Abstract
Liver resection is still the most commonly used therapeutic treatment for hepatocellular carcinoma (HCC), and liver regeneration promotes HCC growth in the regenerating liver. The high recurrence/metastasis of HCC is the main cause of death for HCC patients after liver resection. However, how the augmented growth and metastasis of residual HCC induced by the promoted liver regeneration following liver resection can be abolished remains unclear. In this study, a rat model with liver cirrhosis and diffused HCC was established by administration of diethylnitrosamine. Recombinant miR‐203 adenovirus was administered to induce hepatic miR‐203 overexpression and 30% partial hepatectomy (PH) followed. The effect of miR‐203 on the proliferation, invasion and metastasis of the residual HCC in the remnant cirrhotic liver with promoted regeneration was investigated. We found that the basic spontaneous regeneration of the non‐tumorous liver by 30% PH promoted proliferation, invasion and lung metastasis of the hepatic residual HCC. miR‐203 overexpression further promoted the regeneration of the non‐tumorous liver by upregulating Ki67 expression and enhancing IL‐6/SOCS3/STAT3 pro‐proliferative signals. Importantly, miR‐203 overexpression markedly inhibited the proliferation, invasion and metastasis of hepatic residual HCC through suppressing expression of Ki67, CAPNS1 and lung metastasis. Moreover, it was found that miR‐203 overexpression reversed the epithelial–mesenchymal transition induced by hepatectomy through targeting IL‐1β, Snail1 and Twist1. In conclusion, our results suggested that miR‐203 overexpression inhibited the augmented proliferation and lung metastasis of the residual HCC induced by the promoted liver regeneration following PH partly by regulating epithelial–mesenchymal transition.
Keywords: Hepatectomy, hepatocellular carcinoma, liver regeneration, miR‐203 overexpression, proliferation and metastasis
Hepatocellular carcinoma (HCC), a high mortality disease, is the third leading cause of cancer‐related death worldwide, ranked only after gastric cancer and esophagus cancer.1, 2, 3 Liver resection is the most commonly used therapeutic approach for HCC at present. However, the high postoperative recurrence is an important risk factor of poor prognosis, and liver regeneration promotes HCC growth in the regenerating liver.4 However, liver regeneration is usually initiated after partial hepatectomy (PH), and so the proliferation of the possible residual HCC in the regenerated remnant liver can be accelerated. Unfortunately, how the augmented growth and metastasis of residual HCC induced by the promoted liver regeneration following liver resection can be abolished remains unclear. Therefore, the exploration of new treatments that could inhibit the augmented proliferation and metastasis of HCC induced by liver regeneration after liver resection is urgently needed.
Cumulative evidence indicates that miRNA serve as tumor promoters or suppressors, regulating a wide range of biologic processes, such as proliferation, apoptosis, invasion and metastasis. In particular, it has been demonstrated that miR‐203 serves as a potential tumor suppressor in HCC.5, 6, 7, 8, 9, 10 In investigating the clinical implications and biological relevance of aberrant expression of miR‐203 in HCC, higher miR‐203 expression was suggested to predict better prognosis.7, 9 In HCC cell line experiments, miR‐203 overexpression inhibited growth and metastasis of HCC, with potential targets including surviving, EZH2, Bmi‐1 and ATP binding cassette E1.5, 6, 8 da Silva et al.11 report that miR‐203 acted as a positive intermediary in A20‐enhanced IL‐6/STAT3 pro‐proliferative signals by downregulating SOCS3, which may promote liver regeneration and repair after PH. Our previous study also confirmed that miR‐203 overexpression could directly promote the initiation of cirrhotic liver regeneration by targeting SOCS3 and then enhancing IL‐6/STAT3 signaling pathway after 70% PH in CCl4 induced liver cirrhosis in an animal model.12 Whether miR‐203 can simultaneously inhibit HCC progression while promoting liver regeneration following liver resection for HCC needs to be investigated.
In the present study, we established a rat model with liver cirrhosis and diffused HCC to explore the effect of miR‐203 on proliferation, invasion and metastasis of the residual HCC in the remnant non‐tumorous cirrhotic liver with promoted regeneration after hepatectomy.
Materials and Methods
Animal model
Male Wistar rats (obtained from Chengdu Dossy Experimental Animals, Jianyang, China), weighing 180−220 g, were housed with a 12‐h light–dark cycle and provided a standard laboratory diet and clean water ad libitum before and after surgery. The present study was approved by the Animal Studies Committee of West China Hospital, Sichuan University (Chengdu, China), and all procedures were performed following the Guidelines for the Care and Use of Laboratory Animals of Sichuan University and the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. All operations were performed under ether anesthesia.
Before surgery, 70 rats were allowed to acclimatize for 1 week. The hind limbs of rats were injected subcutaneously (0.3 mL/100 g of body weight, twice a week) with CCl4 (compound of neutral rapeseed oil and CCl4 at 1:1 ratio) with a rest for 2 weeks after four injections. Diethylnitrosamine (DEN, Sigma, Los Angeles, CA, USA; formulated using DEN and sterile water at a concentration of 95 μg/mL, and refreshed once a day) was freely available from the 15th day post the final injection of CCl4 for 12 weeks. One week after the final administration of DEN, 10 rats were randomly selected and sacrificed with ether anesthesia before checking for liver cirrhosis and HCC. The venous blood was harvested for liver function analysis and alpha‐fetoprotein (AFP) detection, and the liver tissues were fixed with 4% paraformaldehyde for HE staining.
Following confirmation of the liver cirrhosis and HCC (Fig. 1a), 10 days after the final administration of DEN, rats with high activity were selected and randomly divided into four groups: an miR‐203 transfection and PH group (n = 12), an miR‐negative control (miR‐NC) transfection and PH group (n = 12), a PH group (n = 12), and a sham group (n = 16). Rats of miR‐203+PH group were injected adenovirus containing miR‐203 overexpression (bought from Neuron Biotech Co., Ltd., Shanghai, China) through the penis vein (volume of adenovirus is 50 μL, concentration of adenovirus is 5 × 109). Recombinant miR‐203 adenovirus was labeled by GFP. Rats in the miR‐NC+PH group were injected with equal volume of negative control adenovirus. The rats in the other two groups were injected with equal volume of saline. Four days after adenovirus transfection, a 30% partial hepatectomy was performed in rats in the miR‐203+PH group, the miR‐NC+PH group and the PH group. PH was performed as described previously.13 In brief, hilar vessels of the left lobe of the liver were ligated and the ligated lobe was subsequently excised. Portions of the hepatic parenchyma equal to 30% of the total liver were removed in this manner. Liver tissues of the left lobe were harvested as specimens of postoperative day 0 (POD 0). Eight rats from the sham group were killed and liver tissues of the left lobe were harvested; the rest of the rats from the sham group only underwent abdominal incision and gentle manipulation of the liver (Fig. 1b).
Figure 1.
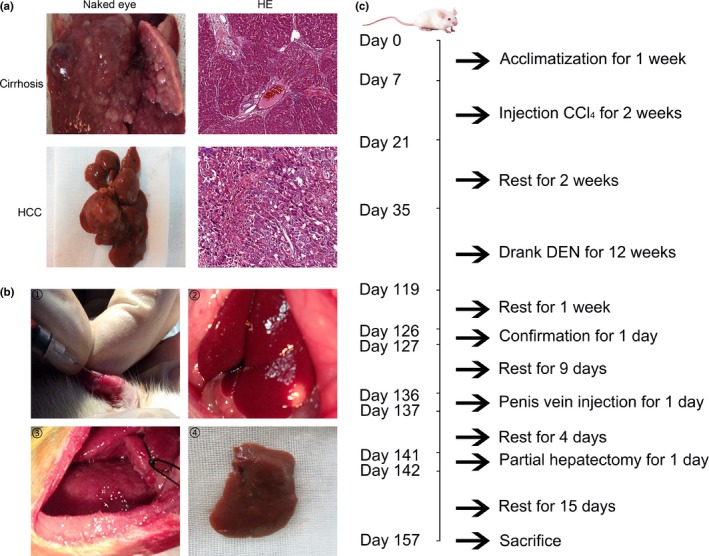
Confirmation of rat model with cirrhosis and hepatocellular carcinoma (HCC) and experimental manipulation. (a) Images of hepatic cirrhosis and HCC confirmed by naked eye. HE from representative cases are shown (HE original magnification, 200×). (b) Interference of adenovirus injection and partial hepatectomy were performed. ① Rats of the miR‐203+PH group were injected with recombinant miR‐203 overexpression adenovirus through the penis vein. ② HCC nodules were observed after abdominal incision. ③ Partial hepatectomy was performed in the rats of the miR‐203+PH, miR‐NC+PH, and PH groups. ④ Liver specimen of left lobe resected by partial hepatectomy. (c) Time course of the animal model experiment.
All rats were killed on the 15th day after partial hepatectomy (POD 15); blood samples of abdominal aorta were harvested, partial tumor tissues from liver and lung were fixed with 4% paraformaldehyde for HE and immunohistochemistry (IHC) staining, the other tumor tissues from liver were harvested for qRT‐PCR assay and western blot check, and partial non‐tumor cirrhotic tissues were fixed with 4% paraformaldehyde for IHC staining. The time course of the animal model experiment is illustrated in Figure 1c.
Analysis of liver function, alpha‐fetoprotein and HE
Liver function was tested using a Fully Automatic Biochemical Analyzer (BS‐120; Mindray, Shenzhen, China). The AFP of peripheral blood was tested by ELISA (SEA153Ra 96T assay kit; Cloud‐Clone, Shanghai, China). Pathological examination for hepatic cirrhosis and HCC was performed by staining with HE. Pathological grades of HCC were evaluated by Edmondson–Steiner stage.14
Detection of hepatic adenovirus transfection
Hepatic miR‐203 expression on the POD 0 and POD 15 was analyzed with qRT‐PCR. Liver tissue of POD 0 were embedded in frozen blocks and cut into 4‐μm sections. DAPI (Sigma), at the concentration of 1 g/mL, was included for the identification of nuclei, and GFP was examined with a fluorescence microscope (ECLIPSE 80i; Nikon, Sendai, Japan).
Quantitative RT‐PCR analysis for miR‐203 and mRNA expression
Total RNA was isolated from the liver tissues using the TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions, and the first strand cDNA was synthesized using a miRNA 1st‐Strand cDNA Synthesis Kit (Shanghai Newgene Biosciences, Shanghai, China) according to the manufacturer's instructions. Quantitative real‐time PCR analyses for mRNA of miR‐203 and β‐actin were conducted with Power SYBR Master Mix (Invitrogen). miRNA levels were quantified using ViiA7 (Applied Biosystems, Foster City, CA, USA) and the 2−ΔΔct relative quantification method. All primers used are listed in Table 1.
Table 1.
Primer sequence used in this study
| Name | Sequence |
|---|---|
| miR‐203 | 5′‐CAGCGGGTGAAATGTTTAGGAC‐3′ (forward) |
| 5′‐AGTGCAGGGTCCGAGGT‐3′ (reverse) | |
| E‐cadherin | 5′‐AAGGTCAGCCTGGTATAGTAAG‐3′ (forward) |
| 5′‐TTCCCATCAGAGTGGTGTTT‐3′ (reverse) | |
| Vimentin | 5′‐ATGTCCGCCAGCAGTATGA‐3′ (forward) |
| 5′‐CACTTCGCAGGTGAGTGA‐3′ (reverse) | |
| N‐cadherin | 5′‐GGCAATCCCACTTACGGC‐3′ (forward) |
| 5′‐GTTGGCTACAATGACATCCACT‐3′ (reverse) | |
| Snail1 | 5′‐CTGATGGAAGGCAGAGTGC‐3′ (forward) |
| 5′‐CCAGTGGGTTGGCTTTAGTT‐3′ (reverse) | |
| Twist1 | 5′‐AGGTCTTGCCAATCAGTCA‐3′ (forward) |
| 5′‐AGTTTGATCCCAGCGTTTT‐3′ (reverse) | |
| ZEB1 | 5′‐AAAGTGGCTGTAGATGGTAA‐3′ (forward) |
| 5′‐GAAGACTGATGGCAGAAAT‐3′ (reverse) | |
| ZEB2 | 5′‐ACCAGCGGAAACAAGGAT‐3′ (forward) |
| 5′‐CGTATTTATGTCGCAGAAGG‐3′ (reverse) |
Immunohistochemistry for detection of protein expression in situ
The tissue samples were cut into 4‐μm sections. After antigen retrieval, the sections were incubated overnight with the first antibody (PCNA, 2586, CST, 1:1000; Ki67, AB9260, millipore, 1:20; Cyclin D1, 2978, CST, 1:100; CAPNS1, HPA006872, Atlas Antibodies, 1:200; P‐STAT3, ab68653, Abcam, 1:100; SOCS3, ab16030, Abcam, 1:100). A standard two‐step immunoperoxidase‐labeled protocol with goat anti‐rat HRP (EnVision, Dako, Glostrup, Denmark) was applied stringently on all slides. Immunohistochemical analysis results was evaluated by immunohistochemistry score (IHS), HIS = A × B (A: positive cell number; B: grading of color intensity of positive cells, 0 [negative], 1 [weak positive], 2 [positive], 3 [strong positive]). Two pathologists assessed the slides without knowledge of subgroups of rats and were blinded to each other's evaluation.
Western blot for detection of protein expression
Protein expression levels were assessed by western blot. In brief, tissues (20–50 mg) were lysed in lysis buffer containing 1 PBS, 1% NP‐40, 01% SDS, 5 mmol/L EDTA, 0.5% sodium deoxycholate and 1 mmol/L of sodium orthovanadate and protease inhibitors. The target protein was then probed using antibodies (1:1000, Santa Cruz, CA, USA), and rabbit anti‐tubulin‐alpha (1:500, Santa Cruz) antibody was used to detect tubulin‐alpha, which served as an internal control.
Detection of lung metastasis
Lung metastatic tumors were confirmed by HE examination, and the incidence of lung metastasis was calculated in each group of rats.
ELISA for detection of hepatic IL‐6 level
Supernatant samples from the liver tissues were analyzed for IL‐6 using ELISA according to the manufacturer's instructions.
Detection of IL‐1β, IL‐6 and TNF‐α of rat serum
Serum levels of IL‐1β, IL‐6 and TNF‐α were tested by Magpix (RECYTMAG‐65K, Millipore, Billerica, MA, USA). In brief, all reagents were allowed to warm to room temperature (20–25°C) before use in the assay. After adding all reagents and samples into each well of the plate, run plate on MAGPIX with xPONENT software was initiated to work. The median fluorescent intensity data were saved and analyzed using a 5‐parameter logistic or spline curve‐fitting method for calculating analyte concentrations in samples.
Statistical analysis
The differences in numeric variables between groups were analyzed using one‐way analysis of variance. χ2 or Fisher's exact tests were performed to determine the ratio of Edmondson–Steiner stage and the ratio of lung metastasis. Statistical analyses and graphing were conducted using SPSS 20.0 (IBM, Chicago, IL, USA), and GraphPad Prism 5 software (GraphPad Company, La Jolla, CA, USA). P < 0.05 was considered a significant difference.
Results
Confirmation of rat model with cirrhosis and diffused hepatocellular carcinoma
Hepatic HE staining showed characteristics of cirrhosis, including disordered structure of the hepatic lobules, and formation of fibrous septum and pseudo‐lobuli. Hepatic HE staining also showed characteristics of HCC, including formation of tumor nodules, tumor cells arranged in a funicular or crumb, and pathological nuclear fission (Fig. 1a). The AFP of peripheral blood was significantly higher than normal (167.713 ± 35.629 ng/mL) above characteristics of HE examination and AFP level in peripheral blood of rat model with cirrhosis and HCC were confirmed. Liver function examination showed slightly elevated transaminases (ALT: 56.5 ± 10.9 IU/L; AST; 231.8 ± 150.6 IU/L), normal total bilirubin level (1.8 ± 0.7 μmol/L) and normal albumin level (40.2 ± 1.7 g/L). The normal range of the laboratory data is: AFP, <8 ng/mL; ALT, 33.70–98.70 IU/L; AST, 69.70–322.90 IU/L; total bilirubin, 0.07–2.04 μmol/L; albumin, 31.70–43.70 g/L. These results suggested that the liver function of rats was still in the compensatory stage.
Hepatic miR‐203 overexpression was induced by miR‐203 adenovirus transfection
Expression of GFP of liver tissues was detected by fluorescence microscope at POD 0. GFP in the miR‐203+PH group rats was significantly higher than that of the other three groups (Fig. 2a). qPCR was performed to detect the relative hepatic levels of miR‐203 at POD 0 and POD 15. Hepatic miR‐203 expression in the miR‐203+PH group rats was significantly higher than that of the other three groups, and this effect lasted to POD 15 (Fig. 2b).
Figure 2.
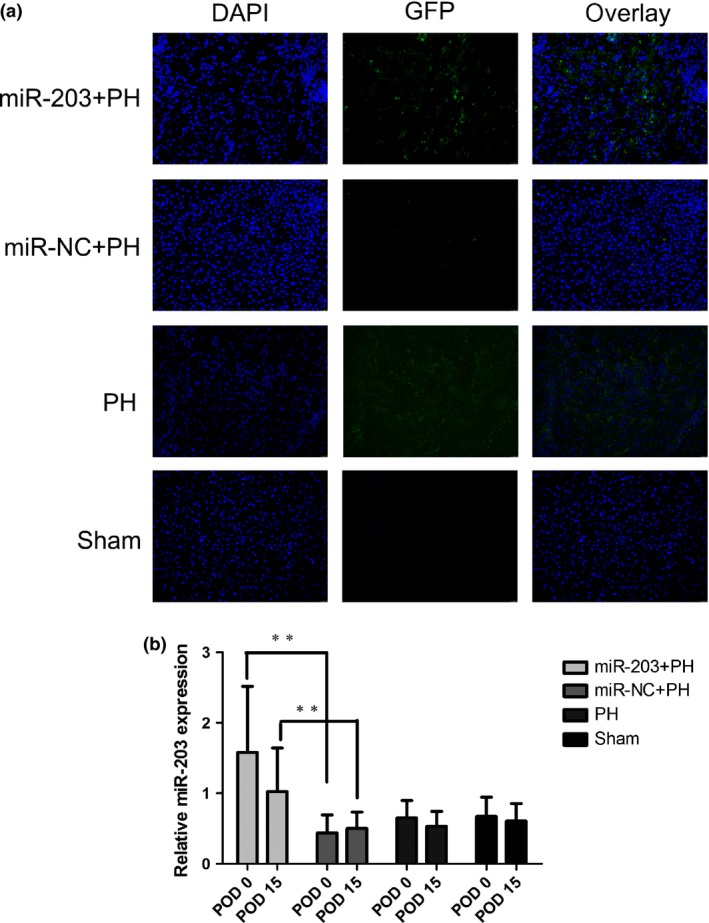
Validation of hepatic miR‐203 overexpression. (a) Expression of GFP of liver tissues of each group was detected at POD0; images from representative cases are shown. (b) qPCR was performed to detect the hepatic relative levels of miR‐203 of liver tissues of each group at POD0 and POD15 (**P<0.01).
miR‐203 overexpression promoted regeneration of the non‐tumorous remnant cirrhotic liver
PCNA, Ki67 and cyclin D1 (biomarkers of proliferation) expression were detected with IHC in the non‐tumorous remnant cirrhotic liver. Ki67 expression in the cirrhotic non‐tumor tissues was significantly higher in the PH group compared to the sham group. Ki67 expression in cirrhotic non‐tumor tissues was significantly higher in the miR‐203+PH group compared to the other three groups. Expressions of PCNA and cyclin D1 were not significantly different in the four groups (Fig. 3a,b). Hepatic IL‐6 level in cirrhotic tissues was significantly higher in the PH group than in the sham group, and reached the highest level in the miR‐203+PH group (Fig. 3c). Expression of SCOS3 in cirrhotic tissues was significantly lower in the miR‐203+PH group than in the other three groups, and expression of P‐STAT3 in cirrhotic tissues was significantly higher in the miR‐203+PH group than in the other three groups. Expressions of SCOS3 and P‐STAT3 were not significantly different between the PH and sham groups (Fig. 3d,e).
Figure 3.
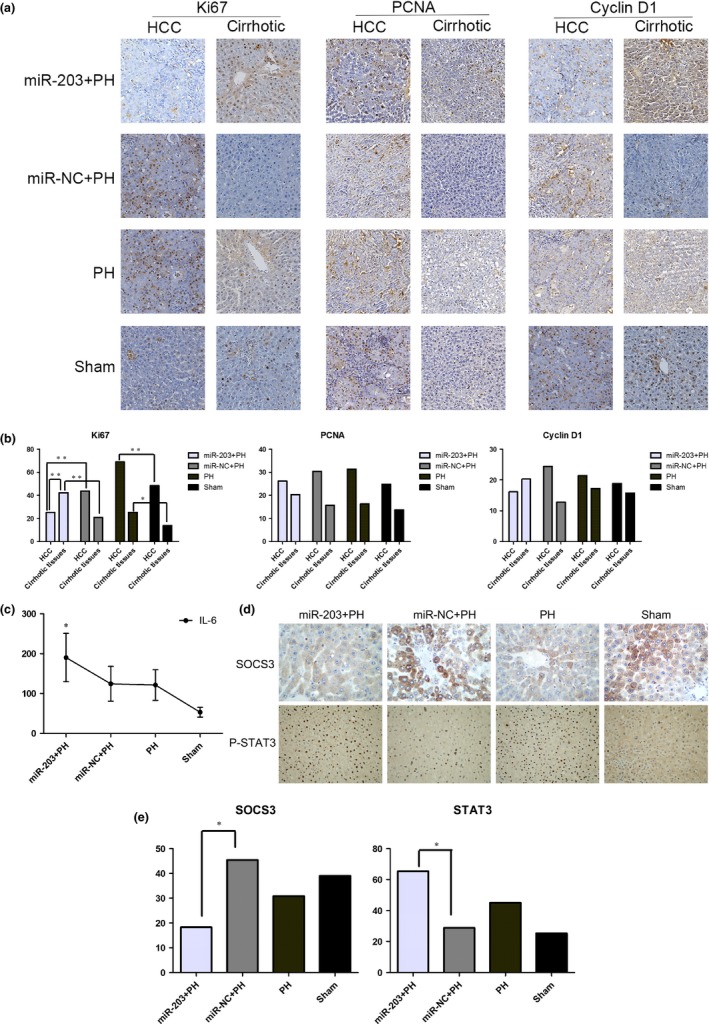
miR‐203 overexpression with two‐sided effect in hepatic residual hepatocellular carcinoma (HCC) and cirrhosis tissues. (a) Immunohistochemistry (IHC) was performed to detect PCNA, Ki67 and cyclin D1 expression in HCC and cirrhotic tissues. Images from representative cases are shown (IHC, ×200). (b) IHC scores were calculated to detect the level of PCNA, Ki67 and cyclin D1 expression (*P < 0.05; **P < 0.01). (c) Hepatic IL‐6 expression was measured by ELISA (*P < 0.05). (d) IHC was performed to detect SCOS3 and P‐STAT3 expression in cirrhotic tissues; images from representative cases are shown (SCOS3, ×400; P‐STAT3, ×100). (e) IHC scores were calculated to detect the level of SCOS3 and P‐STAT3 expression (*P < 0.05).
miR‐203 overexpression suppressed proliferation of the hepatic residual hepatocellular carcinoma
PCNA, Ki67 and cyclin D1 expression were detected in HCC with IHC. Ki67 expression in hepatic residual HCC was significantly higher in the PH group compared to the sham group. Ki67 expression in HCC tissues was significantly lower in the miR‐203+PH group compared to the other three groups. Moreover, further analysis of the miR‐203+PH group revealed that Ki67 expression was significantly higher in cirrhotic non‐tumor tissues compared to HCC tissues. Expressions of PCNA and cyclin D1 were not significantly different in the four groups (Fig. 3a,b).
miR‐203 overexpression inhibited residual hepatocellular carcinoma invasion and lung metastasis after partial hepatectomy
CAPNS1 (a biomarker of HCC invasiveness) expression was detected with IHC. CAPNS1 expression level was significantly higher in the PH group rats compared to the sham group, and significantly lower in the miR‐203+PH group compared to the miR‐NC+PH group (Fig. 4a,b). Tumor stage was judged according to Edmondson–Steiner stage. The ratios of stage I + II and stage III + IV were calculated to detect pathological progression. The ratio of the Edmondson–Steiner stage III + IV in the PH group rat was significantly higher than that of the sham group, and there was no significant difference between the miR‐203+PH group and the miR‐NC+PH group (Fig. 4c,d). Multiple tumor nodules in the lung were observed by naked eye in rats with lung metastasis. Different sized lung tumor cells accompanied by irregular hyperchromatic nuclei located in the central and abundant cytoplasm were detected by HE. The ratio of lung metastasis was significantly higher in the PH group rats compared to the sham group, whereas the ratio of lung metastasis was significantly lower in the miR‐203+PH group rat compared to the miR‐NC+PH group and the sham group (Fig. 4e,f).
Figure 4.
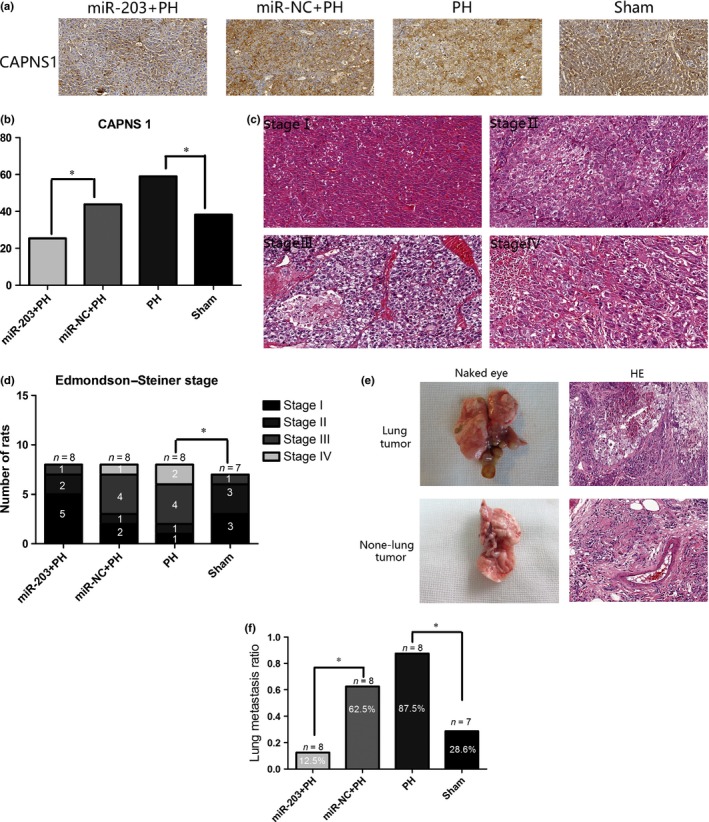
miR‐203 overexpression inhibited invasion and lung metastasis after partial hepatectomy (PH). (a) Immunohistochemistry (IHC) was performed to detect CAPNS1 expression in hepatocellular carcinoma (HCC) tissues; images from representative cases are shown (IHC, ×200). (b) IHC scores were calculated to detect the level of CAPNS1 expression (*P < 0.05). (c) Images of every stage of HCC from representative cases are shown (HE, ×200). (d) Numbers of rat divided by Edmondson–Steiner stage were counted, then the ratios of stage I + II and stage III + IV of each group were calculated (*P < 0.05). (e) Lung metastasis was confirmed by pathological examination of naked eye and HE. Images from representative cases are shown (HE, ×200). (f) Ratio of lung metastasis of each group was calculated (*P < 0.05).
miR‐203 overexpression inhibited epithelial–mesenchymal transition of the residual hepatocellular carcinoma after partial hepatectomy
The expression of E‐cadherin, vimentin and N‐cadherin were examined by qPCR and western blot. E‐cadherin is significantly lower in the PH group rats compared to the sham group and significantly higher in the miR‐203+PH group rats compared to the miR‐NC+PH group rats. Expressions of vimentin and N‐cadherin were significantly higher in the PH group compared to the sham group and significantly lower in the miR‐203+PH group compared to the miR‐NC+PH group (Fig. 5a,b).
Figure 5.
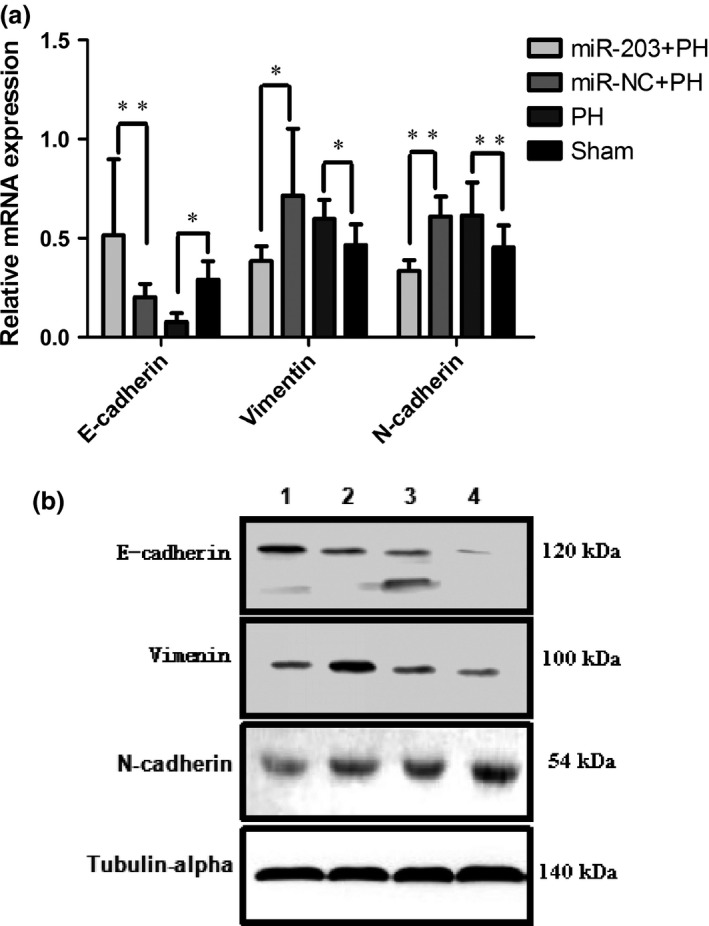
miR‐203 overexpression inhibited epithelial–mesenchymal transition (EMT) of residual hepatocellular carcinoma (HCC) after partial hepatectomy (PH). (a) qPCR was performed to detect the relative levels of E‐cadherin, vimentin and N‐cadherin of liver tissues (*P < 0.05; **P < 0.01). (b) The expression of E‐cadherin, vimentin and N‐cadherin were examined by western blot (1, miR‐203+PH; 2, miR‐NC+PH; 3, PH; 4, sham).
miR‐203 downregulated IL‐1β, Snail1 and Twist1 to inhibit epithelial–mesenchymal transition
Serum levels of IL‐1β, IL‐6 and TNF‐α were measured by Magpix. IL‐1β and IL‐6 were significantly higher in the PH group than the sham group, whereas IL‐1β was significantly lower in the miR‐203+PH group than the miR‐NC+PH group. TNF‐α was not significantly different among the four groups (Fig. 6a). The expressions of Snail1, Twist1, ZEB1 and ZEB2 were examined by qPCR and western blot. Snail1 levels were significantly higher in the PH group rat than the sham group, whereas levels of Snail1 and Twist1 were significantly lower in the miR‐203+PH group than in the miR‐NC+PH group. Levels of ZEB1 and ZEB2 were not significantly different among the four groups (Fig. 6b,c).
Figure 6.
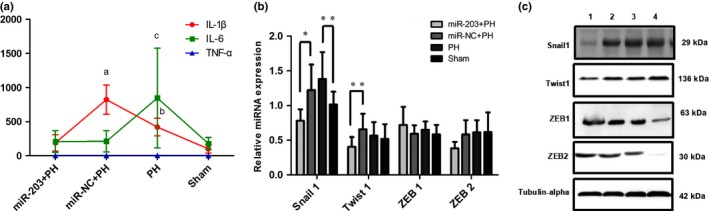
miR‐203 downregulated IL‐1β, Snail1 and Twist1 to inhibit epithelial–mesenchymal transition (EMT). (a) IL‐1β, IL‐6 and TNF‐α were measured by Magpix (a, P = 0.000, miR‐203+PH vs miR‐NC+PH; b, P = 0.000, partial hepatectomy (PH) vs sham; c, P = 0.003, PH vs sham). (b) qPCR was performed to detect the relative levels of Snail1, Twist1, ZEB1 and ZEB2 of liver tissues (*P < 0.05; **P < 0.01). (c) The expression of Snail1, Twist1, ZEB1 and ZEB2 were examined by western blot (1, miR‐203+PH; 2, miR‐NC+PH; 3, PH; 4, sham).
Discussion
In this study, rat livers were subject to a sequential process of toxic injury, cirrhosis and tumorigenesis. This pathological process is similar to the carcinogenesis of human liver cancer induced by hepatitis B virus infection. Growth and metastasis of residual HCC following whole left lobe resection in this experiment were similar to the clinical progression of HCC following radical liver resection. Given the high similarity of this rat model with the clinical progression of HCC patients, our study aims to investigate the effect of miR‐203 on the proliferation and metastasis of residual HCC following liver resection. We found that the basic spontaneous regeneration of the non‐tumorous liver by 30% PH promoted proliferation, invasion and lung metastasis of the hepatic residual HCC. miR‐203 overexpression further promoted the regeneration of the non‐tumorous liver by upregulating Ki67 expression and enhancing IL‐6/SOCS3/STAT3 pro‐proliferative signals. Importantly, miR‐203 overexpression markedly inhibited the proliferation, invasion and metastasis of hepatic residual HCC through suppressing the expression of Ki67 and CAPNS1 and lung metastasis. Moreover, it was also found that miR‐203 overexpression reversed the epithelial–mesenchymal transition (EMT) induced by hepatectomy through targeting IL‐1β, Snail1 and Twist1. Our results suggested that miR‐203 overexpression inhibited the augmented proliferation and lung metastasis of the residual HCC induced by the promoted liver regeneration after PH partly by regulating EMT. miR‐203 could acts both an inhibitor of the augmented proliferation and metastasis of residual HCC as well as a promoter of liver regeneration after liver resection.
Liver regeneration after hepatectomy has been reported to be involved in recurrence and metastasis of HCC.4 In this study, expressions of Ki67 and IL‐6 in non‐tumor cirrhotic tissues were higher in the PH group compared to the sham group; this indicated that elevated cytokines induced by hepatectomy initiated liver regeneration. Moreover, we found that hepatectomy promoted the progression of Edmondson–Steiner stage and lung metastasis, and elevated the expression of CAPNS1. These results indicated that the basic spontaneous regeneration of the non‐tumorous liver by 30% PH promoted proliferation, invasion and lung metastasis of the hepatic residual HCC. It is reported that adenovirus could induce immune response to the liver,15 resulting in damage to the liver; our results showed little discrepancy between miR‐NC+PH and PH groups. However, the damage from adenovirus is mild, so the discrepancies between the two groups were not significant.
The IL‐6/STAT3/SOCS3 pathway is a promising target for pro‐regenerative strategies. IL‐6 levels drive hepatocyte entry into cell cycle by enhancing IL‐6/STAT3 pro‐proliferative signals and downregulating SOCS3 expression.16, 17, 18, 19 da Silva et al.11 report that zinc finger protein A20 promoted liver regeneration by downregulating SOCS3 expression, likely in a miR‐203‐dependent manner; their results suggested that miR‐203 regulated A20/IL‐6/STAT3 pro‐proliferative signals in hepatocytes. In this study, the expression of Ki67 in non‐tumor cirrhotic tissues is significantly higher in the miR‐203+PH group than in the other groups, which indicated that miR‐203 accelerated liver regeneration induced by hepatectomy. We further investigated the molecular mechanism by which miR‐203 promotes proliferation activity in non‐tumor cirrhotic tissues. Here, we showed that the levels of IL‐6 and protein expression of STAT3 were upregulated and expression of SOCS3 was downregulated after miR‐203 overexpression. Consistent with the results of da Silva et al.,11 our results also suggest that miR‐203 overexpression promotes non‐tumor cirrhotic tissues regeneration by regulating IL‐6/SOCS3/STAT3 pro‐proliferative signals.
To explore the effect of miR‐203 on invasion and metastasis of the hepatic residual HCC after hepatectomy, we investigated the expression of Ki67 and CAPNS1, Edmondson–Steiner stage, and the incidence of lung metastasis. In this study, although the ratio of the Edmondson–Steiner stage III + IV in the rats in the miR‐203+PH group was not significantly lower than those in the miR‐NC+PH Group, the hepatic expressions of Ki67 and CAPNS1 were significantly inhibited by miR‐203 overexpression. We also found that miR‐203 overexpression significantly suppressed the ratio of lung metastasis. Our results suggested miR‐203 overexpression inhibited proliferation, invasion and metastasis of residual HCC.
The significant inhibitory effect of miR‐203 on residual HCC metastasis prompted us to investigate whether or not miR‐203 plays a suppressive role in EMT. EMT has been confirmed to be involved in invasion, metastasis and chemoresistance of tumors.20, 21, 22, 23, 24 miR‐203 regulates EMT by directly targeting transcriptional factors, such as Snail1, Snail2, ZEB1, ZEB2 and Twist1. Jiang et al. demonstrate a direct negative feedback loop between miR‐203 and ZEB2 participating in tumor stemness and chemotherapy resistance of nasopharyngeal carcinoma.25 Zhang et al. report that miR‐203 was epigenetically silenced, and the silencing promoted tumor cell growth and invasion at least in part by upregulating the Snai2 transcription factor in malignant breast cancer cells.27 In this study, hepatectomy downregulated expression of E‐cadherin and upregulated expression of vimentin and N‐cadherin, whereas miR‐203 overexpression upregulated E‐cadherin expression and downregulated expressions of vimentin and N‐cadherin. Moreover, production of IL‐1β and IL‐6, as well as Snail1 expression, were upregulated by hepatectomy, whereas miR‐203 overexpression decreased IL‐1β, Snail1 and Twist1 expression. Secreted cytokines have been shown to promote invasive phenotypes.28 Our results suggested that inflammation response after hepatectomy might contribute to initiation of EMT, whereas miR‐203 overexpression could reverse EMT by downregulating expression of IL‐1β, Snail1 and Twist1. Of note, although our results showed that miR‐203 inhibited the expression of IL‐1β, Snail1 and Twist1, the inhibitory effect of miR‐203 was modest. Our future research will explore whether there are additional mechanisms underlying the inhibitory effect of miR‐203 on the HCC invasion and metastasis.
In conclusion, our results suggested that miR‐203 overexpression could inhibit the augmented proliferation and lung metastasis of the residual HCC induced by the promoted liver regeneration after PH partly by regulating EMT. To our knowledge, this is the first report on the two‐sided effect of miR‐203. The results demonstrate the feasibility and potential value of its application in clinical cirrhotic HCC patients.
Disclosure Statement
The authors have no conflict of interest to declare.
Acknowledgments
We thank Dr Li Li and Dr Fei Chen (Department of Pathology, West China Hospital, Sichuan University), who reviewed the HE and IHC staining. The authors thank Dr Yun Yang (Department of Biology, West China Hospital, Sichuan University) for his kind support with western blot analysis. This study was supported by grants from the National Natural Science Foundation of China (No. 71673193) and the Key Technology Research and Development Program of Sichuan Province (2015SZ0131).
Cancer Sci 108 (2017) 338–346
Funding Information
This study was supported by grants from the National Natural Science Foundation of China (No. 71673193) and the Key Technology Research and Development Program of Sichuan Province (2015SZ0131).
References
- 1. Fan ST, Lai EC, Lo CM, Ng IO, Wong J. Hospital mortality of major hepatectomy for hepatocellular carcinoma associated with cirrhosis. Arch Surg 1995; 130: 198–203. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 3. Corpechot C, Barbu V, Wendum D et al Hepatocyte growth factor and c‐Met inhibition by hepatic cell hypoxia: a potential mechanism for liver regeneration failure in experimental cirrhosis. Am J Pathol 2002; 160: 613–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shi JH, Huitfeldt HS, Suo ZH, Line PD. Growth of hepatocellular carcinoma in the regenerating liver. Liver Transpl 2011; 17: 866–74. [DOI] [PubMed] [Google Scholar]
- 5. Wei W, Wanjun L, Hui S, Dongyue C, Xinjun Y, Jisheng Z. miR‐203 inhibits proliferation of HCC cells by targeting survivin. Cell Biochem Funct 2013; 31: 82–5. [DOI] [PubMed] [Google Scholar]
- 6. Yang F, Lv LZ, Cai QC, Jiang Y. Potential roles of EZH2, Bmi‐1 and miR‐203 in cell proliferation and invasion in hepatocellular carcinoma cell line Hep3B. World J Gastroenterol 2015; 21: 13268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen HY, Han ZB, Fan JW et al miR‐203 expression predicts outcome after liver transplantation for hepatocellular carcinoma in cirrhotic liver. Med Oncol 2012; 29: 1859–65. [DOI] [PubMed] [Google Scholar]
- 8. Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR‐124 and miR‐203 are epigenetically silenced tumor‐suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis 2010; 31: 766–76. [DOI] [PubMed] [Google Scholar]
- 9. Liu Y, Ren F, Rong M, Luo Y, Dang Y, Chen G. Association between underexpression of microrna‐203 and clinicopathological significance in hepatocellular carcinoma tissues. Cancer Cell Int 2015; 15: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang F, Qiang Y, Zhu L et al MicroRNA‐7 downregulates the oncogene VDAC1 to influence hepatocellular carcinoma proliferation and metastasis. Tumour Biol 2016; 37: 10235–46. [DOI] [PubMed] [Google Scholar]
- 11. da Silva CG, Studer P, Skroch M et al A20 promotes liver regeneration by decreasing SOCS3 expression to enhance IL‐6/STAT3 proliferative signals. Hepatology 2013; 57: 2014–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen XB, Zheng XB, Chai ZX et al MicroRNA‐203 promotes liver regeneration after partial hepatectomy in cirrhotic rats. J Surg Res 2017; 211: 53–63. [DOI] [PubMed] [Google Scholar]
- 13. Aalbers AG, ten Kate M, van Grevenstein WM et al A small mammal model of tumour implantation, dissemination and growth factor expression after partial hepatectomy. Eur J Surg Oncol 2008; 34: 469–75. [DOI] [PubMed] [Google Scholar]
- 14. Zhou L, Rui JA, Ye DX, Wang SB, Chen SG, Qu Q. Edmondson‐Steiner grading increases the predictive efficiency of TNM staging for long‐term survival of patients with hepatocellular carcinoma after curative resection. World J Surg 2008; 32: 1748–56. [DOI] [PubMed] [Google Scholar]
- 15. Kota J, Chivukula RR, O'Donnell KA et al Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009; 137: 1005–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Campbell JS, Prichard L, Schaper F et al Expression of suppressors of cytokine signaling during liver regeneration. J Clin Invest 2001; 107: 1285–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huda KA, Guo L, Haga S et al Ex vivo adenoviral gene transfer of constitutively activated STAT3 reduces post‐transplant liver injury and promotes regeneration in a 20% rat partial liver transplant model. Transpl Int 2006; 19: 415–23. [DOI] [PubMed] [Google Scholar]
- 18. Riehle KJ, Campbell JS, McMahan RS et al Regulation of liver regeneration and hepatocarcinogenesis by suppressor of cytokine signaling 3. J Exp Med 2008; 205: 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wustefeld T, Rakemann T, Kubicka S, Manns MP, Trautwein C. Hyperstimulation with interleukin 6 inhibits cell cycle progression after hepatectomy in mice. Hepatology 2000; 32: 514–22. [DOI] [PubMed] [Google Scholar]
- 20. Eger A, Aigner K, Sonderegger S et al DeltaEF1 is a transcriptional repressor of E‐cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 2005; 24: 2375–85. [DOI] [PubMed] [Google Scholar]
- 21. Kalluri R, Weinberg RA. The basics of epithelial–mesenchymal transition. J Clin Invest 2009; 119: 1420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 2007; 7: 415–28. [DOI] [PubMed] [Google Scholar]
- 23. Winter JM, Ting AH, Vilardell F et al Absence of E‐cadherin expression distinguishes noncohesive from cohesive pancreatic cancer. Clin Cancer Res 2008; 14: 412–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zeisberg M, Neilson EG. Biomarkers for epithelial–mesenchymal transitions. J Clin Invest 2009; 119: 1429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang Q, Zhou Y, Yang H et al A directly negative interaction of miR‐203 and ZEB2 modulates tumor stemness and chemotherapy resistance in nasopharyngeal carcinoma. Oncotarget 2016; 7: 67288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang Z, Zhang B, Li W et al Epigenetic silencing of miR‐203 upregulates SNAI2 and contributes to the invasiveness of malignant breast cancer cells. Genes Cancer 2011; 2: 782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sullivan NJ, Sasser AK, Axel AE et al Interleukin‐6 induces an epithelial–mesenchymal transition phenotype in human breast cancer cells. Oncogene 2009; 28: 2940–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tachibana S, Zhang X, Ito K et al Interleukin‐6 is required for cell cycle arrest and activation of DNA repair enzymes after partial hepatectomy in mice. Cell Biosci 2014; 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]


