Abstract
Internationally, breast cancer is the most common female cancer, and is induced by a combination of environmental, genetic, and epigenetic risk factors. Despite the advancement of imaging techniques, invasive sampling of breast epithelial cells is the only definitive diagnostic procedure for patients with breast cancer. To date, molecular biomarkers with high sensitivity and specificity for the screening and early detection of breast cancer are lacking. Recent evidence suggests that the detection of methylated circulating cell-free DNA in the peripheral blood of patients with cancer may be a promising quantitative and noninvasive method for cancer diagnosis. Methylation detection based on a multi-gene panel, rather than on the methylation status of a single gene, may be used to increase the sensitivity and specificity of breast cancer screening. In this review, the results of 14 relevant studies, investigating the efficacy of cell-free DNA methylation screening for breast cancer diagnosis, have been summarized. The genetic risk factors for breast cancer, the methods used for breast cancer detection, and the techniques and limitations related to the detection of cell-free DNA methylation status, have also been reviewed and discussed. From this review, we conclude that the analysis of peripheral blood or other samples to detect differentially methylated cell-free DNA is a promising technique for use in clinical settings, and may improve the sensitivity of screening for both, early detection and disease relapse, and thus improve the future prognosis of patients with breast cancer.
Keywords: Breast neoplasms, Complementary DNA, Early detection of cancer
INTRODUCTION
Internationally, breast cancer is the most common female cancer, and the second most common cause of death in women. According to the National Cancer Institute, in 2011 more than 200,000 women were diagnosed with breast cancer in the United States [1]. Various treatment options are adopted for patients at various stages, and exhibiting particular molecular subtypes of breast cancer. Surgical operations, including breast-conserving surgery and mastectomy, are definitive treatments for the disease. Adjuvant treatments such as radiation therapy, chemotherapy, targeted therapy, and hormonal therapy, are prescribed to reduce disease recurrence and distant metastasis [2].
In the clinical setting, breast cancer is classified into four different molecular subtypes based on immunohistochemical detection of the expression of either the estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), or mindbomb E3 ubiquitin protein ligase 1 antibody (MIB1) [3,4]. The majority of patients with breast cancer have luminal A or luminal B, hormone-responsive breast cancers, and the standard treatment for which is to prescribe 5 to 10 years of tamoxifen or aromatase inhibitors as hormonal therapy. Of the two types, patients with luminal A breast cancer have a better prognosis than those with luminal B breast cancer [5]. HER2-positive breast cancer affects approximately 25% of all patients with breast cancer, and is treated with targeted therapy, for example, trastuzumab or lapatinib [6]. In contrast, triple negative breast cancer (TNBC) affects approximately 15% to 20% of all patients with breast cancer, and can only be treated systemically with chemotherapy due to a lack of targetable ER, PR, and HER2 expression [7]. Chemotherapy treatment is able to achieve a pathological complete response in patients with TNBC when it is given in a neoadjuvant setting [3]. Although it has been suggested that a targeted therapy such as bevacizumab may be effective in treating TNBC, so far this not been possible [8]. Overall, (i.e., despite the use various therapies), the overall recurrence rate for patients with axillary node-negative breast cancer is approximately 25% after primary treatment, whilst a recurrence rate of up to 70% is observed for patients with axillary node-positive disease [9].
GENETIC SUSCEPTIBILITY TO BREAST CANCER
Over the past decade, several breast cancer susceptibility genes of varying penetrance have been identified by linkage and large-scale, population-based genome-wide association studies [10]. These include such genes as BRCA1, BRCA2, TP53, PTEN, ATM, and CHEK2, all of which are involved in both cell cycle regulation and DNA repair pathways. For example, DNA repair by homologous recombination is activated via phosphorylation of BRCA1 by ataxia telangiectasia mutated kinase (ATM), in response to either ionizing radiation, ultraviolet irradiation, or other DNA-damaging agents [11]. When DNA damage is too severe to be repaired, phosphoryl-ated BRCA1 forms complexes with BRCA2 and Rad51, and these subsequently interact with other proteins such as p53 and CHEK2, which are checkpoint genes that regulate the cell cycle [11]. BRCA1 and BRCA2 mutations are the most frequently identified mutations in hereditary breast cancers [12]. Up to 70% of breast cancers in patients with inherited BRCA1 mutations are TNBC, whilst in contrast, inherited BRCA2 mutations are associated with 87% of HER2 negative breast cancer [13]. Individuals carrying germline BRCA1 or BRCA2 mutations have up to an 80% higher lifetime risk of developing breast cancer compared to individuals without any BRCA mutations [12].
Mutations in TP53 and PTEN are pleiotropic. Germline mutations in TP53 are often found in families with Li-Fraumeni syndrome (LFS), and are associated with several types of neoplasm. Patients with LFS are predisposed to the development of a number of neoplasms, including not only breast cancer, but also brain tumors, soft tissue sarcoma, and other malignancies [14]. Similarly, germline mutations in PTEN are the most commonly observed mutations to cause Cowden syndrome, of which breast cancer may form a clinical feature [12]. The estimated breast cancer risks incurred by carriers of TP53 and PTEN mutations are 50% to 60% by the age of 45 years, and 30% to 50% by the age of 70 years, respectively [12]. Amongst the various breast cancer susceptibility genes, mutations in BRCA1, BRCA2, TP53, and PTEN are rare, but show high penetrance, and thus are included in most gene panels designed to predict breast cancer risk [10,15]. Mutations in moderate penetrance genes such as ATM and CHEK2 (such as 1100decC in exon 10), are present in 2% to 3% of BRCA-negative familial breast cancers [12,16], and may result in an approximately 2-fold increase in the risk of developing breast cancer for female heterozygous carriers [12].
A novel, high-penetrance breast cancer susceptibility gene, RecQ helicase-like (RECQL), was recently identified [17], and confirmed to contribute to familial breast cancer by studies in two independent Chinese populations [18,19]. Although mutation of this gene is very rare, the risk of developing breast cancer for RECQL carriers was found to be 5- and 16-fold higher in unselected Polish, and high-risk cases from Quebec, respectively [20]. Overall, mutations in the known susceptibility genes only account for 50% of hereditary breast cancer cases [17]. The factors responsible for the remaining 50% of hereditary breast cancer, and for the incidence of various sporadic breast cancers, are not clear. Since a complex network of genes is involved in the pathogenesis of breast cancer, many common genetic variations are identified in genome-wide association studies aimed at identifying breast cancer susceptibility genes. Certainly, data from exome-sequencing and genome-wide association studies imply that the pathogenesis of breast cancer may be polygenic [12,21]; however, the synergetic effects of low penetrance genetic variants on the development of the disease is yet to be determined and validated. Thus, a current focus of research in this field is the identification of high-sensitivity, highly specific molecular biomarkers for breast cancer, and the improvement of current genetic testing for breast cancer disease and susceptibility.
CURRENT PROCEDURES FOR BREAST CANCER DETECTION AND DIAGNOSIS
Mammography and ultrasonography (USG) are commonly used imaging modalities for the detection of breast abnormalities. The sensitivity of mammography is only moderate, and moreover is greatly affected by age and breast density [22]. Similarly, the use of USG for breast cancer screening incurs a high rate of false negative results, particularly in women with dense breast tissue. Thus, USG is generally used as a supplement to mammography during screening to improve detection sensitivity, especially for women with dense breast tissue [23]. In contrast, magnetic resonance imaging (MRI) has been shown to have higher sensitivity in breast cancer detection regardless of breast density; however, it is less likely to detect ductal carcinoma in situ, and is much more expensive than detection via mammography [23,24]. Moreover, there is limited evidence to suggest that the use of MRI and/or USG as screening tools can reduce mortality to the same extent as mammography [25]. As a result, mammography remains the standard method for breast cancer screening, whilst USG and MRI are used as supplementary imaging tools for high-risk women with either dense breast tissue, or identified germline mutations [23,24].
Currently, the definitive diagnosis of breast cancer requires the sampling of breast epithelial cells by fine needle aspiration (FNA), and/or core biopsy guided stereotactically, ultrasonically, or where applicable, by MRI [26]. The sensitivity of FNA and core biopsy in detecting breast cancer is 82% and 93% respectively, and this rate is increased to 97% when both FNA and core biopsy are conducted [26]. Due to its high sensitivity, core biopsy with subsequent immunohistochemical assessment to detect breast cancer molecular subtypes is currently the ‘gold standard’ for breast tissue assessment and breast cancer therapy determination [26,27]. After the commencement of treatment, carcinoembryonic antigen (CEA) and CA15-3, which are the most widely used serum tumor biomarkers, are used in the clinical setting to monitor patient therapeutic response and predict prognosis [28]. Although high concentrations of CEA and CA15-3 are associated with poor prognosis and poor treatment response, limited evidence is available to support the use of these serum biomarkers for the routine surveillance for patients with breast cancer [28,29]. CA15-3 is a serum tumor marker not only for breast, but also for colon cancer, and similarly CEA is serum tumor marker for breast, colon, lung, and bladder cancer [30]. In addition, an increased concentration of CA15-3 only occurs in approximately 10% of patients with stage I breast cancer. Thus, early cancer detection is not feasible with these conventional serum tumor biomarkers [28,30], rendering invasive screening and diagnostic procedures unavoidable at present, especially for asymptomatic patients.
DNA METHYLATION STATUS IN BREAST CANCER
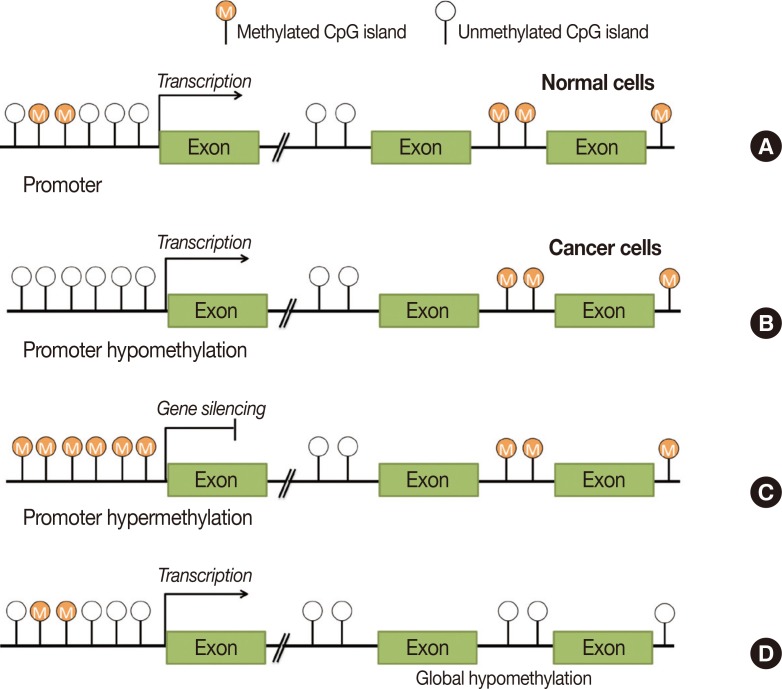
Recent studies have suggested that epigenetic mechanisms such as DNA methylation, microRNA activity, and histone acetylation, that cause gene silencing, may underly the development of breast cancers of varying molecular subtypes. Of these mechanisms, DNA methylation is the predominant epigenetic modification found in human disease, particularly at regions enriched with cytosine and guanine dinucleotides (CpG) [31]. CpG regions are associated with gene promoters, that are located near transcription start sites in 70% of genes [32]. Unlike genetic mutation, DNA methylation is a reversible process that alters gene expression patterns without modifying the DNA sequence. Hypomethylation and hypermethylation states are both associated with human cancer, and have been shown to affect different regions of the genome dependent on the cancer type (Figure 1) [33]. Hypomethylation is often detected in tumorous and metastatic tissues when compared with adjacent normal tissues [33], and generally increases the expression of oncogenes, activates transcription, and alters genome stability. Similarly, hypermethylation of CpG-rich genomic regions represses tumor-suppressing genes, predominantly (i.e., almost 80% of cases) via the most common form of DNA methylation, 5-methylcytosine [33,34, 35]. These epigenetic changes occur in normal tissues during the early stages of carcinogenesis, and eventually lead to the development of breast cancer [36]. Thus, changes to the methylation status of genes related to the epigenetic machinery of carcinogenesis may be useful molecular biomarkers for the early detection of breast cancer.
Figure 1. Hypomethylation and hypermethylation of human genome. (A) In normal cells, cytosine and guanine dinucleotides (CpG) islands in the promoter region are unmethylated. (B) In cancer cells, promoter hypomethylation of oncogenic genes lead to transcription initiation. (C) Promoter hypermethylation of tumor suppressor genes lead to gene silencing. (D) Global hypomethylation affects gene integrity and stability.
The methods used to detect DNA methylation are similar to those used to detect single nucleotide polymorphisms or mutations in genomic DNA, i.e., polymerase chain reaction (PCR) and sequencing, respectively. However, to facilitate the detection of DNA methylation, DNA samples must be pretreated either by enzymatic digestion with restriction enzymes, or by bisulfite conversion [31]. During enzymatic digestion, unmethylated CpG islands are recognized and cleaved by methylation-sensitive restriction enzymes, while methylated CpG islands remain intact. Methylation status can then be assessed via PCR using primers that only amplify regions containing restriction sites [37]. This method is useful for identifying highly methylated genes, for example, those silenced during X-chromosome inactivation [37]. Other methods, such as methylation-sensitive restriction enzyme digestion with real-time quantitative PCR, or combined bisulfite restriction analysis, make use of methylation-sensitive restriction enzymes to digest plasma DNA for subsequent real-time quantitative analysis [38,39]. However, incomplete enzymatic digestion of unmethylated CpG islands can lead to false positive results using these methods [37].
Bisulfite conversion is a chemical modification technique that changes cytosine bases to uracils via three processes: sulphonation, hydrolytic deamination, and alkali desulphonation [38]. Methylated CpG islands are resistant to these chemical modifications, and thus can be identified during PCR-based amplification of the genomic region of interest using primers specific for methylated and unmethylated CpG islands [37]. At unmethylated CpG islands, cytosines are converted to uracils and bind their complementary bases (adenine) during PCR amplification, whereas methylated CpG islands resist conversion and instead bind guanines during amplification [37]. As discussed for enzymatic digestion, incomplete bisulfite conversion may lead to false positive results [38].
Various locus-based methods have been developed to establish the methylation status of bisulfite-treated DNA, including methylation-specific PCR (MSP), quantitative real-time MSP (qMSP), and pyrosequencing [40]. The use of next-generation sequencing-based techniques such as whole genome shotgun bisulfite sequencing have recently become more common [40]. MSP is a qualitative, rapid, and robust detection method with high sensitivity and specificity, in which the amplification of a region of interest is achieved using primers specific to methylated and unmethylated DNA [37], and the resulting PCR-products are then visualized via gel electrophoresis [37]. qMSP was developed from MSP to allow quantification of the real-time methylation status of a given target gene, in addition to MSP-derived qualitative information [38]. Nevertheless, since methylation is both tissue-specific and changes with time, qMSP is unable to generate quantitative information regarding the methylation status of each CpG site in a given target gene. Pyrosequencing is a real-time, quantitative DNA-sequencing technique that detects the light signal generated by pyrophosphate released during the nucleotide incorporation enzymatic cascade [41]. As a result, it is able to obtain not only quantitative information, but can also establish the DNA methylation pattern at individual CpG sites by analyzing the cytosine to thymine ratio in amplified products [41].
These various methods for investigating the DNA methylation status of a given region of interest are particularly useful for studies that employ a candidate-gene approach, i.e., a hypothesis-driven approach to study genes with known functional roles related to a disease of interest [42]. Both enzymatic digestion and bisulfite conversion have recently been adapted for use in array hybridization and next-generation sequencing. This allows for the detection of the genome-wide methylation status of a given individual, and makes the techniques useful in epigenome-wide association studies (EWAS) that evaluate the role of common variants in complex diseases [40,43].
THE DNA METHYLATION STATUS OF CIRCULATING CELL-FREE DNA AS A CANCER DIAGNOSIS TOOL
The discovery of cell-free circulating DNA (cfDNA) in plasma and serum has led to investigation into its potential use as a biomarker for various diseases, including breast cancer. Although the molecular mechanism underlying cfDNA is still unclear, it is likely that a primary source of cfDNA may be the apoptosis of normal nucleated cells. The concentration of cfDNA is generally low in healthy individuals because it is rapidly removed from circulation by phagocytosis [44,45]. During tumorigenesis, excessive amounts of cfDNA are released by necrotic and apoptotic tumor cells, and also by lysed circulating tumor cells in the blood stream, causing a high level of cfDNA to accumulate in patients with cancer [44]. Since cfDNA carries a methylation pattern identical to that of the genomic DNA in tumor cells, detection of cfDNA isolated from plasma or serum represents a potential noninvasive approach for quantifying both genetic and epigenetic information in patients with cancer [45]. Thus, numerous studies investigating the efficacy and validity of using cfDNA for cancer detection have been conducted [44]. A summary of 14 such studies in patients with breast cancer is shown in Table 1.
Table 1. Summary of published studies on investigating diagnosis value of circulating methylated DNA in breast cancer patients.
| Author | Year | Sample size | Type of specimen | Gene | Method | Reference |
|---|---|---|---|---|---|---|
| Chimonidou et al. | 2013 | Control, 60; cancer, 114 | Plasma | SOX17 | qMSP | [46] |
| Chimonidou et al. | 2013 | Control, 37; cancer, 27 (discovery), 46 (metastatic), 123 (validation) | Plasma | CST6 | MSP | [47] |
| Dulaimi et al. | 2004 | Control, 20; benign, 8; cancer, 34 | Serum | RASSF1A, APC, DAP-kinase | MSP | [48] |
| Fu et al. | 2015 | Benign, 60; cancer, 155 | Plasma | SOX17 | MSP | [49] |
| Guerrero-Preston et al. | 2014 | Control, 20 (discovery), 86 (validation); cancer, 20 (discovery), 154 (validation) | Plasma | MAL, KIF1A, FKBP4, VGF, OGDHL | MSP | [50] |
| Hagrass et al. | 2014 | Benign, 100; cancer, 120 | Serum | ESα | MSP | [51] |
| Hoque et al. | 2006 | Control, 76 (discovery), 38 (validation); cancer, 93 (discovery), 47 (validation) | Plasma | APC, GSTP1, RASSF1A, RARβ2 | qMSP | [52] |
| Martínez-Galán et al. | 2008 | Control, 74; benign, 34; cancer, 106; postoperational, 60 | Serum | ESR1, APC, RARB, 14-3-3-σ, E-cadherin | qMSP | [53] |
| Ng et al. | 2011 | Control, 60 (discovery), 20 (validation); breast cancer, 60 (discovery), 38 (vali- dation); gastric cancer, 45 (discovery), 20 (validation) | Plasma | SLC19A3 | MSRED-qPCR | [39] |
| Radpour et al. | 2011 | Control, 30 (plasma, discovery); cancer, 36 (plasma, discovery), 20 (serum, validation) | Plasma, serum | APC, BIN1, BMP6, BRCA1, CST6, ESR-b, GSTP1, p16, p21, TIMP3 | EpiTYPERTM assay | [54] |
| Sharma et al. | 2011 | Control, 30; cancer, 100 | Serum | Maspin (SERPINB5) | MSP | [55] |
| Silva et al. | 1999 | Control, 17; cancer, 35 | Plasma | P16INK4a | MSRE-PCR | [56] |
| Skvortsova et al. | 2006 | Control, 10; benign, 15; cancer, 20 | Plasma | HIC-1, RASSF1A, RARβ2 | MSP | [57] |
| Yamamoto et al. | 2012 | Control, 87; cancer, 159 | Serum | GSTP1, RASSF1A, RARβ2 | One-step MSP | [58] |
Each of these 14 published studies used a candidate-gene approach.
qMSP=quantitative methylation-specific polymerase chain reaction; MSP=methylation-specific polymerase chain reaction; MSRED-qPCR=methylation-sensitive restriction enzyme digestion and real-time quantitative polymerase chain reaction; MSRE-PCR=methylation-sensitive restriction enzyme digestion real-time polymerase chain reaction.
Each of these 14 published studies used a candidate-gene approach to investigate the potential of selected genes as molecular biomarkers for breast cancer detection according to differential methylation status of cfDNA between healthy individuals and patients with breast cancer [39,46,47,48,49,50,51,52,53,54,55,56,57,58]. Seven studies investigated the methylation status of a single candidate gene only between healthy individuals and patients with breast cancer, whilst the remaining studies analyzed between three and nine genes. Amongst all the evaluated candidate genes, adenomatous polyposis coli (APC), Ras association domain family 1 isoform A (RASSF1A), and retinoic acid receptor β2 (RARβ2) were the three most extensively analyzed with regards to breast cancer development. All studies were case-controlled, and included sample sizes ranging from 20 to 159 patients with breast cancer. With the exception of the studies by Hagrass et al. [51] and Fu et al. [49], individuals with benign breast conditions were used as controls. Three studies also included healthy individuals along with patients with benign disease in the control groups, and one study included 45 patients with gastric cancer in this group [39,48,53,57]. Bisulfide conversion-based methods were used in all studies except those by Ng et al. and Silva et al., in which enzymatic digestion-based methods were instead employed [39,46,47,48,49,50,51,52,53,54,55,56,57,58]. Amongst all the bisulfite conversion-based techniques, MSP was the most frequently used to detect the methylation status of cfDNA [47,48,49,51,55,57]. Six studies used quantitative detection methods to assess the cfDNA methylation status, one of which employed a commercially available assay [52,53,54,58]. Eight of the studies analyzed plasma samples, five studies analyzed serum samples, and one study analyzed both sample types in two cohorts of patients with cancer. Five of the fourteen studies used a two-stage study design, including a discovery cohort and independent validation cohort, to assess cfDNA methylation status in the context of breast cancer development [39,46,50,52,54].
The results of the 14 studies promote the use of a multigene panel rather than a single gene as a molecular biomarker, to ensure high sensitivity and specificity during cancer detection. Sex determining region Y box 17 (Sox17) is a transcription factor essential for several developmental processes and for the regulation of stem cell function, and its methylation status with relation to breast cancer development was investigated by both Chimonidou et al. [46] and Fu et al. [49]. Whilst no promoter methylation was detected by Fu et al. in individuals with benign breast conditions, Chimonidou et al. detected SOX17 promoter methylation in 4.3% of control patients. A similar finding was also reported by Hoque et al. [52] regarding the methylation status of RASSF1A, a tumor suppressor gene that regulates cell cycle progression and helps to maintain genetic stability [59]. Inactivation of RASSF1A via promoter methylation has been reported in several primary tumors, including breast tumors [59]. Amongst the four studies that included RASSF1A as a candidate gene, three identified no methylation in the control group, whereas one (by Hoque et al. [52]) found 5% of normal control patients to express methylated RASSF1A in plasma. Another study detected 20% and 49% methylation in at least one gene of a five-gene panel in the control group and cancer patient groups, respectively [50]. Thus, together these results demonstrate that careful consideration must be taken in selecting candidate genes as differential breast cancer biomarkers.
Notably, the sensitivity of methylation-status detection was lower in plasma or serum samples than in primary tumor tissues. For example, the sensitivity of using SOX17 methylation as a breast cancer diagnostic tool was decreased from 73% to 58% when comparing paired tumor and plasma samples [49]. Similarly, the detection sensitivity reported by Dulaimi et al. [48] was 65% in tumor samples and only 56% in serum samples, a finding that studies by both Hoque et al. [52] and Yamamoto et al. [58] supported. The sensitivity of cancer detection using a multi-gene panel comprising RASSF1A and RARβ2 was reported to be as high as 95% for patients with breast cancer, and 100% for healthy controls [57]. Other studies reported similar findings when using multigene panels for cancer diagnosis [52,58]. Therefore, in addition to selecting appropriate candidate genes to increase specificity, the use of a multi-gene panel is recommended to enhance the sensitivity of cancer detection via differential cfDNA methylation levels.
Finally, a minimum of 4 to 40 ng of DNA is recommended per sample input to ensure adequate PCR efficiency during cfDNA analysis, and to prevent the generation of false-negative results due to PCR-inhibition caused by low sample concentration [38]. Notably, of all the cfDNA studies that used bisulfite-based techniques, only two specified the concentration of bisulfite-converted DNA used per reaction in their experiments [47,51].
CONCLUSION
Several recently published studies show promising results in differentiating patients with cancer from healthy individuals according to cfDNA methylation patterns. Nevertheless, future replication studies are required to validate the prediction potentials of the published epigenetic biomarkers with the use of new technologies, for example, next-generation DNA-methylation sequencing to obtain both quantitative and qualitative information. The first EWAS identified 250 differentially methylated CpG islands in the peripheral blood samples of high-risk individuals that developed breast cancer during their post-cancer monitoring, as compared to those that remained cancer-free [60]. Since these high-risk individuals had a family history of breast cancer, genetic, as well as environmental or other factors, may have contributed to these results. In addition, the EWAS analyzed peripheral blood samples only, thus it is not clear whether the evaluation of cfDNA methylation status using plasma or serum samples would have generated the same result. It is also important to note that the patient sample size of published studies thus far, (even including those with a two-stage study design), has been relatively small, limiting their resolution power. Thus, a sample size calculation model specifically designed for future EWAS has been proposed [61]. The sample type used for quantifying cfDNA in patients with cancer will also be critical to improve future studies. Serum has been shown to generally yield a higher concentration of cfDNA than plasma due to the lysis of leukocytes during clotting, such that the results generated using the two sample types are often incomparable. Furthermore, future studies should include not only a large cohort of patients with breast cancer, but also a large control cohort comprising healthy individuals both with and without benign conditions, to more accurately assess the potential of using cfDNA methylation status as an early cancer diagnosis tool. Given that the number of EWAS is increasing, a standardized set of guidelines for conducting these types of studies is needed so as to enable data pooling across studies, thereby overcoming the current limitations of this field of research imposed by the discussed small sample sizes. Thus, it is hoped that continued research into the use of differentially methylated cfDNA to identify breast cancer may lead to the development of a sensitive and specific molecular diagnostic test for use in the clinical setting, to screen for both early detection and patient relapse, and to thus improve prognosis for patients with breast cancer.
Footnotes
CONFLICT OF INTEREST: The authors declare that they have no competing interests.
References
- 1.United States Cancer Statistics Working Group. United States Cancer Statistics: 1999–2011 Incidence and Mortality Web-based Report. Atlanta: U.S.: Department of Health and Human Services, Centers for Disease Control and Prevention, and National Cancer Institute; 2014. [Google Scholar]
- 2.Maughan KL, Lutterbie MA, Ham PS. Treatment of breast cancer. Am Fam Physician. 2010;81:1339–1346. [PubMed] [Google Scholar]
- 3.Brouckaert O, Wildiers H, Floris G, Neven P. Update on triple-negative breast cancer: prognosis and management strategies. Int J Womens Health. 2012;4:511–520. doi: 10.2147/IJWH.S18541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmadeka R, Harmon BE, Singh M. Triple-negative breast carcinoma: current and emerging concepts. Am J Clin Pathol. 2014;141:462–477. doi: 10.1309/AJCPQN8GZ8SILKGN. [DOI] [PubMed] [Google Scholar]
- 5.Schnitt SJ. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod Pathol. 2010;23(Suppl 2):S60–S64. doi: 10.1038/modpathol.2010.33. [DOI] [PubMed] [Google Scholar]
- 6.Jones KL, Buzdar AU. Evolving novel anti-HER2 strategies. Lancet Oncol. 2009;10:1179–1187. doi: 10.1016/S1470-2045(09)70315-8. [DOI] [PubMed] [Google Scholar]
- 7.Gluz O, Liedtke C, Gottschalk N, Pusztai L, Nitz U, Harbeck N. Triple-negative breast cancer: current status and future directions. Ann Oncol. 2009;20:1913–1927. doi: 10.1093/annonc/mdp492. [DOI] [PubMed] [Google Scholar]
- 8.Cameron D, Brown J, Dent R, Jackisch C, Mackey J, Pivot X, et al. Adjuvant bevacizumab-containing therapy in triple-negative breast cancer (BEATRICE): primary results of a randomised, phase 3 trial. Lancet Oncol. 2013;14:933–942. doi: 10.1016/S1470-2045(13)70335-8. [DOI] [PubMed] [Google Scholar]
- 9.Lucci A, Hall CS, Lodhi AK, Bhattacharyya A, Anderson AE, Xiao L, et al. Circulating tumour cells in non-metastatic breast cancer: a prospective study. Lancet Oncol. 2012;13:688–695. doi: 10.1016/S1470-2045(12)70209-7. [DOI] [PubMed] [Google Scholar]
- 10.Fanale D, Amodeo V, Corsini LR, Rizzo S, Bazan V, Russo A. Breast cancer genome-wide association studies: there is strength in numbers. Oncogene. 2012;31:2121–2128. doi: 10.1038/onc.2011.408. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004;95:866–871. doi: 10.1111/j.1349-7006.2004.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ripperger T, Gadzicki D, Meindl A, Schlegelberger B. Breast cancer susceptibility: current knowledge and implications for genetic counselling. Eur J Hum Genet. 2009;17:722–731. doi: 10.1038/ejhg.2008.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mavaddat N, Barrowdale D, Andrulis IL, Domchek SM, Eccles D, Nevanlinna H, et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) Cancer Epidemiol Biomarkers Prev. 2012;21:134–147. doi: 10.1158/1055-9965.EPI-11-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichols KE, Malkin D, Garber JE, Fraumeni JF, Jr, Li FP. Germ-line p53 mutations predispose to a wide spectrum of early-onset cancers. Cancer Epidemiol Biomarkers Prev. 2001;10:83–87. [PubMed] [Google Scholar]
- 15.Easton DF, Pharoah PD, Antoniou AC, Tischkowitz M, Tavtigian SV, Nathanson KL, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372:2243–2257. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turnbull C, Seal S, Renwick A, Warren-Perry M, Hughes D, Elliott A, et al. Gene-gene interactions in breast cancer susceptibility. Hum Mol Genet. 2012;21:958–962. doi: 10.1093/hmg/ddr525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cybulski C, Carrot-Zhang J, Kluźniak W, Rivera B, Kashyap A, Wokołorczyk D, et al. Germline RECQL mutations are associated with breast cancer susceptibility. Nat Genet. 2015;47:643–646. doi: 10.1038/ng.3284. [DOI] [PubMed] [Google Scholar]
- 18.Sun J, Wang Y, Xia Y, Xu Y, Ouyang T, Li J, et al. Mutations in RECQL gene are associated with predisposition to breast cancer. PLoS Genet. 2015;11:e1005228. doi: 10.1371/journal.pgen.1005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong A, Shin VY, Cheuk IW, Chen J, Au CH, Ho DN, et al. Germline RECQL mutations in high risk Chinese breast cancer patients. Breast Cancer Res Treat. 2016;157:211–215. doi: 10.1007/s10549-016-3784-1. [DOI] [PubMed] [Google Scholar]
- 20.Akbari MR, Cybulski C. RECQL: a DNA helicase in breast cancer. Oncotarget. 2015;6:26558–26559. doi: 10.18632/oncotarget.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevens KN, Vachon CM, Couch FJ. Genetic susceptibility to triple-negative breast cancer. Cancer Res. 2013;73:2025–2030. doi: 10.1158/0008-5472.CAN-12-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Srivastava S. Strategic approach to validating methylated genes as biomarkers for breast cancer. Cancer Prev Res (Phila) 2010;3:16–24. doi: 10.1158/1940-6207.CAPR-09-0098. [DOI] [PubMed] [Google Scholar]
- 23.Lee CH, Dershaw DD, Kopans D, Evans P, Monsees B, Monticciolo D, et al. Breast cancer screening with imaging: recommendations from the Society of Breast Imaging and the ACR on the use of mammography, breast MRI, breast ultrasound, and other technologies for the detection of clinically occult breast cancer. J Am Coll Radiol. 2010;7:18–27. doi: 10.1016/j.jacr.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 24.Morrow M, Waters J, Morris E. MRI for breast cancer screening, diagnosis, and treatment. Lancet. 2011;378:1804–1811. doi: 10.1016/S0140-6736(11)61350-0. [DOI] [PubMed] [Google Scholar]
- 25.Shetty MK. Screening for breast cancer with mammography: current status and an overview. Indian J Surg Oncol. 2010;1:218–223. doi: 10.1007/s13193-010-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieske B, Ravichandran D, Wright D. Role of fine-needle aspiration cytology and core biopsy in the preoperative diagnosis of screen-detected breast carcinoma. Br J Cancer. 2006;95:62–66. doi: 10.1038/sj.bjc.6603211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eccles SA, Aboagye EO, Ali S, Anderson AS, Armes J, Berditchevski F, et al. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res. 2013;15:R92. doi: 10.1186/bcr3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duffy MJ. Serum tumor markers in breast cancer: are they of clinical value? Clin Chem. 2006;52:345–351. doi: 10.1373/clinchem.2005.059832. [DOI] [PubMed] [Google Scholar]
- 29.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 30.Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210–219. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- 31.Dahl C, Guldberg P. DNA methylation analysis techniques. Biogerontology. 2003;4:233–250. doi: 10.1023/a:1025103319328. [DOI] [PubMed] [Google Scholar]
- 32.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 34.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 35.Bock C. Analysing and interpreting DNA methylation data. Nat Rev Genet. 2012;13:705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 36.Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. 2016;17:284–299. doi: 10.1038/nrg.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark SJ, Statham A, Stirzaker C, Molloy PL, Frommer M. DNA methylation: bisulphite modification and analysis. Nat Protoc. 2006;1:2353–2364. doi: 10.1038/nprot.2006.324. [DOI] [PubMed] [Google Scholar]
- 39.Ng EK, Leung CP, Shin VY, Wong CL, Ma ES, Jin HC, et al. Quantitative analysis and diagnostic significance of methylated SLC19A3 DNA in the plasma of breast and gastric cancer patients. PLoS One. 2011;6:e22233. doi: 10.1371/journal.pone.0022233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 41.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 42.Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet. 2002;3:391–397. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- 43.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwarzenbach H, Pantel K. Circulating DNA as biomarker in breast cancer. Breast Cancer Res. 2015;17:136. doi: 10.1186/s13058-015-0645-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swarup V, Rajeswari MR. Circulating (cell-free) nucleic acids: a promising, non-invasive tool for early detection of several human diseases. FEBS Lett. 2007;581:795–799. doi: 10.1016/j.febslet.2007.01.051. [DOI] [PubMed] [Google Scholar]
- 46.Chimonidou M, Strati A, Malamos N, Georgoulias V, Lianidou ES. SOX17 promoter methylation in circulating tumor cells and matched cell-free DNA isolated from plasma of patients with breast cancer. Clin Chem. 2013;59:270–279. doi: 10.1373/clinchem.2012.191551. [DOI] [PubMed] [Google Scholar]
- 47.Chimonidou M, Tzitzira A, Strati A, Sotiropoulou G, Sfikas C, Malamos N, et al. CST6 promoter methylation in circulating cell-free DNA of breast cancer patients. Clin Biochem. 2013;46:235–240. doi: 10.1016/j.clinbiochem.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 48.Dulaimi E, Hillinck J, Ibanez de Caceres I, Al-Saleem T, Cairns P. Tumor suppressor gene promoter hypermethylation in serum of breast cancer patients. Clin Cancer Res. 2004;10(18 Pt 1):6189–6193. doi: 10.1158/1078-0432.CCR-04-0597. [DOI] [PubMed] [Google Scholar]
- 49.Fu D, Ren C, Tan H, Wei J, Zhu Y, He C, et al. Sox17 promoter methylation in plasma DNA is associated with poor survival and can be used as a prognostic factor in breast cancer. Medicine (Baltimore) 2015;94:e637. doi: 10.1097/MD.0000000000000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guerrero-Preston R, Hadar T, Ostrow KL, Soudry E, Echenique M, Ili-Gangas C, et al. Differential promoter methylation of kinesin family member 1a in plasma is associated with breast cancer and DNA repair capacity. Oncol Rep. 2014;32:505–512. doi: 10.3892/or.2014.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagrass HA, Pasha HF, Ali AM. Estrogen receptor alpha (ER alpha) promoter methylation status in tumor and serum DNA in Egyptian breast cancer patients. Gene. 2014;552:81–86. doi: 10.1016/j.gene.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 52.Hoque MO, Feng Q, Toure P, Dem A, Critchlow CW, Hawes SE, et al. Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. J Clin Oncol. 2006;24:4262–4269. doi: 10.1200/JCO.2005.01.3516. [DOI] [PubMed] [Google Scholar]
- 53.Martínez-Galán J, Torres B, Del Moral R, Muñoz-Gámez JA, Martín-Oliva D, Villalobos M, et al. Quantitative detection of methylated ESR1 and 14-3-3-sigma gene promoters in serum as candidate biomarkers for diagnosis of breast cancer and evaluation of treatment efficacy. Cancer Biol Ther. 2008;7:958–965. doi: 10.4161/cbt.7.6.5966. [DOI] [PubMed] [Google Scholar]
- 54.Radpour R, Barekati Z, Kohler C, Lv Q, Bürki N, Diesch C, et al. Hypermethylation of tumor suppressor genes involved in critical regulatory pathways for developing a blood-based test in breast cancer. PLoS One. 2011;6:e16080. doi: 10.1371/journal.pone.0016080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharma G, Mirza S, Parshad R, Srivastava A, Gupta SD, Pandya P, et al. Clinical significance of Maspin promoter methylation and loss of its protein expression in invasive ductal breast carcinoma: correlation with VEGF-A and MTA1 expression. Tumour Biol. 2011;32:23–32. doi: 10.1007/s13277-010-0087-8. [DOI] [PubMed] [Google Scholar]
- 56.Silva JM, Dominguez G, Villanueva MJ, Gonzalez R, Garcia JM, Corbacho C, et al. Aberrant DNA methylation of the p16INK4a gene in plasma DNA of breast cancer patients. Br J Cancer. 1999;80:1262–1264. doi: 10.1038/sj.bjc.6690495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skvortsova TE, Rykova EY, Tamkovich SN, Bryzgunova OE, Starikov AV, Kuznetsova NP, et al. Cell-free and cell-bound circulating DNA in breast tumours: DNA quantification and analysis of tumour-related gene methylation. Br J Cancer. 2006;94:1492–1495. doi: 10.1038/sj.bjc.6603117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamamoto N, Nakayama T, Kajita M, Miyake T, Iwamoto T, Kim SJ, et al. Detection of aberrant promoter methylation of GSTP1, RASSF1A, and RARbeta2 in serum DNA of patients with breast cancer by a newly established one-step methylation-specific PCR assay. Breast Cancer Res Treat. 2012;132:165–173. doi: 10.1007/s10549-011-1575-2. [DOI] [PubMed] [Google Scholar]
- 59.Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120(Pt 18):3163–3172. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- 60.Xu Z, Bolick SC, DeRoo LA, Weinberg CR, Sandler DP, Taylor JA. Epigenome-wide association study of breast cancer using prospectively collected sister study samples. J Natl Cancer Inst. 2013;105:694–700. doi: 10.1093/jnci/djt045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsai PC, Bell JT. Power and sample size estimation for epigenome-wide association scans to detect differential DNA methylation. Int J Epidemiol. 2015;44:pii: dyv041. doi: 10.1093/ije/dyv041. [DOI] [PMC free article] [PubMed] [Google Scholar]