Abstract
To investigate the genetic variation and molecular evolution of potato virus X (PVX), 87 coat protein (CP) gene sequences were retrieved from GenBank and analyzed. Of the PVX isolates studied, one recombinant isolate (X3) was detected from South America population of the virus. The other isolates belonged to two lineages, Eurasia and America, with the significant FST value (0.60). Non-synonymous nucleotide diversity to synonymous nucleotide diversity (ω = dN/dS) was less than 1 indicating that the CP gene has been under negative selection or neutral evolution. Further analysis showed that all of the codons in both lineages were under negative selection pressure. No significant genetic differentiation was found between Chinese, Indian, Iranian, Japanese, and the UK populations whereas South America population was distinctly differentiated from other populations. Different evolutionary constraints found for the two lineages suggest that possible mutations and genetic drift were important evolutionary forces driving the genetic diversification of PVX population.
Electronic supplementary material
The online version of this article (doi:10.1007/s13337-017-0362-z) contains supplementary material, which is available to authorized users.
Keywords: Evolutionary force, Genetic drift, Negative selection, Recombination
Introduction
Error-prone nature of RNA-depended RNA polymerase, large population size and short generation time are important factors in existence of high mutation rate in RNA viruses [5]. In addition, RNA recombination is a significant evolutionary factor involved in genetic variation and evolution of plant viruses [8]. On the opposite site, viruses maintain their genomic stability by purifying (negative) selection. Negative selection helps virus populations to eliminate mutants with non-coding vital genes [6]. Negative, neutral and directional (positive) are three types of selection which are determined by the ratio of diversity of non-synonymous sites to synonymous sites (ω = dN/dS). This ratio is <1, =1 and >1 for negative, neutrality and positive selection, respectively [9]. A better knowledge of genetic diversity and how it is generated in viruses helps us to gain insight into trend of virus population, develop reliable diagnostic methods in clean-plant program and to establish best strategy for virus management.
Potato virus X (PVX) is a damaging plant pathogen and spread world-wide in potato growing areas, infecting Solanaceous plants. PVX with flexuous filaments viroins belongs to the genus Potexvirus in the family Alphaflexiviridae. The virus genome is composed of a positive sense, single-stranded RNA molecule which is polyadenylated at its 3′ end, and has a methylguanosine cap at its 5′ end [17]. Five proteins are encoded by the RNA genome which are from 5′ to 3′ direction, RNA-depended RNA polymerase (RdRp; 166 kDa), triple gene block (TGB1; 25KDa, TGB2; 12KDa and TGB3; 8KDa) and coat protein (CP; 25 kDa) [25, 40]. It is transmitted mechanically via infected sap and infected seed potato tubers, but not by insect vectors [35].
Based on amino acid sequences of the CP and ability to induce symptoms on the potato genotypes carrying resistant gene, PVX strains are classified into two types, type X and type B [32]. Previous studies have shown that the PVX isolates can be classified into two lineages, Eurasia and America, which correspond to their geographical origin [22, 24, 44, 45]. PVX is well-known as a model for understanding pathogen-host interaction [4, 14, 20] and efforts in recent years have established good knowledge about genetic basis of resistance and the genetic system controlling behavior of the pathogen [2, 21, 32].
It is accepted that PVX strains in respect to the Nx, Nb and Rx host resistance genes show classical “gene-for-gene” type resistance interactions [20, 29] and CP is avirulence (resistance sensitivity) determinants of both the Nx and Rx-mediate resistances [2] whereas movement protein is avirulence determinants of Rx-mediated resistance [15]. Currently, more than one-hundred fifty sequences of the CP gene of PVX and 25 complete genomes are available in GenBank database. Despite world-wide distribution of this virus, population genetic structure and molecular evolution are poorly understood and needs to be further explored.
Although there are several studies on phylogenetic analysis, selection pressure operating on different genes, and estimation of genetic variability on different coding regions of PVX [24, 44, 45], there still remained a lack of information on evolutionary constraints for the two virus lineages, molecular diversity patterns, population differentiation and recombination event(s). Here, population genetic analysis of PVX isolates from different countries was done based on the CP sequences because conserved nature of the CP gene makes it a suitable candidate for such studies. The CP gene in plant viruses is commonly used for investigation on genetic variation and molecular evolution [10, 27, 41, 43] and was therefore selected for this study.
Materials and methods
Virus sequences
Multiple sequence alignment of 87 nucleotide (nt) sequences of the CPs was performed using the ClustalW algorithm [38] implemented in MEGA6 [37] with the default parameters. For the population genetic analysis, each country or geographical region with at least five PVX CP gene sequences was considered as a population, including Chinese, Indian, Iranian, Japanese, South America and the United Kingdom (UK) populations. Table 1 shows the origins, hosts and accession numbers of PVX isolates used in this study.
Table 1.
Origins, hosts and accession numbers of Potato virus X isolates/strains analyzed in this study
| Origin | Isolate/straina | Hostb | Accession number |
|---|---|---|---|
| China | pWC18, SL104, Ur2-PVXCP1, Ur2-PVXCP3, Guizhou CP01, Shandong, -, FX21 | Ns, Ns, Potato, Potato, Potato, Ns, Potato | U19790, EF063709, HM036193, HM036195, KF225469, AF528555, EU571480, EF423572 |
| India | OOT-5, DEE-58, O-MAS-18, JAL-2, MOD-S1, N-NAL-86, -, O-MAS-26, N-PAR-117, KUF-5, KUF-3, N-NAG-34, PAT-4, O-GEG-44, PAT-38, N-CHA-60, N-MEH-95, KHA-1, SHI-1, O-KOK-37, ptDel-9 | Potato | KR605343, KR605392, KR605379, KR605396, KR605360, KR605372, GU256064, KR605380, KR605373, KR605397, KR605330, KR605370, KR605367, KR605384, KR605366, KR605376, KR605369, KR605359, KR605348, KR605382, JF430080 |
| Iran | KER.LA.1, ZA.WE.UR.1, KER.LA.2, KH.SH.1, ARD.GI.1, ARD.GR.2, KER.ZA.2, AZA.ES.TAB.1, Iran | Potato | KF568905, KF568900, KF575175, KF568904, KF568897, KF568898, KF568906, KF537623, FJ461343 |
| Japan | BS, BH, OS, TO, -, OG | Potato, Potato, Ns, Potato, Potato | AB056719, AB195999, AB056718, AB196001, D87962, AB196000 |
| Peru | GAF225 EuA, GAF225 SA-CIP, SA-CIP, X3, XC, CP4, GAF226, GAF224, - | Potato | KJ534604, KJ534603, KJ534601, M31541, X12804, AF172259, KJ534605, KJ534602, M63141 |
| UK | Sam-15-PVX, Sam-18-PVX, WARB, Sam-23-PVX, KP, Sam-13-PVX, Sam-12-PVX, -, Sam-14-PVX, WATom, COPR, XS, RQTH1, Sam-16-PVX, EX, EX2, DY | Potato | GU144351, GU144349, GU384727, GU144346, X88783, GU144353, GU144354, AF493949, GU144352, GU384735, M95516, X88788, AF111193, GU144350, X88782, GU384737, X88781 |
| Estonia | S6111, PSS1, CP2 | Ns | Z29333, Z29334, Z29335 |
| USA | PVX2.7, PVX2.7b, WS2 | Potato | HQ450387, HQ450388, X887786 |
| Bolivia | Harpenden, HB | Potato | Z23256, X72214 |
| Netherlands | X3, N14 | Ns, Potato | D00344, X88785 |
| South Korea | KO1, KO2 | Potato | AF260640, AF260641 |
| Spain | SP1 | Tomato | AJ505748 |
| Australia | QLD1970, QLD1974, VPRI 31683b | Potato | GU384733, GU384732, GU384729 |
| Russia | Tula, - | Potato | EU021215, X05198 |
| Colombia | PVX-Gooseberry | Gooseberry | KM659859 |
| Taiwan | Taiwan | Potato | AF272736 |
Ns non stated
aNames of isolates are written in an order respective to their hosts and accession numbers
bThe same host for all isolates in an origin is written only once
- Unknown isolate name
Recombination analysis
Recombination detection program RDP4 [23] was applied to detect and analyze recombination signal in the 87 aligned PVX CP unique sequences using several methods (Chimaera, Geneconv, MaxChi, RDP, Siscan and 3Seq), with the default settings, and a P value threshold of 0.05. First, auto-masking was performed and 30 isolates were masked. Then, position of the masking isolates was ascertained by the use of “trees” menu to generate UPGMA phylogenetic tree. Finally, to find the recombinant isolates all methods implemented in the program were run with default settings.
Phylogenetic analysis and population genetic parameters
Phylogenetic tree were constructed by the neighbor-joining (NJ) method implemented in MEGA6 without root and branch support was computed using the bootstrap method based on 1000 replicates. Genetic distance within and between lineages of PVX CP were estimated using Kimura two-parameter method [16] implemented in MEGA6. Aligned PVX sequences were assessed using DnaSP v.5.10.01 [31] to estimate number of haplotypes (H), number of polymorphic sites (S), total number of mutation (η), the average pairwise nucleotide diversity (π = Pi), average number of nucleotide differences (K) and dN/dS. To identify individual codon positions evolving under natural selection, alignment of amino-acid coding sequences was performed in MEGA6 and four different codon-based maximum-likelihood algorithms including single likelihood ancestor counting (SLAC), fixed effects likelihood (FEL), internal fixed effects likelihood (IFEL) and fast unbiased bayesien approximation (FUBAR) all implemented in HyPhy software package [28] and available in Datamonkey server (www.datamonkey.org) were used with significance level set at P value <0.05.
Neutrality test and tests of population differentiation
Tajima’s D [36], Fu and Li’s D&F [7] statistical tests, implemented in DnaSP v.5.10.01, were used to examine the hypothesis of neutral selection operating on the CP between lineages and populations. Independent statistical tests of population differentiation (KS*, KST*, Z*, Snn and FST), were done using DnaSP v.5.10.01 between geographical origins of isolates (China, India, Iran, Japan, South America and the UK). Under the null hypothesis (no genetic differentiation), Kst* is expected to be near zero, but if Ks*, Kst* P value is supported by small P value (<0.05), the null hypothesis is rejected. Z* statistic is a logarithmic variant of Z statistic and if it is too small and supported by P value (<0.05) the null hypothesis of no genetic differentiation is rejected [11]. A Snn (the nearest neighbors of sequences) test statistic value is ranged between one, when population are distinctly differentiated, to 1/2 in the case of panmixia [13]. Finally, FST value (fixation index) shows the amount of inter-population diversity and the value ranges between zero (indicating no differentiation between populations) and one (indicating full differentiation between populations) [12]. The coefficient of FST was used to estimate the extent of genetic differentiation between populations [39]. Normally, an absolute value of FST > 0.33 suggests gene flow is negligible, while absolute value of FST < 0.33 indicates gene flow is frequent [30].
Results
Recombination analysis
One recombination event was detected in CP gene of a PVX isolate, X3 (Accession number: M31541) with five independent methods (RDP, MaxChi, Chimaera, Siscan and 3Seq) [23]. This isolate from Peru appeared to be a recombinant parented by a local isolate Xc (Accession number: X12804) and an unknown isolate (Table 2).
Table 2.
Recombination analysis of potato virus X sequences
| Recombinant isolate/origin | Parents Major/minor |
Breakpointsa (begin-end) | RDP-implemented methodb (P value) |
|---|---|---|---|
| X3/Peru | Xc/Peru × unknown | 1–159/314–753, 160–315 | R (3.577 × 10−2), M (2.276 × 10−06), C (1.117 × 10−02), S (3.464 × 10−11), 3s (6.415 × 10−04) |
aPosition in alignment
b R, RDP; M, MaxChi; C, Chimaera; S, Siscan; 3 s, 3seq
Phylogenetic positions of populations
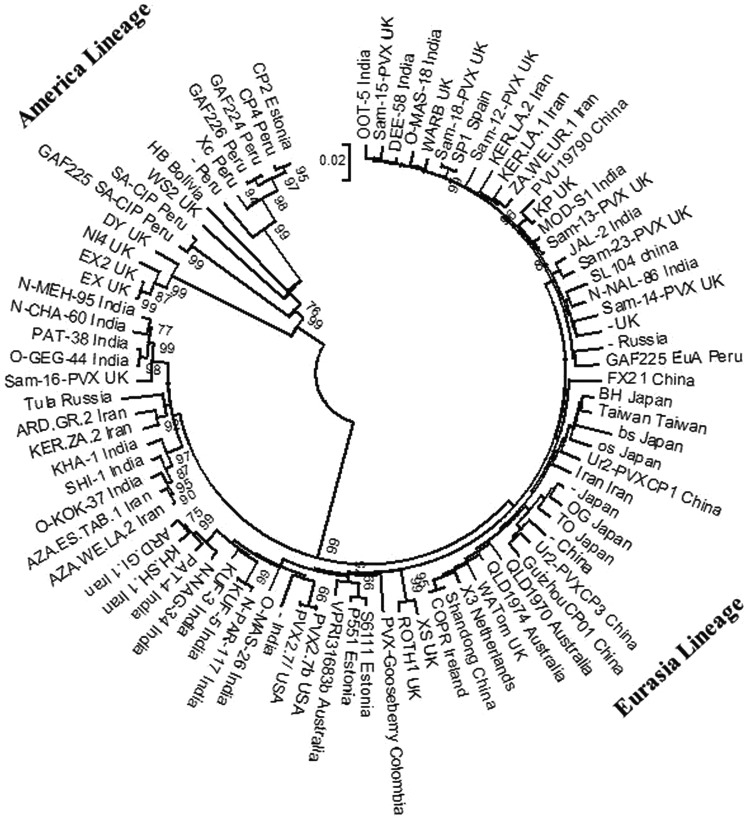
The 86 recombinant–free PVX CP gene sequences from China, India, Iran, Japan, South America and UK were used to reconstruct the phylogeny tree. The results revealed the phylogenetic tree with similar topologies based on the nucleotides and amino acids (Data not shown). In consistent with a previous studies [18, 24, 44, 45], as shown in Fig. 1, the 86 recombination-free PVX isolates fell into two well-defined, namely Eurasia and America lineages. Surprisingly, Isolates GAF-225EuA and PVX-Gooseberry from South America were not clustered with America lineages and there were grouped with Eurasia isolates. All PVX isolates originated from Asia were clustered with Eurasia lineage and PVX isolates from Europa, North and South America were clustered with both lineages suggesting lack of correlation between isolates’ origins and phylogenetic positions.
Fig. 1.
Phylogenetic relationships of potato virus X isolates constructed based on the virus coat protein gene sequence data without root using the neighbor-joining method implemented in MEGA6. Branch support was computed using the bootstrap method based on 1000 replicates. Bootstrap values over 60% are given at the nodes. Isolates are indicated in the tree by isolate name and country of origin, respectively
Pairwise comparisons showed that the genetic distances within variants belonging to Eurasia lineage and America lineage were 0.039 ± 0.003 and 0.120 ± 0.008, respectively, and the genetic diversity between variants from these two lineages was 0.202 ± 0.012. In details, South America population had a highest genetic distance (0.127 ± 0.008), followed by the UK (0.107 ± 0.007), Iran (0.040 ± 0.005), India (0.039 ± 0.004), China (0.035 ± 0.004), and Japan population (0.030 ± 0.004). In addition, the highest and lowest genetic distances between populations were obtained between South America and Japan populations (0.171 ± 0.011), and China and Japan populations (0.034 ± 0.003), respectively (Table S1).
Population characteristic and polymorphism
The overall nucleotide diversity (π) value for all PVX isolates was 0.0619 ± 0.005, which was higher than 0.005, indicating a high genetic diversity in PVX isolates. In details, the largest average number of differences (86 nt) between the sequences from the same group (K) and the greatest π value (0.127) were obtained for the South America population whereas the largest number of segregating sites (S) (216), and mutation within the segregating sites (η) (287) were found for the UK population (Table 3).
Table 3.
Genetic characterization of potato virus X coat protein gene from different lineages and populations
| Lineage and population | N | Hd | S | η | K | π | dS | dN | ω |
|---|---|---|---|---|---|---|---|---|---|
| All | 86 | 0.999 | 283 | 429 | 58.399 | 0.0861 | 0.3033 | 0.0188 | 0.0619 |
| Eurasia | 72 | 0.999 | 262 | 346 | 29.720 | 0.0428 | 0.1583 | 0.0067 | 0.0423 |
| America | 14 | 1.000 | 197 | 254 | 81.066 | 0.1197 | 0.4286 | 0.0227 | 0.0530 |
| Geographic origin | |||||||||
| China | 8 | 1.000 | 75 | 77 | 24.714 | 0.0348 | 0.1288 | 0.0052 | 0.0404 |
| India | 20 | 1.000 | 114 | 117 | 27.542 | 0.0387 | 0.1484 | 0.0041 | 0.0276 |
| Iran | 10 | 1.000 | 80 | 88 | 28.400 | 0.0399 | 0.1482 | 0.0060 | 0.0405 |
| Japan | 6 | 1.000 | 55 | 57 | 21.667 | 0.0305 | 0.1119 | 0.0050 | 0.0447 |
| South America | 10 | 1.000 | 206 | 253 | 86.200 | 0.1271 | 0.4334 | 0.0315 | 0.0727 |
| UK | 19 | 1.000 | 216 | 287 | 75.444 | 0.1067 | 0.3687 | 0.0239 | 0.0648 |
N, sample size; H, number of haplotypes/isolates; S, number of polymorphic (Segregating) sites; η (Eta), total number of mutations; k, average number of nucleotide differences between sequences; p, nucleotide diversity; SS, total number of synonymous sites analyzed; NS, total number of non-synonymous sites analyzed; dS, synonymous nucleotide diversity; dN, non-synonymous nucleotide diversity
Maximum respective values between groups are in bold
The ω estimates for Eurasia and America lineages and each population were smaller than 1 (Table 3). These results suggested that CP gene of PVX in the both lineages and all PVX groups were under negative selection. The results also showed that Eurasia lineage (ω = 0.042) was subjected to a more intense negative selection than America lineage (ω = 0.053). The largest and smallest ω values were obtained from South America and India, 0.073 and 0.028, respectively. In both lineages, there were no codons identified as being under positive selection by four methods (SLAC, FEL, IFEL and FUBAR), using HyPhy software implemented in Datamonkey sever. These results indicated that CP gene in both lineages is under strong negative evolutionary constraints.
Neutrality test on the PVX CP gene
The molecular diversity patterns from segregated sites in both lineages were calculated using Tajima’D and Fu and Li D*&F* statistical tests (Table 4). The results showed that the values of Tajima’D and Fu and Li D*&F* were significantly negative in the Eurasia lineage indicating that the population maintained low frequency polymorphism caused either by genetic hitchhiking, background selection or recent expansion. When the tests were done using the “sliding window” option with 100 and 25 as the window and step size, respectively, no positive values were found in the subsections of CP gene indicating that demographic forces are acting on the members of Eurasia lineage. Based on this, the values were negative across the CP gene indicating the occurrence of recent PVX population expansions. In contrast, Tajima’D, Fu and Li D*&F* values were non-significantly positive in the America lineage indicating an excess of high frequency polymorphism, which suggested that expansion or balancing selection has occurred.
Table 4.
Representation of estimated parameters and test statistics for demographic trends in potato virus X lineages
| Phylogenetic Lineage | π | Tajima’D | Fu&Li D* | Fu&Li F* |
|---|---|---|---|---|
| All | 0.0861 | −1.0853 ns | −0.2566 ns | −0.7333 ns |
| Eurasia | 0.0385 | −2.0372* | −3.2895 | −3.3299** |
| America | 0.1197 | 0.0675 ns | 0.2998 ns | 0.2710 ns |
ns not significant
* 0.01 < P value > 0.05; ** 0.001 < P value > 0.01; *** P value < 0.001
Differentiation of populations
Analysis of PVX population differentiation showed that Eurasia and America lineages with significantly three independent tests were completely distinct. The FST value between the two lineages was 0.60 (> 0.33) indicating a high genetic differentiation between phylogenetic lineages of PVX isolates. Among PVX populations, tests of population differentiation showed that the South America PVX population was distinct from the other populations based on the related Kst*, Z*, Snn and FST values, where Kst* values were not near zero, Z* values were too small, Snn values were significantly high (mostly near 1.000) and FST values were higher than 0.33. The highest FST value (0.54) was obtained between Japanese and South America populations, and the lowest FST value (0.008) was obtained when comparing the India and Iran populations (Table 5).
Table 5.
Genetic differentiation estimates for lineages and geographical populations of potato virus X
| Comparisons | aKS* | aKST* | P value | aZ* | P value | Snn | P value | bFST |
|---|---|---|---|---|---|---|---|---|
| Eurasia (n = 74) versus America (n = 12) | 3.4212 | 0.0915 | 0.000*** | 6.9268 | 0.000*** | 0.967 | 0.000*** | 0.597 |
| China (n = 8) versus India (n = 20) | 3.2697 | 0.0160 | 0.005** | 4.7932 | 0.004** | 0.875 | 0.000*** | 0.103 |
| China (n = 8) versus Iran (n = 10) | 3.2779 | 0.0194 | 0.014* | 3.8530 | 0.017* | 0.713 | 0.049* | 0.101 |
| China (n = 8) versus Japan (n = 6) | 3.1598 | 0.0066 | 0.159 ns | 3.467 | 0.149 ns | 0.607 | 0.228 ns | 0.034 |
| China (n = 8) versus South America (n = 10) | 3.7462 | 0.1124 | 0.002** | 3.5245 | 0.001** | 0.861 | 0.003** | 0.525 |
| China (n = 8) versus UK (n = 19) | 3.7164 | 0.0108 | 0.074 ns | 4.7774 | 0.029* | 0.796 | 0.008** | 0.129 |
| India (n = 20) versus Iran (n = 10) | 3.2953 | 0.0039 | 0.204 ns | 5.0568 | 0.255 ns | 0.750 | 0.023* | 0.008 |
| India (n = 20) versus Japan (n = 6) | 3.2447 | 0.0211 | 0.004** | 4.6140 | 0.005** | 1.000 | 0.000*** | 0.158 |
| India (n = 20) versus South America (n = 10) | 3.5405 | 0.1090 | 0.000*** | 4.6859 | 0.000*** | 0.933 | 0.000*** | 0.506 |
| India (n = 20) versus UK (n = 19) | 3.5789 | 0.0200 | 0.003** | 5.5184 | 0.006** | 0.833 | 0.000*** | 0.124 |
| Iran (n = 10) versus Japan (n = 6) | 3.2340 | 0.0279 | 0.005** | 3.5562 | 0.006** | 0.9375 | 0.003** | 0.150 |
| Iran (n = 10) versus South America (n = 10) | 3.7274 | 0.1124 | 0.000*** | 3.7622 | 0.000*** | 0.900 | 0.002** | 0.504 |
| Iran (n = 10) versus UK (n = 19) | 3.7072 | 0.0180 | 0.039* | 4.8908 | 0.013* | 0.862 | 0.003** | 0.135 |
| Japan (n = 6) versus South America (n = 10) | 3.7796 | 0.1107 | 0.004** | 3.2934 | 0.001** | 0.875 | 0.013* | 0.543 |
| Japan (n = 6) versus UK (n = 19) | 3.7299 | 0.0182 | 0.063 ns | 4.5272 | 0.006** | 0.1000 | 0.000*** | 0.182 |
| South America (n = 10) versus UK (n = 19) | 3.9545 | 0.0683 | 0.000*** | 4.7406 | 0.000*** | 0.8965 | 0.000*** | 0.295 |
ns not significant
* 0.01 < P value > 0.05; ** 0.001 < P value > 0.01; *** P value < 0.001
aK*, Kst*, Z* and Snn are test statistics of genetic differentiation
bFST, coefficient of gene differentiation, which measures inter-population diversity
Discussion
In this study, genetic variation and molecular evolution of 86 recombinant-free PVX isolates based on the CP gene were determined. Recombination is an important source of genetic variation in plant viruses [9]. Recombination at the CP gene in many RNA viruses such as Sugercane mosaic virus [19], Turnip mosaic virus [26], Grpevine fanleaf virus [41], Banan bract mosaic virus [1] has been detected. In this study, one recombination event in one virus isolate (X3) was detected with five methods (Table 2). Recombinant events have not been reported before in PVX isolates. This might be due to the fact that PVX is not transmitted by vectors and, therefore, there has been an unlikely event of being in a mixed infection with a different PVX variant. Recombination was usually parented by the isolates from the same geographic region due to more chances to be exposed each other [33], recombinant isolate (X3) was parented by a local isolates (Xc) and an unknown isolate (Table 2).
Phylogenetic analysis of the CP gene sequences showed that PVX isolates from Eurasia lineage were the most common occurring isolates confirming previous results [24, 44]. The amino acids K at position 20, A at 64, M at 128 and T at 227 were conserved in the CP of all members of Eurasia lineage and they have been reported to be avirulent on potato cultivars carrying the Nx resistance gene as described for the type X of PVX [20, 24]. Accordingly, although members of America lineage were also conserved in the same positions they possessed A at position 20 (gap in some isolates), V at 64, I at 128 and A at position 227, and they are known as being capable of overcoming the Nx resistant gene. Thus, the phylogenetic analysis showed clear relationships between the phylogeny and pathogenicity of PVX isolates on potato cultivars carrying the resistance genes [20]. However, it was difficult to find a clear correlation between phylogeny and geographical origin suggesting that PVX variants have been spread from one region to other geographical regions. Because the virus variants from China, India, Iran, Japan and the UK were placed in several subclades (Fig. 1), it is suggested that the infections may have resulted from founder effect (genetic drift), i.e., different PVX genotypes have been transmitted via propagated material and have gone through bottlenecks at different places and times.
America lineage seemed to have older isolates compared to that of Eurasia lineage which is in agreement with origination of potato from America [34]. The deep-branching with America variants might also point to lack of bottlenecks with these isolates whereas with Eurasia isolates it seems that because PVX has spread from America it has gone through bottlenecks and resulted in founder effects. Accordingly, Eurasia isolates distantly related to the main node and bear shorter branches. This finding also suggests co-evolution of PVX with potato plant.
Isolates GAF-225EuA and PVX-Goosberry within Eurasia lineage seems to be the oldest isolates giving rise to this lineage. This suggests that these isolates as the variants were produced from America variants after a bottleneck event.
Comparison of nt sequence diversity (π) among Eurasia and America lineages (Table 3) showed that the isolates of Eurasia lineage (π = 0.043) were less diverse than that of America lineage (π = 0.119) and overall nt sequence diversity was 0.086. There are three possible explanations for low genetic diversity between isolates of Eurasia lineage. One is that viruses transmitted by infected propagative materials would not have higher nt diversity than aphid-borne or seed-borne viruses (Yu et al. 2010). The second explanation is that frequent gene flow within this lineage (Table 5) and the other might be due to strong negative selection (ω = 0.042). In regard to higher nt diversity within the America lineage, it appeared that the Peruvian and Bolivian PVX isolates have not diversified recently and mutations have accumulated over a long time because their isolates appear as a deep-branching cluster (Fig. 1), which is in agreement with the large number of segregating sites (206) and the large number of mutations (253) in this lineage (Table 3). It seems that primarily originated potatoes growing in Peru and Bolivia [34] have played a role in this scenario. An isolate from Estonia (CP2) and another from USA (WS2) were clustered with Peruvian and Bolivian isolates (Fig. 1) suggesting that the PVX isolates from South America have been transported to USA and Europe by human. This may serve as an example in which human activity has played a role in virus evolution.
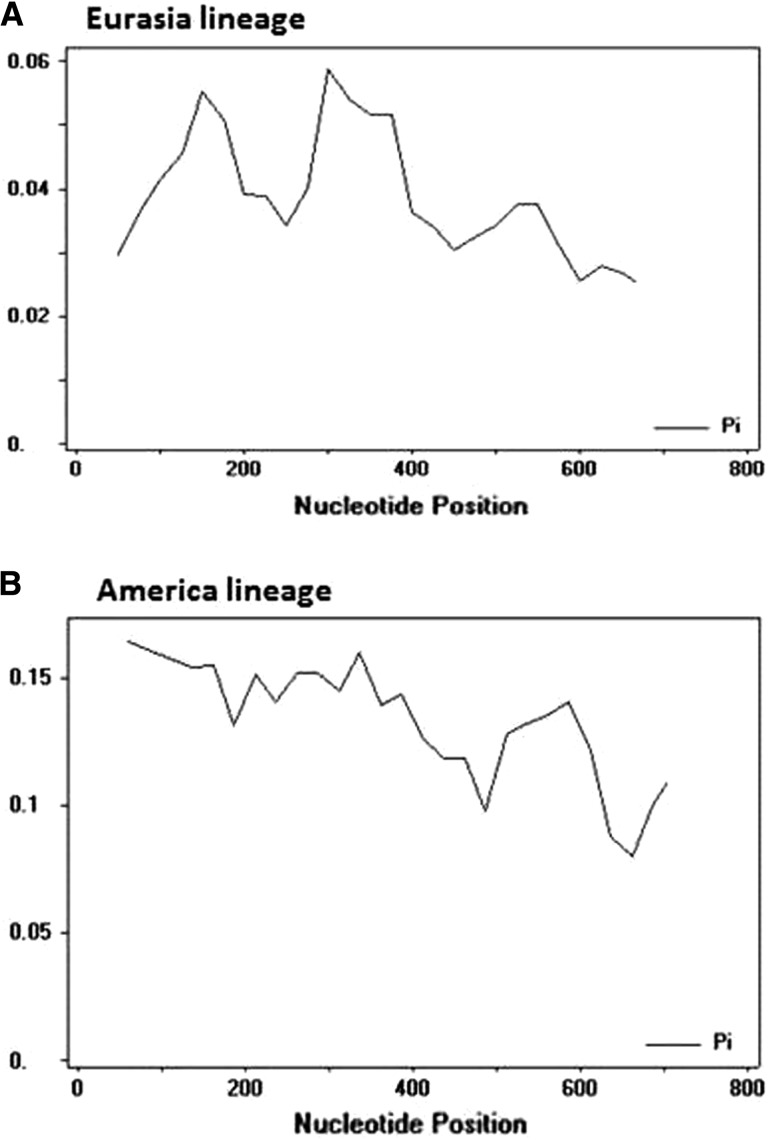
Analysis of nucleotide diversity by “sliding window” option with 100 and 25 as the window and step size, respectively, revealed high polymorphism at middle region and 5′ part of CP gene within Eurasia and America lineages, respectively (Fig. 2). This information helps to select regions of viral genome targeted to design specific primers for virus detection and the occurrence of mutations in the primer-targeted sites may limit the suitability of RT-PCR for viral detection.
Fig. 2.
Trend of nucleotide diversity along the coat protein gene in the potato virus X lineages, A: Eurasia lineage and B: America lineage. The curves were generated by “sliding windows” with 50 and 25 as the window and step sizes, respectively
The direction and degree of selective constraints operating on PVX CP were estimated by calculating ω values [42]. The estimated ω ratios were less than one for the both lineages and the populations (Table 3) suggesting that the negative selection against amino acid changes serves as a driving force for PVX evolution which is consistent with previous results [44, 45]. Overall, the CP gene is going through negative selection as the ω ratio value for the whole PVX population of 86 recombinant-free isolates was 0.062 indicating that the CP is under functional constraint due to its crucial role in virulence, cell-to-cell movement and multiplication [3]. Several studies on viral CP genes in other RNA viruses have revealed that this protein can be conserved under negative selection [8, 42].
All the PVX populations examined, except for the South America population, are non-differentiated from each other because the Kst* values were near zero and supported by none or weak significant P-values. No differentiations between these populations were also confirmed by Z* and Snn values. To estimate the degree of virus migration (Gene flow) between geographical origins (two by two), the statistic FST was calculated (Table 5) showing that gene flow of the PVX populations between Asian and European countries were fairly frequent (FST < 0.19). In addition, the results showed that gene flow between Asian countries, India/Iran and China/Japan populations were frequent with FST values of 0.008 and 0.034, respectively. A possible explanation for this result is relatively close distances between these countries. On the other hand, the highest FST value between South America and Japanese populations (0.54) could be due to long distances between these geographic regions, indicative of a correlation between geographical position and genetic distances. Results from the PVX population genetic differential analysis agreed with those of the phylogenetic analysis. For instance, the lowest FST was found between China and Japan (FST = 0.034) which clustered together in a subclade (Fig. 1).
In agreement with a high genetic diversity between South America population with other populations (Table S1), low gene flow values were estimated (>0.29) (Table 5). No vector transmission is known for PVX and it can be transmitted by infected propagated materials which are exchanged frequently between countries. This result is further emphasizing the use of virus- free plant materials in preventing the dissemination of PVX.
In conclusion, global PVX isolates analyzed in this study could be divided into two distinct lineages with lack of correlation between the phylogeny and the geographical origin of PVX isolates suggesting that there might have been a convergent evolution. This study also revealed that negative selection and genetic drift may be the potential drivers of the dynamics of PVX molecular evolution in Eurasia lineage (π = 0.043) whereas negative selection and recombination may have involved in PVX molecular evolution in the members of America lineage (π = 0.12). To the best of our knowledge, this is the first extensive study on the genetic variability of global PVX isolates and first report of a recombination event in the PVX population. It also seemed that exchange of infected seed potato tubers have played a role in shaping the PVX population structure.
Electronic supplementary material
Below is the link to the electronic supplementary material.
References
- 1.Balasubramainan V, Selvarajan R. Genetic diversity and recombination analysis in the coat protein gene of Banana bract mosaic virus. Virus Genes. 2016;48:509–517. doi: 10.1007/s11262-014-1056-x. [DOI] [PubMed] [Google Scholar]
- 2.Baulcombe DC, Kohm B, Goulden MG, Santa Cruz S, Forsyth A, Chapman S, Kavanagh T. In: Bills D, Owens R, Baker CJ, Hutcheson S, Straney D, Fischoff D, Shah editors. Proceedings of the fifth international symposium on biotechnology and plant protection. New Jersey: University of Maryland, World Scientific Publishing Co. 1993. pp. 101–110.
- 3.Callaway A, Giesman-Cookmeyer D, Gillock ET, Sit TL, Lommel SA. The multifunctional capsid proteins of plant RNA viruses. Annu Rev Phytopathol. 2001;39:419–460. doi: 10.1146/annurev.phyto.39.1.419. [DOI] [PubMed] [Google Scholar]
- 4.Cox BA, Jones RA. Genetic variability in the coat protein gene of Potato virus X and the current relationship between phylogenetic placement and resistance groupings. Arch Virol. 2010;155:1349–1356. doi: 10.1007/s00705-010-0711-3. [DOI] [PubMed] [Google Scholar]
- 5.Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 6.Elena SF, Agudelo-Romero P, Carrasco P, Codoñer FM, Martín S, Torres-Barceló C, Sanjuán R. Experimental evolution of plant RNA viruses. Heredity. 2008;100:478–483. doi: 10.1038/sj.hdy.6801088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao F, Lin W, Shen J, Liao F. Genetic diversity and molecular evolution of arabis mosaic virus based on the CP gene sequence. Arch Virol. 2016;161:1047–1051. doi: 10.1007/s00705-015-2729-z. [DOI] [PubMed] [Google Scholar]
- 9.García-Arenal F, Fraile A, Malpica JM. Variability and genetic structure of plant virus populations. Annu Rev Phytopathol. 2001;39:157–186. doi: 10.1146/annurev.phyto.39.1.157. [DOI] [PubMed] [Google Scholar]
- 10.He Z, Yasaka R, Lie W, Lia S, Ohshimac K. Genetic structure of populations of Sugarcane streak mosaic virus in China: comparison with the populations in India. Virus Res. 2016;211:103–116. doi: 10.1016/j.virusres.2015.09.020. [DOI] [PubMed] [Google Scholar]
- 11.Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting population subdivision. Mol Biol Evol. 1992;9:138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- 12.Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hudson RR. A new statistic for detecting genetic differentiation. Genetics. 2000;155:2011–2014. doi: 10.1093/genetics/155.4.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karpova O, Zayakina O, Arkhipenko M, Sheval E, Kiselyova O, Poljakov V, Yaminsky IV, Rodionova NP, Atabekov JG. Potato virus X RNA-mediated assembly of single-tailed ternary ‘coat protein–RNA–movement protein complexes. J Gen Virol. 2006;87:2731–2740. doi: 10.1099/vir.0.81993-0. [DOI] [PubMed] [Google Scholar]
- 15.Kavanagh T, Goulden M, Santa-Cruz S, Chapman S, Barker I, Baulcombe DC. Molecular analysis of a resistance breaking strain of Potato virus X. Virology. 1992;189:609–617. doi: 10.1016/0042-6822(92)90584-C. [DOI] [PubMed] [Google Scholar]
- 16.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 17.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Virus taxonomy-ninth report of the International Committee on Taxonomy of viruses. London: Academic Press; 2011. [Google Scholar]
- 18.Kutanjak D, Silvestre R, Cuellar W, Perez W, Muller G, Ravinkar M, Kreuze J. Complete genome sequence of new divergent Potato virus X isolates and discrimination between strains in a mixed infection using small RNAs sequencing approach. Virus Res. 2014;191:45–50. doi: 10.1016/j.virusres.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Liu R, Zhou T, Fan Z. Genetic diversity and population structure of Sugarcane mosaic virus. Virus Res. 2013;171:242–246. doi: 10.1016/j.virusres.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 20.Malcuit I, Jong WD, Baulcombe DC, Shields DC, Kavangah TA. Acquisition of multiple virulence/avirulence determinants by Potato virus X (PVX) has occurred through convergent evolution rather than through recombination. Virus Genes. 2000;20:165–172. doi: 10.1023/A:1008178800366. [DOI] [PubMed] [Google Scholar]
- 21.Malcuit IM, Marano MR, Kavanagh TA, De Jong W, Forsyth A, Baulcombe DC. The 25-kDa movement protein of PVX elicits Nb-mediated hypersensitive cell death in potato. Mol Plant Microbe Interact. 1999;12:536–543. doi: 10.1094/MPMI.1999.12.6.536. [DOI] [Google Scholar]
- 22.Mandal B, Kumar A, Rani P, Jain RK. Complete genome sequence, phylogenetic relationships and molecular diagnosis of an Indian isolate of Potato virus X. J Phytopath. 2012;160:1–5. doi: 10.1111/j.1439-0434.2011.01848.x. [DOI] [Google Scholar]
- 23.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massumi H, Poormohammadi S, Pishyar S, Maddahian M, Heydarnejad J, Hosseini-pour A, Bysterveldt K, Varsani A. Molecular characterization and field survey of Iranian Potato virus X isolates. VirusDisease. 2014;25:338–344. doi: 10.1007/s13337-014-0222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morozov S, Solovyev AG. Triple gene block: modular design of a multifunctional machine for plant virus movement. J Gen Virol. 2003;84:1351–1366. doi: 10.1099/vir.0.18922-0. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen HC, Tran HTN, Oshima K. Genetic variation of the Turnip mosaic virus population of Vietnam: a case study of founder, regional and local influences. Virus Res. 2013;171:138–149. doi: 10.1016/j.virusres.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa T, Tomitaka Y, Nakagawa A, Ohshima K. Genetic structure of a population of Potato virus Y inducing potato tuber necrotic ring spot disease in Japan; comparison with North American and European populations. Virus Res. 2008;131:199–212. doi: 10.1016/j.virusres.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 29.Querci M, Baulcombe DC, Goldbach RW, Salazar LF. Analysis of the resistance-breaking determinant of Potato virus X (PVX) strain HB on different potato genotype expressing extreme resistance to PVX. Phytopathol. 1995;85:1003–1010. doi: 10.1094/Phyto-85-1003. [DOI] [Google Scholar]
- 30.Rozas J, Sanchez-De I, Barrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 31.Rozas J. DNA sequence polymorphism analysis using DnaSP. In: Posada D, editor. Bioinformatics for DNA sequence analysis; methods in molecular biology series. Humana Press, New York. 2009; 537:337–350. [DOI] [PubMed]
- 32.Santa-Cruz S, Baulcombe D. Analysis of Potato virus X coat protein genes in relation to resistance conferred by the genes Nx, Nb and Rx1 of potato. J Gen Virol. 1995;76:2057–2061. doi: 10.1099/0022-1317-76-8-2057. [DOI] [PubMed] [Google Scholar]
- 33.Sokhandan-Bashir N, Melcher U. Population genetic analysis of grapevine fanleaf vius. Arch Virol. 2012;157:1919–1929. doi: 10.1007/s00705-012-1381-0. [DOI] [PubMed] [Google Scholar]
- 34.Spooner DM, McLean K, Ramsay G, Waugh R, Bryan GJ. A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. PNAS. 2005;102:14694–14699. doi: 10.1073/pnas.0507400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevenson WR, Loria R, Franc GD, Weingartner DP. Compendium of Potato diseases. 2. St. Paul: APS Press; 2001. [Google Scholar]
- 36.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsompana M, Abad J, Purugganan M, Moyer JW. The molecular population genetics of the Tomato spotted wilt virus (TSWV) genome. Mol Ecol. 2005;14:53–66. doi: 10.1111/j.1365-294X.2004.02392.x. [DOI] [PubMed] [Google Scholar]
- 40.Verchot-Lubicz J. A new cell-to-cell transport model for potexviruses. Mol Plant-Microbe Interact. 2005;18:283–290. doi: 10.1094/MPMI-18-0283. [DOI] [PubMed] [Google Scholar]
- 41.Vigne H, Bergdoll M, Guyader S, Fuchs M. Population structure and genetic variability within isolates of Grapevine fanleaf virus from a naturally infected vineyard in France: evidence for mixed infection and recombination. J Gen Virol. 2004;85:2435–2445. doi: 10.1099/vir.0.79904-0. [DOI] [PubMed] [Google Scholar]
- 42.Yang Z, Bielawski JP. Statistical methods for detecting molecular adaption. Trends Ecol Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon JY, Joa HJ, Choi KS, Do KS, Lim HC, Chung BN. Genetic diversity of a natural population of Apple stem pitting virus isolated from apple in Korea. Plant Pathol J. 2014;30:95–99. doi: 10.5423/PPJ.NT.02.2014.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu XQ, Jia JL, Zhang C, Li XD, Wang YJ. Phylogenetic analyses of an isolate obtained from potato in 1985 revealed Potato virus X was introduced to China via multiple events. Virus Genes. 2010;40:447–451. doi: 10.1007/s11262-010-0468-5. [DOI] [PubMed] [Google Scholar]
- 45.Yu XQ, Wang HY, Lan YF, Zhu XR, Li XD, Fan ZF, Li HF, Wang YY. Complete genome of a Chinese isolate of Potato virus X and analysis of genetic diversity. J Phytopathol. 2008;156:346–351. doi: 10.1111/j.1439-0434.2007.01365.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.