Abstract
Genomic analyses of cutaneous melanoma (CM) have yielded biological and therapeutic insights, but understanding of non-ultraviolet (UV)-derived CMs remains limited. Deeper analysis of acral lentiginous melanoma (ALM), a rare sun-shielded melanoma subtype associated with worse survival than CM, is needed to delineate non-UV oncogenic mechanisms. We thus performed comprehensive genomic and transcriptomic analysis of 34 ALM patients. Unlike CM, somatic alterations were dominated by structural variation and absence of UV-derived mutation signatures. Only 38% of patients demonstrated driver BRAF/NRAS/NF1 mutations. In contrast with CM, we observed PAK1 copy gains in 15% of patients, and somatic TERT translocations, copy gains, and missense and promoter mutations, or germline events, in 41% of patients. We further show that in vitro TERT inhibition has cytotoxic effects on primary ALM cells. These findings provide insight into the role of TERT in ALM tumorigenesis and reveal preliminary evidence that TERT inhibition represents a potential therapeutic strategy in ALM.
Comprehensive sequencing of large cancer cohorts has revealed genomic landscapes of common malignant diseases, enabling deduction of tumorigenic pathways and clinically actionable information. Such genomic analyses of more than 600 cutaneous melanomas (CM), the most common melanoma subtype, have identified a small number of frequent driver mutations (impacting BRAF, NRAS, and NF1), a large number of low-frequency mutations, and a complex array of genotypes reflecting diverse paths to tumorigenesis (for review, see Zhang et al. (2016)). Although all melanomas arise from transformed melanocytes, the genetics, biology, and pathology of melanoma subtypes are shaped by anatomic location (cutaneous or acral skin, mucosal surfaces, or the eye), host-tumor microenvironment, and the unique mutational processes at work in each location. Mapping the relatively uncharted landscapes of melanoma in noncutaneous sites will have broad relevance for understanding melanoma biology and clinical management.
Acral lentiginous melanoma (ALM), first described in Reed (1976), is a rare subtype of non-CM. Although there is debate about ALM's distinctive histologic features, it is characterized by occurrence in sun-shielded skin sites on palms, soles, or nail beds (subungual). It comprises 2%–3% of melanoma cases in the United States and is more prevalent than CM in heavily pigmented ethnic populations with an age-adjusted incidence rate of 1.8 per 1,000,000 persons/year (Bradford et al. 2009). ALM has a worse prognosis than CM, demonstrating inferior 10-yr and overall survival rates (Bradford et al. 2009; Bello et al. 2013). Unfortunately, understanding of ALM's genomic landscape remains limited. Analysis of small ALM cohorts by next-generation or targeted sequencing has revealed a low single-nucleotide variant (SNV) and high structural variant (SV) burden (Curtin et al. 2005; Turajlic et al. 2012; Furney et al. 2014). Notably, CM bears the highest mutation burden of any cancer due to DNA damage from ultraviolet (UV) radiation, an established melanoma risk factor. UV induces characteristic C > T transitions at dipyrimidines that occur in >76% of CMs with variability based on anatomic site and degree of sun exposure (The Cancer Genome Atlas Network 2015). A UV signature is present when C > T transitions at dipyrimidine sites account for >60%, or CC > TT mutations account for >5%, of the total mutation burden. UV signatures are less frequent in ALM in keeping with lower SNV burden and occurrence in sun-shielded sites, although exceptions have been reported (Turajlic et al. 2012; Furney et al. 2014). Driver mutations implicated in ALM include mutations at BRAF (B-Raf proto-oncogene) V600 (reported in ∼15% of ALMs) (The Cancer Genome Atlas Network 2015) and in KIT (KIT proto-oncogene receptor tyrosine kinase; reported in ∼11% of ALMs) (Curtin et al. 2006) in addition to focal amplifications involving CCND1 (cyclin D1), CDK4 (cyclin dependent kinase 4), and GAB2 (GRB2-associated binding protein 2) (Sauter et al. 2002; Curtin et al. 2005; Chernoff et al. 2009; Krauthammer et al. 2012). Finally, TERT (telomerase reverse transcriptase) promoter mutations driving TERT overexpression occur as an early tumorigenic event in >70% of CMs (Horn et al. 2013; Huang et al. 2013; Griewank et al. 2014), resulting from C > T transitions reflective of UV mutagenesis, and often in cooperation with BRAF mutations. These events are more rare in ALM with a recent report identifying TERT promoter mutations in 9.3% of 48 ALMs (de Lima Vazquez et al. 2016). The unique etiology of non-UV and non-CM melanoma still remains to be understood. Further, targeted treatment for ALM is limited. To date, KIT and BRAF mutations, although infrequent, have been the focus of targeted therapy for ALM. It is thus critical to define additional targetable genomic or transcriptomic alterations in ALM. To address these needs, we performed comprehensive, integrated genomic and transcriptomic analysis of 38 ALMs from 34 patients with detailed clinical annotation, along with downstream in vitro analyses based on our findings. These data establish a foundation for understanding ALM's genetic etiology with the ultimate goal to inform ALM clinical management.
Results
The genomic and transcriptomic landscape of ALM
Patient information is shown in Supplemental Tables S1A,B, and overall survival and progression-free survival analyses are shown in Supplemental Figure S1A,B. Paired tumor/constitutional exome sequencing was performed across 33 patients, and tumor-only exome sequencing was performed for the 34th patient, for whom constitutional DNA was not available (Methods). For 32 patients, sufficient amounts of DNA were extracted to support additional analysis using long-insert whole-genome (LIWG) sequencing of ∼900 bp inserts to identify breakpoints that reflect structural variants (SVs) in the form of copy number variants (CNVs) and translocations (Liang et al. 2014). RNA-seq was additionally performed for 33 patients. A summary of assays performed and estimated tumor cellularities (median = 50%) are listed in Supplemental Table S2, and sequencing metrics are listed in Supplemental Table S3.
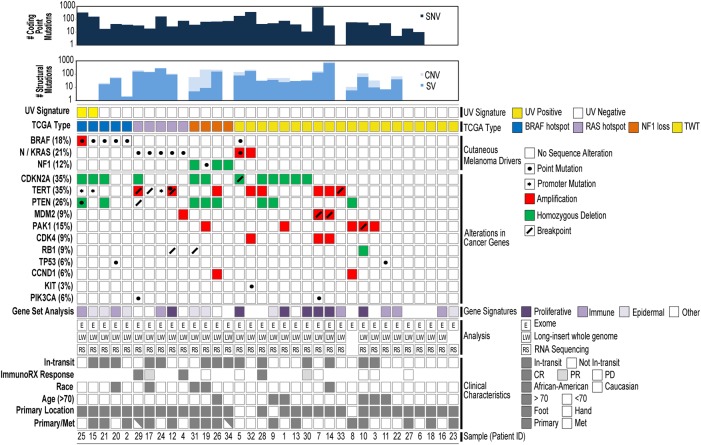
Paired-exome sequencing across 34 patients led to the identification of 9522 somatic SNVs and 72 somatic small indels (SNV/small indel median = 116, range = 3–2278), including intronic, UTR, loss of function, missense, splice site (splicing altered or splice site loss), and synonymous events. With respect to coding somatic mutations (missense, nonsense, splice site, indels), a median of 42 mutations (range 0–869) across all tumors was observed (SNV mutation burden) (Fig. 1). These findings are consistent with prior next-generation sequencing studies that have identified a relatively low ALM coding mutation burden with averages ranging from 9–80 somatic coding mutations per tumor dependent on platform, analysis, and cohort characteristics (Berger et al. 2012; Hodis et al. 2012; Krauthammer et al. 2012; Turajlic et al. 2012; Furney et al. 2014). Notably, the treatment status of sequenced tumors may also impact mutation burden as the tumor demonstrating the highest SNV burden in our study (patient 7; 867 coding SNVs) was collected following treatment with ipilimumab, followed by pembrolizumab (Supplemental Table S1B). Pretreated tumors were also sequenced for patients 4, 5, and 28, and the number of identified coding SNVs also trended toward higher levels for these patients (median = 75, range = 42–177). As secondary confirmation of somatic coding point mutations identified in our study, 12% of these events were confirmed by LIWG data in tumors with both exome and LIWG data, noting the lower coverage for LIWG sequencing. Furthermore, 90% of somatic coding point mutations were confirmed in RNA-seq data in tumors with both exome and RNA-seq data.
Figure 1.
The mutational landscape of ALM. A summary of somatic alterations is shown. Patients for whom multiple tumors were sequenced are shown with partially shaded rectangles on the bottom row.
Overall, C > T transitions, which have a 75% probability of occurring in dipyrimidine sequences (Brash 2015), were the most common base substitution in the ALMs analyzed here, making up 55.0% of all identified somatic point mutations (Supplemental Fig. S2). Across all patients, 39.4% of C > T transitions occurred in dipyrimidine sequences, which falls short of the 60% threshold that characterizes the presence of a dominant UV signature (Brash 2015; The Cancer Genome Atlas Network 2015) observed in CMs, such that a more minor signature may be present. However, individual analysis of each tumor revealed the presence of a UV signature in two ALMs (Fig. 1), with patient 7 demonstrating a trend toward the presence of a signature with 59% of C > T transitions occurring at dipyrimidines.
SNVs identified through exome sequencing, along with respective sequence contexts, were used to characterize somatic mutational signatures across all tumors (Methods). The distribution of signatures across tumors and the context of each identified signature are shown in Supplemental Figure S3, A and B, respectively. Analysis of variance (ANOVA) indicated that differences in the frequency of signatures across all samples was not statistically significant (P = 0.699). Correlation of the identified signatures against previously reported somatic cancer signatures (Alexandrov et al. 2013) was also performed (Supplemental Table S4). Overall, the most common ALM signature that was identified is S1, with an incidence of 0.658 across samples. ALM signature S1 (cosine similarity value [CSV] = 0.981) correlated with the Alexandrov UV light signature (Alexandrov signature S7) and was also the dominant signature for samples 7, 15, 24, 25a, and 25b. Notably, these patients overlap with the patients for whom UV signatures were previously identified (patients 25 and 15) from analysis of C > T transitions in dipyrimidines. These findings provide evidence of a putative UV signature in a small subset of patients. We additionally observed limited correlation between ALM signature S4 and Alexandrov signature S1A (CSV = 0.846), as well as between ALM signatures S7 and S10 with Alexandrov signature S1B (CSV = 0.710–0.761). Alexandrov signatures S1A and S1B were previously reported across 83% of cancer classes analyzed and may be associated with increased spontaneous deamination of 5-methyl-cytosine events resulting in C > T transitions in both normal and cancerous cells (Alexandrov et al. 2013).
Analysis of somatic coding SNVs and small indels revealed NRAS and BRAF as the most significant putative drivers in ALM. Three patients demonstrated the well-recognized activating NRAS (neuroblastoma RAS viral oncogene homolog) Q61K (9%) hotspot mutation that has been reported in CMs (The Cancer Genome Atlas Network 2015), which was validated in two of three patients based on available DNA, and a fourth demonstrated an A59G event. With respect to BRAF alterations, four patients each demonstrated V600E (12%), a fifth demonstrated a G466E mutation, and a sixth demonstrated two separate BRAF mutations (V600K, R462K). The NRAS and BRAF events were mutually exclusive (Fig. 1), and these putative driving events occurred in 10 of the 34 assayed patients (29%), indicating that the remaining 24 patients garner unique driving alterations. Previous studies also reported the presence of mutually exclusive mutant BRAF and mutant NRAS in ALMs (Curtin et al. 2005; Krauthammer et al. 2012), in addition to KIT mutations in 11% of ALMs (Curtin et al. 2006). However, in our analysis, we identified only one patient with a KIT L576P mutation, that was exclusive of driving BRAF/NRAS events, and that was previously reported in ALM patients (Krauthammer et al. 2012). No correlation was observed between mutation burden and BRAF or NRAS mutation status.
In addition to these driving events, we also observed low incidence of somatic mutations in other key genes, including NF1 (neurofibromin 1), EGFR (epidermal growth factor receptor), KRAS (KRAS proto-oncogene, GTPase), and TP53 (tumor protein p53) (Supplemental Table S5). We observed homozygous loss of NF1 in 9% of patients (Fig. 1), and in a fourth patient (19), loss of heterozygosity (LOH) of NF1 was accompanied by a nonsense mutation (E2578*) on the second allele. Additional somatic mutations were identified in EGFR (R334C, V726M), KRAS (V14L with a CNV gain in patient 5), TP53 (R248W), and ERBB3 (erb-b2 receptor tyrosine kinase 3; S1119C). Two nonsynonymous events were also detected (patients 11 [S1167T] and 24 [A355T]) in PREX2 (phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2), which has been reported to be frequently mutated in CM (Berger et al. 2012). These two point mutations have not been described in melanoma, but the S1167 position was found to also be mutated in a CM metastasis (S1167N) (Krauthammer et al. 2012).
We additionally performed mutational landscape analyses by segregating primary from metastatic tumors. Although no novel drivers emerged, BRAF remained significant (IntOgen; Q-value < 0.05) in both groups. UV signatures were also absent in each group, and mutation burden differences trended toward significance (P = 0.06) with a higher number of mutations in metastases (mean = 113) compared to primary ALMs (mean = 50).
Structural alterations in ALM
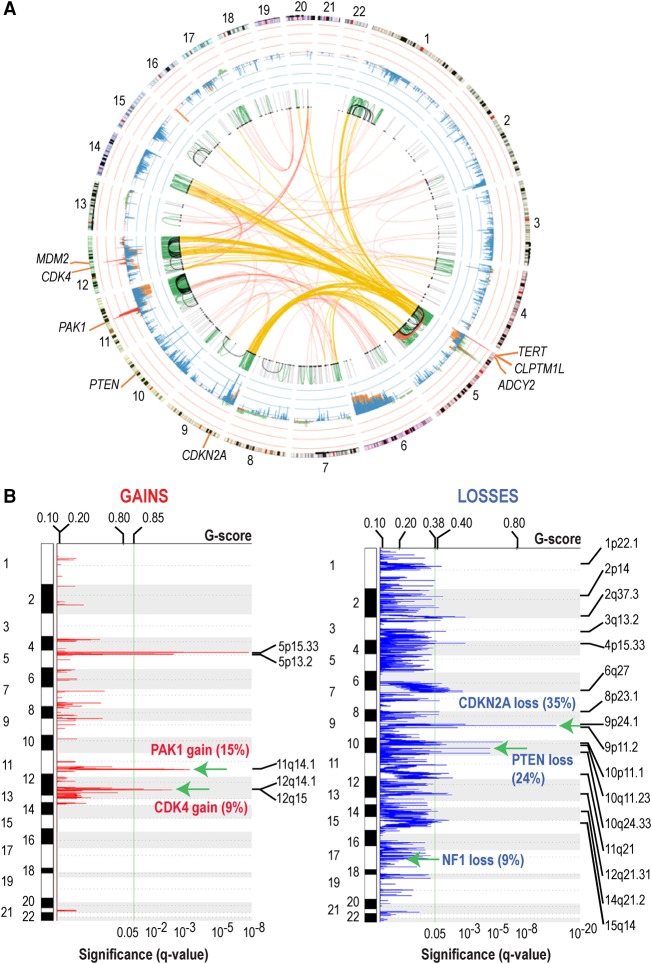
Across 31 patients with LIWG data, a total of 2490 somatic breakpoints, in the form of inversions, large indels, or translocations, were identified across 74% of patients (25 ALMs; median = 31, range = 0–683) (Figs. 1, 2; genic breakpoints listed in Supplemental Table S6; tumor-specific SVs are shown in Supplemental Fig. S3). Notably, patient 14 showed evidence of chromothripsis with 503 breakpoints on Chromosome 12. Patients 24 and 29c additionally demonstrated a high number (more than 200), with 242 total events falling within Chromosome 11 for patient 24 and 135 breakpoints falling within Chromosome 5 for patient 29c. Overall, two genes were found to be most highly impacted across the cohort—these include ADCY2 (adenylate cyclase 2), for which 32 breakpoints were identified across seven samples (21% of patients), and CLPTM1L (CLPTM1 like), for which 22 breakpoints were identified across six samples (15% of patients). ADCY2 translocation partners include CLPTM1L, HECTD4 (HECT domain containing E3 ubiquitin protein ligase 4), TERT, and UBE2QL1 (ubiquitin conjugating enzyme E2Q family-like 1). CLPTM1L partners include ADCY2, PDZD2 (PDZ domain containing 2), RAI14 (retinoic acid induced 14), and TRIO (trio Rho guanine nucleotide exchange factor). These breakpoints all occurred in BRAF wild-type tumors.
Figure 2.
Consensus somatic SVs and CNVs. (A) Summary of somatic SVs and CNVs. Intrachromosomal SVs: (gray) <1 Mb; (green) ≥1 Mb and <50 Mb; (black) ≥50 Mb and <100 Mb; (red) ≥100 Mb. Interchromosomal SVs are shown in red; interchromosomal SVs impacting Chr 5 are in yellow. Consensus CNVs are shown in the inner circle adjacent to chromosomes: (red) exome CNV gain; (green) LIWG CNV gain; (blue) exome CNV loss; (orange) LIWG CNV loss. (B) Consensus CNVs. Selected common gains and losses are indicated by green arrows. The percentage of impacted tumors is shown in parentheses. The plot shows the Q-values; Benjamini and Hochberg FDR (bottom) and G-score (top), with the copy number gains (left) indicated in red and copy number losses (right) in blue. Chromosome positions are indicated along the y-axis.
Overall, a total of 1115 somatic focal CNVs (median = 12, range = 0–211) (Fig. 2A,B; Supplemental Fig. S3, tumor-specific CNVs) were identified. To identify statistically significant consensus CNVs across all samples, CNVs detected using either exome or LIWG data were integrated. As a result, 48 total CNVs were identified (95% confidence interval). CNVs include 40 deleted regions and eight amplified regions (Supplemental Table S7). Key events include TERT and CLPTM1L gains on Chr 5, loss of CDKN2A on Chr 9, and a gain in a region on Chr 12 encompassing CDK4. TERT gains were separately validated by real-time PCR for available samples (six of eight ALMs; 75%). Following segregation of samples based on primary or metastasis status, the TERT and CLPTM1L gains and the CDKN2A losses remained statistically significant even when the primary (Q = 1.28 × 10−4) or metastatic lesions (Q = 1.98 × 10−7) were analyzed separately. However, the CDK4 gain only retained significance in metastases. Notably, relevant CNVs that were not previously observed in the analysis of all samples include PTEN (10q23.31) and NF1 (17q11.2) deletions in only the primary tumors and gain of MDM2 (MDM2 proto-oncogene; 12q15) in only metastases.
Focal copy gains impacting PAK1 were also observed in five (15%) patients, with one of these patients also demonstrating multiple SVs impacting PAK1, including evidence of multiple inversions and a large indel. PAK1 gains were separately validated by real-time PCR for available samples (four of five ALMs; 80%). Elevated expression of PAK1 was also observed in two of five patients (1, 10). These patients were exclusive of characteristic CM BRAF and RAS subtypes, with one patient demonstrating the NF1 subtype and the remaining four demonstrating the triple-wild-type (TWT) subtype, providing evidence that alternate routes toward dysregulation of MAPK signaling may be present in the TWT context.
RNA fusions
Using RNA-seq data, we detected 106 RNA fusions across 74% of patients (median = 2) (Supplemental Table S8). Thirteen of these fusions (13%) were also supported by the detection of a corresponding DNA breakpoint. Additional selected fusions include two MDM2 events (MDM2:GNS, MDM2:CCT2), and PTEN:RPL11, PAK2:LOC646214, and MAP3K8:DEK fusions (each observed in a single tumor). MDM2 structural breakpoints and overexpression were additionally observed in the respective patients with the reported fusions. However, no additional alterations in PTEN and PAK2 were identified in the tumors demonstrating PTEN to PAK2 fusions, but patient 28, who garners the MAP3K8 fusion, also demonstrated a breakpoint adjacent to the fusion boundary.
Perturbed biological processes in ALM
In order to evaluate the impact of identified mutations on pathways, we assessed somatic genomic alterations according to a manually curated set of commonly altered melanoma pathways (Supplemental Fig. S4A). These pathways include MAPK/PI3K (proliferation/survival, altered in 66% of patients), TERT (telomere maintenance, altered in 37% of patients), CDK4/CDKN2A (cell-cycle progression, altered in 51% of patients), and MDM2/TP53 (apoptosis and senescence, altered in 17% of patients). Next, we integrated gene expression data from 24 samples in order to identify transcriptionally dysregulated pathways in ALM and to pinpoint associations with genomic and clinical characteristics of these samples. Through unsupervised hierarchical clustering of the 421 genes with the greatest differential expression, we identified three dominant sample clusters (Supplemental Fig. S4B). Of note, these 421 genes did not include the melanogenesis associated transcription factor (MITF), which demonstrates key roles in CMs through regulation of melanocyte development and function. The three dominant clusters trended toward segregation of primary from metastatic samples, but did not significantly correlate with mutational profiles or other clinical features including age, gender, race, primary location, in-transit status, and response to immunotherapy. Five dominant gene clusters were present, including gene sets associated with (1) protein translation; (2) the ER stress response; (3) antigen presentation and immune processes; (4) keratinization, chemotaxis, and intermediate filaments; and (5) G2/M cell-cycle, pigmentation, and nonsense-mediated decay. Notably, cluster 4 containing keratin genes was down-regulated in metastases and overexpressed in primary tumors to parallel previous reports in CM (The Cancer Genome Atlas Network 2015).
Neo-antigen burden
Although response to immune checkpoint inhibitors has been well demonstrated in CM (Snyder et al. 2014; Van Allen et al. 2015), it has not been well studied in ALM. In a previous retrospective analysis, 11% of ALM cases responded to ipilimumab, but the response rate to anti-PD1 has not been reported (Johnson et al. 2015). In this cohort, 22 patients received immune checkpoint inhibitors, with 10 receiving only anti-CTLA4 and 10 receiving both anti-CTLA4 and anti-PD1. In order to help inform utility of checkpoint blockade in ALM, we assessed neo-antigen burden and HLA expression in patients for whom both RNA and DNA data were available (Supplemental Fig. S5). Neo-antigen burden was associated with mutation burden (0.89, Pearson's correlation) as shown in studies of CM (Snyder et al. 2014; Van Allen et al. 2015; Hugo et al. 2016). Mutation and neo-antigen burden has been previously reported to be associated with response to immune checkpoint blockade (Snyder et al. 2014; Van Allen et al. 2015). Although we did not observe that trend here, the number of samples limits our power to detect such a correlation. Interestingly, we observed that two patients with complete response to anti-PD1, and one with complete response to anti-CTLA4 treatments had lower mutation (less than 75) and neo-antigen (less than 60) burdens.
TERT alterations in ALM
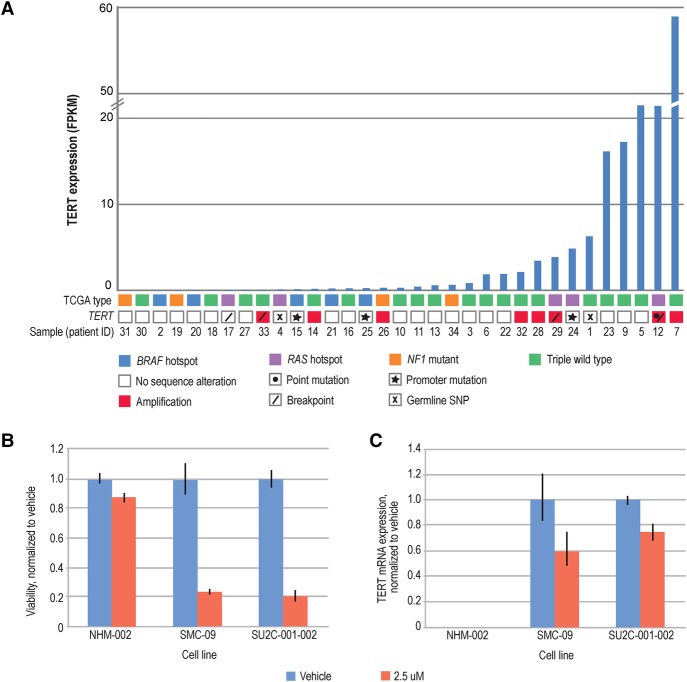
In addition to TERT gains, additional TERT events were also detected with a total of 14 (41%) patients demonstrating either somatic or germline aberrations in this gene. TERT promoter mutations were not initially identified because of limited coverage of the promoter region by exome baits. However, targeted Sanger sequencing of TERT promoters led to the identification of promoter mutations in 9% of patients (four of 28 ALMs; 15, 24, 25a, 25b) with available DNA (Chr 5: 1,295,113: G>A) (Fig. 1) to parallel previous reports (Liau et al. 2014; de Lima Vazquez et al. 2016). From exome sequencing, one nonsynonymous mutation in patient 12 (F919L) was also identified; in patient 24, an intronic SNV (Chr 5: 1,293,410: C > A), which is localized to TERT's RNA-interacting domain and a region required for oligomerization, was observed. Notably, two rare germline polymorphisms were identified in patients 1 and 4 (A1062T) and 8 (T1110M) and were mutually exclusive of patients with somatic TERT events. Both these events are predicted to be damaging by FATHMM (functional analysis through hidden Markov models). One case, patient 1, additionally displayed LOH in tumor DNA.
LIWG also supported the identification of TERT breakpoints in four patients (12, 17, 29a and 29c, 33). In patient 12, a breakpoint was identified at −:5:36,583,500|−:5:1,288,500. Patient 12's tumor demonstrated a complex rearrangement at the Chr 5p locus (Supplemental Fig. S6). This event encompasses an inversion in intron 11 of TERT, directly 3′ of exon 11, as well as a translocation impacting exon 11. The inversion is evidenced by TERT intron 11 reads with mates mapping to intron 3 of NIPBL (cohesion loading factor), whereas the translocation suggests loss of exon 11 of TERT. No RNA reads supporting a TERT:NIPBL fusion were found. However, RNA reads supporting an intra-exon translocation in exon 11 of TERT were observed. Because of the complexity of these events, it is unclear what the functional impact is, but in the same tumor, TERT expression is elevated with an FPKM of 32.5 (Fig. 3) to suggest the possibility of TERT activation. Assembly of RNA-seq data of the same tumor also revealed the presence of a TERT:ADCY2 RNA fusion. In patient 17, a breakpoint was also called in intron 2 of TERT, but closer evaluation indicated that this event falls in a GC-rich region with reads demonstrating low mapping quality. RNA-seq data indicated that this event was not expressed, coinciding with lower TERT expression in this tumor (FPKM < 1). In both metastatic lesions from patient 29, a TERT:PDCD1LG2 (PDL2; programmed cell death 1 ligand 2) interchromosomal rearrangement was identified (29a: −:5:1,272,000|+:9:5,560,500; 29c: −:5:1,271,200|+:9:5,560,800). Manual evaluation led to the observation that the breakpoint in TERT falls within Chr 5: 1,272,200–1,272,400, a region which encompasses exon 7 of the gene and which corresponds to the reverse transcriptase domain of TERT. This region is linked to intron 5 of PDCD1LG2. Although both TERT and PDCD1LG2 were expressed in both tumors, expression of an RNA fusion was not observed. In both tumors, a possible inversion impacting TERT and FER (FER tyrosine kinase; −:5:108,410,400|+:5:1,276,800) was also detected. The breakpoints disrupt intron 6 of TERT and intron 16 of FER. Although both TERT and FER (29a FPKM = 6.7, 29c FPKM = 6.4) were expressed in both tumors, an RNA fusion resulting from this event was not detected. Lastly, a possible large indel was detected in patient 33. Upon closer inspection, this was determined to be an intrachromosomal rearrangement (−:5:7,752,000| +:5:1,294,500) between the 5′ UTR and exon 1 of TERT to intron 15 of ADCY2. A corresponding RNA fusion was not detected, and both expression of TERT and ADCY2 were low in this patient's tumor (FPKM < 1). Variable levels of expression of TERT were, however, observed across all patients (Fig. 3).
Figure 3.
TERT in ALM. (A) TERT aberrations in ALM. 41% of patients demonstrated TERT alterations (somatic, germline) and TERT expression. (B,C) TERT inhibition is selectively cytotoxic in ALM cell lines and reduces TERT expression. (B) Cell lines were treated with DMSO vehicle or Telomerase Inhibitor IX, and after 72 h cell viability was assessed by CellTiterGlo. Viability was reduced by at least 75% in ALM cell lines, but only 12% in normal melanocyte controls. (C) Cells were treated with DMSO vehicle or Telomerase Inhibitor IX, and after 72 h, TERT mRNA was quantified by reverse transcription and qPCR. Expression was reduced by at least 25% in ALM cell lines. NHM-002 TERT expression was undetectable.
TERT inhibition in ALM
To evaluate the impact of TERT inhibition in ALM, we performed viability assays on two primary ALM cell lines, one with a TERT CNV gain (SMC-09) and one with a homozygous TERT promoter mutation (SU2C-001-002; Chr 5: 1,295,113: G > A [hg38]), as well as on a normal melanocyte line (NHM-002). Testing was performed using Telomerase inhibitor IX (Fig. 3B,C) or vehicle. After 72 h of drug treatment, we observed at least a 75% decrease in cell viability with 2.5 µM of Telomerase Inhibitor IX, and not in normal melanocytes (Fig. 3B). To explore the effect of telomerase inhibition on TERT gene expression, we treated the same ALM cells with 2.5 µM Telomerase Inhibitor IX, and measured TERT mRNA by qPCR after 72 h of drug treatment. TERT mRNA expression was also reduced at 72 h by at least 25% in the ALM cell lines and was undetected in normal melanocytes (Fig. 3C). These data thus demonstrate that TERT inhibition may be an effective approach to reduce cell viability in TERT-dependent ALMs.
Discussion
Our analysis of 38 ALMs across 34 patients confirms a number of findings of previous smaller analyses, but also sheds new light on the molecular foundations of human ALM. As others have, we observed lower SNV and higher SV burdens in ALMs (Furney et al. 2014) compared to CM. Importantly, in our study, pretreated tumors demonstrated elevated SNV burden, with one such tumor demonstrating the highest SNV burden in this cohort. UV signature analysis of the entire cohort based on the frequency of C > T transitions in dipyrimidines revealed the absence of a dominant UV signature. However, two cases, both of which are BRAF mutant, individually demonstrated the presence of a UV signature, with one of these two cases also being the tumor with the greatest SNV burden. Secondary somatic signature analysis of SNVs, and sequence contexts of these events, additionally led to the identification of another two patients, in addition to the two previously identified cases, that demonstrate correlations with the previously defined cancer UV signature (Alexandrov et al. 2013) to provide evidence of the presence of this signature in a small subset of ALMs.
BRAF/NRAS mutant tumors did not demonstrate statistically significant differences from wild-type tumors with respect to SNV, CNV, or SV burden. These findings contrast with a previous analysis of sun-shielded melanomas, which reported a high number of CNVs and lower mutation load in BRAF/NRAS wild-type versus mutant melanomas (Krauthammer et al. 2012). We additionally observed a low number of BRAF mutations in ALMs to parallel previous reports of a lower incidence of BRAF alterations in sun-protected melanomas (Curtin et al. 2006). On the other hand, we did not observe any somatic SNVs or indels in DYNC1I1 (dynein, cytoplasmic 1, intermediate chain 1), ARID1A (AT rich interaction domain 1A), and APC (APC, WNT signaling pathway regulator) (Furney et al. 2014), all of which have been reported to be recurrently mutated in ALM. Additional key findings include identification of PAK1 copy gains in a subset of patients, all in the BRAF/NRAS wild-type context. PAK1 has been proposed as a therapeutic target in BRAF wild-type melanoma (Ong et al. 2013) based on its activation in multiple tumors (Radu et al. 2014) as demonstrated by increased cell proliferation, survival, invasion, and metastasis pathways.
In 41% of acral melanoma patients, we observed TERT aberrations encompassing promoter regions and point mutations, breakpoints, copy gains, and coding germline mutations. All ALMs with TERT copy gains were also all BRAF wild type, but overlapped with N/KRAS and NF1 alterations, and trended toward higher levels of somatic alterations. We also identified potentially damaging germline point mutations in TERT. Although TERT promoter germline mutations have been described in melanoma (Horn et al. 2013), nonsynonymous coding germline mutations have not been reported. TERT breakpoints were observed in 12% of patients. Although one tumor demonstrated elevated TERT expression, it remains to be clarified if and how these genomic events may result in TERT activation. In addition to observed somatic TERT SVs, breakpoints were also identified in CLPTM1L, which is 5′ to TERT on Chr 5. Breakpoints impacting CLPTM1L have been observed in two ALM patients (Berger et al. 2012; Furney et al. 2014), and a CLPTM1L:ADCY2 translocation has also been reported in one ALM patient (Furney et al. 2014). Notably, single nucleotide polymorphisms (SNPs) within the TERT-CLPTM1L locus have been reported to influence risk of developing melanoma (Law et al. 2012) and other cancers (Carvajal-Carmona et al. 2015; Liu et al. 2015; Zhang et al. 2015; Bei et al. 2016). Furthermore, SVs identified on Chr 5p are adjacent to, or fall within, reported super enhancers (Khan and Zhang 2016; Wei et al. 2016) and may thus impact transcription. In our analyses, the same patients who garnered a CLPTM1L SV also demonstrate at least one somatic TERT aberration (SV or copy gain) to provide evidence that TERT may be impacted by events proximal to the gene. Notably, corresponding expressed RNA fusions were not detected for identified SVs impacting TERT, with the exception that patient 12, whose tumor demonstrated a complex rearrangement encompassing TERT, also expressed a TERT:ADCY2 fusion. Overall, all patients with somatic TERT events showed expression of TERT, which is reported to be overexpressed in >90% of cancers (Kim et al. 1994; Shay and Bacchetti 1997). Importantly, normal cells typically lack telomerase activity and bear 0.004 RNA molecules on average, whereas in tumor cells, 0.2 TERT transcripts can lead to activation, although tumors may express hundreds of TERT transcripts per cell (Yi et al. 2001; Akıncılar et al. 2015). Lastly, our observation that TERT inhibition (in the context of TERT promoter mutation or CNV gain) successfully decreases ALM cell viability provides evidence that targeting TERT under aberrant conditions may be efficacious in ALM. This novel finding is particularly relevant given that TERT has been described as a driver in CM but has not been appreciated as a potential driver in ALM.
Although ALM is defined as occurring on sun-shielded plantar locations, its etiological relationship to sun-shielded CM has been unclear. Characterization of ALM thus simultaneously establishes a foundation for understanding tumorigenic mechanisms in this rare subtype as well as supports the development and identification of efficacious treatment options for patients. In doing so, the unveiling of non-UV-derived drivers and oncogenic mechanisms may lend insight into other cancers, beyond sun-shielded CM, and may ultimately also benefit these patients. Based on our findings of TERT alterations in nearly half of ALMs analyzed here—and while further functional studies are necessary to verify the impact of such aberrations on TERT activity—TERT inhibitors represent a putative therapeutic strategy in ALM. This finding parallels recent work showing that pharmacological repression of TERT expression in melanoma cells leads to cell death (Kang et al. 2016). This is especially important since ALM has a limited number of targeted treatment options. Continued characterization of ALM and evaluation of the functional implications of TERT aberrations hold promise for paving an avenue toward improving outcomes for ALM patients.
Methods
Patient enrollment and consent
Patients in this study were enrolled from either Vanderbilt University or the Memorial Sloan-Kettering Cancer Center (MSKCC). At Vanderbilt and MSKCC, all patients were consented on a protocol approved by the institutional review board (IRB). Samples were obtained in accordance with standard biopsy or surgical procedures. Tissue was selected by the pathologist to limit the amount of necrotic tissue and placed into a vial and submerged into a liquid nitrogen container.
Sample collection
For MSKCC cases, adjacent normal tissue was collected, and for Vanderbilt cases, a tube of blood was collected during routine blood drawing, for DNA extraction of germline DNA. An H+E slide was made to confirm normal or malignant tissue and <50% necrosis, which was reviewed by the pathologist.
For three patients, multiple tumors were collected for analysis—for patient 25, two metastases were collected; for patient 29, the primary (29b) and two metastases (29a, 29c) were collected; and for patient 34, a primary (34b) and metastasis (34a) were collected. All specimens were fresh frozen with the exception of 34b, which was formalin-fixed and paraffin embedded (FFPE).
Next-generation sequencing and analysis
Supplemental Methods provides details on sample preparation, sequencing, data analysis, and experimental validations. In brief, paired tumor/normal whole exomes and long-insert whole-genome, as well as tumor RNA, libraries were constructed and sequenced on the Illumina HiSeq using V3 reagents. FASTQs were aligned to build 37 of the human genome using BWA (Burrows–Wheeler Aligner) (Li and Durbin 2009). Somatic variants were identified by requiring detection by two of three callers (Seurat; quality score>30) (Christoforides et al. 2013), MuTect (Cibulskis et al. 2013), and Strelka (Saunders et al. 2012). LIWG data were utilized for copy number and breakpoint detection analyses (Liang et al. 2014). A minimum tumor allele ratio of 0.10 and a minimum quality score (depth) of 20 is required for an SV to be called. For CNV detection, normalized log2 fold-changes between tumor and normal are calculated, and a smoothing window is applied. In addition, we used allele frequencies in the tumor of known heterozygous germline SNPs identified within the normal to both evaluate potential false positives and correct biases. Lastly, we applied a circular binary segmentation (CBS) algorithm to corrected log2 fold-changes using the Bioconductor DNAcopy implementation (https://bioconductor.org/packages/release/bioc/html/DNAcopy.html). RNA reads were aligned to build 37 of the human genome using STAR (Dobin et al. 2013), and differential analysis against a universal RNA control was performed using Cufflinks v2.2.1 Cuffdiff (Q-value < 0.05) (Trapnell et al. 2010, 2013) and DESeq2 (P-adjusted < 0.05) (Love et al. 2014). RNA fusions were detected using TopHat-Fusion (quality score > 100) (Kim and Salzberg 2011).
Clinical and pathologic characteristics were collected from disease-specific databases. Survival curves were generated using the date of the pathologic diagnosis until death from melanoma or date of last follow-up. Analysis was performed using SAS software (v9.4) (Supplemental Fig. S1A,B).
TERT inhibitor viability experiments
Cell lines were plated in 96-well microplates, in 100 µL culture medium. Twenty-four hours after plating cells, dimethyl sulfoxide (DMSO) vehicle, or Telomerase Inhibitor IX (EMD Millipore) was diluted in culture medium and added to wells at 100 µL/well. Cell Titer Glo (Promega) was added to cells after 72 h of drug treatment. Plates were incubated at 37°C, and luminescence was measured using a FlexStation 3 microplate reader (Molecular Devices). The Supplemental Methods provide details on quantitative PCR methods. In brief, qPCR was performed using the Kapa Fast qPCR master mix (Kapa Biosystems) and TaqMan probes.
Data access
The sequencing data from this study have been submitted to the NCBI Database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap) under accession number phs001036.v1.p1.
Supplementary Material
Acknowledgments
This publication is based on research supported by the Melanoma Research Alliance (MRA) – Hidary Foundation Team Science Award for Acral Melanoma Genomics (J. Sosman, C.E. Ariyan, and J. Trent, PI’s). Additionally, the research was supported in part by TGen, the TGen Foundation, and a Stand Up To Cancer (SU2C) – Melanoma Research Alliance Melanoma Dream Team Translational Cancer Research Grant (#SU2C-AACR-DT0612). Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research. The authors thank the patients and their families for participating in this study. We additionally thank TGen's Seungchan Kim for RNA-seq analysis feedback; Jessica Aldrich, Daniel Enriquez, and Fatima Naveed for assistance with bioinformatics; and Cassandra Lucas, Cynthia Lechuga, and Kati Koktavy (TGen) for administrative support.
Footnotes
[Supplemental material is available for this article.]
Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.213348.116.
References
- Akıncılar SC, Low KC, Liu CY, Yan TD, Oji A, Ikawa M, Li S, Tergaonkar V. 2015. Quantitative assessment of telomerase components in cancer cell lines. FEBS Lett 589: 974–984. [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bei JX, Su WH, Ng CC, Yu K, Chin YM, Lou PJ, Hsu WL, McKay JD, Chen CJ, Chang YSD, et al. 2016. A GWAS meta-analysis and replication study identifies a novel locus within CLPTM1L/TERT associated with nasopharyngeal carcinoma in individuals of Chinese ancestry. Cancer Epidemiol Biomarkers Prev 25: 188–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello DM, Chou JF, Panageas KS, Brady MS, Coit DG, Carvajal RD, Ariyan CE. 2013. Prognosis of acral melanoma: a series of 281 patients. Ann Surg Oncol 20: 3618–3625. [DOI] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, et al. 2012. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 485: 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford PT, Goldstein AM, McMaster ML, Tucker MA. 2009. Acral lentiginous melanoma: incidence and survival patterns in the United States, 1986-2005. Arch Dermatol 145: 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brash DE. 2015. UV signature mutations. Photochem Photobiol 91: 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. 2015. Genomic classification of cutaneous melanoma. Cell 161: 1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Carmona LG, O'Mara TA, Painter JN, Lose FA, Dennis J, Michailidou K, Tyrer JP, Ahmed S, Ferguson K, Healey CS, et al. 2015. Candidate locus analysis of the TERT–CLPTM1L cancer risk region on chromosome 5p15 identifies multiple independent variants associated with endometrial cancer risk. Hum Genet 134: 231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff KA, Bordone L, Horst B, Simon K, Twadell W, Lee K, Cohen JA, Wang S, Silvers DN, Brunner G, et al. 2009. GAB2 amplifications refine molecular classification of melanoma. Clin Cancer Res 15: 4288–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforides A, Carpten JD, Weiss GJ, Demeure MJ, Von Hoff DD, Craig DW. 2013. Identification of somatic mutations in cancer through Bayesian-based analysis of sequenced genome pairs. BMC Genomics 14: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Brocker EB, LeBoit PE, et al. 2005. Distinct sets of genetic alterations in melanoma. N Engl J Med 353: 2135–2147. [DOI] [PubMed] [Google Scholar]
- Curtin JA, Busam K, Pinkel D, Bastian BC. 2006. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol 24: 4340–4346. [DOI] [PubMed] [Google Scholar]
- de Lima Vazquez V, Vicente AL, Carloni A, Berardinelli G, Soares P, Scapulatempo C, Martinho O, Reis RM. 2016. Molecular profiling, including TERT promoter mutations, of acral lentiginous melanomas. Melanoma Res 26: 93–99. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furney SJ, Turajlic S, Stamp G, Thomas JM, Hayes A, Strauss D, Gavrielides M, Xing W, Gore M, Larkin J, et al. 2014. The mutational burden of acral melanoma revealed by whole-genome sequencing and comparative analysis. Pigment Cell Melanoma Res 27: 835–838. [DOI] [PubMed] [Google Scholar]
- Griewank KG, Murali R, Puig-Butille JA, Schilling B, Livingstone E, Potrony M, Carrera C, Schimming T, Möller I, Schwamborn M, et al. 2014. TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. J Natl Cancer Inst 106: dju246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. 2012. A landscape of driver mutations in melanoma. Cell 150: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. 2013. TERT promoter mutations in familial and sporadic melanoma. Science 339: 959–961. [DOI] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. 2013. Highly recurrent TERT promoter mutations in human melanoma. Science 339: 957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G. 2016. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165: 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DB, Peng C, Abramson RG, Ye F, Zhao S, Wolchok JD, Sosman JA, Carvajal RD, Ariyan CE. 2015. Clinical activity of ipilimumab in acral melanoma: a retrospective review. Oncologist 20: 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Cui Y, Yin H, Scheid A, Hendricks WP, Schmidt J, Sekulic A, Kong D, Trent JM, Gokhale V, et al. 2016. A pharmacological chaperone molecule induces cancer cell death by restoring tertiary DNA structures in mutant hTERT promoters. J Am Chem Soc 5: 5. [DOI] [PubMed] [Google Scholar]
- Khan A, Zhang X. 2016. dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res 44: D164–D171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Salzberg SL. 2011. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 12: R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015. [DOI] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, et al. 2012. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet 44: 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law MH, Montgomery GW, Brown KM, Martin NG, Mann GJ, Hayward NK, MacGregor S. 2012. Meta-analysis combining new and existing data sets confirms that the TERT–CLPTM1L locus influences melanoma risk. J Invest Dermatol 132: 485–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Aldrich J, Tembe W, Kurdoglu A, Cherni I, Phillips L, Reiman R, Baker A, Weiss GJ, Carpten JD, et al. 2014. Long insert whole genome sequencing for copy number variant and translocation detection. Nucleic Acids Res 42: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liau JY, Tsai JH, Jeng YM, Chu CY, Kuo KT, Liang CW. 2014. TERT promoter mutation is uncommon in acral lentiginous melanoma. J Cutan Pathol 41: 504–508. [DOI] [PubMed] [Google Scholar]
- Liu SG, Ma L, Cen QH, Huang JS, Zhang JX, Zhang JJ. 2015. Association of genetic polymorphisms in TERT-CLPTM1L with lung cancer in a Chinese population. Genet Mol Res 14: 4469–4476. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CC, Jubb AM, Jakubiak D, Zhou W, Rudolph J, Haverty PM, Kowanetz M, Yan Y, Tremayne J, Lisle R, et al. 2013. P21-activated kinase 1 (PAK1) as a therapeutic target in BRAF wild-type melanoma. J Natl Cancer Inst 105: 606–607. [DOI] [PubMed] [Google Scholar]
- Radu M, Semenova G, Kosoff R, Chernoff J. 2014. PAK signalling during the development and progression of cancer. Nat Rev Cancer 14: 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, ed. 1976. New concepts in surgical pathology of the skin. John Wiley, New York. [Google Scholar]
- Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. 2012. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics 28: 1811–1817. [DOI] [PubMed] [Google Scholar]
- Sauter ER, Yeo UC, von Stemm A, Zhu W, Litwin S, Tichansky DS, Pistritto G, Nesbit M, Pinkel D, Herlyn M, et al. 2002. Cyclin D1 is a candidate oncogene in cutaneous melanoma. Cancer Res 62: 3200–3206. [PubMed] [Google Scholar]
- Shay JW, Bacchetti S. 1997. A survey of telomerase activity in human cancer. Eur J Cancer 33: 787–791. [DOI] [PubMed] [Google Scholar]
- Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS. 2014. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371: 2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turajlic S, Furney SJ, Lambros MB, Mitsopoulos C, Kozarewa I, Geyer FC, Mackay A, Hakas J, Zvelebil M, Lord CJ, et al. 2012. Whole genome sequencing of matched primary and metastatic acral melanomas. Genome Res 22: 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM. 2015. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350: 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Zhang S, Shang S, Zhang B, Li S, Wang X, Wang F, Su J, Wu Q, Liu H, et al. 2016. SEA: a super-enhancer archive. Nucleic Acids Res 44: D172–D179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Shay JW, Wright WE. 2001. Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res 29: 4818–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Zhao J, Xu J, Liu F, Xu Y, Bu X, Dai C, Song C. 2015. Genetic variations in the TERT and CLPTM1L gene region and gastrointestinal stromal tumors risk. Oncotarget 6: 31360–31367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Dutton-Regester K, Brown KM, Hayward NK. 2016. The genomic landscape of cutaneous melanoma. Pigment Cell Melanoma Res 29: 266–283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.