Abstract
Phosphoinositides and soluble inositol phosphates are essential components of a complex intracellular chemical code that regulates major aspects of lipid signaling in eukaryotes. These involvements span a broad array of biological outcomes and activities, and cells are faced with the problem of how to compartmentalize and organize these various signaling events into a coherent scheme. It is in the arena of how phosphoinositide signaling circuits are integrated and, and how phosphoinositide pools are functionally defined and channeled to privileged effectors, that phosphatidylinositol (PtdIns) transfer proteins (PITPs) are emerging as critical players. As plant systems offer some unique advantages and opportunities for study of these proteins, we discuss herein our perspectives regarding the progress made in plant systems regarding PITP function. We also suggest interesting prospects that plant systems hold for interrogating how PITPs work, particularly in multi-domain contexts, to diversify the biological outcomes for phosphoinositide signaling.
Keywords: phosphatidylinositol transfer proteins, Sec14, phosphoinositides
Introduction
Phosphatidylinositol is a minor phospholipid in most eukaryotic cells. It is primarily, and perhaps exclusively, produced in the endoplasmic reticulum (ER) via the action of PtdIns synthase. This enzyme uses inositol and cytidine-diphospho-diacylglycerol (CDP-DAG) as substrates to produce PtdIns and cytidine-monophosphate. CDP-DAG is produced by another primarily ER-localized enzyme, the CDP-DAG synthase, which uses phosphatidic acid (PtdOH) and cytidine-trisphosphate as substrates (1,2). The minor contribution of PtdIns to bulk membrane mass notwithstanding, PtdIns is an essential phospholipid in eukaryotic cells. The essential nature of PtdIns in no small part reflects the fact that it is the immediate metabolic precursor of phosphoinositides – an essential cohort of chemically distinct phosphorylated derivatives of PtdIns produced by dedicated lipid kinases that modify the inositol headgroup of PtdIns, or even of other phosphoinositides, at specific positions.
The biological importance of phosphoinositides rests in their utility as versatile signaling molecules in eukaryotic cells. These lipids are not only precursors for other soluble and lipid second messengers, but phosphoinositides collectively form the chemical basis for an important and diverse signaling code (1–4). Their intrinsic signaling properties take several forms. For example, phosphoinositides contribute to the assembly of chemically distinct platforms on membrane surfaces that allow spatial and temporal regulation of protein activities recruited from the cytoplasm. Phosphoinositides are also critical allosteric regulators of numerous enzymes (such as GTPases and phospholipase D; 5,6) and ion channels (7,8). In terms of serving as metabolic precursors for other second messenger signaling molecules, the best documented example is the hydrolysis by phospholipases C of phosphatidylinositol-4,5-bisphosphate (PtdIns-4,5-P2) to generate diacylglycerol (a lipid) and soluble inositol polyphosphates that subsequently launch a protein kinase cascade and a wave of calcium signaling, respectively (9,10). Soluble inositol-polyphosphates are also recognized to play a number of other interesting roles such as serving as allosteric regulators of protein activities (11–14), and even as essential structural cofactors for protein folding (15,16)
Plant systems engage in robust pathways for lipid signaling, including phosphoinositide signaling, and this review is focused on novel mechanisms by which phosphatidylinositol transfer proteins regulate phosphoinositide signaling and the fascinating plant biology associated with such mechanisms. A number of excellent reviews that more broadly discuss lipid and phosphoinositide signaling in plants have recently been published, and we refer the interested reader to them for general information (17–19). The subject of lipid trafficking and the potential roles of other types of lipid transfer proteins in such processes is also an active field in plant biology, and outstanding reviews have been produced regarding various aspects of lipid transfer mechanisms and lipid transfer proteins in plants (20–22). Because this review has a specific focus that does not encompass those important mechanisms and pathways, the reader is again referred to those discussions for additional information.
Setting the conceptual stage for phosphatidylinositol transfer proteins
The experimental linkage of phospholipase C-mediated cleavage of PtdIns-4,5-P2 to calcium signaling was forecast some forty years ago in a remarkable synthesis produced by Robert Michell (Birmingham, UK; 23). Of relevance to this discussion, the Michell review crystallized a topological problem associated with phosphoinositide signaling–that is, how are phosphoinositides replenished at the plasma membrane when PLC activity is highly stimulated? Michell’s conjecture was that phosphoinositide resynthesis at the mammalian plasma membrane demands PtdIns be transported from its compartment of synthesis (the ER) to the plasma membrane where PtdIns kinases can regenerate phosphoinositide pools. It is this ER to plasma membrane PtdIns transfer concept that led to the prognostication that a set of cytoplasmic soluble PtdIns carriers, the PtdIns transfer proteins (PITPs), exist for just such a purpose. In a case of ‘seek and you shall find’, proteins satisfying the biochemical criteria for PITPs were subsequently purified from multiple sources and biochemically characterized (24,25). In lock step with the Michell conjecture, there is to this day a school of thought that interprets PITP cellular activities as intracellular lipid carriers that resupply membranes engaged in phosphoinositide signaling with PtdIns from the ER (26–28). Direct evidence to support such an interpretation is lacking, however.
One issue that frustrated lipid transfer protein studies for many years was the complete lack of knowledge regarding the biological functions of these proteins, and the lack of systems from which to address biological questions–a drought that was broken with the recognition that the yeast Sec14 protein, essential for membrane trafficking through late Golgi/endosomal compartments, is a PITP (29). While studies in multiple genetically tractable systems are now drawing back the curtain to reveal the biological functions of PITPs, central details for how these proteins execute these functions remain unclear. For the Sec14-like PITPs, it is no longer the case that the simplest interpretations of the data are consistent with lipid transfer mechanisms for these proteins. Other ideas now demand consideration. Moreover, the biological functions of multi-domain proteins that harbor Sec14-like modules, and the mechanisms that underlie such functions, are poorly understood. It is in these areas of inquiry that plant systems offer new and exciting avenues for studying the biological and biochemical activities of Sec14-like PITPs. This review summarizes the sparse, but increasingly exciting, progress in understanding mechanisms of plant PITP function. It is abundantly clear that plants employ PITPs in interesting ways, and we endeavor to highlight the unique opportunities provided by plant systems for studying Sec14 - like PITPs in physiologically relevant contexts.
Sec14-like proteins and the coupling of lipid metabolism to phosphoinositide signaling
Recent work demonstrates that PITPs play novel roles in regulating PtdIns kinase activities via mechanisms that do not involve intermembrane PtdIns transport (30,31). This concept derives from a body of work on arguably the best understood of the PITPs -- Sec14, the major yeast PITP, and other Sec14-like proteins (29,31–35). The body of evidence is consistent with Sec14 and other Sec14-like proteins executing an interfacial ‘presentation’ of PtdIns to PtdIns 4-OH kinases. That is, PtdIns 4-OH kinases are biologically inadequate enzymes because of their inability to efficiently engage membrane-incorporated PtdIns substrates. The poor ability of the enzyme to produce PtdIns-4-P is of insufficient power to counter the repressive action of cellular ‘brakes’ that antagonize PtdIns-4-P signaling -- such as the Sac1 phosphoinositide phosphatase and the PtdIns-4-P binding protein Kes1/Osh4 (36–43). Sec14 is proposed to help the PtdIns 4-OH kinase to overcome that threshold by executing abortive PtdIns exchange events, when bound to (i.e. primed with) PtdCho, that expose PtdIns in a neither fully incorporated membrane-bound or Sec14-bound states. Thus, for a brief window in time, a frustrated PtdIns molecule foiled in its entry into the Sec14 hydrophobic pocket is a superior substrate for PtdIns 4-OH kinases. It is via such an abortive exchange mechanism that the stimulated enzyme can generate sufficient PtdIns-4-P to overcome the threshold set by negative regulators of PtdIns-4-P signaling (30,31,41).
There are two attractive corollaries to this mechanism. The first is that the PITP presentation activity exerts an essential level of control that not only stimulates the biologically inadequate activity of PtdIns 4-OH kinases, but does so in response to binding of a second lipid ligand (PtdCho in the case of Sec14). This circuit is suggested to define a common strategy for how diverse cohorts of Sec14-like PITPs (that presumably bind diverse cohorts of second ligands) integrate the activities of multiple lipid metabolic pathways with phosphoinositide signaling (30,31,44,45). Powerful examples in support of this concept include: (i) the biologically essential role of Sec14 in coordinating metabolic flux through the CDP-choline pathway for PtdCho biosynthesis with PtdIns 4-OH kinase activity to regulate the interface between PtdIns-4-P, PtdCho and diacylglycerol metabolism and vesicle trafficking through the yeast trans-Golgi/endosomal pathway (29,33,34,36,39–41,46), and (ii) the role of the Sec14-like PITP Sfh4 in regulating an endosomal pathway for phosphatidylserine (PtdSer) decarboxylation (47–49). The second is that productive activity of the lipid kinase requires a vicinal PITP to present substrate, thereby defining a precise site on a membrane surface where signaling is initiated. As discussed in greater detail below, when coupled to appropriate pre-existing protein assemblies, such local control of PtdIns-4-P production can be channeled directly to unique sets of effector molecules–thereby creating a highly compact and tightly integrated signaling ‘pixel’. This engineering strategy holds the prospect for enabling execution of phosphoinositide signaling at ‘point’ resolution. Such capabilities give birth to powerful new ideas for how cells might prosecute high definition phosphoinositide signaling, and plant systems offer unique models for thorough examination of both of these corollaries given the clear phenotypes associated with specific plant PITP defects that are ripe for productive exploitation.
The plant Sec14-like protein family
Higher eukaryotes express a large number of Sec14 proteins and most of these define interesting modular proteins with Sec14-domains. The dizzying breadth of the Sec14 superfamily is obvious upon examination of even individual plant genomes. For example, Arabidopsis alone expresses 32 distinct members of this superfamily and these proteins span an interesting spectrum of domain arrangements–ranging from the plant-specific Sec14-nodulin two-domain proteins, to Sec14-GOLD proteins also found in other multicellular eukaryotes, to single-domain Sec14 proteins (Figure 1). This dramatic amplification of the plant Sec14 family, as exemplified by Arabidopsis, is a common feature throughout the plant kingdom, and highlights the difficulties with kingdom-wide treatments of the subject. So, this discussion focuses on members of the plant Sec14 superfamily for which functional information already exists, or cases where functional information is almost certain to translate to areas of biological impact. The Sec14-like proteins of other plant species that will be discussed are highlighted in Figure 2 for reference. We expect the concepts gleaned from those case studies to apply generally to PITPs of plant species not discussed here.
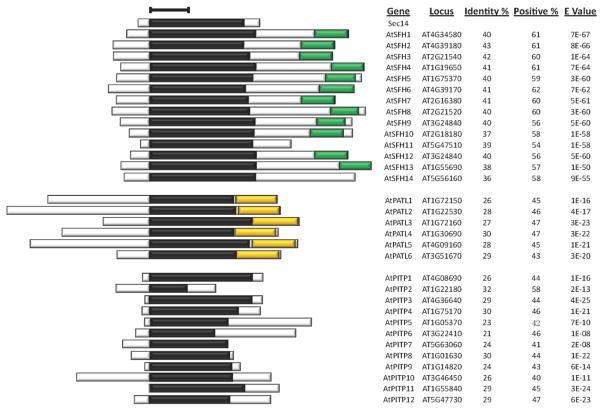
Figure 1. Arabidopsis Sec14-homology proteins.
Sec14-like proteins were identified by interrogating NCBI (http://www.ncbi.nlm.nih.gov/), greenphyl (http://www.greenphyl.org/cgi-bin/index.cgi), and phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) databases with yeast Sec14 as query primary sequence and BlastP as search tool using default parameters (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome). For confirmed the Sec14-homology proteins, the nodulin domain was further screened using Nlj16 sequence as query primary sequence and BlastP as the search tool with default parameters. A hit score of 40 was set as the BlastP cutoff value. The GOLD domain was scanned by the InterPro tool (http://www.ebi.ac.uk/interpro/search/sequence-search) with default parameters. Sec14 domains are indicated by black boxes; nodulin domains are in green, and GOLD domains are in gold. Bar = 100 amino acids. Gene designations and locus identifiers, primary sequence identities and positive similarities of corresponding Sec14-domains to yeast Sec14, and corresponding E-values are listed at right.
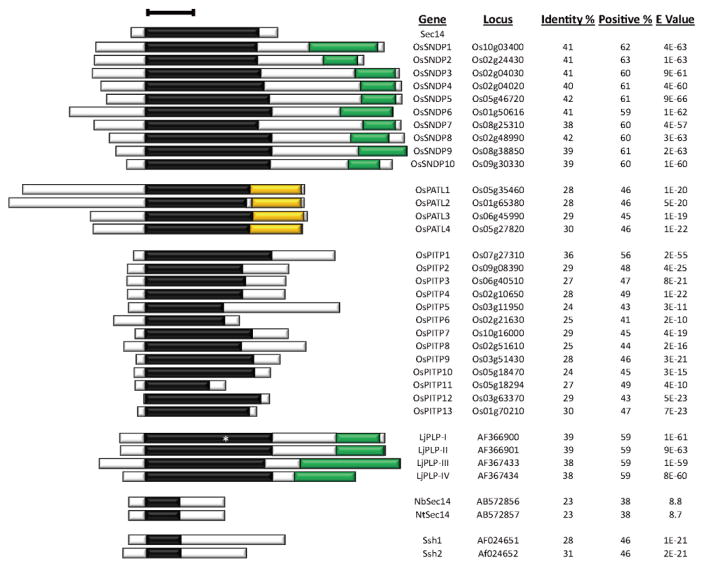
Figure 2. Sec14-homology proteins of rice, birdsfoot trefoil, tobacco, and soybean.
Database searches were as described in the legend to Figure 1, and Figure labeling is also as described in Figure 1. The asterisk in the LjPLP-I denotes an amber stop in the Sec14-domain coding sequence (55), and the Lotus japonicus genome assembly build 3.0 tool (http://www.kazusa.or.jp/lotus/blast.html) predicts two annotated genes (Lj4g3v0988870.1 and Lj4g3v0988870.2, rather than a single Sec14-nodulin gene as shown here. The .1 and .2 suffixes indicate alternative splicing from the same pre-mRNA. The predicted Lj4g3v0988870.1 translation product is a 384 polypeptide containing the N-terminal 159 residues of the Sec14 domain, while the Lj4g3v0988870.2 is predicted to encode a 314 amino acid polypeptide consisting of a 79 residue C-terminus from the Sec14-domain joined to the nodulin-domain. As translational read-through of translation stop codons is known, we list the full-length LjPLP-I polypeptide (550 amino acids) as such read-through could potentially generate such a translation product at some low frequency. Thus, the Sec14-nodulin protein family of Lotus consists of at least three, and perhaps four, proteins. Abbreviations: Os, Oryza sativa; Lj, Lotus japonicus; Nb, Nicotiana benthamiana; Ssh, soybean Sec14 homolog.
In this review, particular emphasis is invested in discussion of plant multi-domain Sec14 proteins. The Arabidopsis and rice cohorts of Sec14-like proteins amply demonstrate that these structurally complex Sec14-like proteins are not only prevalent (at least 18 of the 32 are confidently classified as multi-domain proteins in Arabidopsis), but that some of these arrangements are also more broadly conserved throughout the multicellular Eukaryota. Yet, little information exists regarding in vivo functions for multi-domain Sec14-like proteins. Plant experimental systems offer unique prospects for future research in this arena of lipid metabolism. The free-standing Sec14-like proteins are also numerous (as many as 14 Arabidopsis Sec14-like proteins may fall into this category depending on how stringently one defines a domain), and these too hold their own interesting properties and potentials for new insight into how the phosphoinositide signaling landscape is managed in plants.
Sec14-nodulin two-domain proteins
An impressive feature of the Arabidopsis and rice Sec14-protein families is the large number of Sec14-nodulin proteins. Functional analyses of the Sec14-domains from Arabidopsis Sec14-nodulin proteins demonstrate that all share intrinsic Sec14-like activities (50). These two-domain proteins are of particular interest because these are plant-specific constructions, and the path for how Sec14-nodulins came onto the scientific radar is a curious one. That path deserves a brief summary as it has its origins in the study of the bacterial/legume symbiosis nitrogen fixation and forecasts a large biological significance for these proteins.
The genesis of Sec14-nodulin research came from expression profiling studies that catalogued the genes/proteins of leguminous plants whose expression is elevated during the nodulation phase of legume infection by Rhizobium. While the tractability of Rhizobia to genetic approaches spearheaded progress in understanding the bacterial functions required for productive nodulation, the intractability of legumes to such approaches has retarded progress in understanding the symbiosis problem from the perspective of the plant (51–53). Of the plant proteins expressed during nodulation (hence the name nodulins), one class represents a family of small 14–16Kda proteins for which a cDNA was isolated from the leguminous plant Lotus japonicus. That specific cDNA was derived from an mRNA expressed exclusively in N2-fixing root nodules during late stages of the nodulation program (54). A family of four related nodulin genes, referred to as the Nlj16-like nodulin family, was subsequently identified in Lotus (Figure 2). Curiously, these nodulins are genetically encoded as two-domain Sec14-nodulin proteins with the Sec14 domain having intrinsic PITP activity (LjPLP-IV in the specific case of the Nlj16-nodulin; 55). Nodule-specific expression of the Nlj16 nodulin domain was determined to be driven by a complex and developmentally-regulated reprogramming of gene transcription that activated nodulin domain expression at the expense of the full-length Sec14-nodulin protein (55). The proliferation of Sec14-nodulin proteins in
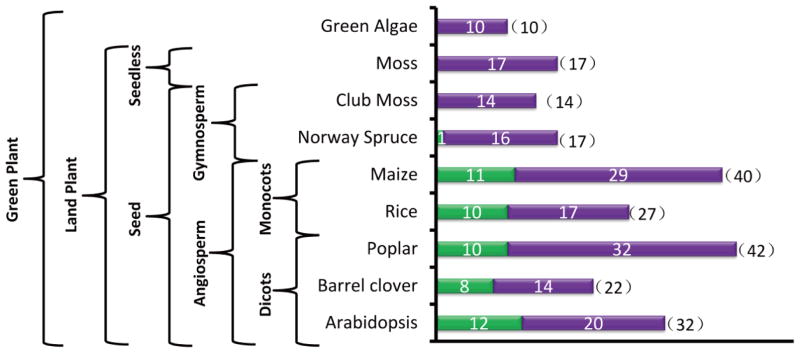
Arabidopsis indicates this modular arrangement is not restricted to legumes. However, survey of plant genomes for Sec14-nodulin genes also indicates these two-domain proteins are restricted to the most evolutionarily advanced plants. Whereas Norway spruce potentially encodes for only one Sec14-nodulin protein, and the presumptive nodulin domain is significantly diverged from other Nlj16-family sequences, individual flowering plant species encode many Sec14-nodulin proteins and the arrangement is conserved across the angiosperms (Figure 3). By far the best studied Sec14-nodulin to date is the Arabidopsis AtSfh1 protein, and it is the product of the gene defined by the can-of-worms (cow1) complementation group identified in studies of short root hair Arabidopsis mutants (50,56,57). We refer to this gene and its product in AtSFH1 (AtSfh1) nomenclature as that designation transmits both functional and gene relatedness information that the COW1 designation does not. Arabidopsis mutants devoid of AtSfh1 are fertile but, consistent with the root-specific expression profile of the AtSFH1 gene, produce short and stubby root hairs that often exhibit two and sometimes even three growing tips. In wild-type plants, AtSfh1 is most concentrated at the tip of the developing root hair root where it is most heavily concentrated on vesicles in the tip cytoplasm, the tip plasma membrane, and in a surprisingly intricate spiraling distribution along the root hair cortical plasma membrane (50). The root hair phenotype of AtSfh1-deficient plants is the consequence of the collapse of a tip-directed PtdIns(4,5)P2 (and perhaps a PtdIns-4-P) gradient in the growing root hair in Arabidopsis (58,59), loss of membrane vesicles from the tip cytoplasm, and derangements in the tip actin microfilament network (a phosphoinositide-responsive system) and in the cortical microtubule cytoskeleton. These derangements are impressively local. That is, the root hair cortical actin cytoskeleton appears substantially normal, and the microtubule cytoskeleton is also apparently normal in the body of the parent trichoblast cell from which the root hair emanates.
Figure 3. A survey of Sec14-nodulin proteins across the plant kingdom.
BlastP searches using the yeast Sec14 as query primary sequence against NCBI: http://www.ncbi.nlm.nih.gov/, greenphyl: http://www.greenphyl.org/cgi-bin/index.cgi, and phytozome: https://phytozome.jgi.doe.gov/pz/portal.html databases were performed. For confirmed the Sec14-homology proteins, the nodulin domain was further screened using Nlj16 sequence as query primary sequence and BlastP as the search tool with default parameters. A hit score of 40 was set as the BlastP cutoff value. Indicated plant species were analyzed. Numbers of total Sec14-like proteins and Sec14-nodulin proteins are plotted. Green bars indicate numbers of Sec14-nodulin proteins, and purple bars indicate numbers of Sec14-like polypeptides without nodulin domains. Numbers in parentheses indicate total Sec14-like proteins produced by the indicated plant species. Taxonomic hierarchy of the selected plant species surveyed is shown at left.
The cumulative data indicate that, in AtSfh1-deficient mutants, a highly polarized system of membrane trafficking collapses into an isotropic mode of deposition of membrane contents into the plasma membrane–including such cargo as ion channels and other receptors. Indeed, rather than exhibiting a strong tip-directed cytoplasmic Ca++ gradient typical of growing root hairs (60), AtSfh1-deficient root hairs present precocious ‘hot-spots’ of high cytoplasmic Ca++ (>600 nM) along the cortical plasma membrane fueled by robust Ca++ influx from the extracellular milieu (~ 4–8pmol Ca++/cm2/sec). This derangement of what is typically a tightly spatially organized pattern of Ca++ uptake is consistent with inappropriate distribution of Ca++-channels across the root hair plasma membrane (50). It is therefore almost certain that other ion channels and solute transporters are similarly mis-distributed in AtSfh1-deficient root hair plasma membrane with the result that root hair ion and solute gradients are generally disrupted.
The detailed functional analyses of AtSfh1 suggest some members of the Sec14-nodulin protein family represent a novel class of polarized membrane trafficking regulators. This inference is supported by tissue expression profiling data indicating that ten of the Arabidopsis Sec14-nodulin proteins are preferentially expressed in tissues that undergo programs of extreme polarized membrane growth -- i.e. root and pollen (www.ncbi.nlm.nih.gov/geo/; summarized in 61). Independent studies also find that the Sec14-nodulin proteins AtSfh3 and AtSfh12 are most highly expressed in flowers (for AtSfh3), or exclusively in flowers for AtSfh12. More detailed analyses show that AtSfh3 is expressed in pollen grains both prior to and after fertilization while AtSfh12 is expressed only in mature and germinating pollen grains (62).
While the expression profiling data are suggestive of general roles for Sec14-nodulin proteins in polarized membrane trafficking, more convincing data that speak directly to this issue are forthcoming from other plant systems. Forward genetic screens for short root hair mutants in rice, coupled to map-based cloning and sequencing approaches, demonstrated that the OsSNDP1 gene is required for proper root hair biogenesis, that the gene product is a plasma membrane-localized Sec14-nodulin protein that exhibits high homology throughout its amino acid sequence to AtSfh1, and that OsSNDP1 expression rescued short root hair phenotypes in Atsfh1 null mutant Arabidopsis plants (63). The inactivating allele identified in the screen evokes a single Gly → Val missense substitution in the Sec14-domain of the protein that does not alter protein localization to the plasma membrane, but presumably inactivates Sec14 domain activity. Moreover, the short-root hair phenotype of Ossndp1 loss-of-function mutants closely mirrors that of Atsfh1 null mutants. OsSNDP1 transcriptional expression is highest in roots and in flowers, particularly anthers, suggesting a role for this Sec14-nodulin protein in pollen tube formation as well (63).
The data obtained from Arabidopsis and rice directly demonstrate that Sec14-nodulin domain proteins play critical roles in regulating extreme modes of polarized membrane growth (i.e. tip-growth) in higher plants. It is also worth emphasizing yet again that rice encode 27 Sec14-like proteins, of which 10 are Sec14-nodulin proteins, and that one of those ten Sec14-nodulin proteins plays a critical role in root hair biogenesis (63; Figure 3). The emerging theme from these early studies is that, the impressive amplification of the Sec14-like protein and the Sec14-nodulin domain families notwithstanding, these proteins channel their respective activities to surprisingly specific physiological contexts (e.g. even specific aspects of membrane polarity programs?).
Functional diversification is also determined by the nodulin domain
Emerging data report that diversification of Sec14-nodulin protein functions does not rest solely with diversification of the biochemical properties of Sec14 domains (see below), but that the nodulin domains also play determining roles. While there exists no structural information for the 16kDa Nlj16-like nodulin unit, both ab initio structure prediction and limited templated structural threading routines consistently project nodulins to assume coiled-coil structures–most probably three-helix coiled coils (61). The Nlj16-like nodulin domains share significant sequence homology across their entire primary sequences, but these modules can be subdivided into three classes based on the nature of their extreme C-terminal sequences. The Class I sequences are marked by an uninterrupted stretch of seven basic residues with vicinal aromatic residues. While the Class II nodulins also have basic C-termini with aromatic residues, the basic residue tracts are shorter than those of the Class I nodulins and Cys residues (potential sites for palmitoylation or prenylation) are more prevalent. The Class III nodulin C-termini also exhibit these features but the sequences are more divergent (61).
The classification translates to biochemical differences. Class I nodulin domains represent phosphoinositide-binding units whereas neither Class II nor Class III have detectable phosphoinositide or other anionic phospholipid binding activity–at least not in the assay systems employed (61). Comprehensive analyses of the AtSfh1 nodulin domain (and indeed all Class I nodulins tested) demonstrate it to be a highly specific PtdIns(4,5)P2-binding unit, that this binding is mediated by the C-terminal run of basic amino acids that distinguish Class I nodulins, and that PtdIns(4,5)P2-binding by this domain is required for AtSfh1 function in promoting polarized tip-directed membrane trafficking in growing root hairs. Several other biochemical features of the nodulin domain are similarly noteworthy (61). These are as follows:
First, these units form stable homo-oligomers and this oligomerization lends high avidity to what is otherwise a highly specific, but low affinity, PtdIns(4,5)P2-binding activity. AtSfh1 nodulin domain mutants that fail to homo-oligomerize fail to bind PtdIns(4,5)P2 in biological membranes and are non-functional proteins in plants.
Second, the extent to which AtSfh1 nodulins self-assemble into larger order oligomers is promoted by neutralizing the basic charge of their C-termini–as would happen upon PtdIns(4,5)P2-binding. Since AtSfh1 localization to the plant plasma membrane is not dependent on the phosphoinositide-binding activity of the nodulin domain, that unit is concluded to function in lateral organization/sequestration of PtdIns(4,5)P2.
Third, AtSfh1 nodulin domains exhibit other functionally important properties that are independent of PtdIns(4,5)P2-binding. That is, AtSfh1 nodulin domain missense mutants were obtained that preserve the nodulin domain’s ability to homo-oligomerize and bind PtdIns(4,5)P2, but are nevertheless non-functional as AtSfh1 nodulin units in the plant. What the activities identified by these particular mutants may be remain unclear. However, one attractive possibility is that the responsible amino acid substitutions break critical protein::protein interactions.
The functional principles gleaned from analyses of AtSfh1 will likely apply to many (if not all) of the Sec14-like proteins with Class I nodulin domains. In this regard, Sec14-nodulin proteins offer interesting possibilities for a lipid signaling nanoreactor that couples phosphoinositide synthesis with spatial organization of the product (61,64). Using AtSfh1 as conceptual template, the phosphoinositide lateral organization principle is envisioned to reside in the nodulin domain, while synthesis of the PtdIns(4,5)P2 ‘pool’ is potentiated by the Sec14-domain. That locally produced pool of phosphoinositide is efficiently captured by electrostatic interactions with the AtSfh1 nodulin domain–thereby enabling a nanopatterning of a specific PtdIns(4,5)P2 pool on root hair membrane surfaces with the potential to channel the phosphoinositide to vicinal effector molecules. AtSfh1 and PtdIns(4,5)P2 confocal imaging data in growing root hairs show that AtSfh1 and areas of PtdIns(4,5)P2 enrichment adopt similar patterns (50). Although it is yet to be demonstrated that these patterns are coincident, the imaging data are consistent with AtSfh1 promoting PtdIns(4,5)P2 synthesis and lateral organization in root hair membrane ‘nano-domains’. This concept is further supported by the demonstration that loss of AtSfh1 function results in loss of the PtdIns(4,5)P2 arrangement typical of growing wild-type root hairs.
The notion that Class I nodulin domains interact physically (and perhaps transiently) with appropriate PtdIns 4-OH and PtdIns4-P 5-OH kinase isozymes is now an attractive one as such a design would consolidate the cardinal features of AtSfh1 biochemistry and cell biology. In that regard, the question of whether AtSfh1 interacts with the PtdIns 4-OH kinase beta-1 and/or PIP5K3 PtdIns4-P 5-OH kinase isozymes is an especially interesting one given that PtdIns 4-OH kinase beta-1 and PIP5K3 deficiencies evoke short root hair phenotypes similar to those associated with AtSfh1 deficiencies (59,65). Moreover, the fact that biological outcomes of PtdIns(4,5)P2 signaling in pollen tubes can be altered by PtdIns-4-P 5-OH kinase domain-swap indicates the determining role played by lipid kinase interaction partners (66). If AtSfh1 does play a role in PtdIns(4,5)P2 nanopatterning, an important question that remains is how the nodulin domain:: PtdIns(4,5)P2 clamp is released. While the data indicate high local Ca++ is insufficient to do so by itself, a Ca++-calmodulin-dependent release mechanism remains a possibility, given the high affinity of Ca++-calmodulins for basic peptides, and post-translational modifications of the nodulin domain may also be involved (61). The effectors of AtSfh1-dependent PtdIns(4,5)P2 signaling remain to be identified. But root hair phospholipase D isoforms (e.g. PLDζ; 67) are potential candidates of lipid signaling import to consider, as is the Agd1 Arf GTPase activating protein given Agd1 shows genetic interactions with AtSfh1 and PIP5K3 (68).
Some of the functional principles gleaned from study of AtSfh1 will translate to Class II and Class III nodulin-containing Sec14 nodulin domains as well. For example, all of the Arabidopsis nodulin domains homo-oligomerize when expressed as recombinant proteins (61). But, their protein interaction partners and small molecule ligands (if any) will almost certainly be different. What biological functions these Sec14-nodulins play, and what the underlying biochemical principles are, define questions for the future. We suggest that these early returns already make a compelling case that detailed study of the biology of Sec14-nodulin proteins is warranted. For plant biologists, the first wave of scientific developments should hold an added attraction as the question defines an area of inquiry that is uniquely of plants.
The Sec14-GOLD patellins
Another structural arrangement favored by evolution is the coupling of Sec14 domains to what is referred to as a GOLD (Golgi dynamic) domain–a module originally identified as a family of primary sequences related to the luminal portion of the p24 protein superfamily of secretory cargo receptors using bioinformatics approaches (69,70). Crystallography shows the GOLD domain of the mammalian Sec14-GOLD protein Sec14L2 adopts an eight-stranded jelly-roll β-barrel structural fold (71). It remains to be determined whether this structural fold generally applies to all GOLD domains. It is also unclear whether the GOLD domain is a functional signature for membrane trafficking-associated proteins, or not – the suggestive designation notwithstanding. Nonetheless, the Sec14-GOLD arrangement is on display in worms, flies, mammals, and plants, and the Sec14-module is consistently placed N-terminal to the GOLD domain. Unlike the nodulins, that are genetically encoded as fusions to Sec14 domains (see above), Sec14-GOLD proteins comprise only one subset of a total of six distinct sub-families of GOLD-containing proteins. Moreover, genetically fused Sec14-GOLD domains do not seem to be uncoupled via altered transcriptional schemes. As is the case with Sec14-nodulin proteins, the patellins are rather late additions to the plant repertoire of Sec14-like proteins as these Sec14-GOLD proteins first appear in gymnosperms.
Arabidopsis expresses six Sec14-GOLD proteins referred to as the patellins PATL1-PATL6 (Figure 1), and the rice genome sequence encodes four patellins (Figure 2). This gene family was discovered by Peterman and co-workers who leveraged a zucchini tryptic peptide sequence obtained from a protein that co-purified with a membrane-associated actin-binding protein to isolation of the cognate Arabidopsis and zucchini genes and subsequent recognition of their other homologs (72,73). The patellin moniker given to these proteins acknowledges the recruitment of AtPATL1 from cytoplasmic and/or endosomal reservoirs to the expanding cell plate of actively dividing cells where this Sec14-GOLD protein is proposed to be involved in maturation of the cell plate during the late telophase stage of cytokinesis (72). This is an intriguing result on two counts. First, cell plate biogenesis and subsequent maturation relies on a complex course of membrane trafficking to build the initial structure and intensive membrane remodeling to complete it. Second, the Sec14-GOLD construction itself combines a Sec14 lipid-binding unit demonstrated to play an essential role in yeast membrane trafficking with a putative protein interaction domain recognized in the p24 class of membrane trafficking cargo receptors.
At this point, the precise role of AtPATL1 in cell plate maturation remains unclear. The timing of AtPATL1 recruitment to the cell plate suggests the protein is not involved in the early membrane trafficking events associated with cell plate biogenesis. Rather, on the basis of several observations, it is inferred that AtPATL1 more likely participates in clathrin-dependent endocytic events that aid in the remodeling and completion of the cell plate (72). This inference is an interesting one given the modular engineering of AtPATL1 as a lipid signaling unit. Its C - terminal GOLD domain exhibits a lysine rich 551KX10K/RK3MQ2-3YR573 motif that resembles known PtdIns(4,5)P2 binding motifs of proteins directly involved in endocytosis. Furthermore, in vitro vesicle sedimentation experiments indicate that AtPATL1 is a phosphoinositide binding protein with some positional specificity of binding (72). Thus, as has been amply demonstrated for the Class 1 Sec14-nodulin proteins discussed above, the Sec14-GOLD proteins may also be built as tightly integrated signaling circuits where stimulated phosphoinositide synthesis is coupled to lateral organization of phosphoinositide via direct channeling to a binding module–in this case the GOLD domain. The presence of a ‘YGEFQ’ motif suggests a AtPATL1 recognition sequence for γ-ear domains of clathrin adaptor proteins (72), and raises the intriguing possibility that this specific patellin potentiates local phosphoinositide synthesis and organization by channeling newly synthesized phosphoinositide to lipid-regulated components of the clathrin endocytic machinery.
The fact that AtPATL1 is functionally the best characterized patellin emphasizes just how little is known about these intriguing proteins in plants. This is not exceptional in and of itself as such gaps in functional understanding are the case for the Sec14-GOLD proteins in general. Yet, the broad conservation of Sec14-GOLD proteins in multicellular eukaryotes forecasts an important physiological function for these proteins. Thus, Sec14-GOLD proteins are compelling subjects for future study, and plant systems are poised for exploitation as attractive models systems in this area of investigation. With regard to efforts in plant systems, other avenues for investigating patellin biological functions are emerging that promise new insights and opportunities. For example, brassinosteroid signaling in plants regulates many developmental processes that are influenced by cellular functions that could reasonably involve PATL proteins, such as membrane trafficking, cytoskeleton dynamics, lipid signaling, etc. In that regard, 2D-DIGE proteomic profiling efforts document, and follow-up experiments confirm, that AtPATL1, AtPATL2 and AtPATL4 protein expression is rapidly elevated upon exposure of Arabidopsis to brassinosteroid (74). Moreover, AtPATL1 and AtPATL2 exhibit behaviors suggesting these patellins are core members of this steroid hormone response pathway given that mutants deranged for brassinosteroid signaling are also coherently deranged for AtPATL1 and AtPATL2 expression responses to challenge with hormone (74). One interesting possibility is that the AtPATL Sec14-domains bind brassinosteroid as ‘second-ligand’ in a version of the Sec14 PtdIns-presentation mechanism for stimulation of PtdIns-kinase activity as discussed above. Such an idea may hold considerable merit given that the mammalian Sec14-GOLD protein Sec14L2 stimulates activity of the cholesterol biosynthetic pathway enzyme squalene epoxidase, and that its Sec14 domain binds tocopherols, tocotrienols, squalene, and oxidized sterols (75–77).
Recent work is featuring patellins on another exciting front, and that is in plant host defense mechanisms to viral infection. Protein interaction screens identified AtPATL3 and AtPATL6 as direct interactors with the Alfalfa mosaic virus movement protein that plays a critical role in systemic spread of virus from the site of initial infection. Interestingly, both patellins antagonize movement protein activity by interfering with its targeting to the plasmodesmata of infected plants. This interference is of biological relevance as both single and double mutant plants showed increased viral RNA loads and increased susceptibility to systemic dissemination of virus (78). The roles of Sec14-domain proteins in host/pathogen contexts and in plant immunity are also gaining traction in other systems as well (see below), and this idea identifies an open area of impactful research.
Free-standing Sec14-domain proteins in plants
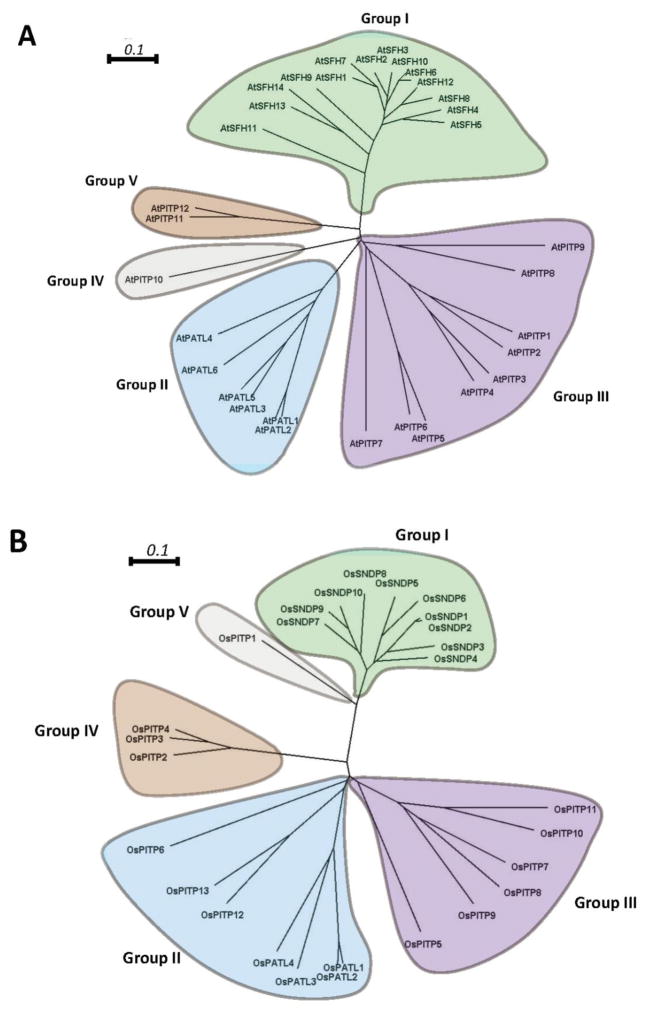
Phylogenetic analyses that interrogate the relatedness of isolated Arabidopsis Sec14-like proteins/domains distribute these primary sequences into five clades (Figure 4A). Strikingly, the Sec14-domains of Sec14-nodulins form one clade of their own (Group I), and the patellin Sec14-domains also distribute into their own exclusive clade (Group II). The stand-alone Sec14 proteins are more diverse and are classified into three distinct clades (Groups III, IV and V). This phylogenetic structure is recapitulated in the rice Sec14-homology protein family (Figure 4B). Again, the Sec14 domains of Sec14-nodulins and patellins define their own exclusive clades while the free-standing Sec14-like proteins fall into three others. One interpretation of these patterns is that Arabidopsis and rice Sec14-domains define at least five distinct biochemical categories of Sec14-like protein/domain. As described below, these distinctions might lie primarily in the nature of their ‘second-ligand’ lipid-binding properties.
Figure 4. Molecular phylogenetic analysis of Arabidopsis and rice Sec14-like domains by the neighbor-joining method.
Arabidopsis (A) and rice (B) Sec14-domain primary sequences were aligned using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) with the default parameters except order = input. Aligned sequence files were the process using the ClustalW2 phylogeny tool with default parameters (http://www.ebi.ac.uk/Tools/phylogeny/clustalw2_phylogeny/). The phylogenetic tree files were plotted and edited using SplitsTree4. For both the Arabidopsis and the rice analyses, the Sec14-domains were classified into 5 clades. These clades are designated Group I - Group V and are highlighted by differential coloring. Bars = 0.1 represent amino acid genetic changes of 0.1 (http://www.ebi.ac.uk/Tools/phylogeny/clustalw2_phylogeny/).
Although our discussion of the plant single-domain Sec14-like proteins follows that of the multi-domain Sec14-like proteins, the former were the first to appear on the plant PITP scene. Their discovery was facilitated by construction of cDNA libraries from various plant sources for the purpose of supporting phenotypic rescue screens designed to identify functional plant paralogs of yeast activities of interest for which convenient mutant alleles were available. The temperature sensitive for growth sec14-1ts yeast strain was one that was exploited in this fashion to identify presumptive plant PITP genes, and the first progress on that front came from the isolation of two genes from soybean and one from Arabidopsis (79,80). All three genes encode single-domain Sec14-like proteins. The Arabidopsis gene was originally designated as AtSEC14 on the basis of its ability to rescue Sec14 defects in yeast but, as it is a more distant Sec14 paralog, we designate it as AtPITP11. This gene is ubiquitously expressed throughout the plant, and the AtPITP11 exhibits PtdIns-transfer activity, but no PtdCho-transfer activity, in vitro (78). Functional analyses of AtPITP11 have yet to be reported, but we note that Arabidopsis encodes a close paralog (AtPITP12) that likely shares at least some functional redundancy with AtPITP11–thereby potentially complicating such analyses (Figure 4A).
The two soybean genes (Soybean Sec14 Homologs 1 and 2) encode what are almost certainly functionally distinct Sec14-like proteins (79). Whereas the SSH1 gene is transcribed most robustly in leaves and roots, SSH2 expression is highest in developing seeds. The proteins are also biochemically distinct. Ssh2 exhibits PtdIns-transfer activity in vitro, but not PtdCho-transfer activity, while Ssh1 does not exhibit either lipid transfer activity–at least not in the assay systems employed. More interestingly, photo-labeling and competitive binding experiments identified both Ssh1 and Ssh2 as phosphoinositide-binding proteins albeit with distinct binding specificities (79). Whereas the significance of those activities remains to be determined, the ability of the osmotic stress-induced phosphoinositide PtdIns(3,5)P2 to bind Ssh1 is of particular interest given that hyperosmotic stress, particularly salt stress, evokes rapid phosphorylation of Ssh1 (but not of Ssh2) both in a heterologous yeast system and in plants. The net result of this modification is to release Ssh1 from membranes–suggesting that this Sec14-like polypeptide is mobilized from membrane stores upon challenge with osmotic stress so as to launch at least one phosphoinositide signaling arm of a larger cellular osmoregulatory response pathway (79,81).
The phosphorylation of Ssh1 in plants is a strikingly local phenomenon (81). The response is tightly confined to the tissue region in direct contact with the osmotic stressor, and the modification is induced in response to several different types of hyperosmotic stress (e.g. dehydration, high sugar, high salt). Phosphorylation occurs exclusively on Ser and Thr (predominantly Thr) residues, is directly mediated by the highly homologous pair of soybean stress-activated kinases SPK1 and SPK2, and these kinases are activated rapidly upon challenge with stressor (81). Whereas the sites of Ser/Thr modification have not been precisely determined, and functional studies of Ssh1 and Ssh2 in the plant remain to be done, the collective Ssh1 data forecast that Sec14-like proteins will play important stress-response roles in plants. In many cases, it may ultimately prove that it is under specific stress conditions that the physiological roles for individual Sec14-like proteins will be revealed. In that regard, we note that AtPITP11/AtPITP12 represent the pair of Arabidopsis proteins most related to soybean Ssh1 by primary sequence–suggesting that one or both of these proteins prosecute Ssh1-like functions in Arabidopsis.
The Nicotiana benthamiana SEC14 gene (NbSEC14) is another, more distantly-related, single-domain Sec14-like protein whose discovery was driven by differential display approaches (82). NbSec14 too has the capacity, when expressed in yeast, to phenotypically rescue the growth and membrane trafficking defects that accompany shift of sec14-1ts mutants to restrictive temperatures, and the polypeptide exhibits both PtdIns- and PtdCho-transfer activity in vitro. These properties are quite surprising given the significant divergence of this PITP from Sec14. The differential display parameter that revealed NbSEC14 involved challenge of Nicotiana with a virulent soil pathogen (Ralstonia solanacearum) in a screen for genes whose products are involved in the plant immune response to this gram-negative bacterium. The data confirm that NbSec14 expression is induced upon stimulation of either transmembrane protein pattern recognition receptor or effector-triggered immunity pathways. This infection-induced expression pattern is also physiologically relevant as plants silenced for NbSEC14 expression, although viable, are markedly sensitized to Ralstonia infection (82). NbSec14 reduces plant vulnerability to infection by stimulating activity of the jasmonic acid pathway for plant defense responses, and stimulating activities for multiple lipid signaling pathways involving phospholipases C and D and diacylglycerol kinase–all of which are linked to innate immunity in plants (83). Thus, NbSec14 plays an important role in coordinating the lipid signaling interface with innate plant immunity responses. We interpret this to be yet another example where a Sec14-like protein is involved in a specialized plant stress response pathway.
Phospholipid-binding bar codes in plant Sec14-like proteins
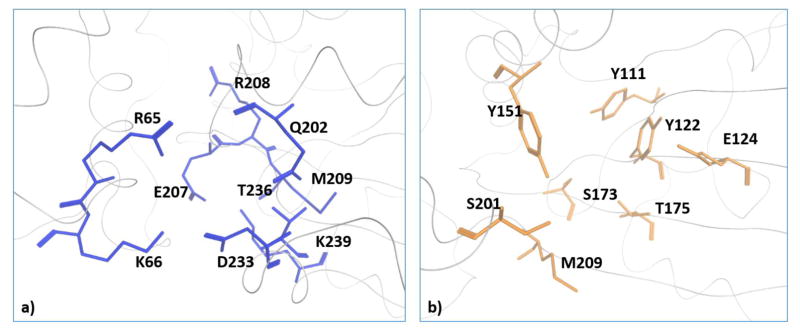
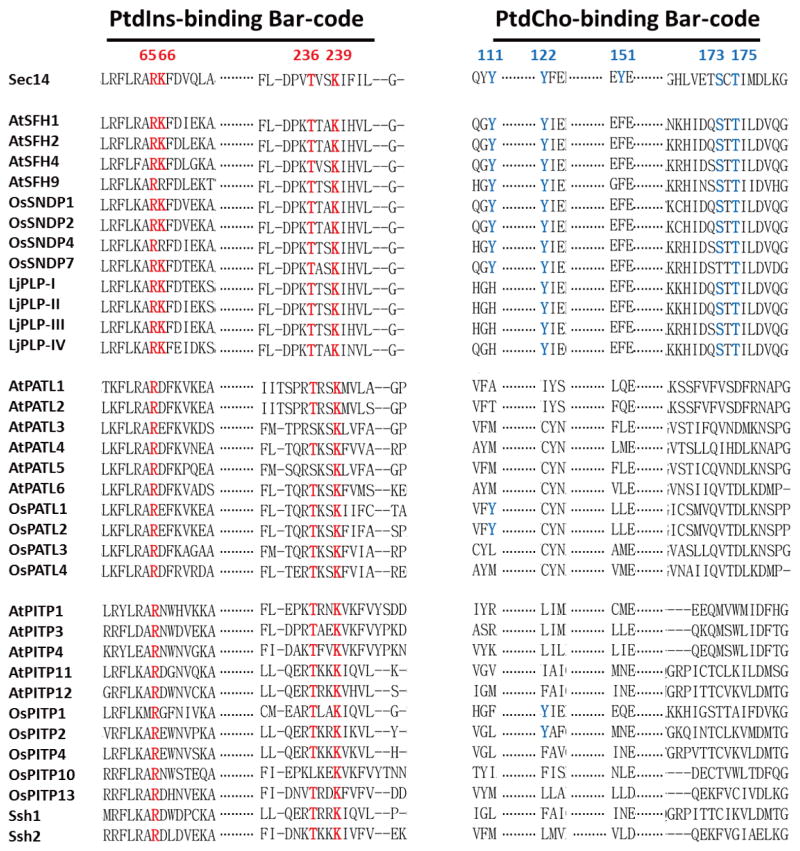
X-Ray crystallography studies demonstrate that yeast Sec14-like PtdIns/PtdCho-transfer proteins bind PtdCho and PtdIns headgroups at physically distinct sites, and identify structural signatures (bar-codes) for PtdIns - and PtdCho-binding that can be recognized at the primary sequence level (30). These bar codes translate 3-dimensional information into the language of primary sequence, and structural depictions of the PtdIns and PtdCho-binding bar-codes of Sec14 are shown in Figure 5. Whereas the PtdIns headgroup is coordinated by an extensive H-bond network and one electrostatic interaction, the PtdCho headgroup is coordinated by two H-bonds from Ser173/Thr175 residues interacting with the headgroup phosphate and cation-π interactions involving two Tyr residues (Tyr122,Tyr151) that form a ‘tyrosine cage’ surrounding the choline headgroup.
Figure 5. Structural description of Sec14 PtdIns- and PtdCho-binding bar-codes.
The Sec14 PtdIns and PtdCho bar-code residues are rendered in blue stick and orange stick mode, respectively. The phospholipids are not included in these models. See ref 30 for those structural details.
Primary sequence comparisons of plant Sec14 domains show that the PtdIns-binding bar code is conserved in the Sec14 superfamily proteins, particularly in the cases of higher plant Sec14-like proteins, whereas the PtdCho-binding bar code is not (Figure 5). Those divergences predict that the plant Sec14 superfamily conserves PtdIns-binding activity while simultaneously diversifying ‘second ligand’ binding specificity. It is proposed that it is in this way that the plant Sec14-like proteins integrate the metabolism of diverse lipids/lipophilic molecules to stimulated phosphoinositide synthesis. This general mechanism enjoys supporting evidence building in analyses of yeast, mammalian systems, and inherited diseases in humans associated with Sec14-domain protein deficiencies as well (30,31,44,45).
The breakdown of which plant proteins exhibit a PtdCho-binding bar-code vs which proteins do not is intriguing. The Arabidopsis, rice and Lotus Sec14-nodulin proteins all conserve a consensus, or a nearly consensus, PtdCho-binding bar-code. None of the patellins of Arabidopsis or rice do. For those proteins not only is the bar-code degenerate but, throughout, the patellins exhibit substitutions in that motif that are expected to be incompatible with PtdCho-binding (as inferred from mutagenesis experiments of that bar-code in the yeast Sec14-like proteins; 30). Thus, the PtdIns-kinase stimulatory activities of patellin Sec14-domains are projected to be primed by second-ligands that are distinct from those of Sec14-nodulin proteins. Similarly, the single-domain Sec14-like proteins of Arabidopsis and rice also lack recognizable PtdCho-binding bar-codes. The inabilities of Ssh1, Ssh2 and AtPITP11 to transfer PtdCho in vitro (79,80; see above) are fully consistent with those proteins lacking PtdCho-binding bar-codes.
The conformational dynamics wiring scheme of Sec14 domains
As the body of structural information of Sec14-like proteins grows, it is becoming increasingly clear that the general structural principles which underly how Sec14 functions as a molecule will translate to the plant PITPs as well. That is, the nature of the conformational dynamics that accompany the lipid exchange reactions of plant Sec14-like proteins can be inferred from the body of work done with yeast Sec14 and Sfh proteins. The major conformational transition Sec14 undergoes during lipid exchange is from the ‘open’ conformer actively involved in phospholipid exchange on membrane surfaces (84), to a ‘closed’ conformer in which one phospholipid molecule is stably incorporated into the protein interior, is shielded from bulk solvent, and represents the solution structure of the protein (30). The transition between these two conformers is dominated by a large (~ 18 Å) repositioning of a helical gate over the lipid binding pocket, and chemical cross-linking experiments confirm that these helical gate dynamics are essential for phospholipid exchange activity in vitro (85). As far as the phospholipid exchange substrates are concerned, electron spin resonance experiments demonstrate that cycling through the Sec14 protein interior represents a simple partitioning of phospholipid molecules between two chemically equivalent environments. That is, between the Sec14 hydrophobic pocket and the cytoplasmic leaflet of the membrane (86). The lipid-binding pocket of Sec14 is not anhydrous, however (30,84,87). Structural data project that H2O flux in this cavity minimizes the conformational adjustments required for Sec14 to bind PtdCho vs PtdIns. Two ordered H2O molecules fill the phosphocholine headgroup-binding space in PtdIns-bound Sec14/Sfh, whereas five ordered H2O molecules occupy the ‘empty’ phosphoinositol headgroup-binding cleft when Sec14/Sfh1 is bound to PtdCho. These H2O rearrangements in the lipid-binding cavity are proposed to overcome the energy barriers that confront PtdIns/PtdCho-exchange reactions (85).
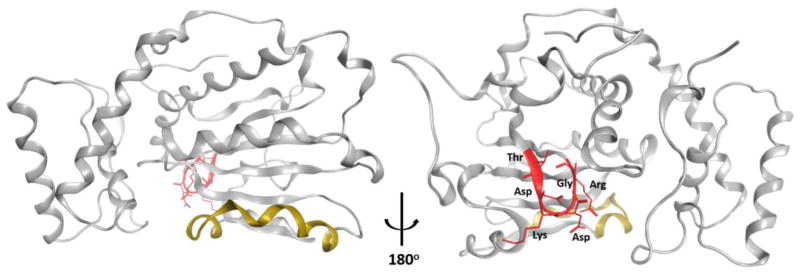
Unrestrained MD simulations that successfully model helical gate closing and opening identify these transitions to be governed by large rigid body motions controlled by a hinge unit that interfaces with the N- and C-terminal ends of the helical gate. MD simulations also identify a conformational switch unit (the G-module) which transduces conformational information to the hinge and, ultimately, to the helical gate (85). The G-module, which is central to control of Sec14 conformational transitions, includes (but is not limited to) a 17 amino acid sequence (residues P108-E125) with a core 115TDKDGR120 motif. Structural representations of this core motif are shown in Figures 7A and 7B, and the G-module connects the interior of the hydrophobic pocket with structural elements at the back-end of the Sec14 molecule required for appropriate helical gate dynamics. The core motif of the G-module is not only conserved in far-flung mammalian members of the Sec14 protein superfamily, but inherited missense mutations associated with this motif are recognized as human disease alleles–thereby suggesting that the conformational dynamics proteins/domains are broadly conserved across the Sec14-like protein superfamily (85). The mechanism by which G-module switch activity is controlled remains an important and outstanding question. However, directed evolution screens point to a key role for H2O fluxes within the lipid-binding cavity in the coupling of G-module activity with structural dynamics of the hydrophobic pocket during lipid exchange (88).
Figure 7. Structural description of the core unit of the Sec14 G-module.
Sec14 α-chain is represented as grey ribbon with the helical substructure that gates entry to the hydrophobic pocket highlighted in yellow. The core TDKDGR motif of the G-module is highlighted in red. Panel at left shows Sec14 with 'helical gate' facing front with TDKDGR motif at the back of the molecule while the panel at right is rotated by 180° to visualize the back face of the Sec14 module and better highlight the core TDKDGR unit.
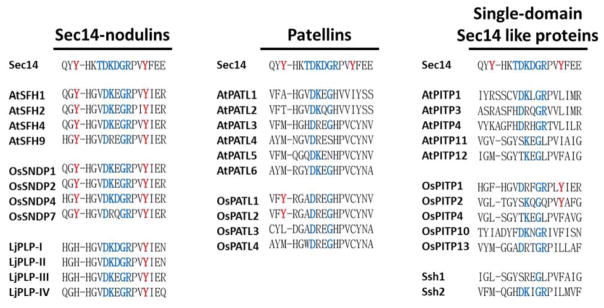
The core element of the G-module is highly conserved in the Sec14-nodulin proteins and the patellins of Arabidopsis, rice and Lotus (Figure 8). While only the identities between the corresponding motifs of these two-domain Sec14 proteins are highlighted in Figure 8, inspection of the alignments show that even the divergences involve conservative amino acid substitutions. The single domain Sec14 homology proteins similarly preserve the core unit of the G-module. Thus, primary sequence alignments forecast that the conformational switch element wiring diagram of the yeast Sec14 will translate broadly to plant Sec14-like proteins.
Figure 8. The core G-module of plant Sec14-homology proteins.
Alignment analyses of the indicated Sec14 homology proteins were performed using ClustalW2 as described in the legend to Figure 6. Precisely conserved core G-module residues (TDKGDR) are highlighted in blue and, for purposes of reference, the Y-residues forming the tyrosine cage of the PtdCho-binding bar code residues are highlighted in red.
Future prospects
Genome sequencing efforts amply demonstrate that plants make robust use of the Sec14 domain and that these organisms do so in the context of both single domain polypeptides and in multi-domain arrangements. Our knowledge of the biological functions of these Sec14-like PITPs is scant, however. In the cases where there is information, the biological context is a significant one. The relationship between plant PITPs and stress-signaling pathways is an area of inquiry that we anticipate will prove an active and fruitful one. We hold the same optimism with regard to plant PITPs and establishment and maintenance of cell polarity and in supporting extreme modes of polarized membrane trafficking. The mechanisms by which these polypeptides are themselves localized and ‘patterned’ raise key questions for future research. To date, most of the progress on plant PITPs has come from functional, physical interaction and expression profiling screens and we expect that trend will continue. However, there have been few efforts targeted at direct functional analyses of individual plant PITPs and this represents a wide open area ripe for exploitation.
A great deal remains to be learned about Sec14-homogy proteins and their activities as molecules. Whereas much of the structural information regarding how the Sec14 domains operate as single molecules can be translated from the yeast proteins to those of plants, many questions remain on that front as well. Prime among these are what are the lipid-binding specificities of plant Sec14-homology proteins, and how do these biochemical properties correlate with phylogenetic relationships? Such information will be invaluable in deciphering what territories of lipid metabolism are integrated with specific phosphoinositide-dependent signaling pathways that channel to specific biological outcomes.
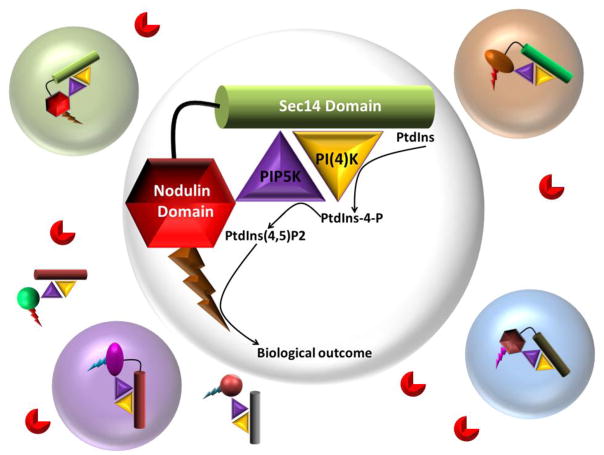
The multi-domain Sec14-like PITPs of plants afford additional opportunities for scientific inquiry. What is the mechanistic relationship between the multi-domain arrangement and biological outcome? What are the downstream effectors to which the phosphoinositide signal is channeled? These questions require a confident knowledge of the compositions of multi-protein complexes of which the PITP is a component. Recent progress made on understanding how Sec14-nodulins function as single molecules argues for more of those kinds of studies. Indeed, ideas for how Sec14-nodulins operate as single molecules set useful conceptual blueprints for interpreting integrated design of phosphoinositide signaling circuits at the molecular level, and how these circuits enable phosphoinositide signaling to be prosecuted at essentially ‘point’ resolution. We note that the patellins are also appropriately configured to participate in analogous circuit designs (Figure 9).
Figure 9. Organization of PITP-dependent phosphoinositide circuits for signaling with point resolution.
Distinct classes of Sec14 multi-domain proteins (Sec14-domains, cylinders; nodulin-domains, hexagons; GOLD-domains ovals) scaffold PtdIns and PtdIns-phosphate kinase (gold and purple triangles, respectively) assemblies with distinct phosphoinositide effectors (bolts). Different colors depict differences in the biochemical properties of Sec14- and associated domains and effectors, and these differences specify the functional channeling of each individual signaling circuit or pixel (open circles). That individual signaling pixels prosecute distinct biological outcomes for phosphoinositide signaling is illustrated by the pixel boundaries being shaded in different colors. Phosphoinositide phosphatases (red PacMan) hydrolyze phosphoinositides that escape pixel boundaries and thereby sharpen those boundaries. Stabilities of these complexes determine their corresponding lifetimes and impose another flexible layer of control to the phosphoinositide signaling landscape.
Finally, there are interesting evolutionary questions. Why is it that the Sec14-domains of Sec14-nodulins, i.e. PITPs of the most evolutionarily advanced plants, share the greatest relatedness to fungal Sec14s? Is this a case of convergent evolution of a Sec14-domain with specific biochemical properties (i.e. PtdCho-binding), or is this the consequence of a fortuitous lateral gene transfer to plants from a fungal pathogen–a circumstance subsequently exploited by plants for organizing and diversifying their phosphoinositide signaling landscapes at what is effectively ‘point’ resolution? We anticipate that studies of plant PITPs in the coming years will make large and unique contributions to a more complete understanding of what is already an exciting biology and a fascinating biochemistry of phosphoinositide signaling. .
Figure 6. Alignment of PtdIns- and PtdCho-binding bar codes in plant Sec14-like proteins.
Alignments of the indicated Arabidopsis and rice Sec14 homology proteins were performed using ClustalW2. As indicated, selected Sec14 homology proteins include Sec14-nodulin proteins (top rows) and GOLD domains (middle rows). Representative single domain Sec14-like proteins discussed in the text are also included (bottom rows). Precisely conserved PtdIns- binding bar code residues (R65T236 and K66K239) are highlighted with red; and conserved PtdCho-binding bar code residues (Y111Y122 Y151S173T175) are highlighted in blue. The Y122 residue is included in the PtdCho-binding bar-code because it is involved in coordinating the binding of other amino-phospholipids (e.g. phosphatidylethanolamine) in the hydrophobic pocket. It is, however, less critical for the binding of PtdCho.
Highlights.
PITPs and the diversification of phosphoinositide signaling outcomes.
Architecture of the plant Sec14-like PITP family.
Biological functions of Sec14-homology proteins in plants.
Functional anatomy for how plant PITP Sec14-homology domains work as molecules.
Acknowledgments
This work was supported by grants from the National Institutes of Health (RO1 GM44530) and the Robert A. Welch Foundation (BE-0017). We thank Ashutosh Tripathi (TAMHSC) for producing Figures 5 and 7. The authors declare no financial conflicts.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Michell RH. Inositol derivatives: evolution and functions. Nat Rev Mol Biol. 2008;9:151–161. doi: 10.1038/nrm2334. [DOI] [PubMed] [Google Scholar]
- 2.Balla T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 4.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 5.Brown FD, Rozelle AL, Yin HL, Balla T, Donaldson JG. Phosphatidylinosito 4,5-bisphosphate and Arf6-regulated membrane traffic. J Cell Biol. 2001;154:1007–1017. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung JK, Sekiya F, Kang HS, Lee C, Han JS, Kim SR, Bae YS, Morris AJ, Rhee SG. Synaptojanin inhibition of phospholipase D activity by hydrolysis of phosphatidylinosito 4,5-bisphosphate. J Biol Chem. 1997;272:15980–15985. doi: 10.1074/jbc.272.25.15980. [DOI] [PubMed] [Google Scholar]
- 7.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 8.Hilgemann DW, Feng S, Nasuhoglu C. The complex and intriguing lives of PIP2 with ion channels and transporters. Science, STKE. 2001:re19. doi: 10.1126/stke.2001.111.re19. [DOI] [PubMed] [Google Scholar]
- 9.Berridge MJ, Irvine RF. Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature. 1984;312:315–321. doi: 10.1038/312315a0. [DOI] [PubMed] [Google Scholar]
- 10.Nishizuka Y. The role of protein kinase C in cell surface signal transduction and tumour promotion. Nature. 1984;308:693–698. doi: 10.1038/308693a0. [DOI] [PubMed] [Google Scholar]
- 11.Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inosito 1,4,5- trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 12.Alcázar-Román AR, Elizabeth J, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- 13.Lee YS, Mulugu S, York JD, O’Shea EK. Regulation of a Cyclin/CDK/CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laha D, Johnen P, Azevedo C, Dynowski M, Weiß M, Capolicchio S, Mao H, Iven T, Steenbergen M, Freyer M, Gaugler P, de Campos MKF, Zhen N, Feussner I, Jessen HJ, Van Wees SC, Saiardi A, Schaaf G. VIH2 regulates the synthesis of inositol pyrophosphate InsP8 and jasmonate-dependent defenses in Arabidopsis. Plant Cell. 2015;27:1082–1097. doi: 10.1105/tpc.114.135160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 17.Thole JM, Nielsen E. Phosphoinositides in plants: novel functions in membrane trafficking. Curr Opin Plant Biol. 2008;11:620–631. doi: 10.1016/j.pbi.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Heilmann M, Heilmann I. Plant phosphoinositides-complex networks controlling growth and adaptation. Biochim Biophys Acta. 2015;1851:759–769. doi: 10.1016/j.bbalip.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Munnik T, Nielsen E. Green light for polyphosphoinositide signals in plants. Curr Opin Plant Biol. 2011;14:489–497. doi: 10.1016/j.pbi.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Kader JC. Lipid-transfer proteins in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:627–654. doi: 10.1146/annurev.arplant.47.1.627. [DOI] [PubMed] [Google Scholar]
- 21.Benning C, Xu C, Awai K. Non-vesicular and vesicular lipid trafficking involving plastids. Curr Opin Plant Biol. 2006;9:241–247. doi: 10.1016/j.pbi.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Benning C. Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu Rev Cell Dev Biol. 2009;25:71–91. doi: 10.1146/annurev.cellbio.042308.113414. [DOI] [PubMed] [Google Scholar]
- 23.Michell RH. Inositol phospholipids and cell surface receptor function. Biochim Biophys Acta. 1975;415:81–147. doi: 10.1016/0304-4157(75)90017-9. [DOI] [PubMed] [Google Scholar]
- 24.Wirtz KWA. Phospholipid transfer proteins. Annu Rev Biochem. 1991;60:73–99. doi: 10.1146/annurev.bi.60.070191.000445. [DOI] [PubMed] [Google Scholar]
- 25.Karel WA, Wirtz KWA. Phospholipid transfer proteins in perspective. FEBS Lett. 2006;580:5436–5441. doi: 10.1016/j.febslet.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 26.Cockcroft S, Carvou N. Biochemical and biological functions of class I phosphatidylinositol transfer proteins. Biochim Biophys Acta. 2007;1771:677–691. doi: 10.1016/j.bbalip.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Shadan S, Holic R, Carvou N, Ee P, Li M, Murray-Rust J, Cockcroft S. Dynamics of lipid transfer by phosphatidylinositol transfer proteins in cells. Traffic. 2008;10:1743–1756. doi: 10.1111/j.1600-0854.2008.00794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prinz WA. Lipid trafficking sans vesicles: where, why, how? Cell. 2010;143:870–874. doi: 10.1016/j.cell.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bankaitis VA, Aitken JR, Cleves AE, Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. doi: 10.1038/347561a0. [DOI] [PubMed] [Google Scholar]
- 30.Schaaf G, Ortlund E, Tyeryar K, Mousley C, Ile K, Woolls M, Garrett T, Raetz CRH, Redinbo M, Bankaitis VA. The functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the Sec14-superfamily. Molecular Cell. 2008;29:191–206. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bankaitis VA, Mousley CJ, Schaaf G. Sec14-superfamily proteins and the crosstalk between lipid signaling and membrane trafficking. Trends in Biochemical Sciences. 2010;35:150–160. doi: 10.1016/j.tibs.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bankaitis VA, Malehorn DE, Emr SD, Greene RR. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J Cell Biol. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cleves AE, McGee TP, Whitters EA, Champion KM, Aitken JR, Dowhan W, Goebl M, Bankaitis VA. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cleves AE, Mcgee T, Bankaitis VA. Phospholipid transfer proteins: a biological debut. Trends Cell Biol. 1991;1:30–34. doi: 10.1016/0962-8924(91)90067-j. [DOI] [PubMed] [Google Scholar]
- 35.Ren J, Lin CPC, Pathak M, Temple BRS, Nile AH, Mousley CJ, Duncan MC, Eckert D, Leiker TJ, Ivanova PT, Milne DS, Murphy RS, Brown HA, Verdaasdonk J, Bloom KS, Ortlund EA, Neiman AM, Bankaitis VA. A phosphatidylinositol transfer protein integrates phosphoinositide signaling with lipid droplet metabolism to regulate a developmental program of nutrient stress-induced membrane biogenesis. Molecular Biology of the Cell. 2014;25:712–727. doi: 10.1091/mbc.E13-11-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cleves AE, Novick PJ, Bankaitis VA. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J Cell Bio. 1989;109:2939–2950. doi: 10.1083/jcb.109.6.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo S, Stolz LE, Lemrow S, York JD. SAC1-like domains of yeast SAC1, INP52 and INP53, and human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- 38.Rivas MP, Kearns BG, Xie Z, Guo S, Sekar MC, Hosaka K, Kagiwada S, York JD, Bankaitis VA. Relationship between altered phospholipid metabolism, DAG, 'bypass Sec14p', and the inositol auxotrophy of yeast sac1 mutants. Mol Biol Cell. 1999;10:2235–2250. doi: 10.1091/mbc.10.7.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang M, Kearns BG, Gedvilaite A, Kagiwada S, Kearns M, Fung MKY, Bankaitis VA. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 1996;15:6447–6459. [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Rivas MP, Fang M, Marchena J, Mehotra B, Chaudhary A, Feng L, Prestwich GD, Bankaitis VA. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J Cell Biol. 2002;157:63–77. doi: 10.1083/jcb.200201037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mousley C, Yuan P, Gaur NA, Trettin KD, Nile AH, Deminoff S, Dewar BJ, Wolpert M, Macdonald JM, Herman PK, Hinnebusch AG, Bankaitis VA. A sterol binding protein integrates endosomal lipid metabolism with TOR signaling and nitrogen sensing. Cell. 2012;148:702–715. doi: 10.1016/j.cell.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G. Osh4p exchanges sterols for phosphatidylinosito 4-phosphate between lipid bilayers. J Cell Biol. 2011;195:965–978. doi: 10.1083/jcb.201104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Filseck JM, Vanni S, Mesmin B, Antonny B, Drin G. A phosphatidylinositol-4-phosphate powered exchange mechanism to create a lipid gradient between membranes. Nature Communications. 2015;6:6671. doi: 10.1038/ncomms7671. [DOI] [PubMed] [Google Scholar]
- 44.Nile AH, Bankaitis VA, Grabon A. Mammalian diseases of phosphatidylinositol transfer proteins and their homologs. Clinical Lipidology. 2010;5:867–897. doi: 10.2217/clp.10.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grabon A, Khan D, Bankaitis VA. Phosphatidylinositol transfer proteins and instructive regulation of lipid kinase biology. Biochim Biophys Acta. 2015;1851:724–735. doi: 10.1016/j.bbalip.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kearns BG, McGee TP, Mayinger P, Gedvilaite A, Phillips SE, Kagiwada S, Bankaitis VA. Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature. 1997;387:101–105. doi: 10.1038/387101a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu WI, Routt S, Bankaitis VA, Voelker D. A new gene involved in transport-dependent metabolism of phosphatidylserine, PSTB2/PDR17, shares sequence similarity with the gene encoding the PI-/PC-TP, Sec14p. J Biol Chem. 2000;275:14446–14456. doi: 10.1074/jbc.275.19.14446. [DOI] [PubMed] [Google Scholar]
- 48.Gulshan K, Shahi P, Moye-Rowley WS. Compartment-specific synthesis of phosphatidylethanolamine is required for normal heavy metal resistance. Mol Biol Cell. 2010;21:443–455. doi: 10.1091/mbc.E09-06-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riekhof W, Wu WI, Jones JL, Nikrad M, Chan MM, Loewen CJR, Voelker DR. An assembly of proteins and lipid-binding domains regulates transport of phosphatidylserine to phosphatidyserine decarboxylas 2 in Saccharomyces cerevisiae. J Biol Chem. 2014;289:5809–5819. doi: 10.1074/jbc.M113.518217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vincent P, Chua M, Nogue F, Fairbrother A, Mekeel H, Xu Y, Allen N, Bibikova TN, Gilroy S, Bankaitis VA. A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hair. J Cell Biol. 2005;168:801–812. doi: 10.1083/jcb.200412074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Long SR. Genes and signals in the Rhizobium-legume symbiosis. Plant Physiology. 2001;125:69–72. doi: 10.1104/pp.125.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gage DJ. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev. 2004;68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oldroyd GE. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol. 2013;11:252–263. doi: 10.1038/nrmicro2990. [DOI] [PubMed] [Google Scholar]
- 54.Kapranov P, de Bruijn FJ, Szczyglowski K. Novel, highly expressed late nodulin gene (LjNOD16) from Lotus japonicas. Plant Physiol. 1997;113:1081–1090. doi: 10.1104/pp.113.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kapranov P, Routt SM, Bankaitis VA, de Bruijn FJ, Szczglowski K. Nodule-specific regulation of phosphatidylinositol transfer protein expression in Lotus japonicas. Plant Cel. 2001;13:1369–1382. doi: 10.1105/tpc.13.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grierson CS, Roberts K, Feldmann KA, Dolan L. The COW1 locus of Arabidopsis acts after RHD2, and in parallel with RHD3 and TIP1, to determine the shape, rate of elongation, and number of root hairs produced from each site of hair formation. 1997;115:981–90. doi: 10.1104/pp.115.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Böhme K, Li Y, Charlot F, Grierson C, Marrocco K, Okada K, Laloue M, Nogué F. The Arabidopsis COW1 gene encodes a phosphatidylinositol transfer protein essential for root hair tip growth. The Plant Journal. 2004;40:686–698. doi: 10.1111/j.1365-313X.2004.02245.x. [DOI] [PubMed] [Google Scholar]
- 58.Braun M, Baluska F, von Witsch M, Menzel D. Redistribution of actin, profilin and phosphatidylinositol-4,5-bisphosphate in growing and maturing root hairs. Planta. 1999;209:435–443. doi: 10.1007/s004250050746. [DOI] [PubMed] [Google Scholar]
- 59.Preuss ML, Schmitz AJ, Thole JM, Bonner HK, Otegui MS, Nielsen E. A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hair cells in Arabidopsis thaliana. J Cell Biol. 2006;172:991–998. doi: 10.1083/jcb.200508116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wymer CL, Bibikova TN, Gilroy S. Cytoplasmic free calcium distributions during the development of root hairs of Arabidopsis thaliana. The Plant Journal. 1997;12:427–439. doi: 10.1046/j.1365-313x.1997.12020427.x. [DOI] [PubMed] [Google Scholar]
- 61.Ghosh R, de Kampos MKF, Huang J, Hur S, Orlowski A, Yang Y, Tripathi A, Nile AH, Lee H-C, Schäfer H, Dynowski M, Rog T, Lete MG, Ahyayauch H, Alonso A, Vattulainen I, Igumenova TI, Schaaf G, Bankaitis VA. Sec14-nodulin proteins and the patterning of phosphoinositide landmarks for developmental control of membrane morphogenesis. Molecular Biology of the Cell. 2015;26:1764–1781. doi: 10.1091/mbc.E14-10-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mo P, Zhu Y, Liu X, Zhang A, Yan C, Wang D. Identification of two phosphatidylinositol/phosphatidylcholine transfer protein genes that are predominantly transcribed in the flowers of Arabidopsis thaliana. J Plant Physiol. 2007;164:478–486. doi: 10.1016/j.jplph.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 63.Huang J, Kim CM, Xuan Y-H, Park SJ, Piao HL, Je BI, Liu J, Kim TH, Kim B-K, Han C-D. OsSNDP1, a Sec14-nodulin domain-containing protein, plays a critical role in root hair elongation in rice. Plant Mol Biol. 2013;82:39–50. doi: 10.1007/s11103-013-0033-4. [DOI] [PubMed] [Google Scholar]
- 64.Ile KE, Schaaf G, Bankaitis VA. Phosphatidylinositol transfer proteins and cellular nanoreactors for lipid signaling. Nature Chem Biol. 2006;2:576–583. doi: 10.1038/nchembio835. [DOI] [PubMed] [Google Scholar]
- 65.Kusano H, Testerink C, Vermeer JE, Tsuge T, Shimada H, Oka A, Munnik T, Aoyama T. The Arabidopsis phosphatidylinositol phosphat 5-kinase PIP5K3 is a key regulator of root hair tip growth. Plant Cell. 2008;20:367–380. doi: 10.1105/tpc.107.056119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stenzel I, Ischebeck T, Quint M, Heilmann I. Variable regions of PI4P 5-kinases direct PtdIns(4,5)P2 toward alternative regulatory functions in tobacco pollen tubes. Front Plant Sci. 2012;2:114. doi: 10.3389/fpls.2011.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohashi Y, Oka A, Rodrigues-Pousada R, Possenti M, Ruberti I, Morelli G, Aoyama T. Modulation of phospholipid signaling by GLABRA2 in root hair pattern formation. Science. 2003;300:1427–1430. doi: 10.1126/science.1083695. [DOI] [PubMed] [Google Scholar]
- 68.Yoo CM, Quan L, Cannon AE, Wen J, Blancaflor EB. AGD1, a clas 1 ARF-GAP, acts in common signaling pathways with phosphoinositide metabolism and the actin cytoskeleton in controlling Arabidopsis root hair polarity. Plant Journal. 2012;69:1064–1076. doi: 10.1111/j.1365-313X.2011.04856.x. [DOI] [PubMed] [Google Scholar]
- 69.Springer S, Chen E, Duden R, Marzioch M, Rowley A, Hamamoto S, Merchant S, Schekman R. The p24 proteins are not essential for vesicular transport in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:4034–4039. doi: 10.1073/pnas.070044097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anantharaman V, Aravind L. The GOLD domain, a novel protein module involved in Golgi function and secretion. Genome Biol. 2002;3:research0023.1–research0023.7. doi: 10.1186/gb-2002-3-5-research0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stocker A, Tomizaki T, Schulze-Briese C, Baumann U. Crystal structure of the human supernatant protein factor. Structur. 2002;10:1533–1540. doi: 10.1016/s0969-2126(02)00884-5. [DOI] [PubMed] [Google Scholar]
- 72.Peterman TK, Ohol YM, McReynolds LJ, Luna EJ. Patelli 1, a novel Sec14-like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiol. 2004;136:3080–3094. doi: 10.1104/pp.104.045369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peterman TK, Sequeira AS, Samia JA, Lunde EE. Molecular cloning and characterization of patellin1, a novel sec14-related protein, from zucchini (Cucurbita pepo) Plant Physiol. 2006;163:1150–1158. doi: 10.1016/j.jplph.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 74.Deng Z, Zhang X, Tang W, Oses-Prieto JA, Suzuki N, Gendron JM, Chen H, Guan S, Chalkley RJ, Peterman TK, Burlingame A, Wang Z-Y. A proteomic study of brassinosteroid response in Arabidopsis. Mol Cell Proteomics. 2007;6:2058–2071. doi: 10.1074/mcp.M700123-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zimmer S, Stocker A, Sarbolouki MN, Spycher SE, Sassoon J, Azzi A. A novel human tocopherol-associated protein: cloning, in vitro expression, and characterization. J Biol Chem. 2000;275:25672–25680. doi: 10.1074/jbc.M000851200. [DOI] [PubMed] [Google Scholar]
- 76.Yamauchi J, Iwamoto T, Kida S, Masushige S, Yamada K, Esashi T. Tocopherol-associated protein is a ligand-dependent transcriptional activator. Biochem Biophys Res Commun. 2001;285:295–299. doi: 10.1006/bbrc.2001.5162. [DOI] [PubMed] [Google Scholar]
- 77.Shibata N, Arita M, Misaki Y, Dohmae N, Takio K, Ono T, Inoue K, Arai H. Supernatant protein factor, which stimulates the conversion of squalene to lanosterol, is a cytosolic squalene transfer protein and enhances cholesterol biosynthesis. Proc Natl Acad Sci USA. 2001;98:2244–2249. doi: 10.1073/pnas.041620398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peiro A, Izquierdo-Garcia AC, Sanchez-Navarro JA, Pallas V, Mulet JM, Aparicio F. Patellin 3 and 6, two members of the plant patellin family, interact with movement protein of alfalfa mosaic virus and interfere with viral movement. Mol Plant Pathol. 2014;15:881–891. doi: 10.1111/mpp.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kearns MA, Monks DE, Fang M, Rivas MP, Courtney PD, Chen J, Prestwich GD, Theibert AB, Dewey RE, Bankaitis VA. Novel developmentally regulated phosphoinositide binding proteins from soybean whose expression bypasses the requirement for an essential phosphatidylinositol transfer protein in yeast. EMBO J. 1998;17:4004–4017. doi: 10.1093/emboj/17.14.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jouannic N, Lepetit M, Vergnolle C, Cantrel C, Gardies AM, Kader JC, Arondel V. Isolation of a cDNA from Arabidopsis thaliana that complements the sec14 mutant of yeast. Eur J Biochem. 1998;258:402–10. doi: 10.1046/j.1432-1327.1998.2580402.x. [DOI] [PubMed] [Google Scholar]
- 81.Monks DE, Aghoram K, Courtney PD, DeWald DB, Dewey RE. Hyperosmotic stress induces the rapid phosphorylation os a soybean phosphatidylinositol protein homolog through activation of the protein kinases SPK1 and SPK2. The Plant Cell. 2001;13:1205–1219. doi: 10.1105/tpc.13.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kiba A, Nakano M, Vincent-Pope P, Takahashi H, Sawasaki T, Endo Y, Ohnishi K, Yoshioka H, Hikichi Y. A novel Sec14 phospholipid transfer protein from Nicotiana benthamania is up-regulated in response to Ralstonia solanacearum infection, pathogen associated molecular patters and effector molecules and involved in plant immunity. J Plant Physiol. 2012;169:1017–1022. doi: 10.1016/j.jplph.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 83.Kiba A, Galis I, Hojo Y, Ohnishi K, Yoshioka H, Hikichi Y. SEC14 phospholipid transfer protein is involved in lipid signaling-mediated plant immune responses in Nicotiana benthamania. PLOS One. 2014;9:e98150. doi: 10.1371/journal.pone.0098150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sha B, Phillips SE, Bankaitis VA, Luo M. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol transfer protein Sec14p. Nature. 1998;391:506–510. doi: 10.1038/35179. [DOI] [PubMed] [Google Scholar]
- 85.Ryan MM, Temple BRS, Phillips SE, Bankaitis VA. Conformational dynamics of the major yeast phosphatidylinositol transfer protein Sec14p: Insights into the mechanisms of phospholipid exchange and diseases of Sec14p-like protein deficiencies. Mol Biol Cell. 2007;18:1928–1942. doi: 10.1091/mbc.E06-11-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smirnova T, Chadwick TG, van Tol J, Ozarowski A, Poluektov O, Schaaf G, Ryan MM, Bankaitis VA. Local polarity and hydrogen bonding inside the Sec14p phospholipid-binding cavity: High-field multifrequency electron paramagnetic studies. Biophys J. 2007;92:3686–3695. doi: 10.1529/biophysj.106.097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smirnova T, Chadwick TG, MacArthur R, Poluekov O, Song L, Ryan M, Schaaf G, Bankaitis VA. The chemistry of PL binding by the Saccharomyces cerevisiae phosphatidylinositol transfer protein Sec14 as determined by electron paramagnetic resonance spectroscopy. J Biol Chem. 2006;281:34897–34908. doi: 10.1074/jbc.M603054200. [DOI] [PubMed] [Google Scholar]
- 88.Schaaf G, Dynowski M, Mousley CJ, Shah SD, Yuan P, Winklbauer E, de Campos MKF, Trettin K, Quinones MC, Smirnova T, Yanagisawa LL, Ortlund E, Bankaitis VA. Resurrection of a functional phosphatidylinositol transfer protein from a pseudo-Sec14 scaffold by directed evolution. Mol Biol Cell. 2011;22:892–905. doi: 10.1091/mbc.E10-11-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]