Significance
Neurotrophic factors are endogenous survival factors for developing neurons during their programmed cell death, and represent therapeutic candidates in several neurodegenerative diseases. Studies in the developing spinal cord suggest that neurotrophic factors promote the survival of motor neurons in a combinatorial manner. To better understand this, we systematically assayed pairwise combinations of neurotrophic factors (NTFs) on highly standardized motor neuron cultures prepared by a unique FACS technique. Our data unravel potent additivity of three neurotrophic factors due to their specific survival effects on distinct classes of motor neurons innervating different targets. Further analyses are required to better understand combinatorial NTF effects in adulthood and to define optimized NTF combinations for degenerative motor neuron diseases.
Keywords: neurotrophic factor, motor neuron, screening, fluorescence-activated cell sorting, Artemin
Abstract
Numerous neurotrophic factors promote the survival of developing motor neurons but their combinatorial actions remain poorly understood; to address this, we here screened 66 combinations of 12 neurotrophic factors on pure, highly viable, and standardized embryonic mouse motor neurons isolated by a unique FACS technique. We demonstrate potent, strictly additive, survival effects of hepatocyte growth factor (HGF), ciliary neurotrophic factor (CNTF), and Artemin through specific activation of their receptor complexes in distinct subsets of lumbar motor neurons: HGF supports hindlimb motor neurons through c-Met; CNTF supports subsets of axial motor neurons through CNTFRα; and Artemin acts as the first survival factor for parasympathetic preganglionic motor neurons through GFRα3/Syndecan-3 activation. These data show that neurotrophic factors can selectively promote the survival of distinct classes of embryonic motor neurons. Similar studies on postnatal motor neurons may provide a conceptual framework for the combined therapeutic use of neurotrophic factors in degenerative motor neuron diseases such as amyotrophic lateral sclerosis, spinal muscular atrophy, and spinobulbar muscular atrophy.
According to the neurotrophic theory (1), populations of developing neurons compete during their period of programmed cell death for limiting amounts of target-derived neurotrophic factors (NTFs), which determines their survival or elimination; this is well illustrated in the embryonic chicken spinal cord where motor neurons show increased cell death after limb bud removal and increased survival after transplantation of a supernumerary limb bud (2). More than 15 NTFs belonging to different protein families and activating distinct receptors and signaling pathways have now been identified for developing motor neurons (3, 4); some are also able to rescue degenerating motor neurons in experimental models of motor neuron diseases, such as amyotrophic lateral sclerosis (ALS) (5).
Mounting evidence suggests that NTFs act in a combinatorial manner. Indeed, knockout of individual NTF genes in mice causes only partial motor neuron loss (3), whereas genetic ablation of cell types releasing multiple NTFs, such as Schwann cells (6) or muscle cells (7), causes almost complete motor neuron loss. Similarly, genetic double knockout of the NTFs IGF1 and LIF (8) or triple knockout of CNTF, CT1, and LIF (9) enhances motor neuron loss compared with respective single or double knockouts. NTFs also synergize to rescue motor neurons after axotomy (10) and in culture (11). Finally, combined administration of the NTFs BDNF + CNTF (12) or NT-3 + CNTF (13) reduces pathologic motor neuron degeneration in wobbler and pmn mice, respectively.
The mechanistic basis for these combinatorial NTF effects remains unclear. One hypothesis postulates the existence of motor neuron subsets with different trophic requirements (3, 4). Testing this hypothesis in a comprehensive manner was hitherto precluded by the plethora of NTFs, the early lethality of many NTF/NTF receptor knockout mouse lines, and the poor characterization of motor neuron subsets in traditional primary cultures of embryonic spinal cord.
To systematically investigate the combinatorial action of NTFs, we screened 66 pairwise combinations of 12 NTFs on pure, highly viable, and perfectly standardized embryonic mouse motor neurons isolated by a unique FACS technique. We demonstrate potent, strictly additive, survival effects of hepatocyte growth factor (HGF), ciliary neurotrophic factor (CNTF), and Artemin (ARTN) due to their selective action on distinct classes of motor neurons.
Results
High-Speed FACS Isolation of Motor Neurons.
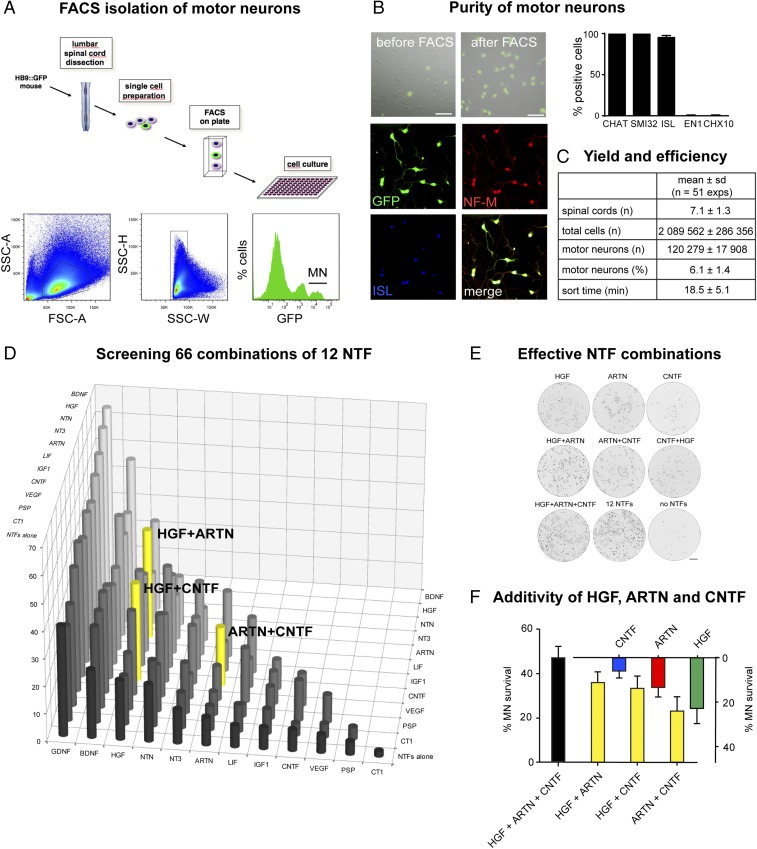
To generate pure, highly viable, and standardized motor neurons, we here set up a unique FACS technique. Using a Becton Dickinson ARIA II FACS sorter, we isolated motor neurons from lumbar spinal cords of embryonic Hb9:GFP mice (14) and seeded them with the built-in AutoClone system directly at predefined numbers on 24-, 96-, or 384-well culture plates (Fig. 1A and SI Appendix, Fig. S1A).
Fig. 1.
Screening neurotrophic factor combinations on FACS-isolated motor neurons. (A) FACS diagram (Upper) depicts FACS-isolation and culture of motor neurons from embryonic E12 Hb9:GFP mice. FACS profiles (Lower) show sequential gating of cells through forward-scatter area (FSC-A)/side scatter area (SSC-A), side scatter width (SSC-W)/side scatter height (SSC-H), and GFP fluorescence to isolate motor neurons (MN) (Right) from bulk cells (Left) and interneurons (Center). (B) Purity of FACS-isolated motor neurons. GFP/DIC images (Top) show cells before and after FACS. 20× objective. (Scale bar: 50 µm.) Immunofluorescence images (Middle and Bottom) show FACS-isolated motor neurons positive for GFP, neurofilament-M (NF-M), and ISL1/2. The diagram indicates that FACS-isolated cells are positive for motor neuron markers CHAT, SMI32, and ISL1/2 but negative for interneuron markers EN1 and CHX10 (mean ± SD, n = 4 replicates). (C) Yield and efficiency of FACS-based MN isolation. (D) Testing 66 NTF combinations on motor neuron survival reveals potentiated effects of pairwise combinations among HGF, ARTN, and CNTF. P < 0.0001 (HGF + CNTF, HGF + ARTN) and P < 0.04 (ARTN + CNTF) by Kruskal–Wallis test and Dunn’s post hoc test, n = 12 wells each, compared with the individual NTFs. Motor neuron survival at 3 DIV is expressed relative to the values for 12 NTFs (100%) and no NTF (0%). (E) Whole-well images showing motor neurons cultured for 3 DIV in the presence of the indicated NTFs. (F) Diagram showing strictly additive survival effects of HGF, ARTN, and CNTF in pairwise and triple combination (mean ± SD). Statistical significance was tested by Kruskal–Wallis test and Dunn’s post hoc test.
We show that the FACS-isolated cells are all strongly GFP positive and large sized, in contrast to the heterogeneous cell population before FACS (Fig. 1B); they express the motor neuron markers CHAT (100%), SMI 32 (100%), and ISL (95.2 ± 2.4%), but not the interneuron markers EN1 or CHX10 (Fig. 1B), and thus represent bona fide motor neurons of exquisite purity.
On a routine basis, we obtain ∼120,000 lumbar motor neurons per typical mouse litter within less than 90 min, including <20 min high-speed FACS at maximum flow rate (Fig. 1C and SI Appendix, Fig. S1B). The FACS-isolated motor neurons survive and grow well and in a highly reproducible manner in culture despite the rapid cell acceleration/deceleration and the strong electromagnetic fields encountered during FACS (Fig. 1B and SI Appendix, Fig. S1 A and B). These data attest the high yield, rapidity, and standardization of this technique.
Combinatorial Screening of Neurotrophic Factors.
To study the combinatorial effects of NTFs, we selected 12 commercially available NTFs belonging to different protein families, including the neurotrophins BDNF and NT3; the GDNF family members GDNF, Neurturin (NTN), ARTN, and Persephin (PSPN); and the cytokines CNTF, CT1, and LIF, as well as HGF, IGF1, and VEGF (3).
We first verified that all NTFs were fully biologically active by comparing side-by-side batches from different suppliers (SI Appendix, Fig. S2A). Each NTF was tested at its reported optimal concentration in chemically defined medium. FACS-isolated lumbar motor neurons were directly seeded into 96-well plates and their survival assessed by automated imaging analysis. Under these conditions, all NTFs significantly enhanced motor neuron survival. BDNF and GDNF were most effective, whereas PSPN, IGF1, and VEGF were least effective (SI Appendix, Fig. S2A). For each NTF, there was little variation between batches from different commercial suppliers, suggesting full biological activity of all NTFs (SI Appendix, Fig. S2A).
In mouse lumbar spinal cord, motor neurons undergo programmed cell death (PCD) between embryonic days E12.5 and E14.5 (15). To assay combinatorial NTF effects on lumbar motor neuron PCD, we isolated them before their PCD at E12, seeded them at a low density of 1,000 cells per well, and monitored their survival over 3 d in vitro (DIV). In the absence of any NTFs, the cultured motor neurons died very rapidly (SI Appendix, Fig. S2B); by contrast, they survived at a high rate in the presence of all NTFs, suggesting potentiation between some NTFs.
To screen the effects of the 66 pairwise NTF combinations, we designed a strict flowchart for experimentation and data analysis (SI Appendix, Fig. S3 A and B) (16). Three pairwise combinations of NTFs turned out to significantly potentiate motor neuron survival (mean ± SD): HGF + ARTN (40 ± 5%), HGF + CNTF (36 ± 6%), and CNTF + ARTN (22 ± 6%), in comparison with HGF (22 ± 7%), ARTN (10 ± 4%), and CNTF (8 ± 3%), all 12 NTFs (set 100%) or no NTFs (set 0%) (Fig. 1 D and E and SI Appendix, Fig. S3 C–E). Interestingly, no such potentiation was seen for HGF, ARTN, or CNTF in combination with the other nine factors (Fig. 1D and SI Appendix, Fig. S3 C–E).
We then found that the triple combination of HGF + ARTN + CNTF is even more effective than the three pairwise combinations and keeps alive 46 ± 5% of motor neurons (Fig. 1 E and F). Strikingly, this percentage is equivalent to the numerical sum of the effects by the three individual factors (Fig. 1F); it is also equivalent to the sum of the effects by one factor and the two others combined (Fig. 1F). These data indicate that HGF, ARTN, and CNTF act in a strictly additive manner, suggesting their action on distinct subsets of motor neurons.
Motor Neuron Subsets in Lumbar Spinal Cord.
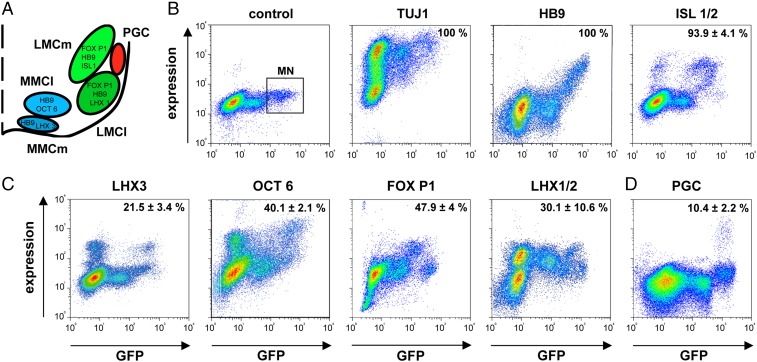
The lumbar spinal cord contains three major subsets of motor neurons that differ in their position, targets, and molecular markers (Fig. 2A) (17). Motor neurons of the medial motor column (MMC-MN) innervate axial body muscles and express the transcription factors OCT-6/SCIP-1 and LHX3 (MMCm). Motor neurons of the lateral motor column (LMC-MN) innervate hindlimb muscles and express FOX P1 and LHX 1/2 (LMCl) (18). Preganglionic motor neurons (PGC-MN) of the sympathetic and parasympathetic nervous system ensure the innervation of ganglionic neurons in pelvic organs and express nNOS, HNF-6, and low levels of FOX P1 (19).
Fig. 2.
Quantitation of motor neuron subsets by double-color flow cytometry. (A) Schematic depicting position and molecular markers of motor neuron subsets in mouse E12 lumbar spinal cord. (B) Density plots indicating the percentage of neurons (TUJ1) and motor neurons (HB9, ISL1/2) in lumbar ventral spinal cord of HB9:GFP mice analyzed by double-color flow cytometry. A representative control (Left) showing labeling with isotype-matched primary antibody and indicating the motor neuron population (MN). (C) Density plots indicating the percentage of MMC motor neurons (LHX3, OCT6) and LMC motor neurons (FOXP1, LHX1/2) among motor neurons. Note overlap of some markers in motor neuron subsets and expression of LHX3, OCT6, and LHX1/2 in subsets of interneurons. (D) Density plot indicating percentage of preganglionic (PGC) neurons after bulk retrograde labeling with tetramethylrhodamine-dextran from bladder. Percentages represent means ± SD of 3–4 independent experiments.
Before testing the potentially distinct effects of HGF, ARTN, and CNTF on these motor neuron subsets, we verified their percentages in the FACS single-cell suspensions. Double-color flow cytometry analysis confirmed that all GFP-positive motor neurons express the generic MN markers HB9 (100%) and ISL 1/2 (94 ± 4%, mean ± SD) (Fig. 2B), and 40 ± 2% of them express the MMC marker OCT 6 and 22 ± 3% the MMCm marker LHX3 (Fig. 2C), in line with the fraction of LHX3/HB9 MN in mouse spinal cord (20). However, 30.1 ± 10.6 are positive for the LMCl marker LHX1/2, and 48 ± 4% for the LMC/PGC marker FOX P1 (Fig. 2C).
Because reported PGC-specific antibodies were not suitable for flow cytometry, we identified PGC-MN by bulk retrograde labeling from bladder using fluorescent tetramethylrhodamine-dextran, yielding ∼10 ± 2% labeled cells (Fig. 2D), in keeping with the 1:4 ratio of lumbar PGC-MN to LMC-MN (18).
These data demonstrate that LMC-MN, MMC-MN, and PGC-MN are present in single-cell FACS preparations at similar percentages as in vivo, warranting analysis of their responses to neurotrophic factors.
Distinct Effects of HGF, CNTF, and ARTN on Motor Neuron Subsets.
There are currently no established methods to specifically isolate and culture MMC-MN, LMC-MN, or PGC-MN from rodents. To overcome this issue, we isolated lumbar MMC-MN from LMC-MN and PGC-MN by anatomic dissection and FACS (Experimental Procedures). Using gene expression profiling on microarrays, we demonstrated that MMC-MN show strong up-regulation of the MMC markers Lhx3, Lhx4, and Oct6/Pou3f1, whereas LMC/PGC-MN show strong up-regulation of the LMC markers Raldh2 and Lhx1, and the PGC markers Nos1 (nNos), Hnf6, and Smad1 (SI Appendix, Fig. S4A). Global gene expression profiles were highly correlated between independent replicate samples (MMC-MN: R = 0.9924; LMC/PGC-MN: R = 0.9899; SI Appendix, Fig. S4B). We further verified that in culture, MMC-MN and LMC/PGC-MN have a similar survival rate in the presence of all 12 NTFs and undergo rapid indistinguishable cell death in the absence of any NTF (SI Appendix, Fig. S5A).
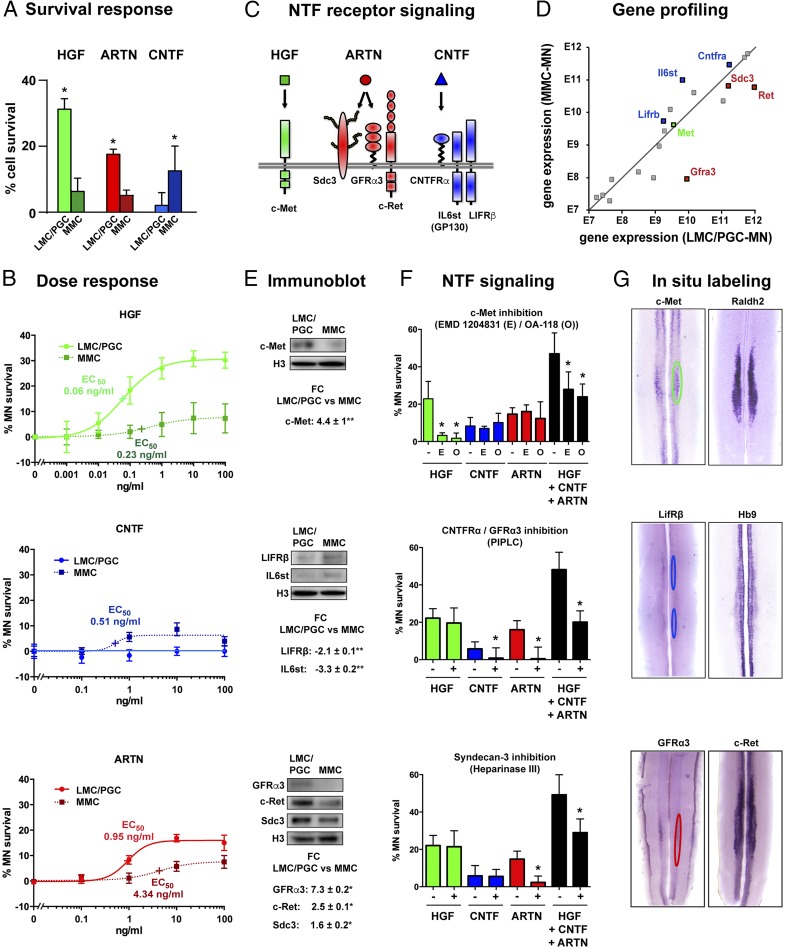
The strong enrichment, high standardization, and similar global survival response of FACS-isolated MMC-MN and LMC/PGC-MN enabled us to compare the effects of individual NTFs. We found that HGF and ARTN support the survival of a large fraction of LMC/PGC-MN (HGF: 30 ± 3%; ARTN: 17 ± 1%) but have a minor effect on MNC-MN (Fig. 3A). By contrast, CNTF supports the survival of 12 ± 7% of MMC-MN but has no effect on LMC/PGC-MN (Fig. 3A). These differential effects of HGF, CNTF, and ARTN were confirmed by dose–response curves showing significant differences in their semimaximal effective concentrations (EC50) for each MN subset (Fig. 3B). No differential effects were seen with the other nine NTFs (SI Appendix, Fig. S5B). HGF and ARTN thus preferentially promote the survival of LMC/PGC-MN, whereas CNTF selectively promotes the survival of MMC-MN.
Fig. 3.
Characterization of motor neurons responsive to HGF, CNTF, and ARTN. (A) Distinct survival responses of LMC/PGC-MN and MMC-MN to HGF, ARTN, and CNTF (mean ± SD, *P < 0.05, Mann–Whitney test). (B) Dose–response curves. Semimaximal effective concentrations (EC50) of HGF and ARTN are 3.8× and 4.5× lower, respectively, for LMC/PGC-MN than for MMC-MN. EC50 95% confidence intervals do not overlap (not shown). (C) Schematic showing HGF, ARTN, and CNTF receptor complexes. (D) Gene expression profiling of MN subsets. Note up-regulation of Lifrb (1.4-fold, P < 0.03) and Il6st/Gp130 (2.3-fold, P < 0.0003) in MMC-MN and of Gfra3 (fourfold, P < 0.0006), Ret (2.3-fold, P < 0.0003), and Sdc3 (1.3-fold, P < 0.008) in LMC/PGC-MN. No differential gene expression was seen for Met (in gray) and the receptors of the nine other NTFs (in gray). Student’s t test, n = 3 independent sample pairs. (E) Immunoblot showing differential expression (FC, fold change) of HGF, CNTF, and ARTN receptor components in LMC/PGC-MN vs. MMC-MN. Histone H3 indicates equal loading. **P < 0.0003, *P < 0.03, Mann–Whitney test, n = 4 independent blots each. (F) NTF signaling. Inhibition of c-Met (Top) with the c-Met kinase inhibitor EMD 1204831 (E, 100 nM) reduces survival of HGF-responsive MN by 21% and of HGF + CNTF + ARTN responsive MN by 19%. The c-Met neutralizing antibody OA-118 (O, 0.5 µg/mL) has similar effects. Inhibition of CNTFRα and GFRα3 signaling (Middle) with PIPLC specifically blocks the effects of CNTF and ARTN. Inhibition of the ARTN coreceptor Sdc3 (Bottom) with Heparinase III specifically affects survival of ARTN-responsive MN. Mean ± SD, six wells per condition. *Kruskal–Wallis and Dunn’s post hoc test. Cell survival is expressed relative to values for all NTFs (100%) or no NTF (0%). (G) In situ labeling in E12 lumbar spinal cord. c-Met–positive MN (Top) are located in the LMC labeled by Raldh2. Lifrβ-positive MN (Middle) are located in the MMC identified by strong Hb9 mRNA expression. Gfrα3-positive MN (Bottom) are positioned in the lower lumbar and sacral spinal cord and represent a fraction of c-Ret-positive neurons.
Differential Expression of the HGF, CNTF, and ARTN Receptors in Motor Neuron Subsets.
The differential survival response of LMC/PGC-MN vs. MMC-MN to HGF, ARTN, and CNTF prompted us to analyze their expression of NTF receptors (Fig. 3C) by microarray and Western blot analyses.
HGF is known to trigger survival through the transmembrane receptor c-Met (Fig. 3C). We found that c-Met is not differentially regulated at the mRNA level (Fig. 3D) but increased by 4.4 ± 0.54-fold (mean ± SD) at the protein level in LMC/PGC-MN compared with MMC-MN (Fig. 3E, Top), in agreement with the preferential response of LMC/PGC-MN to HGF.
The CNTF receptor comprises the CNTFRα receptor and the two signal-transducing components LIFRβ and IL6st (GP130) (Fig. 3C). LIFRβ and GP130 are significantly up-regulated in MMC-MN both at the mRNA (Fig. 3D) and the protein level (Fig. 3E, Middle), in line with the specific response of MMC-MN to CNTF (Fig. 3 A and B). Interestingly, CNTFRα is not differentially regulated, potentially reflecting the presence of soluble forms signaling in trans (21).
ARTN signals through a receptor complex composed by GFRα3, the proteoglycan coreceptor Syndecan-3 (Sdc3), and the signal transducing tyrosine kinase c-Ret (Fig. 3C). We found that GFRα3, Sdc3, and c-Ret are all strongly up-regulated in LMC/PGC-MN at the mRNA (Fig. 3D) and the protein level (Fig. 3E, Bottom), which matches the preferential response of LMC/PGC-MN to ARTN. No such differential regulation was seen for the receptors of the other NTFs tested (Fig. 3D).
HGF/c-Met Signaling Promotes Survival of Limb-Innervating LMC Motor Neurons.
To characterize the motor neuron subsets supported by HGF, ARTN, and CNTF, we used specific inhibitors to block NTF receptor-mediated survival signaling in culture (Fig. 3F) and analyzed the expression of the corresponding NTF receptors in E12 lumbar spinal cord (Fig. 3G).
To analyze HGF/c-Met survival signaling in lumbar motor neurons, we used EMD 1204831, a c-Met tyrosine kinase inhibitor with minimal off-target effects (22). We found that EMD 1204831 (100 nM) completely abrogates the survival effects of HGF and partially those of HGF + ARTN + CNTF but not those of ARTN or CNTF alone (Fig. 3F, Top). Similar effects were seen with OA-118, a monovalent antibody that neutralizes c-Met signaling (Fig. 3F, Top and SI Appendix, Supplemental Material and Methods).
To localize the HGF-responsive lumbar motor neurons, we performed whole-mount in situ hybridizations and detected the c-Met positive motor neurons almost exclusively in the LMC identified by the LMC marker Raldh2 (Fig. 3G, Top). The c-Met–positive MN form well-delineated groups potentially corresponding to individual motor pools innervating limb muscles (Fig. 3G, Top). Few c-Met–positive motor neurons are also present in the adjacent thoracic and sacral spinal cord, in line with an earlier report (23). Taken together, these data indicate that HGF specifically promotes the survival of a fraction of hindlimb-innervating LMC motor neurons through activation of the c-Met tyrosine kinase receptor.
CNTF/CNTFRα Signaling Supports Subsets of Axial Muscle-Innervating MMC Motor Neurons.
We then blocked CNTF signaling by using phosphoinositol phospholipase C (PIPLC), an enzyme that cleaves the phosphoinositol side chain of the CNTFRα receptor necessary for its plasma membrane insertion (24); this completely blocks the survival of CNTF-dependent motor neurons but has no effect on the survival of HGF-dependent motor neurons (Fig. 3F, Middle).
We localized the CNTF-responsive motor neurons by whole-mount in situ hybridization of Lifrβ (Fig. 3G, Middle), which revealed two distinct small MN subsets in the MMC identified by strong Hb9 mRNA expression. We conclude that CNTF specifically promotes the survival of two MMC-MN subsets by activating survival signaling triggered by CNTFRα and, presumably, LIFRβ/GP130.
ARTN Acts as a Survival Factor for Parasympathetic Preganglionic Motor Neurons Through GFRα3/Sdc3 Signaling.
ARTN has not been previously recognized as a neurotrophic factor for motor neurons. To analyze ARTN survival signaling, we used Heparinase III, which cleaves the proteoglycan side chains of the ARTN coreceptor Sdc3 (25). We show that Heparinase III almost completely blocks the survival of motor neurons depending on ARTN (Fig. 3F, Bottom). Similar effects were seen with PIPLC (Fig. 3F, Middle), which cleaves the phosphoinositol chain of GFRα3 (26), and with antibodies against the extracellular domain of GFRα3 (SI Appendix, Fig. S6). ARTN-mediated survival effects were not inhibited with neutralizing antibodies against GFRα1 (SI Appendix, Fig. S6), the preferred α-receptor of GDNF (26), indicating specificity.
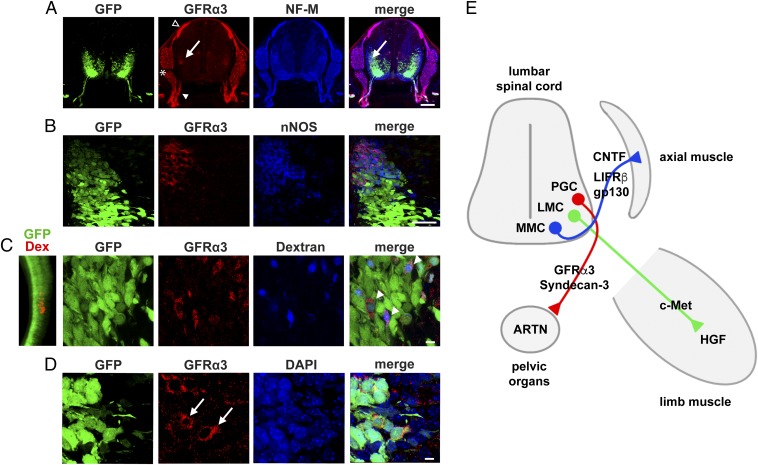
To localize the ARTN-responsive motor neurons, we performed whole-mount immunolabeling, demonstrating that GFRα3-positive cells represent a fraction of c-Ret–positive cells (Fig. 3G, Bottom). The GFRα3-positive cells are localized in a tiny column that extends from lumbar L5 to sacral S2 segments (Fig. 3G, Bottom) at a lateral spinal cord position (Fig. 4A) reminiscent of parasympathetic preganglionic (PGC) motor neurons innervating urogenital sphincter organs; to confirm this, we demonstrated that GFRα3-positive cells coexpress the PGC marker nNOS (Fig. 4B) and fluorescent dextrans retrogradely bulk injected into bladder (Fig. 4C). As expected for an NTF receptor, GFRα3 is expressed in PGC cell bodies (Fig. 4D) and ventral root axons labeled by neurofilament M (Fig. 4A).
Fig. 4.
GFRα3 expression in parasympathetic preganglionic motor neurons. (A) Overview images showing position of GFRα3-positive motor neurons colabeled for NF-M in lower lumbar spinal cord of E12 Hb9:GFP mice. Cell bodies are indicated by arrow and ventral root axons by filled arrowhead. GFRα3 is also strongly expressed in some DRG neurons (asterisk) and the dorsal root entry zone (open arrowhead). (B) Images showing GFRα3 expression in PGC motor neurons identified by nNOS. (C) (Left) Lumbosacral spinal cord after retrograde bulk labeling with tetramethylrhodamine-dextran from developing bladder. (Right) GFRα3-positive cells retrogradely labeled from bladder with Alexa 647-dextran (arrowheads in merge). (D) GFRα3 expression in cell bodies of PGC motor neurons (arrows). Nuclei are labeled with DAPI. (Scale bars: A, 100 µm; B, 50 µm; C and D, 10 µm.) (E) Schematic indicating a close match between the expression of NTF receptors in lumbar LMC, MMC, and parasympathetic PGC motor neurons and the expression of the NTF ligands in the corresponding target tissues and organs.
Taken together, these data identify ARTN as a survival factor for parasympathetic preganglionic motor neurons through activation of GFRα3/Sdc3 signaling.
Discussion
In sum, using a combinatorial screening on highly standardized motor neuron cultures, we here identify previously unrecognized survival effects of the three neurotrophic factors HGF, CNTF, and ARTN on distinct subsets of motor neurons in the lumbar spinal cord.
We demonstrate that HGF almost exclusively supports hindlimb-innervating LMC motor neurons through activation of c-Met kinase (Fig. 3 D–F). Remarkably, the strong up-regulation of the receptor c-Met in hindlimb motor neurons (Fig. 3G) perfectly matches the restricted expression of its ligand HGF in its target muscles in the limb (27, 28), which nicely illustrates the neurotrophic theory. The effects of HGF on limb-innervating motor neurons may also underlie its strong therapeutic potency in mutant SOD1 ALS model mice, where it reduces limb muscle paralysis and increases limb muscle weight (29).
The specific action of CNTF on subsets of axial MMC-MN was more surprising but is in line with its reported potent effects on MMC-MN in spinal cord slice cultures (ref. 30, figure 8) and its weak effects on axotomized neonatal LMC-MN (10). According to our data, the CNTF-responsive MMC motor neurons represent less than 10% of total at embryonic stage E12 (Figs. 1D and 3 A and B). This finding contrasts with the massive loss of motor neurons (33–40%) in knockout mice for the CNTF receptor genes Lifrb, gp130, or Cntfra around birth (3), suggesting additional roles for CNTF or CNTF-like cytokines during late embryonic development.
ARTN has been previously shown to support peripheral sympathetic neurons (26, 31). We here identify ARTN as a survival factor for parasympathetic preganglionic motor neurons in the lumbosacral spinal cord (Fig. 4 A–D), which connect to ganglionic neurons in distal colon, bladder, and genital sphincter organs (32, 33). Consistent with this novel role, ARTN is expressed in these target organs (34); its receptor GFRα3 is expressed in the axons and cell bodies of PGC-MN during development (Fig. 4 A and D) and adulthood (33); and transgenic ARTN causes hyperplasia of parasympathetic nerves (34).
Our findings on HGF, CNTF, and ARTN leave open potential combinatorial effects of other NTFs. Indeed, several NTF combinations, including BDNF + GDNF, tended to be effective in our screen without, however, reaching statistical significance (SI Appendix, Fig. S3 C–E). Furthermore, all NTFs were assayed here at their reported optimal concentration, which favors additive rather than synergistic effects. This may explain why GDNF + CT1, which synergize on motor neurons at low concentration (11), failed to do so at high concentration (SI Appendix, Fig. S3 C–E). Similarly, the design of our screen may have missed the detection of sequential NTF effects—in particular, of NTFs with short half-lives, because NTFs were only added once at the start of the cultures. Finally, several of the NTFs tested here not only promote motor neuron survival but also favor axon and dendrite growth as well as synapse formation (3, 4). These considerations warrant further analyses on combinatorial NTF effects in developing motor neurons.
Are screens on cultured motor neurons to some extent predictive for clinical trials in human motor neuron disease? Published studies with small molecule compounds provide first hints. Indeed, both Olesoxime (TRO19622) (35) and Kenpaullone (36) have been identified in survival screens on neurotrophic factor-deprived motor neurons. Olesoxime, a cholesterol-like molecule, showed positive effects in human spinal muscular atrophy (SMA) (37) but was ineffective in ALS (38). Kenpaullone, an inhibitor of the phospho c-Jun–mediated apoptotic pathway, was reported to be effective on motor neurons carrying mutations in the human ALS genes SOD1 (36) or FUS (39).
Most NTFs have failed in past clinical trials (40), and this failure was widely attributed to delivery problems of the recombinant proteins. In preclinical ALS and SMA models, however, several NTFs have provided substantial therapeutic benefit when delivered by gene- or cell-based systems (13, 41, 42). Importantly, a metaanalysis of 226 studies in mutant SOD1 mice (5) has identified four NTFs among the six most-effective treatments (apart from SOD1 modifiers), which prolonged the animal’s survival by >30%.
It should however be stressed that the response of postnatal motor neurons to NTFs is largely unknown and potentially differs from that of developing motor neurons (our study). Ongoing research efforts thus aim to isolate disease-relevant motor neuron subsets at a postnatal stage to determine their NTF receptor profile and to compare their survival response to NTFs (43, 44). These types of studies may help to tailor future experimental NTF therapies to those motor neurons that are preferentially affected in each form of motor neuron disease, i.e., limb-innervating motor neurons in classical ALS, proximal motor neurons in SMA, and brainstem motor neurons in bulbar ALS or spinobulbar muscular atrophy (4, 45).
Experimental Procedures
Mice.
Transgenic Hb9:GFP mice [line mHB9-Gfp1b (14), kindly provided by T. Jessell] were maintained on a C57/BL6 background and mated with female CD1 mice (Charles River). All experiments with mice were performed in strict compliance with institutional and national guidelines and approved by Marseille’s Ethics Committee No. 71.
FACS and Cell Culture.
Lumbar spinal cords were dissected from E12 Hb9:GFP embryos, and longitudinal ventral spinal cord segments Th13 to S1 isolated under a Leica MZ16FA fluorescence stereomicroscope. Single-cell suspensions were prepared as described (46), resuspended in a 1:1 (vol/vol) mix of L15 medium:Facsflow, and subjected to FACS with an ARIA II SORP (Becton Dickinson). Cells were isolated at a sheath pressure of 45 pounds/psi through an 85-µm nozzle, seeded with the AutoClone system in culture wells coated with polyornithin/laminin, and cultured in supplemented Neurobasal medium without riboflavin.
Screening NTF Combinations.
Effects of pairwise NTF combinations were assayed in a two-step test/retest procedure (SI Appendix, Fig. S3 A and B). Survival of calcein-positive motor neurons was assessed by imaging entire wells with an inverted microscope (DMI 4000B, Leica, 1.25× objective, DX360 camera). Images were processed with ImageJ software and analyzed by an experimenter masked to the experimental conditions.
Flow Cytometry.
Single-cell suspensions were taken up in DMEM/F12, fixed for 15 min on ice in 2% (wt/vol) formaldehyde in PBS, washed twice in PBS 0.1% BSA, and permeabilized for 15 min on ice in PBS 0.5% saponin (wt/vol). Flow cytometry was performed essentially as described (47). Cell suspensions were incubated overnight at 4 °C with primary antibodies against Hb9, Isl 1/2, Lhx 1/2, Lhx3, Foxp1, Oct6 (SI Appendix, Table 2) or with control antibodies, washed twice, and incubated for 45 min at 4 °C with secondary antibodies. After two washes, cells were resuspended in PBS 1% BSA, analyzed with a FACS ARIA II SORP or a FACS ARIA III and data plotted with FlowJo software (Tree Star).
Statistical Analyses.
Each experiment was performed with several biological replicates (see figure legends) and repeated at least twice. Data were analyzed with GraphPad Prism 6 (GraphPad). Data from two groups showing Gaussian distribution were analyzed with Student's t test two-tailed; otherwise, the Mann–Whitney test was used. Data from more than two groups were analyzed with the Kruskal–Wallis test and Dunn post hoc test. Dose–response curves, EC50 values, and 95% confidence intervals were computed using a nonlinear fit model with variable slope. Cytometry data were tested for significance with the χ2 test using FlowJo software.
SI Appendix provides experimental details on gene expression profiling, immunoblot analyses, whole-mount labeling, immunohistochemistry, retrograde labeling, and signaling studies.
Supplementary Material
Acknowledgments
We acknowledge the contribution of A. Gonté and E. Buhler as well as Drs. D. Bohl and S. Blanchard to an early stage of this work. We thank Drs. F. Tell for expert advice on statistical data analysis; S. Zaffran for logistic help with in situ hybridization; Prof. Michael Schaefer for expert help with gene expression analyses; Drs. P. Zimmermann, S. Driegen, and F. Clotman for providing antibodies; F. Bladt for providing the c-Met tyrosine kinase inhibitor EMD 1204831; Drs. F. Morel and J. L. Boisneault for continuous support; Drs. G. P. Schiavo and C. Rabouille for critical reading of the manuscript; and the reviewers for their incisive comments. This study was financed by grants from Association Française contre les Myopathies (AFM), Agence Nationale pour la Recherche (ANR), ERANET Neuron, INSERM, and Aix-Marseille University. S.S. received salary support from ANR; D.B. received a PhD fellowship from Fondation pour la Recherche Médicale (FRM), AFM, and ANR; A.A. received salary support from ANR; and A.J. was supported by PhD fellowships from INSERM/Région Provence Alpes Côtes d’Azur and FRM.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data of FACS-isolated motor neurons reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE86478).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615372114/-/DCSupplemental.
References
- 1.Purves D. Body and Brain: A Trophic Theory of Neural Development. Harvard Univ Press; Cambridge, MA: 1986. [Google Scholar]
- 2.Hamburger V. The developmental history of the motor neuron. The F.O. Schmitt lecture in neuroscience. Neurosci Res Program Bull. 1977;15(Suppl.):1–37. doi: 10.1007/978-1-4899-6743-5_5. [DOI] [PubMed] [Google Scholar]
- 3.Gould TW, Enomoto H. Neurotrophic modulation of motor neuron development. Neuroscientist. 2009;15(1):105–116. doi: 10.1177/1073858408324787. [DOI] [PubMed] [Google Scholar]
- 4.Kanning KC, Kaplan A, Henderson CE. Motor neuron diversity in development and disease. Annu Rev Neurosci. 2010;33:409–440. doi: 10.1146/annurev.neuro.051508.135722. [DOI] [PubMed] [Google Scholar]
- 5.Turner BJ, Talbot K. Transgenics, toxicity and therapeutics in rodent models of mutant SOD1-mediated familial ALS. Prog Neurobiol. 2008;85(1):94–134. doi: 10.1016/j.pneurobio.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Riethmacher D, et al. Severe neuropathies in mice with targeted mutations in the ErbB3 receptor. Nature. 1997;389(6652):725–730. doi: 10.1038/39593. [DOI] [PubMed] [Google Scholar]
- 7.Grieshammer U, Lewandoski M, Prevette D, Oppenheim RW, Martin GR. Muscle-specific cell ablation conditional upon Cre-mediated DNA recombination in transgenic mice leads to massive spinal and cranial motoneuron loss. Dev Biol. 1998;197(2):234–247. doi: 10.1006/dbio.1997.8859. [DOI] [PubMed] [Google Scholar]
- 8.Vicario-Abejón C, Fernández-Moreno C, Pichel JG, de Pablo F. Mice lacking IGF-I and LIF have motoneuron deficits in brain stem nuclei. Neuroreport. 2004;15(18):2769–2772. [PubMed] [Google Scholar]
- 9.Holtmann B, et al. Triple knock-out of CNTF, LIF, and CT-1 defines cooperative and distinct roles of these neurotrophic factors for motoneuron maintenance and function. J Neurosci. 2005;25(7):1778–1787. doi: 10.1523/JNEUROSCI.4249-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vejsada R, Sagot Y, Kato AC. Quantitative comparison of the transient rescue effects of neurotrophic factors on axotomized motoneurons in vivo. Eur J Neurosci. 1995;7(1):108–115. doi: 10.1111/j.1460-9568.1995.tb01025.x. [DOI] [PubMed] [Google Scholar]
- 11.Arce V, et al. Synergistic effects of Schwann- and muscle-derived factors on motoneuron survival involve GDNF and cardiotrophin-1 (CT-1) J Neurosci. 1998;18(4):1440–1448. doi: 10.1523/JNEUROSCI.18-04-01440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitsumoto H, et al. Arrest of motor neuron disease in wobbler mice cotreated with CNTF and BDNF. Science. 1994;265(5175):1107–1110. doi: 10.1126/science.8066451. [DOI] [PubMed] [Google Scholar]
- 13.Haase G, et al. Gene therapy of murine motor neuron disease using adenoviral vectors for neurotrophic factors. Nat Med. 1997;3(4):429–436. doi: 10.1038/nm0497-429. [DOI] [PubMed] [Google Scholar]
- 14.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110(3):385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto Y, Henderson CE. Patterns of programmed cell death in populations of developing spinal motoneurons in chicken, mouse, and rat. Dev Biol. 1999;214(1):60–71. doi: 10.1006/dbio.1999.9413. [DOI] [PubMed] [Google Scholar]
- 16.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24(2):167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 17.Dasen JS, Jessell TM. Hox networks and the origins of motor neuron diversity. Curr Top Dev Biol. 2009;88:169–200. doi: 10.1016/S0070-2153(09)88006-X. [DOI] [PubMed] [Google Scholar]
- 18.Rousso DL, Gaber ZB, Wellik D, Morrisey EE, Novitch BG. Coordinated actions of the forkhead protein Foxp1 and Hox proteins in the columnar organization of spinal motor neurons. Neuron. 2008;59(2):226–240. doi: 10.1016/j.neuron.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francius C, Clotman F. Dynamic expression of the Onecut transcription factors HNF-6, OC-2 and OC-3 during spinal motor neuron development. Neuroscience. 2010;165(1):116–129. doi: 10.1016/j.neuroscience.2009.09.076. [DOI] [PubMed] [Google Scholar]
- 20.Jung H, et al. Evolving Hox activity profiles govern diversity in locomotor systems. Dev Cell. 2014;29(2):171–187. doi: 10.1016/j.devcel.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis S, et al. Released form of CNTF receptor alpha component as a soluble mediator of CNTF responses. Science. 1993;259(5102):1736–1739. doi: 10.1126/science.7681218. [DOI] [PubMed] [Google Scholar]
- 22.Bladt F, et al. EMD 1214063 and EMD 1204831 constitute a new class of potent and highly selective c-Met inhibitors. Clin Cancer Res. 2013;19(11):2941–2951. doi: 10.1158/1078-0432.CCR-12-3247. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto Y, et al. Hepatocyte growth factor (HGF/SF) is a muscle-derived survival factor for a subpopulation of embryonic motoneurons. Development. 1997;124(15):2903–2913. doi: 10.1242/dev.124.15.2903. [DOI] [PubMed] [Google Scholar]
- 24.Davis S, et al. The receptor for ciliary neurotrophic factor. Science. 1991;253(5015):59–63. doi: 10.1126/science.1648265. [DOI] [PubMed] [Google Scholar]
- 25.Bespalov MM, et al. Heparan sulfate proteoglycan Syndecan-3 is a novel receptor for GDNF, Neurturin, and Artemin. J Cell Biol. 2011;192(1):153–169. doi: 10.1083/jcb.201009136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baloh RH, et al. Artemin, a novel member of the GDNF ligand family, supports peripheral and central neurons and signals through the GFRalpha3-RET receptor complex. Neuron. 1998;21(6):1291–1302. doi: 10.1016/s0896-6273(00)80649-2. [DOI] [PubMed] [Google Scholar]
- 27.Ebens A, et al. Hepatocyte growth factor/scatter factor is an axonal chemoattractant and a neurotrophic factor for spinal motor neurons. Neuron. 1996;17(6):1157–1172. doi: 10.1016/s0896-6273(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 28.Sonnenberg E, Meyer D, Weidner KM, Birchmeier C. Scatter factor/hepatocyte growth factor and its receptor, the c-met tyrosine kinase, can mediate a signal exchange between mesenchyme and epithelia during mouse development. J Cell Biol. 1993;123(1):223–235. doi: 10.1083/jcb.123.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun W, Funakoshi H, Nakamura T. Overexpression of HGF retards disease progression and prolongs life span in a transgenic mouse model of ALS. J Neurosci. 2002;22(15):6537–6548. doi: 10.1523/JNEUROSCI.22-15-06537.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rakowicz WP, Staples CS, Milbrandt J, Brunstrom JE, Johnson EM., Jr Glial cell line-derived neurotrophic factor promotes the survival of early postnatal spinal motor neurons in the lateral and medial motor columns in slice culture. J Neurosci. 2002;22(10):3953–3962. doi: 10.1523/JNEUROSCI.22-10-03953.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honma Y, et al. Artemin is a vascular-derived neurotropic factor for developing sympathetic neurons. Neuron. 2002;35(2):267–282. doi: 10.1016/s0896-6273(02)00774-2. [DOI] [PubMed] [Google Scholar]
- 32.Park MJ, Chung K. Endogenous lectin (RL-29) expression of the autonomic preganglionic neurons in the rat spinal cord. Anat Rec. 1999;254(1):53–60. doi: 10.1002/(SICI)1097-0185(19990101)254:1<53::AID-AR7>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 33.Forrest SL, Payne SC, Keast JR, Osborne PB. Peripheral injury of pelvic visceral sensory nerves alters GFRα (GDNF family receptor alpha) localization in sensory and autonomic pathways of the sacral spinal cord. Front Neuroanat. 2015;9:43. doi: 10.3389/fnana.2015.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolon B, et al. The candidate neuroprotective agent Artemin induces autonomic neural dysplasia without preventing peripheral nerve dysfunction. Toxicol Pathol. 2004;32(3):275–294. doi: 10.1080/01926230490431475. [DOI] [PubMed] [Google Scholar]
- 35.Bordet T, et al. Identification and characterization of cholest-4-en-3-one, oxime (TRO19622), a novel drug candidate for amyotrophic lateral sclerosis. J Pharmacol Exp Ther. 2007;322(2):709–720. doi: 10.1124/jpet.107.123000. [DOI] [PubMed] [Google Scholar]
- 36.Yang YM, et al. A small molecule screen in stem-cell-derived motor neurons identifies a kinase inhibitor as a candidate therapeutic for ALS. Cell Stem Cell. 2013;12(6):713–726. doi: 10.1016/j.stem.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finkel R, Bertini E, Muntoni F, Mercuri E. 209th ENMC International Workshop: Outcome Measures and Clinical Trial Readiness in Spinal Muscular Atrophy 7-9 November 2014, Heemskerk, The Netherlands. Neuromuscul Disord. 2015;25(7):593–602. doi: 10.1016/j.nmd.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 38.Lenglet T, et al. Mitotarget study group A phase II-III trial of olesoxime in subjects with amyotrophic lateral sclerosis. Eur J Neurol. 2014;21(3):529–536. doi: 10.1111/ene.12344. [DOI] [PubMed] [Google Scholar]
- 39.Liu ML, Zang T, Zhang CL. Direct lineage reprogramming reveals disease-specific phenotypes of motor neurons from human ALS patients. Cell Reports. 2016;14(1):115–128. doi: 10.1016/j.celrep.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gould TW, Oppenheim RW. Motor neuron trophic factors: Therapeutic use in ALS? Brain Res Brain Res Rev. 2011;67(1-2):1–39. doi: 10.1016/j.brainresrev.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sendtner M, et al. Ciliary neurotrophic factor prevents degeneration of motor neurons in mouse mutant progressive motor neuronopathy. Nature. 1992;358(6386):502–504. doi: 10.1038/358502a0. [DOI] [PubMed] [Google Scholar]
- 42.Lesbordes JC, et al. Therapeutic benefits of cardiotrophin-1 gene transfer in a mouse model of spinal muscular atrophy. Hum Mol Genet. 2003;12(11):1233–1239. doi: 10.1093/hmg/ddg143. [DOI] [PubMed] [Google Scholar]
- 43.Comley L, et al. Motor neurons with differential vulnerability to degeneration show distinct protein signatures in health and ALS. Neuroscience. 2015;291:216–229. doi: 10.1016/j.neuroscience.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 44.Ozdinler PH, Macklis JD. IGF-I specifically enhances axon outgrowth of corticospinal motor neurons. Nat Neurosci. 2006;9(11):1371–1381. doi: 10.1038/nn1789. [DOI] [PubMed] [Google Scholar]
- 45.Swinnen B, Robberecht W. The phenotypic variability of amyotrophic lateral sclerosis. Nat Rev Neurol. 2014;10(11):661–670. doi: 10.1038/nrneurol.2014.184. [DOI] [PubMed] [Google Scholar]
- 46.Raoul C, et al. Motoneuron death triggered by a specific pathway downstream of Fas: Potentiation by ALS-linked SOD1 mutations. Neuron. 2002;35(6):1067–1083. doi: 10.1016/s0896-6273(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 47.Toli D, et al. Modeling amyotrophic lateral sclerosis in pure human iPSc-derived motor neurons isolated by a novel FACS double selection technique. Neurobiol Dis. 2015;82:269–280. doi: 10.1016/j.nbd.2015.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.