Abstract
Background: Giant-Skipper butterflies from the genus Megathymus are North American endemics. These large and thick-bodied Skippers resemble moths and are unique in their life cycles. Grub-like at the later stages of development, caterpillars of these species feed and live inside yucca roots. Adults do not feed and are mostly local, not straying far from the patches of yucca plants.
Methods: Pieces of muscle were dissected from the thorax of specimens and genomic DNA was extracted (also from the abdomen of a specimen collected nearly 60 years ago). Paired-end libraries were prepared and sequenced for 150bp from both ends. The mitogenomes were assembled from the reads followed by a manual gap-closing procedure and a phylogenetic tree was constructed using a maximum likelihood method from an alignment of the mitogenomes.
Results: We determined mitogenome sequences of nominal subspecies of all five known species of Megathymus and Agathymus mariae to confidently root the phylogenetic tree. Pairwise sequence identity indicates the high similarity, ranging from 88-96% among coding regions for 13 proteins, 22 tRNAs and 2 rRNA, with a gene order typical for mitogenomes of Lepidoptera. Phylogenetic analysis confirms that Giant-Skippers (Megathymini) originate within the subfamily Hesperiinae and do not warrant a subfamily rank. Genus Megathymus is monophyletic and splits into two species groups. M. streckeri and M. cofaqui caterpillars feed deep in the main root system of yucca plants and deposit frass underground. M. ursus, M. beulahae and M. yuccae feed in the yucca caudex and roots near the ground, and deposit frass outside through a "tent" (a silk tube projecting from the center of yucca plant). M. yuccae and M. beulahae are sister species consistently with morphological similarities between them.
Conclusions: We constructed the first DNA-based phylogeny of the genus Megathymus from their mitogenomes. The phylogeny agrees with morphological considerations.
Keywords: phylogeny, mitochondria, sequence assembly, Hesperiidae, Megathymini
Giant-Skippers (Lepidoptera: Hesperiidae: Megathymini) are large, fat-bodied butterflies endemic to the North American continent 1– 3. Their caterpillars adapted to feeding inside large roots and fleshy leaves of Yucca and Agave plants and relatives. Protected from many predators living within their nutrition-rich food sources, Megathymini are larger in size than most other skippers, and don't feed as adults. Genus Megathymus is characterized by root-feeding caterpillars, mostly in Yucca plants, that build a "tent" (a silk tube projecting above the ground) at least prior to pupation. Caterpillars of the genus Agathymus live inside Agave leaves and make a "trap-door" (a round, hardened disk of silk) to close the entrance to their leaf chamber before pupation.
To better understand the evolution and phylogeny of Megathymus, we sequenced complete mitogenomes of all five known species from the genus: M. yuccae, M. beulahae, M. ursus, M. streckeri, and M. cofaqui ( http://www.butterfliesofamerica.com/L/Hesperiidae.htm). For most species, nominotypical subspecies from or near the type localities were used (see Figure 1 for specimen data; collected under the permit #08-02Rev). M. beulahae specimen, male paratype, was from the National Museum of Natural History collection (Smithsonian Institution, Washington, DC, USA). To confidently root the Megathymus tree, we also sequenced a complete mitogenome of Agathymus mariae as an outgroup. Methods for genomic DNA extraction, library construction, next-generation sequencing, and computational procedures followed those we reported previously 4– 14. The sequences have been deposited in GenBank and received accessions KY630500–KY630505.
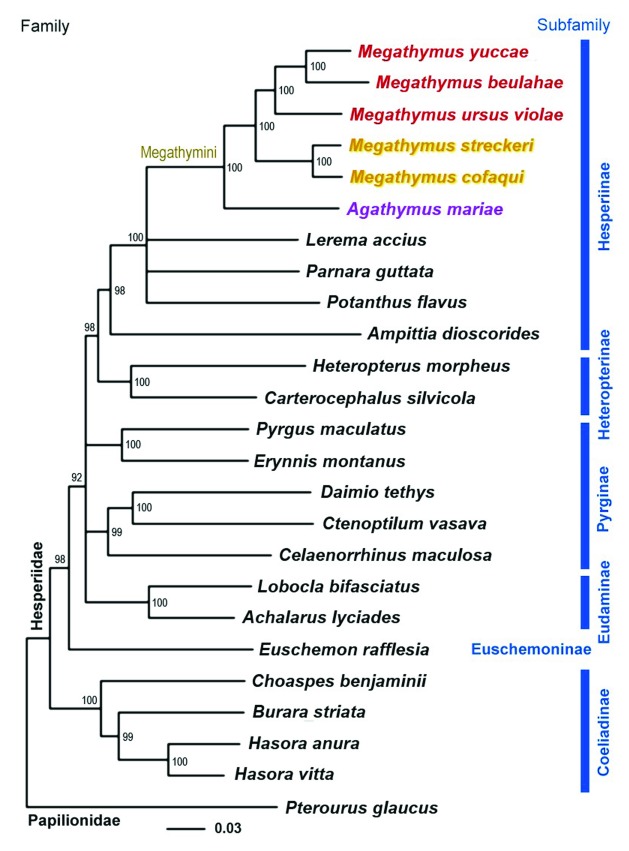
Figure 1. Maximum likelihood tree of complete mitogenomes of 24 Hesperiidae species rooted with Pterourus glaucus (Papilionidae).
Species names for mitogenome reported here are colored red. Numbers by the nodes show bootstrap support values and branches; bootstraps less than 70% are collapsed. GenBank accessions for sequences and data for specimens with mitogenomes reported here are: Achalarus lyciades NC_030602.1; Agathymus mariae mariae KY630504, voucher NVG-1647, female, USA: New Mexico, Eddy County, 22-Sep-2013; Ampittia dioscorides KM102732.1; Burara striata KY524446; Carterocephalus silvicola NC_024646.1; Celaenorrhinus maculosa NC_022853.1; Choaspes benjaminii NC_024647.1; Ctenoptilum vasava NC_016704.1; Daimio tethys NC_024648.1; Euschemon rafflesia KY513288; Erynnis montanus NC_021427.1; Hasora anura NC_027263.1; Hasora vitta NC_027170.1; Heteropterus morpheus NC_028506.1; Lerema accius NC_029826.1; Lobocla bifasciatus NC_024649.1; Megathymus beulahae beulahae KY630505, voucher 11-BOA-13385G05, paratype, male, Mexico, Hidalgo, near Ixmiquilpan, highway 85, klm. 176, 19-Aug-1957; Megathymus cofaqui cofaqui KY630503, voucher NVG-1536, female, USA: Georgia, Burke County, 2-Aug-2013; Megathymus streckeri streckeri KY630501, voucher NVG-1461, male, USA: Arizona, Apache County, southeast of Holbrook, 19-May-2013; Megathymus ursus violae KY630502, voucher NVG-1504, male, USA: Texas, Pecos County, Glass Mountains, 7-Jun-2013; Megathymus yuccae yuccae KY630500, voucher NVG-1185, male, USA: South Carolina, Aiken County, 25-Feb-2013; Papilio glaucus NC_027252; Parnara guttata NC_029136.1; Potanthus flavus NC_024650.1; Pyrgus maculatus NC_030192.1.
All specimens, but one, were collected in 2013 and pieces of their muscles cut out of the thorax were preserved in 100% ethanol to ensure best DNA quality. However, M. beulahae paratype specimen was collected in 1957 15 and stored pinned, spread and dry in a museum drawer for 60 years. DNA was extracted from its abdomen prior to genitalia dissection and produced good quality genomic reads resulting in a complete mitogenome assembly. Similarly to the results reported previously 16, we see that dry insect collections are an invaluable source of specimens for DNA studies; DNA can be extracted from Lepidoptera without damaging specimens beyond standard genitalia dissection procedure; and good quality DNA sequences can be obtained from specimens collected many decades ago.
Sequence comparison revealed that mitogenomes of all six species of Megathymini were very similar, about 15K base pairs in length, coding for 13 proteins, 22 transfer RNAs and 2 ribosomal RNA with gene order typical for mitogenomes of Lepidoptera. The A+T-rich control region is most variable in sequence and length and contains several direct repeats of about 360 bp present in all six species. Among Hesperiidae with available mitogenomes 6, 10, 11, 13, 14, 17– 24 these repeats are unique to Megathymini. The repeats cause difficulty with mitogenome assembly and their number remains uncertain.
To obtain the first DNA-based phylogeny of Megathymus, we constructed RAxML 25 (version 8.2.8, model GTRGAMMA, 100 bootstrap replicates) maximum likelihood tree from available high-quality mitogenomes of Hesperiidae 6, 10, 11, 13, 14, 17– 24 rooted with Pterourus glaucus (Papilionidae) sequence 10 ( Figure 1). While not giving confident resolution to the relationships between subfamilies Eudaminae and Pyrginae, the tree confirms the placement of Megathymini within the subfamily Hesperiinae 26– 28 and argues against historical treatment of Giant-Skippers at subfamily level. The tree resolves the Megathymini phylogeny with 100% bootstrap, supports monophyly of the genus Megathymus and suggests a split between the two species groups. The first group is formed by M. streckeri and M. cofaqui. Caterpillars of these species feed deep in the main root system of yucca plants and deposit frass underground 2, 3. They build a tent only prior to pupation and the tent usually projects from the ground surface. Males of these two species possess hair-like scales, particularly prominent on dorsal hindwing. M. streckeri and M. cofaqui are the closest sister species among Megathymus ( Figure 1). Due to their apparently close relationship and allopatric distribution, Scott has suggested that M. streckeri and M. cofaqui may be subspecies of the same biological species 2. However, the COI barcode sequences we obtained show about 4% divergence between them, revealing significant differences and supporting the two taxa as distinct species. COI barcode divergence in different populations of the same species mostly falls within 2% 29.
The second species group consists of M. yuccae, M. beulahae and M. ursus. Caterpillars of these species feed in Yucca caudex and in roots close to the ground, maintaining the tent throughout development and depositing frass outside the tent 2, 3. Males of these three species lack hair-like scales. M. yuccae and M. beulahae are sister species, as expected from their close morphological similarities. However, their COI barcodes show a very large divergence of 9%. This pronounced divergence was unexpected, because the two species are quite similar in appearance and some males are difficult to tell apart ( http://www.butterfliesofamerica.com/L/t/Megathymus_a.htm). The most noticeable difference between M. yuccae and M. beulahae is the larger white ventral hindwing spots in the latter species, frequently fused to form a bad, especially in females. However, these spots may be significantly reduced in males, frequently in the northern populations. Interestingly, M. beulahae is the only Megathymus species that feeds in yucca-like Agave plant 1, 15, but it is a confident sister of Yucca-feeding M. yuccae. M. ursus is a sister to these two Megathymus. M. ursus has rather different wing shape and patterns. The wings are narrower with more extended apex, forewing spots well-separated in M. yuccae form a band-like arrangement, and hindwings lack spots that females M. yuccae and M. beulahae possess.
In conclusion, we sequenced mitochondrial genomes of all five known species of Megathymus and one species of Agathymus as an outgroup, and constructed the first DNA-based phylogeny of Megathymus. The phylogeny is fully consistent with morphological and behavioral similarities between species. Our results support phylogenetic placement of Megathymini within the subfamily Hesperiinae and clarify the relationships between Megathymus species. In particular, the major phylogenetic split is between the shallow and deep yucca root feeders, and significant mitochondrial DNA divergences between M. yuccae and M. beulahae and between M. streckeri and M. cofaqui support the species status of these allopatric and similar in appearance taxa.
Acknowledgements
We acknowledge Texas Parks and Wildlife Department (Natural Resources Program Director David H. Riskind) for the permit #08-02Rev that makes research based on material collected in Texas State Parks possible. We are grateful to Robert K. Robbins, John M. Burns, and Brian Harris (National Museum of Natural History, Smithsonian Institution, Washington, DC) for granting access to the collections under their care. We thank Lisa N. Kinch for critical suggestions and proofreading of the manuscript. This work was supported in part by the National Institutes of Health (GM094575 to NVG) and the Welch Foundation (I-1505 to NVG).
Funding Statement
This work was supported by National Institutes of Health [GM094575] and the Welch Foundation [I-1505].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved]
References
- 1. Freeman HA: Systematic review of the Megathymidae. J Lepid Soc. 1969;23:1–62. Reference Source [Google Scholar]
- 2. Scott JA: The Butterflies of North America: A Natural History and Field Guide.(Standford University Press);1986. Reference Source [Google Scholar]
- 3. Roever K: The Butterflies of North America.(ed W. H. Howe); (Doubleday and Co.).1975;411–422. [Google Scholar]
- 4. Cong Q, Borek D, Otwinowski Z, et al. : Tiger Swallowtail Genome Reveals Mechanisms for Speciation and Caterpillar Chemical Defense. Cell Rep. 2015. pii: S2211-1247(15)00051-0. 10.1016/j.celrep.2015.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cong Q, Borek D, Otwinowski Z, et al. : Skipper genome sheds light on unique phenotypic traits and phylogeny. BMC Genomics. 2015;16:639. 10.1186/s12864-015-1846-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cong Q, Grishin NV: The complete mitochondrial genome of Lerema accius and its phylogenetic implications. PeerJ. 2016;4:e1546. 10.7717/peerj.1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cong Q, Shen J, Borek D, et al. : When COI barcodes deceive: complete genomes reveal introgression in hairstreaks. Proc Biol Sci. 2017;284(1848): pii: 20161735. 10.1098/rspb.2016.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cong Q, Shen J, Borek D, et al. : Complete genomes of Hairstreak butterflies, their speciation, and nucleo-mitochondrial incongruence. Sci Rep. 2016;6: 24863. 10.1038/srep24863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cong Q, Shen J, Warren AD, et al. : Speciation in Cloudless Sulphurs Gleaned from Complete Genomes. Genome Biol Evol. 2016;8(3):915–931. 10.1093/gbe/evw045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shen J, Cong Q, Grishin NV: The complete mitochondrial genome of Papilio glaucus and its phylogenetic implications. Meta Gene. 2015;5:68–83. 10.1016/j.mgene.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen J, Cong Q, Grishin NV: The complete mitogenome of Achalarus lyciades (Lepidoptera: Hesperiidae). Mitochondrial DNA B Resources. 2016;1(1):581–583. 10.1080/23802359.2016.1197070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shen J, Cong Q, Kinch LN, et al. : Complete genome of Pieris rapae, a resilient alien, a cabbage pest, and a source of anti-cancer proteins [version 1; referees: 2 approved]. F1000Res. 2016;5:2631. 10.12688/f1000research.9765.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J, Cong Q, Shen J, et al. : The complete mitogenome of Euschemon rafflesia (Lepidoptera: Hesperiidae). Mitochondrial DNA B Resources. 2017;2(1):136–138. 10.1080/23802359.2017.1292478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang J, Cong Q, Shen J, et al. : The complete mitochondrial genome of a skipper Burara striata (Lepidoptera: Hesperiidae). Mitochondrial DNA B Resources. 2017; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stallings DB, Turner JR: A review of the Megathymidae of Mexico, with a synopsis of the classification of the family. The Lepidopterists' News. 1958;11:113–137. Reference Source [Google Scholar]
- 16. Timmermans MJTN, Viberg C, Martin G, et al. : Rapid assembly of taxonomically validated mitochondrial genomes from historical insect collections. Biol J Linn Soc. 2016;117(1):83–95. 10.1111/bij.12552 [DOI] [Google Scholar]
- 17. Cao L, Wang J, James John Y, et al. : The complete mitochondrial genome of Hasora vitta (Butler, 1870) (Lepidoptera: Hesperiidae). Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27(4):3020–3021. 10.3109/19401736.2015.1063048 [DOI] [PubMed] [Google Scholar]
- 18. Hao J, Sun Q, Zhao H, et al. : The Complete Mitochondrial Genome of Ctenoptilum vasava (Lepidoptera: Hesperiidae: Pyrginae) and Its Phylogenetic Implication. Comp Funct Genomics. 2012;2012: 328049. 10.1155/2012/328049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim MI, Baek JY, Kim MJ, et al. : Complete nucleotide sequence and organization of the mitogenome of the red-spotted apollo butterfly, Parnassius bremeri (Lepidoptera: Papilionidae) and comparison with other lepidopteran insects. Mol Cells. 2009;28(4):347–363. 10.1007/s10059-009-0129-5 [DOI] [PubMed] [Google Scholar]
- 20. Kim MJ, Wang AR, Park JS, et al. : Complete mitochondrial genomes of five skippers (Lepidoptera: Hesperiidae) and phylogenetic reconstruction of Lepidoptera. Gene. 2014;549(1):97–112. 10.1016/j.gene.2014.07.052 [DOI] [PubMed] [Google Scholar]
- 21. Shao L, Sun Q, Hao J: The complete mitochondrial genome of Parara guttata (Lepidoptera: Hesperiidae). Mitochondrial DNA. 2015;26(5):724–725. 10.3109/19401736.2013.845759 [DOI] [PubMed] [Google Scholar]
- 22. Wang AR, Jeong HC, Han YS, et al. : The complete mitochondrial genome of the mountainous duskywing, Erynnis montanus (Lepidoptera: Hesperiidae): a new gene arrangement in Lepidoptera. Mitochondrial DNA. 2014;25(2):93–94. 10.3109/19401736.2013.784752 [DOI] [PubMed] [Google Scholar]
- 23. Wang J, James John Y, Xuan S, et al. : The complete mitochondrial genome of the butterfly Hasora anura (Lepidoptera: Hesperiidae). Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27(6):4401–4402. 10.3109/19401736.2015.1089543 [DOI] [PubMed] [Google Scholar]
- 24. Wang K, Hao J, Zhao H: Characterization of complete mitochondrial genome of the skipper butterfly, Celaenorrhinus maculosus (Lepidoptera: Hesperiidae). Mitochondrial DNA. 2015;26(5):690–1. 10.3109/19401736.2013.840610 [DOI] [PubMed] [Google Scholar]
- 25. Stamatakis A: RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 26. Warren AD, Ogawa JR, Brower AVZ: Phylogenetic relationships of subfamilies and circumscription of tribes in the family Hesperiidae (Lepidoptera: Hesperioidea). Cladistics. 2008;24(5):642–676. 10.1111/j.1096-0031.2008.00218.x [DOI] [Google Scholar]
- 27. Warren AD, Ogawa JR, Brower AVZ: Revised classification of the family Hesperiidae (Lepidoptera: Hesperioidea) based on combined molecular and morphological data. Syst Entomol. 2009;34(3):467–523. 10.1111/j.1365-3113.2008.00463.x [DOI] [Google Scholar]
- 28. Yuan X, Gao K, Yuan F, et al. : Phylogenetic relationships of subfamilies in the family Hesperiidae (Lepidoptera: Hesperioidea) from China. Sci Rep. 2015;5:11140. 10.1038/srep11140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huemer P, Mutanen M, Sefc KM, et al. : Testing DNA barcode performance in 1000 species of European lepidoptera: large geographic distances have small genetic impacts. PLoS One. 2014;9(12):e115774. 10.1371/journal.pone.0115774 [DOI] [PMC free article] [PubMed] [Google Scholar]