Summary
How eukaryotic chromosomes fold inside the nucleus is an age-old question that remains unanswered today. Early biochemical and microscopic studies revealed the existence of chromatin domains and loops as a pervasive feature of interphase chromosomes, but the biological implications of such organizational features were obscure. Genome-wide analysis of pair-wise chromatin interactions using chromatin conformation capture (3C)-based techniques has shed new light on the organization of chromosomes in interphase nuclei. Particularly, the finding of cell type invariant, evolutionarily conserved topologically associating domains (TADs) in a broad spectrum of cell types has provided a new molecular framework for study of animal development and human diseases. Here we review recent progress in characterization of such chromatin domains and delineation of mechanisms of their formation in animal cells.
Introduction
It has been more than a century since Rabl and Boveri proposed that chromosomes in eukaryotic cells exist in discrete chromosome territories (CT) in interphase nuclei (Boveri, 1909; Rabl, 1885). After a period when the CT concept had been abandoned in favor of an alternative model of intermingled chromatin fibers, recent technological advances have led to accumulation of compelling evidence that allowed this model to be fully established and extended (Cremer and Cremer, 2010). It is now clear that CT is a major feature of the chromosome architecture. According to this model, each chromosome resides in a separate territory in the nucleus. So far, no clear rules have been identified to govern the positions of CTs in the nucleus, and it appears that during each cell division the CTs are re-shuffled randomly. However, some preferences for radial positions by chromosomes have been noted (Cremer and Cremer, 2010).
While the territorial model of chromosomal organization has now been generally accepted, the internal structure of the chromosomes is much less understood. Recent studies suggest that each chromosome is comprised of many distinct chromatin domains, referred to variably as topological domains or topologically associating domains (TADs), that are hundreds of kilobases to several million bases in length (Dixon et al., 2012; Jackson and Pombo, 1998; Ma et al., 1998; Nora et al., 2012; Sexton et al., 2012). These chromatin domains are stable for many cell divisions, invariant across diverse cell types, and evolutionarily conserved in related species. Because of the high degree of conservation, these chromatin domains have been considered the basic units of chromosome folding, and regarded as an important secondary structure in chromosome organization (Cremer and Cremer, 2010; Dekker and Heard, 2015; Sexton and Cavalli, 2015).
The discovery, characterization and function of chromatin domains have been covered recently by a number of excellent reviews (de Laat and Duboule, 2013; Dekker and Heard, 2015; Gorkin et al., 2014; Rowley and Corces, 2016; Sexton and Cavalli, 2015), but important questions remain. For example, the exact nature of the chromatin domains has not been clarified, leading to inconsistent definitions of domain boundaries in the genome by different groups (Dixon et al., 2012; Filippova et al., 2014). The lack of a clear definition of chromatin domains has also led to them being referred to sometimes as domains, and sometimes as loops (Rao et al., 2014). Most importantly, the mechanisms of their formation continue to be unresolved, though a number of different models have been proposed over the years (Dekker and Mirny, 2016; Rowley and Corces, 2016; Sanborn et al., 2015; Vietri Rudan and Hadjur, 2015). In this review, we attempt to provide a unified definition of chromatin domains, by considering the multiple orthogonal lines of evidence that support this concept. We propose here that the essential feature of the chromatin domains is that they perpetuate through mitosis and are conserved among different cell lineages in animals. We discuss the physical and biochemical models that have been put forward to explain such properties of the chromatin domains.
Converging evidence of chromatin domains in animal cells
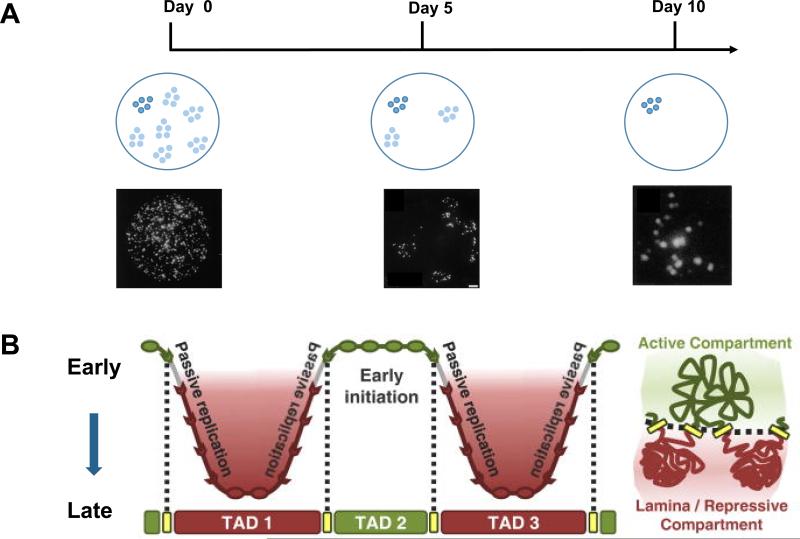
With 146-bp DNA wrapped around a histone octamer in about 1 and ½ turns, the nucleosome is the basic structural unit of chromatin. Nucleosomes assemble into 10nm chromatin fibers as beads on a string, which in turn fold into higher order structures, the details of which have remained unresolved. In mammalian cells, some of the clearest early evidence for domain partitioning of the genome in interphase comes from studies of DNA replication timing (Ferreira et al., 1997; Sparvoli et al., 1994) (See review by Rivera-Mulia and Gilbert, this issue). Using bromo-deoxy-uridin (BrdU) pulse labeling of early S phase nuclei followed by 3D fluorescence imaging, Ma et al found nearly 1,100 sites of DNA replication, called Replication Sites (RS), in NIH 3T3 cells (Ma et al., 1998). They found that each RS contains 5-6 replicons, and the replicon clusters persist after many cell divisions. They also found that the average size of the RS was about one million bases. Independently, Jackson and Pombo made similar observations in the HeLa cells (Jackson and Pombo, 1998). Both studies showed that the megabase-sized chromatin domains remain unchanged after many cell divisions (Figure 1a).
Figure 1. Converging evidence of chromatin domains in animal cells.
A) BrdU pulse-chase labeling followed by immunofluorescence imaging showed that replicon clusters are stable over many cell divisions. A schematic of replication clusters in the nucleus as visualized immediately or 5 days and 10 days after BrdU labeling are shown. The replication clusters remain together even after multiple cell divisions. Image data from Jackson & Pombo (1998). B) Maps of replication domains and chromatin interactions by Repli-chip and Hi-C, respectively, show one-to-one correspondence between replication domains and TADs. A schematic of TADs is on the right. Figure adopted from Pope et al. (2014).
Development of genome-wide chromosome conformation capture techniques (3C, 4C, 5C and Hi-C) has led to further support for the concept of chromatin domains in recent years (de Wit and de Laat, 2012; Dekker et al., 2002; Dixon et al., 2012; Nora et al., 2012; Sexton et al., 2012). Using 5C and Hi-C, three groups independently showed that chromatin interactions are spatially restricted into repeated chromatin domains (Dixon et al., 2012; Nora et al., 2012; Sexton et al., 2012). For example, we determined that chromatin interactions in human and mouse cells occur predominantly within domains with an average size of 880Kb, similar to that of RS observed under microscope. These chromatin interaction domains have come to be widely known as Topologically Associated Domains, or TADs. Like RS, the TADs are generally stable, with diverse cell types such as ES and fibroblasts sharing the majority of the TADs defined in each cell type.
The similar sizes of RS and TADs in mammalian cells, coupled with the apparent stability of these chromatin domains through cell divisions, strongly suggest that these two are the same chromatin structures. Evidence in support of this notion came when Gilbert and colleagues used a method that combine BrdU pulse labeling with DNA microarrays to determine the early and late replication domains in multiple human and mouse tissues and cell types. They found that the replication domains defined in each tissue/cell type were highly consistent with TADs (Pope et al., 2014) (Figure 1b). In sum, the available data strongly suggest that chromatin is organized into repeating domain-like units of chromatin organization in the interphase nuclei. Each domain likely contains hundreds of kilobases of DNA, and the composition of these domains appears to vary little between cell types. Importantly, it is now clear that these chromatin domains have functional implications, such as the regulation of genes within each domain, timing of DNA replication, and propagation of chromatin state along the DNA. Therefore, it is crucial to understand the nature of this fundamental feature of chromatin organization, as well as its mechanism of formation.
It is must be noted that the field has yet to settle on a unifying definition of these chromatin domains, or even a single term to refer to them. For the remainder of this review we will refer to these domains as TADs, because this seems to be the most widely used term in the literature at present. We include a further discussion of terminology below.
Nature of TADs
At their most fundamental level, TADs represent a physical compartmentalization of the genome. The critical observation is that two regions within a TAD associate on average more frequently with each other than with regions outside of the TAD (Dixon et al., 2012; Nora et al., 2012; Sexton et al., 2012). This suggests two basic features of TAD organization: first, a “self association” property of regions within the TAD, and second, an “insulation” property between regions in neighboring TADs. These properties are essentially what the various computational algorithms used to identify TADs attempt to resolve (Dixon et al., 2012; Filippova et al., 2014; Hou et al., 2012; Rao et al., 2014; Sexton et al., 2012; Shin et al., 2016). Interestingly, the properties of self-association and insulation from neighboring regions are also predicted using polymer physics models to be the consequence of forming a “loop” between monomers in a polymer (Dekker and Mirny, 2016; Doyle et al., 2014; Sanborn et al., 2015). In this regard, an obvious question facing the field is to what degree TADs are a reflection of looping events or “loop domains” in the genome. Contributing to the confusion is the recognition that TADs are hierarchical in nature (Figure 2a). TADs themselves may contain smaller “sub-TADs” (Phillips-Cremins et al., 2013; Rao et al., 2014), and ultimately at their most local level may contain individual “loops” (Rao et al., 2014) or “insulation neighborhoods” (Dowen et al., 2014; Ji et al., 2016). Like TADs themselves, sub-TADs, loops, or insulation neighborhoods, are regions that display both the self-associative and insulation properties ascribed to TADs. In this regard, a question currently facing the field is whether TADs are fundamentally different than sub-TADs, loops, or insulation neighborhoods, or are these all the same features of the genome at different length scales? Certainly, some extent of the differences in these structures may reflect differences in the computational algorithms used to identify them, or the type and quality of the data used for analysis. Indeed, there are a variety of algorithms that have been developed to identify domains in chromatin interaction data without a clear indication of which methods provide the “best” set of domain calls. However, going beyond algorithmic or data quality differences, there are several lines of evidence that suggest that TADs are functionally distinct from sub-TADs, loops, and insulation neighborhoods. Perhaps the best current insights into these issues relate to how each of these features are conserved between different cell types. We and others have found little evidence that TADs vary between cell types in a given organism, suggesting that they are a largely invariant feature of genome organization (Dixon et al., 2015; Dixon et al., 2012; Nora et al., 2012). On the contrary, sub-TADs, loops, and insulation neighborhoods all appear to differ, at least partially, between different cell lineages (Dowen et al., 2014; Ji et al., 2016; Phillips-Cremins et al., 2013; Rao et al., 2014). The cell type specific organization of sub-TADs, loops, and insulation neighborhoods appear to be related to cell type specific regulatory events. In this regard, we suspect that TADs represent a larger, more invariant feature of genome organization within which cell type specific structures can form to play roles in lineage specific genome regulation. A second indication that TADs may be functionally distinct from sub-TADs and loops comes from comparisons of TADs and DNA replication timing. In examining replication timing profiles across a variety of cell types, Pope and colleagues demonstrated that TADs correspond to the “units” of replication timing that switch between lineages(Pope et al., 2014). In this regard, these authors suggest that TADs are themselves equivalent to replication domains. This does not, however, appear to apply as well to sub-TADs and individuals loops in the genome, suggesting that these are functionally distinct in their role in regulating DNA replication.
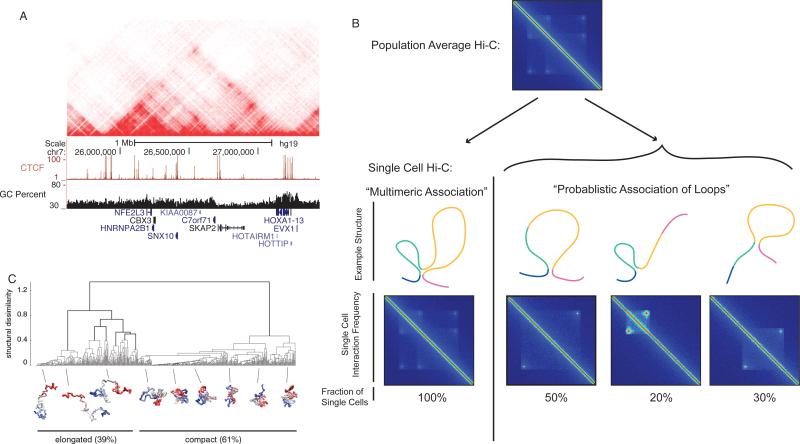
Figure 2. Hierarchical organization of TADS.
A) Interaction heat map of one TAD located near the HoxA locus in GM12878 lymphoblastoid cells from Rao et al. (2014) at 10kb resolution. Also shown below are ChIP-seq tracks from ENCODE and the GC content over the region. Note that the interaction heat map within the TAD has multiple points where the signal is enriched locally, indicative of local “looping” structures. Within the larger TAD there appears to be the presence of multiple smaller sub-TADs and individual loops. B) Diagram of how hierarchical organization within TADs may occur using simulated data. On the top is a heat map of a single TAD with several smaller internal sub-TADs or loops. In the “multimeric association” example, the TAD in each individual cells represents a complex structure consisting of multiple loops or subdomains that would individually largely reflect the population average. In the “probabilistic association of loops example,” individual cells would variable contain single loops or subdomains, and that the population average would reflex a mixture of these single cell structures. In this case, a TAD would represent a region of the genome with increased probability of forming loops or domains within single cells. C) Polymer models of internal TAD structure from Giorgetti et al (2014) indicate that at least in some TADs there is variability in the internal TAD organization from cell to cell.
Given the observation that TADs are hierarchical in nature, a related question is whether TADs represent a “population average” of individual loops that may differ on a cell-to-cell basis. In this regard, a TAD could represent a set of more probabilistic loops that may be variably present in single cells (Figure 2b). Indeed, the extent of possible hierarchical structures in certain TADs would suggest that at least some degree of cell-to-cell variability must be present. Further, some of the highest resolution FISH studies of TAD organization indicate that there can be variability in the internal organization of TADs (Figure 2c) (Giorgetti et al., 2014). However, the sole single cell Hi-C study to date indicated that TADs appear to be present in individual cells (Nagano et al., 2013). This would suggest that TADs are indeed present in individual cells and that their observed hierarchy may reflect multimeric associations between individual regions within the TAD. It should be noted that the current resolution of single cell Hi-C is coarse, and improvements in the resolution of single cell methods will be necessary to conclusively resolve this issue.
Function of TADs
The chromatin contact maps from diverse cell types and tissues indicate that TADs are relatively cell-type invariant, in contrast to other chromatin organization features. Furthermore, comparison of chromatin contact maps in related species also showed that TADs are preserved in syntenic sequences (Dixon et al., 2012; Vietri Rudan et al., 2015). These initial observations have led to the proposal that TADs serve as the basic unit of chromosome folding (Dekker, 2014). As such, TADs may be involved in a broad set of nuclear processes that involve chromatin organization. Indeed, a growing body of evidence has shown that TADs play important roles in transcriptional regulation, DNA replication, and VDJ recombination (Hu et al., 2015; Lucas et al., 2014; Nora et al., 2012; Pope et al., 2014; Sanborn et al., 2015).
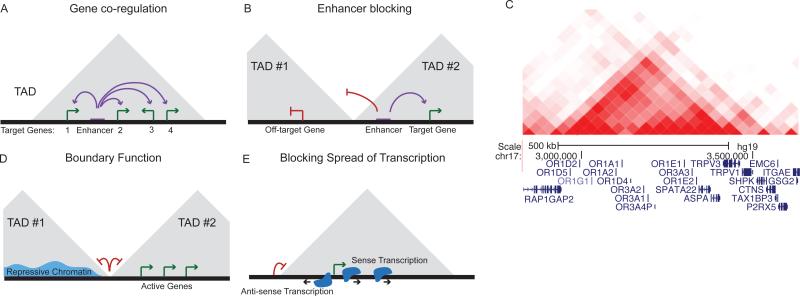
As TADs represent physically isolated units of genome organization, perhaps it should come as no surprise that they also represent functionally isolated units of the genome. This appears to manifest in two distinct phenomena: first, the “co-regulation” of genes within TADs, and second, the blocking of the “spread” of activity between neighboring TADs. With regard to gene co-regulation, by constraining the chromatin interactions among loci within each TAD to the same domain, they effectively create autonomous gene regulatory domains (Figure 3a). Supporting this notion, it has been observed that genes within the same TADs share coordinated gene expression profiles across different cell types and tissues (Flavahan et al., 2016; Nora et al., 2012; Shen et al., 2012). We would note, however, that this effect is not absolute. TADs do not function similarly to a bacterial operon, with all genes within a TAD being activated simultaneously. Instead, co-regulation may affect only a subset of genes within any individual TAD, and is likely more prevalent at certain genomic loci (Nora et al., 2012; Shen et al., 2012). For example, there is clear evidence that gene clusters, such as cytochrome genes, olfactory receptors, and protocadherin genes, are organized into individual TADs, indicating that genes that have functional needs for co-regulation tend to be associated in the same TAD (Figure 3c). Likewise, Grubert et al. and Waszak et al. found evidence of quantitative trait loci (QTL) that appear to coordinately affect the activity of multiple regulatory elements in genomic loci spanning >100kb (Grubert et al., 2015; Waszak et al., 2015). These genetically co-regulated loci were restricted within the same TAD, suggesting that sequence variants that affect activity of regulatory elements may ultimately affect the activity of multiple loci within a TAD. A rather remarkable example of such co-regulation within TADs comes from Symmons and co-workers, who generated mice with a reporter construct inserted at different positions along the chromosome. The reporter was driven by a weak promoter and would only become active under the control of endogenous regulatory elements. Remarkably, the reporter reflected the expression patterns of nearby genes, but only of genes within the same TAD (Symmons et al., 2014). One implication of this finding is that distal regulatory interactions have the potential for being “non-specific” within a given TAD in the genome. This restriction of co-regulation to genes within a domain is likely maintained by “boundary” activity between neighboring TADs (Figure 3b). Indeed, deletion of boundaries has been shown to lead to ectopic gene activation in cultured cells and in vivo (Lupianez et al., 2015; Narendra et al., 2015). For example, deletion of a TAD boundary on X chromosome in mouse ES cells led to enhanced expression of several genes located next to the deleted boundary (Nora et al., 2012). Further, deletion of boundaries has been attributed as the cause for ectopic expression of several developmental regulator genes during limb formation that lead to polydactyly in several families (Lupianez et al., 2015).
Figure 3. Function of TADs in genome regulation.
A) Example diagram showing the co-regulation of multiple genes by a single regulatory element within a TAD. B) Interaction heat map showing a single TAD encompassing a cluster of olfactory receptor genes (data from Dixon et al. 2015) C) Diagram of the potential for TAD boundaries to serve an enhancer blocking role that restricts enhancers to target genes within the same TAD. D) Diagram of the potential for TAD boundaries to restrict the spread of repressive chromatin into active domains and vice versa. E) Diagram of the role of TADs in forming a barrier to divergent transcriptional “noise” in the genome.
Related to their ability to restrict regulatory element interactions with target genes, TADs also appear to play a role in restricting recombination events during VDJ recombination during B-cell maturation (Hu et al., 2015). In mapping high-resolution breakpoints using a RAG mediated “bait,” Hu and colleagues demonstrated that the vast majority of recombination events occur within the same TAD as the bait sequence. This suggests that in this context TADs either facilitate proper recombination during maturation or suppress potential for erroneous breaks and deletions during this process. Remarkably, these authors also observed a clear orientation bias in the generation of these breaks, implying a potential underlying “linear tracking” mechanism. This has important implications in terms of TAD formation that will be discussed in more depth later in this review.
Similarly, there is also growing evidence that the boundaries between TADs can serve as “barriers” to the spread of activity within the genome. A striking example of this comes from a recent study examining “transcriptional noise” in the genome. Many promoter and enhancers display bi-directional transcription, which is actively suppressed but has also been suggested to contribute to function of regulatory elements. Natoli and colleagues showed that upon knock-down of the Set complex protein WDR82, there is a dramatic increase in the amount of non-coding divergent transcription from promoters and enhancers (Austenaa et al., 2015). Remarkably, these authors also showed that such non-coding transcription appears to end abruptly at the boundaries of TADs. This suggests that the boundaries of TADs can form a physical barrier that prevents the linear tracking of molecules along a chromosome (Figure 3d). Similar findings have been observed in studies where the potential boundary elements of TADs are deleted using CRISPR. Reinberg and colleagues showed that in mouse embryonic-stem-cell derived motor neurons, the HoxA locus is divided in two separate TADs, one that is active, and one that is repressed by polycomb group proteins (Narendra et al., 2015). Upon deletion of a CTCF binding site that likely represents the boundary of these TADs, they observed the genes in the polycomb repressed domain nearest to the boundary element become activated. In light of a long history of boundary elements being thought of as regions that would restrict the spread of repressive heterochromatin, these results were somewhat surprising, as the evidence clearly showed the active marks encroaching into the repressed domain. This suggests that ability of TAD boundaries to functionally restrict the genome applies to both active and repressive regions of the genome (Figure 3e).
Given their emerging role in serving as potential functional units of the genome, it may be expected that these domains are conserved in evolution. Our original study demonstrated that TADs are well conserved over syntenic regions of the genome between mice and humans, species separated by ~80 million years of evolution (Dixon et al., 2012). Similar observations of TAD conservation have been made in macaques and dogs (Vietri Rudan et al., 2015). Remarkably, these authors also showed was that breaks in synteny between species, where chromosomes are broken and rearranged, are greatly enriched for proximity to the boundaries between TADs. This is perhaps some of the strongest evidence to date for the notion that TADs are functional units of the genome, as these studies suggest that mutations that would “break” a TAD in evolution appear to be strongly selected against. Theoretically, TADs afford genes with the potential to be controlled by a large set of DNA sequences within the same domain. Given the average size of the TADs in the range of close to a million basepairs, the space for the possibilities of distinct regulatory elements could be very large. This has the potential to be beneficial to an organism's adaptation to the changing environment and add new competitive traits. On the other hand, TADs also place constraints on gene regulatory sequences, limiting those regulatory interactions to the same TAD, but not beyond.
Despite their apparent presence in humans, macaques, dogs, mice, and Drosophila, the organization of chromosomes into TADs does appear different in C. elegans (Crane et al., 2015). Remarkably, while TADs appear to be present on the X-chromosome in C. elegans, they are markedly less frequent on the somatic chromosomes (Crane et al., 2015). Further, depletion of the Condensin complex reduces the appearance of TADs on the X-chromosome but does not appear to affect the structure of autosomes. The origin of the discrepancy in the appearance of TADs between C. elegans and other metazoans is not clear. Comparative evolution studies have indicated a loss of both CTCF (Heger et al., 2009) and a loss of the enrichment of the CTCF motif (Heger et al., 2012) in C. elegans and some other nematodes. Indeed, the presence of CTCF and the enrichment of its motif appears to have arisen early in metazoan evolution and been restricted to bilaterian organisms, but with some apparent loss of CTCF amongst certain metazoan lineages (Heger et al., 2012). It is tempting to speculate that the origin of TADs in evolution coincided with the origin of CTCF, and that subsequent losses of CTCF may alter the presence of TADs in certain organisms.
Given the role of TADs in restricting cis regulatory sequences to their target genes, it is not surprising that an increasing number of diseases have been attributed to disruption of TAD structure. The clearest examples of this so far have come from Mendelian diseases affecting limb development and from cancer. Mundlos and colleagues demonstrated that several families displaying limb malformation can be accounted for by the deletion, inversion, or duplication of TAD boundaries, resulting in ectopic expression of genes in the developing limb bud (Lupianez et al., 2015). With regards to cancer cells, several recent reports have indicated that structural alteration to cancer genomes can result in “enhancer hijacking”, in which an enhancer acts on a gene other than its normal target due the elimination of a barrier such as a TAD boundary. This is known to occur as a the result of a recurrent inversion on chromosome 3 in AML, as well as recurrent deletions and duplications in Meduloblastoma (Groschel et al., 2014; Northcott et al., 2014). Likewise, deletions of CTCF binding sites at “insulation neighborhoods” appears to result in aberrant activation of oncogenes by altering their relationship with nearby enhancers (Hnisz et al., 2016). Similarly, gliomas with aberrant DNA methylation patterns as the result of IDH1 mutations appear to have altered binding of CTCF, resulting in a similar “enhancer hijacking” phenomenon (Flavahan et al., 2016). Given the plethora of structural variations that can occur in cancer genomes, we suspect that alterations which affect 3D genome organization and ultimately contribute to oncogenesis are not likely to be limited to the above mentioned examples.
Mechanisms of TAD Formation
The most prominent feature of TADs is that they are to a large degree cell-type invariant (Dixon et al., 2015; Dixon et al., 2012; Nora et al., 2012). By contrast, the designation of compartment A and B from Hi-C datasets tends to vary widely between cell types and during development (Dixon et al., 2015; Lieberman-Aiden et al., 2009). Additionally, the chromatin loops between promoters and distal enhancers also are known to be cell type dependent (Dixon et al., 2012; Ji et al., 2016; Jin et al., 2013; Kieffer-Kwon et al., 2013; Li et al., 2012; Rao et al., 2014; Sanyal et al., 2012; Tang et al., 2015; Zhang et al., 2013). Given the relatively stable nature of TADs during development, it is important to understand the mechanisms that could account for such remarkable consistency between different cell types.
To understand the mechanisms of TAD formation, one may need to distinguish the different physical forces that underlie chromatin contacts in cells. These forces are not necessarily specific to TAD structure alone, but are a reflection of the processes that can shape a chromatin polymer in the nucleus. To date, several different processes have been envisioned to contribute to chromatin organization: (a) chromatin fiber movement that can be described using polymer physics (Dekker and Mirny, 2016), such as fractional Langevin motion (Lucas et al., 2014) or fractal globule (Lieberman-Aiden et al., 2009), (b) attraction within chromatin domains due to presence of multiple binding sites for certain diffusible molecules, which could be either a large protein complex, or a chromatin-associated RNA, or an RNA/protein complex (Barbieri et al., 2012) (See review by Mele and Rinn in this issue on the role of RNA in shaping nuclear structure) (Figure 4); (c) insulation due to specific sequence binding proteins factors or RNAs that introduce topological constraints to the local chromatin fiber (Phillips-Cremins and Corces, 2013; Vietri Rudan and Hadjur, 2015).
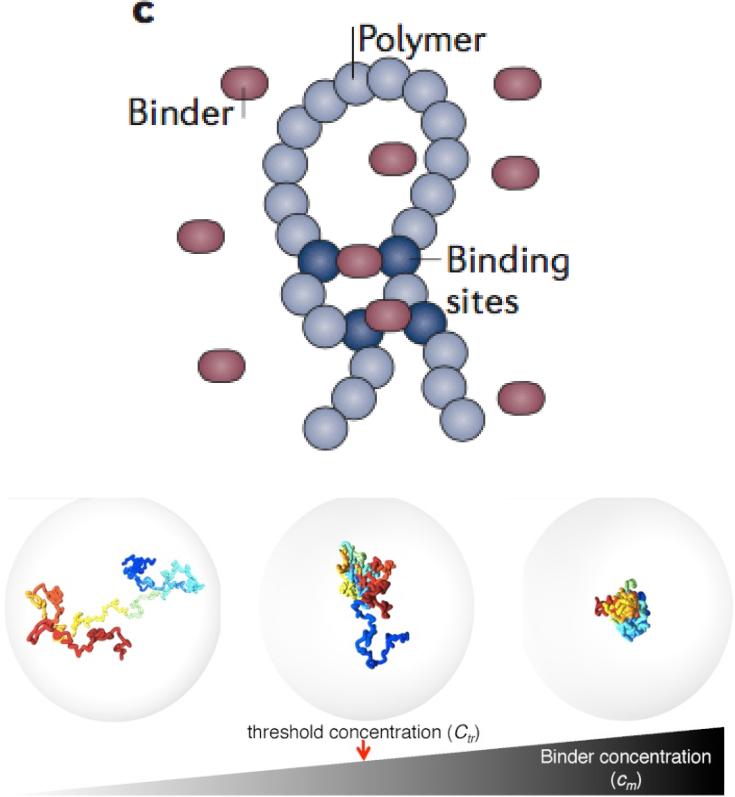
Figure 4. A Strings and Binders Switch model to describe the processes shaping chromatin organization.
Chromatin fiber is modeled as a self-avoiding polymer (blue) with binding sites for Brownian binder particles (red) with concentration (cm) and multiple binding sites on the polymer. Mont Carlo simulation of the polymer and binders leads to three stable states of organization reflecting an open conformation, a fractal conformation and a compact conformation. Figure adopted from Barbieri et al. (2012) and Pombo & Dillon (2015).
One physical process that helps TAD formation is an “attractive” force and likely involves the Cohesin complex (Kagey et al., 2010; Phillips-Cremins et al., 2013; Seitan et al., 2013; Sofueva et al., 2013; Vietri Rudan and Hadjur, 2015; Watrin et al., 2016; Zuin et al., 2014). Cohesin is a ring-shaped protein complex consisting of SMC1, SMC3, and RAD21 (Watrin et al., 2016). Its role in cell cycle has been well documented and characterized. It can encircle two chromatin fibers with its 30-40nm ring structure and hold the two sister chromatids together after DNA replication until the onset of anaphase, when the ring is removed from the chromosomes and the two sister chromatids segregate into the two daughter cells. Overwhelming evidence now suggests that Cohesin complex not only contributes to sister chromatid cohesion, but also plays an active role in mediating long-range chromatin interactions between enhancers and promoters (Kagey et al., 2010; Phillips-Cremins et al., 2013; Seitan et al., 2013; Sofueva et al., 2013; Zuin et al., 2014). In mouse ES cells, Cohesin is found to be associated with the Mediator complex and localized to active enhancers (Kagey et al., 2010). Dynamic occupancy of the enhancers by the Cohesin complex correlates with cell type specific enhancer/promoter interactions in the mouse ES cells and MEF cells. More recent studies in human HEK293 cells, mouse thymocytes, mouse ES cell-derived neuron stem cells and astrocytes showed that loss of Cohesin complex leads to decreased enhancer/promoter interactions within TADs (Seitan et al., 2013; Sofueva et al., 2013; Zuin et al., 2014). Using a combination of Hi-C experiments and FISH, it was shown that intra-TAD chromatin interactions decreases and the TADs volume increases in the cells deleted of the Cohesin subunit Rad21 (Sofueva et al., 2013). Finally, TADs have been shown to be disrupted in mitosis, and then quickly reestablish accompanying the initiation of DNA replication, correlating with the dynamics of Cohesin loading at the end of telophase (Naumova et al., 2013). These observations taken together strongly suggest an active role for Cohesin complex in the formation of TAD interactions.
One consequence of this “attractive” property of TADs that has emerged is that these may serve to restrict chromatin interactions in 3D space (Figure 4). Live cell imaging approaches have shown that individual loci in the genome undergo sub-diffusive motion, best described as Fractional Langevin motion (Lucas et al., 2014). Essentially, this indicates that as a region of the genome begins to diffuse in the nucleus, the most likely next place this region will go is to return back to its original position. This suggests that individual genomic loci are restricted in the nuclear space they have to “explore,” at least over short time scales. The physical determinants of this property remain unclear, as is the mechanism by which two regions, if originally spatially isolated, ultimately find each other to form more stable structures. These questions will likely be addressed as improved live cell imaging, in particular super-resolution live cell imaging, becomes more readily available. Another interesting principle of how chromatin domains are organized in the genome comes from super-resolution imaging of Drosophila cells (Boettiger et al., 2016), which indicated that the internal organization of domains depends on the chromatin state of the regions within the domain. Remarkably, domains marked by polycomb group proteins show a drastically different organization than “inactive” or “active” domains, which are associated with distinct chromatin states (Boettiger et al., 2016). These polycomb bound domains appear to be highly compacted, suggesting that the internal chromatin state of a domain may also influence the presence of “attractive” forces that contribute to domain formation. The same study also indicated that while epigenetic domains are spatially isolated from each other, there can be degrees of intermixing between neighboring regions. However, the degree of intermixing depends on the activity of the two domains in question. This likely indicates that when considering what physical domains “look like” in the genome, it is important to take into account the chromatin state of regions within the domain. Attractive forces alone, however, cannot explain TAD structure. For example, attractive forces have difficulty explaining the depletion of contacts between neighboring domains, or the observation that upon deletion, TAD boundaries tend to “shift” along chromatin rather than resulting in a complete merging of neighboring domains (Narendra et al., 2015; Nora et al., 2012; Sanborn et al., 2015).
In addition to the above factors that enhance chromatin interactions within TADs, a process exists to actively suppress (or “insulate”) chromatin interactions between TADs. A primary protein factor involved in this process is likely to be CTCF, a Zinc-Finger containing DNA binding protein that has been long known to play a key role at insulator elements (Ghirlando and Felsenfeld, 2016; Phillips-Cremins and Corces, 2013). Binding of the insulator-binding proteins to DNA is proposed to prevent enhancers from interacting with inappropriate gene promoters, and separate the euchromatin from heterochromatin in the genome (Ghirlando and Felsenfeld, 2016; Phillips-Cremins and Corces, 2013). TAD borders are enriched for CTCF binding sites in mammalian cells (Dixon et al., 2012). In mammalian cells, ChIP-seq analyses of CTCF, the only known insulator protein with a DNA binding domain characterized by 11 zinc-fingers, showed that CTCF binding sites demarcate nearly 90% of the TAD boundaries (Dixon et al., 2012), a more than 2-fold enrichment over that expected by chance (Dixon et al., 2012; Phillips-Cremins et al., 2013). While CTCF appears to be a key insulating factor in mammalian cells, it is probably not alone. Recently, it was shown that ZNF143 could play a role in chromatin organization (Bailey et al., 2015; Heidari et al., 2014). This zinc-finger protein is essential for ES cell pluripotency (Chia et al., 2010). It co-localizes with CTCF to the genome of embryonic stem cells (Bailey et al., 2015; Heidari et al., 2014). Depletion of ZNF143 leads to alteration of chromatin interactions (Bailey et al., 2015). Thus, ZNF143 could be another architecture protein that either together with CTCF or complement CTCF's function in TAD formation. In Drosophila, multiple insulator proteins have been identified, besides the CTCF homolog protein dCTCF (Phillips-Cremins and Corces, 2013). These include su(Hw), BEAF-32, CP190. These insulator proteins were found to be highly enriched at the borders of chromatin domains defined in Drosophila cells (Hou et al., 2012; Sexton et al., 2012). It was observed that multiplicity of these factors’ binding to DNA scales with the insulation strengths of TAD boundaries, thus the more types of boundary binding proteins present on DNA, the higher the insulation score and more stable the TAD (Van Bortle et al., 2014). This observation suggests a common mechanism by the non-CTCF factors in mediating TAD formation in the Drosophila cells. Recently, TFIIIC was also found to reside at TAD boundaries (Van Bortle et al., 2014; Van Bortle et al., 2012). This is likely to be true in mammalian cells, since TFIIIC also are found to be localized to a subset of TAD boundaries
A plethora of studies have now provided compelling evidence that CTCF directly mediates TAD formation in mammalian cells. First, depletion of the CTCF protein in a human cell line using siRNA increased chromatin interactions between neighboring TADs but reduced chromatin interactions within the same TADs (Zuin et al., 2014). Second, deletion of a CTCF motif at a TAD boundary within the HoxA locus in the mouse genome resulted in expansion of the TAD containing the HoxA1-A5 genes to the adjacent TAD and ectopic activation of HoxA7 gene in mESC-derived motor neurons (Narendra et al., 2015). Similarly, deletion of a CTCF motif in the homologous region in a human embryonal carcinoma cell line also resulted in alterations of chromatin organization and dramatic changes in expression of genes in the locus (Xu et al., 2014). Third, deletion of CTCF binding sites at the borders of a CFTR-containing TAD in a human epithelial colorectal adenocarcinoma cell line (Caco2) altered chromatin organization and enhanced chromatin interaction between the CFTR promoter and sequences outside of the TAD (Yang et al., 2015). Fourth, comparative analysis of chromatin organization in several mammalian species revealed evolutionarily conserved TAD boundaries that are associated with conserved CTCF binding sites across species, as well as a strong correlation between the turn over of CTCF binding sites and loss or gain of TAD boundaries in these species (Vietri Rudan et al., 2015). Thus, loss of function studies, sequence perturbation experiments and evolutionary conservation analyses all point to CTCF as a key player in mediating the TAD organization in mammalian cells and likely other species.
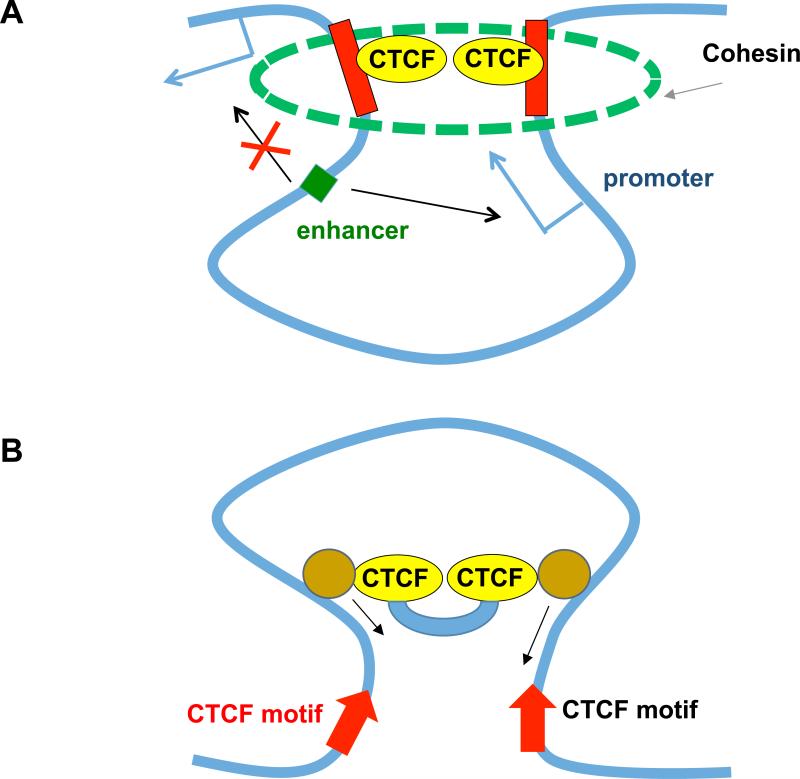
How exactly CTCF “insulates” chromatin interactions between TADs has been a question of intensive research. While a complete, clear answer is still not forthcoming, two sets of models have been proposed (Figure 5). The first set argues that CTCF-containing multimeric complexes simultaneously bind to two ends of a TAD to create a chromatin loop that separates the TAD sequences from the neighboring DNA. This could be achieved by two potential mechanisms. The first model, referred to as the “hand-cuff model” (Vietri Rudan and Hadjur, 2015), posits that the two ends of a TAD are brought together in 3D space by the CTCF proteins, which bind to each boundary via the DNA motifs and recruit the Cohesin complex via protein-protein interactions between CTCF and Rad21, a component of the Cohesin complex (Vietri Rudan and Hadjur, 2015) (Figure 5a). This model is supported by many lines of experimental evidence: a) The Cohesin complex generally co-localizes with CTCF throughout the mammalian genome (Parelho et al., 2008; Wendt et al., 2008); b) Chromatin immunoprecipitation of CTCF bound chromatin followed by proximity ligation and paired-end sequencing (ChIA-PET) showed that chromatin interactions between TAD borders are generally bridged by CTCF and Cohesin (Ji et al., 2016; Tang et al., 2015); c) Dynamics of CTCF binding at the borders correlate with gains or losses of chromatin loop interactions involving TAD boundaries (Narendra et al., 2015; Sanborn et al., 2015). This is a fairly simple model to explain several observations regarding the relationship between CTCF, Cohesin, and 3D chromatin structure. However, there are some difficulties with this model. First, the number of CTCF binding sites along the mammalian genome far exceeds the number of TAD boundaries. A critical question is, why are not TADs more abundant and what confers the specificity of CTCF interactions? A second, perhaps related issue, is how CTCF binding sites on one TAD border find another TAD border in space to interact and form a TAD? In addition, the handcuff model does not provide an explicit definition of the processes used to form a domain in space and time.
Figure 5. Two models for TAD formation.
(A) A hand-cuff model describing the formation of TADs by CTCF ad Cohesin complex connecting the two boundary sequences together. (B) The “extrusion” model involves a pair of tethered CTCF proteins bound to chromatin motors that propel the extrusion of chromatin fiber while the two CTCF molecules slide the chromatin fiber in opposite directions before pausing at converging CTCF DNA binding motifs (Red).
A second model, termed the “extrusion model”, builds upon the “handcuff” model and aims to account for CTCF's role in setting up TADs and conferring specificity to their interactions (Figure 5b). This model proposes that a DNA loop can be generated dynamically by a pair of tethered DNA binding units that can load to the DNA and travel along the chromatin fiber in opposite directions (Alipour and Marko, 2012; Dekker and Mirny, 2016; Fudenberg et al., 2015; Nasmyth, 2001; Sanborn et al., 2015). While the two DNA binding units move along the DNA, loops would form, until each unit reach a landing pad where they will reside a short while before dropping off. In one rendition of this model, two complexes of CTCF and Cohesin interact and bind together to DNA, tethered by heterodimerization between CTCF proteins (Figure 5b). The two complexes would move in opposite directions along the DNA, with the DNA between the two Cohesin rings extruding out in the process. The extrusion stops when the two tethered CTCF proteins reached a pair of convergent binding motifs. This extrusion model explains several phenomena: 1) TAD boundaries frequently contain pairs of CTCF binding sites with convergent CTCF motif orientations (Guo et al., 2015; Rao et al., 2014; Sanborn et al., 2015; Vietri Rudan et al., 2015). 2) The extrusion model would also explain how deletion of one CTCF binding motif does not lead to complete collapse of TAD structure, as the hand-cuff model would predict (Sanborn et al., 2015). Instead, the TAD boundary simply shifts to the next available CTCF binding site with the matching orientation. Indeed, using physical simulations based on the extrusion model, it was possible to predict the formation of new loops after deletion of a CTCF binding site (Dekker and Mirny, 2016; Sanborn et al., 2015). The “extrusion model” depends on several currently unproven assumptions: a) there exists a large protein complex with two motors that can traverse the chromatin fiber in opposite directions and sufficient cellular energy to facilitate such a process; b) there exists a mechanism to clear up the supercoils built up due to the movement of such protein complex along hundreds of kilobases of chromatin; c) the movement of such “chromatin topology machine” will be compatible with other nuclear processes – transcription, DNA replication – and can occur in both euchromatin and heterochromatin compartments. It is also unclear based on the extrusion model how multimeric chromatin loops may form, and the model largely aims to define how to establish pairwise CTCF/Cohesin based interactions.
Both the “hand cuff” model and the “extrusion” model recognize that CTCF and Cohesin work together to mediate the formation of TAD. This would predict that loss of CTCF or Cohesin complex would have the same effect on chromatin organization and TAD. However, when Zuin et al. performed Hi-C experiments and compared the effects of depletion of Cohesin and CTCF on chromatin architecture in human HEK293 cells, they found loss of CTCF and Cohesin complex affected chromatin organization differently (Zuin et al., 2014). While depletion of CTCF resulted in an increase of inter-TAD interactions, deletion of Cohesin complex led to a decrease in inter-TAD interactions (Zuin et al., 2014). Similar results were independently obtained using the post-mitotic thymocytes, where loss of Cohesin resulted in no alteration of the TADs in these cells (Seitan et al., 2013). Therefore, Cohesin complex plays somewhat distinct roles from CTCF in chromatin organization.
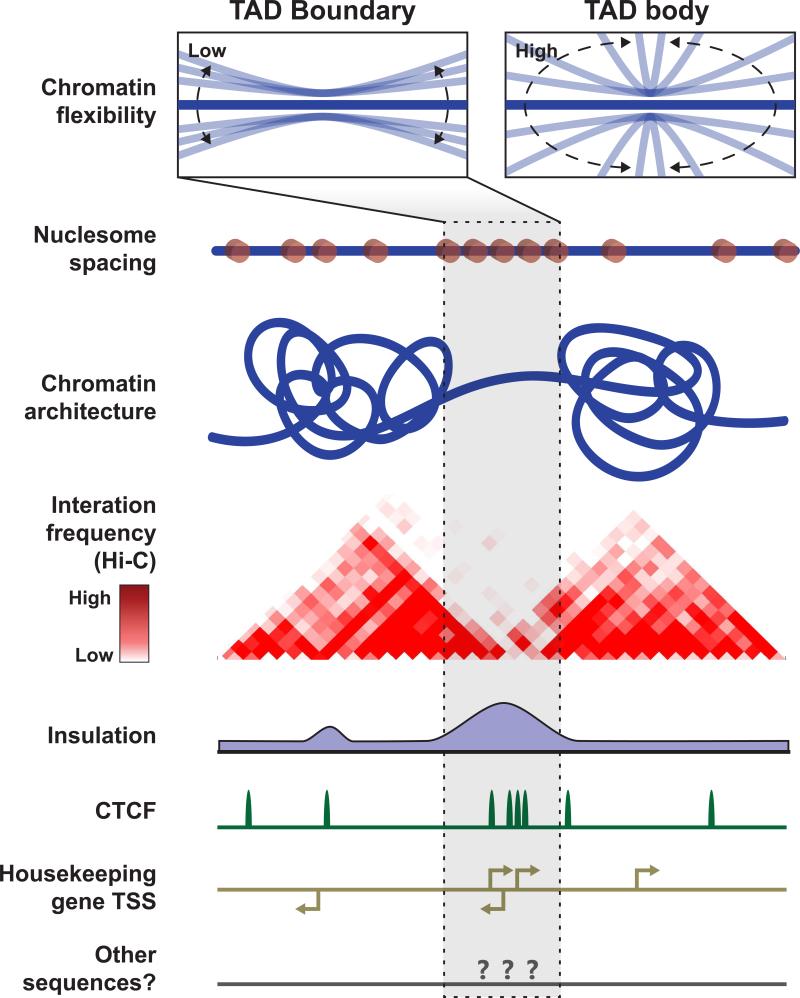
We can envision a second set of models in which the insulating forces at TAD boundaries are initiated independent of loop formation (Figure 6). One clue about the mechanism(s) that create insulating forces at TAD boundaries may come from a common property of CTCF binding sites and housekeeping (highly active) TSSs, which are highly enriched at TAD boundaries: both types of functional elements create arrays of closely-spaced nucleosomes in adjacent chromatin (Fu et al., 2008; Gaffney et al., 2012; Valouev et al., 2011). CTCF positions roughly 20 nucleosomes around its binding site, spaced at ~185 bp intervals (Gaffney et al., 2012). Likewise, highly active TSS like those of housekeeping genes create arrays of well positioned nucleosomes adjacent to the relatively small nucleosome free region that overlaps the TSS. Notably, nucleosome spacing decreases with increasing transcriptional activity. In fact, average nucleosome spacing around active promoters (~178-187bp) is shorter than in other regions including heterochromatin domains marked by H3K9me3 or H3K27me3 (~205bp) (Valouev et al., 2011). Importantly, in vitro studies have shown that spacing between adjacent nucleosomes impacts chromatin flexibility (Correll et al., 2012). More specifically, the evidence suggests that short spacing between adjacent nucleosomes can inhibit higher order chromatin folding. Further, in silico modeling of chromatin dynamics suggests that regions of local polymer “stiffness” can create local insulation in contact frequencies, though this fails to create an entire “TAD-like” structure in some simulations (Fudenberg et al., 2015). We hypothesize that the insulation observed at TAD boundaries results from a lack of flexibility (i.e. stiffness) of the chromatin fiber, which may be caused by functional elements including CTCF binding sites, highly active TSSs, and perhaps other functional elements that position arrays of tightly-spaced nucleosomes in adjacent chromosomes(Figure 6). Following the establishment of insulation forces at the boundary, “attractive” forces within the domain can then confer specific local chromatin interactions, yielding a joint “insulation-attraction” model of TAD formation.
Figure 6. Chromatin stiffness may create insulation at TAD boundaries.
Diagrammatic representation of a chromatin fiber (blue) spanning two adjacent TADs and the boundary region between them. According to this model, chromatin at the TAD boundary region is less flexible than chromatin in the TAD body, as depicted in the two boxes above the chromatin fiber. The low flexibility of chromatin at the TAD boundary could inhibit interaction between regions on opposite sides of the boundary. Chromatin flexibility may be modulated by nucleosome spacing, as dictated by functional elements including CTCF binding sites and gene promoters. However, chromatin flexibility may be modulated by other properties in addition to or instead of nucleosome spacing. Nucleosomes are represented as orange circles on the chromatin fiber. They are not drawn to scale, as they are intended only to convey that variation in nucleosome spacing occurs along the chromatin fiber. Additional tracks are shown below the chromatin fiber for conceptual representation: “Interaction frequency” shows example Hi-C data from a region with a distinct TAD boundary. The “Insulation” track reflects a defining property of TAD boundaries – there are relatively few interactions that cross TAD boundaries, and thus insulation at TAD boundaries is high. “CTCF” and “Housekeeping gene TSS” tracks depict features typically found at TAD boundaries. The “other sequences?” track is meant to convey the idea that unknown sequence features may also contribute to TAD formation.
This “insulation-attraction” model is speculative, but it does explain several experimental observations that are not well explained by the handcuff or extrusion models. 1) It has been observed that many domain boundaries do not form loops, suggesting that in at least some cases boundary regions do not have to form a loop in order to function as boundaries (Rao et al., 2014). The “insulation-attraction” model predicts that any factor which reduces the flexibility of chromatin could in theory produce a TAD boundary, and thus boundary formation does not require boundary looping. Boundary looping may serve as an attractive force to further encourage intra-TAD interactions, which could explain the prevalence of convergent CTCF motifs at adjacent boundaries, but boundary looping would not be required to establish or maintain a boundary. It should be noted that according to computer simulations the extrusion model does not require stable looping interactions between boundaries as long as there is an active loop extrusion factor (Fudenberg et al., 2015). However, according to this model, loss of Cohesin – the posited extrusion factor – should abrogate TAD boundaries at non-looping boundaries. On the contrary, multiple studies have found that loss of Cohesin has only a subtle impact on TAD boundary formation (Seitan et al., 2013; Sofueva et al., 2013; Zuin et al., 2014). 2) Loss of CTCF has a more profound impact on boundary function than loss of Cohesin (Zuin et al., 2014). If nucleosome spacing is an important mediator of chromatin stiffness (as postulated above), then loss of Cohesin would not be predicted to have a major impact on boundary formation. In contrast, loss of CTCF would be predicted to have a stronger impact on boundary formation because loss of CTCF binding would presumably disrupt the nucleosome arrays normally formed in CTCF-adjacent chromatin. 3) At least 10% of the TAD boundaries lack detectable CTCF binding, suggesting that CTCF-independent mechanisms may exist to mediate the formation of TADs in mammalian cells (Dixon et al., 2012). According to the “insulation-attraction” model CTCF is not strictly necessary for TAD boundary formation, as any factor that shortens nucleosome spacing, or perhaps impacts the flexibility of a chromatin fiber in other ways, could lead to boundary formation. We suspect that in most cases the formation of a TAD boundary requires more than one functional element – for example, the combination of several CTCF binding sites, housekeeping TSSs, and/or repetitive elements, spread over several kb.
Future Perspectives
The improvement in methods for studying 3D genome organization led to the identification of TADs as well as other features of higher order chromatin structure. Further advancements in our understanding of what TADs are will be similarly aided by future technological developments. One of the outstanding questions in the field is, what do TADs actually look like in cells? A variety of potential models have been suggested based on existing experimental data (Giorgetti et al., 2014; Sanborn et al., 2015; Tang et al., 2015; Vietri Rudan and Hadjur, 2015), yet we still have a rather poor understanding of what the structure of TADs are like inside cells. Super-resolution microscopy will likely be critical in resolving these issues. There have already been some studies characterizing “epigenetic” domains from a structural level using super resolution imaging (Boettiger et al., 2016). Improvements in both resolution and the number of possible dyes will provide key insights into the organization of TADs and the elements within them. Likewise, the adoption of imaging approaches using dye based barcoding strategies to study chromosomal structures may also greatly increase our understanding of the structure of chromosomes in single cells (Beliveau et al., 2015; Chen et al., 2015; Lee et al., 2014; Lubeck et al., 2014). Similarly, in order to understand the dynamics of TAD organization, improvement in imaging techniques will also be essential. These can include super-resolution imaging, CRISPR-Cas9 based imaging (Chen et al., 2013; Tanenbaum et al., 2014), and improvements in multi-color live cell imaging. Lastly, there is currently a revolution occurring in cryo-EM based characterization of large macromolecular complexes(Bai et al., 2015). Whether these methods will be able to be extended to study what are possibly 200nM or greater sized chromosomal domains inside of cells is unclear. However, these methods will likely be essential for unraveling the structure of the protein complexes that are critical for TAD organization.
Single cell methods also show great promise for helping to unravel the degree of cellular variability in various biological processes. From even relatively low resolution FISH studies, it is clear that chromosome structure can show high variability between cells, so it will be crucial to characterize this degree of variability more comprehensively. Remarkably, single cell Hi-C methods have already been developed (Nagano et al., 2013; Nagano et al., 2015). These have shown that TAD structure is present even in individual cells, but that contacts between TADs are highly variable. However, these initial single cell Hi-C methods suffered from relatively poor resolution and low sample throughput. Improvements in either of these aspects of the methodology will potentially have major impact on unraveling the variability of internal TAD structure between individual cells.
From a mechanistic perspective, the clearest outstanding question in the field of chromosome organization relates to how CTCF manages to define chromatin domains. The recent identification of the preference of binding between regions containing convergent CTCF motifs provides a critical clue (Rao et al., 2014), but it also raises many important questions. These have lent credence to the possibility of either “linear tracking” or “loop extrusion” models, which are also not mutually exclusive. If either of these models are indeed true, it will be critical to understand what are the molecular forces that may control loop extrusion, or what molecules, such as polymerases, helicases, non-coding RNAs, may be mediating linear tracking. The holy grail to address these questions would be to observe these processes taking place inside of a cell, likely either through sophisticated live cell imaging or clever time course experiments.
From a functional perspective, the emerging appreciation of the impact of disruptions to higher order chromatin structure in human disease will only continue to grow. Several studies have recently shown the impact of genomic structural variations that alter 3D genome structure in human Mendelian diseases and in cancer. Given the number large number of uncharacterized structural variants in the human genome as well as the dramatic and extensive alterations seen to the genome in cancer cells (Zack et al., 2013), characterizing how these structural variations impact TAD structure will be a Herculean effort. However, given the profound effects seen in the few studies that have characterized the impact of structural variations on 3D genome structure (Flavahan et al., 2016; Groschel et al., 2014; Hnisz et al., 2016), this line of research will likely provide a rich amount of information about the mechanisms by which structural changes to the linear sequence of our genome can impact human disease.
ACKNOWLEDGEMENTS
We apologize to those authors whose work could not be represented here due to limitation in space. This work is supported by funding from Ludwig Institute for Cancer Research (BR), A.P. Giannini Foundation (DG), NIH IRACDA K12 GM068524 (DG), NIH (U54HG006997, 1R01ES024984, P50GM085764, and 1U54DK107977) to BR. JD is supported by the Leona M. and Harry B. Helmsley Charitable Trust Grant 2012-PG-MED002.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES CITED
- Alipour E, Marko JF. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012;40:11202–11212. doi: 10.1093/nar/gks925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340:1234167. doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- Austenaa LM, Barozzi I, Simonatto M, Masella S, Della Chiara G, Ghisletti S, Curina A, de Wit E, Bouwman BA, de Pretis S, et al. Transcription of Mammalian cis-Regulatory Elements Is Restrained by Actively Enforced Early Termination. Mol Cell. 2015;60:460–474. doi: 10.1016/j.molcel.2015.09.018. [DOI] [PubMed] [Google Scholar]
- Bai XC, McMullan G, Scheres SH. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 2015;40:49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Bailey SD, Zhang X, Desai K, Aid M, Corradin O, Cowper-Sal Lari R, Akhtar-Zaidi B, Scacheri PC, Haibe-Kains B, Lupien M. ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nat Commun. 2015;2:6186. doi: 10.1038/ncomms7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri M, Chotalia M, Fraser J, Lavitas LM, Dostie J, Pombo A, Nicodemi M. Complexity of chromatin folding is captured by the strings and binders switch model. Proc Natl Acad Sci U S A. 2012;109:16173–16178. doi: 10.1073/pnas.1204799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau BJ, Boettiger AN, Avendano MS, Jungmann R, McCole RB, Joyce EF, Kim-Kiselak C, Bantignies F, Fonseka CY, Erceg J, et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat Commun. 2015;6:7147. doi: 10.1038/ncomms8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Bintu B, Moffitt JR, Wang S, Beliveau BJ, Fudenberg G, Imakaev M, Mirny LA, Wu CT, Zhuang X. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016;529:418–422. doi: 10.1038/nature16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T. Die blastomerenkerne von Ascaris meglocephala und die theorie der chromosomenindividualitaet. Archiv für Zellforschung. 1909;3:181–268. [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Correll SJ, Schubert MH, Grigoryev SA. Short nucleosome repeats impose rotational modulations on chromatin fibre folding. EMBO J. 2012;31:2416–2426. doi: 10.1038/emboj.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. Two ways to fold the genome during the cell cycle: insights obtained with chromosome conformation capture. Epigenetics Chromatin. 2014;7:25. doi: 10.1186/1756-8935-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Heard E. Structural and functional diversity of Topologically Associating Domains. FEBS Lett. 2015;589:2877–2884. doi: 10.1016/j.febslet.2015.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Mirny L. The 3D Genome as Moderator of Chromosomal Communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle B, Fudenberg G, Imakaev M, Mirny LA. Chromatin loops as allosteric modulators of enhancer-promoter interactions. PLoS Comput Biol. 2014;10:e1003867. doi: 10.1371/journal.pcbi.1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira J, Paolella G, Ramos C, Lamond AI. Spatial organization of large-scale chromatin domains in the nucleus: a magnified view of single chromosome territories. J Cell Biol. 1997;139:1597–1610. doi: 10.1083/jcb.139.7.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova D, Patro R, Duggal G, Kingsford C. Identification of alternative topological domains in chromatin. Algorithms Mol Biol. 2014;9:14. doi: 10.1186/1748-7188-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suva ML, Bernstein BE. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of Chromosomal Domains by Loop Extrusion. bioRxiv. 2015 doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney DJ, McVicker G, Pai AA, Fondufe-Mittendorf YN, Lewellen N, Michelini K, Widom J, Gilad Y, Pritchard JK. Controls of nucleosome positioning in the human genome. PLoS Genet. 2012;8:e1003036. doi: 10.1371/journal.pgen.1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirlando R, Felsenfeld G. CTCF: making the right connections. Genes Dev. 2016;30:881–891. doi: 10.1101/gad.277863.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Galupa R, Nora EP, Piolot T, Lam F, Dekker J, Tiana G, Heard E. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014;157:950–963. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorkin DU, Leung D, Ren B. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell. 2014;14:762–775. doi: 10.1016/j.stem.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BA, Erpelinck C, van der Velden VH, Havermans M, Avellino R, van Lom K, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- Grubert F, Zaugg JB, Kasowski M, Ursu O, Spacek DV, Martin AR, Greenside P, Srivas R, Phanstiel DH, Pekowska A, et al. Genetic Control of Chromatin States in Humans Involves Local and Distal Chromosomal Interactions. Cell. 2015;162:1051–1065. doi: 10.1016/j.cell.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger P, Marin B, Bartkuhn M, Schierenberg E, Wiehe T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc Natl Acad Sci U S A. 2012;109:17507–17512. doi: 10.1073/pnas.1111941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger P, Marin B, Schierenberg E. Loss of the insulator protein CTCF during nematode evolution. BMC Mol Biol. 2009;10:84. doi: 10.1186/1471-2199-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari N, Phanstiel DH, He C, Grubert F, Jahanbani F, Kasowski M, Zhang MQ, Snyder MP. Genome-wide map of regulatory interactions in the human genome. Genome Res. 2014;24:1905–1917. doi: 10.1101/gr.176586.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Zhang Y, Zhao L, Frock RL, Du Z, Meyers RM, Meng FL, Schatz DG, Alt FW. Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes. Cell. 2015;163:947–959. doi: 10.1016/j.cell.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Dadon DB, Powell BE, Fan ZP, Borges-Rivera D, Shachar S, Weintraub AS, Hnisz D, Pegoraro G, Lee TI, et al. 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell Stem Cell. 2016;18:262–275. doi: 10.1016/j.stem.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grontved L, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SS, Li C, Amamoto R, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonfat N, Montavon T, Darbellay F, Gitto S, Duboule D. Convergent evolution of complex regulatory landscapes and pleiotropy at Hox loci. Science. 2014;346:1004–1006. doi: 10.1126/science.1257493. [DOI] [PubMed] [Google Scholar]
- Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas JS, Zhang Y, Dudko OK, Murre C. 3D trajectories adopted by coding and regulatory DNA elements: first-passage times for genomic interactions. Cell. 2014;158:339–352. doi: 10.1016/j.cell.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng PC, Meng C, Berezney R. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J Cell Biol. 1998;143:1415–1425. doi: 10.1083/jcb.143.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Yaffe E, Wingett SW, Dean W, Tanay A, Fraser P. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat Protoc. 2015;10:1986–2003. doi: 10.1038/nprot.2015.127. [DOI] [PubMed] [Google Scholar]
- Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, Reinberg D. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347:1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Lee C, Zichner T, Stutz AM, Erkek S, Kawauchi D, Shih DJ, Hovestadt V, Zapatka M, Sturm D, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Corces VG. Chromatin insulators: linking genome organization to cellular function. Mol Cell. 2013;50:461–474. doi: 10.1016/j.molcel.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl C. Über Zellteilung. Morphologisches Jahrbuch. 1885;10:214–330. [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014 doi: 10.1016/j.cell.2014.11.021. published online December 12, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Corces VG. The three-dimensional genome: principles and roles of long-distance interactions. Curr Opin Cell Biol. 2016;40:8–14. doi: 10.1016/j.ceb.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, Ing-Simmons E, Lenhard B, Giorgetti L, Heard E, Fisher AG, et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013;23:2066–2077. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Cavalli G. The role of chromosome domains in shaping the functional genome. Cell. 2015;160:1049–1059. doi: 10.1016/j.cell.2015.02.040. [DOI] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shi Y, Dai C, Tjong H, Gong K, Alber F, Zhou XJ. TopDom: an efficient and deterministic method for identifying topological domains in genomes. Nucleic Acids Res. 2016;44:e70. doi: 10.1093/nar/gkv1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Yaffe E, Chan WC, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparvoli E, Levi M, Rossi E. Replicon clusters may form structurally stable complexes of chromatin and chromosomes. J Cell Sci. 1994;107(Pt 11):3097–3103. doi: 10.1242/jcs.107.11.3097. [DOI] [PubMed] [Google Scholar]
- Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, Ettwiller L, Spitz F. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24:390–400. doi: 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K, Nichols MH, Li L, Ong CT, Takenaka N, Qin ZS, Corces VG. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014;15:R82. doi: 10.1186/gb-2014-15-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K, Ramos E, Takenaka N, Yang J, Wahi JE, Corces VG. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res. 2012;22:2176–2187. doi: 10.1101/gr.136788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M, Hadjur S. Genetic Tailors: CTCF and Cohesin Shape the Genome During Evolution. Trends Genet. 2015;31:651–660. doi: 10.1016/j.tig.2015.09.004. [DOI] [PubMed] [Google Scholar]
- Waszak SM, Delaneau O, Gschwind AR, Kilpinen H, Raghav SK, Witwicki RM, Orioli A, Wiederkehr M, Panousis NI, Yurovsky A, et al. Population Variation and Genetic Control of Modular Chromatin Architecture in Humans. Cell. 2015;162:1039–1050. doi: 10.1016/j.cell.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Watrin E, Kaiser FJ, Wendt KS. Gene regulation and chromatin organization: relevance of cohesin mutations to human disease. Curr Opin Genet Dev. 2016;37:59–66. doi: 10.1016/j.gde.2015.12.004. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Xu M, Zhao GN, Lv X, Liu G, Wang LY, Hao DL, Wang J, Liu DP, Liang CC. CTCF controls HOXA cluster silencing and mediates PRC2- repressive higher-order chromatin structure in NT2/D1 cells. Mol Cell Biol. 2014;34:3867–3879. doi: 10.1128/MCB.00567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Kerschner JL, Gosalia N, Neems D, Gorsic LK, Safi A, Crawford GE, Kosak ST, Leir SH, Harris A. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng CZ, Wala J, Mermel CH, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wong CH, Birnbaum RY, Li G, Favaro R, Ngan CY, Lim J, Tai E, Poh HM, Wong E, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IWF, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]