Abstract
Laccases have gained significant attention due to their emerging applications including bioremediation, biomass degradation and biofuel cells. One of the prerequisites for the industrial application of laccases is their sufficient availability. However, expression levels of recombinantly expressed laccases are often low. In this study Mrl2, a new laccase from the basidiomycete Moniliophthora roreri, was cloned in Pichia pastoris and produced in an optimized fed-batch process at an exceptionally high yield of 1.05 g l−1. With a redox potential of 0.58 V, Mrl2 belongs to mid-redox potential laccases. However, Mrl2 demonstrated high kcat values of 316, 20, 74, and 36 s−1 towards 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS), syringaldazine (SGZ), 2,6-dimethoxyphenol (2,6-DMP) and guaiacol, respectively. Mrl2 remained stable above pH 6 and in the presence of many metal ions, which is important for application in bioremediation. Mrl2 was investigated for the ability to degrade endocrine disrupting chemicals (EDCs) and non-steroidal anti-inflammatory drugs (NSDAIs) at neutral pH value. The enzyme accepted and converted estrone, 17β-estradiol, estriol, the synthetic contraceptive 17α-ethinyl estradiol and bisphenol A at pH 7 faster than high-potential laccases from Trametes versicolor. For example, within 30 min Mrl2 removed more than 90% bisphenol A, 17ß-estradiol, 17α-ethinyl estradiol and estriol, respectively. The concentration of the recalcitrant drug diclofenac dropped by 56% after 20 h incubation with Mrl2.
Electronic supplementary material
The online version of this article (doi:10.1186/s13568-017-0368-3) contains supplementary material, which is available to authorized users.
Keywords: Laccase, Expression, Pichia pastoris, Micropollutant degradation
Introduction
Laccases (EC 1.10.3.2) are multi-copper oxidases which catalyze the oxidation of various electron-rich organic and inorganic molecules like mono- and diphenols, polyphenols, diamines, aminophenols, aromatic or aliphatic amines with the four-electron reduction of molecular oxygen to water (Thurston 1994; Xu 1996; Xu et al. 1996). Small compounds, so called redox mediators, can expand the substrate range to non-phenolic lignin derivatives (Bourbonnais and Paice 1990; Eggert et al. 1996) or recalcitrant dyes (Soares et al. 2001). Laccases contain four copper ions. Substrates are oxidized at the mononuclear type 1 (T1) copper site and electrons are transferred to the trinuclear site comprising one T2 and two T3 copper ions, where the reduction of oxygen to water takes place (Solomon et al. 2008). Laccases are ubiquitous and found in plants, fungi, insects (Mayer and Staples 2002), yeast (Kalyani et al. 2015) and bacteria (Dwivedi et al. 2011). The redox potential of laccases is quite different and range from 0.36 to 0.8 V. It is widely accepted that the axial ligand of the T1 copper as well as a tripeptide in the T1 copper site (LEA) roughly indicate the redox potential of laccases (Mot and Silaghi-Dumitrescu 2012; Xu et al. 1998). Low-potential laccases, mainly found in bacteria, with a redox potential from 0.36 to 0.46 V exhibit a methionine at the axial position, while middle-potential laccases, mainly from ascomycete origin, with a redox potential of 0.46–0.71 V usually have a leucine. High-potential laccases with a redox potential >0.71 V are mainly found in basidiomycetes. They display a phenylalanine as a non-coordinating axial ligand (Mot and Silaghi-Dumitrescu 2012). High-potential laccases typically possess a broader substrate spectrum than low- or middle-potential laccases and are even able to oxidize substrates with a redox potential of up to 1.2 or 1.4 V (Tadesse et al. 2008). Regarding their broad substrate spectrum while only relying on water as a cosubstrate, laccases often are called “green” biocatalysts (Pardo and Camarero 2015). Therefore, these enzymes, and high-potential laccases in particular, are attractive for industrial applications as in the pulp and paper, food and textile industry, nanobiotechnology, synthetic chemistry or bioremediation (Rodriguez Couto and Toca Herrera 2006).
Micropollutants comprising endocrine disrupting, toxic, persistent and bioaccumulative substances, like EDCs and NSAIDs (Table 1), are one of the key problems facing humanity (Schwarzenbach et al. 2006). They are ubiquitous in aquatic environments, since e.g. human or veterinary pharmaceuticals are only partially metabolized and eventually end up in the wastewater system (Hamid and Eskicioglu 2012). In an aging society it is very likely that more and more pharmaceuticals will be released in the environment. Human hormones as 17β-estradiol or the contraceptive 17α-ethinyl estradiol have been reported to lead to feminization or sexual disruption of fish (Chen et al. 2016a; Orn et al. 2016). Other estrogens as estrone which is predominant in menopausal women and estriol, a metabolite of estrone and 17β-estradiol, are also found in wastewaters (Auriol et al. 2007a, b). The diphenylmethane derivatives bisphenol A and bisphenol S used in plastics have endocrine mimicking properties (Ji et al. 2013; Vinas and Watson 2013). Diclofenac and naproxen are NSAIDs which show analgesic, antipyretic and anti-inflammatory effects. Because of more and more emerging concerns about the fate of EDCs and NSAIDs much effort has been made to develop alternative strategies to remove those micropollutants since conventional wastewater treatment processes do not meet the demands (Schroder et al. 2016). Among others laccases are capable of reducing estrogenic activity in wastewater (Suzuki et al. 2003; Tsutsumi et al. 2001). However, most high-redox potential laccases demonstrate low activity under neutral or basic pH conditions found in wastewater (Baldrian 2006). Another limitation for a broader laccase application is their rather low expression level in recombinant hosts.
Table 1.
Chemical structures of endocrine disrupting chemicals and nonsteroidal anti-inflammatory drugs investigated in this study
| Endocrine disrupting chemicals (EDCs) | |||
| E1 |
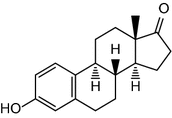
|
E2 |
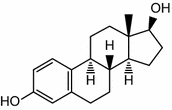
|
| EE2 |
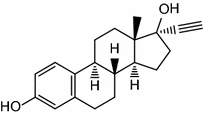
|
E3 |
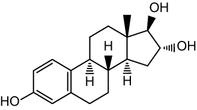
|
| BPA |
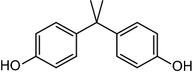
|
BPS |
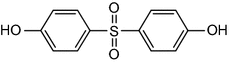
|
| Nonsteroidal anti-inflammatory drugs (NSAIDs) | |||
| Diclofenac | Naproxen | ||
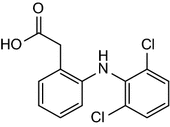
|
|
||
E1 estrone, E2 17ß-estradiol, EE2 17α-ethinyl estradiol, E3 estriol, BPA bisphenol A, BPS bisphenol S
In this study, we identified and characterized a new laccase, Mrl2, from Moniliophthora roreri. Mrl2 was produced at exceptionally high levels in Pichia pastoris (1.05 g l−1) in a 3 l fed-batch fermentation process. High stability above pH 6 and resistance to many metal ions make this enzyme suitable for application in wastewater treatment. Amongst others, 17β-estradiol, 17α-ethinyl estradiol and diclofenac, which are listed by the European Union (EU) as dangerous compounds which should be monitored by the EU members (Schroder et al. 2016) could be degraded by Mrl2.
Materials and methods
Materials
Unless specified otherwise, all chemicals (of analytical grade or higher) and commercial proteins were acquired from AppliChem. (Darmstadt, Germany), Thermo Fisher Scientific Inc. (Waltham, Massachusetts, USA), Sigma-Aldrich (Schnelldorf, Germany), VWR (Darmstadt, Germany), Fermentas (St. Leon-Rot, Germany) or New England Biolabs (Ipswich, Massachusetts, USA).
Cloning of mrl2
The gene for Mrl2 (NCBI Reference Sequence: XP_007855001.1) was codon optimized (GenBank accession number: KY111767) with JCat for expression in yeast (http://www.jcat.de/) and synthesized by Eurofins (Ebersberg Germany). The gene was cloned in the pPICZαA vector from Invitrogen™ (Carlsbad, California, USA) with both the native signal peptide and the α-factor secretion signal from Saccharomyces cerevisiae. Mrl2 was amplified with primers mrl2_BstBI_fw (GATAttcgaaATGGCTAGATTGCAATTC) and mrl2_XbaI_rev (CAagatctTTACAAGTCGTCGTCAG) and mrl2_XhoI_fw (GTATctcaga AAAAGATCTATCGGTCCAATCG) and mrl2_XbaI_rev for the native signal sequence and the α-factor construct, respectively. Recognition sites for the endonucleases are underlined. The forward primer contained the Kex2 signal cleavage site (highlighted in bold), thus mrl2 starts right behind Kex2 at the end of the alpha factor pre pro leader sequence. The vector and the amplified genes were subjected to cleavage with respective restriction enzymes and ligated into pPICZαA to generate pPICZαAmrl2 and pPICZAmrl2. The sequence of the constructs was confirmed by sequencing (GATC Biotech, Konstanz, Germany). pPICZαAmrl2 and pPICZAmrl2 were linearized with PmeI and inserted in P. pastoris X-33 (Invitrogen, Carlsbad, California, USA) by electroporation. After transformation, cells were selected at 30 °C on Yeast Extract Peptone Dextrose medium with sorbitol (YPDS; 10 g l−1 yeast extract; 182.2 g l−1 sorbitol; 20 g l−1 peptone; 15 g l−1 agar) agar plates with 100 µg ml−1 Zeocin™. For further assessment clones were streaked out on 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS)-Buffered Minimal Methanol (BMM; 13.4 g l−1 Yeast nitrogen base w/o amino acids; 15 g l−1 agar; 0.00004% biotin, 0.5% methanol; 100 mM potassium phosphate buffer pH 6; 0.3 mM CuSO4; 0.2 mM ABTS) agar plates and incubated at 30 °C. Clones which formed greenish halos were chosen for further experiments.
Expression of Mrl2 in shaking flasks
Pichia pastoris transformants were grown in 10 ml Buffered Complex Glycerol medium (BMGY; 10 g l−1 yeast extract; 20 g l−1 peptone; 100 mM potassium phosphate buffer pH 6; 1% glycerol; 0.00004% biotin) over night at 30 °C and 180 rpm. Overnight cultures were used for inoculation of 50 ml BMM medium in baffled flasks to an OD600 of 0.1. The cultures were shaken at 30 °C and 180 rpm for 3 days. The medium was supplemented with methanol every day to 0.5% (v/v) final concentration. Samples were taken daily for cell density and laccase activity monitoring.
Expression of Mrl2 in a 7.5 l bioreactor
Pichia pastoris transformants were grown in 10 ml BMGY (preculture) over night at 30 °C. A starter culture of 200 ml BMGY medium was inoculated with the preculture to an OD600 of 0.01–0.05 and grown over night at 30 °C and 180 rpm. The starter culture was used to inoculate a 7.5 l bioreactor (Infors, Bottmingen, Switzerland) containing 3 l fermentation basal salt medium (0.47 g l−1 CaSO4·2 H2O, 9.1 g l−1 K2SO4, 7.5 g l−1 MgSO4·7 H2O, 4.2 g l−1 KOH, 8 ml H3PO4 (85%), 50 g l−1, glycerol (87%), 0.87 mg l−1 biotin, 4.35 ml l−1 Pichia trace metals (PTM1, 6 g l−1 CuSO4·5 H2O, 0.08 g l−1 NaI, 3 g l−1 MnSO4·H2O, 0.5 g l−1 CoCl2, 20 g l−1 ZnCl2, 0.02 g l−1 H3BO3, 0.2 g l−1 Na2Mo4·2 H2O, 65 g l−1 FeSO4·7 H2O, 0.2 g l−1 biotin, 5 ml l−1 H2SO4) to an OD600 of 0.5. pH 6 was adjusted with 10% phosphoric acid and 25% ammonium hydroxide. In the first growth phase on glycerol the temperature was maintained at 30 °C. When glycerol was used up (noticeable in pO2 increase) expression was induced by addition of methanol (0.5% (w/v) containing 12 g l−1 trace metal salts PTM1) and temperature was shifted to 25 °C. Methanol was added automatically when the C-source was depleted, indicated by a sharp increase in pO2 value. 0.9 ml 1 M CuSO4 was added daily to the fermentation broth. Samples were taken daily for monitoring OD600, laccase activity and determining protein concentration by the Bradford assay using bovine serum albumin (BSA) as standard.
Purification and characterization of Mrl2
The fermentation broth was centrifuged (10,000g; 15 min; 4 °C) and the supernatant was concentrated by Crossflow ultra-filtration with a cut-off membrane of 10 kDa (Pall, East Hills, NY, USA). The concentrated supernatant was centrifuged (22,000g; 20 min; 4 °C) and filtered through a 0.22 µm pore size filter. 5–10 ml of the concentrated supernatant was purified by DEAE FF anion exchange chromatography with an Äkta FPLC (GE Healthcare, Chalfont St Giles, Buckinghamshire, UK). After protein application the column was washed with 50 mM potassium phosphate buffer pH 6 and 200 mM NaCl. Mrl2 was eluted with 50 mM potassium phosphate buffer pH 6 and 250 mM NaCl. Blueish, active fractions (towards ABTS as substrate) were pooled and concentrated with a MILLIPORE Amicon® stirred ultrafiltration cell 8200 (Bedford, Maine, USA) with a cut-off membrane of 10 kDa and desalted with a PD Midi Trap G-25 desalting column (GE Healthcare, Chalfont St Giles, Buckinghamshire, UK). Mrl2 was stored in 50 mM potassium phosphate buffer pH 6 at −20 °C until further use.
Protein concentration was estimated by the Bradford Assay with BSA as standard. Deglycosylation was conducted with PNGase F as described in the manual instruction. SDS-PAGE was conducted according to Laemmli (1970). Samples used for the zymogram were not heated at 95 °C. For activity staining the SDS-PAGE gel was incubated in 100 mM sodium acetate buffer pH 5 supplemented with 0.5 mM ABTS.
Copper content of Mrl2 was determined by atomic absorption spectroscopy on a Perkin Elmer AAnalyst 100 (Waltham, USA) equipped with an air-acetylene burner. Mrl2 was diluted in water for copper measurement. The same sample was used for protein concentration determination by Bradford assay and at 280 nm. For determining the protein concentration the molar extinction coefficient was calculated with http://web.expasy.org/protparam/as 80,580 M−1 cm−1.
Redox potential measurements were performed in a nitrogen flushed glove box under the absence of oxygen. For the titration redox mediator couples K3[Fe(CN)6]/K4[Fe(CN)6] and [Fe(bipy)2]3+/[Fe(bipy)2]2+ were used with a standard redox potential of 0.433 V (O’Reilly 1973) and 0.78 V versus the standard hydrogen electrode, respectively. For the bipyridine complex solutions, iron(III) chloride hexahydrate and iron(II) chloride were mixed in 1:2 ratio with 2,2′-bipyridine. The reduction status of the T1 copper was monitored at 600–750 nm. Measurements were performed in 50 mM potassium phosphate buffer, pH 7.5 at room temperature in duplicate. The normalized absorbance was plotted against the redox potential with the software Igor Pro (Wavemetrics; https://www.wavemetrics.com/) and the midpoint was determined using the Nernst equation for a one electron redox reaction: with n = 1, F = 96,486 C mol−1, R = 8.314 J mol−1 K−1, T = 293 K, Em = midpoint potential.
Enzyme activity determination and kinetic parameters
Enzyme activity was determined in 100 mM sodium acetate buffer, pH 5 with 0.5 mM ABTS at room temperature. The increase of absorbance resulting from ABTS oxidation was monitored at 420 nm (ε = 36,000 M−1 cm−1). Kinetic parameters of Mrl2 with the substrates ABTS, 2,6-dimethoxyphenol (2,6-DMP; 468 nm; ε = 49,600 M−1 cm−1), guaiacol (470 nm; ε = 26,000 M−1 cm−1) and syringaldazine (SGZ; 530 nm; ε = 65,000 M−1 cm−1) were determined in 100 mM citrate phosphate buffer at their optimal pH. The substrate concentration ranges were 0.00781–1 mM, 0.000976–0.5 mM, 0.001–10 mM and 0.0001–10 mM for ABTS, SGZ, 2,6-DMP, and guaiacol, respectively. One Unit is defined as the amount of enzyme that converts 1 µmol substrate per minute.
Effect of pH, temperature, metal ions and inhibitors on enzyme activity
For determining the optimal pH the assay was conducted at different pH values: pH 2 and 8–10 in 100 mM Britton Robinson buffer and pH 3–7 in 100 mM citrate phosphate buffer. For temperature stability investigations the enzyme was diluted to 500 µg ml−1 in 50 mM potassium phosphate buffer pH 7 in 0.2 ml reaction tubes. The tubes were incubated at 20, 30, 40, 50, 60, 70 and 80 °C in a PCR Cycler and the residual activity was measured with the standard ABTS activity assay. For pH stability study Mrl2 was diluted in 100 mM Britton Robinson buffer at pH 2-10 and incubated at room temperature. Aliquoted samples were used directly for measuring residual activity with the ABTS assay. The activity in the presence of metal ions as indicated in Table 4 was determined with ABTS as substrate at pH 3 in 100 mM citrate phosphate buffer. The enzyme was incubated with the metal ions for 5 min before the addition of ABTS. The metal ions were added as sulfates or in the case of calcium as nitrate. To assess the metal chelating effect of the citrate buffer used, enzyme activity was tested also in 100 mM HEPES buffer, pH 3 in the presence of 100 mM calcium, nickel, cobalt or zinc. Enzyme stability in the presence of 10 and 100 mM glycerin, acetonitrile, methanol, ethanol, 2-propanol, acetone, dimethyl sulfoxide (DMSO), and dimethylformamide (DMF) was determined as described for metal ion tolerance but in sodium acetate buffer pH 5. For determining activity in the presence of potential inhibitors the protein was incubated in 100 mM sodium acetate buffer pH 5 including 10–100 mM NaCl, 0.1–1% SDS or 0.1% sodium azide for 5 min at room temperature. After incubation time ABTS was added to the reaction mixture and oxidation was followed at 420 nm.
Table 4.
Residual activity of Mrl2 towards ABTS in presence of different metal ions, cosolvents and inhibitors
| Metal ions | Residual activity (%) | Cosolvent | Residual activity (%) | Inhibitor | Concentration | Residual activity (%) | ||
|---|---|---|---|---|---|---|---|---|
| 10 mM | 100 mM | 10% | 20% | |||||
| Mn2+ | 99.3 ± 1.9 | 119.4 ± 3.8 | Ethanol | 81.9 ± 1.5 | 21.7 ± 2.4 | SDS | 0.1% | 100.3 ± 0.7 |
| Co2+ | 100.9 ± 0.9 | 104.6 ± 2.9 | Methanol | 76.5 ± 2.7 | 39 ± 0.8 | 1% | 77.7 ± 1.1 | |
| Ni2+ | 82.7 ± 1.5 | 26.2 ± 0.9 | 2-Propanol | 61.9 ± 2.5 | 5.5 ± 2.5 | |||
| Ca2+ | 85.4 ± 1 | 50.4 ± 1.9 | ACN | 52 ± 2.8 | 0.7 ± 0.4 | Sodium azide | 0.1% | 3.3 ± 1.9 |
| Cu2+ | 104.9 ± 2.5 | 95.2 ± 2.7 | DMSO | 22.9 ± 2.2 | 26.1 ± 3.6 | |||
| Na+ | 99 ± 2.8 | 105.1 ± 4.4 | Acetone | 22.2 ± 2 | 16.6 ± 2.3 | Cl− | 10 mM | 37.3 ± 0.9 |
| Mg2+ | 101.5 ± 1.5 | 80.1 ± 1.7 | DMF | 40.2 ± 4.3 | 16.6 ± 0.4 | 50 mM | 12.2 ± 0.15 | |
| Zn2+ | 104.9 ± 2.1 | 97.0 ± 1.2 | Glycerol | 85.7 ± 0.5 | 89.7 ± 1.9 | 100 mM | 6.7 ± 0.04 | |
ACN acetonitrile, DMSO dimethyl sulfoxide, DMF dimethylformamid
EDC and NSDAI degradation
Stock solutions of 100 mM estrone, 17ß-estradiol, 17α-ethinyl estradiol, estriol, bisphenol A, bisphenol S, naproxen and diclofenac, respectively, were made in DMF. Degradation studies were carried out in 2 ml tubes at room temperature with a total reaction volume of 750 µl. The reaction solution contained 50 mM potassium phosphate buffer, pH 7 containing 10% ethanol (Sei et al. 2008), 100 µM EDCs or NSDAIs and 20 U ml−1 Mrl2. The same samples but containing heat-inactivated Mrl2 were used as a control. The reaction tubes were shaken in a rotating over-head incubator at room temperature. 100 µl samples were taken after 0, 0.5, 1, and 20 h and mixed with 3 µl 6 M HCl to stop the reaction. 100 µl Methanol was added, samples were centrifuged (12,300 g, 10 min) and analyzed by HPLC. Laccases from Trametes versicolor (Sigma Aldrich) were used for comparative degradation studies. The laccases were dissolved in 50 mM potassium phosphate buffer, pH 7, and protein concentration was determined with the Bradford assay using BSA as standard. The solution was stored at −20 °C until use. Degradation reactions with 30 µg ml−1 laccase (Mrl2 and from T. versicolor, respectively) were conducted in a total volume of 500 µl 50 mM potassium phosphate buffer, pH 7 containing 10% ethanol, 100 µM EDCs or NSDAIs. For HPLC analysis samples were prepared as described above. Samples were analyzed immediately.
HPLC analysis
Samples were analyzed on a Shimadzu HPLC (Shimadzu, Duisburg, Germany) with a Chromolith® C18 100 mm, 4.6 mm reversed phase column (Merck, Darmstadt, Germany). As mobile phase a mixture of 100% methanol (mobile phase A) and water supplemented with 0.1% formic acid (v/v) (mobile phase B) was used. 10–25 µl of sample were injected for analysis. Steroids were analyzed with an isocratic elution method. The methanol concentration was 55% for estrone, 60% for 17ß-estradiol and 17α-ethinyl estradiol and 33% for estriol. For the NSAIDs the following gradients were used: for bisphenol A in 10 min from 50% mobile phase A to 75% mobile phase A, from 10 to 12 min 100% mobile phase A, from 12 to 15 min 50% mobile phase A; for diclofenac in 10 min from 70% mobile phase A to 85% mobile phase A, from 10 to 12 min 100% mobile phase A, from 12 to 15 min 50% mobile phase A; for bisphenol A in 10 min from 25% mobile phase A to 75% mobile phase A, from 10 to 12 min 100% mobile phase A, from 12 to 15 min 50% mobile phase A and for naproxen in 10 min from 50% mobile phase A to 75% mobile phase A, from 10 to 12 min 100% mobile phase A, from 12 to 15 min 50% mobile phase A. The flow of the mobile phase was 1–2 ml min−1. The compounds were evaluated at 280 nm. Stock solutions of EDCs and NSAIDs dissolved in 50% methanol to a concentration of 100 mM were used as standards. Degradation of the chemical compounds was calculated as percentage of the initial (time point 0 h) peak area size.
Results
Identification of a putative high-potential laccase
The catalytic activity of laccases is influenced by the difference in the redox potential of a substrate and the type 1 copper (Xu 1996; Xu et al. 1996). Thus, in most cases, high-potential laccases accept a broader range of substrates than low-potential laccases (Tadesse et al. 2008). To identify a laccase with a high redox potential, a BLAST search was done with the motif HCHIDWHLEAGF, containing the coordinating histidines of the T1 and T3 copper ions, the LEA tripeptide and the non-coordinating axial ligand phenylalanine, that is highly conserved within high-potential laccases (Mot and Silaghi-Dumitrescu 2012). A gene with the NCBI Reference Sequence XP_007855001.1 was identified as a putative laccase from a fungus Moniliophthora roreri causing frosty pod rot in cacao. The new gene was designated mrl2.
Cloning and recombinant expression of Mrl2 in shaking flasks
The gene mrl2 was cloned with either its native signal sequence or with the α-mating factor signal sequence from Saccharomyces cerevisiae for secretion into the vector pPICZαA (Invitrogen, Carlsbad, California, USA) resulting in pPICZAmrl2 (nMrl2) and pPICZαAmrl2 (αMrl2), respectively. The constructs were used to transform P. pastoris X-33. Transformants were screened for laccase production on buffered minimal methanol agar plates containing ABTS. After overnight incubation at 30 °C transformants of both constructs formed strong green halos. Selected Pichia transformants were cultivated in buffered minimal methanol medium in shaking flasks at 30 °C. Activity of Mrl2 (towards ABTS) increased over 3 days of expression and reached 1850 U l−1 for nMrl2 and 2630 U l−1 for αMrl2. Since Mrl2, secreted via the α-factor, provided higher activity towards ABTS, this construct was used for laccase production in a 3 l fed-batch fermentation process.
Fermentation and purification of Mrl2
Laccase production was evaluated in a 3 l high cell-density fed-batch fermentation process. The highest volumetric activity of 281,000 U l−1 was reached on day seven after induction with methanol. The fermentation broth was harvested on day eight after induction when a volumetric activity of about 272,800 U l−1 was measured. Cell-density reached OD600 = 460. Mrl2 was purified from concentrated supernatant by DEAE FF anion exchange chromatography to a specific activity of 248 U mg−1 towards ABTS which resulted in 1.33-fold purification. The yield of laccase calculated on the basis of the specific activity was 1.05 g l−1.
SDS-PAGE of purified Mrl2 revealed a strong band around 90 kDa (Fig. 1). Deglycosylation with PNGase F resulted in a shift of the 90 kDa band to around 60 kDa. The calculated molecular weight of Mrl2 is 54 kDa. The discrepancy to the calculated molecular weight might be due to additional O-glycosylation of Mrl2, which is not cleaved off by PNGase F. Laccase zymogram in the gel supplemented with ABTS revealed a strong band at ~55 kDa (Fig. 1). Migration of Mrl2 in the zymogram at a molecular weight of ~55 kDa might result from faster migration of properly folded, unboiled Mrl2 sample loaded onto the gel. Boiled Mrl2 samples completely lost their activity in the zymogram.
Fig. 1.
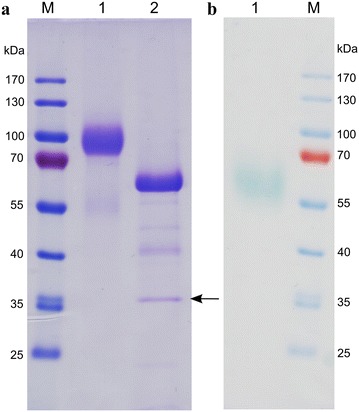
12.5% SDS-PAGE of Mrl2 purification. 5 µg total protein was loaded in each lane; M pre-stained protein markers; a Coomassie staining; 1 Mrl2 after DEAE FF purification, 2 Mrl2 after DEAE FF purification and PNGase F (indicated by an arrow) treatment, b zymogram with ABTS 1 purified Mrl2
Biochemical characterization
Atomic absorption spectroscopy revealed fully copper loaded Mrl2 (4.8 copper ions per molecule). The pH optimum of Mrl2 was determined with four typical laccase substrates: ABTS, SGZ, 2,6-DMP and guaiacol. Mrl2 was rather active at acidic pH values (Fig. 2). For ABTS oxidation the optimal pH was 2, for SGZ and 2,6-DMP oxidation the pH optimum was 4 and the highest guaiacol oxidation was found at pH 5 (Fig. 2). Kinetic parameters were determined for all four substrates at their optimal pH value (Table 2). Mrl2-catalyzed reactions followed Michaelis–Menten kinetics. The highest kcat value was determined with ABTS as substrate (316 s−1) and the lowest with SGZ (21 s−1). Km values ranged from 12.45 µM for syringaldazine to 2235 µM for guaiacol.
Fig. 2.
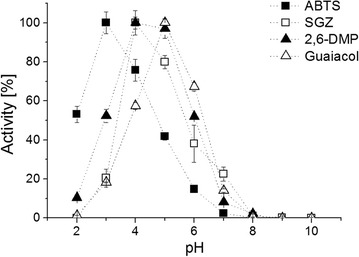
pH optimum of Mrl2 in reactions with the substrates ABTS, SGZ, 2,6-DMP and guaiacol. The highest measured activity for each substrate was set to 100%
Table 2.
Km and kcat values of Mrl2 with ABTS, SGZ, 2,6-DMP and guaiacol at their corresponding pH optima
| Km (µM) | kcat (s−1) | kcat K−1m (µM−1 s−1) | |
|---|---|---|---|
| ABTS | 24.13 ± 1.9 | 316 | 13.076 |
| SGZ | 12.45 ± 1.6 | 21 | 1.684 |
| 2,6-DMP | 358 ± 58.5 | 74 | 0.207 |
| Guaiacol | 2235 ± 117.5 | 37 | 0.016 |
Stability of Mrl2 was determined at pH 2–10 and was found to be higher at basic pH values than at acidic ones. After 1 h incubation at pH 2 no activity of Mrl2 was detectable (Fig. 3). At pH 3–6 after 1 h incubation residual activity of 45–75% was found, while after 24 h the activity was hardly detectable. At pH 7 the activity was still around 50% after 24 h incubation and at pH 8–10 residual activity was highest with around 70% after 24 h (Fig. 3).
Fig. 3.
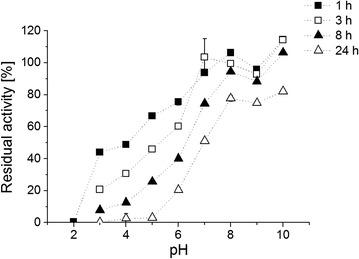
Stability of Mrl2 at different pH values. After 1 h (black square), 3 h (white square), 8 h (black triangle) and 24 h (white triangle) incubation the residual activity (indicated in % of initial activity) was determined
Half-life of Mrl2 at temperatures between 20 and 80 °C was determined at pH 7. Mrl2 was quite stable at 20 °C and pH 7 with a half-life of 13 days (Table 3). At elevated temperature the stability of Mrl2 was quite low with a half-life of 59 min at 50 °C and less than 1 min at 60 °C and above.
Table 3.
Half-life of 500 µg ml−1 Mrl2 after incubation in 50 mM potassium phosphate buffer, pH 7 at different temperatures
| Temperature (°C) | Half-life |
|---|---|
| 20 | 13 days |
| 30 | 3 days |
| 40 | 12.5 h |
| 50 | 59 min |
| 60, 70, 80 | <1 min |
The effect of different metal ions, water-miscible organic solvents as well as some inhibitors on Mrl2 was examined by using ABTS as substrate. The metal ions were applied as sulfates, except Ca2+, which was applied as Ca(NO3)2 due to low solubility of CaSO4. Activity of Mrl2 was hardly or not affected by Mn2+, Co2+, Cu2+, Na2+, Zn2+ and K+ even at 100 mM concentration. At a concentration of 10 mM only Ni2+ and Ca2+ led to a slight inhibition of Mrl2 of 15–18%. At 100 mM metal ion concentration again Ni2+ and Ca2+ ions led to the highest inhibition of Mrl2 with residual activities towards ABTS of 26.2 and 50.4%, respectively. NO3 − was not responsible for the inhibitory effect of Ca(NO3)2 since 100 mM KNO3 had no effect on the activity of Mrl2 (data not shown). 100 mM Mg2+ ions diminished the activity by about 20%, while the other tested metal ions hardly reduced the activity but even increased it in the case of Mn2+ (Table 4).
Water-miscible organic solvents more severely affected Mrl2 activity than metal ions. Already with 10% DMSO or acetone residual activity of Mrl2 dropped by about 80% (Table 4). 10% of ethanol and methanol slightly affected activity of Mrl2, while at a concentration of 20% residual activity of Mrl2 was 22 and 39%, respectively. 20% acetonitrile and 2-propanol inhibited Mrl2 almost completely. Activity of Mrl2 was hardly impaired by glycerol with about 90% residual activity at 20% glycerol.
Low concentrations of SDS had little effect on Mrl2 while 0.1% sodium azide almost completely inhibited Mrl2. Cl− ions had a strong inhibitory effect on Mrl2. In the presence of 10 mM Cl− Mrl2 showed 37% residual activity and at 50 and 100 mM concentration only 12.2 and 6.74% residual activity towards ABTS could be determined.
Redox potential
For redox potential determination we used [Fe(CN)6]3−/[Fe(CN)6]4–and [Fe(bipy)2]3+/Fe[(bipy)2]2+ redox couples with a standard redox potential of 0.433 V (O’Reilly 1973) and 0.78 V, respectively. In first titrations with the [Fe(bipy)2]3+/Fe[(bipy)2]2+ redox mediator couple Mrl2 could not be fully reduced (data not shown), therefore the [Fe(CN)6]3–/[Fe(CN)6]4– couple with the lower standard potential was applied. The redox titration with ferro- and ferricyanide could not fully oxidize Mrl2. For this reason the sample was aerated and the potential was set to an arbitrary high potential of 1.23 V. Redox titrations with iron(II/III) bipyridine indicated that the redox potential must be lower than 0.7 V. Titrations with the ferri/ferrocyanides confirmed these findings and revealed a redox potential of 0.58 V (Additional file 1: Figure S1).
Degradation of EDCs and NSAIDs
In the next step we examined the degradation of several micropollutants (Table 1) by Mrl2 at pH 7. Mrl2 degraded bisphenol A and the steroids 17ß-estradiol, 17α-ethinyl estradiol and estriol within 30 min below 10% residual concentration. Estrone seemed to be the most recalcitrant among the tested estrogens. After 1 h incubation a residual concentration of about 55% could be determined. After 20 h concentration of estrone decreased below the detection limit. Diclofenac was the most recalcitrant compound that could be degraded. After a reaction time of 20 h still about 42% diclofenac remained. Bisphenol S and naproxen were not affected by Mrl2 treatment within 20 h (Fig. 4).
Fig. 4.
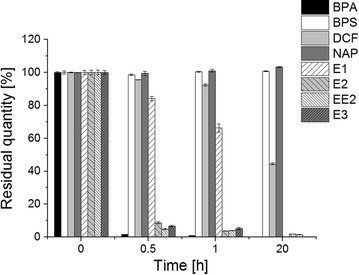
Residual quantity of EDCs and NSAIDs (%) after 0.5, 1 and 20 h of incubation with 20 U ml−1 Mrl2 at room temperature in 50 mM potassium phosphate buffer, pH 7. Measurements were carried out in triplicate
A comparative EDCs and NSAIDs degradation study was conducted with commercially available laccases preparation from T. versicolor (TvL). 30 µg ml−1 Mrl2 and TvL were used to degrade 100 µM micropollutants. After 1 h treatment, Mrl2 degraded all four estrogens and bisphenol A to a higher extent than TvL, while for diclofenac hardly any difference could be noticed between the two laccases. The reaction time for diclofenac was extended to 24 h. In this case TvL showed approximately twice the activity of Mrl2 (Fig. 5).
Fig. 5.
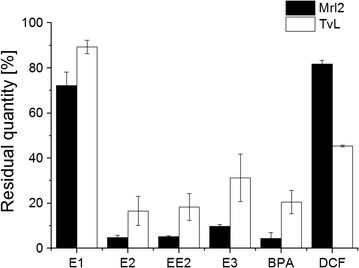
Residual quantity of EDCs and NSAIDs (%) after incubation with 30 µg ml−1 Mrl2 or TvL. The reaction time was set to 1 h except for DCF, for which it was 24 h. Measurements were carried out in triplicate
Discussion
Laccases have gained significant attention due to emerging applications including biomass degradation, biofuel cells, biocatalysis and bioremediation (Cannatelli and Ragauskas 2016; Kudanga et al. 2011; Mikolasch and Schauer 2009; Senthivelan et al. 2016; Strong and Claus 2011; Suzuki et al. 2003). However, low expression levels have been recognized as a major obstacle to industrial use of these enzymes (Piscitelli et al. 2010). Our results demonstrate that Mrl2 was expressed at exceptionally high levels in P. pastoris in a 3 l fed-batch fermentation process. To our knowledge Mrl2 expression achieved the highest level of a fungal laccase in P. pastoris reported so far with 281,000 U l−1, which corresponds to 1.05 g Mrl2/l medium at a specific activity of 248 U mg−1. Similar laccase yield in P. pastoris was only reported for a bacterial laccase from Thermus thermophilus with 1.2 g l−1 (Liu et al. 2015a). Specific activity of this enzyme (1.12 U mg−1) was, however, much lower compared to Mrl2. Comparable laccase yields of 0.8–1 g l−1 of the heterologously expressed fungal laccase T. versicolor could only be reached using the filamentous fungal host Trichoderma reesei (Baker and White 2001). Higher yields were only reported for homologous expression of laccases in basidiomycetes (Table 5). Explanations for the exceptional high expression of Mrl2 in P. pastoris remain elusive. Both the α-factor and the native secretion signal sequence resulted in high secretion of Mrl2, indicating that high expression yield can rather be traced back to the laccase sequence than signal peptide used.
Table 5.
Comparison of reported laccase yields and volumetric activity in different expression hosts
| Source | Expression host | Laccase | Yield (mg l−1) | Vol. activity (U l−1) | Reference |
|---|---|---|---|---|---|
| Heterologous expression in yeast | |||||
| Trametes versicolor | Pichia pastoris | 2.8 | Bohlin et al. (2006) | ||
| Cryphonectria parasitica | Saccharomyces cerevisiae | lac3 | 41.5 | Kim et al. (2010) | |
| Pleurotus eryngii | Saccharomyces cerevisiae | 146 | Bleve et al. (2008) | ||
| Melanocarpus albomyces | Saccharomyces cerevisiae | 3 | 168 | Kiiskinen and Saloheimo (2004) | |
| Bacillus licheniformis | Pichia pastoris | 227.9 | Lu et al. (2013) | ||
| Moniliophthora perniciosa FA553 | Pichia pastoris | LacMP | 232 | Liu et al. (2015b) | |
| Melanocarpus albomyces | Saccharomyces cerevisiae | MaL | 7 | 270 | Andberg et al. (2009) |
| Ganoderma lucidum | Pichia pastoris | 580 | 685.8 | You et al. (2014) | |
| Pycnoporus cinnabarinus | Yarrowia lipolytica | 19.84 | 1024 | Madzak et al. (2005) | |
| Yarrowia lipolytica | Pichia pastoris | YlLac | 1290 | Kalyani et al. (2015) | |
| Bacillus subtilis | Pichia pastoris | CotA | 1648.15 | Wang et al. (2015) | |
| Ganoderma weberianum TZC-1 | Pichia pastoris GS115 | GwLac1 | 2260 | Zhou et al. (2014) | |
| Trametes trogii | Pichia pastoris | Lcc1 | 17 | 2520 | Colao et al. (2006) |
| Botrytis aclada | Pichia pastoris | 517 | 3220 | Kittl et al. (2012a) | |
| Ganoderma fornicatum 814 | Pichia pastoris | rLac1 | 3460 | Huang et al. (2011) | |
| Lenzites gibbosa | Pichia pastoris | 5406 | Zheng et al. (2014) | ||
| Trametes sp. AH 28-2 | Pichia pastoris | lacA | 4 | 5470 | Hong et al. (2006) |
| Rigidoporus microsporus (Fomes lignosus) | Pichia pastoris | 9.03 | 5950 | Liu et al. (2003) | |
| Thermus thermophilus SG0.5JP17-16 | Pichia pastoris | LacTT | 1200 | 6130 | Liu et al. (2015a) |
| Cerrena sp. HYB07 | Pichia pastoris | Lac1 | 19.3 | 6300 | Yang et al. (2015) |
| Pleurotus sajor-caju | Pichia pastoris | lac4 | 110 | 10,200 | Soden et al. (2002) |
| Trametes versicolor | Pichia methanolica | 12,600 | Guo et al. (2006) | ||
| Trametes versicolor | Pichia pastoris | lccA | 18,123 | Li et al. (2014) | |
| Botrytis aclada | Pichia pastoris | 495 | 51,000 | Kittl et al. (2012b) | |
| Trametes versicolor | Pichia pastoris | 140,000 | Hong et al. (2002) | ||
| Trametes sp. 420 | Pichia pastoris | 136 | 239,000 | Zhou et al. (2007) | |
| Pycnoporus cinnabarinus | Pichia pastoris | 8 | Otterbein et al. (2000) | ||
| Myceliophthora thermophila | Saccharomyces cerevisiae | 18 | Bulter et al. (2003) | ||
| Heterologous expression in filamentous fungi | |||||
| Trametes versicolor | Aspergillus niger | 2700 | Bohlin et al. (2006) | ||
| Pycnoporus coccineus | Aspergillus oryzae | 3000 | Hoshida et al. (2005) | ||
| Trametes hirsute | Penicillium canescens | 3000 | Abianova et al. (2010) | ||
| Myceliophthora thermophila | Aspergillus oryzae | r-MtL | 19 | Berka et al. (1997) | |
| Phlebia radiate | Trichoderma reesei | 20 | Saloheimo and Nikupaavola (1991) | ||
| Pycnoporus cinnabarinus | Aspergillus niger | 70 | Record et al. (2002) | ||
| Melanocarpus albomyces | Trichoderma reesei | 920 | Kiiskinen et al. (2004) | ||
| Trametes versicolor | Trichoderma reesei | 800–1000 | Baker and White (2001) | ||
| Heterologous expression in bacteria | |||||
| Bacillus pumilus W3 | Bacillus subtilis WB600 | CotA | 373,100 | Guan et al. (2015) | |
| Natural production host | |||||
| Trametes versicolor 1017 | Trametes versicolor 1017 | Tvlac | 10,000 | Chen et al. (2016b) | |
| Trametes sp. AH 28-2 | Trametes sp. AH 28-2 | rLacB | 31.6 | 32,000 | Li et al. (2007) |
| Trametes pubescens MB 89 | Trametes pubescens MB 89 | 65,000 | Galhaup and Haltrich (2001) | ||
| Trametes multicolor MB 49 | Trametes multicolor MB 49 | 85,000 | Hess et al. (2002) | ||
| Cerrena sp. WR1 | Cerrena sp. WR1 | Lcc3 | 200 | 202,000 | Chen et al. (2012) |
| Cerrena sp. HYB07 | Cerrena sp. HYB07 | LacA | 108 | 210,800 | Yang et al. (2014) |
| White rot fungus WR-1 | White rot fungus WR-1 | 692,000 | Revankar and Lele (2006) | ||
Mrl2 possesses a phenylalanine as an axial ligand of the T1 copper. A positive correlation between hydrophobicity of this ligand and the redox potential E° has been reported (Marshall et al. 2009). Phenylalanine at this position implicates a high redox potential. Moreover, the hydrogen bond between serine at position 113 and glutamate at position 455 contributes to a long T1 Cu-His ligand distance which, according to Piontek and colleagues, strongly anticipates a high redox potential (Piontek et al. 2002). With a redox potential of 0.58 V, Mrl2, however, belongs to the middle-potential laccases. Besides the nature of the axial ligand and neighboring amino acids, redox potential is obviously influenced by other factors including hydrogen bonding of H(N)backbone and the coordinating SCys as well as protein and solvent dipoles and solvent accessibility (Hong et al. 2011). The reason why Mrl2 has a middle redox potential is under further investigation.
Important prerequisites for application of laccases in bioremediation for example in wastewater treatment are besides their sufficient availability, high stability at pH 7 or higher and the ability of degrading micropollutants including pharmaceuticals under neutral or slightly alkaline conditions even without adding redox mediators. In respect of these factors, Mrl2 looks very promising. Moreover, Mrl2 demonstrated high stability in the presence of metal ions which are often present in wastewater. Even at 100 mM concentration Mn2+, Co2+, Cu2+, Na2+, Zn2+ and K+ had almost no effect on laccase activity, examined in citrate phosphate buffer pH 3. A chelating effect of citrate on metal ions and thus their weaker effect on the laccase activity could be excluded since measurements in HEPES buffer provided comparable results (data not shown).
Despite middle redox potential, Mrl2 was able to degrade bisphenol A to 100% and all tested estrogens to more than 97% at neutral pH, with estrone being the most recalcitrant one. Diclofenac that is considered as poorly degradable in studies dealing with its removal (Barbosa et al. 2016), was degraded to 56%. Several reports describe effective removal of EDCs and NSAIDs by other laccases as well. Those experiments were carried out at acidic pH values, due to higher laccase activities at these pH values (Asadgol et al. 2014; Garcia-Morales et al. 2015; Macellaro et al. 2014; Sei et al. 2008; Tsutsumi et al. 2001). However, wastewater of plant effluents, which are considered the main source of estrogens (Snyder et al. 2001), usually show a neutral or basic pH. For this reason we used pH 7 for removal experiments with Mrl2. Despite the fact that fungal laccases usually demonstrate very low activities at that pH, Mrl2 still showed reasonable activity, thus representing an effective biocatalyst for the use in bioremediation. For example, Saito et al. (2004) used 50,000 U l−1 purified laccase from an ascomycete fungus belonging to Chaetomiaceae family to degrade 93.7% bisphenol A within 1 h, whereas 20,000 U l−1 Mrl2 degraded 98% bisphenol A within 30 min. To degrade the recalcitrant NSAID diclofenac Lloret et al. (2010, 2013) used 2000 U l−1 mid-redox potential laccase MtL from Myceliophthora thermophila and 2000 U l−1 (147 mg l−1) of high-redox potential T. versicolor. After 8 h incubation with MtL and 24 h incubation with T. versicolor laccase 2 and 27% diclofenac was removed, respectively, while 20,000 U l−1 (80 mg l−1) Mrl2 decreased diclofenac concentration by 8 and 56% after 1 and 20 h incubation, respectively. Lloret et al. (2010) also tested 2000 U l−1 MtL with the estrogens estrone, 17ß-estradiol and 17α-ethinyl estradiol at pH 7. While removal of 17ß-estradiol with Mrl2 and MtL was comparable (96 and 99% decrease for Mrl2 and MtL after 1 and 8 h, respectively), the contraceptive 17α-ethinyl estradiol was degraded by Mrl2 (95%, 1 h) much faster than by MtL (85%, 8 h).
Yet comparison of different micropollutant removal experiments might be misleading in regard to different specific activities of the laccases and thereby quite different amounts of applied enzyme as e.g. Mrl2 has an exceptionally high specific activity towards ABTS. This is why we applied commercial TvL and Mrl2 in a degradation experiment of several micropollutants at equal enzyme amounts. Though TvL possess a high redox potential (Reinhammar 1972), Mrl2 showed faster degradation of estrogens and bisphenol A within 1 h. Besides laccases’ redox potential, amino acids involved in substrate binding near the T1 copper site contribute to high substrate turnover rates. Our findings suggest better binding of hydroxylated compounds like estrogens and bisphenol A by Mrl2 than TvL. With the more recalcitrant, non-hydroxylated diclofenac, TvL displayed about 2.5-fold higher activity than Mrl2. In this case the higher redox potential of TvL might afford faster degradation of diclofenac compared to Mrl2. Another aspect might be different stability of the enzymes during 24 h incubation.
In concordance with previous studies, bisphenol S and naproxen were not degraded by laccase alone (Lloret et al. 2010). Laccase-mediated removal of bisphenol S and naproxen seems to require addition of small redox mediators, applied to expand the substrate spectrum of laccases to e.g. compounds with higher redox potentials like non-phenolic substances. These mediators are, however, mostly expensive and often not environmentally friendly, and their application in wastewater treatment thus doubtful.
In conclusion, Mrl2, a new laccase from M. roreri was identified and showed exceptionally high expression levels during fed-batch fermentation of recombinant P. pastoris (280,000 U l−1; 1.05 mg l−1) matching those of filamentous fungi. The enzyme is stable up to 30 °C, at alkaline pH values and active in the presence of several water-miscible organic solvents and metal ions. Despite its redox potential of 0.58 V Mrl2 degrades estrogens like estrone, 17β-estradiol, estriol and the contraceptive 17α-ethinyl estradiol and an estrogenic active substance bisphenol A faster than TvL at neutral pH. This makes Mrl2 a promising candidate for application in wastewater treatment.
Authors’ contributions
AB designed and conducted the experiments, evaluated the results and drafted the manuscript. KK drafted the manuscript and helped in research design and data analysis. PLH helped in redox potential titrations, evaluated the redox potential titration results, and revised the manuscript. VBU gave advices in the research work, helped in drafting the manuscript, and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Annette Ricken (Institute for Bioinorganic Chemistry, Heinrich-Heine University Düsseldorf) and Anna Olbrich (Institute of Biochemistry II, Heinrich-Heine University Düsseldorf) for copper content analysis.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The data on which the conclusions are made are all presented in this paper.
Ethical approval and consent to participate
This paper does not contain any studies with human participants or animals performed by any of the authors.
Funding
The scientific activities of the Bioeconomy Science Center were financially supported by the Ministry of Innovation, Science and Research within the framework of the NRW-Strategieprojekt BioSC (No. 313/323-400-002 13, EnZip and BioDeg projects).
Abbreviations
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- kDa
kilo Dalton
- PNGase F
peptide-N-Glycosidase F
- HPLC
high performance liquid chromatography
- BMGY
buffered complex glycerol medium
- BMM
buffered minimal methanol
- DMSO
dimethyl sulfoxide
- DMF
dimethylformamide
- TvL
laccases preparation from Trametes versicolor
- PTM1
Pichia trace metal salts
- ABTS
2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid)
- SGZ
syringaldazine
- 2,6-DMP
2,6-dimethoxyphenol
- HEPES
2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid
- DEAE FF
diethylaminoethyl-Sepharose Fast Flow
- BPA
bisphenol A
- BPS
bisphenol S
- DCF
diclofenac
- NAP
naproxen
- E1
estrone
- E2
17ß-estradiol
- EE2
17α-ethinyl estradiol
- E3
estriol
- EDC
endocrine disrupting compound
- NSAID
non-steroidal anti-inflammatory drug
Additional file
Additional file 1: Figure S1. Additional information.
Contributor Information
Agathe Bronikowski, Email: agathe.bronikowski@uni-duesseldorf.de.
Peter-Leon Hagedoorn, Email: p.l.hagedoorn@tudelft.nl.
Katja Koschorreck, Email: katja.koschorreck@uni-duesseldorf.de.
Vlada B. Urlacher, Email: vlada.urlacher@uni-duesseldorf.de
References
- Abianova AR, Chulkin AM, Vavilova EA, Fedorova TV, Loginov DS, Koroleva OV, Benevolenskii SV. A heterologous production of the Trametes hirsuta laccase in the fungus Penicillium canescens. Prikl Biokhim Mikrobiol. 2010;46(3):342–347. [PubMed] [Google Scholar]
- Andberg M, Hakulinen N, Auer S, Saloheimo M, Koivula A, Rouvinen J, Kruus K. Essential role of the C-terminus in Melanocarpus albomyces laccase for enzyme production, catalytic properties and structure. FEBS J. 2009;276(21):6285–6300. doi: 10.1111/j.1742-4658.2009.07336.x. [DOI] [PubMed] [Google Scholar]
- Asadgol Z, Forootanfar H, Rezaei S, Mahvi AH, Faramarzi MA. Removal of phenol and bisphenol-A catalyzed by laccase in aqueous solution. J Environ Health Sci Eng. 2014;12:93. doi: 10.1186/2052-336X-12-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auriol M, Filali-Meknassi Y, Tyagi RD, Adams CD. Laccase-catalyzed conversion of natural and synthetic hormones from a municipal wastewater. Water Res. 2007;41(15):3281–3288. doi: 10.1016/j.watres.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Auriol M, Filali-Meknassi Y, Tyagi RD, Adams CD. Oxidation of natural and synthetic hormones by the horseradish peroxidase enzyme in wastewater. Chemosphere. 2007;68(10):1830–1837. doi: 10.1016/j.chemosphere.2007.03.045. [DOI] [PubMed] [Google Scholar]
- Baker CJO, White TC. Expression of laccase I and IV genes from Trametes versicolor in Trichoderma reesei. In: Argyropoulos D, editor. Oxidative delignification chemistry. Washington, DC: ACS Symposium Series; 2001. pp. 413–426. [Google Scholar]
- Baldrian P. Fungal laccases—occurrence and properties. FEMS Microbiol Rev. 2006;30(2):215–242. doi: 10.1111/j.1574-4976.2005.00010.x. [DOI] [PubMed] [Google Scholar]
- Barbosa MO, Moreira NF, Ribeiro AR, Pereira MF, Silva AM. Occurrence and removal of organic micropollutants: an overview of the watch list of EU decision 2015/495. Water Res. 2016;94:257–279. doi: 10.1016/j.watres.2016.02.047. [DOI] [PubMed] [Google Scholar]
- Berka RM, Schneider P, Golightly EJ, Brown SH, Madden M, Brown KM, Halkier T, Mondorf K, Xu F. Characterization of the gene encoding an extracellular laccase of Myceliophthora thermophila and analysis of the recombinant enzyme expressed in Aspergillus oryzae. Appl Environ Microbiol. 1997;63(8):3151–3157. doi: 10.1128/aem.63.8.3151-3157.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleve G, Lezzi C, Mita G, Rampino P, Perrotta C, Villanova L, Grieco F. Molecular cloning and heterologous expression of a laccase gene from Pleurotus eryngii in free and immobilized Saccharomyces cerevisiae cells. Appl Microbiol Biotechnol. 2008;79(5):731–741. doi: 10.1007/s00253-008-1479-1. [DOI] [PubMed] [Google Scholar]
- Bohlin C, Jonsson LJ, Roth R, van Zyl WH. Heterologous expression of Trametes versicolor laccase in Pichia pastoris and Aspergillus niger. Appl Biochem Biotechnol. 2006;129–132:195–214. doi: 10.1385/ABAB:129:1:195. [DOI] [PubMed] [Google Scholar]
- Bourbonnais R, Paice MG. Oxidation of non-phenolic substrates. An expanded role for laccase in lignin biodegradation. FEBS Lett. 1990;267(1):99–102. doi: 10.1016/0014-5793(90)80298-W. [DOI] [PubMed] [Google Scholar]
- Bulter T, Alcalde M, Sieber V, Meinhold P, Schlachtbauer C, Arnold FH. Functional expression of a fungal laccase in Saccharomyces cerevisiae by directed evolution. Appl Environ Microbiol. 2003;69(2):987–995. doi: 10.1128/AEM.69.2.987-995.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannatelli MD, Ragauskas AJ. Conversion of lignin into value-added materials and chemicals via laccase-assisted copolymerization. Appl Microbiol Biotechnol. 2016;100(20):8685–8691. doi: 10.1007/s00253-016-7820-1. [DOI] [PubMed] [Google Scholar]
- Chen SC, Wu PH, Su YC, Wen TN, Wei YS, Wang NC, Hsu CA, Wang AH, Shyur LF. Biochemical characterization of a novel laccase from the basidiomycete fungus Cerrena sp. WR1. Protein Eng Des Sel. 2012;25(11):761–769. doi: 10.1093/protein/gzs082. [DOI] [PubMed] [Google Scholar]
- Chen L, Jiang X, Feng H, Shi H, Sun L, Tao W, Xi Q, Wang D. Simultaneous exposure to estrogen and androgen resulted in feminization and endocrine disruption. J Endocrinol. 2016;228(3):205–218. doi: 10.1530/JOE-15-0432. [DOI] [PubMed] [Google Scholar]
- Chen L, Yi X, Deng F, Fang W, Zhang X, Wang X, Fang Z, Xiao Y. A novel ethanol-tolerant laccase, Tvlac, from Trametes versicolor. Biotechnol Lett. 2016;38(3):471–476. doi: 10.1007/s10529-015-1994-y. [DOI] [PubMed] [Google Scholar]
- Colao MC, Lupino S, Garzillo AM, Buonocore V, Ruzzi M. Heterologous expression of lcc1 gene from Trametes trogii in Pichia pastoris and characterization of the recombinant enzyme. Microb Cell Fact. 2006;5:31. doi: 10.1186/1475-2859-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi UN, Singh P, Pandey VP, Kumar A. Structure-function relationship among bacterial, fungal and plant laccases. J Mol Catal B-Enzym. 2011;68(2):117–128. doi: 10.1016/j.molcatb.2010.11.002. [DOI] [Google Scholar]
- Eggert C, Temp U, Dean JF, Eriksson KE. A fungal metabolite mediates degradation of non-phenolic lignin structures and synthetic lignin by laccase. FEBS Lett. 1996;391(1–2):144–148. doi: 10.1016/0014-5793(96)00719-3. [DOI] [PubMed] [Google Scholar]
- Galhaup C, Haltrich D. Enhanced formation of laccase activity by the white-rot fungus Trametes pubescens in the presence of copper. Appl Microbiol Biotechnol. 2001;56(1–2):225–232. doi: 10.1007/s002530100636. [DOI] [PubMed] [Google Scholar]
- Garcia-Morales R, Rodriguez-Delgado M, Gomez-Mariscal K, Orona-Navar C, Hernandez-Luna C, Torres E, Parra R, Cardenas-Chavez D, Mahlknecht J, Ornelas-Soto N. Biotransformation of endocrine - disrupting compounds in groundwater: bisphenol A, nonylphenol, ethynylestradiol and triclosan by a laccase cocktail from CS43. Water Air Soil Pollut. 2015;226(8):251. doi: 10.1007/s11270-015-2514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan ZB, Shui Y, Song CM, Zhang N, Cai YJ, Liao XR. Efficient secretory production of CotA-laccase and its application in the decolorization and detoxification of industrial textile wastewater. Environ Sci Pollut Res Int. 2015;22(12):9515–9523. doi: 10.1007/s11356-015-4426-6. [DOI] [PubMed] [Google Scholar]
- Guo M, Lu F, Du L, Pu J, Bai D. Optimization of the expression of a laccase gene from Trametes versicolor in Pichia methanolica. Appl Microbiol Biotechnol. 2006;71(6):848–852. doi: 10.1007/s00253-005-0210-8. [DOI] [PubMed] [Google Scholar]
- Hamid H, Eskicioglu C. Fate of estrogenic hormones in wastewater and sludge treatment: a review of properties and analytical detection techniques in sludge matrix. Water Res. 2012;46(18):5813–5833. doi: 10.1016/j.watres.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Hess J, Leitner C, Galhaup C, Kulbe KD, Hinterstoisser B, Steinwender M, Haltrich D. Enhanced formation of extracellular laccase activity by the white-rot fungus Trametes multicolor. Appl Biochem Biotechnol. 2002;98–100:229–241. doi: 10.1385/ABAB:98-100:1-9:229. [DOI] [PubMed] [Google Scholar]
- Hong F, Meinander NQ, Jonsson LJ. Fermentation strategies for improved heterologous expression of laccase in Pichia pastoris. Biotechnol Bioeng. 2002;79(4):438–449. doi: 10.1002/bit.10297. [DOI] [PubMed] [Google Scholar]
- Hong Y, Xiao Y, Zhou H, Fang W, Zhang M, Wang J, Wu L, Yu Z. Expression of a laccase cDNA from Trametes sp. AH28-2 in Pichia pastoris and mutagenesis of transformants by nitrogen ion implantation. FEMS Microbiol Lett. 2006;258(1):96–101. doi: 10.1111/j.1574-6968.2006.00209.x. [DOI] [PubMed] [Google Scholar]
- Hong G, Ivnitski DM, Johnson GR, Atanassov P, Pachter R. Design parameters for tuning the type 1 Cu multicopper oxidase redox potential: insight from a combination of first principles and empirical molecular dynamics simulations. J Am Chem Soc. 2011;133(13):4802–4809. doi: 10.1021/ja105586q. [DOI] [PubMed] [Google Scholar]
- Hoshida H, Fujita T, Murata K, Kubo K, Akada R. Copper-dependent production of a Pycnoporus coccineus extracellular laccase in Aspergillus oryzae and Saccharomyces cerevisiae. Biosci Biotechnol Biochem. 2005;69(6):1090–1097. doi: 10.1271/bbb.69.1090. [DOI] [PubMed] [Google Scholar]
- Huang W-T, Tai R, Hseu R-S, Huang C-T. Overexpression and characterization of a thermostable, pH-stable and organic solvent-tolerant Ganoderma fornicatum laccase in Pichia pastoris. Process Biochem. 2011;46(7):1469–1474. doi: 10.1016/j.procbio.2011.03.020. [DOI] [Google Scholar]
- Ji K, Hong S, Kho Y, Choi K. Effects of bisphenol s exposure on endocrine functions and reproduction of zebrafish. Environ Sci Technol. 2013;47(15):8793–8800. doi: 10.1021/es400329t. [DOI] [PubMed] [Google Scholar]
- Kalyani D, Tiwari MK, Li J, Kim SC, Kalia VC, Kang YC, Lee JK. A highly efficient recombinant laccase from the yeast Yarrowia lipolytica and its application in the hydrolysis of biomass. PLoS ONE. 2015;10(3):e0120156. doi: 10.1371/journal.pone.0120156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiiskinen LL, Saloheimo M. Molecular cloning and expression in Saccharomyces cerevisiae of a laccase gene from the ascomycete Melanocarpus albomyces. Appl Environ Microbiol. 2004;70(1):137–144. doi: 10.1128/AEM.70.1.137-144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiiskinen LL, Kruus K, Bailey M, Ylosmaki E, Siika-Aho M, Saloheimo M. Expression of Melanocarpus albomyces laccase in Trichoderma reesei and characterization of the purified enzyme. Microbiology. 2004;150(Pt 9):3065–3074. doi: 10.1099/mic.0.27147-0. [DOI] [PubMed] [Google Scholar]
- Kim JM, Park SM, Kim DH. Heterologous expression of a tannic acid-inducible laccase3 of Cryphonectria parasitica in Saccharomyces cerevisiae. BMC Biotechnol. 2010;10:18. doi: 10.1186/1472-6750-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittl R, Gonaus C, Pillei C, Haltrich D, Ludwig R. Constitutive expression of Botrytis aclada laccase in Pichia pastoris. Bioengineered. 2012;3(4):232–235. doi: 10.4161/bioe.20037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittl R, Mueangtoom K, Gonaus C, Khazaneh ST, Sygmund C, Haltrich D, Ludwig R. A chloride tolerant laccase from the plant pathogen ascomycete Botrytis aclada expressed at high levels in Pichia pastoris. J Biotechnol. 2012;157(2):304–314. doi: 10.1016/j.jbiotec.2011.11.021. [DOI] [PubMed] [Google Scholar]
- Kudanga T, Nyanhongo GS, Guebitz GM, Burton S. Potential applications of laccase-mediated coupling and grafting reactions: a review. Enzyme Microb Technol. 2011;48(3):195–208. doi: 10.1016/j.enzmictec.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li JF, Hong YZ, Xiao YZ, Xu YH, Fang W. High production of laccase B from Trametes sp in Pichia pastoris. World J Microb Biot. 2007;23(5):741–745. doi: 10.1007/s11274-006-9286-2. [DOI] [Google Scholar]
- Li Q, Pei J, Zhao L, Xie J, Cao F, Wang G. Overexpression and characterization of laccase from Trametes versicolor in Pichia pastoris. Prikl Biokhim Mikrobiol. 2014;50(2):163–170. [PubMed] [Google Scholar]
- Liu W, Chao Y, Liu S, Bao H, Qian S. Molecular cloning and characterization of a laccase gene from the basidiomycete Fome lignosus and expression in Pichia pastoris. Appl Microbiol Biotechnol. 2003;63(2):174–181. doi: 10.1007/s00253-003-1398-0. [DOI] [PubMed] [Google Scholar]
- Liu H, Cheng Y, Du B, Tong C, Liang S, Han S, Zheng S, Lin Y. Overexpression of a novel thermostable and chloride-tolerant laccase from Thermus thermophilus SG0.5JP17-16 in Pichia pastoris and its application in synthetic dye decolorization. PLoS ONE. 2015;10(3):9833. doi: 10.1371/journal.pone.0119833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Tong C, Du B, Liang S, Lin Y. Expression and characterization of LacMP, a novel fungal laccase of Moniliophthora perniciosa FA553. Biotechnol Lett. 2015;37(9):1829–1835. doi: 10.1007/s10529-015-1865-6. [DOI] [PubMed] [Google Scholar]
- Lloret L, Eibes G, Lú-Chau TA, Moreira MT, Feijoo G, Lema JM. Laccase-catalyzed degradation of anti-inflammatories and estrogens. Biochem Eng J. 2010;51(3):124–131. doi: 10.1016/j.bej.2010.06.005. [DOI] [Google Scholar]
- Lloret L, Eibes G, Moreira MT, Feijoo G, Lema JM. On the use of a high-redox potential laccase as an alternative for the transformation of non-steroidal anti-inflammatory drugs (NSAIDs) J Mol Catal B-Enzym. 2013;97:233–242. doi: 10.1016/j.molcatb.2013.08.021. [DOI] [Google Scholar]
- Lu L, Wang TN, Xu TF, Wang JY, Wang CL, Zhao M. Cloning and expression of thermo-alkali-stable laccase of Bacillus licheniformis in Pichia pastoris and its characterization. Bioresour Technol. 2013;134:81–86. doi: 10.1016/j.biortech.2013.02.015. [DOI] [PubMed] [Google Scholar]
- Macellaro G, Pezzella C, Cicatiello P, Sannia G, Piscitelli A. Fungal laccases degradation of endocrine disrupting compounds. Biomed Res Int. 2014;2014:614038. doi: 10.1155/2014/614038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madzak C, Otterbein L, Chamkha M, Moukha S, Asther M, Gaillardin C, Beckerich JM. Heterologous production of a laccase from the basidiomycete Pycnoporus cinnabarinus in the dimorphic yeast Yarrowia lipolytica. FEMS Yeast Res. 2005;5(6–7):635–646. doi: 10.1016/j.femsyr.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Marshall NM, Garner DK, Wilson TD, Gao YG, Robinson H, Nilges MJ, Lu Y. Rationally tuning the reduction potential of a single cupredoxin beyond the natural range. Nature. 2009;462(7269):113–116. doi: 10.1038/nature08551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer AM, Staples RC. Laccase: new functions for an old enzyme. Phytochemistry. 2002;60(6):551–565. doi: 10.1016/S0031-9422(02)00171-1. [DOI] [PubMed] [Google Scholar]
- Mikolasch A, Schauer F. Fungal laccases as tools for the synthesis of new hybrid molecules and biomaterials. Appl Microbiol Biotechnol. 2009;82(4):605–624. doi: 10.1007/s00253-009-1869-z. [DOI] [PubMed] [Google Scholar]
- Mot AC, Silaghi-Dumitrescu R. Laccases: complex architectures for one-electron oxidations. Biochemistry (Mosc) 2012;77(12):1395–1407. doi: 10.1134/S0006297912120085. [DOI] [PubMed] [Google Scholar]
- O’Reilly JE. Oxidation-reduction potential of the ferro-ferricyanide system in buffer solutions. Biochim Biophys Acta. 1973;292(3):509–515. doi: 10.1016/0005-2728(73)90001-7. [DOI] [PubMed] [Google Scholar]
- Orn S, Holbech H, Norrgren L. Sexual disruption in zebrafish (Danio rerio) exposed to mixtures of 17alpha-ethinylestradiol and 17beta-trenbolone. Environ Toxicol Pharmacol. 2016;41:225–231. doi: 10.1016/j.etap.2015.12.010. [DOI] [PubMed] [Google Scholar]
- Otterbein L, Record E, Longhi S, Asther M, Moukha S. Molecular cloning of the cDNA encoding laccase from Pycnoporus cinnabarinus I-937 and expression in Pichia pastoris. Eur J Biochem. 2000;267(6):1619–1625. doi: 10.1046/j.1432-1327.2000.01166.x. [DOI] [PubMed] [Google Scholar]
- Pardo I, Camarero S. Laccase engineering by rational and evolutionary design. Cell Mol Life Sci. 2015;72(5):897–910. doi: 10.1007/s00018-014-1824-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontek K, Antorini M, Choinowski T. Crystal structure of a laccase from the fungus Trametes versicolor at 1.90-A resolution containing a full complement of coppers. J Biol Chem. 2002;277(40):37663–37669. doi: 10.1074/jbc.M204571200. [DOI] [PubMed] [Google Scholar]
- Piscitelli A, Pezzella C, Giardina P, Faraco V, Giovanni S. Heterologous laccase production and its role in industrial applications. Bioeng Bugs. 2010;1(4):252–262. doi: 10.4161/bbug.1.4.11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record E, Punt PJ, Chamkha M, Labat M, van Den Hondel CA, Asther M. Expression of the Pycnoporus cinnabarinus laccase gene in Aspergillus niger and characterization of the recombinant enzyme. Eur J Biochem. 2002;269(2):602–609. doi: 10.1046/j.0014-2956.2001.02690.x. [DOI] [PubMed] [Google Scholar]
- Reinhammar BRM. Oxidation-reduction potentials of the electron acceptors in laccases and stellacyanin. BBA-Bioenergetics. 1972;275(2):245–259. doi: 10.1016/0005-2728(72)90045-X. [DOI] [PubMed] [Google Scholar]
- Revankar MS, Lele SS. Enhanced production of laccase using a new isolate of white rot fungus WR-1. Process Biochem. 2006;41(3):581–588. doi: 10.1016/j.procbio.2005.07.019. [DOI] [Google Scholar]
- Rodriguez Couto S, Toca Herrera JL. Industrial and biotechnological applications of laccases: a review. Biotechnol Adv. 2006;24(5):500–513. doi: 10.1016/j.biotechadv.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Saito T, Kato K, Yokogawa Y, Nishida M, Yamashita N. Detoxification of bisphenol A and nonylphenol by purified extracellular laccase from a fungus isolated from soil. J Biosci Bioeng. 2004;98(1):64–66. doi: 10.1016/S1389-1723(04)70243-1. [DOI] [PubMed] [Google Scholar]
- Saloheimo M, Nikupaavola ML. Heterologous production of a ligninolytic enzyme - expression of the Phlebia Radiata laccase gene in Trichoderma reesei. Nat Biotechnol. 1991;9(10):987–990. doi: 10.1038/nbt1091-987. [DOI] [Google Scholar]
- Schroder P, Helmreich B, Skrbic B, Carballa M, Papa M, Pastore C, Emre Z, Oehmen A, Langenhoff A, Molinos M, Dvarioniene J, Huber C, Tsagarakis KP, Martinez-Lopez E, Pagano SM, Vogelsang C, Mascolo G. Status of hormones and painkillers in wastewater effluents across several European states-considerations for the EU watch list concerning estradiols and diclofenac. Environ Sci Pollut Res Int. 2016;23(13):12835–12866. doi: 10.1007/s11356-016-6503-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach RP, Escher BI, Fenner K, Hofstetter TB, Johnson CA, von Gunten U, Wehrli B. The challenge of micropollutants in aquatic systems. Science. 2006;313(5790):1072–1077. doi: 10.1126/science.1127291. [DOI] [PubMed] [Google Scholar]
- Sei K, Takeda T, Soda SO, Fujita M, Ike M. Removal characteristics of endocrine-disrupting chemicals by laccase from white-rot fungi. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2008;43(1):53–60. doi: 10.1080/10934520701750397. [DOI] [PubMed] [Google Scholar]
- Senthivelan T, Kanagaraj J, Panda RC. Recent trends in fungal laccase for various industrial applications: an eco-friendly approach—a review. Biotechnol Bioprocess Eng. 2016;21(1):19–38. doi: 10.1007/s12257-015-0278-7. [DOI] [Google Scholar]
- Snyder SA, Villeneuve DL, Snyder EM, Giesy JP. Identification and quantification of estrogen receptor agonists in wastewater effluents. Environ Sci Technol. 2001;35(18):3620–3625. doi: 10.1021/es001254n. [DOI] [PubMed] [Google Scholar]
- Soares GMB, de Amorim MTP, Costa-Ferreira M. Use of laccase together with redox mediators to decolourize Remazol Brilliant Blue R. J Biotechnol. 2001;89(2–3):123–129. doi: 10.1016/S0168-1656(01)00302-9. [DOI] [PubMed] [Google Scholar]
- Soden DM, O’Callaghan J, Dobson AD. Molecular cloning of a laccase isozyme gene from Pleurotus sajor-caju and expression in the heterologous Pichia pastoris host. Microbiology. 2002;148(Pt 12):4003–4014. doi: 10.1099/00221287-148-12-4003. [DOI] [PubMed] [Google Scholar]
- Solomon EI, Augustine AJ, Yoon J. O2 reduction to H2O by the multicopper oxidases. Dalton Trans. 2008;30(30):3921–3932. doi: 10.1039/b800799c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong PJ, Claus H. Laccase: a review of its past and its future in bioremediation. Crit Rev Env Sci Technol. 2011;41(4):373–434. doi: 10.1080/10643380902945706. [DOI] [Google Scholar]
- Suzuki K, Hirai H, Murata H, Nishida T. Removal of estrogenic activities of 17β-estradiol and ethinylestradiol by ligninolytic enzymes from white rot fungi. Water Res. 2003;37(8):1972–1975. doi: 10.1016/S0043-1354(02)00533-X. [DOI] [PubMed] [Google Scholar]
- Tadesse MA, D’Annibale A, Galli C, Gentili P, Sergi F. An assessment of the relative contributions of redox and steric issues to laccase specificity towards putative substrates. Org Biomol Chem. 2008;6(5):868–878. doi: 10.1039/b716002j. [DOI] [PubMed] [Google Scholar]
- Thurston CF. The structure and function of fungal laccases. Microbiology. 1994;140:19–26. doi: 10.1099/13500872-140-1-19. [DOI] [Google Scholar]
- Tsutsumi Y, Haneda T, Nishida T. Removal of estrogenic activities of bisphenol A and nonylphenol by oxidative enzymes from lignin-degrading basidiomycetes. Chemosphere. 2001;42(3):271–276. doi: 10.1016/S0045-6535(00)00081-3. [DOI] [PubMed] [Google Scholar]
- Vinas R, Watson CS. Bisphenol S disrupts estradiol-induced nongenomic signaling in a rat pituitary cell line: effects on cell functions. Environ Health Perspect. 2013;121(3):352–358. doi: 10.1289/ehp.1205826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TN, Lu L, Wang JY, Xu TF, Li J, Zhao M. Enhanced expression of an industry applicable CotA laccase from Bacillus subtilis in Pichia pastoris by non-repressing carbon sources together with pH adjustment: recombinant enzyme characterization and dye decolorization. Process Biochem. 2015;50(1):97–103. doi: 10.1016/j.procbio.2014.10.009. [DOI] [Google Scholar]
- Xu F. Oxidation of phenols, anilines, and benzenethiols by fungal laccases: correlation between activity and redox potentials as well as halide inhibition. Biochemistry. 1996;35(23):7608–7614. doi: 10.1021/bi952971a. [DOI] [PubMed] [Google Scholar]
- Xu F, Shin WS, Brown SH, Wahleithner JA, Sundaram UM, Solomon EI. A study of a series of recombinant fungal laccases and bilirubin oxidase that exhibit significant differences in redox potential, substrate specificity, and stability. Biochim Biophys Acta Protein Struct Mol Enzymol. 1996;1292(2):303–311. doi: 10.1016/0167-4838(95)00210-3. [DOI] [PubMed] [Google Scholar]
- Xu F, Berka RM, Wahleithner JA, Nelson BA, Shuster JR, Brown SH, Palmer AE, Solomon EI. Site-directed mutations in fungal laccase: effect on redox potential, activity and pH profile. Biochem J. 1998;334(Pt 1):63–70. doi: 10.1042/bj3340063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lin Q, Ng TB, Ye X, Lin J. Purification and characterization of a novel laccase from Cerrena sp. HYB07 with dye decolorizing ability. PLoS ONE. 2014;9(10):e110834. doi: 10.1371/journal.pone.0110834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Ng TB, Lin J, Ye X. A novel laccase from basidiomycete Cerrena sp.: cloning, heterologous expression, and characterization. Int J Biol Macromol. 2015;77:344–349. doi: 10.1016/j.ijbiomac.2015.03.028. [DOI] [PubMed] [Google Scholar]
- You LF, Liu ZM, Lin JF, Guo LQ, Huang XL, Yang HX. Molecular cloning of a laccase gene from Ganoderma lucidum and heterologous expression in Pichia pastoris. J Basic Microbiol. 2014;54(Suppl 1):S134–S141. doi: 10.1002/jobm.201200808. [DOI] [PubMed] [Google Scholar]
- Zheng M, Chi Y, Yi H, Shao S. Decolorization of Alizarin Red and other synthetic dyes by a recombinant laccase from Pichia pastoris. Biotechnol Lett. 2014;36(1):39–45. doi: 10.1007/s10529-013-1323-2. [DOI] [PubMed] [Google Scholar]
- Zhou HM, Hong YZ, Xiao YZ, Cui TJ, Wang XT, Pu CL. High output of a Trametes laccase in Pichia pastoris and characterization of recombinant enzymes. Sheng wu gong cheng xue bao = Chin J Biotechnol. 2007;23(6):1055–1059. doi: 10.1016/S1872-2075(07)60063-6. [DOI] [PubMed] [Google Scholar]
- Zhou YP, Chen QH, Xiao YN, Ke DS, Tian CE. Gene cloning and characterization of a novel laccase from the tropical white-rot fungus Ganoderma weberianum TZC-1. Appl Biochem Microbiol. 2014;50(5):500–507. doi: 10.1134/S0003683814050147. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data on which the conclusions are made are all presented in this paper.


