Abstract
Metabolomics generates a profile of small molecules that are derived from cellular metabolism and can directly reflect the outcome of complex networks of biochemical reactions, thus providing insights into multiple aspects of cellular physiology. Technological advances have enabled rapid and increasingly expansive data acquisition with samples as small as single cells; however substantial difficulties remain. In this primer, we provide an overview of metabolomics, especially mass spectrometry based metabolomics that uses liquid chromatography for separation, and discuss its utilities and limitations. We identify several areas at the frontier of metabolomics technology development. Our goal is to give the reader a sense of what might be accomplished when conducting a metabolomics experiment, now and in the near future.
Keywords: Metabolomics, mass spectrometry, metabolic network, metabolic biology, quantitative biology
Metabolomics-overview
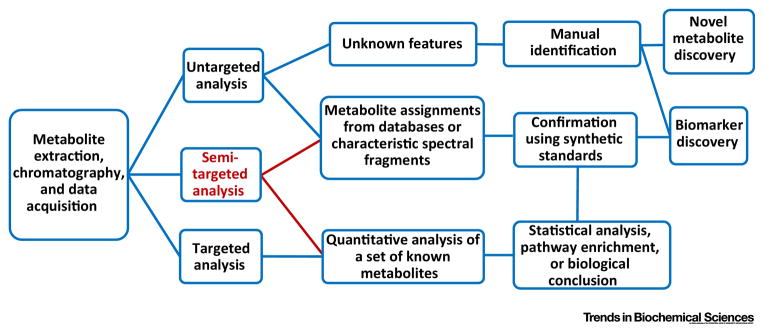
Metabolomics investigates the activity and status of cellular and organismal metabolism, on global- or network-scale, to delineate the end points of physiology and pathophysiology [1–5]. It involves the measurement of small molecule compounds, including endogenous and exogenous molecules, that are the products and substrates of chemical reactions within biological systems. A metabolomics experiment directly reflects the activity of the metabolic network that leads to the production of these metabolites and yields essential information about the underlying biological status of the system in question. Thus, metabolomics is not defined by any particular experiment but reflects the study of metabolism in a comprehensive way. It can involve an “untargeted” screen where thousands of unknown features are profiled and the relative differences in two conditions or across a population (semi-quantitation) are measured (Figure 1). Such a screening experiment could be useful in identifying a new metabolite that may be present in a genetic condition or a newly engineered metabolic pathway [6–12]. However, a semi-targeted metabolomics experiment is often more useful; here, a large number of molecules are unambiguously identified and quantified [13, 14]. This process allows the user to generate data that are otherwise obtained from hundreds of separate biochemical assays, to characterize the properties of a network or pathway[12, 15–20]. These experiments are considered semi-targeted because, while the list of metabolites is defined, the hypothesesis may not be. Targeted experiments often provide deeper insights by testing a specific hypothesis because the absolute concentrations of molecules are measured (absolute quantitation) [16, 21–23] or the rates or fluxes of the conversion of one molecule to another can be obtained [22, 24, 25]. Thus, a targeted metabolomics analysis requires substantial pre-existing knowledge and its success depends on strength of the hypothesis being tested.
Figure 1. Targeted, semi-targeted and untargeted analysis.
General scheme of different workflows that are available for metabolomics studies.
Both nuclear magnetic resonance (NMR) and mass spectrometry (MS) are effective tools that analyze the molecular composition of a sample (Table 1) [26]. NMR detects molecular features by measuring an intrinsic magnetic property of atomic nuclei (i.e. the “spin”) that encodes information about the chemical environment and thus its molecular structure. MS is more commonly used for semi-targeted or untargeted metabolomics because it is more sensitive, higher-throughput, and can measure more molecules in a complex biological sample. However, one advantage to NMR is that it is quantitative in that the number of molecules in the sample corresponds to the number of nuclei in that sample although this limitation in MS can be overcome by incorporating internal standards before extraction. Both liquid chromatography (LC) and gas chromatography (GC) are used for metabolite separatation (Table 1). Efforts have also been made to combine these different instruments (LC-NMR-MS) to advance strucuture elucidation[33]. In addition, the amount of material required for metabolomics is also getting smaller, which allows for single cell metabolite profiling or spatial resolution within cells or tissues. However, data collection is merely the first step of metabolomics, and metabolomics is ultimately an integration of instrumentation, chemistry, statistics, and computer science with a biological problem [4, 39, 40]. In this article, we provide a primer on metabolomics discuss its process, the type of data that are obtained, and its applications.
Table 1.
Metabolomics platforms
| Pros | Cons | Special Applicationsa | |
|---|---|---|---|
| LC-MS |
|
|
Comprehensive (broad coverage) metabolomics analysis |
| GC-MS |
|
|
Petrol chemical analysis |
| NMR |
|
|
|
unique applications (besides metabolomics) compared to other platforms.
From experiment to MS data – sample preparation and instrumentation
There is no limitation of the sample type that is suitable for a metabolomics study. However, the sample type and metabolites of interest determine the appropriate sample preparation procedures [41–43]. Furthermore, data interpretation is markedly different depending on the biological system that the metabolites originated. For example, to compare health and disease states or to study drug actions, measuring the level of a metabolite in the serum may be a reasonable proxy for a physiological function [44], since serum metabolome is the net effect of diet, environment and whole-body response to a disease or a drug; however, measuring that same metabolite level in a tissue would have a different interpretation, because it reflects more the cell autonomous effect. In sample preparation procedures, a general principle is to preserve the original state of the biological system as much as possible by minimizing the amount of enzyme activity and chemical reactivity that occurs during metabolite extraction, a process to isolate or purify metabolites from original biological matrix (cells, serum, tissue, etc).
For small molecule metabolites, protein precipitation or liquid-liquid extraction are the most commonly used methods. Polar organic solvents, such as methanol, acetonitrile or isopropanol, are used to extract mostly polar metabolites, whereas relatively non-polar solvents, such as hexane, chloroform, or methyl tertiary butyl ether, or the combination of polar and non-polar solvents, are used to extract lipids [45, 46]. Occasionally, acid is added to the extraction solvent to preserve the stability of certain compounds, such as acyl-Coenzyme A compounds [47]. However, although acidic solvents may stabilize one class of metabolites, they may also simultaneously cause degradation of other types of metabolites and possibly cause an overall reduction in the sensitivity of the experiment due to ion suppression [48, 49]. Despite the diverse chemical properties of metabolites, protocols have been developed that allow for a broad coverage of metabolites while maintaining, at a tolerable level, the inherent tradeoffs [34, 50–52]. For instance, these general protocols may not offer the best sensitivity for every single metabolite, and therefore, unstable or low abundant metabolites may require special care.
MS involves first ionizing the molecules (i.e., adding a positive or negative charge to) and then moving these molecules through electric fields where they are eventually analyzed. At each time point, the data are recorded as mass spectra, composed of mass to charge ratio for each intact ion and corresponding intensity. Each ion has a retention time and mass spectra, the values of which are dependent on instrument setup.
Two commonly used approaches for MS analysis are Multiple Reaction Monitoring (MRM) and high resolution MS (HRMS). MRM experiments are usually conducted on a triple quadrupole mass spectrometer [21, 32, 52]. The first quadrupole filters ions (parent ion) within a defined molecular weight (usually with a resolution of 1 atomic mass unit). The second quadrupole fragments the molecules that have been selected and the third quadrupole selects characteristic fragments. Therefore, before any data acquisition, the parent and fragement ions must be defined, and the optimized energy for the fragmentation and retention time for each metabolite is needed.
Another increasingly common method for MS is high resolution MS (HRMS). These approaches rely on the high mass resolution of the mass analyzer. One commonly used mass analyzer is an OrbitrapTM which records the oscillation of the ions, the frequency of which provides information of the molecular mass [53]. Another is a Time of Flight (TOF) instrument, which records the time it take for an ion to traverse through an electric field [54, 55]. These mass analyzers greatly simplify compound identification compared to lower resolution methods. When high resolution mass analyzers are coupled to collision cells that can fragment ions before they are sent to the mass analyzer, the fragment pattern of ions provide additional structural information. Then at the data processing stage, metabolite assignments are made.
From mass spectrometry data to metabolite profiling
There are numerous open source and commercial software available for raw mass spectrometry data analysis (Table 2). The output data includes features (peaks with specific retention time and mass to charge ratio m/z); peak area, which is usually the preferred parameter to represent relative abundance of each metabolite in different samples. These software packages typically involve chromatographic alignment, peak selection and compound identification by searching against metabolomics databases. This untargeted experiment is relatively thorough and unbiased, but it usually contains thousands of features, and unfortunately, these features do not directly reflect the metabolite identity (Figure 1). To characterize metabolites, it starts from matching of selected features with that of known metabolites. These databases are publically available at several online servers [56–60]. For metabolomics data generated from HRMS, m/z is often used as the only criteria for feature identification, and therefore, many features often return multiple metabolite identities, which is caused by isomers or ion source fragmentation [61]. A further complication is that chromatographic retention times (RT) are highly dependent on the LC or GC setup, are difficult to reproduce from external databases, and also vary over time even within a given lab. Lots of efforts have been made to advance untargeted analysis and feature identification, including new MS/MS workflow or network integration (Table 2) [62, 63]. A more detailed discussion of unknown metabolite identification is contained in a recent article [4]. Overall, untargeted analysis could be challenging and the result could be hard to interpret.
Table 2.
Metabolomics data processing software
| Software | Data Format | Statistics | Pathwaysb | Data Visualization | Isotope tracing | MS/MSc | Semi-targeted analysisd | Multi-omics integration | Source |
|---|---|---|---|---|---|---|---|---|---|
| XCMS | Alla | √ | √ | √ | √ | xcmsonline.scripps.edu | |||
| 13C XCMS | All | √ | √ | pattilab.wustl.edu/x13cms | |||||
| MAVEN | All | √ | √ | √ | √ | maven.princeton.edu | |||
| MsXelerator | All | √ | √ | √ | √ | msmetrix.com | |||
| MetaboAnalyst | All | √ | √ | √ | metaboanalyst.ca | ||||
| MetAlign | All | √ | √ | √ | metalign.nl | ||||
| MZmine | All | √ | √ | √ | mzmine.sourceforge.net | ||||
| SIEVE | .raw | √ | √ | √ | √ | Thermo | |||
| Compound discover | .raw | √ | √ | √ | √ | √ | √ | Thermo | |
| Mass Profiler | All | √ | √ | √ | √ | Agilent | |||
| MarkerLynx | .raw | √ | √ | Waters | |||||
| MarkerView | .d | √ | √ | AB Sciex | |||||
| MS-DIAL | All | √ | √ | √ | prime.psc.riken.jp/Meta bolomics_Software/MS-DIAL |
“All” means LC-MS data generated from all instruments could be either directly used as input data or it can be transferred to mzXML format before loading into the software.
“Pathway” means identified metabolites can be linked to corresponding pathways and used for pathway enrichment analysis.
“MS/MS” means the software can automatically extract MS/MS spectra and search against databases for metabolite identification.
“Semi-targeted analysis” involves both untargeted feature extraction, and a list of targeted metabolites (including m/z and retention time) with semi-quantitative values from raw data.
Therefore, a more targeted or semi-targeted data analysis is also performed simultaneously (Figure 1). An internal reference library including both m/z and RT, or even MS/MS spectra, is constructed in-house either by using pure chemical standards or by generating them “on the fly” by spiking in reference compounds into the metabolite extract. For a typical semi-targeted analysis, dozens to hundreds of metabolites can be assigned with high confidence from various biological samples and 3000–10000 other features are present in the spectra and remain unidentified [64, 65]. Nevertheless, this semi-targeted analysis provides a time- and cost-effective yet informative metabolic profile and allows researchers to either test multiple hypotheses at once or investigate systems biology-level questions.
Converting ion intensity to metabolite concentration is complex, which depends on variables such as percent of the compound recovered from the original material, column binding capacity, ionization efficiency, and transmission efficiency through the mass spectrometer. However, MS based metabolomics data are often semi-quantitative, which means that although the signal itself (metabolite peak area) does not reflect the absolute concentration, differences in peak area do scale linearly with metabolite concentration. A differential analysis provides the relevant biological information.
Normalization may be required in certain cases. For example, inconsistent sample preparation or extraction from different sources can result in varying ion suppression and cause nonlinear shifts of MS intensities of metabolites in different samples. In these situations isotopically labelled standards (internal standards) are added to each sample before the extraction for normalization [50]. However, applying internal standards is challenging due to the wide diversity of chemical properties and wide concentration ranges of metabolites in biological samples. When carrying out analyses in the absence of these standards, we therefore recommend making comparisons using material from a similar origin (compare serum A to serum B or tumor A to tumor B) and as similiar an amount of material as possible. Alternatively, pooled quality control samples can be used to reduce variation due to batch effects[66] . When applying these principles, most MRM and HRMS methods have yielded a linear range of quantitation for 3 to 4 orders of magnitude.
Analysis of a metabolomics experiment
Since metabolomics experiments typically contain information that could otherwise be obtained from hundreds of separate biochemical assays, usually some pre-existing biological knowledge helps with interpretation of a metabolomics experiment. Under this framework one can simply use the data to ask biologically relevant questions and make conclusions following the standard scientific method. Such questions could be: Does the energy status change under this condition? How about the redox status? Are the nucleotide levels maintained when this gene is overexpressed? Treating a metabolomics experiment under this mindset often allows for the one to reach conclusions about a biological mechanism in a highly expedited fashion as opposed to pursuing these hypotheses one by one with separate assays.
Nevertheless it is not humanely possible to process the entirety of the data from intuition alone. Computational tools are needed for further analysis. Software for feature extraction (Table 2) often include additional data analysis functions, such as principal components analysis, hierarchical clustering, and numerous statistical tests and data visualization plots to identify the largest changing features and specific signatures in the data. Pathway enrichment analyses, which are commonly used in gene expression analysis [67], can be also used with metabolomics data to identify affected metabolic pathways. However, metabolite annotation and pathway demarcations are not as well developed as in the genomics field thus often times these analyses produce results that are hard to interpret. Nevertheless, pathway enrichment analyses still give some useful insights into groups of metabolites of interest. For example, in a recent study pathway enrichment analysis of metabolomics data showed that the methionine cycle was altered before other parts of the network were affected [20]. Further, network mappings can contribute to systems level analysis upon integration with other ‘omics’ data sets [68, 69]. One commonly used network mapping tool is Cytoscape and the metabolism plugin for Cytoscape, called Metscape [70, 71]. There are other software that employ the Cytoscape platform, such as MetaMapR and GAM, which may generate metabolic networks based on enzymatic transformations, metabolite structural similarity, mass spectral similarity or empirical associations [68, 72]. A recently developed software PIUMet facilitates unknown metabolite identification by network integration of untargeted metabolomics [63]. These tools become more powerful upon integration with other omics data and allow the user to find regions of the metabolic network that correspond to a phenotype or are altered in a condition.
Isotope tracing, flux analysis, and computational modeling
The overall metabolite profile is very informative in many cases as discussed above, but for metabolites involved in multiple catabolic and anabolic pathways, metabolite levels reflect some complicated conglomeration of each individual pathway contribution. An appropriately designed isotopic tracing study is then used to identify the activity within each pathway of that metabolite and is the phenotypic readout of metabolism. The isotopic labeling pattern of downstream metabolites is used to represent the metabolic flux from different sources. Typical protocols involve incubating a stable isotope labeled nutrient (such as glucose, an amino acid, a lipid, or other molecules) at the same concentration as the original experiment (different concentrations will induce changes to metabolism from the original condition of interests) and waiting for the metabolic flux to reach steady state [74, 76]. Note that ‘steady state’ means that the isotopic labeling pattern of metabolites of interest is no longer time-dependent.
Much of the time, comparing the labeling pattern of a certain metabolite in different experimental conditions or performing a simple calculation of the labeled nutrient’s contribution to downstream metabolites is sufficient to interpret a metabolic flux qualitatively. However, model-based flux calculations often provide more accurate and sometimes comprehensive views of metabolism that can not be obtained from intuition alone[74, 76–79]. These techniques are collectively termed “Metabolic Flux Analysis” and involve taking a series of isotope measurements of different metabolites, overlaying them onto a metabolic network, and fitting a mathematical model of the fluxes onto a the network that best fits the data. This approach is very powerful but at the moment the implementation of the technology has been limited and most computations involving flux estimation and statistical analysis of the results are typically carried out on an ad hoc basis. Future directions will involve successfully implementing these approaches to serve a broader audience. More detailed discussions about flux calculations have been covered by previous reviews and we refer the reader to these[74, 76, 77, 79].
Recent biological insights obtained by metabolomics
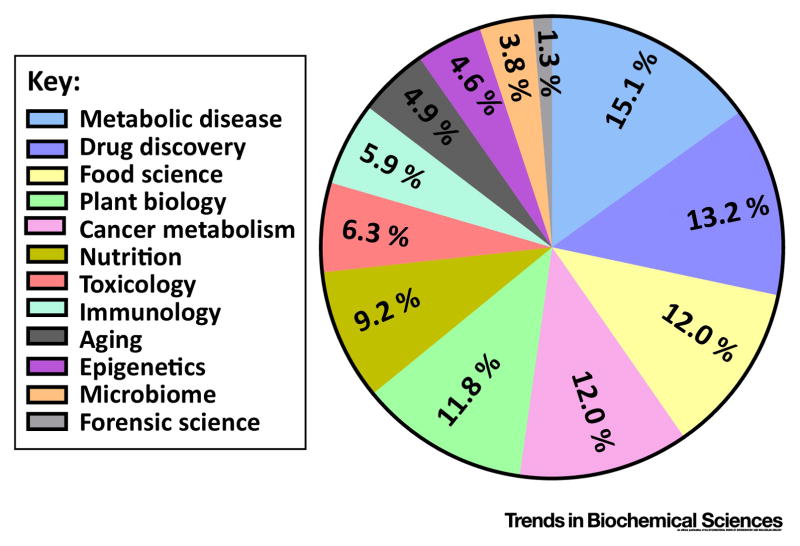
In recent years, metabolomics has been applied in multiple fields to make new discoveries and confirm hypotheses (Figure 2). A seminal advance in cancer biology used untargeted LC-HRMS to discovery that cancer cells with mutant IDH1 produce 2-hydroxyglutarate, a metabolite that was found in the spectra of the mutants but not the wild type cells after untargeted metabolomics using LC-HRMS. This metabolite was later shown to provide a link between metabolism and epigenetics[80, 81].
Figure 2. Distribution of recent publications on applications of metabolomics by area.
Number of publications were obtained by searching the corresponding keywords (e.g., drug discovery & metabolomics) in Google Scholar, with time range of 2015 – current (Nov 2016), and the fraction was calculated by dividing the number obtained from each area by the total number of publications in metabolomics.
Metabolomics has also been widely used in drug discovery and drug action[3, 82]. An extensive drug database, DrugBank (http://www.drugbank.ca/) now exists [83]. Over 8000 drug entries are recorded in this database, and the related information, such as known drug metabolism and known or proposed drug target, is also provided. Meanwhile, metabolomics is helping researchers to gain new knowledge of drug action. For example, through a metabolomics study, Ser et al. showed that 5-FU (5-fluorouracil), a commonly used chemotherapy drug, caused overproduction of the nucleotide deoxyuridine, which was also measured in 5-FU treated mouse serum. The overflow of this nucleotide could be potentially used as a biomarker for positive response to 5-FU treatment[84].
Semi-targeted metabolomics analyses led to a deeper understanding of the etiology of aging, cardiometabolic disease, and cancer, where metabolomics data not only tests existing hypotheses that can come from transcriptional or proteomics data but also provides insights when there is lack of hypothesis or when original hypothesis is rejected. For example, a study on Drosophila employed HRMS and demonstrated a critical role for specific clock genes in modulating the effects of nutrient manipulation on fat metabolism and aging[85]. In a separate study, MRM-based analyses have identified signatures of the onset of pancreatic cancer and diabetes [86]. Elevated plasma levels of branched-chain amino acids (BCAAs) were found to be associated increased risk of diabetes and other cardiometabolic diseases[87, 88]. These studies spawned further mechanistic analyses, which have pointed to the role of amino acid oxidation in the mitochondria as a key player in the etiology of these diseases. This same signature was also associated with an increased risk of future pancreatic cancer diagnosis[88]. These findings led to the mechanistic insight that the cause of this elevation was an increase in tissue breakdown, at least in the case of tumor development. Semi-targeted metabolomics analyses have also lent insights into human population genetics and the origins of metabolic traits. Genome-wide association studies (GWAS) studies for variation in metabolite levels were carried out in human blood from 2820 individuals[89]. Surprisingly, numerous genetic loci associated with blood metabolite concentrations were discovered and the locations of these loci, for the most part, were in promixity to a gene encoding a metabolic enzyme involved the production or consumption of the given metabolite. This analysis provided evidence that some of the metabolome variation observed across individuals may be directly encoded in the genome. Such studies may be valuble for understanding personalized nutrition, which is still in its infant stages[90].
Flux analyses have also led to important advances in cancer biology. One example was the discovery of the diversion of glycolytic flux into de novo serine metabolism as an important process in some cancers. Further efforts to trace the fate of serine using stable isotopes, HRMS and mathematical modeling revealed how the network downstream of serine is coordinated, leading to the identification of a potential cancer therapeutic[91]. These studies have also identified systems-level properties that show how gene expression levels can be used to sometimes predict metabolic flux, in which case then biomarkers for metabolic flux could be obtained from human specimens, from which mRNA is often more readily available[92]. Other flux analyses have identified previously unappreciated pathways that are important in cancer, leading to new drug targets[93–95]. As another example, acetate has been identified as an alternative fuel source for cancer metabolism[96–98]. Mashimo et al. first performed in vivo tracing in patients and showed the increased oxidation of acetate in the brains of patients with glioblastoma and brain metastases using 13C-NMR, and furthermore, Comerfold and Schug et al. demonstrated the utilization of acetate in tumors required the enzyme acetyl-CoA synthetase 2 which is now an important drug target in cancer. Thus is these cases metabolomics has led to novel findings that have advanced key areas of biomedical research.
Challenges and future directions in metabolomics
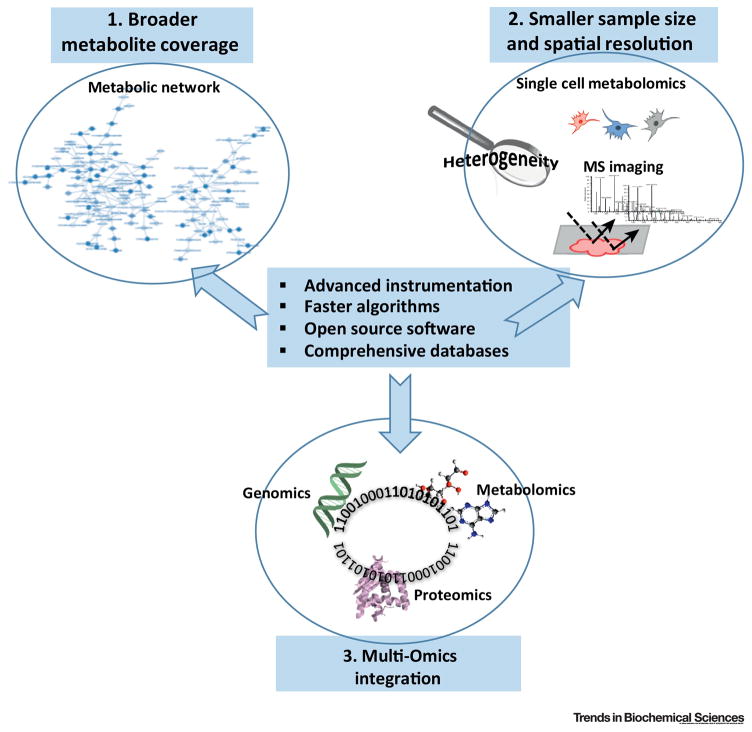
In summary, much progress in metabolomics has been made and obtaining a metabolite profile or measuring metabolic flux is now standard practice. Efforts are also being made to advance the field by covering more metabolites with less materials or efforts, achieving spatial resolution, and integration of multi-omics (Figure 3). Nevertheless, challenges remain in multiple areas.
Figure 3. Trends in metabolomics.
Trends include broader metabolite coverage from smaller sample sizes, achieving spatial resolution, and the integration of multi-omics data.
The greatest challenge for metabolomics is how to best obtain biological insight with the appropriate experimental design. Success often achieves a balance of biological intuition that is supplemented with computation. Choices of experimental models are an essential component in this process. For example, ex vivo systems that can successfully model in vivo metabolism are important[99–101]. Conclusions drawn from cell culture have substantially advanced biomedical knowledge but must be assessed with respect to the assumptions in the model such as the nutrient rich environements that typical cultures conditions consist of. Another major challenge is subcellular compartmentalization as metabolomics data reflects the sum of metabolites in different cellular organelles. Although it would be preferable to assay metabolites from specific organelles, it is very challenging, if currently impossible, to separate organelles while preserving the metabolic state of these structures. Recent studies have used clever isotope tracing strategies to probe cofactors that occur in a specific compartment such as the cytosol[102]. Metabolic flux analysis can be also used to estimate fluxes that involve metabolites that are shared across compartments [91, 95].
Metabolic heterogeneity is another issue in understanding metabolism and efforts to carry out metabolomics at the single cell level are underway[103–106]. This particularly exciting frontier in metabolomics research involves the analysis of single cells to obtain information that is masked in bulk studies. There are various approaches developed and successfully applied to plant, neuron, yeast, bacteria and animal cells in the last decade [105, 107]. Single cells could be isolated with a needle, or through microfluidic devices, followed by metabolite extraction and further analysis with HRMS or MS/MS[108–110]. Intact cells or tissue samples can also directly interact with an ionization source to generate ions, which are then analyzed by MS [111–113]. These technologies in princible can achieve spatial resolution of sub micrometer, and therefore, metabolomics imaging and analysis of subcellular organelles can be achieved. However, with this extremely fine spatial resolution, the data acquisition time and data size are also dramatically increased, which makes it difficult to perform single cell metabolomics in a high throughput fasion. In addition, efforts are also required to improve the robustness, number of metabolites covered and accuracy. Therefore, standard metabolomics techniques that average cells are still the mainstay.
Better approaches to standardize metabolomics data are also essential to advance the field. Thus, efforts are underway to create better normalization procedures and better protocols to rapidly obtain absolute metabolite concentration values [114, 115]. For flux analysis, the mathematical frameworks and algorithms are well beyond what is typically implemented and assessable to larger a community. Such an effort to further disseminate these capabilities will prove valuable. Nevertheless, current metabolomics technologies coupled with interesting questions already allow rapid inroads to be made.
Box 1. Outstanding Questions.
How does one best extract meaningful biological information from massive amounts of metabolomics data?
How can in vitro models more accurately reflect in vivo metabolism?
How can intracellular compartmentalization be resolved in metabolomics experiments?
What are the best ways forward in conducting single cell metabolomics experiments?
What are the best ways to make metabolic flux analysis more broadly used in the biomedical community?
Trends.
Extraction from a biological a sample followed by chromatographic separation and mass spectrometry allows for the simultaneous measurement of hundreds of metabolites.
The application of metabolomics to address numerous biological questions has been successfully demonstrated.
Advances in instrumentation and computational tools make metabolomics.
Emerging technologies are further advancing metabolomics.
Acknowledgments
Work was funded by (R00CA168997) and (R01CA193256) to JWL from National Cancer Institute at the National Institutes of Health. We thank the anonymous reviewers for helpful suggestions in improving this manuscript.
Glossary
- Absolute quantitation
a measurement of concentration of a metabolite.
- Chromatography
techniques used to separate metabolites based on chemical properties.
- Features
signal patterns recorded in metabolomics data (defined by the pair of retention time and mass to charge ratio in LC- or GC-MS based metabolomics data).
- In source fragmentation
fragmentation of molecules that occurs during ionization of the sample.
- Ion suppresion
the reduction of sensitivy due to the difference in the ability to ionize a molecule as a result of the surrounding composition of that molecule.
- Mass resolution
the ability to distinguish a molecule due to its mass to charge ratio from another molecular with a different mass to charge ratio.
- Pooled quality control samples
a mixture of samples to be analyzed with similar matrix to provide a representative “mean” sample, which is used to check the performance variation among individuals or over time.
- Retention time
the time taken for a metabolite to be eluted from a column.
- Semi-quantitation
the measurement of the relative level of a molecule in two conditions or across a population.
- Semi-targeted metabolomics
a system for quantitation of hundreds of known metabolites, and simultaneous detection of thousands of unknown features which are tentatively identified or not identied.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cajka T, Fiehn O. Toward Merging Untargeted and Targeted Methods in Mass Spectrometry-Based Metabolomics and Lipidomics. Anal Chem. 2016;88(1):524–45. doi: 10.1021/acs.analchem.5b04491. [DOI] [PubMed] [Google Scholar]
- 2.Johnson CH, et al. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17(7):451–9. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wishart DS. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 2016;15(7):473–84. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- 4.Zamboni N, et al. Defining the metabolome: size, flux, and regulation. Mol Cell. 2015;58(4):699–706. doi: 10.1016/j.molcel.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guma M, et al. Metabolomics in rheumatic diseases: desperately seeking biomarkers. Nat Rev Rheumatol. 2016;12(5):269–81. doi: 10.1038/nrrheum.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doroghazi JR, et al. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol. 2014;10(11):963–8. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denzel MS, et al. Hexosamine pathway metabolites enhance protein quality control and prolong life. Cell. 2014;156(6):1167–78. doi: 10.1016/j.cell.2014.01.061. [DOI] [PubMed] [Google Scholar]
- 8.Barkal LJ, et al. Microbial metabolomics in open microscale platforms. Nat Commun. 2016;7:10610. doi: 10.1038/ncomms10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen W, et al. Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat Commun. 2014;5:3438. doi: 10.1038/ncomms4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang C, et al. A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nat Med. 2016;22(4):421–6. doi: 10.1038/nm.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson CH, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015;21(6):891–7. doi: 10.1016/j.cmet.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin SY, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46(6):543–50. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breitling R, et al. Ab initio prediction of metabolic networks using Fourier transform mass spectrometry data. Metabolomics. 2006;2(3):155–164. doi: 10.1007/s11306-006-0029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gika HG, et al. Protocol for quality control in metabolic profiling of biological fluids by U(H)PLC-MS. J Chromatogr B Analyt Technol Biomed Life Sci. 2016;1008:15–25. doi: 10.1016/j.jchromb.2015.10.045. [DOI] [PubMed] [Google Scholar]
- 15.Hensley CT, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164(4):681–94. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Link H, et al. Real-time metabolome profiling of the metabolic switch between starvation and growth. Nat Methods. 2015;12(11):1091–7. doi: 10.1038/nmeth.3584. [DOI] [PubMed] [Google Scholar]
- 17.Yun J, et al. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science. 2015;350(6266):1391–6. doi: 10.1126/science.aaa5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Overmyer KA, et al. Maximal oxidative capacity during exercise is associated with skeletal muscle fuel selection and dynamic changes in mitochondrial protein acetylation. Cell Metab. 2015;21(3):468–78. doi: 10.1016/j.cmet.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartiala JA, et al. Genome-wide association study and targeted metabolomics identifies sex-specific association of CPS1 with coronary artery disease. Nat Commun. 2016;7:10558. doi: 10.1038/ncomms10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mentch SJ, et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab. 2015;22(5):861–73. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett BD, et al. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat Protoc. 2008;3(8):1299–311. doi: 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park JO, et al. Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat Chem Biol. 2016;12(7):482–9. doi: 10.1038/nchembio.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain M, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336(6084):1040–4. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shlomi T, et al. Network-based prediction of human tissue-specific metabolism. Nat Biotechnol. 2008;26(9):1003–10. doi: 10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- 25.Alves TC, et al. Integrated, Step-Wise, Mass-Isotopomeric Flux Analysis of the TCA Cycle. Cell Metab. 2015;22(5):936–47. doi: 10.1016/j.cmet.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunn WB, et al. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev. 2011;40(1):387–426. doi: 10.1039/b906712b. [DOI] [PubMed] [Google Scholar]
- 27.Lisec J, et al. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1(1):387–96. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y, et al. Evaluation of accurate mass and relative isotopic abundance measurements in the LTQ-orbitrap mass spectrometer for further metabolomics database building. Anal Chem. 2010;82(13):5490–501. doi: 10.1021/ac100271j. [DOI] [PubMed] [Google Scholar]
- 29.Brown SC, et al. Metabolomics applications of FT-ICR mass spectrometry. Mass Spectrom Rev. 2005;24(2):223–31. doi: 10.1002/mas.20011. [DOI] [PubMed] [Google Scholar]
- 30.Michalski A, et al. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics. 2011;10(9):M111011015. doi: 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson AC, et al. Development of a GC/Quadrupole-Orbitrap mass spectrometer, part I: design and characterization. Anal Chem. 2014;86(20):10036–43. doi: 10.1021/ac5014767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitteringham NR, et al. Multiple reaction monitoring for quantitative biomarker analysis in proteomics and metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(13):1229–39. doi: 10.1016/j.jchromb.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 33.Godejohann M. Hydrophilic interaction chromatography coupled to nuclear magnetic resonance spectroscopy and mass spectroscopy--a new approach for the separation and identification of extremely polar analytes in bodyfluids. J Chromatogr A. 2007;1156(1–2):87–93. doi: 10.1016/j.chroma.2006.10.053. [DOI] [PubMed] [Google Scholar]
- 34.Lu W, et al. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem. 2010;82(8):3212–21. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frey AJ, et al. LC-quadrupole/Orbitrap high-resolution mass spectrometry enables stable isotope-resolved simultaneous quantification and (13)C-isotopic labeling of acyl-coenzyme A thioesters. Anal Bioanal Chem. 2016;408(13):3651–8. doi: 10.1007/s00216-016-9448-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mackay GM, et al. Analysis of Cell Metabolism Using LC-MS and Isotope Tracers. Methods Enzymol. 2015;561:171–96. doi: 10.1016/bs.mie.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 37.Boudah S, et al. Annotation of the human serum metabolome by coupling three liquid chromatography methods to high-resolution mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;966:34–47. doi: 10.1016/j.jchromb.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 38.Raro M, et al. Untargeted metabolomics in doping control: detection of new markers of testosterone misuse by ultrahigh performance liquid chromatography coupled to high-resolution mass spectrometry. Anal Chem. 2015;87(16):8373–80. doi: 10.1021/acs.analchem.5b02254. [DOI] [PubMed] [Google Scholar]
- 39.Patti GJ, et al. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263–9. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sud M, et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016;44(D1):D463–70. doi: 10.1093/nar/gkv1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dietmair S, et al. Towards quantitative metabolomics of mammalian cells: development of a metabolite extraction protocol. Anal Biochem. 2010;404(2):155–64. doi: 10.1016/j.ab.2010.04.031. [DOI] [PubMed] [Google Scholar]
- 42.Ser Z, et al. Extraction parameters for metabolomics from cultured cells. Anal Biochem. 2015;475:22–8. doi: 10.1016/j.ab.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Want EJ, et al. Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry. Anal Chem. 2006;78(3):743–52. doi: 10.1021/ac051312t. [DOI] [PubMed] [Google Scholar]
- 44.Psychogios N, et al. The human serum metabolome. PLoS One. 2011;6(2):e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Folch J, et al. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226(1):497–509. [PubMed] [Google Scholar]
- 46.Reis A, et al. A comparison of five lipid extraction solvent systems for lipidomic studies of human LDL. J Lipid Res. 2013;54(7):1812–24. doi: 10.1194/jlr.M034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Basu SS, Blair IA. SILEC: a protocol for generating and using isotopically labeled coenzyme A mass spectrometry standards. Nat Protoc. 2012;7(1):1–12. doi: 10.1038/nprot.2011.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X, et al. High-Resolution Metabolomics with Acyl-CoA Profiling Reveals Widespread Remodeling in Response to Diet. Mol Cell Proteomics. 2015;14(6):1489–500. doi: 10.1074/mcp.M114.044859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Annesley TM. Ion suppression in mass spectrometry. Clin Chem. 2003;49(7):1041–4. doi: 10.1373/49.7.1041. [DOI] [PubMed] [Google Scholar]
- 50.Bain JR, et al. Metabolomics applied to diabetes research: moving from information to knowledge. Diabetes. 2009;58(11):2429–43. doi: 10.2337/db09-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X, et al. Development and quantitative evaluation of a high-resolution metabolomics technology. Anal Chem. 2014;86(4):2175–84. doi: 10.1021/ac403845u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan M, et al. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7(5):872–81. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olsen JV, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4(12):2010–21. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Plumb R, et al. Ultra-performance liquid chromatography coupled to quadrupole-orthogonal time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2004;18(19):2331–7. doi: 10.1002/rcm.1627. [DOI] [PubMed] [Google Scholar]
- 55.Zhu ZJ, et al. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat Protoc. 2013;8(3):451–60. doi: 10.1038/nprot.2013.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith CA, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27(6):747–51. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 57.Wishart DS, et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41(Database issue):D801–7. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanehisa M, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42(Database issue):D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horai H, et al. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45(7):703–14. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 60.Fahy E, et al. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009;50(Suppl):S9–14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu YF, et al. Avoiding misannotation of in-source fragmentation products as cellular metabolites in liquid chromatography-mass spectrometry-based metabolomics. Anal Chem. 2015;87(4):2273–81. doi: 10.1021/ac504118y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsugawa H, et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods. 2015;12(6):523–6. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pirhaji L, et al. Revealing disease-associated pathways by network integration of untargeted metabolomics. Nat Methods. 2016 doi: 10.1038/nmeth.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nygren H, et al. Liquid chromatography-mass spectrometry (LC-MS)-based lipidomics for studies of body fluids and tissues. Methods Mol Biol. 2011;708:247–57. doi: 10.1007/978-1-61737-985-7_15. [DOI] [PubMed] [Google Scholar]
- 65.Liu X, et al. A strategy for sensitive, large scale quantitative metabolomics. J Vis Exp. 2014;(87) doi: 10.3791/51358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wehrens R, et al. Improved batch correction in untargeted MS-based metabolomics. Metabolomics. 2016;12:88. doi: 10.1007/s11306-016-1015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jha AK, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42(3):419–30. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 69.Wanichthanarak K, et al. Genomic, Proteomic, and Metabolomic Data Integration Strategies. Biomark Insights. 2015;10(Suppl 4):1–6. doi: 10.4137/BMI.S29511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gao J, et al. Metscape: a Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks. Bioinformatics. 2010;26(7):971–3. doi: 10.1093/bioinformatics/btq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grapov D, et al. MetaMapR: pathway independent metabolomic network analysis incorporating unknowns. Bioinformatics. 2015;31(16):2757–60. doi: 10.1093/bioinformatics/btv194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang X, et al. X13CMS: global tracking of isotopic labels in untargeted metabolomics. Anal Chem. 2014;86(3):1632–9. doi: 10.1021/ac403384n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buescher JM, et al. A roadmap for interpreting (13)C metabolite labeling patterns from cells. Curr Opin Biotechnol. 2015;34:189–201. doi: 10.1016/j.copbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Higashi RM, et al. Stable isotope-labeled tracers for metabolic pathway elucidation by GC-MS and FT-MS. Methods Mol Biol. 2014;1198:147–67. doi: 10.1007/978-1-4939-1258-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Niedenfuhr S, et al. How to measure metabolic fluxes: a taxonomic guide for (13)C fluxomics. Curr Opin Biotechnol. 2015;34:82–90. doi: 10.1016/j.copbio.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 77.Shestov AA, et al. Computational approaches for understanding energy metabolism. Wiley Interdiscip Rev Syst Biol Med. 2013;5(6):733–50. doi: 10.1002/wsbm.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weitzel M, et al. 13CFLUX2--high-performance software suite for (13)C-metabolic flux analysis. Bioinformatics. 2013;29(1):143–5. doi: 10.1093/bioinformatics/bts646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Antoniewicz MR, et al. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006;8(4):324–37. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 80.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–8. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaddurah-Daouk R, et al. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–83. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 83.Wishart DS, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Database issue):D668–72. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ser Z, et al. Targeting One Carbon Metabolism with an Antimetabolite Disrupts Pyrimidine Homeostasis and Induces Nucleotide Overflow. Cell Rep. 2016;15(11):2367–76. doi: 10.1016/j.celrep.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Katewa SD, et al. Peripheral Circadian Clocks Mediate Dietary Restriction-Dependent Changes in Lifespan and Fat Metabolism in Drosophila. Cell Metab. 2016;23(1):143–54. doi: 10.1016/j.cmet.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mayers JR, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med. 2014;20(10):1193–8. doi: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Newgard CB, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–26. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang TJ, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448–53. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Suhre K, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477(7362):54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zeevi D, et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell. 2015;163(5):1079–94. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 91.Mehrmohamadi M, et al. Characterization of the usage of the serine metabolic network in human cancer. Cell Rep. 2014;9(4):1507–19. doi: 10.1016/j.celrep.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481(7381):380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481(7381):385–8. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fan J, et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510(7504):298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mashimo T, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159(7):1603–14. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schug ZT, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015;27(1):57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Comerford SA, et al. Acetate dependence of tumors. Cell. 2014;159(7):1591–602. doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boj SF, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160(1–2):324–38. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kenny HA, et al. Use of a novel 3D culture model to elucidate the role of mesothelial cells, fibroblasts and extra-cellular matrices on adhesion and invasion of ovarian cancer cells to the omentum. Int J Cancer. 2007;121(7):1463–72. doi: 10.1002/ijc.22874. [DOI] [PubMed] [Google Scholar]
- 101.Hirayama A, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69(11):4918–25. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 102.Lewis CA, et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell. 2014;55(2):253–63. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hu J, et al. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat Biotechnol. 2013;31(6):522–9. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cairns RA, et al. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 105.Zenobi R. Single-cell metabolomics: analytical and biological perspectives. Science. 2013;342(6163):1243259. doi: 10.1126/science.1243259. [DOI] [PubMed] [Google Scholar]
- 106.Rubakhin SS, et al. Profiling metabolites and peptides in single cells. Nat Methods. 2011;8(4 Suppl):S20–9. doi: 10.1038/nmeth.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rubakhin SS, et al. Progress toward single cell metabolomics. Curr Opin Biotechnol. 2013;24(1):95–104. doi: 10.1016/j.copbio.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lapainis T, et al. Capillary electrophoresis with electrospray ionization mass spectrometric detection for single-cell metabolomics. Anal Chem. 2009;81(14):5858–64. doi: 10.1021/ac900936g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fujii T, et al. Direct metabolomics for plant cells by live single-cell mass spectrometry. Nat Protoc. 2015;10(9):1445–56. doi: 10.1038/nprot.2015.084. [DOI] [PubMed] [Google Scholar]
- 110.Gong X, et al. Single cell analysis with probe ESI-mass spectrometry: detection of metabolites at cellular and subcellular levels. Anal Chem. 2014;86(8):3809–16. doi: 10.1021/ac500882e. [DOI] [PubMed] [Google Scholar]
- 111.Phelps MS, et al. Nanomanipulation-Coupled Matrix-Assisted Laser Desorption/Ionization-Direct Organelle Mass Spectrometry: A Technique for the Detailed Analysis of Single Organelles. J Am Soc Mass Spectrom. 2016;27(2):187–93. doi: 10.1007/s13361-015-1232-9. [DOI] [PubMed] [Google Scholar]
- 112.Shrestha B, Vertes A. In situ metabolic profiling of single cells by laser ablation electrospray ionization mass spectrometry. Anal Chem. 2009;81(20):8265–71. doi: 10.1021/ac901525g. [DOI] [PubMed] [Google Scholar]
- 113.Guenther S, et al. Spatially resolved metabolic phenotyping of breast cancer by desorption electrospray ionization mass spectrometry. Cancer Res. 2015;75(9):1828–37. doi: 10.1158/0008-5472.CAN-14-2258. [DOI] [PubMed] [Google Scholar]
- 114.Veselkov KA, et al. Optimized preprocessing of ultra-performance liquid chromatography/mass spectrometry urinary metabolic profiles for improved information recovery. Anal Chem. 2011;83(15):5864–72. doi: 10.1021/ac201065j. [DOI] [PubMed] [Google Scholar]
- 115.De Livera AM, et al. Normalizing and integrating metabolomics data. Anal Chem. 2012;84(24):10768–76. doi: 10.1021/ac302748b. [DOI] [PubMed] [Google Scholar]