Abstract
Cognitive impairments, uncontrolled drinking, and neuropathological cortical changes characterize alcohol use disorder. Dysfunction of the orbitofrontal cortex (OFC), a critical cortical subregion that controls learning, decision-making, and prediction of reward outcomes, contributes to executive cognitive function deficits in alcoholic individuals. Electrophysiological and quantitative synaptomics techniques were used to test the hypothesis that heavy drinking produces neuroadaptations in the macaque OFC. Integrative bioinformatics and reverse genetic approaches were used to identify and validate synaptic proteins with novel links to heavy drinking in BXD mice. In drinking monkeys, evoked firing of OFC pyramidal neurons was reduced, whereas the amplitude and frequency of postsynaptic currents were enhanced compared with controls. Bath application of alcohol reduced evoked firing in neurons from control monkeys, but not drinking monkeys. Profiling of the OFC synaptome identified alcohol-sensitive proteins that control glutamate release (e.g., SV2A, synaptogyrin-1) and postsynaptic signaling (e.g., GluA1, PRRT2) with no changes in synaptic GABAergic proteins. Western blot analysis confirmed the increase in GluA1 expression in drinking monkeys. An exploratory analysis of the OFC synaptome found cross-species genetic links to alcohol intake in discrete proteins (e.g., C2CD2L, DIRAS2) that discriminated between low- and heavy-drinking monkeys. Validation studies revealed that BXD mouse strains with the D allele at the C2cd2l interval drank less alcohol than B allele strains. Thus, by profiling of the OFC synaptome, we identified changes in proteins controlling glutamate release and postsynaptic signaling and discovered several proteins related to heavy drinking that have potential as novel targets for treating alcohol use disorder.
SIGNIFICANCE STATEMENT Clinical research identified cognitive deficits in alcoholic individuals as a risk factor for relapse, and alcoholic individuals display deficits on cognitive tasks that are dependent upon the orbitofrontal cortex (OFC). To identify neurobiological mechanisms that underpin OFC dysfunction, this study used electrophysiology and integrative synaptomics in a translational nonhuman primate model of heavy alcohol consumption. We found adaptations in synaptic proteins that control glutamatergic signaling in chronically drinking monkeys. Our functional genomic exploratory analyses identified proteins with genetic links to alcohol and cocaine intake across mice, monkeys, and humans. Future work is necessary to determine whether targeting these novel targets reduces excessive and harmful levels of alcohol drinking.
Keywords: alcohol, electrophysiology, genetics, orbitofrontal cortex, proteomics
Introduction
Alcohol abuse disorder (AUD) is a detrimental worldwide public health problem with increased prevalence over the past 20 years (Whiteford et al., 2013). Accordingly, alcohol ranks higher than 20 abused substances, including heroin, cocaine, and methamphetamine, as the most harmful drug (Nutt et al., 2010). AUD is a chronic relapsing disease characterized by functional and metabolic tolerance, a distinctive withdrawal syndrome, recurrent and heavier alcohol use, and cognitive impairments. Importantly, compelling evidence from studies that have examined the neurobiology of cognitive deficits in alcoholic individuals has demonstrated that poor cognitive function is a risk factor for increased vulnerability to relapse (Charlet et al., 2014), suggesting that improving cognitive performance during abstinence may prevent relapse. However, current pharmacotherapies for treating AUD do not target alcohol-induced cognitive impairments.
Subregions of the prefrontal cortex (PFC) are key regulators of executive functions that are often critically impaired following heavy alcohol use. Among these, the orbitofrontal cortex (OFC) contributes to outcome-based decision-making related to the expected rewards and the pleasurable properties of rewarding stimuli (Stalnaker et al., 2015). Imaging and behavioral studies show that patients with AUD have greater activation of the OFC and reduced performance when completing OFC-dependent tasks compared with healthy control subjects, demonstrating that prolonged drinking can impair OFC function (Fortier et al., 2008, 2009). Consistent with deficits reported in individuals with AUD, rodent models of alcohol dependence induce OFC-dependent behavioral deficits and adaptations in synaptic physiology and intrinsic excitability in OFC neurons (Crews and Boettiger, 2009; Badanich et al., 2011; McGuier et al., 2015; Nimitvilai et al., 2016). Although rodent studies on the effects of alcohol on cortical function are valuable, they are limited by the reduced complexity of the rodent cortex present in primates (Carmichael and Price, 1994; Ongür and Price, 2000). To circumvent these limitations, a nonhuman primate alcohol drinking model was used that closely mimics the range of consumption patterns reported in humans and generates a proportion of monkeys self-administering alcohol in a chronic, heavy manner (Baker et al., 2014). Using this model, we combined comprehensive functional measurements of synaptic physiology with integrated synaptic profiling to identify neuroadaptations and biomarkers in the OFC of heavy-drinking monkeys. Because the nonhuman primate self-administration model is highly translational but is limited by small cohort sizes, additional drinking and integrative genomic experiments were performed using BXD recombinant inbred (RI) strains of mice to validate the novel targets identified from the monkey studies.
Materials and Methods
Alcohol-self administration.
Monkey care and all procedures were approved by the Oregon National Primate Research Center Animal Care and Use Committee at Oregon Health & Science University and performed according to the National Institutes of Health (NIH) Guidelines for the Care and Use of Mammals in Neuroscience and Behavioral Research. Two cohorts of adult male cynomolgus monkeys (n = 21; Macaca fascicularis; age, 67–84 months; weight, 6.02–9.12 kg; World Wide Primates) were used for these studies, and 16 of the monkeys were trained to self-administer alcohol using a schedule-induced polydipsia technique as described previously (Vivian et al., 2001; Grant et al., 2008). Following the 3-month induction phase, monkeys had open access to 4% alcohol (v/v) for 22 h/d for 6 months. Blood samples were obtained by femoral blood sampling at 7 h into the daily 22 h access period every 4–7 d. At the completion of the drinking paradigm, control and drinking monkeys were anesthetized at a time when they would normally begin a drinking session. Brains were rapidly removed following routine procedures (Davenport et al., 2014), and tissue blocks of area 13L were prepared for electrophysiology or protein analysis. Ten monkeys (2 controls and 8 drinkers; MATRR.org INIA Cohort 13) were used for functional assays, and 11 monkeys (3 controls and 8 drinkers; MATRR.org INIA Cohort 9) were used for quantitative proteomics and Western blot analyses. Because of the individual differences in average daily intake in this voluntary alcohol-drinking procedure (Baker et al., 2014, 2017) that may preclude identifying functional or proteomic neuroadaptations, alcohol-drinking monkeys were oversampled in these studies.
Preparation of brain slices.
Brain slices containing area 13L were prepared for whole-cell patch-clamp electrophysiology experiments with the experimenter blind to the treatment conditions. Following the necropsy procedure and rapid removal of the brain, the tissue was blocked coronally for the frontal cortex and mounted in a Vibroslicer (Leica) containing ice-cold oxygenated (95% O2, 5% CO2) sucrose containing buffer, and coronal sections (300 μm) were cut. Slices containing area 13L were immediately placed in a holding chamber containing oxygenated artificial CSF (aCSF) at 34°C for 45 min and kept at room temperature for at least 30 min before transferring to the recording chamber. The glutamate NMDA receptor blocker AP5 (50 μm) was added to the aCSF during incubation to prolong the viability of neurons, and slices were washed with regular oxygenated aCSF for at least 20 min before the whole-cell recordings were performed. Although AP5 is reported to wash out of acute slice preparations (Dozmorov et al., 2004; Herman et al., 2011), application of AP5 for ≥12 h enhances evoked firing (Ishikawa et al., 2009; Lee and Chung, 2014), suggesting that there may be a compensatory increase in intrinsic excitability under these recording conditions. However, all groups were treated identically, so this is unlikely to explain a difference in firing between treatment groups.
The composition of the sucrose-containing cutting solution used was as follows (in mm): 194 sucrose, 30 NaCl, 4.5 KCl, 1.2 NaH2PO4, 1 MgCl2, 10 glucose, and 26 NaHCO3, adjusted to 305–315 mOsm. The composition of the aCSF was as follows (in mm): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 1.3 MgCl2, 2.0 CaCl2, 0.4 ascorbate, 10 glucose, and 25 NaHCO3, adjusted to 290–310 mOsm. Both solutions were saturated with 95% O2/5% CO2, pH 7.4.
Whole-cell patch-clamp electrophysiology.
An individual slice was placed in the recording chamber fixed to the stage of an upright microscope (Axio scope examiner D1, Zeiss) and perfused with 29–31°C oxygenated aCSF. Recordings were localized to area 13L of the OFC under infrared optics using a 40× water-immersion objective. Thin-wall borosilicate glass electrodes (outer diameter = 1.5 mm; inner diameter = 1.17 mm) were pulled on a Sutter Instrument P97 Micropipette Puller and had tip resistances ranging from 1.9 to 5.5 MΩ. Patch pipettes filled with an internal solution were slowly lowered onto the layer V/VI pyramidal neurons to obtained a seal (>1 GΩ) followed by breakthrough to gain whole-cell access. All the whole-cell recordings were performed using an Axon MultiClamp 700B amplifier (Molecular Devices) and analyzed with pClamp software (Molecular Devices). Events were filtered at 2 kHz and digitized at a sampling rate of 10 kHz. To measure the intrinsic excitability of layer V/VI area 13L pyramidal neurons, current-clamp recordings were performed. Spike firing was induced by direct current injection (−40 to 240 pA) through patch pipettes filled with a potassium gluconate internal solution [composition as follows (in mm): 120 K-gluconate, 10 KCl, 10 HEPES, 2 MgCl2, 1 EGTA, 2 NaATP, and 0.3 NaGTP]. Recordings were analyzed off-line in AxographX (Axograph) for the number of spikes in response to each current step, resting membrane potential (in millivolts), action potential threshold (in millivolts), height (in millivolts), half-width (in milliseconds), rise time (in milliseconds), interspike interval (ISI; ms), frequency (in hertz), and after-hyperpolarization (AHP; in millivolts). The spike frequency adaptation ratio was calculated as first ISI/last ISI. To monitor spontaneous postsynaptic currents (sPSCs), OFC pyramidal neurons were voltage clamped at a membrane potential of −70 mV with patch pipettes filled with a cesium methanesulfonate internal solution [with composition as follows (in mm): 125 CsMeSO3, 10 CsCl, 5 NaCl, 10 HEPES, 1 EGTA, 2 MgCl2, 5 MgATP, and 0.3 NaGTP]. To maximize the chances of observing changes in synaptic activity in the limited number of OFC slices available, no blockers of synaptic activity were used in these recordings. However, under the recording conditions used, the estimated reversal potentials are biased in favor of detecting AMPA-mediated conductances (reversal potential, ∼0 mV) over those generated by chloride-permeable GABAA receptors (reversal potential, −51.3 mV). Differences in the number of spikes were assessed with two-way ANOVA, and differences in other electrophysiological characteristics of OFC neurons were analyzed with unpaired t test. Spontaneous events were analyzed off-line with AxographX software using a sliding template-matching algorithm. Differences in sPSC parameters were assessed with unpaired t test using GraphPad Prism (GraphPad Software). Comparisons were considered significantly different at p < 0.05, and the experimenter was blind to the treatment conditions for data acquisition of the electrophysiology experiments.
Preparation of iTRAQ-labeled peptides.
For proteomic analysis, Triton X-100 insoluble membrane fractions that are enriched in postsynaptic density (PSD) proteins (Uys et al., 2016) were prepared from 11 monkeys (3 controls and 8 drinkers) following our routine methods (Mulholland et al., 2011; Uys et al., 2016). Due to the limited number of isobaric tags, 8 (3 controls and 5 drinkers) of the samples were used for mass spectrometry (MS), while all 11 samples were used for Western blotting, as described below. Isobaric Tag for Relative and Absolute Quantitation (iTRAQ; AB SCIEX) uses covalent attachment of isobaric tags to individual samples, allowing for qualitative and quantitative analysis of eight protein samples in a single solution by liquid chromatography-tandem MS (LC-MS/MS; Ross et al., 2004). An aliquot of each sample was taken for determination of protein concentration by the bicinchoninic acid assay (Pierce Biotechnology). Each sample (50 μg of protein in equal volumes) was reduced in 5 mm tris-(2-carboxyethyl)phosphine for 1 h at 60°C, alkylated in 9 mm methyl methane-thiosulfonate for 30 min at room temperature, diluted to 0.1% in PPS Silent surfactant (Agilent Technologies) with 50 mm triethylammonium bicarbonate, and digested overnight in trypsin (1:10, enzyme/protein ratio; Promega Corporation) at 37°C. Tryptic peptides were labeled with iTRAQ 8-plex reagents for 2 h at room temperature, and the contents of all digested samples were combined after quenching the labeling reaction with 50 mm ammonium bicarbonate. Control monkeys were labeled with tags 113, 114, and 116, whereas monkeys with a history of alcohol self-administration were labeled with tags 115, 117, 118, 119, and 121.
Strong cationic exchange of labeled peptides and LC-MS/MS.
Lyophilized labeled peptides were solubilized, fractionated by strong cation exchange, and mass analyzed following our previously published methods (Uys et al., 2016). Briefly, eluting peptides were mass analyzed by data-dependent acquisition on the Orbitrap Elite mass spectrometer (Thermo Scientific) with Xcalibur 2.2 software (Thermo Scientific). The top seven most intense ions in the Fourier Transform Mass Spectrometry survey scan were selected for fragmentation by alternating higher-energy collisional dissociation (HCD) at a normalized collision energy of 40% and collision-induced dissociation (CID) at 35%. Full-scan MS spectra were acquired at a target value of 1 × 106 and a resolution of 60,000, and CID MS/MS spectra were acquired in the ion trap at a target value of 1 × 104; the HCD MS/MS spectra were recorded at a target value of 5 × 104 with a resolution of 15,000. Ions with a +1 charge were excluded from selection. Dynamic exclusion was enabled with a repeat count of 2, a duration of 30 s, an exclusion list size of 500, and an exclusion duration of 80 s.
Database searching, peptide identification, quantitation, and functional annotation.
A recent study demonstrated that the human Uniprot database is an excellent alternative strategy to replace deficient nonhuman primate databases for proteomic profiling of monkey tissue samples (Lee et al., 2015). Thus, the raw files were searched using Mascot and SequestHT in Proteome Discoverer 2.1 (Thermo Scientific) against a human database (Swiss-Prot; 20,186 reviewed sequences containing isoforms; downloaded in February 2016). Parameters for peptide identification were as follows: precursor mass tolerance of 10 ppm, fragment mass tolerance was set at 0.8 Da for the CID spectra and 0.1 Da for the HCD spectra, fully tryptic peptides with a maximum of two missed cleavages, static modifications of peptide N termini, and lysines with the iTRAQ reagent and cysteine alkylation with methylthio; methionine oxidation was included as a variable modification. The search results were filtered using Percolator 2.04 (Käll et al., 2007) to yield peptides with a false discovery rate of <1%. The raw data for unique peptides were log transformed and normalized (central mean tendency), and fold changes were calculated using InfernoRDN software (Polpitiya et al., 2008). The log-transformed, normalized data were exported for statistical evaluation using mixed linear modeling (PROC MIXED, SAS software version 9.4). This analytical method has been used previously to analyze MS data (Wildburger et al., 2015) and was selected because of the capacity to handle unbalanced and complex repeated-measures data and the ability to model the variance and correlation structure of repeated-measures experimental designs (Littell et al., 1998). The spectra were nested at the peptide level, and the peptide data were further nested at the protein level. Proteins that were identified by a single peptide were excluded, and a protein was considered differentially expressed if the p value was <0.05. Mascot scores and the percentage of coverage were calculated in Proteome Discover 2.1 software. Although only differentially expressed proteins are reported here, the full raw and normalized datasets are archived in the Monkey Alcohol Tissue Research Resource (www.MATRR.com). Functional annotation of the synaptomics data was performed using Ingenuity Pathway Analysis (QIAGEN) following our previously reported methods (Uys et al., 2016).
Western blotting.
Western blotting procedures for GluA1 followed our routine methods (Padula et al., 2015). Briefly, PSD-enriched fractions from three controls and all eight drinking monkeys were diluted with NuPAGE 4× LDS sample loading buffer (Invitrogen; pH 8.5) containing 50 mm dithiothreitol, and samples were denatured for 10 min at 70°C. Ten micrograms of each sample (controls, n = 3; drinkers, n = 8) was separated using the Bis-Tris (375 mm resolving buffer and 125 mm stacking buffer, pH 6.4; 7.5% acrylamide) discontinuous buffer system with MOPS (3-(N-morpholino)propanesulfonic acid) electrophoresis buffer (50 mm MOPS, 50 mm Tris, 0.1% SDS, 1 mm EDTA, pH 7.7). Protein was then transferred to Immobilon-P PVDF membranes (Millipore) using a semidry transfer apparatus (Bio-Rad). After transfer, blots were washed with PBS containing 0.1% Tween 20 (PBST) and then blocked with PBST containing 5% nonfat dried milk (NFDM) for 1 h at room temperature with agitation. The membranes were then incubated overnight at 4°C with primary antibody against GluA1 (catalog #AB1504, Millipore; RRID: AB_2113602) diluted 1:4000 in PBST containing 0.5% NFDM and washed in PBST before a 1 h incubation at room temperature with horseradish peroxidase-conjugated secondary antibody diluted 1:2000 in PBST. Membranes received a final wash in PBST, and the antigen–antibody complex was detected by enhanced chemiluminescence using a ChemiDoc MP Imaging system (Bio-Rad). Before running the experimental samples, Western blots were performed using different titrations of sample and antibody to establish the linear range for GluA1 in cynomolgus macaque tissue. Bands were quantified by mean optical density using Image Lab software (version 4.0.1; Bio-Rad) and were normalized to a total protein stain (Gürtler et al., 2013). Normalized protein expression data were analyzed with a one-tailed t test, and standard linear regression was used to analyze protein expression levels with optical density values and average daily alcohol intake. The specificity of this antibody for Western blots has been confirmed in GluA1 knock-out mice (Zamanillo et al., 1999) and characterized using human, macaque, and rodent tissue from frontal cortex (Tucholski et al., 2014).
Partial least-squares discriminant analysis regression.
To identify proteins that best discriminate between control and drinking monkeys, we used a partial least-squares discriminant analysis (PLS-DA) regression approach (SOLO, Eigenvector Research). PLS-DA is a multivariate technique that maximizes the separation of the between-groups covariance matrix in large datasets and has been validated as an exploratory approach to identify potential biomarkers in MS data (Jonsson et al., 2005; Rajalahti et al., 2009a). PLS is an extension of multiple regression that is used as a means of exploratory analysis and variable selection when faced with a large number of independent variables and often a relatively small number of dependent variables. For this analysis, monkeys were classified according to their drinking history on the y matrix. The x matrix consisted of log-transformed, normalized (mean centered) spectra data that were rolled up to the protein level using Inferno software. We used the selectivity ratio, calculated as the ratio between explained and residual variance, to determine the proteins with the highest proportion of variance that best discriminate phenotypes, as previously described for identifying biomarkers in MS profiles (Rajalahti et al., 2009a,b). Proteins are reported that have selectivity ratios above the 95% confidence limit that is based on the Hotelling T2 limit, scaled by the eigenvalue of the given principle components.
Drinking-in-the-dark mouse model.
Fifteen female BXD RI strains were obtained from the University of Tennessee Health Science Center and were individually housed in temperature- and humidity-controlled environments. Adult mice (n = 73 total mice; 8–10 weeks old at the start of the experiment) were kept on a 12 h light/dark cycle, and food and water were available ad libitum during the experiment except when alcohol was presented in their home cages. The Pennsylvania State University Institutional Animal Care and Use Committee approved all procedures in accordance with NIH guidelines for the humane care and use of laboratory animals. Binge-like ethanol consumption was induced using a standard 4 d drinking-in-the-dark (DID) protocol (Rhodes et al., 2005; Rinker et al., 2016a). During test week 1, water bottles were replaced with 20% alcohol (v/v) for a 2 h period beginning 3 h into the dark cycle for 3 consecutive days. On the fourth day, alcohol was available for an extended 4 h period. This paradigm was repeated for two additional weeks. Due to an unanticipated increase in the temperature (>30°C) and humidity of the colony room during test week 1 that caused a spike in alcohol drinking, these data were excluded from analysis. Drinking values on day 4 were averaged for each strain and then averaged across test weeks 2 and 3. A two-tailed, unpaired t test was used for analysis in GraphPad software. Strains were genotyped at the C2cd2l interval in GeneNetwork, and all drinking data are available in GeneNetwork.
Bioinformatics analyses.
To provide additional evidence for the alcohol-sensitive proteins and the proteins that best discriminate between low- and heavy-drinking monkeys, we performed two additional open source integrative bioinformatics analyses following our previously reported methods (Padula et al., 2015; McGuier et al., 2016; Rinker et al., 2017). First, we queried the GeneWeaver software system (www.geneweaver.org), a database containing major curated repositories as well as functional genomics results obtained from experiments across nine species (Baker et al., 2012), to retrieve data that implicate the genes that encode the identified proteins in alcohol-related phenomena. Next, we performed targeted analyses using existing genetic and phenotypic data in GeneNetwork (www.genenetwork.org). We correlated PFC robust multi-array average levels in male and female alcohol-naive BXD RI strains of mice [GeneNetwork dataset: VCU BXD PFC Sal M430 2.0 (Dec06) RMA] for C2cd2l (also known as Tmem24; probe set: 1458713_at and 1428095_a_at), Diras2 (1455436_at), and Pycr2 (1448315_a_at) with published phenotypic data, including data for intravenous cocaine self-administration and cocaine-induced conditioned place preference in BXD RI strains (Philip et al., 2010; Dickson et al., 2016). To match the monkey-drinking paradigm, only significant Spearman correlations related to alcohol consumption or preference in two-bottle choice (alcohol vs water) voluntary models were reported. All datasets generated on alcohol and cocaine for this article can be queried on GeneWeaver and GeneNetwork.
Results
Functional studies of OFC pyramidal neurons from heavy drinking macaques
As an initial measure of functional alterations, we recorded passive and active membrane properties of OFC pyramidal neurons in brain slices from control and drinking monkeys. The majority of monkeys used for electrophysiological studies consumed ∼3 g/kg/d (equivalent to ≥12 daily human drinks) and achieved blood alcohol concentrations (BACs) >80 mg/dl that correlated with alcohol intake (Fig. 1A–C). Consistent with known membrane properties of cortical pyramidal neurons in primates (Povysheva et al., 2006), the spike frequency adaptation ratio of deep-layer area 13L pyramidal neurons in controls was 0.45 ± 0.04 (n = 5 cells from two monkeys). All measures of passive membrane (i.e., cell capacitance, resting membrane potential, input resistance) and biophysical properties (i.e., threshold, peak, rise time, half-width, decay, adaptation ratio, AHP amplitude) of the evoked action potentials (APs) were similar between controls and drinkers (Table 1). However, there was a significant reduction in the number of evoked APs (two-way repeated-measures ANOVA, F(14,308) = 2.49, p = 0.002; Sidak's post hoc test, *p < 0.05 vs controls; Fig. 1F,G) and a corresponding increase in rheobase current in OFC neurons from drinking monkeys [controls, n = 5/2 (cells/monkeys); drinkers, n = 19/8; unpaired two-tailed t test, t(22) = 2.42, *p = 0.0239 vs controls; Fig. 1H]. Bath application of 22 (∼100 mg/dl) and 66 mm (∼300 mg/dl) alcohol significantly reduced evoked AP firing in pyramidal neurons in controls (two-way repeated-measures ANOVA, F(28,56) = 1.70, p = 0.046; Sidak's post hoc test, *p < 0.05; n = 3/1; Fig. 1I), but not in drinking monkeys (two-way repeated-measures ANOVA, F(2,16) = 0.131; p = 0.88, n = 9/7; Fig. 1J).
Figure 1.
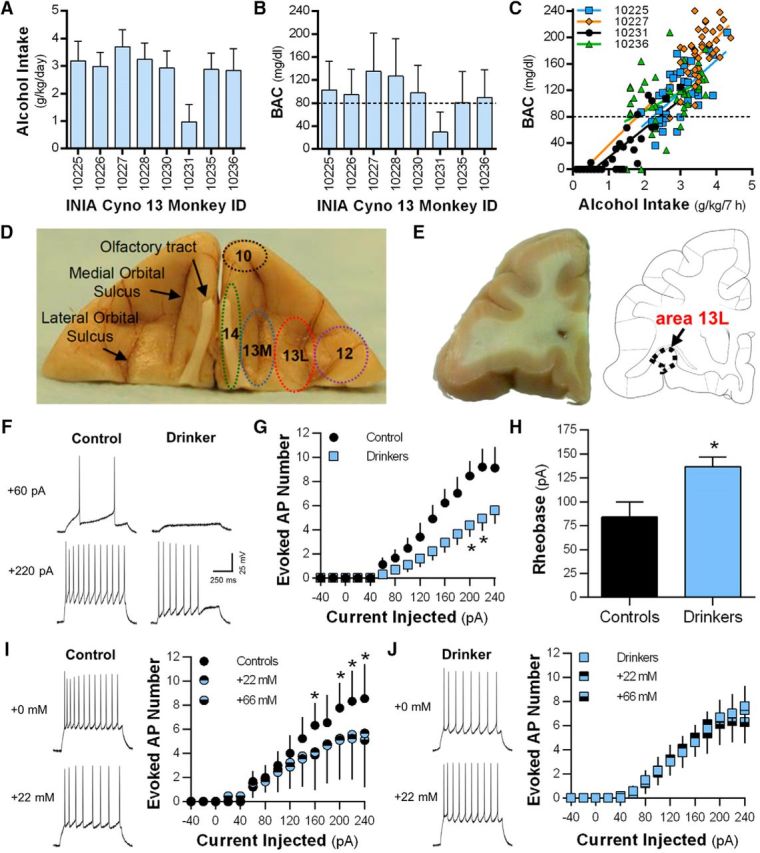
Long-term alcohol drinking reduces evoked spiking in area 13L pyramidal neurons. A, B, Alcohol consumption levels (A; mean ± SD) and BACs (B; mean ± SD) during the 6 months of 22 h access to alcohol self-administration in the INIA Cyno 13 cohort of monkeys used for electrophysiological analysis. C, Representative correlations between alcohol intake and BACs. D, Example image of the ventral aspect of a cynomolgus macaque brain showing prominent anatomical markers and areas 10, 12, 13L, 13M, and 14. E, A representative example of a coronal section of the OFC-containing area 13L that was used for functional and biochemical analyses. F, Representative traces showing reduced evoked spiking in a control and a drinking monkey. G, Number of action potentials at current injections ranging from −40 to −240 pA in control and drinking monkeys. H, Rheobase was increased in the drinking monkeys. I, J, Representative traces and average number of evoked firing in the absence and presence of 22 mm alcohol bath application in control (I) and drinking (J) monkeys. Electrophysiological data are expressed as the mean ± SEM.
Table 1.
Measures of passive membrane and biophysical properties of the evoked APs in deep-layer OFC pyramidal neurons from control and long-term drinking monkeys
| Cell capacitance (pF) | AP threshold (mV) | Input resistance (ΩM) | RMP (ΩM) | AP height (mV) | AP rise time (μs) | AP half-width (ms) | AP decay (ms) | AHP amplitude (mV) | |
|---|---|---|---|---|---|---|---|---|---|
| Controls | 31.10 ± 3.87 | −46.35 ± 3.17 | 250.0 ± 37.05 | −72.44 ± 0.43 | 65.40 ± 5.26 | 768.8 ± 105.20 | 2.083 ± 0.28 | 2.099 ± 0.34 | 11.46 ± 3.53 |
| Drinkers | 53.40 ± 6.26 | −41.50 ± 2.01 | 186.9 ± 33.38 | −70.91 ± 0.88 | 73.39 ± 3.77 | 667.7 ± 46.96 | 1.789 ± 0.09 | 1.616 ± 0.09 | 17.26 ± 1.22 |
| t value (df = 22) | 1.777 | 1.095 | 0.921 | 0.864 | 1.014 | 0.955 | 1.354 | 1.961 | 1.964 |
| p value | 0.089 | 0.285 | 0.370 | 0.397 | 0.3218 | 0.350 | 0.190 | 0.063 | 0.062 |
Data are expressed as the mean ± SEM, unless otherwise indicated.
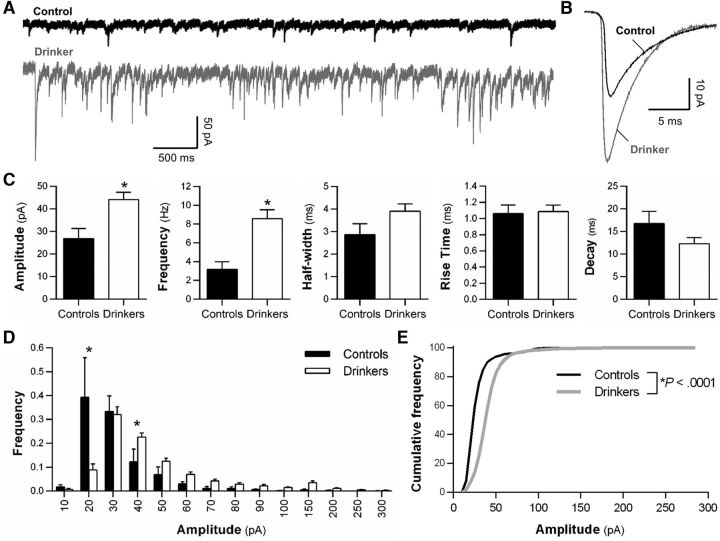
Because plasticity of intrinsic excitability coincides with postsynaptic adaptations during learning or alcohol and drug exposure (Sehgal et al., 2013; Kourrich et al., 2015; Nimitvilai et al., 2016), we next determined whether there were functional synaptic adaptations in drinking monkeys. Representative traces of sPSCs obtained from a drinking (gray) and a control (black) monkey are shown in Figure 2, A and B. The average frequency, amplitude, and rise time of sPSCs in OFC pyramidal neurons from control monkeys (Fig. 2C) are consistent with postsynaptic events recorded from pyramidal neurons in monkey prefrontal cortex (Amatrudo et al., 2012; Medalla and Luebke, 2015). Analysis of the sPSCs shows that chronic alcohol self-administration significantly increased amplitude and frequency without altering half-width, rise time, or decay time constant [amplitude: t test, t(20) = 2.057, *p = 0.027; frequency: t test, t(20) = 2.24, *p = 0.037; half-width: t test, t(20) = 1.24, p = 0.11; rise time: t test, t(20) = 1.12, p = 0.45; decay: t test, t(20) = 1.27, p = 0.11; controls, n = 3/2 (cells/monkeys); drinkers, n = 19/5; Fig. 2C]. Frequency histograms for binned (two-way repeated-measures ANOVA, F(13,260) = 5.46, p < 0.0001; Fisher LSD post hoc, *p < 0.007; Fig. 2D) and cumulative (Kolmogorov–Smirnov test, *p < 0.0001; Fig. 2E) distribution revealed rightward shifts of sPSCs amplitudes in pyramidal neurons recorded from drinkers. Together, these functional analyses of OFC pyramidal neurons indicate that alcohol self-administration produces opposing effects on evoked action potential firing and postsynaptic signaling.
Figure 2.
Long-term alcohol self-administration enhances the amplitude and frequency of sPSCs in deep-layer area 13L pyramidal neurons. A, B, Representative traces of sPSCs from a control (black trace) and a drinking (gray trace) monkey. C, Average amplitude, frequency, half-width, rise time, and decay in controls and drinkers. Data are expressed as the mean ± SEM. D, E, Frequency histograms for binned and cumulative sPSC amplitude distributions in controls and drinking monkeys.
Synaptomic analysis in drinking monkeys
An unbiased quantitative synaptomics approach was then used to identify synaptic proteins that were altered by drinking. A separate cohort of cynomolgus macaques was used for this study, and their drinking data are shown in Figure 3A–C. After 6 months of free access to alcohol, brains were extracted before the start of a drinking session and PSD-enriched fractions were prepared from area 13L for proteomic analysis. LC/MS-MS analysis identified 976 proteins with isoforms (879 unique proteins and 3809 unique peptides). A normality test of the normalized data fit the expression ratio frequency distribution (R2 = 0.988; Fig. 3D,E). Functional annotation revealed that 33.4% of the unique proteins localize to the plasma membrane or were identified as ion channels or membrane-bound transporters. Analysis showed that long-term drinking significantly altered the expression of a diverse group of 57 proteins (Table 2), of which 29 are transmembrane proteins and 47 were significantly upregulated. We then analyzed all differentially expressed OFC proteins to identify prominent biological networks that are affected by long-term drinking. Proteins linked to cell-to-cell signaling and interaction, cell signaling, and nucleic acid metabolism were the top three biological function annotations of the alcohol-sensitive proteins (Fig. 3F). Data were filtered using a conservative threshold of p < 0.01 (−log(B-H corrected p value) >2). Because Triton X-100 fractions are enriched in excitatory synaptic proteins (Uys et al., 2016), it is possible that these three annotations are also highly ranked when considering the entire dataset. However, analysis showed that these biological annotations were ranked 1st, 6th, and 23rd, respectively, demonstrating that alcohol self-administration affects subsets of OFC proteins. A closer inspection of the significantly dysregulated proteins reveals apparent coordinated adaptations in 23 proteins that control presynaptic glutamate release and postsynaptic glutamatergic signaling (Fig. 3G, Table 2). In addition to excitatory synaptic proteins, GABAA receptors (Centanni et al., 2014) and other key scaffolding and signaling proteins expressed in GABAergic inhibitory synapses (Sassoè-Pognetto et al., 2011) are also present in the Triton X-100 insoluble fraction. However, in contrast with adaptations in excitatory proteins, GABAergic inhibitory synaptic proteins present in the dataset were unaltered by long-term alcohol consumption (Table 3).
Figure 3.
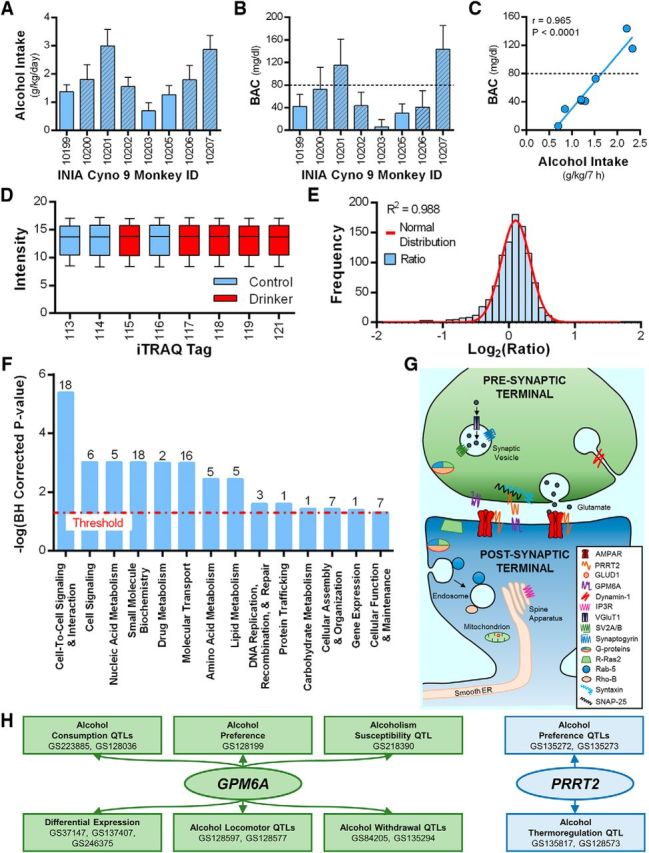
Synaptomic profiling of PSD-enriched fractions from control and drinking cynomolgus macaques identifies novel alcohol-sensitive proteins (controls, n = 3; drinkers, n = 5). A, B, Average daily alcohol intake (A) and BACs (B) during the 6 months of self-administration. The monkeys used for MS/MS analysis are shown in the hatched bars. Two of the monkeys (10201 and 10207) reached criteria for heavy drinking (≥3 g/kg on 48% and 38% of the open-access drinking days), and the remaining monkeys fit the criteria for low drinkers achieving ≤3 g/kg intake on <1% of the open access days. Data are the mean ± SD. C, Correlation analysis of average BAC and alcohol intake (g/kg) at 7 h into the daily session for all eight drinking monkeys. D, E, Frequency distribution of the normalized log-transformed median expression ratios of the 976 proteins. The histogram was fit by a normal distribution. Frequency distribution of the ratios is shown in blue bars, and a red line shows the results of the normal fit. F, IPA Core analysis identified highly ranked biological functions of the Triton X-100 insoluble proteins in the area 13L that were differentially expressed by alcohol self-administration. The red line denotes threshold, and the number above each bar shows the number of proteins contained within that annotation. G, Presynaptic and postsynaptic proteins related to glutamatergic signaling that were significantly different between control and drinking monkeys. H, GeneWeaver search results of alcohol-related gene sets (GS) containing GPM6A and PRRT2.
Table 2.
Long-term drinking induces expression changes in proteins in the macaque OFC
| Protein | Gene | Accession # | Peptides/spectra | F value | p value | Fold change | Coverage % | Mascot score |
|---|---|---|---|---|---|---|---|---|
| Cytoplasm | ||||||||
| cAMP-dependent PKA C-α, Isoform 2 | PRKACA | P17612-2 | 2/2 | 17.52 | 0.006 | 1.09 | 26.5 | 250 |
| D-3-phosphoglycerate dehydrogenase | PHGDH | O43175 | 4/5 | 7.72 | 0.032 | 1.26 | 14.1 | 457 |
| Dynamin-1* | DNM1 | Q05193 | 6/7 | 25.79 | 0.002 | 0.95 | 12.0 | 103 |
| FAS-associated factor 2 | FAF2 | Q96CS3 | 4/5 | 6.94 | 0.039 | 1.19 | 24.3 | 263 |
| Glial fibrillary acidic protein | GFAP | P14136 | 10/17 | 7.01 | 0.038 | 0.62 | 49.1 | 866 |
| G(I) protein subunit α-1, Isoform 2* | GNAI1 | P63096-2 | 6/19 | 20.35 | 0.004 | 1.20 | 51.4 | 2977 |
| G(I) protein subunit α-2, Isoform 3* | GNAI2 | P04899-3 | 6/15 | 8.91 | 0.025 | 1.13 | 55.5 | 2755 |
| G(I)/G(S)/G(O) protein subunit γ-3* | GNG3 | P63215 | 4/17 | 8.11 | 0.029 | 1.16 | 72.0 | 908 |
| G(I)/G(S)/G(T) protein subunit β-2* | GNB2 | P62879 | 2/2 | 11.8 | 0.014 | 1.24 | 44.7 | 11,797 |
| G(k) protein subunit α* | GNAI3 | P08754 | 2/2 | 13.38 | 0.011 | 1.20 | 34.5 | 1566 |
| Hemoglobin subunit β | HBB | P68871 | 2/3 | 7.24 | 0.036 | 0.31 | 34.7 | 353 |
| MICOS complex subunit MIC19 | CHCHD3 | Q9NX63 | 10/14 | 6.65 | 0.042 | 0.78 | 53.3 | 543 |
| Myosin light polypeptide 6 | MYL6 | P60660 | 4/6 | 8.63 | 0.026 | 0.55 | 13.9 | 161 |
| Protein NipSnap homolog 3A | NIPSNAP3A | Q9UFN0 | 2/2 | 15.47 | 0.008 | 1.24 | 15.0 | 105 |
| Endoplasmic reticulum membrane | ||||||||
| Inositol 1,4,5-trisphosphate receptor type 1, Isoform 5* | ITPR1 | Q14643-5 | 3/3 | 7.21 | 0.036 | 1.16 | 4.2 | 417 |
| Oxysterol-binding protein-related protein 8, Isoform 2 | OSBPL8 | Q9BZF1-2 | 3/3 | 6.92 | 0.039 | 0.85 | 6.1 | 176 |
| Very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase 3 | HACD3 | Q9P035 | 2/3 | 7.81 | 0.031 | 0.79 | 14.9 | 343 |
| α-Soluble NSF attachment protein | NAPA | P54920 | 3/3 | 6.82 | 0.040 | 1.19 | 44.4 | 271 |
| Mitochondria | ||||||||
| Amine oxidase (flavin-containing) B, Isoform 2 | MAOB | P27338-2 | 8/43 | 6.58 | 0.043 | 1.30 | 36.0 | 3199 |
| Calcium uptake protein 3, mitochondrial | MICU3 | Q86XE3 | 2/3 | 41.1 | 0.001 | 0.82 | 7.4 | 233 |
| cGMP-dependent phosphodiesterase, isoform PDE2A1 | PDE2A | O00408-2 | 9/18 | 37.48 | 0.001 | 1.07 | 26.9 | 1345 |
| Cytochrome b–c1 complex subunit Rieske | UQCRFS1 | P47985 | 2/10 | 6.6 | 0.042 | 3.08 | 21.9 | 1305 |
| Cytochrome c oxidase subunit 1 | MT-CO1 | P00395 | 2/2 | 7.51 | 0.034 | 1.20 | 4.9 | 127 |
| Dihydrolipoyl dehydrogenase, mitochondrial, Isoform 3 | DLD | P09622-3 | 6/19 | 7.91 | 0.031 | 1.16 | 25.9 | 1481 |
| Glutamate dehydrogenase 1, mitochondrial* | GLUD1 | P00367 | 6/9 | 7.33 | 0.035 | 1.15 | 35.8 | 601 |
| NAD(P) transhydrogenase, mitochondrial | NNT | Q13423 | 21/53 | 6.76 | 0.041 | 1.31 | 32.7 | 3472 |
| Prohibitin-2 | PHB2 | Q99623 | 12/95 | 6.52 | 0.043 | 1.35 | 51.2 | 6861 |
| Sideroflexin-1 | SFXN1 | Q9H9B4 | 8/14 | 7.01 | 0.038 | 1.30 | 51.9 | 1155 |
| Solute carrier family 25 member 46 | SLC25A46 | Q96AG3 | 3/5 | 6.71 | 0.041 | 1.17 | 11.7 | 287 |
| Tricarboxylate transport protein, mitochondrial | SLC25A1 | P53007 | 3/5 | 6.96 | 0.039 | 0.82 | 29.3 | 225 |
| Nucleus | ||||||||
| Histone H4 | HIST1H4A | P62805 | 2/2 | 9.89 | 0.020 | 0.40 | 40.8 | 68 |
| Plasma membrane | ||||||||
| 4F2 cell-surface antigen heavy chain, Isoform 3 | SLC3A2 | P08195-3 | 3/6 | 6.23 | 0.047 | 1.23 | 8.2 | 231 |
| Dipeptidyl aminopeptidase-like protein 6, Isoform DPPX-S | DPP6 | P42658-2 | 2/4 | 9.86 | 0.020 | 1.14 | 7.4 | 163 |
| FXYD domain-containing ion transport regulator 6 | FXYD6 | Q9H0Q3 | 2/7 | 6.99 | 0.038 | 1.28 | 50.5 | 280 |
| Glutamate receptor 1, Isoform 4* | GRIA1 | P42261-4 | 4/4 | 9.23 | 0.023 | 1.23 | 13.2 | 442 |
| Neuronal membrane glycoprotein M6-a* | GPM6A | P51674 | 2/3 | 12.11 | 0.013 | 1.38 | 12.9 | 120 |
| Neurotrimin, Isoform 4 | NTM | Q9P121-4 | 6/19 | 7.25 | 0.036 | 1.13 | 34.9 | 776 |
| Proline-rich transmembrane protein 2* | PRRT2 | Q7Z6L0 | 2/3 | 6.71 | 0.041 | 1.49 | 5.8 | 194 |
| Ras-related protein Rab-5C | RAB5C | P51148 | 2/2 | 8.76 | 0.025 | 1.17 | 30.9 | 94 |
| Ras-related protein R-Ras2* | RRAS2 | P62070 | 2/2 | 16.79 | 0.006 | 1.33 | 13.3 | 155 |
| Reticulon-4 receptor-like 2 | RTN4RL2 | Q86UN3 | 3/4 | 7.17 | 0.037 | 1.18 | 13.1 | 258 |
| Rho-related GTP-binding protein RhoB* | RHOB | P62745 | 2/3 | 10.54 | 0.018 | 1.33 | 47.4 | 368 |
| Na+/Ca2+ exchanger 2 | SLC8A2 | Q9UPR5 | 6/8 | 10.69 | 0.017 | 1.37 | 18.3 | 491 |
| Na+/K+-transporting ATPase subunit α-1, isoform 2 | ATP1A1 | P05023-2 | 12/26 | 6.14 | 0.048 | 1.32 | 34.9 | 4315 |
| Na+/K+-transporting ATPase subunit α-3, isoform 3 | ATP1A3 | P13637-3 | 13/84 | 6.55 | 0.043 | 1.27 | 41.4 | 7233 |
| Na+/K+-transporting ATPase subunit β-2 | ATP1B2 | P14415 | 3/3 | 6.07 | 0.049 | 1.35 | 19.0 | 222 |
| Solute carrier family 12 member 5, Isoform 2 | SLC12A5 | Q9H2X9-2 | 10/15 | 9.56 | 0.021 | 1.20 | 16.6 | 965 |
| Synaptic vesicle membrane | ||||||||
| Sodium-dependent neutral amino acid transporter* | SLC6A17 | Q9H1V8 | 2/2 | 13.42 | 0.011 | 1.30 | 8.0 | 288 |
| Synaptic vesicle glycoprotein 2A, Isoform 2* | SV2A | Q7L0J3-2 | 3/3 | 6.74 | 0.041 | 1.21 | 11.6 | 346 |
| Synaptic vesicle glycoprotein 2B* | SV2B | Q7L1I2 | 4/4 | 6.35 | 0.045 | 1.23 | 11.4 | 305 |
| Synaptogyrin-1, isoform 1B* | SYNGR1 | O43759-2 | 2/2 | 8.44 | 0.027 | 1.35 | 10.3 | 95 |
| Synaptogyrin-3* | SYNGR3 | O43761 | 2/5 | 8.92 | 0.024 | 1.27 | 11.8 | 337 |
| Synaptosomal-associated protein 25, Isoform 2* | SNAP25 | P60880-2 | 2/2 | 16.45 | 0.007 | 1.28 | 64.1 | 1772 |
| Syntaxin-12* | STX12 | Q86Y82 | 2/3 | 8.68 | 0.026 | 1.21 | 16.7 | 521 |
| Syntaxin-1A, isoform 2* | STX1A | Q16623-2 | 8/18 | 9.24 | 0.023 | 1.26 | 47.2 | 1468 |
| Syntaxin-1B* | STX1B | P61266 | 10/15 | 6.53 | 0.043 | 1.17 | 41.7 | 901 |
| Vesicular glutamate transporter 1, Isoform 3* | SLC17A7 | Q9P2U7-3 | 2/5 | 6.27 | 0.046 | 1.21 | 12.7 | 249 |
*Denotes synaptic proteins depicted in Figure 3G.
Table 3.
Long-term alcohol self-administration does not affect expression levels of key proteins found in GABAergic inhibitory synapses
| Protein | Gene | Accession # | Peptides/spectra | F value | p value | Fold change | Coverage % | Mascot score |
|---|---|---|---|---|---|---|---|---|
| GABAA receptor subunit α-1 | GABRA1 | P14867 | 2/2 | 0.52 | 0.499 | 0.93 | 12.9 | 73 |
| GABAB receptor subunit 2 | GABBR2 | O75899 | 3/3 | 0.02 | 0.890 | 0.99 | 14.9 | 200 |
| Gephyrin | GPHN | Q9NQX3 | 3/3 | 1.74 | 0.235 | 0.69 | 15.1 | 260 |
| Neurofascin | NFASC | O94856 | 7/7 | 3.56 | 0.108 | 1.17 | 8.2 | 351 |
| Neuroligin-2 | NLGN2 | Q8NFZ4 | 3/3 | 3.70 | 0.103 | 1.12 | 12.9 | 198 |
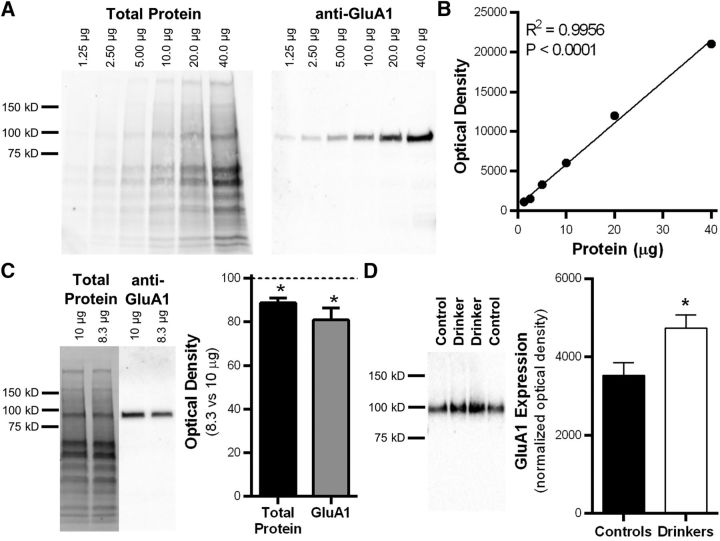
The increase in GluA1 expression found in the synaptomics screen (Table 2) and demonstration of enhanced postsynaptic currents from the electrophysiology experiments suggests an increase in postsynaptic AMPA receptor function (Fig. 2) in drinking monkeys. Because AMPA receptor expression is increased in the lateral OFC of alcohol-dependent mice (Nimitvilai et al., 2016), GluA1 expression levels in this cohort of monkeys were measured using Western blotting to validate cross-species effect of alcohol on AMPA receptors. Characterization of GluA1 immunoreactivity in monkey OFC samples revealed a linear dynamic range across twofold dilutions between 1.25 and 40 μg of protein (R2 = 0.9956; Fig. 4A,B). We also detected significant differences in total protein intensity and the raw GluA1 signal when there was an intentional 17% difference in control sample load (total protein stain: t test, t(14) = 2.85, *p = 0.013, n = 8/group; anti-GluA1: t test, t(14) = 2.32, *p = 0.036; n = 8/group; Fig. 4C), further demonstrating that this quantitative approach is reliable and sensitive (Gürtler et al., 2013). Consistent with results from the proteomics and functional studies, GluA1 level was significantly elevated in drinking monkeys compared with nondrinking controls (one-tailed t test, t(9) = 2.00, *p = 0.039; controls, n = 3; drinkers, n = 8; Fig. 4D).
Figure 4.
Western blot characterization and analysis of synaptic GluA1 expression in the OFC of drinking monkeys. A, Characterization of GluA1 Western blot in macaque cortical tissue (protein loading range, 1.25–40 μg). B, Positive correlation between the amount of protein loaded and GluA1 optical density values. C, Representative images and quantitation of an intentional 17% decrease in total protein load. D, Representative GluA1 blot and quantitation of normalized GluA1 expression in controls and drinkers. Data are the mean ± SEM.
Although these data validate cross-species effects of alcohol on GluA1 expression, a primary goal of synaptic profiling was to identify novel proteins that underpin drinking-induced neuroadaptations. To our knowledge, this is the first study to link alcohol with two synaptic plasma membrane proteins: proline-rich transmembrane protein 2 (PRRT2) and neuronal membrane glycoprotein M6-a (GPM6A). Since these membrane proteins showed the largest fold change, we further explored their relationship with drinking by querying the GeneWeaver.org database (Baker et al., 2012) for functional genomic experiments related to alcohol intake and other alcohol-related behaviors across species. GPM6A is found in 11 published quantitative trait loci (QTLs) for alcohol drinking and other alcohol-related behaviors in humans, rats, and mice (Fig. 3H), while PRRT2 is found in four alcohol-related gene sets (Fig. 3H). PRRT2 is an outer core AMPA receptor complex protein (Schwenk et al., 2012), and GRIA1, the gene encoding the GluA1 subunit of AMPA receptors, is found in 19 alcohol-related gene sets that cross multiple species (GeneWeaver.org analysis not shown). Thus, the synaptomic and functional genomic analyses identified novel alcohol-sensitive synaptic proteins with cross-species genetic links to alcohol consumption phenotypes.
Identification and validation of heavy-drinking synaptic biomarkers
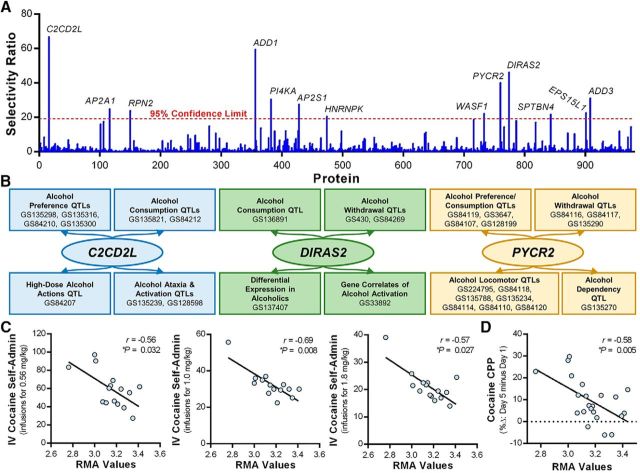
While a recent study has identified a biomarker in blood and a subcortical brain region that predicted heavy alcohol consumption in monkeys (Cervera-Juanes et al., 2016), the identity of synaptic proteins that may drive excessive drinking remains largely unknown. We thus conducted a final exploratory analysis of the synaptomic data using partial least squares discriminant analysis (PLS-DA) to determine which key proteins or groups of proteins best discriminate between control and drinking monkeys. Despite marked changes in functional measures of synaptic excitability in drinking monkeys, PLS-DA only identified two proteins [myosin light chain 6B (MYL6B) and neuronal cell adhesion molecule (NRCAM)] that were above the selectivity ratio criteria that discriminates between controls and drinkers. The low number of discriminatory proteins identified in this analysis suggests unexplained variance in the proteomic dataset, possibly stemming from the allelic diversity reported in alcohol-naive cynomolgus macaques (Kanthaswamy et al., 2013). An equally plausible hypothesis is that individual differences in alcohol intake may contribute to the unexplained variance. To account for this variance, monkeys were classified into low, binge, or heavy drinkers using previously described criteria (Baker et al., 2014). Two of the monkeys (Fig. 3) reached the criteria for heavy drinking (≥3 g/kg on 48% and 38% of the open-access drinking days), and the remaining monkeys fit the criteria for low drinkers achieving ≤3 g/kg intake on <1% of the open access days. PLS-DA of these two groups identified 13 proteins that best discriminated between low and high drinking phenotypes (Fig. 5A). Subsets of the family of 13 proteins fall into functional classes, as 4 proteins have biological processes involving actin cycling (ADD1, ADD3, WASF1, and SPTBN4) and 3 proteins control clathrin-mediated endocytosis (AP2S1, AP2A1, and EPS15L1). Two of the five top proteins are addiction-related proteins in the adducin family (Jung et al., 2013; Han et al., 2015), thus supporting the use of this approach to identify novel brain biomarkers.
Figure 5.
Exploratory analysis of the orbitofrontal cortex synaptome identified proteins that best discriminate between low- and high-drinking cynomolgus monkeys and genes with cross-species links to alcohol- and cocaine-related phenotypes. A, Selectivity ratio plot for all proteins identified in the synaptomics screen. Proteins with selectivity ratios above the 95% confidence limit are identified by gene name. B, GeneWeaver search results of alcohol-related gene sets (GS) containing C2CD2L, DIRAS2, and PYCR2. C, D, Correlations between C2cd2l transcript levels in the PFC and cocaine self-administration and cocaine-conditioned place preference in BXD RI strains.
Interestingly, genes encoding the remaining three top proteins (C2CD2L, DIRAS2, PYCR2) are located within published human and rodent QTLs for alcohol drinking and other alcohol-related behaviors (Fig. 5B). To further validate these initial genetic findings, we experimentally tested associations between PFC transcript levels for these genes and voluntary alcohol drinking in BXD RI strains of mice using the GeneNetwork.org software system (Baker et al., 2012). BXD RI strains generated by ethanol-preferring C57BL/6J (B) and ethanol-avoiding DBA/2J (D) mice are valuable for studying genetic drivers of variation in alcohol consumption (Philip et al., 2010) and our previous work used this approach to identify candidate genes as pharmacological targets for reducing heavy drinking (Padula et al., 2015; Rinker et al., 2017). In alcohol-naive BXD strains, PFC transcript levels for C2cd2l, Diras2, and Pycr2 significantly correlate with alcohol consumption in a standard two-bottle choice (alcohol vs water) drinking model (Table 4). C2cd2l was negatively correlated with 12 datasets for alcohol consumption and preference in male and female BXD RI strains across a range of alcohol concentrations (3–15%, v/v). Because of the relationship between C2CD2L and drinking in monkeys and mice, we then exploited the DBA/2J sequence (Keane et al., 2011; Wang et al., 2016) to extract all sequence variants in and around C2cd2l (e.g., rs32609282). While there are no high-impact C2cd2l variants (Wang et al., 2016), C2cd2l is associated with cis-acting expression QTLs (logarithm of odds scores >3; Pandey and Williams, 2014) in multiple addiction-related brain regions. As neurocircuitry and mechanisms of drug-seeking are shared across drug classes (Mulholland et al., 2016), PFC transcript levels for C2cd2l were then correlated with behaviors related to cocaine experience (Philip et al., 2010; Dickson et al., 2016). Comparable to the relationship between C2cd2l and alcohol intake, there were negative correlations between C2cd2l and cocaine self-administration, as well as cocaine-conditioned place preference (Fig. 5C,D). C2cd2l is also found in QTLs for alcohol-, cocaine-, methamphetamine-, morphine-, and nicotine-related phenotypes (GeneWeaver.org analysis not shown), suggesting that C2CD2L may be associated with genetic vulnerability associated with multiple abused substances.
Table 4.
Correlations between PFC transcript levels in male and female alcohol-naive BXD recombinant inbred strains of mice and alcohol consumption and preference in two-bottle choice (alcohol vs water) drinking models
| Gene | Record ID | Phenotype | PubMed ID | Sex | r | Number of strains | p value |
|---|---|---|---|---|---|---|---|
| Pycr2 | 10156 | Consumption (10% alcohol, g/kg/d) | 8651451 | ♀ | 0.98 | 9 | 4.94−09 |
| 10159 | Consumption (10% alcohol, g/kg/d) | 8651451 | ♂, ♀ | 0.83 | 9 | 0.003 | |
| 10155 | Consumption (10% alcohol, g/kg/d) | 8651451 | ♂ | 0.80 | 9 | 0.007 | |
| 12964 | Consumption (15% alcohol, g/kg/2 h) | 27793543 | ♂, ♀ | 0.69 | 11 | 0.023 | |
| 10581 | Consumption (10% alcohol, g/kg/d) | 7695038 | ♀ | 0.49 | 19 | 0.030 | |
| 10154 | Preference (10% alcohol) | 8651451 | ♀ | 0.95 | 9 | 7.23−06 | |
| 10153 | Preference (10% alcohol) | 8651451 | ♂ | 0.93 | 9 | 5.18−05 | |
| 10158 | Preference (10% alcohol) | 8651451 | ♂, ♀ | 0.85 | 9 | 0.002 | |
| 10583 | Preference (10% alcohol) | 7695038 | ♀ | 0.50 | 19 | 0.027 | |
| C2cd2l | 10582 | Consumption (10% alcohol, g/kg/d) | 7695038 | ♂ | −0.47 | 19 | 0.044 |
| 10475 | Consumption (3% alcohol, g/kg/d) | 7978106 | ♀ | −0.56 | 17 | 0.019 | |
| 10474 | Consumption (3% alcohol, g/kg/d) | 7978106 | ♀ | −0.56 | 17 | 0.017 | |
| 12745 | Consumption (15% alcohol, g/kg/2 h) | 27793543 | ♂, ♀ | −0.65 | 11 | 0.030 | |
| 10155 | Consumption (10% alcohol, g/kg/d) | 8651451 | ♂ | −0.67 | 9 | 0.049 | |
| 10477 | Consumption (10% alcohol, g/kg/d) | 7978106 | ♀ | −0.67 | 17 | 0.004 | |
| 12964 | Consumption (15% alcohol, g/kg/2 h) | 27793543 | ♂, ♀ | −0.71 | 11 | 0.012 | |
| 10159 | Consumption (10% alcohol, g/kg/d) | 8651451 | ♂, ♀ | −0.73 | 9 | 0.022 | |
| 10584 | Preference (10% alcohol) | 7695038 | ♀ | −0.47 | 19 | 0.043 | |
| 10588 | Preference (10% alcohol) | 7695038 | ♂, ♀ | −0.52 | 19 | 0.022 | |
| 10583 | Preference (10% alcohol) | 7695038 | ♀ | −0.55 | 19 | 0.013 | |
| 10153 | Preference (10% alcohol) | 8651451 | ♂ | −0.81 | 9 | 0.005 | |
| Diras2 | 12623 | Consumption (10% alcohol, g/kg/d) | n/a | ♂ | −0.70 | 13 | 0.009 |
Data were obtained from GeneNetwork.org. n/a, Not available.
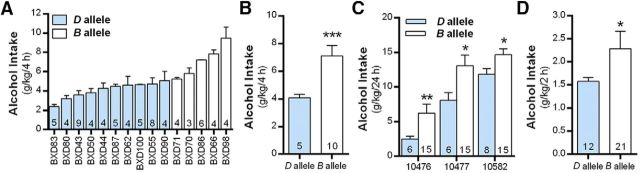
As a final validation for the role of C2cd2l in alcohol drinking, we used a reverse genetic approach (Wang et al., 2016) that can evaluate the spectrum of phenotypes directly linked to the C2cd2l interval and compared alcohol intake in BXD strains that inherited the D allele in comparison with the B allele. Fifteen strains of female BXD mice were allowed to consume alcohol in a binge-like DID model that promotes high levels of alcohol intake. As expected, average alcohol intake in BXDs during the 4 h test drinking sessions varied across strains (range, 2.4–9.5 g/kg/4 h; n = 73 mice; Fig. 6A), and strains that inherited the D allele at C2cd2l on average drank markedly less than strains that inherited the B allele (GeneNetwork record IDs: 13568 and 13571; t test: t(13) = 4.75, ***p < 0.001; Fig. 6B). Consistent with these results, examination of the C2cd2l interval using additional existing datasets in GeneNetwork.org shows that male and female strains that inherited the D allele at C2cd2l consumed significantly lower amounts of alcohol in standard two-bottle choice drinking models [24 h model, 10% alcohol: GeneNetwork record IDs: 10476 (t test, t(19) = 3.58, **p < 0.002), 10477 (t test, t(19) = 2.49, *p < 0.05), and 10582 (t test, t(21) = 2.15, *p = 0.043); 2 h model, 15% alcohol: GeneNetwork record ID 12745 (t test, t(31) = 2.33, *p < 0.027); Fig. 6C,D). Together, these converging analyses corroborate C2CD2L as a high-priority candidate for differential alcohol consumption across species.
Figure 6.
Alcohol consumption in BXD RI strains of mice that inherited the B or D allele at the C2cd2l interval. A, Alcohol (20%, v/v) intake across 15 BXD RI strains in a single-bottle, 4 h DID model. The number within each bar for A shows the number of BXD RI mice within each strain. B–D, BXD mice that inherited the D allele at C2cd2l consumed significantly less alcohol in the DID model (B) and two-bottle choice (C) 24 and 2 h access (D) models. The number within each bar shows the number of strains with the B or D alleles for the C2cd2l interval for B–D.
Discussion
A major finding of this study is that long-term drinking produces functional adaptations in OFC pyramidal neurons of macaques. The electrophysiological analysis revealed functional alterations, while the synaptome screen identified novel proteins altered by alcohol self-administration including presynaptic and postsynaptic proteins that regulate glutamatergic, but not GABAergic, signaling. Because tissue samples for all of these studies were acquired before the start of a daily drinking session, these neuroadaptations may reflect an abnormal state of OFC pyramidal neurons that is present in nonabstinent, actively drinking monkeys. Exploratory integrative bioinformatic approaches unveiled a group of proteins that best discriminate between low and heavy alcohol-drinking subjects. The characterization of these proteins and their contribution to the neurophysiological mechanisms of heavy alcohol drinking is an important step in the development of potentially clinically effective therapeutic strategies to target individuals with AUD.
Long-term drinking and functional adaptations
In this study, we show opposing actions of long-term drinking on evoked action potential firing and spontaneous synaptic activity of monkey OFC pyramidal neuron physiology. Drinking reduced evoked firing and markedly enhanced the amplitude and frequency of sPSCs recorded from OFC pyramidal neurons. These findings are in opposition to intrinsic and synaptic plasticity in models of learning that report learning-related enhanced dendritic and somatic excitability as an early mechanism that facilitates synaptic plasticity (Sehgal et al., 2013). However, consistent with our findings, opposing changes in intrinsic and synaptic plasticity have been observed in the nucleus accumbens shell of rats that self-administered cocaine and may reflect a homeostatic response to normalize global neuronal activity or a shift in the signal-to-noise ratio of behaviorally relevant inputs (Kourrich et al., 2015). Regardless of which hypothesis is correct, these findings suggest that abused substances produce aberrant plasticity of somatic and dendritic excitability of neurons in the corticostriatal circuitry. While enhanced postsynaptic excitability and loss of acute alcohol inhibition of firing are consistent with our previously published study in mice (Nimitvilai et al., 2016), OFC neurons in alcohol-dependent mice show enhanced firing following withdrawal. This apparent discrepancy may reflect the use of different withdrawal times between the mouse (3–14 d) and monkey (<6 h) studies. To directly examine this question, we measured evoked firing of OFC neurons prepared from alcohol-dependent mice within 2 h of their last ethanol exposure, similar to the time of the monkey OFC recordings. Compared with air-exposed control mice, there was no significant effect of long-term intermittent alcohol exposure on OFC neuron firing (two-way repeated-measures ANOVA, F(1,29) = 0.624, p = 0.449). These results are consistent with those results from other mouse studies showing withdrawal time-dependent differences in markers of cortical neuron excitability (Holmes et al., 2012; Kroener et al., 2012; McGuier et al., 2015; Pleil et al., 2015; Nimitvilai et al., 2016). With regard to the effects of firing rate changes on behavior, in vivo recordings in mice (Bissonette et al., 2015) and monkeys (Chang et al., 2005) show that both low and high firing rates are related to poor performance on OFC- and PFC-dependent tasks, suggesting that disruption of optimal firing of OFC pyramidal neurons by long-term alcohol consumption may underlie cognitive decline in alcoholic individuals.
Long-term alcohol consumption and AMPA receptors
A notable result from profiling the OFC synaptome was the finding that long-term drinking enhances GluA1 expression, a result confirmed by Western blotting and consistent with the increase in the amplitude of putative AMPA-mediated sPSCs in deep-layer OFC pyramidal neurons observed in drinking monkeys. While we cannot rule out that increases in sPSC amplitude reflect changes in GABA-mediated signaling, GRIA1 levels are also increased in the superior frontal cortex of alcoholic individuals (Lewohl et al., 2000), and increases in AMPA receptor function and expression have been reported in the lateral OFC, but not prelimbic PFC, of alcohol-dependent mice (Kroener et al., 2012; Hu et al., 2015; Nimitvilai et al., 2016). Our recent chemogenetic and lesion study has implicated the OFC in controlling escalated drinking in dependent mice (den Hartog et al., 2016), and there is evidence from studies with rats showing that AMPA receptors affect operant self-administration of alcohol (Wang et al., 2012; Cannady et al., 2016). Interestingly, sPSC frequency was also elevated in the drinking monkeys, and the proteomics screen identified multiple upregulated presynaptic proteins that control glutamate release from synaptic vesicles. A recent study has reported that SV2A, an alcohol-responsive presynaptic protein, regulates dendritic spine density and postsynaptic AMPA receptor expression (Cohen et al., 2011). Although we did not examine dendritic spine changes in drinking monkeys, alcohol dependence enhanced dendritic spine density in the mouse OFC (McGuier et al., 2015). Together, this converging evidence reveals a cross-species effect of long-term alcohol consumption on the GluA1 subunit of AMPA receptors in the OFC and suggests that presynaptic adaptations produced by drinking may be molecular mechanisms that drive functional and morphological postsynaptic adaptations.
Novel alcohol-sensitive synaptic proteins
An advantage of our approach over previous proteomic studies in brain tissue is the ability to solubilize and quantify transmembrane synaptic proteins. Our profiling of the macaque OFC synaptome identified novel alcohol-sensitive transmembrane proteins, and the two synaptic transmembrane proteins that were most altered by long-term drinking (i.e., GPM6A and PRRT2) localize to presynaptic sites where they control glutamate release (Roussel et al., 1998; Valente et al., 2016). In addition, postsynaptic GPM6A has been reported to promote dendritic spine formation (Alfonso et al., 2005), whereas PRRT2 functions in spines to modulate AMPA receptor gating properties and surface expression (Schwenk et al., 2012; Li et al., 2013). Recent studies suggest a central role for GPM6A and PRRT2 in cognitive impairments (Najmabadi et al., 2011; Gregor et al., 2014), and our bioinformatic analysis links the genes encoding these proteins to alcohol drinking and other alcohol-related behaviors. Loss-of-function PRRT2 mutations are a causative factor for familial paroxysmal kinesigenic dyskinesia (PKD; Heron and Dibbens, 2013) and PKD patients with an insertion mutation in PRRT2 show enhanced spontaneous low-frequency fluctuations in corticostriatal circuitry (Luo et al., 2013). Neuroimaging studies in alcoholic individuals reveal aberrant executive control network activity and altered brain connectivity (Müller-Oehring et al., 2015; Zhu et al., 2015) that could reflect disruptions in GPM6A/PRRT2 expression or function. Future studies are needed to validate the importance of these proteins in mediating the altered cognition, glutamatergic signaling, and corticostriatal circuitry observed in individuals with AUD.
Our final query of the OFC synaptome was an exploratory analysis that identified functional groups of proteins that are discriminators of a heavy-drinking phenotype. The subset of identified proteins that best discriminated between high and low drinkers is the known regulators of actin cycling within dendritic spines and has been previously implicated in drug addiction (Jung et al., 2013; Han et al., 2015). Adaptations in dendritic spines on neurons within the corticostriatal circuitry are a shared feature of drug-induced plasticity (Mulholland et al., 2016). Additional bioinformatic analysis revealed that C2CD2L, DIRAS2, and PYCR2 are found within multiple published QTLs for alcohol drinking and dependence in human and rodent studies, suggesting that they may be relevant brain biomarkers for a heavy-drinking phenotype. In support of this, PFC transcript levels for these three genes in BXD strains correlate with voluntary drinking. Like Diras2, several other genes encoding proteins in the RAS family are associated with excessive drinking (Repunte-Canonigo et al., 2010; Ben Hamida et al., 2012; Stacey et al., 2012; Ojelade et al., 2015). Finally, strains with the C2cd2l D allele drink markedly less alcohol across multiple paradigms, including a binge-like drinking model. Thus, these findings suggest that DIRAS2 and C2CD2L are brain biomarkers of heavy drinking and should be explored and validated as potential targets in preclinical models of excessive drinking.
Although the findings reported here uncovered adaptations and potential pharmacotherapeutic targets for the treatment of AUD, there are some limitations to consider. First, sample sizes for the functional measures were unequal due to a limited number of monkeys assigned to the control condition. During these studies, the experimenter was blind to the treatment groups so no adjustments could be made to account for this difference. While the limited number of control monkeys is potentially a concern, the robust effects of drinking on sPSCs and the congruence of the proteomic changes in presynaptic and postsynaptic glutamatergic signaling machinery help to mitigate this limitation. Second, the overrepresentation of high drinkers used for these studies precludes correlations between functional adaptations and individual differences in alcohol intake. Nonetheless, the key proteins that discriminated between low and high male drinkers were validated in multiple alcohol-drinking models using both male and female mice. In addition, as previously discussed, the recording conditions used to monitor sPSCs, although biased in favor of AMPA events, do not rule out the possibility of changes in inhibitory synaptic transmission. Last, because data were collected at a single time point from long-term drinking monkeys, it is unknown at what stage the observed functional neuroadaptations occurred and whether they would persist. These are important considerations as such changes may be long-lasting and could contribute to relapse to alcohol-seeking behaviors even after long periods of abstinence. Future studies are needed to address these questions.
In summary, long-term alcohol self-administration is associated with complex neuroadaptations in deep-layer OFC pyramidal neurons in heavy-drinking macaques. The congruence of alcohol effects across species suggests that aberrant adaptations in glutamatergic signaling are key mechanisms underpinning alcohol-induced OFC dysfunction. Finally, the novel proteins (e.g., C2CD2L) that show links to a heavy-drinking phenotype in rodents, monkeys, and humans have potential as targets for the treatment of AUD. Further clinical and preclinical studies are necessary to determine whether targeting these biomarkers prevents excessive intake and reduces the incidence of relapse.
Footnotes
This work was supported by National Institutes of Health (NIH) Grants AA-020930, AA-023288, RR-024485, AA-024426, AA-019431, AA-013541, AA-016662, AA-013499, AA-010761, AA-009986, AA-021951, and OD-11092. The Medical University of South Carolina Mass Spectrometry Facility receives support from the SC COBRE in Oxidants, Redox Balance, and Stress Signaling (Grant P20 GM103542) and the Office of the Provost. The Orbitrap Elite Mass Spectrometer was acquired through NIH/National Center for Research Resources Grant S10-OD-010731. We thank Susana Comte-Walters. We also thank Dr. James B. Daunais for the macaque frontal cortex images.
The authors declare no competing financial interests.
References
- Alfonso J, Fernández ME, Cooper B, Flugge G, Frasch AC (2005) The stress-regulated protein M6a is a key modulator for neurite outgrowth and filopodium/spine formation. Proc Natl Acad Sci U S A 102:17196–17201. 10.1073/pnas.0504262102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatrudo JM, Weaver CM, Crimins JL, Hof PR, Rosene DL, Luebke JI (2012) Influence of highly distinctive structural properties on the excitability of pyramidal neurons in monkey visual and prefrontal cortices. J Neurosci 32:13644–13660. 10.1523/JNEUROSCI.2581-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badanich KA, Becker HC, Woodward JJ (2011) Effects of chronic intermittent ethanol exposure on orbitofrontal and medial prefrontal cortex-dependent behaviors in mice. Behav Neurosci 125:879–891. 10.1037/a0025922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EJ, Jay JJ, Bubier JA, Langston MA, Chesler EJ (2012) GeneWeaver: a web-based system for integrative functional genomics. Nucleic Acids Res 40:D1067–D1076. 10.1093/nar/gkr968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EJ, Farro J, Gonzales S, Helms C, Grant KA (2014) Chronic alcohol self-administration in monkeys shows long-term quantity/frequency categorical stability. Alcohol Clin Exp Res 38:2835–2843. 10.1111/acer.12547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EJ, Walter NA, Salo A, Rivas Perea P, Moore S, Gonzales S, Grant KA (2017) Identifying future drinkers: behavioral analysis of monkeys initiating drinking to intoxication is predictive of future drinking classification. Alcohol Clin Exp Res 41:626–636. 10.1111/acer.13327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Hamida S, Neasta J, Lasek AW, Kharazia V, Zou M, Carnicella S, Janak PH, Ron D (2012) The small G protein H-Ras in the mesolimbic system is a molecular gateway to alcohol-seeking and excessive drinking behaviors. J Neurosci 32:15849–15858. 10.1523/JNEUROSCI.2846-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissonette GB, Schoenbaum G, Roesch MR, Powell EM (2015) Interneurons are necessary for coordinated activity during reversal learning in orbitofrontal cortex. Biol Psychiatry 77:454–464. 10.1016/j.biopsych.2014.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannady R, Fisher KR, Graham C, Crayle J, Besheer J, Hodge CW (2016) Potentiation of amygdala AMPA receptor activity selectively promotes escalated alcohol self-administration in a CaMKII-dependent manner. Addict Biol. Advance online publication. Retrieved March 4, 2017. doi: 10.1111/adb.12357. 10.1111/adb.12357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael ST, Price JL (1994) Architectonic subdivision of the orbital and medial prefrontal cortex in the macaque monkey. J Comp Neurol 346:366–402. 10.1002/cne.903460305 [DOI] [PubMed] [Google Scholar]
- Centanni SW, Teppen T, Risher ML, Fleming RL, Moss JL, Acheson SK, Mulholland PJ, Pandey SC, Chandler LJ, Swartzwelder HS (2014) Adolescent alcohol exposure alters GABAA receptor subunit expression in adult hippocampus. Alcohol Clin Exp Res 38:2800–2808. 10.1111/acer.12562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera-Juanes R, Wilhem LJ, Park B, Lee R, Locke J, Helms C, Gonzales S, Wand G, Jones SR, Grant KA, Ferguson B (2016) MAOA expression predicts vulnerability for alcohol use. Mol Psychiatry 21:472–479. 10.1038/mp.2015.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YM, Rosene DL, Killiany RJ, Mangiamele LA, Luebke JI (2005) Increased action potential firing rates of layer 2/3 pyramidal cells in the prefrontal cortex are significantly related to cognitive performance in aged monkeys. Cereb Cortex 15:409–418. 10.1093/cercor/bhh144 [DOI] [PubMed] [Google Scholar]
- Charlet K, Beck A, Jorde A, Wimmer L, Vollstädt-Klein S, Gallinat J, Walter H, Kiefer F, Heinz A (2014) Increased neural activity during high working memory load predicts low relapse risk in alcohol dependence. Addict Biol 19:402–414. 10.1111/adb.12103 [DOI] [PubMed] [Google Scholar]
- Cohen JE, Lee PR, Chen S, Li W, Fields RD (2011) MicroRNA regulation of homeostatic synaptic plasticity. Proc Natl Acad Sci U S A 108:11650–11655. 10.1073/pnas.1017576108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews FT, Boettiger CA (2009) Impulsivity, frontal lobes and risk for addiction. Pharmacol Biochem Behav 93:237–247. 10.1016/j.pbb.2009.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport AT, Grant KA, Szeliga KT, Friedman DP, Daunais JB (2014) Standardized method for the harvest of nonhuman primate tissue optimized for multiple modes of analyses. Cell Tissue Bank 15:99–110. 10.1007/s10561-013-9380-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hartog C, Zamudio-Bulcock P, Nimitvilai S, Gilstrap M, Eaton B, Fedarovich H, Motts A, Woodward JJ (2016) Inactivation of the lateral orbitofrontal cortex increases drinking in ethanol-dependent but not non-dependent mice. Neuropharmacology 107:451–459. 10.1016/j.neuropharm.2016.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson PE, Miller MM, Calton MA, Bubier JA, Cook MN, Goldowitz D, Chesler EJ, Mittleman G (2016) Systems genetics of intravenous cocaine self-administration in the BXD recombinant inbred mouse panel. Psychopharmacology 233:701–714. 10.1007/s00213-015-4147-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozmorov M, Li R, Xu HP, Jilderos B, Wigström H (2004) Slowly developing depression of N-methyl-D-aspartate receptor mediated responses in young rat hippocampi. BMC Neurosci 5:26. 10.1186/1471-2202-5-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier CB, Steffen EM, Lafleche G, Venne JR, Disterhoft JF, McGlinchey RE (2008) Delay discrimination and reversal eyeblink classical conditioning in abstinent chronic alcoholics. Neuropsychology 22:196–208. 10.1037/0894-4105.22.2.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier CB, Maksimovskiy AL, Venne JR, LaFleche G, McGlinchey RE (2009) Silent trace eliminates differential eyeblink learning in abstinent alcoholics. Int J Environ Res Public Health 6:2007–2027. 10.3390/ijerph6072007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant KA, Leng X, Green HL, Szeliga KT, Rogers LS, Gonzales SW (2008) Drinking typography established by scheduled induction predicts chronic heavy drinking in a monkey model of ethanol self-administration. Alcohol Clin Exp Res 32:1824–1838. 10.1111/j.1530-0277.2008.00765.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor A, Kramer JM, van der Voet M, Schanze I, Uebe S, Donders R, Reis A, Schenck A, Zweier C (2014) Altered GPM6A/M6 dosage impairs cognition and causes phenotypes responsive to cholesterol in human and Drosophila. Hum Mutat 35:1495–1505. 10.1002/humu.22697 [DOI] [PubMed] [Google Scholar]
- Gürtler A, Kunz N, Gomolka M, Hornhardt S, Friedl AA, McDonald K, Kohn JE, Posch A (2013) Stain-free technology as a normalization tool in Western blot analysis. Anal Biochem 433:105–111. 10.1016/j.ab.2012.10.010 [DOI] [PubMed] [Google Scholar]
- Han L, Liu P, Wang C, Zhong Q, Fan R, Wang L, Duan S, Zhang L (2015) The interactions between alcohol consumption and DNA methylation of the ADD1 gene promoter modulate essential hypertension susceptibility in a population-based, case-control study. Hypertens Res 38:284–290. 10.1038/hr.2014.172 [DOI] [PubMed] [Google Scholar]
- Herman MA, Nahir B, Jahr CE (2011) Distribution of extracellular glutamate in the neuropil of hippocampus. PLoS One 6:e26501. 10.1371/journal.pone.0026501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heron SE, Dibbens LM (2013) Role of PRRT2 in common paroxysmal neurological disorders: a gene with remarkable pleiotropy. J Med Genet 50:133–139. 10.1136/jmedgenet-2012-101406 [DOI] [PubMed] [Google Scholar]
- Holmes A, Fitzgerald PJ, MacPherson KP, DeBrouse L, Colacicco G, Flynn SM, Masneuf S, Pleil KE, Li C, Marcinkiewcz CA, Kash TL, Gunduz-Cinar O, Camp M (2012) Chronic alcohol remodels prefrontal neurons and disrupts NMDAR-mediated fear extinction encoding. Nat Neurosci 15:1359–1361. 10.1038/nn.3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Morris B, Carrasco A, Kroener S (2015) Effects of acamprosate on attentional set-shifting and cellular function in the prefrontal cortex of chronic alcohol-exposed mice. Alcohol Clin Exp Res 39:953–961. 10.1111/acer.12722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M, Mu P, Moyer JT, Wolf JA, Quock RM, Davies NM, Hu XT, Schlüter OM, Dong Y (2009) Homeostatic synapse-driven membrane plasticity in nucleus accumbens neurons. J Neurosci 29:5820–5831. 10.1523/JNEUROSCI.5703-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson P, Bruce SJ, Moritz T, Trygg J, Sjöström M, Plumb R, Granger J, Maibaum E, Nicholson JK, Holmes E, Antti H (2005) Extraction, interpretation and validation of information for comparing samples in metabolic LC/MS data sets. Analyst 130:701–707. 10.1039/B501890K [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y, Mulholland PJ, Wiseman SL, Chandler LJ, Picciotto MR (2013) Constitutive knockout of the membrane cytoskeleton protein beta adducin decreases mushroom spine density in the nucleus accumbens but does not prevent spine remodeling in response to cocaine. Eur J Neurosci 37:1–9. 10.1111/ejn.12037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods 4:923–925. 10.1038/nmeth1113 [DOI] [PubMed] [Google Scholar]
- Kanthaswamy S, Ng J, Satkoski Trask J, George DA, Kou AJ, Hoffman LN, Doherty TB, Houghton P, Smith DG (2013) The genetic composition of populations of cynomolgus macaques (Macaca fascicularis) used in biomedical research. J Med Primatol 42:120–131. 10.1111/jmp.12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellåker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, et al. (2011) Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477:289–294. 10.1038/nature10413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourrich S, Calu DJ, Bonci A (2015) Intrinsic plasticity: an emerging player in addiction. Nat Rev Neurosci 16:173–184. 10.1038/nrn3877 [DOI] [PubMed] [Google Scholar]
- Kroener S, Mulholland PJ, New NN, Gass JT, Becker HC, Chandler LJ (2012) Chronic alcohol exposure alters behavioral and synaptic plasticity of the rodent prefrontal cortex. PLoS One 7:e37541. 10.1371/journal.pone.0037541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JG, McKinney KQ, Lee YY, Chung HN, Pavlopoulos AJ, Jung KY, Kim WK, Kuroda MJ, Han DK, Hwang S (2015) A draft map of rhesus monkey tissue proteome for biomedical research. PLoS One 10:e0126243. 10.1371/journal.pone.0126243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Chung HJ (2014) NMDA receptors and L-type voltage-gated Ca(2)(+) channels mediate the expression of bidirectional homeostatic intrinsic plasticity in cultured hippocampal neurons. Neuroscience 277:610–623. 10.1016/j.neuroscience.2014.07.038 [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA (2000) Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res 24:1873–1882. 10.1111/j.1530-0277.2000.tb01993.x [DOI] [PubMed] [Google Scholar]
- Li KW, Chen N, Smit AB (2013) Interaction proteomics of the AMPA receptor: towards identification of receptor sub-complexes. Amino Acids 44:1247–1251. 10.1007/s00726-013-1461-9 [DOI] [PubMed] [Google Scholar]
- Littell RC, Henry PR, Ammerman CB (1998) Statistical analysis of repeated measures data using SAS procedures. J Anim Sci 76:1216–1231. 10.2527/1998.7641216x [DOI] [PubMed] [Google Scholar]
- Luo C, Chen Y, Song W, Chen Q, Gong Q, Shang HF (2013) Altered intrinsic brain activity in patients with paroxysmal kinesigenic dyskinesia by PRRT2 mutation: altered brain activity by PRRT2 mutation. Neurol Sci 34:1925–1931. 10.1007/s10072-013-1408-7 [DOI] [PubMed] [Google Scholar]
- McGuier NS, Padula AE, Lopez MF, Woodward JJ, Mulholland PJ (2015) Withdrawal from chronic intermittent alcohol exposure increases dendritic spine density in the lateral orbitofrontal cortex of mice. Alcohol 49:21–27. 10.1016/j.alcohol.2014.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuier NS, Griffin WC 3rd, Gass JT, Padula AE, Chesler EJ, Mulholland PJ (2016) Kv7 channels in the nucleus accumbens are altered by chronic drinking and are targets for reducing alcohol consumption. Addict Biol 21:1097–1112. 10.1111/adb.12279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medalla M, Luebke JI (2015) Diversity of glutamatergic synaptic strength in lateral prefrontal versus primary visual cortices in the rhesus monkey. J Neurosci 35:112–127. 10.1523/JNEUROSCI.3426-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland PJ, Becker HC, Woodward JJ, Chandler LJ (2011) Small conductance calcium-activated potassium type 2 channels regulate alcohol-associated plasticity of glutamatergic synapses. Biol Psychiatry 69:625–632. 10.1016/j.biopsych.2010.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland PJ, Chandler LJ, Kalivas PW (2016) Signals from the fourth dimension regulate drug relapse. Trends Neurosci 39:472–485. 10.1016/j.tins.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Oehring EM, Jung YC, Pfefferbaum A, Sullivan EV, Schulte T (2015) The resting brain of alcoholics. Cereb Cortex 25:4155–4168. 10.1093/cercor/bhu134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, Hosseini M, Behjati F, Haas S, Jamali P, Zecha A, Mohseni M, Püttmann L, Vahid LN, Jensen C, Moheb LA, Bienek M, Larti F, Mueller I, Weissmann R, et al. (2011) Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478:57–63. 10.1038/nature10423 [DOI] [PubMed] [Google Scholar]
- Nimitvilai S, Lopez MF, Mulholland PJ, Woodward JJ (2016) Chronic intermittent ethanol exposure enhances the excitability and synaptic plasticity of lateral orbitofrontal cortex neurons and induces a tolerance to the acute inhibitory actions of ethanol. Neuropsychopharmacology 41:1112–1127. 10.1038/npp.2015.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt DJ, King LA, Phillips LD, Independent Scientific Committee on D (2010) Drug harms in the UK: a multicriteria decision analysis. Lancet 376:1558–1565. 10.1016/S0140-6736(10)61462-6 [DOI] [PubMed] [Google Scholar]
- Ojelade SA, Jia T, Rodan AR, Chenyang T, Kadrmas JL, Cattrell A, Ruggeri B, Charoen P, Lemaitre H, Banaschewski T, Büchel C, Bokde AL, Carvalho F, Conrod PJ, Flor H, Frouin V, Gallinat J, Garavan H, Gowland PA, Heinz A, et al. (2015) Rsu1 regulates ethanol consumption in Drosophila and humans. Proc Natl Acad Sci U S A 112:E4085–E4093. 10.1073/pnas.1417222112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongür D, Price JL (2000) The organization of networks within the orbital and medial prefrontal cortex of rats, monkeys and humans. Cereb Cortex 10:206–219. 10.1093/cercor/10.3.206 [DOI] [PubMed] [Google Scholar]
- Padula AE, Griffin WC 3rd, Lopez MF, Nimitvilai S, Cannady R, McGuier NS, Chesler EJ, Miles MF, Williams RW, Randall PK, Woodward JJ, Becker HC, Mulholland PJ (2015) KCNN genes that encode small-conductance Ca2+-activated K+ channels influence alcohol and drug addiction. Neuropsychopharmacology 40:1928–1939. 10.1038/npp.2015.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey AK, Williams RW (2014) Genetics of gene expression in CNS. Int Rev Neurobiol 116:195–231. 10.1016/B978-0-12-801105-8.00008-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, Hamre KM, Lariviere WR, Matthews DB, Mittleman G, Goldowitz D, Chesler EJ (2010) High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav 9:129–159. 10.1111/j.1601-183X.2009.00540.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleil KE, Lowery-Gionta EG, Crowley NA, Li C, Marcinkiewcz CA, Rose JH, McCall NM, Maldonado-Devincci AM, Morrow AL, Jones SR, Kash TL (2015) Effects of chronic ethanol exposure on neuronal function in the prefrontal cortex and extended amygdala. Neuropharmacology 99:735–749. 10.1016/j.neuropharm.2015.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polpitiya AD, Qian WJ, Jaitly N, Petyuk VA, Adkins JN, Camp DG 2nd, Anderson GA, Smith RD (2008) DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics 24:1556–1558. 10.1093/bioinformatics/btn217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povysheva NV, Gonzalez-Burgos G, Zaitsev AV, Kröner S, Barrionuevo G, Lewis DA, Krimer LS (2006) Properties of excitatory synaptic responses in fast-spiking interneurons and pyramidal cells from monkey and rat prefrontal cortex. Cereb Cortex 16:541–552. 10.1093/cercor/bhj002 [DOI] [PubMed] [Google Scholar]
- Rajalahti T, Arneberg R, Berven FS, Myhr KM, Ulvik RJ, Kvalheim OM (2009a) Biomarker discovery in mass spectral profiles by means of selectivity ratio plot. Chemometr Intell Lab Syst 95:35–48. 10.1016/j.chemolab.2008.08.004 [DOI] [Google Scholar]
- Rajalahti T, Arneberg R, Kroksveen AC, Berle M, Myhr KM, Kvalheim OM (2009b) Discriminating variable test and selectivity ratio plot: quantitative tools for interpretation and variable (biomarker) selection in complex spectral or chromatographic profiles. Anal Chem 81:2581–2590. 10.1021/ac802514y [DOI] [PubMed] [Google Scholar]
- Repunte-Canonigo V, van der Stap LD, Chen J, Sabino V, Wagner U, Zorrilla EP, Schumann G, Roberts AJ, Sanna PP (2010) Genome-wide gene expression analysis identifies K-ras as a regulator of alcohol intake. Brain Res 1339:1–10. 10.1016/j.brainres.2010.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes JS, Best K, Belknap JK, Finn DA, Crabbe JC (2005) Evaluation of a simple model of ethanol drinking to intoxication in C57BL/6J mice. Physiol Behav 84:53–63. 10.1016/j.physbeh.2004.10.007 [DOI] [PubMed] [Google Scholar]
- Rinker JA, Marshall SA, Mazzone CM, Lowery-Gionta EG, Gulati V, Pleil KE, Kash TL, Navarro M, Thiele TE (2016) Extended amygdala to ventral tegmental area corticotropin-releasing factor circuit controls binge ethanol intake. Biol Psychiatry. Advance online publication. Retrieved March 4, 2017. doi: 10.1016/j.biopsych.2016.02.029. 10.1016/j.biopsych.2016.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinker JA, Fulmer DB, Trantham-Davidson H, Smith ML, Williams RW, Lopez MF, Randall PK, Chandler LJ, Miles MF, Becker HC, Mulholland PJ (2017) Differential potassium channel gene regulation in BXD mice reveals novel targets for pharmacogenetic therapies to reduce heavy alcohol drinking. Alcohol 58:33–45. 10.1016/j.alcohol.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3:1154–1169. 10.1074/mcp.M400129-MCP200 [DOI] [PubMed] [Google Scholar]
- Roussel G, Trifilieff E, Lagenaur C, Nussbaum JL (1998) Immunoelectron microscopic localization of the M6a antigen in rat brain. J Neurocytol 27:695–703. 10.1023/A:1006924400768 [DOI] [PubMed] [Google Scholar]
- Sassoè-Pognetto M, Frola E, Pregno G, Briatore F, Patrizi A (2011) Understanding the molecular diversity of GABAergic synapses. Front Cell Neurosci 5:4. 10.3389/fncel.2011.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Brechet A, Zolles G, Berkefeld H, Müller CS, Bildl W, Baehrens D, Hüber B, Kulik A, Klöcker N, Schulte U, Fakler B (2012) High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron 74:621–633. 10.1016/j.neuron.2012.03.034 [DOI] [PubMed] [Google Scholar]
- Sehgal M, Song C, Ehlers VL, Moyer JR Jr (2013) Learning to learn—intrinsic plasticity as a metaplasticity mechanism for memory formation. Neurobiol Learn Mem 105:186–199. 10.1016/j.nlm.2013.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey D, Bilbao A, Maroteaux M, Jia T, Easton AC, Longueville S, Nymberg C, Banaschewski T, Barker GJ, Büchel C, Carvalho F, Conrod PJ, Desrivières S, Fauth-Bühler M, Fernandez-Medarde A, Flor H, Gallinat J, Garavan H, Bokde AL, Heinz A, et al. (2012) RASGRF2 regulates alcohol-induced reinforcement by influencing mesolimbic dopamine neuron activity and dopamine release. Proc Natl Acad Sci U S A 109:21128–21133. 10.1073/pnas.1211844110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalnaker TA, Cooch NK, Schoenbaum G (2015) What the orbitofrontal cortex does not do. Nat Neurosci 18:620–627. 10.1038/nn.3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucholski J, Pinner AL, Simmons MS, Meador-Woodruff JH (2014) Evolutionarily conserved pattern of AMPA receptor subunit glycosylation in Mammalian frontal cortex. PLoS One 9:e94255. 10.1371/journal.pone.0094255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uys JD, McGuier NS, Gass JT, Griffin WC 3rd, Ball LE, Mulholland PJ (2016) Chronic intermittent ethanol exposure and withdrawal leads to adaptations in nucleus accumbens core postsynaptic density proteome and dendritic spines. Addict Biol 21:560–574. 10.1111/adb.12238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente P, Castroflorio E, Rossi P, Fadda M, Sterlini B, Cervigni RI, Prestigio C, Giovedì S, Onofri F, Mura E, Guarnieri FC, Marte A, Orlando M, Zara F, Fassio A, Valtorta F, Baldelli P, Corradi A, Benfenati F (2016) PRRT2 is a key component of the Ca(2+)-dependent neurotransmitter release machinery. Cell Rep 15:117–131. 10.1016/j.celrep.2016.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian JA, Green HL, Young JE, Majerksy LS, Thomas BW, Shively CA, Tobin JR, Nader MA, Grant KA (2001) Induction and maintenance of ethanol self-administration in cynomolgus monkeys (Macaca fascicularis): long-term characterization of sex and individual differences. Alcohol Clin Exp Res 25:1087–1097. 10.1111/j.1530-0277.2001.tb02321.x [DOI] [PubMed] [Google Scholar]
- Wang J, Ben Hamida S, Darcq E, Zhu W, Gibb SL, Lanfranco MF, Carnicella S, Ron D (2012) Ethanol-mediated facilitation of AMPA receptor function in the dorsomedial striatum: implications for alcohol drinking behavior. J Neurosci 32:15124–15132. 10.1523/JNEUROSCI.2783-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Pandey AK, Mulligan MK, Williams EG, Mozhui K, Li Z, Jovaisaite V, Quarles LD, Xiao Z, Huang J, Capra JA, Chen Z, Taylor WL, Bastarache L, Niu X, Pollard KS, Ciobanu DC, Reznik AO, Tishkov AV, Zhulin IB, et al. (2016) Joint mouse-human phenome-wide association to test gene function and disease risk. Nat Commun 7:10464. 10.1038/ncomms10464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford HA, Degenhardt L, Rehm J, Baxter AJ, Ferrari AJ, Erskine HE, Charlson FJ, Norman RE, Flaxman AD, Johns N, Burstein R, Murray CJ, Vos T (2013) Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. Lancet 382:1575–1586. 10.1016/S0140-6736(13)61611-6 [DOI] [PubMed] [Google Scholar]
- Wildburger NC, Lichti CF, LeDuc RD, Schmidt M, Kroes RA, Moskal JR, Nilsson CL (2015) Quantitative proteomics and transcriptomics reveals metabolic differences in attracting and non-attracting human-in-mouse glioma stem cell xenografts and stromal cells. EuPA Open Proteomics 8:94–103. 10.1016/j.euprot.2015.06.006 [DOI] [Google Scholar]
- Zamanillo D, Sprengel R, Hvalby O, Jensen V, Burnashev N, Rozov A, Kaiser KM, Köster HJ, Borchardt T, Worley P, Lübke J, Frotscher M, Kelly PH, Sommer B, Andersen P, Seeburg PH, Sakmann B (1999) Importance of AMPA receptors for hippocampal synaptic plasticity but not for spatial learning. Science 284:1805–1811. 10.1126/science.284.5421.1805 [DOI] [PubMed] [Google Scholar]
- Zhu X, Cortes CR, Mathur K, Tomasi D, Momenan R (2017) Model-free functional connectivity and impulsivity correlates of alcohol dependence: a resting-state study. Addict Biol 22:206–217. 10.1111/adb.12272 [DOI] [PMC free article] [PubMed] [Google Scholar]