Abstract
The brown planthopper (BPH) is the most serious insect pest of rice in Asia. The indica rice cultivar ADR52 carries two BPH resistance genes, BPH26 (BROWN PLANTHOPPER RESISTANCE 26) and BPH25. Map-based cloning of BPH26 revealed that BPH26 encodes a coiled-coil-nucleotide-binding-site–leucine-rich repeat (CC–NBS–LRR) protein. BPH26 mediated sucking inhibition in the phloem sieve element. BPH26 was identical to BPH2 on the basis of DNA sequence analysis and feeding ability of the BPH2-virulent biotype of BPH. BPH2 was widely incorporated in elite rice cultivars and was well-cultivated in many Asian countries as a favorable gene resource in rice breeding against BPH. However, BPH2 was rendered ineffective by a virulent biotype of BPH in rice fields in Asia. In this study, we suggest that BPH2 can be reused by combining with other BPH resistance genes, such as BPH25, to ensure durable resistance to BPH.
Plants have various mechanisms to afford protection against herbivorous insects1. However, varieties of highly productive crops suitable for multi-fertilizer cultivation lose their inherent protective mechanisms against insect-feeding. Development and utilization of resistant crop varieties will promote environment-friendly agricultural practices involving limited use of pesticides. Many insect-resistant crop species have been screened, and their application in breeding programs is being investigated2.
Rice is one of the most widely grown crops worldwide. The brown planthopper (BPH), Nilaparvata lugens Stål (Homoptera: Delphacidae), is the most serious insect pest of rice in Asia. BPH is a monophagous sucking insect that mainly feeds on rice (Oryza sativa L.)3,4. These insects cause direct damage by sucking sap from the phloem of susceptible rice varieties and indirect damage by transmitting viral diseases5. When the density of BPH remarkably increases, many rice plants die and, eventually, “hopperburn” appears in the rice paddy. Large amounts of insecticides are used to control BPH. However, BPH showing resistance to insecticides such as imidacloprid have emerged6, requiring the development of other control methods as an alternative to insecticide use.
Rice varieties resistant to BPH have been screened on a large scale at the International Rice Research Institute (IRRI)7. The resistance gene from the indica cultivar Mudgo was named Bph1 and that from the indica cultivar ASD7 was named bph2. The indica cultivar IR26 harboring the Bph1 gene was bred at IRRI and used as the first commercial cultivar for controlling BPH. However, a Bph1-vilurent biotype of BPH gradually rended the resistance gene Bph1 ineffective8. Next, some cultivars such as IR42 harboring bph2 were released for BPH control. However, they were also rendered ineffective by the appearance of a new virulent BPH biotype9.
BPH resistance genes were screened from not only O. sativa L., which is the main host plant of BPH but also other Oryza species, and currently, more than 30 BPH resistance loci have been reported10. Breeding of resistant rice cultivars for controlling BPH necessitates the discovery of new gene sources and the development of control methods to better retain the effect of resistance genes. Myint et al.11 found two BPH resistance genes in the indica cultivar ADR52 and named them BPH25 and BPH26. BPH25 is located on the short arm of chromosome 6, and BPH26 is located on the long arm of chromosome 12. Many BPH resistance genes, such as Bph1, bph2, Bph9, Bph10, Bph18(t), and Bph21(t), have been mapped to the long arm of chromosome 1210. Bph112,13, bph212,14,15, Bph916,17,18, and BPH26 were found in O. sativa L. varieties; Bph10, in O. australiensis19 and O. officinalis20; Bph18(t), in O. australiensis21; and Bph21(t), in O. minuta22. The long arm of chromosome 12 is considered to be one of the most important locations for conferring resistance against BPH in Oryza species. However, there are no reports on the molecular cloning of the BPH resistance gene in this region. In this study, we isolated BPH26 from the long arm of chromosome 12 of the indica cultivar ADR52 (O. sativa L.). Till date, only a few resistance genes against sap-sucking insects, such as the aphid resistance gene Mi (Mi-1.2) from tomato23,24 and the BPH resistance gene Bph1425, have been cloned and their nucleotide sequences have been reported. Although Bph14 originated from the wild rice variety O. officinalis on chromosome 3 has already been isolated as a BPH resistance gene using introgression lines25,26,27, none of the BPH resistance genes has been isolated from O. sativa L., which is the main host plant of BPH.
Furthermore, we confirmed that BPH26 is identical to BPH2, which was rendered ineffective by a virulent biotype of BPH in the Asian rice fields. Herein, we discuss the possibility for reusing BPH2.
Results
Delimitation of a candidate genomic region of BPH26
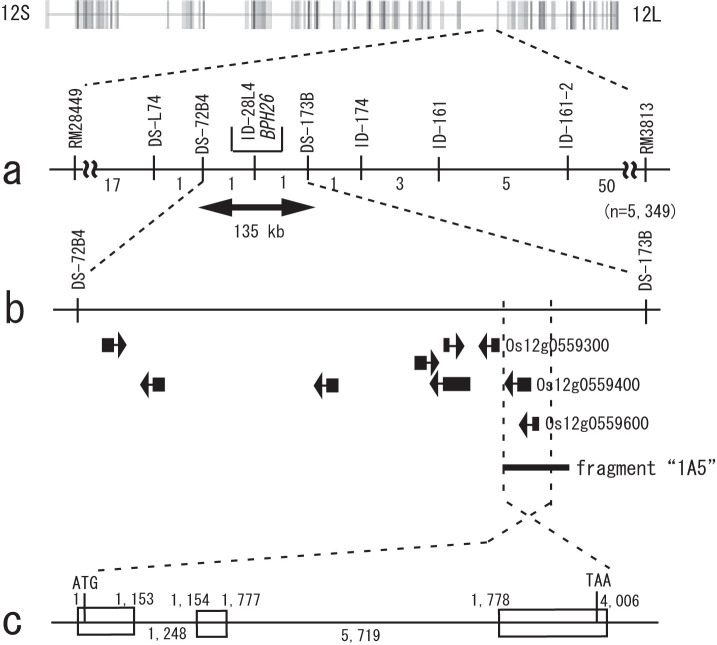
High-resolution genetic maps of BPH26 were constructed using 5,349 BC6F3 individuals (Fig. 1a). Seventy-nine plants showing recombination between two markers (RM28449 and RM3813) were selected, and the BPH resistance of these plants was analyzed based on the survival rate of BPH in a test tube. Eventually, we determined the candidate genomic region of BPH26 in a 135-kb region between two flanking markers, DS-72B4 and DS-173B (Fig. 1a). Nine putative genes were predicted in the corresponding candidate region of Nipponbare by using a rice annotation project database (RAP-DB; Fig. 1b). Among them, the amino acid sequences of Os12g0559300, Os12g0559400, and Os12g0559600 were homologous to those of NBS-LRR type disease resistance proteins (Supplementary Table S1 online). Two putative genes (Os12g0559400 and Os12g0559600) were predicted to encode parts of one nucleotide-binding site–leucine-rich repeat (NBS-LRR) protein, although one putative gene (Os12g0559300) was considered to be incomplete and was non-functional. One bacterial artificial chromosome (BAC) clone 14H01 (153 kb), containing Os12g0559400 and Os12g0559600, was selected from a BAC genomic library of ADR52, and the corresponding region was sequenced (Supplementary Fig. S1 online). One gene that was expected to code the NBS-LRR protein was also present on the corresponding sequences of the BAC clone 14H01, as revealed by the rice genome automated annotation system (RiceGAAS). Rapid amplification of cDNA ends (RACE)-PCR was performed, and cDNA encoding coiled coil-nucleotide binding site-leucine rich repeat (CC-NBS-LRR) protein was cloned and sequenced, using primers designed from the sequence data of the BAC clone 14H01A.
Figure 1. High-resolution genetic map of BPH26.
(a) Genetic linkage map of 5,349 BC6F3 plants showing the relative position of BPH26 with the single-sequence repeat, insertion-deletion, and single nucleotide polymorphism markers on chromosome 12. The numbers under the linkage map indicate the number of recombinants in the adjacent marker intervals. Framework maps are quoted from Harushima et al58. (b) Putative genes in the candidate region whose presence is predicted by the Rice Annotation Project Database. A fragment 1A5 shows the genomic DNA of ADR52 which was used for the functional complementation test. (c) The structure of BPH26 on the genomic DNA of ADR52.
Functional complementation with the candidate gene in transgenic rice
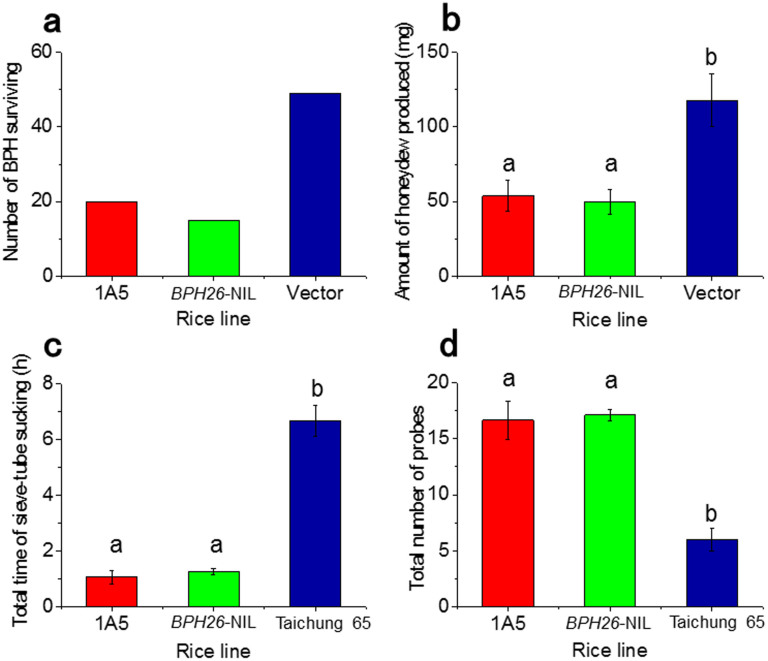
A genomic sub-clone library of about 20 kb was constructed from the BAC clone 14H01. A clone containing a 15.6-kb DNA fragment (fragment 1A5) with the full length of the candidate gene was selected (Fig. 1b). There was only one gene predicted on the fragment 1A5, as revealed by BLASTN search. The clone was transformed into the susceptible cultivar Taichung 65. Of the five independent transgenic lines tested, all transgenic lines with fragment 1A5 showed resistance to BPH (Supplementary Fig. S2 online). The result of the complementation test using one of those five lines (line name, “1A5”) was compared to those of the near isogenic line carrying BPH26 (line name, “BPH26-NIL”) and vector control (line name, “Vector”; Fig. 2). The BPH survival was significantly decreased on the transgenic “1A5” in comparison with that on the “Vector” control, and was equivalent to that on the “BPH26-NIL” (chi-square test28; Fig. 2a). Similarly, the amount of honeydew excreted on “1A5” and “BPH26-NIL” was smaller than that on “Vector” control (Fig. 2b). The feeding activities of BPH were investigated using an alternating current electric penetration graph (AC-EPG). The total hours of sucking sieve-tube on “1A5” and “BPH26-NIL” were suppressed around to only 1 h compared to 7 h of sucking on the susceptible cultivar Taichung 65 during the 10-h exposure to BPH (Fig. 2c). Furthermore, the numbers of probes on “1A5” and “BPH26-NIL” were higher than those on the susceptible cultivar Taichung 65 (Fig. 2d). These data suggest that phloem ingestion was inhibited and probing behavior was repeated on “1A5” and “BPH26-NIL.” Therefore, the gene on fragment 1A5 was identified as the resistance gene BPH26, because the "1A5" line exhibited the same resistance as the original line "BPH26-NIL.”
Figure 2. Functional complementation test of candidate genes.
T1 lines that have been PCR validated to harbor the gene were used in the bioassays. The BPH2-avirulent biotype was used in these experiments. (a) The survival number of brown planthopper (BPH) on the three rice lines. The 1A5 line is the transgenic plant with the predicted gene on the DNA fragment 1A5 in the background of Taichung 65. The vector line is the transgenic plant with only vector fragment in the background of Taichung 65. BPH26-NIL is the near isogenic line with BPH26 in the genetic background of Taichung 65. In all, sixty last instar larvae that had been fasted for 24 h were released on each rice line for 5 days. (b) The amount of honeydew excreted by BPH females on the three rice lines. Values represent mean and SE (n = 10). Means labeled with the same letters are not significantly different by Tukey–Kramer test (P < 0.05). (c) The total duration of phloem ingestion on the three rice lines. The feeding behavior of BPH females was measured using an AC-EPG system for 10 h. Values represent mean and SE (n = 10). Means labeled with the same letters are not significantly different by Tukey–Kramer test (P < 0.05). (d) The total number of probes. The feeding behavior of BPH females was measured using an AC-EPG system for 10 h. Values represent mean and SE (n = 10). Means labeled with the same letters are not significantly different by Tukey–Kramer test (P < 0.05).
Sequence analysis of the BPH26 gene
Comparison of the cDNA and genomic DNA sequences revealed that there are three exons and two introns (Fig. 1c). The deduced open reading frame (ORF) of BPH26 encoded a protein of 1,218 amino acids. The cDNA and amino acid sequences are shown in Supplementary Fig. S3 online and Fig. 3, respectively. Motifs present in the protein encoded by BPH26 were assessed on the basis of the report of Zhou et al29. BPH26 also contained many kinds of motifs, e.g., CC motif; motifs characteristic of the NBS domain; and LRR motifs, which are found in other CC-NBS-LRR proteins (Fig. 3).
Figure 3. The deduced amino acid sequence of BPH26.
Typical motifs with rice R proteins are shown in blue letters, and the leucine-rich repeat (LRR) domain is underlined in red.
Next, the deduced amino acid sequence of BPH26 was compared with those in other rice varieties (Supplementary Fig. S4 online). There was a nonsense mutation in the allele of Nipponbare and Taichung 65, which are both susceptible japonica varieties. Indica rice variety Babawee and Rathu Heenati do not possess the resistance gene on chromosome 1230,31,32. The nucleotide sequence was identical between Rathu Heenati and Babawee, and there was no nonsense mutation in their sequences. There were many substitutions and losses of amino acid sequences when the alleles of ADR52 were compared with those of Rathu Heenati. Amino acid substitutions tended to be mainly found in the LRR domain (Supplementary Fig. S4 online).
Identification of BPH26 as BPH2
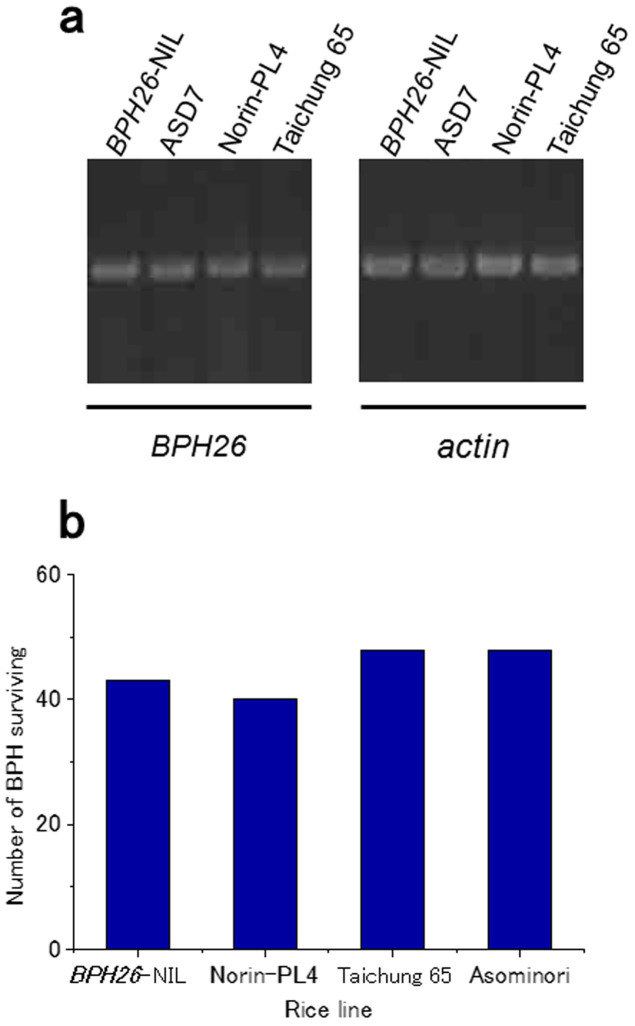
The map positions of both BPH26 and BPH2 seemed to overlap with each other on the long arm of chromosome 12. Nucleotide and amino acid sequences of BPH26 were entirely identical to those of ASD7 and IR1154–243 (Supplementary Fig. S4 online), which are known to harbor the BPH2 gene12,14,15. The expression analysis by reverse transcriptase (RT)-PCR was conducted using BPH26-NIL, Taichung 65, ASD7, and Norin-PL4. Norin-PL4 included a BPH2 originated from IR1125-243 in the background of the japonica cultivar Asominori15. Actin primers were used in the control amplification. The expressions of the BPH26 gene were observed in all the varieties tested (Fig. 4a). Taichung 65 was considered to have lost resistance because the normal BPH26 protein had not been translated due to a nonsense mutation, although gene expression was observed. Gene expressions were also observed in ASD7 and Norin-PL4, both of which have the BPH2 gene. The feeding abilities of BPH on BPH26-NIL and Norin-PL4 were investigated using a BPH2-virulent biotype of BPH. The chi-square test28 revealed that there was no difference between the numbers of survivors on BPH26-NIL and Norin-PL4 (Fig. 4b). This result indicated that the feeding capability of the BPH2-virulent biotype on the plants harboring BPH26 was almost the same as that on the plants harboring BPH2. Thus, BPH26 could be considered to be identical to BPH2 on the basis of the results of map positioning, gene sequence analysis, gene expression analysis, and feeding ability of the BPH2-virulent biotype.
Figure 4. The comparisons of BPH26 and BPH2.
(a) Gene expression analysis of BPH26 in some rice varieties by using reverse transcriptase-PCR. Actin primers were used in the control amplification. The gels have been run under the same experimental conditions. Full-length gel is presented in Supplementary Figure S7. (b) The feeding abilities of the BPH2-virulent biotype of brown planthopper (BPH) on BPH26-near isogenic line and Norin-PL4 harboring the BPH2 gene. Total of sixty last instar larvae that had been starved for 24 h were released on each rice line for 5 days.
Phylogenetic analysis
Phylogenetic relationships were investigated among the CC-NBS-LRR proteins, such as BPH14 and the proteins encoded by blast-disease-resistance genes in Oryza species (Fig. 5). BPH26 was more closely related with some disease-resistance proteins than with BPH14. The amino acid sequences of the main motifs in BPH26, BPH14, and PIB were compared. In CC-NBS domain, the amino acid sequences of the main motifs were well-conserved, although none of the motifs had the same amino acid sequences between BPH26 and BPH14 (Supplementary Fig. S5 online). The structures of LRR domains of BPH26, BPH14, and PIB were more different than those of CC-NBS domains (Supplementary Fig. S6 online).
Figure 5. Phylogenic tree of some disease and pest resistance proteins in Oryza species.
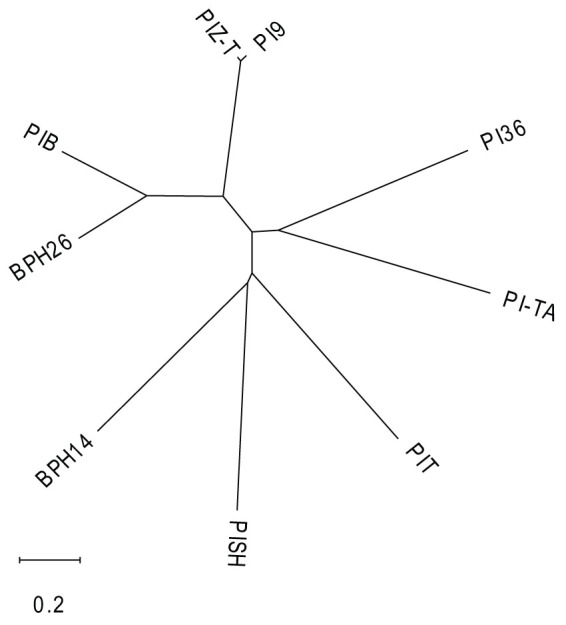
Sequences were aligned by MEGA6 program51, and a full-length amino acid sequence was used for clustering.
Histochemical detection of BPH26 expression
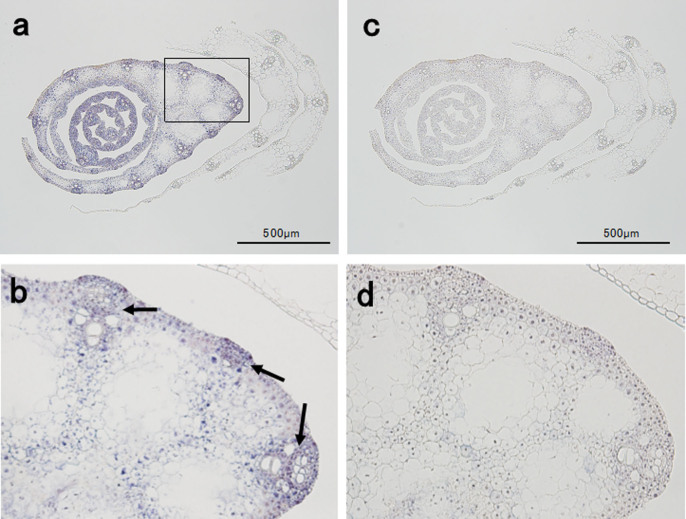
The expression site of BPH26 was analyzed using the in situ hybridization method (Fig. 6). BPH26 showed relatively stronger expression in the inner leaf than in the outer leaf of the leaf sheath (Fig. 6a). Furthermore, BPH26 showed relatively stronger expression in vascular bundles than in other tissues in the leaves of the middle layer of the leaf sheath (Fig. 6b).
Figure 6. The results of in situ hybridization of BPH26.
Near isogenic lines with BPH26 at the 5- to 6-leaf stages were used for in situ hybridization analysis. Cross-sections of the leaf sheath are shown. (a): antisense probe of BPH26 m-RNA, (b): enlarged photograph of a square in (a), (c): sense probe of BPH26 m-RNA (blank sample), (d): enlarged photograph of (c).
Discussion
The host-plant resistance gene BPH2, which conferred resistance to BPH, was widely incorporated in elite rice cultivars, and was well-cultivated in many Asian countries as a favorable gene resource in rice breeding against BPH33. The BPH resistance gene BPH26 was found in the O. sativa L. ssp. indica cultivar ADR52 and mapped on the rice chromosome 12. In the present study, map-based cloning of the BPH resistance gene BPH26 showed that BPH26 is identical to BPH2 on the basis of DNA sequence analysis and feeding ability of the BPH2-virulent biotype of BPH. The protein encoded by BPH26 was a CC-NBS-LRR protein. The sequence of BPH26 derived from ADR52 was completely identical to one from ASD7, which carried the resistance allele of BPH2. On the other hand, there was a nonsense mutation in the sequence of the susceptible cultivar Taichung 65. The aphid resistance gene Mi from tomato23,24 and BPH resistance gene BPH14 from wild rice25 have been reported to encode a CC-NBS-LRR-receptor kinase-like protein. Although only a few resistance genes against sap-sucking insects have been cloned, genes belonging to the NBS-LRR family might confer resistance to sap-sucking insects, in addition to conferring disease resistance.
The R genes of the NBS-LRR family, such as BPH26, are considered to be receptors related to signal perception and transduction. BPH26 plants showed resistance by inhibiting sucking from the phloem, as revealed by the analysis of AC-EPG. Many studies also indicated that the inhibition of sucking occurred in the phloem of the resistant O. sativa L. variety34,35,36. In the case of a non-host barnyard grass, probing of BPH was interrupted before the stylets reached the sieve elements of the phloem because the barnyard grass constitutively contains trans-aconitic acid, which acts as an antifeedant of BPH, in the non-phloem tissues such as parenchyma37,38,39. BPH14 originated from the wild rice O. officinalis mediated sucking inhibition in the phloem25. O. sativa L. and O. officinalis seem to have the same mechanism of resistance, i.e., defense in the phloem, against BPH. The R genes of the NBS-LRR family against BPH might mediate sucking inhibition in the phloem sieve element.
The sieve element can be plugged to prevent the erosion of the phloem fluid after damage40, and sieve tube occlusion in the phloem was suggested to contribute to the inhibition against sucking insects41. In legumes, crystalline P-protein (forisome), which is contained in the sieve elements, is responsible for the plugging of sieve elements40,42. In the case of monocotyledons, sieve element plastids are suggested to promote occlusion in sieve-plate pores, similar to the P-protein43,44. BPH14 has been inferred to mediate the deposition of callose (β-1,3 glucan polymer) in phloem cells25,27. Further studies are required to confirm the defense mechanism mediated by BPH26 in the phloem of O. sativa L.
There are about 480 predicted NBS-LRR genes in the rice genome29, of which some have been identified as disease-resistance genes. Phylogenetic analysis of BPH26, BPH14, and some blast-resistance proteins revealed that BPH26 was closely related to many blast-resistance proteins rather than BPH14. The indica variety Engkatek that harbored the blast-resistance gene PYRICULARIA ORYZAE RESISTANCE B (Pib) did not show resistance to BPH (data not shown). When the amino acid sequences of BPH26, BPH14, and other blast-resistance proteins were compared, typical consensus sequences between the two BPH resistance proteins (BPH26 and BPH14) were not found in the main motifs of the CC-NBS domain and LRR domain (Supplementary Figs. S5 and S6 online). It is necessary to analyze in more detail not only the amino acid sequence but also their three-dimensional structures that separate blast resistance and BPH resistance. BPH26 showed relatively strong expression in the vascular bundle of young leaves inside the leaf sheath, as revealed by in situ hybridization analysis. This suggests that BPH26 is especially required in the vascular bundles where BPH often attacks to feed. BPH14 has been suggested to be remarkably expressed in the vascular bundles via the β-glucuronidase (GUS) reporter system25. Expression of R gene in the vascular bundles might be important for BPH resistance.
Initially, BPH2 was reported as a recessive gene12. However, Murai et al. reported that BPH2 was a dominant gene by analyzing Norin-PL4 that harbored BPH2 from IR1154-243 in the genetic background of a susceptible japonica cultivar. BPH26 has also been reported as a dominant gene11. Cultivars containing the resistance allele of BPH2, such as IR42, were bred in Southeast Asia to control BPH. However, a virulent biotype of BPH appeared, and the resistance afforded by BPH2 was gradually rendered ineffective9. Every year, BPH populations that propagated in Southeast Asia such as northern Vietnam and the Philippines migrate via south China and Taiwan to Japan in early summer45,46 and cannot overwinter in Japan. Therefore, the strains of BPH that migrate to Japan are affected by the resistant cultivars cultivated in Southeast Asia. Myint et al.11 reported that BPH26-NIL showed resistance to the BPH strain collected in Japan in 1989 (strain-Chikugo-89). However, it did not show resistance to the strain collected in 2006 (strain-Japan-KG-06). “Japan-KG-06” is considered to be a BPH2-virulent biotype, because cultivars with the BPH2 gene have been used in Southeast Asia, and the virulent biotype has been dominated in these regions. Nevertheless, BPH26 showed resistance to BPH “Japan-KG-06” because of the coexistence of BPH25, although neither BPH26 nor BPH25 showed resistance to this strain11. Thus, the BPH2 gene, which has lost its resistance, can be reused along with other genes, such as BPH25.
Of more than 30 BPH-resistance loci reported, some genes are likely to be located in similar positions10. Controlling BPH by using BPH resistance genes might become difficult if all BPH resistance genes are rendered ineffective by respective virulent biotypes, because of the limited number of BPH resistance genes. Our findings might encourage the breeding of rice varieties containing insect-resistance genes.
Methods
Gene nomenclature
We follow the new gene nomenclature system for rice47. The gene full name of resistance to BPH is BROWN PLANTHOPPER RESISTANCE, and a gene symbol is an abbreviation of the gene full name such as BPH and written in italics and all capital letters.
Plant materials
The BPH-resistant indica cultivar ADR52 and the susceptible japonica cultivar Taichung 65 were used as parental lines for obtaining a BPH26 mapping population. The F1 plants from this cross were backcrossed for 5 rounds with Taichung 65 through marker-assisted selection (MAS), and BC6F2 population was obtained to develop NILs carrying BPH26 gene in a Taichung 65 genetic background48. Heterozygous individuals in the BPH26 region on chromosome 12 were selected from the BC6F2 population and self-pollinated to collect a large-scaled segregating population (BC6F3). The BC6F3 population was used for fine-scaled linkage mapping. Recombinant BC6F3 individuals were selected by MAS and self-pollinated to confirm the phenotype in the progeny. The BC6F6 seeds carrying BPH26 gene in a Taichung 65 genetic background (BPH26-NIL; line name, "TBPH101") were used for molecular analysis and bioassays.
The rice cultivar, ADR52, was originally obtained from the International Rice Research Institute, the Philippines and has been maintained in Kyushu University. The rice varieties, ASD7, IR1154–243, Rathu Heenati, Babawee, Norin-PL4, and Asominori (which are stocked at the Rice Genome Resource Center), were used for sequencing the exon regions of BPH26 and for bioassays.
BPH (N. lugens Stål) rearing
BPH adults (BPH2-avirulent biotype) were collected in Osaka Prefecture, Japan, in 1973. A stock colony of BPH was reared successively on rice seedlings at our institute at 25 ± 2°C and 70 ± 5% relative humidity under a 16:8 h light/dark photoperiod. The BPH was reared on the sensitive cultivar Koshihikari. BPH collected from Shimane Prefecture, Japan, in 1983, were used for selecting a BPH2-virulent biotype that can attack ASD7 carrying BPH2. The BPH2-virulent biotype was also reared on the sensitive cultivar Koshihikari, and its ability to feed on ASD7 was monitored at regular intervals.
Bioassays
Survival rate of BPH. A germinating seedling was placed in a test tube (diameter, 1 cm; height, 18 cm) with absorbent cotton moistened with liquid fertilizer, and the test tube was secured with a cotton plug. It was incubated for 13 days at 25°C under a 16-h light:8-h dark cycle. The two individuals of last instar larvae that had been fasted for 24 h on a moist filter paper were released in each test tube, and the numbers of live BPH were recorded 5 days after releasing. The tests were repeated 30 times (the total number of BPH was 60), and the number of living BPH was summed accordingly. Chi-square tests28 were used to compare the number of individuals surviving in the two different groups.
Electronic monitoring of feeding
The feeding activities of BPH on some rice lines were investigated using an AC-EPG as previously described39. An oscillator (IEM-04; Tsukuba Rica Seiki Inc.) was set at 0.5 V and 500 Hz, and all recordings were acquired over 12 h. The waveforms during probing behavior of each insect for 10 h from the start of the first probe were analyzed. The waveforms were interpreted on the basis of a previous report39. Each variety of plant was tested 10 times by using fresh insects and fresh plants. The data on the total time of phloem ingestion and the numbers of probe were analyzed using Tukey–Kramer test28.
Honeydew assays
For quantitative analysis of honeydew, modified Parafilm sachet technique was used49. A Parafilm sachet was attached to the leaf sheath of each seedling at the 5– to 6-leaf stage. A female BPH within 2 days after emergence was then enclosed in a Parafilm sachet. After 48 h of incubation at 25°C under a 16-h light:8-h dark cycle, each insect was removed, and the sachet was weighed before and after the removal of honeydew. Each line of plants was tested 10 times by using fresh insects and fresh plants. The data was analyzed using Tukey–Kramer test28.
DNA markers for mapping
The candidate region of BPH26 was elucidated using single sequence repeat (SSR) makers reported by the International Rice Genome Sequencing Project50. Furthermore, newly developed insertion-deletion (InDel) and single nucleotide polymorphism (SNP) markers in the candidate region were used to determine regions having a tight linkage to the resistance gene. These new markers were developed using the direct sequences of both Taichung 65 and ADR52 genomic DNA. SNP was detected by direct sequencing of the DNA amplified by PCR. The primers for markers are shown in Supplementary Table S2 online.
RNA extraction and analysis
Total RNA was extracted from the shoots and leaf blades by using a NucleoSpin RNA II (Macherey-Nagel). Genomic DNA was removed using TURBO DNA-freeTM Kit. RT-PCR assay was conducted using one-step RT-PCR kit (Quick Master Mix; TOYOBO) according to the manufacturer's instructions. PCR analysis consisted of 40 cycles by using primers shown in Supplementary Table S2 online. RNA extraction and RT-PCR were repeated three times for each sample. To isolate the cDNA of BPH26 from ADR52, ASD7, and Norin-PL4, RACE-PCR was performed using a SMARTer RACE cDNA Amplification Kit (Clontech). RACE-PCR products were cloned and sequenced.
Sequence comparisons of BPH26 among some rice varieties
Three exon regions of BPH26 from some rice varieties were amplified by PCR using the genomic DNA as template. PCR analyses were conducted using KOD-Plus neo (TOYOBO) and the primers shown in Supplementary Table S2 online. The PCR products were cloned using Target cloneTM Plus (TOYOBO) and sequenced to compare the alleles from some rice varieties. In addition to the genomic sequences, cDNA sequences from ADR52, ASD7, and Norin-PL4 were also used for sequence comparisons. Amino acid sequences were deduced from these nucleotide sequences.
Phylogenetic analysis
The phylogenetic analysis was conducted using the MEGA6 program51. A neighbor-joining (NJ) method was then applied to produce a phylogenetic tree52. The evolutionary distances were computed using the Poisson correction method53 and are in the units of the number of amino acid substitutions per site.
Complementation test
A BAC library of ADR52 genomic DNA was constructed as described previously54, and a BAC clone (14H01) covering the BPH26 gene was selected using DNA markers tightly linked to BPH26. A genomic sub-clone library of about 20 kb was constructed from 14H01. Restriction fragments of 14H01 after digestion with Sau3AI were gel purified and cloned into the BamHI site of binary plasmid vector pPZP2H-lac55 by ligation. Recombinants were cloned into the Escherichia coli DH5α strain, and the clones that contained a fragment with the candidate BPH26 gene were selected by colony PCR and sequenced. The selected recombinants were purified and introduced into Agrobacterium strain EHA10156 to infest calluses of susceptible cultivar Taichung 65. Plants regenerated from hygromycin-resistant calluses were grown in growth chambers. DNA from T1 plants was collected from leaf tips. T1 plants that were PCR validated to harbor the candidate BPH26 gene were selected using primers complementary to the insertion site of the pPZP2H-lac (Supplementary Table S2 online) and used in the biological assays. For analysis of the expression levels and copy numbers of Bph26 in the transgenic lines, real-time quantitative RT-PCR were performed using a Light Cycler 480 Instrument (Roche) in accordance with the manufacturer's instructions. Total RNA for the analysis of gene expression was extracted from the leaf sheath of 14-d-old seedlings by using NucleoSpin RNA II (Macherey-Nagel), and a PrimeScript RT reagent Kit (Takara) was used for reverse transcription. DNA for analysis of the copy number was extracted from the leaves of the same 14-d-old seedlings used for RNA extraction. The gene-specific primers used for Bph26 and an internal standard gene are shown in Supplementary Table S2 online.
In situ hybridization analysis
NIL plants carrying BPH26 in a Taichung 65 background at the 5- to 6-leaf stages were used for in situ hybridization analysis of BPH26 m-RNA. NIL plants that did not experience sucking from BPH were used for this analysis. In situ hybridization was conducted according to the methods described previously57. The BPH26 antisense probe was synthesized by in vitro transcription by using SP6 RNA polymerase from a 250-bp fragment that was a part of the BPH26-specific 3′UTR sequences amplified using PCR (Supplementary Table S2 online).
Supplementary Material
Supplementary imformation
Acknowledgments
We thank Yano, M. for helpful advice and technical support; Kimura, Y., Hashino, K., Shiba, J. & Watanabe, M. for laboratory assistant; Hasegawa, T., Matsumoto, Y., Asano, T. & Kobayashi, T. for helpful discussion; Tagiri, A. & Hara, N. for technical support on in situ hybridization analysis; Ando, T. for technical support on molecular analysis; Sugimoto, K. & Ono, K. for helpful advice on plant transformation technics; Uchiyama, T. & Nojiri, M. in Hokko Chemical Industry co. Ltd for giving BPH colony collected in 1972; Yoshimura, A. & Yamagata, Y. for supporting material development. This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation QTL-2001 and Genomics-based Technology for Agricultural Improvement LCT-0011).
Footnotes
The authors declare no competing financial interests.
Author Contributions Y.T. designed and performed the experiments and wrote the manuscript. H.Y. developed the materials, provided advice on the experiments, and improved the manuscript. M.H. supported bioassays and cloning of cDNA. H.Y. and M.Y. analyzed the structure of genes. A.T. provided advice on the experiments and phylogenetic analysis. J.W. designed BAC clone analysis. N.S. designed in situ hybridization analysis. All authors reviewed the manuscript.
References
- Schoonhoven L. M., van Loon J. J. A. & Dicke M. Insect-Plant Biology, 2nd Ed. (Oxford University Press, New York, 2005). [Google Scholar]
- Clement S. L. & Quisenberry S. S. Global Plant Genetic Resources for Insect-Resistant Crop. (CRC Press LLC, New York, 1999). [Google Scholar]
- Nasu S. Rice leafhoppers. In: The Major Insect Pests of the Rice Plant. 493–523 (Johns Hopkins Press, Baltimore, Maryland, 1964). [Google Scholar]
- Kisimoto R. Studies on polymorphism and its role playing in the population growth of the brown planthopper. Bull. Shikoku Agric. Exp. Stn. 13, 1–106 (1965). [Google Scholar]
- Dyck V. A. & Thomas B. The Brown planthopper problem. In: Brown Planthopper: Threat to Rice Production in Asia. 3–17 (International Rice Research institute, Manila, Philippines, 1979). [Google Scholar]
- Matsumura M. et al. Current status of insecticide resistance in rice planthoppers in Asia. In: Planthoppers: New Threats to the Sustainability of Intensive Rice Production Systems in Asia. 233–244 (International Rice Research Institute, Manila, Philippines, 2009). [Google Scholar]
- Pathak M. D. & Khush G. S. Studies of varietal resistance in rice to the brown planthopper at the International Rice Research Institute. In: Brown Planthopper: Threat to Rice Production in Asia. 285–301 (International Rice Research Institute, Manila, Philippines, 1979). [Google Scholar]
- Sogawa K. The rice brown planthopper: feeding physiology and host plant interactions. Ann. Rev. Entomol. 27, 49–73 (1982). [Google Scholar]
- Sogawa K. & Kilin D. Biotype shift in a brown planthopper (BPH) population on IR42. Int. Rice Res. Newslett. 12, 40 (1987). [Google Scholar]
- Fujita D., Kohli A. & Horgan F. G. Rice resistance to planthoppers and leafhoppers. Criti. Rev. Plant Sci. 32, 162–191 (2013). [Google Scholar]
- Myint K. K. M. et al. Mapping and pyramiding of two major genes for resistance to the brown planthopper (Nilaparvata lugens [Stål]) in the rice cultivar ADR52. Theor. Appl. Genet. 124, 495–504 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athwal D. S., Pathak M. D., Bacalangco E. H. & Pura C. D. Genetics of resistance to brown planthopper and green leafhoppers in Oryza sativa L. Crop Sci. 11, 747–750 (1971).
- Hirabayashi H. & Ogawa T. RFLP mapping of Bph1 (Brown planthopper resistance gene) in rice. Breeding Sci. 45, 369–371 (1995). [Google Scholar]
- Martinez C. R. & Khush G. S. Sources and inheritance of resistance to brown planthopper in some breeding lines of rice. Crop Sci. 14, 264–267 (1974). [Google Scholar]
- Murai H. et al. Construction of a high-resolution linkage map of a rice brown planthopper (Nilaparvata lugens Stål) resistance gene bph2. Theor. Appl. Genet. 103, 526–532 (2001). [Google Scholar]
- Nemoto H., Ikeda R. & Kaneda C. New genes for resistance to brown planthopper Nilaparvata lugens Stål in rice. Jpn. J. Breed. 39, 23–28 (1989). [Google Scholar]
- Murata K. et al. Bph9, a dominant brown planthopper resistance gene, is located on the long arm of rice chromosome 12. Rice Genet. Newslett. 17, 84–86 (2000). [Google Scholar]
- Su C. C., Zhai H. Q., Wang C. M., Sun L. H. & Wan J. M. SSR mapping of brown planthopper resistance gene Bph9 in Kaharamana, an Indica rice (Oryza sativa L.). Acta Genet. Sinica 33, 262–268 (2006). [DOI] [PubMed] [Google Scholar]
- Ishii T., Brar D. S., Multani D. S. & Khush G. S. Molecular tagging of genes for brown planthopper resistance and earliness introgressed from Oryza australiensis into cultivated rice, O. sativa L. Genome 37, 217–221 (1994). [DOI] [PubMed] [Google Scholar]
- Lang N. T. & Buu B. C. Genetic and physical maps of gene Bph10 controlling brown plant hopper resistance in rice (Oryza sativa L.). Omonrice. 11, 35–41 (2003). [Google Scholar]
- Jena K. K., Jeung J. U., Lee J. H., Choi H. C. & Brar D. S. High-resolution mapping of a new brown planthopper (BPH) resistance gene, Bph18 (t), and marker-assisted selection for BPH resistance in rice (Oryza sativa L.). Theor. Appl. Genet. 112, 288–297 (2006). [DOI] [PubMed] [Google Scholar]
- Rahman M. L. et al. High-resolution mapping of two rice brown planthopper resistance genes, Bph20 (t) and Bph21 (t), originating from Oryza minuta. Theor. Appl. Genet. 119, 1237–1246 (2009). [DOI] [PubMed] [Google Scholar]
- Rossi M. et al. The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. U. S. A. 95, 9750–9754 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P. et al. The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat. Biotechnol. 16, 1365–1369 (1998). [DOI] [PubMed] [Google Scholar]
- Du B. et al. Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc. Natl. Acad. Sci. U. S. A. 106, 22163–22168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., He G., Shu L., Li X. & Zhang Q. Identification and mapping of two brown planthopper resistance genes in rice. Theor. Appl. Genet. 102, 929–934 (2001). [Google Scholar]
- Hao P. et al. Herbivore-induced callose deposition on the sieve plates of rice: an important mechanism for host resistance. Plant Physiol. 146, 810–1820 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute Inc. JMP User's Guide, Version 5.1. (SAS Institute Inc., Cary, North Carolina, 2003). [Google Scholar]
- Zhou T. et al. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genomics. 271, 402–415 (2004). [DOI] [PubMed] [Google Scholar]
- Kawaguchi M. et al. Assignment of a brown planthopper (Nilaparvata lugens Stål) resistance gene bph4 to the rice chromosome 6. Breeding Sci. 51, 13–18 (2001). [Google Scholar]
- Sun L., Su C., Wang C., Zhai H. & Wan J. (2005) Mapping of a major resistance gene to the brown planthopper in the rice cultivar Rathu Heenati. Breeding Sci. 55, 391–396. [Google Scholar]
- Jairin J., Phengrat K., Teangdeerith S., Vanavichit A. & Toojinda T. Mapping of a broad-spectrum brown planthopper resistance gene, Bph3, on rice chromosome 6. Mol. Breeding. 19, 35–44 (2007). [Google Scholar]
- Khush G. S. & Virk P. S. IR Varieties and Their Impact. (International Rice Research Institute, Manila, Philippines, 2005). [Google Scholar]
- Velusamy R. & Heinrichs E. A. Electronic monitoring of feeding behavior of Nilaparvata lagens (Homoptera: Delphacidae) on resistant and susceptible rice cultivars. Environ. Entomol. 15, 678–682 (1986). [Google Scholar]
- Khan Z. R. & Saxena R. C. Probing behavior of three biotypes of Nilaparvata lugens (Homoptera: Delphacidae) on different resistant and susceptible rice varieties. J. Econ. Entomol. 81, 1338–1345 (1988). [Google Scholar]
- Kimmis F. M. Electrical penetration graphs from Nilaparvata lugens on resistant and susceptible rice varieties. Entomol. Exp. Appl. 50, 69–79 (1989). [Google Scholar]
- Kim M., Koh H.-S., Obata T., Fukami H. & Ishii S. Isolation and identification of trans-aconitic acid as the antifeedant in barnyard grass against the brown planthopper, Nilaparvata lugens (Stål) (Homoptera: Delphacidae). Appl. Ent. Zool. 11, 53–57 (1976). [Google Scholar]
- Katsuhara M., Sakano K., Sato M., Kawakita H. & Kawabe S. Distribution and production of trans-aconitic acid in barnyard grass (Echinochloa crus-galli var. oryzicola) as putative antifeedant against brown planthoppers. Plant Cell Physiol. 34, 251–254 (1993). [Google Scholar]
- Hattori M. Probing behavior of the brown planthopper, Nilaparvata lugens Stål (Homoptera: Delphacidae) on a non-host barnyard grass, and resistant and susceptible varieties of rice. Appl. Entomol. Zool. 36, 83–89 (2001). [Google Scholar]
- Knoblauch M. & Mullendore D. Sieve element occlusion. In: Phloem: Molecular Cell Biology, Systemic Communication, Biotic Interactions. 141–153 (Wiley-Blackwell, Iowa, 2013). [Google Scholar]
- Shinoda T. Callose reaction induced in melon leaves by feeding of melon aphid, Aphis gossypii GLOVER, as possible aphid-resistant factor. Jpn. J. Appl. Entomol. Zool. 37, 145–152 (1993). [Google Scholar]
- Knoblauch M., Peters W. S., Ehlers K. & van Bel A. J. Reversible calcium-regulated stopcocks in legume sieve tubes. Plant Cell. 13, 1221–1230 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M. A. & Melarangno J. E. Structural evidence for plastid inclusions as a possible ‘sealing' mechanism in the phloem of monocotyledons. J. Exp. Bot. 32, 311–320 (1981). [Google Scholar]
- Paivaa É. A. S. & Machado S. R. Can sieve-element plastids in Panicum maximum (Poaceae) leaves act in the blockage of injured sieve-tube elements? Flora Morphol. Distribut. Funct. Ecol. Plants. 203, 327–331 (2008). [Google Scholar]
- Otuka A. et al. A migration analysis of the rice planthopper Nilaparvata lugens from the Philippines to East Asia with three-dimensional computer simulations. Popul. Ecol. 47, 143–150 (2005). [Google Scholar]
- Otuka A., Matsumura M., Watanabe T. & Dinh T. V. A migration analysis for rice planthoppers, Sogatella furcifera (Horváth) and Nilaparvata lugens (Stål) (Homoptera: Delphacidae), emigrating from northern Vietnam from April to May. Appl. Entomol. Zool. 43, 527–534 (2008). [Google Scholar]
- McCouch S. R. & CGSNL. Gene nomenclature system for rice. Rice 1, 72–84 (2008). [Google Scholar]
- Yara A., Phi C. N., Matsumura M., Yoshimura A. & Yasui H. Development of near-isogenic lines for BPH25(t) and BPH26(t), which confer resistance to the brown planthopper, Nilaparvata lugens (Sål.) in indica rice ‘ADR52'. Breeding Sci. 60, 639–647 (2010). [Google Scholar]
- Pathak P. K., Saxena R. C. & Heinrichs E. A. Parafilm sachet for measuring honeydew excretion by Nilaparvata lugens on rice. J. Econ. Entomol. 75, 194–195 (1982). [Google Scholar]
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature 436, 793–800 (2004). [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N. & Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E. & Pauling L. Evolutionary divergence and convergence in proteins. In: Evolving Genes and Proteins. 97–166 (Academic Press, New York, 1965). [Google Scholar]
- Hayashi N. et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 64, 498–510 (2010). [DOI] [PubMed] [Google Scholar]
- Fuse T., Sasaki T. & Yano M. Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol. 18, 219–222 (2001). [Google Scholar]
- Toki S. Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol. Biol. Rep. 15, 16–21 (1997). [Google Scholar]
- Komatsuda T. et al. Six-rowed barley originated from a mutation in a homeodomain–leucine zipper I–class homeobox gene. Proc. Natl. Acad. Sci. U. S. A. 104, 1424–1429 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima Y. et al. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148, 479–494 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary imformation