Abstract
Background
Children with risk alleles at the 17q21 genetic locus who wheeze during rhinovirus illnesses have a greatly increased likelihood of developing childhood asthma. In mice, overexpression of the 17q21 gene ORMDL3 leads to airway remodeling and hyper-responsiveness. However, the mechanisms by which ORMDL3 predisposes to asthma are unclear. Previous studies have suggested that ORMDL3 induces endoplasmic reticulum (ER) stress and production of the type I interferon (IFN) regulated chemokine CXCL10.
Objective
The purpose of this study was to determine the relationship between ORMDL3 and rhinovirus-induced ER stress and type I IFN in human leukocytes.
Methods
ER stress was monitored by measuring HSPA5, CHOP and spliced XBP1 gene expression, and type I IFN by measuring IFNB1 (IFN-β) and CXCL10 expression in human cell lines and primary leukocytes following treatment with rhinovirus. Requirements for cell contact and specific cell type in ORMDL3 induction were examined by transwell assay and depletion experiments, respectively. Finally, the effects of 17q21 genotype on the expression of ORMDL3, IFNB1, and ER stress genes were assessed.
Results
THP-1 monocytes overexpressing ORMDL3 responded to rhinovirus with increased IFNB1 and HSPA5. Rhinovirus-induced ORMDL3 expression in primary leukocytes required cell-cell contact, and induction was abrogated by plasmacytoid dendritic cell depletion. The degree of rhinovirus induced ORMDL3, HSPA5, and IFNB1 expression varied by leukocyte type and 17q21 genotype, with the highest expression of these genes in the asthma-associated genotype.
Conclusions & Clinical Relevance
Multiple lines of evidence support an association between higher ORMDL3 and increased rhinovirus-induced HSPA5 and type I IFN gene expression. These associations with ORMDL3 are cell-type specific, with the most significant 17q21 genotype effects on ORMDL3 expression and HSPA5 induction evident in B cells. Together, these findings have implications for how the interaction of increased ORMDL3 and rhinovirus may predispose to asthma.
Introduction
Association with polymorphisms at the 17q21 locus remains one of the strongest and most widely replicated GWAS findings in childhood asthma [1–3]. The 17q21 asthma risk genotype is associated with increased expression of two co-regulated genes at that locus, GSDMB (gasdermin B) and ORMDL3 (orosomucoid like-3) in peripheral blood cells [1, 4, 5]. Furthermore, exposure of PBMC to rhinovirus (RV), a common trigger of wheezing in early childhood and exacerbations in established asthma, upregulates GSDMB and ORMDL3 expression [4]. In the high risk COAST (Childhood Origins of ASThma) birth cohort, children with the TT genotype at rs7216389 who wheezed with RV in the first three years of life had significantly greater prevalence of asthma later in childhood compared to children who did not wheeze, providing evidence for a gene-pathogen interaction [4].
The strong linkage disequilibrium between the nucleotide polymorphisms (SNPs) at 17q21, and the correlation of asthma-associated SNPs with altered expression of both ORMDL3 and GSDMB, have complicated the assignment of causality in asthma risk [4, 6]. Recently, human ORMDL3 over-expressing transgenic mice have presented compelling evidence for ORMDL3 as a pathogenic factor. These mice manifest spontaneous airway remodeling, increased fibrosis, mucus production, inflammation, and methacholine-induced hyper-responsiveness [7]. ORMDL3 is an endoplasmic reticulum (ER) resident protein that appears to have two major functions in cells: 1) inhibition of SPT (serine palmitoyl CoA transferase), reducing de novo sphingolipid synthesis and 2) inhibition of the sarco/endoplasmic reticulum Ca2+ ATPase (SERCA2b) pump that maintains the calcium gradient between ER and cytoplasm [8, 9]. Both of these functions could be related to abnormal airway physiology in asthma. By regulating ER calcium stores, ORMDL3 influences a stress response pathway known as the “Unfolded Protein Response” (UPR).
In immune cells such as macrophages, UPR induction dramatically increases production of type I IFN (with obvious relevance for anti-viral responses), and pro-inflammatory cytokines such as the IL-17-inducing cytokine IL-23 [10–13]. In murine studies, ORMDL3 has been linked to increased production of the type I IFN regulated chemokine CXCL10 [7, 14]. However, it is unclear how ORMDL3 alters the function of human leukocytes, particularly vis-a-vis asthma associated infections such as RV. We hypothesize that the increased ORMDL3 expression associated with the high risk 17q21 genotype may predispose to asthma by regulating RV induced ER stress and type I IFN production.
During a respiratory infection, RV primarily infects and replicates in airway epithelium. However, RV also stimulates responses such as cytokine and chemokine production, proliferation and antigen presentation in different peripheral blood cell types including monocytes, CD4 and CD8 T cells, B cells, dendritic cells and eosinophils [15–21]. In airway tissue from human asthmatic subjects, RV co-localizes with monocytes and macrophages [22]. Furthermore, interactions between RV stimulated monocytes and epithelium dramatically increases CXCL10 production [23]. Therefore, in this study we investigated the relationship between ORMDL3 expression and RV-induced ER stress and type I IFN production in multiple primary human leukocyte cell types, including cells from individuals with the high risk and low risk genotypes at the 17q21 asthma locus. Together our results revealed associations between elevated ORMDL3 expression and increased HSPA5 (BiP) induction and type I IFN responses in response to RV.
Materials and Methods
Human subjects
Participants were recruited for studies of eosinophil function. All participants provided written informed consent in a University of Wisconsin-Madison Institutional Review Board-approved blood donation protocol (approval #2013-1570). Individuals ranged in age from 19–55 and most had either self-reported allergies (94%), and/or mild asthma (56%). For 17q21 genotyping, whole blood was processed for DNA using the Qiagen Gentra Puregene Blood kit according to manufacturer protocols. SNP rs7216389 genotype within the 17q21 locus was assessed using a TaqMan SNP genotyping assay (Applied Biosystems, Foster City, CA, Life Technologies product number: C 29062108 10). Donors with the homozygous TT (high risk) or CC (low risk) genotype were selected for analysis to maximize differences in our samples. Although TT donors had higher eosinophil counts (manuscript submitted), the rest of their cell differentials did not differ from CC donors at the time of blood collection.
Cells, reagents, and stimulations
THP1 (American Type Culture Collection, Rockville, MD) and BJAB (gift of Shannon Kenney, University of Wisconsin-Madison) cell lines were maintained in DMEM/high glucose (Mediatech, Manassas, VA) with 10% FBS (Hyclone, Logan, UT) and antibiotic-antimycotic solution (Mediatech). Comparisons of PBMC, leukocyte fractions and eosinophil subsets, as well as comparisons of cell subsets isolated before or after RV stimulation involved cells from the same donors as cell yields permitted. Percoll enabled separation of eosinophils and PBMC from the same blood donation: For eosinophil isolation, whole blood was centrifuged over Percoll, and eosinophils purified from the granulocyte layer by negative selection using anti-CD16 beads (Miltenyi Biotech, Auburn, CA). Peripheral blood mononuclear cells (PBMC) were isolated from blood by centrifugation over Percoll or Lymphocyte Seperation Medium (Fisher, Waltham, MA). CD4, CD8, CD14, CD19 and plasmacytoid dentritic cells (pDC) were separated or depleted from PBMC by positive selection with magnetic microbeads (Miltenyi) according to the manufacturer’s instructions. The CD303/BDCA-2 kit was used for pDC depletion. “CD19−CD8−” cells represent column flow through from sequential CD19+ and CD8+ cell removal. Cell fraction purification studies were carried out on ice with ice-cold solutions to minimize effects on gene expression. For RV stimulations, all primary cells were maintained in DMEM supplemented with 10% FBS (not heat-inactivated; Invitrogen) and antibiotic-antimycotic solution. For plasmid transfection studies, THP1 cells were transiently transfected with plasmids of control-GFP and ORMDL3-GFP (GeneCopoeia, Rockville, MD) using AMAXA electroporation (Lonza, Walkersville, MD) as follows: 4×106 cells were combined with 5 μg DNA in 100 μL Nucleofector solution (Kit V) and then electroporated using program Y-006 one day prior to stimulation with virus. HRV16A was purified by sucrose gradient and added at the concentration of 2–10 pfu per cell for 24 hrs unless otherwise indicated. For the untreated cells, a vehicle control was added for the same duration. IFN-β protein was detected in supernatant using ProcartaPlex Human IFN-beta Simplex kit (eBioscience, San Diego, CA). The magnetic anti-IFN-β beads were detected on a Luminex 100 machine (Austin, TX). 12-well 0.4 μM pore size transwell dishes were obtained from Corning (Toledo, OH). These pores are large enough for diffusion of 30 nM rhinovirus. For “co-culture” conditions, CD19+ B cells were positively isolated from PBMC using Miltenyi magnetic beads according to the manufacturer’s directions. 1×106 CD19+ cells were placed in the insert with 12×106 CD19− PBMC (flow-through) in the well below. The same amount of total RV was used for transwell and whole PBMC stimulations.
Quantitative PCR
Cells were lysed with TRIzol (Invitrogen, Waltham, MA) and processed according to the manufacturer’s instructions. Briefly, RNA was extracted with chloroform and precipitated with isopropanol. RNA quality (260/280 ratio) and quantity was assessed by NanoDrop 2000 (Thermo Scientific, Wilmington, DE). Total RNA was treated with DNase I (Invitrogen) prior to reverse transcription using an Iscript kit (Bio-Rad, Hercules, CA). Gene expression in cDNA was quantitated by SYBR Green (Bio-Rad, Hercules, CA) fluorescence detected on a CFX96 machine (Bio-Rad). Reaction efficiency was assessed using a serially diluted standard curve and product by melting curve. Relative mRNA expression was normalized to the 18S rRNA housekeeping gene using the standard ΔCt/ΔCt method. Primers were designed using Beacon design software (Premier Biosoft, Palo Alto, CA): 18S rRNA, F:5′-GGACACGGACAGGATTGACAG3; R:ATCGCTCCACCAACTAAGAACG; IFNB1, F: TGGCTAATGTCTATCATCA; R: CTTCAGTTTCGGAGGTAA; HSPA5, F:GTGGAATGACCCGTCTGTG; R:TTGGTTGCTTGGCGTTGG; CHOP, F:CTAGTGCCAATGATGTGA; R:ATATACAAGCTGAGACCTT; CXCL10, F:GCT GCT ACT ACT CCT GTA; R:GGT CAG AAC ATC CAC TAA G; ORMDL3, F:ATCCCGTTTGTGAGTGTCC; R: CTTAGTGTAGAAGCTGGTGAGG; spliced XBP1, F: 5′-TGCTGAGTCCGCAGCAGGTG-3′, R:5′-GCTGGCAGGCTCTGGGGAAG-3′.
Western blotting
Cells were lysed with RIPA buffer containing protease and phosphatase inhibitor cocktails (Sigma-Aldrich, St. Louis, MO), and whole-cell lysates resolved by 12% SDS-PAGE. Samples were transferred to polyvinylidene difluoride membrane (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK) and immunoblotted with primary Abs anti-ORMDL3 Ab (Millipore, Bedford, MA), anti-β-Actin Ab (Santa Cruz, Dallas, TX), followed by a flourescence-conjugated secondary Ab (Licor, Lincoln, NE). Proteins were visualized with the Odyssey system (Licor).
Statistical analysis
Gene expression in different cell types and comparisons of genotype differences (CC vs TT at rs7216389) with or without RV treatment were assessed using two way repeated measures ANOVA with treatment, genotype and their interactions as fixed effects. Different PBMC cell types purified before or after simulating with either RV or vehicle control were compared using one way repeated measures ANOVA. All responses were log transformed to better approximate normal distributions. Associations between the expressions of each gene were assessed using Pearson’s correlation coefficient in a log transformed scale. P < 0.05 was the criterion for statistical significance. Analyses were performed in SAS version 9.4 (SAS Institute, Cary, NC). Error bars in bar graphs denote standard errors of the mean. In all figures, *p≤0.05 **p≤0.01 ***p≤0.001 unless otherwise indicated.
Results
ORMDL3 enables RV upregulation of HSPA5 and type I IFN gene expression in human monocytes
The UPR encompasses three major signaling cascades initiated by activation of ER resident proteins inositol requiring kinase 1 (IRE1), PKR-like endoplasmic reticulum kinase (PERK) and activating transcription factor (ATF6). IRE1 is both a signaling kinase and an endonuclease that splices the XBP1 mRNA to generate the active XBP1 transcription factor. PERK phosphorylates eIF2α, resulting in transient global translational attenuation, apart from specific target genes. Triggering of the UPR also activates the ATF6 transcription factor. Together, these three pathways regulate the transcription of UPR target genes aimed at restoring ER homeostasis [24, 25]. Interestingly, although one study has linked ORMDL3 to eIF2α phosphorylation (PERK pathway) and BiP induction (ATF6 target), another group has reported unique activation of the ATF6 pathway by ORMDL3 over-expression [7, 9, 14].
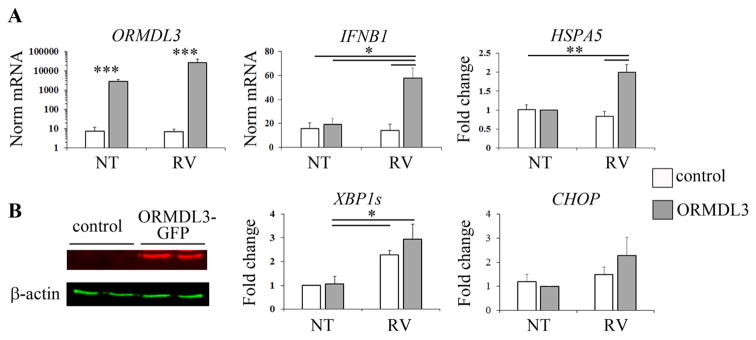
The effect of RV stimulation on the UPR was initially assessed in human cell lines: Bjab (non-EBV transformed B cells) and monocytic THP-1 cells were incubated with virus and then activation of the three UPR axes was monitored via the expression of pathway-specific targets: spliced XBP1 mRNA for the IRE1 pathway, HSPA5 (encoding BiP/GRP78) for ATF6, and CHOP for the PERK pathway [25, 26]. In Bjab B cells RV stimulation induced IFNB1 and CXCL10 mRNA, and also upregulated ORMDL3 and HSPA5 expression (Fig. 1). Spliced XBP-1 was not significantly up regulated, and CHOP expression was unchanged with 2pfu (Fig. 1) and inhibited by 10pfu (Supplemental Fig. S1).
Figure 1. RV induces type I IFN, HSPA5 and ORMDL3 gene expression in Bjab cells.
B cells were stimulated with RV (gray bars) or untreated (NT, open bars). RNA was reverse transcribed and examined for gene expression by quantitative PCR (qPCR). 18S normalized mRNA or fold changes (NT=1) are shown (note different y-axes). N=7 independent experiments for IFNB1, 6 for HSPA5 and ORMDL3, 5 for CHOP and 3 for CXCL10. *p≤0.02 comparing geometric mean expression.
In THP1 monocytes, RV induced expression of spliced XBP1, but not IFNB1 or other UPR target genes (Fig. 2, open bars). As native ORMDL protein was not readily detectable in THP1 monocytes, these cells were transfected with an ORMDL3 expression plasmid. Following ORMDL3 transfection, RV stimulation of THP1 cells increased expression of HSPA5 and IFNB1, but not spliced XBP1 or CHOP. Transfected ORMDL3 also resulted in higher IFN-β production (Supplemental Fig. S2). Together these results suggest that RV induces cell type specific IFN-β and UPR responses, and that ORMDL3 expression enhances RV-induced upregulation of HSPA5 and IFNB1 expression.
Figure 2. Synergistic IFNB1 and HSPA5 expression in ORMDL3 over-expressing monocytes stimulated with RV.
THP-1 monocytes were transfected with a control GFP plasmid (control, open bars) or plasmid containing ORMDL3-GFP (gray bars). A) Transfectants were untreated or stimulated with RV as indicated, and then RNA reverse transcribed and analyzed by qPCR for gene expression. Expression was normalized to the 18S housekeeping gene as indicated. Fold change is compared to unstimulated (NT) ORMDL3 transfected cells or control unstimulated cells. N=6 independent experiments for IFNB1 and CHOP, 5 for ORMDL3, 4 for HSPA5 and 3 for XBP1. B) Whole cell lysates (duplicate samples) from control plasmid or ORMDL3-GFP transfectants were resolved by SDS PAGE and blots probed with antibodies specific for actin or ORMDL as indicated.
ORMDL3 expression and HSPA5 upregulation varies in different primary human leukocyte cell types
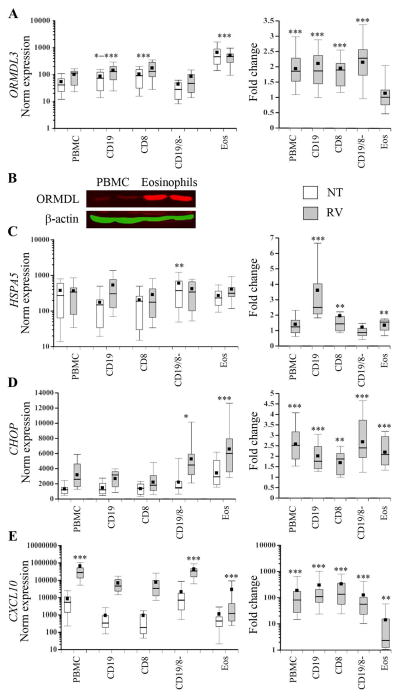
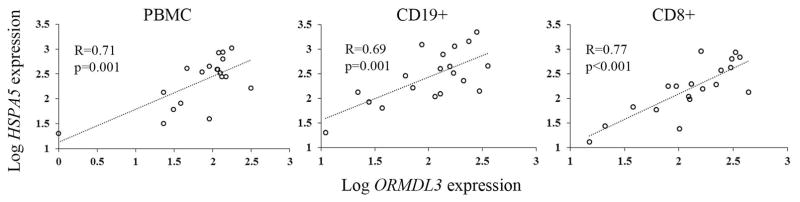
Caliskan et al. described a 2-fold increase in ORMDL3 expression following RV stimulation of PBMC, a mixed cell population [4]. Recent analysis of ORMDL3 and GSDMB in different unstimulated PBMC cell types revealed elevated gene expression in CD8+ and possibly CD19+ cells, respectively [6]. One study in a mouse model described high ORMDL3 expression in eosinophils [27]. Therefore, to determine which primary human leukocyte cell types upregulate ORMDL3 expression after RV stimulation, we incubated eosinophils or PBMC with RV, and then isolated CD19 B cells, CD8 T cells, or CD8−CD19− (mostly monocytes, CD4 T cells, and fewer NK cells and rare dendritic cells) cell fractions. ORMDL3 expression was highest in eosinophils (Fig. 3) for both untreated and RV stimulated cells, with untreated mean levels 4.7–11 fold higher than other leukocyte populations (p<0.001). Eosinophil ORMDL3 mRNA was also >18-fold higher than that observed in monocytes (Fig. S3). These differences in mRNA levels correlated with elevated ORMDL protein detected by Western blot. Currently available “ORMDL3” antibodies also recognize ORMDL1 and ORMDL2 related to the high homology between these proteins. However, ORMDL1 and ORMDL2 mRNA were not readily detected in freshly isolated human eosinophils (data not shown). A previous report also described greater ormdl3 expression in murine eosinophils compared to ormdl1 and ormdl2, so the detected band in eosinophils most likely represents primarily ORMDL3 [14]. B cell and CD8 T cells also expressed more ORMDL3 mRNA than either whole PBMC (p=0.02 to <0.001) or the CD19−CD8− (p<0.001) populations. RV induced ORMDL3 expression in all leukocyte cell types (ranging from 1.7–2.0 fold increase (p≤0.001)) except eosinophils. Eosinophils failed to upregulate ORMDL3 expression in response to RV, despite a modest induction of the UPR indicator HSPA5, and comparable upregulation of CHOP (1.6–2.5 fold increase in all leukocytes, p≤0.002). Likewise, eosinophils responded with much less CXCL10 induction (4.1-fold, p=0.01) compared to other cell types (130–149-fold increase in CD19+ and CD8+ cells and 54–79 fold in PBMC and CD19−C8− cells respectively, p<0.001). In response to RV, HSPA5 upregulation was greatest in B-cells (mean 2.9-fold, p<0.001) and greater than in other cell types, particularly PBMC and CD19−CD8− populations (p<0.001), which did not significantly upregulate HSPA5. Thus, the cells that increased ORMDL3 expression in response to RV tended to also upregulate CXCL10 and HSPA5. Furthermore, in PBMC, CD8, and CD19 B cells, RV-induced HSPA5 gene expression significantly correlated with RV-stimulated ORMDL3 gene expression (Fig. 4). RV-induced CXCL10 expression also correlated with ORMDL3 expression in CD8 T cells and HSPA5 expression in CD8 T cells, PBMC and CD8−CD19− cells (Fig. S4).
Figure 3. ORMDL3, HSPA5, CHOP and CXCL10 gene expression in primary leukocytes.
A, C–E) Eosinophils or PBMC were unstimulated (NT, open bars) or stimulated with RV (gray bars). After PBMC stimulation, CD19+ (B cells), CD8+ (T cells) or CD19−CD8− (CD19/8−) fractions were isolated prior to RNA extraction. Gene expression was analyzed by qPCR and normalized to 18S (left graphs) and further normalized to NT for ratio of stimulation (Fold change, right graphs). Standard box plots denote 25th percentile, median and 75th percentiles with whiskers at 10th and 90th percentiles and black boxes indicating means. For ORMDL3, HSPA5 and CXCL10, data are from 19 donors (PBMC, CD19, CD8), 17 donors (CD19−CD8−) or 12 donors (eosinophils). For CHOP, data are from 11 donors. B) Western blots of whole cell lysates from PBMC or eosinophils (duplicate samples) probed with fluorescence-tagged antibodies specific for β −actin or ORMDL as indicated. Results are representative of 3 experiments showing elevated ORMDL protein in eosinophils. A) ORMDL3 expression: CD19 vs. PBMC *p≤0.02, CD19 vs. CD19−CD8− ***p<0.001. CD8s vs both PBMC and CD19−CD8− ***p<0.001. Eosinophils vs. all others, ***p<0.001. C) HSPA5 expression: untreated CD19−CD8 vs. other cell types, **p≤0.003. D) CHOP expression: *p<0.03 comparing CD19−CD8− vs. PBMC, CD19 and CD8. ***P<0.001 for eosinophils vs PBMC, CD19 and CD8. E) CXCL10 expression: ***p<0.001 for PBMC and CD19−CD8− vs. CD19, CD8 and Eos. Eosinophil expression lower than other cell types ***p<0.001. Fold change asterisks refer to comparisons of RV stimulated and untreated samples for those cell fractions.
Figure 4. Correlation of RV-induced ORMDL3 with BiP gene expression.
CD19+ and CD8+ cells were isolated from PBMC following stimulation with RV. Gene expression was determined by qPCR. Statistical evaluation of association was performed on log transformed data as shown. Data are from 19 donors.
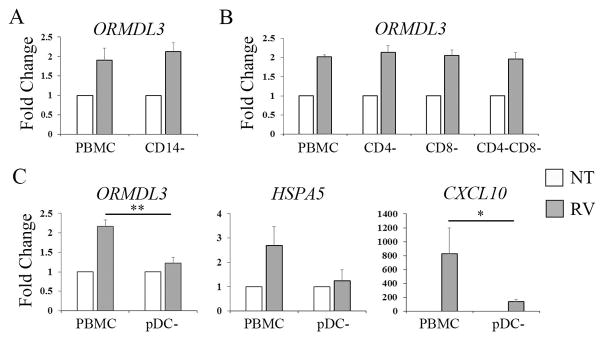
RV upregulation of ORMDL3 in primary human leukocytes requires cell-cell contact and the presence of plasmacytoid dendritic cells (pDC)
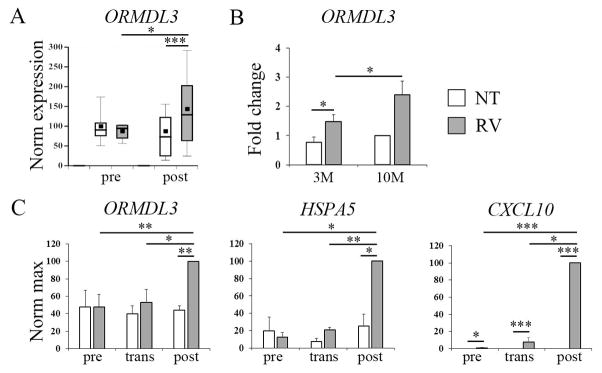
In contrast to transformed cell lines, or B cells isolated following RV stimulation of whole PBMC, isolated primary human B cells failed to upregulate ORMDL3 in response to RV (Fig. 5A). This suggested that either a soluble factor generated during whole PBMC stimulation or cell-cell contact between B cells and other cells was required. Previous studies had suggested STAT6 (induced by IL-4 or IL-13 signaling) was sufficient for ORMDL3 regulation [14, 28]. However, incubation of PBMC with exogenous IL-4 or IL-13 failed to induce ORMDL3 expression (Fig. S5). Increasing the cell number per well (and thereby cell-cell contact) during RV stimulation enhanced ORMDL3 upregulation (Fig. 5B). To more directly address the need for cell-cell contact, transwell assays were performed in which the B cells were present in one compartment, PBMC in another, and soluble factors (such as cytokines and RV) could freely diffuse (Fig. 5C). B cells co-cultured with PBMC by transwell did not significantly upregulate ORMDL3 or HSPA5 expression. Further, maximal induction of HSPA5 and CXCL10 expression required whole PBMC culture. These results indicate a requirement for cell-cell contact.
Figure 5. Upregulation of ORMDL3 by RV in B cells requires cell-cell contact.
A) B cells were isolated prior to stimulation (pre), or after PBMC stimulation (post) with media (NT, open bars) or RV (gray bars) for 24h. Gene expression was examined by qPCR with normalization to 18S. Data is from 6 pre donors and 19 post donors. *p=0.02, ***p<0.001. B) PBMC were incubated with 2 pfu/cell RV for 24h at a concentration of 3×106 cells per well (3M) or 10×106 cells per well (10M). Fold change ORMDL3 (vs. 10M non-treated) expression was determined by qPCR. N= 5 independent experiments (5 donors).*p≤0.04. C) B cells were isolated prior to stimulation (pre), incubated in a transwell with PBMC (trans), or isolated following stimulation with RV (post). RNA expression was normalized to post stimulation (=100). Data are from 4–5 independent experiments. ORMDL3 *p=0.01, **p≤0.005; HSPA5 *p≤0.04, **p=0.005; CXCL10 *p≤0.02, ***p<0.001.
To determine which cell types in PBMC were critical for ORMDL3 upregulation by RV, monocytes (Fig. 6A), CD4, CD8 T cells or both T cell subsets (Fig. 6B), or plasmacytoid dendritic cells (pDC) were depleted (Fig. 6C) prior to stimulation with RV. Only depletion of pDC abrogated the otherwise reliable 2-fold increase in RV-induced ORMDL3 expression. pDC were required but not sufficient for ORMDL3 upregulation, as isolated B cells incubated with pDC did not increase ORMDL3 (data not shown), suggesting the need for a more complex cell-cell interaction during the PBMC cultures.
Figure 6. RV-induced ORMDL3 in PBMC requires pDC.
Individual populations of cells including CD14 monocytes (A), T cells (B) and pDC (C) were depleted from PBMC prior to stimulation with RV. Gene expression was determined by qPCR with normalization to unstimulated (NT). A) N=4 experiments. B) N=3 experiments. C) N= 6 experiments for ORMDL3 and HSPA5 and 4 for CXCL10. *p=0.03, **p=0.003.
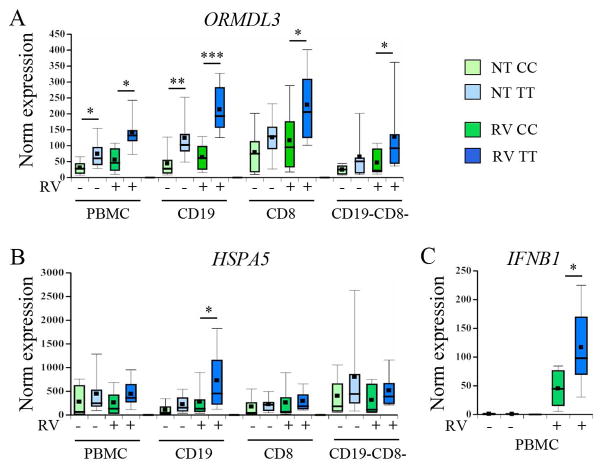
17q21 genotypic differences in RV-induced ORMDL3 and HSPA5 are most pronounced in CD19+ B cells
The ORMDL3 transfection studies, transwell assays and cell depletion studies supported an association between ORMDL3 upregulation, HSPA5 and type I IFN gene expression. We next determined whether 17q21 genotype affects ORMDL3 expression and upregulation of UPR genes and type I IFN in response to RV. Multiple asthma-associated SNPs in 17q21 display significant linkage disequilibrium [4, 29]. As the rs7216389 SNP has been reported to correlate with differential ORMDL3 expression in whole PBMC, rs7216389 was chosen as a surrogate genotypic marker [4]. The homozygous CC and TT genotypes were chosen to maximize observable differences in a small sample size. Participant characteristics for each genotype are described in Table 1. There was a non-significant trend towards lower age of onset in the TT genotype. ORMDL3 expression levels were higher in both untreated PBMC and B cells with the asthma risk TT genotype. Following RV-stimulation, the TT genotype was associated with increased ORMDL3 expression in all PBMC cell fractions (Fig. 7A). The differences in ORMDL3 expression between CC and TT subjects were most striking and significant (p≤0.004) in the CD19+ B cell subset, with or without RV stimulation. Interestingly, only RV-stimulated B cells showed increased HSPA5 expression in TT cells compared to CC cells (p=0.03, Fig 7B); although there were trends towards increased RV-stimulated HSPA5 in TT vs. CC PBMCs and CD19−CD8− cells (p=0.08 and 0.09 respectively). CHOP and CXCL10 expression induction did not differ between genotypes (Fig. S6) and PBMCs failed to increase XBP1 splicing in response to RV (not shown). However, RV-induced IFNB1 expression in PBMCs was significantly higher in cells with the asthma risk-associated TT genotype (Fig. 7C).
Table 1. Demographics and clinical characteristics of genotyped study participants.
CC and TT represent genotypes at the childhood asthma-associated SNP rs7216389. SD=standard deviation.
| Characteristics | CC (n=9) | TT (n=10) | p-value |
|---|---|---|---|
| Sex (% male) | 33 | 30 | 0.80 |
| Race (%White, Non-Hispanic) | 89 | 100 | 0.47 |
| Asthma (%) | 67 | 90 | 0.30 |
| Age (mean years (SD)) | 43.9 (10.3) | 43.9 (10.7) | 1.00 |
| Age asthma onset (mean years (SD)) | 25.3 (16.8) | 14.1 (10.6) | 0.15 |
Figure 7. 17q21 genotypic differences in ORMDL3, HSPA5 and IFNB1 gene expression.
PBMC from donors with CC (low risk) or TT (high risk) genotypes at the 17q21 locus were stimulated with media (pale boxes) or RV (dark boxes) prior to isolation of different cell subsets. Gene expression was determined by qPCR with normalization to 18S. CC genotypes are green and TT are in blue. For ORMDL3 and HSPA5 expression, PBMC, CD19 and CD8 cells were obtained from 9 CC and 10 TT donors. CD19−CD8− data are from 9 CC and 8 TT donors. For IFNB1, PBMC data were from 9 CC and 8 TT donors. A) ORMDL3 *p≤0.03, **p=0.004, ***p<0.001. B) HSPA5 and C) IFN-β *p=0.03.
Discussion
Genotype at the 17q21 locus is strongly linked to childhood asthma, especially in children who experience an RV wheezing episode during the first three years of life [4]. The goal of our study was to investigate cell type-specific relationships between RV infection and 17q21 genotype, ORMDL3 expression, and the UPR. Our results support the hypothesis that ORMDL3 regulates both ATF6 pathway activity and type I IFN responses to RV in specific blood leukocytes. In monocytic cells, exogenous ORMDL3 synergized with RV to induce HSPA5 and IFNB1 gene expression. Furthermore, RV coordinately induced ORMDL3 and HSPA5 expression in PBMC and lymphocytes. Cells with the rs7216389 TT genotype at the 17q21 locus expressed higher levels of ORMDL3 expression and more RV-induced HSPA5 and IFNB1 mRNA. Finally, cell to cell contact appeared to be required for both ORMDL3 and HSPA5 upregulation in PBMCs.
The requirement for cell-cell interaction for RV-induced upregulation of ORMDL3 expression in PBMC contrasts with the sufficiency of STAT6-dependent cytokine stimulation reported in murine and other in vitro systems [14]. In particular, the cell type depletion studies suggest that virus-responsive IFN-producing pDC are critical for upregulating ORMDL3 expression (Figure 6). Moreover, the results of the transwell assays suggest that direct interaction with pDC rather than soluble factors (e.g. IFNs) are necessary to upregulate ORMDL3 expression upon exposure to RV. The requisite role that pDC are playing in ORMDL3 regulation will require further study.
Cell-specific patterns of ORMDL3 expression were highly variable and may provide clues that link this gene to asthma pathogenesis. Our results examining ORMDL3 expression in different leukocyte populations are consistent with a previous study showing higher expression of this gene in CD8 T cells and B cells and lower expression in monocytes [6]. The strikingly high levels of ORMDL3 mRNA and ORMDL protein in human eosinophils had not been previously reported, and suggest the possibility that ORMDL3 might affect eosinophil inflammatory functions. In this study, we found that B cells had the greatest genotype-specific differences in ORMDL3 expression, in both untreated and RV-stimulated cells. B cells have been implicated in asthma exacerbations, asthma severity, and are key effector cells in allergic diseases through the production of allergen specific-IgE [30–32]. Although the 17q21 genotype has not been associated with allergic sensitization, it has recently been associated with allergic rhinitis, suggesting a link between this locus and allergic disease in the presence of asthma [4, 33].
Our studies also demonstrate that RV infection can induce the UPR in a cell-type specific manner. An increasing body of literature has described both induction and inhibition of the UPR by viral infections [13, 34]. Viruses may utilize the pro-survival anti-apoptotic arms of the UPR, as well as increased ER capacity to enhance viral replication in otherwise stressed cells. On the other hand, the transient translational inhibition that decreases protein production during the UPR could hamper viral replication. Results from this study suggest that depending upon cell type (e.g. monocytes vs B-cells or PBMC) RV stimulation may induce all three arms of the UPR, as evident from increases in HSPA5 (encoding BiP), CHOP, and spliced XBP1 mRNA. In primary human cells, HSPA5 induction by RV stimulation was particularly sensitive to cell type, being greatest in CD19 B cells. One possible explanation is that B cells are already “primed” to be ER-stress sensitive, given the central role of the UPR in both the generation and differentiation of B-cells into plasma cells. However, most studies have pointed to XBP1 and the IRE1 pathway in B cell development and function, rather than the ATF6 pathway [35, 36]. As another possibility, one study has suggested that RV replicates and produces infectious virions in B-cells, in contrast to other circulating blood cell types. Viral replication may thus increase ER stress in B cells [19].
To date, the mechanisms by which ORMDL3 influences ER stress are not entirely clear. ORMDL3 has been reported to induce ER calcium depletion and thus ER stress via blockade of the SERCA pump, as seen with the well-known pharmacologic UPR inducer and SERCA inhibitor thapsigargin [9, 37]. Although thapsigargin was first described as an inducer of BiP, one would predict an effect on all three UPR axes [38]. In the Bjab B cells, THP1 monocytes and genotyped primary cells used in this study, ORMDL3 induction by RV was primarily associated with HSPA5 expression, and not increases in RV-induced spliced XBP1 or CHOP. These results are consistent with previous studies in the murine system whereby over-expressed ORMDL3 primarily influenced ATF6 activation [7, 14]. The discrepancies with reports supporting a wider effect of ORMDL3 on UPR may reflect differences in experimental system (e.g. stimulus and cell type or species) [9].
It is not clear if the induction of the type I IFN gene IFNB1 and HSPA5 are separate or related outcomes of RV stimulation. Our previous studies have described synergistic augmentation of type I IFN genes by the UPR, both through XBP1 binding of an Ifnb1 enhancer as well as activation of IRF3, a transcription factor essential for IFN-β expression [10, 39]. Further, blockade of ATF6 activation prevented IRF3 phosphorylation by pharmacologic UPR inducers tunicamycin and 2-deoxyglucose [39]. Another study has also implicated ATF6 in the activation of the pro-inflammatory transcription factor NF-κB, which enhances IFN-β production [40]. Thus, increased ATF6 pathway activation could directly impact RV-induced IFNB1 expression.
Currently, it is unclear how increased IFN-β would contribute to asthma. Indeed, one trial suggested IFN-β may ameliorate virus-induced exacerbations, especially in individuals with more severe asthma [41]. On the other hand, both children and adults who wheeze in response to virus display increased interferon responses, and it is possible that exuberant interferon responses add to airway inflammation and obstruction [42, 43]. Subject phenotype, timing, and magnitude of IFN production in response to viral infections may be critical. Alternatively, ER stress augments the production of other pro-inflammatory cytokines, including TNF-α and IL-23 [11, 12]. These pro-inflammatory cytokines could promote the neutrophilic inflammation seen in acute exacerbations of asthma.
One limitation to this study was the use of cells from adult subjects with allergies or mild asthma. The enrichment for adults with allergies and asthma facilitated procurement of eosinophils, which are relatively rare cell types in peripheral blood from non-allergic individuals. However, these characteristics of our sample limit the generalizability of our results, which will require validation in healthy subjects and in children. Although our small sample sizes may have reduced the power to detect some differences, genotype-specific differences in ORMDL3 expression were highly significant, particularly in CD19+ B cells, attesting to the robust and significant effects of 17q21 genotype on ORMDL3 expression. Another caveat to interpretation comes from our use of positive selection to obtain multiple cell fractions. Positive selection could inhibit responses to RV. However, untreated populations isolated before (“pre”) or following PBMC culture (“post”) did not display any differences in ORMDL3 expression (Figure 5A). Also, 17q21 genotype comparisons (Figure 7) and the associations between differing levels of CXCL10, ORMDL3 and HSPA5 expression (Figure 4 and S4) were all from similarly treated cell populations, so positive selection should not have been an issue for these results.
In summary, our work describes a striking association between elevated ORMDL3 expression and increased HSPA5 and type I IFN production in response to RV. Together with other published studies, our results support an association between the risk genotype at the 17q21 asthma locus and elevated ORMDL3 expression [1, 4, 6, 29, 44]. Our results suggest that immune cells with higher ORMDL3 (the 17q21 risk genotype) respond to RV with augmented ER stress (at least ATF6 pathway activation) and IFNB1 expression. The link between ER stress and type I IFN may be direct, for instance through increased activation of the IFN-β regulatory transcription factor IRF3, or indirect. One implication of our results is that increased ER stress could promote increased IFN responses or other pro-inflammatory consequences that predispose to the development of asthma. Alterations in ER stress and type I IFN production may be of particular importance during early childhood, when exposure to viruses is high and anti-viral immunity has not yet matured. Future studies teasing apart the mechanistic relationships between ER stress, ORMDL3 expression, and type I IFN responses to RV are clearly warranted.
Supplementary Material
Acknowledgments
This work was funded by NIH-NHLBI P01 HL070831, P01HL088584, and NIH-NIAID R21 AI121808.
Footnotes
Conflict of Interest: The authors have no conflicts of interest to declare.
References
- 1.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448(7152):470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 2.Ono JG, Worgall TS, Worgall S. 17q21 locus and ORMDL3: an increased risk for childhood asthma. Pediatr Res. 2014;75(1–2):165–70. doi: 10.1038/pr.2013.186. [DOI] [PubMed] [Google Scholar]
- 3.Moffatt MF, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363(13):1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caliskan M, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368(15):1398–407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy A, et al. Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum Mol Genet. 2010;19(23):4745–57. doi: 10.1093/hmg/ddq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Acevedo N, et al. Risk of childhood asthma is associated with CpG-site polymorphisms, regional DNA methylation and mRNA levels at the GSDMB/ORMDL3 locus. Hum Mol Genet. 2015;24(3):875–90. doi: 10.1093/hmg/ddu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller M, et al. ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J Immunol. 2014;192(8):3475–87. doi: 10.4049/jimmunol.1303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breslow DK, et al. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463(7284):1048–53. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantero-Recasens G, et al. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010;19(1):111–21. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 10.Smith JA, et al. Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN-beta induction via X-box binding protein 1. Eur J Immunol. 2008;38(5):1194–203. doi: 10.1002/eji.200737882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinon F, et al. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010 doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLay ML, et al. HLA-B27 misfolding and the unfolded protein response augment interleukin-23 production and are associated with Th17 activation in transgenic rats. Arthritis Rheum. 2009;60(9):2633–43. doi: 10.1002/art.24763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith JA. A new paradigm: innate immune sensing of viruses via the unfolded protein response. Front Microbiol. 2014;5:222. doi: 10.3389/fmicb.2014.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller M, et al. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc Natl Acad Sci U S A. 2012;109(41):16648–53. doi: 10.1073/pnas.1204151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gern JE, et al. Rhinovirus enters but does not replicate inside monocytes and airway macrophages. J Immunol. 1996;156(2):621–7. [PubMed] [Google Scholar]
- 16.Saba TG, et al. Rhinovirus-induced macrophage cytokine expression does not require endocytosis or replication. Am J Respir Cell Mol Biol. 2014;50(5):974–84. doi: 10.1165/rcmb.2013-0354OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilarraza R, et al. Rhinovirus has the unique ability to directly activate human T cells in vitro. J Allergy Clin Immunol. 2013;131(2):395–404. doi: 10.1016/j.jaci.2012.11.041. [DOI] [PubMed] [Google Scholar]
- 18.Gern JE, et al. Rhinovirus produces nonspecific activation of lymphocytes through a monocyte-dependent mechanism. J Immunology. 1996;157(4):1605–12. [PubMed] [Google Scholar]
- 19.Aab A, et al. Human Rhinoviruses Enter and Induce Proliferation of B Lymphocytes. Allergy. 2016 doi: 10.1111/all.12931. [DOI] [PubMed] [Google Scholar]
- 20.Pritchard AL, et al. Innate IFNs and plasmacytoid dendritic cells constrain Th2 cytokine responses to rhinovirus: a regulatory mechanism with relevance to asthma. J Immunol. 2012;188(12):5898–905. doi: 10.4049/jimmunol.1103507. [DOI] [PubMed] [Google Scholar]
- 21.Handzel ZT, et al. Eosinophils bind rhinovirus and activate virus-specific T cells. J Immunol. 1998;160(3):1279–84. [PubMed] [Google Scholar]
- 22.Bentley JK, et al. Rhinovirus colocalizes with CD68- and CD11b-positive macrophages following experimental infection in humans. J Allergy Clin Immunol. 2013;132(3):758–761 e3. doi: 10.1016/j.jaci.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korpi-Steiner NL, et al. Human monocytic cells direct the robust release of CXCL10 by bronchial epithelial cells during rhinovirus infection. Clin Exp Allergy. 2010;40(8):1203–13. doi: 10.1111/j.1365-2222.2010.03546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–89. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 25.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–6. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto K, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13(3):365–76. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 27.Ha SG, et al. ORMDL3 promotes eosinophil trafficking and activation via regulation of integrins and CD48. Nat Commun. 2013;4:2479. doi: 10.1038/ncomms3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu R, et al. Signal transducer and activator of transcription 6 directly regulates human ORMDL3 expression. FEBS J. 2013;280(9):2014–26. doi: 10.1111/febs.12225. [DOI] [PubMed] [Google Scholar]
- 29.Schedel M, et al. Polymorphisms related to ORMDL3 are associated with asthma susceptibility, alterations in transcriptional regulation of ORMDL3, and changes in TH2 cytokine levels. J Allergy Clin Immunol. 2015;136(4):893–903 e14. doi: 10.1016/j.jaci.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Korsgren M, et al. Allergic eosinophil-rich inflammation develops in lungs and airways of B cell-deficient mice. J Exp Med. 1997;185(5):885–92. doi: 10.1084/jem.185.5.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamelmann E, et al. Development of eosinophilic airway inflammation and airway hyperresponsiveness requires interleukin-5 but not immunoglobulin E or B lymphocytes. Am J Respir Cell Mol Biol. 1999;21(4):480–9. doi: 10.1165/ajrcmb.21.4.3659. [DOI] [PubMed] [Google Scholar]
- 32.Drake LY, et al. B cells play key roles in th2-type airway immune responses in mice exposed to natural airborne allergens. PLoS One. 2015;10(3):e0121660. doi: 10.1371/journal.pone.0121660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuertes E, et al. Associations between the 17q21 region and allergic rhinitis in 5 birth cohorts. J Allergy Clin Immunol. 2015;135(2):573–6. doi: 10.1016/j.jaci.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 34.Li S, Kong L, Yu X. The expanding roles of endoplasmic reticulum stress in virus replication and pathogenesis. Crit Rev Microbiol. 2015;41(2):150–64. doi: 10.3109/1040841X.2013.813899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reimold AM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412(6844):300–7. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 36.Hu CC, et al. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. EMBO J. 2009;28(11):1624–36. doi: 10.1038/emboj.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13(10):1211–33. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 38.Price BD, Mannheim-Rodman LA, Calderwood SK. Brefeldin A, thapsigargin, and AIF4- stimulate the accumulation of GRP78 mRNA in a cycloheximide dependent manner, whilst induction by hypoxia is independent of protein synthesis. J Cell Physiol. 1992;152(3):545–52. doi: 10.1002/jcp.1041520314. [DOI] [PubMed] [Google Scholar]
- 39.Liu YP, et al. Endoplasmic reticulum stress regulates the innate immunity critical transcription factor IRF3. J Immunol. 2012;189(9):4630–9. doi: 10.4049/jimmunol.1102737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao J, et al. ATF6 mediates a pro-inflammatory synergy between ER stress and TLR activation in the pathogenesis of liver ischemia-reperfusion injury. Am J Transplant. 2014;14(7):1552–61. doi: 10.1111/ajt.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Djukanovic R, et al. The effect of inhaled IFN-beta on worsening of asthma symptoms caused by viral infections. A randomized trial. Am J Respir Crit Care Med. 2014;190(2):145–54. doi: 10.1164/rccm.201312-2235OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller EK, et al. A mechanistic role for type III IFN-lambda1 in asthma exacerbations mediated by human rhinoviruses. Am J Respir Crit Care Med. 2012;185(5):508–16. doi: 10.1164/rccm.201108-1462OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwantes EA, et al. Interferon gene expression in sputum cells correlates with the Asthma Index Score during virus-induced exacerbations. Clin Exp Allergy. 2014;44(6):813–821. doi: 10.1111/cea.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lluis A, et al. Asthma-associated polymorphisms in 17q21 influence cord blood ORMDL3 and GSDMA gene expression and IL-17 secretion. J Allergy Clin Immun. 2011;127(6):1587–94 e6. doi: 10.1016/j.jaci.2011.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.