Abstract
The advantages of the model organism Drosophila melanogaster, including low genetic redundancy, functional simplicity, and the ability to conduct large-scale genetic screens, have been essential for understanding the molecular nature of circadian (∼24 hr) rhythms, and continue to be valuable in discovering novel regulators of circadian rhythms and sleep. In this review, we discuss the current understanding of these interrelated biological processes in Drosophila and the wider implications of this research. Clock genes period and timeless were first discovered in large-scale Drosophila genetic screens developed in the 1970s. Feedback of period and timeless on their own transcription forms the core of the molecular clock, and accurately timed expression, localization, post-transcriptional modification, and function of these genes is thought to be critical for maintaining the circadian cycle. Regulators, including several phosphatases and kinases, act on different steps of this feedback loop to ensure strong and accurately timed rhythms. Approximately 150 neurons in the fly brain that contain the core components of the molecular clock act together to translate this intracellular cycling into rhythmic behavior. We discuss how different groups of clock neurons serve different functions in allowing clocks to entrain to environmental cues, driving behavioral outputs at different times of day, and allowing flexible behavioral responses in different environmental conditions. The neuropeptide PDF provides an important signal thought to synchronize clock neurons, although the details of how PDF accomplishes this function are still being explored. Secreted signals from clock neurons also influence rhythms in other tissues. SLEEP is, in part, regulated by the circadian clock, which ensures appropriate timing of sleep, but the amount and quality of sleep are also determined by other mechanisms that ensure a homeostatic balance between sleep and wake. Flies have been useful for identifying a large set of genes, molecules, and neuroanatomic loci important for regulating sleep amount. Conserved aspects of sleep regulation in flies and mammals include wake-promoting roles for catecholamine neurotransmitters and involvement of hypothalamus-like regions, although other neuroanatomic regions implicated in sleep in flies have less clear parallels. Sleep is also subject to regulation by factors such as food availability, stress, and social environment. We are beginning to understand how the identified molecules and neurons interact with each other, and with the environment, to regulate sleep. Drosophila researchers can also take advantage of increasing mechanistic understanding of other behaviors, such as learning and memory, courtship, and aggression, to understand how sleep loss impacts these behaviors. Flies thus remain a valuable tool for both discovery of novel molecules and deep mechanistic understanding of sleep and circadian rhythms.
Keywords: FlyBook: Drosophila, circadian rhythms, molecular neuroscience, neuroscience, sleep
Part 1: Circadian Rhythms
Genetics of circadian rhythms
Circadian rhythms are daily rhythms in behavior or physiology that reoccur approximately every 24 hr. Circadian rhythms can be entrained by external environmental cues (i.e., light and temperature), but persist in the absence of these cues, with free-running periods that deviate slightly from the expected 24 hr in constant environmental conditions. Circadian rhythms had been observed in living organisms for centuries (de Mairan 1729), but it was not until the advent of Drosophila research in the 1960s that we began to understand the genetic and molecular nature of this phenomenon. Circadian rhythms are evident in many different aspects of Drosophila behavior and physiology, but the major assays used to discover the key molecules driving circadian rhythms measured either eclosion (emergence of adult flies from pupae) or locomotor activity. Eclosion is a one-time event in a single fly, but it can be monitored as a rhythm in a population, with peaks of emerging flies typically observed in the early daytime hours. The first genetic studies of circadian rhythms began with the observation of heritable early or late eclosion times in D. pseudoobscura (Pittendrigh 1967), but it was the historic genetic screen of D. melanogaster conducted by R. Konopka in Seymour Benzer’s laboratory that led to the identification of single gene mutants, which were mapped and eventually cloned (Konopka and Benzer 1971; Bargiello et al. 1984; Zehring et al. 1984). Konopka identified mutants with shortened (to 19 hr) or lengthened (to 28 hr) periodicity of rhythms, or completely eliminated rhythms, in constant conditions, and found that all these mutations mapped to the same gene, which he named period (per). For a long time per was the only circadian gene known, but rapid progress in the circadian field in the 1990s led to the identification of timeless (tim) (Sehgal et al. 1994), Clock (Clk) (Allada et al. 1998), and cycle (cyc) (Rutila et al. 1998), which, together with per, make up the core transcriptional feedback loop that drives circadian rhythms. The late 1990s and early 2000s saw the identification of a circadian photoreceptor, cryptochrome (cry) (Emery et al. 1998; Stanewsky et al. 1998), the kinases double-time (dbt) (Kloss et al. 1998; Price et al. 1998) and shaggy (sgg) (Martinek et al. 2001), and a transcriptional activator involved in a second feedback loop, vrille (vri) (Blau and Young 1999). Following the identification of tim in 1994, which was identified using the same eclosion assay used by Konopka, the circadian field switched to locomotor activity as the assay of choice for circadian rhythms.
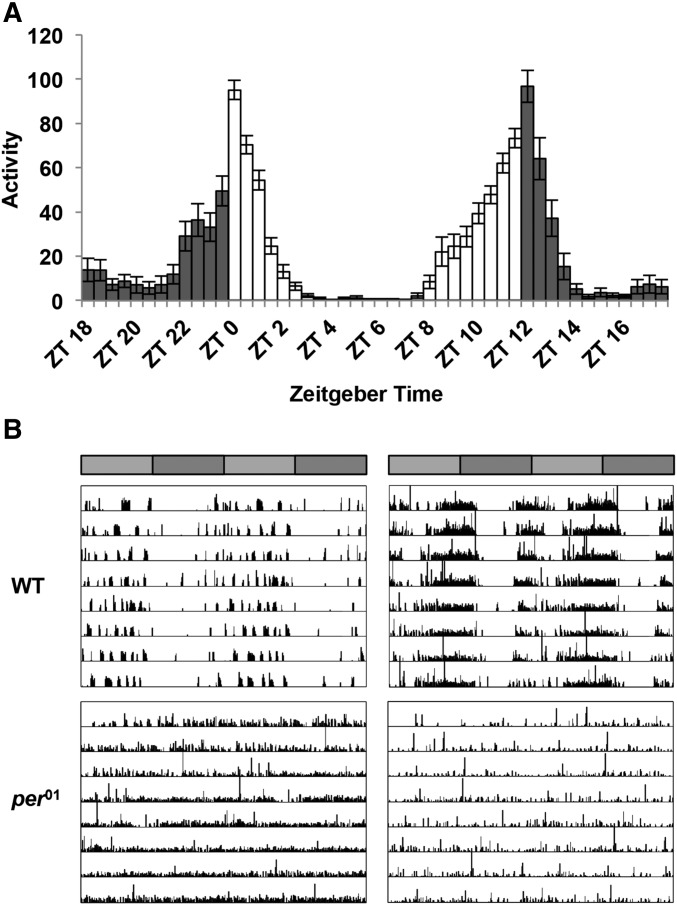
Locomotor activity in Drosophila is organized such that, in a 12:12 hr light:dark (LD) cycle, flies exhibit peaks of activity during dawn and dusk (morning and evening peaks), with increases in activity typically occurring slightly before (anticipating) the lights-on and lights-off transitions (Figure 1A), although under more naturalistic conditions, an additional afternoon peak of activity can be observed (Vanin et al. 2012; Green et al. 2015). In constant dark conditions (DD), the morning peak shrinks and only the evening peak persists, reoccurring with a period of ∼23.8 hr (Figure 1B). In addition to eclosion and locomotor activity, circadian rhythms also drive other aspects of physiology and behavior, including sleep and an increasingly appreciated role in metabolism. Circadian control of all of these processes relies not only on the intracellular clock, but also on networks of cells that interact to influence circadian outputs. In part 1 of this review, we will focus on how intracellular rhythms are sustained and how the network of clock-expressing cells in the brain drives rest:activity behavior and other outputs. The role of specific clock neurons in driving sleep behavior will be discussed in part 2.
Figure 1.
(A) Activity for a group of wild-type (WT) male flies in a 12:12 hr light:dark (LD) cycle at 25°. Flies anticipate lights-off, and under these conditions also lights-on, with increased activity in advance of these transitions. (B) Double-plotted activity in constant darkness for two individual WT and per01 male flies after entrainment in standard LD conditions. Data are double-plotted for ease of interpretation, with each day of data plotted in a new row concatenated with the data from the subsequent day, such that Row 1 displays data for Day 1 and Day 2, Row 2 displays data for Day 2 and Day 3, etc. Activity is concentrated in the subjective day in a pattern that recurs with a period of slightly <24 hr in WT flies.
Molecular mechanism of the clock
Transcription-based feedback loops:
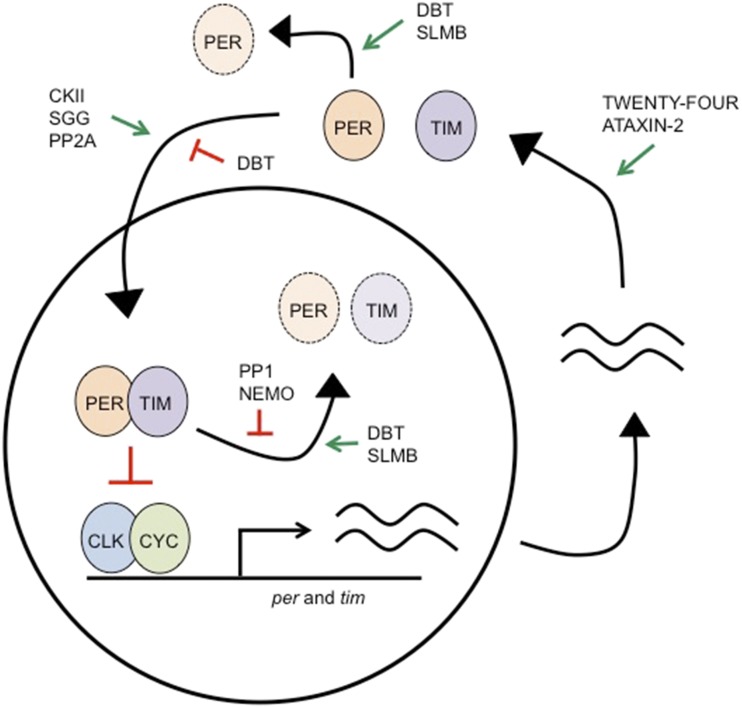
The finding that per RNA and protein are expressed cyclically and rising levels of protein are associated with declining levels of the mRNA led to the postulate that the PER protein negatively regulates its own transcription to generate an autoregulatory circadian loop (Siwicki et al. 1988; Hardin et al. 1990; Zerr et al. 1990). Subsequent studies with tim supported this idea, showing that the two mRNAS cycle in phase and the PER and TIM proteins interact directly and affect their own transcription (Gekakis et al. 1995; Sehgal et al. 1995). The negative feedback loop thus generated constitutes the basis of overt rhythms in Drosophila. As described below, maintenance of the loop requires the activity of additional components (Figure 2).
Figure 2.
The molecular feedback loop is formed by the negative feedback of Period (PER) and Timeless (TIM) on their own transcription. Delays exist between transcription of per and tim mRNA and the localization of these proteins in the nucleus, where they can interact with transcriptional activators Clock (CLK) and Cycle (CYC). These delays are thought to be important for allowing the molecular clock to cycle with a period of ∼24 hr. Critical regulators have been identified at several steps of the cycle that are necessary for accurate timing and strength of molecular rhythms. Degradation of PER and TIM allows the cycle to start anew. Not pictured is the second feedback loop formed by PDP1 and Vrille, which produces cycling of Clk mRNA. This secondary feedback loop is thought to reinforce molecular oscillations, although cycling of the CLK protein is not necessary for rhythms. CKII, Casein Kinase II; SGG, shaggy; PP2A, Protein Phosphatase 2A; PP1, Protein Phosphatase 1; DBT, doubletime.
Levels of per and tim mRNA rise during the day and are highest in the early evening. At this time, the two proteins start to accumulate, initially in the cytoplasm, and then around the middle of the night in the nucleus (reviewed in Zheng and Sehgal 2012). TIM stabilizes PER in the cytoplasm, and is required to transport it to the nucleus. Nuclear localization of the two proteins is also regulated by specific importins (Jang et al. 2015) and appears to be temporally regulated (Curtin et al. 1995; Meyer et al. 2006). Nuclear localization of PER and TIM coincides with the decline of their mRNA levels due to negative autoregulatory feedback by the proteins. PER and TIM cannot bind DNA, but regulate transcription by inhibiting their transcriptional activators, Clock and Cycle. Although the mechanisms of negative feedback are not completely understood, biochemical data support sequestration of the CLK–CYC complex from DNA by PER and a model where PER recruits the kinase DBT (a CK1ε homolog) to promote CLK phosphorylation (Lee et al. 1999; Kim and Edery 2006; Kim et al. 2007; Nawathean et al. 2007). The role of TIM in negative feedback is less clear. In the early morning, TIM levels decline sharply, and, by midafternoon, PER expression is also greatly diminished, lifting the negative feedback and allowing a new cycle of transcription.
In addition to the major feedback loop described above, the Drosophila clock contains a second loop interlocked with the first. In this loop, the CLK–CYC complex drives sequential expression of a transcriptional activator, PDP1, and a repressor, Vrille (VRI), of Clk expression (Cyran et al. 2003; Glossop et al. 2003). By feeding back on Clk expression in a rhythmic fashion, PDP1 and VRI maintain rhythmic expression of Clk mRNA. However, given that CLK protein levels do not cycle, the purpose of the mRNA cycling is unclear (Houl et al. 2006). It is thought that the second loop stabilizes the system and provides greater precision (Cyran et al. 2003; Glossop et al. 2003). Other proteins, such as Clockwork Orange, KAYAK-α, and E75, are also implicated in transcriptional control (Kadener et al. 2007; Lim et al. 2007; Matsumoto et al. 2007; Ling et al. 2012; Kumar et al. 2014).
Post-translational regulation within the clock:
It is clear that a molecular cycle as described above can be only maintained through the incorporation of critical delays. For instance, negative feedback likely has to be delayed or the system might reach an equilibrium between mRNA synthesis and mRNA repression, which would be incompatible with cyclic expression. As noted, expression of the PER and TIM proteins is delayed by several hours relative to expression of the mRNAs. Indeed, the timely appearance and disappearance of the proteins is very important, and is controlled in large part by post-transcriptional mechanisms. Translation of PER is regulated by a complex consisting of the Twenty-four and Ataxin-2 proteins, but this complex does not appear to act in a time-specific manner (Lim et al. 2011; Lim and Allada 2013; Zhang et al. 2013). There is more evidence for temporal regulation of protein stability, which is mediated to a large extent by phosphorylation. Both PER and TIM are cyclically phosphorylated, and many of the relevant kinases and phosphatases have been identified. Thus, Casein Kinase 1ε (called doubletime in Drosophila) and NEMO phosphorylate PER to control its stability (Kloss et al. 1998; Price et al. 1998; Chiu et al. 2011), while Casein Kinase II and GSK3b (sgg) target PER alone or TIM and PER, respectively, to promote the nuclear entry of PER and TIM (Martinek et al. 2001; J. Lin et al. 2002; Lin et al. 2005; Akten et al. 2003; Ko et al. 2010). However, phosphorylation is not always a positive regulator of nuclear localization. Protein Phosphatase 2A dephosphorylates PER, and yet is also a positive regulator of nuclear localization and stability (Sathyanarayanan et al. 2004). Protein Phosphatase 1 also dephosphorylates and stabilizes TIM and PER, and effects on PER appear to be regulated by TIM (Fang et al. 2007). DBT may also delay nuclear entry of PER (Bao et al. 2001; Cyran et al. 2005; Zheng et al. 2014) in addition to its other roles in the clock.
In general, the mechanism by which phosphorylation impacts nuclear entry is not known. We have a somewhat better understanding of how phosphorylation affects protein stability. Phosphorylation of PER by DBT at S47 and likely other nearby sites promotes its recognition by the F-box protein SLMB, a component of the SCF E3 ubiquitin ligase complex, which targets PER for degradation (Grima et al. 2002; Ko et al. 2002; Chiu et al. 2008). Mutation of S47 to a residue that cannot be phosphorylated lengthens circadian period, most likely because PER expression persists longer than normal, while a phospho-mimetic S47D mutation shortens it (Chiu et al. 2008). On the other hand, phosphorylation of PER by the kinase NEMO at S596, along with DBT-dependent phosphorylation at neighboring sites, inhibits phosphorylation at S47 to stabilize PER, and mutations at these sites therefore shorten the circadian period (Chiu et al. 2011). A mutation in one of the DBT-dependent phosphorylation sites near S596 is, in fact, the basis of the 19 hr period in the original pershort mutant isolated by Konopka (Baylies et al. 1987; Yu et al. 1987). Phosphorylated residues S610 and S613 also cooperatively regulate PER stability and lengthen the period when mutated, and genetic evidence suggests S613 may be upstream of the phosphor-cluster around S596, although the relevant kinase for these sites is not known (Garbe et al. 2013). Thus, an interplay of phosphorylation sites on PER tightly regulates its stability, and, ultimately, circadian period. It is important to note, though, that while a number of period-altering mutations have been identified in per, tim, and the relevant kinases, we still do not have a complete understanding of how the ∼24 hr period is generated.
O-GlcNAcylation of serine/threonine residues on PER and CLK provides yet another level of post-translational control (Kim et al. 2012; Kaasik et al. 2013). Knockdown or overexpression of the enzyme that confers this modification, OGlcNAc transferase (OGT), alters circadian period, perhaps by affecting phosphorylation of PER. The latter is based on the finding that O-GlycNAcylation of human Per2 competes with phosphorylation in critical regions (Kaasik et al. 2013).
It is important to note that mechanisms of the circadian clock elucidated in Drosophila are conserved through evolution. Inherent to clock function in flies is a feedback loop in which the per and tim genes are expressed cyclically and negatively regulated by their own protein products (Hardin et al. 1990; Sehgal et al. 1995). The Neurospora clock is likewise comprised of a negative feedback loop generated through cyclic activity of the frequency gene product (Aronson et al. 1994). Even the cyanobacterial clock, which can be reconstituted in vitro through the cyclic action of clock proteins (Nakajima et al. 2005), includes a transcriptional feedback mechanism in vivo (Ishiura et al. 1998).
Insects and mammals share not only the regulatory logic of a transcriptional feedback loop, but the functions of many clock genes are also conserved. Mammalian Per2 and CK1δ, both homologs of genes first linked to circadian rhythms in the fruit fly, have been implicated in the human circadian disorder Advanced Sleep Phase Syndrome (Toh et al. 2001; Xu et al. 2005). Homologs of CLK, CYC (called BMAL1 in mammals), and PER serve similar functions in mammals as they do in Drosophila (reviewed in Partch et al. 2014). The closest mammalian CRY homolog, on the other hand, does not act as a photoreceptor—mammalian light input to the clock is delivered non-cell-autonomously via intrinsically photosensitive retinal ganglion cells (Güler et al. 2008)—but instead appears to have taken the function of TIM, acting with PER to repress its own transcription. Other insects, such as honeybees and butterflies, appear to have a mammalian-like CRY that can act as a negative regulator of transcription in addition to, or instead of, a Drosophila-like photosensitive CRY, suggesting that the mammalian CRY is evolutionary old, even though it seems to have been lost in Drosophila (Zhu et al. 2005; Rubin et al. 2006). Mammals also have a second transcriptional feedback loop analogous to the PDP1/VRI loop in flies (Partch et al. 2014).
The transcriptional mechanisms discussed earlier do not just maintain rhythmic expression of per and tim, but also drive cycling of many output genes that contain enhancer elements recognized by CLK-CYC. Transcriptomic analysis has identified many cyclically expressed genes in Drosophila heads, brains, clock neurons, and peripheral tissues (Claridge-Chang et al. 2001; McDonald and Rosbash 2001; Ceriani et al. 2002; Ueda et al. 2002; Y. Lin et al. 2002; Wijnen et al. 2006; Keegan et al. 2007; Kula-Eversole et al. 2010; Xu et al. 2011; Hughes et al. 2012). Cycling transcripts that are direct targets of CLK-CYC can then drive broader changes in physiology. For example, cycling of the Nlf-1 transcript results in time-of-day dependent changes in sodium leak current that, in turn, drive rhythmic neuronal activity in a subset of clock neurons (Flourakis et al. 2015). NLF-1 is thought to act by stabilizing and promoting trafficking of a sodium leak channel, Narrow Abdomen (na). This example shows how clock-dependent cycling transcription can be the basis for rhythms in physiology that, in this case, likely contribute to behavior. In addition to these cell autonomous effects of the clock on cell physiology, however, clocks can also drive rhythms in physiology of other cells non-cell-autonomously, by controlling signaling through neuronal circuits and release of secreted factors (Jaramillo et al. 2004; Cavey et al. 2016; Erion et al. 2016). The next section will discuss how clock neurons function as a network to orchestrate rest:activity rhythms.
The clock cell network
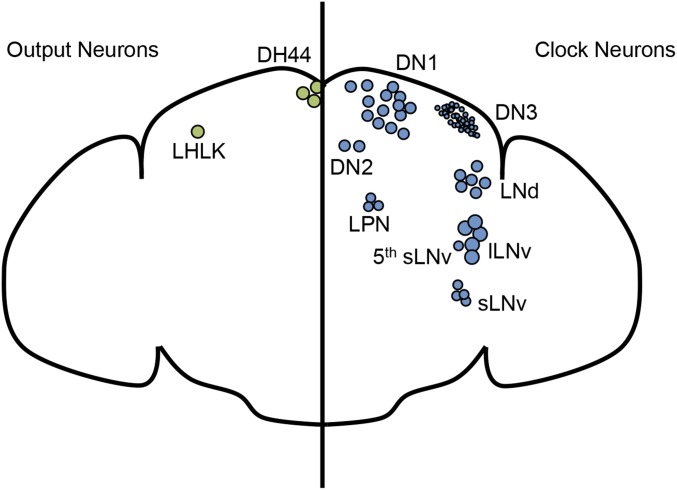
Behavioral rhythms of rest:activity require clock activity in the brain, which was suggested even by early experiments of Konopka in which he restored short period rhythms in per null flies by transplanting brains of pershort mutants into the abdomen—an experiment that suggests some secreted humoral signal from the brain as a final output from the clock, which has not yet been identified (Handler and Konopka 1979). In Drosophila, the core molecular clock components are coexpressed only in a restricted set of ∼150 neurons, which serve a function similar to the mammalian superchiasmatic nucleus (SCN) in regulating circadian rhythms in behavioral activity. These neurons are clustered into discrete groups: specifically three groups of dorsal neurons (DN1, 2, and 3) and four groups of lateral neurons (LNs) (Figure 3). The lateral neurons consist of two ventral clusters—large and small ventrolateral neurons (lLNvs and sLNvs)—one group of dorsolateral neurons (LNds), and the lateral posterior neurons (LPNs). There is heterogeneity both between and within clusters; like cells in the mammalian SCN (reviewed in Welsh et al. 2010; Bedont and Blackshaw 2015), these cells express different neurotransmitters, neuropeptides, and receptors, and serve different functional roles, as discussed below. Clock cells also exhibit cycling neuronal activity (Cao and Nitabach 2008; Sheeba et al. 2008b; Flourakis et al. 2015), and the peak of neuronal activity, as reflected by intracellular calcium levels, occurs at different phases for different groups (Liang et al. 2016). However, in wild-type flies kept in conditions that approximate the natural world (including LD and early DD), the core molecular clocks in nearly all groups of clock neurons cycle approximately in phase with each other (Yoshii et al. 2009a; Roberts et al. 2015). The mechanisms through which the cycling of the molecular clock generates complex and flexible behavioral outputs are thus probably related to these other properties of the clock circuit, rather than to differences in the cycling of the molecular clock itself.
Figure 3.
Clock cells, which express the core components of the molecular clock, are depicted on the right. These cells are interconnected and heterogeneous between and within clusters, allowing cells to serve different functions, and respond to different environmental conditions. On the left, two groups of output neurons that do not express the molecular components of the clock, but have cycling neuronal activity and are important for behavioral activity rhythms, suggesting clock input. DN, dorsal neurons; LN, lateral neurons (lLNv, large ventral lateral neurons; sLNv, small ventral lateral neurons; LNd, dorsal lateral neurons); LPN, lateral posterior neurons; LHLK, lateral horn leucokinin neuron; DH44, Diuretic Hormone 44.
Pigment dispersing factor (PDF) and “morning” cells:
An important signaling molecule necessary for keeping clock cells synchronized with each other and orchestrating behavioral activity is the neuropeptide PDF. This peptide is expressed in all LNvs, with the exception of a single bilaterally represented PDF-negative cell referred to as “the 5th sLNv” (Helfrich-Förster 1995; 5th sLNv first characterized in adults by Rieger et al. 2006). The PDF receptor, PDFR, is expressed in a subset of clock neurons distributed throughout the network (Im and Taghert 2010). Without PDF, the molecular clock in some groups of clock neurons run fast, while others dampen as individual cells fall out of phase with each other, and yet others may slow (Klarsfeld et al. 2004; Lin et al. 2004; Yoshii et al. 2009b; L. Zhang et al. 2010). Unsurprisingly, then, flies with mutations in either PDF or the PDF receptor (PDFR) have pronounced behavioral phenotypes, with no morning peak of activity and an early evening peak of activity in LD, and short-period rhythms that dampen quickly in DD (Renn et al. 1999; Lin et al. 2004; Hyun et al. 2005; Lear et al. 2005; Mertens et al. 2005).
Although both the lLNvs and sLNvs express PDF, the sLNvs are particularly important for behavioral activity rhythms and have robust molecular rhythms that persist many days into constant darkness (Grima et al. 2004; Stoleru et al. 2005; Roberts et al. 2015). The PDF+ LNvs are necessary and rescue of clock functions in PDF+ LNvs cells is sufficient for free-running behavior in DD and morning anticipation of lights-on in LD (Grima et al. 2004; Stoleru et al. 2004). The evening peak of activity in LD, on the other hand, is not affected by PDF+ LNv ablation or clock rescue, and, because of this apparent specificity, PDF+ sLNvs are often referred to as “morning” or “M” cells. Although convenient, this classification is widely recognized as imperfect. While the cells themselves are necessary, a functional molecular clock is not actually required in PDF+ LNvs for morning anticipation, and manipulating the pace of the clock in these cells does not change the phase of morning behavior (Stoleru et al. 2004; Guo et al. 2014). Moreover, there is evidence to suggest that additional clock cells, including the so-called “evening,” or “E,” cells (the PDF-negative 5th sLNv and LNds, discussed below), as well as the DN1s, are also important for morning activity, with roles that can change in different environmental conditions (Stoleru et al. 2007; L. Zhang et al. 2010a, b; Guo et al. 2014). Calcium imaging of these cells, on the other hand, somewhat reinforces the “M” and “E” roles; for both groups of cells, peak neuronal activity as reflected by intracellular calcium precedes the respective peak of behavior associated with each set of neurons by ∼2–4 hr after entrainment to different light cycles (Liang et al. 2016).
In DD, PDF+ sLNvs are notable in their ability to drive both the speed of molecular cycling in other clock neurons, and, in large part, the speed of behavior in a PDF-dependent manner (Stoleru et al. 2005; Guo et al. 2014; Yao and Shafer 2014). If the period of molecular rhythms in PDF+ sLNvs is accelerated or slowed with genetic manipulations, the speed of molecular clocks in a subset of LNds and some or all DN1s and DN3s will follow suit, suggesting that, in the absence of external cues, PDF+ sLNvs provide a signal that sets the pace of the clock throughout this network of cells (Stoleru et al. 2005; Yao and Shafer 2014). If the difference in the period of rhythms between PDF+ sLNvs and other clock cells is sufficiently large, some flies will exhibit two peaks of activity running with different period lengths, one apparently set by the PDF+ sLNvs and the cells that follow the PDF+ sLNv rhythms, the other apparently driven by the cells that are unaffected by the manipulation in PDF+ sLNvs (Yao and Shafer 2014). The ability of the PDF+ sLNvs to drive the pace of behavior is completely dependent on PDF signaling, suggesting that PDF is an essential part of this coupling mechanism (Yao and Shafer 2014). A similar coupling exists where the molecular clock in DN2s sets the pace of the molecular clock in the lLNvs, although the relevant intercellular signals involved have not been identified, and these cells do not seem to have profound effects on rhythmic rest:activity behavior (Stoleru et al. 2005). It is also important to note that these coupling relationships are likely different in LD cycles, and may also change in light cycles that mimic different seasonal conditions (Stoleru et al. 2007; Guo et al. 2014).
Although it is clear that PDF provides an important signal to synchronize clocks, the exact mechanisms through which this signal impinges on the molecular clock are still being explored. Activation of PDFR with bath application of PDF in brain explants results in increases in cAMP throughout the clock cell network, although this receptivity is gated by time of day such that it is greatest in early morning (Shafer et al. 2008; Klose et al. 2016). Biochemical and genetic evidence suggests that increases in cAMP produced by PDF signaling may act through PKA to stabilize PER, and, indeed, dbt, a regulator of PER stability, is necessary for clock neuron synchrony (Li et al. 2014; Zheng et al. 2014). TIM has also been proposed as a target of PDF and/or PDF+ LNv firing, based in one case on non-cell-autonomous degradation of TIM throughout the clock cell network induced by PDF+ LNv firing, and in the other case by restoration of rhythmic TIM expression in DN1s with per expression solely in PDF+ LNvs, although these authors propose that PDF signaling stabilizes TIM (Guo et al. 2014; Seluzicki et al. 2014). Finally, PDF acutely promotes firing of its target neurons, which may feed back onto the molecular clock (Nitabach et al. 2002, 2004; Seluzicki et al. 2014). In addition to its function as a synchronizing signal, PDF signaling is also required for “E” neuron activity at the proper phase in the circadian cycle (Liang et al. 2016). These proposed mechanisms for PDF action are not mutually exclusive, and may work together or in context-dependent ways to mediate the functions of PDF in the circadian clock circuit.
“Evening” cells:
Based on the ability of the PDF+ sLNvs to set the pace of behavioral and molecular rhythms in DD, these cells are often referred to as the “master pacemakers,” but LNds are also crucial for many aspects of rhythmic rest:activity behavior. Like sLNvs, LNds have sustained molecular rhythms days into constant darkness (Grima et al. 2004; Roberts et al. 2015). LNds, together with the single bilateral PDF-negative sLNv often referred to as the “5th sLNv,” are often referred to as “evening,” or “E,” cells because they are necessary for evening anticipation in LD cycles, and rescuing the clock in both “E” and “M” cells produces normal anticipation at both times of day, while rescuing in “M” cells alone only promotes the morning peak (Grima et al. 2004; Stoleru et al. 2004). The pace of the clock in the “E” cells sets the pace of the peak of evening activity in LD cycles, suggesting both that the “E” cells govern evening activity and also that the pace of the molecular clock of “E” cells is not governed by “M” cells in LD as it is in DD (Stoleru et al. 2007; Guo et al. 2014). As with “M” cells, however, this classification is likely an oversimplification; recent work has suggested that disrupting synaptic transmission from “E” cells disrupts both the morning and evening peaks of activity and weakens activity rhythms in constant darkness (Guo et al. 2014). A functional clock in “E” cells is also sufficient for rhythmic behavior under high light conditions, such as experimental conditions where mutations in photoreceptive pathways allow flies exposed to bright light 24 hr a day (LL) to express rhythms (wild-type flies are arrhythmic under these conditions) or where darkness is replaced by low-intensity “moonlight” (Picot et al. 2007; Stoleru et al. 2007; Rieger et al. 2009). The LNds are quite heterogeneous, with only a subset of cells expressing the photoreceptor CRY or PDFR, and with subsets of LNds expressing different neuropeptides (Benito et al. 2008; Yoshii et al. 2008; Johard et al. 2009; Im and Taghert 2010; Im et al. 2011). In some cases, it has been shown that manipulating only a subset of LNds, along with the 5th sLNv, is sufficient for the phenotypes described (Picot et al. 2007; Rieger et al. 2009; Guo et al. 2014; Schlichting et al. 2016).
DN1s and nonclock output cells:
DN1s are downstream synaptic partners of sLNvs that are also important for morning anticipation under LD conditions (L. Zhang et al. 2010; Y. Zhang et al. 2010; Cavanaugh et al. 2014; Seluzicki et al. 2014). A molecular clock in a subset of DN1 is sufficient to drive morning anticipatory activity in LD cycles, and, in certain temperature conditions, can drive evening anticipation as well (Y. Zhang et al. 2010). Unlike sLNv and LNd clocks, DN1 clocks dampen quickly in DD (Yoshii et al. 2009b; Roberts et al. 2015). However, DN1s make synaptic contact with some “output” (i.e., nonclock) cells in the pars intercerebralis (PI) that are necessary for strong activity rhythms in constant darkness, suggesting that they may act as a relay point between the sLNvs and clock output (Cavanaugh et al. 2014). Thus, it is possible that these cells maintain rhythms in neuronal activity despite a dampened molecular clock in DD conditions. Recent evidence also suggests that DN1s provide inhibitory feedback to both “M” and “E” cells (Guo et al. 2016), a finding similar to previous observations made in the somewhat simplified larval clock neuron network (Collins et al. 2014). A role for DN1s in sleep regulation is discussed below (Kunst et al. 2014; Guo et al. 2016).
The cells of the PI, thought to be the Drosophila equivalent of the hypothalamus (de Velasco et al. 2007), do not express components of the molecular clock, but rhythmic neuronal activity in at least some of these cells suggests that these cells receive time of day signals from the clock circuitry (Cavey et al. 2016). The PI secretes several neuropeptides, of which Diuretic Hormone 44 (DH44) is a critical regulator of robust activity rhythms in DD (Cavanaugh et al. 2014). In addition to the PI cells, two cells in the lateral horn expressing the neuropeptide leucokinin (LHLK cells) also show rhythms of neuronal activity and receive input from clock cells, most likely sLNvs (Cavey et al. 2016). Like DH44, leucokinin is necessary for robust activity rhythms in DD. However, it remains unclear if either of these molecules, or PDF, represents the secreted humoral factor(s) proposed by Konopka capable of regulating rest:activity.
Although not all clock cells have strong functional roles as mediators of rest:activity rhythms under the conditions typically used in the laboratory, these other clock neurons may be important for entrainment to different environmental cues, as discussed below, or in mediating other behavioral or nonbehavioral outputs from the clock. Rhythms in temperature preference in adult flies, for example, are not dependent on PDF or PDFR, but are driven by the DN2s (Kaneko et al. 2012). The timing of sleep after lights-off is regulated by rhythms in GABA receptivity in lLNvs, and DN1s promote both sleep during the day and wakefulness just prior to lights-on (Kunst et al. 2014; Liu et al. 2014; Guo et al. 2016). Secreted signals from different groups of clock neurons also influence clocks and cycling transcripts in other tissues, discussed more below.
Entrainment
To act as reliable time-keepers, clocks must not only maintain a 24 hr rhythm, but also entrain to environmental cues. Light is the dominant entraining stimulus (zeitgeber) for circadian rhythms, although temperature can also entrain clocks in both cold-blooded animals and in mammals. Clock adaptation to zeitgebers can be studied by measuring behavioral and molecular adaptation to a shift in the timing of a zeitgeber, or by studying the effect of a brief pulse of a zeitgeber at an unexpected time of day. The latter approach, used with light or heat, will either accelerate or delay the phase of clock cycling and behavior, depending on when the pulse is delivered. These responses can be summarized by a phase-response curve (PRC), which shows the magnitude of phase shifting when the zeitgeber is delivered at different times of day. Clocks can be entrained to environmental zeitgebers either cell-autonomously or through neural circuits. TIM is rapidly degraded in response to light pulses, and is thought to confer environmental cues to the rest of the molecular clock (Hunter-Ensor et al. 1996; Myers et al. 1996; Zeng et al. 1996). Interestingly, there is now evidence that both cell autonomous and non-cell-autonomous mechanisms for entrainment in Drosophila converge on TIM degradation as a mechanism of resetting the clock.
Light:
Entrainment to pulses of light requires a dedicated circadian photoreceptor, crytochrome (CRY) (Emery et al. 1998; Stanewsky et al. 1998). CRY is a flavin-based light sensor similar to photolyase molecules originally found in bacteria; in flies, light that penetrates the cuticle is sensed directly by CRY. CRY binds directly to TIM upon light treatment, and promotes its turnover through the F-box SCF ubiquitin ligase component JETLAG (JET) (Ceriani et al. 1999; Koh et al. 2006b). This entire light response can be reconstituted in cultured cells by transfecting cry, tim, and jet (Koh et al. 2006b). CRY and JET are required for proper phase delays and advances in response to light pulses and also prevent flies from expressing behavioral rhythms in constant light, presumably because TIM is continuously degraded in these conditions (Stanewsky et al. 1998; Emery et al. 2000; Koh et al. 2006b; Lamba et al. 2014). CRY, however, is expressed in only a subset of clock cells (Benito et al. 2008; Yoshii et al. 2008), and is not required for synchronization to normal light:dark cycles because the visual system provides an additional mechanism of entrainment (Stanewsky et al. 1998; Helfrich-Förster et al. 2001; Rieger et al. 2003; Klarsfeld et al. 2004; Veleri et al. 2007). Although the mechanisms of visual-mediated entrainment are less clear, LNvs have processes in the optic lobe, and are well-positioned to mediate transmitted visual signals to other cells of the clock network (Schlichting et al. 2016; Yoshii et al. 2016). Indeed, genetic manipulations to drive neuronal activity in PDF+ LNvs for 2- or 3-hr bursts in the early night (ZT15) produce a behavioral phase delay, while driving activity late at night (ZT21) produces a phase advance, mimicking the effects of light pulses during these times (Guo et al. 2014; Eck et al. 2016). The firing-mediated phase shifts at both time points are dependent on Pdfr and coincide with TIM degradation across the clock cell network, suggesting that non-cell-autonomous responses may also work through TIM degradation (Guo et al. 2014). The firing-mediated early night phase delays and degradation of TIM are also dependent on the E3 ubiquitin ligase component Cullin 3, potentially representing an alternative mechanism for TIM degradation that does not rely on jet (Grima et al. 2012; Guo et al. 2014). CRY is also capable of directly modulating neuronal activity in response to light, although this has not yet been linked to circadian entrainment (Fogle et al. 2011, 2015).
There is some evidence to support a model where LNds integrate cell autonomous and non-cell-autonomous light signals, and then provide a re-entrainment signal for other clock neurons. First, while both LNvs and LNds are required for phase shifts in response to light pulses, TIM is not actually degraded in PDF+ LNvs during early night light pulses that induce phase delays, suggesting that these cells are not the integrators, at least at this time of day (Tang et al. 2010). Likewise, SGG, which promotes TIM stability, disrupts phase-response curves when overexpressed in PDF-negative clock cells but not PDF-positive clock cells (Stoleru et al. 2007). After a phase-advancing light pulse in late night, ex vivo brain imaging of PER transcription shows that LNds are the first to shift and resynchronize, consistent with a role in re-entraining other clock neurons (Roberts et al. 2015). Finally, CRY is required in LNds but not in other clock cells for quick behavioral re-entrainment to a shifted light cycle, and, even in the absence of CRY, the molecular clock in these cells will partially re-entrain within a day, whereas other clock cells do not show clear re-entrainment on the same time scale (Yoshii et al. 2015). This suggests that both CRY-dependent and vision-dependent signals have rapid effects on the clock in these cells, and that LNds are functionally important for re-entrainment. However, this remains an area of active research, and a conclusive integrator role for LNds has not yet been shown.
Temperature:
Like light, temperature can be sensed both cell autonomously and non-cell-autonomously. Non-cell-autonomous mechanisms are mediated by heat-sensitive ionotropic receptors on the chordotonal organ (ChO), a peripheral sensory structure, and mediate behavioral and, in some clock neurons, molecular entrainment to temperature cycles with differences in temperature as low as 2° (Wheeler et al. 1993; Sehadova et al. 2009; Wolfgang and Simoni 2013; Chen et al. 2015). Cell- or tissue-autonomous mechanisms allow Drosophila peripheral, and possibly brain, molecular clocks to entrain to temperature cycles independently from non-cell-autonomous temperature inputs, although these experiments use higher amplitude temperature cycles (Glaser and Stanewsky 2005; Sehadova et al. 2009; Tataroglu et al. 2015). LNvs and LNds are not necessary for rest:activity adaptations to temperature cycles, although PDF+ LNvs are necessary for free-running activity in constant conditions after temperature entrainment, and multiple groups of clock cells appear to be capable of mediating temperature entrainment (Yoshii et al. 2005; Busza et al. 2007). While most cells, including clock neurons, are capable of entraining to temperature cycles in a wild-type fly, it has been suggested that DNs and LPNs are particularly sensitive to temperature cycles based on molecular cycling when conflicting light and temperature cues are given, and because the DN3s and LPNs display robust molecular cycling in LL temperature cycles even in the absence of LNvs and LNds (Yoshii et al. 2005, 2009a; Miyasako et al. 2007). In the absence of ChO input, molecular clock entrainment to low-amplitude temperature cycles in a subset of clock neurons is disrupted, although other clock neurons are still able to cycle, and the particular neurons disrupted are different for DD and LL conditions (Chen et al. 2015). Blocking output from DN1s or DN2s has some effects on entrainment to low-amplitude temperature cycles.
Recent work in S2 cells and live flies suggests that sustained high intracellular calcium induced by heat promotes degradation of TIM through a calmodulin/calpain (sol)-dependent mechanism (Tataroglu et al. 2015). SOL was found to mediate both molecular and behavioral phase-shifting and adaptation to temperature, and its mammalian homolog, SOLH, likewise promotes degradation of Per2. This work not only provides a mechanism for cell-autonomous entrainment of clocks to temperature cycles, but also might provide a mechanism for resetting in response to other stimuli that would produce sustained increases in intracellular calcium, such as non-cell-autonomous entrainment signals transmitted by neuronal circuits.
Peripheral clocks
Rhythms are observed not just in locomotor behavior, but in many physiological processes throughout the body, and these rhythms rely on the brain clock to various extents. Rhythms of eclosion are mediated by a clock in the prothoracic gland (PG), located in the thoracic region of the developing pupa (Myers et al. 2003). This gland produces the hormone ecdysone, which declines in levels, presumably in response to circadian signals, just prior to eclosion. Although weak autonomous oscillations of clock genes in PG cells can be seen in the absence of brain input, input from the central nervous system (dependent on PDF and LNs) dramatically amplifies and synchronizes oscillations in this tissue (Myers et al. 2003; Morioka et al. 2012). On the other hand, a clock in the Malphigian Tubules, equivalent to the kidney, appears to be entirely autonomous and directly entrained by light, presumably by CRY expression in this tissue (Giebultowicz et al. 2000). The Drosophila fat body, which serves functions of the liver and adipose tissue, does not require the brain clock for molecular clock cycling in LD cycles, but dampens rapidly in constant darkness in the absence of PDF (Erion et al. 2016). Providing an additional layer of complexity, only some cycling transcripts in the fat body rely on the fat body clock, while others are driven by secreted signals from neuronal clocks. Finally, PDF also influences the phase and period of the clock in oenocytes, cells responsible for sex pheromone production in insects, although, curiously, PDF and PDFR have opposite effects on the speed of the clock in these cells (Krupp et al. 2013).
Conclusion
Drosophila has been essential for identifying the key clock molecules and the negative feedback loop mechanism that produces 24 hr cycles of gene expression and overt rhythms. This negative feedback loop is conserved in mammals, and drives rhythms in behavior, metabolism, and other physiological processes, making it a powerful regulator of virtually any process where time-of-day regulation is advantageous with profound implications for human health. Although we know the identities of the molecules involved in driving intracellular rhythms, it is still unclear exactly how the pace of this negative feedback loop is set. Several points of regulation involving phosphorylation of PER have been identified, but further work is needed to fully understand how these phosphorylation events, as well as perhaps other critical delays, set the 24 hr period.
Although the core negative feedback loop provides an elegant mechanism for timekeeping in both clock neurons and peripheral tissues, studying outputs of the circadian clock has revealed that the regulation of physiology by the intracellular clock is not straightforward. In the case of rest:activity rhythms, a heterogeneous network of clock neurons interact with each other to ensure that activity occurs at the appropriate time of day in a variety of different environmental contexts. Studying how signaling within this network of clock cells allows clocks to entrain to cues in the environment, maintain synchronized molecular rhythms, and adapt to changes such as seasons should provide important insights into how the analogous group of neurons in the SCN accomplish these tasks. Work in Drosophila has also provided several different models for how circadian regulation in nonbrain peripheral tissues is produced, with various degrees of dependence on the “master pacemakers” in the brain. Importantly, even at the level of output from the circadian clock, there is evidence to suggest direct homology between mechanisms in flies and mammals (Liu et al. 2014; Flourakis et al. 2015; Erion et al. 2016). There is also evidence for functional homology even when the exact molecules are not conserved, as both the Drosophila and mammalian clocks are made up of a heterogeneous network of neurons where peptidergic signaling plays a fundamental role in synchronizing clocks and producing behavioral output (Vosko et al. 2007; Welsh et al. 2010). The relative simplicity and ease of manipulation of Drosophila position it to continue to be a valuable tool to the circadian community in the discovery of novel mechanisms that control circadian processes.
Part 2: Sleep
Introduction
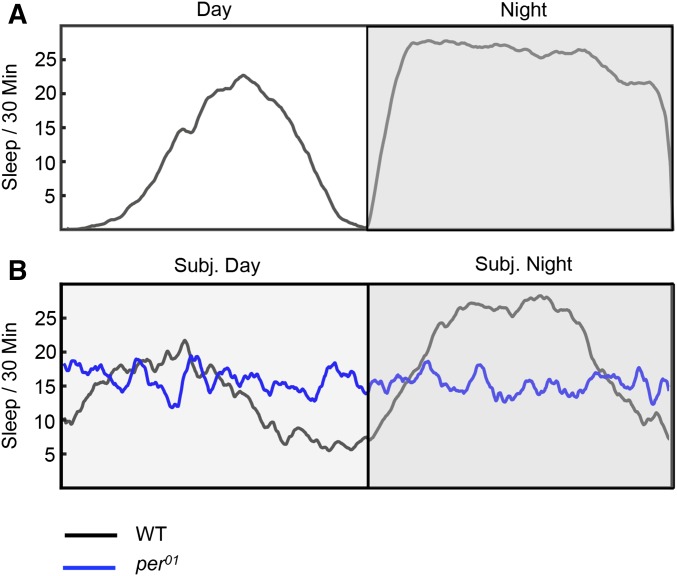
Compared to the long history of circadian rhythm research, sleep research in D. melanogaster has been relatively recent, originating with two studies published in 2000 (Hendricks et al. 2000; Shaw et al. 2000). In these studies, it was found that Drosophila periodically enter a state of quiescence that meets a series of criteria for sleep: (1) this quiescent state is characterized by an increased arousal threshold (decreased responsiveness to sensory stimuli), but (2) it can be distinguished from coma or anesthesia by its rapid reversibility with a stimulus that is sufficiently strong. (3) The timing of sleep is regulated by the circadian clock, although these two processes can also be separated; flies with mutations in the core clock genes have fragmented sleep across the day, but can have normal overall sleep amounts (Hendricks et al. 2003) (Figure 4), and flies with mutations that result in very low total sleep amounts can still show robust circadian activity rhythms. (4) Sleep is also homeostatically regulated, such that when flies are deprived of sleep using a mechanical stimulus, they compensate with longer and deeper sleep the following day. This suggests that sleep serves an important restorative function rather than simply reflecting ecologically advantageous periods of inactivity. (5) Flies experience broad changes in neuronal activity during sleep. Although rhythmic neuronal activity, like that observed with the EEG in mammals, has not been observed in flies, local field potential recordings of the protocerebral area and imaging with the optical calcium indicator GCaMP in the mushroom body show that sleep is a state of reduced neuronal activity and blunted neuronal responses to sensory stimuli (Nitz et al. 2002; Bushey et al. 2015). Importantly, many genetic and molecular regulators of sleep are conserved across species (reviewed in Crocker and Sehgal 2010). Thus, sleep in flies closely resembles sleep in other organisms, and researchers can take advantage of the benefits of this small, genetically tractable model organism to advance our understanding of the molecular neuroscience of sleep.
Figure 4.
(A) Sleep behavior for a group of wild-type (WT) female flies in a 12:12 hr light:dark cycle. Flies have short bouts of siesta sleep in the middle of the day (more pronounced in males) and a relatively consolidated period of sleep at night. (B) Sleep behavior for WT and per01 male flies in constant darkness (DD). per01 flies, which do not display circadian rhythms of activity, spend approximately the same amount of time in sleep, but have sleep that is fragmented across the day. Data appear slightly noisier as fewer flies are represented compared to (A).
At the center of much sleep research is the enigmatic question: what is the function of sleep? We know that in flies, as well as in mammals, important brain processes like learning and memory suffer when animals are sleep deprived and can be recovered by allowing sleep to occur. However, we do not yet know what, at a molecular level, is being depleted and restored. A related line of thought presumes that if we can understand the regulatory factors that underlie the sleep homeostasis, this will lead to a better understanding of sleep function. Gene expression studies have revealed interesting molecular signatures of sleep across the animal kingdom (Mackiewicz et al. 2009), and this has led to a number of interesting hypotheses about sleep function: that it is a time for particular synaptic plasticity processes (Tononi and Cirelli 2006), or specific metabolic activities (Mackiewicz et al. 2007), but evidence supporting these hypotheses is mixed (Scharf et al. 2008; Frank and Cantera 2014; Tononi and Cirelli 2014). An additional physiological correlate of sleep in mammals is greater influx of cerebral spinal fluid into the brain, which may also have a functional role, but has not been directly connected to behavior (Xie et al. 2013). Likewise, research in mammalian systems has uncovered at least some of the relevant neural circuitry for sleep regulation, and a flip-flop switch model for how sleep and wake states are stabilized (Saper et al. 2010; Weber and Dan 2016), but has not revealed satisfying mechanisms to explain what forces cause this switch to flip.
Small model organisms have great potential to reveal single genes and molecules with large impacts on sleep regulation or function, potentially providing answers to these big questions. However, work in model organisms over the past 16 years has also revealed the complexity of even evolutionarily early sleep states. Circadian and homeostatic regulation of sleep were important for establishing similarities between Drosophila sleep and sleep in mammals, but, in addition to regulation by the circadian clock and homeostatic system, sleep in Drosophila can be modulated by diverse environmental factors (Zimmerman et al. 2012) such as social experience (Ganguly-Fitzgerald et al. 2006; Bushey et al. 2011; Chi et al. 2014; Liu et al. 2015; Lone et al. 2016), mating (Isaac et al. 2009), light (Shang et al. 2008), temperature (Parisky et al. 2016), feeding (Keene et al. 2010; Thimgan et al. 2010), age (Koh et al. 2006a; Seugnet et al. 2011a; Kayser et al. 2014; Metaxakis et al. 2014), infection (Kuo et al. 2010; Kuo and Williams 2014), and stress (Lenz et al. 2015). Some of these environmental factors act on the circadian and homeostatic circuitry, but many of these environmental modulators also employ independent mechanisms that do not seem to interfere with circadian timekeeping, sleep amount when animals are undisturbed, or the homeostatic response to sleep loss. In C. elegans, two different sleep-like states have been described that meet nearly all the criteria above, but instead of regulation by the circadian clock, these sleep states are induced by either the molting phase of worm development or by stress (Raizen et al. 2008; Hill et al. 2014). Thus, complex regulation of sleep by diverse environmental factors is likely a general principal of sleep that can be extracted from evolutionarily primitive organisms like insects and nematodes. The picture that emerges from this work, then, is not of a uniform state with simple regulatory mechanisms, but rather of a state that is subject to regulation by a variety of external and internal forces, which may serve different molecular functions in different neuronal or environmental contexts.
Measuring sleep
Based on initial studies of arousal threshold, sleep in Drosophila is commonly defined as a period of inactivity lasting 5 min or longer (Shaw et al. 2000; Huber et al. 2004; Andretic and Shaw 2005). Sleep is typically monitored through the same Drosophila Activity Monitoring System (DAMS) used to analyze circadian behavior. This system relies on an active fly crossing the center of the locomotor tube to break the infrared beam passed across the middle, but this system is generally sufficient to differentiate sleep from activity in young, healthy flies, where activity levels are high enough that it is unlikely that a 5-min or greater period of inactivity would be recorded by chance.
In old or sick flies with reduced overall activity, it may be useful to use a more sensitive method of evaluating sleep behavior. There is also the possibility that extended feeding behavior, in which a fly would dwell at the end of the tube with food and fail to cross the center beam, could be misconstrued as sleep (discussed in Cavanaugh et al. 2016). There are two alternatives to traditional single-beam DAMS monitors that can be used to address these concerns. Multi-beam DAMS monitors, where 17 infrared beams along the length of a locomotor tube are used to monitor activity, provide a similar environment to the traditional locomotor tube set up but offer increased sensitivity (Garbe et al. 2015). Video monitoring systems have also been set up to monitor the activity of individual flies (Zimmerman et al. 2008; Donelson et al. 2012; Gilestro 2012; Faville et al. 2015; Garbe et al. 2015). Video monitoring systems, while potentially offering increased sensitivity, also present a difficulty in that no standard for the sensitivity to motion for these systems has been agreed upon. A very sensitive system may detect leg twitches or imaging artifacts during sleep and inappropriately read these as activity. Video monitoring could also introduce another potential confounding factor if it uses small arenas instead of the typical locomotor tube, as this can result in differences in behavior (Garbe et al. 2015). Thus, while different results can sometimes be observed between video systems and traditional DAMS monitors, these results should be interpreted with caution.
When observing a fly with reduced or elevated overall levels of sleep, it can be conceptually useful to determine how sleep bout architecture is changed (Andretic and Shaw 2005). For example, short-sleeping mutants may initiate fewer bouts of sleep, or may be unable to maintain sleep over long bouts, which implies different mechanisms of action for these genes. Most software used for automated analysis of sleep behavior allows for study of sleep bout architecture in addition to total sleep time.
Sleep depth is an additional dimension of sleep that DAMS monitoring alone does not detect, although automated systems to probe sleep depth have been developed (Faville et al. 2015). While initial characterizations of sleep depth suggested that sensory unresponsiveness plateaus after 5 min of inactivity, subsequent studies have demonstrated that sleep depth varies predictably over longer bouts of sleep as well. Troughs in arousability have been observed after 15 and 30 min of sleep, and protocerebral local field potential recordings show variation in neuronal activity based on length of sleep bouts, in some ways resembling the changes in sleep depth (“sleep stages”) that occur during bouts of sleep in mammals (van Alphen et al. 2013). Depth of sleep also differs between day and night, such that daytime “siesta” sleep in flies is generally a lighter sleep state. Increased sleep depth is also a component of the homeostatic response to sleep deprivation (Huber et al. 2004; van Alphen et al. 2013; Dubowy et al. 2016), and mutations can affect sleep depth in ways that would not be predicted by changes in sleep amount (Faville et al. 2015).
Genetic tools for sleep research
Sleep research in Drosophila, like a lot of molecular neuroscience in this model organism, has drawn heavily on both the study of mutations that lead to aberrant sleep behavior, as well as the use of a genetic toolkit for manipulating neuronal activity. One successful strategy for identifying novel regulators of sleep is to conduct forward genetic screens for mutants with very extreme phenotypes. Another strategy is manipulation of different neuroanatomic loci labeled by Gal4 drivers by activating or suppressing neuronal firing. Researchers can use a variety genetic tools to manipulate neuronal activity. The bacterial sodium channel NaChBac (Luan et al. 2006; Nitabach et al. 2006) and the potassium channel Kir2.1 (Baines et al. 2001) can be driven either throughout fly development or in a time-restricted manner using inducible binary expression systems to activate or silence cells, respectively. Thermogenetic tools, such as the heat-activated depolarizing channel TrpA1 (Hamada et al. 2008; Parisky et al. 2008), or the temperature-sensitive dominant negative allele of shibire used to block synaptic transmission (Kitamoto 2001), as well as optogenetic tools, such as the light-activated depolarizing CsChrimson channel (Klapoetke et al. 2014) are also frequently used for conditional manipulation of neurons.
In many cases, gene- and circuit-based approaches intersect. Many, though not all, genes that regulate sleep have been shown to function in specific neuroanatomic loci, in some cases identifying novel sleep regulating areas of the fly brain. Important advances have also come from studying interactions between genes that regulate sleep, as well as studying genes that produce sleep phenotypes and have unknown or unappreciated roles in controlling neuronal activity. Studying sleep in Drosophila then not only leads to insight into sleep-regulatory mechanisms that may extend to mammals, but also identifies novel regulators of neuronal function, and provides new insight into brain signaling and metabolism. In this review, we present a thorough discussion of the genetics and neuroanatomy of sleep, with an emphasis on how sleep-regulating genes act in the context of sleep-regulating brain regions, and how different sleep regulating genes and brain areas interact with each other.
Sleep regulation through global modulation of neuronal activity
The Shaker potassium channel (Cirelli et al. 2005; Bushey et al. 2007) and its modulator sleepless (Koh et al. 2008) were two early hits with extreme short-sleeping phenotypes from large-scale genetic screens. Both genes are expressed throughout the fly brain (Wu et al. 2009), and neither of these phenotypes has been fully mapped to specific neuroanatomic loci, suggesting that they exert widespread effects on brain activity or metabolism that feed back onto sleep regulation. Shaker is a voltage-gated potassium channel involved in membrane repolarization. sleepless is a Ly6 neurotoxin-like molecule that, in the years since its discovery as a sleep regulator, has been found to promote Shaker expression and activity and inhibit nicotinic acetylcholine (nAChR) function, such that loss of sleepless might lead to increased neuronal activity through multiple mechanisms (Wu et al. 2009; Shi et al. 2014; Wu et al. 2014). The molecular functions of these genes therefore suggest a mechanism of sleep regulation where wakefulness is produced by broadly increasing neuronal excitability. Indeed, broadly inhibiting cholinergic transmission partially suppresses both the Shaker and sleepless phenotypes, and RNAi knockdown of the nAChRα3 subunit suppresses the sleepless phenotype (Wu et al. 2014). However, recent work has revealed a more complicated role for these genes. Although it has typically been assumed that the Shaker phenotype results from increased neuronal activity of wake-promoting cells, a recent study found that knocking down Shaker in sleep-promoting cells actually lengthens the inter-spike interval and reduces neuronal activity in these populations to favor wake (Pimentel et al. 2016). Another study found that, in contrast with the generally wake-promoting effects of cholinergic neurotransmission in the fly brain (Wu et al. 2014; Seidner et al. 2015), a specific nAChR subunit, redeye, is strongly sleep-promoting (Shi et al. 2014). Genetic evidence suggests that sleepless also interacts with the redeye subunit, in this case acting as a wake-promoting rather than sleep-promoting molecule, consistent with sleepless inhibiting nAChRs regardless of subunit composition. Recent studies of sleepless have also suggested that it in part regulates sleep by non-cell-autonomously promoting metabolism of GABA in glia, perhaps also through its effect on neural activity (Chen et al. 2014; Maguire et al. 2015). Shaker and sleepless thus both seem to interact in a nonstraightforward way with sleep-regulatory genes and cells in the nervous system, and work with sleepless suggests a potential connection between neuronal activity and metabolism of neurotransmitters, although the details of this connection remain unclear.
The mushroom body
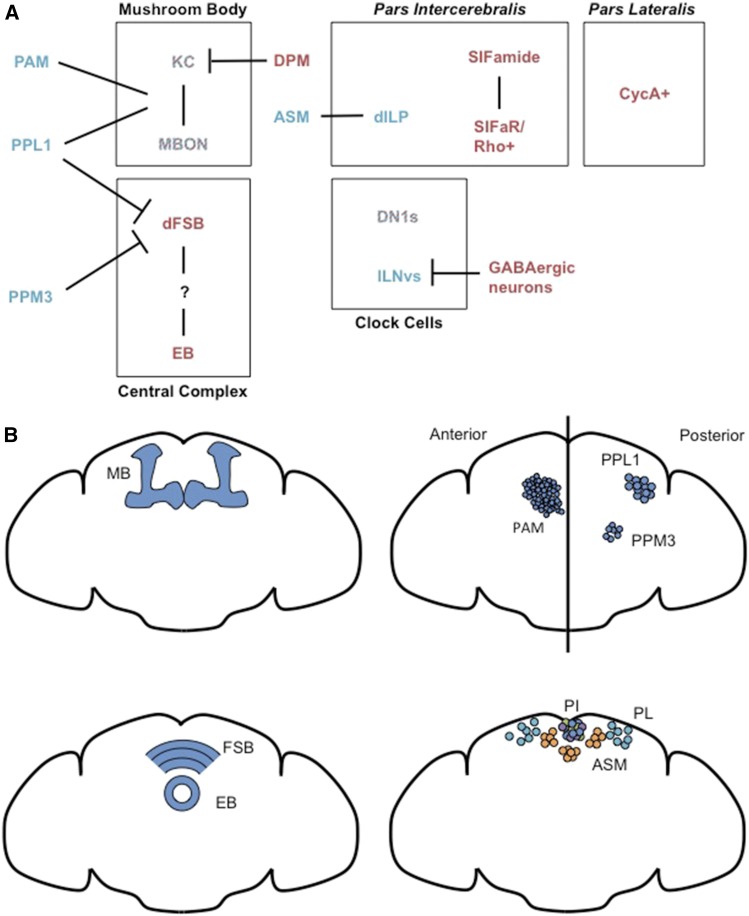
The mushroom body is the center of olfactory memory in the fly brain and as a result of years of intense research, there is detailed anatomic and functional data available for mushroom body circuits (reviewed in Guven-Ozkan and Davis 2014; Owald and Waddell 2015). The mushroom body consists primarily of ∼2000 Kenyon cells, most of which receive input from an average of six stochastically connected projection neurons, with each projection neuron encoding input from a single type of odorant receptor neuron. Each Kenyon cell projects axons to a subset of lobes of the mushroom body, forming three classes: those that project to the α and β lobes, the α’ and β’ lobes, or the γ lobe. Within each lobe there exist several compartments, defined by the dendrites of different mushroom body output neurons (MBONs) and axonal projections of different dopaminergic neurons, which respond to aversive or appetitive unconditioned stimuli like electric shock or sugar. At least some MBONs have an inherent valence, which is correlated with neurotransmitter expression; flies will act to avoid optogenetic activation of aversive glutamtergic MBONs, and act to prolong activation of attractive MBONs, which can be cholinergic or GABAergic (Aso et al. 2014b). A simple model of learning and memory in the mushroom body posits that within a mushroom body compartment, the strength of the synapses between Kenyon cells, which encode odor, and MBONs, which encode valence, is modulated by dopaminergic neurons in response to pairing of an odor with an aversive or appetitive stimulus (Owald and Waddell 2015). MBONs project to largely uncharacterized protocerebral areas of the fly brain (Aso et al. 2014a). In addition to the neurons described above, mushroom bodies also receive octopaminergic input, and are innervated by the dorsal paired medial (DPM), anterior paired lateral (APL), and dorsal anterior lateral (DAL) pairs of neurons (Guven-Ozkan and Davis 2014). These neurons may serve functions in memory consolidation or in fine-tuning olfactory coding.
The mushroom body was also the first neuroanatomic structure identified as a regulator of sleep in Drosophila (Joiner et al. 2006; Pitman et al. 2006). Conditional approaches were used to block synaptic transmission, perturb PKA signaling, or manipulate excitability of mushroom body neurons, primarily using the relatively broad Gal4 drivers that were available at the time, but also taking advantage of methods that could target the mushroom body specifically. These approaches suggested that the mushroom body contains both sleep-promoting and wake-promoting cells: for example, flies slept less when hydroxyurea was used to ablate α/β and α’/β’ mushroom body lobes, suggesting a sleep-promoting role for these cells, but slept more when a relatively restricted mushroom body GeneSwitch line was used to silence specific cells in adulthood.
Later work using more restricted split-Gal4 lines identified specific mushroom body circuits that underlie both wake- and sleep-promoting effects (Aso et al. 2014b; Sitaraman et al. 2015a). Several MBONs are capable of regulating sleep behavior, and interestingly these same MBONs are also necessary for certain types of learning and memory (Aso et al. 2014b). The sleep- and wake-promoting characteristics of MBONs seem to correlate with their aversive or appetitive nature, such that the two identified wake-promoting groups of MBONs are aversive and glutamatergic, whereas two identified sleep-promoting MBONs are appetitive and cholinergic or GABAergic; an unusual MBON with dendritic projections in the calyx and no identified neurotransmitter or valence is also sleep-promoting (Aso et al. 2014b). Wake- and sleep-promoting characteristics of different Kenyon cell populations then seem to reflect which of the sleep-controlling MBONs the Kenyon cells in question target most prominently (Sitaraman et al. 2015a). For example, neural epistasis experiments suggest that the wake-promoting glutamatergic MBONs (γ5β’2a/β’2mp’/β’2mp_bilateral) are downstream of a wake-promoting α’β’ KC driver and a broad wake-promoting KC driver that encompasses γ-dorsal, γ-main, and α/β KCs (γ-dorsal KCs are sleep-promoting on their own, but γ-main KCs are wake-promoting). On the other hand, blocking the sleep-promoting cholinergic γ2α’1 MBONs makes the wake-promoting effects of the broad KC driver that encompasses γ-dorsal, γ-main, and α/β KCs even stronger, suggesting that γ2α’1 MBONs receive sleep-promoting input from these cells, even if the net effect of the KC driver conferred by other downstream MBONs is wake-promoting. Likewise, the DPM neurons, which are proposed to provide inhibitory input to α’β’ KCs via GABA and/or serotonin, are strongly sleep-promoting when activated, consistent with an overall wake-promoting effect of α’β’ KCs (Haynes et al. 2015). Loss of the d5-HT1 serotonin receptor in mushroom bodies also produces a weak short-sleep phenotype, which can be rescued with expression driven by the MB-GeneSwitch driver (Yuan et al. 2006). This finding is consistent with serotonin released from DPMs acting to inhibit wake-promoting α’β’ KCs.
Dopamine and the dorsal fan-shaped body
Perhaps the strongest parallel between mammalian and Drosophila sleep regulation identified so far is the strong wake-promoting effects of the monoamine neurotransmitters dopamine and octopamine (the insect homolog of norepinephrine, discussed in the next section). fumin, one of the first short-sleeping mutants identified, is a mutation in a dopamine transporter that presumably results in elevated dopamine levels throughout the fly brain (Kume et al. 2005), and dopaminergic neurons are strongly wake-promoting when activated (Shang et al. 2011; Liu et al. 2012). Conversely, mutants deficient for the CNS-specific isoform of tyrosine hydroxylase (TH), the rate-limiting enzyme for dopamine synthesis, have increased sleep throughout the day (Riemensperger et al. 2011).
One site of dopaminergic action is the central complex, an area of the brain that has been hypothesized to serve a basal ganglia-like function in action selection based in part on the input it receives from protocerebral areas and its functional role in motor output (Strausfeld and Hirth 2013). Thermogenetic activation of the ExF/2 neurons in the dorsal fan-shaped body, a region of the central complex, is strongly sleep-promoting (Donlea et al. 2011). Sleep deprivation changes the electrophysiologic properties of these neurons to favor activity, suggesting they may play a role in output of homeostatic sleep signals (Donlea et al. 2014). Dopamine provides a wake-promoting stimulus by silencing these neurons, although there is some disagreement regarding the relevant cluster of dopaminergic neurons as well as the relevant D1-like dopamine receptor. A MARCM approach to target single dopaminergic neurons indicated that individual PPM3s with projections to the fan-shaped body exert small but significant effects on sleep behavior, while a separate study comparing expression of wake-promoting and non-wake-promoting Gal4 drivers suggested that PPL1s with projections to the fan-shaped body provide wake-promoting input (Liu et al. 2012; Ueno et al. 2012). It is possible that both groups of cells provide wake-promoting dopaminergic input to this brain area. Likewise, it was initially thought that Dop1R1 was the relevant receptor for wake-promoting dopaminergic signaling in the brain. Dop1R1 mutations suppress the fumin phenotype, and, unlike wild-type flies, Dop1R1 mutants do not experience severe sleep reduction when fed L-DOPA (Liu et al. 2012; Ueno et al. 2012). These effects can be rescued with Dop1R1 expression driven by the relatively specific fan-shaped body driver 104y-Gal4. However, more recent work shows that RNAi knockdown of the related receptor Dop1R2 in the dorsal fan-shaped body ExF/2 neurons is sufficient to prevent both short-term hyperpolarization and longer-term silencing of these cells by dopamine, and this manipulation also produces a long-sleep phenotype (Pimentel et al. 2016).
Dopaminergic neurons with projections to the mushroom body also have wake-promoting effects (Sitaraman et al. 2015b; Nall et al. 2016). Neurons of the PAM cluster, as well as a subset of neurons of the PPL1 cluster distinct from those that project to the fan-shaped body, project to specific compartments of the mushroom body (MB). Recent work has suggested that MB-PAM neurons and MB-PPL1 neurons can be wake-promoting when thermogenetically activated. The wake-promoting effects of caffeine are also mediated by the PAM cluster of neurons (Nall et al. 2016). However, although Split-Gal4 drivers have been used to elegantly identify specific mushroom body circuits that control sleep, the PAM and PPL1 neurons that promote wake do not seem to neatly correspond to these circuits (Sitaraman et al. 2015b). It is possible that diffusion of dopamine or functional interconnectivity between dopaminergic neurons (Cohn et al. 2015) contributes to these results.
Genetic knock-outs and experiments silencing dopaminergic neurons show that endogenous dopamine plays an important role in daily sleep regulation; however, it is interesting that increases in global dopamine levels can be compensated with loss of the Dop1R1 receptor to achieve approximately normal amounts of daily sleep (Ueno et al. 2012). Thermogenetic activation of dopaminergic neurons produces a sleep rebound once activation is stopped, suggesting that these wake-promoting neurons are upstream of neuronal machinery capable of producing homeostatic responses to extended wakefulness (Seidner et al. 2015; Dubowy et al. 2016). Alterations in dopamine signaling are also implicated in sleep regulation by developmental or environmental cues; the increased sleep amounts that young flies experience have been attributed to decreased dopaminergic input to ExF/2 neurons, and dopamine has also been proposed to play a role in the adaptation of sleep amount to changing social environments (Ganguly-Fitzgerald et al. 2006; Kayser et al. 2014). In addition to inhibitory, wake-promoting input from dopamine, ExF/2 neurons may also receive input from unidentified sleep-promoting neurons labeled by the 201y-Gal4 driver (Cavanaugh et al. 2016). Thus, the dorsal fan-shaped body is well-positioned to act as an integrator and output for many sleep-regulatory signals.
In addition to fumin, other short-sleeping mutants also appear to depend on dopamine or the fan-shaped body for their mechanisms of action. The Rho-GAP crossveinless c is a sleep-promoting molecule that disrupts the physiological membrane properties of the ExF/2 neurons when mutated, resulting in reduced sleep (Donlea et al. 2014). The 2-pore potassium channel Sandman is necessary for dopamine-mediated silencing of these neurons, and knockdown of this channel in these neurons also produces a short sleep phenotype (Pimentel et al. 2016). The spatial requirements for the sleep-promoting ubiquitin ligase component Cullin 3, and its interacting BTB adaptor insomniac have not been established (Stavropoulos and Young 2011), but pharmacologically blocking dopamine synthesis blocks the short-sleeping phenotypes of these mutants, suggesting that Cullin 3-mediated protein turnover and dopamine signaling may interact to regulate sleep (Pfeiffenberger and Allada 2012).
Octopamine, the pars intercerebralis, and the pars lateralis
Octopamine, the insect homolog of norepinephrine, is another wake-promoting monoaminergic neurotransmitter (Crocker and Sehgal 2008). Mutating the enzymes responsible for octopamine synthesis or silencing octopaminergic neurons increases daily sleep amount, while activating octopaminergic neurons or feeding flies octopamine decreases sleep (Crocker and Sehgal 2008; Seidner et al. 2015). Although octopamine provides input to the mushroom body, the wake-promoting effects of octopamine do not appear to be mediated by this structure. Instead, a MARCM approach identified the octopaminergic ASM neurons, which project to the pars intercerebralis (PI), as sufficient to drive increased wake when chronically activated, and the PI insulin-like peptide (ILP)-secreting neurons as downstream mediators of octopamine signaling (Crocker et al. 2010). The effect sizes observed when ASM or ILP-secreting neurons are manipulated are somewhat smaller than those observed with manipulation of all octopaminergic neurons, so it is possible that other neurons important for the wake-promoting effects of octopamine have not yet been found. Unlike dopaminergic neurons, activating octopaminergic neurons produces strong sleep loss without an apparent rebound the next day, suggesting that octopaminergic neurons provide a wake-promoting stimulus that is able to circumvent sleep homeostasis (Seidner et al. 2015). This work suggests that octopaminergic neurons may be a neural substrate for environmental factors that promote wake without any apparent homeostatic compensation.
In addition to the ILP-expressing neurons, distinct sets of neurons in the PI expressing EGFR ligands and SIFamide are also sleep-promoting (Foltenyi et al. 2007; Park et al. 2014). rhomboid (rho), an enzyme necessary for the production of EGFR ligands, is expressed prominently in the PI, and manipulating its activity using Gal4 drivers with expression in the PI produces sleep when rho is overexpressed, and wake when rho is knocked down (Foltenyi et al. 2007). SIFamide is a sleep-promoting insect neuropeptide expressed in four PI neurons; ablation of these neurons or knockdown of the peptide with RNAi decreases sleep (Park et al. 2014). Both rho and the SIFamide receptor (SIFaR) are required in c767-Gal4 labeled neurons for normal sleep amounts, suggesting that SIFamide acts through inter-PI signaling, and implicating EGFR ligands as a possible output from this circuit (Foltenyi et al. 2007; Park et al. 2014). However, c767-Gal4 also drives expression outside the PI, and so a function of these molecules outside the PI cannot be excluded. EGFR signaling in clock neurons may mediate the effects of social enrichment on sleep, although it is not clear if this is functionally related to the release of EGFR ligand from the PI (Donlea et al. 2009).
A separate, but related neuroendocrine structure, the pars lateralis (PL) (de Velasco et al. 2007), was recently identified as a site of action for cell cycle regulators that modulate sleep in adult postmitotic neurons. Two cell cycle regulators, Rca1 (Regulator of Cyclin A) and taranis (tara, a Trip-Br family transcriptional coregulator), were independently identified in genetic screens for short-sleeping mutants, and, following the identification of Rca1, it was found that knocking down Cyclin A (CycA) itself in neurons produces an equally strong short-sleeping phenotype (Rogulja and Young 2012; Afonso et al. 2015). CycA and tara genetically interact, and TARA binds to and post-transcriptionally promotes stable expression of Cyclin A in PL neurons (Afonso et al. 2015). Postmitotic expression of Cyclin A is relatively restricted in the fly brain, but includes ∼14 neuroendocrine cells in the pars lateralis, and knocking down tara in this structure partially recapitulates the short sleeping phenotype of tara mutants. Experimentally activating and silencing these neurons supports a wake-promoting role. No mechanism has yet been identified for the involvement of these cell cycle regulators in neuronal activity or sleep, but this will be an interesting area of future research.
Clock regulation of sleep
The circadian clock is essential for restricting sleep to environmentally advantageous times of day. A role for the circadian clock has been established in flies in both putting flies to sleep at night once the dark period has begun, and waking them up in advance of dawn (Kunst et al. 2014; Liu et al. 2014). Interestingly, these pathways seem to mechanistically diverge, suggesting that circadian regulation of sleep is not driven by continuous oscillation of a single sleep- or wake-promoting factor, but is rather driven by time-of-day specific modulation of distinct sleep- and wake-promoting mechanisms. Clock cells also have broader noncircadian roles in sleep regulation as mediators of the effects of temperature and social enrichment on sleep (Donlea et al. 2009; Parisky et al. 2016).
One particularly well-studied mechanism of circadian sleep regulation regulates sleep around the time of lights-off, and is driven by cyclic expression of a gene that regulates responsiveness to neuronal signals in a specific set of clock neurons. The large ventral lateral neurons (lLNvs) are a wake-promoting population of neurons with neuronal activity that fluctuates over the course of the day, such that firing frequency is reduced around the time of lights-off as well as later in the night in an LD cycle (Cao and Nitabach 2008; Parisky et al. 2008; Shang et al. 2008; Sheeba et al. 2008a,b; Liu et al. 2014). Manipulations of lLNv activity produce broad effects on sleep and wake throughout the day, but effects are particularly pronounced at night, with clear effects on length to sleep onset (sleep latency) after lights-off. Genetic and pharmacological studies suggest that the silencing of these neurons during this time is mediated by GABA-A receptor Rdl, and, indeed, broadly silencing GABAergic neurons in the brain substantially lengthens the sleep latency after lights-off in flies (Agosto et al. 2008; Parisky et al. 2008; Chung et al. 2009). The positive and negative arms of the molecular clock also oppositely regulate sleep latency, such that Clock and cyc mutants have increased sleep latency after lights-off, while per and tim mutants have shortened sleep latency after lights-off (Liu et al. 2014). Thus, changes in activity in this circuit seem to drive sleep in response to time of day around the transition to darkness.
A key molecular mediator of these changes in activity was initially discovered in a genetic screen for short-sleeping mutants. Although wide awake (wake) mutant flies were found to have reduced sleep across the day and night, which may be due to activity of wake in other neuroanatomic loci, the increased latency to sleep in wake mutants could be anatomically mapped to the lLNvs (Liu et al. 2014). Transcription of wake was found to cycle in the lLNvs, with increased transcription and protein levels at dusk, and WAKE physically and genetically interacts with the Rdl GABA-A receptor. Crucially, wake mutants did not display rhythms in lLNv firing frequency, and GABA-induced inhibitory currents in lLNvs were dampened. Thus, clock-driven expression of wake in lLNvs appears to be the key time-of-day driven regulator that induces sleep after lights-off.
A distinct mechanism in a different set of clock neurons is invoked to promote wake at the end of the night, just before lights-on. Diuretic Hormone 31 (DH31) is a wake-promoting neuropeptide expressed in the DN1 clock neurons, and manipulating its expression in these cells produces sleep phenotypes specifically from ZT21-24; knockdown of the peptide in DN1s increases sleep during this time period, while overexpression of the peptide in these cells decreases it (Kunst et al. 2014). Expressing a tethered PDF peptide in the DN1s, which should produce PDFR activation in these cells, also reduces sleep specifically during late night, as does pan-neuronal expression of tethered DH31. The time-specific effects of DH31 might therefore be gated both by time-specific PDF responsiveness in DN1s, and by time-specific DH31 responsiveness in downstream neurons.
However, DN1s are sleep-promoting at other times of day; optogenetically or thermogenetically activating these cells increases daytime sleep, suppressing the normal “evening” peak of activity, while silencing them decreases sleep during early night (Guo et al. 2016). The sleep-promoting effects of DN1s during the day can be blocked with RNAi targeting mGluR in “E” cells, suggesting “E” cells might also have a role in sleep regulation. The role of sleep-promoting signals from DN1s in normal daily sleep regulation remains unclear, although the authors propose that variations in activity of DN1s may explain sexually dimorphic sleep patterns and regulation of sleep by high temperature.
Metabolic regulation of sleep
Food availability is a potent environmental regulator of sleep in fruit flies. Starvation strongly suppresses sleep, perhaps so that flies can devote more time to foraging for food (Keene et al. 2010; Thimgan et al. 2010). Mechanisms that regulate sleep at baseline and in response to food availability have some overlap with pathways that regulate metabolic energy storage, but these pathways are ultimately separable, such that sleep phenotypes do not depend on differences in metabolic stores (Erion et al. 2012; Masek et al. 2014; Murakami et al. 2016). Pharmacological evidence suggests that the suppression of sleep in response to starvation can be mimicked by feeding flies a glycolysis inhibitor, but not an inhibitor of fatty acid β-oxidation, suggesting that the suppression of sleep with starvation is related to reduced metabolic mobilization of glucose, not the taste of sugars or to lipid metabolism (Murakami et al. 2016).
An essential molecular mediator for these effects was recently identified in the nucleotide binding protein translin (Murakami et al. 2016). translin is highly upregulated upon starvation, and translin knockdown completely prevents starvation-induced sleep loss in flies. However, other sleep and starvation-related behaviors, such as sleep at baseline, sleep after sleep deprivation, preference for sucrose or yeast after starvation, and the proboscis extension reflex following starvation, are completely unaffected, and there is no evidence that translin knockdown alters energy stores. The effects of this molecular mediator were mapped to neurons expressing the neuropeptide leucokinin. Like translin knockdown, silencing leucokinin neurons prevented sleep suppression in response to starvation.
Although pharmacology suggests that sleep suppression in response to starvation is related to glucose metabolism, and is mechanistically distinct from the response to mechanical sleep deprivation, which induces a homeostatic response, a separate body of work suggests that genes involved in lipid metabolism can specifically modulate the rebound response to mechanical sleep deprivation (Thimgan et al. 2010, 2015). However, a mechanism through which lipid metabolism modulates sleep following sleep deprivation and neuronal substrates of this process remain unknown, and it is still unclear whether lipid metabolic stores are directly related to these phenotypes, or whether lipid metabolism and sleep homeostasis share common pathways.
Homeostatic response to sleep deprivation
Sleep homeostasis ensures that flies sleep the proper amount by recovering lost sleep after periods of extended wakefulness. Sleep homeostasis is often conceptualized as a continuous build-up of sleep need over periods of wakefulness and dissipation over periods of sleep, such that the same mechanisms should be invoked both when flies are spontaneously waking and during periods of forced wakefulness (sleep deprivation). However, recent work in Drosophila has called this view into question.
A disconnect between regulation of sleep following spontaneous wakefulness and sleep following sleep deprivation is supported by a number of observations. While it is true that many short-sleeping mutants have impaired sleep rebound, these phenotypes may arise from the general inability to initiate or maintain sleep in these flies, such that even high sleep pressure cannot overcome these deficits; it is also difficult to interpret rebound data from short-sleeping flies because their habitual short sleep means they have less sleep to lose. Indeed, the converse relationship does not seem to hold: a number of genetic perturbations have been identified that specifically affect sleep after sleep deprivation with little to no effect on baseline sleep, suggesting that sleep following sleep deprivation is produced by an independent mechanism (Seugnet et al. 2011b; Seidner et al. 2015; Thimgan et al. 2015; Dubowy et al. 2016; Liu et al. 2016).
Likewise, it seems that the nature of sleep deprivation matters for the homeostatic response. Activating different populations of wake-promoting neurons in the fly brain produces varying amounts of rebound the following day, ranging from no rebound response at all, as is seen with activation of octopaminergic neurons, to a rebound that in some cases far exceeds the amount of sleep lost (Seidner et al. 2015; Dubowy et al. 2016). Different environmental stimuli used to keep flies awake can also produce the varying effects. Particularly strikingly, one group has reported that starving flies produces equivalent amounts of sleep loss as mechanical sleep deprivation without producing any observable sleep rebound (Thimgan et al. 2010). It is possible that even different mechanical sleep deprivation approaches invoke different neural pathways to keep flies awake, which may explain why so few mutants with impaired sleep rebound have been validated across laboratories.
Despite these challenges, there is a picture emerging about the relevant circuitry for sleep homeostasis. Groups of wake-promoting cells that do produce sleep rebound after activation include dopaminergic neurons, as well as at least one restricted set of cholinergic cells, which produce a particularly strong rebound with even short periods of activation (Seidner et al. 2015; Dubowy et al. 2016). In addition, electrophysiology suggests that the sleep-promoting dorsal fan-shaped body neurons have reduced input resistance and reduced membrane time constants, suggesting greater activity, following sleep deprivation (Donlea et al. 2014); as discussed previously, this brain area is well-positioned to act as an integrator or output for multiple sleep regulatory signals, including, it seems, the response to sleep deprivation. It has also been suggested that silencing MBON-γ2α’1 neurons can block sleep rebound, although the data do not exclude the possibility that this is due to a general wake-promoting effect of silencing MBON-γ2α’1 neurons during the early day when rebound occurs (Sitaraman et al. 2015a).
A recently identified element of sleep-regulatory circuitry with an apparently specific role in sleep homeostasis is the ellipsoid body R2 neurons (Liu et al. 2016). These neurons were initially of interest because they produce a persistent sleep-promoting signal when thermogenetically activated; while no changes in sleep are reported at the time of activation, which can be as short as 30 min, a dramatic rebound-like increase in sleep is observed for the next 12 hr. Structural plasticity in the R2 neurons seems to underlie the phenotype, as circuit-specific analysis of Bruchpilot expression showed greater synapse number and size for R2 neurons after sleep deprivation, and genetic manipulations that block this plasticity partially block sleep rebound. A neuronal epistasis experiment suggests that these cells are upstream (although not necessarily directly connected to) the dorsal fan-shaped body. The manipulations of R2 neurons that affect sleep rebound have no effect on sleep at baseline, however, again supporting the idea that regulation of the homeostatic response to sleep deprivation is mechanistically different from the regulation of baseline sleep.
Function of sleep
Sleep affects neurobehavioral performance across the animal kingdom, and flies are no exception. Sleep has a bidirectional relationship with learning and memory; sleep deprivation in adult flies has been shown to interfere with both short- and long-term memory, while inducing sleep allows memories to form in contexts where an experience would ordinarily be forgotten (Ganguly-Fitzgerald et al. 2006; Seugnet et al. 2008; Donlea et al. 2011; Berry et al. 2015; Dissel et al. 2015). Sleep loss also has consequences for social behavior in flies; in adult flies, acute sleep loss results in impaired aggressive behavior (Kayser et al. 2015). There also appears to be a critical window during development where sleep loss produces long-lasting deficits in courtship behavior and short-term memory that persist into adulthood (Seugnet et al. 2011a; Kayser et al. 2014). Precisely how these deficits arise, however, remains an outstanding question in the field.
One general line of thought supposes that there are brain-wide molecular pathways that are different during sleep and wake, and perturbed by sleep loss, that underlie these behavioral changes. Indeed, molecular characterization comparing sleeping, spontaneously waking, and sleep-deprived brains has found widespread differences in gene expression between different behavioral states (Cirelli 2006; Zimmerman et al. 2006; Williams et al. 2007). The types of changes observed appear to be conserved across organisms: broadly, genes involved in synaptic plasticity/function, cellular stress, and metabolism are affected by sleep and wake across species studied (Mackiewicz et al. 2009).
One hypothesis based on this data, put forth by Tononi and Cirelli (2006), proposes that global synaptic downscaling occurs during sleep to counteract overpotentiation that might occur during wake. Work from these authors shows evidence that, broadly and within specific circuits of the adult fly brain, synaptic markers increase after wake or sleep deprivation and decrease following sleep, suggesting changes in the number or size of synapses (Gilestro et al. 2009; Bushey et al. 2011). Several shared regulators of learning, synaptic plasticity, and sleep have been identified, but a direct link between synaptic plasticity and either sleep regulation or neurobehavioral performance has been difficult to establish (Bushey et al. 2009; Bai and Sehgal 2015; Robinson et al. 2016). In some cases, it seems that the effects of sleep and synaptic plasticity can in fact be separated; for example, in the learning-impaired mutant dunce, inducing sleep improves learning even though the global changes in synaptic markers typically associated with sleep are not observed (Dissel et al. 2015).
Another hypothesis states that sleep is a time where metabolic functions such as macromolecule biosynthesis can be carried out in the brain in the absence of the more urgent metabolic demands of waking. This may also explain why extended sleep loss results in induction of cellular stress genes. As with learning and synaptic plasticity, shared regulators of metabolic or cellular stress and sleep regulation or function have been identified (Shaw et al. 2002; Naidoo et al. 2007; Thimgan et al. 2010, 2015; Maguire et al. 2015), and flies increase sleep following a heat pulse that induces a cellular stress response (Lenz et al. 2015), but a direct link that would establish cellular metabolism as an essential function of sleep has not yet been shown.
Some progress in understanding neurobehavioral changes with sleep comes from examining specific neurotransmitter systems or circuits that are perturbed by sleep loss. In the case of learning deficits with sleep loss, performance can be restored by overexpressing Dop1R1 or pharmacologically promoting dopamine signaling (Seugnet et al. 2008). In the case of loss of aggression after sleep loss, feeding flies the dopamine precursor L-DOPA does not improve behavior, but instead an octopamine agonist is effective at restoring aggression (Kayser et al. 2015). Studying the mechanisms that allow increased sleep to promote memory have also yielded insights; in particular, recent work suggests that inducing sleep may promote the formation of an aversive olfactory memory by suppressing a dopamine-dependent “active forgetting” process that occurs when flies are awake and moving (Berry et al. 2015). Whether these changes in neurotransmitter pathways are downstream of global metabolic or plasticity pathways that are altered during sleep will be an interesting area of future research.
Conclusions
The study of sleep in Drosophila has allowed us to harness the power of forward genetics to make significant advances in the study of sleep and neuroscience more broadly. The neuroanatomy of sleep in Drosophila, while not comprehensive, has identified a diverse set of neurons in the fly brain that can regulate sleep (Figure 5). We are also moving toward a better understanding of how these circuits interact with each other, which will enable us to build models for how sleep regulation works that can be applied to mammalian brains. The neuroanatomy and neurochemistry of sleep in Drosophila includes many parallels between flies and mammals. Disruptions of potassium channel function have profound effects on sleep in flies and are also linked to human sleep phenotypes (Allebrandt et al. 2011; Cornelius et al. 2011). The wake-promoting effects of catecholamines and the sleep-regulatory roles of hypothalamus-like structures are strong parallels between flies and mammals, and the direction of sleep regulation for most neurotransmitters is preserved across evolution (Crocker and Sehgal 2010). The insect mushroom body and the central complex, on the other hand, have less clear parallels to mammalian sleep-regulatory neuroanatomy, but may still share functional homology to mammalian sleep-regulatory circuits. A better understanding of protocerebral areas of the fly brain, many of which are relatively unexplored but have connections to both the mushroom body and the central complex, may also lend insights into sleep function and regulation.
Figure 5.
(A) Schematic of sleep-promoting (red), and sleep-inhibiting (blue), neurons in the fly brain. Sleep-regulating neurons are identified by neurotransmitter, neuropeptide, or molecular marker expression, and/or neuroanatomic location. Dopaminergic neurons: PAM, protocerebral anterior lateral; PPL1, protocerebral posterior lateral; and PPM3, protocerebral posterior medial. Mushroom body (MB) neurons: KC, Kenyon cells; MBON, mushroom body output neurons. Central complex: dFSB, dorsal fan-shaped body; EB, ellipsoid body. Pars intercerebralis (PI): SIFaR, SIFamide Receptor; Rho, Rhomboid; and dILP, Drosophila insulin-like peptide. Octopaminergic neurons: ASM, anterior superior medial. Pars lateralis (PL): CycA, CyclinA. Clock cells: DN, dorsal neurons; lLNvs, large ventral lateral neurons. (B) Location of sleep-regulating neurons in the fly brain.
An important lesson already apparent from studying Drosophila is that sleep regulation is orchestrated by a complex set of genes, neurons, and environmental conditions. Although there is a tendency in the field to reduce sleep regulation to a homeostatic and a circadian component, this thinking has not been sufficient to understand sleep regulation in flies, and perhaps also in other systems. Instead, there appear to be different sets of genes and cells that regulate basal sleep drive, sleep in response to environmental cues, as well as sleep in response to forced wakefulness. Likewise, the circadian component is comprised of different cell groups and different circadian output molecules regulating sleep and wake at specific times of day, not a single oscillating signal.
Sleep also has profound affects on waking behavior in Drosophila, making flies suitable model organisms to study the function of sleep. Excitingly, we are learning more and more about how complex behaviors are orchestrated in flies, providing more power to examine specifically how sleep and wake impinge on these processes. As we enter an era where identifying more precise mechanisms for the effects of sleep on biological functions is possible, we can begin finding commonalities across different behaviors and processes influenced by sleep, and use these findings to make general statements about what sleep does to make it necessary across the animal kingdom.
Acknowledgments
We thank Sehgal laboratory members for critical reading of the manuscript and helpful comments. The laboratory is supported by National Institutes of Health (NIH) grant R37NS048471. C.D. was supported by T32-MH017168 and T32-GM007229.
Footnotes
Communicating editor: J. Truman
Literature Cited
- Afonso D. J. S., Liu D., Machado D. R., Pan H., Jepson J. E. C., et al. , 2015. TARANIS functions with cyclin A and Cdk1 in a novel arousal center to control sleep in Drosophila. Curr. Biol. 25: 1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosto J., Choi J. C., Parisky K. M., Stilwell G., Rosbash M., et al. , 2008. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat. Neurosci. 11: 354–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akten B., Jauch E., Genova G. K., Kim E. Y., Edery I., et al. , 2003. A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosci. 6: 251–257. [DOI] [PubMed] [Google Scholar]
- Allada R., White N. E., So W. V., Hall J. C., Rosbash M., 1998. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 93: 791–804. [DOI] [PubMed] [Google Scholar]
- Allebrandt K. V., Amin N., Müller-Myhsok B., Esko T., Teder-Laving M., et al. , 2011. A KATP channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Mol. Psychiatry 18: 122–132. [DOI] [PubMed] [Google Scholar]
- Andretic R., Shaw P. J., 2005. Essentials of sleep recordings in Drosophila: moving beyond sleep time. Methods Enzymol. 393: 759–772. [DOI] [PubMed] [Google Scholar]
- Aronson B., Johnson K., Loros J., Dunlap J., 1994. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science 263: 1578–1584. [DOI] [PubMed] [Google Scholar]
- Aso Y., Hattori D., Yu Y., Johnston R. M., Iyer N. A., et al. , 2014a The neuronal architecture of the mushroom body provides a logic for associative learning. eLife 3: e04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y., Sitaraman D., Ichinose T., Kaun K. R., Vogt K., et al. , 2014b Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife 3: e04580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L., Sehgal A., 2015. Anaplastic lymphoma kinase acts in the Drosophila mushroom body to negatively regulate sleep. PLoS Genet. 11: e1005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines R. A., Uhler J. P., Thompson A., Sweeney S. T., Bate M., 2001. Altered electrical properties in Drosophila neurons developing without synaptic transmission. J. Neurosci. 21: 1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S., Rihel J., Bjes E., Fan J. Y., Price J. L., 2001. The Drosophila double-timeS mutation delays the nuclear accumulation of period protein and affects the feedback regulation of period mRNA. J. Neurosci. 21: 7117–7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargiello T. A., Jackson F. R., Young M. W., 1984. Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature 312: 752–754. [DOI] [PubMed] [Google Scholar]
- Baylies M. K., Bargiello T. A., Jackson F. R., Young M. W., 1987. Changes in abundance or structure of the per gene product can alter periodicity of the Drosophila clock. Nature 326: 390–392. [DOI] [PubMed] [Google Scholar]
- Bedont J. L., Blackshaw S., 2015. Constructing the suprachiasmatic nucleus: a watchmaker’s perspective on the central clockworks. Front. Syst. Neurosci. 9: 350–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito J., Houl J. H., Roman G. W., Hardin P. E., 2008. The blue-light photoreceptor CRYPTOCHROME is expressed in a subset of circadian oscillator neurons in the Drosophila CNS. J. Biol. Rhythms 23: 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J. A., Cervantes-Sandoval I., Chakraborty M., Davis R. L., 2015. Sleep facilitates memory by blocking dopamine neuron-mediated forgetting. Cell 161: 1656–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau J., Young M. W., 1999. Cycling vrille expression is required for a functional Drosophila clock. Cell 99: 661–671. [DOI] [PubMed] [Google Scholar]
- Bushey D., Huber R., Tononi G., Cirelli C., 2007. Drosophila Hyperkinetic mutants have reduced sleep and impaired memory. J. Neurosci. 27: 5384–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D., Tononi G., Cirelli C., 2009. The Drosophila fragile X mental retardation gene regulates sleep need. J. Neurosci. 29: 1948–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D., Tononi G., Cirelli C., 2011. Sleep and synaptic homeostasis: structural evidence in Drosophila. Science 332: 1576–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D., Tononi G., Cirelli C., 2015. Sleep- and wake-dependent changes in neuronal activity and reactivity demonstrated in fly neurons using in vivo calcium imaging. Proc. Natl. Acad. Sci. USA 112: 4785–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busza A., Murad A., Emery P., 2007. Interactions between circadian neurons control temperature synchronization of Drosophila behavior. J. Neurosci. 27: 10722–10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G., Nitabach M. N., 2008. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J. Neurosci. 28: 6493–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh D. J., Geratowski J. D., Wooltorton J. R. A., Spaethling J. M., Hector C. E., et al. , 2014. Identification of a circadian output circuit for rest: activity rhythms in Drosophila. Cell 157: 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh D. J., Vigderman A. S., Dean T., Garbe D. S., Sehgal A., 2016. The Drosophila circadian clock gates sleep through time-of-day dependent modulation of sleep-promoting neurons. Sleep 39: 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavey M., Collins B., Bertet C., Blau J., 2016. Circadian rhythms in neuronal activity propagate through output circuits. Nat. Neurosci. 19: 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani M. F., Darlington T. K., Staknis D., Mas P., Petti A. A., et al. , 1999. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science 285: 553–556. [DOI] [PubMed] [Google Scholar]
- Ceriani M. F., Hogenesch J. B., Yanovsky M., Panda S., Straume M., et al. , 2002. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J. Neurosci. 22: 9305–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Buhl E., Xu M., Croset V., Rees J. S., et al. , 2015. Drosophila Ionotropic Receptor 25a mediates circadian clock resetting by temperature. Nature 527: 516–520. [DOI] [PubMed] [Google Scholar]
- Chen W.-F., S. Maguire, M. Sowcik, W. Luo, K. Koh et al., 2014 A neuron-glia interaction involving GABA transaminase contributes to sleep loss in sleepless mutants. Mol. Psychiatry 20: 240–251. [DOI] [PMC free article] [PubMed]
- Chi, M., L. Griffith, and C. Vecsey, 2014 Larval population density alters adult sleep in wild-type Drosophila melanogaster but not in Amnesiac mutant flies. Brain Sci. 4: 453–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu J. C., Vanselow J. T., Kramer A., Edery I., 2008. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 22: 1758–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu J. C., Ko H. W., Edery I., 2011. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell 145: 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B. Y., Kilman V. L., Keath J. R., Pitman J. L., Allada R., 2009. The GABAA receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr. Biol. 19: 386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C., 2006. Cellular consequences of sleep deprivation in the brain. Sleep Med. Rev. 10: 307–321. [DOI] [PubMed] [Google Scholar]
- Cirelli C., Bushey D., Hill S., Huber R., Kreber R., et al. , 2005. Reduced sleep in Drosophila Shaker mutants. Nature 434: 1087–1092. [DOI] [PubMed] [Google Scholar]
- Claridge-Chang A., Wijnen H., Naef F., Boothroyd C., Rajewsky N., et al. , 2001. Circadian regulation of gene expression systems in the Drosophila head. Neuron 32: 657–671. [DOI] [PubMed] [Google Scholar]
- Cohn R., Morantte I., Ruta V., 2015. Coordinated and compartmentalized neuromodulation shapes sensory processing in Drosophila. Cell 163: 1742–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B., Kaplan H. S., Cavey M., Lelito K. R., Bahle A. H., et al. , 2014. Differentially timed extracellular signals synchronize pacemaker neuron clocks. PLoS Biol. 12: e1001959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelius J. R., Pittock S. J., McKeon A., Lennon V. A., Aston P. A., et al. , 2011. Sleep manifestations of voltage-gated potassium channel complex autoimmunity. Arch. Neurol. 68: 733–738. [DOI] [PubMed] [Google Scholar]
- Crocker A., Sehgal A., 2008. Octopamine regulates sleep in Drosophila through protein kinase A-dependent mechanisms. J. Neurosci. 28: 9377–9385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker A., Sehgal A., 2010. Genetic analysis of sleep. Genes Dev. 24: 1220–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker A., Shahidullah M., Levitan I. B., Sehgal A., 2010. Identification of a neural circuit that underlies the effects of octopamine on sleep: wake behavior. Neuron 65: 670–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin K. D., Huang Z. J., Rosbash M., 1995. Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron 14: 365–372. [DOI] [PubMed] [Google Scholar]
- Cyran S. A., Buchsbaum A. M., Reddy K. L., Lin M.-C., Glossop N. R. J., et al. , 2003. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112: 329–341. [DOI] [PubMed] [Google Scholar]
- Cyran S. A., Yiannoulos G., Buchsbaum A. M., Saez L., Young M. W., et al. , 2005. The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J. Neurosci. 25: 5430–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mairan J., 1729. Observation botanique. Hist Acad Roy Sci 1729: 35.
- de Velasco B., Erclik T., Shy D., Sclafani J., Lipshitz H., et al. , 2007. Specification and development of the pars intercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev. Biol. 302: 309–323. [DOI] [PubMed] [Google Scholar]
- Dissel S., Angadi V., Kirszenblat L., Suzuki Y., Donlea J., et al. , 2015. Sleep restores behavioral plasticity to Drosophila mutants. Curr. Biol. 25: 1270–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelson N., Kim E. Z., Slawson J. B., Vecsey C. G., Huber R., et al. , 2012. High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the “Tracker” program. PLoS One 7: e37250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea J. M., Ramanan N., Shaw P. J., 2009. Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 324: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea J. M., Thimgan M. S., Suzuki Y., Gottschalk L., Shaw P. J., 2011. Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332: 1571–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea J. M., Pimentel D., Miesenböck G., 2014. Neuronal machinery of sleep homeostasis in Drosophila. Neuron 81: 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubowy C., Moravcevic K., Yue Z., Wan J. Y., Van Dongen H. P. A., et al. , 2016. Genetic dissociation of daily sleep and sleep following thermogenetic sleep deprivation in Drosophila. Sleep 39: 1083–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eck S., Helfrich-Förster C., Rieger D., 2016. The timed depolarization of morning and evening oscillators phase shifts the circadian clock of Drosophila. J. Biol. Rhythms 31: 428–442. [DOI] [PubMed] [Google Scholar]
- Emery P., So W. V., Kaneko M., Hall J. C., Rosbash M., 1998. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95: 669–679. [DOI] [PubMed] [Google Scholar]
- Emery P., Stanewsky R., Hall J. C., Rosbash M., 2000. A unique circadian-rhythm photoreceptor. Nature 404: 456–457. [DOI] [PubMed] [Google Scholar]
- Erion R., DiAngelo J. R., Crocker A., Sehgal A., 2012. Interaction between sleep and metabolism in Drosophila with altered octopamine signaling. J. Biol. Chem. 287: 32406–32414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erion R., King A. N., Wu G., Hogenesch J. B., Sehgal A., 2016. Neural clocks and Neuropeptide F/Y regulate circadian gene expression in a peripheral metabolic tissue. eLife 5: pii: e13552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Sathyanarayanan S., Sehgal A., 2007. Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1). Genes Dev. 21: 1506–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faville R., Kottler B., Goodhill G. J., Shaw P. J., van Swinderen B., 2015. How deeply does your mutant sleep? probing arousal to better understand sleep defects in Drosophila. Sci. Rep. 5: 8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flourakis M., Kula-Eversole E., Hutchison A. L., Han T. H., Aranda K., et al. , 2015. A conserved bicycle model for circadian clock control of membrane excitability. Cell 162: 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogle K. J., Parson K. G., Dahm N. A., Holmes T. C., 2011. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science 331: 1409–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogle K. J., Baik L. S., Houl J. H., Tran T. T., Roberts L., et al. , 2015. CRYPTOCHROME-mediated phototransduction by modulation of the potassium ion channel β-subunit redox sensor. Proc. Natl. Acad. Sci. USA 112: 2245–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltenyi K., Greenspan R. J., Newport J. W., 2007. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat. Neurosci. 10: 1160–1167. [DOI] [PubMed] [Google Scholar]
- Frank M. G., Cantera R., 2014. Sleep, clocks, and synaptic plasticity. Trends Neurosci. 37: 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly-Fitzgerald I., Donlea J., Shaw P. J., 2006. Waking experience affects sleep need in Drosophila. Science 313: 1775–1781. [DOI] [PubMed] [Google Scholar]
- Garbe D. S., Fang Y., Zheng X., Sowcik M., Anjum R., et al. , 2013. Cooperative interaction between phosphorylation sites on PERIOD maintains circadian period in Drosophila. PLoS Genet. 9: e1003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe D. S., Bollinger W. L., Vigderman A., Masek P., Gertowski J., et al. , 2015. Context-specific comparison of sleep acquisition systems in Drosophila. Biol. Open 4: 1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekakis N., Saez L., Delahaya-Brown A.-M., Myers M. P., Sehgal A., et al. , 1995. Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science 370: 811–815. [DOI] [PubMed] [Google Scholar]
- Giebultowicz J. M., Stanewsky R., Hall J. C., Hege D. M., 2000. Transplanted Drosophila excretory tubules maintain circadian clock cycling out of phase with the host. Curr. Biol. 10: 107–110. [DOI] [PubMed] [Google Scholar]
- Gilestro G. F., 2012. Video tracking and analysis of sleep in Drosophila melanogaster. Nat. Protoc. 7: 995–1007. [DOI] [PubMed] [Google Scholar]
- Gilestro G. F., Tononi G., Cirelli C., 2009. Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science 324: 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser F. T., Stanewsky R., 2005. Temperature synchronization of the Drosophila circadian clock. Curr. Biol. 15: 1352–1363. [DOI] [PubMed] [Google Scholar]
- Glossop N. R. J., Houl J. H., Zheng H., Ng F. S., Dudek S. M., et al. , 2003. VRILLE feeds back to control circadian transcription of clock in the Drosophila circadian oscillator. Neuron 37: 249–261. [DOI] [PubMed] [Google Scholar]
- Green E. W., O’Callaghan E. K., Hansen C. N., Bastianello S., Bhutani S., et al. , 2015. Drosophila circadian rhythms in seminatural environments: summer afternoon component is not an artifact and requires TrpA1 channels. Proc. Natl. Acad. Sci. USA 112: 8702–8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B., Lamouroux A., Chélot E., Papin C., Limbourg-Bouchon B., et al. , 2002. The F-box protein Slimb controls the levels of clock proteins period and timeless. Nature 420: 178–182. [DOI] [PubMed] [Google Scholar]
- Grima B., Chélot E., Xia R., Rouyer F., 2004. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431: 869–873. [DOI] [PubMed] [Google Scholar]
- Grima B., Dognon A., Lamouroux A., Chélot E., Rouyer F., 2012. CULLIN-3 controls TIMELESS oscillations in the Drosophila circadian clock. PLoS Biol. 10: e1001367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güler A. D., Ecker J. L., Lall G. S., Haq S., Altimus C. M., et al. , 2008. Melanopsin cells are the principal conduits for rod–cone input to non-image-forming vision. Nature 453: 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Cerullo I., Chen X., Rosbash M., 2014. PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. eLife 3: e02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Yu J., Jung H. J., Abruzzi K. C., Luo W., et al. , 2016. Circadian neuron feedback controls the Drosophila sleep–activity profile. Nature 536: 292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T., Davis R. L., 2014. Functional neuroanatomy of Drosophila olfactory memory formation. Learn. Mem. 21: 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada F. N., Rosenzweig M., Kang K., Pulver S. R., Ghezzi A., et al. , 2008. An internal thermal sensor controlling temperature preference in Drosophila. Nature 454: 217–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler A. M., Konopka R. J., 1979. Transplantation of a circadian pacemaker in Drosophila. Nature 279: 236–238. [DOI] [PubMed] [Google Scholar]
- Hardin P. E., Hall J. C., Rosbash M., 1990. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343: 536–540. [DOI] [PubMed] [Google Scholar]
- Haynes P. R., Christmann B. L., Griffith L. C., 2015. A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. eLife 4: e03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C., 1995. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 92: 612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C., Winter C., Hofbauer A., Hall J. C., Stanewsky R., 2001. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron 30: 249–261. [DOI] [PubMed] [Google Scholar]
- Hendricks J. C., Finn S. M., Panckeri K. A., Chavkin J., Williams J. A., et al. , 2000. Rest in Drosophila is a sleep-like state. Neuron 25: 129–138. [DOI] [PubMed] [Google Scholar]
- Hendricks J. C., Lu S., Kume K., Yin J. C. P., Yang Z., et al. , 2003. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J. Biol. Rhythms 18: 12–25. [DOI] [PubMed] [Google Scholar]
- Hill A. J., Mansfield R., Lopez J. M. N. G., Raizen D. M., Van Buskirk C., 2014. Cellular stress induces a protective sleep-like state in C. elegans. Curr. Biol. 24: 2399–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houl J. H., Yu W., Dudek S. M., Hardin P. E., 2006. Drosophila CLOCK is constitutively expressed in circadian oscillator and non-oscillator cells. J. Biol. Rhythms 21: 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R., Hill S. L., Holladay C., Biesiadecki M., Tononi G., et al. , 2004. Sleep homeostasis in Drosophila melanogaster. Sleep 27: 628–639. [DOI] [PubMed] [Google Scholar]
- Hughes M. E., Grant G. R., Paquin C., Qian J., Nitabach M. N., 2012. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res. 22: 1266–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter-Ensor M., Ousley A., Sehgal A., 1996. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell 84: 677–685. [DOI] [PubMed] [Google Scholar]
- Hyun S., Lee Y., Hong S.-T., Bang S., Paik D., et al. , 2005. Drosophila GPCR han is a receptor for the circadian clock neuropeptide PDF. Neuron 48: 267–278. [DOI] [PubMed] [Google Scholar]
- Im S. H., Taghert P. H., 2010. PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J. Comp. Neurol. 518: 1925–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im S. H., Li W., Taghert P. H., 2011. PDFR and CRY signaling converge in a subset of clock neurons to modulate the amplitude and phase of circadian behavior in Drosophila. PLoS One 6: e18974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac R. E., Li C., Leedale A. E., Shirras A. D., 2009. Drosophila male sex peptide inhibits siesta sleep and promotes locomotor activity in the post-mated female. Proc. Biol. Sci. 277: 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiura M., Kutsuna S., Aoki S., Iwasaki H., Andersson C. R., et al. , 1998. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281: 1519–1523. [DOI] [PubMed] [Google Scholar]
- Jang A. R., Moravcevic K., Saez L., Young M. W., Sehgal A., 2015. Drosophila TIM binds importin α1, and acts as an adapter to transport PER to the nucleus. PLoS Genet. 11: e1004974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo A. M., Zheng X., Zhou Y., Amado D. A., Sheldon A., et al. , 2004. Pattern of distribution and cycling of SLOB, Slowpoke channel binding protein, in Drosophila. BMC Neurosci. 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johard H. A. D., Yoishii T., Dircksen H., Cusumano P., Rouyer F., et al. , 2009. Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J. Comp. Neurol. 516: 59–73. [DOI] [PubMed] [Google Scholar]
- Joiner W. J., Crocker A., White B. H., Sehgal A., 2006. Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441: 757–760. [DOI] [PubMed] [Google Scholar]
- Kaasik K., Kivimäe S., Allen J. J., Chalkley R. J., Huang Y., et al. , 2013. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 17: 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S., Stoleru D., McDonald M., Nawathean P., Rosbash M., 2007. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 21: 1675–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H., Head L. M., Ling J., Tang X., Liu Y., et al. , 2012. Circadian rhythm of temperature preference and its neural control in Drosophila. Curr. Biol. 22: 1851–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M. S., Yue Z., Sehgal A., 2014. A critical period of sleep for development of courtship circuitry and behavior in Drosophila. Science 344: 269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M. S., Mainwaring B., Yue Z., Sehgal A., 2015. Sleep deprivation suppresses aggression in Drosophila. eLife 4: e07643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan K. P., Pradhan S., Wang J.-P., Allada R., 2007. Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Comput. Biol. 3: e208–e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene A. C., Duboué E. R., McDonald D. M., Dus M., Suh G. S. B., et al. , 2010. Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr. Biol. 20: 1209–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. Y., Edery I., 2006. Balance between DBT/CKIε kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc. Natl. Acad. Sci. USA 103: 6178–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. Y., Ko H. W., Yu W., Hardin P. E., Edery I., 2007. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol. Cell. Biol. 27: 5014–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. Y., Jeong E. H., Park S., Jeong H. J., Edery I., et al. , 2012. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 26: 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto T., 2001. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J. Neurobiol. 47: 81–92. [DOI] [PubMed] [Google Scholar]
- Klapoetke N. C., Murata Y., Kim S. S., Pulver S. R., Birdsey-Benson A., et al. , 2014. Independent optical excitation of distinct neural populations. Nat. Methods 11: 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarsfeld A., Malpel S., Michard-Vanhée C., Picot M., Chélot E., et al. , 2004. Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila. J. Neurosci. 24: 1468–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose M., Duvall L. B., Li W., Liang X., Ren C., et al. , 2016. Functional PDF signaling in the Drosophila circadian neural circuit is gated by Ral A-Dependent modulation. Neuron 90: 781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B., Price J. L., Saez L., Blau J., Rothenfluh A., et al. , 1998. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell 94: 97–107. [DOI] [PubMed] [Google Scholar]
- Ko H. W., Jiang J., Edery I., 2002. Role for Slimb in the degradation of Drosophila period protein phosphorylated by doubletime. Nature 420: 673–678. [DOI] [PubMed] [Google Scholar]
- Ko H. W., Kim E. Y., Chiu J., Vanselow J. T., Kramer A., et al. , 2010. A hierarchical phosphorylation cascade that regulates the timing of PERIOD nuclear entry reveals novel roles for proline-directed kinases and GSK-3 /SGG in circadian clocks. J. Neurosci. 30: 12664–12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K., Evans J. M., Hendricks J. C., Sehgal A., 2006a A Drosophila model for age-associated changes in sleep: wake cycles. Proc. Natl. Acad. Sci. USA 103: 13843–13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K., Zheng X., Sehgal A., 2006b JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science 312: 1809–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K., Joiner W. J., Wu M. N., Yue Z., Smith C. J., et al. , 2008. Identification of SLEEPLESS, a sleep-promoting factor. Science 321: 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka R. J., Benzer S., 1971. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 68: 2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp J. J., Billeter J.-C., Wong A., Choi C., Nitabach M. N., et al. , 2013. Pigment-dispersing factor modulates pheromone production in clock cells that influence mating in Drosophila. Neuron 79: 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kula-Eversole E., Nagoshi E., Shang Y., Rodriguez J., Allada R., 2010. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc. Natl. Acad. Sci. USA 107: 13497–13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Chen D., Jang C., Nall A., Zheng X., et al. , 2014. An ecdysone-responsive nuclear receptor regulates circadian rhythms in Drosophila. Nat. Commun. 5: 5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K., Kume S., Park S. K., Hirsh J., Jackson F. R., 2005. Dopamine is a regulator of arousal in the fruit fly. J. Neurosci. 25: 7377–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst M., Hughes M. E., Raccuglia D., Felix M., Li M., et al. , 2014. Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Curr. Biol. 24: 2652–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T.-H., Williams J. A., 2014. Acute sleep deprivation enhances post-infection sleep and promotes survival during bacterial infection in Drosophila. Sleep 37: 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T.-H., Pike D. H., Beizaeipour Z., Williams J. A., 2010. Sleep triggered by an immune response in Drosophila is regulated by the circadian clock and requires the NFκB Relish. BMC Neurosci. 11: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba P., Bilodeau-Wentworth D., Emery P., Zhang Y., 2014. Morning and evening oscillators cooperate to reset circadian behavior in response to light input. Cell Rep. 7: 601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear B. C., Merrill C. E., Lin J.-M., Schroeder A., Zhang L., et al. , 2005. A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron 48: 221–227. [DOI] [PubMed] [Google Scholar]
- Lee C., Bae K., Edery I., 1999. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol. Cell. Biol. 19: 5316–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz O., Xiong J., Nelson M. D., Raizen D. M., Williams J. A., 2015. FMRFamide signaling promotes stress-induced sleep in Drosophila. Brain Behav. Immun. 47: 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Guo F., Shen J., Rosbash M., 2014. PDF and cAMP enhance PER stability in Drosophila clock neurons. Proc. Natl. Acad. Sci. USA 111: E1284–E1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Holy T. E., Taghert P. H., 2016. Synchronous Drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science 351: 976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C., Allada R., 2013. ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science 340: 875–879. [DOI] [PubMed] [Google Scholar]
- Lim C., Chung B. Y., Pitman J. L., McGill J. J., Pradhan S., et al. , 2007. clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Curr. Biol. 17: 1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C., Lee J., Choi C., Kilman V. L., Kim J., et al. , 2011. The novel gene twenty-four defines a critical translational step in the Drosophila clock. Nature 470: 399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Stormo G. D., Taghert P. H., 2004. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J. Neurosci. 24: 7951–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.-M., Schroeder A., Allada R., 2005. In vivo circadian function of casein kinase 2 phosphorylation sites in Drosophila PERIOD. J. Neurosci. 25: 11175–11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. M., Kilman V. L., Keegan K., Paddock B., Emery-Le M., et al. , 2002. A role for casein kinase 2α in the Drosophila circadian clock. Nature 420: 816–820. [DOI] [PubMed] [Google Scholar]
- Lin Y., Han M., Shimada B., Wang L., Gibler T. M., et al. , 2002. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 99: 9562–9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J., Dubruille R., Emery P., 2012. KAYAK-alpha modulates circadian transcriptional feedback loops in Drosophila pacemaker neurons. J. Neurosci. 32: 16959–16970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., P. R. Haynes, N. C. Donelson, S. Aharon, and L. C. Griffith, 2015 Sleep in populations of Drosophila melanogaster. eNeuro 2: pii: ENEURO.0071-15.2015. [DOI] [PMC free article] [PubMed]
- Liu Q., Liu S., Kodama L., Driscoll M. R., Wu M. N., 2012. Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr. Biol. 22: 2114–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Lamaze A., Liu Q., Tabuchi M., Yang Y., et al. , 2014. WIDE AWAKE mediates the circadian timing of sleep onset. Neuron 82: 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Liu Q., Tabuchi M., Wu M. N., 2016. Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell 165: 1347–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone S. R., Potdar S., Srivastava M., Sharma V. K., 2016. Social experience is sufficient to modulate sleep need of Drosophila without increasing wakefulness. PLoS One 11: e0150596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H., Lemon W. C., Peabody N. C., Pohl J. B., Zelensky P. K., et al. , 2006. Functional dissection of a neuronal network required for cuticle tanning and wing expansion in Drosophila. J. Neurosci. 26: 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz M., Shockley K. R., Romer M. A., Galante R. J., Zimmerman J. E., et al. , 2007. Macromolecule biosynthesis: a key function of sleep. Physiol. Genomics 31: 441–457. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M., Zimmerman J. E., Shockley K. R., Churchill G. A., Pack A. I., 2009. What are microarrays teaching us about sleep? Trends Mol. Med. 15: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire S. E., Rhoades S., Chen W.-F., Sengupta A., Yue Z., et al. , 2015. Independent effects of GABA transaminase (GABAT) on metabolic and sleep homeostasis. J. Biol. Chem. 290: 20407–20416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S., Inonog S., Manoukian A. S., Young M. W., 2001. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105: 769–779. [DOI] [PubMed] [Google Scholar]
- Masek P., Reynolds L. A., Bollinger W. L., Moody C., Mehta A., et al. , 2014. Altered regulation of sleep and feeding contributes to starvation resistance in Drosophila melanogaster. J. Exp. Biol. 217: 3122–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A., Ukai-Tadenuma M., Yamada R. G., Houl J., Uno K. D., et al. , 2007. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 21: 1687–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald M. J., Rosbash M., 2001. Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107: 567–578. [DOI] [PubMed] [Google Scholar]
- Mertens I., Vandingenen A., Johnson E. C., Shafer O. T., Li W., et al. , 2005. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron 48: 213–219. [DOI] [PubMed] [Google Scholar]
- Metaxakis A., Tain L. S., Grönke S., Hendrich O., Hinze Y., et al. , 2014. Lowered insulin signalling ameliorates age-related sleep fragmentation in Drosophila. PLoS Biol. 12: e1001824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P., Saez L., Young M. W., 2006. PER-TIM interactions in living Drosophila cells: an interval timer for the circadian clock. Science 311: 226–229. [DOI] [PubMed] [Google Scholar]
- Miyasako Y., Umezaki Y., Tomioka K., 2007. Separate sets of cerebral clock neurons are responsible for light and temperature entrainment of Drosophila circadian locomotor rhythms. J. Biol. Rhythms 22: 115–126. [DOI] [PubMed] [Google Scholar]
- Morioka E., Matsumoto A., Ikeda M., 2012. Neuronal influence on peripheral circadian oscillators in pupal Drosophila prothoracic glands. Nat. Commun. 3: 909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Yurgel M. E., Stahl B. A., Masek P., Mehta A., et al. , 2016. Translin is required for metabolic regulation of sleep. Curr. Biol. 26: 972–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers E. M., Yu J., Sehgal A., 2003. Circadian control of eclosion: interaction between a central and peripheral clock in Drosophila melanogaster. Curr. Biol. 13: 526–533. [DOI] [PubMed] [Google Scholar]
- Myers M. P., Wager-Smith K., Rothenfluh-Hilfiker A., Young M. W., 1996. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science 271: 1736–1740. [DOI] [PubMed] [Google Scholar]
- Naidoo N., Casiano V., Carter J., Zimmerman J., Pack A. I., 2007. A role for the molecular chaperone protein BiP/GRP78 in Drosophila sleep homeostasis. Sleep 30: 557–565. [DOI] [PubMed] [Google Scholar]
- Nakajima M., Imai K., Ito H., Nishiwaki T., Murayama Y., et al. , 2005. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308: 414–415. [DOI] [PubMed] [Google Scholar]
- Nall A. H., Shakhmantsir I., Cichewicz K., Birman S., Hirsh J., et al. , 2016. Caffeine promotes wakefulness via dopamine signaling in Drosophila. Sci. Rep. 6: 20938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawathean P., Stoleru D., Rosbash M., 2007. A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization, and transcriptional repressor activity. Mol. Cell. Biol. 27: 5002–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach M. N., Blau J., Holmes T. C., 2002. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell 109: 485–495. [DOI] [PubMed] [Google Scholar]
- Nitabach M. N., Sheeba V., Vera D. A., Blau J., Holmes T. C., 2004. Membrane electrical excitability is necessary for the free-running larval Drosophila circadian clock. J. Neurobiol. 62: 1–13. [DOI] [PubMed] [Google Scholar]
- Nitabach M. N., Wu Y., Sheeba V., Lemon W. C., Strumbos J., et al. , 2006. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J. Neurosci. 26: 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitz D. A., van Swinderen B., Tononi G., Greenspan R. J., 2002. Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr. Biol. 12: 1934–1940. [DOI] [PubMed] [Google Scholar]
- Owald D., Waddell S., 2015. Olfactory learning skews mushroom body output pathways to steer behavioral choice in Drosophila. Curr. Opin. Neurobiol. 35: 178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisky K. M., Agosto J., Pulver S. R., Shang Y., Kuklin E., et al. , 2008. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60: 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisky K. M., Agosto Rivera J. L., Donelson N. C., Kotecha S., Griffith L. C., 2016. Reorganization of sleep by temperature in Drosophila requires light, the homeostat, and the circadian clock. Curr. Biol. 26: 882–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Sonn J. Y., Oh Y., Lim C., Choe J., 2014. SIFamide and SIFamide receptor define a novel neuropeptide signaling to promote sleep in Drosophila. Mol. Cells 37: 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch C. L., Green C. B., Takahashi J. S., 2014. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 24: 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffenberger C., Allada R., 2012. Cul3 and the BTB adaptor insomniac are key regulators of sleep homeostasis and a dopamine arousal pathway in Drosophila. PLoS Genet. 8: e1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot M., Cusumano P., Klarsfeld A., Ueda R., Rouyer F., 2007. Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol. 5: 2513–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel D., Donlea J. M., Talbot C. B., Song S. M., Thurston A. J. F., et al. , 2016. Operation of a homeostatic sleep switch. Nature 536: 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman J. L., McGill J. J., Keegan K. P., Allada R., 2006. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441: 753–756. [DOI] [PubMed] [Google Scholar]
- Pittendrigh C. S., 1967. Circadian systems. I. The driving oscillation and its assay in Drosophila pseudoobscura. Proc. Natl. Acad. Sci. USA 58: 1762–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J. L., Blau J., Rothenfluh A., Abodeely M., Kloss B., et al. , 1998. Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94: 83–95. [DOI] [PubMed] [Google Scholar]
- Raizen D. M., Zimmerman J. E., Maycock M. H., Ta U. D., You Y.-J., et al. , 2008. Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451: 569–572. [DOI] [PubMed] [Google Scholar]
- Renn S. C., Park J. H., Rosbash M., Hall J. C., Taghert P. H., 1999. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99: 791–802. [DOI] [PubMed] [Google Scholar]
- Rieger D., Stanewsky R., Helfrich-Förster C., 2003. Cryptochrome, compound eyes, Hofbauer-Buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila Melanogaster. J. Biol. Rhythms 18: 377–391. [DOI] [PubMed] [Google Scholar]
- Rieger D., Shafer O. T., Tomioka K., Helfrich-Förster C., 2006. Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J. Neurosci. 26: 2531–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger D., Wulbeck C., Rouyer F., Helfrich-Förster C., 2009. Period gene expression in four neurons is sufficient for rhythmic activity of Drosophila melanogaster under dim light conditions. J. Biol. Rhythms 24: 271–282. [DOI] [PubMed] [Google Scholar]
- Riemensperger T., Isabel G., Coulom H., Neuser K., Seugnet L., et al. , 2011. Behavioral consequences of dopamine deficiency in the Drosophila central nervous system. Proc. Natl. Acad. Sci. USA 108: 834–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts L., Leise T. L., Noguchi T., Galschiodt A. M., Houl J. H., et al. , 2015. Light evokes rapid circadian network oscillator desynchrony followed by gradual phase retuning of synchrony. Curr. Biol. 25: 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. E., Paluch J., Dickman D. K., Joiner W. J., 2016. ADAR-mediated RNA editing suppresses sleep by acting as a brake on glutamatergic synaptic plasticity. Nat. Commun. 7: 10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja D., Young M. W., 2012. Control of sleep by cyclin a and its regulator. Science 335: 1617–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin E. B., Shemesh Y., Cohen M., Elgavish S., Robertson H. M., et al. , 2006. Molecular and phylogenetic analyses reveal mammalian-like clockwork in the honey bee (Apis mellifera) and shed new light on the molecular evolution of the circadian clock. Genome Res. 16: 1352–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutila J. E., Suri V., Le M., So W. V., Rosbash M., et al. , 1998. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93: 805–814. [DOI] [PubMed] [Google Scholar]
- Saper C. B., Fuller P. M., Pedersen N. P., Lu J., Scammell T. E., 2010. Sleep state switching. Neuron 68: 1023–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayanan S., Zheng X., Xiao R., Sehgal A., 2004. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell 116: 603–615. [DOI] [PubMed] [Google Scholar]
- Scharf M. T., Naidoo N., Zimmerman J. E., Pack A. I., 2008. The energy hypothesis of sleep revisited. Prog. Neurobiol. 86: 264–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting M., Menegazzi P., Lelito K. R., Yao Z., Buhl E., et al. , 2016. A neural network underlying circadian entrainment and photoperiodic adjustment of sleep and activity in Drosophila. J. Neurosci. 36: 9084–9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehadova H., Glaser F. T., Gentile C., Simoni A., Giesecke A., et al. , 2009. Temperature entrainment of Drosophila’s circadian clock involves the gene nocte and signaling from peripheral sensory tissues to the brain. Neuron 64: 251–266. [DOI] [PubMed] [Google Scholar]
- Sehgal A., Price J. L., Man B., Young M. W., 1994. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science 263: 1603–1606. [DOI] [PubMed] [Google Scholar]
- Sehgal A., Rothenfluh-Hilfiker A., Hunter-Ensor M., Chen Y., Myers M. P., et al. , 1995. Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation. Science 270: 808–810. [DOI] [PubMed] [Google Scholar]
- Seidner G., Robinson J. E., Wu M., Worden K., Masek P., et al. , 2015. Identification of neurons with a privileged role in sleep homeostasis in Drosophila melanogaster. Curr. Biol. 25: 2928–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluzicki A., Flourakis M., Kula-Eversole E., Zhang L., Kilman V., et al. , 2014. Dual PDF signaling pathways reset clocks via TIMELESS and acutely excite target neurons to control circadian behavior. PLoS Biol. 12: e1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L., Suzuki Y., Vine L., Gottschalk L., Shaw P. J., 2008. D1 receptor activation in the mushroom bodies rescues sleep-loss-induced learning impairments in Drosophila. Curr. Biol. 18: 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L., Suzuki Y., Donlea J. M., Gottschalk L., Shaw P. J., 2011a Sleep deprivation during early-adult development results in long-lasting learning deficits in adult Drosophila. Sleep 34: 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L., Suzuki Y., Merlin G., Gottschalk L., Duntley S. P., et al. , 2011b Notch signaling modulates sleep homeostasis and learning after sleep deprivation in Drosophila. Curr. Biol. 21: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer O. T., Kim D. J., Dunbar-Yaffe R., Nikolaev V. O., Lohse M. J., et al. , 2008. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron 58: 223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Griffith L. C., Rosbash M., 2008. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc. Natl. Acad. Sci. USA 105: 19587–19594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Haynes P., Pírez N., Harrington K. I., Guo F., et al. , 2011. Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat. Neurosci. 14: 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P. J., Cirelli C., Greenspan R. J., Tononi G., 2000. Correlates of sleep and waking in Drosophila melanogaster. Science 287: 1834–1837. [DOI] [PubMed] [Google Scholar]
- Shaw P. J., Tononi G., Greenspan R. J., Robinson D. F., 2002. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature 417: 287–291. [DOI] [PubMed] [Google Scholar]
- Sheeba V., Fogle K. J., Kaneko M., Rashid S., Chou Y.-T., et al. , 2008a Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr. Biol. 18: 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V., Gu H., Sharma V. K., O’Dowd D. K., Holmes T. C., 2008b Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J. Neurophysiol. 99: 976–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Yue Z., Kuryatov A., Lindstrom J. M., Sehgal A., 2014. Identification of redeye, a new sleep-regulating protein whose expression is modulated by sleep amount. eLife 3: e01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman D., Aso Y., Jin X., Chen N., Felix M., et al. , 2015a Propagation of homeostatic sleep signals by segregated synaptic microcircuits of the Drosophila mushroom body. Curr. Biol. 25: 2915–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman D., Aso Y., Rubin G. M., Nitabach M. N., 2015b Control of sleep by dopaminergic inputs to the Drosophila mushroom body. Front. Neural Circuits 9: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwicki K. K., Eastman C., Petersen G., Rosbash M., Hall J. C., 1988. Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron 1: 141–150. [DOI] [PubMed] [Google Scholar]
- Stanewsky R., Kaneko M., Emery P., Beretta B., Wager-Smith K., et al. , 1998. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95: 681–692. [DOI] [PubMed] [Google Scholar]
- Stavropoulos N., Young M. W., 2011. Insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron 72: 964–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D., Peng Y., Agosto J., Rosbash M., 2004. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431: 862–868. [DOI] [PubMed] [Google Scholar]
- Stoleru D., Peng Y., Nawathean P., Rosbash M., 2005. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature 438: 238–242. [DOI] [PubMed] [Google Scholar]
- Stoleru D., Nawathean P., Fernández M. P., Menet J. S., Ceriani M. F., et al. , 2007. The Drosophila circadian network is a seasonal timer. Cell 129: 207–219. [DOI] [PubMed] [Google Scholar]
- Strausfeld N. J., Hirth F., 2013. Deep homology of arthropod central complex and vertebrate basal Ganglia. Science 340: 157–161. [DOI] [PubMed] [Google Scholar]
- Tang C.-H. A., Hinteregger E., Shang Y., Rosbash M., 2010. Light-mediated TIM degradation within Drosophila pacemaker neurons (s-LNvs) is neither necessary nor sufficient for delay zone phase shifts. Neuron 66: 378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tataroglu O., Zhao X., Busza A., Ling J., O’Neill J. S., et al. , 2015. Calcium and SOL protease mediate temperature resetting of circadian clocks. Cell 163: 1214–1224. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Thimgan M. S., Seugnet L., Turk J., Shaw P. J., 2015. Identification of genes associated with resilience/vulnerability to sleep deprivation and starvation in Drosophila. Sleep 38: 801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimgan M. S., Suzuki Y., Seugnet L., Gottschalk L., Shaw P. J., 2010. The perilipin homologue, lipid storage droplet 2, regulates sleep homeostasis and prevents learning impairments following sleep loss. PLoS Biol. 8: e1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh K., Jones C. R., He Y., Eide E. J., Hinz W. A., et al. , 2001. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291: 1040–1043. [DOI] [PubMed] [Google Scholar]
- Tononi G., Cirelli C., 2006. Sleep function and synaptic homeostasis. Sleep Med. Rev. 10: 49–62. [DOI] [PubMed] [Google Scholar]
- Tononi G., Cirelli C., 2014. Sleep and the price of plasticity: from synaptic and cellular homeostasis to memory consolidation and integration. Neuron 81: 12–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H. R., Matsumoto A., Kawamura M., Iino M., Tanimura T., et al. , 2002. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J. Biol. Chem. 277: 14048–14052. [DOI] [PubMed] [Google Scholar]
- Ueno T., Tomita J., Tanimoto H., Endo K., Ito K., et al. , 2012. Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat. Neurosci. 15: 1516–1523. [DOI] [PubMed] [Google Scholar]
- van Alphen B., Yap M. H. W., Kirszenblat L., Kottler B., van Swinderen B., 2013. A dynamic deep sleep stage in Drosophila. J. Neurosci. 33: 6917–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanin S., Bhutani S., Montelli S., Menegazzi P., Green E. W., et al. , 2012. Unexpected features of Drosophila circadian behavioural rhythms under natural conditions. Nature 484: 371–375. [DOI] [PubMed] [Google Scholar]
- Veleri S., Rieger D., Helfrich-Förster C., Stanewsky R., 2007. Hofbauer-buchner eyelet affects circadian photosensitivity and coordinates TIM and PER expression in Drosophila clock neurons. J. Biol. Rhythms 22: 29–42. [DOI] [PubMed] [Google Scholar]
- Vosko A. M., Schroeder A., Loh D. H., Colwell C. S., 2007. Vasoactive intestinal peptide and the mammalian circadian system. Gen. Comp. Endocrinol. 152: 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., Dan Y., 2016. Circuit-based interrogation of sleep control. Nature 538: 51–59. [DOI] [PubMed] [Google Scholar]
- Welsh D. K., Takahashi J. S., Kay S. A., 2010. Suprachiasmatic nucleus: cell autonomy and network properties. Annu. Rev. Physiol. 72: 551–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D. A., Hamblen-Coyle M. J., Dushay M. S., Hall J. C., 1993. Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J. Biol. Rhythms 8: 67–94. [DOI] [PubMed] [Google Scholar]
- Wijnen H., Naef F., Boothroyd C., Claridge-Chang A., Young M. W., 2006. Control of daily transcript oscillations in Drosophila by light and the circadian clock. PLoS Genet. 2: e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. A., Sathyanarayanana S., Hendricks J. C., Sehgal A., 2007. Interaction between sleep and the immune response in Drosophila: a role for the NFkB elish. Sleep 30: 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang W., Simoni A., Gentile C., Stanewsky R., 2013. The Pyrexia transient receptor potential channel mediates circadian clock synchronization to low temperatures cycles in Drosophila melanogaster. Proc. Biol. Sci. 280: 20130959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. N., Joiner W. J., Dean T., Yue Z., Smith C. J., et al. , 2009. SLEEPLESS, a Ly-6/neurotoxin family member, regulates the levels, localization and activity of Shaker. Nat. Neurosci. 13: 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Robinson J. E., Joiner W. J., 2014. SLEEPLESS is a bifunctional regulator of excitability and cholinergic synaptic transmission. Curr. Biol. 24: 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Kang H., Xu Q., Chen M. J., Liao Y., et al. , 2013. Sleep drives metabolite clearance from the adult brain. Science 342: 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., DiAngelo J. R., Hughes M. E., Hogenesch J. B., Sehgal A., 2011. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab. 13: 639–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Padiath Q. S., Shapiro R. E., Jones C. R., Wu S. C., et al. , 2005. Functional consequences of a CKIδ mutation causing familial advanced sleep phase syndrome. Nature 434: 640–644. [DOI] [PubMed] [Google Scholar]
- Yao Z., Shafer O. T., 2014. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 343: 1516–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T., Hermann-Luibl C., Helfrich-Förster C., 2016. Circadian light-input pathways in Drosophila. Commun. Integr. Biol. 9: e1102805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T., Heshiki Y., Ibuki-Ishibashi T., Matsumoto A., Tanimura T., et al. , 2005. Temperature cycles drive Drosophila circadian oscillation in constant light that otherwise induces behavioural arrhythmicity. Eur. J. Neurosci. 22: 1176–1184. [DOI] [PubMed] [Google Scholar]
- Yoshii T., Todo T., Wülbeck C., Stanewsky R., Helfrich-Förster C., 2008. Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J. Comp. Neurol. 508: 952–966. [DOI] [PubMed] [Google Scholar]
- Yoshii T., Vanin S., Costa R., Helfrich-Förster C., 2009a Synergic entrainment of Drosophila’s circadian clock by light and temperature. J. Biol. Rhythms 24: 452–464. [DOI] [PubMed] [Google Scholar]
- Yoshii T., Wulbeck C., Sehadova H., Veleri S., Bichler D., et al. , 2009b The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J. Neurosci. 29: 2597–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T., Hermann-Luibl C., Kistenpfennig C., Schmid B., Tomioka K., et al. , 2015. Cryptochrome-dependent and -independent circadian entrainment circuits in Drosophila. J. Neurosci. 35: 6131–6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Jacquier A. C., Citri Y., Hamblen M., Hall J. C., et al. , 1987. Molecular mapping of point mutations in the period gene that stop or speed up biological clocks in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 84: 784–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q., Joiner W. J., Sehgal A., 2006. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr. Biol. 16: 1051–1062. [DOI] [PubMed] [Google Scholar]
- Zehring W. A., Wheeler D. A., Reddy P., Konopka R. J., Kyriacou C. P., et al. , 1984. P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell 39: 369–376. [DOI] [PubMed] [Google Scholar]
- Zeng H., Qian Z., Myers M. P., Rosbash M., 1996. A light-entrainment mechanism for the Drosophila circadian clock. Nature 380: 129–135. [DOI] [PubMed] [Google Scholar]
- Zerr D. M., Hall J. C., Rosbash M., Siwicki K. K., 1990. Circadian fluctuations of period protein lmmunoreactivity in the CNS and the visual system of Drosophila. J. Neurosci. 10: 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ling J., Yuan C., Dubruille R., Emery P., 2013. A role for Drosophila ATX2 in activation of PER translation and circadian behavior. Science 340: 879–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chung B. Y., Lear B. C., Kilman V. L., Liu Y., et al. , 2010. DN1p circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr. Biol. 20: 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu Y., Bilodeau-Wentworth D., Hardin P. E., Emery P., 2010. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr. Biol. 20: 600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Sehgal A., 2012. Speed control: cogs and gears that drive the circadian clock. Trends Neurosci. 35: 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Sowcik M., Chen D., Sehgal A., 2014. Casein kinase 1 promotes synchrony of the circadian clock network. Mol. Cell. Biol. 34: 2682–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Yuan Q., Froy O., Casselman A., Reppert S. M., 2005. The two CRYs of the butterfly. Curr. Biol. 15: R953–R954. [DOI] [PubMed] [Google Scholar]
- Zimmerman J. E., Rizzo W., Shockley K. R., Raizen D. M., Naidoo N., et al. , 2006. Multiple mechanisms limit the duration of wakefulness in Drosophila brain. Physiol. Genomics 27: 337–350. [DOI] [PubMed] [Google Scholar]
- Zimmerman J. E., Raizen D. M., Maycock M. H., Maislin G., Pack A. I., 2008. A video method to study Drosophila sleep. Sleep 31: 1587–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman J. E., Chan M. T., Jackson N., Maislin G., Pack A. I., 2012. Genetic background has a major impact on differences in sleep resulting from environmental influences in Drosophila. Sleep 35: 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]