Abstract
The high overall genetic homology between human and rhesus macaques, coupled with the phenotypic conservation of lymphocyte populations, highlights the potential utility of non-human primates (NHPs) for the preclinical evaluation of vaccine candidates. For HIV-1, experimental models are needed to identify vaccine regimens capable of eliciting desired immune responses, such as broadly neutralizing antibodies. One important neutralization target on the HIV-1 envelope glycoproteins (Env) is the conserved primary CD4 receptor binding site (CD4bs). The isolation and characterization of CD4bs-specific neutralizing monoclonal antibodies (MAbs) from HIV-1 infected individuals has provided insights into how broadly reactive antibodies target this conserved epitope. In contrast, and for reasons that are not understood, current Env immunogens elicit CD4bs-directed antibodies with limited neutralization breadth. To facilitate the use of the NHP model to address this and other questions relevant to human humoral immunity, we defined features of the rhesus macaque immunoglobulin (Ig) loci and compared these to the human Ig loci. We then studied Env immunized rhesus macaques, identified single B-cells expressing CD4bs-specific antibodies, and sequenced and expressed a panel of functional MAbs. Comparison of vaccine-elicited MAbs with HIV-1 infection-induced MAbs revealed differences in the degree of somatic hypermutation of the Abs, as well as in the fine specificities targeted within the CD4bs. These data support the use of the preclinical NHP model to characterize vaccine-induced B cell responses at high resolution.
INTRODUCTION
The extreme variation of the HIV-1 envelope glycoproteins (Env) provides a major hurdle for developing a protective vaccine. So far, clinical Env immunization trials have resulted in the elicitation of antibodies with limited neutralization breadth (1, 2). In contrast, some chronically HIV-infected individuals develop remarkably potent and broad serum neutralizing activity (reviewed in (3)), suggesting that the human immune system is capable of generating such responses if exposed to Env during years of active viral replication. Once broad and potent neutralizing responses appear, a single or a few specificities can account for much of the neutralizing capacity present in the polyclonal serum of these individuals (4–6).
The ontogeny of neutralizing Ab responses elicited during chronic HIV-1 infection is under active investigation (7–11) and multiple monoclonal antibodies (MAbs) from individuals displaying broadly neutralizing serological activity are now described (9, 10, 12, 13). Several of these MAbs are directed against the CD4 binding site (CD4bs), a functionally conserved region spanning the inner and outer domains of gp120. However, CD4bs-directed Abs vary in their capacity to mediate broadly neutralizing responses; for example MAb b12 displays considerably broader neutralizing activity than MAb b13, despite recognizing overlapping epitopes (14). Recent studies reveal that extensive somatic hypermutation (SHM) of CD4bs-directed MAbs, which are as high as 30% divergent from the germline sequence, is required for efficient neutralization of primary HIV-1 isolates. Furthermore, many of the broadly neutralizing CD4bs-directed Abs display a restricted variable heavy chain (VH) gene usage (VH1-2*02) (9, 10, 15). Whether these features are required for broadly neutralizing CD4bs-directed Ab activity is not known. In contrast, the origin and evolution of CD4bs-directed Ab responses elicited by subunit Env vaccination have not been elucidated. Since existing HIV-1 vaccine candidates do not elicit broadly neutralizing Abs, it is important to define the Ab specificities elicited by candidate Env immunogens to inform the design of regimens that more successfully promote Ab responses against relevant neutralizing determinants, such as the conserved CD4bs.
The CD4bs is shielded by highly immunogenic elements that tolerate extreme variability designated variable regions V1–V5 (16). The V regions not only dominate the humoral immune response during natural infection, but also during Env immunization (17). Clearly, HIV-1 has evolved mechanisms to occlude critical functional elements of Env from Ab recognition. We recently demonstrated that CD4bs-directed Abs capable of neutralizing selected HIV-1 isolates are elicited in rhesus macaques immunized with soluble gp140-F trimers (18). This provided an opportunity to isolate CD4bs-directed MAbs from macaque memory B cells and to investigate the genetic and functional properties of such Abs to gain insights into how the HIV-1 Env CD4bs is seen by the host immune system in the context of vaccination.
Here, we report the isolation of a panel of Env vaccine-elicited CD4bs-directed macaque MAbs and the definition of genetic and functional features that distinguish these Abs from broadly neutralizing CD4bs-directed MAbs produced during chronic HIV-1 infection. Furthermore, we describe a comparative analysis of the human and rhesus macaque Ig loci to illustrate their close genetic relationship as a basis for further B cell studies in rhesus macaques. The similarities of the immunogenetics and B cell biology between humans and macaques suggest that the information gained from the approaches described here can be used to guide specific modifications to current Env immunization regimens to accelerate clinical HIV-1 vaccine studies as well as to study B cell repertoires and affinity maturation of vaccine-elicited B cell responses in general.
RESULTS
The rhesus and human heavy chain Ig loci are highly homologous
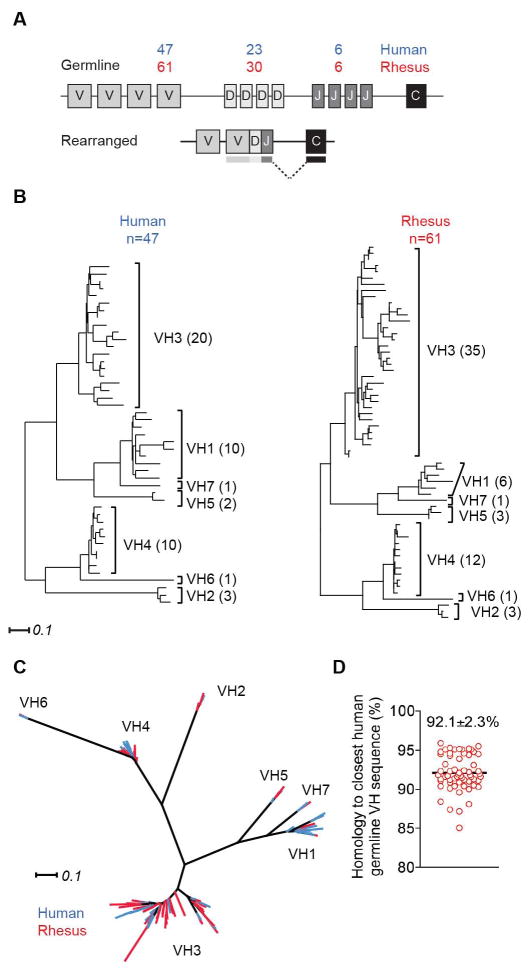
The full genome of an Indian rhesus macaque was published in 2007, but the assembly and annotation of the Ig loci are so far incomplete (19). To mine the rhesus Ig loci for possible V(D)J segments, we used the online IMGT®/LIGMotif tool developed by the international immunogenetics information system® (IMGT®). IMGT®/LIGMotif searches the submitted sequences for patterns of short defined conserved sequences specific for immunoglobulin or T cell receptor genes. We also used previously published rhesus germline sequences (20–25) to blast the genome for potential V(D)J exons. We determined that the Ig heavy chain locus covering the rhesus VDJ genes was situated on the distal part of the long arm of chromosome 7 from base pair 169,100,000 to the telomere. An additional 10 contigs of less certain chromosomal location containing variable (VH)-like genes were also identified. In total, the IMGT®/LIGMotif tool found 124 proposed rhesus macaque VH genes, mainly distributed on chromosome 7 and a contig labeled MMUL_1:1099548049584 (Ensemble). However, as these regions contain sequence gaps, some VH genes may be missing. This interpretation is supported by the high level of diversity observed between some of the previously isolated rhesus macaque VH genes and those reported here (fig. S1A).
Additionally we identified 30 diversity (DH) segments and 6 joining (JH) segments in the rhesus macaque genome, all situated on chromosome 7. The DH and JH genes have been described previously (25). By further analyzing the proposed VH genes for intact leader sequences and complete exons, we identified and annotated 61 possible open reading frames (ORFs) (table S1). In comparison, the human genome has been reported to contain 47 functional VH genes, 23 D segments and 6 J segments (IMGT®) (fig. 1A). The phylogenetic relationship of the rhesus macaque VH ORFs was evaluated using MUSCLE and PhyML and was compared to previously published functional human VH genes (fig. 1B). The VH gene family organization of the rhesus ORFs closely mimicked that of the human genes with most members belonging to the VH3 family, followed by the VH4 and VH1 families. Upon performing a joined sequence analysis with both rhesus and human VH regions, the genes clustered according to family distribution rather than by species, indicating a high level of gene family conservation between human and rhesus macaques VH genes (fig. 1C). The average homology between the rhesus VH and the corresponding functional human VH genes was 92.1±2.3% (SD; n=61) (fig. 1D), closely matching the overall ~93% homology between the rhesus and human genomes (19). We utilized the same approach for the light chain loci and identified and annotated 50 lambda chain variable gene (VL) ORFs, mainly distributed on chromosome 10 (fig. S2 and table S2), as well as 62 kappa chain variable gene (VK) ORFs, distributed over two separate regions on chromosome 13 (fig. S3 and table S3).
Figure 1. Human and rhesus macaque VH gene sequence homology.
(A) Number of functional heavy chain V, D, J genes in humans and rhesus macaques. (B) Comparison of human and rhesus VH families and family members. (C) Joint phylogenetic relationships of rhesus and human VH genes. (D) Average homology between the rhesus VH ORFs and corresponding human VH genes (n=61).
Total and antigen-specific rhesus memory B cells were isolated for single-cell analysis of Ab sequences
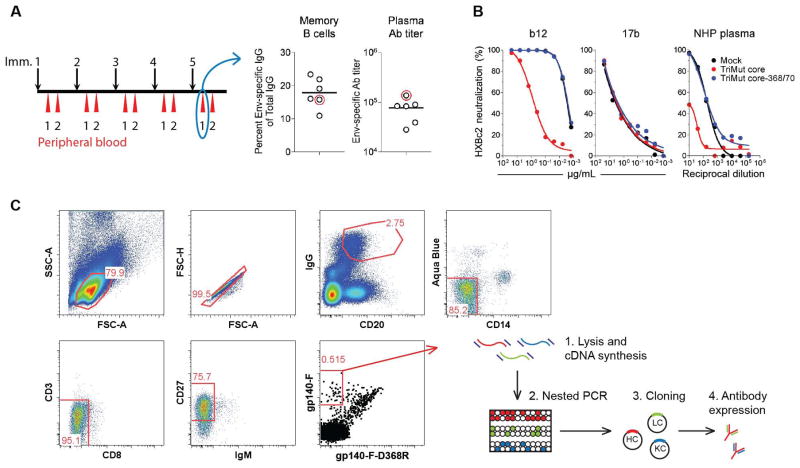
To isolate Env vaccine-elicited CD4bs-directed MAbs, we selected peripheral blood mononuclear cells (PBMCs) recovered following five Env trimer immunization of monkey F128 (26), which possessed high anti-Env Ab titers and memory B cell responses (fig. 2A, red circles). Our previous study shows that F128 was also one of the animals that displayed the most potent neutralizing titers against a panel of virus tested among the animals included in that study (26). The plasma from F128 was evaluated for the presence of CD4bs-directed Abs using a differential competition neutralization assay based on a pair of probes, referred to as TriMut core and TriMut core-368/70 (27). The TriMut probes are based on the gp120 core V3S protein with three additional mutations in the gp120 bridging sheet region, I423M, N425K and G431E, which eliminate CD4 binding without affecting recognition by any of the known CD4bs antibodies (28). The TriMut core-368/70 probe contains two additional mutations in the CD4 binding loop (D368R and E370F), which are known to specifically eliminate recognition by most CD4bs-directed antibodies. MAbs b12 (CD4bs-directed) and 17b (co-receptor binding site (CoRbs)-directed) were used to confirm the specificity of the assay (fig. 2B). Assessment of the plasma from animal F128 with these probes demonstrated that most of the neutralization (>92%) of HXBc2 was absorbed by TriMut core, but not by TriMut core-368/70, confirming the presence of CD4bs-directed antibodies in the plasma.
Figure 2. Isolation of vaccine-induced NHP memory B cells specific for the CD4bs of HIV-1 gp120.
(A) Schematic representation of the immunization schedule (left panel). Inoculations (black arrows) were given five times at monthly intervals to six rhesus macaques (26). Blood and cells were collected one and two weeks after each inoculation (red arrows). The frequency of Env-specific memory B cells one week after the fifth immunization (middle panel) and Env-specific IgG Ab titers two weeks after the fifth immunization, presented as Log10 OD50 titers (right panel) (n=6). Circled in red are values for macaque F128, which was used for the single-cell sorts. (B) Verification of probe specificity to detect CD4bs-directed neutralization using b12 (left panel), and 17b (middle panel). Right panel shows the presence of CD4bs-directed antibodies in the NHP plasma at the time of sort. (C) Gating scheme for sorting IgG+, CD4bs-specific memory B cells from immunized rhesus macaques: CD20+, IgG+, CD27+, CD3−, CD8−, CD14−, IgM−, gp140-F+, gp140-F-D368R- cells were sorted at single cell density into 96-well plates containing lysis buffer. The frequency (percentage) of gated cells is listed in red color. Upon cDNA synthesis, the heavy, lambda light and kappa light chains were amplified by nested PCR and the V(D)J regions were cloned into expression vectors.
To differentially isolate CD4bs-directed memory B cells from the PBMCs of monkey F128, we used trimeric gp140-F and gp140-F-D368R probes and antibodies against cell surface markers to sort CD20+IgG+CD27+gp140-F+gp140-D368R- cells at single cell density into 96-well plates (fig. 2C). Following reverse transcription of the RNA, nested PCR of heavy and light chains and cloning into expression vectors (15, 29, 30) a panel of MAbs were produced. The number of stained and sorted cells, the number of wells that yielded positive antibody heavy and light chains (HC+LC) and the number of expressed MAbs are summarized in table S4. To enable a general evaluation of the NHP memory B cell compartment, in addition to isolating CD4bs-directed memory B cells, we single-cell sorted total Env-specific memory B cells using the gp140-F probe only (CD20+IgG+CD27+gp140-F+), as well as total (non-antigen-specific) memory B cells (CD20+IgG+CD27+) (fig. 3). The total memory B cells were sorted from a different monkey (F133) only immunized with adjuvant as control (26). The RNA was reverse transcribed and, following nested heavy chain PCR the amplified uncloned VH sequences were evaluated for gene family usage, SHM, and Complementary-Determining Region 3 (CDR3) length.
Figure 3. Evaluation of Ig heavy chain genetics in sorted memory B cell populations.
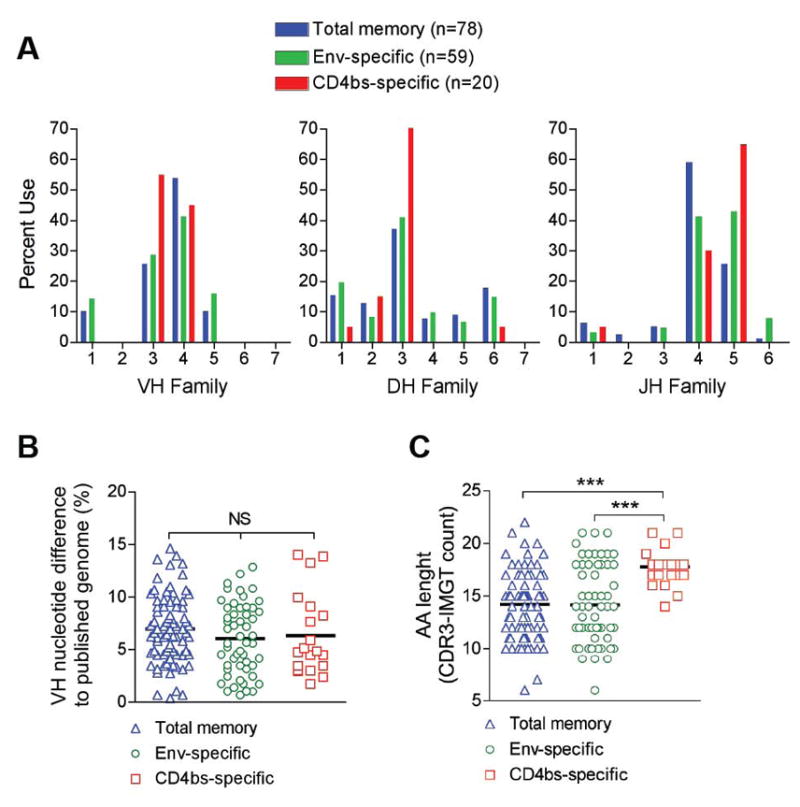
Three memory B cell populations were sorted; Total memory (Blue; CD20+CD27+IgG+; n=78), total Env-specific (Green; CD20+CD27+IgG+gp140-F+; n=53), and CD4bs-specific (Red; CD20+CD27+IgG+gp140-F+gp140-F-D368R-; n=20). (A) V(D)J family distribution was evaluated using IMGT®/V-Quest and Joinsolver. (B) SHM in single-cell sorted memory B cells (Total, Env-specific and CD4bs-specific) is shown as % nucleotide difference calculated by aligning VH regions to the corresponding germline sequences of the Indian rhesus reference genome. (C) Evaluation of heavy chain CDR3 regions based on the CDR3-IMGT definition. Statistical differences were evaluated using the non-parametric Kruskal-Wallis test followed by Dunn’s post-test for comparison of specific samples. Significance was determined as; *p≤0.05, **p≤0.01, and ***p≤0.001
The NHP Ab sequences are genetically diverse and moderately mutated
The genetic composition of the NHP sequences was analyzed using IMGT®/V-Quest and Joinsolver®. As these tools use human V(D)J sequences for comparative analysis, the degree of SHM and specific V(D)J genes cannot be definitively determined using these tools alone. However, as human and rhesus V(D)J genes are highly homologous, they are valuable for assigning the gene families from which the NHP Abs are comprised (fig. 1C). We determined the heavy chain V(D)J family distribution in three sorted memory B cell populations (fig. 3A): total memory (blue), total Env-specific (green), and CD4bs-specific (red). All three populations showed similar gene family distributions, where the majority of the VH genes belonged to the VH3 and VH4 families, the DH genes to DH3, and the JH genes to JH4 and JH5. When examining nine isolated CD4bs-directed MAbs, annotated GE121, GE125, GE136, GE137, GE140, GE143, GE145, GE147 and GE148, four were composed of VH genes belonging to the VH3 family and five to the VH4 family (table 1). By phylogenetic analysis and sequence alignment, we determined that GE140 and GE145 were identical clones. We could also determine that some of the other MAbs utilized the same germline VH gene, but combined with different D and/or J genes or paired to different light chains, demonstrating that they were of unique clonal origins. (table 1 and fig. S1B).
Table 1.
Vaccine- and infection-induced CD4bs-directed MAb Ig heavy chain genetics
| MAb | VH familya | VH Mutation rateb | CDR3 sequence | CDR3 length | DH family | JH family | |
|---|---|---|---|---|---|---|---|
| (bp) | (AA) | (AA) | |||||
| Vaccine-induced NHP MAbs | |||||||
|
| |||||||
| GE121 | 3.8 | 4.5% | 8.2% | CAKGPMTIFGLFIIRFDVW | 17 | 3 | 5 |
| GE125 | 3.8 | 3.4% | 8.2% | CAKGRITIFGLVITYFDSW | 17 | 3 | 4 |
| GE136 | 4.11 | 5.4% | 14.3% | CVREGIVLVNLAVKNWFDVW | 18 | 2 | 5 |
| GE137 | 3.18 | 4.6% | 7.1% | CTRDPRITVFGLINGGDKWFDVW | 21 | 3 | 5 |
| GE140* | 4.11 | 4.4% | 12.2% | CARHRGTIFGLVIFNWFDVW | 18 | 3 | 5 |
| GE143 | 4.40 | 8.4% | 13.3% | CARHATSPYYSISIRSQNWFDVW | 21 | 3 | 5 |
| GE147 | 3.24 | 1.4% | 1% | CTRGRIAGNIFVWFDVW | 15 | 1 | 5 |
| GE148 | 4.57 | 6.3% | 13% | CARVQNIVVVFTIKEFFELW | 18 | 2 | 1 |
|
| |||||||
| Infection-induced human MAbs | |||||||
|
| |||||||
| Non-bNAbs | |||||||
|
| |||||||
| b13 | 3–30 | ND | 7.1% | CARDIGLKGEHYDILTAYGPDYW | 21 | ND | ND |
| F105 | 4–59 | ND | 8.2% | CARGPVPAVFYGDYRLDPW | 17 | ND | ND |
|
| |||||||
| bNAbs | |||||||
|
| |||||||
| VRC01 | 1–2 | 32% | 41.8% | CTRGKNCDYNWDFEHW | 14 | 3–16 | 1 |
| b12 | 1–3 | 13% | 20.4% | CARVGPYSWDDSPQDNYYMDVW | 20 | 3–10 | 6 |
ND, Not determined
Highest scoring germline sequence as determined by AlignX and annotated according to table S1.
Divergence to highest scoring germline sequence
GE140 and GE145 were identical clones
For SHM analysis, we first asked if there were any significant sequence differences in germline VH genes between the Chinese origin rhesus macaques, used here, and the Indian rhesus macaque reference sequences. Germline VH genes from Chinese origin macaques were sequenced and aligned to Indian origin macaque VH sequences and then compared to the allelic diversity observed in humans (fig. S1C). We found the divergence between Chinese and Indian rhesus macaques to be within the same range as that seen for human allelic variation (p=0.65) and we could therefore use the Indian rhesus macaque Ig germline to calculate SHM for the NHP antibody sequences isolated in the current study. The three sorted populations displayed a similar level of SHM, where total memory displayed an average SHM of 7.0±3.2% (blue; n=78), Env-specific 6.1±3.3% (green; n=53), and CD4bs-specific 6.3±3.9% (red; n=20) (fig. 3B), indicating that the immunization regimen used here stimulated SHM of antigen-specific antibody genes to similar levels as those observed in the general B cell memory compartment. For the isolated NHP MAbs, we identified the average SHM to be 4.8±2% (range 1.4–8.4%) at the nucleotide level and 9.7±4.4% (range 1–14.3%) at the amino acid level. This is similar to the levels previously reported for human memory B cells (15, 31) and consistent with our analysis of the sequences from the total memory B cells analyzed here (fig. 3B). Examination of the heavy chain CDR3 region (by IMGT®/V-Quest analysis) revealed no differences between total memory and Env-specific memory B cells, while interestingly the CDR3 regions from CD4bs-directed antibody gene segments were significantly longer (fig. 3C). This observation was confirmed when we analyzed CD4bs-specific memory B cells from a second monkey (F125) that had received the same immunization regimen as monkey F128 (26). The VDJ family usage, degree of SHM, and CDR3 length were highly comparable between the two monkeys (fig S4).
The vaccine-elicited CD4bs-specific MAbs display neutralizing activity and high affinity to Env ligands
The isolated NHP MAbs were characterized for their capacity to bind a panel of Env ligands including gp120, gp120-D368R, core (V3S), stabilized core (2CC) (32) and TriMut core (27). The rationale for examining this panel of Env variants was as follows. All MAbs are expected to bind gp120 but most CD4bs-directed MAbs do not efficiently recognize the D368R variant. All CD4bs MAbs should recognize gp120 core independently of the deleted variable regions, while the stabilized core, which is mutagenically induced in the CD4-bound conformation of gp120, is efficiently recognized only by broadly neutralizing CD4bs MAbs. The TriMut core contains mutations in the gp120 bridging sheet that eliminates efficient binding by CD4, but retains recognition by all known CD4bs MAbs. All NHP MAbs bound gp120 with high efficiency, while six of the eight NHP MAbs, GE121, GE136, GE140, GE143, GE147 and GE148 displayed no detectable binding to gp120-D368R (fig. S5A). Only two MAbs, GE125 and GE137, exhibited some binding to gp120-D368R, although with reduced efficiency. Among the human MAbs, VRC01 bound gp120-D368R weakly, while b12 and F105 binding to gp120 was completely abrogated by the D368R mutation, consistent with previous results. All NHP MAbs, except GE147, bound core, while only one NHP MAb, GE148, and the broadly neutralizing human MAbs, VRC01 and b12, exhibited detectable binding to the stabilized core. Similarly to the human MAbs, all NHP MAbs, except GE147, bound TriMut core (fig. S5A). The epitope specificity of the NHP MAbs to the CD4 binding region of gp120 was confirmed by competition ELISA. Binding to gp120 by biotinylated NHP MAbs was competed with a panel of CD4bs ligands, including b6, b12, VRC01 and CD4-Ig while no competition was observed for MAb 2G12 (fig. S5B). Binding of the NHP MAbs to gp120 was moderately competed by the CoRbs Ab, 17b, consistent with previous observations that CD4bs-directed Abs compete with CoRbs-directed (33).
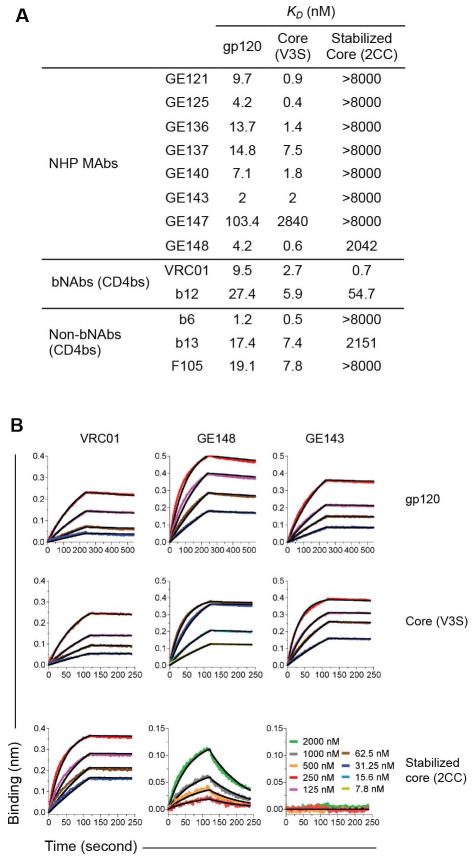
The binding characteristics of the NHP MAbs to selected gp120 Envs variants were further analyzed by Bio-Layer Interferometry which, besides the relative half maximal binding approximated by ELISA, can determine the precise kinetic parameters of Ab association (on-rate) and dissociation (off-rate) with its epitope on a given antigen. All NHP MAbs, except GE147, demonstrated high binding affinities for gp120, with dissociation constant KD value ranging from 2–15 nM, similar to that of the known human CD4bs MAbs (fig. 4A). Consistent with the ELISA results, the binding affinities of all NHP MAbs to gp120-D368R were below detection, with the exception of GE125 and GE137, which demonstrated weak affinities (KD values in the μM range) (table S5). Furthermore, the NHP MAbs and all human CD4bs MAbs bound core with similar affinities, while in contrast, only the broadly neutralizing antibodies, VRC01 and b12, were capable of binding stabilized core with high affinity (fig 4). Of the non-broad MAbs, only the vaccine-induced MAb GE148 and the infection-induced MAb b13 exhibited weak affinity for stabilized core (fig. 4 and table S5).
Figure 4. NHP MAb affinity to Env.
(A) The binding affinities of the NHP MAbs to gp120, core and stabilized core were analyzed by Bio-Layer Interferometry using an Octet Red96 system with the MAbs immobilized on the chip surface and the Env ligands in the analyte. Selected human CD4bs-specific MAbs were examined for comparison. The dissociation constants (KD) between the different MAbs and ligands are shown in nM. (B) The kinetics of binding of VRC01, GE148 and GE143 to gp120, core and stabilized core are shown at different concentrations of the ligand.
We next examined the neutralizing capacity of the NHP MAbs compared to the human CD4bs-directed MAbs using a standardized neutralization assay (34). The panel included clade B isolates representing neutralization-sensitive viruses (Tier 1) and isolates representing more neutralization-resistant primary viruses (Tier 2), a designation based upon the capacity of these viruses to be neutralized by HIV-1+ immune-sera. All NHP MAbs, except GE147, displayed neutralizing activity against several Tier 1 viruses including HXBc2, a virus known to be sensitive to CD4bs-directed Abs (table 2). Five of the NHP MAbs (GE125, GE136, GE140 and GE143) also neutralized SF162 and MN. MN was neutralized by VRC01, b12, b6 and b13, but not by F105, F91 and 1.5E. Several NHP MAbs (GE125, GE136, GE137 and GE143) neutralized either or both of the non-clade B viruses DJ263 and MW965, demonstrating that these MAbs were capable of recognizing the CD4bs of diverse Env spanning several HIV-1 clades (table 2). These results demonstrate the potential of the vaccine-elicited Abs to recognize this functionally conserved, antigenic surface across diverse HIV-1 clades.
Table 2.
Neutralizing properties of vaccine-elicited CD4bs-directed MAbs
| MN.3 | HXBc2 | SF162 | Clade B BaL.26 |
JRFL | 89.6 | YU2 | Clade A DJ263. 8 |
Clade C MW965.26 |
||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| Vaccine-induced NHP MAbs | Plasma | 7044 | 131 | 183 | ND | <10 | 22 | 93 | 39 | 3898 |
| GE121 | >50 | 0.216 | 10.5 | >50 | >50 | >50 | >50 | >50 | >50 | |
| GE125 | 0.109 | 0.348 | 1.95 | 46.6 | >50 | >50 | >50 | 10.4 | >50 | |
| GE136 | 0.227 | 4.04 | 6.85 | >50 | >50 | >50 | >50 | >50 | 28.2 | |
| GE137 | >50 | 10.3 | >50 | >50 | >50 | >50 | >50 | 27.4 | 0.142 | |
| GE140 | 0.438 | 1.79 | 3.88 | >50 | >50 | >50 | >50 | >50 | >50 | |
| GE143 | 0.759 | 0.976 | 2.66 | >50 | >50 | >50 | >50 | 28.3 | 17 | |
| GE147 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | |
| GE148 | 0.788 | 0.404 | 1.53 | 11.9 | >50 | >50 | >50 | >50 | >50 | |
|
|
||||||||||
| Infection-induced Human MAbs | VRC01 | 0.02 | 0.013 | 0.106 | 0.027 | 0.032 | 0.685 | 0.115 | 0.083 | 0.056 |
| b12 | 0.003 | 0.007 | 0.07 | 0.051 | 0.022 | 0.14 | 2.18 | 0.812 | 0.19 | |
| F105 | >50 | 0.266 | 1.15 | 9.4 | >50 | 50 | >50 | >50 | >50 | |
| b6 | 0.118 | 0.138 | 0.8 | 11.4 | >50 | >50 | >50 | 0.67 | >50 | |
| b13 | 0.945 | 0.165 | 1 | 4.18 | 15.8 | 49.2 | >50 | >50 | 4.31 | |
| F91 | >50 | 0.321 | 0.893 | 22.9 | >50 | >50 | >50 | >50 | >50 | |
| 1.5E | >50 | 3.28 | 2.36 | >50 | >50 | >50 | >50 | >50 | >50 | |
ND, Not determined
| <1 | 1–10 | 10–50 | >50 | μg/mL (MAb) |
| >1000 | 100–1000 | 10–100 | <10 | ID50 (Plasma) |
Fine mapping of the vaccine-elicited MAbs reveals distinct binding specificities to the CD4bs
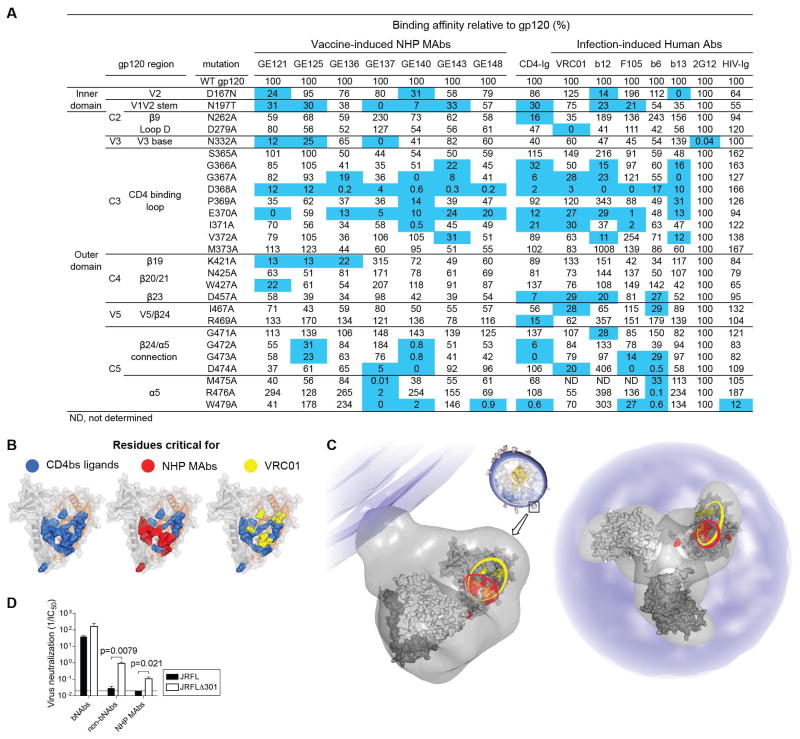
To fine-map the sub-specificities of the NHP MAbs within the CD4 binding region, we selected 27 JRCSF gp120 mutants containing single alanine (Ala) point mutations in gp120 to perform a limited Ala scan (35). Residues were selected based on the criteria that they were known contact residues for CD4, CD4bs-directed Abs or the structurally defined CD4bs Abs in complex with gp120 (14, 36–39). As expected, recognition by all NHP and human MAbs and CD4-Ig was dramatically changed by mutations in CD4 binding loop (residues 365–373) (fig. 5A). Mutations in β24/α5 connections (residues 471–474) affected only some NHP MAbs, while the CD4bs-directed infection-induced MAbs were more frequently affected. Beyond these regions, mutations in loop D, β23, and V5/β24 affected recognition by VRC01 or/and CD4-Ig binding but not the NHP MAbs or the non-broad human MAbs except b6, while mutations in the V3 base and β19 affected binding by the NHP MAbs but not the infection-induced human MAbs. These data suggest that the CD4bs specificities elicited by YU2 trimer immunization overlap with those of the non-broad neutralizing CD4bs MAbs, but markedly less so with the HIV-1 broadly neutralizing MAbs. The sub-specificities within the CD4bs recognized by the vaccine-elicited MAbs, shown as a composite of all residues that affect recognition by the eight NHP MAbs, compared to residues that affect the broadly neutralizing, infection-induced CD4bs MAb, VRC01, are dramatically revealed when the Ala scan data are mapped onto the molecular surface of the gp120 core crystal structure (fig. 5B, individual footprints in fig. S6). The difference in the approximate NHP MAb and VRC01 footprints on gp120 are even more apparent when the regions identified in the Ala scan are modeled onto the proposed Env trimer and fitted into the density of the previously described cryo-EM structure of the functional HIV-1 spike (40). Collectively, this analysis shows that the vaccine-elicited MAbs map closer to the trimer axis, while VRC01 maps more distally on the assembled spike, perhaps accounting for the better access of VRC01 to the functional Env spike and its much greater neutralization capacity (fig. 5C).
Figure 5. Alanine scanning of NHP MAb binding sites.
(A) A panel of JRCSF Env mutants containing single alanine (Ala) point mutations in gp120 was used for a limited Ala scan of the CD4 binding region for the NHP MAbs, CD4-Ig and selected human CD4bs-directed MAbs. MAb 2G12 and HIV-Ig were used as controls. The effect of a given Ala mutation on ligand binding is shown relative to wild-type (WT) gp120. Mutations resulting in a three-fold reduction in binding of CD4-Ig and the CD4bs MAbs relative to WT gp120 are highlighted in blue. (B) The gp120 core molecular surface was used to highlight the residues affecting recognition by CD4-Ig and previously known CD4bs MAbs (blue). Of these, the residues affecting binding of VRC01 are marked in yellow and those affecting the NHP MAbs are marked in red. The alpha carbon backbone of the inner domain and bridging sheet, depicted in ribbon, are in gray and the outer domain is highlighted in salmon. (C) Side and top views depict the cryo-EM density map with the trimeric cores fit into the density. Circled in red is the approximate location of residues affecting binding of the NHP MAbs while circled in yellow is the approximate footprint of VRC01. (D) Neutralization of JRFL and JRFLΔ301 by the CD4bs-directed MAbs shown in (A): human bNAbs (n=2), human non-bNAbs (n=5) and NHP MAbs (n=8). Statistical significance between inverse virus neutralization IC50 values was evaluated with a Mann-Whitney test using Graph Pad Prism software.
Based on this model, we hypothesized that the lack of neutralizing activity against resistant Tier 2 isolates, such as JRFL, might be due to quaternary packing constraints occluding access to the CD4bs by the vaccine-elicited MAbs. We therefore assessed the neutralizing activity of an engineered variant of JRFL, which lack an N-linked glycan at the base of V3, JRFLΔ301. Elimination of this glycan renders JRFL sensitive to non-broad human CD4bs-directed MAbs, presumably due to the removal of quaternary conformational constraints (41). We found that similar to the non-broad human CD4bs-directed MAbs, the NHP CD4bs-directed MAbs also neutralized JRFLΔ301, but not the unmodified JRFL virus (fig. 5D), consistent with a limited capacity of these MAbs to efficiently access the unmodified JRFL spike due to conformational constrains. In contrast, the more broadly neutralizing human MAbs, VRC01 and b12, potently neutralized both the JRFL and JRFLΔ301 viruses.
DISCUSSION
In this study, we established an approach for high-resolution definition of humoral immune responses in non-human primates and demonstrate its use for genetic and functional analyses of B cell responses elicited by clinical vaccine candidates. Specifically, we investigated antibodies harbored in the IgG-switched memory B cell pool induced by a highly relevant vaccine antigen, HIV-1 Env. This line of investigation breaks new territory in defining vaccine-induced B cell responses to Env, a heavily glycosylated antigen that is considered a critical component of an effective HIV-1 vaccine. Our analysis of the genetic organization and sequence homologies between human and rhesus macaque antibody heavy and light chains (fig. 1 and fig S1–S3) provides a foundation for future efforts in this area. Having conducted a comprehensive analysis of the human and rhesus immunoglobulin loci, we amplified antibody heavy and light chain sequences from total, Env-specific and CD4bs-specific memory B cells. Analysis of these sequences revealed similar V(D)J gene distribution in these populations and, intriguingly, a clear increase in the CDR3 length in the CD4bs-specific populations compared to total memory or total Env-specific cells.
Recent studies describe the isolation of HIV-1 infection-induced MAbs targeting the CD4bs that are considerably more potent and more broadly neutralizing than the prototype CD4bs-directed MAb b12 (9, 15), demonstrating that highly effective Ab responses are elicited with detectable frequency during chronic HIV-1 infection. In contrast, broadly neutralizing antibodies directed against the CD4bs, or against any broadly neutralizing epitope on HIV-1 Env, have so far not been elicited by subunit Env immunization. To establish a baseline of the types of Abs elicited against the CD4bs and to begin to understand the limitations of Env vaccine-induced Ab responses, we isolated eight clonally distinct CD4bs-directed MAbs and characterized them by ELISA, cross-competition analysis and binding kinetics compared to a panel of infection-induced, CD4bs-directed MAbs. In contrast to the infection-induced MAbs, VRC01 and b12, the vaccine-elicited CD4bs MAbs were modestly mutated compared to closest matching germline sequence, around 5%, which was similar to that observed in the total memory pool. The marked difference in SHM between vaccine-elicited and infection-elicited MAbs is not surprising given the persistent antigen load and chronic immune activation that typifies chronic HIV-1 infection. The average time for development of Abs displaying broadly neutralizing activity in naturally infected persons is approximately 2.5 years and breadth only develops in a minority of infected individuals. When rare but effective neutralizing responses do appear, the Abs mediating this activity are highly mutated, suggesting that multiple rounds of SHM and B cell affinity maturation against the target epitope are required to elicit such neutralizing activity.
When the epitopes of the vaccine-elicited MAbs were examined by Alanine scanning and modeled onto the structure of the gp120 core, we found that their composite binding footprint was more proximal to the trimer axis and overlapped more with that of a non-broadly CD4bs-directed neutralizing infection-induced MAb, F105 (38) than that of the broadly neutralizing MAb VRC01, which was more dependent on residues located distally in the outer domain of gp120 (37). These data and analysis provide important information as they suggest that despite the high affinity of the vaccine-induced MAbs to soluble Env ligands, their limited capacity to neutralize diverse HIV-1 isolates is due to inefficient recognition of their cognate epitopes in the context of the functional spike, perhaps due to steric occlusion by quaternary packing. The differential footprints between the vaccine-elicited NHP CD4bs MAbs and VRC01 are striking and may be a result of the recombinant trimeric immunogen used here as well as glycan-influenced shifting of B cell recognition, or greater immunogenicity distal to the glycan-shielded outer domain, as previously suggested (37, 42). Importantly, the differential footprints suggest avenues for improved immunogen design. For example, approaches to occlude access to the bridging sheet-proximal regions of the CD4bs while maintaining access to the outer domain of gp120 might shift the sub-specificities of elicited responses and result in Abs with improved neutralizing potential. The observation that GE148 weakly recognized the stabilized core (2CC) suggests that, perhaps, with a properly focused antigenic drive and additional affinity maturation, improved antibody responses against the neutralization-relevant sub-region of the CD4bs might be elicited. Such improvements in immunogen design and elicitation in NHPs would have a direct route of translation into human clinical studies. Structural analysis of selected NHP CD4bs-directed MAbs described here and the elucidation of their precise footprints on gp120 would provide valuable information to complement the epitope mapping performed in this study.
In conclusion, the current study establishes an approach for the iterative analysis of vaccine-induced B cell responses in an experimental model system, which is highly relevant to humans at the molecular level. While the panel of expressed MAbs and the immunoglobulin sequences analyzed here represents a small sample of the total CD4bs-directed memory B cell repertoire, we believe that the results obtained provide an important foundation for the design of improved vaccine candidates that target the HIV-1 gp120 CD4bs to be accelerated into clinical studies. Furthermore, characterization of immune responses in NHPs permits the sampling of cellular compartments not easily accessed in humans, such as bone marrow and lymph nodes, and facilitates longitudinal studies of vaccine-induced B cell responses in a given vaccine-recipient, providing translational opportunities for Env-based immunogenicity testing in humans.
MATERIALS AND METHODS
Animals and sampling
Three year old rhesus macaques (Macaca Mulatta) of Chinese origin where housed at the animal facility Astrid Fagraeus Laboratory at the Swedish Institute for Infectious Disease Control. Housing and care procedures were in compliance with the guidelines of the Swedish Board of Agriculture. The facility has been assigned an Animal Welfare Assurance number by the Office of Laboratory Animal Welfare (OLAW) at NIH. All procedures were approved by the Local Ethical Committee on Animal Experiments. A detailed description of the immunization experiment was reported previously (26). Essentially, it consists of purified gp140-F trimers in adjuvant inoculated five times at monthly intervals.
Expression and purification of Env glycoproteins
The soluble gp140-F trimers (43) used as the immunogen were produced by transient transfection of a CMV-driven expression plasmid into Freestyle 293F suspension cells (Invitrogen) as previously described (44). Env ligands used in binding studies, gp120, gp120-D368R, gp120 core (core, previously referred to as V3S) and stabilized gp120 core (stabilized core, previously referred to as 2CC) were purified by a 17b MAb-coupled protein A-Sepharose column (45). TriMut core and TriMut core-368/70 were purified by lentil-lectin and gel filtration chromatography. The biotinylated gp140-F and gp140-F-D368R probes used for Env-specific cell sorting and enrichment of CD4bs-directed memory B cells flow cytometry were purified by lectin chromatography and IMAC. Both probes carried an Avi-tag signal for site-specific biotinylation to the C-terminus of gp140-F (46) and biotinylation was performed with biotin ligase Bir A (Avidity). All gp120 and gp140 proteins were on the YU2 background, while the core proteins were based on HXBc2.
Rhesus Ab heavy chain gene analysis
The rhesus macaque genome (19) was mined for possible V(D)J genes using IMGT®/LIGMotif version 4.1.1 (47). Proposed V(D)J genes where further evaluated using IMGT®/V-Quest (48) and Joinsolver® (49) to determine gene family usage and possible exon truncations. An intact exon and leader sequence defined the sequence as an ORF. To determine the VH gene similarity between Chinese and Indian rhesus macaques, we sequenced proposed VH gene units, including the leader sequence and VH exon. The sequences (average 3–4 per proposed VH gene) were assembled in ContigExpress (VectorNTI; Invitrogen) and aligned to the closest matching Indian rhesus macaque sequence to determine homology (fig. S1C). As a reference for normal allelic deviation, human VH allelic variants (IMGT®) were used. For phylogenetic analysis, germline VH genes from human (IMGT®) and rhesus macaques where aligned using MUSCLE (version 3.7) iterated 50 times. Alignment gaps were removed using Gblocks. Phylogeny was calculated using PhyML (version 3.0) and maximal likelihood trees were rendered using the HKY85 substitution model and bootstrapping (200–500 replicates) with TreeDyn (version 198.3) (50). The trees where then graphically edited using Dendroscope (51).
SHM calculations were performed on the VH region between codon 1 and ~104 (IMGT® count; conserved cysteine, marking the end of the V region) through aligns to the previously published rhesus genome (19). CDR3 lengths were determined via IMGT®/V-Quest.
Single cell sorting by flow cytometry
Cell sorting of memory B cells from NHPs F133 (Total memory), F125 and F128 following immunization 5 (Total Env-specific and CD4bs-specific) was performed as follows. Frozen monkey PBMCs were re-suspended in 10 ml RPMI 1640 medium (Invitrogen) pre-warmed to 37°C containing 10% fetal bovine serum (FBS) and 10,000 unit/ml DNase I (Roche). After centrifugation, the cells were re-suspended in 50 μl pre-chilled PBS. 5 μl of aqua fluorescent reactive dye (Invitrogen) reconstituted in DMSO and 40-fold diluted in distilled water was added to the suspension for 20 min at 4°C in the dark. The cells were then incubated with a cocktail of fluorescently labeled antibodies to cell surface markers (table S6). The Ab-Env probe cocktail was prepared in 50 μl of PBS including antibodies specific for the T cell markers CD3 and CD8 and B cell markers CD20, CD27, IgG and IgM. CD14- was used for negative selection of monocytes. Probes to enrich for CD4bs-specific B cells were also included in the cocktail and added to stain the cells for 1 h at 4°C. The gp140-F-biotin trimers were conjugated to streptavidin-allophycocyanin (SA-APC) (Invitrogen) and the gp140-F-biotin-D368R trimers to extravidin-phycoerythrin (SA-PE) (Sigma) to yield gp140-F-APC and gp140-F-D368R-PE (15). Following conjugation, each Env-probe was added to the Ab-cell cocktail at a final concentration of 4 μg/ml. The stained cells were washed with 10 ml of pre-chilled PBS and resuspended in 500 μl of PBS, passed through a 70 μm cell mesh (BD Biosciences) and sorted using a modified 3-laser or 4-laser FACS Aria cell sorter (BD Biosciences). CD4bs-specific memory B cells were defined as CD3-, CD8−, Aqua blue-, CD14−, CD20+, IgG+, CD27+, IgM−, gp140-F+ and gp140F-D368R- and were collected into single wells of 96-well PCR plate containing 20 μl of cell lysis buffer (15). The numbers of cells in the sorted populations are indicated in table S4.
RT-PCR of IgG genes, cloning and expression
The IgG heavy and the corresponding light chain gene transcripts of single sorted NHP memory B cells were amplified by RT-PCR and cloned into eukaryotic expression vectors to produce full IgG1 antibodies, using immunoglobulin-specific PCR conditions, primers and expression vectors as described (15, 30). For expression, equal amounts of heavy chain and light chain plasmid DNAs (250 μg of each) were transfected with 1 ml of 293Fectin into 50 ml FreeStyle 293F cells at a density of 1.2 million cells/ml. The cell culture supernatant containing the secreted IgG was harvested 4 days following transfection and purified by protein A Sepharose columns (GE Healthcare). The efficiency of the RT-PCR and cloning is indicated in table S4.
Characterization of Env binding and cross-competition ELISA
The MAbs where tested for binding using Reacti-bind® (Pierce) ELISA plates coated at 2 μg/ml with gp120, gp120-D368R, core (V3S), stabilized core (2CC), TriMut core or TriMut core-368/70 in PBS at 4°C over night. After blocking in B3T buffer (150 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 3.3% fetal bovine serum, 2% bovine albumin, 0.07% Tween 20), the MAbs were added and incubated for 1h at 37°C. Binding was detected by secondary HRP-conjugated anti-human Fcγ (Jackson ImmunoResearch) at 1:10 000 for 1h. The signal was developed by addition of TMB substrate (SureBlue; KPL) for 10 minutes. Reactions were terminated with 1N sulfuric acid and the optical density (OD) read at 450 nm. Between each incubation step, the plates were washed 6 times with PBS containing 0.05% Tween 20.
Competition ELISA was performed as previously described (15). Briefly, the NHP MAbs were biotin-labeled with Peirce EZ-Link NHS-Biotin reagent (ThermoScientific) per manufacturer instruction. The gp120 was captured in the ELISA wells pre-coated with the sheep anti-gp120 C5 region-specific Ab, D7324 (Aalto Bio Reagents). The unlabelled competitor human MAbs or CD4-Ig were diluted in blocking buffer and plated at 10-fold serial dilution, starting at 50 μg/ml. Following 30 minutes incubation at 37°C, biotin-labeled antibodies at a single concentration were added to the wells. This concentration was determined by previous titration experiments to give OD450 nm value in the range of 1 to 2. The binding signal was developed by incubation with streptavidin-HRP (Sigma) and TMB substrate (Invitrogen).
Antibody binding kinetics analysis
The kinetics of NHP and human MAb binding to a panel of Env variants were assessed with an Octet Red96 system (ForteBio) using Bio-Layer Interferometry (BLI) in a 96-well format following manufacturer instructions. Prior to use, all Env variants were subjected to size-exclusion chromatography to remove undesired oligomeric forms where applicable. Antibodies at 10 μg/ml diluted in PBS/0.2% Tween 20 were captured on the surface of the anti-huIgG Fc capture biosensors (ForteBio) for 1 minute. Following capture, a 1 minute wash in buffer removed excess unbound Ab and to establish new baseline signal. The biosensor tip was then immersed in wells containing the Env variants in solution. The glycoproteins were 2-fold serially diluted from an initial starting concentration of 250 nM. In the case of low affinity, Env protein concentrations starting from 8 μM or 1 μM were used to obtain higher initial binding signal. Ab-Env association rate (on-rate) was measured over a 2 minute interval, followed by placing the sensors in wells containing buffer to measure dissociation rate (off-rate) over a 2 minute interval. KD values (in nM) were calculated as off-rate/on-rate. The interaction of Ab and Env on the tip of the sensor was assessed with a wavelength shift, Δλ due to the change in thickness of the sensor tip if a ligand-ligand interaction had occurred. The sensograms were corrected with the blank reference and fit with the software, ForteBio Data Analysis 6.4, using 1:1 binding model using the global fitting function (grouped by color, Rmax).
Alanine scanning
A panel of 27 gp120 Ala mutants containing alterations known to affect CD4 binding or recognition by a set of CD4bs-directed MAbs was selected. The Ala mutations were generated previously in the context of the full-length JRCSF gp160 expression plasmid (35). The Env plasmids were individually co-transfected into 293T cells together with a plasmid containing the remaining HIV structural genes to produce Env-pseudoviruses as described previously (34). For binding analysis, the gp120 was released from the pseudovirus by detergent lysis and then captured by the sheep anti-gp120 C5 Ab, D7324 (Aalto Bio Reagents) previously coated into ELISA wells of the 96-well plate. Following washing, binding to gp120 was assessed for the human and NHP MAbs using an anti-Fc HRP secondary Ab and the TMB substrate as described previously. The level of binding by the human MAb 2G12 was used to normalize the variant gp120 expression levels. The effect of a given Ala mutation on Ab binding was represented by apparent affinity (avidity) relative to a binding level to wild-type (WT) gp120, calculated with the formula ([(EC50_WT/EC50_mutant)/(EC50_WT for 2G12/EC50_mutant for 2G12)]*100) as previously described (39).
Virus neutralization assays
Neutralization assays were performed using an HIV-1 Env pseudovirus assay and TZM-bl target cells (34). To determine the serum dilution that resulted in a 50% reduction in Relative luciferace units (RLU) serial dilutions of sera were performed and the neutralization dose-response curves were fit by non-linear regression using a 4 parameter hill slope equation programmed into JMP statistical software (JMP 5.1, SAS Institute Inc.). The results are reported as the serum neutralization ID50, which is the reciprocal of the serum dilution producing 50% virus neutralization. Diverse HIV-1 virus isolates, including viruses from clades A, B and C were used in the neutralization assays. The sources of the Env-encoding plasmids were described previously (26).
Molecular modeling
Amino acid residues affecting the binding of the NHP MAbs to gp120 were highlighted on the gp120 core structure using the PDB entry code: 2NY3. To position the affected residues in the context of the HIV-1 spike, the gp120 cores were superimposed on the gp120 core protomers already fitting the density map of the unliganded HIV-1 spike (PDB: 3DNN, EMDB: EMD-5019). The molecular modeling, SSM superimposition and presentation were done using the molecular graphics program, CCP4mg (52).
Human monoclonal antibodies
The MAb 2G12 was purchased from Polymun Scientific Inc. The anti-CD4bs MAb F105 was provided by Marshall Posner (Dana Farber Cancer Institute) and the CD4bs MAbs b6, b12, and b13 were provided by Dennis Burton (The Scripps Research Institute). The co-receptor-directed mAbs 17b was provided by James Robinson (Tulane University). HIV immune globulin (HIVIG) was obtained from the NIH AIDS Research and Reference Reagent Program. Soluble CD4 (sCD4 with CD4 D1–D4 domains) was purchased from Progenics (Tarrytown). The CD4-Ig plasmid expression construct was provided by Joseph Sodroski (Dana Farber Cancer Institute). VRC01 was previously described (15).
Statistical analysis
Statistical evaluation between individual groups was performed with a Mann-Whitney test Evaluation of ≥3 groups was done using the non-parametric Kruskal-Wallis test followed by Dunn’s post-test for comparison of specific samples. Statistical significance was determined as: *p≤0.05, **p≤0.01, and ***p≤0.001. All statistics were evaluated using GraphPad prism version 4 software.
Supplementary Material
Acknowledgments
We thank M. Roederer and his team at the Vaccine Research Center (VRC) FACS core for fluorescent Ab production and cell sorting, Y. Xia and B. Beutler at the Scripps Research Center for sequencing VH genes from Chinese rhesus macaques and M. Nussenzweig at the Rockefeller University for antibody expression plasmids.
Funding: This study was supported by grants from the Swedish Research Council, Sida/SAREC, International AIDS Vaccine Initiative (IAVI), the Bill and Melinda Gates Foundation and the intramural program of the VRC at the National Institutes of Health and by the IAVI Center at Scripps. We are also grateful to the Fulbright Commission, The Carlsberg Foundation and Karolinska Institutet for scholarships to NH, CP and CS respectively.
Footnotes
Author contributions: CS and YL performed experiments, participated in experimental design, analyzed the data and wrote the paper. NH, CP, RW, SO and YF performed experiments. JRM, RTW, and GBKH participated in experimental design, analyzed the data and wrote the paper.
Competing interests: The authors have no conflict of interests.
Data and Material: The MAbs characterized in the current study have been submitted to the GenBank database under accession numbers: JQ885990-JQ886005.
References
- 1.Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, van Griensven F, Hu D, Tappero JW, Choopanya K. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 2.Spearman P, Lally MA, Elizaga M, Montefiori D, Tomaras GD, McElrath MJ, Hural J, De Rosa SC, Sato A, Huang Y, Frey SE, Sato P, Donnelly J, Barnett S, Corey LJ. A trimeric, V2-deleted HIV-1 envelope glycoprotein vaccine elicits potent neutralizing antibodies but limited breadth of neutralization in human volunteers. J Infect Dis. 2011;203:1165–1173. doi: 10.1093/infdis/jiq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009;15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 4.Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, Decker JM, Wycuff D, Harris L, Hawkins N, Wood B, Nathe C, Richman D, Tomaras GD, Bibollet-Ruche F, Robinson JE, Morris L, Shaw GM, Montefiori DC, Mascola JR. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82:11651–11668. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, Wu X, Shaw GM, Connors M, Wyatt RT, Mascola JR. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, Sinangil F, Pancera M, Yongping Y, Zhang B, Zhu J, Kwong PD, O’Dell S, Mascola JR, Wu L, Nabel GJ, Phogat S, Seaman MS, Whitesides JF, Moody MA, Kelsoe G, Yang X, Sodroski J, Shaw GM, Montefiori D, Kepler TB, Tomaras GD, Alam SM, Liao HX, Haynes BF. Analysis of a Clonal Lineage of HIV-1 Envelope V2/V3 Conformational Epitope-Specific Broadly Neutralizing Antibodies and Their Inferred Unmutated Common Ancestors. J Virol. 2011 doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 9.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Olivera TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton D, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. Sequence and Structural Convergence of Broad and Potent HIV Antibodies That Mimic CD4 Binding. Science. 2011 doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O’Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose N, Connors M, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. Focused Evolution of HIV-1 Neutralizing Antibodies Revealed by Structures and Deep Sequencing. Science. 2011 doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao X, Chen W, Feng Y, Zhu Z, Prabakaran P, Wang Y, Zhang MY, Longo NS, Dimitrov DS. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun. 2009;390:404–409. doi: 10.1016/j.bbrc.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011 doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, Kwon YD, Zhou T, Wu X, O’Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O’Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 17.Dosenovic P, Chakrabarti B, Soldemo M, Douagi I, Forsell MN, Li Y, Phogat A, Paulie S, Hoxie J, Wyatt RT, Karlsson Hedestam GB. Selective expansion of HIV-1 envelope glycoprotein-specific B cell subsets recognizing distinct structural elements following immunization. J Immunol. 2009;183:3373–3382. doi: 10.4049/jimmunol.0900407. [DOI] [PubMed] [Google Scholar]
- 18.Douagi I, Forsell MN, Sundling C, O’Dell S, Feng Y, Dosenovic P, Li Y, Seder R, Lore K, Mascola JR, Wyatt RT, Karlsson Hedestam GB. Influence of novel CD4 binding-defective HIV-1 envelope glycoprotein immunogens on neutralizing antibody and T-cell responses in nonhuman primates. J Virol. 2010;84:1683–1695. doi: 10.1128/JVI.01896-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, Batzer MA, Bustamante CD, Eichler EE, Hahn MW, Hardison RC, Makova KD, Miller W, Milosavljevic A, Palermo RE, Siepel A, Sikela JM, Attaway T, Bell S, Bernard KE, Buhay CJ, Chandrabose MN, Dao M, Davis C, Delehaunty KD, Ding Y, Dinh HH, Dugan-Rocha S, Fulton LA, Gabisi RA, Garner TT, Godfrey J, Hawes AC, Hernandez J, Hines S, Holder M, Hume J, Jhangiani SN, Joshi V, Khan ZM, Kirkness EF, Cree A, Fowler RG, Lee S, Lewis LR, Li Z, Liu YS, Moore SM, Muzny D, Nazareth LV, Ngo DN, Okwuonu GO, Pai G, Parker D, Paul HA, Pfannkoch C, Pohl CS, Rogers YH, Ruiz SJ, Sabo A, Santibanez J, Schneider BW, Smith SM, Sodergren E, Svatek AF, Utterback TR, Vattathil S, Warren W, White CS, Chinwalla AT, Feng Y, Halpern AL, Hillier LW, Huang X, Minx P, Nelson JO, Pepin KH, Qin X, Sutton GG, Venter E, Walenz BP, Wallis JW, Worley KC, Yang SP, Jones SM, Marra MA, Rocchi M, Schein JE, Baertsch R, Clarke L, Csuros M, Glasscock J, Harris RA, Havlak P, Jackson AR, Jiang H, Liu Y, Messina DN, Shen Y, Song HX, Wylie T, Zhang L, Birney E, Han K, Konkel MK, Lee J, Smit AF, Ullmer B, Wang H, Xing J, Burhans R, Cheng Z, Karro JE, Ma J, Raney B, She X, Cox MJ, Demuth JP, Dumas LJ, Han SG, Hopkins J, Karimpour-Fard A, Kim YH, Pollack JR, Vinar T, Addo-Quaye C, Degenhardt J, Denby A, Hubisz MJ, Indap A, Kosiol C, Lahn BT, Lawson HA, Marklein A, Nielsen R, Vallender EJ, Clark AG, Ferguson B, Hernandez RD, Hirani K, Kehrer-Sawatzki H, Kolb J, Patil S, Pu LL, Ren Y, Smith DG, Wheeler DA, Schenck I, Ball EV, Chen R, Cooper DN, Giardine B, Hsu F, Kent WJ, Lesk A, Nelson DL, O’Brien WE, Prufer K, Stenson PD, Wallace JC, Ke H, Liu XM, Wang P, Xiang AP, Yang F, Barber GP, Haussler D, Karolchik D, Kern AD, Kuhn RM, Smith KE, Zwieg AS. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 20.Andris JS, Miller AB, Abraham SR, Cunningham S, Roubinet F, Blancher A, Capra JD. Variable region gene segment utilization in rhesus monkey hybridomas producing human red blood cell-specific antibodies: predominance of the VH4 family but not VH4-21 (V4-34) Mol Immunol. 1997;34:237–253. doi: 10.1016/s0161-5890(97)00021-7. [DOI] [PubMed] [Google Scholar]
- 21.Bible JM, Howard W, Robbins H, Dunn-Walters DK. IGHV1, IGHV5 and IGHV7 subgroup genes in the rhesus macaque. Immunogenetics. 2003;54:867–873. doi: 10.1007/s00251-003-0536-2. [DOI] [PubMed] [Google Scholar]
- 22.Helmuth EF, Letvin NL, Margolin DH. Germline repertoire of the immunoglobulin V(H)3 family in rhesus monkeys. Immunogenetics. 2000;51:519–527. doi: 10.1007/s002510000170. [DOI] [PubMed] [Google Scholar]
- 23.Howard WA, Bible JM, Finlay-Dijsselbloem E, Openshaw S, Dunn-Walters DK. Immunoglobulin light-chain genes in the rhesus macaque II: lambda light-chain germline sequences for subgroups IGLV1, IGLV2, IGLV3, IGLV4 and IGLV5. Immunogenetics. 2005;57:655–664. doi: 10.1007/s00251-005-0020-2. [DOI] [PubMed] [Google Scholar]
- 24.Howard WA, Bible JM, Finlay-Dijsselbloem E, Openshaw S, Dunn-Walters DK. Immunoglobulin light-chain genes in the rhesus macaque I: kappa light-chain germline sequences for subgroups IGKV1, IGKV and IGKV3. Immunogenetics. 2005;57:210–218. doi: 10.1007/s00251-005-0794-2. [DOI] [PubMed] [Google Scholar]
- 25.Link JM, Hellinger MA, Schroeder HW., Jr The Rhesus monkey immunoglobulin IGHD and IGHJ germline repertoire. Immunogenetics. 2002;54:240–250. doi: 10.1007/s00251-002-0468-2. [DOI] [PubMed] [Google Scholar]
- 26.Sundling C, Forsell MN, O’Dell S, Feng Y, Chakrabarti B, Rao SS, Lore K, Mascola JR, Wyatt RT, Douagi I, Karlsson Hedestam GB. Soluble HIV-1 Env trimers in adjuvant elicit potent and diverse functional B cell responses in primates. J Exp Med. 2010;207:2003–2017. doi: 10.1084/jem.20100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Y, McKee K, Tran K, O’Dell S, Schmidt SD, Phogat A, Forsell MN, Karlsson Hedestam GB, Mascola JR, Wyatt RT. Biochemically defined HIV-1 envelope glycoprotein variant immunogens display differential binding and neutralizing specificities to the CD4-binding site. The Journal of biological chemistry. 2012;287:5673–5686. doi: 10.1074/jbc.M111.317776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, Wang L, Hendrickson WA, Doyle ML, Sodroski J. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol. 2002;76:9888–9899. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheid JF, Mouquet H, Feldhahn N, Walker BD, Pereyra F, Cutrell E, Seaman MS, Mascola JR, Wyatt RT, Wardemann H, Nussenzweig MC. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods. 2009;343:65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, Mehta A, Razavi B, Del Rio C, Zheng NY, Lee JH, Huang M, Ali Z, Kaur K, Andrews S, Amara RR, Wang Y, Das SR, O’Donnell CD, Yewdell JW, Subbarao K, Marasco WA, Mulligan MJ, Compans R, Ahmed R, Wilson PC. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dey B, Svehla K, Xu L, Wycuff D, Zhou T, Voss G, Phogat A, Chakrabarti BK, Li Y, Shaw G, Kwong PD, Nabel GJ, Mascola JR, Wyatt RT. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 2009;5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore JP, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pantophlet R, Ollmann Saphire E, Poignard P, Parren PW, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J Virol. 2003;77:642–658. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, O’Dell S, Walker LM, Wu X, Guenaga J, Feng Y, Schmidt SD, McKee K, Louder MK, Ledgerwood JE, Graham BS, Haynes BF, Burton DR, Wyatt RT, Mascola JR. Mechanism of Neutralization by the Broadly Neutralizing HIV-1 Monoclonal Antibody VRC01. J Virol. 2011;85:8954–8967. doi: 10.1128/JVI.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koch M, Pancera M, Kwong PD, Kolchinsky P, Grundner C, Wang L, Hendrickson WA, Sodroski J, Wyatt R. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology. 2003;313:387–400. doi: 10.1016/s0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- 42.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 43.Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76:4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forsell MN, Dey B, Morner A, Svehla K, O’Dell S, Hogerkorp CM, Voss G, Thorstensson R, Shaw GM, Mascola JR, Karlsson Hedestam GB, Wyatt RT. B cell recognition of the conserved HIV-1 co-receptor binding site is altered by endogenous primate CD4. PLoS Pathog. 2008;4:e1000171. doi: 10.1371/journal.ppat.1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dey B, Pancera M, Svehla K, Shu Y, Xiang SH, Vainshtein J, Li Y, Sodroski J, Kwong PD, Mascola JR, Wyatt R. Characterization of human immunodeficiency virus type 1 monomeric and trimeric gp120 glycoproteins stabilized in the CD4-bound state: antigenicity, biophysics, and immunogenicity. J Virol. 2007;81:5579–5593. doi: 10.1128/JVI.02500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doria-Rose NA, Klein RM, Manion MM, O’Dell S, Phogat A, Chakrabarti B, Hallahan CW, Migueles SA, Wrammert J, Ahmed R, Nason M, Wyatt RT, Mascola JR, Connors M. Frequency and Phenotype of HIV Envelope-specific B Cells From Patients With Broadly Cross-Neutralizing Antibodies. J Virol. 2008 doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lane J, Duroux P, Lefranc MP. From IMGT-ONTOLOGY to IMGT/LIGMotif: the IMGT standardized approach for immunoglobulin and T cell receptor gene identification and description in large genomic sequences. BMC Bioinformatics. 2010;11:223. doi: 10.1186/1471-2105-11-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Souto-Carneiro MM, Longo NS, Russ DE, Sun HW, Lipsky PE. Characterization of the human Ig heavy chain antigen binding complementarity determining region 3 using a newly developed software algorithm, JOINSOLVER. J Immunol. 2004;172:6790–6802. doi: 10.4049/jimmunol.172.11.6790. [DOI] [PubMed] [Google Scholar]
- 50.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;1:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460–465. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McNicholas S, Potterton E, Wilson KS, Noble ME Principal Investigators PG. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.