ABSTRACT
The increase in public online databases dedicated to fungal identification is noteworthy. This can be attributed to improved access to molecular approaches to characterize fungi, as well as to delineate species within specific fungal groups in the last 2 decades, leading to an ever-increasing complexity of taxonomic assortments and nomenclatural reassignments. Thus, well-curated fungal databases with substantial accurate sequence data play a pivotal role for further research and diagnostics in the field of mycology. This minireview aims to provide an overview of currently available online databases for the taxonomy and identification of human and animal-pathogenic fungi and calls for the establishment of a cloud-based dynamic data network platform.
KEYWORDS: databases, taxonomy, nomenclature, fungi, pathogenic
INTRODUCTION
Fungi are ubiquitous microorganisms with a profound influence on agriculture and human and animal life. The global health and socioeconomic impacts of fungal diseases are underrecognized and increasing. Worldwide, on an annual scale, at least 11.5 million deaths are attributed to cases of life-threatening invasive fungal disease (IFD) (http://www.gaffi.org/); this exceeds deaths from malaria and tuberculosis (1). IFDs now cause ∼10% of all nosocomial infections, with high mortality rates (40 to 100%) (1, 2), despite modern therapies. Furthermore, the health impact of chronic respiratory, mucocutaneous, and allergic fungal diseases is enormous. Current expenses are appraised at $2.6 billion/year in the United States alone, with a projected annual increase of 2 to 3% (3). With a burgeoning population at risk for IFDs (4), impacts of natural disasters, and predicted effects of climate change (5), fungal diseases continue to inflict human and animal health with a huge economic burden (1, 4, 5); as such, they are subject to extensive studies, making correct taxonomic identification paramount.
Fungal identification has been based traditionally on subjective morphological and phenotypic characteristics, often leading to multiple names for a single species or, conversely, a single name for distinct species, resulting in erroneous species identifications. Traditional methods based on morphological or biochemical characteristics enable, in most cases, genus- and species-level identifications, but they are slow and can be inaccurate. Diagnostic approaches, such as matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), require fungal culture, and although this can rapidly identify common yeasts, mold identification is more complex and requires access to validated purpose-built databases of reference spectra (6, 7). Undeniably delayed treatment of fungal infections leads to adverse outcomes. However, the prerequisite for early targeted therapy is accurate and timely fungal species identification (ID) (8), which currently is only partially been realized by robust accessible diagnostic platforms (9). Molecular biology now allows for a more objective approach to fungal phylogeny and subsequent correct identification, but at the same time, it produces ever-increasing amounts of sequence data. This creates a new challenge, that of ensuring prompt provision of the vast amount of sequence data available to the end user. With advancements in computational technology and bioinformatics tools, large volumes of data can now be easily stored, annotated, and accessed remotely with relative ease. As a result of this, a superfluity of databases for fungal studies exists, which requires a detailed understanding of each.
ESSENTIALS OF AN ONLINE SEQUENCE DATABASE
In general, an online fungal database comprises sequence information for genes (mainly from the ribosomal DNA [rDNA] gene cluster and protein-coding genes), polyphasic characters, and other associated metadata. Exploring a database usually involves the following steps: (i) accessing a database, (ii) planning a strategy for a particular search, (iii) performing the search, and (iv) retrieving data. The prominence of a database lies in the relative ease of end-user accessibility to deposit, store, annotate, and retrieve data. Any database has an inherent propensity to become obsolete over a period of time. To overcome this and to maintain effective databases relevant to both in diagnostic mycology and in research, it is imperative that a constant and consistent curation effort by a dedicated team of experts is in place.
Over the last decade, a large number of online fungal databases have been established for the mycology research community. This minireview attempts a holistic overview of the more widely used repositories, such as Aspergillus Genome Database (AspGD), Aspergillus & Aspergillosis Website, BOLD, Broad Institute databases, CBS-KNAW, Candida Genome Database (CGD), Doctor Fungus, FungiDB, Fusarium Database, Fusarium MLST (MLST, multilocus sequence typing), Index Fungorum, Institut Pasteur-FungiBank, International Society for Human and Animal Mycology-Internal Transcribed Spacer (ISHAM-ITS), ISHAM-MLST, Mycology Online, MycoBank, NCBI GenBank, NCBI RefSeq, and UNITE, with a focus on the nomenclature, taxonomy, identification, and genotyping of pathogenic fungi.
APPROACHES WITH ITS AS DNA BARCODE, MLST, AND OTHER GENE LOCI
For molecular species identification, an increasingly popular concept of utilizing short DNA sequences, called DNA barcodes, was recommended (10). A DNA barcode constitutes of short conserved (500- to 800-bp) regions containing species-specific genome diversity. It easily enables comparisons with well-identified reference collections of DNA sequence signatures of the species currently in a database, requiring limited fungus-specific identification expertise (11). However, the reliability of a universal gene to achieve an accurate delineation of fungal species and identity still remains a key constraint to its application.
Currently, there is consensus among the mycology community to use the internal transcribed spacer (ITS) sequences of rDNA as the primary barcode for fungi (12). Initially, due to spurious identifications and incomplete ITS sequence deposits, sequence-matched approaches for molecular identification and phylogenetic studies in the public databases of the International Nucleotide Sequence Database Collaboration (INSDC) were often flawed (13). This has been recently overcome through the development of quality-controlled public databases, such as NCBI RefSeq, BOLD, UNITE, and the ISHAM-ITS database (12, 14–16).
Recently, Delgado-Serrano et al. (17) applied a machine learning-based open-source algorithm approach (Mycofier) for classification of the fungal ITS sequence data. They demonstrated that a large pool of ITS1 sequence data could be classified with accuracy comparable to that of the Basic Local Alignment Search Tool (BLAST) algorithm.
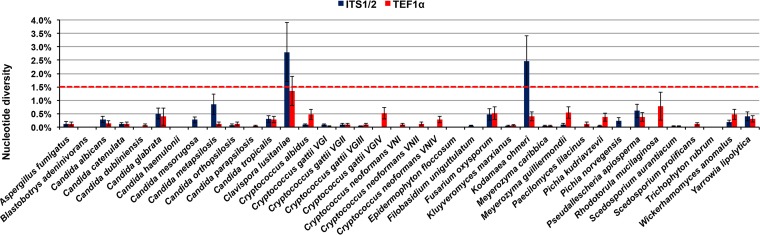
However, for many taxa, additional barcodes are necessary. These secondary and even tertiary barcodes, usually based on sequences of housekeeping genes, are needed for accurate species identification, e.g., the β-tubulin (βTUB) gene for Aspergillus spp. (18) and Scedosporium spp. (19), the translation elongation factor 1α (TEF1α) for Fusarium spp. (20), the intergenic spacer 1 (IGS1) for Trichosporon spp. (21), and the D1/D2 region of the rDNA gene cluster for Lichtheimia spp. (22) Currently, there is no consensus about these supplementary barcodes, since for many taxa, they are genus dependent. Initial work by Robert et al. (23) and Stielow et al. (24) assessed different protein-coding genes from fungi, with the aim of finding an appropriate candidate for a secondary fungal DNA barcode with high phylogenetic resolution, and proposed the TEF1α gene. Preliminary studies showed that the intraspecies variation dropped below 1.5%, enabling a more accurate species delineation (Fig. 1).
FIG 1.
Examples of intraspecies variation in the primary fungal DNA barcode, the ITS1/2 regions compared with the secondary fungal DNA barcode (blue bars), the elongation factor TEF1α, showing the superiority of the TEF1α locus (red bars), with all so-far-tested species having less than 1.5% intraspecies variation, the cutoff needed for accurate species identification. Clavispora lusitaniae (anamorph: Candida lusitaniae) and Kodamaea ohmeri are examples of too-high intraspecies variation in the ITS1/2 locus.
Other molecular approaches, such as multilocus sequence typing (MLST), utilize four or more gene loci for strain typing to aid investigations on epidemiology and population genetics with consistency and reproducibility. The discriminatory power of a specific MLST scheme is dependent on the choice of the gene loci and accuracy of sequences.
ONLINE DATABASES FOR TAXONOMY AND IDENTIFICATION OF PATHOGENIC FUNGI
A vast number of online databases ranging from morphological, biochemical, and clinical to gene- and genome sequence-based databases are now available (Table 1). On the basis of diagnostic convenience and the data elements delimited in each, they can be broadly grouped as (i) clinical-biochemical, morphological, and taxonomy-based; (ii) gene-based; (iii) strain typing-based; and (iv) genome-based, databases.
TABLE 1.
Salient features of currently available fungal databases in public domain
| Database (abbreviation) | URL | Yr instituted | Foundation/host organization | Curation status | Reference |
|---|---|---|---|---|---|
| Aspergillus and Aspergillosis Website | http://www.aspergillus.org.uk/ | 1998 | National Aspergillosis Centre (NAC) and University of Manchester-Fungal Infection Trust | Yes | |
| Aspergillus Genome Database (AspGD) | http://www.aspgd.org/ | 2008 | Board of Trustees, Leland Stanford Junior University, and Broad Institute | Yes | 44 |
| Assembling the Fungal Tree of Life (AFTOL) | https://aftol.umn.edu/ | 2006 | Assembling the Fungal Tree of Life (AFTOL) project, U.S. National Science Foundation | Yes | 29 |
| Barcode of Life Data Systems (BOLD) | http://v4.boldsystems.org/ | 2007 | Canadian Centre for Biodiversity Genomics | Yes | 16 |
| Broad Institute databases | http://www.broadinstitute.org/scientific-community/data/ | 2003 | Broad Institute of Harvard and MIT | Yes (collaborative) | |
| Candida Genome Database (CGD) | http://www.candidagenome.org/ | 2004 | U.S. National Institutes of Health | Yes (except Ensembl Fungi gene annotations) | 47 |
| Centraalbureau voor Schimmelcultures databases (CBS-KNAW) | http://www.cbs.knaw.nl/collections/ | 1989 | CBS-KNAW Fungal Biodiversity Centre, Royal Netherlands Academy of Arts and Sciences | Yes | |
| Doctor Fungus | http://mycosesstudygroup.org/aboutdrf/index.htm | 2007 | Mycoses study group (MSG)–Doctor Fungus Corporation | Yes | |
| EzFungi | http://www.ezbiocloud.net/ezfungi | Seoul National University and ChunLab, Inc. | Yes | 58 | |
| FungiDB | http://fungidb.org/fungidb/ | 2012 | The Eukaryotic Pathogen Database Project Team | Yes | 48 |
| Fusarium databases | http://www.cbs.knaw.nl/fusarium/, http://isolate.fusariumdb.org/ | 2005 | CBS-KNAW Fungal Biodiversity Centre, Royal Netherlands Academy of Arts and Sciences, U.S. Department of Agriculture, Peoria, Illinois, and Penn State University | Yes | 31, 32 |
| Index Fungorum | http://www.indexfungorum.org/names/names.asp | 2000 | Royal Botanic Gardens, Kew, Landcare Research and Institute of Microbiology at Chinese Academy of Sciences | Yes | |
| Institut Pasteur FungiBank | http://fungibank.pasteur.fr/ | 2015 | French National Reference Center for Invasive Mycoses and Antifungals | Yes | |
| International Society for Human and Animal Mycology (ISHAM-ITS) | http://its.mycologylab.org/ | 2011 | DNA Barcoding Working Group of International Society for Human and Animal Mycology | Yes (dedicated curator) | 14 |
| International Society for Human and Animal Mycology (ISHAM-MLST) | http://mlst.mycologylab.org/ | 2011 | MLST Working Group of International Society for Human and Animal Mycology | Yes (dedicated curators) | 35–38 |
| Multi Locus Sequence Typing (MLST.net) | http://www.mlst.net/databases/ | 2005 | Wellcome Trust, Imperial College, UK | Yes | 39–43, 59 |
| MycoBank | http://www.mycobank.org/ | 2004 | International Mycological Association | Yes (registered mycologist) | 26 |
| Mycology Online | http://www.mycology.adelaide.edu.au/ | NAa | Department of Molecular & Cellular Biology, School of Biological Sciences, University of Adelaide, Australia | Yes | |
| National Center for Biotechnology Information (NCBI) GenBank | http://www.ncbi.nlm.nih.gov/GenBank/ | 1982 | National Center for Biotechnology Information, USA | No | 27 |
| NCBI RefSeq database | http://www.ncbi.nlm.nih.gov/refseq/ | 2014 | National Center for Biotechnology Information | Yes (manual curation) | 12 |
| NCBI Whole Genome Database | https://www.ncbi.nlm.nih.gov/genome/ | National Center for Biotechnology Information | No | 27 | |
| The Fungal Genomics Resource (MycoCosm) | http://genome.jgi.doe.gov/programs/fungi/index.jsf | 2013 | 1000 Fungal Genome Project, U.S. Department of Energy Joint Genome Institute | Yes | 49 |
| Unified system for the DNA-based fungal species linked to the classification (UNITE) | https://unite.ut.ee/ | 2003 | UNITE board | Yes (user curated) | 15 |
NA, not applicable.
(i) Clinical, biochemical, morphology, and taxonomy-based databases.
Doctor Fungus (http://www.mycosesstudygroup.org/), Mycology Online (http://www.mycology.adelaide.edu.au/), and the Aspergillus and Aspergillosis Website (http://www.aspergillus.org.uk/) are some of the most extensively used online educational databases providing images and describing morphological characteristics for fungal species and information pertaining to fungal infections in humans and animals across the globe. These online databases, when routinely updated, could be valuable tools for the routine clinical laboratories, which currently rely on online textbooks.
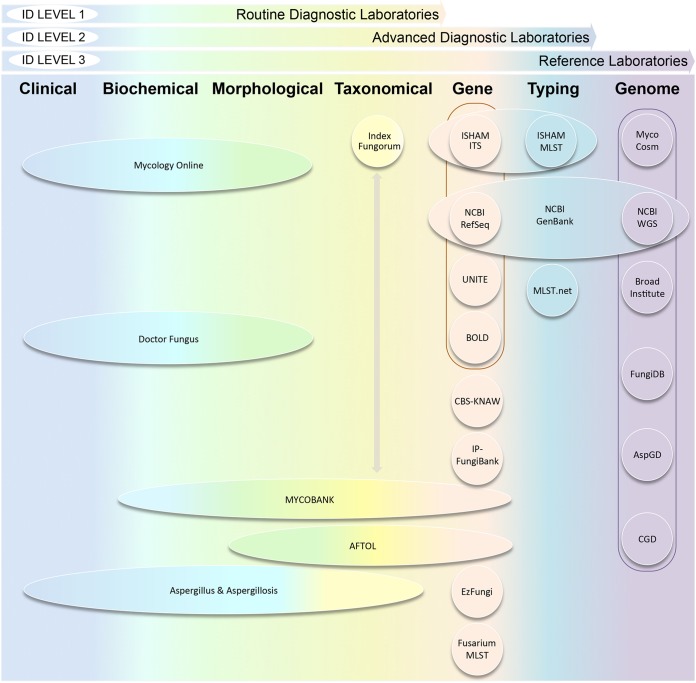
In 2004, the MycoBank Database initiative was started at the CBS-KNAW Fungal Biodiversity Centre at the Royal Netherlands Academy of Arts and Sciences, which was later transferred to the International Mycological Association (IMA). MycoBank is a dedicated online fungal nomenclature and taxonomic database, which is remotely curated and widely used by the mycological community (25, 26). It documents fungal nomenclature and other associated data, such as descriptions and illustrations from all known fungal species. This centralized deposit of fungal taxa provides synonyms, basionyms, teleomorph-anamorph equivalence names, taxonomic classifications, as well as wide-ranging reference links for external resources and pertaining bibliography. Moreover, it comprehensively searches for pairwise sequence alignments against numerous curated reference databases, such as ISHAM-ITS, UNITE, GenBank, CBS, and Institut Pasteur-FungiBank (IP-FungiBank) databases (Fig. 2). In contrast to most other currently available fungal databases for pathogenic fungi, which mainly have provisions for gene and genomic data alone, MycoBank offers the most comprehensive search options on molecular data alone or using a combination of morphological, physiological, and molecular criteria in a polyphasic approach.
FIG 2.
Illustration of predominant online databases for pathogenic fungi with a gradient from clinical, biochemical, morphological, taxonomical, to gene- to genome-based data resources and the identification level achieved according to the kind of diagnostic laboratory. Outlines indicate existing linkages between the databases. Arrows indicate data existing data exchange between databases.
Index Fungorum (http://www.indexfungorum.org/) is an open-access database totally dedicated to fungal nomenclature. It is supported by the collective partnerships of The Royal Botanic Gardens Kew, Landcare Research-NZ, and the Institute of Microbiology of the Chinese Academy of Sciences. In addition, the data elements were sourced from professional mycology consortia, such as the Centre for Agriculture and Biosciences International (CABI) publications, Index of Fungi, and the MycoBank databases.
(ii) Gene sequence-based databases.
Erroneous or mismatched sequences are an ongoing problem faced by many public databases. The most commonly used public database, GenBank (27), hosted at the National Center for Biotechnology Information (NCBI), contains over 10% flawed ITS sequences, annotations, and species definitions (28). To overcome this limitation, the arbitrator-curated NCBI RefSeq Targeted Loci (RTL) database has been established by the NCBI (12). RefSeq is a curated database of fully annotated sequences from type and expert-verified materials. It also facilitates cross-platform multiple-sequence markers and whole-genome searches (12).
The Assembling the Fungal Tree of Life (AFTOL) project set up yet another valuable online resource for fungi, data matrices, and phylogenetic reconstructions (29). The data elements are curated from published literature and include post-taxonomic assessments for inclusion in the AFTOL database, combining character illustrations and molecular data.
The Barcode of Life Data Systems (BOLD) represents the first barcode database system since the initiation of the barcoding concept (16). The analytical platform of this database was developed in the Canadian Centre for Biodiversity Genomics with cloud-based data storage. It has strict guidelines for the submission of barcodes and is curated with a focus on animals and plants. Recent efforts are in place to incorporate more data sets from fungi. In 2015, the ISHAM-ITS database (see below) was incorporated into BOLD (10) (Fig. 2).
The Centraalbureau voor Schimmelcultures Collection and associated Databases (CBS-KNAW) hosts the largest living collection of fungal strains, with more than 80,000 strains in the public collection belonging to almost 15,000 different species. The ITS and LSU loci have been sequenced for most of the strains, including almost all known pathogenic species. The CBS-KNAW website offers the possibility to perform pairwise DNA sequence alignments, as well as polyphasic identifications based on a combination of morphological, physiological, and molecular characteristics for a number of different fungal groups (dermatophytes, Fusarium, medical fungi, Penicillium, Phaeoacremonium, Scedosporium, and yeasts). Like for MycoBank, ISHAM-ITS, ISHAM-MLST, and IP-FungiBank, which all use the same software system, pairwise DNA alignments can be performed simultaneously using several remote reference databases, and the best results are combined into a single matching list.
The EzFungi database is a sister database of the EzTaxon database and contains manually selected and verified ITS sequences to facilitate routine identification of fungal pathogens. The EzFungi database is a result of the collaboration between Seoul National University and ChunLab, Inc. and is maintained and curated by the EzFungi Team.
The French National Reference Center for Invasive Mycoses and Antifungals (NRCMA) hosts the IP-FungiBank, a restricted database for pathogenic fungi. This database allows pairwise gene sequence alignments for medically important yeasts and molds. The polygenic sequence database has been derived from strains systematically identified on the basis of morphology, MALDI-TOF MS, and DNA sequencing. The IP-FungiBank provides updated nomenclature and DNA sequence information in addition to ITS sequences for species-specific identification of Fusarium, Aspergillus, and Trichosporon species viz. the TEF1α gene, βTUB, and IGS1, respectively (http://fungibank.pasteur.fr/).
The taxonomy of the genus Fusarium is inherently complex and has relied upon automated molecular approaches and multiple gene sequences for reliable molecular identification (30). To provide the Fusarium community with reliable identification tools, two widely used databases for this important plant and human pathogen have been established: the Fusarium ID Database, hosted at Pennsylvania State University (31, 32), and the Fusarium MLST database, a dedicated online tool jointly instituted by the CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands, the USDA (Peoria, IL, USA), and the Pennsylvania State University, College Township, PA, USA (33). The Fusarium MLST database enables single-sequence and multisequence alignments for unknown sequence queries (33). This database is linked with GenBank and the CBS-KNAW sequence databases and remains one of the most extensively used databases for Fusarium research groups, soliciting contributions from the user community for continuous development and additions (Fig. 2).
In 2011, the International Society for Human and Animal Mycology-Internal Transcribed Sequences (ISHAM-ITS) database for human- and animal-pathogenic fungi was instituted under the aegis of the ISHAM international working group on “DNA barcoding of human and animal pathogenic fungi.” This widely accessed curated quality-controlled online database currently comprises >3,750 ITS sequences associated with various types of metadata derived from over 500 human- and animal-pathogenic fungal species (14). Most of the species can be reliably identified by ITS sequences, but some of them, such as dermatophytes, Aspergillus, Fusarium, Penicillium, and emerging pathogenic yeasts, require additional molecular methods/gene loci (14, 34). Apart from being well curated, the ISHAM-ITS database is seamlessly integrated into the NCBI RefSeq, UNITE, and BOLD databases through direct linkouts and unique flagging (Fig. 2).
UNITE was first created in 2003, mainly focusing on the ITS sequences of ectomycorrhizal fungi (15). The database has undergone significant changes over the years to provide reliable molecular identification for a large group of fungi. The clustering of fungal species mainly from environmental habitats was achieved by introducing the “species hypothesis” concept. A comprehensive workbench (PlutoF) is also available for the molecular identification, taxonomy, and analysis of sequences derived from metagenome analysis, including Geographic Information System (GIS) (15). UNITE is extensively linked by means of cross-references and linkouts with the NCBI GenBank, NCBI RefSeq, and ISHAM-ITS databases (Fig. 2), and as a result, it holds various fungal ITS sequences of pathogenic fungi.
(iii) Fungal strain genotyping databases.
The International Society for Human and Animal Mycology-multilocus sequence typing (ISHAM-MLST) database provides access to a curated MLST scheme for the following pathogenic fungal species: (i) Cryptococcus neoformans and Cryptococcus gattii (35); (ii) Scedosporium apiospermum and Scedosporium boydii (36); (iii) Scedosporium aurantiacum; (iv) Bipolaris australiensis, Bipolaris hawaiiensis, and Bipolaris spicifera (37); and (v) Pneumocystis jirovecii (38) (Table 2).
TABLE 2.
MLST schemes for pathogenic fungi
| Species | Locia | URL | Reference |
|---|---|---|---|
| Cryptococcus neoformans | CAP59, GPD1, IGS1, LAC1, PLB1, SOD1, URA5 | http://mlst.mycologylab.org/cneoformans/ | 35 |
| Cryptococcus gattii | http://mlst.mycologylab.org/cgattii/ | ||
| Scedosporium apiospermum | ACT, βTUB, CAL, RPB2, SOD2 | http://mlst.mycologylab.org/sapiospermum/ | 36 |
| Scedosporium boydii | http://mlst.mycologylab.org/sboydii/ | ||
| Scedosporium aurantiacum | ACT, βTUB, CAL, RPB2, SOD2, TEF1α | http://mlst.mycologylab.org/saurantiacum/ | |
| Bipolaris australiensis | BRN1, GPD1, RPB1, RPB2, SAL1, TEF1α | http://mlst.mycologylab.org/baustraliensis/ | 37 |
| Bipolaris hawaiiensis | http://mlst.mycologylab.org/bhawaiiensis/ | ||
| Bipolaris spicifera | http://mlst.mycologylab.org/bspicifera/ | ||
| Pneumocystis jirovecii | βTUB, DHPS, ITS1/2, mtLSU | http://mlst.mycologylab.org/pjirovecii/ | 38 |
| Candida albicans | AAT1a, ACC, ADP1, MPIb, SYA1, VPS13, ZWF1b | http://calbicans.mlst.net/ | 40 |
| Candida glabrata | FKS, LEU2, NMT1, TRP1, UGP1, URA3 | http://cglabrata.mlst.net/ | 41 |
| Candida krusei | ADE2, HIS3, LEU2, LYS2D, NMT1, TRP1 | http://pubmlst.org/ckrusei/ | 42 |
| Candida tropicalis | ICL1, MDR1, SAPT2, SAPT4, XYR1, ZWF1a | http://pubmlst.org/ctropicalis/ | 43 |
CAP59, capsule polysaccharide; GPD1, glycerol 3-phosphate dehydrogenase; IGS1, intergenic spacer 1; LAC1, laccase 1; PLB1, phospholipase B1; SOD1, superoxide dismutase; URA5, orotidine monophosphate pyrophosphorylase; ACT, actin; βTUB, β-tubulin; CAL, calmodulin; RPB2, second largest subunit of RNA polymerase II; SOD2, manganese superoxide dismutase; TEF1α, translation elongation factor 1α; BRN1, melanin reductase; RPB1, largest subunit of RNA polymerase I; SAL1, scytalone dehydratase; DHPS, dihydropteroate synthase; ITS1/2, internal transcribed spacer 1 and 2 regions and 5.8S rRNA gene of the nuclear rRNA gene cluster; mtLSU, mitochondrial large subunit rRNA; AAT1a, aspartate aminotransferase; ACC, acetyl-coenzyme A carboxylase; ADP1, ATP-dependent permease; MPIb, mannose phosphate isomerase; SYA1, alanyl-RNA synthetase; VPS13, vacuolar protein sorting protein; ZWF1b, glucose-6-phosphate dehydrogenase; FKS, 1,3-β-Glucan synthase; LEU2, 3-isopropylmalate dehydrogenase; NMT1, myristoyl-CoA:protein N-myristoyltransferase; TRP1, phosphoribosyl-anthranilate isomerase; UGP1, UTP-glucose-1-phosphate uridylyltransferase; URA3, orotidine-5-phosphate decarboxylase; ADE2, adenylosuccinate synthetase; HIS3, imidazole glycerol-phosphate dehydratase; LYS2D, l-aminoadipate-semialdehyde dehydrogenase; ICL1, isocitrate lyase 1; MDR1, major facilitator transporter; SAPT2, secreted aspartic protease 2; SAPT4, secreted aspartic protease 4; XYR1, xylanase regulator 1; ZWF1a, glucose-6-phosphate dehydrogenase.
Similarly, the MLST.Net Database hosted at the Imperial College, London, UK, enables a discriminatory typing system applicable to a number of the pathogenic yeast species useful for epidemiological purposes (39), including MLST schemes for (i) Candida albicans (46), (ii) Candida glabrata (41), (iii) Candida krusei (Pichia kudriavzevii) (42), and (iv) Candida tropicalis (43) (Table 2).
(iv) Genome-based databases.
The Aspergillus Genome Database (AspGD) collects biologically important information of genomic records, proteins, subcellular localizations, and functions of the genus Aspergillus, predominantly for the A. nidulans, A. fumigatus, A. flavus, A. niger, and A. oryzae species complexes. In addition to analytical tools and multispecies comparison, it provides annotation updates and literature links (44).
The Broad Institute databases have exhaustive sequence repositories for fungal data, with specific links to dermatophytes, dimorphic fungal pathogens, and medically important yeasts (https://www.broadinstitute.org/scientific-community/data). The Dermatophyte Comparative, hosted at the Broad Institute, utilizes an expressed sequence tag (EST) approach and contains genome assemblies and annotations for dermatophytes of the genera Trichophyton and Microsporum (45). This database is exceptionally useful for the zoophilic, geophilic, and anthropophilic dermatophytes, viz. Trichophyton rubrum, Trichophyton tonsurans, Trichophyton equinum, Microsporum canis, and Microsporum gypseum, and has features for comparative genome studies which are specific to this group, including gain or loss of gene functions and mating competencies (45). This database is supplemented by the T. rubrum Expression Database (TrED) for specialized analysis of sequence data sets for the aforementioned superficial fungi (46).
The Candida Genome Database (CGD) is a Candida-specific database for sequences of genome and protein data for C. albicans and other Candida species and is funded and hosted by U.S. National Institutes of Health (47). It uses multigenome BLAST for the gene annotations from Ensembl Fungi. However, the gene annotation of the Candida strains is not curated (47).
The FungiDB (48), a constituent of the EuPathDB Bioinformatics Resource Center, provides multiple genome analysis data sets, gene records, data downloads, and diverse data mining tools. It has a sustainable model for curation from the user community with PubMed ID updates and supports with comments, phenotypes, and images (44).
In 2013, MycoCosm, a fungal genomics gateway envisioning the documentation of 1,000 fungal genomes, was initiated. This database was established to integrate and analyze fungal genome data to achieve a better phylogenetic placement of all fungi by the U.S. Department of Energy Joint Genome Institute, and it solicits the user community to partake in the proposal of new species for sequencing, annotation, and data analysis (49).
The NCBI Whole Genome Database serves as a common platform for the deposit of whole fungal genomes from a broad range of genome centers (e.g., the Broad Institute), and as such may serve as the umbrella platform to host and unite these diverse databases in the future.
APPLICATION OF ONLINE FUNGAL DATABASES FOR IDENTIFICATION OF HUMAN- AND ANIMAL-PATHOGENIC FUNGI
An accurate diagnosis (or exclusion) of fungal disease impacts both clinical outcomes and the use and timing of empirical, preemptive, or targeted antifungal therapy. Judicious use of appropriate antifungal drugs is essential in improving outcomes, reducing unnecessary drug toxicity, minimizing health costs, and in delaying the emergence of drug resistance.
A holistic awareness of the specific utility of available online databases for pathogenic fungi with a gradient from clinical-biochemical, morphological-taxonomical, to gene- and genome-based data resources enables identification for a given purpose (Fig. 2). Routine mycology labs, which heavily rely on databases for conventional identifications and related information, will profit from accessing clinical-biochemical-morphological databases. Further, to keep updated with nomenclatural changes, synonyms, basionyms, and obsolete names, taxonomy-specific databases should be accessed. For epidemiological purposes, strain-typing databases (e.g., MLST) should be employed. In reference and advanced research labs, gene- and genome-based databases will aid in the highest taxonomic resolution and discriminatory power for accurate pathogen identification. The ISHAM-ITS database, followed by either of NCBI RefSeq, UNITE, or BOLD, will achieve this purpose. While whole-genome sequencing (WGS), with its highest discriminatory power and accuracy, is potentially attractive, per se, it is not currently feasible for fungi due to the large costs and annotation efforts associated with their large genomes (15 to 40 Mbp), the lack of reference genomes, and the impossibility of providing routine ID within the 48-h window required for early antifungal therapy to greatly improve outcome. Focused-group databases enable genus- or species-specific elements for specific purposes related to diagnostic and research needs. However, a combinatorial approach of the aforementioned databases will obviate tardiness and systematize the required stringency for fast identification purposes.
IMPORTANCE OF ONLINE FUNGAL DATABASES IN THE RAPIDLY EVOLVING GENOMICS ERA
Online fungal databases in the emerging genomics era have been immensely valuable tools. The International Code of Nomenclature for algae, fungi, and plants (ICN) has put forth the mandatory requirement of fungal nomenclature registration, wherein any taxonomical novelties will be assigned a MycoBank ID and scrutinized by experts for its validity and legitimacy in the process of making it available in the online databases (26).
The advent of WGS and metagenomics has revolutionized the field of biology and bioinformatics. These rapidly developing fields in molecular and DNA-based methodologies for fungal identification have generated a whirlwind of data and pose a tremendous challenge for storage, sharing, and ongoing curation. Additionally, the concept of MLST for epidemiological typing purposes has been gradually shifting from using only a couple of loci to WGS. Such efforts have led to large data sets, which demand massive database structures, which are currently lacking. There is an increasing taxonomic restructuring and reshuffling following the post-Amsterdam Amsterdam Declaration on Fungal Nomenclature of one fungus = one name (IF = IN) (50, 51). Further, metagenomics approaches, which rely largely on data mining for understanding biological systems and their interaction with other life forms in a specific niche (52), mean more members of the fungal kingdom are going to be discovered as research in this direction progresses.
LIMITATIONS AND CURRENT CHALLENGES IN COMPREHENSIVE ONLINE FUNGAL DATABASE MANAGEMENT
Although many of the databases listed are in one or another way curated, the frequency and their monitoring are unclear, since some of them have usability limitations or are not updated (e.g., Doctor Fungus or AFTOL) due to a lack of ongoing maintenance.
In most cases, clinical mycology laboratories that use Sanger sequencing, according to the recommendations of the Clinical Laboratory Standards International (CLSI) MM18-A guideline, to the genus level might report filamentous molds as “species complexes,” which in most cases may be sufficient for appropriate clinical care (60). However, the utility of multiple databases discussed herein might enable a further identification to the species level, which may be necessary in specific cases to allow an even greater impact on clinical care and treatment.
Limited data sharing options available among the preexisting databases is one of the major challenges currently being faced in comprehensive online fungal database management. To meet the demands and requisites for advancement in the fungal genomic studies, an option to resolve this would be to reconsider data sharing options and enhancing integrated connectivity. A growing database has the intrinsic chance of becoming redundant and accumulating bias and errors, which could be mitigated by collective efforts for a robust integrated data curation program. This would enable minimization of the potential to accumulate errors and their prompt rectification. In addition, real-time data set sharing and annotations would be an ideal proposition for improving the operational sustainability of databases.
Yet another constraint is the inconsistency in grant support, which impedes the advancement and development of accurately curated and networked databases. This limits the database utility for molecular identification, epidemiology, population genetics, and gene locus studies. It is imperative that a long-term vision and priority by grant agencies with steadfast funding support programs for intensive database curation and its operative sustenance is in place. In addition, there is a great need for funding to extend the current efforts of whole-fungal-genome sequencing to establish a broad platform for studies of the mycobiome, compared with the efforts under way on the bacterial microbiome.
The restraints in the fluctuations in funding support for focused single-pathogen databases, which serve relatively small research groups, could be overcome by database integration. One such successful model network is AspGD, which was revitalized via integration with FungiDB, a component of the EuPathDB Bioinformatics Resource Centre (44). This has maximized the potential of the single-pathogen-focused AspGD database to the general user community. However, different strategies are being adopted for databases, such as Index Fungorum or MycoBank, which ascertain that in the event of discontinuation of their support, coordination, and curation, custodianship is or could be vested to the International Mycological Association (IMA) or International Union of the Microbiological Societies (IUMS) to ensure the continued availability to the community (http://www.indexfungorum.org/). Additionally, interlinking of databases of morphological, biochemical, and sequence data of fungi has been attempted to enable synchronization of molecular data to the fungus-specific attribute, making them relevant and useful for the scientific community (53).
PROPOSAL FOR A CLOUD-BASED DYNAMIC DATABASE NETWORK PLATFORM AND INTEGRATION AMONG SPECIFIC FOCUSED-GROUP DATABASES
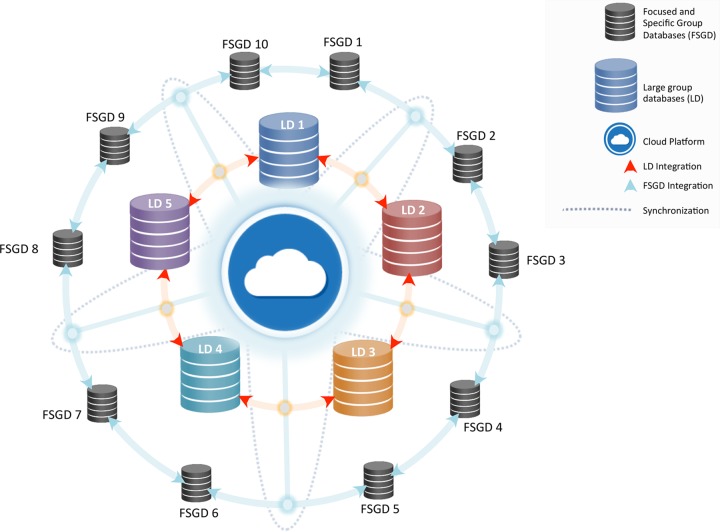
To circumvent the aforementioned crippling challenges for comprehensive data management, we propose a dynamic database network environment by integration among specific focused-group databases with maximum access and functional features for the end-user community. Linkage of standalone specific focused-group databases (e.g., those for Fusarium and dermatophytes) to larger databases will enable better search results and comprehensive understanding of important members of those fungal groups. One of the best ways to achieve integrated data networks would be the utilization of the emerging cloud computing platforms (Fig. 3). Cloud platforms refer to Internet-based shared processing of a large composite of data and digital resources with secured access to machines in public domains enabled by means of virtual private networks (VPNs) (54). These cloud platforms offer considerable potential in further developments of online databases with metadata pertinent to strain, genomics, and taxonomy, along with associated and supportive data sets for pathogenic fungi. In addition, the lack of a unique strain ID for multiple entries of the same strain leads to confusion. This can be avoided when interlinking the databases by having a Unified Strain ID (US-ID), which will improve tracing and indexing of the fungal metadata records for each strain. Future trends may also be a foreseeable application of quantum computing (QBITS) with advanced processing abilities for an aptly manageable, interlinked, and easily retrievable online fungal database (55).
FIG 3.
Architectural flow diagram for the proposed dynamic integrated fungal databases network link.
With Sanger sequencing data generation being limited to the isolated strains or the amplification of a single sequence target with specific primers, the use of next-generation sequencing (NGS) in routine diagnostics will be a game changer, as clinical samples can be directly sequenced, and the number of sequences generated can grow to several thousand to millions of reads that need to be analyzed and compared against reference databases. While a single sequence alignment against a reference database of a few million sequences may take a few milliseconds to 2 to 3 s on a given computer, the comparison of millions of sequences against millions of references will cause serious scalability problems and challenges. Faster heuristic comparative methods (56, 57) will have to be developed and implemented in a cloud- or grid-based environment.
CONCLUSION
Future research toward clinically and agriculturally important fungi demands high-quality and maximum-utility databases for the research community. Given that exotic invasive fungal infections are often zoonotic, i.e., encountered from diverse environmental habitats, especially plant pathogens, interlinking of focused and specific group databases enables a highly dynamic integrated network. Thus, at this juncture, with vast expanses of biodiversity habitats under exploration and newer fungal species being defined, linking fungal databases in a virtually connected environment will enhance better search strategies, improve taxonomic resolution toward identification of pathogenic fungi, and contribute to significant fungal infectious disease research ramifications.
ACKNOWLEDGMENTS
This work was supported by a National Health and Medical Research Council of Australia (NH&MRC) grant APP1121936 to W.M., S.C., and V.R. P.Y.P. is recipient of Australian Government's Department of Education and Training Endeavor Scholarships and Fellowships 2016 program and is supported by the University of Sydney.
P.Y.P., L.I., and W.M. conducted the statement literature search; P.Y.P., L.I., and W.M. created the figures; P.Y.P., L.I., C.H., S.C., V.R., and W.M. designed the study; P.Y.P., L.I., and W.M. collected the data; P.Y.P., L.I., V.R., and W.M. analyzed the data; P.Y.P., L.I., C.H., S.C., V.R., and W.M. interpreted the data; and P.Y.P., L.I., C.H., S.C., V.R., and W.M wrote the manuscript. All authors participated sufficiently in this work and took part in the final approval of the published version.
We declare no potential conflicts of interest.
REFERENCES
- 1.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Pfaller MA, Pappas PG, Wingard JR. 2006. Invasive fungal pathogens: current epidemiological trends. Clin Infect Dis 43:S3–S14. doi: 10.1086/504490. [DOI] [Google Scholar]
- 3.Panackal AA. 2011. Global climate change and infectious diseases: invasive mycoses. J Earth Sci Climate Change 1:108. [Google Scholar]
- 4.Armstrong-James D, Meintjes G, Brown GD. 2014. A neglected epidemic: fungal infections in HIV/AIDS. Trends Microbiol 22:120–127. doi: 10.1016/j.tim.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. 2012. Emerging fungal threats to animal, plant and ecosystem health. Nature 484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sleiman S, Halliday CL, Chapman B, Brown M, Nitschke J, Lau AF, Chen SC. 2016. Performance of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of Aspergillus, Scedosporium, and Fusarium spp. in the Australian clinical setting. J Clin Microbiol 54:2182–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizzato C, Lombardi L, Zoppo M, Lupetti A, Tavanti A. 2015. Pushing the Limits of MALDI-TOF mass spectrometry: beyond fungal species identification. J Fungi 1:367. doi: 10.3390/jof1030367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen SC, Sorrell TC, Chang CC, Paige EK, Bryant PA, Slavin MA. 2014. Consensus guidelines for the treatment of yeast infections in the haematology, oncology and intensive care setting, 2014. Intern Med J 44:1315–1332. doi: 10.1111/imj.12597. [DOI] [PubMed] [Google Scholar]
- 9.Westblade LF, Jennemann R, Branda JA, Bythrow M, Ferraro MJ, Garner OB, Ginocchio CC, Lewinski MA, Manji R, Mochon AB, Procop GW, Richter SS, Rychert JA, Sercia L, Burnham CA. 2013. Multicenter study evaluating the Vitek MS system for identification of medically important yeasts. J Clin Microbiol 51:2267–2272. doi: 10.1128/JCM.00680-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hebert PD, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proc Biol Sci 270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frézal L, Leblois R. 2008. Four years of DNA barcoding: current advances and prospects. Infect Genet Evol 8:727–736. doi: 10.1016/j.meegid.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Schoch CL, Robbertse B, Robert V, Vu D, Cardinali G, Irinyi L, Meyer W, Nilsson RH, Hughes K, Miller AN, Kirk PM, Abarenkov K, Aime MC, Ariyawansa HA, Bidartondo M, Boekhout T, Buyck B, Cai Q, Chen J, Crespo A, Crous PW, Damm U, De Beer ZW, Dentinger BT, Divakar PK, Dueñas M, Feau N, Fliegerova K, García MA, Ge ZW, Griffith GW, Groenewald JZ, Groenewald M, Grube M, Gryzenhout M, Gueidan C, Guo L, Hambleton S, Hamelin R, Hansen K, Hofstetter V, Hong SB, Houbraken J, Hyde KD, Inderbitzin P, Johnston PR, Karunarathna SC, Kõljalg U, Kovács GM, Kraichak E, et al. 2014. Finding needles in haystacks: linking scientific names, reference specimens and molecular data for fungi. Database (Oxford) 2014:bau061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura Y, Cochrane G, Karsch-Mizrachi I, International Nucleotide Sequence Database Collaboration . 2013. The international nucleotide sequence database collaboration. Nucleic Acids Res 41:D21–D24. doi: 10.1093/nar/gks1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irinyi L, Serena C, Garcia-Hermoso D, Arabatzis M, Desnos-Ollivier M, Vu D, Cardinali G, Arthur I, Normand AC, Giraldo A, da Cunha KC, Sandoval-Denis M, Hendrickx M, Nishikaku AS, de Azevedo Melo AS, Merseguel KB, Khan A, Parente Rocha JA, Sampaio P, da Silva Briones MR, e Ferreira RC, de Medeiros Muniz M, Castanon-Olivares LR, Estrada-Barcenas D, Cassagne C, Mary C, Duan SY, Kong F, Sun AY, Zeng X, Zhao Z, Gantois N, Botterel F, Robbertse B, Schoch C, Gams W, Ellis D, Halliday C, Chen S, Sorrell TC, Piarroux R, Colombo AL, Pais C, de Hoog S, Zancope-Oliveira RM, Taylor ML, Toriello C, de Almeida Soares CM, Delhaes L, Stubbe D, et al. 2015. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database–the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med Mycol 53:313–337. doi: 10.1093/mmy/myv008. [DOI] [PubMed] [Google Scholar]
- 15.Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson K-H. 2013. Towards a unified paradigm for sequence-based identification of Fungi. Mol Ecol 22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 16.Ratnasingham S, Hebert PD. 2007. BOLD: the Barcode of Life Data system (http://www.barcodinglife.org). Mol Ecol Notes 7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delgado-Serrano L, Restrepo S, Bustos JR, Zambrano MM, Anzola JM. 2016. Mycofier: a new machine learning-based classifier for fungal ITS sequences. BMC Res Notes 9:1–8. doi: 10.1186/s13104-015-1837-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samson RA, Visagie CM, Houbraken J, Hong SB, Hubka V, Klaassen CHW, Perrone G, Seifert KA, Susca A, Tanney JB, Varga J, Kocsubé S, Szigeti G, Yaguchi T, Frisvad JC. 2014. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol 78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilgado F, Cano J, Gené J, Guarro J. 2005. Molecular phylogeny of the Pseudallescheria boydii species complex: proposal of two new species. J Clin Microbiol 43:4930–4942. doi: 10.1128/JCM.43.10.4930-4942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Short DP, O'Donnell K, Thrane U, Nielsen KF, Zhang N, Juba JH, Geiser DM. 2013. Phylogenetic relationships among members of the Fusarium solani species complex in human infections and the descriptions of F. keratoplasticum sp. nov. and F. petroliphilum stat. nov. Fungal Genet Biol 53:59–70. doi: 10.1016/j.fgb.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Chagas-Neto TC, Chaves GM, Melo AS, Colombo AL. 2009. Bloodstream infections due to Trichosporon spp.: species distribution, Trichosporon asahii genotypes determined on the basis of ribosomal DNA intergenic spacer 1 sequencing, and antifungal susceptibility testing. J Clin Microbiol 47:1074–1081. doi: 10.1128/JCM.01614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Hermoso D, Hoinard D, Gantier JC, Grenouillet F, Dromer F, Dannaoui E. 2009. Molecular and phenotypic evaluation of Lichtheimia corymbifera (formerly Absidia corymbifera) complex isolates associated with human mucormycosis: rehabilitation of L. ramosa. J Clin Microbiol 47:3862–3870. doi: 10.1128/JCM.02094-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robert V, Szöke S, Eberhardt U, Cardinali G, Seifert KA, Lévesque CA, Lewis CT, Meyer W. 2011. The quest for a general and reliable fungal DNA barcode. Open Appl Inform J 5:45–61. doi: 10.2174/1874136301105010045. [DOI] [Google Scholar]
- 24.Stielow JB, Lévesque CA, Seifert KA, Meyer W, Iriny L, Smits D, Renfurm R, Verkley GJM, Groenewald M, Chaduli D, Lomascolo A, Welti S, Lesage-Meessen L, Favel A, Al-Hatmi AMS, Damm U, Yilmaz N, Houbraken J, Lombard L, Quaedvlieg W, Binder M, Vaas LAI, Vu D, Yurkov A, Begerow D, Roehl O, Guerreiro M, Fonseca A, Samerpitak K, van Diepeningen AD, Dolatabadi S, Moreno LF, Casaregola S, Mallet S, Jacques N, Roscini L, Egidi E, Bizet C, Garcia-Hermoso D, Martín MP, Deng S, Groenewald JZ, Boekhout T, de Beer ZW, Barnes I, Duong TA, Wingfield MJ, de Hoog GS, Crous PW, Lewis CT, et al. 2015. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia 35:242–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crous PW, Gams W, Stalpers D, Robert V, Stegehuis G. 2004. MycoBank: an online initiative to launch mycology into the 21st century. Stud Mycol 50:19–22. [Google Scholar]
- 26.Robert V, Vu D, Amor AB, van de Wiele N, Brouwer C, Jabas B, Szoke S, Dridi A, Triki M, Ben Daoud S, Chouchen O, Vaas L, de Cock A, Stalpers JA, Stalpers D, Verkley GJ, Groenewald M, Dos Santos FB, Stegehuis G, Li W, Wu L, Zhang R, Ma J, Zhou M, Gorjon SP, Eurwilaichitr L, Ingsriswang S, Hansen K, Schoch C, Robbertse B, Irinyi L, Meyer W, Cardinali G, Hawksworth DL, Taylor JW, Crous PW. 2013. MycoBank gearing up for new horizons. IMA Fungus 4:371–379. doi: 10.5598/imafungus.2013.04.02.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2014. GenBank. Nucleic Acids Res 42:D32–D37. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsson R, Ryberg M, Kristiansson E, Abarenkov K, Larsson K, Kõljalg U. 2006. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS One 1:e59. doi: 10.1371/journal.pone.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Celio GJ, Padamsee M, Dentinger BT, Bauer R, McLaughlin DJ. 2006. Assembling the Fungal Tree of Life: constructing the structural and biochemical database. Mycologia 98:850–859. doi: 10.3852/mycologia.98.6.850. [DOI] [PubMed] [Google Scholar]
- 30.O'Donnell K, Ward TJ, Robert VARG, Crous PW, Geiser DM, Kang S. 2015. DNA sequence-based identification of Fusarium: current status and future directions. Phytoparasitica 43:583–595. doi: 10.1007/s12600-015-0484-z. [DOI] [Google Scholar]
- 31.Geiser DM, del Mar Jiménez-Gasco M, Kang S, Makalowska I, Veeraraghavan N, Ward TJ, Zhang N, Kuldau GA, O'Donnell K. 2004. FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur J Plant Pathol 110:473–479. doi: 10.1023/B:EJPP.0000032386.75915.a0. [DOI] [Google Scholar]
- 32.Park B, Park J, Cheong KC, Choi J, Jung K, Kim D, Lee YH, Ward TJ, O'Donnell K, Geiser DM, Kang S. 2011. Cyber infrastructure for Fusarium: three integrated platforms supporting strain identification, phylogenetics, comparative genomics and knowledge sharing. Nucleic Acids Res 39:D640–D646. doi: 10.1093/nar/gkq1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Donnell K, Sutton DA, Rinaldi MG, Sarver BA, Balajee SA, Schroers HJ, Summerbell RC, Robert VA, Crous PW, Zhang N, Aoki T, Jung K, Park J, Lee YH, Kang S, Park B, Geiser DM. 2010. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J Clin Microbiol 48:3708–3718. doi: 10.1128/JCM.00989-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visagie CM, Houbraken J, Frisvad JC, Hong SB, Klaassen CHW, Perrone G, Seifert KA, Varga J, Yaguchi T, Samson RA. 2014. Identification and nomenclature of the genus Penicillium. Stud Mycol 78:343–371. doi: 10.1016/j.simyco.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer W, Aanensen DM, Boekhout T, Cogliati M, Diaz MR, Esposto MC, Fisher M, Gilgado F, Hagen F, Kaocharoen S, Litvintseva AP, Mitchell TG, Simwami SP, Trilles L, Viviani MA, Kwon-Chung J. 2009. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med Mycol 47:561–570. doi: 10.1080/13693780902953886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernhardt A, Sedlacek L, Wagner S, Schwarz C, Wurstl B, Tintelnot K. 2013. Multilocus sequence typing of Scedosporium apiospermum and Pseudallescheria boydii isolates from cystic fibrosis patients. J Cyst Fibros 12:592–598. doi: 10.1016/j.jcf.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 37.Pham CD, Purfield AE, Fader R, Pascoe N, Lockhart SR. 2015. Development of a multilocus sequence typing system for medically relevant Bipolaris species. J Clin Microbiol 53:3239–3246. doi: 10.1128/JCM.01546-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phipps LM, Chen SC, Kable K, Halliday CL, Firacative C, Meyer W, Wong G, Nankivell BJ. 2011. Nosocomial Pneumocystis jirovecii pneumonia: lessons from a cluster in kidney transplant recipients. Transplantation 92:1327–1334. doi: 10.1097/TP.0b013e3182384b57. [DOI] [PubMed] [Google Scholar]
- 39.Aanensen DM, Spratt BG. 2005. The multilocus sequence typing network: mlst.net. Nucleic Acids Res 33:W728–W733. doi: 10.1093/nar/gki415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bougnoux ME, Tavanti A, Bouchier C, Gow NA, Magnier A, Davidson AD, Maiden MC, D'Enfert C, Odds FC. 2003. Collaborative consensus for optimized multilocus sequence typing of Candida albicans. J Clin Microbiol 41:5265–5266. doi: 10.1128/JCM.41.11.5265-5266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dodgson AR, Pujol C, Denning DW, Soll DR, Fox AJ. 2003. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J Clin Microbiol 41:5709–5717. doi: 10.1128/JCM.41.12.5709-5717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobsen MD, Gow NA, Maiden MC, Shaw DJ, Odds FC. 2007. Strain typing and determination of population structure of Candida krusei by multilocus sequence typing. J Clin Microbiol 45:317–323. doi: 10.1128/JCM.01549-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tavanti A, Davidson AD, Johnson EM, Maiden MC, Shaw DJ, Gow NA, Odds FC. 2005. Multilocus sequence typing for differentiation of strains of Candida tropicalis. J Clin Microbiol 43:5593–5600. doi: 10.1128/JCM.43.11.5593-5600.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cerqueira GC, Arnaud MB, Inglis DO, Skrzypek MS, Binkley G, Simison M, Miyasato SR, Binkley J, Orvis J, Shah P, Wymore F, Sherlock G, Wortman JR. 2014. The Aspergillus genome database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res 42:D705–D710. doi: 10.1093/nar/gkt1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White TC, Oliver BG, Graser Y, Henn MR. 2008. Generating and testing molecular hypotheses in the dermatophytes. Eukaryot Cell 7:1238–1245. doi: 10.1128/EC.00100-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, Chen L, Wang L, Zhang W, Liu T, Jin Q. 2007. TrED: the Trichophyton rubrum Expression Database. BMC Genomics 8:250. doi: 10.1186/1471-2164-8-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inglis DO, Arnaud MB, Binkley J, Shah P, Skrzypek MS, Wymore F, Binkley G, Miyasato SR, Simison M, Sherlock G. 2011. The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res 40:D667–D674. doi: 10.1093/nar/gkr945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS, Kissinger JC, Li W, Nayak V, Pinney DF, Stoeckert CJ Jr, Roos DS. 2012. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res 40:D675–D681. doi: 10.1093/nar/gkr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor JW. 2011. One fungus = one name: DNA and fungal nomenclature twenty years after PCR. IMA Fungus 2:113–120. doi: 10.5598/imafungus.2011.02.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hawksworth DL, Crous PW, Redhead SA, Reynolds DR, Samson RA, Seifert KA, Taylor JW, Wingfield MJ, Abaci Ö, Aime C, Asan A, Bai F-Y, de Beer ZW, Begerow D, Berikten D, Boekhout T, Buchanan PK, Burgess T, Buzina W, Cai L, Cannon PF, Crane JL, Damm U, Daniel H-M, van Diepeningen AD, Druzhinina I, Dyer PS, Eberhardt U, Fell JW, Frisvad JC, Geiser DM, Geml J, Glienke C, Gräfenhan T, Groenewald JZ, Groenewald M, de Gruyter J, Guého-Kellermann E, Guo L-D, Hibbett DS, Hong S-B, de Hoog GS, Houbraken J, Huhndorf SM, Hyde KD, Ismail A, Johnston PR, Kadaifciler DG, Kirk PM, Kõljalg U, et al. 2011. The Amsterdam Declaration on Fungal Nomenclature. IMA Fungus 2:105–112. doi: 10.5598/imafungus.2011.02.01.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjøller R, Kõljalg U, Pennanen T, Rosendahl S, Stenlid J, Kauserud H. 2013. Fungal community analysis by high-throughput sequencing of amplified markers–a user's guide. New Phytol 199:288–299. doi: 10.1111/nph.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat J, Buyck B, Cai L, Dai Y-C, Abd-Elsalam KA, Ertz D, Hidayat I, Jeewon R, Jones EBG, Bahkali AH, Karunarathna SC, Liu J-K, Luangsa-ard JJ, Lumbsch HT, Maharachchikumbura SSN, McKenzie EHC, Moncalvo J-M, Ghobad-Nejhad M, Nilsson H, Pang K-L, Pereira OL, Phillips AJL, Raspé O, Rollins AW, Romero AI, Etayo J, Selçuk F, Stephenson SL, Suetrong S, Taylor JE, Tsui CKM, Vizzini A, Abdel-Wahab MA, Wen T-C, Boonmee S, Dai DQ, Daranagama DA, Dissanayake AJ, Ekanayaka AH, Fryar SC, Hongsanan S, Jayawardena RS, Li W-J, Perera RH, Phookamsak R, de Silva NI, Thambugala KM, et al. 2015. The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers 74:3–18. doi: 10.1007/s13225-015-0351-8. [DOI] [Google Scholar]
- 54.Saxena V, Doddavula SK, Jain A. 2012. Implementation of a secure genome sequence search platform on public cloud-leveraging open source solutions. J Cloud Comp Adv Syst Appl 1:1–14. doi: 10.1186/2192-113X-1-1. [DOI] [Google Scholar]
- 55.Boixo S, Ronnow TF, Isakov SV, Wang Z, Wecker D, Lidar DA, Martinis JM, Troyer M. 2014. Evidence for quantum annealing with more than one hundred qubits. Nat Phys 10:218–224. doi: 10.1038/nphys2900. [DOI] [Google Scholar]
- 56.Yu YW, Daniels NM, Danko DC, Berger B. 2015. Entropy-scaling search of massive biological data. Cell Syst 1:130–140. doi: 10.1016/j.cels.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vu D, Szoke S, Wiwie C, Baumbach J, Cardinali G, Rottger R, Robert V. 2014. Massive fungal biodiversity data re-annotation with multi-level clustering. Sci Rep 4:6837. doi: 10.1038/srep06837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. 2012. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 59.Jolley KA, Chan M-S, Maiden MC. 2004. mlstdbNet–distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:1–8. doi: 10.1186/1471-2105-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petti CA, Bosshard PP, Brandt ME, Clarridge JE, Feldblyum TV, Foxall P, Furtado MR, Pace N, Procop G. 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline–vol 28, no. 12. CLSI document MM18-A Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]