Abstract
Aberrant activation of telomerase occurs in 85–90% of all cancers and underpins the ability of cancer cells to bypass their proliferative limit, rendering them immortal. The activity of telomerase is tightly controlled at multiple levels, from transcriptional regulation of the telomerase components to holoenzyme biogenesis and recruitment to the telomere, and finally activation and processivity. However, studies using cancer cell lines and other model systems have begun to reveal features of telomeres and telomerase that are unique to cancer. This review summarizes our current knowledge on the mechanisms of telomerase recruitment and activation using insights from studies in mammals and budding and fission yeasts. Finally, we discuss the differences in telomere homeostasis between normal cells and cancer cells, which may provide a foundation for telomere/telomerase targeted cancer treatments.
Keywords: shelterin, telomere length homeostasis, replicative senescence, Hayflick limit, t-stumps, S. pombe
1. Introduction: chromosome maintenance and cell proliferation
1.1. Telomere homeostasis in normal and cancer cells
All dividing eukaryotic cells require telomeres to maintain the ends of the chromosomes and sustain chromosome stability. To protect the genetic information contained within the chromosomes, telomeres sacrifice their non-coding DNA sequences in the erosion that occurs during DNA replication in each cell cycle [1,2]. Most somatic cells that have undergone sufficient cell divisions to cause critical telomere shortening enter into replicative senescence. However, some cells, including lymphocytes, germ cells, stem cells and unicellular eukaryotes like yeast, express the enzyme telomerase, which has the ability to replenish telomeres and allow further replicative potential [3–7].
Telomerase is a ribonucleoprotein complex, composed of a reverse transcriptase enzyme catalytic subunit and a long non-coding RNA that contains the template sequence for telomere synthesis [6]. Whereas expression of the telomerase components is tightly regulated in differentiated cells, the vast majority of human cancers express active telomerase, effectively rendering them immortal [8–10]. A direct correlation between telomere maintenance and indefinite cell division was demonstrated in vitro by ectopic expression of telomerase in somatic cell culture [11]. However, while cancer cells stably maintain telomeres, they tend to be short [12,13]. In particular, some of them are critically short, termed ‘t-stumps’ [14], resulting in immortal cells that sustain a high risk of chromosome instability. This is strikingly different from our understanding of telomerase action in normal cells, in which telomerase preferentially elongates shorter telomeres until they are no longer short [15–17]. Why telomerase acts differently in cancer cells remains a mystery. In this review, we summarize our current knowledge of fundamental telomerase action, and highlight the phenotypes uniquely observed in cancer cells.
1.2. Proliferation and protection: the problems faced by telomeres
Progressive telomere shortening occurs each time a cell divides owing to incomplete replication of linear chromosome ends by the conventional DNA polymerases. This shortening is termed the end replication problem [18]. Nucleases also trim the telomeres to shape the chromosome ends for protection, thereby causing loss of telomeric DNA after S phase [19]. DNA replication-associated telomere shortening limits the number of divisions a cell can undergo, known as the Hayflick limit, before triggering the cessation of growth [20]. Once telomeres become critically short, the DNA damage response machinery is activated, and cells enter replicative senescence or undergo programmed cell death [21,22]. Telomere shortening leading to programmed cell death is a major tumour suppressor mechanism, and as such, most cancer cells require telomerase to be active in order to survive.
In the absence of the senescence checkpoint per se, critically short telomeres become ‘uncapped’; they lose their end protection ability. One essential role of the telomeres is the differentiation of bona fide chromosomal ends from damaged DNA double-stranded breaks [23,24]. This function is indispensable for maintaining chromosome integrity, as illicit repair of chromosome ends could result in chromosome fusions. Such fusions would induce mitotic arrest and cell death [25] or cause breakage–fusion–bridge cycles in subsequent cell divisions, leading to translocations, aneuploidy and eventually genomic instability [26]. This is called the end protection problem.
2. The structure of telomeres and telomerase
2.1. The shelterin complex and telomere conformation
Telomeres are specialized DNA–protein complexes found at the ends of all linear chromosomes. Telomeric DNA is composed of arrays of short guanine-rich tandem repeats, and while most of the telomere is double-stranded (ds), they terminate in a single-stranded (ss) G-rich 3′ overhang called the G-tail [24,27–29]. These telomeric ds and ssDNA repeats are covered by a specialized protein complex, termed shelterin, to evade recognition of the chromosome ends by the DNA damage response machinery. Together with the shelterin complex, telomeres establish a heterochromatin structure that packages up the ends of the chromosomes and prevents them from being aberrantly recognized as DNA double-stranded breaks (DSBs) [30]. The G-tail is thought not to be exposed, but rather hidden within the dsDNA by forming a displacement loop (D-loop). Telomeres are further compacted into a lasso-like structure called a t-loop with the aid of the proteins in the shelterin complex [31,32]. In addition, the shelterin complex interacts with DNA damage response factors, preventing induction of their downstream pathways. Thus, failure of the chromosome ends to interact with shelterin can expose the DNA ends and elicit the DNA damage response [23,24,33].
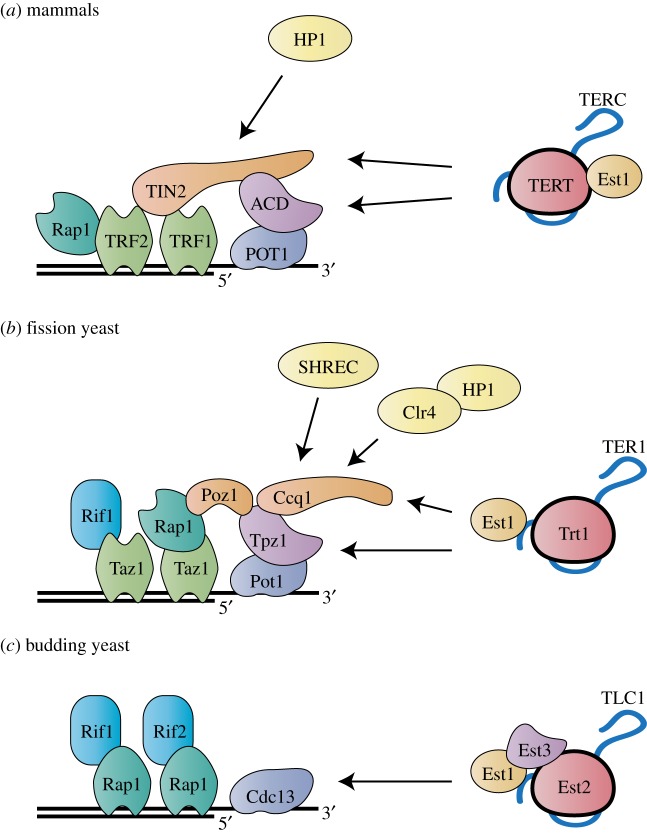
In mammals, shelterin is composed of six proteins: TRF1, TRF2, RAP1, TIN2, ACD (previously known as TPP1) and POT1 (figure 1a) [34]. TRF1 and TRF2 bind the telomeric dsDNA and recruit TIN2, which associates with ACD. POT1 forms a heterodimer with ACD and directly binds to the telomeric ssDNA at the D-loop and the G-tail. Thus, the shelterin complex formation bridges the telomeric ds and ssDNA and stabilizes the telomeric proteins. It is also thought to negatively regulate telomere lengthening [35–40]. TRF2 is required for the formation of t-loops [31,32] and for suppressing ATM activation and non-homologous end-joining (NHEJ) of chromosome ends [41]. These functions are promoted by TRF2-dependent topological changes [42]. RAP1, which binds to TRF2, is also thought to play a role in inhibiting NHEJ and homology directed repair but its actual function remains debatable [43,44]. TRF2 and RAP1 may be redundantly required for telomere protection even though RAP1 localization is dependent on TRF2. Interestingly, TRF2 and RAP1 also bind to internal telomere sequences and modulate transcription [45]. TRF1 is dispensable for end protection, but is required for lagging strand synthesis during DNA replication [46]. POT1 blocks the binding of replication protein A (RPA) to the telomeric ssDNA, thereby preventing recruitment of ATR [23]. Among the members of the shelterin complex, TIN2 and ACD are responsible for recruiting telomerase to the telomeres (discussed in a later section).
Figure 1.
Shelterin conservation at the telomere from yeast to mammals. Schematic diagrams depict the proteins of the shelterin complexes and telomerase complex in (a) mammalian cells and (b) fission and (c) budding yeasts. Orthologous proteins are shaded in the same colour. Known interactions with the telomerase complexes and chromatin modifying proteins are indicated. Other interaction proteins are omitted from these diagrams.
The shelterin complex is well conserved between fission yeast and mammals (figure 1b) [34,47,48]. The telomeric dsDNA binding protein Taz1 (orthologue of TRF1 and TRF2 in mammals) supports the replication of telomeric DNA and inhibits the NHEJ pathway [49,50]. Pot1 directly binds the telomeric ssDNA to inhibit degradation of chromosome ends [51]. Like mammalian shelterin, Taz1 forms the shelterin bridging structure with Rap1, Poz1 (TIN2 orthologue), Tpz1 (ACD orthologue) and Pot1 [48]. Thus, fission yeast shelterin also connects the ds and ss telomeric DNA, and maintenance of this connection has been shown to negatively regulate telomerase activity [48,52–55]. Tpz1 also interacts with Ccq1, which recruits telomerase [48,56]. We believe that mammalian TIN2 may be a bifunctional protein orthologue of fission yeast Poz1 and Ccq1 (discussed later). While each protein has a distinct role, the overall formation of the shelterin complex is crucial for telomere maintenance and telomerase regulation.
Although a shelterin-like protein complex has not been found, budding yeast has been the best-studied model system for telomere biology. In this organism, Rap1 directly binds the telomeric dsDNA and, with its associated proteins Rif1 and Rif2, negatively controls telomerase action [57,58]. Cdc13 solely binds the telomeric ssDNA to recruit and stabilize telomerase [59–61]. Although these proteins do not associate to bridge the ds and ss telomeric DNA (figure 1c), each protein functions to maintain telomere structure and protect the chromosome ends. Importantly, the mechanisms and principles of telomeric protein-mediated telomere homeostasis observed in budding yeast appear to be largely conserved to fission yeast and mammals (conservation of the structure and function of telomeres and telomerase has been reviewed [24,47,62,63]).
2.2. Telomerase structure and accessory proteins in fission yeast and mammals
The core components of telomerase comprise the telomerase RNA (TER1 in fission yeast and TR or TERC in mammals) and the catalytic reverse transcriptase protein (Trt1 in fission yeast and TERT in mammals) [64]. The reverse transcriptase subunit has been well conserved throughout evolution [65]. It can be divided into three major structural and functional domains: a telomerase essential amino-terminal domain (TEN), a telomerase RNA-binding domain (TRBD) and a reverse transcriptase domain (reviewed in [66]). In contrast, the telomerase RNA varies widely in length and sequence between different organisms [67–69], and accommodates distinct RNA recognition proteins. However, some conserved functional elements exist, including the template domain, a template boundary element to limit the extent of reverse transcription, and a pseudo-knot domain important for binding to the telomerase catalytic protein [70]. While these two core components alone are required for in vitro telomere synthesis [71], the telomerase accessory proteins contribute to the assembly, stabilization and trafficking of telomerase (reviewed in [72]).
In fission yeast, the Sm family of proteins associate with the TER1 RNA, contributing to telomerase maturation and stability. Subsequent replacement of Sm with the Lsm2–8 complex promotes Trt1–TER1 interaction [73]. Est1 directly binds TER1 and directs telomerase to telomeres through an interaction between its 14-3-3-like domain and the shelterin component Ccq1 [74,75].
In mammals, telomerase RNA maturation uses ribosomal RNA biogenesis (reviewed in [76]). The telomerase RNA, TERC, is part of a group of RNAs called H/ACA and binds to a tetrameric complex, composed of the dyskerin, NAF1, NHP2 and NOP10 proteins [77,78]. This H/ACA ribonucleoprotein complex stabilizes TERC and ensures the localization of telomerase to small sub-nuclear organelles called Cajal bodies where NAF1 is replaced by GAR1 [79–81]. Once inside the Cajal bodies, TERC associates with TERT to form the mature telomerase complex. After the assembly of a minimal telomerase complex containing TERT, TR and dyskerin, interaction with a protein called TCAB1 facilitates trafficking of telomerase to the telomeres [82].
Mammalian telomerase biogenesis additionally requires the molecular chaperones heat shock protein 90 (HSP90) and P23, which bind TERT for assembly with TERC. They are also thought to provide a binding site for proteins which link to the dynein–dynactin motor, thereby promoting the transport of hTERT to the nucleus along microtubules [83]. In mammals, the yeast Est1 orthologue, EST1A (SMG6), interacts with TERT and can bind to the telomeric ssDNA [84]. However, Est1A is not directly involved in telomerase recruitment but rather telomere protection and maintenance [85]. It also plays a role in nonsense mediated-mRNA decay and appears to affect the abundance of telomeric RNA transcripts called TERRA [86], which contribute to the regulation of telomere length homeostasis (reviewed in [87]).
3. Fundamental mechanisms of telomerase action in yeasts and mammals
3.1. Telomerase expression and cellular proliferation
The level of functional telomerase enzyme expressed in a wide range of different cell types has been characterized using the telomeric repeat amplification protocol assay. This method essentially allows a measure of the telomerase activity contained within a cell lysate in vitro [9]. Using this assay, it has been well documented that most differentiated somatic cells lack detectable telomerase activity [9,10], explaining the propensity for telomere shortening through successive cell divisions [11–13,88].
Telomerase is, however, highly expressed in adult testes and ovaries, allowing consistently longer telomeres to be inherited by the next generation [4,13]. Telomerase remains active during early embryonic development but expression declines after the blastocyst stage and can no longer be detected in neonatal somatic cells [4,89,90]. Nevertheless, most stem cell populations possess weak telomerase activity [3,5,9,10], which is not sufficient to immortalize cells but does extend the proliferative ability of these self-renewal tissues (reviewed in [91,92]). Notably, the Hayflick limit of somatic cells can be indefinitely evaded when telomere length is maintained by high ectopic expression of telomerase [11]. Therefore, the level of telomerase expression defines telomere length homeostasis and proliferative capacity.
3.2. Common mechanisms for telomerase recruitment
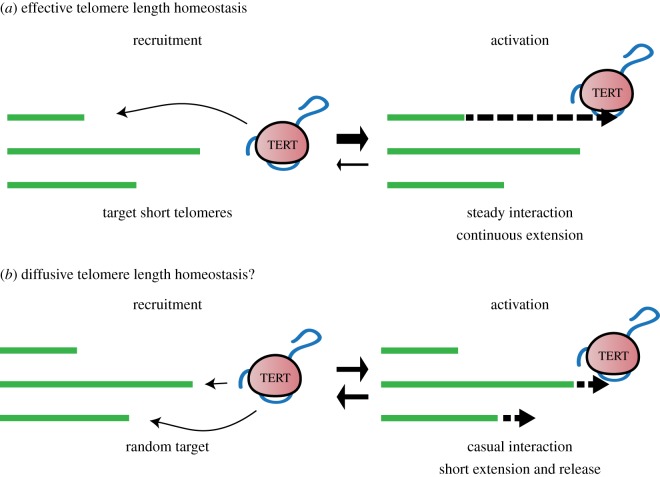
To maintain telomere length homeostasis, active telomerase needs to be efficiently recruited to every short telomere. Although telomeric proteins and telomerase components differ between budding and fission yeasts and mammals, on-going studies reveal that fundamental common features operate in telomerase action. In yeasts, telomerase is specifically recruited to shortened telomeres and the shorter telomeres are elongated the most during S phase [17]. As shorter telomeres can accommodate fewer telomeric DNA binding proteins, a quantitative negative regulation effect is thought to define the frequency of telomerase recruitment [93,94]. This system allows telomeric DNA to be retained at every chromosome end despite the presence of only a few molecules of active telomerase (figure 2a). This model has also been indirectly demonstrated in mammals using TERT heterozygous mouse cells [15].
Figure 2.
Models of telomere maintenance in normal and cancer cells. Schematic diagrams depicting two mode of telomere length homeostasis by distinct telomerase action. In normal cells (a), telomerase is recruited to the shortest telomere and it is extended until it is no longer short. Theoretically, such action would require a steady interaction between the telomere and telomerase, resulting in continuous extension. In this mode, distribution of telomere length would be the Gaussian like. In contrast, in cancer cells (b), telomerase recruitment appears to be random as it is independent of telomere length. Furthermore, telomeres are extended only a small amount, perhaps indicating an inability to maintain a steady interaction with telomerase, resulting in short extension and release. Such casual interaction permits repetitive recruitment of telomerase to new telomeres. In this mode, distribution of telomere length would be dispersal, resulting in retention of some short telomeres.
The mechanism of telomerase recruitment was originally best defined in budding yeasts. Recruitment of telomerase during S phase is mediated by association of Est1 and the single-stranded telomeric DNA binding protein Cdc13 [95–99]. This interaction is triggered via phosphorylation of Cdc13 by the DNA damage response kinases Tel1/Mec1 (ATM/ATR orthologues) and the cell cycle coordinator Cdk1 [61,99–103]. As mentioned, telomerase recruitment occurs preferentially at short telomeres owing to a quantitative negative regulation effect of telomere-bound Rap1 [93,94].
Several studies have shown that recruitment of telomerase to the telomere in fission yeast occurs via direct interaction of the telomerase accessory protein Est1 with Ccq1, the telomeric Tpz1 (ACD orthologue) binding protein. This occurs after phosphorylation of Ccq1 by the Rad3 and Tel1 kinases (ATR and ATM orthologues respectively) [68,69,75,104,105]. However, recent work indicates that the Ccq1–Est1 interaction is likely to be transient, as association of Est1 with TER1 and Ccq1 is mutually exclusive [75,106]. Further stable association of telomerase at the telomere is achieved via interactions between Tpz1, Ccq1 and Trt1 [106,107]. The telomeric dsDNA binding protein Taz1 (TRF1/2 orthologue) restricts Rad3/Tel1 activation and telomerase recruitment at the telomere, limiting telomerase recruitment to S phase of the cell cycle [108,109]. Similar to the quantitative negative regulation effect observed in budding yeast, Taz1 suppresses activation of Rad3/Tel1 in a telomere length-dependent manner. Therefore, Rad3 and telomerase are efficiently recruited to short telomeres.
Mammalian telomerase recruitment also occurs in S phase of the cell cycle. The rest of the time it appears to be concentrated in Cajal bodies or elsewhere in the nucleus [110,111]. The POT1–ACD complex is known to associate with telomerase [112–114]. TIN2, the shelterin protein that bridges TRF1/2 and ACD, is crucial for ACD localization at telomeres and is also involved in telomerase recruitment [30,114–116]. Similar to yeasts, phosphorylation of a telomeric protein, ACD, occurs during S phase by a kinase, Cdk1, which is thought to increase the stability of its interaction with TERT [117]. However, this phosphorylation-mediated interaction remains debatable [118]. Therefore, how the interaction of ACD and telomerase is achieved remains to be elucidated. It is possible that TIN2 may function like Ccq1 in fission yeast and initiate the interaction between ACD and TERT. In support of this hypothesis, the TIN2-R282H mutation, which is found in patients with dyskeratosis congenita, impairs telomerase recruitment, whereas ACD recruitment and shelterin formation are unaffected [116,119]. Another similarity to the yeast recruitment process is the contribution of ATM/ATR signalling to telomere elongation. Phosphorylation of the shelterin component TRF1 at an ATM/ATR target site (S367) has been shown to increase telomerase recruitment [120,121]. As with yeasts, mammalian telomeres are also thought to have a quantitative negative regulatory effect, with proteins such as TRF1 and TRF2 negatively regulating telomere extension by telomerase [122,123]. However, quantification of TRF1 and TRF2 molecules on the telomeres suggests the number of TRF proteins is limited and long telomeres may not possess more [124,125]. Indeed, an alternative telomeric DNA binding protein, TZAP, binds to long telomeres in a manner that is mutually exclusive to TRF protein binding [126]. TZAP functions to trim the telomeres by excising the t-loop, thereby making long telomeres short. As TZAP counteracts the possession of very long telomeres, we anticipate that it may also function in restricting telomerase activity.
The three model systems described above highlight common fundamental features of telomerase recruitment. The DNA damage checkpoints appear to monitor and flag the shorter telomeres that harbour fewer numbers of the telomere dsDNA binding proteins. In fission yeast and mammals, a direct interaction between the OB (oligosaccharide/oligonucleotide)-fold domain of Tpz1 and ACD, especially the so-called TEL patch on the surface [127], and the TEN domain of the telomerase catalytic subunit is necessary for both telomerase retention and processivity [106,107,114,128–130]. Notably, mammalian TIN2 and fission yeast Ccq1, which bind to the C-terminus of ACD and Tpz1 respectively, are also required for telomerase recruitment, as demonstrated by loss of function mutations [104,116]. TIN2 and Ccq1 also recruit and associate with the heterochromatin proteins to control the status of condensation or cohesion at telomeres [131–135]. Owing to the similarities in function between TIN2 and Ccq1 in terms of end protection, telomerase recruitment and interaction with chromatin modifying proteins, it is tempting to speculate that Ccq1 may be the functional equivalent of mammalian TIN2. The connection between telomere architecture and telomerase accessibility is slowly being uncovered [30]. Both fission yeast and mammalian shelterin formation controls the extendible, non-extendible and extending states [52]. Thus, although the structural similarities of fission yeast and mammalian shelterin proteins are not great, their functions and roles are highly conserved.
3.3. Current models of telomerase activation
Once telomerase is at the telomere, the single-stranded 3′ overhang at the distal end of the telomeric DNA forms the substrate for telomerase. This anneals with the template region on the telomerase RNA to form a DNA/RNA hybrid, and one repeat of telomeric DNA can be added to the end of the 3′ tail using the complementary RNA sequence as a template. Telomerase then repositions its RNA template on the 3′ end of the substrate and adds another telomeric repeat. The ability of a single telomerase complex to add multiple repeats in a single cell cycle without dissociation is termed repeat addition processivity (RAP) [136,137]. The exact mechanism by which telomerase can reposition itself on the template remains to be elucidated. Single molecule imaging of telomerase revealed that rearrangement of the telomerase RNA molecule is coupled with catalytic action, posing a possible model for resolution of the DNA/RNA hybrid and translocation of the RNA template after synthesis [138]. Thus, it is becoming increasingly clear that multiple factors affect the processivity or activity (how fast nucleotides are added) of telomerase after it has been recruited to the telomere.
In mammals, stable association of telomerase with the POT1–ACD complex occurs after recruitment to telomeres. Binding of POT1–ACD to the telomeric ssDNA has been shown to decrease the rate of RNA primer dissociation, aid template translocation and enhance telomerase processivity in vitro [112,139]. Indeed, mutations within the TEL patch in the OB fold of ACD, which impair association with TERT, have been shown to decrease processivity of telomerase by POT1–ACD in vitro, compared with wild-type ACD [127,140]. Interestingly, however, another recent study found that the POT1–ACD complex increases not only telomerase processivity but also activity, resulting in more rapid dissociation from the primer [141]. Thus, ACD-mediated retention of TERT improves telomerase processivity and activity.
The TEN domain of mammalian TERT has dual functions. In addition to binding the OB-fold domain of ACD for telomerase recruitment and RAP stimulation, the TEN domain supports the stable formation of the RNA–DNA duplex in the active site of the enzyme [142–144]. The reverse transcriptase and C-terminal domains of telomerase have also been proposed to interact with the telomeric DNA substrate to help promote RAP [145,146]. Thus, while the TEN domain of TERT associates with ACD for recruitment, it also promotes stable association of TERT with the telomeric ssDNA for repeat synthesis. How these two activities of the TEN domain are coordinated remains to be elucidated.
Recent studies have begun to demonstrate that recruitment of telomerase to the telomere in S/G2 phase does not equate to activation of the enzyme. In mammals, a residue within ACD (L104), found on the opposite face of the OB fold to the TEL patch, has been implicated in regulating telomerase activity. Mutation of this residue causes short telomeres despite the mutant protein binding a similar amount of telomerase to mutant ACD proteins in cells with longer telomeres [118,127]. Intriguingly, a mutant form of the ACD orthologue in fission yeast, Tpz1 (K75A), also cannot maintain telomere length despite being fully capable of recruiting telomerase to telomeres [106,107]. Nevertheless, a stable interaction between Tpz1 and Trt1 is required for the processive activity of telomerase, as the Tpz1 (K75A) mutation could be overcome by fusion of Tpz1 directly to Trt1 [106]. However, telomerase activity requires Ccq1 association with the telomerase-bound Tpz1. It has been proposed that recruitment of telomerase by Ccq1 might temporally and locally resolve shelterin formation to allow access to the telomeric 3′ end [52]. Thus, these studies demonstrate that association of telomerase with telomeres is not sufficient to regulate telomere length and a subsequent activation step must exist.
In budding yeast, another telomerase component, Est3, complexes with Est1 and Est2 at the telomere in S/G2 phase to activate telomerase [96,97,147,148] (figure 1c). Est3 is composed of the OB-fold domain and interacts with the TEN domain of Est2 with the aid of Est1 [149]. It has been proposed that the surface residues on Est3 required for telomerase activation might be comparable to residue L104 in ACD [150]. Thus, in all three model organisms, telomerase activation requires a stable association with the OB-fold ACD family of proteins as well as conformational changes in telomere structure to provide telomerase with access to the ssDNA end.
3.4. Termination of telomerase activity
To understand the processivity of telomerase, we also need to know how telomerase action is terminated. Several factors at the telomeres can inhibit, rather than stimulate, telomerase processivity. The CST (CTC1, STN1 and TEN1) complex, of which homologues for STN1 and TEN1 exist in fission and budding yeast, is thought to terminate telomere elongation by recruiting DNA polymerase alpha to the ssDNA overhang, thereby displacing telomerase [151–153]. Studies in budding yeast have shown that Stn1 replaces Est1 as the Cdc13 binding partner and blocks further telomerase recruitment after S phase [154,155]. However, because telomerase can repeatedly access the same telomere for further extension during S phase, we may need to separately consider termination of processivity and inhibition of telomerase recruitment.
The lack of coupling between telomere extension and telomere lagging strand synthesis may itself lead to inhibition or termination of telomerase processivity. The presence of a long G-tail can encourage the formation of certain structural conformations in the telomeric DNA, such as G-quadruplexes, which could potentially inhibit the access of telomerase to the telomere [32,52,156]. The POT1–ACD complex plays a role in preventing the formation of such secondary structures [112,139], as does RPA in fission yeast [157]. As such, the extent of telomere extension may well be monitored/controlled by the amount of ssDNA binding proteins recruited to the telomere. Further investigations will be needed to define these differences and to reveal how telomerase is temporally released from the 3′ telomeric overhang to terminate processivity.
4. Telomere biology in cancer
4.1. Unique features of telomerase action in cancer
Telomerase reactivation or upregulation is a critical feature in the vast majority of cancers. While the mechanisms controlling hTERT expression are not fully understood, they are thought to include hTERT promoter mutations, alterations in alternative splicing of hTERT pre-mRNA, hTERT gene amplification, epigenetic changes and disruption of the telomere position effect machinery (reviewed in [158]). Many studies carried out in human cells in actual fact use established cancer cell lines, because primary cells senesce in cell culture without the induction of telomerase expression. The characterization of telomeres in cancer cells has revealed that activated telomerase can largely maintain telomere length homeostasis as well as cell proliferation. The average length at which telomeres are maintained directly correlates with the expression level of telomerase [159–161]. Nevertheless, despite the fact that many cancer cells express highly active telomerase [9,10], their telomeres are shorter than in paired differentiated normal tissue [162,163]. Strikingly, a subset of telomeres are left very short (t-stumps) in cancer cells [14]. Thus, although activated telomerase maintains chromosome ends overall, the manner in which the telomeres are maintained appears to differ from that in normal tissue, such as germ cells.
The reason for the persistent presence of short telomeres in cancer cells might originate from some modified action of telomerase. Our understanding of telomerase from studies in yeasts and murine embryonic stem cells is that it preferentially elongates shorter telomeres until they are no longer short [15,17]. In contrast, in cancer cells, the majority of telomeres are elongated, but they are only extended a short length [164]. A recent single cell live imaging study using HeLa cells demonstrated that human telomerase forms short dynamic interactions with the majority of telomeres, probing each chromosome end multiple times during S phase [165]. Thus, we predict that cancer telomerase targets every telomere but only extends them a little (figure 2b). This model would explain how some telomeres can be left at a short length or lost but the overall mean telomere length reflects/correlates with the amount of active telomerase.
Curiously, telomere extension by telomerase is not coupled with synthesis of the complementary C-rich strand by polymerase alpha [164], leading to long G-tail extensions during S phase. It has been proposed that telomerase recruitment/retention (and hence processivity) is terminated by recruitment of the CST complex, which associates with the polymerase alpha complex [151,152,166]. However, the probing interactions described in HeLa cells are only rarely converted into static interactions long enough to allow telomere elongation [165]. Thus, it is possible that telomerase might associate with telomeres in an unstable manner in cancer cells, and therefore it exhibits low processivity and dissociates before C-strand synthesis.
The hallmark of irregular telomerase action in cancer cells might also/otherwise stem from the presence of alternatively spliced hTERT mRNA isoforms. Alternative splicing events are commonly observed in the majority of cancer cells, and can both control transcript abundance and contribute to proteome diversity [158]. hTERT mRNA is alternatively spliced in a wide range of species [167], and a number of variants are co-expressed at significant levels in tumour and stem cells [168–170]. However, the regulation and function of these splice variants is not well understood. Expression of a major splice variant lacking most of the RT domain has been correlated with low telomerase activity in cancers [171,172], and a recent study has shown that the translated protein product can bind the telomerase RNA and suppress telomerase activity [173], presumably by competing with the fully functional hTERT isoform for TERC binding. Therefore, such deletions or substitutions of other key residues and domains may well affect the association of telomerase with shelterin components or the telomeric ssDNA overhang.
The shelterin complex proteins are important not only for telomerase recruitment but also for control of the DNA damage response and cell cycle control machineries at telomeres. These machineries are impaired or altered in cancer cells. Several mutations in genes encoding the components of the shelterin complex have also been identified in cancers. These mutations might affect telomere status and telomerase accessibility or processivity. For example, in familial cases of chronic lymphocytic leukaemia (CLL) loss-of-function mutations in POT1, affecting either its interaction with the telomeric ssDNA or with ACD, were found to co-segregate with CLL [174]. Furthermore, in melanoma patients, elongated telomeres were associated with mutations that impair telomeric ssDNA binding and Pot1–ACD–TIN2 interactions [175–178]. Such mutations would impair the complete formation of the shelterin bridge between the ds and ss telomeric DNA, disrupting the telomerase non-extendible state. Finally, in patients with myeloproliferative neoplasms, significantly reduced telomere length has been associated with elevated levels of POT1 and TIN2 expression, and downregulation of ACD and RAP1 compared with healthy controls [179]. It is not clear whether these alterations in the genes encoding the shelterin components are a cause or a consequence of the cancers. However, abnormal expression and/or function of the shelterin proteins is likely to contribute to telomere dysfunction, thereby driving genetic aberrations and cancer pathogenesis. At present, the factors contributing to the hallmarks of telomeres unique to cancer remain a mystery. However, growing data indicate that cancer cells may harbour impairments affecting the later stages of telomerase activity regulation, such as the targeting and processivity of telomerase, thereby leaving some telomeres short.
On top of aberrant telomerase action, overall shortening of telomeres could be consequences of selection. Random addition of telomeric repeats should lead to some very long telomeres. As the amount of telomeric proteins are limited, insufficient binding of the TRF proteins can cause fragile telomeres and constitutive DNA damage [46]. Hence, cells containing long telomeres are sensitive to further DNA damage and could be selectively eliminated by cell death [180]. Alternatively, homologous recombination or TZAP may become highly active in cancer cells, thereby actively trimming long telomeres. Further investigation into the functions of the telomeric proteins and their potential aberrations in cancer would benefit our understanding of how telomere homeostasis is differently maintained in cancer cells.
4.2. Targeting cancer telomeres and telomerase
Telomerase is an attractive potential drug target in the fight against cancer owing to its low/absent expression levels in normal somatic cells and high expression in cancer. Robust hTERT inhibition can lead to progressive telomere shortening and eventually cancer cell death. Thus, it should be possible to target cancer cells reasonably selectively, while the effect on normal cells should be minimal [181]. Several different compounds that directly target telomerase activity are currently under development, for example antisense oligonucleotides such as imetelstat/GRN163L (reviewed in [8]) and small molecules targeting hTR or hTERT such as BIBR1532 [182]. However, there is a long lag time between administration and clinical response for therapies that target telomerase activity, as telomeres must shorten before the effect is seen [8]. As such, therapies that target the non-canonical functions of telomerase, or which induce a DNA-damage response (DDR) at telomeres (i.e. 6-thio-dG, G-quadruplex stabilizers and oligonucleotides homologous to the 3′ telomeric overhang) may present a better treatment option [8,183].
Overexpression of an hTERT variant, which lacks most of the RT domain, was found to confer cells with a protective advantage against cisplatin-induced apoptosis, indicating a telomere homeostasis independent role for hTERT in cancer pathophysiology [173]. Thus, elucidating the connection between telomerase regulation and the regulation of RNA splicing has a great deal of potential to provide new insights into cancer biology [8]. Similarly, inhibition of the functions of the shelterin proteins could present a viable therapeutic option. For example, TRF1 is overexpressed in many types of cancer and plays an important role in telomeric DNA replication. Loss of TRF1 leads to uncapping of telomeres regardless of telomere length, and has been shown to impair lung tumour growth in mouse models [184]. However, it is not clear what effects targeting the shelterin proteins would have on normal cells.
It has been demonstrated that hTERT is also involved in upregulation of tRNAs [185] and WNT/β-catenin signalling [186] by interacting with their promoter regions. This non-canonical function of TERT appears to be TERC and RT activity-independent. Such aberrant transcription owing to TERT reactivation contributes to carcinogenesis. Therefore, inhibitors against multifunctional tankyrase, which is involved in telomere homeostasis, mitotic spindle formation and WNT/β-catenin signalling, and HSP90, which is involved in signal transduction, intracellular transport and protein degradation, have been explored to selectively kill cancer cells [183]. Finally, a number of immunotherapies are being tested in clinical trials, which aim to sensitize the immune system to tumour cells expressing protein fragments or peptides of telomerase on their cell surface. These are among the most promising telomerase targeting therapeutics, with hTERT specific immune responses being seen in telomerase positive tumours, minimal effects in normal cells and no autoimmunity (reviewed in [8]). Thus, there is a lot of potential in anti-telomerase therapeutics for cancer treatment, and a greater understanding of the regulation of telomerase expression, functions and activity can only serve to further enlighten the search for safe and effective treatments.
5. Conclusions and future perspectives
Until fairly recently, the activity of telomerase was thought to be controlled by limiting access to the telomeres. However, the collective data illustrate that telomerase recruitment and activation are separate events. Such a two-step mechanism is likely to be conserved from yeast to humans. However, the structural biology and biochemistry underlying the process of telomerase activation remains largely unknown and presents an important area for future research. In many human cancer cells, telomerase is highly expressed and recruited indiscriminately to all telomeres. Nevertheless, processivity is low, resulting in the maintenance of short telomeres. As such, both the preferential targeting of short telomeres and the processivity/activity of telomerase may be altered in cancer cells. We believe that t-stumps and altered telomerase regulation, such unique feature of telomeres in cancer, would be an ideal target for cancer therapeutics. Further investigation of telomerase regulation and action would benefit our understanding of the differences in telomere homeostasis between cancer and normal cells, and hopefully lead to the development of effective and safe anti-cancer treatments.
6. Take home messages
— Telomere function and action of telomerase are largely conserved between yeasts and mammals.
— The structural biology and biochemistry underlying the process of telomerase activation remains largely unknown, but emerging studies indicate evolutionary conservation of the mechanisms of telomere homeostasis from yeast to mammals.
— Telomere homeostasis differs between normal cells and cancer cells.
— The maintenance of short telomeres in cancer cells is thought to predispose to genomic instability, and indicates that the targeting and processivity of telomerase may be impaired in cancer.
— Telomeres and telomerase are attractive targets for anti-cancer therapeutics owing to its uniqueness in cancer cells, allowing selective targeting of cancer cells whilst having minimal effects on normal tissue.
Acknowledgements
The authors thank their laboratory members for comments on the manuscript.
Authors' contributions
The authors designed and wrote the manuscript.
Competing interests
We have no competing interests on this manuscript.
Funding
This work is supported by Cancer Research UK (C36439/A12097) and the European Research Council (281722-HRMCB).
References
- 1.Szostak JW, Blackburn EH. 1982. Cloning yeast telomeres on linear plasmid vectors. Cell 29, 245–255. (doi:10.1016/0092-8674(82)90109-X) [DOI] [PubMed] [Google Scholar]
- 2.Zakian VA. 1995. Telomeres: beginning to understand the end. Science 270, 1601–1607. (doi:10.1126/science.270.5242.1601) [DOI] [PubMed] [Google Scholar]
- 3.Hiyama K, Hirai Y, Kyoizumi S, Akiyama M, Hiyama E, Piatyszek MA, Shay JW, Ishioka S, Yamakido M. 1995. Activation of telomerase in human lymphocytes and hematopoietic progenitor cells. J. Immunol. 155, 3711–3715. [PubMed] [Google Scholar]
- 4.Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. 1996. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 18, 173–179. (doi:10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3) [DOI] [PubMed] [Google Scholar]
- 5.Ramirez RD, Wright WE, Shay JW, Taylor RS. 1997. Telomerase activity concentrates in the mitotically active segments of human hair follicles. J. Invest. Dermatol. 108, 113–117. (doi:10.1111/1523-1747.ep12285654) [DOI] [PubMed] [Google Scholar]
- 6.Greider CW, Blackburn EH. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43, 405–413. (doi:10.1016/0092-8674(85)90170-9) [DOI] [PubMed] [Google Scholar]
- 7.Lingner J, Cech TR, Hughes TR, Lundblad V. 1997. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl Acad. Sci. USA 94, 11 190–11 195. (doi:10.1073/pnas.94.21.11190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jafri MA, Ansari SA, Alqahtani MH, Shay JW. 2016. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 8, 69 (doi:10.1186/s13073-016-0324-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim NW, et al. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015. (doi:10.1126/science.7605428) [DOI] [PubMed] [Google Scholar]
- 10.Shay JW, Bacchetti S. 1997. A survey of telomerase activity in human cancer. Eur. J. Cancer 33, 787–791. (doi:10.1016/S0959-8049(97)00062-2) [DOI] [PubMed] [Google Scholar]
- 11.Bodnar AG, et al. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279, 349–352. (doi:10.1126/science.279.5349.349) [DOI] [PubMed] [Google Scholar]
- 12.Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC. 1990. Telomere reduction in human colorectal carcinoma and with ageing. Nature 346, 866–868. (doi:10.1038/346866a0) [DOI] [PubMed] [Google Scholar]
- 13.de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE. 1990. Structure and variability of human chromosome ends. Mol. Cell. Biol. 10, 518–527. (doi:10.1128/MCB.10.2.518) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu L, Blackburn EH. 2007. Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol. Cell 28, 315–327. (doi:10.1016/j.molcel.2007.10.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Kha H, Ungrin M, Robinson MO, Harrington L. 2002. Preferential maintenance of critically short telomeres in mammalian cells heterozygous for mTert. Proc. Natl Acad. Sci. USA 99, 3597–3602. (doi:10.1073/pnas.062549199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samper E, Goytisolo FA, Menissier-de Murcia J, Gonzalez-Suarez E, Cigudosa JC, de Murcia G, Blasco MA. 2001. Normal telomere length and chromosomal end capping in poly(ADP-ribose) polymerase-deficient mice and primary cells despite increased chromosomal instability. J. Cell Biol. 154, 49–60. (doi:10.1083/jcb.200103049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teixeira MT, Arneric M, Sperisen P, Lingner J. 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117, 323–335. (doi:10.1016/S0092-8674(04)00334-4) [DOI] [PubMed] [Google Scholar]
- 18.Olovnikov AM. 1996. Telomeres, telomerase, and aging: origin of the theory. Exp. Gerontol. 31, 443–448. (doi:10.1016/0531-5565(96)00005-8) [DOI] [PubMed] [Google Scholar]
- 19.Lingner J, Cooper JP, Cech TR. 1995. Telomerase and DNA end replication: no longer a lagging strand problem? Science 269, 1533–1534. (doi:10.1126/science.7545310) [DOI] [PubMed] [Google Scholar]
- 20.Hayflick L, Moorhead PS. 1961. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25, 585–621. (doi:10.1016/0014-4827(61)90192-6) [DOI] [PubMed] [Google Scholar]
- 21.Lundblad V, Szostak JW. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57, 633–643. (doi:10.1016/0092-8674(89)90132-3) [DOI] [PubMed] [Google Scholar]
- 22.Artandi SE, Attardi LD. 2005. Pathways connecting telomeres and p53 in senescence, apoptosis, and cancer. Biochem. Biophys. Res. Commun. 331, 881–890. (doi:10.1016/j.bbrc.2005.03.211) [DOI] [PubMed] [Google Scholar]
- 23.Denchi EL, de Lange T. 2007. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448, 1068–1071. (doi:10.1038/nature06065) [DOI] [PubMed] [Google Scholar]
- 24.Palm W, de Lange T. 2008. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334. (doi:10.1146/annurev.genet.41.110306.130350) [DOI] [PubMed] [Google Scholar]
- 25.Hayashi MT, Cesare AJ, Rivera T, Karlseder J. 2015. Cell death during crisis is mediated by mitotic telomere deprotection. Nature 522, 492–496. (doi:10.1038/nature14513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClintock B. 1939. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl Acad. Sci. USA 25, 405–416. (doi:10.1073/pnas.25.8.405) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blackburn EH, Gall JG. 1978. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 120, 33–53. (doi:10.1016/0022-2836(78)90294-2) [DOI] [PubMed] [Google Scholar]
- 28.Hiraoka Y, Henderson E, Blackburn EH. 1998. Not so peculiar: fission yeast telomere repeats. Trends Biochem. Sci. 23, 126 (doi:10.1016/S0968-0004(98)01176-1) [DOI] [PubMed] [Google Scholar]
- 29.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. 1988. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA 85, 6622–6626. (doi:10.1073/pnas.85.18.6622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canudas S, Houghtaling BR, Bhanot M, Sasa G, Savage SA, Bertuch AA, Smith S. 2011. A role for heterochromatin protein 1γ at human telomeres. Genes Dev. 25, 1807–1819. (doi:10.1101/gad.17325211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. 1999. Mammalian telomeres end in a large duplex loop. Cell 97, 503–514. (doi:10.1016/S0092-8674(00)80760-6) [DOI] [PubMed] [Google Scholar]
- 32.Doksani Y, Wu JY, de Lange T, Zhuang X. 2013. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 155, 345–356. (doi:10.1016/j.cell.2013.09.048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carneiro T, Khair L, Reis CC, Borges V, Moser BA, Nakamura TM, Ferreira MG. 2010. Telomeres avoid end detection by severing the checkpoint signal transduction pathway. Nature 467, 228–232. (doi:10.1038/nature09353) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Lange T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110. (doi:10.1101/gad.1346005) [DOI] [PubMed] [Google Scholar]
- 35.van Steensel B, de Lange T. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385, 740–743. (doi:10.1038/385740a0) [DOI] [PubMed] [Google Scholar]
- 36.Li B, Oestreich S, de Lange T. 2000. Identification of human Rap1: implications for telomere evolution. Cell 101, 471–483. (doi:10.1016/S0092-8674(00)80858-2) [DOI] [PubMed] [Google Scholar]
- 37.Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. 2000. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 20, 1659–1668. (doi:10.1128/MCB.20.5.1659-1668.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houghtaling BR, Cuttonaro L, Chang W, Smith S. 2004. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 14, 1621–1631. (doi:10.1016/j.cub.2004.08.052) [DOI] [PubMed] [Google Scholar]
- 39.Ye JZ, Donigian JR, van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T. 2004. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 279, 47 264–47 271. (doi:10.1074/jbc.M409047200) [DOI] [PubMed] [Google Scholar]
- 40.Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. 2004. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 18, 1649–1654. (doi:10.1101/gad.1215404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Steensel B, Smogorzewska A, de Lange T. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92, 401–413. (doi:10.1016/S0092-8674(00)80932-0) [DOI] [PubMed] [Google Scholar]
- 42.Benarroch-Popivker D, et al. 2016. TRF2-mediated control of telomere DNA topology as a mechanism for chromosome-end protection. Mol. Cell 61, 274–286. (doi:10.1016/j.molcel.2015.12.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarthy J, Bae NS, Scrafford J, Baumann P. 2009. Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J. 28, 3390–3399. (doi:10.1038/emboj.2009.275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. 2010. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 327, 1657–1661. (doi:10.1126/science.1185100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye J, Renault VM, Jamet K, Gilson E. 2014. Transcriptional outcome of telomere signalling. Nat. Rev. Genet. 15, 491–503. (doi:10.1038/nrg3743) [DOI] [PubMed] [Google Scholar]
- 46.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138, 90–103. (doi:10.1016/j.cell.2009.06.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jain D, Cooper JP. 2010. Telomeric strategies: means to an end. Annu. Rev. Genet. 44, 243–269. (doi:10.1146/annurev-genet-102108-134841) [DOI] [PubMed] [Google Scholar]
- 48.Miyoshi T, Kanoh J, Saito M, Ishikawa F. 2008. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 320, 1341–1344. (doi:10.1126/science.1154819) [DOI] [PubMed] [Google Scholar]
- 49.Miller KM, Rog O, Cooper JP. 2006. Semi-conservative DNA replication through telomeres requires Taz1. Nature 440, 824–828. (doi:10.1038/nature04638) [DOI] [PubMed] [Google Scholar]
- 50.Ferreira MG, Cooper JP. 2001. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell 7, 55–63. (doi:10.1016/S1097-2765(01)00154-X) [DOI] [PubMed] [Google Scholar]
- 51.Baumann P, Cech TR. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292, 1171–1175. (doi:10.1126/science.1060036) [DOI] [PubMed] [Google Scholar]
- 52.Jun HI, Liu J, Jeong H, Kim JK, Qiao F. 2013. Tpz1 controls a telomerase-nonextendible telomeric state and coordinates switching to an extendible state via Ccq1. Genes Dev. 27, 1917–1931. (doi:10.1101/gad.219485.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooper JP, Nimmo ER, Allshire RC, Cech TR. 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385, 744–747. (doi:10.1038/385744a0) [DOI] [PubMed] [Google Scholar]
- 54.Chikashige Y, Hiraoka Y. 2001. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr. Biol. 11, 1618–1623. (doi:10.1016/S0960-9822(01)00457-2) [DOI] [PubMed] [Google Scholar]
- 55.Kanoh J, Ishikawa F. 2001. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 11, 1624–1630. (doi:10.1016/S0960-9822(01)00503-6) [DOI] [PubMed] [Google Scholar]
- 56.Tomita K, Cooper JP. 2008. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev. 22, 3461–3474. (doi:10.1101/gad.498608) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wotton D, Shore D. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11, 748–760. (doi:10.1101/gad.11.6.748) [DOI] [PubMed] [Google Scholar]
- 58.Shore D, Bianchi A. 2009. Telomere length regulation: coupling DNA end processing to feedback regulation of telomerase. EMBO J. 28, 2309–2322. (doi:10.1038/emboj.2009.195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nugent CI, Hughes TR, Lue NF, Lundblad V. 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274, 249–252. (doi:10.1126/science.274.5285.249) [DOI] [PubMed] [Google Scholar]
- 60.Lin JJ, Zakian VA. 1996. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl Acad. Sci. USA 93, 13 760–13 765. (doi:10.1073/pnas.93.24.13760) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pennock E, Buckley K, Lundblad V. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104, 387–396. (doi:10.1016/S0092-8674(01)00226-4) [DOI] [PubMed] [Google Scholar]
- 62.Hug N, Lingner J. 2006. Telomere length homeostasis. Chromosoma 115, 413–425. (doi:10.1007/s00412-006-0067-3) [DOI] [PubMed] [Google Scholar]
- 63.Lewis KA, Wuttke DS. 2012. Telomerase and telomere-associated proteins: structural insights into mechanism and evolution. Structure 20, 28–39. (doi:10.1016/j.str.2011.10.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beattie TL, Zhou W, Robinson MO, Harrington L. 1998. Reconstitution of human telomerase activity in vitro. Curr. Biol. 8, 177–180. (doi:10.1016/S0960-9822(98)70067-3) [DOI] [PubMed] [Google Scholar]
- 65.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277, 955–959. (doi:10.1126/science.277.5328.955) [DOI] [PubMed] [Google Scholar]
- 66.Nandakumar J, Cech TR. 2013. Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 14, 69–82. (doi:10.1038/nrm3505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blasco MA, Funk W, Villeponteau B, Greider CW. 1995. Functional characterization and developmental regulation of mouse telomerase RNA. Science 269, 1267–1270. (doi:10.1126/science.7544492) [DOI] [PubMed] [Google Scholar]
- 68.Leonardi J, Box JA, Bunch JT, Baumann P. 2008. TER1, the RNA subunit of fission yeast telomerase. Nat. Struct. Mol. Biol. 15, 26–33. (doi:10.1038/nsmb1343) [DOI] [PubMed] [Google Scholar]
- 69.Webb CJ, Zakian VA. 2008. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat. Struct. Mol. Biol. 15, 34–42. (doi:10.1038/nsmb1354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin J, Ly H, Hussain A, Abraham M, Pearl S, Tzfati Y, Parslow TG, Blackburn EH. 2004. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc. Natl Acad. Sci. USA 101, 14 713–14 718. (doi:10.1073/pnas.0405879101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276, 561–567. (doi:10.1126/science.276.5312.561) [DOI] [PubMed] [Google Scholar]
- 72.Schmidt JC, Cech TR. 2015. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev. 29, 1095–1105. (doi:10.1101/gad.263863.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang W, Kannan R, Blanchette M, Baumann P. 2012. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature 484, 260–264. (doi:10.1038/nature10924) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beernink HT, Miller K, Deshpande A, Bucher P, Cooper JP. 2003. Telomere maintenance in fission yeast requires an Est1 ortholog. Curr. Biol. 13, 575–580. (doi:10.1016/S0960-9822(03)00169-6) [DOI] [PubMed] [Google Scholar]
- 75.Webb CJ, Zakian VA. 2012. Schizosaccharomyces pombe Ccq1 and TER1 bind the 14-3-3-like domain of Est1, which promotes and stabilizes telomerase–telomere association. Genes Dev. 26, 82–91. (doi:10.1101/gad.181826.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.MacNeil DE, Bensoussan HJ, Autexier C. 2016. Telomerase regulation from beginning to the end. Genes (Basel) 7, 64 (doi:10.3390/genes7090064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mitchell JR, Cheng J, Collins K. 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3' end. Mol. Cell. Biol. 19, 567–576. (doi:10.1128/MCB.19.1.567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT. 2006. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 173, 207–218. (doi:10.1083/jcb.200601105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pogacic V, Dragon F, Filipowicz W. 2000. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 20, 9028–9040. (doi:10.1128/MCB.20.23.9028-9040.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Egan ED, Collins K. 2012. An enhanced H/ACA RNP assembly mechanism for human telomerase RNA. Mol. Cell. Biol. 32, 2428–2439. (doi:10.1128/MCB.00286-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jady BE, Bertrand E, Kiss T. 2004. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 164, 647–652. (doi:10.1083/jcb.200310138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. 2009. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323, 644–648. (doi:10.1126/science.1165357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jeong YY, Her J, Oh SY, Chung IK. 2016. Hsp90-binding immunophilin FKBP52 modulates telomerase activity by promoting the cytoplasmic retrotransport of hTERT. Biochem. J. 473, 3517–3532. (doi:10.1042/BCJ20160344) [DOI] [PubMed] [Google Scholar]
- 84.Snow BE, Erdmann N, Cruickshank J, Goldman H, Gill RM, Robinson MO, Harrington L. 2003. Functional conservation of the telomerase protein Est1p in humans. Curr. Biol. 13, 698–704. (doi:10.1016/S0960-9822(03)00210-0) [DOI] [PubMed] [Google Scholar]
- 85.Redon S, Reichenbach P, Lingner J. 2007. Protein RNA and protein protein interactions mediate association of human EST1A/SMG6 with telomerase. Nucleic Acids Res. 35, 7011–7022. (doi:10.1093/nar/gkm724) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801. (doi:10.1126/science.1147182) [DOI] [PubMed] [Google Scholar]
- 87.Wang C, Zhao L, Lu S. 2015. Role of TERRA in the regulation of telomere length. Int. J. Biol. Sci. 11, 316–323. (doi:10.7150/ijbs.10528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Harley CB, Futcher AB, Greider CW. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345, 458–460. (doi:10.1038/345458a0) [DOI] [PubMed] [Google Scholar]
- 89.Gilchrist GC, Kurjanowicz P, Mereilles FV, King WA, LaMarre J. 2015. Telomere length and telomerase activity in bovine pre-implantation embryos in vitro. Reprod. Domest. Anim. 50, 58–67. (doi:10.1111/rda.12449) [DOI] [PubMed] [Google Scholar]
- 90.Turner S, Wong HP, Rai J, Hartshorne GM. 2010. Telomere lengths in human oocytes, cleavage stage embryos and blastocysts. Mol. Hum. Reprod. 16, 685–694. (doi:10.1093/molehr/gaq048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Greenwood MJ, Lansdorp PM. 2003. Telomeres, telomerase, and hematopoietic stem cell biology. Arch. Med. Res. 34, 489–495. (doi:10.1016/j.arcmed.2003.07.003) [DOI] [PubMed] [Google Scholar]
- 92.Hiyama E, Hiyama K. 2007. Telomere and telomerase in stem cells. Br. J. Cancer 96, 1020–1024. (doi:10.1038/sj.bjc.6603671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marcand S, Gilson E, Shore D. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275, 986–990. (doi:10.1126/science.275.5302.986) [DOI] [PubMed] [Google Scholar]
- 94.Bianchi A, Shore D. 2007. Increased association of telomerase with short telomeres in yeast. Genes Dev. 21, 1726–1730. (doi:10.1101/gad.438907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin JJ, Zakian VA. 1995. An in vitro assay for Saccharomyces telomerase requires EST1. Cell 81, 1127–1135. (doi:10.1016/S0092-8674(05)80017-0) [DOI] [PubMed] [Google Scholar]
- 96.Evans SK, Lundblad V. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286, 117–120. (doi:10.1126/science.286.5437.117) [DOI] [PubMed] [Google Scholar]
- 97.Taggart AK, Teng SC, Zakian VA. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297, 1023–1026. (doi:10.1126/science.1074968) [DOI] [PubMed] [Google Scholar]
- 98.Bianchi A, Negrini S, Shore D. 2004. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell 16, 139–146. (doi:10.1016/j.molcel.2004.09.009) [DOI] [PubMed] [Google Scholar]
- 99.Wu Y, Zakian VA. 2011. The telomeric Cdc13 protein interacts directly with the telomerase subunit Est1 to bring it to telomeric DNA ends in vitro. Proc. Natl Acad. Sci. USA 108, 20 362–20 369. (doi:10.1073/pnas.1100281108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tseng SF, Lin JJ, Teng SC. 2006. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 34, 6327–6336. (doi:10.1093/nar/gkl786) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sabourin M, Tuzon CT, Zakian VA. 2007. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol. Cell 27, 550–561. (doi:10.1016/j.molcel.2007.07.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW. 2007. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol. Cell 27, 851–858. (doi:10.1016/j.molcel.2007.08.007) [DOI] [PubMed] [Google Scholar]
- 103.Li S, Makovets S, Matsuguchi T, Blethrow JD, Shokat KM, Blackburn EH. 2009. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell 136, 50–61. (doi:10.1016/j.cell.2008.11.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moser BA, Chang YT, Kosti J, Nakamura TM. 2011. Tel1ATM and Rad3ATR kinases promote Ccq1–Est1 interaction to maintain telomeres in fission yeast. Nat. Struct. Mol. Biol. 18, 1408–1413. (doi:10.1038/nsmb.2187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yamazaki H, Tarumoto Y, Ishikawa F. 2012. Tel1ATM and Rad3ATR phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev. 26, 241–246. (doi:10.1101/gad.177873.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Armstrong CA, Pearson SR, Amelina H, Moiseeva V, Tomita K. 2014. Telomerase activation after recruitment in fission yeast. Curr. Biol. 24, 2006–2011. (doi:10.1016/j.cub.2014.07.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hu X, Liu J, Jun HI, Kim JK, Qiao F. 2016. Multi-step coordination of telomerase recruitment in fission yeast through two coupled telomere–telomerase interfaces. eLife 5, e15470 (doi:10.7554/eLife.15470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dehe PM, Rog O, Ferreira MG, Greenwood J, Cooper JP. 2012. Taz1 enforces cell-cycle regulation of telomere synthesis. Mol. Cell 46, 797–808. (doi:10.1016/j.molcel.2012.04.022) [DOI] [PubMed] [Google Scholar]
- 109.Moser BA, Subramanian L, Khair L, Chang YT, Nakamura TM. 2009. Fission yeast Tel1ATM and Rad3ATR promote telomere protection and telomerase recruitment. PLoS Genet. 5, e1000622 (doi:10.1371/journal.pgen.1000622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jady BE, Richard P, Bertrand E, Kiss T. 2006. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol. Biol. Cell 17, 944–954. (doi:10.1091/mbc.E05-09-0904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP. 2006. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell 17, 955–965. (doi:10.1091/mbc.E05-09-0903) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. 2007. The POT1–TPP1 telomere complex is a telomerase processivity factor. Nature 445, 506–510. (doi:10.1038/nature05454) [DOI] [PubMed] [Google Scholar]
- 113.Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. 2007. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature 445, 559–562. (doi:10.1038/nature05469) [DOI] [PubMed] [Google Scholar]
- 114.Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. 2010. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol. Cell. Biol. 30, 2971–2982. (doi:10.1128/MCB.00240-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim SH, Kaminker P, Campisi J. 1999. TIN2, a new regulator of telomere length in human cells. Nat. Genet. 23, 405–412. (doi:10.1038/70508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Frank AK, Tran DC, Qu RW, Stohr BA, Segal DJ, Xu L. 2015. The Shelterin TIN2 subunit mediates recruitment of telomerase to telomeres. PLoS Genet. 11, e1005410 (doi:10.1371/journal.pgen.1005410) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang Y, Chen LY, Han X, Xie W, Kim H, Yang D, Liu D, Songyang Z. 2013. Phosphorylation of TPP1 regulates cell cycle-dependent telomerase recruitment. Proc. Natl Acad. Sci. USA 110, 5457–5462. (doi:10.1073/pnas.1217733110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sexton AN, et al. 2014. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev. 28, 1885–1899. (doi:10.1101/gad.246819.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Savage SA, Giri N, Baerlocher GM, Orr N, Lansdorp PM, Alter BP. 2008. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am. J. Hum. Genet. 82, 501–509. (doi:10.1016/j.ajhg.2007.10.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee SS, Bohrson C, Pike AM, Wheelan SJ, Greider CW. 2015. ATM kinase is required for telomere elongation in mouse and human cells. Cell Rep. 13, 1623–1632. (doi:10.1016/j.celrep.2015.10.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tong AS, Stern JL, Sfeir A, Kartawinata M, de Lange T, Zhu XD, Bryan TM. 2015. ATM and ATR signaling regulate the recruitment of human telomerase to telomeres. Cell Rep. 13, 1633–1646. (doi:10.1016/j.celrep.2015.10.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feuerhahn S, Chen LY, Luke B, Porro A. 2015. No DDRama at chromosome ends: TRF2 takes centre stage. Trends Biochem. Sci. 40, 275–285. (doi:10.1016/j.tibs.2015.03.003) [DOI] [PubMed] [Google Scholar]
- 123.Ancelin K, Brunori M, Bauwens S, Koering CE, Brun C, Ricoul M, Pommier JP, Sabatier L, Gilson E. 2002. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 22, 3474–3487. (doi:10.1128/MCB.22.10.3474-3487.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. 2010. In vivo stoichiometry of shelterin components. J. Biol. Chem. 285, 1457–1467. (doi:10.1074/jbc.M109.038026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Grolimund L, Aeby E, Hamelin R, Armand F, Chiappe D, Moniatte M, Lingner J. 2013. A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nat. Commun. 4, 2848 (doi:10.1038/ncomms3848) [DOI] [PubMed] [Google Scholar]
- 126.Li JS, Miralles Fuste J, Simavorian T, Bartocci C, Tsai J, Karlseder J, Lazzerini Denchi E. 2017. TZAP: a telomere-associated protein involved in telomere length control. Science 355, 638–641. (doi:10.1126/science.aah6752) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. 2012. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492, 285–289. (doi:10.1038/nature11648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE. 2012. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150, 481–494. (doi:10.1016/j.cell.2012.07.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sexton AN, Youmans DT, Collins K. 2012. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J. Biol. Chem. 287, 34 455–34 464. (doi:10.1074/jbc.M112.394767) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schmidt JC, Dalby AB, Cech TR. 2014. Identification of human TERT elements necessary for telomerase recruitment to telomeres. eLife 3, e03563 (doi:10.7554/eLife.03563) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Canudas S, Smith S. 2009. Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J. Cell Biol. 187, 165–173. (doi:10.1083/jcb.200903096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Houghtaling BR, Canudas S, Smith S. 2012. A role for sister telomere cohesion in telomere elongation by telomerase. Cell Cycle 11, 19–25. (doi:10.4161/cc.11.1.18633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang J, Cohen AL, Letian A, Tadeo X, Moresco JJ, Liu J, Yates JR III, Qiao F, Jia S. 2016. The proper connection between shelterin components is required for telomeric heterochromatin assembly. Genes Dev. 30, 827–839. (doi:10.1101/gad.266718.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Reyes C, Serrurier C, Gauthier T, Gachet Y, Tournier S. 2015. Aurora B prevents chromosome arm separation defects by promoting telomere dispersion and disjunction. J. Cell Biol. 208, 713–727. (doi:10.1083/jcb.201407016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Motwani T, Doris R, Holmes SG, Flory MR. 2010. Ccq1p and the condensin proteins Cut3p and Cut14p prevent telomere entanglements in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 9, 1612–1621. (doi:10.1128/EC.00339-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Greider CW. 1991. Telomerase is processive. Mol. Cell. Biol. 11, 4572–4580. (doi:10.1128/MCB.11.9.4572) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yang W, Lee YS. 2015. A DNA-hairpin model for repeat-addition processivity in telomere synthesis. Nat. Struct. Mol. Biol. 22, 844–847. (doi:10.1038/nsmb.3098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Parks JW, Kappel K, Das R, Stone MD. 2017. Single-molecule FRET-Rosetta reveals RNA structural rearrangements during human telomerase catalysis. RNA 23, 175–188. (doi:10.1261/rna.058743.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Latrick CM, Cech TR. 2010. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 29, 924–933. (doi:10.1038/emboj.2009.409) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dalby AB, Hofr C, Cech TR. 2015. Contributions of the TEL-patch amino acid cluster on TPP1 to telomeric DNA synthesis by human telomerase. J. Mol. Biol. 427, 1291–1303. (doi:10.1016/j.jmb.2015.01.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hwang H, Opresko P, Myong S. 2014. Single-molecule real-time detection of telomerase extension activity. Sci. Rep. 4, 6391 (doi:10.1038/srep06391) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Robart AR, Collins K. 2011. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol. Cell 42, 308–318. (doi:10.1016/j.molcel.2011.03.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu RA, Collins K. 2014. Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. EMBO J. 33, 921–935. (doi:10.1002/embj.201387205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Akiyama BM, Parks JW, Stone MD. 2015. The telomerase essential N-terminal domain promotes DNA synthesis by stabilizing short RNA–DNA hybrids. Nucleic Acids Res. 43, 5537–5549. (doi:10.1093/nar/gkv406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Huard S, Moriarty TJ, Autexier C. 2003. The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity. Nucleic Acids Res. 31, 4059–4070. (doi:10.1093/nar/gkg437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xie M, Podlevsky JD, Qi X, Bley CJ, Chen JJ. 2010. A novel motif in telomerase reverse transcriptase regulates telomere repeat addition rate and processivity. Nucleic Acids Res. 38, 1982–1996. (doi:10.1093/nar/gkp1198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tuzon CT, Wu Y, Chan A, Zakian VA. 2011. The Saccharomyces cerevisiae telomerase subunit Est3 binds telomeres in a cell cycle- and Est1-dependent manner and interacts directly with Est1 in vitro. PLoS Genet. 7, e1002060 (doi:10.1371/journal.pgen.1002060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Talley JM, DeZwaan DC, Maness LD, Freeman BC, Friedman KL. 2011. Stimulation of yeast telomerase activity by the ever shorter telomere 3 (Est3) subunit is dependent on direct interaction with the catalytic protein Est2. J. Biol. Chem. 286, 26 431–26 439. (doi:10.1074/jbc.M111.228635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee J, Mandell EK, Tucey TM, Morris DK, Lundblad V. 2008. The Est3 protein associates with yeast telomerase through an OB-fold domain. Nature Struct. Mol. Biol. 15, 990–997. (doi:10.1038/nsmb.1472) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rao T, Lubin JW, Armstrong GS, Tucey TM, Lundblad V, Wuttke DS. 2014. Structure of Est3 reveals a bimodal surface with differential roles in telomere replication. Proc. Natl Acad. Sci. USA 111, 214–218. (doi:10.1073/pnas.1316453111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. 2009. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 36, 193–206. (doi:10.1016/j.molcel.2009.08.009) [DOI] [PubMed] [Google Scholar]
- 152.Chen LY, Redon S, Lingner J. 2012. The human CST complex is a terminator of telomerase activity. Nature 488, 540–544. (doi:10.1038/nature11269) [DOI] [PubMed] [Google Scholar]
- 153.Wu P, Takai H, de Lange T. 2012. Telomeric 3' overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell 150, 39–52. (doi:10.1016/j.cell.2012.05.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Grandin N, Damon C, Charbonneau M. 2000. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol. Cell. Biol. 20, 8397–8408. (doi:10.1128/MCB.20.22.8397-8408.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chandra A, Hughes TR, Nugent CI, Lundblad V. 2001. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15, 404–414. (doi:10.1101/gad.861001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zaug AJ, Podell ER, Cech TR. 2005. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl Acad. Sci. USA 102, 10 864–10 869. (doi:10.1073/pnas.0504744102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Audry J, Maestroni L, Delagoutte E, Gauthier T, Nakamura TM, Gachet Y, Saintome C, Geli V, Coulon S. 2015. RPA prevents G-rich structure formation at lagging-strand telomeres to allow maintenance of chromosome ends. EMBO J. 34, 1942–1958. (doi:10.15252/embj.201490773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Avin BA, Umbricht CB, Zeiger MA. 2016. Human telomerase reverse transcriptase regulation by DNA methylation, transcription factor binding and alternative splicing (review). Int. J. Oncol. 49, 2199–2205. (doi:10.3892/ijo.2016.3743) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vera E, Canela A, Fraga MF, Esteller M, Blasco MA. 2008. Epigenetic regulation of telomeres in human cancer. Oncogene 27, 6817–6833. (doi:10.1038/onc.2008.289) [DOI] [PubMed] [Google Scholar]
- 160.Cristofari G, Lingner J. 2006. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 25, 565–574. (doi:10.1038/sj.emboj.7600952) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cao Y, Huschtscha LI, Nouwens AS, Pickett HA, Neumann AA, Chang AC, Toouli CD, Bryan TM, Reddel RR. 2008. Amplification of telomerase reverse transcriptase gene in human mammary epithelial cells with limiting telomerase RNA expression levels. Cancer Res. 68, 3115–3123. (doi:10.1158/0008-5472.CAN-07-6377) [DOI] [PubMed] [Google Scholar]
- 162.Park KH, et al. 1998. Telomerase activity and telomere lengths in various cell lines: changes of telomerase activity can be another method for chemosensitivity evaluation. Int. J. Oncol. 13, 489–495. (doi:10.3892/ijo.13.3.489) [DOI] [PubMed] [Google Scholar]
- 163.Rha SY, et al. 1999. Changes of telomerase and telomere lengths in paired normal and cancer tissues of breast. Int. J. Oncol. 15, 839–845. (doi:10.3892/ijo.15.4.839) [DOI] [PubMed] [Google Scholar]
- 164.Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE. 2009. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138, 463–475. (doi:10.1016/j.cell.2009.05.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Schmidt JC, Zaug AJ, Cech TR. 2016. Live cell imaging reveals the dynamics of telomerase recruitment to telomeres. Cell 166, 1188–1197.e9 (doi:10.1016/j.cell.2016.07.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gu P, Min JN, Wang Y, Huang C, Peng T, Chai W, Chang S. 2012. CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 31, 2309–2321. (doi:10.1038/emboj.2012.96) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sykorova E, Fajkus J. 2009. Structure–function relationships in telomerase genes. Biol. Cell/Eur. Cell Biol. Organ. 101, 375–392. (doi:10.1042/BC20080205) [DOI] [PubMed] [Google Scholar]
- 168.Ulaner GA, Hu JF, Vu TH, Giudice LC, Hoffman AR. 1998. Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res. 58, 4168–4172. [PubMed] [Google Scholar]
- 169.Ulaner GA, Hu JF, Vu TH, Oruganti H, Giudice LC, Hoffman AR. 2000. Regulation of telomerase by alternate splicing of human telomerase reverse transcriptase (hTERT) in normal and neoplastic ovary, endometrium and myometrium. Int. J. Cancer 85, 330–335. (doi:10.1002/(SICI)1097-0215(20000201)85:3<330::AID-IJC6>3.0.CO;2-U) [PubMed] [Google Scholar]
- 170.Saeboe-Larssen S, Fossberg E, Gaudernack G. 2006. Characterization of novel alternative splicing sites in human telomerase reverse transcriptase (hTERT): analysis of expression and mutual correlation in mRNA isoforms from normal and tumour tissues. BMC Mol. Biol. 7, 26 (doi:10.1186/1471-2199-7-26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lincz LF, Mudge LM, Scorgie FE, Sakoff JA, Hamilton CS, Seldon M. 2008. Quantification of hTERT splice variants in melanoma by SYBR green real-time polymerase chain reaction indicates a negative regulatory role for the beta deletion variant. Neoplasia 10, 1131–1137. (doi:10.1593/neo.08644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu Y, Wu BQ, Zhong HH, Tian XX, Fang WG. 2012. Quantification of alternative splicing variants of human telomerase reverse transcriptase and correlations with telomerase activity in lung cancer. PLoS ONE 7, e38868 (doi:10.1371/journal.pone.0038868) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Listerman I, Sun J, Gazzaniga FS, Lukas JL, Blackburn EH. 2013. The major reverse transcriptase-incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis. Cancer Res. 73, 2817–2828. (doi:10.1158/0008-5472.CAN-12-3082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Speedy HE, et al. 2016. Germline mutations in shelterin complex genes are associated with familial chronic lymphocytic leukemia. Blood 128, 2319–2326. (doi:10.1182/blood-2016-01-695692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Aoude LG, et al. 2015. Nonsense mutations in the shelterin complex genes ACD and TERF2IP in familial melanoma. J. Natl. Cancer Inst. 107, dju408. (doi:10.1093/jnci/dju408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Shi J, et al. 2014. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat. Genet. 46, 482–486. (doi:10.1038/ng.2941) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Robles-Espinoza CD, et al. 2014. POT1 loss-of-function variants predispose to familial melanoma. Nat. Genet. 46, 478–481. (doi:10.1038/ng.2947) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Liu J, Yu C, Hu X, Kim JK, Bierma JC, Jun HI, Rychnovsky SD, Huang L, Qiao F. 2015. Dissecting fission yeast shelterin interactions via MICro-MS links disruption of shelterin bridge to tumorigenesis. Cell Rep. 12, 2169–2180. (doi:10.1016/j.celrep.2015.08.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dahlstrom J, Zhang X, Ghaderi M, Hultcrantz M, Bjorkholm M, Xu D. 2015. Dysregulation of shelterin factors coupled with telomere shortening in Philadelphia chromosome negative myeloproliferative neoplasms. Haematologica 100, e402–e405. (doi:10.3324/haematol.2015.125765) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Fairlie J, Harrington L. 2015. Enforced telomere elongation increases the sensitivity of human tumour cells to ionizing radiation. DNA Repair (Amst) 25, 54–59. (doi:10.1016/j.dnarep.2014.11.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Buseman CM, Wright WE, Shay JW. 2012. Is telomerase a viable target in cancer? Mutat. Res. 730, 90–97. (doi:10.1016/j.mrfmmm.2011.07.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bryan C, Rice C, Hoffman H, Harkisheimer M, Sweeney M, Skordalakes E. 2015. Structural basis of telomerase inhibition by the highly specific BIBR1532. Structure 23, 1934–1942. (doi:10.1016/j.str.2015.08.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ruden M, Puri N. 2013. Novel anticancer therapeutics targeting telomerase. Cancer Treat. Rev. 39, 444–456. (doi:10.1016/j.ctrv.2012.06.007) [DOI] [PubMed] [Google Scholar]
- 184.Garcia-Beccaria M, et al. 2015. Therapeutic inhibition of TRF1 impairs the growth of p53-deficient K-RasG12 V-induced lung cancer by induction of telomeric DNA damage. EMBO Mol. Med. 7, 930–949. (doi:10.15252/emmm.201404497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Khattar E, et al. 2016. Telomerase reverse transcriptase promotes cancer cell proliferation by augmenting tRNA expression. J. Clin. Invest. 126, 4045–4060. (doi:10.1172/JCI86042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Park JI, et al. 2009. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 460, 66–72. (doi:10.1038/nature08137) [DOI] [PMC free article] [PubMed] [Google Scholar]