A new small RNA overexpression system could highly overexpress not only artificial and endogenous miRNAs but also endogenous siRNAs in Nicotiana benthamiana.
Abstract
With experimental and bioinformatical methods, numerous small RNAs, including microRNAs (miRNAs) and short interfering RNAs (siRNAs), have been found in plants, and they play vital roles in various biological regulation processes. However, most of these small RNAs remain to be functionally characterized. Until now, only several viral vectors were developed to overexpress miRNAs with limited application in plants. In this study, we report a new small RNA overexpression system via viral satellite DNA associated with Tomato yellow leaf curl China virus (TYLCCNV) vector, which could highly overexpress not only artificial and endogenous miRNAs but also endogenous siRNAs in Nicotiana benthamiana. First, we constructed basic TYLCCNV-amiRPDS(319L) vector with widely used AtMIR319a backbone, but the expected photobleaching phenotype was very weak. Second, through comparing the effect of backbones (AtMIR319a, AtMIR390a, and SlMIR159) on specificity and significance of generating small RNAs, the AtMIR390a backbone was optimally selected to construct the small RNA overexpression system. Third, through sRNA-Seq and Degradome-Seq, the small RNAs from AtMIR390a backbone in TYLCCNV-amiRPDS(390) vector were confirmed to highly overexpress amiRPDS and specifically silence targeted PDS gene. Using this system, rapid functional analysis of endogenous miRNAs and siRNAs was carried out, including miR156 and athTAS3a 5′D8(+). Meanwhile, through designing corresponding artificial miRNAs, this system could also significantly silence targeted endogenous genes and show specific phenotypes, including PDS, Su, and PCNA. These results demonstrated that this small RNA overexpression system could contribute to investigating not only the function of endogenous small RNAs, but also the functional genes in plants.
Small RNAs, especially microRNAs (miRNAs) and transacting siRNAs (tasiRNAs), play vital roles in different kinds of biological regulation processes, including leaf development, phase transition, and abiotic and biotic stresses resistance (Byrne, 2012; Khraiwesh et al., 2012; Teotia and Tang, 2015; Wang and Chekanova, 2016). In recent years, as a hotspot in the field of plant physiology, small RNAs have been largely found in various plants (Zhang et al., 2014; Yi et al., 2015).
miRNAs, a class of 20- to 24-nucleotide small RNAs, widely regulate the expression of their target genes in almost all plants, animals, and even viruses (Voinnet, 2009; Chávez Montes et al., 2014; Taylor et al., 2014; Xie et al., 2015). In plants, mature miRNAs originate from primary miRNAs, which are transcribed by RNA polymerase II and contain an imperfect intramolecular stem loop and are formed by the cleavage of DICER-like1 (DCL1) endonuclease (Rogers and Chen, 2013). According to previous studies, plant miRNAs can directly cleave or inhibit translation of their targets to induce gene silencing via ARGONAUTE proteins (Rogers and Chen, 2013; Bologna and Voinnet, 2014). Generally, plant miRNAs become involved in massive biological processes by mediating plant transcriptional factors, such as miR165/miR166-class III homeodomain-Leu zipper and miR164-cup-shaped cotyledon modules in leaf development (Byrne, 2012), and miR156-squamosa promoter-binding protein-like (SPL) and miR172-apetala2 modules in flowering-determining (Teotia and Tang, 2015). Moreover, many miRNAs are involved in phytohormone signaling pathways (Curaba et al., 2014) and abiotic and biotic stresses (Sunkar, 2010; Khraiwesh et al., 2012).
As for short interfering RNAs (siRNAs), they are divided into secondary siRNAs (including tasiRNAs and phased siRNAs), heterochromatic siRNAs, and natural antisense transcript siRNAs (Axtell, 2013). In general, tasiRNAs are mainly found in plants, such as Arabidopsis (Arabidopsis thaliana) and tomato (Solanum lycopersicum; Allen et al., 2005; Zhang et al., 2014). Combined with miRNA and siRNA pathways, tasiRNAs are synthesized from TAS transcripts (Fei et al., 2013), and they play vital roles in leaf morphology (Adenot et al., 2006), lateral root growth (Marin et al., 2010), and regulation of ARF (Allen et al., 2005). Generally, tasiRNAs from TAS3 can target ARF3 and ARF4, such as Arabidopsis TAS3a 5′D7(+) and 5′D8(+) (Allen et al., 2005) and rice (Oryza sativa) TAS3a Osta-siR2141 (Wang et al., 2010).
As a special and efficient gene-silencing technology, artificial miRNA (amiRNA) technology (Schwab et al., 2006), has been widely utilized for functional characterization of plant-specific miRNAs in many kinds of plants, such as Arabidopsis, rice, and tomato, through several amiRNA backbones, including Arabidopsis MIR319a (AtMIR319a; Schwab et al., 2006), Arabidopsis MIR390a (AtMIR390a; Carbonell et al., 2014), and tomato MIR159 (SlMIR159; Kravchik et al., 2014). Moreover, amiRNA technology could be also applied to overexpress tasiRNAs in rice and Arabidopsis (Wang et al., 2010).
With the rapid development of next-generation sequencing technology, more and more small RNAs were identified in various plants, following experimental and functional identification of miRNAs with transformation technology (Sunkar and Jagadeeswaran, 2008; Baksa et al., 2015). However, as we all know, the genetic transformation technology is so laborious and time consuming, which leads to the functions of most small RNAs still remaining unknown. Thus, there is an urgency for simple, rapid, and highly effective tools to clarify the exact functions of those small RNAs. For small RNAs with high expression in plants, we used virus-based miRNA silencing/suppression systems to silence those small RNAs by at least five virus vectors, such as Tobacco rattle virus (TRV; Sha et al., 2014; Yan et al., 2014), Cotton leaf crumple virus (CLCrV; Gu et al., 2014), Cucumber mosaic virus (Du et al., 2014; Liao et al., 2015), Barley stripe mosaic virus (Jiao et al., 2015), and Potato virus X (PVX; Zhao et al., 2016). As for small RNAs with low expression in plants, virus-induced gene silencing (VIGS) using amiRNAs (MIR VIGS) has provided a simple and effective tool to overexpress these miRNAs in plants (Tang et al., 2010). Until now it has been reported that MIR VIGS was only caused by four plant virus vectors, including two DNA viruses in the genus Begomovirus, family Geminiviridae, of Cabbage leaf curl virus (CaLCuV) (Tang et al., 2010) and CLCrV (Gu et al., 2014), and two RNA viruses in genus Tobravirus, family Virgaviridae, of TRV and in genus Potexvirus, family Alphaflexiviridae of PVX (Chen et al., 2015). Moreover, these vectors were only used to overexpress miRNAs in plant. In this study, we reported a new versatile system based on a viral satellite associated with Tomato yellow leaf curl China virus (TYLCCNV) vector for rapidly functional characterization of small RNAs, including miRNAs and siRNAs.
TYLCCNV, like CaLCuV and CLCrV, belongs to the genus Begomovirus, family Geminiviridae, and replicates in the nucleus (Zhou, 2013), so we inferred the virus could also be used as a viral promoter to drive miRNA precursor to form mature miRNA. TYLCCNV consists of DNA-A and DNA-β. DNA-A, as a helper virus, is an about 2.7-kilonucleotide circular single-stranded DNA (ssDNA) and encodes six genes, including AC1, AC2, AC3, AC4, AV1, and AV2, and DNA-β, as a beta satellite, is an about 1.3-kilonucleotide ssDNA and encodes one βC1 gene (Tao and Zhou, 2004). TYLCCNV vector (βC1 deletion), which was applied as VIGS and virus-induced gene transcriptional silencing (VITGS) vector, has already been used to induce transcriptional gene silencing or posttranscriptional gene silencing without causing any obvious viral symptoms in plants (Tao and Zhou, 2004; Tao et al., 2006; Cai et al., 2007; Ju et al., 2016). Here, we reported a new TYLCCNV vector, which could efficiently overexpress not only amiRNAs but also endogenous small RNAs to study their biological functions in N. benthamiana.
RESULTS
Establishment of a TYLCCNV-Based Small RNA Overexpression System
TYLCCNV vector has been used for VIGS and VITGS by respectively inserting the fragment of genes or promoters to regulate specific plant gene expression (Tao and Zhou, 2004; Tao et al., 2006; Ju et al., 2016). The currently available TYLCCNV vector, p1.7A+2mβ, which carried both viral DNA-A and DNA-mβ (βC1 gene deletion but retains βC1 promoter), was reconstructed in our previous study (Ju et al., 2016). To determine whether the TYLCCNV vector could be used for overexpressing specific small RNA, the amiRPDS sequence, targeting N. benthamiana PDS gene (NCBI:DQ469932) as a reporter, was designed and inserted in the common AtMIR319a backbone according to Tang’s study (Tang et al., 2010). We replaced the native miR319a with amiRPDS via traditionally tedious three-step PCR and cloned it into TYLCCNV in the same direction of the βC1 gene to produce TYLCCNV-amiRPDS(319L) vector, which represented long primary AtMIR319a sequence as backbone. In the nucleus, the amiRNA precursors could be transcribed by βC1 promoter and were expected to be processed to mature amiRNAs, which could silence corresponding target genes.
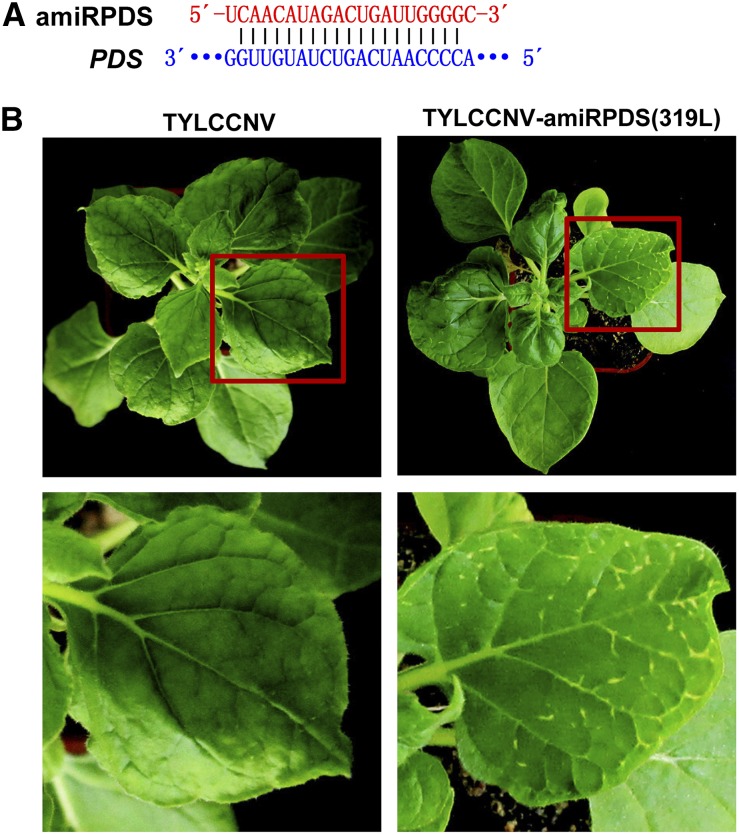
According to previous studies, silencing of PDS gene could result in photobleached leaves (Tao and Zhou, 2004; Tao et al., 2006; Ju et al., 2016). Consistently, most N. benthamiana plants (12/18) infiltrated with Agrobacterium tumefaciens containing TYLCCNV-amiRPDS(319L) developed a photobleached phenotype in veins at about 2 to 3 weeks postinfiltration, while all TYLCCNV-infected control plants had no apparent symptoms (Fig. 1; Supplemental Fig. S1; the silencing efficiency is shown in Supplemental Table S1). These data indicated that TYLCCNV-amiRPDS(319L) infiltration could induce PDS gene silencing phenotype in N. benthamiana, which might be attributed to amiRPDS overexpression resulting from TYLCCNV-amiRPDS(319L). However, only spots in the vein could be observed and could not be confirmed by real-time quantitative PCR (RT-qPCR; Fig. 1). Thus, the small RNA overexpression system was further optimized to strengthen the PDS silencing phenotype in this research below.
Figure 1.
Silence of PDS by TYLCCNV-amiRPDS(319L) in N. benthamiana. A, Scheme of amiRPDS and its target mRNA sequence. B, PDS-silencing phenotype in N. benthamiana plants inoculated with TYLCCNV-amiRPDS(319L). TYLCCNV empty vector-inoculated plants were used as negative control. Photographs were taken at 30 dpi.
Optimization of TYLCCNV-Based Small RNA Overexpression System
To optimize our overexpressing amiRNA system, different amiRNA backbones and strategies of constructing amiRNA precursors were considered to generate high-abundance amiRPDS. Among the amiRNA backbones that had been used to overexpress miRNA via transgenic technology (Schwab et al., 2006; Khraiwesh et al., 2008; Li et al., 2014b), we chose three kinds of amiRNA backbones, including AtMIR319a and SlMIR159, which were processed as loop-to-base miRNA processing mechanism, and AtMIR390a, which was synthesized according to base-to-loop miRNA processing mechanism. AtMIR319a precursor was widely used to overexpress miRNA in many plants, such as N. benthamiana, Arabidopsis, and tomato, via transgenic technology or virus inducing miRNA overexpression method with CaLCuV and CLCrV (Tang et al., 2010; Gu et al., 2014; Li et al., 2014b). SlMIR159 precursor has been previously reported in tomato to induce up-regulation of specific amiRSlDCL3, which markedly raised 21- and 22-nucleotide sRNA levels and significantly downregulated 24-nucleotide sRNA levels (Kravchik et al., 2014). As for AtMIR390a precursor, it was found to be processed accurately and to produce more miRNAs than AtMIR319a precursor in Arabidopsis (Carbonell et al., 2014).
First, we designed specific primers to simplify the process of constructing TYLCCNV-amiRPDS vector (primers shown in Supplemental Table S2), which replaced common three-step PCR with only one-step PCR (Supplemental Figs. S2 and S3); for example, the TYLCCNV-amiRPDS(319) vector was constructed by using short preAtMIR319a structure instead of long primary AtMIR319a backbone. In the one-step process, long primers in Supplemental Table S2 contained prolonged 15-nucleotide bases, which were needed in base-to-loop processing mechanisms, such as MiR390, while pre-MIRNA structure only was needed in loop-to-base processing mechanisms, such as MIR319 and MiR159.
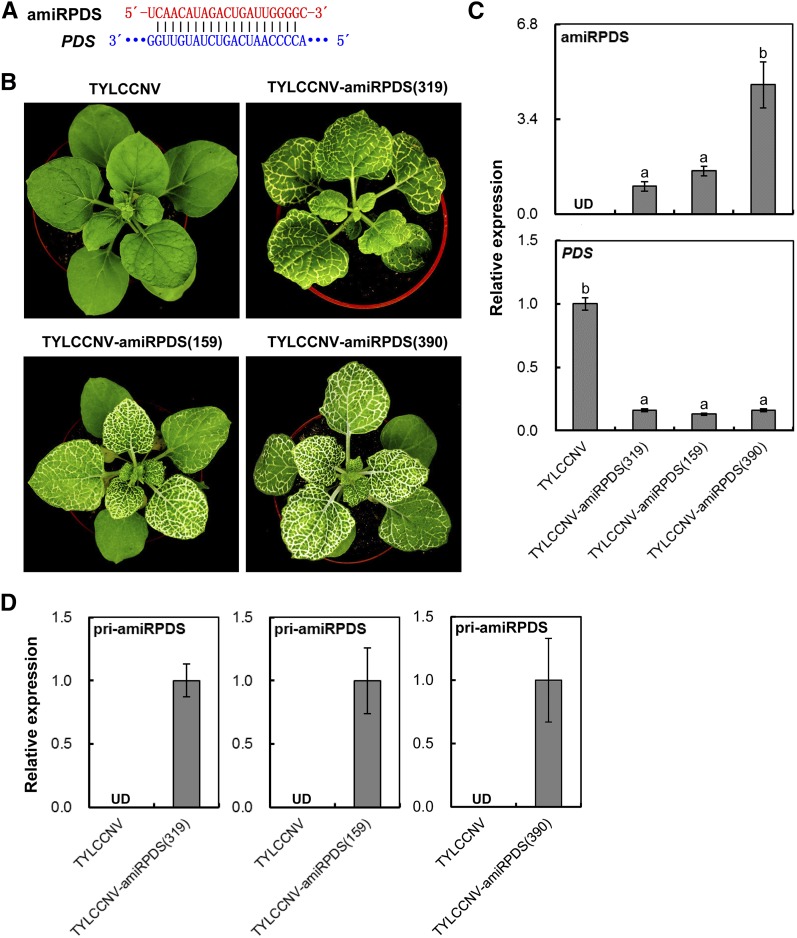
Second, those three kinds of amiRPDS precursors were respectively cloned into TYLCCNV to form TYLCCNV-amiRPDS(319), TYLCCNV-amiRPDS(159), and TYLCCNV-amiRPDS(390) vector to silence PDS gene (Fig. 2A). In about 2 to 3 weeks postinfiltration, all of TYLCCNV-amiRPDS(319)-, TYLCCNV-amiRPDS(159)-, and TYLCCNV-amiRPDS(390)-infected N. benthamiana could induce significantly plant photobleached phenotype in veins of leaf, stem, and flower tissues (Fig. 2B; pooled phenotype in Supplemental Figs. S4 and S5; the silencing efficiency is shown in Supplemental Table S3). Interestingly, TYLCCNV-amiRPDS(390)-infiltrated plants exhibited more severe phenotype than that of other vectors. Further RT-qPCR and northern-blot analysis also showed that TYLCCNV-amiRPDS(390) produced more amiRPDS than other vectors with different backbones (Fig. 2C; Supplemental Fig. S6A). Moreover, pri-amiRPDS could be detected in all of TYLCCNV-amiRPDS(319)-, TYLCCNV-amiRPDS(159)-, and TYLCCNV-amiRPDS(390)-infected N. benthamiana, but not in TYLCCNV-infected control plants (Fig. 2D; Supplemental Fig. S6A). In addition, compared to TYLCCNV-infected control plants, the mRNA levels of PDS were significantly reduced in these three TYLCCNV-amiRPDS-infiltrated plants (Fig. 2C; Supplemental Fig. S6B). Taken together, those results suggested that TYLCCNV-based vectors can be used to overexpress specific amiRNA to induce efficient gene silencing in N. benthamiana plants, and AtMIR390a backbone was optimally selected to further studies.
Figure 2.
Silence of PDS by TYLCCNV-amiRPDS vectors in different miRNA backbones in N. benthamiana. A, Scheme of amiRPDS and its related target mRNA sequence. B, PDS-silencing phenotype in N. benthamiana plants inoculated with TYLCCNV-amiRPDS(319), TYLCCNV-amiRPDS(159), and TYLCCNV-amiRPDS(390). TYLCCNV empty vector-inoculated plants were used as negative control. Photographs were taken at 25 dpi. C, Relative expressions of amiRPDS and PDS in plants inoculated with TYLCCNV or TYLCCNV-amiRPDS vectors in different miRNA backbones by RT-qPCR. Data were presented as the means of three independent experiments. Different lowercase letters indicate a statistical significance using ANOVA followed by Duncan’s test (P < 0.05). eIF4α was used as an internal control. D, Relative expression levels of pri-amiRPDS in plants inoculated with TYLCCNV or TYLCCNV-amiRPDS vectors in different miRNA backbones by RT-qPCR. Data are presented as the mean of three independent experiments. The error bars indicate mean ± sd. UD, Undetectable. Asterisks indicate statistical significance (Student’s t test, P < 0.01). eIF4α was used as an internal control.
Validation of amiRNA Sequences from AtMIR390a-Based Backbone through sRNA-Seq and Degradome-Seq
In order to confirm that the small RNAs generated by TYLCCNV vector could specifically overexpress amiRPDS and cleave targeted PDS transcripts in infiltrated leaves, we performed sRNA-Seq and Degradome-Seq separately.
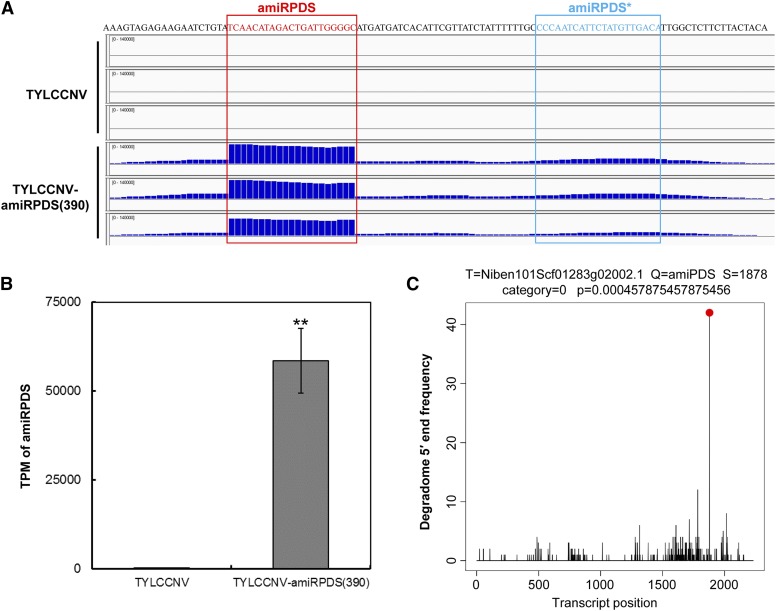
To more precisely evaluate the processing and accumulation of the amiRNA, small RNA libraries of three TYLCCNV-amiRPDS(390) and three TYLCCNV control-infiltrated leaf samples were compared in IGV genome browser (Fig. 3A). The result showed that higher abundance of small RNA reads was accumulated specifically in the amiRPDS backbone location than in control libraries (2943 folds in transcripts per million, P = 1.83e-4; Fig. 3B). This result corresponded to correctly processed 21-nucleotide amiRNA, while reads from the amiRNA* strands were always comparatively insufficient, indicating that the AtMIR390a precursor is processed precisely (Fig. 3A).
Figure 3.
Analysis of sRNA-Seq and Degradome-Seq data to show overexpression of specific small RNAs by TYLCCNV-amiRPDS(390) vector and cleavage of targeted PDS gene mRNA. A, amiRPDS distribution shown in IGV by analysis of sRNA-Seq. B, Transcripts per million of amiRPDS reads in plants inoculated with TYLCCNV or TYLCCNV-amiRPDS(390). Data are presented as the mean of three independent experiments. The error bars indicate mean ± sd. Asterisks indicate statistical significance (Student’s t test, P < 0.01). C, Target plots showed cleavage features in PDS mRNA by amiRPDS from the TYLCCNV-amiRPDS(390)-inoculated N. benthamiana degradome library.
The Degradome-Seq data were bioinformatically analyzed using the CleaveLand tool (Addo-Quaye et al., 2009a) with all small RNAs mapped to the amiRPDS(390) insert against the N. benthamiana cDNA library. The results showed that the degraded RNA reads were only accumulated and overrepresented on the predicted amiRPDS cleavage site (Fig. 3C), proving that amiRPDS induced the correct cleavage of its targeted PDS transcripts and was not off target. Taken together, the TYLCCNV-amiRPDS(390) was validated to specifically overexpress amiRPDS and exert its biological role in plants, suggesting the TYLCCNV-amiR(390) vector could be utilized in further study.
Overexpression of Plant Endogenous miRNAs via TYLCCNV Vector
Generally, overexpression of miRNAs is one of the most popular methods to explore the function of miRNAs in plants (Schwab et al., 2006; Khraiwesh et al., 2008; Ossowski et al., 2008; Warthmann et al., 2008). In order to estimate whether the TYLCCNV-based miRNA-overexpressing vector could be used to characterize the function of plant endogenous miRNAs, we constructed TYLCCNV-amiR156 vector to overexpress miR156b in N. benthamiana plants (Fig. 4A; Supplemental Fig. S7). MiR156 is a conservative miRNA in plants and target SPL transcription factors, such as in Arabidopsis, rice, and cotton (Gossypium hirsutum; Xie et al., 2012; Xing et al., 2013; Gu et al., 2014).
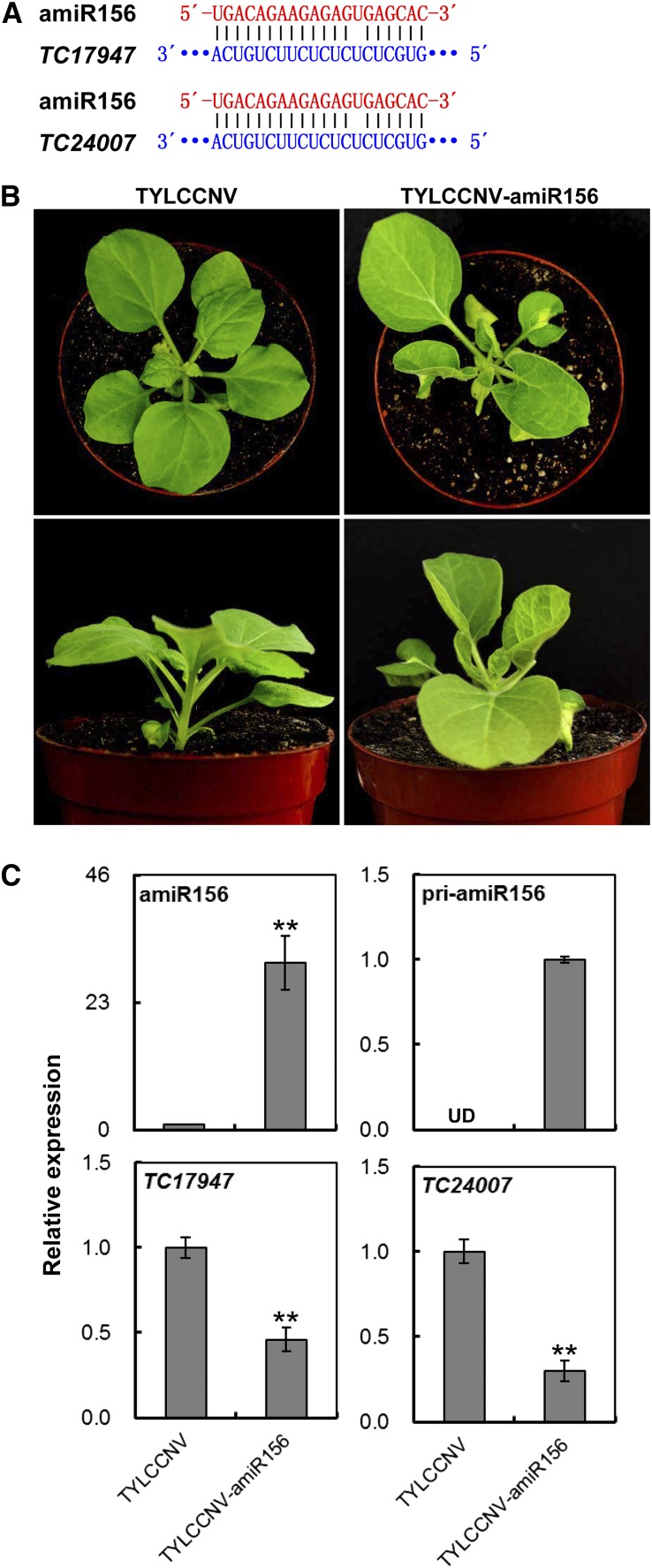
Figure 4.
Overexpression of miR156 by TYLCCNV-amiR156 in N. benthamiana. A, Scheme of amiR156 and its related target mRNA sequences. B, The phenotype of overexpressing miR156 in N. benthamiana plants inoculated with TYLCCNV-amiR156. TYLCCNV empty vector-inoculated plants were used as negative control. Photographs were taken at 14 dpi. C, Relative expressions of amiR156, pri-amiR156, and miR156 targeted TC17947 and TC24007 genes in plants inoculated with TYLCCNV or TYLCCNV-amiR156 by RT-qPCR. Data are presented as the mean of three independent experiments. The error bars indicate mean ± sd. UD, Undetectable. Asterisks indicate statistical significance (Student’s t test, P < 0.01). eIF4α was used as an internal control.
In as early as 14 d postinfiltration (dpi), a total 13 of 18 plants infiltrated with A. tumefaciens carrying TYLCCNV-amiR156 showed typical leaf-curl phenotype, while plants infected by TYLCCNV control appeared normal (Fig. 4B; pooled phenotype in Supplemental Fig. S8; the silencing efficiency is shown in Supplemental Table S4). In TYLCCNV-amiR156 infiltrated plants, the mRNA levels of miR156 were remarkably increased about 30 times than that in control (TYLCCNV; Fig. 4C). Furthermore, pri-amiR156 abundance was also detected in TYLCCNV-amiR156-infiltrated plants, while undetected in control plants. These data indicated amiR156 biogenesis was largely promoted in N. benthamiana plants infected with TYLCCNV-amiR156 vector.
In N. benthamiana, miR156 had at least five target genes, including two putative SPL genes (TC17947 and TC24007), which were found with the miRNA target search tool included in Web MicroRNA Designer (Ossowski et al., 2008). In order to know the molecular effect of overexpressing amiR156, the relative expressions of TC17947 and TC24007 were analyzed to evaluate the silencing effect of miR156 on its targets. Compared to TYLCCNV-infected control plants, the mRNA levels of TC17947 and TC24007 were significantly reduced in TYLCCNV-amiR156 infiltrated plants (Fig. 4C). These results suggested that this vector could be utilized to effectively study the functions of plant endogenous miRNAs.
Overexpression of Plant Endogenous tasiRNAs through TYLCCNV Vector
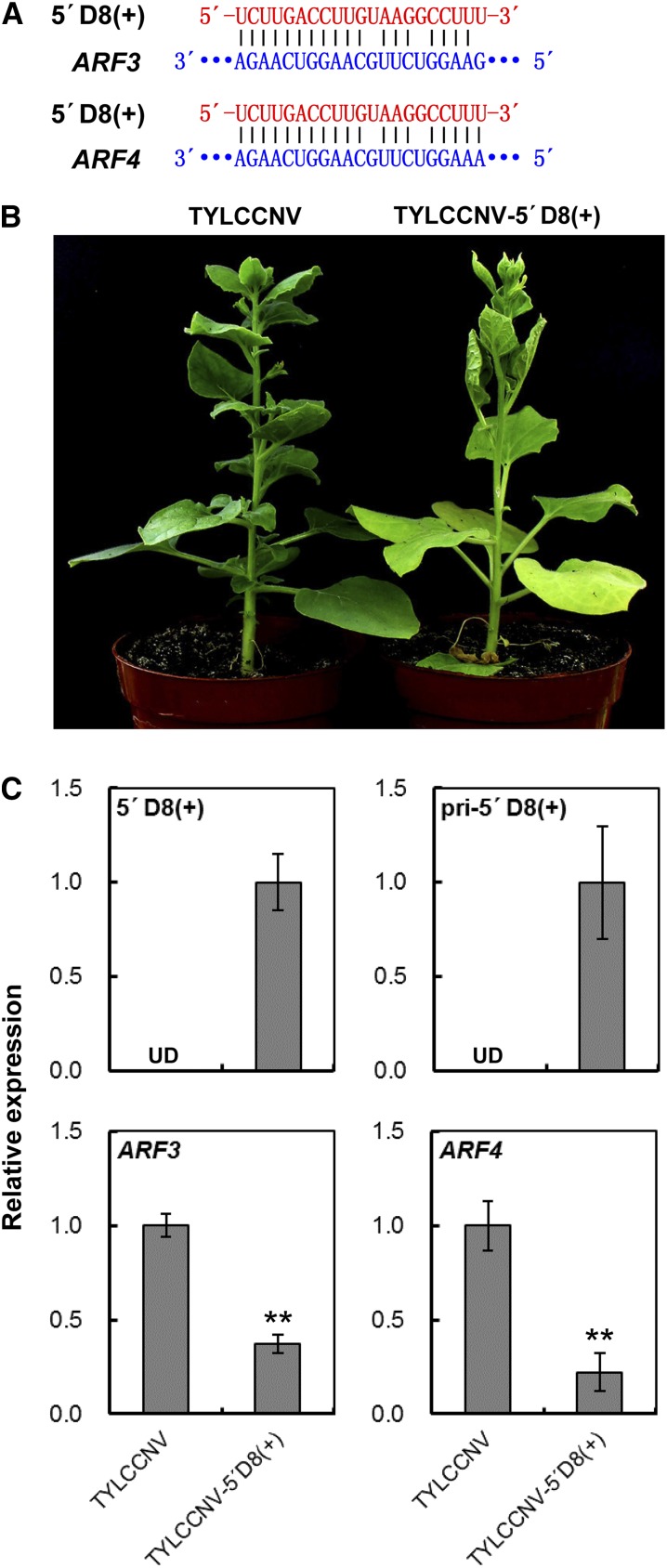
In order to determine whether our TYLCCNV-based small RNA-overexpressing vector could be used to study the functions of plant endogenous siRNAs, the Arabidopsis TAS3a-5′D8(+) tasiRNA sequence was constructed into TYLCCNV-amiR(390) vector to produce 5′D8(+) (Fig. 5A; Supplemental Fig. S7). Corresponding to the effect of overexpressing TAS3a-5′D8(+) in a previous study (Wang et al., 2010), 14 of 18 plants infiltrated with A. tumefaciens carrying TYLCCNV-5′D8(+) showed typical leaf-curl phenotype at 30 dpi (Fig. 5B; Supplemental Fig. S9; the silencing efficiency is shown in Supplemental Table S5). In contrast, all of plants infected by TYLCCNV control still appeared normal (Fig. 5B; Supplemental Fig. S9).
Figure 5.
Overexpression of 5′D8(+) by TYLCCNV-5′D8(+) in N. benthamiana. A, Scheme of 5′D8(+) and its related target mRNA sequences. B, The phenotype of overexpressing 5′D8(+) in N. benthamiana plants inoculated with TYLCCNV-5′D8(+). TYLCCNV empty vector-inoculated plants were used as negative control. Photographs were taken at 30 dpi. C, Relative expressions of 5′D8(+), pri-5′D8(+), and 5′D8(+) targeted ARF4 and ARF3 genes in plants inoculated with TYLCCNV or TYLCCNV-5′D8(+) by RT-qPCR. Data are presented as the mean of three independent experiments. The error bars indicate mean ± sd. UD, Undetectable. Asterisks indicate statistical significance (Student’s t test, P < 0.01). eIF4α was used as an internal control.
To assess the accumulation of tasiRNA in TYLCCNV-5′D8(+)-inoculated plants, systemic leaves were harvested, and we performed RT-qPCR. The result showed that the corresponding mature 5′D8(+) was distinctly detected in TYLCCNV-5′D8(+)-infiltrated plants, but not in plants infiltrated with empty TYLCCNV, as well as the result of pri-5′D8(+) (Fig. 5C). These results suggested that the expression of tasiRNA 5′D8(+) is specifically increased by using TYLCCNV-5′D8(+) vector.
As for the targets of tasiRNA 5′D8(+), there were at least five genes, including two putative ARF genes (ARF3 and ARF4) predicted by searching the N. benthamiana cDNA library with the psRNATarget tool (Dai and Zhao, 2011), which was similar to that in Arabidopsis (Wang et al., 2010). Then the relative expressions of ARF3 and ARF4 were assayed by RT-qPCR to assess the silencing effect of 5′D8(+). In TYLCCNV-5′D8(+)-infected plants, ARF3 and ARF4 expressed obviously higher than in TYLCCNV-infected control plants (Fig. 5C). Taken together, these results indicate that the promotion of specific endogenous tasiRNA expression using TYLCCNV-amiR(390) vector could effectively induce expected molecular and morphological changes in N. benthamiana, suggesting that TYLCCNV-amiR(390) vector could prospectively be applied to evaluate the functions of plant endogenous tasiRNAs.
Silence of Functional Gene with TYLCCNV amiRNA Vector
The above experiment of amiRPDS overexpression with TYLCCNV vector has proven that this vector also resulted in PDS gene silencing in N. benthamiana plant. To further test whether the TYLCCNV vector was a useful artificial tool with a 21-nucleotide short sequence for inducing functional gene silencing in plant, Su and PCNA genes were chosen to design and construct TYLCCNV-amiRSu and TYLCCNV-amiRPCNA vectors (Figs. 6 and 7; Supplemental Figs. S7, S10, and S11). According to previous studies, silencing the Su gene in N. benthamiana could result in yellow-colored leaves, while silencing the PCNA gene leads to misshapen newly produced leaves with truncated basipetal growth at the shoot tips (Tao and Zhou, 2004; Tang et al., 2010).
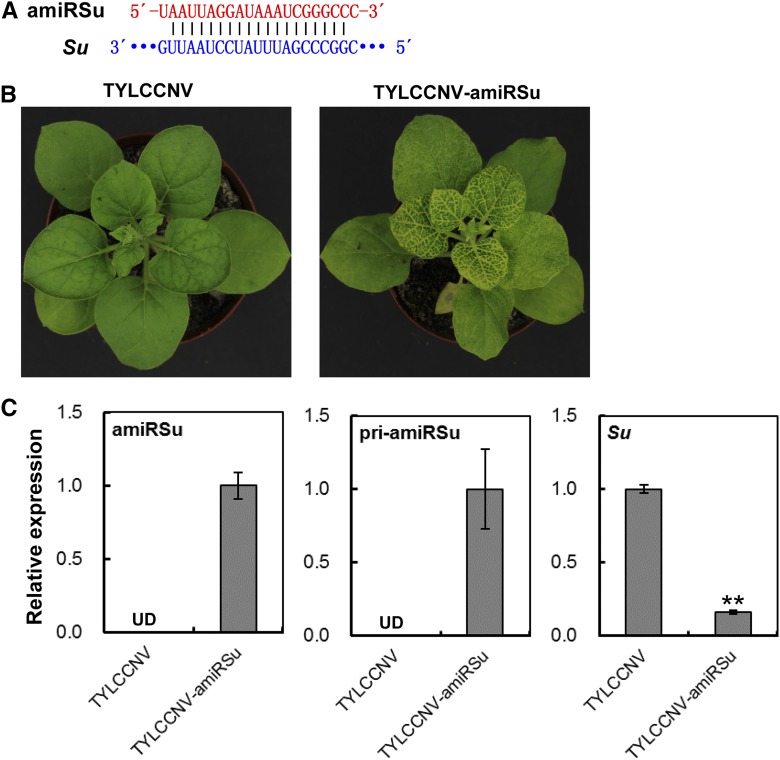
Figure 6.
Silence of Su by TYLCCNV-amiRSu in N. benthamiana. A, Scheme of amiRSu and its related target mRNA sequence. B, Su-silencing phenotype in N. benthamiana plants inoculated with TYLCCNV-amiRSu. TYLCCNV empty vector-inoculated plants were used as negative control. Photographs were taken at 25 dpi. C, Relative expressions of amiRSu, pri-amiRSu, and Su in plants inoculated with TYLCCNV or TYLCCNV-amiRSu by RT-qPCR. Data are presented as the mean of three independent experiments. The error bars indicate mean ± sd. UD, Undetectable. Asterisks indicate statistical significance (Student’s t test, P < 0.01). eIF4α was used as an internal control.
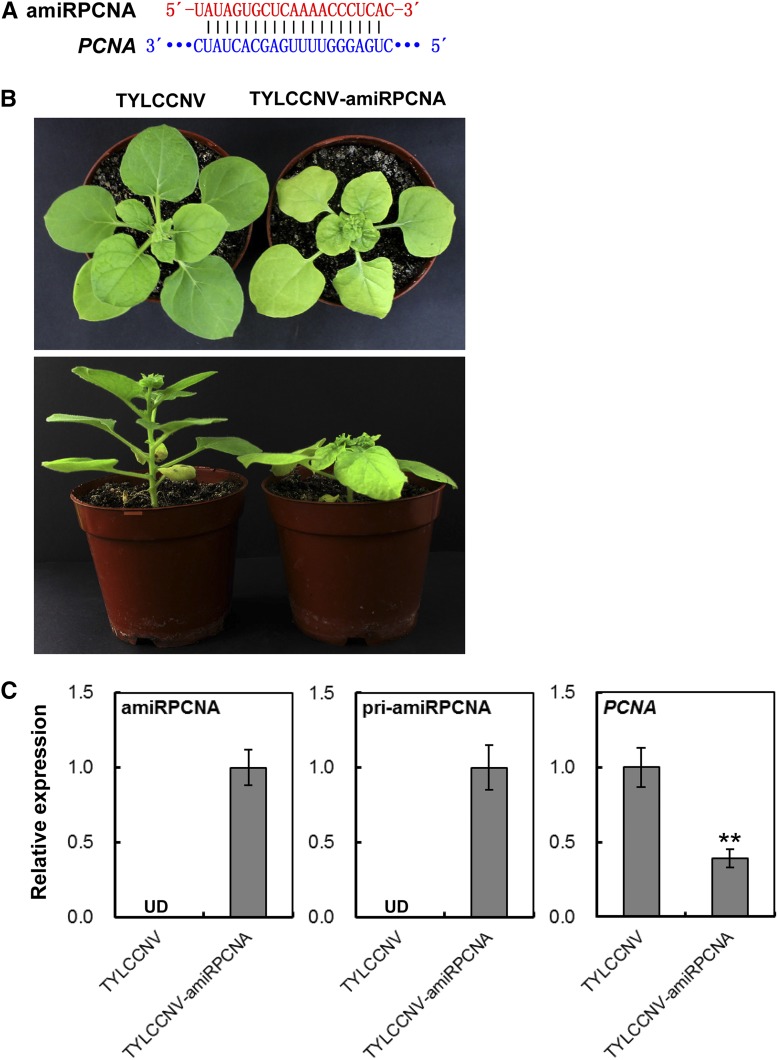
Figure 7.
Silence of PCNA by TYLCCNV-amiRPCNA in N. benthamiana. A, Scheme of amiRPCNA and its related target mRNA sequence. B, PCNA-silencing phenotype in N. benthamiana plants inoculated with TYLCCNV-amiRPCNA. TYLCCNV empty vector-inoculated plants were used as negative control. Photographs were taken at 25 dpi. C, Relative expressions of amiRPCNA, pri-amiRPCNA, and PCNA in plants inoculated with TYLCCNV or TYLCCNV-amiRPCNA by RT-qPCR. Data are presented as the mean of three independent experiments. The error bars indicate mean ± sd. UD, Undetectable. Asterisks indicate statistical significance (Student’s t test, P < 0.01). eIF4α was used as an internal control.
Similarly, all plants (18/18) infiltrated with TYLCCNV-amiRSu produced phenotype of yellow leaves in vein as early as 25 dpi (Fig. 6B; Supplemental Fig. S10; the silencing efficiency is shown in Supplemental Table S6), and all plants (18/18) infected with TYLCCNV-amiRPCNA showed obviously abnormal growth at 25 dpi (Fig. 7B; Supplemental Fig. S11; the silencing efficiency is shown in Supplemental Table S7). In contrast, the TYLCCNV-infiltrated control plants appeared normal, and none of them showed any silencing phenotypes (Fig. 6B and Supplemental Fig. S10; Fig. 7B and Supplemental Fig. S11). These distinct phenotypes proved that 21-nucleotide artificial specific sequences constructed in TYLCCNV-amiRSu and TYLCCNV-amiRPCNA vectors sufficiently induced corresponding gene-silencing phenotype.
Furthermore, the corresponding mature amiRSu and amiRPCNA were respectively detected in TYLCCNV-amiRSu- and TYLCCNV-amiRPCNA-infiltrated plants, but not in plants infected with empty TYLCCNV vector, which was similar to the result of pri-amiRSu and pri-amiRPCNA (Figs. 6C and 7C). These data showed that the artificial amiRNAs designed from functional genes were highly accumulated in N. benthamiana plants after inoculating with A. tumefaciens containing TYLCCNV-amiRSu and TYLCCNV-amiRPCNA vector. Moreover, the mRNA levels of Su and PCNA in TYLCCNV-amiRSu and TYLCCNV-amiRPCNA-infiltrated leaf tissues were all obviously lower than in TYLCCNV-infected control plants, respectively (Fig. 6C and 7C). Overall, these results proved that the TYLCCNV-based amiRNA vector can be utilized to overexpress specific 21-nucleotide amiRNAs for sufficiently inducing target gene silencing in N. benthamiana plants.
DISCUSSION
TYLCCNV Vector Was a Versatile and Efficient Vector for Small RNA Overexpression
Similar to other important virus vectors, traditional TYLCCNV vector has been utilized as a useful tool to cause transcriptional gene silencing and posttranscriptional gene silencing in plants, namely VITGS and VIGS (Tao and Zhou, 2004; Tao et al., 2006; Ju et al., 2016). In this study, this vector was also proved to up-regulate the expressions of small RNAs in plants and avoided the time-consuming and laborious process of plant genetic transformation. Moreover, an optimized amiRNA backbone based on AtMIR390a precursor was designed and constructed into TYLCCNV vector, which finally formed TYLCCNV-amiRNA vector. After being infiltrated in plant tissues with A. tumefaciens mediation, this vector largely induced the accumulation of specific 21-nucleotide short small RNAs artificially designed in the manner of miRNA biogenesis.
Theoretically, miRNAs or siRNAs, even probably 21-nucleotide small RNAs like miRNAs and siRNAs, could be overexpressed via TYLCCNV-amiRNA vector. Hence, in this study, following experimental validation and bioinformatical approaches have demonstrated the above inference and confirmed that this new TYLCCNV vector could be a highly efficient small RNA-overexpressing vector. Using this system, we successfully overexpressed endogenous miR156 and tasiRNA TAS3a-5′D8(+) in N. benthamiana, respectively. In addition, in the manner of amiRNAs, we separately silenced several endogenous functional genes in N. benthamiana plants, including PDS, Su, and PCNA. Besides, TYLCCNV-amiRPDS(390)-inoculated tomato (Micro-Tom) plants showed the PDS-silencing phenotype (Supplemental Fig. S12). These results provided us a comprehensive and versatile tool for characterization of small RNAs and functional genes, which have several distinct characteristics and advantages.
First, unlike previous reported successful vectors, including CaLCuV (Tang et al., 2010), CLCrV (Gu et al., 2014), TRV, and PVX (Chen et al., 2015), the viral satellite DNA associated with TYLCCNV could be applied to high-efficiency investigation of the physiological roles of several kinds of small RNAs, including endogenous miRNAs, siRNAs, and even new 21-nucleotide artificial short RNAs (especially siRNAs). Up to date, there was still no report of using viral vectors to explore the roles of endogenous tasiRNAs in plants, so our research would help to promote the progress of siRNA characterization in this field. Secondly, TYLCCNV virus was well known for its mild pathogenicity and wide adaptability to its host plants, such as N. benthamiana, tomato, Nicotiana glutinosa, and petunia (Petunia hybrida; Tao et al., 2006; Tao and Zhou, 2004), predicting that TYLCCNV amiRNA(390) vector would be applied in a wide range of plant kingdom for small RNA functional characterization in the future. Finally, three miRNA backbones, including AtMIR319a, SlMIR159, and AtMIR390a, were utilized in this study to compare their effect on small RNA-specific biosynthesis, and the AtMIR390a backbone was considered as optimal selection, indicating that TYLCCNV vector could facilitate another potential purpose to estimate the influence of different backbones and their structures of miRNA precursors on miRNA biogenesis.
Strategy of Small RNA Overexpression for Functional Characterization
In general, there are two strategies to overexpressing small RNAs for artificially regulating small RNAs in plant. One is amiRNA technology (Schwab et al., 2006), and another is synthetic transacting small interfering RNAs (syn-tasiRNAs) technology (Montgomery et al., 2008).
As an efficient and specific gene-silencing technology, amiRNA has been widely applied in functional genomics in many kinds of plants (Li et al., 2014b), such as Arabidopsis, rice, and tomato. This technology was largely developed through kinds of amiRNA backbones, such as AtMIR319a (Schwab et al., 2006; Tang et al., 2010; Gu et al., 2014), AtMIR390a (Carbonell et al., 2014), and SlMIR159 (Kravchik et al., 2014). Additionally, amiRNA technology was also reported to overexpress tasiRNA OstasiR2141 via genetic transformation in rice and Arabidopsis (Wang et al., 2010).
As for syn-tasiRNAs technology, it could efficiently induce gene silencing through replacing 5′D7(+) and 5′D8(+) with designated sequences in Arabidopsis TAS3a or substituting 3′D3(+) and 3′D4(+) in Arabidopsis TAS1c (Montgomery et al., 2008; Carbonell et al., 2014). The syn-tasiRNA sequence was similar to miRNA sequences such as atasiRPDS (Montgomery et al., 2008); thus, it was assumed that this technology could also substitute syn-tasiRNA with miRNA sequences, which would cause overexpression of specific miRNAs.
However, these two strategies in the previous reports mainly depended on laborious and time-consuming genetic transformation process, so high-throughput and rapid methods were urgently needed to identify the functions of small RNAs largely predicted by bioinformatics analysis and next-generation sequencing (Sunkar and Jagadeeswaran, 2008; Baksa et al., 2015).
Until now, only the first strategy was chosen to develop amiRNA-overexpression virus vectors, such as reported CaLCuV (Tang et al., 2010) and CLCrV (Gu et al., 2014) virus, which drove the transcription of amiRNA backbone in nucleus under respective promoter and were subsequently processed to mature miRNAs by DCL1 (Xie et al., 2015). In this study, the amiRNA strategy was also applied to construct TYLCCNV amiRNA-overexpressing vectors, which would expand the pool of potential amiRNA-overexpression tools, and the classes of small RNAs needed identification, especially due to retained βC1 promoter to produce amiRNA precursors in nucleus.
miRNA Backbone Design According to miRNA Processing Mechanism
Generally, the original miRNA sequence in the backbone was replaced with amiRNA sequence by three-step PCR with six primers, such as AtMIR319a backbone in transgenic technology (Schwab et al., 2006) and MIR VIGS (Tang et al., 2010; Gu et al., 2014). To simplify the construction processes in this study, we replaced long pri-miRNA (300–500 nucleotides) with short pri-miRNA structure (only 100–200 nucleotides) to form mature miRNA (Supplemental Figs. S2 and S3), which only needs one-step PCR with two primers, and finally caused satisfactorily expected specific amiRNA accumulation.
There are two kinds of miRNA-processing mechanisms: one is base-to-loop, which processes pri-miRNA from the base-proximal side of the stem and needs ≥15-nucleotide length stem sequences below the miRNA/miRNA* duplex to identify the right initial processing position of DCL1, such as MIR172 and MIR390 (Mateos et al., 2010; Song et al., 2010; Werner et al., 2010; Cuperus et al., 2011; Carbonell et al., 2014), and the other is loop-to-base, which processes pri-miRNA from the loop-proximal side of the stem and does not need the ≥15-nucleotide length stem sequences below the miRNA/miRNA* duplex, such as the deeply conserved MIR159 and MIR319 family (Addo-Quaye et al., 2009b; Bologna et al., 2009; Cuperus et al., 2011). According to those two miRNA processing mechanisms, only two special primers, both containing ≥15 nucleotides or not, needed to be designed, which simplified the construction of amiRNA precursor via one-step PCR (Supplemental Fig. S2).
miR390 Precursor as Good Candidate Backbone for amiRNA Overexpression
Although there were several famous backbones in amiRNA overexpression, such as AtMIR319a (Schwab et al., 2006) and SlMIR159 (Kravchik et al., 2014), the AtMIR390a backbone was recently reported with many advantages, including deep conservation in plants, accurate process, relatively short distal stem loop, high-expression 21- or 22-nucleotide amiRNA, and so on (Carbonell et al., 2014). In addition, the Carbonell study showed AtMIR390a backbone was processed more accurately than AtMIR319a backbone and produced more amiRNA (Carbonell et al., 2014), which was consistent with our result of TYLCCNV-amiRPDS inducing specific amiRNA accumulation targeting PDS in N. benthamiana (Fig. 2C; Supplemental Fig. S6A). Similarly, OsMIR390 and OsMIR390-AtL backbones (chimeric OsMIR390-based precursor including distal stem-loop sequences from AtMIR390a backbone), could highly express amiRNA in Brachypodium distachyon plants via transgenic technology (Carbonell et al., 2015). Thus, in our overexpressing small RNA system, we would attempt to use OsMIR390 and OsMIR390-AtL backbones in the future. Moreover, several MIR390 backbones from other plants or chimeric MIR390 backbones, even backbones by artificial design, would be considered to unceasingly optimize the structure of MIR390 backbone in our further studies.
MATERIALS AND METHODS
Plant Growth and Agroinfiltration
Nicotiana benthamiana were cultured in a growth chamber at 22°C under a 16 h light/8 h dark cycle. All viral infectious clones were separately introduced into Agrobacterium tumefaciens strain GV3101. Agroinfiltration was performed as described in our recent study (Ju et al., 2016). Plant tissues were collected, frozen instantly in liquid nitrogen, and stored in the −80°C freezer for further studies.
Plasmid Construction
The amiRPDS sequence was selected according to Tang’s report (Tang et al., 2010), the amiRNAs targeting Su and PCNA genes were designed by using Web MicroRNA Designer (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi), and the N. benthamiana mature miR156 sequence as well as Arabidopsis (Arabidopsis thaliana) TAS3a 5′D8(+) were respectively chosen based on previous studies (Allen et al., 2005). All sequences of amiRNA, endogenous miRNA and tasiRNA were shown in Supplemental Text S1.
The amiRPDS precursor of TYLCCNV-amiRPDS(319L) vector, using AtMIR319a (At4g23713) as a backbone, was formed via three-step PCR with the six primers referred to Tang’s report, and the detailed protocol can be found in his supplemental data (Supplemental Materials and Methods S1; Tang et al., 2010). Then, the PCR product containing amiRPDS was digested with XbaI and SmaI and subsequently cloned into p1.7A+2mβ vector to produce p1.7A+2mβ-amiRPDS(319L) [TYLCCNV-amiRPDS(319L)].
The amiRPDS precursors of TYLCCNV-amiRPDS(319) vector and TYLCCNV-amiRPDS(159) vector, separately adopting Arabidopsis AtMIR319a (At4g23713) and tomato (Solanum lycopersicum) endogenous SlMIR159 (NCBI: HG975515.1) as a backbone, were formed via only one-step PCR with two primers, which both obeyed loop-to-base miRNA process mechanisms. Then, the PCR products containing amiRPDS were digested by XbaI and SmaI and cloned into p1.7A+2mβ to produce p1.7A+2mβ-amiRPDS(319) [TYLCCNV-amiRPDS(319)] and p1.7A+2mβ-amiRPDS(159) [TYLCCNV-amiRPDS(159)], respectively.
The precursors of TYLCCNV-amiRPDS(390), TYLCCNV-amiRSu, TYLCCNV-amiRPCNA, TYLCCNV-amiR156, and TYLCCNV-5′D8(+) vector, all using Arabidopsis endogenous AtMIR390a as backbone, were amplified via one-step PCR with two specific primers, according to base-to-loop miRNA process mechanisms. Subsequently, the PCR products containing amiRPDS, amiRSu, amiRPCNA, N. benthamiana mature miR156, and Arabidopsis TAS3a 5′D8(+) were digested by XbaI and SmaI and introduced into p1.7A+2mβ to respectively produce p1.7A+2mβ-amiRPDS(390) [TYLCCNV-amiRPDS(390)], p1.7A+2mβ-amiRSu (TYLCCNV-amiRSu), p1.7A+2mβ-amiRPCNA (TYLCCNV-amiRPCNA), p1.7A+2mβ-amiR156 (TYLCCNV-amiR156), and p1.7A+2mβ-5′D8(+) [TYLCCNV-5′D8(+)].
All primers for plasmid construction are presented in Supplemental Table S2.
DNA and RNA Isolation, PCR Detection, and RT-qPCR Analysis
Total DNA was extracted from N. benthamiana leaves using an EasyPure Plant Genome DNA Kit (Transgen). Primers DNA-A-F and DNA-A-R were used for detecting TYLCCNV DNA-A, and DNA-mβ-F and DNA-mβ-R for TYLCCNV DNA-mβ.
Total RNA was extracted from agroinoculated N. benthamiana leaves using a modified Trizol method (Simms et al., 1993). First-strand cDNA was synthesized using 2 μg total RNA, random 6-mers or oligo(dT) primer by TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgen).
Transcriptional expression of target genes and miRNA precursors by RT-qPCR were performed as described previously using TransStart Top Green qPCR SuperMix (Transgen; Cao et al., 2014), and the expression levels of miRNA and tasiRNA using RT-qPCR were implemented in accordance with the method described by Varkonyi-Gasic et al. (2007). NbeIF4α (NCBI, KX247369.1) was the internal control gene (Sha et al., 2014), and the 2−∆∆Ct method was used to calculate the relative expressions of genes (Livak and Schmittgen, 2001). Each gene expression assay was repeated three times biologically. All primers for RT-qPCR were designed using Beacon Designer 8.13 (http://www.premierbiosoft.com/crm/jsp/com/pbi/crm/clientside/ProductList.jsp).
All primers used for PCR detection and RT-qPCR analysis are listed in Supplemental Table S2.
Northern-Blot Analysis
For northern-blot assays of amiRPDS and pri-amiRPDS, 20 μg of total RNA extracted from TYLCCNV-inoculated leaves and from TYLCCNV-amiRPDS-inoculated leaves were separately fractionated on 15% denaturing polyacrylamide gels containing 7 m urea in 1× Tris-Borate-EDTA (TBE) buffer and then transferred to Hybond-N+ membranes (Amersham) using the semidry transfer apparatus (Bio-Rad) in 0.5× TBE at 250 mA for 1.5 h. The transferred RNAs were cross-linked to the membrane by 0.16 m 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (Sigma-Aldrich) at 60°C for 1.5 h (Pall and Hamilton, 2008). Two DNA oligonucleotides complementary to N. benthamiana U6 RNA (Li et al., 2014a) and amiRPDS and separately labeled one digoxin (DIG) at 3′ were synthesized (TSINGKE) and used as probes for hybridization (Supplemental Table S2). Hybridization was carried out in the PerfectHyb Plus Hybridization buffer (Sigma-Aldrich) at 42°C overnight in a LF-III molecular hybridization furnace (Scientz), and hybridization signals were detected with DIG Luminescent Detection Kit (Roche) and the MiniChemi imager (SageCreation).
For northern-blot assay of PDS, 15 μg of total RNA extracted from TYLCCNV-inoculated leaves and from TYLCCNV-amiRPDS-inoculated leaves were respectively fractionated on 1.2% denaturing agarose gels containing 2.4 m formaldehyde in 1× MOPS buffer and then transferred to Hybond-N+ membranes (Amersham) by capillary transfer in 20× SSC overnight. The transferred RNAs were UV cross-linked to the membrane four times in a Xinyi03-II UV cross-linker (Scientz). In order to obtain PDS probe, 717-bp fragment of N. benthamiana PDS was constructed to pEasy-Blunt vector (Transgen) and confirmed by sequencing (TSINGKE). Then, the 717-bp PDS fragment was added to the T7 promoter at 3′ using PDS-PF and PDS-PT7R primers by PCR and was transcription-labeled with DIG using T7 RNA polymerase (Roche) and DIG RNA Labeling Mix (Roche) to produce the PDS RNA probe. Hybridization was carried out in the PerfectHyb Plus Hybridization buffer (Sigma-Aldrich) at 68°C overnight, and hybridization signals were detected as described above for northern-blot assays of amiRPDS and pri-amiRPDS.
All primers used for northern-blot analysis were listed in Supplemental Table S2.
Preparation of Small RNA Libraries and Sequencing Analysis
Small RNA libraries were constructed using the methods described previously with some modifications (Lu et al., 2007). The quality and purity of total RNA were analyzed by Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (Agilent) with RIN value ≥ 7.0. Approximately 1 μg of total RNA, respectively, from three control samples from TYLCCNV-inoculated leaves and three samples from TYLCCNV-amiRPDS(390)-inoculated leaves were used to prepare the small RNA library based on the protocol of TruSeq Small RNA Sample Prep Kits (Illumina). The 18- to 30-nucleotide small RNAs were separated by polyacrylamide gel electrophoresis and ligated to 5′ and 3′ RNA adapter by T4 RNA ligase. Subsequently, the adapter-ligated RNAs were transcribed to single-stranded cDNAs by Superscript II reverse transcriptase (Promega). Thereafter, the cDNAs were used as templates to produce double-stranded cDNAs by PCR amplification using primers that anneal to adapters. The cDNA libraries were obtained by 16% TBE gel. Finally, these six small RNA libraries were submitted to LC-Bio for the single-end sequencing on an Illumina HiSeq 2500, at least 10 million reads depth per library. The raw data of the sRNA-Seq are available at NCBI Short Read Archive (SRP096568).
The raw reads of sRNA-Seq were preprocessed with the Fastx-toolkit (v0.013.2) pipeline (http://hannonlab.cshl.edu/fastx_toolkit/) to remove the adapter sequences, low-quality sequences, and repetitive reads. Reads larger than 30 nucleotides and smaller than 18 nucleotides were discarded. Sequencing reads were parsed to identify library-specific barcodes were collapsed to a unique set with read counts. Unique sequences were aligned to a TYLCCNV genome [TYLCCNB-Y10 DNA-A (AJ319675); TYLCCNB-Y10 DNAβ (AJ421621)] index database containing the sequence of AtMIR390a-based amiRNA using BOWTIE version 1.0.0 (Langmead et al., 2009), with only perfect matches settings (-f -v 0 -a -S). Small RNA alignments were saved in sequence alignment/map format and were queried using SAMtools version 1.2 (Li et al., 2009). The bowtie result was sorted using SAMtools, finally exhibited and visualized in IGV genome browser (Thorvaldsdóttir et al., 2013).
Preparation of Degradome Library and Sequencing Analysis
To investigate the target specificity of amiRPDS, one degradome cDNA library was constructed using sliced ends of polyadenylated transcripts isolated from the TYLCCNV-amiRPDS(390) agroinoculated leaves (mixture of three samples for small RNA sequencing). Approximately 20 μg of total RNA from TYLCCNV-amiRPDS-inoculated leaves was used to prepare degradome library. The method was based on the description of Ma et al. (2010) with some modifications: (1) About 150 ng of poly(A)+ RNA was used as input RNA and annealing with biotinylated random primers; (2) strapavidin capture of RNA fragments was performed through biotinylated random primers; (3) 5′ adaptor ligation occurred in only those RNAs containing 5′-monophosphates; and (4) reverse transcription and PCR were used to obtain the degradome library. Finally, the degradome library was submitted to LC-Bio (Hangzhou) for the single-end sequencing on an Illumina HiSeq 2500 at a depth of ∼20 million reads. The raw data of Degradome-Seq is available at NCBI Short Read Archive (SRP096568).
Degradome clean reads were acquired using Fastx-toolkit to trim 5′ adaptor sequences and low-quality reads. Only 20- and 22-nucleotide reads perfectly matching to the amiRPDS backbone in the above clean small RNA libraries and reads with >5 clones in degradome library were collected for potentially cleaved target identification. The amiRPDS target cleavage was analyzed against N. benthamiana cDNA library (Niben101_annotation.transcripts) using CleaveLand v4.3 (Addo-Quaye et al., 2009a) with parameters -e, -u, -n -d, -g. Finally, miRNA targets and cleavage T-plot with P value ≤ 0.01 cutoff were kept.
Accession Numbers
Raw sequencing data from the sRNA-Seq and Degradome-Seq experiment are stored at NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under accession number SRP096568.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Silence of PDS by TYLCCNV-amiRPDS(319L) in N. benthamiana.
Supplemental Figure S2. Diagram of constructing amiRNA precursor by three-step and one-step PCR.
Supplemental Figure S3. Secondary structures of pri-amiRPDS(319), pri-amiRPDS(159), and pri-amiRPDS(390).
Supplemental Figure S4. Silence of PDS by TYLCCNV-amiRPDS vectors in different miRNA backbones in N. benthamiana.
Supplemental Figure S5. Phenotype of amiRPDS precursors in different miRNA backbones at 45 dpi.
Supplemental Figure S6. Northern-blot analysis of amiRPDS, pri-amiRPDS, and PDS in plants inoculated with TYLCCNV or TYLCCNV-amiRPDS vectors in different miRNA backbones.
Supplemental Figure S7. Secondary structures of pri-amiR156, pri-5′D8(+), pri-amiRSu, and amiRPCNA.
Supplemental Figure S8. Overexpression of miR156 by TYLCCNV-amiR156 in N. benthamiana.
Supplemental Figure S9. Overexpression of 5′D8(+) by TYLCCNV-5′D8(+) in N. benthamiana.
Supplemental Figure S10. Silence of Su by TYLCCNV-amiRSu in N. benthamiana.
Supplemental Figure S11. Silence of PCNA by TYLCCNV-amiRPCNA in N. benthamiana.
Supplemental Figure S12. PDS-silencing phenotype in tomato plants inoculated with TYLCCNV-amiRPDS(390).
Supplemental Table S1. The silencing efficiency of PDS gene by TYLCCNV-amiRPDS(319L) vector in N. benthamiana plants at 30 dpi.
Supplemental Table S2. Lists of primers used in this study.
Supplemental Table S3. The silencing efficiency of PDS gene by TYLCCNV-amiRPDS(319), TYLCCNV-amiRPDS(159), and TYLCCNV-amiRPDS(390) vectors in N. benthamiana plants at 25 dpi.
Supplemental Table S4. The silencing efficiency of overexpressing miR156 by TYLCCNV-amiR156 vectors in N. benthamiana plants at 14 dpi.
Supplemental Table S5. The silencing efficiency of overexpressing 5′D8(+) by TYLCCNV-5′D8(+) vectors in N. benthamiana plants at 30 dpi.
Supplemental Table S6. The silencing efficiency of Su gene by TYLCCNV-amiRSu vectors in N. benthamiana plants at 25 dpi.
Supplemental Table S7. The silencing efficiency of PCNA gene by TYLCCNV-amiRPCNA vectors in N. benthamiana plants at 25 dpi.
Supplemental Text. Sequences of pri-amiRNAs (5′−3′).
Supplementary Material
Acknowledgments
We thank Prof. Xueping Zhou (Zhejiang University) for providing pGEM-1.7A and pBinPLUS-2mβ-Sub vectors, Prof. Yingwu Yang (Chongqing University) for providing technologic help in northern-blot analysis of miRNA, and Prof. Zhiping Deng (Zhejiang Academy of Agricultural Sciences) for polishing this article.
Glossary
- tasiRNA
transacting siRNA
- amiRNA
artificial miRNA
- VIGS
virus-induced gene silencing
- VITGS
virus-induced transcriptional gene silencing
- dpi
days postinfiltration
- syn-tasiRNA
synthetic transacting small interfering RNA
Footnotes
This work was supported by the National Nature Science Foundation of China (grant nos. 31271959 and 31571894) and by the DBN Education Foundation from the China Agricultural University Education Foundation (grant no. 1061-2415002).
References
- Addo-Quaye C, Miller W, Axtell MJ (2009a) CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25: 130–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C, Snyder JA, Park YB, Li YF, Sunkar R, Axtell MJ (2009b) Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA 15: 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouché N, Gasciolli V, Vaucheret H (2006) DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16: 927–932 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Axtell MJ. (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64: 137–159 [DOI] [PubMed] [Google Scholar]
- Baksa I, Nagy T, Barta E, Havelda Z, Várallyay É, Silhavy D, Burgyán J, Szittya G (2015) Identification of Nicotiana benthamiana microRNAs and their targets using high throughput sequencing and degradome analysis. BMC Genomics 16: 1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna NG, Mateos JL, Bresso EG, Palatnik JF (2009) A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J 28: 3646–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna NG, Voinnet O (2014) The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol 65: 473–503 [DOI] [PubMed] [Google Scholar]
- Byrne ME. (2012) Making leaves. Curr Opin Plant Biol 15: 24–30 [DOI] [PubMed] [Google Scholar]
- Cai X, Wang C, Xu Y, Xu Q, Zheng Z, Zhou X (2007) Efficient gene silencing induction in tomato by a viral satellite DNA vector. Virus Res 125: 169–175 [DOI] [PubMed] [Google Scholar]
- Cao D, Ju Z, Gao C, Mei X, Fu D, Zhu H, Luo Y, Zhu B (2014) Genome-wide identification of cytosine-5 DNA methyltransferases and demethylases in Solanum lycopersicum. Gene 550: 230–237 [DOI] [PubMed] [Google Scholar]
- Carbonell A, Fahlgren N, Mitchell S, Cox KL Jr, Reilly KC, Mockler TC, Carrington JC (2015) Highly specific gene silencing in a monocot species by artificial microRNAs derived from chimeric miRNA precursors. Plant J 82: 1061–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A, Takeda A, Fahlgren N, Johnson SC, Cuperus JT, Carrington JC (2014) New generation of artificial microRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol 165: 15–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez Montes RA, de Fátima Rosas-Cárdenas F, De Paoli E, Accerbi M, Rymarquis LA, Mahalingam G, Marsch-Martínez N, Meyers BC, Green PJ, de Folter S (2014) Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat Commun 5: 3722. [DOI] [PubMed] [Google Scholar]
- Chen W, Kong J, Lai T, Manning K, Wu C, Wang Y, Qin C, Li B, Yu Z, Zhang X, et al. (2015) Tuning LeSPL-CNR expression by SlymiR157 affects tomato fruit ripening. Sci Rep 5: 7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curaba J, Singh MB, Bhalla PL (2014) miRNAs in the crosstalk between phytohormone signalling pathways. J Exp Bot 65: 1425–1438 [DOI] [PubMed] [Google Scholar]
- Dai X, Zhao PX (2011) psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res 39: W155–W159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Chen A, Chen W, Westwood JH, Baulcombe DC, Carr JP (2014) Using a viral vector to reveal the role of microRNA159 in disease symptom induction by a severe strain of Cucumber mosaic virus. Plant Physiol 164: 1378–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Xia R, Meyers BC (2013) Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25: 2400–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Huang C, Li F, Zhou X (2014) A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol J 12: 638–649 [DOI] [PubMed] [Google Scholar]
- Jiao J, Wang Y, Selvaraj JN, Xing F, Liu Y (2015) Barley stripe mosaic virus (BSMV) induced microRNA silencing in common wheat (Triticum aestivum L.). PLoS One 10: e0126621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Z, Wang L, Cao D, Zuo J, Zhu H, Fu D, Luo Y, Zhu B (2016) A viral satellite DNA vector-induced transcriptional gene silencing via DNA methylation of gene promoter in Nicotiana benthamiana. Virus Res 223: 99–107 [DOI] [PubMed] [Google Scholar]
- Khraiwesh B, Ossowski S, Weigel D, Reski R, Frank W (2008) Specific gene silencing by artificial microRNAs in Physcomitrella patens: An alternative to targeted gene knockouts. Plant Physiol 148: 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B, Zhu J, Zhu J (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta 1819: 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchik M, Damodharan S, Stav R, Arazi T (2014) Generation and characterization of a tomato DCL3-silencing mutant. Plant Sci 221-222: 81–89 [DOI] [PubMed] [Google Scholar]
- Langmead B, Schatz MC, Lin J, Pop M, Salzberg SL (2009) Searching for SNPs with cloud computing. Genome Biol 10: R134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Huang C, Li Z, Zhou X (2014a) Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin-like protein to repress RDR6 expression. PLoS Pathog 10: e1003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Zhang D, Sheen J (2014b) Epitope-tagged protein-based artificial miRNA screens for optimized gene silencing in plants. Nat Protoc 9: 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q, Tu Y, Carr JP, Du Z (2015) An improved cucumber mosaic virus-based vector for efficient decoying of plant microRNAs. Sci Rep 5: 13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−Δ ΔC(T) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu C, Meyers BC, Green PJ (2007) Construction of small RNA cDNA libraries for deep sequencing. Methods 43: 110–117 [DOI] [PubMed] [Google Scholar]
- Ma Z, Coruh C, Axtell MJ (2010) Arabidopsis lyrata small RNAs: Transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell 22: 1090–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A (2010) miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos JL, Bologna NG, Chorostecki U, Palatnik JF (2010) Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr Biol 20: 49–54 [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Yoo SJ, Fahlgren N, Gilbert SD, Howell MD, Sullivan CM, Alexander A, Nguyen G, Allen E, Ahn JH, et al. (2008) AGO1-miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci USA 105: 20055–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ (2008) Improved northern blot method for enhanced detection of small RNA. Nat Protoc 3: 1077–1084 [DOI] [PubMed] [Google Scholar]
- Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha A, Zhao J, Yin K, Tang Y, Wang Y, Wei X, Hong Y, Liu Y (2014) Virus-based microRNA silencing in plants. Plant Physiol 164: 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms D, Cizdziel PE, Chomczynski P (1993) TRIzol: A new reagent for optimal single-step isolation of RNA. Focus 15: 532–535 [Google Scholar]
- Song L, Axtell MJ, Fedoroff NV (2010) RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr Biol 20: 37–41 [DOI] [PubMed] [Google Scholar]
- Sunkar R. (2010) MicroRNAs with macro-effects on plant stress responses. Semin Cell Dev Biol 21: 805–811 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Jagadeeswaran G (2008) In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol 8: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Wang F, Zhao J, Xie K, Hong Y, Liu Y (2010) Virus-based microRNA expression for gene functional analysis in plants. Plant Physiol 153: 632–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Qian Y, Zhou X (2006) Modification of a viral satellite DNA-based gene silencing vector and its application to leaf or flower color change in Petunia hybrida. Chin Sci Bull 51: 2208–2213 [Google Scholar]
- Tao X, Zhou X (2004) A modified viral satellite DNA that suppresses gene expression in plants. Plant J 38: 850–860 [DOI] [PubMed] [Google Scholar]
- Taylor RS, Tarver JE, Hiscock SJ, Donoghue PC (2014) Evolutionary history of plant microRNAs. Trends Plant Sci 19: 175–182 [DOI] [PubMed] [Google Scholar]
- Teotia S, Tang G (2015) To bloom or not to bloom: Role of microRNAs in plant flowering. Mol Plant 8: 359–377 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang HL, Chekanova JA (2016) Small RNAs: Essential regulators of gene expression and defenses against environmental stresses in plants. Wiley Interdiscip Rev RNA 7: 356–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gao X, Li L, Shi X, Zhang J, Shi Z (2010) Overexpression of Osta-siR2141 caused abnormal polarity establishment and retarded growth in rice. J Exp Bot 61: 1885–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warthmann N, Chen H, Ossowski S, Weigel D, Hervé P (2008) Highly specific gene silencing by artificial miRNAs in rice. PLoS One 3: e1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Wollmann H, Schneeberger K, Weigel D (2010) Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr Biol 20: 42–48 [DOI] [PubMed] [Google Scholar]
- Xie K, Shen J, Hou X, Yao J, Li X, Xiao J, Xiong L (2012) Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice. Plant Physiol 158: 1382–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Zhang S, Yu B (2015) microRNA biogenesis, degradation and activity in plants. Cell Mol Life Sci 72: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Salinas M, Garcia-Molina A, Höhmann S, Berndtgen R, Huijser P (2013) SPL8 and miR156-targeted SPL genes redundantly regulate Arabidopsis gynoecium differential patterning. Plant J 75: 566–577 [DOI] [PubMed] [Google Scholar]
- Yan F, Guo W, Wu G, Lu Y, Peng J, Zheng H, Lin L, Chen J (2014) A virus-based miRNA suppression (VbMS) system for miRNA loss-of-function analysis in plants. Biotechnol J 9: 702–708 [DOI] [PubMed] [Google Scholar]
- Yi X, Zhang Z, Ling Y, Xu W, Su Z (2015) PNRD: A plant non-coding RNA database. Nucleic Acids Res 43: D982–D989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Li G, Zhu S, Zhang S, Fang J (2014) tasiRNAdb: A database of ta-siRNA regulatory pathways. Bioinformatics 30: 1045–1046 [DOI] [PubMed] [Google Scholar]
- Zhao J, Liu Q, Hu P, Jia Q, Liu N, Yin K, Cheng Y, Yan F, Chen J, Liu Y (2016) An efficient Potato virus X-based microRNA silencing in Nicotiana benthamiana. Sci Rep 6: 20573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. (2013) Advances in understanding begomovirus satellites. Annu Rev Phytopathol 51: 357–381 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.