ABSTRACT
Insects and other animals commonly form symbioses with heritable bacteria, which can exert large influences on host biology and ecology. The pea aphid, Acyrthosiphon pisum, is a model for studying effects of infection with heritable facultative symbionts (HFS), and each of its seven common HFS species has been reported to provide resistance to biotic or abiotic stresses. However, one common HFS, called X-type, rarely occurs as a single infection in field populations and instead typically superinfects individual aphids with Hamiltonella defensa, another HFS that protects aphids against attack by parasitic wasps. Using experimental aphid lines comprised of all possible infection combinations in a uniform aphid genotype, we investigated whether the most common strain of X-type provides any of the established benefits associated with aphid HFS as a single infection or superinfection with H. defensa. We found that X-type does not confer protection to any tested threats, including parasitoid wasps, fungal pathogens, or thermal stress. Instead, component fitness assays identified large costs associated with X-type infection, costs which were ameliorated in superinfected aphids. Together these findings suggest that X-type exploits the aphid/H. defensa mutualism and is maintained primarily as a superinfection by “hitchhiking” via the mutualistic benefits provided by another HFS. Exploitative symbionts potentially restrict the functions and distributions of mutualistic symbioses with effects that extend to other community members.
IMPORTANCE Maternally transmitted bacterial symbionts are widespread and can have major impacts on the biology of arthropods, including insects of medical and agricultural importance. Given that host fitness and symbiont fitness are tightly linked, inherited symbionts can spread within host populations by providing beneficial services. Many insects, however, are frequently infected with multiple heritable symbiont species, providing potential alternative routes of symbiont maintenance. Here we show that a common pea aphid symbiont called X-type likely employs an exploitative strategy of hitchhiking off the benefits of a protective symbiont, Hamiltonella. Infection with X-type provides none of the benefits conferred by other aphid symbionts and instead results in large fitness costs, costs lessened by superinfection with Hamiltonella. These findings are corroborated by natural infections in field populations, where X-type is mostly found superinfecting aphids with Hamiltonella. Exploitative symbionts may be common in hosts with communities of heritable symbionts and serve to hasten the breakdown of mutualisms.
KEYWORDS: Hamiltonella defensa, PAXS, entomopathogenic fungi, heat stress, heritable symbionts, parasitoids, pea aphid resistance
INTRODUCTION
Heritable bacterial symbionts are ubiquitous in arthropod species, especially insects occupying terrestrial ecosystems (1). These heritable symbionts are usually maternally transmitted and cannot survive outside host tissues and consequently have limited strategies for reaching new hosts (2–4). Given that host fitness and symbiont fitness are intimately linked, symbionts can invade host populations by providing net fitness benefits, often by mediating resource acquisition or providing protection against natural enemies (5). Alternatively, heritable symbionts can invade populations by manipulating host reproduction to favor infected females (6, 7). Moreover, individual hosts are often infected with multiple inherited symbionts and antagonistic or synergistic interactions between superinfecting symbionts may enhance or reduce mutualistic services or costs, influencing symbiont spread within the host population (8–10). Instances of heritable pathogens are expected to be exceedingly rare (1), yet under some conditions, a heritable symbiont that was neither providing conditional benefits nor manipulating host reproduction could spread by “hitchhiking” alongside a beneficial symbiont (11). A hitchhiking symbiont could be maintained in host populations even when superinfection causes harm to host fitness as long as the conditional benefits provided by the mutualist in the superinfection context outweigh these costs. Such symbionts can act as third-party exploiters and potentially interfere with the services conferred by beneficial symbionts, altering spatial and temporal dynamics of the symbioses and possibly hastening the breakdown of the mutualism (12).
The pea aphid Acyrthosiphon pisum (Hemiptera: Aphididae) has emerged as a model organism for studying phenotypic effects of infection with heritable facultative symbionts (HFS) (3). This is due to the diverse assemblage of heritable symbiont species infecting this aphid combined with its clonal reproduction and the relative ease in manipulating heritable symbiont communities (3). All pea aphids carry the obligate symbiont Buchnera aphidicola, which provides essential nutrients not found in sufficient quantities in their phloem diet (13, 14). In addition, most individual pea aphids are variably infected with up to seven species of HFS, which confer diverse protective benefits to the aphid host (3, 8). For example, Hamiltonella defensa confers resistance to the dominant parasitoid of the pea aphid, Aphidius ervi (15, 16). The mechanism of resistance is not known but requires that the bacterial symbiont be infected with the bacteriophage APSE, which encodes eukaryotic toxins hypothesized to harm A. ervi larvae developing within the aphid (17–19). Other HFS commonly found infecting pea aphids and associated with protective roles include Serratia symbiotica, which confers resistance to thermal stress, and Regiella insecticola, Spiroplasma, Rickettsia, and Rickettsiella viridis, which protect against an aphid-specific fungal pathogen, Pandora neoaphidis (8, 20–24).
Despite the pea aphid's status as a model for studying HFS, relatively little is known about the consequences of infection with some species, including the informally named X-type (also known as PAXS, for pea aphid X-type symbiont [25]). This common HFS often infects up to 40% of individuals in some North American populations (8, 11). Sequences of the X-type 16S rRNA gene place this HFS in the Enterobacteriaceae (Gammaproteobacteria), along with other aphid symbionts, including Buchnera and H. defensa, as well as notable human pathogens, such as Salmonella (25). Interestingly, population-level studies show that X-type occurs uncommonly as a single infection but rather is typically found “superinfecting” individual pea aphids with the defensive symbiont H. defensa (8). Here we define superinfection as an infection with >1 HFS species in an individual aphid (e.g., aphid with H. defensa plus X-type) and excluding Buchnera, which is present in all individuals. While HFS superinfections are common in pea aphids, most HFS species also occur frequently as single infections (8, 26). Host-level selection can result in the spread of particular combinations of synergistic HFS (e.g., multiple distinct benefits conferred) or remove combinations exhibiting increased costs (relative to singly infected and uninfected aphids); costs to the symbiont may occur over competition for space or resources within the host (4, 8, 10, 27–29). Nonselective factors may also be important in HFS maintenance, as vertical transmission rates are generally expected to be lower in superinfections than in single infections (8–10, 27, 30), although transmission rates of either partner may vary for specific HFS pairings. The propensity of X-type to occur only as a superinfection suggests that it may act synergistically by improving the performance of superinfected aphids, either by providing distinct benefits or by enhancing those of H. defensa. Alternatively, X-type may deploy a cheating strategy and spread by “hitchhiking” with the defensive mutualist H. defensa (4, 9).
Building on prior research indicating that H. defensa-conferred protection against the parasitoid A. ervi likely fails at warmer temperatures (31), a subsequent study identified a possible synergistic role for X-type, as symbiont-based protection was not lost at higher temperatures in aphids infected with both X-type and H. defensa (25). These studies did not control for variation in aphid and symbiont genotypes, which potentially contributed to the observed results, especially given the later findings that some pea aphid genotypes are resistant to A. ervi and that endogenous aphid resistance does not fail at high temperatures (32, 33). Furthermore, aphids were dissected prior to parasitoid pupation such that effects caused by strains of H. defensa (e.g., those with APSE2) that kill wasps late in development (32) may not have been observed. Indeed, in a separate study which controlled for aphid genotypic effects, we found no evidence that X-type rescued loss of H. defensa-based protection at high temperatures, nor did we find that X-type provided any protection against the wasp A. ervi as a single infection (33). However, several studies with aphids, including pea aphids, show that symbiont-based protection against parasitoids can be specialized to particular parasitoid species (34–37), rendering it possible that X-type provides protection specifically against the only other parasitoid, Praon pequodorum, commonly attacking the pea aphid in North America (38). In European pea aphids, some X-type strains have been implicated in providing protection from thermal stress, as well as challenges by the wasp A. ervi and the fungal pathogen P. neoaphidis (39). In this previous study, X-type could not be isolated from the superinfecting symbiont Spiroplasma, but lines with X-type and Spiroplasma generally received more protection than lines with only Spiroplasma, which has also been shown to provide protection against P. neoaphidis (24). In North American pea aphid populations, Spiroplasma is less common and does not frequently partner with X-type (8, 33).
In summary, several reports indicate that X-type provides diverse protective roles to pea aphids, yet no benefits have been established for X-type as a single infection, and these other studies have not fully separated effects from coinfecting symbionts and aphid genotypes. Here we examined roles of X-type as a single infection and superinfection with its common partner H. defensa. This was accomplished by establishing experimental lines sharing aphid genotype, yet differing in the four possible symbiont infection types: uninfected with HFS (UC), infected with H. defensa only (H), infected with X-type only (X), and superinfected (XH). Using these lines, we examined the constitutive cost of harboring the X-type symbiont, both as a single infection and as a superinfection. We then investigated whether X-type infection results in increased host protection against two natural enemies—P. pequodorum, a species of parasitoid wasp native to North America, and the fungal pathogen P. neoaphidis—as well as thermal stress. We also estimated symbiont titers using quantitative PCR (qPCR) to investigate possible interactions between these two symbionts. Finally, we analyzed strain variation using a multilocus typing approach and assessed the phylogenetic placement of North American X-type based on several housekeeping genes of related bacteria. In doing so, we hope to deliver a robust account of X-type-induced effects on the aphid phenotype in an effort to determine the propagation strategy of this common heritable symbiont.
RESULTS
X-type infection is highly costly as a single infection and as a superinfection.
We found substantial reductions in aphid fecundity for aphids harboring X-type relative to uninfected controls (Fig. 1; Table 1). Surprisingly, while H. defensa infection resulted in significant increases in fecundity compared to that of controls, superinfections with X-type not only eliminated this benefit but also significantly decreased fecundity, almost to levels seen in X-type single infections (Fig. 1). X-type-infected aphids (X and XH) reproduced at rates similar to those in controls for the first 3 to 6 days of adulthood, at which point reproduction was abruptly curtailed (Fig. 1). This cessation of reproduction is not due to shortened life span, as X-type-infected aphids exhibited an estimated time to 0.5 survival (i.e., time point at which 50% of aphids were still alive) similar to those of H. defensa-infected aphids and uninfected aphids (Table 1). Interestingly, this reproduction cessation (3 to 6 days postadulthood) coincides with a severe phenotypic change seen in X-type-infected aphids (both X and XH), in which the aphids become extremely engorged and occasionally produce small, developmentally incomplete and inviable offspring (Fig. 2A). Dissections of these large, bloated aphids revealed only a few malformed offspring, while uninfected aphids of the same age contained ∼15 to 20 properly developed offspring (Fig. 2B and C).
FIG 1.
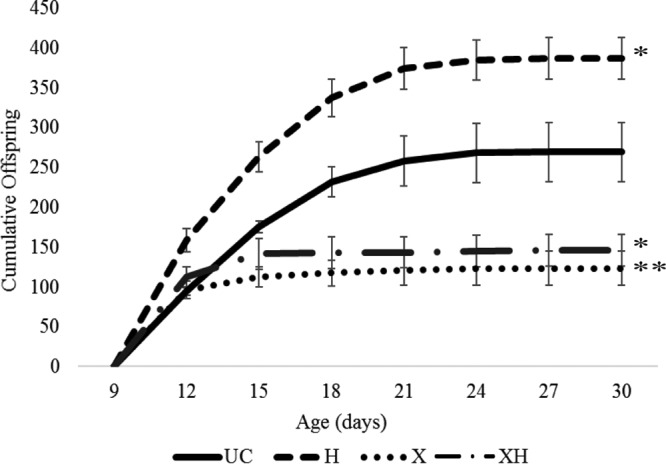
Cumulative fecundity in the absence of natural enemies. Bars represent standard errors. UC, uninfected with HFS; H, infected with H. defensa only; X, infected with X-type only; XH, infected with X-type plus H. defensa. Total fecundity at day 30 was compared among lines using Dunnett's test with the UC line as control. Significant differences between lines are marked in Table 1. *, P < 0.01; **, P < 0.001.
TABLE 1.
Aphid development and fecundity at 20°Ca
| Line | Time to first reproduction (h), mean ± SE (P value) | Fresh wt (mg), mean ± SE (P value) | Cumulative 30-day fecundity (no. of offspring/female), mean ± SE (P value) | Time (days) to 0.5 survival (n = 30) |
|---|---|---|---|---|
| UC | 237.13 ± 2.49 | 3.13 ± 0.16 (1.0) | 268.5 ± 37.23 (1.0) | 19.15 |
| H | 246.66 ± 3.61 (0.2166) | 3.11 ± 0.092 (0.9983) | 386.17 ± 26.07 (0.0153) | 19.58 |
| X | 227.62 ± 2.36 (0.0196) | 3.63 ± 0.10 (0.0163) | 123 ± 21.44 (0.003) | 18.91 |
| XH | 230.67 ± 2.54 (0.0399) | 3.65 ± 0.14 (0.0111) | 145.83 ± 19.79 (0.0118) | 16.87 |
Analyzed with multiple Wilcoxon rank sum tests (with Benjamini-Hochberg correction; α = 0.0214); only comparisons to the uninfected control line are included. Fresh weight, body size, and fecundity were compared using Dunnett's test with the uninfected line as the control. Survival data were fit to a lognormal distribution for estimates of 0.5 survivorship; α = 0.05.
FIG 2.

(A) X-type-infected adults exhibiting the “puffy” phenotype (∼12 days old). The red arrow indicates aborted offspring. (B and C) Embryos from an ∼12-day-old dissected uninfected aphid (B) and embryos from an ∼12-day-old dissected X-type-infected aphid (C). The live-aphid photo (A) is courtesy of Laura Kraft (reprinted with permission); dissection photos (B and C) are by Matthew R. Doremus.
Single infection with X-type resulted in significantly faster aphid development than for both uninfected and H. defensa-infected lines (Table 1). Aphids infected with X-type (both X and XH) were significantly larger and heavier at adulthood (measured as period from birth to first reproduction [TFR]) than uninfected aphids (UC and H) (Table 1). H. defensa-only-infected aphids did not differ significantly in development time, weight, or body size from the uninfected control (Table 1).
X-type infection does not protect aphids from heat stress, but heat shock ameliorates costs in superinfections.
To account for the increased mortality that can affect fecundity at later time points, reproductive output was analyzed in terms of both cumulative fecundity and reproduction rate (offspring of each living aphid/day). Surprisingly, within-line comparisons (heat shock versus 20°C control) revealed that exposure to heat shock generally increased total fecundity (Student's t test: TX = 1.81 and P = 0.0384; TXH = 4.28 and P = 0.0001) and rate of reproduction (TX = 2.24 and P = 0.0148; TXH = 3.14 and P = 0.0015) (Table 2) of aphids infected with X-type (X) or superinfected with both X-type and H. defensa (XH). However, heat shock had no effect on total fecundity (TUC = 0.332 and P = 0.742; TH = 0.11 and P = 0.91) or rate of reproduction (TUC = 0.395 and P = 0.695; TH = −0.361 and P = 0.72) (Table 2) of uninfected (UC) and H. defensa-infected (H) aphids compared to 20°C controls. In among-line comparisons to the uninfected control, X-type-only-infected aphids again exhibited greatly reduced fecundity and reproductive rate; thus, despite increased reproductive output post-heat shock, these aphids still suffered a severe reduction in fecundity relative to that of uninfected controls (Table 2). In contrast, H. defensa infection did not impose a cost to total fecundity or reproductive rate in either treatment compared to the uninfected control (Table 2), but we also did not observe fecundity increases as seen in the first assay. Finally, superinfected aphids also exhibited a reduction in fecundity and reproductive rate at 20°C, but interestingly, this cost was eliminated in the heat shock assay, in which the reproduction of superinfected aphids was on par with that of controls (Table 2).
TABLE 2.
Aphid development and fecundity following heat shocka
| Line | Temp treatment | Time to first reproduction (h), mean ± SE (P value) |
Daily fecundity (no. of offspring/female), mean ± SE (P value) |
||
|---|---|---|---|---|---|
| Compared to UC | Compared to 20°C | Compared to UC | Compared to 20°C | ||
| UC | Control (20°C) | 242 ± 3.18 (1.0) | (0.1626) | 5.00 ± 0.31 (1.0) | (0.6951) |
| Heat shock (37.5°C) | 251.9 ± 4.52 (1.0) | 5.16 ± 0.29 (1.0) | |||
| H | Control (20°C) | 243.35 ± 4.6 (0.782) | (0.1555) | 4.65 ± 0.34 (0.4638) | (0.7202) |
| Heat shock (37.5°C) | 251.44 ± 3.82 (0.8125) | 4.50 ± 0.20 (0.1081) | |||
| X | Control (20°C) | 228.03 ± 5.57 (0.0063) | (0.0055) | 1.99 ± 0.22 (<0.0001) | (0.0296) |
| Heat shock (37.5°C) | 245.93 ± 4.69 (0.4075) | 2.64 ± 0.18 (<0.0001) | |||
| XH | Control (20°C) | 234.56 ± 3.64 (0.1338) | (0.8738) | 3.89 ± 0.29 (0.0097) | (0.0029) |
| Heat shock (37.5°C) | 236.74 ± 2.92 (0.0083) | 5.10 ± 0.26 (0.9960) | |||
Analyzed under control and heat shock conditions with multiple Wilcoxon rank sum tests (Bonferroni corrected; α = 0.0083), with comparisons to the control line (among lines) and 20°C (within-line comparison; P values only) included. Daily fecundity was compared using Dunnett's test with UC; α = 0.05.
Consistent with our first fitness assay, conducted only at 20°C, aphids infected singly with X-type reached developmental maturity faster than uninfected or H. defensa singly infected aphids in both the presence and absence of heat stress (Table 2). Superinfected aphids also developed faster than uninfected control aphids when exposed to heat stress (Table 2). Heat shock significantly delayed reproduction of X-type singly infected aphids compared to that of their 20°C counterparts, while the other aphid lines (UC, H, and XH) experienced only a minor delay in reproduction. Interestingly, the delay was slightest for superinfected aphids. Infection with HFS (X-type or H. defensa) as a single infection or superinfection, as well as heat shock, had no effect on aphid survival to reproduction, although there was a nonsignificant reduction in survival in all aphid lines in the heat shock treatment and in singly infected aphids (both HFS species) in both temperature treatments (logistic regression equation [LRE], Y = −0.059 + 0.085XH − 0.299X − 0.152H − 0.154temp; likelihood ratio test [LRT], FXH = 0.202 and P = 0.653; FX = 2.27 and P = 0.132; FH = 0.611 and P = 0.435; Ftemp = 1.13 and P = 0.287).
X-type infection does not confer protection against the parasitoid P. pequodorum.
Single infection with X-type did not reduce P. pequodorum parasitism success (Fig. 3) (LRE, Y = −0.17 − 0.03XH − 0.03H − 0.28X; LRT, FX = 0.935 and P = 0.3335), nor did aphids superinfected with X-type and H. defensa-APSE8 receive any protection against P. pequodorum (Fig. 3) (LRT, FXH = 0.0061 and P = 0.9376). As seen in prior studies using H. defensa carrying different APSE variants, including APSE8, single infection with the present study's strain of H. defensa with APSE8 did not confer any protection against attack by the parasitoid P. pequodorum (Fig. 3) (LRT, FH = 0.011 and P = 0.9155 [36]).
FIG 3.
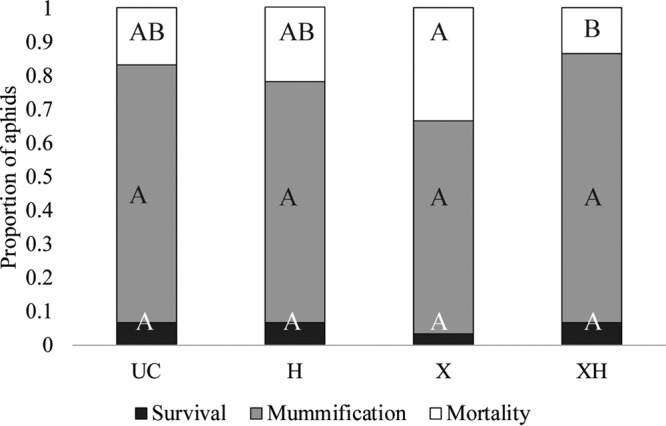
Mean aphid survival, mummification, and mortality after parasitism by P. pequodorum. Different letters represent significant differences determined by ANOVA with Tukey's HSD test after arcsin transformation of data (α = 0.05).
X-type infection does not confer protection against specialized fungal pathogens.
Neither single infection with X-type nor superinfection with both HFS increased aphid survival when exposed to P. neoaphidis (Fig. 4) (LRE, Y = −1.10 + 0.09H + 0.07X + 0.07XH +0.79R; LRT, FX = 0.67 and P = 0.4118; LRT, FXH = 0.69, P = 0.4062). Infection with H. defensa also did not significantly increase aphid survival against attack by P. neoaphidis (LRT, FH = 1.07 and P = 0.3004); however, a significant increase in survival was seen with R. insecticola infection (LRT, FR = 10.76 and P = 0.001).
FIG 4.
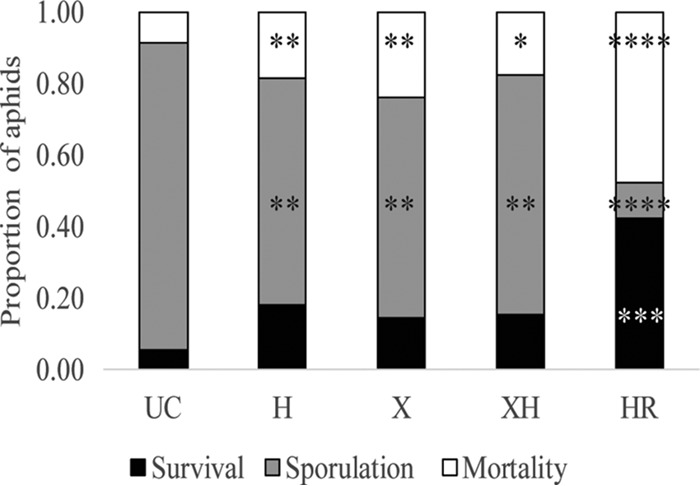
Mean aphid survival, sporulation, and mortality after exposure to P. neoaphidis. Asterisks represent significant differences compared to the uninfected control line (UC) determined by logistic regression with a likelihood ratio test using arcsin-transformed data (α = 0.05). HR, H. defensa plus R. insecticola. *, 0.05 > P > 0.01; **, 0.01 > P > 0.001; ***, 0.001 > P > 0.0001; ****, 0.0001 > P > 0.00001.
Both X-type and H. defensa single infections resulted in decreased fungal sporulation compared to that of the uninfected aphid line (Fig. 4) (LRE, Y = 0.27 − 0.637H − 0.571X − 0.561XH − 1.28HR; LRT, FH = 11.56 and P = 0.0007; LRT, FX = 8.33 and P = 0.0039). Additionally, superinfection resulted in a similar, minor decrease in sporulation compared to that of the uninfected line, suggesting that superinfection with X-type and H. defensa does not provide any additive benefits in this interaction (LRT, FXH = 8.53 and P = 0.0035). In all three of these cases (X, H, and XH) reductions in sporulation were small relative to that observed for aphids infected with R. insecticola (Fig. 4) (LRT, FR = 29.55 and P ≤ 0.0001 [24, 39]). HFS-infected lines also showed increased host mortality after pathogen exposure, which may be responsible for the observed reduction in sporulation (Fig. 4) (LRE, Y = −0.29 + 0.43H + 0.51X + 0.34XH + 0.84R; LRT, FH = 6.92 and P = 0.0085; FX = 10.61 and P = 0.0011; FXH = 4.22 and P = 0.0401; FR = 18.53 and P ≤ 0.0001). Additionally, average time of sporulation was delayed in all lines infected with HFS compared to that of the line lacking HFS (see Fig. S1 in the supplemental material; multiple Wilcoxon test, P ≤ 0.001 for all comparisons, and Bonferroni-corrected α = 0.0125).
No clear indication of antagonistic interactions between superinfecting HFS based on estimates of within-aphid symbiont abundance.
H. defensa titers increased during nymphal development after single infection or superinfection (48 to 144 h) but decreased or leveled off upon adulthood (Fig. 5A). In adult aphids (e.g., 192 and 288 h) there were significantly fewer H. defensa cells in superinfected aphids relative to those infected with H. defensa only, suggesting that superinfection with X-type negatively impacts H. defensa abundance. X-type titers increased through adulthood (Fig. 5B) and were substantially higher in 12-day (288-h)-old adult aphids, suggesting that X-type begins to overproliferate in older aphids. The increase, however, was not as large in superinfected aphids, which may explain why costs to these aphids are not as severe. Buchnera genomic copies varied over time within each experimental line, with no consistent effects of single or superinfections with HFS on Buchnera titers, although titers were generally higher in the superinfected line (Fig. 6). In prior studies (29, 40), Buchnera copy number increased during aphid development into early adulthood; however, we observed consistent decreases across all experimental lines in middle-instar nymphs. The reason for this is unclear but may involve stage-specific amplification inhibitors.
FIG 5.
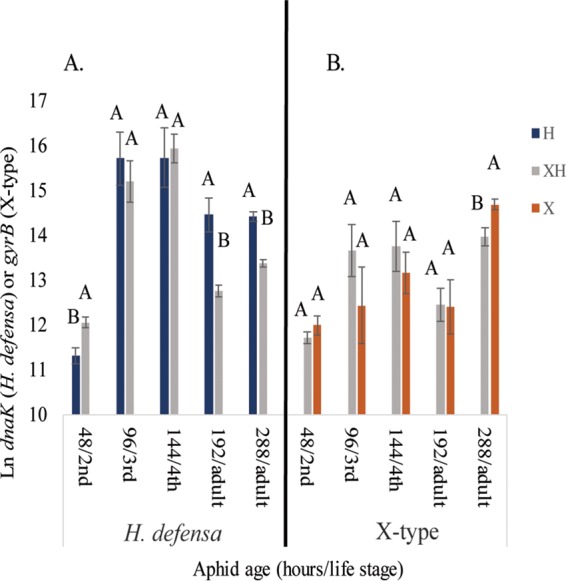
Natural log values of mean estimated gene copies within aphids across five time points. (A) Mean H. defensa dnaK copies in single infections and superinfections, compared using Student's t test. (B) Mean X-type gyrB copies in single infections and superinfections, compared using Student's t test. Letters represent significant differences; α = 0.05 for all comparisons.
FIG 6.
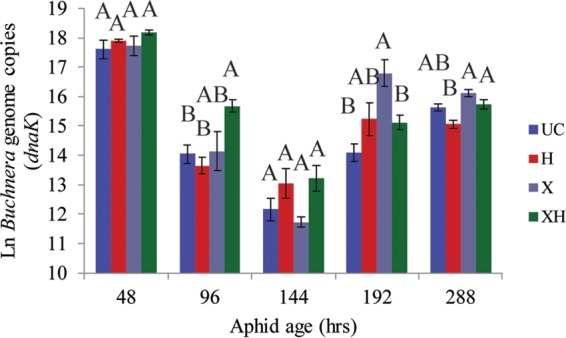
Natural log values of estimated Buchnera genome copy number in differentially infected aphids across five time points. Different letters represent significant differences in the number of Buchnera genome copies between aphid lines, determined by ANOVA with Tukey's HSD test; α = 0.05.
X-type natural occurrence, strain variation and phylogenetic placement.
Surveys of HFS from field-collected pea aphids collected from 2013 to 2015 found that 27% of aphids (totaling 485) were infected with X-type. The vast majority of these (n = 449 [93%]) occurred as superinfections, with most involving X-type plus H. defensa (n = 401 [82.7%]); a subset of these (n = 95) also carried a third HFS (most often Rickettsiella). Only 6% of superinfected aphids carried X-type and an HFS other than H. defensa.
A maximum likelihood phylogeny of 3 concatenated loci (gyrB plus accD plus 16S rRNA genes) revealed that X-type resides within the Enterobacteriaceae and is closely related to H. defensa and R. insecticola (Fig. 7). We also identified virtually no strain variation in the X-type symbiont. In two housekeeping genes (hrpA and rpoS) we found 100% nucleotide similarity sequenced across samples from Wisconsin and reference sequences from earlier studies (8, 41). Among 146 X-type infected aphids collected across a 4-year span from Wisconsin, we found a single nucleotide polymorphism (SNP) in the accD sequence from a single sample. While the sampling is most intense in Wisconsin, comparisons with sequences of samples from New York and Pennsylvania identified no additional variation and strongly suggest that X-type diversity is very limited in North America. Importantly, it identifies the strain used in this study as the dominant strain in Wisconsin and likely North America (see Table S2 in the supplemental material).
FIG 7.
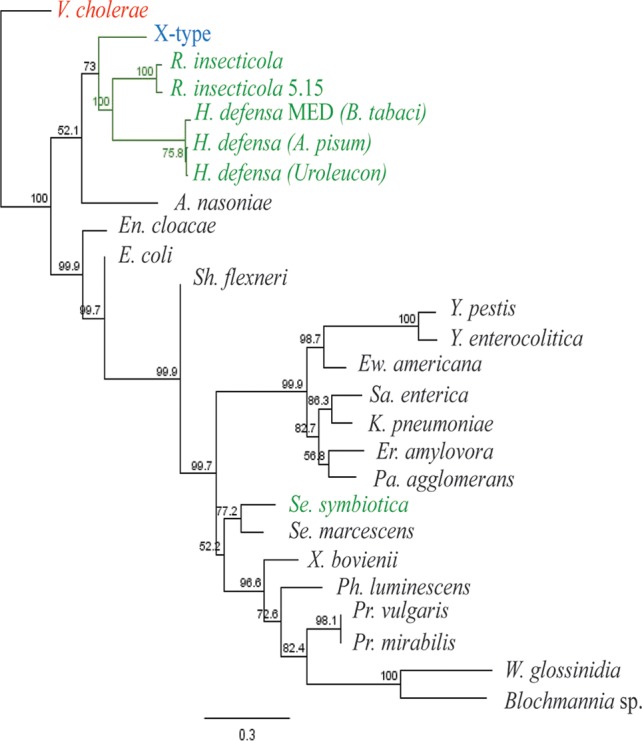
PHYML TN93+G+I 1000 bootstrap phylogenetic tree showing the placement of X-type within the Enterobacteriaceae, based on concatenated sequences of gyrB plus accD plus 16S rRNA genes with Vibrio cholerae as the outgroup. X-type is shown in blue, and other A. pisum HFS are shown in green.
As X-type is typically found superinfecting aphids with H. defensa, we also sequenced two H. defensa housekeeping genes (ptsI and recJ [Table S2]). Remarkably, we found that 58 of the 65 total sequenced superinfected samples contained a single H. defensa strain. Thus, X-type primarily associates with a single strain of H. defensa, suggesting the presence of more complex interactions between not only HFS species but also strains within those species (Fisher's exact test, P < 0.0001).
DISCUSSION
Pea aphids are commonly infected with one or more of seven HFS, most of which have been experimentally shown to confer protection against thermal stress and attack by specialized fungal pathogens and parasitic wasps (3). One common HFS, X-type, has been implicated in providing thermal tolerance and enhancing protection against parasitoid wasps and fungal pathogens (25, 39). X-type is unusual among pea aphid HFS, as it occurs primarily as a superinfection with a second symbiont, H. defensa (8; this study); however, no studies have disentangled the impact of X-type on the aphid protective phenotype in single-infection and superinfection contexts. After creating a full complement of experimental aphid lines with all X-type–H. defensa infection combinations in a shared aphid genotype, we examined the effects of X-type infection on aphid fitness at normal temperatures and under thermal stress and when challenged by specialized fungal pathogens and parasitoid wasps. In total, we found that as a single infection, X-type confers no protective services against thermal stress, fungal pathogens, or parasitoids, and it neither substantially enhances nor degrades protection against parasitoids or other threats when co-occurring with H. defensa (Table 2; Fig. 3 and 4). Instead, we identified large fecundity costs for aphids infected with only X-type (Table 1; Fig. 1). These costs are partially ameliorated in superinfections with H. defensa at cool temperatures and are entirely recovered in superinfections following heat shock (Table 2). That this strain of X-type confers none of the benefits hitherto attributed to pea aphid HFS yet confers large costs, especially in the single-infection context, suggests that X-type is maintained in pea aphid populations by exploiting the mutualism between pea aphids and H. defensa and essentially hitchhiking via the protective benefits conferred by its coinfector. This hypothesis is bolstered by the aforementioned field surveys showing that X-type mostly co-occurs with H. defensa and by our finding that H. defensa's protective benefits are not substantially reduced, while costs are ameliorated, in superinfections.
Two related parasitoid species, A. ervi and P. pequodoroum, commonly attack alfalfa-associated pea aphids in North America. A prior study found that X-type infection did not impact interactions with the dominant parasitoid A. ervi (33), and here we show that X-type, as a single infection or superinfection with H. defensa, does not contribute significant protection to the wasp P. pequodorum. Together these studies indicate that this common X-type strain is not directly involved in providing protection against either wasp species that attacks this aphid. So far, no identified aphid defenses, either of endogenous or symbiont origin, provide any protection against P. pequodorum, which may partially explain why this native wasp persists while other competitors have been excluded (37).
Infection with X-type also did not protect pea aphids from challenge by the specialized aphid fungal pathogen P. neoaphidis. Although X-type single infections and superinfections did decrease P. neoaphidis sporulation compared to that of uninfected controls, these infections did not result in significantly increased aphid survival, rendering a direct antifungal benefit unlikely (Fig. 4). Rather, the reduction of sporulation in X-type-infected lines correlated with increased aphid mortality prior to fungal sporulation. We also found that X-type did not provide thermal protection in our experimental lines. Aphids infected only with X-type exhibited high fecundity costs at constant, cool temperatures and under thermal stress. Interestingly, superinfection mitigated the cost of harboring X-type under conditions of heat stress, where we found no difference in fecundity between super- and uninfected aphids (Tables 1 and 2). The cause of this mitigation is unclear but may involve reduction of symbiont titers (23). Based on these findings, we may expect that superinfections involving X-type plus H. defensa will be more common in warmer regions or seasons, although the loss of H. defensa-mediated resistance to parasitism at warmer temperatures (31, 33) obscures this hypothesis. Our results contrast with a previous study finding that European pea aphids superinfected with X-type and Spiroplasma showed decreased susceptibility to numerous stressors relative to those infected with Spiroplasma only (39). Given that this previous study did not isolate X-type single infections and that Spiroplamsa's only associated benefit is fungal protection (24), indirect effects may account for the enhanced protection in superinfected hosts. The presence of an additional HFS (X-type) may have amplified the protective effect of Spiroplasma. Also, variation between North American and European X-type strains potentially contributed to the different findings. For example, strains of H. defensa can vary from completely protective to nonprotective depending on the presence of its associated phage, APSE (19).
While we examined only a single strain of X-type in our assays, sequence typing to characterize strain variation revealed that this was the dominant strain found across several sampled North American populations over a 4-year period. Moreover, this dominant X-type strain was most often found coinfecting pea aphids with the specific strain of APSE8-bearing H. defensa used in this study. Together, these findings indicate that our results likely generalize to many, if not most, North American X-type infections, with the caveat that aphid genotype by X-type interactions may introduce variation. Unfortunately, numerous attempts to place this X-type strain into other aphid genotypes were unsuccessful. Many successful transfections resulted in unstable infections in aphid lines that produced so few offspring that we unable to use them in bioassays, indicating that only some aphid genotypes are capable of tolerating infection with X-type. The presence of genotypic tolerance suggests that we are generally underestimating costs to infection with X-type via selection of lines which produce enough offspring to experimentally examine. These findings make it even more surprising that X-type can infect up to 40% of individuals in pea aphid populations (9).
While X-type may yet provide some untested benefit, such as resistance to bacterial or viral pathogens or modification of host dietary range (42, 43), pea aphid HFS are not known to provide resistance to such pathogens and European X-type strains do not improve aphid performance when aphids are fed on alfalfa or clover (39). The lack of expected benefits identified in this study suggests that North American X-type is not a conditional mutualist like the other common aphid HFS. It is also unlikely that X-type invades pea aphid populations as a reproductive manipulator, as pea aphids primarily reproduce asexually, although some reproductive manipulation has been seen with Spiroplasma-infected pea aphids during their sexual phase (44). Preliminary transmission experiments in the lab found high vertical transmission rates (>99%) and no instances of horizontal (i.e., infectious) transmission, together indicating that X-type is primarily maternally transmitted, similar to other common pea aphid HFS (45). Reduced costs and increased transmission rates of superinfections may allow for the maintenance of H. defensa and X-type superinfections when parasitism pressure from A. ervi is high.
Of course, there are numerous cases of heritable symbionts carrying high infection costs, including strains of H. defensa in black bean aphids (46), but these costs are offset by conditional benefits, with balancing selection maintaining intermediate frequencies (47). There are also virulent Wolbachia strains, most notably wMelPop, yet these HFS also likely spread via protective benefits and/or reproductive manipulation (48, 49). The phenotype conferred by X-type infection shares particular similarity with that of wMelPop; both carry severe costs that develop in adult hosts, possibly due to overreplication of the virulent symbiont (48, 50, 51). In this study, we found large increases in X-type titers only in older aphids (Fig. 5B), but other genotypes less tolerant of X-type infection may exhibit more dramatic increases in symbiont abundance. Indeed, preliminary titer estimations of X-type in a second aphid genotype (WI-12) are suggestive of overreplication (see Fig. S2).
Theory and empirical work have revealed two primary strategies for the spread and maintenance of heritable symbioses: providing net fitness benefits and manipulating host reproduction (1–4). However, given the mostly parthenogenetic reproduction of the aphid host, the findings that X-type does not confer any of the benefits attributed to other aphid HFS, imposes large fitness costs in single infections, and occurs primarily as a superinfection with defensive symbionts indicate that X-type likely persists in pea aphid populations by exploiting the protective services of H. defensa. This appears to be a tenuous strategy; X-type requires both a beneficial partner (H. defensa) and pressure from a specific natural enemy to provide selection for the partner symbiont. Yet aphids singly infected with H. defensa should still be expected to outperform superinfected aphids, implying that susceptible hosts are also required for the maintenance of this superinfection. In such a scenario, aphids superinfected with X-type and H. defensa should increase in the population relative to susceptible aphids (uninfected or infected with HFS other than H. defensa) during parasitism pressure. However, spread of this superinfection is ultimately impeded as single H. defensa infections concomitantly increase due to wasp pressure. Alternative explanations for X-type maintenance include improved rates of vertical transmission in superinfections relative to single infections or frequent lateral transfer, but neither of these was supported in preliminary lab trials. While X-type appears to be an atypical case, superinfections are common in many insect species (8, 10, 52) and this hitchhiking strategy, or similar scenarios, may occur in other systems, even among conditional beneficial symbionts not under direct selection (e.g., coinfecting antifungal HFS spread with H. defensa under parasitism pressure [11]). Of course, confirming a purely exploitative hitchhiking status of symbionts will remain troublesome, due to the difficulty in “proving a negative,” as there is always the possibility that the hitchhiker confers some unknown conditional benefit.
In a broad sense, facultative defensive mutualisms, such as those involving protective HFS in arthropods, are maintained only in the presence of a third, nonmutualist species (12). In pea aphids, for example, the proportion of aphids infected with H. defensa increases in cage studies in the presence of parasitoids but decreases in their absence (47). However, third-party participants can also exploit and impede mutualisms (12, 53, 54). For instance, when communities of HFS occur within individuals, exploitative HFS potentially obtain benefits supplied by other players in the interaction, without contributing any benefits themselves. In our case, X-type, which is vertically transmitted at high rates and presumably cannot live outside arthropod hosts, receives nutrient-provisioned housing and increased access to new hosts under conditions of high parasitism pressure when H. defense-infected aphids spread relative to aphids lacking H. defensa. While we did not find that protective services were reduced in superinfected aphids, costs in the absence of enemy challenge were much higher, indicating that symbiont-based defenses are likely to be removed from the population faster under conditions of low parasitism pressure. Thus, identifying potential exploitative symbionts is important, as they can be expected to degrade heritable mutualisms by depressing the benefit of the inherited mutualists. Given that defensive heritable symbionts appear to be common and often occur in communities (5), such interactions potentially restrict the distributions and function of these widespread mutualisms with effects that cascade to trophic levels above and below.
MATERIALS AND METHODS
Establishment of pea aphid experimental lines.
Pea aphids are phloem-feeding herbivores that consume a variety of herbaceous legumes. Due to their cyclically parthenogenetic life cycle, asexual clonal lines can be maintained in the laboratory indefinitely via continued exposure to a long-day-length photoperiod. All pea aphid experimental lines were maintained in small cages containing a fava bean plant, Vicia fava, within Percival biological incubators at 20°C with 70% humidity under 16-h light/8-h dark conditions, unless otherwise noted. The primary aphid line (5D) used in these experiments was collected on alfalfa (Medicago sativa) from Dane County, WI, in 2012. A single parthenogenetic female was placed in a petri dish with V. fava leaves to establish the line. A subset of her offspring were screened for all known pea aphid HFS using diagnostic PCR (primers and reaction conditions found in references 32 and 41) as well as any “unexpected” symbionts using universal primers with denaturing gradient gel electrophoresis (DGGE) (32). Line 5D was found to be naturally superinfected with H. defensa and the X-type symbiont. The H. defensa strain used in this study carries the phage APSE8, which encodes the cdtB2 toxin; this strain is functionally very similar to better-studied APSE2-H. defensa strains, and both provide a moderate level of protection against A. ervi (16, 32, 37, 55).
Using the 5D aphid genotype, we created a full complement of differentially infected aphid experimental lines for use in this study, consisting of the naturally superinfected line (XH), a line singly infected with H. defensa-APSE8 (H), a line singly infected with X-type (X), and a line uninfected with either symbiont (UC [uninfected control]). To create the X-type-only and uninfected lines, we selectively cured aphids by allowing aphid nymphs to feed on aphid artificial diet spiked with an antibiotic cocktail modified from that described in reference 56. We were unable to generate the H. defensa-only line through selective symbiont curing and we instead used transinfection to create this line. Briefly, hemolymph containing H. defensa was collected by homogenizing several superinfected 5D aphids in 40 μl of 1× phosphate-buffered saline (PBS) in a 0.5-ml tube; the hemolymph was then centrifuged at 7,000 × g for 15 s and transferred via pulled glass capillary needles to our UC line cured of both symbionts (16, 37). After producing the four experimental lines, we confirmed the expected symbiont profile of each line using diagnostic PCR protocols over multiple aphid generations and again just prior to use in experimental assays; experimental lines were maintained for at least six generations prior to use in experiments (18; see supplemental material for full details). We also performed microsatellite genotyping (32), using four loci (Ap-02, Ap-03, Ap-05, and Aph10M), to confirm that all four experimental lines were aphid genotype 5D (Table S1). We made numerous attempts to establish X-type single infections in other aphid genotypes but were unable to generate stable infections that produced sufficient offspring to conduct bioassays.
Pea aphid natural enemies examined in this study.
After its introduction in 1959 to control pea aphids in North America, the parasitoid A. ervi competitively displaced numerous native and introduced parasitoids attacking this aphid, except for the native aphidiine braconid (Hymenoptera) P. pequodorum (38). Thus, P. pequodorum, which also has a standard solitary endoparasitoid life cycle, is typically the only primary parasitoid other than A. ervi attacking pea aphids in North America. The P. pequodorum wasps in this study were field-caught wasps reared on a mixture of susceptible pea aphid genotypes. Adult wasps were provided honey and water, and females were at least 3 days old and mated prior to all parasitism assays.
The fungal pathogen Pandora neoaphidus (Zygomycota: Entomophthorales) is another specialized, widespread, and important natural enemy of the pea aphid acting as an effective natural biological control of pea aphid populations (57, 58). The P. neoaphidis (genotype ARSEF 2588) used in this study originated from the USDAARS Collection of Entomopathogenic Fungal Cultures. P. neoaphidis was maintained on the susceptible aphid line WI-48 (no HFS), with desiccated fungal cadavers held at 4°C with low humidity for no more than 2 months prior to use in experiment. Prior to experimental use, cadavers were rehydrated to induce sporulation.
Fitness assays at 20°C.
Casual observations of X-type-infected aphids, as well as the difficulty we experienced in creating and maintaining X-type single infections, suggested that this symbiont imposed significant constitutive costs on the aphid host, beyond that typically seen with other pea aphid HFS (22, 27). Component fitness assays were conducted to compare single infections and superinfections relative to uninfected controls. Assays were modified from the parameters reported in reference 27 and designed to measure aphid survival, fecundity, and developmental time, defined as the period from birth to first reproduction (TFR), as well as fresh weight and dorsal body size at TFR. Six replicates of newborn aphids were used for a total of 30 aphids/line (=120 aphids) in each assay. Aphid survival and fecundity were checked every 3 days from the onset of reproduction (day 9) to day 30, at which point almost all aphids had died or ceased reproduction; total output of offspring was summed as lifetime fecundity. To measure the effects of HFS infection on development, we monitored aphids every 2 days until day 8 (near adulthood), when we then monitored aphids every 3 h until first reproduction. Upon reproduction, the new mothers were removed and weighed using a microscale. Aphid body size was then estimated using dorsal view photographs generated with an Olympus SZX16. The dorsal circumference of the body, excluding antennae and legs, was traced to estimate the dorsal body area of the aphid from each photo using the measurement function in Olympus cellSens Standard software version 1.4.1.
Heat shock assay.
The heat shock assays consisted of a modified version of the previous fitness assays, except that cohorts consisted of 12 replicates of 10 newborn aphids per line which were divided into one of two temperature treatments, modified from those described in reference 22. The control treatment consisted of four replicates (=160 aphids) kept at the control temperature of 20°C for the duration of the experiment, which lasted for 20 days in total. The remaining eight replicates (=320 aphids) were exposed to a heat shock regimen in which aphids were maintained at 20°C for 2 days (∼2nd instar) and then the temperature was gradually increased over the span of 2 h to 37.5°C (±1°C) and held for 2 h before being reduced gradually first to 30°C (±1°C) and held for 1 h and then reduced gradually back to 20°C for the remainder of the experiment. Additionally, heat-shocked aphids were transferred to fresh plants the day after heat shock to maintain consistent host plant quality between the heat shock and control treatments. TFR was measured as described above, while fecundity was measured as described in reference 22.
P. pequodorum parasitism assays.
To test if X-type confers protection against P. pequodorum, either as a single infection or as a superinfection with H. defensa, we conducted a parasitism assay at 20°C. Cohorts of 15 2nd- to 3rd-instar pea aphids infected with either X-type (X), H. defensa-APSE8 (H), or X-type plus H. defensa-APSE8 (XH) or lacking both symbionts (UC) were placed onto a fresh V. fava plant and given 2 h to settle. A mated, female P. pequodorum wasp was then introduced into the cage and, after visual confirmation that the wasp would readily attack aphids, was allowed to parasitize aphids for 24 h, after which point it was removed. Nine days later, the proportions of aphids surviving (resistant), mummified (susceptible), and dead (dual mortality of aphid and parasitoid) were determined.
Fungal assays.
To determine if X-type confers protection against a second natural enemy, the fungal pathogen P. neoaphidis, we performed assays modified from those described in references 24, 59, and 60). Briefly, 16 cohorts of 10 apterous adult aphids (=160 aphids, 10 ± 1 days old) from each line (5D-X, H, XH, UC; 640) were placed in a 35-mm-diameter petri dish with an agar plate inoculated with two sporulating P. neoaphidis-infested cadavers inverted over them for 90 min, thus mimicking a natural spore shower. Additionally, four replicates (=40) of an aphid line (WI27-HR) superinfected with H. defensa and Regiella insecticola and highly resistant to P. neoaphidis were included as a control in this assay. A slide coverslip was placed under the aphids to ensure that sporulation occurred postexposure, and fungal plates were rotated every 15 min between similarly infected replicates to normalize sporulation. Each cohort was then placed onto a fresh V. fava plant at 20°C with 100% humidity (via an unvented cup lid) and normal long-day light conditions for 24 h. At this point, the unvented lid was then replaced with a vented lid (24, 59, 60; see supplemental material for additional setup methods). Aphids were checked every 24 h for 10 days postexposure for aphid survival, dual mortality (aphid and pathogen), and fungal sporulation (P. neoaphidis survival); any offspring produced were removed.
Symbiont titers.
Symbiont titers in uninfected (only B. aphidicola), singly infected (H or X), and superinfected (XH) aphids were estimated from whole extractions of aphids collected at five time points representing the 2nd through 4th instars, as well as early and older adults (i.e., 48 h [2nd instar]), 96 h [3rd instar], 144 h [4th instar], 192 h [early adult], and 288 h [older adult]) in replicates of eight aphids per time point. DNA was extracted as previously described, except in the case of the 288-h-old aphids, for which DNA was extracted using the E.Z.N.A. tissue DNA kit (OMEGA Bio-Tek) by following the manufacturer's protocol. Fragments of the single-copy bacterial genes dnaK from H. defensa and gyrB from X-type, along with the dnaK gene from polyploid B. aphidicola, were amplified using quantitative PCR (32, 61–63). The X-type primers new to this study were ACG GAG GTG AGT ACC CGA AAA (forward) and ATC AGC GTT CAT CTC TCC CA (reverse). PCRs were performed as described in reference 63; symbiont titers were estimated using external standard curves of each gene, and the aphid Ef-1 α gene was amplified in all samples to correct for differing extraction efficiencies between DNA samples (27).
X-type natural occurrence, strain variation, and phylogeny.
To investigate infection frequencies and patterns of X-type in natural populations, we extracted DNA from field-caught pea aphids collected on alfalfa from Dane County, WI, from 2013 to 2015 using the E.Z.N.A. tissue DNA kit (OMEGA Bio-Tek). DNA was stored at −80°C until tested for the presence of pea aphid HFS using the protocols described above (see “Establishment of pea aphid experimental lines”).
To examine strain variation of North American X-type, we PCR amplified and Sanger sequenced (using Eurofins MWG) five X-type housekeeping genes (accD, hrpA, murE, recJ, and rpoS) (41) from two pea aphid samples collected from 2012 to 2013 from Wisconsin, after enzyme cleaning with 0.3 μl of exonuclease I and 0.5 μl of FastAP under the following protocol: 35°C for 15 min, 85°C for 15 min, and holding at 10°C. Sequences, including references, were aligned with ClustalW using MUSCLE, trimmed manually using Geneious v 8.1.5, and compared to European X-type sequences obtained from GenBank (Table S2) (41). Preliminary screening revealed variation (a single, uncommon SNP) only at the accD locus; hence, we sequenced a total of 146 aphid clones collected from 2012 to 2105 from Dane Country, WI, at this locus. A subset of these samples was also used to generate additional rpoS (=19) and hrpA (=21) sequences to reduce the likelihood that we were missing variation. The same protocol was used to investigate the strain diversity of H. defensa found superinfecting with X-type, focusing on the ptsI and recJ genes of H. defensa (Table S2; primers and reaction from reference 18). A total of 89 samples (65 from superinfections with X-type) collected from Wisconsin from 2011 to 2015 were sequenced and aligned as previously described.
To create a phylogeny of X-type within the Enterobacteriaceae, two X-type housekeeping genes (accD and gyrB) and the 16S rRNA gene were amplified from strain 5D. We also obtained representative sequences of these same housekeeping genes and 16S rRNA genes of several other Enterobacteriaceae, including other pea aphid HFS and free-living bacteria downloaded from GenBank (Table S3). Concatenated sequences were aligned in Geneious as described above, and the Akaike information criteria (AIC) were calculated using MEGA v6.0 and used to predict the best-fit model of evolution. Additionally, concatenated sequences underwent DualBrothers recombination analysis. The best-fit model, Tamura-Nei with gamma distribution and transition/transversion bias, was used along with PHYML in Geneious to generate maximum likelihood trees, which were run with 1,000 bootstrap iterations.
Statistical analyses.
All analyses were performed using the statistical package SAS JMP Pro version 11 (SAS Institute Inc., Cary, NC). In the fitness assays, lifetime fecundity, aphid body size, and fresh weight upon reproduction were compared using Dunnett's test with the UC line as the control. TFR was analyzed using the nonparametric multiple Wilcoxon rank sum tests with Benjamini-Hochberg-corrected significance values, as the TFR data had a nonnormal distribution even after transformation attempts. Aphid survival was fit to a lognormal distribution to estimate 0.5 survival time. The effects of heat shock were analyzed as in the first fitness assay, except that the numbers of aphids surviving to adulthood were compared between aphid line and temperature treatment using logistic regression. To compare TFRs, the nonparametric Wilcoxon rank sum test with a Bonferroni-corrected significance level was used to compare developmental times between lines and temperature treatments. To compensate for adult mortality at the later time points, fecundity data were treated both as rate of reproduction (offspring/day) and summed as total fecundity and compared using analysis of variance (ANOVA) with Tukey's honestly significant difference (HSD) test. Aphid survival, mummification (or sporulation), and mortality after P. pequodorum parasitism and P. neoaphidis exposure, for each line, were arcsin transformed and the distributions were checked for normality using the goodness-of-fit test. The results were then compared between aphid lines and the UC line using logistic regression, as well as ANOVA with Tukey's HSD post hoc test to compare means. Additionally, a lognormal distribution was used to estimate 0.5 survival time after exposure to P. neoaphidis as in the fecundity assay. Symbiont titers in differentially infected hosts were log transformed, and the distributions of symbiont titers in each experimental line at each time treatment were checked for normality using the goodness-of-fit test. Transformed titers of each symbiont were then compared (when applicable) between aphid lines and time using ANOVA with Tukey's HSD test. Finally, the tendency for a particular strain of H. defensa to associate with X-type was analyzed using Fisher's exact test with Bonferroni correction.
Supplementary Material
ACKNOWLEDGMENTS
Thanks are due to Adam Martinez, Stephanie Weldon, Clesson Higashi, Kyungsun Kim, Nancy Miorelli, and Jordan Harris for assistance in setting up and completing assays. Thanks are due to Nicole Gerardo (Emory) for providing P. neoaphidis-infected cadavers, Jason Harmon (North Dakota State) for providing P. pequodorum, and Jacob Russell for providing sequences.
This project was supported by NSF-DEB award 1240892 to K.M.O.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03276-16.
REFERENCES
- 1.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 2.Werren JH, O'Neill SL. 1997. The evolution of heritable symbionts. Oxford University Press, New York, NY. [Google Scholar]
- 3.Oliver KM, Smith AH, Russell JA. 2014. Defensive symbiosis in the real world—advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct Ecol 28:341–355. doi: 10.1111/1365-2435.12133. [DOI] [Google Scholar]
- 4.Jaenike J. 2012. Population genetics of beneficial heritable symbionts. Trends Ecol Evol 27:226–232. doi: 10.1016/j.tree.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Oliver KM, Martinez AJ. 2014. How resident microbes modulate ecologically-important traits of insects. Curr Opin Insect Sci 4:1–7. doi: 10.1016/j.cois.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Duron O, Bouchon D, Boutin S, Bellamy L, Zhou LQ, Engelstadter J, Hurst GD. 2008. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol 6:27. doi: 10.1186/1741-7007-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 8.Russell JA, Weldon S, Smith AH, Kim KL, Hu Y, Łukasik P, Doll S, Anastopoulos I, Novin M, Oliver KM. 2013. Uncovering symbiont-driven genetic diversity across North American pea aphids. Mol Ecol 22:2045–2059. doi: 10.1111/mec.12211. [DOI] [PubMed] [Google Scholar]
- 9.Chiel E, Gottlieb Y, Zchori-Fein E, Mozes-Daube N, Katzir N, Inbar M, Ghanim M. 2007. Biotype-dependent secondary symbiont communities in sympatric populations of Bemisia tabaci. Bull Entomol Res 97:407–413. doi: 10.1017/S0007485307005159. [DOI] [PubMed] [Google Scholar]
- 10.Toju H, Fukatsu T. 2011. Diversity and infection prevalence of endosymbionts in natural populations of the chestnut weevil: relevance of local climate and host plants. Mol Ecol 20:853–868. doi: 10.1111/j.1365-294X.2010.04980.x. [DOI] [PubMed] [Google Scholar]
- 11.Smith AH, Lukasik P, O'Connor MP, Lee A, Mayo G, Drott MT, Doll S, Tuttle R, Disciullo RA, Messina A, Oliver KM, Russell JA. 2015. Patterns, causes and consequences of defensive microbiome dynamics across multiple scales. Mol Ecol 24:1135–1149. doi: 10.1111/mec.13095. [DOI] [PubMed] [Google Scholar]
- 12.Bronstein JL, Barbosa P. 2002. Multitrophic/multispecies mutualistic interactions: the role of non-mutualists in shaping and mediating mutualisms, p 44–66. In Tscharntke T, Hawkins BA (ed), Multitrophic level interactions. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 13.Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience Publishers, New York, NY. [Google Scholar]
- 14.Douglas A. 1992. Requirement of pea aphids (Acyrthosiphon pisum) for their symbiotic bacteria. Entomol Exp Appl 65:195–198. doi: 10.1111/j.1570-7458.1992.tb01643.x. [DOI] [Google Scholar]
- 15.Oliver KM, Russell JA, Moran NA, Hunter MS. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci U S A 100:1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliver KM, Moran NA, Hunter MS. 2005. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc Natl Acad Sci U S A 102:12795–12800. doi: 10.1073/pnas.0506131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moran NA, Russell JA, Koga R, Fukatsu T. 2005. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. J Appl Environ Microbiol 71:3302–3310. doi: 10.1128/AEM.71.6.3302-3310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Degnan PH, Moran NA. 2008. Diverse phage-encoded toxins in a protective insect endosymbiont. J Appl Environ Microbiol 74:6782–6791. doi: 10.1128/AEM.01285-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliver KM, Degnan PH, Hunter MS, Moran NA. 2009. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325:992–994. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montllor CB, Maxmen A, Purcell AH. 2002. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol 27:189–195. doi: 10.1046/j.1365-2311.2002.00393.x. [DOI] [Google Scholar]
- 21.Scarborough CL, Ferrari J, Godfray HCJ. 2005. Aphid protected from pathogen by endosymbiont. Science 310:1781. doi: 10.1126/science.1120180. [DOI] [PubMed] [Google Scholar]
- 22.Russell JA, Moran NA. 2006. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc R Soc Lond B Biol Sci 273:603–610. doi: 10.1098/rspb.2005.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burke G, Fiehn O, Moran N. 2010. Effects of facultative symbionts and heat stress on the metabolome of pea aphids. ISME J 4:242–252. doi: 10.1038/ismej.2009.114. [DOI] [PubMed] [Google Scholar]
- 24.Łukasik P, van Asch M, Guo H, Ferrari J, Godfray CJ. 2013. Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol Lett 16:214–218. doi: 10.1111/ele.12031. [DOI] [PubMed] [Google Scholar]
- 25.Guay JF, Boudreault S, Michaud D, Cloutier C. 2009. Impact of environmental stress on aphid clonal resistance to parasitoids: role of Hamiltonella defensa bacterial symbiosis in association with a new facultative symbiont of the pea aphid. J Insect Physiol 55:919–926. doi: 10.1016/j.jinsphys.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Ferrari J, West JA, Via S, Godfray HCJ. 2012. Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution 66:375–390. doi: 10.1111/j.1558-5646.2011.01436.x. [DOI] [PubMed] [Google Scholar]
- 27.Oliver KM, Moran NA, Hunter MS. 2006. Costs and benefits of a superinfection of facultative symbionts in aphids. Proc R Soc Lond B Biol Sci 273:1273–1280. doi: 10.1098/rspb.2005.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vautrin E, Vavre F. 2009. Interactions between vertically transmitted symbionts: cooperation or conflict? Trends Microbiol 17:95–99. doi: 10.1016/j.tim.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Koga R, Tsuchida T, Fukatsu T. 2003. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc R Soc Lond B Biol Sci 270:2543–2550. doi: 10.1098/rspb.2003.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandström JP, Russell JA, White JP, Moran NA. 2001. Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol Ecol 10:217–228. doi: 10.1046/j.1365-294X.2001.01189.x. [DOI] [PubMed] [Google Scholar]
- 31.Bensadia F, Boudreault S, Guay J-F, Michaud D, Cloutier C. 2006. Aphid clonal resistance to a parasitoid fails under heat stress. J Insect Physiol 52:146–157. doi: 10.1016/j.jinsphys.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 32.Martinez AJ, Weldon SR, Oliver KM. 2014. Effects of parasitism on aphid nutritional and protective symbioses. Mol Ecol 23:1594–1607. doi: 10.1111/mec.12550. [DOI] [PubMed] [Google Scholar]
- 33.Doremus MR. 2016. Friend or foe: characterization of the X-type symbiont in the pea aphid, Acyrthosiphon pisum. Master's thesis University of Georgia, Athens, GA. [Google Scholar]
- 34.Asplen MK, Bano N, Brady CM, Desneux N, Hopper KR, Malouines C, Oliver KM, White JA, Heimpel GE. 2014. Specialisation of bacterial endosymbionts that protect aphids from parasitoids. Ecol Entomol 39:736–739. doi: 10.1111/een.12153. [DOI] [Google Scholar]
- 35.Cayetano L, Vorburger C. 2015. Symbiont-conferred protection against Hymenopteran parasitoids in aphids: how general is it? Ecol Entomol 40:85–93. doi: 10.1111/een.12161. [DOI] [Google Scholar]
- 36.McLean AHC, Godfray HCJ. 2015. Evidence for specificity in symbiont-conferred protection against parasitoids. Proc R Soc Lond B Biol Sci 282:20150977. doi: 10.1098/rspb.2015.0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez AJ, Kim KL, Harmon JP, Oliver KM. 2016. Specificity of multi-modal aphid defenses against two rival parasitoids. PLoS One 11:e0154670. doi: 10.1371/journal.pone.0154670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schellhorn NA, Kuhman TR, Olson AC, Ives AR. 2002. Competition between native and introduced parasitoids of aphids: nontarget effects and biological control. Ecology 83:2745–2757. [Google Scholar]
- 39.Heyworth ER, Ferrari J. 2015. A facultative endosymbiont in aphids can provide diverse ecological benefits. J Evol Biol 28:1753–1760. doi: 10.1111/jeb.12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simonet P, Duport G, Gaget K, Weiss-Gayet M, Colella S, Febvay G, Charles H, Viñuelas J, Heddi A, Calevro F. 2016. Direct flow cytometry measurements reveal a fine-tuning of symbiotic cell dynamics according to the host developmental needs in aphid symbiosis. Sci Rep 6:19967. doi: 10.1038/srep19967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henry LM, Peccoud J, Simon JC, Hadfield JD, Maiden MJ, Ferrari J, Godfray HC. 2013. Horizontally transmitted symbionts and host colonization of ecological niches. Curr Biol 23:1713–1717. doi: 10.1016/j.cub.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamilton PT, Perlman SJ. 2013. Host defense via symbiosis in Drosophila. PLoS Pathog 9:e1003808. doi: 10.1371/journal.ppat.1003808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner SM, Martinez AJ, Ruan Y-M, Kim KL, Lenhart PA, Dehnel AC, Oliver KM, White JA. 2015. Facultative endosymbionts mediate dietary breadth in a polyphagous herbivore. Funct Ecol 29:1402–1410. doi: 10.1111/1365-2435.12459. [DOI] [Google Scholar]
- 44.Simon J-C, Boutin S, Tsuchida T, Koga R, Le Gallic J-F, Frantz A, Outreman Y, Fukatsu T. 2011. Facultative symbiont infections affect aphid reproduction. PLoS One 6:e21831. doi: 10.1371/journal.pone.0021831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moran NA, Dunbar HE. 2006. Sexual acquisition of beneficial symbionts in aphids. Proc Natl Acad Sci U S A 103:12803–12806. doi: 10.1073/pnas.0605772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vorburger C, Gouskov A. 2011. Only helpful when required: a longevity cost of harbouring defensive symbionts. J Evol Biol 24:1611–1617. doi: 10.1111/j.1420-9101.2011.02292.x. [DOI] [PubMed] [Google Scholar]
- 47.Oliver KM, Campos J, Moran NA, Hunter MS. 2008. Population dynamics of defensive symbionts in aphids. Proc R Soc Lond B Biol Sci 275:293–299. doi: 10.1098/rspb.2007.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Min K-T, Benzer S. 1997. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc Natl Acad Sci U S A 94:10792–10796. doi: 10.1073/pnas.94.20.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chrostek E, Marialva MSP, Esteves SS, Weinert LA, Martinez J, Jiggins FM, Teixeira L. 2013. Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLoS Genet 9:e1003896. doi: 10.1371/journal.pgen.1003896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reynolds KT, Thomson LJ, Hoffmann AA. 2003. The effects of host age, host nuclear background and temperature on phenotypic effects of the virulent Wolbachia strain popcorn in Drosophila melanogaster. Genetics 164:1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chrostek E, Teixeira L. 2015. Mutualism breakdown by amplification of Wolbachia genes. PLoS Biol 13:e1002065. doi: 10.1371/journal.pbio.1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gottlieb Y, Ghanim M, Gueguen G, Kontsedalov S, Vavre F, Fleury F, Zchori-Fein E. 2008. Inherited intracellular ecosystem: symbiotic bacteria share bacteriocytes in whiteflies. FASEB J 22:2591–2599. doi: 10.1096/fj.07-101162. [DOI] [PubMed] [Google Scholar]
- 53.Little AEF, Currie CR. 2008. Black yeast symbionts compromise the efficiency of antibiotic defenses in fungus-growing ants. Ecology 89:1216–1222. doi: 10.1890/07-0815.1. [DOI] [PubMed] [Google Scholar]
- 54.Vidal MC, Sendoya SF, Oliveira PS. 2016. Mutualism exploitation: predatory drosophilid larvae sugar-trap ants and jeopardize facultative ant-plant mutualism. Ecology 97:1650–1657. doi: 10.1002/ecy.1441. [DOI] [PubMed] [Google Scholar]
- 55.Weldon SR. 2015. Matryoshka mutualisms: developing the bacteriophage APSE-Hamiltonella defensa-pea aphid systems as a model for tripartite symbiosis. PhD dissertation University of Georgia, Athens, GA. [Google Scholar]
- 56.Douglas AE, François CLMJ, Minto LB. 2006. Facultative “secondary” bacterial symbionts and the nutrition of the pea aphid, Acyrthosiphon pisum. Physiol Entomol 31:262–269. doi: 10.1111/j.1365-3032.2006.00516.x. [DOI] [Google Scholar]
- 57.Pickering J, Dutcher JD, Ekbom BS. 1989. An epizootic caused by Erynia neoaphidis and E. radicans (Zygomycetes, Entomophthoraceae) on Acyrthosiphon pisum (Aphididae) on legumes under overhead irrigation. J Appl Entomol 107:331–333. doi: 10.1111/j.1439-0418.1989.tb00265.x. [DOI] [Google Scholar]
- 58.Roy HE, Brodie EL, Chandler D, Goettel MS, Pell JK, Wajnberg E, Vega FE. 2010. Deep space and hidden depths: understanding the evolution and ecology of fungal entomopathogens. Biocontrol 55:1–6. doi: 10.1007/s10526-009-9244-7. [DOI] [Google Scholar]
- 59.Ferrari J, Muller CB, Kraaijeveld AR, Godfray HC. 2001. Clonal variation and covariation in aphid resistance to parasitoids and a pathogen. Evolution 55:1805–1814. [DOI] [PubMed] [Google Scholar]
- 60.Parker BJ, Spragg CJ, Altincicek B, Gerardo NM. 2013. Symbiont-mediated protection against fungal pathogens in pea aphids: a role for pathogen specificity? Appl Environ Microbiol 79:2455–2458. doi: 10.1128/AEM.03193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moran NA, Degnan PH, Santos SR, Dunbar HE, Ochman H. 2005. The players in a mutualistic symbiosis: insects, bacteria, viruses, and virulence genes. Proc Natl Acad Sci U S A 102:16919–16926. doi: 10.1073/pnas.0507029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson AC, Dunbar HE, Davis GK, Hunter WB, Stern DL, Moran NA. 2006. A dual-genome microarray for the pea aphid, Acyrthosiphon pisum, and its obligate bacterial symbiont, Buchnera aphidicola. BMC Genomics 7:50. doi: 10.1186/1471-2164-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weldon S, Strand M, Oliver K. 2013. Phage loss and the breakdown of a defensive symbiosis in aphids. Proc R Soc Lond B Biol Sci 280:20122103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


