Abstract
A cataract is a pathological condition characterized by the clouding of the normally clear eye lens brought about by deposition of crystallin proteins in the lens fiber cells. These protein aggregates reduce visual acuity by scattering or blocking incoming light. Chemical damage to proteins of the crystallin family, accumulated over a lifetime, leads to age-related cataract, whereas inherited mutations are associated with congenital or early-onset cataract. The V75D mutant of γD-crystallin is associated with congenital cataract in mice and was previously shown to un/fold via a partially folded intermediate. Here, we structurally characterized the stable equilibrium urea unfolding intermediate of V75D at the ensemble level using solution NMR and small-angle x-ray scattering. Our data show that, in the intermediate, the C-terminal domain retains a folded conformation that is similar to the native wild-type protein, whereas the N-terminal domain is unfolded and comprises an ensemble of random conformers, without any detectable residual structural propensities.
Introduction
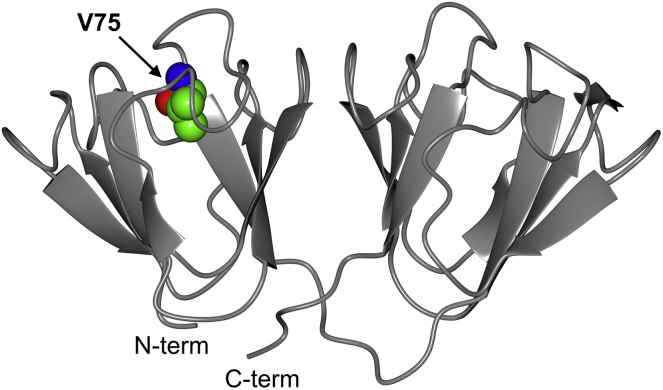
The lens of the eye is subject to stringent functional and structural constraints: it must possess a high refractive index to focus light onto the retina, it needs to be highly transparent so as not to block or scatter incoming light, and it must permit adjustment of its focal length for focusing on objects at variable distances. These requirements are reflected in the unique biology of the lens, the bulk of which is composed of terminally differentiated lens fiber cells filled with a unique set of proteins, the crystallins, that account for ∼90% of the total soluble lens protein. During eye development, the maturing lens cells lose their nuclei and organelles, eliminating cellular structures that constitute potential sources of light scattering (1). As a consequence, no new protein synthesis occurs in mature fiber cells at the core of the lens, and therefore, crystallins must last throughout the lifetime of the organism. Crystallins are classified into α-, β-, and γ-crystallins (2). The α-crystallins are small heat-shock proteins that form large oligomeric structures exhibiting chaperone-like activity (3). The β- and γ-crystallins are two-domain β-strand proteins in which each of the structurally homologous domains comprises two intercalated Greek key motifs (Fig. 1). It is generally assumed that this architecture contributes to the high stability of these proteins and their resistance to damage over their long lifetime (4, 5). The β- and γ-crystallins, which exist as dimers and monomers, respectively, most likely play a predominantly structural role, contributing to lens clarity (6, 7) and refractivity (8).
Figure 1.
Ribbon representation of the x-ray structure of human WT γD-crystallin (PDB: 1HK0). V75, which is mutated to Asp in the cataract-associated variant, is shown in space-filling representation. To see this figure in color, go online.
Despite the thermodynamic stability of the crystallins, the lens is vulnerable to external insults that cause cataract, a pathological opacification resulting in blurred vision and, in advanced cases, blindness. With an estimated 20 million cases, cataract is the leading cause of blindness worldwide, and thus the World Health Organization considers cataract to be a priority eye disease. Cataracts are formed by protein aggregates of damaged or modified proteins that precipitate in the lens. These assemblies reduce visual acuity by scattering incoming light.
Over the lifetime of an organism, various environmental stresses can cause proteins to undergo chemical modifications such as oxidation, deamidation, carbamylation, UV damage to aromatic residues, and cleavage of the peptide backbone (9, 10, 11, 12). In the eye lens, the α-crystallins are believed to mitigate the detrimental effects of damage by sequestering affected β- and γ-crystallins, thereby minimizing the formation of insoluble βγ-crystallin aggregates. Because essentially no new protein synthesis occurs in mature lens fiber cells, the supply of α-crystallins available to bind damaged β- or γ-crystallins will eventually be exhausted, significantly increasing the probability of cataract formation with advancing age.
In addition to age-related cataract, numerous mutations in the β- and γ-crystallins have been identified that are associated with congenital or early-onset cataract (13). Early-onset cataract, which is particularly devastating in those parts of the world that lack routine access to modern surgical techniques for replacing cataractous lenses, implies a lifetime of blindness for those affected. In this work, we focus on γD-crystallin, which makes up ∼11% of the total lens protein in young human lenses (14). γD mutations associated with early-onset cataract include R14C (15), R58H (16), R36S (17), V75D (18), P23T (19, 20), E107A (21), G60C (22), R76S (23), and W42R (24), among others. (Note that two different numbering conventions exist in the γD-crystallin literature: the first starts with an N-terminal Met1, and the second is based on the mature protein sequence starting with Gly1. We use the latter convention in this article and have named all mutants accordingly.) The discovery of mutations associated with congenital and early-onset cataract naturally raised the question as to why these individual mutations lead to cataract so prematurely. Structural analysis of mutants over the last 15 years has revealed that, with the exception of R36S, these mutations cause little or no large-scale conformational perturbation (25, 26, 27, 28, 29, 30, 31, 32, 33), ruling out gross structural disruption or misfolding as the defining cataractogenic feature.
Interestingly, despite the structural similarity of all these mutants to wild-type (WT) γD-crystallin in the native state, two of them, V75D and W42R, exhibit substantially altered folding/unfolding behavior compared to the WT, populating a stable intermediate over a wide range of denaturant concentrations (28, 29). Both mutants possess amino acid changes in the NTD, and both substitutions replace hydrophobic side chains that contribute to the hydrophobic core of the NTD. In this work, we use the term “intermediate” to refer to a stable equilibrium unfolding intermediate in the presence of 4.2 M urea. The urea denaturation profile for V75D is provided in Fig. S1. For V75D, we demonstrate, via sequence-specific NMR resonance assignments, that the intermediate corresponds to a crystallin molecule having an unfolded N-terminal domain (NTD) and a folded C-terminal domain (CTD), consistent with previous observations (29). Although the V75D mutation was originally discovered in mice, a recent study demonstrated that the human mutant appears to be disease relevant: human V75D γD-crystallin transgenically expressed in zebrafish lenses was shown to cause lens defects primarily associated with protein aggregation, whereas human WT γD-crystallin expression resulted in normal lenses (34).
Given the unusual equilibrium unfolding behavior of V75D γD-crystallin, the possibility exists that a particular residual or transient structure in the NTD of this mutant may serve as a specific nucleus for aggregation and cataract formation, rendering this mutant an interesting subject for structural characterization. Furthermore, other studies indicate that V75D is not efficiently recognized by the αA- and αB-crystallin chaperones (35, 36), suggesting that evasion of the innate anti-aggregation system of the lens could also play a role.
Because it is practically difficult to examine and characterize partially unfolded proteins experimentally, structural details of non-native crystallin states are scarce. Here, we report the characterization of the urea-induced, partially un/folded equilibrium structural ensemble of the V75D mutant of human γD-crystallin by combining residue- and atom-specific NMR data and low-resolution small-angle x-ray scattering (SAXS) data with a computational structural ensemble approach. To our knowledge, this is the first experimental structural study of a non-native, partially un/folded state of any mutant crystallin associated with early-onset cataract.
Materials and Methods
Expression and purification of V75D γD-crystallin
A single colony of Escherichia coli BL21 Star (DE3) cells (Life Technologies, Grand Island, NY) bearing a pET14b vector encoding the full-length human V75D γD-crystallin gene was used to inoculate 5 mL of Luria-Bertani (LB) medium supplemented with 100 μg/mL ampicillin. This culture was incubated overnight at 37°C in a rotary shaker, added to 1 L of fresh LB medium containing 100 μg/mL ampicillin, and grown at 37°C in a rotary shaker to an OD600 of ∼0.6, at which point protein expression was induced by the addition of isopropyl β-D-1-thiogalactopyranoside to a final concentration of 0.85 mM. Protein expression was allowed to proceed for 4 h at 37°C, after which the cells were harvested by centrifugation. The cell pellet was re-suspended in ∼40 mL of buffer Q (50 mM Tris (pH 8.0), 1 mM EDTA, and 1 mM dithiothreitol (DTT)) and stored at −80°C until it was thawed for protein purification.
The thawed cell suspension was lysed using a microfluidizer, and cell debris was removed by centrifugation. Nucleic acids were precipitated by adding polyethyleneimine to a final concentration of ∼0.05% w/v and stirring on ice for 25 min, followed by centrifugation. The supernatant was passed over a 5 mL HiTrap Q HP anion exchange column (GE Healthcare Life Sciences, Piscataway, NJ) equilibrated in buffer Q. V75D γD-crystallin was collected in the flow-through, which was dialyzed overnight at 4°C against 4 L of buffer S (25 mM MES (pH 6.0), 1 mM EDTA, 1 mM DTT, and 2% v/v glycerol). After verifying that the pH had equilibrated to 6.0, the dialysate was loaded onto a Mono S 10/100 GL cation exchange column equilibrated in buffer S, and bound proteins were eluted over ∼12 column volumes using a 0–75% gradient of 1 M NaCl in buffer S. The crystallin-containing elution peak was passed over a HiLoad 26/60 Superdex 75 gel filtration column pre-equilibrated in 25 mM MES (pH 6.0), 1 mM EDTA, and 5 mM DTT for final purification. Isotopically labeled protein for NMR studies was produced in the same manner, except that modified M9 minimal medium containing 13C-glucose and 15NH4Cl as the sole carbon and nitrogen sources, respectively, was used for growth.
Preparation of partially unfolded V75D γD-crystallin
Purified V75D at dilute concentration was dialyzed at 4°C against an ∼100-fold excess volume of 25 mM MES (pH 6.0) buffer containing 1 mM EDTA, 5 mM DTT, and 4.2 M urea. This urea concentration was selected because the equilibrium unfolding intermediate is populated over the 4–7 M urea range (Fig. S1). Equilibration was allowed to continue until no further changes were observed in the protein’s 1H-15N heteronuclear single-quantum coherence (HSQC) NMR spectrum, which typically required a period of 1 week. The partially un/folded species was stored at 4°C until it was used in SAXS or NMR experiments, usually within a few days. No visible aggregation occurred at any time between preparing the sample and performing structural experiments.
Small-angle x-ray scattering measurements and analysis
All SAXS measurements were performed at beamline 12-ID-B of the Advanced Photon Source at Argonne National Laboratory (Lemont, IL) using x-rays of energy 14 keV (λ ≈ 0.8856 Å). SAXS data were acquired for both native and partially unfolded V75D. For each protein concentration, 30 x-ray exposures of 1 s each were recorded from the sample under flow to minimize radiation damage. Protein scattering was recorded for several concentrations ranging from 0.5 to 8 mg/mL to check for concentration-dependent effects. Reduction of the raw scattering data was performed using beamline software. The 30 scattering curves for each concentration were checked for outliers using the SAXS Similarity Tool (37), available on the website of the SYBYLS beamline at the Advanced Light Source of Lawrence Berkeley National Laboratory. After removing outliers, the remaining individual scattering curves were averaged to create the final data for each sample.
Buffer scattering for subtraction from native V75D scattering was recorded using protein-free aliquots of the final gel filtration purification buffer, whereas buffer scattering for subtraction from the partially unfolded V75D scattering was recorded from aliquots of the urea-containing dialysis buffer used during unfolding. Subtraction of the buffer scattering for each protein sample was performed using PRIMUS (38) from the ATSAS 2.5.2 package (Biological Small Angle Scattering Group, European Molecular Biology Laboratory, Hamburg, Germany).
NMR spectroscopy
All NMR experiments were carried out at 30°C using Bruker (Billerica, MA) Avance 700 and 800 MHz spectrometers. All samples typically contained ∼300 μM protein. Sequence-specific resonance assignments for the N, HN, Cα, and Cβ atoms of full-length V75D in the presence of 4.2 M urea were obtained from analysis of three-dimensional (3D) HNCACB and HN(CO)CACB spectra in conjunction with the two-dimensional 1H-15N HSQC spectrum. The 3D spectra were acquired non-uniformly using Poisson gap sampling (39) and typically included ∼35% of the points of the total 15N × 13C indirect grid. These spectra were processed using the iterative soft thresholding method as implemented in the istHMS software (40). The 2D 1H-15N HSQC spectrum was sampled uniformly and processed using NMRPipe (41). Spectral analysis was carried out using NMRViewJ (42) and NMRFAM-SPARKY (43).
1DN-HN residual dipolar couplings (RDCs) were measured for the partially unfolded V75D protein using the IPAP method (44) implemented in a 3D HNCO spectrum. Stretched 6% polyacrylamide gels (45) were used as the alignment medium, and RDC values were extracted as the difference between the observed splitting in the aligned and isotropic samples. Spectra were processed using NMRPipe and analyzed using NMRViewJ.
Generation of models of the partially unfolded V75D structure
Because no x-ray or NMR structure of the V75D variant exists, a homology model was generated based on the 1.25 Å resolution x-ray structure of WT γD-crystallin (PDB: 1HK0) (46). The V75D amino acid change was introduced using Coot (47), and the resulting structure was used in the program flexible-meccano (48) to generate a pool of 50,000 models of the partially unfolded state. In the flexible-meccano approach, physically realistic models of unfolded protein segments are generated by randomly assigning residue-specific backbone dihedral angles from a library of non-secondary-structure elements in high-resolution protein x-ray crystal structures. In each generated model, the folded C-terminal domain, residues 87–173, was fixed based on its coordinates in PDB 1HK0, whereas the conformations of residues 1–86 were generated by the flexible-meccano procedure, representing the unfolded N-terminal domain.
Prediction of experimental observables for V75D structural models
The experimental observables for each conformer in the initial pool of conformers were calculated for comparison with the experimental data. N, HN, Cα, and Cβ chemical shifts were predicted using SPARTA (49), 1DN-HN residual dipolar couplings were calculated using PALES (50), and the SAXS curves were predicted using CRYSOL (51).
Modeling of the partially unfolded V75D structural ensemble
The program ASTEROIDS (52) employs a genetic algorithm for selecting conformational ensembles from a large starting pool of possible structures that agree best with the available experimental data. In the first round, ASTEROIDS was used iteratively (53, 54) to select five independent ensembles of 200 structures each, all of which exhibited good agreement with the experimental N, HN, Cα, and Cβ chemical shift data. Subsequently, flexible-meccano was employed to create a pool of 25,000 possible structures using the PDB coordinates of the C-terminal domain and generating the chain of the N-terminal domain using 1,000 residue-specific ϕ/φ values extracted from the earlier ensembles that were selected based on the chemical-shift data. Using the new pool of 25,000 conformers, a second round of ASTEROIDS selections was performed, this time simultaneously selecting based on chemical shift, residual dipolar coupling, and SAXS data together. In each selection, 20,000 iterations of the genetic algorithm were carried out to ensure convergence; in practice, the results ceased to change after at most a few thousand iterations. Ensembles of 50, 100, 200, 300, and 500 structures were generated to explore the effect of ensemble size on the agreement between calculated and observed data. Five independent ASTEROIDS selections were performed per ensemble size to assess the variability of the results for a given ensemble size.
Results
V75D resonance assignments
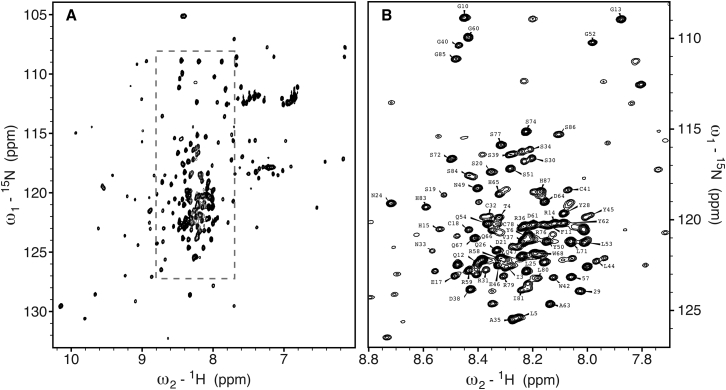
Chemical shift assignments for N, HN, Cα, and Cβ atoms of the V75D equilibrium unfolding intermediate were obtained by analysis of 3D HNCACB and 3D HN(CO)CACB spectra in conjunction with the 2D 1H-15N HSQC spectrum (Fig. 2 A; a superposition of the 1H-15N HSQC spectra of native V75D and V75D in 4.2 M urea is provided in Fig. S2). Resonances associated with the unfolded NTD cluster in the narrow center region of the spectrum (8.8 > δH > 7.8 ppm). As is well known, this lack of spectral dispersion is caused by averaging of the chemical shifts over all possible chemical environments sampled by residues in random coil regions, in contrast to the restricted chemical environments experienced by residues in a folded protein. The boxed region of the 1H-15N HSQC spectrum of Fig. 2 A is expanded in Fig. 2 B, and assignments are shown. The NTD resonances are narrow and intense, given the random-coil, flexible nature of this domain, compared to the folded CTD, for which well dispersed, but less intense, crosspeaks are observed (Fig. 2 A). Of 173 total residues, assignments were obtained for 153 N, 153 HN, 151 Cα, and 136 Cβ resonances. Of the assigned residues, 76 are located in the N-terminal domain. It should be pointed out that using non-uniform sampling for 3D data collection was highly beneficial; the time saved by collecting only 35–40% of the total points in the 2D 15N × 13C indirect dimensions was used to increase the number of both increments and scans per increment. This resulted in improved resolution and signal/noise ratio, mitigating the resonance overlap problem for the random-coil region to some degree.
Figure 2.
1H-15N HSQC spectrum of V75D γD-crystallin in 4.2 M urea. The well-dispersed resonances are associated with residues in the folded C-terminal domain, whereas the sharp and intense resonances, clustered in the center region of the spectrum, belong to residues in the unfolded N-terminal domain. The region boxed in (A) is expanded in (B). In (B), the contour level has been adjusted to optimally show resonances of unfolded residues, rendering the intensities of resonances for the folded residues very small. Resonance assignments are indicated by residue name and number.
Secondary chemical shifts reveal no residual conformational preference in the NTD
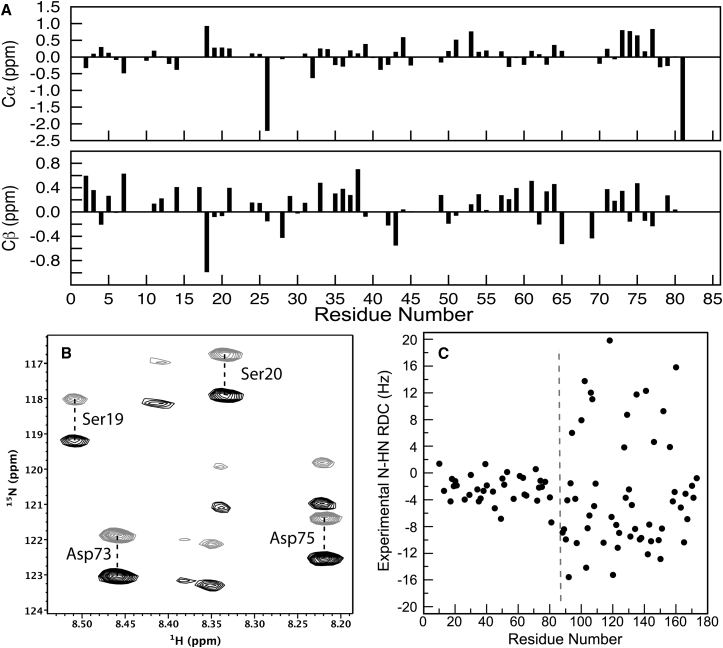
Backbone chemical shifts are exquisitely sensitive to local conformation. Secondary chemical shifts (Δδ), especially those of Cα and Cβ resonances, permit prediction of helical or strand conformations (55, 56, 57). The experimental secondary chemical shifts for the V75D NTD are displayed in Fig. 3 A. As can be noted, only small Δδ values are observed, and no detectable residual secondary structure can be inferred for the NTD of the partially unfolded V75D intermediate.
Figure 3.
NMR data of V75D γD-crystallin in 4.2 M urea. (A) Cα and Cβ secondary chemical shifts of the NTD. (B) Selected region of the 3D IPAP-HNCO spectrum recorded for V75D γD-crystallin, aligned in a stretched polyacrylamide gel in the presence of 4.2 M urea. Superposition of representative upfield (gray) and downfield (black) components for the doublets, recorded in separate sub-spectra, are shown for Ser19, Ser20, Asp73, and Asp75, all in the NTD. The labeled crosspeaks are located at δCO = 174.6 ppm, and the unlabeled weaker crosspeaks are from residues centered on nearby carbonyl planes. (C) Experimentally measured RDC values versus position in the amino acid sequence of V75D γD-crystallin in 4.2 M urea. The dashed gray line marks the boundary between the NTD and CTD.
V75D residual dipolar coupling measurements
RDCs provide global orientation information and inform on possible residual secondary structure. RDC values were extracted as the difference between the isotropic doublet splitting and the doublet splitting in the aligned sample (Fig. 3 B). We decided that only unambiguously determined RDCs were valuable for assessing the NTD structure. Therefore, it was necessary to unequivocally locate the center of each doublet component, which was possible for 33 of the 76 assigned NTD 1H-15N resonances. All NTD residues that exhibited IPAP-HNCO resonances with poorly identifiable centers were excluded from the RDC analysis. Although this conservative approach resulted in accepting approximately only half of all possible RDC values for the V75D NTD, residues for which RDCs could be determined are spread roughly evenly throughout the NTD sequence and, therefore, comprise a representative set with respect to the structure.
Averaging of the NTD RDCs suggests extensive conformational heterogeneity
RDC values contain valuable information about possible long-range order and/or dynamics of the unfolded NTD of V75D. Conformational dynamics results in averaging of the RDCs to small or nearly zero values (58). This is observed here for the V75D NTD. The N-HN RDC values are small, ranging from about −5 to +2 Hz. In contrast, large and diverse RDCs, ranging from −16 to +20 Hz, are measured for the CTD (Fig. 3 C). The small size of the RDCs for residues in the NTD and the large absolute size of the RDCs for CTD residues mirror the stark differences in resonance intensities in the 1H-15N HSQC spectrum. This combined observation is consistent with the notion that an ensemble of interconverting random-coil conformations is present in the NTD, whereas the CTD retains a well-defined structure, similar to the native WT structure.
Our experimental chemical shift and RDC data are consistent with an earlier folding study showing that the V75D mutation destabilizes the NTD, resulting in easier unfolding of the NTD compared to the CTD (29). Furthermore, neither the secondary chemical shifts nor the RDCs of V75D suggest the presence of any residual structure in the NTD of the partially unfolded intermediate. Therefore, if the unfolded NTD served as a seed for protein aggregation, non-specific, random interactions rather than a specific structured aggregation nucleus would be involved.
V75D SAXS
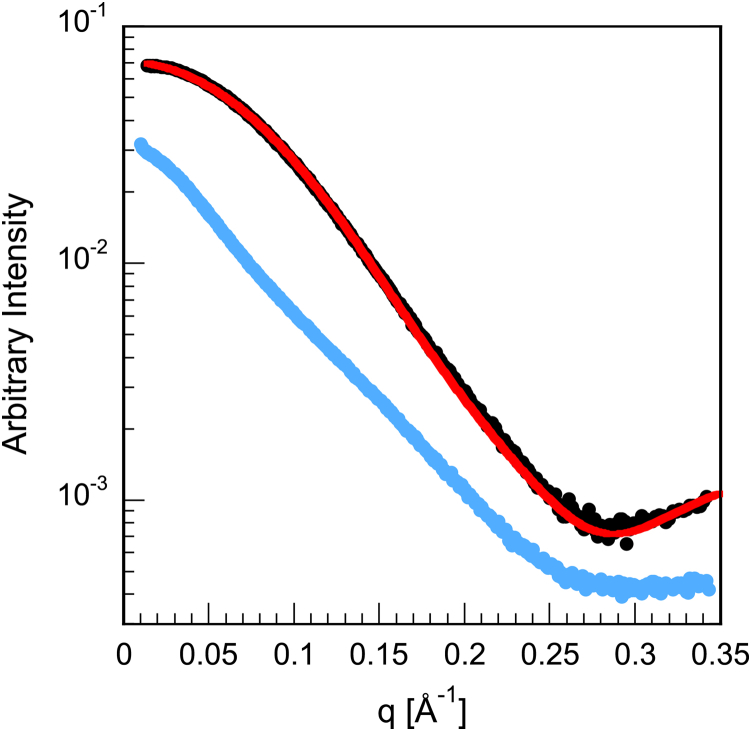
SAXS curves for both native and partially unfolded V75D were recorded. No concentration-dependent anomalies were observed for native V75D, although for V75D in 4.2 M urea, very minor signs of aggregation were present at the lowest scattering angles. We used moderate concentration (200 μM) to collect the SAXS data for partially unfolded V75D, omitting q values <0.017 Å−1 to eliminate from the final analysis the data points most susceptible to corruption by aggregation. Representative scattering curves for native and partially unfolded V75D are shown in Fig. 4, along with the predicted scattering curve for native WT γD. The close agreement between the experimental V75D and theoretical WT γD curves demonstrates the outstanding quality of the experimental data and provides further evidence that the native structure of V75D is not significantly affected by the mutation.
Figure 4.
SAXS curves for V75D human γD-crystallin in the absence (black) and presence (light blue) of 4.2 M urea. The scattering intensity is plotted as a function of the momentum transfer, q (4π sin θ/λ). The red trace is the predicted scattering curve based on the x-ray structure of WT γD (PDB: 1HK0). To see this figure in color, go online.
ASTEROIDS modeling of the partially unfolded V75D ensemble
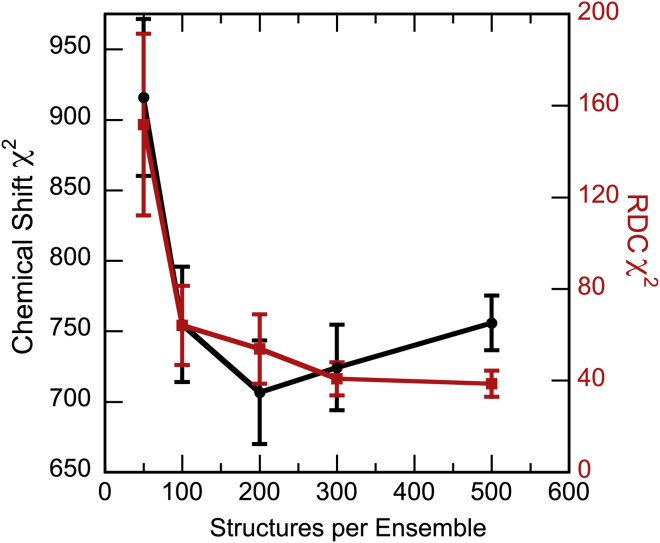
The experimental chemical shift and RDC data indicated that the NTD of V75D in 4.2 M urea is unfolded and devoid of any appreciable residual secondary structure. To gain further insight into the properties of this unfolded ensemble, we used the ASTEROIDS software to select ensembles of model structures that have ensemble-averaged properties that match the experimental data. Starting from an initial pool of 50,000 possible structures (conformation of residues 1–86 generated by flexible-meccano and conformation of residues 87–173 fixed to the crystal coordinates of WT γD), ASTEROIDS was used to select five ensembles of 200 structures that possess ensemble-averaged N, HN, Cα, and Cβ chemical shifts that agree with the experimental chemical shifts (see Materials and Methods). The ϕ/φ propensities for each amino acid were extracted from the selected ensembles and used in flexible-meccano to generate a new pool of 25,000 conformational models. This pool of models was used in additional ASTEROIDS selections in which all chemical shift, RDC, and SAXS data were applied simultaneously. The number of structures required in the ensemble to reproduce all the experimental observations accurately was determined empirically. ASTEROIDS ensembles of 50, 100, 200, 300, and 500 members were generated, and the agreement between the ensemble-averaged and experimental data was examined as a function of ensemble size. Five independent ASTEROIDS selections for each ensemble size were carried out to assess the variability of the selection results for a given ensemble size. As metrics for the agreement between the ensemble-averaged and experimental data, the χ2 goodness of fit was used for the chemical shift and RDC data in each of the five selected ensembles. The χ2 values for each ensemble size were averaged and compared to evaluate whether better agreement between experiment and computational selection was achieved by choosing additional structures. The decrease in χ2 values for increasing ensemble sizes is displayed in Fig. 5. As can be appreciated, an ensemble of 50 structures is clearly insufficient to accurately describe the experimental data, and ensemble sizes of at least 100 structures are necessary. Thus, we chose an ensemble size of 200 structures to ensure that ensemble-averaged properties faithfully reflect the experimental values. This guarantees that sufficient diversity is present for recovering the experimental data without arbitrarily inflating the ensemble size. The discussion presented below is based on one particular selection of 200 structures, but it holds for all independently selected ASTEROIDS ensembles of 200 structures.
Figure 5.
A comparison of the χ2 goodness of fit for the chemical-shift (black) and RDC (red) data as a function of ensemble size. To see this figure in color, go online.
Comparison of ensemble-averaged and experimental data
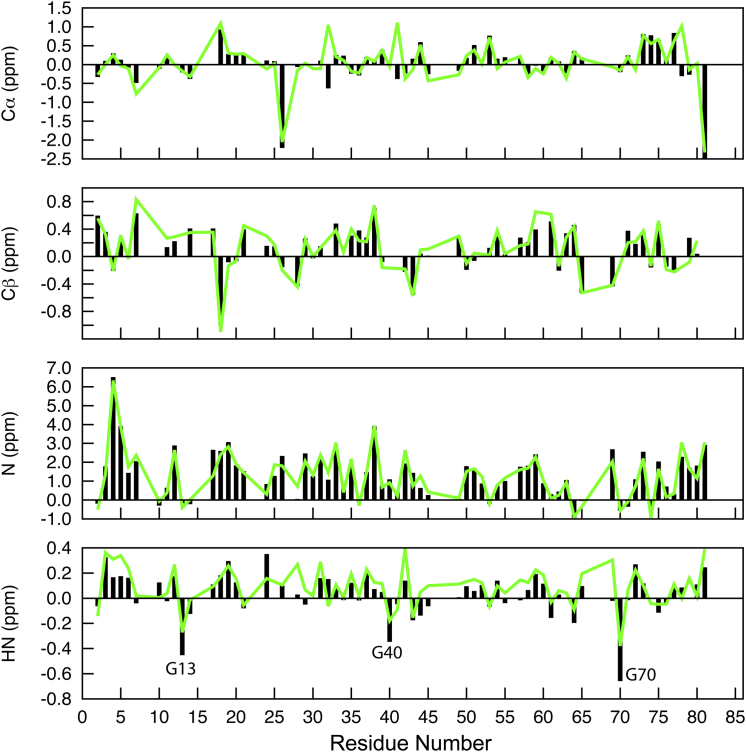
ASTEROIDS uses a genetic algorithm to optimize ensembles of structures that maximally agree with the available experimental data (59). Fig. 6 shows the comparison between the experimental secondary chemical shifts (black bars) with those of the ASTEROIDS ensemble (green lines). As evidenced by the close match between the two sets, the 200-structure ASTEROIDS ensemble reproduces the experimental chemical shifts very well. The largest differences between experimental and calculated values are seen for the HN chemical shifts, with glycine residues 13, 40, and 70 exhibiting the most pronounced deviations. Because it is well known that HN shifts are the most difficult backbone shifts to predict from structure, these differences are not critical.
Figure 6.
Experimental (black bars) and ensemble-averaged (green lines) Cα, Cβ, N, and HN secondary chemical shifts (Δδ) for V75D human γD-crystallin in the presence of 4.2 M urea. The Cα and Cβ secondary shifts do not indicate the presence of any residual secondary structure. Note the excellent agreement between the averaged chemical shifts of the ASTEROIDS-optimized ensemble and the experimental shifts. To see this figure in color, go online.
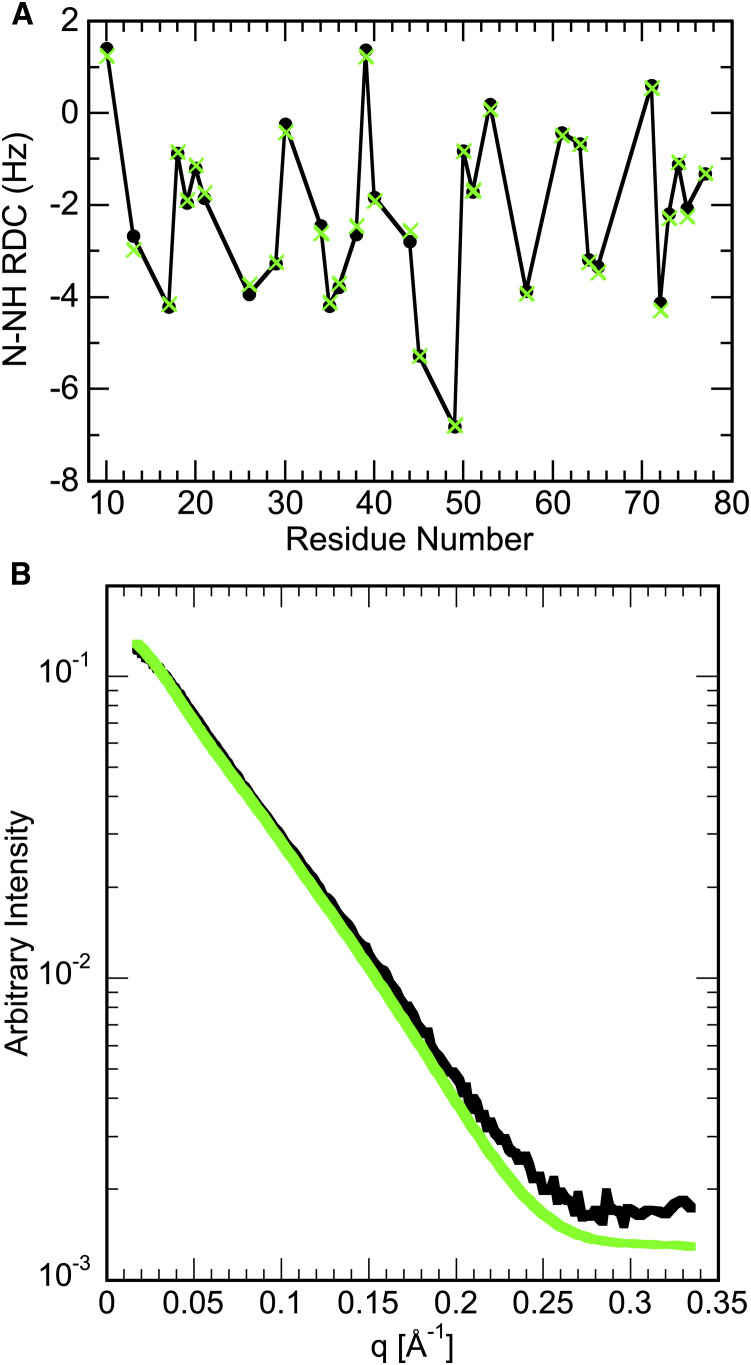
The results of the optimization and selection process for the RDC data are displayed in Fig. 7 A, with the experimental RDC values shown in black and the ensemble-averaged RDC values calculated from the ASTEROIDS-selected ensemble of 200 structures in green. As can be appreciated, the agreement is excellent, with an average difference between experimental and calculated RDCs of 0.1 Hz and a maximal difference of 0.2 Hz.
Figure 7.
Experimental (black) and ensemble-averaged (green) RDC (A) and SAXS (B) data for the NTD of V75D human γD-crystallin in the presence of 4.2 M urea. To see this figure in color, go online.
NMR chemical shifts are exquisitely sensitive to local structure, whereas RDCs report on internuclear vector orientations that provide information on global conformation and long-range angular order. SAXS data permit the generation of overall shapes and were included in the ASTEROIDS calculations as a complement to the NMR data. Because the unfolded NTD exists as an ensemble of dynamically interconverting, random conformers, the overall molecule does not have a single, well-defined global shape as natively folded proteins do. The experimentally obtained SAXS curve and the ensemble-averaged curve for the 200-structure ASTEROIDS ensemble are displayed in Fig. 7 B. The two curves show excellent agreement at low and middle q-values, with some minor divergence at high q-values. Possible reasons for this discrepancy may be related to imperfect buffer subtraction or inaccuracies in modeling a random conformation’s solvent shell by small-angle scattering prediction programs. Indeed, although SAXS prediction works well for native, globular proteins, it is not clear whether the same holds when the protein comprises both a well-packed, globular component and a large unstructured part. Furthermore, the structural models generated by flexible-meccano that were used to calculate the ensemble scattering assume that the CTD structure retains the native conformation, a reasonable approximation given that the CTD transition midpoint lies at a much higher urea concentration than the one used here (Fig. S1) and that resonances from the CTD are well dispersed in the 1H-15N HSQC spectrum and very similar in position to those of the protein without urea, in contrast to the NTD. However, small structural changes in the CTD cannot be excluded, and they may contribute to the discrepancy at the highest scattering angles, which report on shorter-distance structural details. However, because the NMR evidence indicates that the CTD is still folded into a near-native conformation, we believe that the likelier explanation for the modest discrepancy between the ensemble-averaged and experimental curves is sub-optimal buffer subtraction, a problem well known to manifest specifically at high scattering angles.
We also tested whether any subset of the 25,000 starting structures could reproduce the experimental SAXS curve accurately for all q-values. Five independent 200-structure ensembles were selected using only the experimental SAXS data (no chemical shifts or RDCs). The result for one of the ensembles is shown in Fig. S3, but similar outcomes were present for all five ensembles. Only modest improvement was observed in the mid-q range (0.15–0.20 Å−1), and no improvement at high q, compared to ensembles that were optimized using all the experimental data. This indicates that the agreement between experimental and ensemble-averaged SAXS data is not negatively affected when all the experimental data are included in the selection process. Furthermore, the agreement between the ASTEROIDS ensembles and the experimental SAXS data is clearly superior to the agreement between the starting pool of structures and the SAXS data, demonstrating that ASTEROIDS has made meaningful structure selections. Overall, it can be concluded that excellent agreement between the experimental and ensemble-averaged chemical-shift, RDC, and SAXS data is reached, providing confidence that the modeled ensemble accurately reflects the partially unfolded V75D ensemble.
Characterization of structures in the ASTEROIDS-optimized ensemble
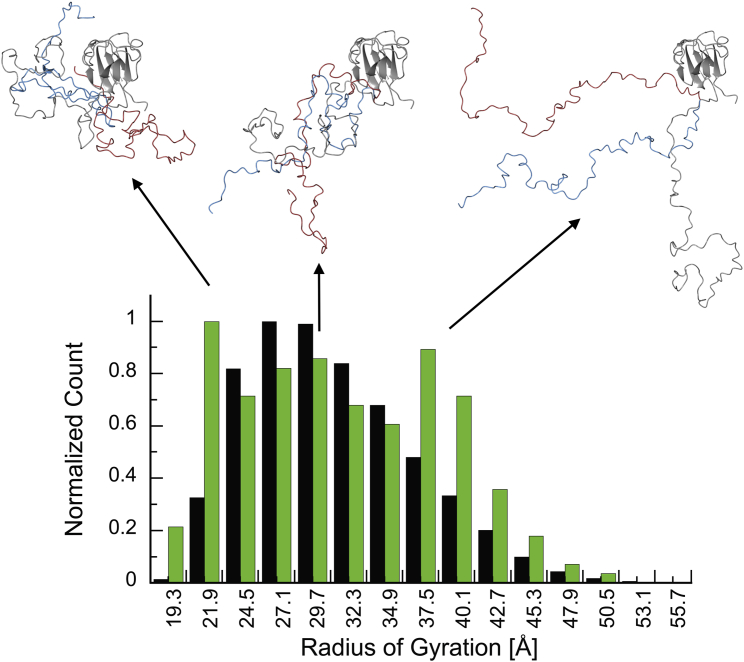
Fig. 8 depicts a comparison between the normalized distribution of the radius of gyration of the 25,000 structures (black bars) in the starting pool and the ASTEROIDS-optimized ensemble of 200 structures (green bars). The starting distribution is unimodal, with most structures exhibiting a radius of gyration of 27–30 Å, which corresponds to a folded CTD with a modestly extended NTD. After selection of an ensemble of 200 structures by ASTEROIDS, based on the experimental data, the distribution becomes trimodal. The first and most populated mode, around Rg ≈ 22 Å, is highly enriched in the ASTEROIDS ensemble compared to the starting pool of conformers. One possible explanation for the abundance of this mode in the selected ensemble is that it may reflect the modest loss of compactness compared to the native state (Rg = 17 Å), which is frequently observed in experimental studies of mono- and multimeric proteins exposed to chaotropic agents (60). The second and third modes represent successively more extended structures, with Rg values of ∼30 and 38–40 Å, respectively. Although the experimental chemical shifts and residual dipolar couplings demonstrate that no residual secondary structure is present, the ensemble’s relative enrichment in extended conformations is itself noteworthy. Whether this is a consequence of using urea as a chaotropic agent or is of any biological significance for promoting aggregation cannot be ascertained at present. Illustrative structures belonging to each of the three modes are displayed above the histogram.
Figure 8.
Structural summary of the ASTEROIDS-optimized V75D human γD-crystallin ensemble in the presence of 4.2 M urea. The histogram displays the radii of gyration in the starting pool of 25,000 structures (black) and the ASTEROIDS-selected ensemble of 200 structures (green). Three models from the ASTEROIDS-optimized ensemble with radii of gyration of ∼22, ∼30, and ∼39 Å, respectively, are displayed. These average radii of gyration correspond to the maxima of the trimodal distribution (green) in the histogram. To see this figure in color, go online.
Validation of the ASTEROIDS ensembles
To assess the quality of our models, we employed a cross-validation strategy. An additional series of ASTEROIDS selections was carried out, in which subsets of the NTD chemical-shift and RDC data were systematically withheld from the ASTEROIDS calculations. The omitted experimental data were then compared with data back-calculated from the ASTEROIDS ensembles. For the chemical-shift validations, the chemical shifts of 16 randomly selected residues distributed throughout the NTD sequence were withheld from ASTEROIDS. The results of this cross-validation are provided in Fig. S4. For all four atom types, (Cα, Cβ, N, and HN), the withheld chemical shifts are accurately predicted by the selected ensemble.
For the RDC validations, a different approach was used. Instead of withholding RDCs randomly throughout the NTD sequence, a series of ASTEROIDS selections was performed in which experimental RDCs were withheld for stretches of 10 consecutive residues (i.e., all available RDCs for residues 20–29, 30–39, 40–49, …, 70–79). This systematic approach allowed us to test whether the distance from the structured CTD plays any role. For the stretches of sequences close to the CTD, the withheld experimental and back-calculated RDC values agree well, whereas for stretches farther away from the CTD, less perfect agreement is noted, a trend that was not observed when validation was carried out using the chemical-shift data. The data for the RDC validations are provided in Fig. S5.
Overall, the agreement between experimental chemical-shift and RDC data omitted from ASTEROIDS and these data back-calculated from the selected ensembles is very good, demonstrating that the ASTEROIDS-optimized ensembles reflect the characteristics of the partially un/folded V75D ensemble.
Discussion
In this study, we performed a comprehensive structural assessment of the folding intermediate of human V75D γD-crystallin in 4.2 M urea, combining experimental and computational methodologies. Although no high-resolution x-ray or NMR structure is available for the folded V75D mutant protein, all existing NMR spectroscopic data suggest that its native structure is very similar to that of the WT and that no gross structural perturbations are present that could explain the mutant’s cataractogenicity. Because previous work established that a partially folded, stable intermediate for this mutant can be observed in the presence of modest chaotrope concentrations, the possibility existed that this partially folded state might promote aggregation and cataract formation. Therefore, we evaluated whether any residual conformational preference exists in the unfolded NTD of the intermediate that may serve as the initiation site for aggregation. Indeed, many proteins, including apomyoglobin (61), barnase (62), CheY (63), and α-lactalbumin (64), are known to retain residual secondary structure under a variety of denaturing conditions. However, as evidenced by all the available data for V75D in 4.2 M urea, no residual structural propensity was detected in the unfolded NTD. At this juncture, we stress that urea-induced unfolding is clearly not identical to unfolding under native conditions. However, the fact that this destabilized mutant can easily populate such a stable unfolding intermediate over a wide range of chaotrope concentrations, in contrast to the WT protein, suggests that this or a similar intermediate could also possibly exist and could contribute to cataractogenesis under physiological conditions.
V75D aggregation
At least three mechanisms for crystallin aggregation and cataract formation have been proposed, each supported by different types of evidence for different crystallins. The first is an amyloid-based mechanism, in which misfolding of part or all of the native crystallin structure leads to the formation of fibrils made from oligomeric β-strand structures. Amyloid fibrils of crystallin have been proposed for WT γD (65, 66, 67, 68), G61C γD (33), and WT γC-crystallin (69), among others. Aggregates of WT γD created in vitro under acidic conditions (pH ≤3) were reported to possess amyloid characteristics; however, such conditions are very different from those found in the eye lens, where the pH lies between 6 and 7, calling into question the physiological relevance of these in vitro crystallin amyloids. A second proposed mechanism for cataract formation involves aggregation via 3D domain swapping. The domain-swapping mechanism of aggregation results in the exchange of one segment of the protein between two or more monomers, resulting in identically structured pseudo-monomers, and in some cases, this mechanism involves complete unfolding and refolding of the protein (70). Such domain-swapped γD-crystallin arrangements have been proposed previously (71, 72). In contrast to domain swapping, amyloid formation generally involves the association of unfolded or misfolded protein segments into non-native intermolecular β-sheet structures, although aggregates can also form directly from reorganization of pre-associated, natively folded monomers (73). Finally, a condensation mechanism has been suggested. For this mechanism, no major unfolding or misfolding is required. Instead, subtle alterations in the physiochemical surface properties of the protein are thought to increase self-association and aggregation. This process has been suggested as the aggregation mechanism for R76S γD (27), P23T γD (74), and V41M γS-crystallin (75).
For the V75D γD variant investigated here, the NTD is thermodynamically destabilized, as evidenced by the observation of a stable folding intermediate (29), and the NMR data demonstrate that the NTD is completely unfolded in 4.2 M urea and lacks any residual structure. Aggregates of V75D γD that form under physiological conditions (pH 7) appear amorphous by negative-stain electron microscopy (unpublished data), suggesting a non-amyloid structure. Therefore, further experimental work is needed to characterize the structure of the aggregate and the molecular mechanism of aggregation. Such studies are ongoing in our laboratory.
Author Contributions
A.M.G. and M.J.W. conceived the study and planned the experiments. M.J.W., Z.X., and J.C.B. expressed and purified the protein. M.J.W., Z.X., and J.C.B. acquired and analyzed the NMR data. M.J.W. collected and analyzed the SAXS data. M.J.W., M.R.J., and M.B. performed the computational selections using ASTEROIDS. M.J.W. and A.M.G. wrote the manuscript. All authors reviewed and edited the manuscript.
Acknowledgments
M.J.W. gratefully acknowledges training received during the 2014 EMBO Practical Course on Solution Scattering from Biological Macromolecules, as well as useful discussions with Dmitri Svergun and his group at the European Molecular Biology Laboratory Hamburg Outstation. The authors declare no conflicts of interest.
This work was funded by the National Institutes of Health (grant EY021193 to A.M.G.) and made use of the SAXS Core Facility of the Center for Cancer Research, National Cancer Institute. Time on the shared scattering beamline 12-ID-B was allocated under the PUP-24152 agreement between the National Cancer Institute and Argonne National Laboratory (ANL). The Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility, is operated by Argonne National Laboratory under contract no. DE-AC02-06CH11357.
Editor: David Eliezer.
Footnotes
Five figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30169-8.
Supporting Material
References
- 1.Bassnett S. On the mechanism of organelle degradation in the vertebrate lens. Exp. Eye Res. 2009;88:133–139. doi: 10.1016/j.exer.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waley S.G. The lens: function and macromolecular composition. In: Davson H., editor. The Eye. Academic Press; New York: 1969. pp. 299–379. [Google Scholar]
- 3.Horwitz J. α-Crystallin can function as a molecular chaperone. Proc. Natl. Acad. Sci. USA. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J., Callis P.R., King J. Mechanism of the very efficient quenching of tryptophan fluorescence in human gamma D- and gamma S-crystallins: the gamma-crystallin fold may have evolved to protect tryptophan residues from ultraviolet photodamage. Biochemistry. 2009;48:3708–3716. doi: 10.1021/bi802177g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaenicke R. Stability and folding of domain proteins. Prog. Biophys. Mol. Biol. 1999;71:155–241. doi: 10.1016/s0079-6107(98)00032-7. [DOI] [PubMed] [Google Scholar]
- 6.Delaye M., Tardieu A. Short-range order of crystallin proteins accounts for eye lens transparency. Nature. 1983;302:415–417. doi: 10.1038/302415a0. [DOI] [PubMed] [Google Scholar]
- 7.Vendra V.P., Agarwal G., Balasubramanian D. Structural integrity of the Greek key motif in βγ-crystallins is vital for central eye lens transparency. PLoS One. 2013;8:e70336. doi: 10.1371/journal.pone.0070336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao H., Brown P.H., Schuck P. The molecular refractive function of lens γ-crystallins. J. Mol. Biol. 2011;411:680–699. doi: 10.1016/j.jmb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hains P.G., Truscott R.J. Post-translational modifications in the nuclear region of young, aged, and cataract human lenses. J. Proteome Res. 2007;6:3935–3943. doi: 10.1021/pr070138h. [DOI] [PubMed] [Google Scholar]
- 10.Hanson S.R., Hasan A., Smith J.B. The major in vivo modifications of the human water-insoluble lens crystallins are disulfide bonds, deamidation, methionine oxidation and backbone cleavage. Exp. Eye Res. 2000;71:195–207. doi: 10.1006/exer.2000.0868. [DOI] [PubMed] [Google Scholar]
- 11.Lampi K.J., Ma Z., David L.L. Age-related changes in human lens crystallins identified by two-dimensional electrophoresis and mass spectrometry. Exp. Eye Res. 1998;67:31–43. doi: 10.1006/exer.1998.0481. [DOI] [PubMed] [Google Scholar]
- 12.Xia Z., Yang Z., Zhou R. UV-radiation induced disruption of dry-cavities in human γD-crystallin results in decreased stability and faster unfolding. Sci. Rep. 2013;3:1560. doi: 10.1038/srep01560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hejtmancik J.F. Congenital cataracts and their molecular genetics. Semin. Cell Dev. Biol. 2008;19:134–149. doi: 10.1016/j.semcdb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson N.E., Lampi K.J., Robinson A.B. Quantitative measurement of young human eye lens crystallins by direct injection Fourier transform ion cyclotron resonance mass spectrometry. Mol. Vis. 2006;12:704–711. [PubMed] [Google Scholar]
- 15.Stephan D.A., Gillanders E., Brownstein M.J. Progressive juvenile-onset punctate cataracts caused by mutation of the γD-crystallin gene. Proc. Natl. Acad. Sci. USA. 1999;96:1008–1012. doi: 10.1073/pnas.96.3.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Héon E., Priston M., Munier F.L. The γ-crystallins and human cataracts: a puzzle made clearer. Am. J. Hum. Genet. 1999;65:1261–1267. doi: 10.1086/302619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kmoch S., Brynda J., Elleder M. Link between a novel human γD-crystallin allele and a unique cataract phenotype explained by protein crystallography. Hum. Mol. Genet. 2000;9:1779–1786. doi: 10.1093/hmg/9.12.1779. [DOI] [PubMed] [Google Scholar]
- 18.Graw J., Löster J., Hrabé de Angelis M. V76D mutation in a conserved gD-crystallin region leads to dominant cataracts in mice. Mamm. Genome. 2002;13:452–455. doi: 10.1007/s00335-002-3021-6. [DOI] [PubMed] [Google Scholar]
- 19.Nandrot E., Slingsby C., Hilal L. Gamma-D crystallin gene (CRYGD) mutation causes autosomal dominant congenital cerulean cataracts. J. Med. Genet. 2003;40:262–267. doi: 10.1136/jmg.40.4.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santhiya S.T., Shyam Manohar M., Graw J. Novel mutations in the γ-crystallin genes cause autosomal dominant congenital cataracts. J. Med. Genet. 2002;39:352–358. doi: 10.1136/jmg.39.5.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messina-Baas O.M., Gonzalez-Huerta L.M., Cuevas-Covarrubias S.A. Two affected siblings with nuclear cataract associated with a novel missense mutation in the CRYGD gene. Mol. Vis. 2006;12:995–1000. [PubMed] [Google Scholar]
- 22.Li F., Wang S., Ma X. Mutation G61C in the CRYGD gene causing autosomal dominant congenital coralliform cataracts. Mol. Vis. 2008;14:378–386. [PMC free article] [PubMed] [Google Scholar]
- 23.Roshan M., Vijaya P.H., Satyamoorthy K. A novel human CRYGD mutation in a juvenile autosomal dominant cataract. Mol. Vis. 2010;16:887–896. [PMC free article] [PubMed] [Google Scholar]
- 24.Wang B., Yu C., Xie L. A novel CRYGD mutation (p.Trp43Arg) causing autosomal dominant congenital cataract in a Chinese family. Hum. Mutat. 2011;32:E1939–E1947. doi: 10.1002/humu.21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banerjee P.R., Pande A., Pande J. Cataract-associated mutant E107A of human γD-crystallin shows increased attraction to α-crystallin and enhanced light scattering. Proc. Natl. Acad. Sci. USA. 2011;108:574–579. doi: 10.1073/pnas.1014653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans P., Wyatt K., Slingsby C. The P23T cataract mutation causes loss of solubility of folded γD-crystallin. J. Mol. Biol. 2004;343:435–444. doi: 10.1016/j.jmb.2004.08.050. [DOI] [PubMed] [Google Scholar]
- 27.Ji F., Jung J., Gronenborn A.M. Structural and biochemical characterization of the childhood cataract-associated R76S mutant of human γD-crystallin. Biochemistry. 2012;51:2588–2596. doi: 10.1021/bi300199d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji F., Jung J., Gronenborn A.M. The human W42R γD-crystallin mutant structure provides a link between congenital and age-related cataracts. J. Biol. Chem. 2013;288:99–109. doi: 10.1074/jbc.M112.416354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreau K.L., King J. Hydrophobic core mutations associated with cataract development in mice destabilize human γD-crystallin. J. Biol. Chem. 2009;284:33285–33295. doi: 10.1074/jbc.M109.031344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pande A., Pande J., Benedek G.B. Molecular basis of a progressive juvenile-onset hereditary cataract. Proc. Natl. Acad. Sci. USA. 2000;97:1993–1998. doi: 10.1073/pnas.040554397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pande A., Pande J., Benedek G.B. Crystal cataracts: human genetic cataract caused by protein crystallization. Proc. Natl. Acad. Sci. USA. 2001;98:6116–6120. doi: 10.1073/pnas.101124798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pande A., Annunziata O., Pande J. Decrease in protein solubility and cataract formation caused by the Pro23 to Thr mutation in human gamma D-crystallin. Biochemistry. 2005;44:2491–2500. doi: 10.1021/bi0479611. [DOI] [PubMed] [Google Scholar]
- 33.Zhang W., Cai H.C., Yan Y.B. The congenital cataract-linked G61C mutation destabilizes γD-crystallin and promotes non-native aggregation. PLoS One. 2011;6:e20564. doi: 10.1371/journal.pone.0020564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu S.Y., Zou P., Mchaourab H.S. Expression of cataract-linked γ-crystallin variants in zebrafish reveals a proteostasis network that senses protein stability. J. Biol. Chem. 2016;291:25387–25397. doi: 10.1074/jbc.M116.749606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra S., Stein R.A., McHaourab H.S. Cataract-linked γD-crystallin mutants have weak affinity to lens chaperones α-crystallins. FEBS Lett. 2012;586:330–336. doi: 10.1016/j.febslet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreau K.L., King J.A. Cataract-causing defect of a mutant γ-crystallin proceeds through an aggregation pathway which bypasses recognition by the α-crystallin chaperone. PLoS One. 2012;7:e37256. doi: 10.1371/journal.pone.0037256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hura G.L., Budworth H., Tainer J.A. Comprehensive macromolecular conformations mapped by quantitative SAXS analyses. Nat. Methods. 2013;10:453–454. doi: 10.1038/nmeth.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konarev P.V., Volkov V.V., Svergun D.I. PRIMUS—a Windows-PC based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 39.Hyberts S.G., Takeuchi K., Wagner G. Poisson-gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J. Am. Chem. Soc. 2010;132:2145–2147. doi: 10.1021/ja908004w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hyberts S.G., Milbradt A.G., Wagner G. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J. Biomol. NMR. 2012;52:315–327. doi: 10.1007/s10858-012-9611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 42.Johnson B.A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 43.Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ottiger M., Delaglio F., Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 45.Chou J.J., Gaemers S., Bax A. A simple apparatus for generating stretched polyacrylamide gels, yielding uniform alignment of proteins and detergent micelles. J. Biomol. NMR. 2001;21:377–382. doi: 10.1023/a:1013336502594. [DOI] [PubMed] [Google Scholar]
- 46.Basak A., Bateman O., Pande J. High-resolution X-ray crystal structures of human γD crystallin (1.25 Å) and the R58H mutant (1.15 Å) associated with aculeiform cataract. J. Mol. Biol. 2003;328:1137–1147. doi: 10.1016/s0022-2836(03)00375-9. [DOI] [PubMed] [Google Scholar]
- 47.Emsley P., Lohkamp B., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ozenne V., Bauer F., Blackledge M. Flexible-meccano: a tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental observables. Bioinformatics. 2012;28:1463–1470. doi: 10.1093/bioinformatics/bts172. [DOI] [PubMed] [Google Scholar]
- 49.Shen Y., Bax A. Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology. J. Biomol. NMR. 2007;38:289–302. doi: 10.1007/s10858-007-9166-6. [DOI] [PubMed] [Google Scholar]
- 50.Zweckstetter M., Bax A. Prediction of sterically induced alignment in a dilute liquid crystalline phase: aid to protein structure determination by NMR. J. Am. Chem. Soc. 2000;122:3791–3792. [Google Scholar]
- 51.Svergun D.I., Barberato C., Koch M.H.J. CRYSOL—a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 52.Nodet G., Salmon L., Blackledge M. Quantitative description of backbone conformational sampling of unfolded proteins at amino acid resolution from NMR residual dipolar couplings. J. Am. Chem. Soc. 2009;131:17908–17918. doi: 10.1021/ja9069024. [DOI] [PubMed] [Google Scholar]
- 53.Jensen M.R., Salmon L., Blackledge M. Defining conformational ensembles of intrinsically disordered and partially folded proteins directly from chemical shifts. J. Am. Chem. Soc. 2010;132:1270–1272. doi: 10.1021/ja909973n. [DOI] [PubMed] [Google Scholar]
- 54.Ozenne V., Schneider R., Blackledge M. Mapping the potential energy landscape of intrinsically disordered proteins at amino acid resolution. J. Am. Chem. Soc. 2012;134:15138–15148. doi: 10.1021/ja306905s. [DOI] [PubMed] [Google Scholar]
- 55.Wishart D.S., Sykes B.D., Richards F.M. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 56.Wishart D.S., Case D.A. Use of chemical shifts in macromolecular structure determination. Methods Enzymol. 2001;338:3–34. doi: 10.1016/s0076-6879(02)38214-4. [DOI] [PubMed] [Google Scholar]
- 57.Redfield C. Using nuclear magnetic resonance spectroscopy to study molten globule states of proteins. Methods. 2004;34:121–132. doi: 10.1016/j.ymeth.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 58.Bernadó P., Blanchard L., Blackledge M. A structural model for unfolded proteins from residual dipolar couplings and small-angle x-ray scattering. Proc. Natl. Acad. Sci. USA. 2005;102:17002–17007. doi: 10.1073/pnas.0506202102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jensen M.R., Zweckstetter M., Blackledge M. Exploring free-energy landscapes of intrinsically disordered proteins at atomic resolution using NMR spectroscopy. Chem. Rev. 2014;114:6632–6660. doi: 10.1021/cr400688u. [DOI] [PubMed] [Google Scholar]
- 60.Dutta S., Bhattacharyya D. Size of unfolded and dissociated subunits versus that of native multimeric proteins. J. Biol. Phys. 2001;27:59–71. doi: 10.1023/A:1011826525684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eliezer D., Chung J., Wright P.E. Native and non-native secondary structure and dynamics in the pH 4 intermediate of apomyoglobin. Biochemistry. 2000;39:2894–2901. doi: 10.1021/bi992545f. [DOI] [PubMed] [Google Scholar]
- 62.Bond C.J., Wong K.-B., Daggett V. Characterization of residual structure in the thermally denatured state of barnase by simulation and experiment: Description of the folding pathway. PNAS. 1997;94:13409–13413. doi: 10.1073/pnas.94.25.13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia P., Serrano L., Bruix M. NMR and SAXS characterization of the denatured state of the chemotactic protein CheY: implications for protein folding initiation. Protein Sci. 2001;10:1100–1112. doi: 10.1110/ps.52701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Higman V.A., Rösner H.I., Smith L.J. Probing the urea dependence of residual structure in denatured human alpha-lactalbumin. J. Biomol. NMR. 2009;45:121–131. doi: 10.1007/s10858-009-9342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moran S.D., Woys A.M., Zanni M.T. Two-dimensional IR spectroscopy and segmental 13C labeling reveals the domain structure of human γD-crystallin amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2012;109:3329–3334. doi: 10.1073/pnas.1117704109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moran S.D., Zhang T.O., Zanni M.T. An alternative structural isoform in amyloid-like aggregates formed from thermally denatured human γD-crystallin. Protein Sci. 2014;23:321–331. doi: 10.1002/pro.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu J.W., Chen M.E., Wang S.S. Comparative analysis of human γD-crystallin aggregation under physiological and low pH conditions. PLoS One. 2014;9:e112309. doi: 10.1371/journal.pone.0112309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Papanikolopoulou K., Mills-Henry I., King J. Formation of amyloid fibrils in vitro by human γD-crystallin and its isolated domains. Mol. Vis. 2008;14:81–89. [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y., Petty S., King J. Formation of amyloid fibrils in vitro from partially unfolded intermediates of human γC-crystallin. Invest. Ophthalmol. Vis. Sci. 2010;51:672–678. doi: 10.1167/iovs.09-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu L., Byeon I.J., Gronenborn A.M. Domain swapping proceeds via complete unfolding: a 19F- and 1H-NMR study of the cyanovirin-N protein. J. Am. Chem. Soc. 2012;134:4229–4235. doi: 10.1021/ja210118w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Das P., King J.A., Zhou R. Aggregation of γ-crystallins associated with human cataracts via domain swapping at the C-terminal β-strands. Proc. Natl. Acad. Sci. USA. 2011;108:10514–10519. doi: 10.1073/pnas.1019152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garcia-Manyes S., Giganti D., Fernández J.M. Single-molecule force spectroscopy predicts a misfolded, domain-swapped conformation in human γD-crystallin protein. J. Biol. Chem. 2016;291:4226–4235. doi: 10.1074/jbc.M115.673871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Plakoutsi G., Bemporad F., Chiti F. Evidence for a mechanism of amyloid formation involving molecular reorganisation within native-like precursor aggregates. J. Mol. Biol. 2005;351:910–922. doi: 10.1016/j.jmb.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 74.Pande A., Ghosh K.S., Pande J. Increase in surface hydrophobicity of the cataract-associated P23T mutant of human γD-crystallin is responsible for its dramatically lower, retrograde solubility. Biochemistry. 2010;49:6122–6129. doi: 10.1021/bi100664s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bharat S.V., Shekhtman A., Pande J. The cataract-associated V41M mutant of human γS-crystallin shows specific structural changes that directly enhance local surface hydrophobicity. Biochem. Biophys. Res. Commun. 2014;443:110–114. doi: 10.1016/j.bbrc.2013.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.