Abstract
Huntington’s disease (HD) is a polyglutamine (polyQ) disease caused by aberrant expansion of the polyQ tract in Huntingtin (HTT). While motor impairment mediated by polyQ-expanded HTT has been intensively studied, molecular mechanisms for nonmotor symptoms in HD, such as psychiatric manifestations, remain elusive. Here we have demonstrated that HTT forms a ternary protein complex with the scaffolding protein DISC1 and cAMP-degrading phosphodiesterase 4 (PDE4) to regulate PDE4 activity. We observed pathological cross-seeding between DISC1 and mutant HTT aggregates in the brains of HD patients as well as in a murine model that recapitulates the polyQ pathology of HD (R6/2 mice). In R6/2 mice, consequent reductions in soluble DISC1 led to dysregulation of DISC1-PDE4 complexes, aberrantly increasing the activity of PDE4. Importantly, exogenous expression of a modified DISC1, which binds to PDE4 but not mutant HTT, normalized PDE4 activity and ameliorated anhedonia in the R6/2 mice. We propose that cross-seeding of mutant HTT and DISC1 and the resultant changes in PDE4 activity may underlie the pathology of a specific subset of mental manifestations of HD, which may provide an insight into molecular signaling in mental illness in general.
Introduction
Huntington’s disease (HD) is a hereditary brain disease characterized by both motor and nonmotor manifestations. Identification of the causal gene for HD, huntingtin (HTT), has facilitated our understanding of the molecular pathways that mediate disease pathology. For instance, it has been demonstrated that structural changes of mutant HTT elicited by the aberrant expansion of polyglutamine (polyQ) lead to the formation of oligomers or insoluble aggregates. The polyQ aggregates, which can recruit various HTT binding partners, impair axonal transport and damage medium spiny neurons, resulting in motor dysfunction in HD (1–7). HD is also characterized by nonmotor dysfunctions including depression (8). Nonetheless, the molecular mechanisms underlying such mental manifestations in HD have not been well studied.
One of the leading mediators for cellular signaling involving higher brain functions is cyclic 3′5′-adenosine monophosphate (cAMP) (9–11). cAMP levels are decreased in cerebrospinal fluid from HD patients and in brains from the HttQ111 knock-in mouse model for HD, which recapitulates the HD polyQ expansion in the murine Htt gene (12, 13). cAMP is degraded by various phosphodiesterases, of which members of the phosphodiesterase 4 (PDE4) family play a major role, including underpinning compartmentalized cAMP signaling in cells (14). Active site-directed small-molecule inhibitors specific for PDE4, such as rolipram, together with targeted gene knockout and genetic studies, have shown that PDE4 regulates a wide range of brain functions — in particular, mental states such as depression and anxiety (15–19). The activity of PDE4 is physiologically regulated at various levels, including intracellular localization, protein interaction, and posttranslational modifications (14, 19). One of the critical direct interactors with PDE4 is the scaffold protein DISC1 (17, 20–23). Physiological roles for DISC1 in biological pathways in brain development and neurosignaling have been intensively studied for the past decade in various animal models (24–26). Yet the significance of the DISC1-PDE4 interaction in pathological contexts is poorly understood. Identification of a DISC1-PDE4 complex in conjunction with dysregulation of cAMP in HD would suggest the possibility that HTT might be involved in the DISC1-PDE4 pathway, but this mechanistic link has not been examined to date. Furthermore, DISC1 was previously shown to form insoluble aggregates (27–30), but the structural basis of DISC1 aggregation and its neuropathological effects have not been well investigated. Here we address a novel molecular mechanism involving the disturbance of DISC1-PDE4 interactions and behavioral changes through aggregation of DISC1 in HD.
Results
Augmentation of PDE4 activity in R6/2 mice representing the polyQ pathology of HD.
The R6/2 mouse model, which expresses the HD-associated human HTT mutant and represents the polyQ pathology of HD, shows that HTT aggregation begins at 3.5 and 4.5 weeks in the cerebral cortex and striatum, respectively, and is clearly detectable at 7 weeks and progressively continues (31). In this model, we first measured PDE4 activity at 4, 8, and 12 weeks. Intriguingly, we observed a profound augmentation of PDE4 activity in both the cerebral cortex and striatum at 8 weeks (Figure 1A). The elevated PDE4 activity was also observed at up to 12 weeks of age, but the increase in PDE4 activity in the cerebral cortex was reduced over time (Figure 1A). This reduction over time in the cerebral cortex may be due to the neuronal cell death in the cortex preceding that in the striatum of R6/2 mice (31). The increased PDE4 activity may provide an explanation why rolipram has beneficial effects in the R6/2 model (32, 33). Consistent with our novel discovery in vivo, when we evaluated a neuro2a cell model that stably expresses an N-terminal 1–67 amino acid fragment (corresponding to exon 1) of Htt with expanded polyQ (HTT67Q150) (34), we found that PDE4 activity was increased more than 3-fold (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI85594DS1). In our assay we used pan-PDE4-selective rolipram (10 μM) at a concentration known to suppress all PDE4 subtypes (35–37).
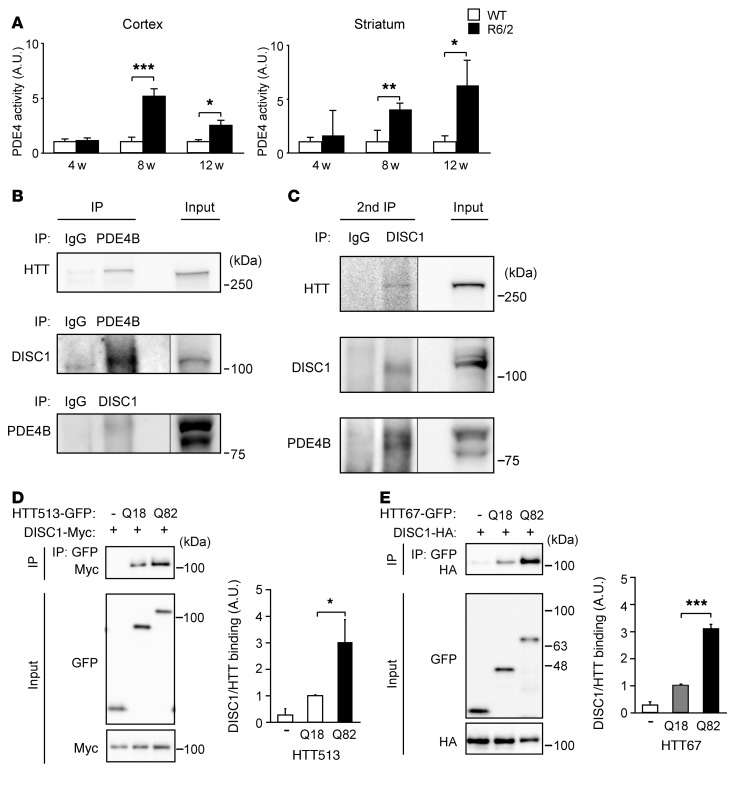
Figure 1. PDE4 activity could be regulated by the newly identified HTT-DISC1-PDE4 ternary protein complex.
(A) PDE4 activities of the homogenates of cerebral cortex (left) and striatum (right) in R6/2 mice were increased compared with those in WT at 8 and 12 weeks (w) of age. Data represent mean + SEM. *P < 0.05, **P < 0.01, ***P < 0.001; unpaired 2-tailed t test. Cortex: 4 weeks, n = 4 per group; 8 weeks, n = 5 per group; 12 weeks, n = 9 (WT), 6 (R6/2). Striatum: 4, 8, and 12 weeks, n = 4 per group. (B) Protein complex of HTT-DISC1-PDE4B in cerebral cortex of WT mice. Brain homogenates were immunoprecipitated by an anti-PDE4B (pan-PDE4B) or anti-DISC1 (mEx3) antibody or IgG control, followed by immunoblotting with an anti-HTT (MAB2144), anti-DISC1 (m317C), or anti-PDE4B (pan-PDE4B) antibody. Representative immunoblots are shown from 3 independent sample sets. (C) Formation of HTT-DISC1-PDE4B tertiary protein complex in cerebral cortex of WT mice was detected with sequential coimmunoprecipitation. Brain homogenates were immunoprecipitated by PDE4B antibody (pan-PDE4B) (1st IP), then the immunoprecipitates were immunoprecipitated by an anti-DISC1 (mEx3) antibody (2nd IP), followed by immunoblotting with an anti-HTT (MAB2166), anti-DISC1 (m317C), or anti-PDE4B (pan-PDE4B) antibody. Negative control was immunoprecipitated by IgG. Representative immunoblots are shown from 3 independent sample sets. (D) Increased binding of HTT513 and DISC1 with expansion of polyQ (Q18 or Q82) in HEK293T cells by coimmunoprecipitation. Data represent mean + SEM (5 independent sample sets). *P < 0.05; 1-way ANOVA followed by Bonferroni post hoc corrections. (E) Increased binding of HTT67 and DISC1 with expansion of polyQ (Q18 or Q82) in HEK293T cells by coimmunoprecipitation. Data represent mean + SEM (3 independent sample sets). ***P < 0.001; 1-way ANOVA followed by Bonferroni post hoc corrections.
Regulation of PDE4 activity by an HTT-DISC1-PDE4 ternary protein complex. DISC1 interacts with dynactin (p150glued), nuclear receptor corepressor (N-CoR), pericentriolar material 1 protein (PCM1), and kalirin-7, all of which also reportedly interact with huntingtin-associated protein 1 (HAP1) and/or HTT (24–26). The frequency of common binding partners of DISC1 and HTT implied that these 2 proteins may interact either directly or indirectly. A recent bioinformatic approach also resulted in a similar proposal (38). As DISC1 and PDE4 proteins interact physically and functionally (17, 20, 39), we hypothesized that DISC1-PDE4, PDE4-HTT, and HTT-DISC1 proteins might interact. To evaluate this hypothesis, we conducted coimmunoprecipitation with the homogenates of whole mouse brains and identified the DISC1-PDE4, PDE4-HTT, and HTT-DISC1 protein interactions (Figure 1B and Supplemental Figure 2). The DISC1-HTT and DISC1-PDE4 binding was more significant in striatum and cerebral cortex than in cerebellum (Supplemental Figure 3A).
Next, we asked whether these 3 proteins might form a ternary complex. Indeed, sequential coimmunoprecipitation demonstrated the formation of such a ternary complex (Figure 1C). One of the PDE4 isoforms, PDE4B, which is relatively abundant in cerebral cortex and striatum interacts directly with DISC1 (20), and has been implicated from genetic association studies to be functionally important DISC1 partners (17, 40). The PDE4B gene encodes several distinct isoforms, of which the major ones are the PDE4B1 and PDE4B3 long forms and the PDE4B2 short form (41). We thus evaluated the binding of DISC1 with each of these isoforms and found that they each bound to DISC1 and to HTT (Supplemental Figure 3, B and C).
Specific effect of polyQ expansion on the protein binding of HTT with DISC1.
Given that PDE4, HTT, and DISC1 form a ternary protein complex, then how does polyQ expansion in HTT affect these protein interactions? We examined how the polyQ expansion affected HTT-DISC1 protein interaction by using an N-terminal 1–513 amino acid fragment of HTT (HTT513) with 18 residues of polyQ, or pathologically expanded 82 residues of polyQ (designated Q18 and Q82, respectively). The HTT513 and DISC1 interaction was augmented more than 2-fold when polyQ was expanded (Figure 1D). The increased binding was also observed when a shorter N-terminal fragment with an equal polyQ expansion (HTT67Q82) was used together with DISC1 (Figure 1E). However, the binding of DISC1 with HTT513 was greater than that with HTT67 (Supplemental Figure 4A). These results indicate that amino acids 1–67 of HTT are sufficient to interact with DISC1, but that additional amino acids between 68 and 513 further facilitate the interaction. The effect of expanded polyQ was not observed in the interaction between PDE4B and HTT513 (Supplemental Figure 4B), implying that the polyQ expansion has unique pathophysiological effects on the HTT-DISC1 interaction.
Selective sequestration of DISC1 into SDS-insoluble HTT aggregates in R6/2 mice and HD brains.
One of the hallmarks of HD is the formation of SDS-resistant aggregates/inclusions of mutant HTT with expanded polyQ (42). DISC1 is also reportedly prone to aggregation (27, 28), but its pathophysiological significance is, as yet, unclear. Thus, we tested whether a pool of DISC1 could be detected in the SDS-resistant fraction of the brains from the R6/2 model, as well as HD patients, by using a filter trap assay. Significant increases in the levels of DISC1 in the SDS-resistant fractions from the cerebral cortex and striatum of R6/2 mice were observed, compared with those of normal mice (Figure 2A). Similar results were observed in the SDS-insoluble fractions from HD patients compared with those from normal controls (Figure 2B). In contrast, we did not find PDE4B immunoreactivity in the SDS-resistant fractions of any brain samples. Furthermore, by Western blotting, we observed immunoreactivity of DISC1, but not of PDE4B, on the top of the gel when we applied brain homogenates from R6/2 mice (Supplemental Figure 5A), further supporting the notion that a pool of DISC1 is insoluble to SDS in the R6/2 brain.
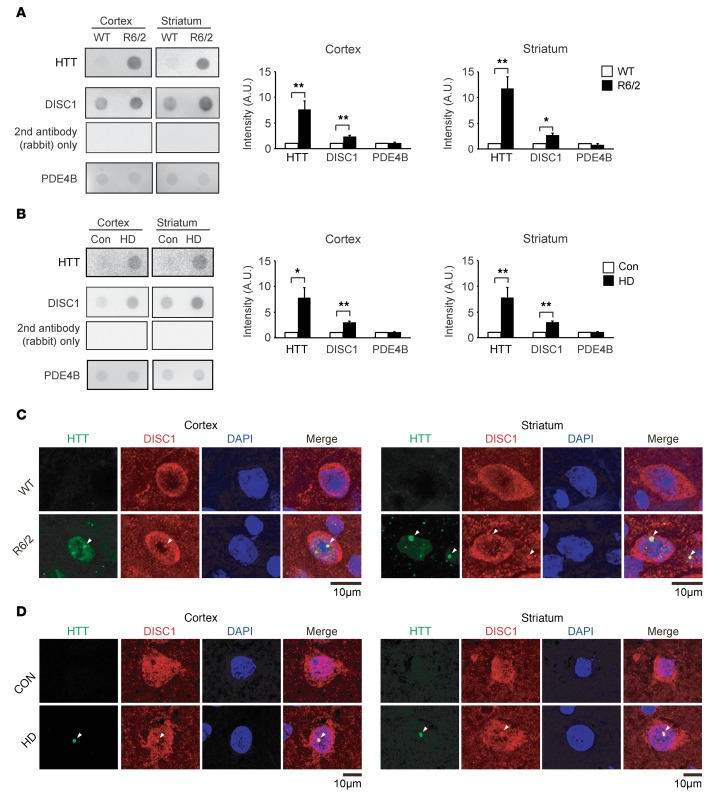
Figure 2. DISC1, but not PDE4B, is sequestered into insoluble HTT aggregates in HD.
(A) Filter trap assay with homogenates of cerebral cortex or striatum from 12-week-old R6/2 mice showed significantly increased levels of HTT (EM48) (top) and DISC1 (m317C) (middle) but not PDE4B (pan-PDE4B) (bottom) in the SDS-resistant fraction, compared with those from WT mice. A negative control with rabbit secondary antibody alone is also shown (middle). Data represent mean + SEM. *P < 0.05, **P < 0.01; unpaired 2-tailed t test. n = 3 per group. (B) Filter trap assay with homogenates of cerebral cortex or striatum showed significantly increased levels of HTT (EM48) (top) and DISC1 (h316C) (middle) but not PDE4B (pan-PDE4B) (bottom) in the SDS-resistant fraction from HD patients (HD), compared with those from controls (Con). A negative control with rabbit secondary antibody alone is also shown (middle). Data represent mean + SEM. *P < 0.05, **P < 0.01; unpaired 2-tailed t test. n = 3 per group. (C) Immunostaining of frozen sections of cerebral cortex (left) or striatum (right) demonstrated that DISC1 was colocalized with intranuclear inclusions of mutant HTT in 12-week-old R6/2 mice. Green, HTT (EM48); red, DISC1 (m317C); blue, DAPI (the nucleus). Arrowheads show intranuclear inclusions. Scale bar: 10 μm. n = 3 per group. (D) Immunostaining of paraffin sections of cerebral cortex (left half) or striatum (right half) in HD patients (HD) and human controls (Con) was performed with anti-HTT (EM48) (green, left) and anti-DISC1 (h316C) (red) antibodies. Nucleus was stained by DAPI (blue). Merged images are shown on the right. Arrowheads show intranuclear inclusions. Scale bar: 10 μm. n = 3 (Con), 6 (HD).
To test whether this insoluble DISC1 is associated with mutant HTT aggregation, we examined immunofluorescent tissue staining. We found that high-intensity DISC1 staining was colocalized with intranuclear aggregates of mutant HTT in both the cerebral cortex and striatum of R6/2 mice (Figure 2C). DISC1 is reportedly localized in both cytosol and nucleus (43–46). Therefore it is likely that the nuclear localization of DISC1 would contribute to the sequestration into intranuclear HTT aggregates. Furthermore, colocalization of DISC1 in the HTT aggregates was also observed in the sections from the cortex and striatum of HD patients (Figure 2D). In contrast, no colocalization of PDE4B in HTT aggregates was detected in R6/2 mouse brains (Supplemental Figure 5B).
Cross-seeding between intrinsically aggregation-prone DISC1 and polyQ-expanded mutant HTT.
The above results suggested that pathologically enhanced interaction of DISC1 and mutant HTT may selectively facilitate the sequestration of DISC1 into the HTT aggregates. Therefore, we set out to evaluate possible biophysical mechanisms underlying the selective aggregation of DISC1 in HD. First, we aimed to reveal structural and physical properties of full-length DISC1 aggregates, which have not yet been fully characterized even though DISC1 has been shown to be aggregation-prone in nature (27, 28). We performed multiple biophysical assays in the cell-free in vitro system and found that DISC1 spontaneously formed aggregates, which reacted to an amyloid-specific dye, thioflavin T, at a level comparable to that seen with the typical amyloid, HTT67Q42 (ref. 42 and Figure 3A). The fibrillar morphology was observed by electron microscopy (Figure 3B). Furthermore, circular dichroism spectra of DISC1 aggregates showed a negative peak at 228 nm, indicating extensive β-sheet–rich structures (ref. 47 and Supplemental Figure 6A). These data show that DISC1 can form fibrillar aggregates in vitro. As DISC1 does not contain glutamine/asparagine-rich domains that could induce aggregation, we examined a core region of DISC1 aggregates by mass spectrometry. Limited proteolysis of DISC1 aggregates with proteinase K treatment followed by MALDI/TOF mass analysis showed a single major peak at 2,012.9 (m/z) in the mass spectrum, which identified the core of DISC1 aggregates as the region consisting of residues 209–227 (Figure 3C).
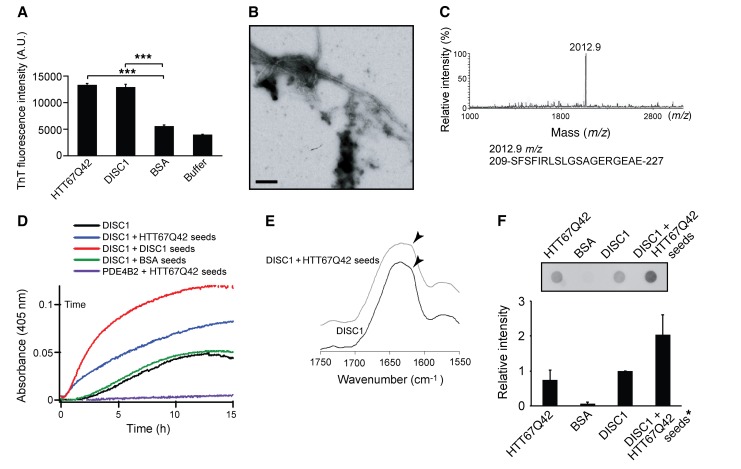
Figure 3. Aggregation of intrinsically aggregation-prone DISC1 is accelerated by cross-seeding with polyQ-expanded mutant HTT.
(A) Thioflavin T (ThT) fluorescent intensities of spontaneous DISC1 and HTT67Q42 aggregates were significantly higher than those of BSA aggregates. Data represent mean + SEM (4 independent sample sets). ***P < 0.001; 1-way ANOVA followed by Bonferroni post hoc corrections. (B) Fibrillar morphology of typical DISC1 aggregates shown by transmission electron microscopy. Scale bar: 100 nm. A representative image is shown from 3 independent samples. (C) MALDI-TOF mass spectrum of DISC1 aggregates digested by proteinase K identified a major mass peak at 2,012.9 m/z corresponding to residues 209–227 in DISC1 as the core of DISC1 aggregates. Representative data are shown from 3 independent samples. (D) Aggregation of DISC1, which was monitored by the absorbance (turbidity) at 405 nm, was significantly accelerated by the HTT67Q42 aggregate seeds but not by BSA aggregates. No aggregation of PDE4B2 was observed in the presence of HTT67Q42 aggregate seeds. Representative data are shown from 3 independent sample sets. (E) FT-IR spectra revealed that the DISC1 aggregates formed in the presence of HTT67Q42 aggregate seeds (gray) contained more intermolecular β-sheet structures (arrowheads) than did spontaneously formed DISC1 aggregates (black). Representative data are shown from 4 independent sample sets. (F) The DISC1 aggregates formed in the presence of HTT67Q42 aggregate seeds showed higher resistance to 2% SDS than spontaneously formed DISC1 aggregates in the filter trap assay with Sypro Ruby staining. The asterisk indicates that 10% of the signal intensity of HTT67Q42 amyloid is subtracted from that of DISC1-HTTQ42 coaggregates for correction of the signal intensity. Data represent mean + SEM (4 independent sample sets). One-way ANOVA was used for statistical analysis.
We then examined the effects of HTT aggregates on the kinetics of DISC1 aggregation. DISC1 spontaneously formed aggregates, but the aggregation was not evident for the initial 2 hours under the conditions tested. This 2-hour lag time was almost completely abolished by the addition of seeds (10% mol/mol) of DISC1 aggregates at the initiation of the reaction, indicating that DISC1 aggregates are self-seeding, a key feature of amyloid fibers (refs. 48–50 and Figure 3D). More importantly, DISC1 aggregation was significantly accelerated with the addition of seeds of HTT67Q42 aggregates, but not with the addition of BSA aggregates, which were used as a control (Figure 3D). Furthermore, we found no aggregation of PDE4B2 or a trigger factor tag in the presence of HTT67Q42 aggregate seeds (Figure 3D and Supplemental Figure 6B). The following immunoprecipitation experiments proved that the accelerated DISC1 aggregation by HTT67Q42 seeds was caused by coaggregation, but not by an increased rate of spontaneous DISC1 aggregation (Supplemental Figure 6C). After soluble DISC1 was incubated with HA-tagged HTT67Q42 amyloid seeds (HTT67 Q42-HA seeds; 10% mol/mol), the resulting HTT67 Q42-HA aggregates contained DISC1, indicating that soluble DISC1 formed coaggregates with HA-HTTQ42 seeds (Supplemental Figure 6B). These data reveal cross-seeding between DISC1 and mutant HTT, which is consistent with the marked colocalization of the 2 proteins in HD brains in vivo (Figure 2D).
The DISC1 aggregates that formed with seeds of HTT67Q42 aggregates showed an increase in the peak at 1,620 cm–1 in Fourier transform infrared (FT-IR) spectra, indicating a greater level of intermolecular β-sheet structures (51), compared with the DISC1 aggregates that were spontaneously formed in the absence of seeds (Figure 3E). Consistently, we found that the DISC1 aggregates seeded by HTT67Q42 amyloid showed the trend of the higher resistance to 2% SDS than spontaneously formed DISC1 aggregates (Figure 3F). The altered structural and physical properties of the DISC1 aggregates also supported the cross-seeding between DISC1 and mutant HTT.
Reduced DISC1-PDE4 complex by decreased soluble DISC1 levels in R6/2 mice.
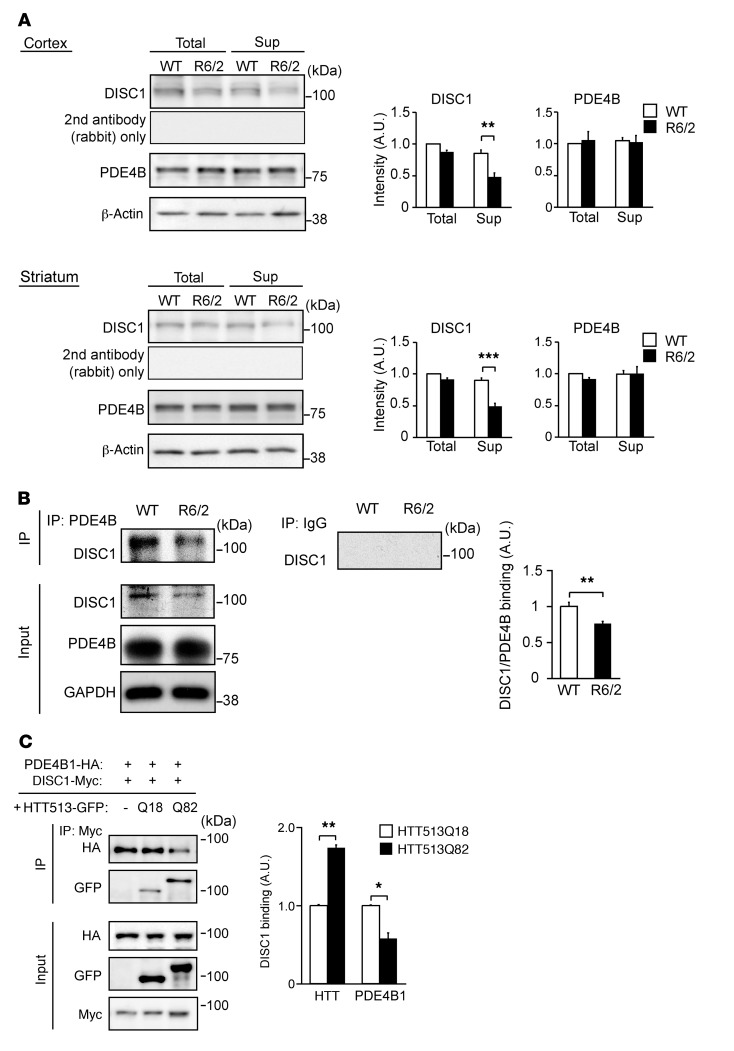
We then asked whether the physiological disposition of DISC1 might be impaired because of the selective sequestration of this protein into the HTT aggregates in HD pathology. First, we used sedimentation analysis with homogenates of the cerebral cortex and striatum, and observed that the level of soluble DISC1, but not that of PDE4B, was reduced in R6/2 mice compared with WT mice (Figure 4A). DISC1 interacts with PDE4B in soluble fractions and regulates the enzymatic activity of PDE4 (17, 39). Thus, reduction in the levels of soluble DISC1 may result in a decrease in the amount of DISC1-PDE4 complex and an increase in the PDE4 activity. Compared with substantial changes of DISC1 in the soluble fractions (detected by Western blotting in Figure 4A) and alteration in PDE4 activity (Figure 1A), the integration of DISC1 staining in the HTT aggregates was more modest (Figure 2, C and D). A portion of, but not entire, insoluble DISC1 with HTT might be visualized as colocalization with large HTT aggregates, since insoluble DISC1 with HTT microaggregates could be diffusible. In mouse brain homogenates, we showed a significant reduction in the amount of DISC1-PDE4 complex in R6/2 compared with that in WT mice (Figure 4B). Furthermore, we addressed whether the reduction of DISC1-PDE4B complex was elicited directly by the presence of mutant HTT. We added lysates of cells overexpressing HTT513Q18 or HTT513Q82 to cell lysates in which DISC1 and PDE4B interact, and examined how exogenous HTT513 proteins impact the DISC1-PDE4B complex. The addition of HTT513Q82 significantly reduced the amount of the preexisting DISC1-PDE4B complex, whereas that of HTT513Q18 did not (Figure 4C). Notably, as described above, augmented binding of HTT to DISC1 was observed with HTT513Q82 compared with HTT513Q18 (Figure 1D). Thus, a relative loss of DISC1’s interaction with PDE4 is likely a major effect of the DISC1 sequestration into HTT aggregates due to the preferred binding of DISC1 with polyQ-expanded mutant HTT.
Figure 4. The amount of DISC1-PDE4 complex was reduced by decreased soluble DISC1 levels in R6/2 mice.
(A) Soluble DISC1 but not soluble PDE4B levels in supernatant fractions (Sup) were reduced in cerebral cortex (top) and striatum (bottom) from 12-week-old R6/2 mice, compared with those from WT mice. Total DISC1 and PDE4B levels in total homogenates (Total) were not changed between WT and R6/2 mice. β-Actin is a loading control. An m317C anti-DISC1 antibody was used to detect DISC1, and a negative control with rabbit secondary antibody alone is also shown. Data represent mean + SEM. **P < 0.01, ***P < 0.001; unpaired 2-tailed t test. n = 3 per group. (B) The amount of DISC1-PDE4B complex in 12-week-old R6/2 mouse brains was decreased compared with that in WT brains. Immunoprecipitation was performed by an anti–pan-PDE4B antibody, followed by immunoblotting with anti-DISC1 antibody (D27). GAPDH is a loading control and used for normalization of the IP data. Data represent mean + SEM. **P < 0.01; unpaired 2-tailed t test. n = 5 per group. (C) Exogenous addition of HTT513Q82, but not HTT513Q18, significantly reduced DISC1-PDE4B interactions in HEK293T cells. Lysates of the cells overexpressing GFP alone (–), HTT513Q18-GFP (Q18), or HTT513Q82-GFP (Q82) were added to lysates of the cells overexpressing DISC1-Myc and PDE4B1-HA. Mixtures were processed for immunoprecipitation with an anti–c-Myc antibody, followed by immunoblotting. Data represent mean + SEM (3 independent sample sets). *P < 0.05, **P < 0.01; unpaired 2-tailed t test.
HTT-DISC1-PDE4 interactions for aberrant enhanced PDE4 activity in R6/2 mice.
We propose that sequestration of DISC1 into HD aggregates may disturb the regulatory effects of DISC1 on PDE4, resulting in aberrant augmentation of PDE4 enzymatic activity, which contributes to HD pathology. If this scenario is correct, supplementing WT DISC1 might ameliorate such increase of PDE4 activity in the models we have studied here. As described above, the overexpression of HTT67Q150 in neuro2a cells, in which substantial levels of endogenous DISC1 were expressed (52), augmented PDE4 activity 3- to 4-fold (Supplemental Figure 1). When we exogenously overexpressed WT DISC1 in the neuro2a cells expressing HTT67Q150, the aberrant augmentation of PDE4 activity was ameliorated (Supplemental Figure 7A). Although these results are promising in terms of protecting cells against pathological PDE4 activation, we were curious why the beneficial effects of exogenous DISC1 were only partial. This may be because exogenously expressed DISC1 was also, at least in part, sequestered into HTT aggregates and could not fully regulate endogenous PDE4 activity.
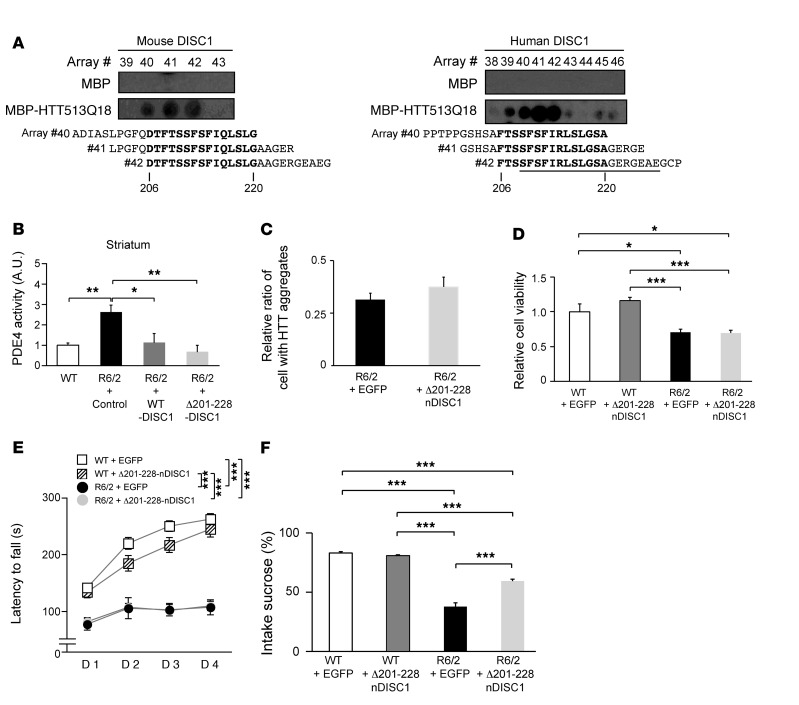
We then set out to identify the region of DISC1 responsible for HTT binding, using a peptide array approach that has been used to successfully map the binding domain of DISC1 with protein interactors and PDE4 with various partner proteins (19, 20). We anticipated that a mutant DISC1 lacking a minimal domain for HTT interaction but maintaining PDE4 binding would not be sequestered into the HTT aggregates and thus would fully regulate PDE4 activity. Analyses of mouse and human DISC1 led to consistent results conserved between the 2 species, in which amino acids 206–220 of both mouse and human DISC1 were crucial for binding with HTT (Figure 5A). Furthermore, in agreement with these observations from peptide arrays, mutant mouse DISC1 lacking amino acids 201–228 (Δ201-228-DISC1) displayed a dramatic reduction in DISC1-HTT513 binding in cells, compared with that in WT DISC1 (Supplemental Figure 7B). On the other hand, this deletion showed no effect on DISC1-PDE4B2 binding, and significant effects on DISC1-PDE4B1 interactions (Supplemental Figure 7B), due to the complexity of the binding between DISC1 and PDE4 isoforms (20, 53). When we expressed WT DISC1 or Δ201-228-DISC1 in neuro2a cells expressing the mutant HTT fragment, we observed that Δ201-228-DISC1 completely normalized aberrantly augmented PDE4 activity. The effect of Δ201-228-DISC1 was greater than that of WT DISC1 (Supplemental Figure 7A).
Figure 5. Aberrantly enhanced PDE4 activity and reduced sucrose preference in R6/2 mice are recovered by exogenous DISC1 expression.
(A) The 25-mer peptides #40–#42 (residues 194–228 for mouse DISC1 [left], residues 196–230 for human DISC1 [right]) in DISC1 showed binding to maltose-binding protein (MBP)–HTT513Q18 (bottom) but not MBP alone (top) on peptide array. A black line corresponds to the core region of aggregates, which was identified by limited proteolysis and mass spectrometry (see Figure 3C). Representative data are shown from 3 and 2 independent samples for mouse and human DISC1, respectively. (B) Aberrant augmentation of PDE4 activity in striatum of R6/2 mice was normalized with WT or Δ201-228-DISC1. Data represent mean + SEM. *P < 0.05, **P < 0.01; 1-way ANOVA followed by Bonferroni post hoc corrections. n = 6, 3, 3, and 3 for WT, R6/2 + Control, R6/2 + WT-DISC1, and R6/2 + Δ201-228-DISC1 mice, respectively. (C) The number of cells with EM48-positive HTT aggregates was not affected by expression of Δ201-228-nDISC1 in striatum of mice at 12 weeks. Data represent mean + SEM. Unpaired 2-tailed t test was used for statistical analysis. n = 3 per group. (D) The number of NeuN-positive neurons was not affected by expression of Δ201-228-nDISC1 in striatum of mice at 12 weeks. Data represent mean + SEM. *P < 0.05, ***P < 0.001; 1-way ANOVA followed by Bonferroni post hoc corrections. n = 4, 4, 6, and 5 for WT + EGFP, WT + Δ201-228-nDISC1, R6/2 + EGFP, and R6/2 + Δ201-228-nDISC1 mice, respectively. (E) The impairment of motor function of R6/2 mice was not rescued by expression of Δ201-228-nDISC1. The latency to fall off the rotarod was examined in mice at 9 weeks. Data represent mean ± SEM. ***P < 0.001; 1-way ANOVA followed by Bonferroni post hoc corrections. n = 17, 16, 8, and 8 for WT + EGFP, WT + Δ201-228-nDISC1, R6/2 + EGFP, and R6/2 + Δ201-228-nDISC1 mice, respectively. (F) Reduced sucrose preference in R6/2 mice was rescued by expression of Δ201-228-nDISC1. The sucrose intake was measured in mice at 9 weeks. Data represent mean + SEM. ***P < 0.001; 1-way ANOVA followed by Bonferroni post hoc corrections. n = 17, 16, 17, and 20 mice for WT + EGFP, WT + Δ201-228-nDISC1, R6/2 + EGFP, and R6/2 + Δ201-228-nDISC1 mice, respectively.
If aggregation of DISC1 is mediating the reduced PDE4 activity in HD brain, then the ectopic introduction of additional DISC1 should rescue the aberrant PDE4 activity seen in HD. Since our observations indicated that PDE4 activity was augmented in the striatum of 8-week-old R6/2 mice (Figure 1A), we next introduced exogenous DISC1 into the striatum of the 2-week-old mice by stereotaxic injection of a lentiviral vector of DISC1 connected with a GFP Venus via IRES (DISC1/Venus). We then collected the DISC1/Venus–expressing cells from the striatum of the mice at 8 weeks by flow cytometry and measured PDE4 activity. Expression of WT DISC1 significantly decreased the aberrant PDE4 activity in R6/2 mice (Figure 5B). Introduction of Δ201-228-DISC1 led to an even greater effect, which is consistent with the observations from the cell model (Figure 5B). These results support the notion that pathological augmentation of PDE4 activity in HD pathology may be predominantly mediated by the partial loss of DISC1 that regulates PDE4 activity. This process is determined by the sequestration of DISC1 from the HTT-DISC1-PDE4 ternary complex as a result of specifically preferred binding and coaggregation between mutant HTT and DISC1.
Significance of HTT-DISC1-PDE4 interactions for nonmotor function in R6/2 mice.
Next, we examined the in vivo consequences of the disturbed DISC1-PDE4 interaction. We generated adeno-associated virus (AAV) encoding an N-terminal 1–316 amino acid fragment of mouse DISC1, which includes most of the binding sites for PDE4 but lacks the binding region for HTT (Δ201-228-nDISC1) (Supplemental Figure 8). We stereotaxically injected AAV encoding control EGFP or Δ201-228-nDISC1 that could interact with PDE4B isoforms but not HTT, into the striatum of 3-week-old R6/2 mice. Immunohistological studies revealed that exogenous expression of Δ201-228-nDISC1 has no effect on HTT aggregation and neuronal viability (Figure 5, C and D). We then investigated motor and nonmotor behaviors of the mice. We confirmed impairment of motor functions in R6/2, evident by deficits in rotarod performance and shorter distance in open field test, which were not altered by exogenous expression of Δ201-228-nDISC1 (Figure 5E and Supplemental Figure 9A). Next, we performed a variety of behavioral tests for nonmotor functions, including light/dark transition, marble burying, nesting, and sucrose preference. Interestingly, in the sucrose preference test, the exogenous expression of Δ201-228-nDISC1, but not expression of a control protein EGFP, significantly restored the reduced preference of R6/2 mice (Figure 5F). In contrast, in marble burying and nesting test, although the deficits existed in the R6/2 mice, the rescue by Δ201-228-nDISC1 was not observed (Supplemental Figure 9, B and C). Note that the R6/2 mice did not show even alterations in light/dark transition and center time in open field test (Supplemental Figure 9, D and E). These results suggest that the DISC1/PDE4/HTT pathway may be involved in a specific subset of nonmotor behaviors in R6/2 mice.
Discussion
Here we demonstrate that the sequestration of intrinsically aggregation-prone DISC1, but not PDE4, into mutant HTT aggregates disturbed the proper formation of an HTT-DISC1-PDE4 ternary protein complex. This generates a PDE4 fraction out of physiological control by DISC1, thereby aberrantly increasing PDE4 activity. Furthermore, our exploratory study in vivo suggests that this molecular pathway may underlie anhedonia in the R6/2 model (Supplemental Figure 10). Our present study uniquely addresses mental manifestation in HD, compared with many previous studies in which coaggregation of mutant HTT with other aggregation-prone proteins was demonstrated to be involved in motor dysfunctions (54–56). Thus, we propose that this mechanism involving the HTT-DISC1-PDE4 ternary protein complex provides a novel molecular basis for the pathology and potentially the psychiatric symptoms underpinning the reduced cAMP levels observed in both HD patients and model mice (12, 13).
Rolipram administration restored motor dysfunction and decreased both HTT aggregation and neuronal death in R6/2 mice (31–33), while we found that Δ201-228-nDISC1 did not affect them (Figure 5, C–E, and Supplemental Figure 9A). These contrasting results underscore that the molecular mechanisms underlying rolipram challenge and ectopic Δ201-228-nDISC1 expression are different. In contrast to the fact that rolipram could target diverse PDE4s (36), Δ201-228-nDISC1 would have specific inhibitory effects on the DISC1-associated PDE4 activity.
We show here that the aggregation of DISC1 is significantly accelerated by the presence of mutant HTT seeds. Importantly, our structural analysis of DISC1 aggregates provided mechanistic insights into the efficient cross-seeding between mutant HTT and DISC1: the core region (residues 209–227) of DISC1 aggregates included the binding site for HTT (residues 206–220). This overlap might increase the chance of interactions between aggregation-prone core regions in distinct DISC1 molecules and thereby facilitate the sequestration of DISC1 into mutant HTT aggregates. In addition, the increased β-sheet structures and SDS resistance of DISC1 aggregates by cross-seeding with mutant HTT may enhance the resistance to degradation and promote further recruitment of soluble DISC1 into mutant HTT aggregates.
This report, with a combined cell biology and biophysical approach that is informed by clinical observations, formulates a novel hypothesis that leads us to suggest 3 questions for future studies. First, it will be important to clarify whether the sequestration of DISC1 into misfolded protein fractions and aggregates is specific to HD or more generally applied to other polyQ diseases. The present data from the R6/2 model that represents the polyQ pathology implies that DISC1 may also be involved in other polyQ diseases. Second, the next study may use a full-length HTT mouse model to address the influence of HTT on broader aspects of HD pathology. Third, it is worthwhile to address how other DISC1 interacting partners could be involved in the PDE4-regulating mechanism controlled by the HTT-DISC1-PDE4 ternary protein complex.
Recent studies have indicated that brain samples from a subgroup of patients with schizophrenia, bipolar disorder, or depression, as well as some preclinical models for such conditions, contain insoluble DISC1 and dysbindin, another protein that might be involved in the biology of mental functions (27, 29, 30, 57, 58). Based on these findings, we propose that a selective network of misfolded proteins that are prone to be insoluble may underlie many aspects of brain dysfunctions, including psychiatric manifestations.
Methods
Plasmids and antibodies
The cDNA of full-length mouse DISC1 was subcloned into a pRK5 vector with an HA or Myc tag at the N-terminus or into a pcDNA4 vector (Invitrogen) with a mys/His6 tag at the C-terminus. The cDNA of full-length human PDE4B1, 4B2, or 4B3 was subcloned into a pRK5 vector with an N-terminal HA tag. The cDNA of HTT67 or HTT513 containing 18 or 82 glutamine repeats was inserted into a pCSII-CMV vector that includes EGFP at the C-terminus. For production of lentivirus, the cDNA of full-length or mutant mouse DISC1 with a C-terminal HA tag was subcloned into a pCSII-CMV-IRES-Venus plasmid. pCSII-CMV, pCAG-HIVgp, and pCAG-VSV-G plasmids were provided by the RIKEN BioResource Center. For production of AAV, the cDNA of full-length or mutant mouse DISC1 was subcloned into a pAAV2 vector. For bacterial expression of DISC1 and HTT67Q42, the cDNA of human DISC1, HTT513Q18, and HTT67Q42 was inserted into pCold-TF (Takara), pMAL (New England Biolabs), and pGEX6P2 (GE Healthcare) vectors including a His tag at the C, N, and N terminus, respectively. For bacterial expression of PDE4B2, the cDNA of human PDE4B2 was introduced into a pET29b vector. Rabbit polyclonal mouse DISC1 (m317C) and human DISC1 (h316C) antibodies were raised against the purified proteins of residues 317–852 in mouse DISC1 and residues 316–854 in human DISC1, respectively, in the Research Resources Center at RIKEN Brain Science Institute (Supplemental Figure 2). A mouse monoclonal antibody against mouse DISC1 (M49) was produced with the purified protein of residues 317–852 as an antigen (MBL) (Supplemental Figure 2). For validation of mouse DISC1 antibodies, Disc1 locus impairment (Disc1 LI) mice, in which a major full-length 100-kDa isoform is depleted (59, 60), were used (Supplemental Figure 2). The following commercial or published antibodies were also used: mouse monoclonal antibodies against HA, Myc, and GFP tags (Nacalai Tesque), against mouse DISC1 (2B3) (59), and against β-actin (Abcam); rabbit polyclonal antibodies against HA, Myc, and GFP tags (MBL), against mouse DISC1 (mEx3, refs. 59–63; and D27, refs. 63–66), against human DISC1 (14F2) (57), and against HTT (EM48 and MAB2144) (Millipore); and a sheep polyclonal anti–pan-PDE4B antibody (41). Mouse, rabbit, or sheep IgG (Santa Cruz Biotechnology) was also used as a control in coimmunoprecipitation experiments.
R6/2 mice
Heterozygous HTT67 (corresponding to HTT exon 1) transgenic R6/2 mice were obtained from Jackson Laboratory and maintained by ovarian transplantation of female R6/2 mice carrying about 130 CAG repeats. R6/2 transgenic and age-matched WT mice were anesthetized and the brains were removed, followed by separation of cerebral cortex and striatum. For measurement of PDE4 activity, storage of brains at –80°C was minimized.
Autopsied brains from HD patients
Autopsied brains (12–32 hours postmortem interval) from 6 HD patients and 3 controls at the age of 37–76 were obtained from Harvard Brain Tissue Resource Center. Frozen and fixed paraffin blocks for cerebral cortex and striatum include the regions from frontal cortex and caudate/putamen/accumbens, respectively.
Measurement of PDE4 activity
PDE4 activity was assayed by our reported protocol (67). Briefly, brains or cells were homogenized in KHEM buffer (50 mM KCl, 10 mM EGTA, 1.92 mM MgCl2, 1 mM DTT, and 50 mM HEPES, pH 7.2) containing protease inhibitor cocktail (Roche Applied Sciences) and centrifuged, and the supernatant fraction was used to measure the PDE4 activity. Total cAMP-specific phosphodiesterase activity was measured with 1 μM/l cAMP substrate concentration using a fluorescence polarization assay (Molecular Devices) under linear conditions with and without the PDE4 inhibitor rolipram at a concentration known to fully and selectively inhibit PDE4 (10 μM) (35).
Western blotting
A standard protocol was used, including 5%–20% gradient acrylamide gels (ATTO). For detection, appropriate peroxidase-conjugated secondary antibodies (GE Healthcare) were used in conjunction with a SuperSignal West Pico or Femto chemiluminescent substrate (Thermo Fisher Scientific) to obtain images with LAS-3000 Imager (GE Healthcare).
Immunoprecipitation
Typically, cells or brains were lysed with RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and a protease inhibitor cocktail [Roche]). Lysates were sonicated, cell debris was cleared by centrifugation at 200 g for 3 minutes at 4°C, and the soluble fraction was subjected to immunoprecipitation with a primary antibody or an appropriate IgG control with Dynabeads Protein G (Invitrogen). Green fluorescent protein (GFP) or monomeric red fluorescent protein was used as a negative control of binding partner. For sequential immunoprecipitation studies, a Crosslink Magnetic IP/Co-IP Kit (Pierce) was used according to the manufacturer’s protocol. The first and second immunoprecipitations were conducted with anti-DISC1 (m317C) and HTT (MAB2166) antibodies, respectively. An appropriate IgG was used as negative control for each reaction. For competitive binding experiments, lysates of the HEK293T cells overexpressing HTT513Q18-GFP or HTT513Q82-GFP were added to lysates of the HEK293T cells overexpressing DISC1-Myc and PDE4B1-HA. The mixture was processed for immunoprecipitation with an anti–c-Myc antibody.
Sedimentation assay
Samples (0.1 g) of cerebral cortex or striatum in WT or R6/2 mice were homogenized with 1 ml of RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitor cocktail [Roche]) by a digital homogenizer at 1,500 rpm for 15 strokes and sonicated for 10 seconds (Branson sonifier, 15%). The homogenates were partially cleared by centrifugation at 200 g for 3 minutes at 4°C, and the supernatant (total fraction) was further centrifuged at 16,000 g for 15 minutes at 4°C. Protein concentration of the total and supernatant fractions was determined by a BCA assay kit (Thermo Fisher), and 60 μg of protein per well of the total and supernatant fractions was used for immunoblotting.
Filter trap assay
Samples (0.1 g) of cerebral cortex or striatum in WT and R6/2 mice or human controls and HD patients were homogenized with 1 ml of PBS including a protease inhibitor cocktail (Roche) by a digital homogenizer at 1,500 rpm for 15 strokes and sonicated for 10 seconds (Branson sonifier, 15%). The homogenates were cleared by centrifugation at 200 g for 3 minutes at 4°C, and protein concentration of the supernatant (total fraction) was determined by a BCA assay kit (Thermo Fisher Scientific). Fifty micrograms of protein per hole of the total brain homogenate was loaded onto a cellulose acetate membrane (0.2 μm; Advantec) in the dot blot apparatus (Bio-Rad), and the membrane was washed with 2% SDS, followed by immunoblotting. Loading of the same protein amounts on the membrane was confirmed by Western blotting of the total fractions.
Immunohistochemistry
Mouse brains were fixed with 4% buffered paraformaldehyde, mounted with Tissu Mount (Shirai Kogyo), and frozen by carbon dioxide. The frozen brains were sectioned with a cryostat at 10 μm (Leica CM3050 S). Paraffin-embedded human brains were sectioned at 5 μm, followed by autoclaving in 10 mM citrate buffer, pH 6.0, at 121°C for 3 minutes. The paraffin sections were treated with TrueBlack (Biotium) for quenching of lipofuscin autofluorescence, and immunostaining was performed with a primary antibody (1:100 to 1:500), followed by Alexa Fluor 488– or 546–conjugated secondary antibodies (1:300) (Invitrogen). Vectashield mounting medium with DAPI (Vector Laboratories) was used to stain the nucleus. Fluorescent images of the serial-cut frozen or paraffin sections were acquired with a confocal microscope TCS SP8 (Leica) or LSM710 (Zeiss).
Preparation of recombinant proteins and aggregates
Expression and purification of HisGST-tagged HTT67Q42 were reported previously (47). His-tagged human PDE4B2 was expressed at 25°C in E. coli BL21 (DE3) Rosetta and purified with Nickel agarose resin (GE Healthcare). DISC1 protein was expressed at 15°C in E. coli BL21 (DE3) Rosetta as a fusion protein with a Hisx7-tag and trigger factor (TF) from a pCold-TF (Takara) vector and was purified with 50 mM Tris-HCl buffer, pH 8.0, with 1 M NaCl and 2 mM β-mercaptoethanol under the native condition with Nickel agarose resin (GE Healthcare). DISC1 protein was eluted with the same buffer containing 250 mM imidazole, concentrated, and passed through a 1000K filter (Millipore) to remove preexisting aggregates. Staining with Coomassie Brilliant Blue showed that DISC1 was more than 90% pure, and the protein concentration was determined at an extinction coefficient of HisTF-DISC1 (85,600 mM–1cm–1). For aggregate formation, typically 1 ml of 5 μM protein solution with 10 units of turbo3C protease (Accelagen) to cleave the tag was rotated in an end-over-end manner in a 2-ml tube at 37°C (47).
Characterization of DISC1 aggregates in vitro
The physical properties of DISC1 aggregates were examined by an amyloid-specific fluorescent dye, thioflavin T (Sigma-Aldrich). The fluorescence of thioflavin T (excitation, 442 nm; emission, 483 nm) in 50 mM Na-glycine buffer (pH 8.0) including 12.5 μM thioflavin T and 5 μM DISC1 aggregates was measured with a fluorescence plate reader (ARVO MX, PerkinElmer) (47). For morphological observation of DISC1 aggregates with transmission electron microscopy (JEM 1200EX, JEOL), an aliquot of 2 μM DISC1 aggregates was adsorbed onto carbon-coated 400-mesh copper grids, followed by 2% paraformaldehyde and 2% glutaraldehyde in 100 mM sodium phosphate buffer. The samples were negatively stained with 2% sodium phosphotungstic acid. Far-UV circular dichroism and FT-IR spectra of DISC1 aggregates were measured as described previously (47, 68). A core region in DISC1 aggregates was identified with a 4800 Plus MALDI-TOF/TOF Analyzer (AB SCIEX) as previously reported (69). The kinetics of DISC1 aggregation was monitored by the absorbance (turbidity) at 405 nm, with 10 μM DISC1 or PDE4B2 protein in 5 mM potassium phosphate buffer containing 150 mM sodium chloride (pH 7.4), and 2 units of turbo3C protease in the absence or presence of seeds of indicated aggregates (10% [mol/mol]) with a plate reader in a 96-well format (SpectraMax M2, Molecular Devices). As a control, TF was also used in the aggregation assay. The aggregate seeds were prepared by sonication (Branson sonifier, 15%) of 5 μM protein aggregates for 10 seconds. The plate reader instrument agitates a 96-well plate for 5 seconds every 2 minutes. A filter trap assay was performed with 10 μg of HTT67Q42, DISC1, or BSA aggregates using a cellulose acetate membrane (0.2 μm; Advantec) in the dot blot apparatus (Bio-Rad). After the protein was allowed to form aggregate for 2 days, the sample was loaded and the membrane was washed with 5 mM potassium phosphate buffer (pH 7.4) with 2% SDS, followed by detection of signals with Sypro Ruby staining (Bio-Rad). For immunoprecipitation experiments, recombinant soluble DISC1 protein was filtered with a 300-kDa cutoff membrane (Sartorius) to remove preexisting oligomers. The recombinant DISC1 or BSA protein was polymerized in the presence or absence of HA-HTT67Q42 amyloid seeds (10% [mol/mol]). After overnight incubation, the resulting aggregates were captured by anti-HA beads (Invitrogen), washed with the buffer containing 0.1% Triton X-100, and processed for Western blotting using anti-HA and h316C DISC1 antibodies.
Peptide array
MBP-HTT513Q18 protein was expressed at 20°C in E. coli BL21 (DE3) Rosetta as a fusion protein with an N-terminal Hisx7 tag and purified as TF-DISC1. Coomassie Brilliant Blue staining showed that MBP-HTT513Q18 was more than 90% pure, and the protein concentration was determined by an extinction coefficient of MBP-HTT513Q18 (37,940 mM–1cm–1). Peptide arrays of mouse and human DISC1 were generated as previously reported (20, 64, 70). One microgram of maltose-binding protein (MBP) alone or MBP-HTT513Q18 protein in PBS was overlaid to the peptide array. The array was washed with PBS, followed by detection as previously reported (20, 64).
Production of lentivirus and AAV, and injection into mice
HEK293T cells were transfected with a pCSII-CMV–mouse DISC1– or Δ201-228-DISC1-IRES-Venus plasmid together with pCAG-HIVgp and pCAG-VSV-G plasmids, and forskolin (Sigma-Aldrich) was added to the culture medium (10 μM). Purification of lentivirus was performed as previously reported (64), except that the lentivirus was concentrated with Lenti-X Concentrator (Clontech) and the pellet was resuspended with MACS Neurobasal media (Miltenyi Biotec) containing MACS B27 plus (Miltenyi Biotec) and penicillin and streptomycin (Nacalai). Stereotaxic injections were bilaterally placed at 0 and 1.0 mm anterior to the bregma, 2.0 mm lateral to the sagittal suture, and 2.0 mm below the skull surface in 2-week-old WT or R6/2 mice. One microliter of lentivirus (200 μl of lentivirus purified from 10 ml culture) was injected at the rate of 0.1 μl/min by a microinjector (Narishige). Immunohistochemical analyses with frozen sections of the mice confirmed the expression of Venus and DISC1 in striatum. At 8 weeks, the striatum of the mice was removed, minced on ice, and treated with a Papain Dissection System (Worthington) to separate cells. The cells were treated with propidium iodide (Nacalai), and the Venus-expressing and propidium iodide–negative cells were collected by flow cytometry (FACSAria, BD), followed by centrifugation at 1,000 g for 15 minutes at 4°C. Preparation of AAV followed a previous report (60). Stereotaxic injections of AAV into mice were performed as described above.
Analysis of HTT aggregates and neuronal viability in AAV-injected mice
The numbers of cells with HTT aggregates and neurons in striatum of AAV-injected mice were examined as previously described (71). An EM48 (Millipore) or NeuN (Millipore) antibody together with Vectashield mounting medium with DAPI (Vector Laboratories) was used for immunostaining of the striatum of AAV-EGFP– or AAV-Δ201-228-nDISC1–injected mice at 12 weeks.
Behavioral analysis of mice
Following injection of AAV in striatum, the behavioral tests were performed for WT and R6/2 mice at the age of 8–9 weeks.
Rotarod performance test.
A mouse was placed on a rotating rod (Muromachi Kikai), and the latency time for which the mouse was able to run on the rod was measured. The speed of rotation was 4 rpm on day 1 and was increased from 4 to 40 rpm over a 4-minute period and then kept at 40 rpm for another 1 minute from day 2 to day 5. Mice were tested in 1 trial for 2 minutes on day 1 and in 4 trials with a maximum time of 300 seconds (intervals between trials were 20–30 seconds) on days 2–5. The latency time of mice on the rotating rod was recorded by the instrument.
Open field test.
The open field test was performed as previously described (72). Briefly, each mouse was placed in the center of the open field (50[width]x50[depth]x40[height]cm) and allowed to move freely for 15 minutes. Distance traveled (centimeters) and time spent (percent) in the center area (30%) of the field or in the 4 corner squares of the 5×5 subdivisions were measured every 1 minute and analyzed by TimeOFCR4 (O’Hara & Co.).
Light/dark transition.
The light/dark transition test was performed as previously described (72). Briefly, mice were individually introduced into the light box, and the door of the tunnel automatically opened immediately after the software detected the mouse. The mice were then allowed to move freely in the light/dark box for 10 minutes. The total distance traveled, the percentage of time spent in the light box, the number of transitions between the light and dark boxes, and the duration of the first latency period before entry to the dark box were measured as indices. Data were collected and analyzed using Time LD4 (O’Hara & Co.).
Sucrose preference test.
Mice were allowed to take water or 2% sucrose from 2 distinct bottles, and consumed water and sucrose were measured every day for 4 days. The positions of the 2 bottles were switched after 2 days. No specificity of the mice to bottle or its position was observed. Sucrose preference was calculated as a percentage of the weight of sucrose intake over the total weight of fluid intake and averaged over the 4 days of testing.
Marble burying test.
A mouse was placed in a cage in the presence of 15 marbles. After 20 minutes, pictures of the cages were taken, and the number of marbles buried in bedding was counted. For nesting behavior test, a mouse was placed in a cage in the presence of cotton square. After 48 hours, pictures of the cages were taken, and efficiency of the nest building was evaluated by a score of 1 (intact) to 5 as previously reported (73).
Statistics
For analyses of 2 groups, P values were determined by unpaired 2-tailed t test. For analyses of 3 or more groups, including all mouse behavioral tests, P values were determined by 1-way ANOVA followed by Bonferroni post hoc corrections. P values of less than 0.05 (*), less than 0.01 (**), and less than 0.001 (***) were considered to be statistically significant.
Study approval
All of the animal experiments were performed in compliance with relevant laws and guidelines issued by RIKEN Brain Science Institute and Johns Hopkins University. Samples of autopsied brains were obtained from the Harvard Brain Tissue Resource Center at McLean Hospital in Belmont, Massachusetts, USA.
Author contributions
MT, K. Ishizuka, and AS designed the experiments. MT, K. Ishizuka, YNM, RE, NT, YK, K. Ishii, KKWH, and SHK performed the cell biology, mouse, and structural biology experiments. HS and ET measured PDE4 activity. AG, EH, and MDH performed peptide array experiments. MK and NN initially provided R6/2 mice. MT, K. Ishizuka, MDH, and AS wrote the paper.
Supplementary Material
Acknowledgments
We thank Pamela Talalay, Mari Kondo, and Charles Yokoyama for critical reading of the manuscript, Yukiko Lema for organizing the figures and manuscript, Mao Yamaguchi and Mizue Honda for help with genotyping of mice and ovarian transplantation, Yumiko Ohhashi for advice on structural analyses of protein aggregates, and Naomi Takahashi for help with subcloning. We thank Carsten Korth for his gift of 14F2 antibody. DNA sequencing, mass spectrometry, flow cytometry, and electron microscopic analyses were performed at RIKEN Brain Science Institute Research Resources Center. This work was supported by US Public Health Service grants MH-084018 (to AS), MH-094268 Silvio O. Conte Center (to AS), MH-069853 (to AS), MH-085226 (to AS), MH-088753 (to AS), MH-092443 (to AS), MH-105660 (to AS and K. Ishizuka), and MH-96208 (to K. Ishizuka); grants from the Stanley Foundation (to AS), Rusk Foundation (to AS), and the S&R Foundation (to AS); Brain & Behavior Research Foundation(to AS and K. Ishizuka); the Maryland Stem Cell Research Fund (to AS and K. Ishizuka); the Next program (LS129 to MT); Grants-in-Aid for Young Scientists (A) 22680030 (to MT), Young Scientists (B) 23700398 (to RE), and Encouragement of Scientists 24930021 (to YNM) from the Ministry of Education, Culture, Sports, Science and Technology–Japan; the Japan Intractable Diseases Research Foundation (to MT); the Medical Research Council (to MDH); and Scottish Enterprise (to MDH).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(4):1438–1450.https://doi.org/10.1172/JCI85594.
Contributor Information
Motomasa Tanaka, Email: motomasa@brain.riken.jp.
Koko Ishizuka, Email: kishizu2@jhmi.edu.
Yoko Nekooki-Machida, Email: y-nekooki@med.teikyo-u.ac.jp.
Ryo Endo, Email: ryo-endo@brain.riken.jp.
Noriko Takashima, Email: norikot@brain.riken.jp.
Hideyuki Sasaki, Email: sasakihideyu@gmail.com.
Yusuke Komi, Email: ykomi@riken.jp.
Amy Gathercole, Email: Amy.gathercole.88@gmail.com.
Elaine Huston, Email: Elaine.Huston@glasgow.ac.uk.
Kazuhiro Ishii, Email: kenkan55672@gmail.com.
Masaru Kurosawa, Email: mskurosa@juntendo.ac.jp.
Sun-Hong Kim, Email: skim37@jhmi.edu.
Nobuyuki Nukina, Email: nnukina@mail.doshisha.ac.jp.
Eiki Takimoto, Email: etakimo1@jhmi.edu.
Miles D. Houslay, Email: miles.houslay@kcl.ac.uk.
Akira Sawa, Email: asawa1@jhmi.edu.
References
- 1.Crook ZR, Housman D. Huntington’s disease: can mice lead the way to treatment? Neuron. 2011;69(3):423–435. doi: 10.1016/j.neuron.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Prospero NA, Fischbeck KH. Therapeutics development for triplet repeat expansion diseases. Nat Rev Genet. 2005;6(10):756–765. doi: 10.1038/nrg1690. [DOI] [PubMed] [Google Scholar]
- 3.Saudou F, Humbert S. The biology of Huntington’s disease. Handb Clin Neurol. 2008;89:619–629. doi: 10.1016/S0072-9752(07)01257-2. [DOI] [PubMed] [Google Scholar]
- 4.Labbadia J, Morimoto RI. Huntington’s disease: underlying molecular mechanisms and emerging concepts. Trends Biochem Sci. 2013;38(8):378–385. doi: 10.1016/j.tibs.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 6.An MC, et al. Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11(2):253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landles C, Bates GP. Huntingtin and the molecular pathogenesis of Huntington’s disease. Fourth in molecular medicine review series. EMBO Rep. 2004;5(10):958–963. doi: 10.1038/sj.embor.7400250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Craufurd D, Snowden J. Neuropsychological and neuropsychiatric aspects of Huntington’s disease. In: Bates G, Harper P, Jones L, eds. Huntington’s Disease. 3rd ed. Oxford, United Kingdom: Oxford University Press; 2002:62–94. [Google Scholar]
- 9.Kandel ER. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol Brain. 2012;5:14. doi: 10.1186/1756-6606-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duman RS. Synaptic plasticity and mood disorders. Mol Psychiatry. 2002;7(suppl 1):S29–S34. doi: 10.1038/sj.mp.4001016. [DOI] [PubMed] [Google Scholar]
- 11.Arnsten AF, Wang MJ, Paspalas CD. Neuromodulation of thought: flexibilities and vulnerabilities in prefrontal cortical network synapses. Neuron. 2012;76(1):223–239. doi: 10.1016/j.neuron.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gines S, et al. Specific progressive cAMP reduction implicates energy deficit in presymptomatic Huntington’s disease knock-in mice. Hum Mol Genet. 2003;12(5):497–508. doi: 10.1093/hmg/ddg046. [DOI] [PubMed] [Google Scholar]
- 13.Cramer H, Warter JM, Renaud B. Analysis of neurotransmitter metabolites and adenosine 3′,5′-monophosphate in the CSF of patients with extrapyramidal motor disorders. Adv Neurol. 1984;40:431–435. [PubMed] [Google Scholar]
- 14.Conti M, Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu Rev Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 15.Menniti FS, Faraci WS, Schmidt CJ. Phosphodiesterases in the CNS: targets for drug development. Nat Rev Drug Discov. 2006;5(8):660–670. doi: 10.1038/nrd2058. [DOI] [PubMed] [Google Scholar]
- 16.O’Donnell JM, Zhang HT. Antidepressant effects of inhibitors of cAMP phosphodiesterase (PDE4) Trends Pharmacol Sci. 2004;25(3):158–163. doi: 10.1016/j.tips.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Millar JK, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310(5751):1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- 18.Burgin AB, et al. Design of phosphodiesterase 4D (PDE4D) allosteric modulators for enhancing cognition with improved safety. Nat Biotechnol. 2010;28(1):63–70. doi: 10.1038/nbt.1598. [DOI] [PubMed] [Google Scholar]
- 19.Houslay MD. Underpinning compartmentalised cAMP signalling through targeted cAMP breakdown. Trends Biochem Sci. 2010;35(2):91–100. doi: 10.1016/j.tibs.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Murdoch H, et al. Isoform-selective susceptibility of DISC1/phosphodiesterase-4 complexes to dissociation by elevated intracellular cAMP levels. J Neurosci. 2007;27(35):9513–9524. doi: 10.1523/JNEUROSCI.1493-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kvajo M, et al. Altered axonal targeting and short-term plasticity in the hippocampus of Disc1 mutant mice. Proc Natl Acad Sci U S A. 2011;108(49):E1349–E1358. doi: 10.1073/pnas.1114113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10(1):40–68; image 5. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 23.Niwa M, et al. DISC1 a key molecular lead in psychiatry and neurodevelopment: No-More Disrupted-in-Schizophrenia 1. Mol Psychiatry. 2016;21(11):1488–1489. doi: 10.1038/mp.2016.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brandon NJ, Sawa A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat Rev Neurosci. 2011;12(12):707–722. doi: 10.1038/nrn3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnstone M, Thomson PA, Hall J, McIntosh AM, Lawrie SM, Porteous DJ. DISC1 in schizophrenia: genetic mouse models and human genomic imaging. Schizophr Bull. 2011;37(1):14–20. doi: 10.1093/schbul/sbq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narayan S, Nakajima K, Sawa A. DISC1: a key lead in studying cortical development and associated brain disorders. Neuroscientist. 2013;19(5):451–464. doi: 10.1177/1073858412470168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leliveld SR, et al. Insolubility of disrupted-in-schizophrenia 1 disrupts oligomer-dependent interactions with nuclear distribution element 1 and is associated with sporadic mental disease. J Neurosci. 2008;28(15):3839–3845. doi: 10.1523/JNEUROSCI.5389-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atkin T, Kittler J. DISC1 and the aggresome: a disruption to cellular function? Autophagy. 2012;8(5):851–852. doi: 10.4161/auto.19979. [DOI] [PubMed] [Google Scholar]
- 29.Hamburg H, et al. Simultaneous effects on parvalbumin-positive interneuron and dopaminergic system development in a transgenic rat model for sporadic schizophrenia. Sci Rep. 2016;6:34946. doi: 10.1038/srep34946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trossbach SV, et al. Misassembly of full-length Disrupted-in-Schizophrenia 1 protein is linked to altered dopamine homeostasis and behavioral deficits. Mol Psychiatry. 2016;21(11):1561–1572. doi: 10.1038/mp.2015.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies SW, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90(3):537–548. doi: 10.1016/S0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 32.DeMarch Z, Giampà C, Patassini S, Bernardi G, Fusco FR. Beneficial effects of rolipram in the R6/2 mouse model of Huntington’s disease. Neurobiol Dis. 2008;30(3):375–387. doi: 10.1016/j.nbd.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 33.Giampà C, et al. Phosphodiesterase type IV inhibition prevents sequestration of CREB binding protein, protects striatal parvalbumin interneurons and rescues motor deficits in the R6/2 mouse model of Huntington’s disease. Eur J Neurosci. 2009;29(5):902–910. doi: 10.1111/j.1460-9568.2009.06649.x. [DOI] [PubMed] [Google Scholar]
- 34.Wang GH, et al. Caspase activation during apoptotic cell death induced by expanded polyglutamine in N2a cells. Neuroreport. 1999;10(12):2435–2438. doi: 10.1097/00001756-199908200-00001. [DOI] [PubMed] [Google Scholar]
- 35.Houslay MD, Adams DR. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem J. 2003;370(pt 1):1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houslay MD, Schafer P, Zhang KY. Keynote review: phosphodiesterase-4 as a therapeutic target. Drug Discov Today. 2005;10(22):1503–1519. doi: 10.1016/S1359-6446(05)03622-6. [DOI] [PubMed] [Google Scholar]
- 37.MacKenzie SJ, Houslay MD. Action of rolipram on specific PDE4 cAMP phosphodiesterase isoforms and on the phosphorylation of cAMP-response-element-binding protein (CREB) and p38 mitogen-activated protein (MAP) kinase in U937 monocytic cells. Biochem J. 2000;347(pt 2):571–578. doi: 10.1042/0264-6021:3470571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boxall R, Porteous DJ, Thomson PA. DISC1 and Huntington’s disease — overlapping pathways of vulnerability to neurological disorder? PLoS One. 2011;6(1):e16263. doi: 10.1371/journal.pone.0016263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlyle BC, Mackie S, Christie S, Millar JK, Porteous DJ. Co-ordinated action of DISC1, PDE4B and GSK3β in modulation of cAMP signalling. Mol Psychiatry. 2011;16(7):693–694. doi: 10.1038/mp.2011.17. [DOI] [PubMed] [Google Scholar]
- 40.Fatemi SH, et al. PDE4B polymorphisms and decreased PDE4B expression are associated with schizophrenia. Schizophr Res. 2008;101(1–3):36–49. doi: 10.1016/j.schres.2008.01.029. [DOI] [PubMed] [Google Scholar]
- 41.Huston E, et al. Molecular cloning and transient expression in COS7 cells of a novel human PDE4B cAMP-specific phosphodiesterase, HSPDE4B3. Biochem J. 1997;328(pt 2):549–558. doi: 10.1042/bj3280549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scherzinger E, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90(3):549–558. doi: 10.1016/S0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 43.Sawamura N, et al. Nuclear DISC1 regulates CRE-mediated gene transcription and sleep homeostasis in the fruit fly. Mol Psychiatry. 2008;13(12):1138–1148, 1069. doi: 10.1038/mp.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sawamura N, Sawamura-Yamamoto T, Ozeki Y, Ross CA, Sawa A. A form of DISC1 enriched in nucleus: altered subcellular distribution in orbitofrontal cortex in psychosis and substance/alcohol abuse. Proc Natl Acad Sci U S A. 2005;102(4):1187–1192. doi: 10.1073/pnas.0406543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soda T, et al. DISC1-ATF4 transcriptional repression complex: dual regulation of the cAMP-PDE4 cascade by DISC1. Mol Psychiatry. 2013;18(8):898–908. doi: 10.1038/mp.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malavasi EL, Ogawa F, Porteous DJ, Millar JK. DISC1 variants 37W and 607F disrupt its nuclear targeting and regulatory role in ATF4-mediated transcription. Hum Mol Genet. 2012;21(12):2779–2792. doi: 10.1093/hmg/dds106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nekooki-Machida Y, Kurosawa M, Nukina N, Ito K, Oda T, Tanaka M. Distinct conformations of in vitro and in vivo amyloids of huntingtin-exon1 show different cytotoxicity. Proc Natl Acad Sci U S A. 2009;106(24):9679–9684. doi: 10.1073/pnas.0812083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harper JD, Lansbury PT. Models of amyloid seeding in Alzheimer’s disease and scrapie: mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 49.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148(6):1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tessier PM, Lindquist S. Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+] Nat Struct Mol Biol. 2009;16(6):598–605. doi: 10.1038/nsmb.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seshadri S, Khurana R, Fink AL. Fourier transform infrared spectroscopy in analysis of protein deposits. Meth Enzymol. 1999;309:559–576. doi: 10.1016/s0076-6879(99)09038-2. [DOI] [PubMed] [Google Scholar]
- 52.Mao Y, et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3β/β-catenin signaling. Cell. 2009;136(6):1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheung YF, et al. PDE4B5, a novel, super-short, brain-specific cAMP phosphodiesterase-4 variant whose isoform-specifying N-terminal region is identical to that of cAMP phosphodiesterase-4D6 (PDE4D6) J Pharmacol Exp Ther. 2007;322(2):600–609. doi: 10.1124/jpet.107.122218. [DOI] [PubMed] [Google Scholar]
- 54.Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11(11):777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- 55.Lee WC, Yoshihara M, Littleton JT. Cytoplasmic aggregates trap polyglutamine-containing proteins and block axonal transport in a Drosophila model of Huntington’s disease. Proc Natl Acad Sci U S A. 2004;101(9):3224–3229. doi: 10.1073/pnas.0400243101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poças GM, Branco-Santos J, Herrera F, Outeiro TF, Domingos PM. α-Synuclein modifies mutant huntingtin aggregation and neurotoxicity in Drosophila. Hum Mol Genet. 2015;24(7):1898–1907. doi: 10.1093/hmg/ddu606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ottis P, et al. Convergence of two independent mental disease genes on the protein level: recruitment of dysbindin to cell-invasive disrupted-in-schizophrenia 1 aggresomes. Biol Psychiatry. 2011;70(7):604–610. doi: 10.1016/j.biopsych.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 58.Nucifora LG, et al. A mutation in NPAS3 that segregates with schizophrenia in a small family leads to protein aggregation. Mol Neuropsychiatry. 2016;2(3):133–144. doi: 10.1159/000447358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shahani N, et al. DISC1 regulates trafficking and processing of APP and Aβ generation. Mol Psychiatry. 2015;20(7):874–879. doi: 10.1038/mp.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seshadri S, et al. Interneuronal DISC1 regulates NRG1-ErbB4 signalling and excitatory-inhibitory synapse formation in the mature cortex. Nat Commun. 2015;6:10118. doi: 10.1038/ncomms10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ishizuka K, et al. Evidence that many of the DISC1 isoforms in C57BL/6J mice are also expressed in 129S6/SvEv mice. Mol Psychiatry. 2007;12(10):897–899. doi: 10.1038/sj.mp.4002024. [DOI] [PubMed] [Google Scholar]
- 62.Saito A, et al. Early postnatal GABAA receptor modulation reverses deficits in neuronal maturation in a conditional neurodevelopmental mouse model of DISC1. Mol Psychiatry. 2016;21(10):1449–1459. doi: 10.1038/mp.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishizuka K, et al. DISC1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature. 2011;473(7345):92–96. doi: 10.1038/nature09859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hayashi-Takagi A, et al. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci. 2010;13(3):327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schurov IL, Handford EJ, Brandon NJ, Whiting PJ. Expression of disrupted in schizophrenia 1 (DISC1) protein in the adult and developing mouse brain indicates its role in neurodevelopment. Mol Psychiatry. 2004;9(12):1100–1110. doi: 10.1038/sj.mp.4001574. [DOI] [PubMed] [Google Scholar]
- 66.Brandon NJ, et al. Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Mol Cell Neurosci. 2004;25(1):42–55. doi: 10.1016/j.mcn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 67.Takimoto E, et al. Chronic inhibition of cyclic GMP phosphodiesterase 5A prevents and reverses cardiac hypertrophy. Nat Med. 2005;11(2):214–222. doi: 10.1038/nm1175. [DOI] [PubMed] [Google Scholar]
- 68.Ohhashi Y, Ito K, Toyama BH, Weissman JS, Tanaka M. Differences in prion strain conformations result from non-native interactions in a nucleus. Nat Chem Biol. 2010;6(3):225–230. doi: 10.1038/nchembio.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science. 2012;336(6079):355–359. doi: 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- 70.Bolger GB, et al. Scanning peptide array analyses identify overlapping binding sites for the signalling scaffold proteins, beta-arrestin and RACK1, in cAMP-specific phosphodiesterase PDE4D5. Biochem J. 2006;398(1):23–36. doi: 10.1042/BJ20060423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanaka M, et al. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat Med. 2004;10(2):148–154. doi: 10.1038/nm985. [DOI] [PubMed] [Google Scholar]
- 72.Takashima N, et al. Impaired cognitive function and altered hippocampal synapse morphology in mice lacking Lrrtm1, a gene associated with schizophrenia. PLoS One. 2011;6(7):e22716. doi: 10.1371/journal.pone.0022716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nollet M, Le Guisquet AM, Belzung C. Models of depression: unpredictable chronic mild stress in mice. Curr Protoc Pharmacol. 2013;Chapter 5:Unit 5.65. doi: 10.1002/0471141755.ph0565s61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.