with extremely high and low EBVs for PP and FP were used for candidate gene identification for milk protein and milk fat. We determined a total of 487 differential CNVRs between the high and low groups, and further found that 235 functional genes were located within these CNVRs. Function analysis showed that the 235 genes were enriched in 163 significant GO terms and 8 significant KEGG pathways. Especially, the processes of long-chain fatty acid binding, protein glycosylation, asymmetric protein localization, glycoprotein biosynthetic process, protein serine/threonine kinase activator activity and negative regulation of protein acetylation were related to protein or lipid metabolism were enriched. Also among the KEGG pathways we detected, insulin/IGF pathway-protein kinase B signaling cascade, prolactin signaling pathway and AMPK signaling pathway are well-known pathways for protein and lipid metabolism [81, 82] and 10 genes involved in the biological process such as cell apoptosis, protein modification, conversion of amino acid and metabolism of fatty acids were included. These were INS, IGF2, FOXO3, TH, SCD5, GALNT18, GALNT16, ART3, SNCA and WNT7A.
The bovine insulin (INS) gene is close to the IGF2 and TH genes. Insulin binding to the insulin receptor (INSR) exerts biological function that maintaining the blood glucose concentration through multiple signaling pathways, such as AMPK, insulin, mTOR and PI3K-Akt signaling pathways, which play critical roles in milk fat and protein synthesis in dairy cattle [83]. The IGF2 gene was related to breast epithelial and stromal cell proliferation in human [84], and over-expression of IGF2 increased breast cancer development [85], thus, it was implied that IGF2 may play an important role in maintaining bovine mammary gland epithelial cell function well. Forkhead box O3 (FOXO3) was also known as FOXO3a, which was considered as a key downstream effector of the well-known signaling pathways for lipid and protein metabolism, i.e. PI3K-Akt, MAPK and Jak-STAT [86, 87]. Tyrosine hydroxylase (TH) is the rate limiting enzyme for converting tyrosine to dopamine which was a crucial regulator of prolactin (PRL) [88]. PRL is essential for mammary gland involution and lactation [88, 89]. The Stearoyl-CoA desaturase 5 (SCD5) gene is located within a known QTL region for milk protein [90] and fat yield [91], but also near to the SNPs significantly associated with milk fat yield, protein yield, fat percentage and protein percentage identified by a previous GWA study [15]. The ADP-ribosyltransferase 3 (ART3) gene encodes the arginine-specific ADP-ribosyltransferase that has impact on cell proliferation and apoptosis etc. [92]. The Wnt family member 7A (WNT7A) gene, as a member of WNT gene family, encodes secreted signaling proteins and is related to suppressing human lung cancer progression [93]. The synuclein alpha (SNCA) gene was found to be associated with Parkinson’s and Alzheimer’s diseases [94, 95]. Among the candidate genes, INS, IGF2, FOXO3, TH, SCD5 were related with milk composition traits according to previous studies, and identification of them in current study confirmed their potential functions. As for the remaining genes, there existed more or less indirect association with milk composition traits. Thereby, to gain further insights into the association of the 10 candidate genes with milk composition traits, we compared the chromosome positions of the 10 genes with the significant SNPs detected by previous GWAS for milk production traits in dairy cattle [4, 5, 7, 10, 15], and found that all genes were located near to multiple significant SNPs for milk protein and fat traits with 0.01 Mb to 9.90 Mb (Additional file 9: Table S9), suggesting their potential associations with milk compositions.
In the study of Xu et al. [13], 34 CNVs were found significantly associated with milk production traits, of which 11 CNVs were included in the differential CNVRs identified in this study, i.e., CNVR45, CNVR46, CNVR47, CNVR189, CNVR190, CNVR200, CNVR201, CNVR202, CNVR203, CNVR399 and CVNR400. Within CNVR400, two candidate genes, INS and IGF2 were enriched. Ben et al. [30] identified two CNVRs associated with milk composition, including one (chr17: 75031000–75158596) with milk fat yield and milk protein yield, and another (chr18: 12381000–12527000) with milk fat yield, and 8 genes were enriched in these two regions, especially the MTHFSD gene within the second CNVR belongs to the folate metabolism gene family and plays critical roles in regulating milk protein synthesis [96]. Although there was no overlap between these CNVRs and ours, two CNVRs in this study were located near to them with 2.51 Mb and 5.83 Mb, respectively. The DEPDC5 gene overlapped with CNVR290 encodes a protein which was a component of the GAP activity toward Rags complex and is involved in mTORC1 pathway [97].
In addition, we found that 95 differential CNVRs detected in this study were overlapped with 75 known QTLs that have been shown to be associated with protein yield, protein percentage, fat yield and fat percentage in dairy cattle (Cattle QTLdb, http://www.animalgenome.org/cgi-bin/QTLdb/BT/index). Eight annotated genes were overlapped with these differential CNVRs (Additional file 7: Table S7).
Conclusions
In conclusion, based on NGS data of 8 Holstein bulls with extremely high and low EBVs for milk PP and FP, we identified a total of 14821 CNVs corresponding to 6015 CNVRs. Of these, 487 differential CNVRs between the high and low groups were obtained. Of note, we further identified 235 annotated genes that were located in or overlapped with these differential CNVRs, including 10 genes significantly enriched for specific biological functions related to protein and lipid metabolism, insulin/IGF pathway-protein kinase B signaling cascade, prolactin signaling pathway and AMPK signaling pathways. These genes included INS, IGF2, FOXO3, TH, SCD5, GALNT18, GALNT16, ART3, SNCA and WNT7A, implying their potential association with milk protein and fat traits.
Additional files
Summary of identified CNVs and CNVRs in the 8 bulls’ genomes. (XLSX 1044 kb)
Primers information and confirmation results of the 11 chosen CNVRs by qPCR analysis. (XLSX 14 kb)
General statistics of 487 differential CNVRs between high and low group based on UMD3.1. (XLSX 28 kb)
The detailed features of genes completely or partial overlapped with differential CNVRs. (XLSX 28 kb)
Functional enrichment of GO and KEGG pathway analysis of genes covered by differential CNVRs. (XLSX 17 kb)
Functional enrichment of GO and KEGG pathway analysis of genes covered by group-specific CNVRs. (XLSX 11 kb)
QTLs covered by differential CNVRs. (XLSX 18 kb)
QTLs covered by group-specific CNVRs. (XLSX 17 kb)
Detailed information on the significant SNPs identified in previous GWAS that are near to the 10 candidate genes identified in this study. (XLSX 17 kb)
Acknowledgements
Not applicable.
Funding
This work was financially supported by the National Science and Technology Programs of China (2013AA102504, 2011BAD28B02, 2014ZX08009-053B), National Natural Science Foundation (31072016, 31472065), Beijing Natural Science Foundation (6152013), the Beijing Dairy Industry Innovation Team (BAIC06-2016), and the Program for Changjiang Scholar and Innovation Research Team in University (IRT1191).
Availability of data and materials
All the re-sequencing data generated in this study have been deposited in National Centre for Biotechnology Information (NCBI) under the BioProject: PRJNA349121, and can be accessed in the Sequence Read Archive (SRA) under the accessions SRX2254497, SRX2254776, SRX2254777, SRX2254778, SRX2254779, SRX2254780, SRX2254813 and SRX2254814.
Authors’ contributions
YG performed bioinformatics and statistical analysis and qPCR experiments, and also was a major contributor to manuscript preparation. JJ and SY performed qRT-PCR experiments and sample collection. YH and GEL participated in data analysis and manuscript preparations. SZ and QZ participated in result interpretation. DS conceived and designed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval
All protocols for collection of the semen samples of Holstein bulls were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at China Agricultural University. Semen samples were collected specifically for this study following standard procedures with the full agreement of the Beijing Dairy Cattle Center who owned the animals.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- Acgh
Array comparative genomic hybridization
- AS
Assembly
- BTF3
Basic transcription factor 3
- BWA
Burrows-wheeler aligner
- CN
Copy number
- CNV
Copy number variation
- CNVR
CNV region
- EBV
Estimated breeding value
- FoSTeS
Fork stalling and template switching
- FP
Fat percentage
- GO
Gene ontology
- GWAS
Genome-wide association analysis
- KEGG
Kyoto encyclopedia of genes and genomes
- LA
Linkage analysis
- NAHR
Non-allelic homologous recombination
- NGS
Next-generation sequencing
- NHEJ
Non-homologous end joining
- PP
Protein percentage
- qPCR
Quantitative PCR
- QTL
Quantitative traits locus
- RD
Read-depth
- RP
Read pair
- SD
Segmental duplications
- SNP
Single nucleotide polymorphisms
- SR
Split-read
Contributor Information
Yahui Gao, Email: 812956123@qq.com.
Jianping Jiang, Email: jiangjianping818@126.com.
Shaohua Yang, Email: yangshaohua309@126.com.
Yali Hou, Email: houyali1210@gmail.com.
George E Liu, Email: george.liu@ars.usda.gov.
Shengli Zhang, Email: zhangslcau@cau.edu.cn.
Qin Zhang, Email: qzhang@cau.edu.cn.
Dongxiao Sun, Email: sundx@cau.edu.cn.
References
- 1.Georges M, Nielsen D, Mackinnon M, Mishra A, Okimoto R, Pasquino AT, et al. Mapping quantitative trait loci controlling milk production in dairy cattle by exploiting progeny testing. Genetics. 1995;139(2):907–20. doi: 10.1093/genetics/139.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daetwyler HD, Schenkel FS, Sargolzaei M, Robinson JA. A genome scan to detect quantitative trait loci for economically important traits in Holstein cattle using two methods and a dense single nucleotide polymorphism map. J Dairy Sci. 2008;91(8):3225–36. doi: 10.3168/jds.2007-0333. [DOI] [PubMed] [Google Scholar]
- 3.Stoop WM, Schennink A, Visker MH, Mullaart E, van Arendonk JA, Bovenhuis H. Genome-wide scan for bovine milk-fat composition. I. Quantitative trait loci for short- and medium-chain fatty acids. J Dairy Sci. 2009;92(9):4664–75. doi: 10.3168/jds.2008-1966. [DOI] [PubMed] [Google Scholar]
- 4.Kolbehdari D, Wang Z, Grant JR, Murdoch B, Prasad A, Xiu Z, et al. A whole genome scan to map QTL for milk production traits and somatic cell score in Canadian Holstein bulls. J Anim Breed Genet. 2009;126(3):216–27. doi: 10.1111/j.1439-0388.2008.00793.x. [DOI] [PubMed] [Google Scholar]
- 5.Jiang L, Liu JF, Sun DX, Ma PP, Ding XD, Yu Y, et al. Genome wide association studies for milk production traits in Chinese Holstein population. PLoS One. 2010;5(10):e13661. doi: 10.1371/journal.pone.0013661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mai MD, Sahana G, Christiansen FB, Guldbrandtsen B. A genome-wide association study for milk production traits in Danish Jersey cattle using a 50 K single nucleotide polymorphism chip. J Anim Sci. 2010;88(11):3522–8. doi: 10.2527/jas.2009-2713. [DOI] [PubMed] [Google Scholar]
- 7.Schopen GC, Visker MH, Koks PD, Mullaart E, van Arendonk JA, Bovenhuis H. Whole-genome association study for milk protein composition in dairy cattle. J Dairy Sci. 2011;94(6):3148–58. doi: 10.3168/jds.2010-4030. [DOI] [PubMed] [Google Scholar]
- 8.Bouwman AC, Bovenhuis H, Visker MHPW, van Arendonk JAM. Genome wide association of milk fatty acids in Dutch dairy cattle. BMC Genet. 2011;12:43. doi: 10.1186/1471-2156-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olsen HG, Hayes BJ, Kent MP, Nome T, Svendsen M, Larsgard AG, et al. Genome-wide association mapping in Norwegian Red cattle identifies quantitative trait loci for fertility and milk production on BTA12. Anim Genet. 2011;42(5):466–74. doi: 10.1111/j.1365-2052.2011.02179.x. [DOI] [PubMed] [Google Scholar]
- 10.Li C, Sun DX, Zhang SL, Wang S, Wu XP, Zhang Q, et al. Genome Wide Association Study Identifies 20 Novel Promising Genes Associated with Milk Fatty Acid Traits in Chinese Holstein. PLoS One. 2014;9(5):e96186. doi: 10.1371/journal.pone.0096186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Littlejohn MD, Tiplady K, Lopdell T, Law TA, Scott A, Harland C, et al. Expression Variants of the Lipogenic AGPAT6 Gene Affect Diverse Milk Composition Phenotypes in Bos taurus. PLoS One. 2014;9(1):e85757. doi: 10.1371/journal.pone.0085757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buitenhuis B, Janss LL, Poulsen NA, Larsen LB, Larsen MK, Sørensen P. Genome-wide association and biological pathway analysis for milk-fat composition in Danish Holstein and Danish Jersey cattle. BMC Genomics. 2014;15:1112. doi: 10.1186/1471-2164-15-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu LY, Cole JB, Bickhart DM, Hou YL, Song JZ, VanRaden PM, et al. Genome wide CNV analysis reveals additional variants associated with milk production traits in Holsteins. BMC Genomics. 2014;15:683. doi: 10.1186/1471-2164-15-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui XG, Hou YL, Yang SH, Xie Y, Zhang SL, Zhang Y, et al. Transcriptional profiling of mammary gland in Holstein cows with extremely different milk protein and fat percentage using RNA sequencing. BMC Genomics. 2014;15:226. doi: 10.1186/1471-2164-15-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole JB, Wiggans GR, Ma L, Sonstegard TS, Lawlor TJ, Jr, Crooker BA, et al. Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary U.S. Holstein cows. BMC Genomics. 2011;12:408. doi: 10.1186/1471-2164-12-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–8. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 18.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–81. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330(6004):641–6. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, et al. Recent segmental duplications in the human genome. Science. 2002;297(5583):1003–7. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 21.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315(5813):848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henrichsen CN, Chaignat E, Reymond A. Copy number variants, diseases and gene expression. Hum Mol Genet. 2009;18(R1):R1–R8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- 23.Bickhart DM, Hou YL, Schroeder SG, Alkan C, Cardone MF, Matukumalli LK, et al. Copy number variation of individual cattle genomes using next-generation sequencing. Genome Res. 2012;22(4):778–90. doi: 10.1101/gr.133967.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang L, Jiang JC, Wang JY, Ding XD, Liu JF, Zhang Q. Genome-wide identification of copy number variations in Chinese Holstein. PLoS One. 2012;7(11):e48732. doi: 10.1371/journal.pone.0048732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jinag L, Jiang JC, Yang J, Liu X, Wang JY, Wang HF, et al. Genome-wide detection of copy number variations using high-density SNP genotyping platforms in Holsteins. BMC Genomics. 2013;14:131. doi: 10.1186/1471-2164-14-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang LZ, Jia SG, Yang MJ, Xu Y, Li CJ, Sun JJ, et al. Detection of copy number variations and their effects in Chinese bulls. BMC Genomics. 2014;15:480. doi: 10.1186/1471-2164-15-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhan BJ, Fadista J, Thomsen B, Hedegaard J, Panitz F, Bendixen C. Global assessment of genomic variation in cattle by genome resequencing and high-throughput genotyping. BMC Genomics. 2011;12:557. doi: 10.1186/1471-2164-12-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stothard P, Choi JW, Basu U, Sumner-Thomson JM, Meng Y, Liao XP, et al. Whole genome resequencing of Black Angus and Holstein cattle for SNP and CNV discovery. BMC Genomics. 2011;12:559. doi: 10.1186/1471-2164-12-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boussaha M, Esquerré D, Barbieri J, Djari A, Pinton A, Letaief R, et al. Genome-Wide Study of Structural Variants in Bovine Holstein, Montbéliarde and Normande Dairy Breeds. PLoS One. 2015;10(8):e0135931. doi: 10.1371/journal.pone.0135931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ben Sassi N, González-Recio Ó, de Paz-Del Río R, Rodríguez-Ramilo ST, Fernández AI. Associated effects of copy number variants on economically important traits in Spanish Holstein dairy cattle. J Dairy Sci. 2016;99(8):6371–80. doi: 10.3168/jds.2015-10487. [DOI] [PubMed] [Google Scholar]
- 31.Hou YL, Bickhart DM, Hvinden ML, Li CJ, Song JZ, Boichard DA, et al. Fine mapping of copy number variations on two cattle genome assemblies using high density SNP array. BMC Genomics. 2012;13:376. doi: 10.1186/1471-2164-13-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou YL, Liu GE, Bickhart DM, Matukumalli LK, Li CJ, Song JZ, et al. Genomic regions showing copy number variations associate with resistance or susceptibility to gastrointestinal nematodes in Angus cattle. Funct Integr Genomics. 2012;12(1):81–92. doi: 10.1007/s10142-011-0252-1. [DOI] [PubMed] [Google Scholar]
- 33.Wu Y, Fan H, Jing S, Xia J, Chen Y, Zhang L, et al. A genome-wide scan for copy number variations using high-density single nucleotide polymorphism array in Simmental cattle. Anim Genet. 2015;46(3):289–98. doi: 10.1111/age.12288. [DOI] [PubMed] [Google Scholar]
- 34.Choi JW, Lee KT, Liao XP, Stothard P, An HS, Ahn S, et al. Genome-wide copy number variation in Hanwoo, Black Angus, and Holstein cattle. Mamm Genome. 2013;24(3-4):151–63. doi: 10.1007/s00335-013-9449-z. [DOI] [PubMed] [Google Scholar]
- 35.Chen WK, Swartz JD, Rush LJ, Alvarez CE. Mapping DNA structural variation in dogs. Genome Res. 2009;19(3):500–9. doi: 10.1101/gr.083741.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicholas TJ, Cheng Z, Ventura M, Mealey K, Eichler EE, Akey JM. The genomic architecture of segmental duplications and associated copy number variants in dogs. Genome Res. 2009;19(3):491–9. doi: 10.1101/gr.084715.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alvarez CE, Akey JM. Copy number variation in the domestic dog. Mamm Genome. 2012;23:144–63. doi: 10.1007/s00335-011-9369-8. [DOI] [PubMed] [Google Scholar]
- 38.Fontanesi L, Beretti F, Martelli PL, Colombo M, Dall’olio S, Occidente M, et al. A first comparative map of copy number variations in the sheep genome. Genomics. 2011;97(3):158–65. doi: 10.1016/j.ygeno.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Fontanesi L, Martelli PL, Beretti F, Riggio V, Dall’Olio S, Colombo M, et al. An initial comparative map of copy number variations in the goat (Capra hircus) genome. BMC Genomics. 2010;11:639. doi: 10.1186/1471-2164-11-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jia XB, Chen SR, Zhou HJ, Li DF, Liu WB, Yang N. Copy number variations identified in the chicken using a 60 K SNP BeadChip. Anim Genet. 2013;44(3):276–84. doi: 10.1111/age.12009. [DOI] [PubMed] [Google Scholar]
- 41.Yi GQ, Qu LJ, Liu JF, Yan YY, Xu GY, Ying N. Genome-wide patterns of copy number variation in the diversified chicken genomes using next-generation sequencing. BMC Genomics. 2014;15:962. doi: 10.1186/1471-2164-15-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramayo-Caldas Y, Castello A, Pena RN, Alves E, Mercade A, Souza CA, et al. Copy number variation in the porcine genome inferred from a 60 k SNP BeadChip. BMC Genomics. 2010;11:593. doi: 10.1186/1471-2164-11-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang JY, Jiang JC, Fu WX, Jiang L, Ding XD, Liu JF, et al. A genome-wide detection of copy number variations using SNP genotyping arrays in swine. BMC Genomics. 2012;13:273. doi: 10.1186/1471-2164-13-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang JC, Wang JY, Wang HF, Zhang Y, Kang HM, Feng XT, et al. Global copy number analyses by next generation sequencing provide insight into pig genome variation. BMC Genomics. 2014;15:593. doi: 10.1186/1471-2164-15-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5(1):e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77(1):78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41(10):1061–7. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinto D, Darvishi K, Shi X, Rajan D, Rigler D, Fitzgerald T, et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol. 2011;29(6):512–20. doi: 10.1038/nbt.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.LaFramboise T. Single nucleotide polymorphism arrays: a decade of biological, computational and technological advances. Nucleic Acids Res. 2009;37(13):4181–93. doi: 10.1093/nar/gkp552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai WR, Johnson MD, Kucherlapati R, Park PJ. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics. 2005;21(19):3763–70. doi: 10.1093/bioinformatics/bti611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winchester L, Yau C, Ragoussis J. Comparing CNV detection methods for SNP arrays. Brief Funct Genomics Proteomics. 2009;8(5):353–66. doi: 10.1093/bfgp/elp017. [DOI] [PubMed] [Google Scholar]
- 52.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–9. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campbell CD, Sampas N, Tsalenko A, Sudmant PH, Kidd JM, Malig M, et al. Population-genetic properties of differentiated human copy-number polymorphisms. Am J Hum Genet. 2011;88(3):317–32. doi: 10.1016/j.ajhg.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teo SM, Pawitan Y, Ku CS, Chia KS, Salim A. Statistical challenges associated with detecting copy number variations with next-generation sequencing. Bioinformatics. 2012;28(21):2711–8. doi: 10.1093/bioinformatics/bts535. [DOI] [PubMed] [Google Scholar]
- 55.Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470(7332):59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6(9):677–81. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hormozdiari F, Alkan C, Eichler EE, Sahinalp SC. Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res. 2009;19(7):1270–8. doi: 10.1101/gr.088633.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiang DY, Getz G, Jaffe DB, O’Kelly MJ, Zhao X, Carter SL, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6(1):99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abyzov A, Gerstein M. AGE: defining breakpoints of genomic structural variants at single-nucleotide resolution, through optimal alignments with gap excision. Bioinformatics. 2011;27(5):595–603. doi: 10.1093/bioinformatics/btq713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie C, Tammi MT. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics. 2009;10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25(21):2865–71. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nijkamp JF, van den Broek MA, Geertman JM, Reinders MJ, Daran JM, de Ridder D. De novo detection of copy number variation by co-assembly. Bioinformatics. 2012;28(24):3195–202. doi: 10.1093/bioinformatics/bts601. [DOI] [PubMed] [Google Scholar]
- 63.Li RQ, Zhu HM, Ruan J, Qian WB, Fang XD, Shi ZB, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20(2):265–72. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40(6):722–9. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21(6):974–84. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang YJ, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464(7289):704–12. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu GE, Hou YL, Zhu B, Cardone MF, Jiang L, Cellamare A, et al. Analysis of copy number variations among diverse cattle breeds. Genome Res. 2010;20(5):693–703. doi: 10.1101/gr.105403.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fadista J, Thomsen B, Holm LE, Bendixen C. Copy number variation in the bovine genome. BMC Genomics. 2010;11:284. doi: 10.1186/1471-2164-11-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bae JS, Cheong HS, Kim LH, NamGung S, Park TJ, Chun JY, et al. Identification of copy number variations and common deletion polymorphisms in cattle. BMC Genomics. 2010;11:232. doi: 10.1186/1471-2164-11-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hou YL, Liu GE, Bickhart DM, Cardone MF, Wang K, Kim ES, et al. Genomic characteristics of cattle copy number variations. BMC Genomics. 2011;12:127. doi: 10.1186/1471-2164-12-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hanson EK, Ballantyne J. A Highly Discriminating 21 Locus Y-STR “Megaplex” System Designed to Augment the Minimal Haplotype Loci for Forensic Casework. J Forensic Sci. 2004;49(1):49–51. doi: 10.1520/JFS2003209. [DOI] [PubMed] [Google Scholar]
- 72.Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7(2):e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H, Durbin R. Fast and accurate short read alignment with burrows wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42(Database issue):D764–70. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xie C, Mao XZ, Huang JJ, Ding Y, Wu JM, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Web Server issue):W316–22. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hu ZL, Fritz ER, Reecy JM. AnimalQTLdb: a livestock QTL database tool set for positional QTL information mining and beyond. Nucleic Acids Res. 2007;35(Database issue):D604–9. doi: 10.1093/nar/gkl946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12(5):363–76. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu GE, Ventura M, Cellamare A, Chen L, Cheng Z, Zhu B, et al. Analysis of recent segmental duplications in the bovine genome. BMC Genomics. 2009;10:571. doi: 10.1186/1471-2164-10-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414(6865):799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 82.Hardie DG. The AMP-activated protein kinase pathway-new players upstream and downstream. J Cell Sci. 2004;117(Pt 23):5479–87. doi: 10.1242/jcs.01540. [DOI] [PubMed] [Google Scholar]
- 83.Bionaz M, Loor JJ. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics. 2008;9:366. doi: 10.1186/1471-2164-9-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cullen KJ, Allison A, Martire I, Ellis M, Singer C. Insulin-like growth factor expression in breast cancer epithelium and stroma. Breast Cancer Res Treat. 1992;22(1):21–9. doi: 10.1007/BF01833330. [DOI] [PubMed] [Google Scholar]
- 85.Pacher M, Seewald MJ, Mikula M, Oehler S, Mogg M, Vinatzer U, et al. Impact of constitutive IGF1/IGF2 stimulation on the transcriptional program of human breast cancer cells. Carcinogenesis. 2007;28(1):49–59. doi: 10.1093/carcin/bgl091. [DOI] [PubMed] [Google Scholar]
- 86.Steelman LS, Abrams SL, Whelan J, Bertrand FE, Ludwig DE, Bäsecke J, et al. Contributions of the Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT pathways to leukemia. Leukemia. 2008;22(4):686–707. doi: 10.1038/leu.2008.26. [DOI] [PubMed] [Google Scholar]
- 87.Poulsen RC, Carr AJ, Hulley PA. Cell proliferation is a key determinant of the outcome of FOXO3a activation. Biochem Biophys Res Commun. 2015;462(1):78–84. doi: 10.1016/j.bbrc.2015.04.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pennacchio GE, Neira FJ, Soaje M, Jahn GA, Valdez SR. Effect of hyperthyroidism on circulating prolactin and hypothalamic expression of tyrosine hydroxylase, prolactin signaling cascade members and estrogen and progesterone receptors during late pregnancy and lactation in the rat. Mol Cell Endocrinol. 2016;442:40–50. doi: 10.1016/j.mce.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 89.Campo Verde Arboccó F, Sasso CV, Actis EA, Carón RW, Hapon MB, Jahn GA. Hypothyroidism advances mammary involution in lactating rats through inhibition of PRL signaling and induction of LIF/STAT3 mRNAs. Mol Cell Endocrinol. 2016;419:18–28. doi: 10.1016/j.mce.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 90.Schrooten C, Bink MC, Bovenhuis H. Whole genome scan to detect chromosomal regions affecting multiple traits in dairy cattle. J Dairy Sci. 2004;87(10):3550–60. doi: 10.3168/jds.S0022-0302(04)73492-X. [DOI] [PubMed] [Google Scholar]
- 91.Harder B, Bennewitz J, Reinsch N, Thaller G, Thomsen H, Kühn C, et al. Mapping of quantitative trait loci for lactation persistency traits in German Holstein dairy cattle. J Anim Breed Genet. 2006;123(2):89–96. doi: 10.1111/j.1439-0388.2006.00577.x. [DOI] [PubMed] [Google Scholar]
- 92.Tan L, Song XD, Sun X, Wang N, Qu Y, Sun ZJ. ART3 regulates triple-negative breast cancer cell function via activation of Akt and ERK pathways. Oncotarget. 2016;7(29):46589–602. doi: 10.18632/oncotarget.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Winn RA, Van Scoyk M, Hammond M, Rodriguez K, Crossno JT, Jr, Heasley LE, et al. Antitumorigenic effect of Wnt 7a and Fzd 9 in non-small cell lung cancer cells is mediated through ERK-5-dependent activation of peroxisome proliferator-activated receptor gamma. J Biol Chem. 2006;281(37):26943–50. doi: 10.1074/jbc.M604145200. [DOI] [PubMed] [Google Scholar]
- 94.Abreu GM, Valença DC, Júnior CM, da Silva CP, Pereira JS, Araujo Leite MA, et al. Autosomal dominant Parkinson's disease: Incidence of mutations in LRRK2, SNCA, VPS35 and GBA genes in Brazil. Neurosci Lett. 2016;635:67–70. doi: 10.1016/j.neulet.2016.10.040. [DOI] [PubMed] [Google Scholar]
- 95.Wang QB, Tian Q, Song XJ, Liu YY, Li W. SNCA Gene Polymorphism may Contribute to an Increased Risk of Alzheimer's Disease. J Clin Lab Anal. 2016;30(6):1092–9. doi: 10.1002/jcla.21986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Menzies KK, Lefèvre C, Macmillan KL, Nicholas KR. Insulin regulates milk protein synthesis at multiple levels in the bovine mammary gland. Funct Integr Genomics. 2009;9(2):197–217. doi: 10.1007/s10142-008-0103-x. [DOI] [PubMed] [Google Scholar]
- 97.D'Gama AM, Geng Y, Couto JA, Martin B, Boyle EA, LaCoursiere CM, et al. Mammalian target of rapamycin pathway mutations cause hemimegalencephaly and focal cortical dysplasia. Ann Neurol. 2015;77(4):720–5. doi: 10.1002/ana.24357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of identified CNVs and CNVRs in the 8 bulls’ genomes. (XLSX 1044 kb)
Primers information and confirmation results of the 11 chosen CNVRs by qPCR analysis. (XLSX 14 kb)
General statistics of 487 differential CNVRs between high and low group based on UMD3.1. (XLSX 28 kb)
The detailed features of genes completely or partial overlapped with differential CNVRs. (XLSX 28 kb)
Functional enrichment of GO and KEGG pathway analysis of genes covered by differential CNVRs. (XLSX 17 kb)
Functional enrichment of GO and KEGG pathway analysis of genes covered by group-specific CNVRs. (XLSX 11 kb)
QTLs covered by differential CNVRs. (XLSX 18 kb)
QTLs covered by group-specific CNVRs. (XLSX 17 kb)
Detailed information on the significant SNPs identified in previous GWAS that are near to the 10 candidate genes identified in this study. (XLSX 17 kb)
Data Availability Statement
All the re-sequencing data generated in this study have been deposited in National Centre for Biotechnology Information (NCBI) under the BioProject: PRJNA349121, and can be accessed in the Sequence Read Archive (SRA) under the accessions SRX2254497, SRX2254776, SRX2254777, SRX2254778, SRX2254779, SRX2254780, SRX2254813 and SRX2254814.


