Abstract
Marine bacteria are the most widely distributed organisms in the ocean environment and produce a wide variety of secondary metabolites. However, traditional screening for bioactive natural compounds is greatly hindered by the lack of a systematic way of cataloguing the chemical profiles of bacterial strains found in nature. Here we present a chemical fingerprint database of marine bacteria based on their secondary metabolite profiles, acquired by high-resolution LC-MS. Till now, 1,430 bacterial strains spanning 168 known species collected from different marine environments were cultured and profiled. Using this database, we demonstrated that secondary metabolite profile similarity is approximately, but not always, correlated with taxonomical similarity. We also validated the ability of this database to find species-specific metabolites, as well as to discover known bioactive compounds from previously unknown sources. An online interface to this database, as well as the accompanying software, is provided freely for the community to use.
Metabolomics is a rapidly growing area of scientific research. Global metabolic profiling employing technological platforms such as nuclear magnetic resonance (NMR) or mass spectrometry (MS) is an increasingly powerful tool for studying biological phenomena1. Thus far, metabolomics research has mostly focused on primary metabolites that have well-known structures and biological functions. On the other hand, much less is known about secondary metabolites, which are defined as organic compounds such as toxins, antibiotics and other outward-directed compounds2 that are not directly involved in normal growth. However, secondary metabolites are rich sources of novel bioactive compounds and drug leads due to their great diversity in chemical structures in terms of carbon skeletons and stereochemistries3,4. In addition, as secondary metabolites often result from the interplay between the genotype and the external environment, the study of these compounds will be important for improving our understanding of how the organism interacts with its environment4.
Marine bacteria are the most widely distributed organisms in the ocean environment (approximately 1029 cells) and produce a great variety of compounds as a result of their unique metabolic and physiological capabilities3,5. Currently, up to 38% of the reported natural products and 20% of new compound entries in AntiBase, a commercial database of natural compounds, are produced by marine bacteria6,7. However, in the past 15 years, efforts of the pharmaceutical industry in natural product research has gradually declined8. The shift of focus to high-throughput screening of synthetic compound libraries is one reason for this decline, but the lack of improvement in traditional screening methods for natural products is another key factor. In such methods, when a positive result is obtained in a preliminary bioactivity screening, the active compounds must be purified in bioassay-guided steps and then identified using various analytical methods. These endeavors are extremely laborious and time-consuming, and often end in disappointing rediscoveries of known active compounds. Although genotyping of the bacteria prior to screening can reduce the chance of unproductive or duplicate efforts, it does not work well for bacteria with highly conserved 16s rRNA sequences. For example, our group recently reported several novel cyclopeptides – thalassospiramides from Thalassospira sp. TrichSKD10 which possessed effective inhibitory activity against human calpain-19. The screening of analogues with better activity and selectivity from other Thalassospira species is worthwhile. However, there are more than 100 Thalassospira samples in our laboratory and most of them were found to have highly conserved 16s rRNA sequences (data published soon). For bacteria that are not clearly identified to the species level, unproductive screening of all bacterial strains and manual examination of crude extracts to exclude rediscovery is unavoidable. Moreover, it is well known that even phylogenetically distant bacteria can sometimes produce similar compounds, mostly due to horizontal gene transfer of secondary metabolite-producing enzymes among bacteria10. In marine bacteria in particular, to ensure resilience in the dynamic marine environment, horizontal gene transfer occurs more frequently as a result of gene transfer agents produced by α-Proteobacteria11. Therefore, efficient methodology improvements are imperative to minimize the risk of such rediscoveries during the screening of marine bacteria, as well as to quickly guide researchers to the more promising bacterial strains.
In current practice, ribotyping, namely, the sequencing of the 16S rRNA gene, is often performed to obtain the assessment of bacterial diversity at the species or genus level12. However, there is ongoing debate regarding how to develop broadly accepted and useful definition of taxonomical levels for bacterial classification. Bacterial genomes are dynamic entities that include large numbers of genes acquired horizontally13, which may result in different phenotypic characteristics, adding new and unexpected levels of complexity to the problem of taxonomical classification. This necessitates more sensitive classification methods that can capture the complex evolutionary and environmental forces14,15. As secondary metabolite profiling is a more direct measurement of the phenotype of the bacteria than ribotyping, it can be viewed as a valuable supplement to the classical method of bacterial classification. Numerous studies have shown that the secondary metabolite profiles can help to distinguish strains within the same species. For example, many Streptomyces strains within the same species can have different secondary metabolite profiles15; nonpathogenic and pathogenic strains of Bacillus cereus can be distinguished by their secondary metabolites16.
To be able to use the secondary metabolite profiles of bacteria as “chemical fingerprints”, a reproducible method for extracting and detecting these compounds, as well as an efficient computational toolkit for managing and matching these fingerprints is required. Ultra-performance liquid chromatography coupled with high-resolution mass spectrometry (UPLC-HRMS) is considered as one of the most powerful tools to analyze metabolites because of the fast separation, high sensitivity, and excellent resolution17,18,19. Here, we present our work establishing a searchable chemical fingerprint database of marine bacteria using UPLC-HRMS, together with a software package that performs LC-MS alignment and fingerprint searching. These methods provide a complementary approach to current methods used for bacterial taxonomy and a useful tool for the screening of promising bioactive compounds in marine bacteria.
Results
Construction of a chemical fingerprint database of marine bacteria
To populate our secondary metabolite LC-MS fingerprint database, bacterial samples were collected from different marine habitats (including the euphotic zone, sediments and water column near deep-sea vents and seeps, and polar seas). In addition, because many antibiotic-producing bacterial strains are derived from eukaryotic hosts20,21,22,23 and some bioactive compounds are produced by bacteria associated with eukaryotic organism21,24,25, our database also included bacteria isolated from sponges, macroalgae, and tunicates, among others. To date, 1,430 bacterial strains collected from the marine environment were successfully cultured and 946 of them had been identified based on 16s rRNA sequences to 168 known species and several new species (for species name and phylogenetic relationship see Supplementary Table 1 and Supplementary Fig. 1). To manage and retrieve the large amount of LC-MS data, tailor-made software (MBMSearcher) was developed and tested. Secondary metabolites are extracted from bacterial lysates by an organic solvent, concentrated and analyzed by LC-MS. MBMSearcher processes the data automatically, integrating steps such as background subtraction, feature detection, and noise reduction. Together with any additional information (e.g. the source of the sample, the species identification by ribotyping, etc.), the profile of each bacterial sample is then stored in a relational database implemented with MySQL 5.5.36 (http://www.mysql.com/) for ready retrieval. Totally, six steps were required to build this database: 1) bacterial isolation and purification from different marine habitats; 2) bacterial identification based on their 16s rRNA gene sequences; 3) bacterial fermentation using standard medium, and extraction and concentration of secondary metabolites; 4) analysis by standard LC-MS; 5) raw data processing by MBMSearcher; and 6) importation into the chemical fingerprint database (flowchart see Supplementary Fig. 2, detailed information of sample preparation and software development see Methods).
Correlation of marine bacterial taxonomy and secondary metabolite profiles
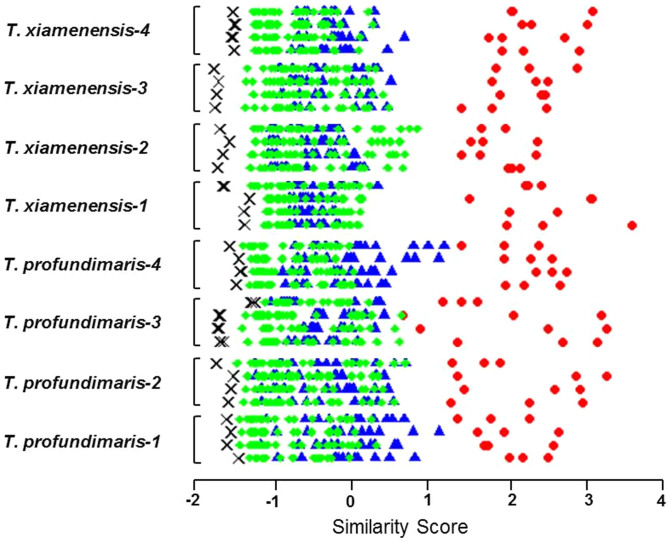
One purpose of developing this database was to study the relationship between the secondary metabolite profiles of marine bacteria and their taxonomy. As we previously discovered a number of novel cyclopeptides from different Thalassospira species (Gram-negative marine bacteria in the Rhodobacteraceae family)9, we first analyzed the metabolite profiles of this genus. We constructed a small test database of 8 Thalassospira strains (4 of Thalassospira xiamenensis and 4 of Thalassospira profundimaris), and one Pseudovibrio denitrificans strain also belonging to the Rhodobacteraceae family, as a negative control. To test the relationship between bacterial secondary metabolites and their taxonomy, 4 biological replicates of each strain were re-cultured and analyzed, and the resulting LC-MS maps were searched against our test database. The results showed that the secondary metabolite profiles of the same bacterial strain had a high similarity, indicating good reproducibility of our culture and analysis methodology (for representative UPLC chromatograms see Supplementary Fig. 3). However, the similarity of the metabolite profiles within a species was relatively low, despite that the similarity of their 16s rRNA gene sequences was over 99%. In some cases, the species-level similarity was even lower than that at the genus-level, most notably for the Thalassospira xiamenensis-2 samples (see Fig. 1). The negative control Pseudovibrio denitrificans showed the lowest similarity score among all of the test queries (see Fig. 1). These results indicate that although the overall trend supports a correlation between the similarity of the secondary metabolite profile and the level of taxonomical differentiation, classification based on the secondary metabolite profile is not in complete agreement with the ribotype defining the species. A similar observation was found using Bacillus subtilis strains, a model species collected from the marine environment, when searched against a much larger database (see Supplementary Fig. 4).
Figure 1. Similarity of secondary metabolite profiles of Thalassospira strains at different taxonomic levels.
A small test database was constructed using 8 Thalassospira strains (4 of T. xiamenensis and 4 of T. profundimaris) and 1 Pseudovibrio denitrificans strain (as negative control). Four biological replicates of each Thalassospira strains were cultured, and its secondary metabolite profiles are searched against the test database. The similarity scores between each biological replicate and all possible answers were plotted as data points on a horizontal line with the following color code: biological replicates of the same strain (red), different Thalassospira strains in the same species (blue), different Thalassospira species in the same genus (green), and the P. denitrificans strain (black). The secondary metabolite profiles of the same strain show high biological reproducibility, and the secondary metabolite profile similarity approximately, but not always, correlates with taxonomical similarity. The profiles of strains differing at the genus level are sometimes more similar than those differing at the species level.
Study of species-specific secondary metabolites
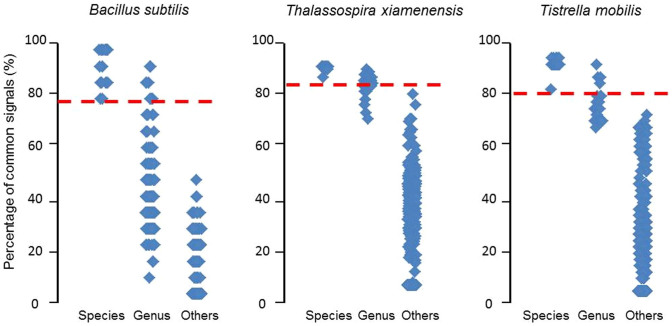
The production of some secondary metabolites is species-specific and can serve as a phenotypic trait associated with broadly distributed bacterial populations26. These metabolites could function as potential indices for bacterial classification. To identify potential species-specific metabolites, 5 reference strains of 3 dominant species in the database (Bacillus subtilis, Thalassospira xiamenensis and Tistrella mobilis) were obtained from the Marine Culture Collection of China, cultured in our laboratory, and analyzed using LC-MS. The common signals of 5 reference LC-MS profiles for each species were extracted (see Supplementary Table 2) and used as a species-specific classification index. All marine bacteria collected from natural environments in our database were then examined to determine how many of these index signals can be found. The results show that indeed, the number of signals detected correlated strongly with the taxonomical similarity (see Fig. 2). This is remarkable given that we employed only 5 reference strains for each species, and the results can likely be further improved with more data. With the expansion of our chemical fingerprint database, more reliable “species-specific signals” for a greater number of bacterial species could be extracted, and potentially identified to individual metabolites, which may reveal interesting biology in addition to being useful phenotypic classification indices.
Figure 2. Using common metabolite signals as a species-specific classification index.
Common metabolite signals were extracted from 5 reference strains of each species (B. subtilis, T. xiamenensis and T. mobilis). Then each LC-MS profile in the database is searched for the presence of these signals, and the percentage found was plotted as a data point. The data points are sorted by taxonomical level: different strains of the same species (Species), different species of the same genus (Genus), and different genera (Others). The results indicate that the number of found “species-specific signals” correlates with the taxonomical similarity, and that one can potentially use these “species-specific signals” as a phenotypic classification index, e.g., percentages above the red line indicate strains from the same species.
Use of this database to facilitate research of natural bioactive compounds
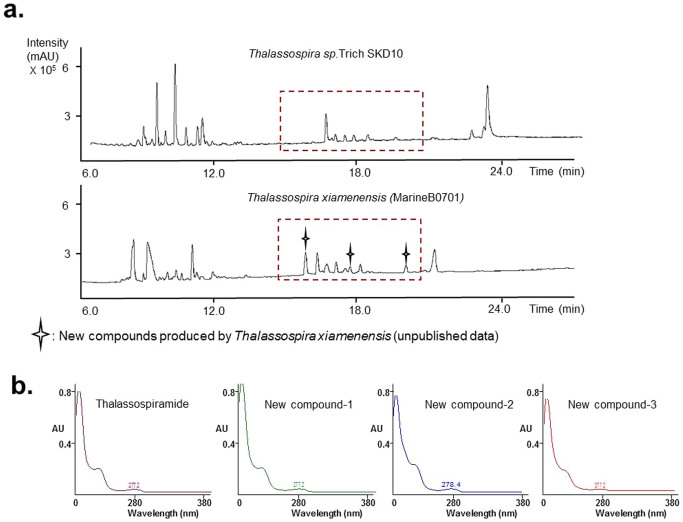
The database can be used to quickly identify relevant strains for bioactive compound screening. For example, as mentioned in the Introduction, we were interested in finding other producers of thalassospiramide analogues which may also have inhibitory activity against human calpain-1. To do so, the secondary metabolite profile of Thalassospira sp. TrichSKD10 (an original thalassospiramides-producing strain), was searched in our database, and the results showed that most of the highly ranked candidates were Thalassospira species (See Supplementary Table 3). The top 5 bacterial strains (marineB0718, marineB0711, marineB0701, marineB0717 and marineB0685) were re-cultured and analyzed by LC-MS. By comparing their LC-MS data, we found, surprisingly, that marine B0701 could produce not only 6 of the known thalassospiramides (thalassospiramide A, A1, A2, B, C and F, chemical structure and retention time see Supplementary Fig. 5) but also 3 new compounds (no hits in AntiBase database) with similar UV absorption and retention times as thalassospiramide (See Fig. 3). Further MSn and NMR analyses confirmed that these 3 compounds were analogues of thalassospiramide (unpublished data). This discovery would not have been possible in such a short time without our chemical fingerprint database, because we would have no way of knowing which of the dozens of Thalassospira strains in our collection would produce secondary metabolites similar to those of TrichSKD10.
Figure 3. Screening for strains with similar secondary metabolite profiles as Thalassospira sp. TrichSKD10 and the discovery of new thalassospiramide analogues.
(a) UPLC comparison of T. sp. TrichSKD10 and MarineB0701 (classified as T. xiamenensis by ribotyping) found by searching our database. The potentially novel compounds are labeled with a star. (b) Comparison of UV absorptions of thalassospiramide and 3 new compounds. The maximum absorption wavelengths of 4 compounds were very similar.
This database can also be used to facilitate novel biosynthetic pathway research and detect horizontal gene transfer among distant species. Once a compound with a unique and previously unknown structure was detected, the database can be searched to find all strains that potentially produce the same compound. As an example, thalassospiramide A, a typical thalassospiramide originally isolated from Thalassospira sp. CNJ328, possesses a 12-membered ring structure which makes it very difficult to synthesize chemically (see Supplementary Fig. 5)27. To examine whether thalassospiramide A is a species-specific compound or whether it can also be synthesized by other marine bacteria, we conducted a search using our software for compounds that satisfy 2 criteria: a signal at m/z 958.55 and retention time (13–14 minutes), with a MS fragment signal for a ring structure (m/z = 390.2) as an additional criterion (see Supplementary Fig. 5). Surprisingly, the search results revealed that MarineB0729 (identified to the species Tistrella bauzanensis by ribotyping) had the highest signal intensity at m/z 958.55; 1 Bacillus circulans strain, 1 Pseudovibrio denitrificans strain, and 4 Tistrella mobilis strains could produce this signal in addition to the Thalassospira strains (see Table 1). To exclude false positive results, all of the identified signals were subjected to MSn analysis, and only Bacillus circulans provided a false result. This finding suggested that the accuracy of the compound searching was reasonably good. The discovery of thalassospiramide A in non-Thalassospira strains suggested that horizontal gene transfer may have occurred among these marine bacteria, which is consistent with the recent report that two homologous gene clusters of thalassospiramides encoding a novel hybrid non-ribosomal peptide synthetase/polyketide synthase were discovered by the complete genome sequence analysis of 2 Tistrella and 2 Thalassospira strains9. Secondary metabolites that are produced by enzymes involved in horizontal gene transfer may allow marine bacteria to better cope with the various environmental changes associated with coastal and oceanic regions, and thus, the biosynthetic pathways for such compounds are of interest for future study.
Table 1. Searching for a specific compound among all strains in the database.
| Strain ID | Strain name | m/z | RT(min) | Intensity (×103) |
|---|---|---|---|---|
| MarineB0729 | Tistrella bauzanensis | 958.54 | 13.3 | 22194 |
| MarineB0492 | Thalassospira sp. | 958.55 | 13.2 | 3442 |
| MarineB0688 | Thalassospira lucentensis | 958.60 | 13.4 | 3437 |
| MarineB0699 | Thalassospira xiamenensis | 958.60 | 13.4 | 3435 |
| MarineB0462 | Thalassospira xiamenensis | 958.56 | 13.2 | 2958 |
| MarineB0493 | Thalassospira xiamenensis | 958.57 | 13.2 | 842 |
| MarineB0439 | Tistrella mobilis | 958.57 | 13.1 | 501 |
| MarineB0453 | Tistrella mobilis | 958.55 | 13.2 | 173 |
| MarineB0446 | Tistrella mobilis | 958.55 | 13.4 | 103 |
| MarineB0450 | Tistrella mobilis | 958.55 | 13.2 | 99 |
| MarineB0487 | Thalassospira sp. | 958.57 | 13.2 | 82 |
| MarineB0486 | Thalassospira lucentensis | 958.56 | 13.2 | 71 |
| Test sample | Thalassospira sp. CNJ328 | 958.55 | 13.5 | 45 |
| MarineB0548 | Bacillus circulans | 958.55 | 13.5 | 43 |
| MarineB0803 | Pseudovibrio denitrificans | 958.55 | 13.1 | 13 |
The compound thalassospiramide A was searched among all strains in the database, based on its m/z (958.55), retention time (13–14 min), and a characteristic fragment (m/z 390.2). The strain from which the compound was first discovered, Thalassospira sp. CNJ328, is labeled in red, and strains from non-Thalassospira species are labeled in blue.
Discussion
This paper reported our ongoing effort to establish a chemical fingerprint database of marine bacteria based on the LC-MS analytical platform. The database is expected to grow rapidly in the next few years, as we continually collect and isolate bacterial samples from different marine habitats. Besides, more information such as genome sequences, bioactivities, and identified metabolites can be added to this platform in the future. As we demonstrated with a few applications, the screening strategy for bioactive natural products from marine bacteria could be substantially improved with this database. An important distinction of our database and other resources for natural compound research is that ours is a strain-specific metabolome database, rather than a metabolite database. Unlike databases that focus only on the compounds and their analytical signatures (such as UV and MS2 spectra)28,29, our approach captures the whole secondary metabolite profiles of bacterial strains. This enables us to use the entire LC-MS profile for chemical fingerprinting, and can quickly determine whether a newly collected bacterium is similar to an existing strain. Besides, by using an effective time alignment algorithm and a longer LC running time (30 minutes), this fingerprint matching strategy greatly benefited from the retention time as a reliable coordinate for peak matching. This is especially useful for trace secondary metabolites because their UV and MS2 spectra are often unreliable. Moreover, our approach captures information of unknown metabolites as well, in the form of signals in LC-MS coordinates, even though most of these signals have yet to be identified to chemical structures. The ability of the database to search for similar signals across many strains allows researchers to focus their attention on potentially interesting molecules.
In summary, we expect that this database will become a useful resource for life science researchers of diverse interests. It should greatly improve the current procedure for natural compound screening, whether for pharmaceuticals or other uses. It should provide an additional dimension for classifying marine bacteria based on their phenotype. It should also help microbiologists discover novel biosynthetic pathways and study the interplay between bacteria and their environments. Lastly, although the database currently focuses on marine bacteria, it can in principle be extended to other organisms such as human pathogens, fungi and plants. The database management software is free and open-source and can be similarly deployed to develop LC-MS fingerprint databases for other realms of biology.
Software Available
The software MBMSearcher can be obtained at https://sourceforge.net/projects/mbmsearcher/ for individual research groups to maintain their own chemical fingerprint database. An online searching service based on this software was also applied (http://mbmsearcher.ust.hk/). Scientists can login, upload LC-MS profiles of unknown bacterial strains to the server and receive a detailed report showing a ranked list of bacteria from the database with similar secondary metabolite profiles, as well as a list of common signals (four standard LC-MS profiles can be downloaded and we will check any new LC-MS profile uploaded by other users). Besides, the distribution of specific metabolite in different bacteria can be listed by inputting its m/z and the range of retention time. (NOTE to editors and reviewers: Please log in using the user name “test” and password “test”. An individualized log-in system will be set up upon publication).
Methods
Isolation of marine bacteria
Each bacterium collected was cultivated on agar plates (yeast: 2 g.L−1, agar: 16 g.L−1, sea salt: 35 g.L−1) in the laboratory. Cultivable strains were purified via a replating process, and the pure bacterial strains were then cultured in a 50-mL falcon tube (tryptone: 10 g.L−1, yeast extract: 5 g.L−1, sea salt: 35 g.L−1, 200 rpm, 37°C, 3–5 days), transferred to glycerin tubes, labelled and stored at −80°C.
16s rRNA sequencing of culturable marine bacteria
The species-level classification was based on the bacterial 16s rRNA sequences. The DNA of individual bacterial strains was extracted according to a established protocol30. Bacterial cells were first lysed in 10 μL of lysozyme (100 mg.mL−1), and the protein was digested using 80 μL of 20% SDS and 8 μL of proteinase K (10 μg.μL−1). The DNA was extracted twice with chloroform:isoamyl alcohol (24:1) and then precipitated with 100% isopropanol, followed by washing with 75% ethanol. The quality and quantity of the DNA were checked using a NanoDrop ND-100 device (Thermo Fisher, USA) and by agarose gel electrophoresis. The V3–V5 region of the 16S rRNA gene was amplified using the following pair of primers: 341F (5′-CCTACGGGAGGCAGCAG-3′) and 907R (5′-CCGTCAATTCCTTTRAGTTT-3′). Each 20 μL of PCR reaction consisted of 4 μL of 5 × Phusion HF Buffer (M0530S New England BioLabs Inc.), 1.6 μL of dNTPs (2.5 μM each), 1 μL of forward and reverse primer (10 μM), 0.6 μL of DMSO, 10 ng of template DNA, 0.2 μL of Phusion® High-Fidelity DNA Polymerase (0.4 units), and 10.6 μL of pure water. The PCR was performed with a thermal cycler (Bio-Rad, USA) using the following program: an initial denaturation at 98°C for 1 min; 25 cycles at 98°C for 10 s, 60°C for 30 s and 72°C for 20 s; and a final extension at 72°C for 5 min. Sequences were assigned according to the Ribosomal Database Project (RDP) Classifier (version 2.2)31,32 of the QIIME pipeline using the Silva108 database32 with a confidence level of 80%.
Fermentation and secondary metabolite extraction
A standardized culture protocol is necessary to enable reproducible fingerprint matching. To obtain sufficient quantities of crude bacterial extracts, each cultivable bacterial strain was fermented in a 250-mL flask. In general, the production of secondary metabolites is highly dependent on the culture medium; the complexity of the culture medium correlates with the chemical diversity of the bacteria2. Therefore, GYPT medium (glucose: 10 g.L−1, yeast extract: 2 g.L−1, tryptone: 2.5 g.L−1, peptone: 2.5 g.L−1, sea salt: 35 g.L−1) was chosen as the standard medium for the culture of the marine bacteria. The bacteria were allowed to grow for 36 ~ 60 hours based on their growth status, until OD600 reached 0.8. As the bacterial broth contained large amounts of sea salt and culture medium, ethyl acetate (EA) was selected to extract the secondary metabolites due to its moderate polarity and low boiling point. The bacterial broth was extracted with twice the volume of EA, and then the suspension was stirred for 10 min. The supernatant was collected, and dried chemical extracts were obtained by complete evaporation of the solvent. In order to reduce noise signal and get a valid LC-MS profile of crude bacterial extract, a flask only containing GYPT medium was also prepared in each fermentation batch and used as a blank sample for subsequent background subtraction by MBMSearcher (the common features shared by the blank sample and each biological sample were removed from the corresponding feature map). Biological replicates were prepared by selecting separate colonies from the agar plate containing the pure bacterial culture, and growing them under the same culture conditions.
LC-MS analysis of crude extracts
The crude extract was analyzed by UPLC (Waters ACQUITY, USA) coupled with the MicroTOF-MS system (Bruker Daltonics GmbH, Bremen, German). Reverse-phase chromatography was conducted in 2.1 × 150 mm columns (Waters, BEH C18, 1.7 μm, USA) at a flow rate of 250 μL min−1, and samples were eluted by gradient mobile phase (5% to 95% acetonitrile with 0.1% formic acid in water) over 30 minutes. The UPLC-separated samples were then analyzed by TOF-MS (mass range: 100–2000 Da) by electrospray ionization (4.5 kV, 0.2 Bar, positive ion mode), and the raw LC-MS data were converted to XML format by CompassXport (http://www.bruker.com) to build the database.
Feature detection and alignment of LC-MS profile data
We developed a software tool, MBMSearcher to perform all data processing and database searching automatically. MBMSearcher automatically detects and extracts signals from the loaded data using the FeatureFinder tool in the OpenMS suite33, and stores the feature maps in its database. The “centroided” mode was chosen for FeatureFinder, and the m/z, retention time (RT), and intensity ranges were (200 Da/e, ∞), (3 min, 25 min) and (300, ∞), respectively. Since the retention time of an analyte depends heavily on the conditions of the chromatographic column in LC-MS, and is often prone to shifts and distortions, an effective time alignment algorithm is an important component of MBMSearcher. This is especially critical considering experiments will be conducted over a period of months, if not years. For efficiency, we adopted a “feature-based” approach to the time alignment problem. That is, we first applied an existing feature-finding algorithm33 to detect “features” in the LC-MS profile data, which were intense signals that were likely to originate from an analyte in the sample (see Supplementary Fig. 6). The detected features in each LC-MS profile formed a feature map, which could then be aligned with other feature maps using a recently developed RT alignment method, LWBMatch34. The time alignment step automatically corrected the RT shifts to match features belonging to the same analyte across different LC-MS runs, based on an efficient, weighted bipartite matching34,35. The parameters for LWBMatch included the retention time and m/z tolerances. If one feature is located outside another feature's retention time or m/z tolerances, LWBMatch will exclude the possibility that this is a matching pair in its algorithm. In our experiments, 60 s and 0.2 Da/e were used. This alignment step was performed in both the building and searching components of the LC-MS fingerprint database. Background subtraction was accomplished by first detecting features in the LC-MS map of a a blank sample, aligning these features with those in LC-MS maps of the bacterial samples, and then excluding any shared features between them.
Consensus map generation
To increase the accuracy and consistency of the fingerprint database, at least 2 technical replicates for each sample were generated and a consensus map was constructed by including only common features shared by these replicates. This procedure was accomplished by first aligning the feature maps of technical replicates using LWBMatch34. Matching feature tables were then generated, and all of the shared features were included in a consensus map, which was used as the reference fingerprint for that bacterial sample (see Supplementary Fig. 7).
Searching the database for similar LC-MS profiles
After completion of the alignment, MBMSearcher would generate a matching table based on the alignment results in which each row was a list of features from each aligned experiment. Next, a similarity score was calculated between the pair of LC-MS profiles (in our case simplified to feature maps) based on the complete feature-to-feature mapping information stored in the matching table. In general, samples that shared a greater number of common compounds had more similar profiles. In our application, we defined the similarity score as a rank-transform dot product36,37,38 between the feature vectors (shown in Supplementary Fig. 8). The detailed algorithm used to calculate similarity scores between two LC-MS feature maps is described in the Supplementary Method and Supplementary Fig. 9. For an unknown sample, the feature map of the query was compared sequentially with all of the feature maps stored in the fingerprint database. For the comparison of each pair, an alignment was first performed using LWBMatch, and then, the similarity score was calculated based on the results of the alignment. Finally, a ranked list was collected as the output, in which the candidate fingerprints in the database were sorted based on their similarity scores (from highest to lowest).
Supplementary Material
supporting information
Acknowledgments
This study was supported by grants from the China Mineral Resources Research and Development Association (COMRRDA12SC01 and COMRRDA12SC02), the Sanya Institute of Deep Sea Science and Engineering (SIDSSE-201206) and the King Abdullah University of Science and Technology (SA-C0040/UK-C0016).
Footnotes
The authors declare no competing financial interests.
Author Contributions L.L., J.W., Y.X., H.L. and P.Q. designed the experiments, L.L., K.W. and Y.L. executed the experiments, J.W., Y.H., H.L. and B.Y. developed analytical tools, L.L., J.W., R.T., Q.L., Z.S. and W.P. collected and analyzed data and L.L. wrote the manuscript with input from all authors.
References
- Schmidt C. Metabolomics takes its place as latest up-and-coming “omic” science. J. Natl. Cancer. Inst. 96, 732–734 (2004). [DOI] [PubMed] [Google Scholar]
- Frisvad J. C., Andersen B. & Thrane U. The use of secondary metabolite profiling in chemotaxonomy of filamentous fungi. Mycol. Res. 112, 231–240 (2008). [DOI] [PubMed] [Google Scholar]
- Whitman W. B., Coleman D. C. & Wiebe W. J. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. USA 95, 6578–6583 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenical W. Chemical studies of marine bacteria - developing a new resource. Chem. Rev. 93, 1673–1683 (1993). [Google Scholar]
- Goodacre R. et al. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 22, 245–252 (2004). [DOI] [PubMed] [Google Scholar]
- Blunt J. W. et al. Marine natural products. Nat. Prod. Rep. 26, 170–244 (2009). [DOI] [PubMed] [Google Scholar]
- Laatsch H. Antibase (2010).
- Li J. W. H. & Vederas J. C. Drug Discovery and Natural Products: End of an Era or an Endless Frontier? Science 325, 161–165 (2009). [DOI] [PubMed] [Google Scholar]
- Ross A. C. et al. Biosynthetic Multitasking Facilitates Thalassospiramide Structural Diversity in Marine Bacteria. J. Am. Chem. Soc. 135, 1155–1162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach M. A. Antibiotics from microbes: converging to kill. Curr. Opin. Microbiol. 12, 520–527 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel L. D. et al. High Frequency of Horizontal Gene Transfer in the Oceans. Science 330, 50–50 (2010). [DOI] [PubMed] [Google Scholar]
- Rossello-Mora R. & Amann R. The species concept for prokaryotes. FEMS. Microbiol. Rev. 25, 39–67 (2001). [DOI] [PubMed] [Google Scholar]
- Ochman H., Lawrence J. G. & Groisman E. A. Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304 (2000). [DOI] [PubMed] [Google Scholar]
- Ochman H., Lerat E. & Daubin V. Examining bacterial species under the specter of gene transfer and exchange. Proc. Natl. Acad. Sci. USA 102, 6595–6599 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P. R. Linking species concepts to natural product discovery in the post-genomic era. J. Ind. Microbiol. Biotechnol. 37, 219–224 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy J. G. et al. Discrimination of pathogenic clinical isolates and laboratory strains of Bacillus cereus by NMR-based metabolomic profiling. FEMS. Microbiol. Lett. 242, 127–136 (2005). [DOI] [PubMed] [Google Scholar]
- Mazzeo J. R., Neue U. D., Kele M. & Plumb R. S. A new separation technique takes advantage of sub-2-mu m porous particles. Anal. Chem. 77, 460A–467A (2005). [Google Scholar]
- Bertrand S. et al. Detection of metabolite induction in fungal co-cultures on solid media by high-throughput differential ultra-high pressure liquid chromatography-time-of-flight mass spectrometry fingerprinting. J. Chromatogr. A 1292, 219–228 (2013). [DOI] [PubMed] [Google Scholar]
- Makarov A. Electrostatic axially harmonic orbital trapping: A high-performance technique of mass analysis. Anal. Chem. 72, 1156–1162 (2000). [DOI] [PubMed] [Google Scholar]
- Wietz M. et al. Wide Distribution of Closely Related, Antibiotic-Producing Arthrobacter Strains throughout the Arctic Ocean. Appl. Environ. Microbiol. 78, 2039–2042 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. W. et al. Soaking it up: the complex lives of marine sponges and their microbial associates. ISME J. 1, 187–190 (2007). [DOI] [PubMed] [Google Scholar]
- Thomas T. et al. Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J. 4, 1557–1567 (2010). [DOI] [PubMed] [Google Scholar]
- Gilturnes M. S., Hay M. E. & Fenical W. Symbiotic marine bacteria chemically defend crustacean embryos form a pathogenic fungus. Science 246, 116–118 (1989). [DOI] [PubMed] [Google Scholar]
- Xu Y. et al. Bacterial Biosynthesis and Maturation of the Didemnin Anti-cancer Agents. J. Am. Chem. Soc. 134, 8625–8632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E. W. & Donia M. S. Life in cellulose houses: symbiotic bacterial biosynthesis of ascidian drugs and drug leads. Curr. Opin. Biotechnol. 21, 827–833 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P. R. et al. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl. Environ. Microbiol. 73, 1146–1152 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D. C. et al. Thalassospiramides A and B, immunosuppressive peptides from the marine bacterium Thalassospira sp. Org. Lett. 9, 1525–1528 (2007). [DOI] [PubMed] [Google Scholar]
- Henrich C. J. & Beutler J. A. Matching the power of high throughput screening to the chemical diversity of natural products. Nat. Prod. Rep. 30, 1284–1298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T. & Masubuchi M. Dereplication of microbial extracts and related analytical technologies. J. Antibiot. 1, 1–8 (2014). [DOI] [PubMed] [Google Scholar]
- Lee O. O., Wong Y. H. & Qian P. Y. Inter- and Intraspecific Variations of Bacterial Communities Associated with Marine Sponges from San Juan Island, Washington. Appl. Environ. Microbiol. 75, 3513–3521 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M. & Cole J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E. et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlbacher O. et al. TOPP - the OpenMS proteomics pipeline. Bioinformatics 23, E191–E197 (2007). [DOI] [PubMed] [Google Scholar]
- Wang J. J. & Lam H. Graph-based peak alignment algorithms for multiple liquid chromatography-mass spectrometry datasets. Bioinformatics 29, 2469–2476 (2013). [DOI] [PubMed] [Google Scholar]
- West D. Introduction to graph theory (second edition). ISBN 0-13-014400-2, Prentice Hall, (2001). [Google Scholar]
- Frank A. M. A ranking-based scoring function for peptide-spectrum matches. J Proteome Res. 8, 2241–2252 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen C. Y., Houel S., Ahn N. G. & Old W. M. Spectrum-to-spectrum searching using a proteome-wide spectral library. Mol. Cell. Proteomics 10, M111.007666 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W., Zhu K. & Lam H. Refining similarity scoring to enable decoy-free validation in spectral library searching. Proteomics 13, 3273–83 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supporting information