ABSTRACT
Rifamycin and its derivatives are particularly effective against the pathogenic mycobacteria Mycobacterium tuberculosis and Mycobacterium leprae. Although the biosynthetic pathway of rifamycin has been extensively studied in Amycolatopsis mediterranei, little is known about the regulation in rifamycin biosynthesis. Here, an in vivo transposon system was employed to identify genes involved in the regulation of rifamycin production in A. mediterranei U32. In total, nine rifamycin-deficient mutants were isolated, among which three mutants had the transposon inserted in AMED_0655 (rifZ, encoding a LuxR family regulator). The rifZ gene was further knocked out via homologous recombination, and the transcription of genes in the rifamycin biosynthetic gene cluster (rif cluster) was remarkably reduced in the rifZ null mutant. Based on the cotranscription assay results, genes within the rif cluster were grouped into 10 operons, sharing six promoter regions. By use of electrophoretic mobility shift assay and DNase I footprinting assay, RifZ was proved to specially bind to all six promoter regions, which was consistent with the fact that RifZ regulated the transcription of the whole rif cluster. The binding consensus sequence was further characterized through alignment using the RifZ-protected DNA sequences. By use of bionformatic analysis, another five promoters containing the RifZ box (CTACC-N8-GGATG) were identified, among which the binding of RifZ to the promoter regions of both rifK and orf18 (AMED_0645) was further verified. As RifZ directly regulates the transcription of all operons within the rif cluster, we propose that RifZ is a pathway-specific regulator for the rif cluster.
IMPORTANCE To this day, rifamycin and its derivatives are still the first-line antituberculosis drugs. The biosynthesis of rifamycin has been extensively studied, and most biosynthetic processes have been characterized. However, little is known about the regulation of the transcription of the rifamycin biosynthetic gene cluster (rif cluster), and no regulator has been characterized. Through the employment of transposon screening, we here characterized a LuxR family regulator, RifZ, as a direct transcriptional activator for the rif cluster. As RifZ directly regulates the transcription of the entire rif cluster, it is considered a pathway-specific regulator for rifamycin biosynthesis. Therefore, as the first regulator characterized for direct regulation of rif cluster transcription, RifZ may provide a new clue for further engineering of high-yield industrial strains.
KEYWORDS: Amycolatopsis mediterranei, rifamycin, rif, RifZ, pathway-specific regulator
INTRODUCTION
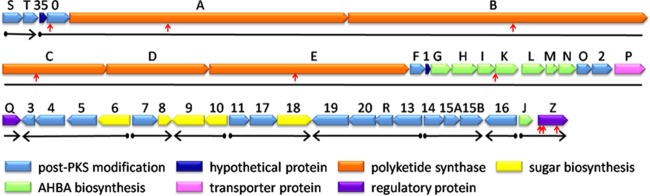
Amycolatopsis mediterranei is well known to produce the commercially important ansamycin antibiotic rifamycin, whose derivatives are particularly effective against pathogenic mycobacteria (1). These years, with the emergence of multidrug-resistant (MDR) and totally drug-resistant (TDR) tuberculosis (2, 3), an in-depth study of rifamycin biosynthesis and the development of new effective derivatives become more and more urgent. Due to their ability to tightly bind to prokaryotic RNA polymerases, rifamycins kill bacteria by blocking the transcription elongation (4, 5). In the past 2 decades, most processes in rifamycin biosynthesis have been characterized, and the nearly 90-kb rifamycin biosynthetic gene cluster (rif cluster) from several A. mediterranei strains has been sequenced (6–9); it starts with AMED_0613 (rifS) and ends with AMED_0655 (the last gene in the rif cluster, thus named rifZ) in A. mediterranei U32 (Fig. 1). The cluster includes genes involved in 3-amino-5-hydroxybenzoic acid (AHBA) biosynthesis (rifG-rifN operon and rifJ), polyketide biosynthesis (rifA-rifE operon), and rifamycin modification and export (6, 10–14). Although two rif genes are annotated as regulatory genes (Fig. 1), little is known about the regulation of rifamycin biosynthesis (9).
FIG 1.
Schematic chart of the rif cluster in A. mediterranei U32. The transposon insertional sites listed in Table 1 are labeled with vertical red arrows. The length and direction of the 10 operons in the rif cluster are indicated by solid horizontal black arrows. Genes listed: S, rifS; T, rifT; 35, orf35 (AMED_0615); 0, orf0 (AMED_0616); A, rifA; B, rifB; C, rifC; D, rifD; E, rifE; F, rifF; 1, orf1 (AMED_0623); G, rifG; H, rifH; I, rifI; K, rifK; L, rifL; M, rifM; N, rifN; O, rifO; 2, orf2 (AMED_0632); P, rifP; Q, rifQ; 3, orf3 (AMED_0635); 4, orf4 (AMED_0636); 5, orf5 (AMED_0637); 6, orf6 (AMED_0638); 7, orf7 (AMED_0639); 8, orf8 (AMED_0640); 9, orf9 (AMED_0641); 10, orf10 (AMED_0642); 11, orf11 (AMED_0643); 17, orf17 (AMED_0644); 18, orf18 (AMED_0645); 19, orf19 (AMED_0646); 20, orf20 (AMED_0647); R, rifR; 13, orf13 (AMED_0649); 14, orf14 (AMED_0650); 15A, orf15A (AMED_0651); 15B, orf15B (AMED_0652); 16, orf16 (AMED_0653); J, rifJ; Z, rifZ (AMED_0655).
Secondary metabolisms can be regulated by either global regulators or pathway-specific regulators. In actinobacteria, GlnR and PhoP are two key global regulators that govern the nitrogen and phosphate metabolisms and then affect the biosynthesis of the secondary metabolites (15–19). In addition, the pathway-specific regulators are also important for the regulation of the biosynthesis of secondary metabolites. The large ATP-binding regulators of the LuxR family (LAL regulators) constitute a large family of pathway-specific regulators, and many regulators in this family have been identified in actinobacteria, such as AveR (20), PikD (21), RapH (22), GdmRI and GdmRII (23, 24), SlnR (25), Dbv3 (26), and Tei16* (27). However, as no LuxI proteins have been found in actinobacteria and no N-acyl-l-homoserine lactone (AHL)-sensing system has been reported for Gram-positive bacteria, LuxR family regulators in actinobacteria are unlikely to be controlled by quorum-sensing signals similar to those in Vibrio fischeri (28).
In A. mediterranei U32, the biosynthesis of rifamycin was influenced after deletion of glnR (29). However, as GlnR is unable to bind the promoter region of the AMED_0615 operon (our unpublished data), the regulation must be conducted via an indirect way. Meanwhile, no pathway-specific regulators have ever been reported for the rif cluster. Therefore, to identify novel regulators for rif transcription, a transposon mutagenesis library was constructed in A. mediterranei U32 and a pathway-specific regulator for the rif cluster was identified and further characterized in this study.
RESULTS AND DISCUSSION
Screening of Tn316 transposon mutants defective in rifamycin production.
An efficient transposon system of Tn316 derived from Nocardia asteroides YP21 (30, 31) was applied to generate a transposon mutant library in A. mediterranei U32 with the efficiency of 1.12 × 102 CFU per microgram Tn316. Tn316 integration sites were determined via plasmid rescue experiments followed by DNA sequencing and were found to be randomly distributed on the U32 chromosome. Based on our previous studies, this transposon system also works efficiently in Streptomyces (32, 33), which therefore suggests that this in vivo transposon system may also be applied in functional genomics study of other actinomycetes.
A library of 20,000 transposon mutants was constructed to screen mutants defective in rifamycin production (see Materials and Methods). In total, 11 mutants defective in rifamycin production were obtained, and 9 of them had the transposon inserted within the rif cluster, including the AHBA formation genes (mutant no. 115C07), polyketide biosynthesis genes (no. 039F10, no. 112C08, no. 110H05, and no. 023A12), a P450 enzyme gene, AMED_0616 (no. 050D05), and a regulatory gene, rifZ (no. 061H02, no. 063H06, and no. 063H07) (Fig. 1 and Table 1).
TABLE 1.
Tn316 insertional locations in A. mediterranei U32
| Transposon mutant no. | Tn316 insertional site (bp) | Inactivated gene | Within rif cluster? |
|---|---|---|---|
| 050D05 | 628514 to 628513 | AMED_0616 | Yes |
| 039F10 | 634881 to 634882 | rifA | Yes |
| 112C08 | 635269 to 635270 | rifB | Yes |
| 110H05 | 660734 to 660735 | rifC | Yes |
| 023A12 | 673636 to 673537 | rifE | Yes |
| 115C07 | 684168 to 684167 | rifK | Yes |
| 061H02 | 716703 to 716704 | rifZ | Yes |
| 063H06 | 715689 to 715690 | rifZ | Yes |
| 063H07 | 715720 to 715721 | rifZ | Yes |
| 109A12 | 2462543 to 2462542 | AMED_2309 | No |
| 050F04 | 5030598 to 5030597 | AMED_4574 | No |
Among these mutants, the one with a mutation in rifZ, which encodes a LuxR family regulator and is located at the end of the rif cluster (9, 14), drew our attention. Bioinformatics analysis revealed a helix-turn-helix (HTH) LuxR-type DNA binding motif in the C terminus of RifZ (see Fig. S2 in the supplemental material). However, no autoinducer binding domain was identified, which indicated that the regulation of RifZ was different from the typical quorum-sensing model in Gram-negative bacteria (e.g., V. fischeri).
RifZ activates the transcription of rif genes in A. mediterranei U32.
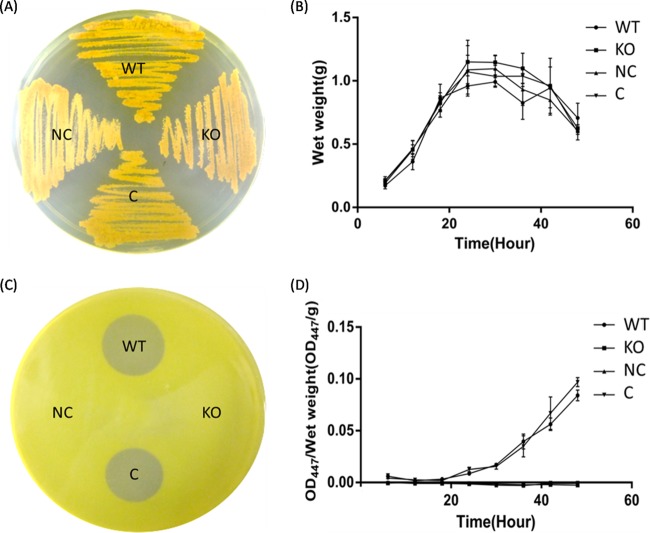
The rifZ gene was further knocked out via homologous recombination to fully uncover its function in A. mediterranei U32. First of all, the deletion of rifZ did not affect bacterial growth either on the plate (Fig. 2A) or in the liquid culture (Fig. 2B), which indicated that rifZ was not related to the cell's growth. What is more, it was noteworthy that there was different pigmentation of the rifZ knockout mutant (KO) and of NC (KO strain integrated with pDZL803, a negative control) with respect to the wild-type strain (WT) and to C (KO strain integrated with pDZL8031, a complementation strain) when growing on the plate. And this may be caused by their different capability for rifamycin biosynthesis, because rifamycins usually appeared golden on the plate. So, the yield of rifamycins was then detected by both the bacterial inhibition test (Fig. 2C) and the chemical method (Fig. 2D), which showed that the KO's yield of rifamycins dropped significantly, just as in the transposon-deactivated rifZ mutants, and subsequent complementation of rifZ with its intact promoter restored the rifamycin production to a level comparable to that of the wild type (Fig. 2C and D). All the above-described results illustrated in Fig. 2 indicated that RifZ positively regulated the biosynthesis of rifamycin.
FIG 2.
RifZ stringently regulates rifamycin biosynthesis in A. mediterranei U32. (A) Growth of A. mediterranei strains on a Bennet plate. (B) Growth curves of different A. mediterranei strains. (C) Bacterial inhibition test of different A. mediterranei strains using Sarcina lutea as the indicator. (D) Rifamycin production by different A. mediterranei strains, detected by a chemical method. Both NC and KO strains lost the ability to produce rifamycin. WT, wild type; KO, rifZ knockout mutant; NC, KO strain integrated with pDZL803, a negative control; C, KO strain integrated with pDZL8031, a complementation strain.
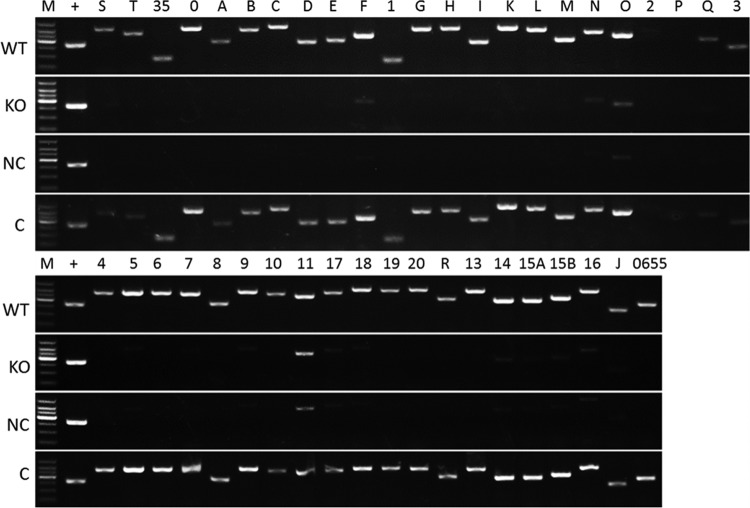
A reverse transcription (RT)-PCR assay was then employed to determine whether RifZ regulated the transcription of the rif cluster. Total RNA was extracted from the wild-type strain, the rifZ knockout mutant, the complementation strain integrated with pDZL8031, and the mutant integrated with the control plasmid pDZL803. The RT-PCR results showed that the levels of transcription of orf2 (AMED_0632) and rifP were very low and even undetectable in the wild type, which was possibly caused by their physical location within the cluster, i.e., near the end of a large operon, or by the independent regulation of unknown factors. Except for these two genes, the transcription of all other genes significantly decreased in the null mutant and the null mutant with the control plasmid, while their transcription was recovered in the complementation strain (Fig. 3). Based on these results, one might conclude that RifZ is a positive regulator for the transcription of the rif cluster.
FIG 3.
RT-PCR analysis of the transcription of genes in the rif cluster in different A. mediterranei strains. No PCR bands were observed when total RNA without reverse transcription was employed as the template, which indicated that the RNA samples were not contaminated by genomic DNA (data not shown). WT, wild type; KO, rifZ knockout mutant; NC, KO strain integrated with pDZL803, a negative control; C, KO strain integrated with pDZL8031, a complementation strain; M, DL1000 DNA ladder; +, the rpoB gene, used as an internal control. The analyzed genes were in the same order as shown in Fig. 1, where “S” stood for rifS, 35 stood for orf35, etc.
RifZ is a pathway-specific regulator for the rif cluster.
To determine whether RifZ was able to directly bind to the promoter regions of its target genes in the rif cluster, overexpression of RifZ was performed in Escherichia coli. However, although different expression conditions that employed different plasmids with distinct promoters and different E. coli hosts were tried, RifZ either failed to be expressed or remained in the form of an insoluble inclusion body. Renaturation of the inclusion body was also attempted but failed (data not shown). We then resorted to overexpressing truncated forms of RifZ that contained the DNA binding domain (amino acids [aa] 346 to 398). Distinct truncated RifZs were tested, and only RifZ-5, which contained the sequence of aa 320 to 404, was soluble in E. coli hosts under the tested conditions (see Fig. S3 and S4 in the supplemental material).
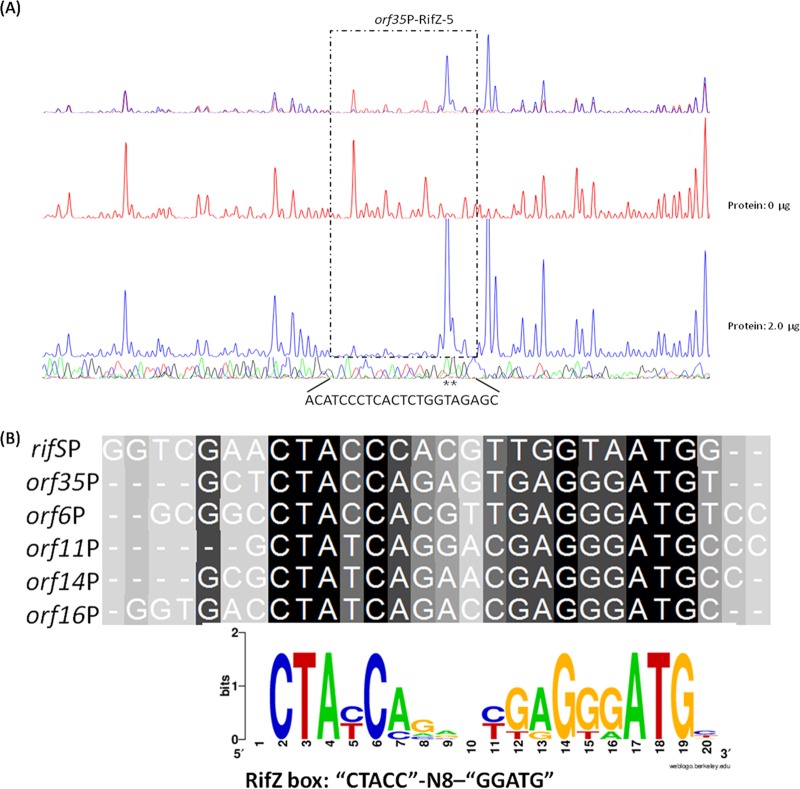
Meanwhile, the cotranscription of rif cluster genes in A. mediterranei U32 was examined by RT-PCR analysis, which indicated that the rif cluster was divided into 10 operons transcriptionally, sharing six promoter regions (Fig. 1; see also Fig. S5 in the supplemental material). Then, all the promoter regions were prepared as 6-carboxyfluorescein (FAM)-labeled probes and were used for electrophoretic mobility shift assay (EMSA) with purified RifZ-5. For those four promoters of divergent operons (promoters of orf6 and orf7, orf10 and orf11, orf13 and orf14, and orf16 and rifJ [Fig. 1]), relatively long probes covering whole intergenic regions were prepared for each promoter region. According to the EMSA results, we found that all these six probes could be specifically bound by RifZ-5 (see Fig. S6A in the supplemental material), which could well explain the RifZ-mediated transcriptional regulation of most genes in the rif cluster. A DNase I footprinting experiment was further employed to determine the RifZ-5 binding sequences in the above-mentioned promoter regions (Fig. 4A; see also Fig. S7 in the supplemental material). Then, a consensus DNA sequence of CTACC-N8-GGATG (RifZ box), which was composed of a pair of imperfect inverted repeats, was identified through DNA alignment analysis (Fig. 4B). As revealed by the DNase I footprinting results, the binding affinity of RifZ-5 to promoter regions of orf35 (AMED_0615) and rifS was the strongest, where 2 μg RifZ-5 could provide obvious protection of target DNA sequences against DNase I digestion, while the binding affinity to the others was relatively weaker, i.e., orf6 (AMED_0638), orf11 (AMED_0643), orf14 (AMED_0650), and orf16 (AMED_0653).
FIG 4.
Characterization of the RifZ-5-protected DNA sequences by DNase I footprinting assay and the speculative RifZ box. (A) Analysis of the RifZ-5-protected DNA sequences in the promoter region of orf35 by DNase I footprinting assay. The electrophoretograms of the control reaction (without RifZ-5, red line) and experimental reaction (with RifZ-5, blue line) are shown in the upper panel, and the lower panel shows the DNA sequencing results. Precise DNA sequences protected by RifZ-5 are indicated at the bottom, and hypersensitive sites are indicated with asterisks. The results of RifZ-5-protected DNA sequences in the promoter regions of other target genes, including rifS, orf6, orf11, orf14, and orf16, are shown in Fig. S7 in the supplemental material. (B) The RifZ binding consensus sequence of CTACC-N8-GGATG was obtained through alignment using the RifZ-5-protected DNA sequences in the promoter regions of all six target genes. The sequence logo at the bottom was generated by web-based WebLogo (version 2.8.2).
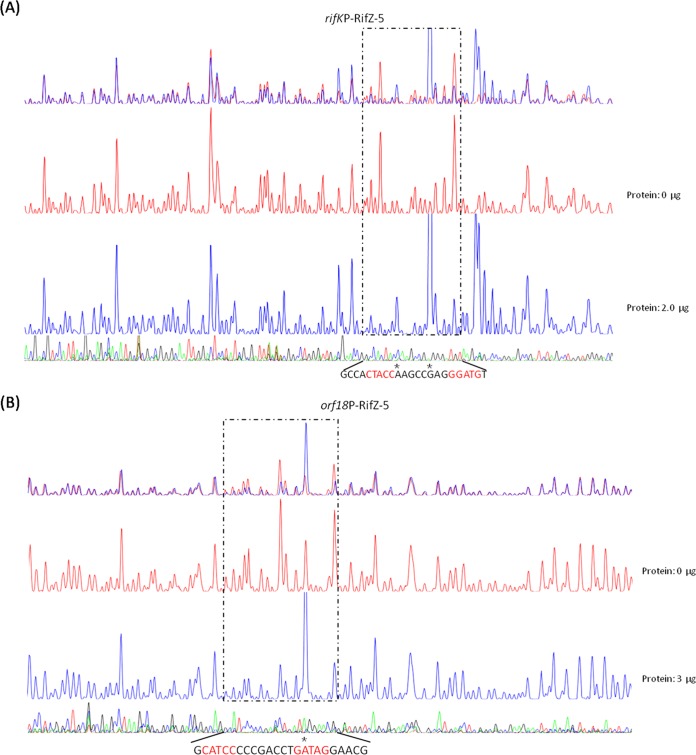
Moreover, the binding motif was used to search for the promoter regions throughout the genome of U32, and five new promoter regions containing the RifZ box were identified (see Table S2 in the supplemental material), among which were rifK and orf18 (AMED_0645), located within the rif cluster. RifK is an AHBA synthase and plays an important role in the AHBA biosynthesis, while Orf18 is a nucleotide diphosphate (NDP)-hexose 2,3-dehydratase and takes part in sugar biosynthesis (9). As both genes transcribed from their operons' promoters and no promoter regions had been previously predicted, the transcriptome data of A. mediterranei U32 (34) were then reanalyzed. Both rifK and orf18 (AMED_0645) were found to transcribe at an obviously higher level than their previous genes (see Fig. S8 in the supplemental material), indicating that both genes might transcribe from their own promoters in addition to their operons' promoters. Both promoter regions of rifK and orf18 were then selected to verify the accuracy of the bioinformatics prediction, and further EMSA (Fig. S6B) and DNase I footprinting (Fig. 5) confirmed the specific binding of RifZ-5 to the promoter regions of rifK and orf18, respectively. As RifZ directly activated the majority of rif genes, it was considered a pathway-specific regulator for the transcription of the rif cluster.
FIG 5.
Verification of the binding of RifZ-5 to the promoter regions of rifK and orf18 by DNase I footprinting assay. rifK (A) and orf18 (B) were identified through searching with the RifZ box in U32 at the whole-genome scale. The DNase I footprinting assay further confirmed that RifZ could specifically bind to both promoter regions, and the two RifZ-5-protected DNA sequences contained the RifZ binding motif, which is highlighted in red.
MATERIALS AND METHODS
Bacterial strains, media, and primers.
Methylation-deficient Escherichia coli JM110 was used to propagate plasmid Tn316. A. mediterranei U32 and its mutants were grown in Bennet medium (35) at 30°C. Sarcina lutea was used as an indicator for rifamycin production (29). When needed, apramycin (50 μg/ml), kanamycin (50 μg/ml), and ampicillin (100 μg/ml) were used. Plasmids used in this study are listed in Table 2, and primers (ordered from Biosune [Shanghai]) are listed in Table S1 in the supplemental material. The DL1000 DNA ladder was ordered from TaKaRa.
TABLE 2.
Plasmids used in this study
| Plasmid name | Relevant characteristics | Source |
|---|---|---|
| Tn316 | Plasmid derived from IS-204 in Nocardia asteroides YP21 | Lab stock |
| pFDZ100 | E. coli-Streptomyces shuttle vector, derivative obtained from pHZ1358, containing attP0 site and attB15 site, resistant to thiostrepton, ampicillin, and chloromycetin | Lab stock |
| pFDZ101 | Derivative obtained from pMD19-T, containing attB0 site and attP6 site, resistant to ampicillin and apramycin | Lab stock |
| pTA0613 | Derivative obtained from pMD19-T, containing attB6 site and attP13 site, resistant to ampicillin and apramycin | Lab stock |
| pFDZ103 | Derivative obtained from pMD19-T, containing attB13 site, attP15 site, and attP0 site, resistant to ampicillin and apramycin | Lab stock |
| pDZL101 | Derivative obtained from pFDZ101, containing the upstream homologous arm of AMED_0655 | This work |
| pDZL103 | Derivative obtained from pFDZ103, containing the downstream homologous arm of AMED_0655 | This work |
| pDZL104 | Derivative obtained from pFDZ100, containing both homologous arms of AMED_0655, resistant to thiostrepton and apramycin | This work |
| pRT801 | E. coli-Streptomyces shuttle plasmid, encoding the ϕBT1-integrase and containing attP, resistant to apramycin | 40 |
| pMD19-hyg | A 1,415-bp hygromycin resistance fragment flanked by two EcoRV sites in pMD19 | Lab stock |
| pDZL803 | EcoRV-digested fragment (1,409 bp) from pMD19-hyg ligated into the SmaI site of pRT801, resistant to apramycin and hygromycin | This work |
| pDZL8031 | AMED_0655 gene with its intact promoter ligated into the EcoRV site of pDZL803 | This work |
| pET-RifZ1 | AMED_0655 gene ligated into pET28a(+) | This work |
| pET-RifZ5 | Coding DNA sequence of RifZ-5 ligated into pET28a(+) | This work |
Screening of the transposon mutants of A. mediterranei U32.
Competent cells for electroporation were prepared according to our previous work (36). Electroporation of A. mediterranei U32 with methylation-free Tn316 was carried out with the Gene Pulser apparatus (Bio-Rad). Specifically, 1 μg DNA was added into 60 μl U32 competent cells and mixed on ice, and the mixture was transferred to a chilled electroporation cuvette (2 mm). The field strength was 9 kV/cm, and the pulse duration was about 13 to 14 ms. After pulsing, 1 ml prewarmed Bennet medium was immediately added to the electroporation cuvette and the mixture was incubated at 30°C for 4 h. Then, the cells were plated on Bennet plates containing apramycin (50 μg/ml) to select out successfully transposed mutants. Proper-size colonies were arrayed into 96-well plates, where each well contained 100 μl 20% glycerol. Finally, these plates were frozen and stored at −80°C.
To screen rifamycin-deficient mutants, frozen mutants were plated onto Bennet plates and further incubated at 30°C for 4 days. Candidate mutants were inoculated into 50 ml liquid Bennet medium and incubated at 30°C for 48 h with shaking at 200 rpm. Then, 2.5 ml culture was further inoculated into 50 ml fresh liquid Bennet medium, followed by continuing incubation for 22 h at 30°C with shaking at 200 rpm.
S. lutea was grown in 10 ml Luria-Bertani (LB) medium and incubated at 37°C for 18 h with shaking at 200 rpm. Then, the culture was mixed with 40 ml LB medium containing 0.8% low-melting agar and then poured onto LB plates containing 1.5% agar. Five microliters of supernatants of mutant cultures (22 h) was added to double plates with S. lutea, which were incubated at 37°C for phenotype observation. Growth of S. lutea was inhibited if the supernatant contained rifamycin.
Characterization of the transposon insertional sites in mutants.
Genomic DNA was extracted using commercial genomic extraction kits and digested with FastDigest ApaI (Thermo Fisher Scientific) for about 3 h. After inactivation of ApaI, the digested mixture was ligated with T4 DNA ligase (Tolo Biotech, Shanghai, China) overnight before being transformed into competent cells of E. coli DH5α. Transformants selected on LB plates containing apramycin (50 μg/ml) were inoculated into 3 ml liquid LB medium and incubated overnight. Rescued plasmids were extracted and sequenced using primer MY-002, and sequences downstream of GAAGAGCC were rescued from the mutant genome. Assisted with alignment analysis, sites and directions of the transposon insertion were determined.
Construction of rifZ knockout mutant and its complementation strain.
To knock out rifZ via homologous recombination, plasmids were constructed as previously described (37). Briefly, primers 0655U-F/0655U-R and 0655D-F/0655D-R were separately used to amplify the upstream and downstream homologous arms of rifZ, and the PCR fragments were introduced into the XcmI site of pFDZ101 and pFDZ103 to obtain pDZL101 and pDZL103 (Table 2), respectively. pDZL101, pDZL103, and pTA0613 were linearized and tandemly assembled with pFDZ100 using φBT1 integrase (38) to produce the knockout plasmid pDZL104, which was further electroporated into U32 to knock out rifZ. Mutants were selected on Bennet plates containing apramycin (50 μg/ml) and verified by PCR employing primers 0655CH-UF/0655CH-UR and 0655CH-DF/0655CH-DR.
The hygromycin resistance cassette from pMD19-hyg was inserted into the SmaI site of pRT801 (39, 40) to obtain plasmid pDZL803. The rifZ gene with its intact promoter region was amplified with primers 0655C-F/0655C-R using Phanta super-fidelity DNA polymerase (Vazyme) and inserted into the EcoRV site of pDZL803, producing plasmid pDZL8031. Plasmid pDZL8031 and the control plasmid pDZL803 were individually introduced into the rifZ null mutant by electroporation, obtaining both the complementation strain and its blank control.
RNA extraction and RT-PCR analysis.
A. mediterranei strains were grown in liquid Bennet medium at 30°C for 48 h with shaking at 200 rpm, and then 2.5 ml culture was inoculated into 50 ml fresh liquid Bennet medium and further incubated for 22 h. Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific) and further treated with RNase-free DNase I (TaKaRa) to prevent contamination of trace genomic DNA. RT was performed using the PrimeScript II 1st Strand cDNA synthesis kit (TaKaRa). Then, PCR was performed with 20 ng cDNA as the template for 28 cycles, using the rpoB gene as the internal control. A negative control was made by following the same procedures except for the omission of reverse transcriptase during RT. Two independent samples were employed for analyses.
Cotranscription assay of all the genes in the rif cluster of A. mediterranei U32.
After being cultured at 30°C in Bennet medium containing 80 mM KNO3 for 24 h, cells were harvested and used to extract total RNA, which was then used for cDNA synthesis. With the cDNA as the template, cotranscription assay of genes (from rifT to rifZ) was achieved by PCR. At the same time, the negative and positive controls were achieved by PCRs with the total RNA and genomic DNA (gDNA) of A. mediterranei U32 as the templates separately. Primers used during PCRs are listed in Table S1. The principle for designing the cotranscription assay primers was as follows: the forward primer was designed at the end of one gene, while the reverse primer was located at the middle of the next gene.
Protein expression in E. coli BL21(DE3).
The amino acid sequence of RifZ was analyzed on NCBI, and the DNA binding domain of RifZ (namely RifZ-5) was predicted to be from aa 346 to 398. To construct plasmids expressing complete and truncated RifZ proteins, the coding DNA sequence was amplified with different primer pairs (from RifZ-1EX-F/RifZ-EX-R to RifZ-5EX-F/RifZ-EX-R [see Fig. S1 in the supplemental material]). PCR amplicons were digested with NdeI and HindIII, and then they were introduced into the same sites in pET-28a(+), generating the expression plasmids pET-RifZ1 to pET-RifZ5.
The expression plasmids were then transformed into E. coli BL21(DE3), and induction of the protein expression was performed by addition of 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and grown at 16°C for 16 h. Cells were harvested, washed twice in potassium phosphate buffer (pH 7.0), and lysed using an ultrasonic cell crusher. In noninduced samples, cells were directly treated with the BugBuster master mix (Novagen) before being loaded for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. To prevent protein degradation, 1 mM (final concentration) phenylmethylsulfonyl fluoride (PMSF) was added. After separation of the cell debris and membrane fractions from the soluble fraction by centrifugation at 15,000 rpm for 30 min using a high-speed refrigerated centrifuge (CR21GIII; Hitachi), His-tagged full-length and truncated RifZ proteins were purified by Ni-nitrilotriacetic acid (Ni-NTA) gravity flow columns and further assessed by SDS-PAGE. Protein concentrations were determined by the Bradford method (41).
Electrophoretic mobility shift assay and DNase I footprinting assay.
Promoter regions were amplified by PCR with Dpx DNA polymerase (Tolo Biotech, Shanghai, China), employing the primer pairs listed in Table S1. Purified PCR amplicons were separately cloned into the TA cloning vector pUC18B-T (Tolo Biotech) and verified through DNA sequencing. Then, plasmids were used as the templates for probe preparation by PCR using primers M13F-47 (FAM labeled) and M13R-48. FAM-labeled probes were purified using Wizard SV gel and the PCR Clean-Up system (Promega) and quantified with a NanoDrop 2000C spectrophotometer (Thermo Fisher Scientific).
The binding of RifZ-5 to probes was carried out in a 20-μl reaction volume containing 50 mM Tris-HCl (pH 8.0), 100 mM KCl, 2.5 mM MgCl2, 1.0 mM dithiothreitol (DTT), 10% glycerol, and 100 ng/μl sheared salmon sperm DNA. FAM-labeled probes (40 ng) and different amounts (0 and 2 μg) of His-tagged RifZ-5 were used for each reaction. After being incubated for 30 min at 30°C, samples were separated by a 6% native polyacrylamide gel in ice-bathed 0.5× Tris-borate-EDTA (TBE) at 150 V. Gels were scanned with an ImageQuant LAS 4000mini imager (GE Healthcare).
DNase I footprinting assays were performed by Tolo Biotech according to the procedures previously described (18). In brief, about 400 ng FAM-labeled probes was incubated with different amounts of His-tagged RifZ-5 in a total volume of 40 μl. After incubation, DNase I digestion was performed for 1 min at room temperature before being stopped by addition of the DNase I stop solution. Digested samples were purified and then loaded into an ABI 3130 sequencer for electrophoresis. The electropherograms were analyzed with PeakScanner v1.0 software (Applied Biosystems).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Key Research Development Program of China (2016YFA0500600), the National Natural Science Foundation of China (no. 31430004, 31670058, and 31300034), and the China National Basic Research Program (973 Program no. 2012CB721102).
We thank Tolo Biotech for their assistance in protein purification, EMSA, and DNase I footprinting assay. We thank Shuangxi Ren for his help in bioinformatics analysis.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03201-16.
REFERENCES
- 1.Calvori C, Frontali L, Leoni L, Tecce G. 1965. Effect of rifamycin on protein synthesis. Nature 207:417–418. doi: 10.1038/207417a0. [DOI] [PubMed] [Google Scholar]
- 2.Atre SR, Murray MB. 2016. Management and control of multidrug-resistant tuberculosis (MDR-TB): addressing policy needs for India. J Public Health Policy doi: 10.1057/jphp.2016.14. [DOI] [PubMed] [Google Scholar]
- 3.Velayati AA, Farnia P, Masjedi MR. 2013. Totally drug-resistant tuberculosis (TDR-TB): a debate on global health communities. Int J Mycobacteriol 2:71–72. doi: 10.1016/j.ijmyco.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Wehrli W, Knusel F, Schmid K, Staehelin M. 1968. Interaction of rifamycin with bacterial RNA polymerase. Proc Natl Acad Sci U S A 61:667–673. doi: 10.1073/pnas.61.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901–912. doi: 10.1016/S0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 6.August PR, Tang L, Yoon YJ, Ning S, Muller R, Yu TW, Taylor M, Hoffmann D, Kim CG, Zhang X, Hutchinson CR, Floss HG. 1998. Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699. Chem Biol 5:69–79. doi: 10.1016/S1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 7.Tang B, Zhao W, Zheng H, Zhuo Y, Zhang L, Zhao GP. 2012. Complete genome sequence of Amycolatopsis mediterranei S699 based on de novo assembly via a combinatorial sequencing strategy. J Bacteriol 194:5699–5700. doi: 10.1128/JB.01295-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verma M, Kaur J, Kumar M, Kumari K, Saxena A, Anand S, Nigam A, Ravi V, Raghuvanshi S, Khurana P, Tyagi AK, Khurana JP, Lal R. 2011. Whole genome sequence of the rifamycin B-producing strain Amycolatopsis mediterranei S699. J Bacteriol 193:5562–5563. doi: 10.1128/JB.05819-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao W, Zhong Y, Yuan H, Wang J, Zheng H, Wang Y, Cen X, Xu F, Bai J, Han X, Lu G, Zhu Y, Shao Z, Yan H, Li C, Peng N, Zhang Z, Zhang Y, Lin W, Fan Y, Qin Z, Hu Y, Zhu B, Wang S, Ding X, Zhao GP. 2010. Complete genome sequence of the rifamycin SV-producing Amycolatopsis mediterranei U32 revealed its genetic characteristics in phylogeny and metabolism. Cell Res 20:1096–1108. doi: 10.1038/cr.2010.87. [DOI] [PubMed] [Google Scholar]
- 10.Absalon AE, Fernandez FJ, Olivares PX, Barrios-Gonzalez J, Campos C, Mejia A. 2007. RifP; a membrane protein involved in rifamycin export in Amycolatopsis mediterranei. Biotechnol Lett 29:951–958. doi: 10.1007/s10529-007-9340-7. [DOI] [PubMed] [Google Scholar]
- 11.Floss HG, Yu TW. 1999. Lessons from the rifamycin biosynthetic gene cluster. Curr Opin Chem Biol 3:592–597. doi: 10.1016/S1367-5931(99)00014-9. [DOI] [PubMed] [Google Scholar]
- 12.Kaur H, Cortes J, Leadlay P, Lal R. 2001. Cloning and partial characterization of the putative rifamycin biosynthetic gene cluster from the actinomycete Amycolatopsis mediterranei DSM 46095. Microbiol Res 156:239–246. doi: 10.1078/0944-5013-00108. [DOI] [PubMed] [Google Scholar]
- 13.Schupp T, Toupet C, Engel N, Goff S. 1998. Cloning and sequence analysis of the putative rifamycin polyketide synthase gene cluster from Amycolatopsis mediterranei. FEMS Microbiol Lett 159:201–207. doi: 10.1111/j.1574-6968.1998.tb12861.x. [DOI] [PubMed] [Google Scholar]
- 14.Yuan H, Zhao W, Zhong Y, Wang J, Qin Z, Ding X, Zhao GP. 2011. Two genes, rif15 and rif16, of the rifamycin biosynthetic gene cluster in Amycolatopsis mediterranei likely encode a transketolase and a P450 monooxygenase, respectively, both essential for the conversion of rifamycin SV into B. Acta Biochim Biophys Sin (Shanghai) 43:948–956. doi: 10.1093/abbs/gmr091. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Garcia A, Sola-Landa A, Apel K, Santos-Beneit F, Martin JF. 2009. Phosphate control over nitrogen metabolism in Streptomyces coelicolor: direct and indirect negative control of glnR, glnA, glnII and amtB expression by the response regulator PhoP. Nucleic Acids Res 37:3230–3242. doi: 10.1093/nar/gkp162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sola-Landa A, Rodriguez-Garcia A, Amin R, Wohlleben W, Martin JF. 2013. Competition between the GlnR and PhoP regulators for the glnA and amtB promoters in Streptomyces coelicolor. Nucleic Acids Res 41:1767–1782. doi: 10.1093/nar/gks1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Zhao GP. 2009. GlnR positively regulates nasA transcription in Streptomyces coelicolor. Biochem Biophys Res Commun 386:77–81. doi: 10.1016/j.bbrc.2009.05.147. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Cen XF, Zhao GP, Wang J. 2012. Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J Bacteriol 194:5237–5244. doi: 10.1128/JB.00989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Wang JZ, Shao ZH, Yuan H, Lu YH, Jiang WH, Zhao GP, Wang J. 2013. Three of four GlnR binding sites are essential for GlnR-mediated activation of transcription of the Amycolatopsis mediterranei nas operon. J Bacteriol 195:2595–2602. doi: 10.1128/JB.00182-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitani S, Ikeda H, Sakamoto T, Noguchi S, Nihira T. 2009. Characterization of a regulatory gene, aveR, for the biosynthesis of avermectin in Streptomyces avermitilis. Appl Microbiol Biotechnol 82:1089–1096. doi: 10.1007/s00253-008-1850-2. [DOI] [PubMed] [Google Scholar]
- 21.Wilson DJ, Xue Y, Reynolds KA, Sherman DH. 2001. Characterization and analysis of the pikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J Bacteriol 183:3468–3475. doi: 10.1128/JB.183.11.3468-3475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aparicio JF, Molnár I, Schwecke T, König A, Haydock SF, Khaw LE, Staunton J, Leadlay PF. 1996. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of the enzymatic domains in the modular polyketide synthase. Gene 169:9–16. doi: 10.1016/0378-1119(95)00800-4. [DOI] [PubMed] [Google Scholar]
- 23.He W, Lei J, Liu Y, Wang Y. 2008. The LuxR family members GdmRI and GdmRII are positive regulators of geldanamycin biosynthesis in Streptomyces hygroscopicus 17997. Arch Microbiol 189:501–510. doi: 10.1007/s00203-007-0346-2. [DOI] [PubMed] [Google Scholar]
- 24.Rascher A, Hu Z, Viswanathan N, Schirmer A, Reid R, Nierman WC, Lewis M, Hutchinson CR. 2003. Cloning and characterization of a gene cluster for geldanamycin production in Streptomyces hygroscopicus NRRL. 3602. FEMS Microbiol Lett 218:223–230. doi: 10.1016/S0378-1097(02)01148-5. [DOI] [PubMed] [Google Scholar]
- 25.Zhu Z, Li H, Yu P, Guo Y, Luo S, Chen Z, Mao X, Guan W, Li Y. 2016. SlnR is a positive pathway-specific regulator for salinomycin biosynthesis in Streptomyces albus. Appl Microbiol Biotechnol doi: 10.1007/s00253-016-7918-5. [DOI] [PubMed] [Google Scholar]
- 26.Lo Grasso L, Maffioli S, Sosio M, Bibb M, Puglia AM, Alduina R. 2015. Two master switch regulators trigger A40926 biosynthesis in Nonomuraea sp. strain ATCC 39727. J Bacteriol 197:2536–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horbal L, Kobylyanskyy A, Truman AW, Zaburranyi N, Ostash B, Luzhetskyy A, Marinelli F, Fedorenko V. 2014. The pathway-specific regulatory genes, tei15* and tei16*, are the master switches of teicoplanin production in Actinoplanes teichomyceticus. Appl Microbiol Biotechnol 98:9295–9309. doi: 10.1007/s00253-014-5969-z. [DOI] [PubMed] [Google Scholar]
- 28.Schaefer AL, Hanzelka BL, Eberhard A, Greenberg EP. 1996. Quorum sensing in Vibrio fischeri: probing autoinducer-LuxR interactions with autoinducer analogs. J Bacteriol 178:2897–2901. doi: 10.1128/jb.178.10.2897-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu H, Yao Y, Liu Y, Jiao R, Jiang W, Zhao GP. 2007. A complex role of Amycolatopsis mediterranei GlnR in nitrogen metabolism and related antibiotics production. Arch Microbiol 188:89–96. doi: 10.1007/s00203-007-0228-7. [DOI] [PubMed] [Google Scholar]
- 30.Yao W, Yang Y, Chiao J. 1994. IS204: an insertion sequence from Nocardia asteroides (mexicana) YP21. Plasmid 32:262–269. doi: 10.1006/plas.1994.1065. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Bao Y, Shi X, Ou X, Zhou P, Ding X. 2012. Efficient transposition of IS204-derived plasmids in Streptomyces coelicolor. J Microbiol Methods 88:67–72. doi: 10.1016/j.mimet.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 32.Ou X, Zhang B, Zhang L, Dong K, Liu C, Zhao G, Ding X. 2008. SarA influences the sporulation and secondary metabolism in Streptomyces coelicolor M145. Acta Biochim Biophys Sin (Shanghai) 40:877–882. doi: 10.1111/j.1745-7270.2008.00466.x. [DOI] [PubMed] [Google Scholar]
- 33.Ou X, Zhang B, Zhang L, Zhao G, Ding X. 2009. Characterization of rrdA, a TetR family protein gene involved in the regulation of secondary metabolism in Streptomyces coelicolor. Appl Environ Microbiol 75:2158–2165. doi: 10.1128/AEM.02209-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao ZH, Ren SX, Liu XQ, Xu J, Yan H, Zhao GP, Wang J. 2015. A preliminary study of the mechanism of nitrate-stimulated remarkable increase of rifamycin production in Amycolatopsis mediterranei U32 by RNA-seq. Microb Cell Fact 14:75. doi: 10.1186/s12934-015-0264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mejia A, Barrios-Gonzalez J, Viniegra-Gonzalez G. 1998. Overproduction of rifamycin B by Amycolatopsis mediterranei and its relationship with the toxic effect of barbital on growth. J Antibiot (Tokyo) 51:58–63. doi: 10.7164/antibiotics.51.58. [DOI] [PubMed] [Google Scholar]
- 36.Ding X, Tian Y, Chiao J, Zhao G, Jiang W. 2003. Stability of plasmid pA387 derivatives in Amycolatopsis mediterranei producing rifamycin. Biotechnol Lett 25:1647–1652. doi: 10.1023/A:1025698824679. [DOI] [PubMed] [Google Scholar]
- 37.Zhang B, Zhang L, Dai R, Yu M, Zhao G, Ding X. 2013. An efficient procedure for marker-free mutagenesis of S. coelicolor by site-specific recombination for secondary metabolite overproduction. PLoS One 8:e55906. doi: 10.1371/journal.pone.0055906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Ou X, Zhao G, Ding X. 2008. Highly efficient in vitro site-specific recombination system based on streptomyces phage phiBT1 integrase. J Bacteriol 190:6392–6397. doi: 10.1128/JB.00777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baltz RH. 2012. Streptomyces temperate bacteriophage integration systems for stable genetic engineering of actinomycetes (and other organisms). J Ind Microbiol Biotechnol 39:661–672. doi: 10.1007/s10295-011-1069-6. [DOI] [PubMed] [Google Scholar]
- 40.Gregory MA, Till R, Smith MC. 2003. Integration site for Streptomyces phage phiBT1 and development of site-specific integrating vectors. J Bacteriol 185:5320–5323. doi: 10.1128/JB.185.17.5320-5323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.