Abstract
Obesity is closely associated with various metabolic disorders. However, little is known about abnormalities in the metabolic change of obese adipose tissue. Here we use static metabolic analysis and in vivo metabolic turnover analysis to assess metabolic dynamics in obese mice. The static metabolic analyses showed that glutamate and constitutive metabolites of the TCA cycle were increased in the white adipose tissue (WAT) of ob/ob and diet-induced obesity mice but not in the liver or skeletal muscle of these obese mice. Moreover, in vivo metabolic turnover analyses demonstrated that these glucose-derived metabolites were dynamically and specifically produced in obese WAT compared with lean WAT. Glutamate rise in obese WAT was associated with down-regulation of glutamate aspartate transporter (GLAST), a major glutamate transporter for adipocytes, and low uptake of glutamate into adipose tissue. In adipocytes, glutamate treatment reduced adiponectin secretion and insulin-mediated glucose uptake and phosphorylation of Akt. These data suggest that a high intra-adipocyte glutamate level potentially relates to adipocyte dysfunction in obesity. This study provides novel insights into metabolic dysfunction in obesity through comprehensive application of in vivo metabolic turnover analysis in two obese animal models.
Keywords: adipocyte, adipose tissue metabolism, glutamate, metabolomics, obesity
Introduction
We and others have demonstrated that adipose tissue produces and secretes a variety of biologically active molecules conceptualized as adipocytokines/adipokines (1–5). Dysregulation of adipocytokines, including low adiponectin secretion, is considered to play a key role in the pathophysiology of metabolic disorders and atherosclerosis in obesity (6, 7).
The hypertrophied adipocytes in obese adipose tissue exhibit hyperlipolytic activity, providing excess free fatty acids and glycerol (8, 9). We reported recently that adipose tissue secretes uric acid and that the production of uric acid is augmented in obese mice (10). Overproduction of uric acid in obese adipose tissue may be associated with enhanced purine metabolism. Several recent reports showed changes in plasma amino acid profiles in obese subjects and obese animal models (11–15). These results suggest that metabolic derangement in obese adipose tissue extends beyond glucose and fatty acid metabolism. To date, the metabolic pathways of glucose, fatty acid, purine, and amino acid metabolism have been examined separately. Only a few studies investigated changes in several metabolic pathways in obese adipose tissues (16, 17). Moreover, there is no report on the metabolic dynamics in in vivo tissues of obesity.
The aim of this study was to systematically determine the static and dynamic metabolic changes in obese adipose tissue compared with lean adipose tissue and to define the association between metabolic changes and characteristics of hypertrophied adipocytes, focusing on adiponectin and insulin actions. For this purpose, we employed an in vivo metabolic turnover analysis (18–20) using a combination of in vivo stable isotope labeling and high-resolution metabolome analysis by time course sampling after intraperitoneal injection of isotope tracers.
Results
Static Metabolic Analysis of Obese Adipose Tissue
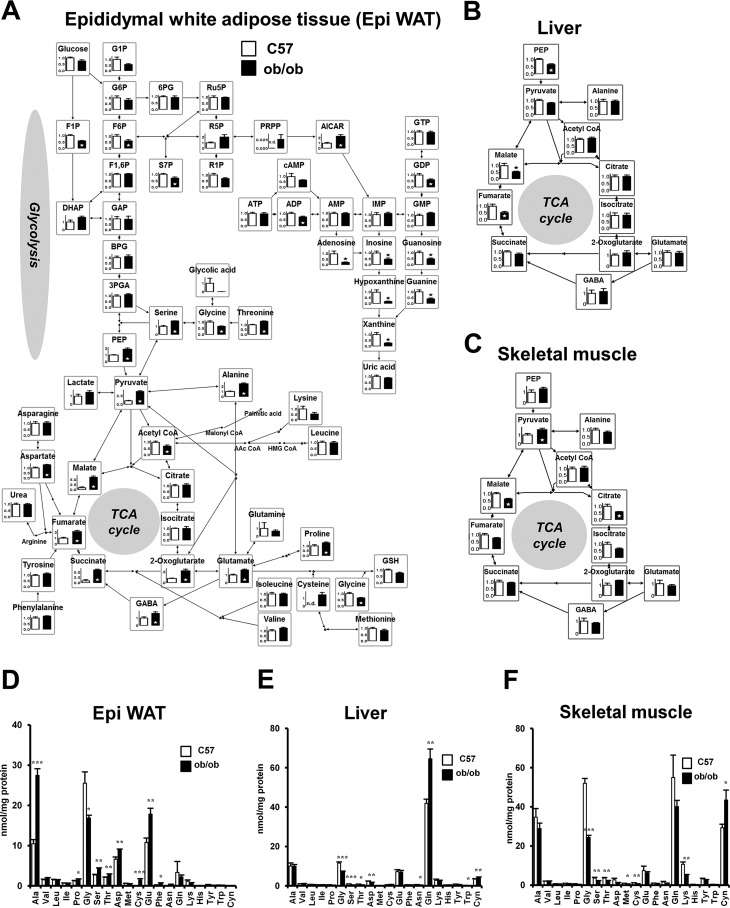
To examine the effect of obesity on adipose tissue metabolism, epididymal white adipose tissues (Epi WAT)2 from 11-week-old lean control male C57BL/6J (C57) mice and 11-week-old male B6.V-Lepob/J (ob/ob) mice were subjected to static metabolic analysis. Tissue metabolites were measured by LC/MS-MS and GC/MS. The relative levels of metabolites involved in glycolysis, the pentose phosphate pathway, TCA cycle, and amino acids are illustrated in the metabolic pathway map in Fig. 1A. Glycolytic intermediates, P-enolpyruvate, and pyruvate, and many constitutive metabolites of the TCA cycle, such as 2-oxoglutarate, succinate, fumarate, and malate, were significantly higher in ob/ob mice than in C57 mice. Among the amino acids, glutamate and alanine were significantly higher in ob/ob mice than in C57 mice. However, no such differences were observed in the liver or skeletal muscle between C57 and ob/ob mice (Fig. 1, B and C, and supplemental Fig. S1, A and B). In the liver, fumarate and malate were significantly lower in ob/ob mice than in C57 mice. Next, using GC/MS, the absolute quantity of amino acids was analyzed in Epi WAT, liver, and skeletal muscle in C57 and ob/ob mice (Fig. 1, D–F). The results showed high concentrations of alanine, glutamate, and glycine in Epi WAT compared with other amino acids. Moreover, the concentrations of alanine and glutamate were significantly higher in Epi WAT of ob/ob mice than of C57 mice.
FIGURE 1.
Static metabolic analysis of central carbon metabolism in Epi WAT, liver, and skeletal muscle of C57 and ob/ob mice. Analysis of 11-week-old male C57 and ob/ob mice (n = 6/group). A, relative levels of Epi WAT metabolites. B, changes in metabolites associated with the TCA cycle in the liver. C, changes in metabolites associated with the TCA cycle in skeletal muscle. The level of each metabolite in C57 mice was set at 1. *, p < 0.05, C57 versus ob/ob mice. D–F, quantitative amino acid analysis of Epi WAT (D), liver (E), and skeletal muscle (F). Data are mean ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001, C57 versus ob/ob mice. G1P, glucose 1-phosphate; G6P, glucose 6-phosphate; 6PG, 6-phosphogluconate; Ru5P, ribulose 5-phosphate; 5P, ribose 5-phosphate; F6P, fructose 6-phosphate; F1P, fructose 1-phosphate; F1,6P, fructose 1,6-bis-phosphate; S7P, sedoheptulose 7-phosphate. DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; BPG, 1,3-bisphosphoglycerate; 3PGA, 3-phosphoglycerate; PEP, P-enolpyruvate; PRPP, phosphoribosyl pyrophosphate; AICAR, 5-aminoimidazole-4-carboxamide ribonucleoside; n.d., not detected.
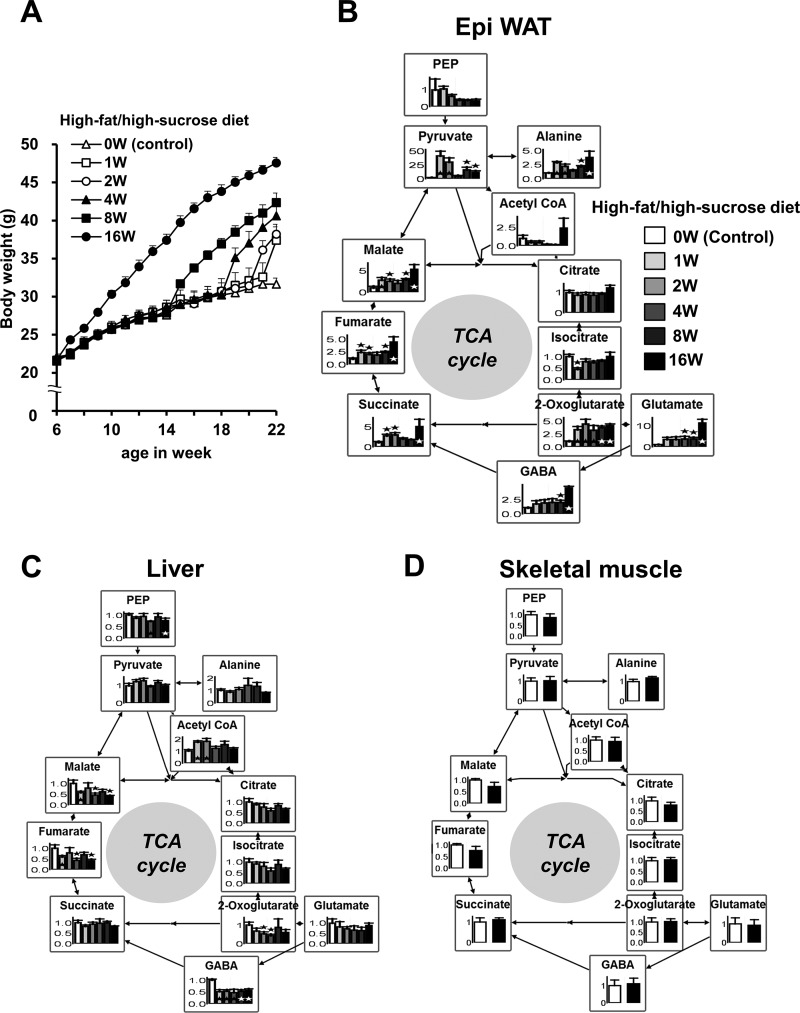
We also analyzed the metabolites in diet-induced obesity (DIO) mice (Fig. 2 and supplemental Fig. S2). Mice of the DIO group were fed a high-fat/high-sucrose diet from 6 weeks of age for 16 weeks, from 14 weeks of age for 8 weeks, from 18 weeks of age for 4 weeks, from 20 weeks of age for 2 weeks, and from 21 weeks of age for 1 week to 22 weeks of age, whereas mice of the control group were fed a regular diet from 6–22 weeks of age (0 weeks). Fig. 2A shows changes in body weight in each group. The relative levels of metabolites associated with the TCA cycle are illustrated in the metabolic pathway map in Fig. 2, B–D. In Epi WAT, glutamate, pyruvate, and many constitutive metabolites of the TCA cycle, such as 2-oxoglutarate, succinate, fumarate, and malate, were significantly higher in DIO mice than in control mice (Fig. 2B and supplemental Fig. S2). Most of these metabolites were elevated with an increase in body weight (Fig. 2, A and B). However, no such changes were noted in the liver or skeletal muscle (Fig. 2, C and D). In the liver, fumarate and malate were lower in DIO mice than in control mice. These differences were similar to those between C57 and ob/ob mice. Thus, the results of the static metabolic analyses suggest the presence of high levels of glutamate and many metabolites associated with the TCA cycle, specifically in adipose tissue but not in the liver or skeletal muscle tissues of obese mice.
FIGURE 2.
Static metabolic analysis of the TCA cycle for Epi WAT, liver, and skeletal muscle of DIO mice. At 6 weeks (W) of age, male C57 mice were randomly divided into six treatment groups (n = 6/group). DIO mice were fed a high-fat/high-sucrose diet from 6 weeks of age for 16 weeks, from 14 weeks of age for 8 weeks, from 18 weeks of age for 4 weeks, from 20 weeks of age for 2 weeks, and from 21 weeks of age for 1 week to 22 weeks of age, whereas mice of the control group were fed a regular diet from 6–22 weeks of age (0 weeks). All mice were analyzed at the age of 22 weeks after 4-h fasting. A, changes in body weight for each group. B, relative levels of metabolites in Epi WAT. C, relative levels of metabolites in the liver. D, relative levels of metabolites in skeletal muscle of control mice (0 weeks) and DIO mice (16 weeks). The level of each metabolite in the control group was set at 1. Data are mean ± S.E. *, p < 0.05, control versus DIO group(s).
Metabolic Turnover Analysis of Obese Adipose Tissue
The above metabolic changes in adipose tissue could be due to two mechanisms: rapid turnover of the TCA cycle resulted in high levels of glutamate and TCA cycle intermediates, or damming the flow of the TCA cycle caused accumulation of glutamate and TCA cycle intermediates. To investigate the differences in metabolic dynamics in each tissue between obese and lean mice, we performed intraperitoneal injection, which allows labeled substrates to reach each tissue through systemic circulation. To address this issue, the use of the isotope tracing method allows determination of not only static metabolite levels but also the dynamics of metabolism by monitoring the change in the distribution of isotopically labeled substrates. The metabolic turnover analysis was applied in this study, which involved intraperitoneal injection of 13C-labeled glucose followed by metabolomic analysis of isotopomers to clarify the metabolic dynamics in obese adipose tissue with a special focus on glutamate and constitutive metabolites of the TCA cycle.
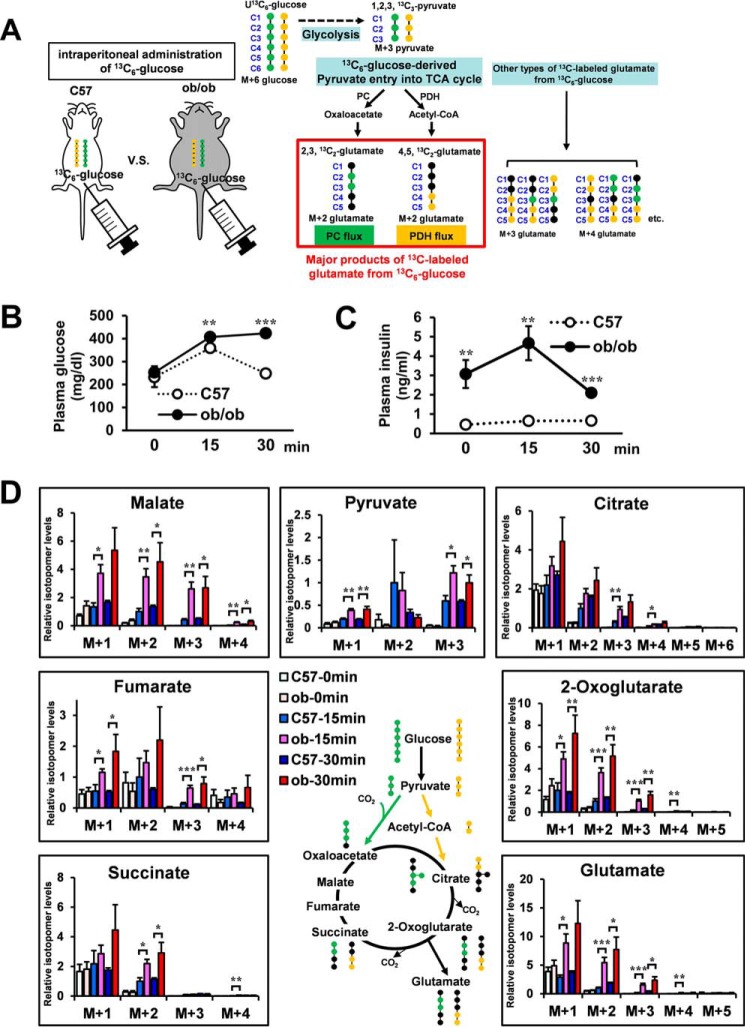
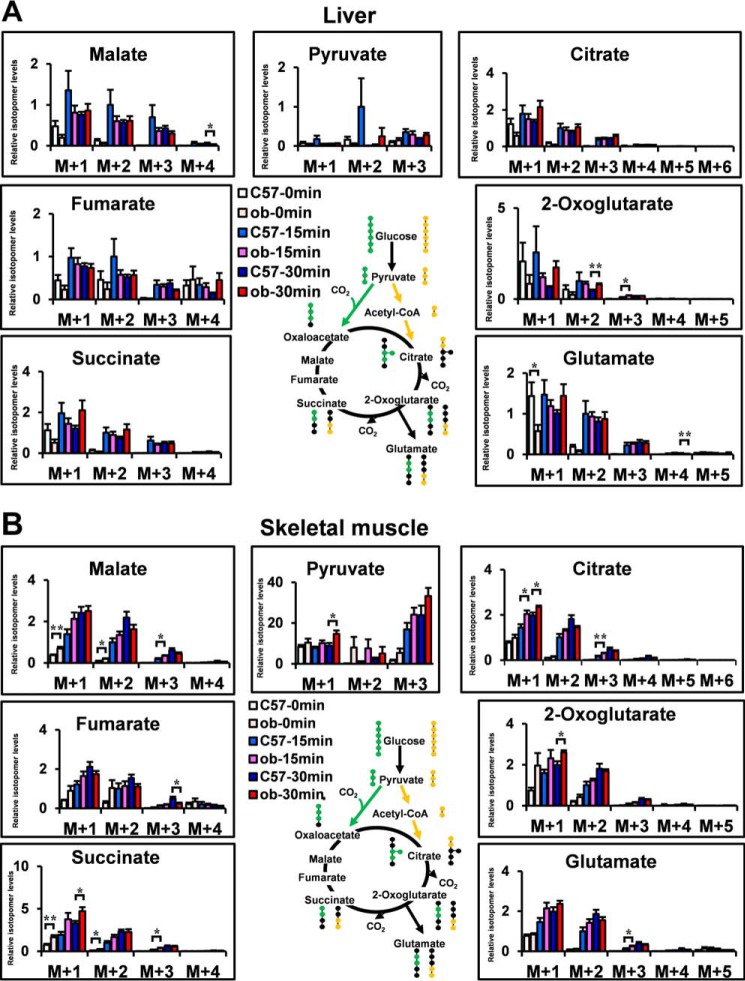
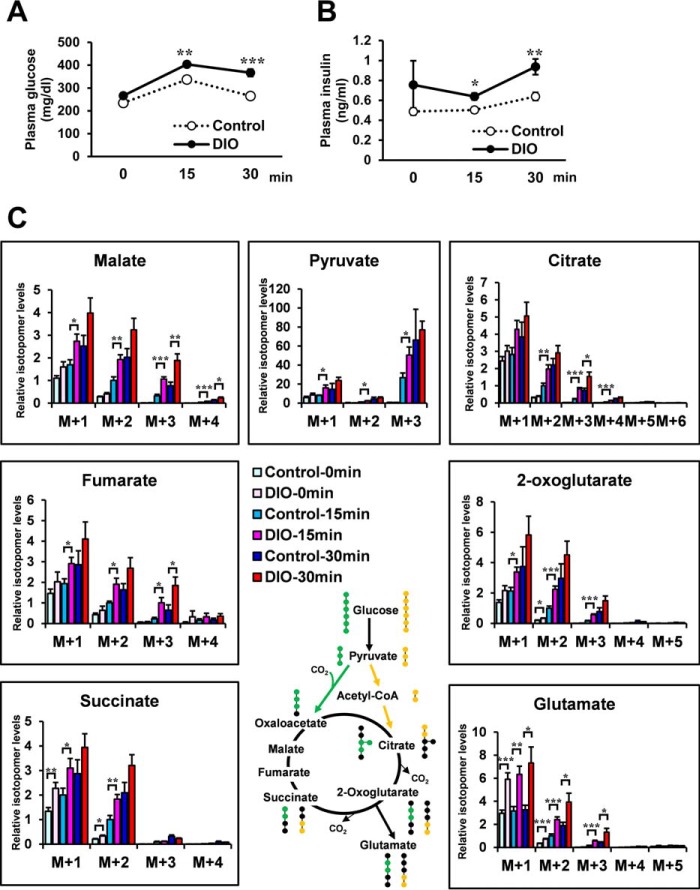
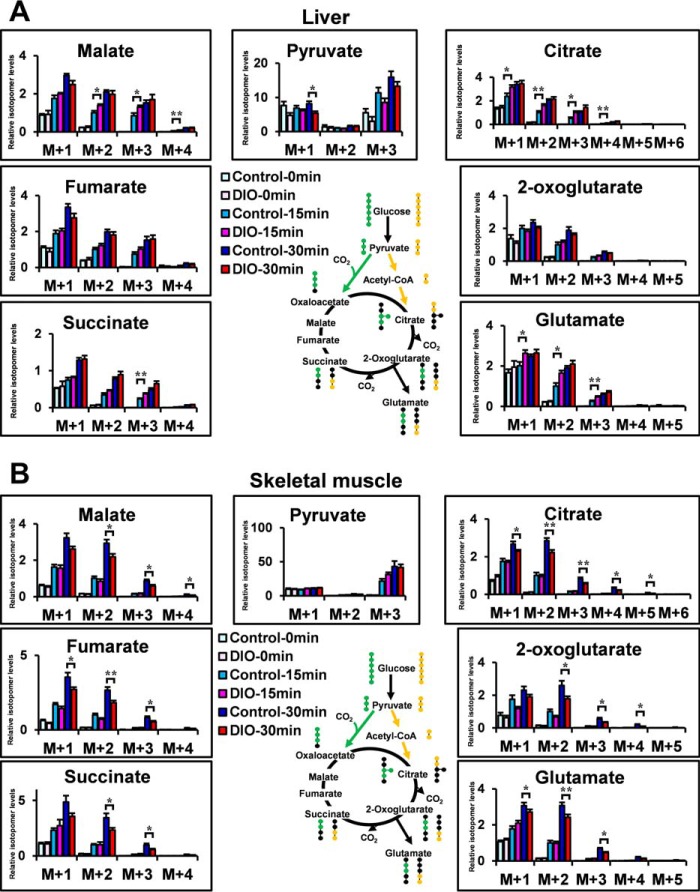
Fig. 3A shows the potential [13C]glutamate derived from [13C6]glucose. The isotopomer levels of metabolites are indicated as molar fractions, where M+1, M+2, M+3, etc. represent the number of 13C atoms in the molecule. The green circles in the compounds represent the fate of the 13C atoms via the pyruvate carboxylase (PC) flux, whereas the yellow circles represent the fate of the 13C atoms via the pyruvate dehydrogenase (PDH) flux (21). Hence, the [13C2]glutamate (M+2) is the isotopomer, which is generated mainly from injected [13C6]glucose through PC or PDH flux. To determine the optimal labeling time, first [13C6]glucose (1 mg/g of body weight) was injected intraperitoneally in 4-h-fasted C57 mice, and we analyzed the mass isotopomer distribution of each metabolite in Epi WAT, liver, and skeletal muscle (supplemental Fig. S3, A–C). Because the increase in the fraction of 13C-labeled isotopomers reached a plateau from 30–60 min after [13C6]glucose administration, we analyzed at 15 and 30 min after [13C6]glucose administration in the following study. M+0, mostly endogenous isotopomer, occupied a great part of each metabolite. To clearly demonstrate the changes in metabolic dynamics, M+1, M+2, and more were shown in the following metabolic turnover analyses. [13C6]glucose (1 mg/g of body weight) was injected intraperitoneally in 4-h-fasted C57 and ob/ob mice. Fig. 3, B and C, shows plasma glucose and plasma insulin of each group. 15 and 30 min after [13C6]glucose administration, Epi WAT, liver, and skeletal muscle were dissected out, and the isotopomer levels of metabolites in C57 and ob/ob mice of the non-administered (C57–0 min and ob-0 min), 15 min-administered (C57–15 min and ob-15 min), and 30 min-administered (C57–30 min and ob-30 min) were analyzed using LC-MS (Figs. 3D and 4, A and B). [13C6-glucose]derived constitutive metabolites of the TCA cycle, pyruvate and glutamate of Epi WAT in C57 and ob/ob mice, are shown in Fig. 3D. [13C3]pyruvate (M+3), [13C2]2-oxoglutarate (M+2), [13C2]succinate (M+2), [13C2]fumarate (M+2), [13C2]malate (M+2), [13C2]glutamate (M+2), [13C3]citrate (M+3), and [13C2]citrate (M+2) were the isotopomers, which were generated mainly from injected [13C6]glucose through PC or PDH flux. Interestingly, the levels of [13C3]pyruvate (M+3), [13C2]2-oxoglutarate (M+2), [13C2]succinate (M+2), [13C2]malate (M+2), [13C2]glutamate (M+2), and [13C3]citrate (M+3) were significantly higher in the ob-15 min group than in the C57–15 min group. The relationship between the C57–30 min group and ob-30 min group was almost similar to that between the C57–15 min group and the ob-15 min group. Thus, the results of the in vivo metabolic turnover analysis showed high levels of glucose-derived constitutive metabolites of the TCA cycle and glutamate biosynthesis of Epi-WAT in ob/ob mice compared with C57 mice. In contrast, no changes in [13C2]glutamate (M+2) were noted in the liver or skeletal muscle of ob/ob mice (Fig. 4, A and B) despite an increase in plasma glucose and insulin after intraperitoneal injection of [13C6]glucose (Fig. 3, B and C).
FIGURE 3.
Metabolic turnover analysis of Epi WAT in C57 and ob/ob mice. Four-hour-fasted 11-week-old male C57 and ob/ob mice were administered [13C6]glucose (0.2 mg/μl in saline, 1 mg/g of body weight) intraperitoneally. 15 and 30 min after [13C6]glucose administration, Epi WAT, liver, and skeletal muscle were harvested for analysis. C57 and ob/ob mice of the non-administered (C57–0 min and ob-0 min), 15 min-administered (C57–15 min and ob-15 min), and 30 min-administered (C57–30 min and ob-30 min) (n = 6 for each of the six groups) were analyzed for isotopomer levels of metabolites using LC/MS. A, representation of potential 13C-labeled glutamate isotopomers derived from [13C6]glucose. Green circles, 13C-labeled carbons in the isotopomers from pyruvate via PC flux; yellow circles, 13C-labeled carbons in the isotopomers from pyruvate via PDH flux. B, changes in plasma glucose levels. C, changes in plasma insulin levels. D, [13C6]glucose-derived constitutive metabolites of the TCA cycle, pyruvate and glutamate, in Epi WAT. The level of each M+2 isotopomer in C57 mice was set at 1. Data are mean ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001; C57 versus ob/ob mice at the same time after [13C6]glucose administration.
FIGURE 4.
Metabolic turnover analysis of the liver and skeletal muscle in C57 and ob/ob mice. A and B, [13C6]glucose-derived constitutive metabolites of the TCA cycle, pyruvate and glutamate of the liver (A) and skeletal muscle (B), in C57 and ob/ob mice. The level of each M+2 isotopomer in C57 mice was set at 1. Data are mean ± S.E. *, p < 0.05; **, p < 0.01; C57 versus ob/ob mice at the same time after [13C6]glucose administration.
We also analyzed the metabolic turnover in DIO mice. Mice of the DIO group were fed a high-fat/high-sucrose diet from 8–12 weeks of age, whereas those of the control group were fed a regular diet. [13C6]glucose (1 mg/g of body weight) was injected intraperitoneally in 4-h-fasted control and DIO mice. Fig. 5, A and B, shows changes in plasma glucose and plasma insulin for each group. 15 and 30 min after [13C6]glucose administration, [13C2]glutamate (M+2) of Epi WAT was significantly higher in DIO mice than in control mice (Fig. 5C). [13C2]glutamate (M+2) of the liver was higher in the DIO-15 min group than in the control-15 min group but not 30 min after [13C6]glucose administration (Fig. 6A). [13C2]glutamate (M+2) of skeletal muscle was slightly lower in the DIO-30 min group than in the control-30 min group (Fig. 6B). Epi WAT levels of [13C3]pyruvate (M+3), [13C2]2-oxoglutarate (M+2), [13C2]succinate (M+2), [13C2]fumarate (M+2), [13C2]malate (M+2), and [13C3]citrate (M+3) were significantly higher in the DIO-15 min group than in the control-15 min group (Fig. 5C). The relationship between the control-30 min group and DIO-30 min group was also similar to that between the control-15 min group and DIO-15 min group.
FIGURE 5.
Metabolic turnover analysis of Epi WAT in DIO mice. Four-hour-fasted 12-week-old male mice were administered [13C6]glucose (0.2 mg/μl in saline, 1 mg/g of body weight) intraperitoneally. DIO group mice were fed a high-fat/high-sucrose diet from 8–12 weeks of age, whereas control group mice were fed regular diet (n = 6/each group). A, changes in plasma glucose levels. B, changes in plasma insulin levels. C, [13C6]glucose-derived constitutive metabolites of the TCA cycle, pyruvate and glutamate, in Epi WAT. The level of each M+2 isotopomer in control mice was set at 1. Data are mean ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001; control mice versus DIO mice at the same time after [13C6]glucose administration.
FIGURE 6.
Metabolic turnover analysis of the liver and skeletal muscle in DIO mice. A and B, [13C6]glucose-derived constitutive metabolites of the TCA cycle, pyruvate and glutamate of the liver (A) and skeletal muscle (B), in DIO mice. The level of each M+2 isotopomer in C57 mice was set at 1. Data are mean ± S.E. *, p < 0.05; **, p < 0.01; control mice versus DIO mice at the same time after [13C6]glucose administration.
The above results suggest that the rise in glutamate level is due to the cataplerotic TCA cycle flux in adipose tissue of DIO mice, similar to ob/ob mice. Gene expression levels associated with the TCA cycle were not different between C57 and ob/ob mice (supplemental Fig. S4A). Gene expression levels and enzyme activities of aspartate transaminase (GOT), alanine transaminase (GPT), and glutamate dehydrogenase (GDH) in Epi WAT, which play a role in glutamate synthesis, were not different between C57 and ob/ob mice (supplemental Fig. S4, A–D).
Glutamate Transporters in Obese Adipose Tissue
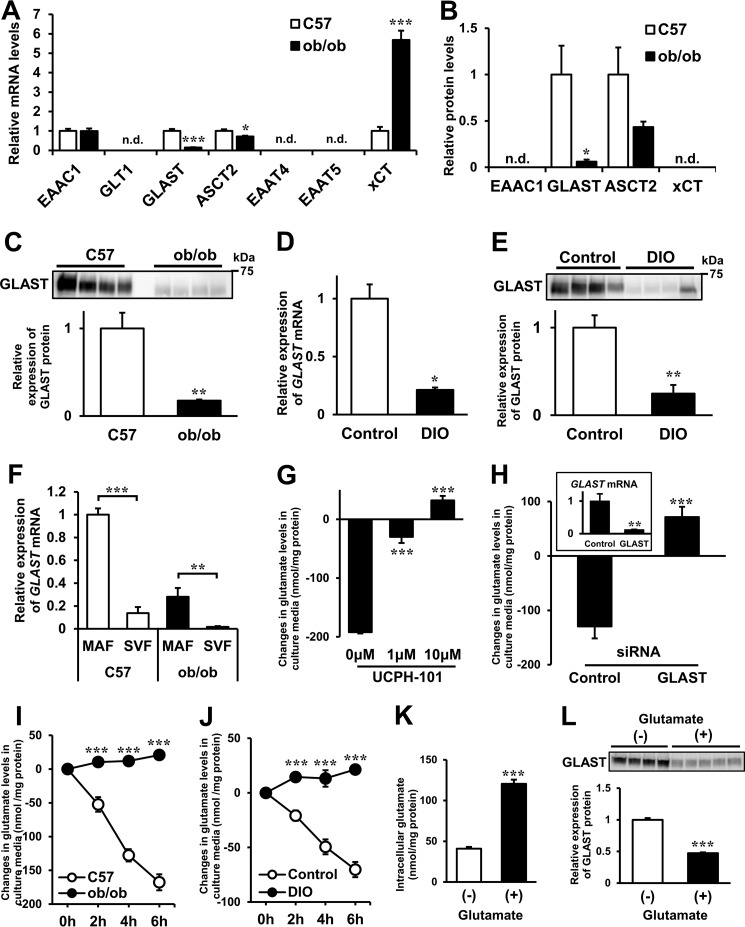
The metabolic turnover analysis of Epi WAT demonstrated that glutamate and constitutive metabolites of the TCA cycle were dynamically produced in obese adipose tissue. Next, to determine the glutamate transfer between the inside and outside of adipose tissue, we analyzed the expression levels and roles of glutamate transporters in adipose tissue. Several amino acid transporters are reported to transport glutamate (22–25). To determine the differences in glutamate transporters of adipose tissue between C57 and ob/ob mice, we examined the gene expression levels of candidate glutamate transporters using RT-PCR (Fig. 7A). The relative expression levels of GLAST and ASCT2 were lower and xCT were higher in ob/ob mice than in C57 mice. We also examined the protein expression levels of glutamate transporters in membrane fractions of Epi WAT by targeted proteomics using LC/MS-MS (Fig. 7B and supplemental Table S1). Protein expression of GLAST and ASCT2 was detected in both species, and the relative expression level of GLAST was markedly lower in ob/ob mice than in C57 mice. Protein expression of EAAC1 and xCT could not be detected. Western blotting analysis confirmed that the GLAST protein level was markedly lower in Epi WAT of ob/ob mice than of C57 mice (Fig. 7C). The mRNA and protein levels of GLAST in Epi WAT were also lower in DIO mice than in control mice (Fig. 7, D and E). GLAST is a sodium-dependent, high-affinity glutamate transporter (26, 27). Because the intracellular sodium concentration is much lower than that of extracellular sodium, GLAST is considered to play a role in glutamate uptake in adipose tissue.
FIGURE 7.
Glutamate transporters and transportation of glutamate in obese adipose tissue. A and B, gene (A, n = 8/group) and protein (B, n = 3/group) expression levels of glutamate transporters in Epi WAT of C57 and ob/ob mice using RT-PCR and LC/MS-MS, respectively. C, protein expression levels of GLAST in Epi WAT of C57 and ob/ob mice using Western blotting analysis (n = 4/group). Top panel, Western blotting analysis of GLAST. Bottom panel, quantitative analysis of GLAST contents in the top panel. Gene and protein levels in Epi WAT of C57 mice were set at 1 (A–C). *, p < 0.05; **, p < 0.01; ***, p < 0.001, C57 versus ob/ob mice; n.d., not detected. D, GLAST mRNA levels in Epi WAT of control and DIO mice (n = 6/group). DIO mice were fed a high-fat/high-sucrose diet from 6–22 weeks of age (16 weeks), whereas mice of the control group were fed a regular diet from 6–22 weeks of age. *, p < 0.05; control versus DIO mice. E, GLAST protein expression levels in Epi WAT of control and DIO mice by Western blotting analysis (n = 4/group) as described in C. Gene and protein levels in Epi WAT of control mice were set at 1. **, p < 0.01; control versus DIO mice. F, GLAST mRNA levels in the MAF and SVF of Epi WAT in C57 and ob/ob mice (n = 5–6/group). The GLAST mRNA level of MAF in C57 mice was set at 1. **, p < 0.01; ***, p < 0.001, MAF versus SVF. G, changes in glutamate levels in culture media from 0- to 2-h incubation of 3T3-L1 mature adipocytes with or without UCPH-101 (n = 3/group). ***, p < 0.001 versus control. H, knockdown of GLAST in 3T3-L1 mature adipocytes (n = 3/group). Shown are changes in glutamate levels in culture media of 3T3-L1 mature adipocytes transfected with siRNAs. GLAST mRNA levels in 3T3-L1 mature adipocytes transfected with siRNAs are shown in the inset. The GLAST mRNA level in 3T3-L1 mature adipocytes transfected with control-siRNA was set at 1. **p < 0.01; ***, p < 0.001 versus control. I, changes in glutamate levels in culture media obtained from organ culture of Epi WAT in C57 and ob/ob mice from 0–6 h over time (n = 6/group). ***, p < 0.001 versus C57 mice. J, changes in glutamate levels in culture media obtained from organ culture of Epi WAT in control and DIO mice from 0–6 h over time (n = 5/group). ***, p < 0.001 versus C57 mice. K and L, intracellular glutamate (K, n = 6/group) and GLAST protein levels (L, n = 4–5/group) in 3T3-L1 adipocytes treated with or without 5 mm glutamate for 6 days from 2 days after differentiation. ***, p < 0.001 versus control. The data in A–L are mean ± S.E.
Inhibition and Knockdown of GLAST in 3T3-L1 Mature Adipocytes
To identify the cell type that contributed to the observed GLAST levels, the mature adipocyte fraction (MAF) and stromal vascular cell fraction (SVF) were isolated from Epi WAT. The gene expression of GLAST in Epi WAT was detected mostly in the MAF (Fig. 7F). Therefore, we conducted the next experiment using 3T3-L1 mature adipocytes to clarify the role of GLAST in glutamate uptake in adipocytes. Addition of UCPH-101, a GLAST-specific inhibitor (28), significantly decreased glutamate uptake, and this effect was dose-dependent (Fig. 7G). To confirm that glutamate uptake in adipocytes was GLAST-dependent, we also performed GLAST gene knockdown experiments. After 48 h of GLAST siRNA transfection, GLAST mRNA levels were decreased by an average of 88% (Fig. 7H, inset). Then the culture medium was changed, and the cells were incubated for 2 h. Glutamate uptake in GLAST siRNA-transfected adipocytes was significantly lower than in control siRNA-transfected adipocytes (Fig. 7H). These results suggest that GLAST plays a key role in glutamate uptake in adipocytes. The organ culture experiment demonstrated continuing reduction of glutamate in the culture medium of adipose tissue derived from control mice but not from obese mice (ob/ob mice and DIO mice), suggesting that glutamate uptake was decreased in the adipose tissue of obese mice (Fig. 7, I and J).
Addition of 5 mm glutamate for 6 days from 2 days after differentiation increased intracellular glutamate in 3T3-L1 adipocytes (Fig. 7K). Under this condition, the GLAST protein expression level was significantly lower in the glutamate-added group than in the control group (Fig. 7L). These results suggest that high intracellular glutamate seems to reduce the GLAST protein expression level, resulting in low uptake of glutamate into adipocytes.
Effects of Glutamate on Adipocyte Function in 3T3-L1 Mature Adipocytes
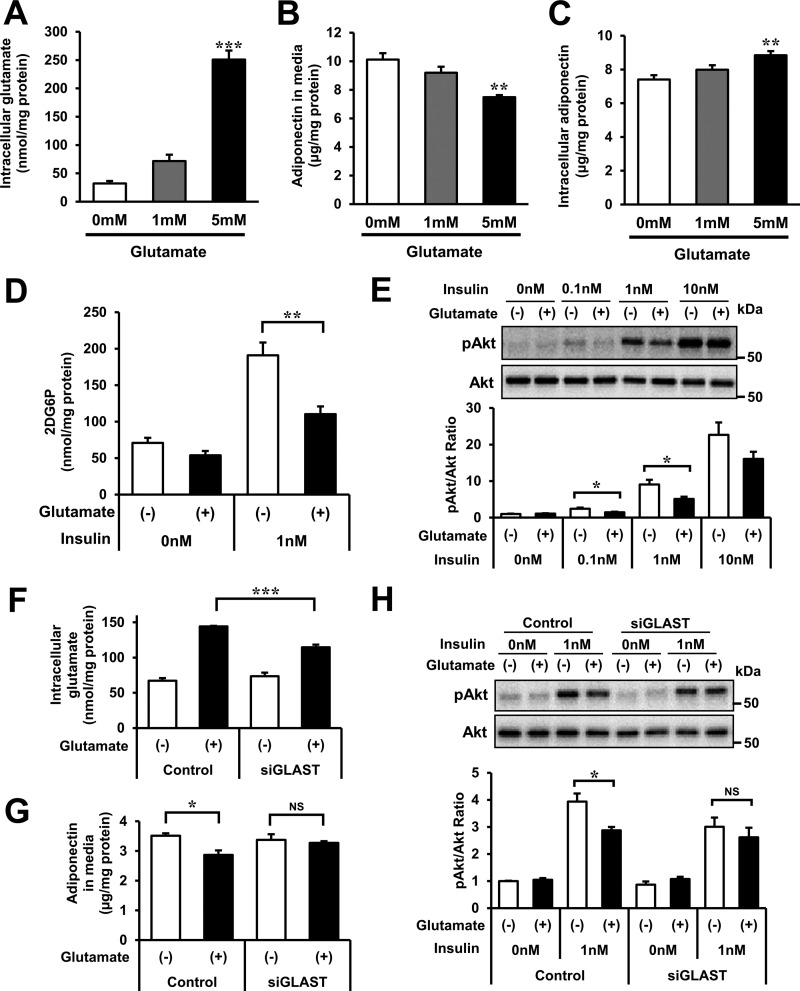
To investigate the effects of high intracellular glutamate on adipocyte function, 3T3-L1 mature adipocytes were incubated with or without 1 and 5 mm glutamate. The addition of 5 mm glutamate allowed intracellular glutamate to increase up to pathophysiological concentrations (Fig. 8A) and significantly reduced the accumulation of adiponectin in the incubation medium (Fig. 8B). The addition of glutamate increased intracellular adiponectin levels (Fig. 8C) with no significant changes of adiponectin mRNA levels (data not shown), suggesting that a glutamate rise in adipocytes inhibited adiponectin secretion.
FIGURE 8.
Effects of glutamate on adipocyte function in 3T3-L1 mature adipocytes. A–D, intracellular glutamate (A), adiponectin protein in culture media (B), and intracellular adiponectin protein (C) in experiments of 3T3-L1 mature adipocytes treated with or without 1 and 5 mm glutamate. In all experiments, cells were exposed to glutamate for 24 h. A–C, n = 4/group. D, glucose uptake as measured by 2DG-6-phosphate accumulation in 3T3-L1 mature adipocytes incubated with or without 5 mm glutamate for 24 h (n = 4/group). 3T3-L1 mature adipocytes were stimulated with or without 1 nm insulin for 10 min after 14 h FBS starvation. E, the ratio of serine-phosphorylated Akt (pAkt) to total Akt in 3T3-L1 mature adipocytes incubated with or without 5 mm glutamate for 24 h. 3T3-L1 mature adipocytes were stimulated with 0, 0.1, 1, or 10 nm insulin for 5 min after 14-h FBS starvation. Top panel, representative Western blotting analysis. Bottom panel, quantitative analysis of pAkt/Akt ratio in the top panel (n = 3/group). The pAkt/Akt ratio of the control group (without glutamate or insulin treatment) was set at 1. F and G, intracellular glutamate (F) and adiponectin protein levels in culture media (G) in gene knockdown experiments of GLAST in 3T3-L1 mature adipocytes treated with or without 5 mm glutamate for 24 h (n = 4/group). H, the ratio of serine-phosphorylated Akt to total Akt in knockdown of GLAST of 3T3-L1 mature adipocytes incubated with or without 5 mm glutamate for 24 h. 3T3-L1 mature adipocytes were stimulated with or without 1 nm insulin for 5 min after 14-h FBS starvation. Top panel, representative Western blotting analysis. Bottom panel, quantitative analysis of the pAkt/Akt ratio in the top panel (n = 3/group). The pAkt/Akt ratio of the control group was set at 1. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control. The data in A–H are mean ± S.E.
Furthermore, to examine the effect of high intracellular glutamate on glucose uptake and insulin signaling, we measured 2-deoxyglucose (2DG) uptake and the ratio of serine-phosphorylated Akt to total Akt in 3T3-L1 mature adipocytes with or without 5 mm glutamate treatment for 24 h (Fig. 8, D and E). The addition of glutamate significantly reduced insulin-mediated 2DG-6-phosphate accumulation, which was generated from 2DG in adipocytes (Fig. 8D). Moreover, glutamate significantly reduced 0.1 and 1 nm insulin-mediated Akt phosphorylation (Fig. 8E). No such effects of glutamate were observed following transfection of GLAST siRNA (Fig. 8, F–H), suggesting that glutamate, which was experimentally given to 3T3-L1 adipocytes, acted in cells by transportation through GLAST. Taken together, the results suggest that high intracellular glutamate decreases adiponectin secretion, insulin-mediated Akt phosphorylation, and glucose uptake in adipocytes.
Discussion
The results of static metabolic analysis showed specific increases in glutamate and several constitutive metabolites of the TCA cycle in obese adipose tissue (both ob/ob mice and DIO mice). These metabolic changes were not found in the liver or skeletal muscle.
Static metabolite levels represent a balance between production and consumption, and, therefore, they do not always reflect the real changes in metabolic dynamics. Several recent studies (29–34) employed isotope tracing analysis of cultured cells or in vivo tissue using 13C-labeled metabolites. In these studies, metabolic dynamics were evaluated by measuring the levels and distributions of 13C-labeled isotopomers. However, no study has comprehensively analyzed the metabolic dynamics in in vivo tissues of obesity. In this study, we examined the metabolic dynamics of obese adipose tissue, focusing on glutamate and constitutive metabolites of the TCA cycle using [13C6]glucose (Figs. 3 and 5). Epi WAT levels of [13C2]glutamate (M+2), [13C3]pyruvate (M+3), [13C2]2-oxoglutarate (M+2), [13C2]succinate (M+2), [13C2]malate (M+2), and [13C3]citrate (M+3) were significantly higher in obese mice (both ob/ob mice and DIO mice) than in control mice after intraperitoneal injection of [13C6]glucose. The results demonstrate increased biosynthesis of glucose-derived glutamate and constitutive metabolites of the TCA cycle in Epi WAT of obese mice compared with control mice. Gene expression levels and enzyme activities of GOT, GPT, and GDH in Epi WAT, which play a role in glutamate synthesis, were not different between C57 and ob/ob mice. These findings suggest that increased dynamics of obese adipose tissue could be related to incorporation of glucose as substrates for the TCA cycle. The TCA cycle in the mitochondrion is the main source of ATP in mammals. Using a 13C-labeled isotope tracing analysis of cultured cells, a recent study demonstrated reduced activity of the TCA cycle in association with mitochondrial dysfunction (34). Adipose tissue expands by energy oversupply (35–38), distinct from skeletal muscle and liver. For this feature, a large amount of ATP should be required for mRNA translation associated with cell proliferation (39–41). Hence, the obese adipose tissue-specific rapid turnover of the TCA cycle may relate to the ATP requirement for adipose expansion. We also found an increase in dihydroxyacetone phosphate (Fig. 1A and supplemental Fig. S2) and increased biosynthesis of citrate (Figs. 3D and 5C), suggesting that these might be associated with glycerolipid biosynthesis and triglyceride storage in obese adipose tissue. It is also possible that other cell types, including macrophages, in obese adipose tissue are involved in these metabolic changes. Further studies focusing on other cell types are also needed in future analyses.
Glutamate transporters play a role in glutamate transfer between the inside and outside of tissues and cells (22). To our knowledge, there are no reports of glutamate transporters in adipose tissue. Among the candidate glutamate transporters, GLAST protein levels in Epi WAT were markedly reduced in ob/ob and DIO mice. The gene expression levels of GLAST in the liver or skeletal muscle were not changed in lean and obese mice (data not shown). GLAST is a sodium-dependent, high-affinity glutamate transporter especially for glutamate uptake (26, 27). The cell experiments suggested that GLAST acted as the main transporter in glutamate uptake in adipocytes. Therefore, down-regulation of GLAST should link to decreased glutamate uptake in obese adipose tissue. In 3T3-L1 adipocytes, GLAST protein was down-regulated with increased intracellular glutamate levels. Taken together, decreased GLAST in obese adipose tissue could be negative feedback compensatory for the buildup of intracellular glutamate.
The results of our in vitro experiments suggest that high intracellular glutamate potentially relates to adipocyte dysfunction, including decreased adiponectin secretion and insulin-mediated glucose uptake. Glutamate treatment of 3T3-L1 adipocytes did not affect the levels of phosphorylated mechanistic target of rapamycin (mTOR) (data not shown). Further studies are needed to elucidate glutamate-dependent changes in specific pathways. From a therapeutic point of view to obesity-based metabolic diseases, further research is needed to elucidate upstream mechanisms of glutamate overproduction in obese adipose tissue and their effects on adiponectin secretion and insulin sensitivity.
In summary, glucose-derived TCA cycle dynamics may induce a rise in glutamate in obese adipose tissue, as confirmed by metabolomics technologies for static and dynamic analyses. Our results also suggest that high glutamate levels in obese adipose cells might disturb adiponectin secretion and insulin sensitivity in adipocytes. This study provides novel insights into metabolic dysfunction in obesity through comprehensive application of in vivo metabolic turnover analysis.
Experimental Procedures
Animals and Experimental Protocol
For metabolic phenotyping of Epi WAT, liver, and skeletal muscle, 11-week-old male C57 and ob/ob mice were used for these analyses. The gastrocnemius muscle was used as the skeletal muscle in all studies. For studies of DIO mice (42), at 6 weeks of age, C57 male mice were randomly divided into six treatment groups and fed either a high-fat/high-sucrose diet from 6 weeks of age for 16 weeks, from 14 weeks of age for 8 weeks, from 18 weeks of age for 4 weeks, from 20 weeks of age for 2 weeks, and from 21 weeks of age for 1 week to 22 weeks of age, whereas mice of the control group were fed a regular diet from 6–22 weeks of age (0 weeks). All mice were used at the age of 22 weeks for these analyses. After 4-h of fasting, tissues were removed and immediately frozen in liquid nitrogen. For metabolic turnover analysis of adipose tissue, 11-week-old male C57, 11-week-old ob/ob mice, and 12 week-old male DIO mice (mice of the DIO group were fed a high-fat/high-sucrose diet from 8–12 weeks of age, whereas control group mice were fed a regular diet) were injected intraperitoneally with [13C6]glucose (0.2 mg/μl in saline, 1 mg/g of body weight) after 4-h fasting. Tissues were dissected out (first, we obtained Epi WAT samples within 15 s, and then we dissected out the liver and skeletal muscle within 15 and 60 s, respectively) and frozen in liquid nitrogen and stored at −80 °C until use. Each sample of Epi WAT, liver, and skeletal muscle for metabolome analysis contained about 200 mg, 50 mg, and 75 mg tissue, respectively, for both lean and obese mice. The mice were housed in rooms set at 22 °C with a 12–12 h dark/light cycle (light cycle, 8 a.m. to 8 p.m.). All experimental protocols described in this study were approved by the Ethics Review Committee for Animal Experimentation of Osaka University School of Medicine.
Sample Preparation for Metabolome Analysis and Metabolic Turnover Analysis
For metabolites extraction of Epi WAT, liver, and skeletal muscle, the Blye-Dyer method was used with modifications (43). To extract compounds, 1 ml of mixed solvent (MeOH/H2O/CHCl3 = 5:2:2, with 0.01 mg/ml of ribitol and 0.01 μg/ml of (1S)-(+)-10-camphersulfonic acid as internal standard) was added to a 2-ml plastic tube containing each tissue and homogenized. The tube was incubated at 1200 rpm at 37 °C for 30 min and centrifuged at 16,000 × g at 4 °C for 3 min. For hydrophilic metabolites, 700 μl of the aqueous layer was transferred to a new plastic tube, mixed with 350 μl of water, and centrifuged at 16,000 × g at 4 °C for 3 min. To remove lipids, 800 μl of the aqueous layer was transferred to a new plastic tube, mixed with 400 μl of chloroform, and centrifuged at 16,000 × g at 4 °C for 3 min. For the GC/MS and LC/MS metabolome analysis, 300 μl and 320 μl of the upper aqueous layer were transferred to new 2-ml plastic tubes, respectively. For GC/MS analysis, the extract was centrifuge-dried for 90 min and freeze-dried overnight. The lyophilizate was suspended in 100 μl of pyridine containing 20 mg/ml methoxyamine hydrochloride and incubated at 1200 rpm at 30 °C for 90 min. Then the sample was mixed with 50 μl of N-methyl-N-(trimethylsilyl) trifluoroacetamide and incubated at 1200 rpm at 37 °C for 30 min. After incubation, the sample was subjected to GC/MS. For the LC/MS analysis, the extract was centrifuge-dried for 1 h. Then the solution was applied to a 5000 molecular weight cutoff ultrafiltration membrane and centrifuged at 16,000 × g until the whole solution was filtered. The filtrate were then freeze-dried. After suspension in 40 μl of water, the sample was subjected to LC/MS. The procedure of metabolic turnover analysis was similar to the one described for LC/MS analysis.
Metabolome Analysis and Metabolic Turnover Analysis
The GC/MS analysis was performed based on previous research using GCMS-QP2010 Ultra (Shimadzu, Kyoto, Japan) (44). For data analysis, metabolites were identified based on retention index and mass spectrum library. Each peak area was expressed relative to the peak area of the internal standard, ribitol, and is described in the text as relative quantity. The absolute quantity of each metabolite was determined using standard curves prepared in the same analytical batch. We used the modified LC/MS method described in detail previously (45). Metabolites were separated by reverse-phase ion pair LC and detected by multiple reaction monitoring by the MS. The conditions were set as follows: instrument, Nexera ultra high performance liquid chromatography (UHPLC) system-equipped LCMS 8030 Plus (Shimadzu); column, Mastro (150 mm length × 2.1 mm inner diameter, 3-μm particle size, Shimadzu GLC, Tokyo, Japan); mobile phase, 10 mm tributylamine and 15 mm acetic acid in water (A) and methanol (B); flow rate, 0.2 ml/min; gradient curve, 0% B at 0–1 min, 0–16% B at 1–2 min, 16–43% B at 4–17 min, 43–100% B at 17–21.5 min, 100–0% B at 23.5–24.5 min and 100–0% B at 13–13.5 min, and 0% B at 24.5–29 min. In the metabolic turnover analysis, LC conditions were similar to those of the metabolome analysis. The MS conditions were described previously (20). In the static metabolic analysis, the final result for each sample was normalized to the respective protein content. In the metabolic turnover analysis, the M+2 isotopomer level of each metabolite in control mice was set at 1 after adjustment for the respective protein content.
Organ Culture of Mouse Adipose Tissue
The adipose tissue organ culture was prepared as described previously (46, 47). Briefly, Epi WAT was obtained from 11-week-old C57 and ob/ob male mice (n = 6 each) and 22-week-old control and DIO male mice (DIO group mice were fed a high-fat/high-sucrose diet from 6–22 weeks of age (16 weeks), whereas mice of the control group were fed a regular diet from 6–22 weeks of age) (n = 5 each) after 4-h fasting. After the whole tissues were minced into small pieces and washed with 6 ml of calcium- and magnesium-free PBS, each tissue was placed onto a 6-well plate filled with 6 ml of DMEM containing 4.5 g/liter glucose and 10% FBS. After 0–2 h, 0–4 h, or 0–6 h culture, the culture media were collected and used for measurement of the glutamate level.
Quantification of mRNA Levels
Total RNA was isolated from mouse tissues as described previously (10) using the RNA STAT-60 (Tel-Test, Inc., Friendswood, TX) according to the protocol supplied by the manufacturer. The quality and quantity of total RNA were determined by using an ND-1000 spectrophotometer (Nano Drop Technologies, Wilmington, DE). First-strand cDNA was synthesized from 160 ng of total RNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science). Real-time quantitative PCR amplification was conducted with the LightCycler 1.5 (Roche Applied Science) using Light Cycler-FastStart DNA Master SYBR Green I (Roche Applied Science) according to the protocol recommended by the manufacturer. The sequences of primers used for real-time PCR were as follows: EAAC1, 5′-AGCCAGAAATCGTGCCAAAG-3′ and 5′-ACGGTCAGTCGGTAGCTTTC-3′; GLT1, 5′-TCTGAGGAGGCCAATACCAC-3′ and 5′-TTCATCCCGTCCTTGAACTC-3′; GLAST, 5′-CTGTTTCGGAATGCCTTCGTT-3′ and 5′-TCACCTCCCGGTAGCTCATTT-3′; ASCT2, 5′-CCAGGTGAAGAACGAGGTGT-3′ and 5′-CCAGTCCATTTCTCCAGCTC-3′; EAAT4, 5′-GGTCATTGGTGGCATGAAGC-3′ and 5′-AGATGATGATGCCCACCAGC-3′; EAAT5, 5′-TCATAGCTGTAGACTGGGCAC-3′ and 5′-AATCCTTCCGGCAGATGTGG-3′; adiponectin, 5′-GATGGCAGAGATGGCACTCC-3′ and 5′-CTTGCCAGTGCTGCCGTCAT-3′; xCT, 5′-CCTGGCATTTGGACGCTACAT-3′ and 5′-TCAGAATTGCTGTGAGCTTGCA-3′ and 36B4, 5′-GGCCAATAAGGTGCCAGCT-3′ and 5′-TGATCAGCCCGAAGGAGAAG-3′; Cs, 5′-AGGCTAGACTGGTCACACAAT-3′ and 5′-AGGACAGGTAAGGGTCTGAAAG-3′; IDH2, 5′-ACGAGCACTTCCTGAACACC-3′ and 5′-GCCATGTACAGAGTACCCACTG-3′; Ogdh, 5′-TCTGAGCCTTCAAGACAATGG-3′ and 5′-ATGCTTAGCTGCCCAAGATG-3′; GOT1, 5′-GCGCCTCCATCAGTCTTTG-3′ and 5′-ATTCATCTGTGCGGTACGCTC-3′; GOT2, 5′-GGACCTCCAGATCCCATCCT-3′ and 5′-GGTTTTCCGTTATCATCCCGGTA-3′; GPT, 5′-TCCAGGCTTCAAGGAATGGAC-3′ and 5′-CAAGGCACGTTGCACGATG-3′; and GDH, 5′-CTGGGATTGGACCTGAGAAC-3′ and 5′-TCGGCTGTGAGCTGTCTATG-3′. The final result for each sample was normalized to the respective 36B4 value.
Preparation of Membrane Fractions for Targeted Proteomics and Western Blotting Analysis
Epi WAT was homogenized at 33 mg of wet tissue/ml in a homogenization buffer (20 mm Tris-HCl (pH 7.6), 250 mm sucrose, 1 mm EDTA, and protease inhibitor mixture (Roche Diagnostics) using Hyscotron (Microtec, Chiba, Japan). The sample was further homogenized by using a Potter-Elvehjem homogenizer and centrifuged at 1000 × g for 5 min at 4 °C. The supernatant was centrifuged at 8000 × g for 5 min at 4 °C. Subsequently, the supernatant was centrifuged at 438,000 × g for 15 min at 4 °C. After ultracentrifugation, the membrane fraction was obtained as a pellet. The pellet of the membrane fraction was suspended in 50 ml of buffer containing 20 mm Tris-HCl (pH 7.6) and 250 mm sucrose. Protein levels were assayed using the BCA protein assay reagent kit (Thermo Fisher Scientific, Waltham, MA).
The membrane fraction was prepared for LC/MS measurement as described previously (48). Briefly, the membrane fraction was washed with 8 m urea and then dissolved in dissolution buffer (6 m urea, 0.1 m Na2CO3, and 0.5% sodium deoxycholate). A 10-μg protein was reduced with 2 mm tris(2-carboxyethyl)phosphine, alkylated with 55 mm iodoacetamide, and diluted with 0.1 m triethylammonium bicarbonate buffer (pH 8.5), followed by digestion with trypsin (trypsin-to-sample ratio = 1:40, Roche) for 3 h at 37 °C. After digestion, 100 fmol of protein digestion standard mixture (Waters, Milford, MA) was added as the internal standard of the sample. Then sodium deoxycholate was removed using the partial transmit sequence method (49). The digested sample was desalted by C18-StageTip (50). The desalted fractions were dried up under a vacuum and dissolved in 20 μl of a measurement buffer (3% acetonitrile and 0.1% formic acid).
LC/MS-MS for Targeted Proteomics
LC/MS measurement was performed using Q Exactive (Thermo Fisher Scientific) with Advance UHPLC (Michrom Bioresources, Auburn, CA) equipped with a trap column (L-column octadecylsilyl, 0.3 × 5 mm, CERI, Tokyo, Japan) and C18 packed tip column (100 μm inner diameter, Nikkyo Technos, Tokyo, Japan). The injection volume was 2 μl, and the flow rate was 400 nl/min. The column temperature was controlled at 60 °C using an ESCO column oven (AMR, Gifu, Japan). The mobile phases were composed of buffer A (0.1% formic acid) and buffer B (100% acetonitrile). The gradient condition was configured as follows: 5–40% buffer B in 100 min. The spray voltage was 2000 V, and the capillary temperature was 275 °C. The targeted MS (49) (parallel reaction monitoring) method was used with the following settings: resolution, 17,500; automatic gain control target, 1 × 105 ions; maximum IT, 200 ms; isolation window, 2.0 m/z; normalized collision energy, 27%. Peptides for the protein of interest in an inclusion list were selected from peptides detected by preliminary comprehensive proteomics. The inclusion list was developed using Pinpoint 1.3 (Thermo Fisher Scientific). All samples were subjected to triplicate analysis.
All raw data obtained by the parallel reaction monitoring method were analyzed using Pinpoint 1.3 (Thermo Fisher Scientific). At least two transitions per peptide from the protein of interest were selected for quantification, and then the quantitative value of the peptide was estimated based on peak areas of selected transitions. The reliability of the quantitative value was confirmed by peak features as described previously (51). The quantitative value was normalized by areas of peptides estimated using internal standard proteins. The quantitative value of the protein represented the mean of the peptide values.
Western Blotting Analysis
Protein extracts from cells were prepared as described previously (52). Preparation of protein extracts from adipose tissue is described under “Preparation of Membrane Fractions for Targeted Proteomics and Western Blotting Analysis.” For measurement of insulin-induced Akt phosphorylation in 3T3-L1 mature adipocytes, 3.3 μg of protein was subjected to a 4–20% gradient SDS-PAGE gel and then transferred onto a nitrocellulose membrane (GE Healthcare). The Western blotting analysis was performed first with anti-Akt and anti-phospho-Akt (Ser-473) antibodies (Cell Signaling Technology, Danvers, MA), followed by incubation with secondary antibody conjugated with horseradish peroxidase. Detection was achieved using the enhanced chemiluminescence kit (GE Healthcare).
For measurement of GLAST in adipose tissue, 30 μg of protein was subjected to 10% SDS-PAGE gel and then transferred onto a PVDF membrane (GE Healthcare). Western blotting analysis was performed first with anti-GLAST antibody (Santa Cruz Biotechnology, Santa Cruz, CA), followed by incubation with secondary antibody conjugated with horseradish peroxidase. Detection was achieved using the enhanced chemiluminescence kit (GE Healthcare).
Fractionation of Mouse Adipose Tissue
Epi WAT from 16-week-old C57 male mice was fractionated using a method described previously (53). Adipose tissues were minced in Krebs-Ringer bicarbonate HEPES buffer containing 120 mmol/liter NaCl, 4 mmol/liter KH2PO4, 1 mmol/liter MgSO4, 1 mmol/liter CaCl2, 10 mmol/liter NaHCO3, 30 mmol/liter HEPES, 20 μmol/liter adenosine, and 4% bovine serum albumin. Tissue suspensions were centrifuged at 500 × g for 5 min to remove erythrocytes and free leukocytes. Collagenase was added to a final concentration of 2 mg/ml, followed by incubation at 37 °C for 30 min under continuous shaking. The cell suspension was filtered through a 250-μm filter and then spun at 300 × g for 1 min to separate the floating MAF from the SVF pellet. The fractionation and washing procedures were repeated twice with Krebs-Ringer bicarbonate HEPES buffer. Finally, both fractions were washed with PBS and used for mRNA analysis.
3T3-L1 Cell Cultures
3T3-L1 cells were differentiated as described previously (54). 3T3-L1 cells were maintained in DMEM with 4.5 g/liter glucose, 100 units/ml penicillin, 100 g/ml streptomycin, and 10% FBS. On day 8 after differentiation, 3T3-L1 mature adipocytes were incubated in the presence or absence of 1 or 10 μm UCPH-101 (Abcam, Cambridge, UK), a GLAST-specific inhibitor (28), dissolved in dimethyl sulfoxide. The cells were harvested 2 h later, and the culture media were analyzed for glutamate levels. In glutamate treatment studies, on day 8 after differentiation, 3T3-L1 mature adipocytes were incubated in the absence or presence of 1 and 5 mm glutamate, and cells were harvested 24 h later. In the insulin-induced Akt phosphorylation study, 3T3-L1 mature adipocytes were stimulated with 0, 0.1, 1, or 10 nm insulin for 5 min after 14 h of FBS starvation. Before the insulin treatment, 3T3-L1 adipocytes were incubated with or without 5 mm glutamate for 24 h (the first 10 h of incubation with FBS, followed by 14 h of incubation without FBS).
Knockdown of GLAST in 3T3-L1 Mature Adipocytes
On day 7 after induction of differentiation, 3T3-L1 mature adipocytes were transfected with GLAST siRNA (catalog no. 10620318 Slc1a3MSS237927, Invitrogen) using Lipofectamine RNAiMAX reagent (Invitrogen) according to the protocol recommended by the manufacturer. After 48-h incubation of the transfected cells, the culture medium was replaced with fresh medium, followed by incubation for another 2 h. Then the cells were harvested, and culture media were collected for measurement of the glutamate level. Allstars negative control siRNA (Qiagen, Valencia, CA) was used as a control.
Glucose Uptake
On day 8 after differentiation, 3T3-L1 mature adipocytes were incubated with or without 5 mm glutamate for 24 h (the first 10 h of incubation with FBS, followed by 14 h of incubation without FBS) and then incubated with Krebs Ringer phosphate HEPES buffer (1.2 mm KH2PO4, 1.2 mm MgSO4, 1.3 mm CaCl2, 118 mm NaCl, 5 mm KCl, and 30 mm HEPES (pH 7.5)) containing 2% bovine serum albumin (Merck Millipore, Darmstadt, Germany) for 30 min with or without 5 mm glutamate. Then cells were treated with insulin solution (final concentration, 0 or 1 nm) for 20 min. Subsequently, the cells were treated with 2DG (Wako Pure Chemical, Osaka, Japan) solution (final concentration, 1 mm) for 10 min. After incubation, the cells were treated with 40 μm cytochalasin B (Wako Pure Chemical) and washed twice with PBS. The cells were lysed with 10 mm Tris-HCl (pH 8.0) and disrupted by microtip sonicator. The cell lysate was heat-treated at 80 °C for 15 min and centrifuged at 15,000 × g for 20 min at 4 °C. Glucose uptake was assayed using a kit (CSR OKP-PMG-K01, Cosmobio, Tokyo, Japan) and the protocol described by the manufacturer.
Biochemical Measurements
Plasma glucose and plasma insulin were measured by glucose CII test (Wako Pure Chemical) and the insulin measurement ELISA kit (Morinaga, Yokohama, Japan), respectively. Glutamate in culture media and intracellular glutamate were analyzed using the Amplex Red glutamic acid/glutamate oxidase assay kit (Invitrogen). Adiponectin in culture media and intracellular adiponectin were assayed using the adiponectin ELISA kit (Otsuka, Tokushima, Japan). Tissue and cultured cells were homogenized in 50 mm Tris-HCl (pH 7.5), 0.1% Triton X-100, and a mixture of complete protease inhibitor (Roche Applied Science) and centrifuged at 20,000 × g for 15 min at 4 °C. Protein concentrations were determined by using the BCA protein assay (Thermo Fisher Scientific).
Statistical Analysis
Results are presented as mean ± S.E. The statistical significance of differences between groups was determined using two-tailed t tests or the Dunnett's test. In all cases, differences were considered significant when the p < 0.05. All analyses were performed using JMP Statistical Discovery Software 11.2.1 (SAS Institute, Cary, NC).
Author Contributions
H. Nagao designed and performed the experiments, analyzed the data, and wrote the manuscript. H. Nishizawa conceived the study, designed the experiments, and wrote the manuscript. T. B. and Y. N. performed the metabolomic analyses, analyzed the data, and wrote the manuscript. N. I., S. N., and Y. K. performed the protein expression analyses for glutamate transporters and wrote the manuscript. Y. T., S. K., S. F., and N. M. performed part of the animal experiments and contributed to the discussions. T. F., E. F., and I. S. conceived the study and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Yu Tsushima for helpful discussions and technical advice. We also thank Kayoko Ohashi (Department of Metabolic Medicine, Graduate School of Medicine, Osaka University), Takanori Kobayashi, and Satori Matsumoto (Department of Bio-system Pharmacology, Graduate School of Medicine, Osaka University) for excellent technical assistance and all members of the IIIrd laboratory (Adiposcience Laboratory), Department of Metabolic Medicine, Osaka University, for helpful discussions.
This work was supported in part by Grant-in-Aid for Scientific Research (C) 15K09412 (to H. Nishizawa), Grant-in-Aid for Scientific Research (B) 26293221 (to T. F.), Grant-in-Aid for Scientific Research Innovative Areas “HD Physiology” 25136714 (to S. N.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and Core Research for Evolutional Science and Technology (CREST) program of Japan Science and Technology Agency (JST) (to E. F and I. S.). S. K., T. F., and N. M. are members of the Department of Metabolism and Atherosclerosis, a sponsored course endowed by Kowa Co. Ltd. The company has a scientific officer who oversees the program.

This article contains supplemental Figs. S1–S4 and Tables S1 and S2.
- Epi WAT
- epididymal white adipose tissue
- DIO
- diet-induced obesity
- PC
- pyruvate carboxylase
- PDH
- pyruvate dehydrogenase
- GLAST
- glutamate aspartate transporter
- GOT
- aspartate transaminase
- GPT
- alanine transaminase
- GDH
- glutamate dehydrogenase
- MAF
- mature adipocyte fraction
- SVF
- stromal vascular fraction
- 2DG
- 2-deoxyglucose.
References
- 1. Flier J. S. (2004) Obesity wars: molecular progress confronts an expanding epidemic. Cell 23, 337–350 [DOI] [PubMed] [Google Scholar]
- 2. Friedman J. M., and Halaas J. L. (1998) Leptin and the regulation of body weight in mammals. Nature 395, 763–770 [DOI] [PubMed] [Google Scholar]
- 3. Hotamisligil G. S., Shargill N. S., and Spiegelman B. M. (1993) Adipose expression of tumor necrosis factor-α: direct role in obesity-linked insulin resistance. Science 259, 87–91 [DOI] [PubMed] [Google Scholar]
- 4. Maeda K., Okubo K., Shimomura I., Funahashi T., Matsuzawa Y., and Matsubara K. (1996) cDNA cloning and expression of a novel adipose specific collagen-like factor, apM1 (AdiPose most abundant gene transcript 1). Biochem. Biophys. Res. Commun. 221, 286–289 [DOI] [PubMed] [Google Scholar]
- 5. Shimomura I., Funahashi T., Takahashi M., Maeda K., Kotani K., Nakamura T., Yamashita S., Miura M., Fukuda Y., Takemura K., Tokunaga K., and Matsuzawa Y. (1996) Enhanced expression of PAI-1 in visceral fat: possible contributor to vascular disease in obesity. Nat. Med. 2, 800–803 [DOI] [PubMed] [Google Scholar]
- 6. Matsuzawa Y., Funahashi T., Kihara S., and Shimomura I. (2004) Adiponectin and metabolic syndrome. Arterioscler. Thromb. Vasc. Biol. 24, 29–33 [DOI] [PubMed] [Google Scholar]
- 7. Rosen E. D., and Spiegelman B. M. (2006) Adipocytes as regulators of energy balance and glucose homeostasis. Nature 444, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smith U., Hammersten J., Björntorp P., and Kral J. G. (1979) Regional differences and effect of weight reduction on human fat cell metabolism. Eur. J. Clin. Invest. 9, 327–332 [DOI] [PubMed] [Google Scholar]
- 9. Mittelman S. D., Van Citters G. W., Kirkman E. L., and Bergman R. N. (2002) Extreme insulin resistance of the central adipose depot in vivo. Diabetes 51, 755–761 [DOI] [PubMed] [Google Scholar]
- 10. Tsushima Y., Nishizawa H., Tochino Y., Nakatsuji H., Sekimoto R., Nagao H., Shirakura T., Kato K., Imaizumi K., Takahashi H., Tamura M., Maeda N., Funahashi T., and Shimomura I. (2013) Uric acid secretion from adipose tissue and its increase in obesity. J. Biol. Chem. 288, 27138–27149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamakado M., Tanaka T., Nagao K., Ishizaka Y., Mitushima T., Tani M., Toda A., Toda E., Okada M., Miyano H., and Yamamoto H. (2012) Plasma amino acid profile is associated with visceral fat accumulation in obese Japanese subjects. Clin. Obes. 2, 29–40 [DOI] [PubMed] [Google Scholar]
- 12. Newgard C. B., An J., Bain J. R., Muehlbauer M. J., Stevens R. D., Lien L. F., Haqq A. M., Shah S. H., Arlotto M., Slentz C. A., Rochon J., Gallup D., Ilkayeva O., Wenner B. R., Yancy W. S. Jr, et al. (2009) A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 9, 311–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakamura H., Jinzu H., Nagao K., Noguchi Y., Shimba N., Miyano H., Watanabe T., and Iseki K. (2014) Plasma amino acid profiles are associated with insulin, C-peptide and adiponectin levels in type 2 diabetic patients. Nutr. Diabetes 4, e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. She P., Van Horn C., Reid T., Hutson S. M., Cooney R. N., and Lynch C. J. (2007) Obesity-related elevations in plasma leucine are associated with alterations in enzymes involved in branched-chain amino acid metabolism. Am. J. Physiol. Endocrinol. Metab. 293, 1552–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. She P., Olson K. C., Kadota Y., Inukai A., Shimomura Y., Hoppel C. L., Adams S. H., Kawamata Y., Matsumoto H., Sakai R., Lang C. H., and Lynch C. J. (2013) Leucine and protein metabolism in obese Zucker rats. PLoS ONE 8, e59443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giesbertz P., Padberg I., Rein D., Ecker J., Höfle A. S., Spanier B., and Daniel H. (2015) Metabolite profiling in plasma and tissues of ob/ob and db/db mice identifies novel markers of obesity and type 2 diabetes. Diabetologia 58, 2133–2143 [DOI] [PubMed] [Google Scholar]
- 17. Cummins T. D., Holden C. R., Sansbury B. E., Gibb A. A., Shah J., Zafar N., Tang Y., Hellmann J., Rai S. N., Spite M., Bhatnagar A., and Hill B. G. (2014) Metabolic remodeling of white adipose tissue in obesity. Am. J. Physiol. Endocrinol. Metab. 307, 262–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harada K., Fukusaki E., Bamba T., Sato F., and Kobayashi A. (2006) In vivo 15N-enrichment of metabolites in suspension cultured cells and its application to metabolomics. Biotechnol. Prog. 22, 1003–1011 [DOI] [PubMed] [Google Scholar]
- 19. Nakayama Y., Putri S. P., Bamba T., and Fukusaki E. (2014) Metabolic distance estimation based on principle component analysis of metabolic turnover. J. Biosci. Bioeng. 118, 350–355 [DOI] [PubMed] [Google Scholar]
- 20. Shimizu R., Dempo Y., Nakayama Y., Nakamura S., Bamba T., Fukusaki E., and Fukui T. (2015) New insight into the role of the Calvin cycle: reutilization of CO2 emitted through sugar degradation. Sci. Rep. 5, 11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Varma V., Boros L. G., Nolen G. T., Chang C. W., Wabitsch M., Beger R. D., and Kaput J. (2015) Metabolic fate of fructose in human adipocytes: a targeted 13C tracer fate association study. Metabolomics 11, 529–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Danbolt N. C. (2001) Glutamate uptake. Prog. Neurobiol. 65, 1–105 [DOI] [PubMed] [Google Scholar]
- 23. Storck T., Schulte S., Hofmann K., and Stoffel W. (1992) Structure, expression, and functional analysis of a Na+-dependent glutamate/aspartate transporter from rat brain. Proc. Natl. Acad. Sci. U.S.A. 89, 10955–10959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Utsunomiya-Tate N., Endou H., and Kanai Y. (1996) Cloning and functional characterization of a system ASC-like Na+-dependent neutral amino acid transporter. J. Biol. Chem. 271, 14883–14890 [DOI] [PubMed] [Google Scholar]
- 25. Sato H., Tamba M., Ishii T., and Bannai S. (1999) Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J. Biol. Chem. 274, 11455–11458 [DOI] [PubMed] [Google Scholar]
- 26. Conradt M., Storck T., and Stoffel W. (1995) Localization of N-glycosylation sites and functional role of the carbohydrate units of GLAST-1, a cloned rat brain L-glutamate/L-aspartate transporter. Eur. J. Biochem. 229, 682–687 [DOI] [PubMed] [Google Scholar]
- 27. Schulte S., and Stoffel W. (1995) UDP galactose:ceramide galactosyltransferase and glutamate/aspartate transporter: copurification, separation and characterization of the two glycoproteins. Eur. J. Biochem. 233, 947–953 [DOI] [PubMed] [Google Scholar]
- 28. Abrahamsen B., Schneider N., Erichsen M. N., Huynh T. H., Fahlke C., Bunch L., and Jensen A. A. (2013) Allosteric modulation of an excitatory amino acid transporter: the subtype-selective inhibitor UCPH-101 exerts sustained inhibition of EAAT1 through an intramonomeric site in the trimerization domain. J. Neurosci. 33, 1068–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gheni G., Ogura M., Iwasaki M., Yokoi N., Minami K., Nakayama Y., Harada K., Hastoy B., Wu X., Takahashi H., Kimura K., Matsubara T., Hoshikawa R., Hatano N., Sugawara K., et al. (2014) Glutamate acts as a key signal linking glucose metabolism to incretin/cAMP action to amplify insulin secretion. Cell Rep. 9, 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fendt S. M., Bell E. L., Keibler M. A., Olenchock B. A., Mayers J. R., Wasylenko T. M., Vokes N. I., Guarente L., Vander Heiden M. G., and Stephanopoulos G. (2013) Reductive glutamine metabolism is a function of the α-ketoglutarate to citrate ratio in cells. Nat. Commun. 4, 2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Metallo C. M., Gameiro P. A., Bell E. L., Mattaini K. R., Yang J., Hiller K., Jewell C. M., Johnson Z. R., Irvine D. J., Guarente L., Kelleher J. K., Vander Heiden M. G., Iliopoulos O., and Stephanopoulos G. (2011) Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Overmyer K. A., Evans C. R., Qi N. R., Minogue C. E., Carson J. J., Chermside-Scabbo C. J., Koch L. G., Britton S. L., Pagliarini D. J., Coon J. J., and Burant C. F. (2015) Maximal oxidative capacity during exercise is associated with skeletal muscle fuel selection and dynamic changes in mitochondrial protein acetylation. Cell Metab. 21, 468–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Le A., Lane A. N., Hamaker M., Bose S., Gouw A., Barbi J., Tsukamoto T., Rojas C. J., Slusher B. S., Zhang H., Zimmerman L. J., Liebler D. C., Slebos R. J., Lorkiewicz P. K., Higashi R. M., et al. (2012) Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 15, 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morita M., Gravel S. P., Chénard V., Sikström K., Zheng L., Alain T., Gandin V., Avizonis D., Arguello M., Zakaria C., McLaughlan S., Nouet Y., Pause A., Pollak M., Gottlieb E., et al. (2013) mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 18, 698–711 [DOI] [PubMed] [Google Scholar]
- 35. Drolet R., Richard C., Sniderman A. D., Mailloux J., Fortier M., Huot C., Rhéaume C., and Tchernof A. (2008) Hypertrophy and hyperplasia of abdominal adipose tissues in women. Int. J. Obes. 32, 283–291 [DOI] [PubMed] [Google Scholar]
- 36. Jo J., Gavrilova O., Pack S., Jou W., Mullen S., Sumner A. E., Cushman S. W., and Periwal V. (2009) Hypertrophy and/or hyperplasia: dynamics of adipose tissue growth. PLoS Comput. Biol. 5, e1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Spalding K. L., Arner E., Westermark P. O., Bernard S., Buchholz B. A., Bergmann O., Blomqvist L., Hoffstedt J., Näslund E., Britton T., Concha H., Hassan M., Rydén M., Frisén J., and Arner P. (2008) Dynamics of fat cell turnover in humans. Nature 453, 783–787 [DOI] [PubMed] [Google Scholar]
- 38. Zeve D., Tang W., and Graff J. (2009) Fighting fat with fat: the expanding field of adipose stem cells. Cell Stem Cell 5, 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Buttgereit F., and Brand M. D. (1995) A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J. 312, 163–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rolfe D. F., and Brown G. C. (1997) Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol. Rev. 77, 731–758 [DOI] [PubMed] [Google Scholar]
- 41. Vander Heiden M. G., Cantley L. C., and Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 22, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maeda N., Shimomura I., Kishida K., Nishizawa H., Matsuda M., Nagaretani H., Furuyama N., Kondo H., Takahashi M., Arita Y., Komuro R., Ouchi N., Kihara S., Tochino Y., Okutomi K., et al. (2002) Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat. Med. 8, 731–737 [DOI] [PubMed] [Google Scholar]
- 43. Bligh E. G., and Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 44. Tsugawa H., Bamba T., Shinohara M., Nishiumi S., Yoshida M., and Fukusaki E. (2011) Practical non-targeted gas chromatography/mass spectrometry-based metabolomics platform for metabolic phenotype analysis. J. Biosci. Bioeng. 112, 292–298 [DOI] [PubMed] [Google Scholar]
- 45. Hashim Z., Teoh S. T., Bamba T., and Fukusaki E. (2014) Construction of a metabolome library for transcription factor-related single gene mutants of Saccharomyces cerevisiae. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 966, 83–92 [DOI] [PubMed] [Google Scholar]
- 46. Frederich R. C. Jr, Kahn B. B., Peach M. J., and Flier J. S. (1992) Tissue-specific nutritional regulation of angiotensinogen in adipose tissue. Hypertension 19, 339–344 [DOI] [PubMed] [Google Scholar]
- 47. Nakagawa Y., Kishida K., Kihara S., Yoshida R., Funahashi T., and Shimomura I. (2011) Nocturnal falls of adiponectin levels in sleep apnea with abdominal obesity and impact of hypoxia-induced dysregulated adiponectin production in obese murine mesenteric adipose tissue. J. Atheroscler. Thromb. 18, 240–247 [DOI] [PubMed] [Google Scholar]
- 48. Uetsuka S., Ogata G., Nagamori S., Isozumi N., Nin F., Yoshida T., Komune S., Kitahara T., Kikkawa Y., Inohara H., Kanai Y., and Hibino H. (2015) Molecular architecture of the stria vascularis membrane transport system, which is essential for physiological functions of the mammalian cochlea. Eur. J. Neurosci. 42, 1984–2002 [DOI] [PubMed] [Google Scholar]
- 49. Masuda T., Tomita M., and Ishihama Y. (2008) Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res. 7, 731–740 [DOI] [PubMed] [Google Scholar]
- 50. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 51. Kitata R. B., Dimayacyac-Esleta B. R., Choong W. K., Tsai C. F., Lin T. D., Tsou C. C., Weng S. H., Chen Y. J., Yang P. C., Arco S. D., Nesvizhskii A. I., Sung T. Y., and Chen Y. J. (2015) Mining missing membrane proteins by high-pH reverse-phase StageTip fractionation and multiple reaction monitoring mass spectrometry. J. Proteome Res. 14, 3658–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Maeda N., Funahashi T., Hibuse T., Nagasawa A., Kishida K., Kuriyama H., Nakamura T., Kihara S., Shimomura I., and Matsuzawa Y. (2004) Adaptation to fasting by glycerol transport through aquaporin 7 in adipose tissue. Proc. Natl. Acad. Sci. U.S.A. 101, 17801–17806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sekimoto R., Kishida K., Nakatsuji H., Nakagawa T., Funahashi T., and Shimomura I. (2012) High circulating levels of S100A8/A9 complex (calprotectin) in male Japanese with abdominal adiposity and dysregulated expression of S100A8 and S100A9 in adipose tissues of obese mice. Biochem. Biophys. Res. Commun. 419, 782–789 [DOI] [PubMed] [Google Scholar]
- 54. Maeda N., Takahashi M., Funahashi T., Kihara S., Nishizawa H., Kishida K., Nagaretani H., Matsuda M., Komuro R., Ouchi N., Kuriyama H., Hotta K., Nakamura T., Shimomura I., and Matsuzawa Y. (2001) PPARγ ligands increase expression and plasma concentrations of adiponectin, an adipose-derived protein. Diabetes 50, 2094–2099 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.