INTRODUCTORY PARAGRAPH
Amniogenesis - the development of amnion - is a critical developmental milestone for early human embryogenesis and successful pregnancy1,2. However, human amniogenesis is poorly understood due to limited accessibility to peri-implantation embryos and a lack of in vitro models. Here we report an efficient biomaterial system to generate human amnion-like tissue in vitro through self-organized development of human pluripotent stem cells (hPSCs) in a bioengineered niche mimicking the in vivo implantation environment. We show that biophysical niche factors act as a switch to toggle hPSC self-renewal versus amniogenesis under self-renewal-permissive biochemical conditions. We identify a unique molecular signature of hPSC-derived amnion-like cells and show that endogenously activated BMP-SMAD signaling is required for the amnion-like tissue development by hPSCs. This study unveils the self-organizing and mechanosensitive nature of human amniogenesis and establishes the first hPSC-based model for investigating peri-implantation human amnion development, thereby helping advance human embryology and reproductive medicine.
MAIN TEXT
During implantation of a human embryo, amnion cells (amnioblasts) are the first differentiated cell group emerging from an expanding pluripotent epiblast population and will give rise to a polarized squamous amniotic epithelium that encloses the amniotic cavity1,2 (Fig. 1a). Despite its basic and clinical significance, amnion development in humans is poorly understood due to limited studies on peri-implantation human embryos2 and drastic differences in amniogenesis between human and other commonly used amniote models1,3. Even with recent progress in developing in vitro systems4,5, including in vitro cultured human embryos6,7, for studying early human embryogenesis, the development of human amnion remains mysterious.
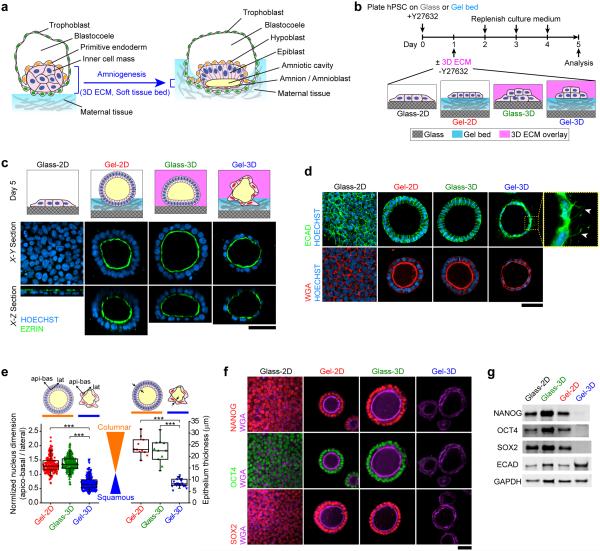
Figure 1. hPSCs form squamous cysts with amnion-like morphology in an implantation-like niche.
(a) Development of amnion/amnioblasts from epiblasts in a peri-implantation human embryo1. Amniogenesis in vivo occurs in a biophysical niche featuring a soft tissue bed (maternal tissue and invading trophoblasts) and a 3D extracellular matrix (ECM) provided by the overlying primitive endoderm/hypoblasts. (b) hPSC amniogenesis assay. (c) Cartoons showing hPSC morphogenesis under different culture conditions (top). Confocal micrographs showing the X-Y (middle) and X-Z (bottom) sections of the hPSC monolayer and cysts formed in indicated conditions at day 5. EZRIN (green) demarcates apical surfaces. HOECHST (blue) counterstains nuclei. (d) Confocal micrographs showing staining of E-CADHERIN (ECAD, green; top), wheat germ agglutinin (WGA, red; bottom), and HOECHST (blue, nucleus) in hPSCs cultured under indicated conditions. Squamous cells exhibit basal ECAD+ protrusions, highlighted by white arrowheads. n = 16 independent experiments. (e) Box charts showing normalized nucleus dimension (left) and epithelium thickness (right) for hPSC cysts in indicated conditions (box: 25% - 75%, bar-in-box: median, and whiskers: 1% and 99%). Cartoons show columnar versus flat, squamous cyst morphologies and the apico-basal (api-bas) and lateral (lat) directions. ncell = 179, 254, and 243, and ncyst = 11, 11, and 23, for Gel-2D, Glass-3D, and Gel-3D, respectively. n = 4 independent experiments. P-values were calculated using unpaired, two-sided Student's t-test. ***: P < 0.001. (f) Confocal micrographs showing NANOG (top), OCT4 (middle), and SOX2 (bottom) immunostaining in hPSCs cultured under indicated conditions. WGA co-staining shows cell morphology. n = 9 independent experiments. (g) Western blot showing protein levels of NANOG, OCT4, SOX2, ECAD, and GAPDH in hPSCs cultured under indicated conditions. n = 3 independent experiments. Scale bars in c, d, and f, 50 μm.
Human pluripotent stem cells (hPSCs), which reside in a developmental state similar to pluripotent epiblasts8,9, have been successfully utilized for modeling post-gastrulation human embryonic development4,10. However, the applicability of hPSCs for modeling peri-implantation, pre-gastrulation developmental events, such as amniogenesis, remains undetermined. Here we adapted a biomimicry approach to engineer a biomaterial-based in vitro hPSC culture system for efficient generation of early human amniotic tissue. Specifically, we constructed a biomimetic implantation-like niche for cultured hPSCs by implementing two major biophysical factors seen in the in vivo amniogenic niche: (a) a three-dimensional (3D) extracellular matrix (ECM) that is provided by the basement membrane surrounding the epiblast during implantation11, and (b) a soft tissue bed provided by the uterine wall and trophoblast to support the developing amnion (Fig. 1a,b). Since amniogenesis initiates from the expanding pluripotent epiblast, we utilized mTeSR1 medium and basement membrane matrix (Geltrex™) to render the culture permissive for pluripotency maintenance.
In this culture system, H9 human embryonic stem cells (hESCs) were plated as single cells at 30,000 cells cm− onto a thick, soft gel bed of Geltrex™ (with thickness ≥ 100 μm, bulk Young's modulus ~900 Pa, coated on glass coverslip), in mTeSR1 medium supplemented with ROCK inhibitor Y2763213 (Fig. 1b). At 24 hr (day 1), medium containing Y27632 was replaced by fresh mTeSR1 supplemented with 4% (v : v) Geltrex™ to establish a 3D implantation-like niche (referred to henceforth as "Gel-3D" condition). To assess the effect of ECM dimensionality and matrix rigidity, respectively, several modifications of this Gel-3D condition were tested (Fig. 1b). First, the Geltrex™ supplement was excluded from the medium, with the gel bed retained (referred to henceforth as "Gel-2D" condition). Second, the soft gel bed was replaced by a 1% Geltrex™-coated glass coverslip (referred to henceforth as "Glass-3D" condition). Finally, a standard 2D culture, using a 1% Geltrex™-coated glass coverslip (referred to henceforth as "Glass-2D" condition), was examined as a control that maintains hPSC self-renewal. Culture medium was replenished daily. Analyses were performed at day 5 unless otherwise noted (Fig. 1b).
In the Glass-2D condition, apico-basally polarized hESC colonies were observed at day 5. Strikingly, in Gel-2D, Glass-3D, and Gel-3D conditions, hESCs formed 3D cysts with EZRIN+ apical surfaces facing inward, reflecting the intrinsic lumenogenic property of hESCs5,7. In both Gel-2D and Glass-3D, > 90% of lumenal cysts are made of tall, columnar E-CADHERIN+ (ECAD+) epithelial cells with apico-basally elongated nuclei and thick epithelium (Fig. 1c-e; Supplementary Fig. 1). In distinct contrast, > 90% of cysts formed in Gel-3D show a squamous epithelial morphology featuring flattened, laterally elongated cell nuclei and reduced epithelium thickness, as well as unique ECAD+ protrusions extending from basal surfaces (Fig. 1c-e; Supplementary Fig. 1).
Notably, all 3D columnar epithelial cysts that formed in Gel-2D and Glass-3D conditions express pluripotency markers NANOG, OCT4, and SOX214, consistent with the well-known association between columnar epithelial morphology and pluripotent epiblast in vivo2,6,9 (Fig. 1f,g). However, in squamous cysts that formed in Gel-3D, expression of NANOG, OCT4, and SOX2 protein is lost, suggesting that they are composed of a differentiated cell type (Fig. 1f,g). qRT-PCR analysis shows reduction in mRNA expression for NANOG and SOX2, but not OCT4 (also known as POU5F1), in Gel-3D (Supplementary Fig. 2a), suggesting a blunted transcriptional regulation of OCT4. Despite loss of pluripotency markers, the squamous cyst maintains an epithelial phenotype, retaining expression of ECAD/CDH1 and CLDN612 (Fig. 1d,g; Supplementary Fig. 2b). This spontaneously differentiated squamous epithelial cystic tissue is morphologically reminiscent of the developing amnion in peri-implantation human embryos1,2.
The development of squamous cysts is characterized by concurrent changes in morphology and cell fate (Supplementary Fig. 3). From days 2-4, the majority of cysts in Gel-3D transitioned from columnar to squamous morphology and lose NANOG and OCT4 expression. Self-organized development of squamous cysts was also observed in two additional hESC lines (UM63-1 and H7) and an hiPSC line (1196a) cultured in Gel-3D, but not under Glass-2D, Gel-2D, or Glass-3D conditions (Supplementary Fig. 4). Thus, uniquely among all conditions examined, the implantation-like Gel-3D biophysical niche is both necessary and sufficient for efficiently inducing self-organized development of hPSCs to a cystic epithelial tissue with amnion-like squamous morphology, under biochemical conditions that permit hPSC self-renewal. Furthermore, the development of squamous cysts in Gel-3D is inhibited as the thickness of the gel bed is decreased to 60 μm or 20 μm, a modulation known to increase apparent substrate rigidity13 (Supplementary Fig. 5). Together, these findings support the notion that the mechanical rigidity and the 3D dimensionality of the ECM are integrated to trigger hPSC differentiation to an amnion-like tissue.
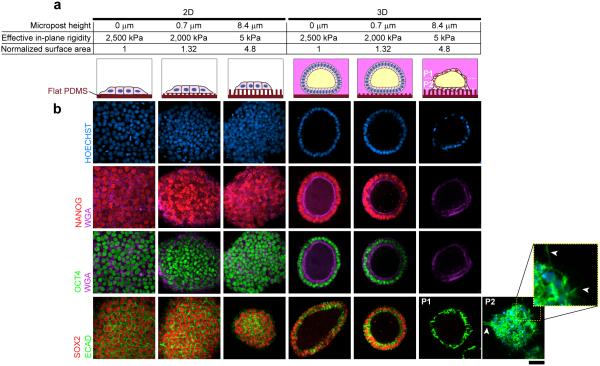
To further confirm the physical niche-dependency of hPSC morphogenic cytodifferentiation, we adopted an artificial matrix, made of a regular array of elastomeric polydimethylsiloxane (PDMS) microposts14 whose height can be precisely modulated to control substrate rigidity and surface area (Fig. 2a; Supplementary Fig. 6a). Consistently, development of squamous cysts occurred only in hPSCs cultured on 8.4 μm tall microposts (a soft matrix), but not on 0.7 μm tall microposts or on flat PDMS surfaces (rigid matrices), even though all included a 3D Geltrex™ overlay (Fig. 2b; Supplementary Fig. 6b). Interestingly, hPSCs cultured on soft 8.4 μm tall microposts in 2D (i.e., without the Geltrex™ overlay) did not form cysts (Fig. 2), in contrast to columnar cysts formed in Gel-2D (Fig. 1c). The mechanism underlying such differential morphogenesis of hPSCs is unclear.
Figure 2. Development of squamous cysts from hPSCs on a synthetic, soft artificial matrix.
(a) Specifics and cartoons of culture conditions using artificial matrices made of PDMS microposts of different post heights. (b) Confocal micrographs showing staining of NANOG (red), OCT4 (green), WGA (purple), ECAD (green), and SOX2 (red) for hPSCs cultured in indicated conditions. HOECHST (blue) counterstains nuclei. For the squamous cyst formed in 3D on 8.4 μm tall microposts, confocal micrographs taken at two planes passing the middle (P1) and bottom (P2) of the cyst (white dashed lines in a) are shown. The yellow square highlights ECAD+ protrusions (white arrowheads) at the interface between the cyst and microposts. n = 3 independent experiments. Scale bar, 50 μm.
We next examined the molecular signature of hPSC-derived squamous cysts and compared it with other embryonic and extraembryonic lineages possibly existing in a peri-implantation embryo, including primitive streak (PS), neuroectoderm, primitive endoderm (PE)/hypoblast, trophectoderm (TE)/trophoblast, primordial germ cells (PGCs), and amnion.
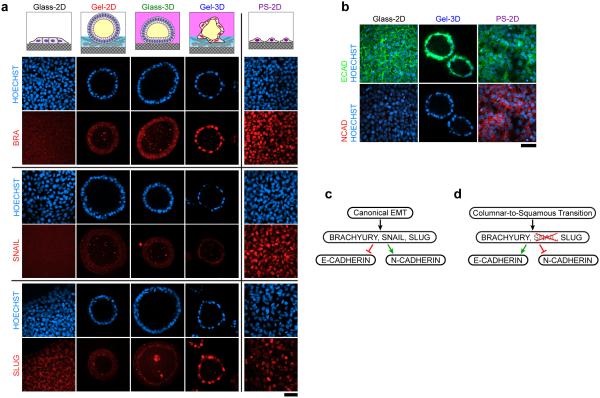
Primitive streak development is associated with an epithelial-to-mesenchymal transition (EMT) accompanied by up-regulation of transcription factors including BRACHYURY (BRA), SNAIL, and SLUG15. Indeed, basal protrusions observed in squamous cysts (Fig. 1d) suggest the possible involvement of EMT. Compared with control hPSCs in Glass-2D, up-regulation of BRA/BRA and SLUG/SNAI2, but not SNAIL/SNAI1, were observed in squamous cysts in Gel-3D; no up-regulation of these transcription factors was seen in Gel-2D or Glass-3D conditions (Fig. 3a; Supplementary Fig. 7a). In contrast, hPSC-derived PS cells (via a 2D culture protocol16; referred to henceforth as PS-2D cells) showed up-regulation of BRA/BRA, SNAIL/SNAI1, and SLUG/SNAI2 (Fig. 3a; Supplementary Fig. 7a). PS-2D cells also showed a decrease in ECAD/CDH1 and loss of ECAD organization, accompanied by increased NCAD/CDH2; none of these changes were seen in squamous cysts in Gel-3D (Fig. 3b; Supplementary Fig. 7b). These data suggest that while PS-2D cells exhibit molecular signatures of canonical EMT (Fig. 3c), squamous cyst development activates a unique subset of EMT-related transcription factors, notably without SNAIL, and elicits a columnar-to-squamous epithelium transition with ECAD/NCAD regulation distinct from that observed in canonical EMT and PS lineage differentiation (Fig. 3d). FOXA2/FOXA2 , a PS/endoderm marker, was undetectable in squamous cysts (Supplementary Fig. 7c,d), further excluding the PS lineage. Absence of SOX2 - a marker of human neuroepithelium17 - in squamous cysts (Fig. 1f,g; Fig. 2) excludes the neuroectodermal lineage. PE/hypoblast markers GATA4 and GATA66 were not up-regulated in squamous cysts, compared with hPSCs, excluding the PE lineage (Fig. 4a). A recent publication suggests that in cynomolgus monkey embryos, PGCs are NANOG+/OCT4+/SOX17+ and emigrate from the amnion by canonical EMT18. However, none of NANOG/OCT4/SOX17, nor canonical EMT, were detected in squamous cysts (Fig. 1f&g, Fig. 3a, and Supplementary Fig. 3c; data not shown for SOX17). Therefore, the squamous cysts do not appear to match the characteristics of PGCs. It remains a future goal to determine whether PGCs can be derived from squamous cysts under suitable conditions.
Figure 3. Squamous cyst development is transcriptionally distinct from canonical EMT or primitive streak.
(a) Confocal micrographs showing immunostaining of BRACHYURY (BRA, red; top), SNAIL (red; middle), and SLUG (red; bottom) in hPSCs cultured under indicated conditions and corresponding staining of PS cells derived from hPSCs under 2D culture16 (PS-2D). HOECHST (blue) counterstains nuclei. (b) Confocal micrographs showing immunostaining of ECAD (green; top) and N-CADHERIN (NCAD, red; bottom) for hPSCs under indicated culture conditions. HOECHST (blue) counterstains the nuclei. Scale bars in a and b, 50 μm. n = 2 independent experiments. (c&d) Schemes summarizing the transcriptional control program for canonical EMT (c) and the columnar-to-squamous transition reminiscent of human amnion development (d).
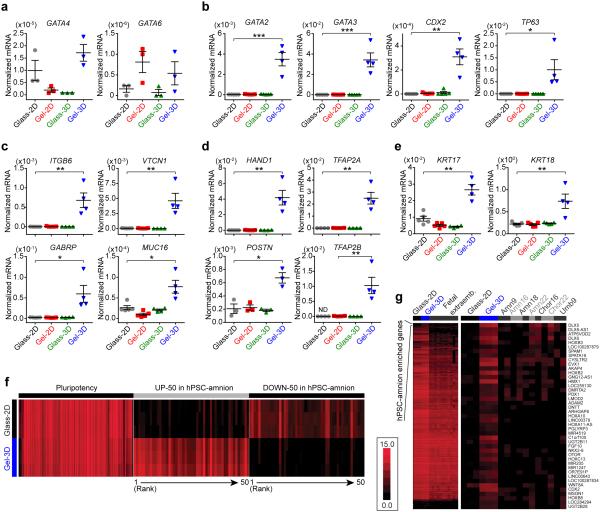
Figure 4. Molecular characterization and identification of the squamous, hPSC-derived amnion-like tissue.
(a-e) qRT-PCR analysis of known markers: (a) primitive endoderm/hypoblast markers GATA4 and GATA6; (b) trophectoderm and trophoblast markers GATA2, GATA3, CDX2, and TP63; (c-e) first trimester human amnion markers ITGB6, VTCN1, GABRP, MUC16, HAND1, POSTN, TFAP2A, TFAP2B, KRT17, and KRT18. All data were normalized against GAPDH and plotted as the mean ± s.e.m, with n = 3 - 5 biological replicates indicated by individual dots, n = 2 independent experiments. P-values were calculated using unpaired, two-sided Student's t-tests. *: P < 0.05; **: P < 0.01; ***: P < 0.001. (f) Heat map showing expression levels of 108 putative pluripotency genes27 (Supplementary Table 2), 50 most up-regulated genes (UP-50), and 50 most down-regulated genes (DOWN-50) (Supplementary Table 3) in hPSC-amnion derived in Gel-3D relative to hPSC colonies in Glass-2D. n = 3 biological replicates. (g) Hierarchical clustering of ~4,000 prospective hPSC-amnion-enriched genes (Supplementary Table 4) among hPSC colonies (Glass-2D), hPSC-amnion (Gel-3D), and published fetal extraembryonic tissues including week 9 amnion and umbilical cord (Amn9, Umb9), week 16 amnion and chorion (Amn16, Chor16), week 18 amnion (Amn18), and week 22 amnion and chorion (Amn22, Chor22)22. The color scale in f and g represents levels of normalized gene expression (Supplementary Methods).
Interestingly, several TE/trophoblast markers - GATA2, GATA3,CDX2, and TP636,19 - were highly up-regulated in squamous cysts compared with hPSC colonies and columnar cysts (Fig. 4b). However, other known trophoblast markers - KRT7, CGA, HLA-G19,20 - were not up-regulated in squamous cysts (Supplementary Fig. 8a). The squamous cysts showed heterogeneous staining of CDX2 and GATA3, which co-localize with BRA (Supplementary Fig. 8b,c), combinations not seen in hPSC-derived trophoblasts19. SSEA-4, a surface antigen associated with the inner cell mass, but not TE21, is also retained in squamous cysts (Supplementary Fig. 8d). Importantly, other studies have reported GATA2, GATA319,22, and SSEA-41,23 expression in human amnion. Together, these results contradict known molecular features of trophoblasts, and suggest that the hPSC-derived squamous cystic tissue resembles early human amnion.
Consistently, mRNA expression of a set of key fate-identifying genes recently reported for first trimester human amnion - ITGB6, VTCN1, GABRP and MUC1622 - are all significantly up-regulated in squamous cysts compared with control hPSCs (Fig. 4c). Additionally, squamous cysts exhibit up-regulated expression of HAND1, POSTN, TFAP2A, and TFAP2B (Fig. 4d); HAND1, POSTN, and TFAP2A are markers for early mouse 27amnion24, and POSTN, TFAP2A, and TFAP2B are reported first trimester human amnion markers22,24,25. The squamous cysts also show up-regulation of KRT17 and KRT18 (Fig. 4e), which are observed in week 10 human amnion26. Together, these data demonstrate that, among all the candidate lineages, the hPSC-derived squamous cysts exhibit a molecular signature most closely matching known aspects of human amnion at the first trimester (e.g., week 9-10), the earliest stage reported so far in the literature.
To establish the transcriptome of the hPSC-derived amnion-like tissue (referred to henceforth as hPSC-amnion), we performed RNA-sequencing (RNA-seq) (Supplementary Table 1). Although the transcriptome of hPSC-amnion differs substantially from that of control hPSCs, expression levels of a cohort of putative pluripotency genes27 are remarkably similar in both; only CUZD1 and CCL26 are substantially down-regulated in hPSC-amnion compared with control hPSCs (Fig. 4f; Supplementary Table 1-3). This observation suggests that hPSC-amnion develops with only slight down-regulation of the transcriptional circuitry maintaining pluripotency, consistent with the emergence of amnion from expanding pluripotent epiblasts in a self-renewal-permissive environment in vivo and here in vitro.
We next selected ~4,000 genes with higher expression in hPSC-amnion than in hPSCs or in previously examined fetal extraembryonic tissues, including amnion, chorion, and umbilical cord (GEO access number GSE66302)22 (Fig. 4g; Supplementary Table 4), and subjected them to hierarchical clustering. This revealed a gene set uniquely enriched in hPSC-amnion and relatively depleted in hPSCs and other extraembryonic tissues (Fig. 4g), probably reflecting the fact that the peri-implantation stage represented by hPSC-amnion is developmentally earlier than previously examined amnion22,25,28 samples19,22,25. We also compared genes enriched in hPSC-amnion with recently reported single cell transcriptomes of 197 non-amniotic cells obtained from post-implantation monkey embryos9. This analysis did not reveal any monkey cell that either displays transcriptomic similarity to hPSC-amnion (data not shown) or is double-positive for hPSC-amnion markers ITGB6 and VTCN1, supporting that hPSC-amnion represents a distinct lineage. We note that hPSC-amnion, which resembles the initial stage of amnion development, is the end-point phenotype in the current culture system (data not shown), suggesting that it may require additional factors for further development of the amnion-like tissue in vitro.
We also performed gene ontology (GO) functional annotation clustering for genes enriched in hPSC-amnion. GO terms for genes enriched in 9-week human amnion as well as human chorion and placenta were similarly clustered. Strikingly, three most enriched annotation clusters in hPSC-amnion were transcription factors, primarily those of the homeobox classes (Supplementary Fig. 9). Interestingly, HOX genes comprised the highest ranked cluster in 9-week amnion as well (Supplementary Fig. 9).
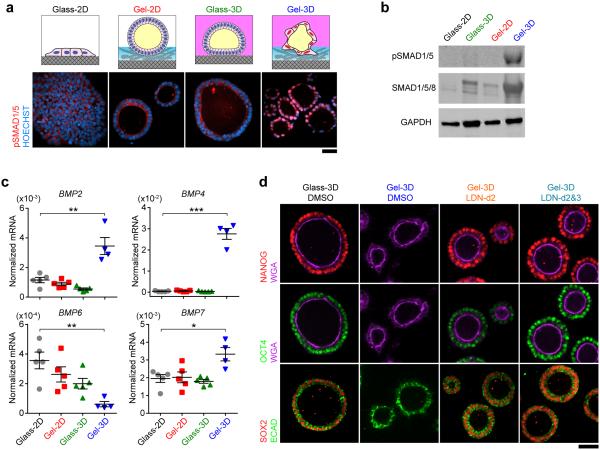
Among genes enriched in hPSC-amnion were several potential BMP targets including DLX5/6 and EVX1 (Fig. 4g). Gene set enrichment analysis (GSEA) also revealed enrichment of genes related to the ALK-pathway, which is associated with BMP signaling, in hPSC-amnion compared with hPSCs (Supplementary Fig. 10; Supplementary Table 5). Indeed, we observe prominent nuclear staining and up-regulated protein level of phosphorylated SMAD1/5 (pSMAD1/5), a downstream target of BMP-SMAD signaling, in hPSC-amnion, but not in other conditions (Fig. 5a,b). It suggests that BMP-SMAD signaling is activated during hPSC-amnion development, consistent with findings in early mouse and primate embryos18,24,28,21. Consistently, both RNA-seq (Supplementary Table 1) and qRT-PCR analysis (Fig. 5c) show up-regulated BMP2/4/7 in hPSC-amnion. Western blotting further confirms increased BMP4 protein level in hPSC-amnion (Supplementary Fig. 11a), suggesting endogenously activated BMP production during the development of hPSC-amnion.
Figure 5. Endogenously activated BMP-SMAD signaling is required for the development of hPSC-amnion.
(a) Confocal micrographs showing immunostaining of phosphorylated SMAD1/5 (pSMAD1/5, red) in hPSCs cultured under indicated conditions. HOECHST (blue) counterstains nuclei. n = 2 independent experiments. (b) Western blot showing protein levels of pSMAD1/5, SMAD1/5/8, and GAPDH for hPSCs cultured under indicated conditions. n = 3 independent experiments. (c) qRT-PCR analysis of BMP2, BMP4, BMP6, and BMP7 for hPSCs cultured under indicated conditions. Data were normalized against GAPDH and plotted as the mean ± s.e.m, with n = 4 - 5 biological replicates indicated by individual dots, n = 2 independent experiments. P-values were calculated using unpaired, two-sided Student's t-tests. *: P < 0.05; **: P < 0.01; ***: P < 0.001. (d) Confocal micrographs showing immunostaining of NANOG (red; top), OCT4 (green; middle), SOX2 (red; bottom), pan-cell membrane marker WGA (purple; top and middle), and basolateral membrane marker ECAD (green; bottom), in hPSC-derived epithelial cysts under Glass-3D and Gel-3D conditions with or without supplementation of BMP inhibitor LDN193189 (LDN) as indicated. LDN was supplemented on either day 2 only (LDN-d2) or on both days 2 and 3 (LDN-d2&3). n = 4 independent experiments. Scale bars in a and d, 50 μm.
To examine whether BMP-SMAD signaling is required for hPSC-amnion development, hPSCs cultured in Gel-3D were treated with a small molecule inhibitor LDN193189 (LDN), which inhibits ALK2/3 receptors that bind to BMP2/4/7. Strikingly, treatments with LDN (on day 2 only or on both days 2 and 3) inhibited hPSC-amnion development (Fig. 5d; Supplementary Fig. 11b&c). The development of hPSC-amnion is also inhibited by treatment with NOGGIN, a protein that antagonizes BMP2/4/7 (Supplementary Fig. 11d&e). These results implicate the requirement of BMP-SMAD signaling for hPSC-amnion development. It remains to be determined how physical signals from the 3D implantation-like niche activate BMP-SMAD signaling to elicit hPSC-amnion development.
In this work, we report a biomimetic implantation-like niche for hPSCs to model human amniogenesis - a key developmental step previously not accessible to study. We demonstrated that amniotic development by hPSCs is a self-organizing process that occurs in the absence of biochemical inductive cues from a maternal or extraembryonic source. Rather, physical signals from the implantation-like niche are necessary and sufficient to trigger the development of amnion-like tissue in a BMP-dependent manner. Interestingly, while the squamous hPSC-amnion reported here closely resembles human amnion tissue from morphological and transcriptomic perspectives, they do not fully recapitulate the patterned tissue morphologies of the peri-implantation human embryo, possibly due to differences in the dynamics of amnion formation in vitro versus in vivo. In addition to advancing fundamental understanding of human amnion development and expanding the application of hPSCs to model peri-implantation human embryogenesis, this efficient hPSC-based 3D amniogenic system could be leveraged for developing high-throughput screening assays to predict human reproductive success, examine the effect of toxins on amniotic development, or provide a therapeutic strategy for in utero treatment of amniotic tears.
METHODS
Cell lines
hPSC lines used in this study included H9 (WA09, P50, WiCell; NIH registration number: 0062), H7 (WA07, P52, WiCell; NIH registration number: 0061), UM63-1 (P25, provided by Dr. Gary D. Smith at the University of Michigan MStem Cell Laboratories; NIH registration number: 0277), and 1196a (an iPSC line, P42, from the University of Michigan Pluripotent Stem Cell Core29). All protocols for the use of hPSC lines were approved by the Human Pluripotent Stem Cell Research Oversight Committee at the University of Michigan. All hPSC lines have been authenticated by the original sources and also authenticated in-house by immunostaining for pluripotency markers and successful differentiation to three germ layer cells. All hPSC lines were maintained in a feeder-free system for at least 10 passages and authenticated as karyotypically normal at the indicated passage number. Karyotype analysis was performed by Cell Line Genetics (Madison, USA). All hPSC lines were tested negative for mycoplasma contamination (LookOut Mycoplasma PCR Detection Kit, Sigma-Aldrich).
Cell culture
hPSCs were maintained in a standard feeder-free culture system using mTeSR1 medium (STEMCELL Technologies) and lactate dehydrogenase-elevating virus (LDEV)-free hESC-qualified reduced growth factor basement membrane matrix Geltrex™ (Thermo Fisher Scientific; derived from Engelbreth-Holm-Swarm tumors similarly as Matrigel®) per the manufacturers' instructions. All cultures were visually examined during every passage to ensure the absence of spontaneously differentiated, mesenchymal-like cells in the culture. All hPSCs were used before reaching P70.
3D amniogenesis assay and BMP inhibition assay
Three different kinds of substrates were used in this study: glass coverslip coated with 1% Geltrex™ solution for 1 hr at room temperature, Geltrex™ beds of different nominal thicknesses (as described below), and artificial matrices of different effective rigidities (as described below). Cultured hPSC colonies were first incubated with Accutase (Sigma-Aldrich) at 37°C for 10 min before triturated and suspended in PBS as single cells. hPSCs were then centrifuged and the cell pellet was resuspended in mTeSR1 containing 10 μM Y27632 (Tocris), a ROCK inhibitor that prevents dissociation-induced apoptosis30. hPSCs were plated as single cells at 30,000 cells cm−2 onto the indicated substrate. After 24 hr (on day 1), culture medium was changed to fresh mTeSR1 without Y27632. For 3D culture conditions, 4% (v : v) Geltrex™ was supplemented in mTeSR1 medium on day 1, as previously described5. Thereafter, mTeSR1 medium was replenished daily, and 4% (v : v) Geltrex™ was supplemented daily for all 3D culture conditions.
In small molecule inhibitor treatment assays, 500 nM BMP inhibitor LDN193189 (LDN; STEMCELL Technologies) was added to the culture medium, either on day 2 only or on both days 2 and 3. DMSO (Sigma-Aldrich) was added to control groups on both days 2 and 3. In experimental groups treated with LDN on day 2 only, DMSO was supplemented on day 3. No LDN or DMSO was added to the culture medium on days other than those specified above.
In the NOGGIN treatment assay, 500 ng ml−1 human recombinant NOGGIN (R&D Systems; reconstituted in PBS) was added to the culture medium for the experimental group from day 0 to day 5. PBS was added to the control group.
Derivation of primitive streak cells
Primitive streak (PS) cells were derived from hPSCs according to a previously published protocol16. In brief, hPSCs (H9 line) were dissociated and plated as single cells at 20,000 cells cm−2 onto glass coverslips coated with 1% Getlrex™ solution in mTeSR1 medium containing 10 μM Y27632. After 24 hr (on day 1), culture medium was replaced by fresh mTeSR1 without Y27632. On day 2, mTeSR1 was replaced by PS-differentiation medium containing Essential 6™ medium (Thermo Fisher Scientific), 20 ng ml−1 FGF2 (Peprotech), and 8 μM CHIR99021 (Tocris). PS cell differentiation was induced for 48 hr before performing downstream assays.
Fabrication of basement membrane matrix gel beds
Fabrication protocol for the Geltrex™ gel bed was based on a "sandwich" setup previously developed for generating polyacrylamide gel substrates31-33. To prepare a substrate that the gel bed could attach to, a 22 × 22 mm2 glass coverslip was first treated with air plasma (Harrick Plasma) for 2 min before coated with 0.1 mg ml−1 poly-(L-lysine) (PLL) solution (Sigma-Aldrich) for 30 min, and 1% glutaraldehyde solution (Electron Microscopy Sciences) for another 30 min. To prepare a substrate that could "sandwich" and release the gel bed, a pre-cleaned glass slide was treated with air plasma for 2 min before coated with 0.1 mg ml−1 poly-(L-lysine)-graft-poly-(ethylene glycol) (PLL-g-PEG; SuSoS) solution for 1 hr. To obtain gel beds with nominal thickness of 20, 60, and 100 μm, undiluted Geltrex™ (10, 30 and 50 μl, respectively) was sandwiched between the pre-treated glass coverslip and glass slide on ice before incubated at 37°C for 30 min for gelation. The gel bed, which was attached to the glass coverslip, was then removed from the glass slide and submerged in DMEM/F12 medium (Thermo Fisher Scientific) and incubated at 37°C overnight before plating cells.
To prepare gel beds with a nominal thickness larger than 100 μm, spacers with the desired thickness were placed between the glass coverslip and the glass slide when preparing the gel bed sandwich. Specifically, spacers were made of polydimethylsiloxane (PDMS; Dow Corning) films. The film was made by spin-coating liquid phase PDMS (mixed at 10:1 of base:curing agent ratio) onto a petri-dish at 500 rpm for 40 s (for generating 150 μm thick film) and cured at 70°C for at least 24 hr before use. A previously published "PDMS film thickness - spin coating parameter" chart34 can be referred to when preparing PDMS films of desired thicknesses.
Fabrication of elastomeric artificial matrices
The elastomeric artificial matrix consisted of a regular PDMS micropost array generated using a microfabrication protocol previously published14. To functionalize inert PDMS surfaces for cell attachment, the PDMS micropost array was treated with air plasma for 2 min and coated with 1% Geltrex™ solution for 2 hr. The Geltrex™ solution was aspirated, and the PDMS micropost array was rinsed with DMEM/F12 before plating hPSCs.
Finite element analysis was performed to determine the nominal spring constant, K, of the PDMS micropost under lateral force applied at the micropost top as previously described35. The nominal spring constant K was converted to an "effective in-plane substrate rigidity Eeff", by Eeff = 9K / 2πD36, where D = 1.83 μm was the micropost diameter.
Cell fixation and immunocytochemistry
hPSCs were fixed in 4% paraformaldehyde (buffered in 1 × PBS) for 30 min, and permeabilized in 0.1% SDS (sodium dodecyl sulfate, dissolved in PBS) solution for another 30 min. Samples were first incubated in 2% goat serum solution (Thermo Fisher Scientific) at 4°C for 24 hr, and then with primary antibody solution prepared in 2% goat serum at 4°C for another 24 hr. Samples were then labeled with goat-raised secondary antibodies (1:500) at 4°C for 24 hr. HOECHST 33342 (Thermo Fisher Scientific) was used for counterstaining the cell nucleus. Alexa-fluor dye-conjugated wheat germ agglutinin (WGA; Thermo Fisher Scientific) was used as a pan-cell membrane marker. All primary antibodies, their sources, and dilutions are listed in Supplementary Table 6.
Confocal microscopy and image analysis
Images were acquired on an Olympus 1X81 fluorescence microscope equipped with a CSU-X1 spinning-disc unit (YOKOGAWA) or a Nikon-A1 laser scanning confocal microscope (Nikon). Fluorescence images acquired from confocal microscopy were reconstructed in 3D using Imaris8.2 (Bitplane).
Scanning electron microscopy
The PDMS micropost array was mounted on a stub and sputter-coated with gold palladium. Samples were observed and photographed using a Philips XL20 scanning electron microscope (Phillips).
Cell and cyst morphology characterization
Measurements of cell nucleus dimensions and cyst epithelial thickness were performed manually using the Measurement tool in ImageJ (NIH)37. The normalized nucleus dimension, defined as the ratio of nucleus dimension along the apico-basal direction to that along the lateral direction, was then calculated. Distance between the apical and basal surfaces (both stained by WGA) of the epithelium was measured at 5 different positions evenly distributed along the circumference of the cyst. The average of these 5 measurements was calculated as the cyst epithelial thickness and was plotted as one data point in Fig. 1e.
Total RNA isolation and qRT-PCR analysis
Samples were collected and prepared for RNA isolation following a protocol commonly used for processing in vivo tissues. In brief, samples were washed with DMEM/F12 to remove ECM overlay. Cysts (ca. 5,000 or more per sample) were then scraped from the culture substrate, before being spun down at 1,000 rpm for 3 min in a conical tube. RNA was isolated from the cell pellet using an RNeasy Micro Kit (QIAGEN) following the manufacturer's instruction. RNA quality and quantity were determined using a NanoDrop 1000 spectrophotometer (Thermo Scientific). Reverse transcription was performed using the iScript™ cDNA synthesis Kit (Bio-Rad). qRT-PCR analysis was performed using Quantitect Sybr Green MasterMix (QIAGEN) and specific primers (as listed in Supplementary Table 7) on a CFX Connect™ Real-Time System (Bio-Rad). qRT-PCR result was considered as "not detected" when no signal was observed within 40 cycles. Human GAPDH was used as an internal control for quantifying relative gene expression by using the 2−ΔCt method38. All analyses were performed with 2 - 3 technical replicates and ≥ 3 biological replicates as described in figure captions. The investigator who performed the qRT-PCR analysis was blinded to the test condition allocation during the experiment. All qRT-PCR results were summarized and tabulated in Supplementary Tables 8&9.
Western blotting
Whole cell lysates were prepared from cells and homogenized by sonication. Proteins in lysates were separated via SDS-polyacrylamide gel electrophoresis (SDS-PAGE) before transferring to PVDF membranes. PVDF membranes were then incubated with blocking buffer (Li-Cor) for 1 hr and then with primary antibodies (Supplementary Table 6) at 4°C overnight with gentle shaking. Blots were then incubated with IRDye secondary antibodies (Li-Cor) for 1 hr before imaging using the Li-Cor Odyssey Sa Infrared Imaging System (Li-Cor). The investigator who performed the Western blotting was blinded to the test condition allocation during the experiment.
RNA-sequencing and data analysis
mRNA-sequencing was performed by the University of Michigan DNA Sequencing Core following Illumina guideline. The investigator who performed the sequencing was blinded to the test condition allocation during the experiment. RNA samples were sequenced on an Illumina HiSeq-2500 High-Output system generating 50 cycle single reads. Samples were barcoded and run on a single lane. Publically available RNA-seq data (GSE66302) were downloaded from GEO. Reads were mapped to the human reference genome (Build hg19) using STAR39 and tabulated using HTSeq40. Differential RNA expression was determined using edgeR41. Normalized levels of gene expression and the color scale (Fig. 4f,g) were determined by scaling raw reads for each gene to account for different total read counts across samples before values were log2 transformed. Raw and processed data files have been deposited in Gene Expression Omnibus (GEO, accession number GSE89479).
Cluster analysis
Hierarchical clustering analysis of transcript expression data was performed within Cluster 3.0 using uncentered correlation with average linkage42. Java Treeview was used for visualizing the clustered data43.
Gene ontology (GO) enrichment analysis
Quality check of the data was conducted using the Trinity toolkit (https://github.com/trinityrnaseq/trinityrnaseq/wiki/QC-Samples-and-Replicates)44. Differential expression analysis was carried out for hPSC-amnion, 9-week human amnion, human chorion, and human placenta, respectively, relative to hPSCs, with the DESeq2 package (https://www.bioconductor.org/packages/release/bioc/html/DESeq2.html)45. The output (.DE_results) were converted to an Excel format, sorted by log2 fold change and used as a unified set for GO enrichment analysis by DAVID, v6.8 (https://david.ncifcrf.gov).
Gene set enrichment analysis (GSEA)
Genes were ranked by fold change in expression level relative to hPSCs. The entire ranked data set shown in Supplementary Table 1 (i.e., genes enriched and depleted in amnion-like cysts) was queried for 35 genes of the ALK-pathway-related gene set, BIOCARTA_ALK_PATHWAY, using the GSEA software (http://www.broad.mit.edu/gsea/) and the Molecular Signature Database (MSigDB)46.
Data availability statement
All data included in this study are available from the corresponding authors upon request. RNA-seq data is also available at the Gene Expression Omnibus (GSE89479; www.ncbi.nlm.nih.gov/geo).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. K. Sue O'Shea, Dr. Sundeep Kalantry, Dr. Toshio Miki, and Dr. William Shawlot for comments on the manuscript. We are grateful to Dr. Michael Czerwinski for help with bioinformatics. We thank Dr. Gary D. Smith at the University of Michigan MStem Cell Laboratories for providing the UM63-1 hESC line and the University of Michigan Pluripotent Stem Cell Core and the Steven Schwartzberg Memorial Fund for the derivation of 1196a hiPSC line. This work is supported by the National Science Foundation (CMMI 1129611 and CBET 1149401, J.F.), the National Institutes of Health (R21 EB017078 and R01 EB019436, J.F.; R01 DK089933, D.G.), and the American Heart Association (12SDG12180025, J.F.). Y.S. is also partially supported by the University of Michigan Rackham Predoctoral Fellowship. The Lurie Nanofabrication Facility at the University of Michigan, a member of the National Nanotechnology Infrastructure Network (NNIN) funded by the National Science Foundation, is acknowledged for support in microfabrication.
Footnotes
AUTHOR CONTRIBUTIONS
Y.S., K.T., D.L.G., and J.F. designed experiments; Y.S., K.T., R.F.T., X.X., K.M.A.Y., and J.S. performed experiments; K.G. processed RNA-seq results and performed hierarchical clustering; Y.S., K.T., K.G., J.R.S., D.L.G., and J.F. analyzed data and wrote the manuscript. D.L.G. and J.F. supervised the project. All authors contributed to the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interest.
REFERENCES
- 1.Dobreva MP, Pereira PN, Deprest J, Zwijsen A. On the origin of amniotic stem cells: of mice and men. Int. J. Dev. Biol. 2010;54:761–777. doi: 10.1387/ijdb.092935md. [DOI] [PubMed] [Google Scholar]
- 2.Luckett WP. The development of primordial and definitive amniotic cavities in early Rhesus monkey and human embryos. Am. J. Anat. 1975;144:149–167. doi: 10.1002/aja.1001440204. [DOI] [PubMed] [Google Scholar]
- 3.Ferner K, Mess A. Evolution and development of fetal membranes and placentation in amniote vertebrates. Respir. Physiol. Neurobiol. 2011;178:39–50. doi: 10.1016/j.resp.2011.03.029. [DOI] [PubMed] [Google Scholar]
- 4.Warmflash A, et al. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods. 2014;11:847–854. doi: 10.1038/nmeth.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taniguchi K, et al. Lumen formation is an intrinsic property of isolated human pluripotent stem cells. Stem Cell Rep. 2015;5:954–962. doi: 10.1016/j.stemcr.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deglincerti A, et al. Self-organization of the in vitro attached human embryo. Nature. 2016;533:251–254. doi: 10.1038/nature17948. [DOI] [PubMed] [Google Scholar]
- 7.Shahbazi MN, et al. Self-organization of the human embryo in the absence of maternal tissues. Nat. Cell Biol. 2016;18:700–708. doi: 10.1038/ncb3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Leary T, et al. Tracking the progression of the human inner cell mass during embryonic stem cell derivation. Nat. Biotechnol. 2012;30:278–282. doi: 10.1038/nbt.2135. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura T, et al. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature. 2016;537:57–62. doi: 10.1038/nature19096. [DOI] [PubMed] [Google Scholar]
- 10.Lancaster M, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enders AC, Schlafke S, Hendrickx AG. Differentiation of the embryonic disc, amnion, and yolk sac in the rhesus monkey. Am. J. Anat. 1986;177:161–185. doi: 10.1002/aja.1001770205. [DOI] [PubMed] [Google Scholar]
- 12.Ben-David U, Nudel N, Benvenisty N. Immunologic and chemical targeting of the tight-junction protein Claudin-6 eliminates tumorigenic human pluripotent stem cells. Nat. Commun. 2013;4:1992. doi: 10.1038/ncomms2992. [DOI] [PubMed] [Google Scholar]
- 13.Buxboim A, Rajagopal K, Brown AEX, Discher DE. How deeply cells feel: Methods for thin gels. J. Phys.: Condens. Mat. 2010;22:194116. doi: 10.1088/0953-8984/22/19/194116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu J, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thiery J, Acloque H, Huang R, Nieto M. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Mendjan S, et al. NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell. 2014;15:310–325. doi: 10.1016/j.stem.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Zhang XQ, et al. Pax6 Is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki K, et al. The germ cell fate of cynomolgus monkeys is specified in the nascent amnion. Dev. Cell. 2016;39:169. doi: 10.1016/j.devcel.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Li YC, et al. BMP4-directed trophoblast differentiation of human embryonic stem cells is mediated through Delta Np63(+) cytotrophoblast stem cell state. Development. 2013;140:3965–3976. doi: 10.1242/dev.092155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee CQE, et al. What is trophoblast? A combination of criteria define human first-trimester trophoblast. Stem Cell Rep. 2016;6:257–272. doi: 10.1016/j.stemcr.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson JK, et al. Preimplantation human embryos and embryonic stem cells show comparable expression of stage-specific embryonic antigens. Stem Cells. 2002;20:329–337. doi: 10.1634/stemcells.20-4-329. [DOI] [PubMed] [Google Scholar]
- 22.Roost MS, et al. KeyGenes, a tool to probe tissue differentiation using a human fetal transcriptional atlas. Stem Cell Rep. 2015;4:1112–1124. doi: 10.1016/j.stemcr.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miki T, Strom SC. Amnion-derived pluripotent/multipotent stem cells. Stem Cell Rev. 2006;2:133–141. doi: 10.1007/s12015-006-0020-0. [DOI] [PubMed] [Google Scholar]
- 24.Dobreva MP, et al. Periostin as a biomarker of the amniotic membrane. Stem Cells Int. 2012;2012:987185. doi: 10.1155/2012/987185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slieker RC, et al. DNA methylation landscapes of human fetal development. Plos Genetics. 2015;11:e1005583. doi: 10.1371/journal.pgen.1005583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Regauer S, Franke WW, Virtanen I. Intermediate filament cytoskeleton of amnion epithelium and cultured amnion epithelial-cells - expression of epidermal cytokeratins in cells of a simple epithelium. J. Cell Biol. 1985;100:997–1009. doi: 10.1083/jcb.100.4.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallon BS, et al. StemCellDB: The human pluripotent stem cell database at the National Institutes of Health. Stem Cell Res. 2013;10:57–66. doi: 10.1016/j.scr.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pereira PN, et al. Amnion formation in the mouse embryo: the single amniochorionic fold model. BMC Dev. Biol. 2011;11:48. doi: 10.1186/1471-213X-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen HM, et al. Transcripts involved in calcium signaling and telencephalic neuronal fate are altered in induced pluripotent stem cells from bipolar disorder patients. Transl. Psychiatry. 2014;4:e375. doi: 10.1038/tp.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 31.Tse J, Engler A. Preparation of hydrogel substrates with tunable mechanical properties. Curr. Protoc. Cell Biol. 2010 doi: 10.1002/0471143030.cb1016s47. Unit 10.16, Ch. 10. [DOI] [PubMed] [Google Scholar]
- 32.Fischer R, Myers K, Gardel M, Waterman C. Stiffness-controlled three-dimensional extracellular matrices for high-resolution imaging of cell behavior. Nat. Protoc. 2012;7:2056–2066. doi: 10.1038/nprot.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buxboim A, Rajagopal K, Brown AEX, Discher DE. How deeply cells feel: Methods for thin gels. J. Phys.: Condens. Mat. 2010;22:194116. doi: 10.1088/0953-8984/22/19/194116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koschwanez JH, Carlson RH, Meldrum DR. Thin PDMS films using long spin times or Tert-Butyl Alcohol as a solvent. PLoS One. 2009;4:e4572. doi: 10.1371/journal.pone.0004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao Y, Mann JM, Chen WQ, Fu JP. Global architecture of the F-actin cytoskeleton regulates cell shape-dependent endothelial mechanotransduction. Integr. Biol. 2014;6:300–311. doi: 10.1039/c3ib40223a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weng S, Fu J. Synergistic regulation of cell function by matrix rigidity and adhesive pattern. Biomaterials. 2011;32:9584–9593. doi: 10.1016/j.biomaterials.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders S, Pyl PT, Huber W. HTSeq - a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saldanha AJ. Java Treeview - extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 44.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data included in this study are available from the corresponding authors upon request. RNA-seq data is also available at the Gene Expression Omnibus (GSE89479; www.ncbi.nlm.nih.gov/geo).