Abstract
The female vaginal environment contains diverse microorganisms, and their interactions play significant roles in health and disease. Lactobacillus species are the predominant vaginal microorganisms in healthy women and relevant as a barrier to defense against pathogens, including Candida albicans. The yeast-to-hyphae transition is believed to be a determinant of C. albicans pathogenesis. In this study, we investigated the effects of vaginal isolates of L. crispatus (seven strains), L. gasseri (six strains), and L. jensenii (five strains) on growth, hyphal formation and virulence-related genes expression of C. albicans ATCC 10231. We found that the L. crispatus showed the most significant antimicrobial activities in microplate-based liquid medium assay (P < 0.05). All seven cell-free supernatants (CFS) from L. crispatus strains reduced the growth of C. albicans by >60%. The effects might be due to their productions of some secretory antimicrobial compounds in addition to H2O2 and organic acids. Furthermore, each of the CFS of Lactobacillus strains was found to significantly suppress the yeast-to-hyphae transition of C. albicans under hyphae-inducing conditions (RPMI 1640 medium supplemented with 10% fetal bovine serum). The hyphae inhibition rates of C. albicans treated by CFS from L. crispatus, L. gasseri, and L. jensenii were 88.3 ± 3.02%, 84.9 ± 6.0%, and 81.9 ± 6.2%, respectively. Moreover, the expression of hyphae-specific genes (ALS3, HWP1, ECE1, EAP1, and SAP5) and transcriptional regulatory genes (EFG1, TEC1, and NRG1) were analyzed using quantitative real-time PCR. The results demonstrated that L. crispatus CFS significantly down-regulated the expression of hyphae-specific genes ALS3 (0.140-fold)), HWP1 (0.075-fold), and ECE1 (0.045-fold), while up-regulated the expression of the negative transcriptional regulator gene NRG1 with 1.911-fold. The antimicrobial compounds from L. crispatus B145 against Candida growth were heat stable and protease resistance, but those against hyphal formation were partially sensitive to the same treatments. Our novel findings suggest that L. crispatus, a dominant Lactobacillus species associated with a healthy vagina, could strongly inhibit C. albicans growth and hyphal formation. L. crispatus might repress the expression of hyphae-specific genes (ALS3, HWP1, and ECE1) in a NRG1-dependent manner. Besides, L. crispatus B145 is highly worthwhile for probiotic investigation.
Keywords: Lactobacillus, Candida albicans, microbiota, vulvovaginal candidiasis, antimicrobial activity, yeast-tohyphae
Introduction
Candida albicans is an opportunistic pathogen and the most prevalent fungal species of the human microbiota (da Silva Dantas et al., 2016). In individuals with healthy immune systems, C. albicans asymptomatically colonizes mucosal surfaces. However, in some conditions this species can cause infections, such as VVC (Nobile and Johnson, 2015). The overgrowth and morphological transition (e.g., yeast-to-hyphae transition) of C. albicans are very important determinants to promote the conversion from a commensal to a pathogen (Jacobsen et al., 2012). VVC is responsible for a great morbidity among women of reproductive-age and significant burden to the health care system due to rising vaginitis-related health care costs. It is estimated that about 75% of all women at the childbearing age are afflicted by VVC at least once in their lifetime with ∼40–50% experiencing at least one additional episode of infection, ∼5–8% of women suffer from at least four recurrent VVC per year (Peters et al., 2014; Cassone, 2015).
It is well accepted that the interactions between C. albicans and other components of the resident bacteria, particularly Lactobacillus spp., play important roles in determining the commensal or pathogenic outcome of this fungus (Förster et al., 2016). Lactobacilli are predominant microorganisms in the normal vaginal microbiota, where the most frequently isolated species are L. crispatus, L. gasseri, and L. jensenii (Petrova et al., 2015). In a healthy status, C. albicans and lactobacilli naturally co-inhabit in the female genitourinary tract, where the former lives as a minority co-habitant. It is reported that lactobacilli could inhibit the overgrowth of C. albicans by producing a wide variety of secondary metabolites with antimicrobial activity, such as lactic acid, hydrogen peroxide (H2O2), bacteriocin-like compounds, and biosurfactants (Borges et al., 2014). For instance, bacteriocin L23 produced by L. fermentum isolate displayed inhibition effect on C. albicans growth (Pascual et al., 2008). However, the mechanisms underlying anti-Candida activity are still not clearly understood.
When the balance of vaginal flora is disturbed by the weakened immune system, antibiotic usage and other risk factors, the lactobacilli biomass would decrease and its species composition might shift, while C. albicans could overgrow and undergo a morphogenetic change from a round-ovoid typical yeast cell to a hyphal growing organism (Ma et al., 2012; Cassone, 2015). The yeast-to-hyphae transition is believed to be a determinant of C. albicans pathogenesis, and it could be triggered by various environmental cues in vitro, including neutral pH, presence of serum or N-acetyl glucosamine, elevated carbon dioxide concentration and physiological temperature (Mayer et al., 2013; Palková and Váchová, 2016). In addition, the morphological transition from yeast to hyphae is regulated by a complex network of signaling pathway in C. albicans. Several virulence-related genes that are specifically expressed in hyphae have been suggested to play essential roles in C. albicans pathogenesis (Sudbery, 2011; Jacobsen et al., 2012). For example, the hyphal wall protein 1 gene (HWP1), agglutinin-like protein gene 3 (ALS3), and extent of cell elongation gene 1 (ECE1) encode cell surface proteins, which are important for hyphal growth and adhesion to host cells (Fan et al., 2013; Vila et al., 2016). Expressions of these hyphae-specific genes are positively regulated by transcription factors Efg1 and Tec1, and are also negatively regulated by the transcriptional repressors Nrg1 (Shapiro et al., 2011).
The use of probiotic lactobacilli against VVC has emerged as an attractive therapeutic option in the era of antibiotic resistance (Matsubara et al., 2016a). Several clinical trials reported that the commercially available probiotic Lactobacillus spp., such as L. rhamnosus GR-1, L. reuteri RC-15, and L. plantarum I1001, are beneficial to women’s urogenital health by decreasing the risk of VVC (Reid et al., 2003; Martinez et al., 2009; Murina et al., 2014; Palacios et al., 2016). However, they are seldom reported as the main resident Lactobacillus species in healthy vagina. Moreover, little is known about the roles and underlying mechanisms of yeast-to-hyphae transition in C. albicans regulated by lactobacilli, especially by those resident Lactobacillus species (Orsi et al., 2014b; Matsubara et al., 2016b; Oliveira et al., 2016).
The aim of this study was to investigate the effects of the most frequently found Lactobacillus spp. in the healthy vagina, i.e., L. crispatus, L. gasseri, and L. jensenii, on growth, hyphal formation and virulence-related genes expression of C. albicans. We found that all 18 Lactobacillus strains tested could inhibit the growth and hyphal formation of C. albicans in a strain-specific manner. In addition, for the first time, we demonstrated that L. crispatus inhibited C. albicans hyphal formation by down-regulating the hyphae-specific genes ALS3, HWP1, and ECE1 possibly through up-regulating the expression of transcriptional repressor gene NRG1. Having the strongest inhibition effects on C. albicans growth and hyphal formation, and being the most commonly found resident Lactobacillus species in the healthy vagina, L. crispatus shows a great potential as a probiotic candidate.
Materials and Methods
Strains and Culture Conditions
Eighteen Lactobacillus strains of three different species (L. crispatus, L. gasseri, and L. jensenii) were originally isolated from human vaginal secretion (Wang et al., 2014, 2015). L. acidophilus ATCC 4356 and C. albicans ATCC 10231 were purchased from the China General Microbiological Culture Collection Center.
All Lactobacillus strains were routinely grown in MRS medium (BD, Sparks, MD, USA) and incubated in an anaerobic atmosphere (BD, Sparks, MD, USA) at 37°C for 48 h. C. albicans was routinely grown for 12 h at 37°C in YPD medium.
Determination of H2O2 Production
The qualitative plate assay for H2O2 production was carried out according to the method of Xu et al. (2008). In brief, following 48 h of anaerobic incubation on the TMB-HRP-MRS plates [MRS agar containing 250 μg/mL 3, 3′, 5, 5′-tetramethylbenzidine (TMB; Amresco, Solon, OH, USA) and 50 μg/mL horseradish peroxidase (HRP; Biodee, Beijing, China)], the colonies of H2O2-producing strains turned blue after 30 min air exposure. Based on the color intensity, the potential of H2O2 production could be divided into four categories: negative (-), weakly positive (+), positive (++), and strongly positive (+++) (Xu et al., 2008). L. acidophilus ATCC 4356 was used as a positive control, and graded as a strong H2O2 producer.
Preparation of Cell-Free Supernatant (CFS) from Lactobacillus Strains and pH Measurement
Lactobacilli were grown in MRS broth (pH 6.5) for 48 h at 37°C in anaerobic condition. CFS was prepared by centrifuging the culture at 8000 rpm for 15 min at 4°C and then filtered through 0.22 μm filters (Millipore, Bedford, MA, USA). The pH value of CFS was measured by using a pH meter (PHS-3E, INESA Scientific Instrument, Shanghai, China). CFS was stored as single-use aliquots at -20°C until needed.
Antimicrobial Activity of Lactobacilli against C. albicans
The modified agar overlay method was used to determine the antimicrobial activity of lactobacilli against C. albicans (do Carmo et al., 2016). Ten microliter of Lactobacillus spp. suspensions (∼106 CFU/mL) were spotted onto MRS agar plates, and incubated anaerobically for 48 h. The MRS agar plates containing the growth of lactobacilli (≈8 mm in diameter) were overlayed with a C. albicans suspension (∼107 CFU/mL) in 10 mL of YPD soft agar (0.5% agar). The inhibitory effect of MRS was tested as a negative control on each plate. After aerobic incubation for 24 h, inhibition zones were read. The GIA was calculated as follows: GIA (mm) = (IZD – CD)/2, where IZD was inhibition zone diameter and CD was colony diameter of a Lactobacillus spot. A GIA grading system was applied, in which GIA < 0.5 mm was recorded as negative (–), (0.5, 2) mm as weak positive (+), (2, 3.5) mm as intermediate positive (++), and ≥ 3.5 mm as strong positive (+++).
Antimicrobial Activity of CFS against C. albicans
A microplate assay was used to evaluate the antimicrobial activity of the CFS against C. albicans according to Parolin et al. (2015) with some modifications. A mixture of 100 μL C. albicans suspensions in YPD broth (∼106 CFU/mL) and 100 μL tested CFS was added to each well of a sterile 96-well microplate (Costar 3799, Corning, NY, USA). MRS broth was used instead of CFS as a positive growth control. The plate was incubated at 37°C aerobically for 24 h, and the growth of C. albicans was recorded as OD at 630 nm wavelength using a microplate reader (Model 680, Bio-Rad, Hercules, CA, USA). The growth inhibition rate (%) was expressed as: (ODcontrol – ODCFS)/ODcontrol × 100.
Effect of CFS on C. albicans Hyphal Formation
Hyphal growth assay of C. albicans was performed in RPMI 1640 medium supplemented with 10% FBS (Sun et al., 2015). We used both solid and liquid medium assays to evaluate the effects of CFS on C. albicans hyphal formation. In these two assays, MRS broth instead of CFS was used as control. All assays were repeated for five times.
For solid medium assay, C. albicans cells from an overnight culture were washed with PBS and spread (∼100 colonies per plate) on plates of solidified medium supplemented with 50 μL of CFS. Plates were aerobically incubated at 37°C for 5 days. Images of colony edges were obtained using a stereomicroscope (SZ66, Optec, Chongqing, China).
To assess the effect of CFS in liquid medium, a mixture of 900 μL C. albicans suspensions (∼106 CFU/mL) in medium and 100 μL CFS was added into each well of a 48-well microplate, and aerobically incubated at 37°C for 4 h. Quantification of inhibition of the yeast-to-hyphae transition was accomplished by counting the number of individual yeast cells versus the number of hyphae in the population under a light microscope (AxioCam MRc5; Carl Zeiss, Jena, Germany). More than 100 cells were counted for each well in duplicate, and all assays were repeated for five times. The hyphae inhibition rate (%) was calculated as: (Hyphae%control – Hyphae%CFS)/Hyphae%control × 100.
Quantitative Real-Time Reverse Transcription PCR (qRT-PCR)
To determine the effects of CFS in RPMI 1640 + 10% FBS broth on the transcription of C. albicans genes related to yeast-to-hyphae transition, gene expression levels of GSP1, NRG1, TEC1, EFG1, EAP1, SAP5, ALS3, HWP1, and ECE1 were evaluated by two-step qRT-PCR (Sun et al., 2015; James et al., 2016). Total RNA of C. albicans was extracted using a Yeast RiboPure RNA Purification kit (CoWin Biosciences, Beijing, China). Concentration, purity, and quality of the isolated RNA samples were determined using a Nano Drop One Spectrophotometer (Thermo Scientific, Waltham, MA, USA). RNA (1 μg) from each sample was immediately reverse transcribed into cDNA using a Prime Script RT reagent kit (Takara, Tokyo, Japan) according to the manufacturer’s instructions. The conditions for reverse transcription were 15 min at 37°C, 5 s at 85°C. qRT-PCR was performed with 80 ng cDNA template and SYBR Premix Ex Taq (Takara) in ABI StepOne Real Time PCR System (Applied Biosystems). Primer sequences used for amplification of specific genes were shown in Table 1. GSP1, which was not transcriptionally regulated in the morphogenesis switch, served as the internal control (Sun et al., 2015). The following parameters were used for qRT-PCR: one cycle at 95°C for 15 s, 40 cycles at 95°C for 5 s and then 60°C for 30 s. Specificity of the primers was confirmed by melting curve analysis. The generated CT values of target genes were normalized to the CT value of reference gene GSP1. Relative expression fold changes were evaluated by ΔΔCT method using 2-ΔΔCT formula (Livak and Schmittgen, 2001).
Table 1.
List of primers used for qRT-PCR experiments.
| Gene | Function | Primer sequence (5′–3′) |
|---|---|---|
| GSP1 | GTP-binding protein (Reference gene) | F: TGAAGTCCATCCATTAGGAT |
| R: ATCTCTATGCCAGTTTGGAA | ||
| NRG1 | Negative transcriptional regulator | F: CACCTCACTTGCAACCCC |
| R: GCCCTGGAGATGGTCTGA | ||
| TEC1 | Conserved filamentation activator | F: AGGTTCCCTGGTTTAAGTG |
| R: ACTGGTATGTGTGGGTGAT | ||
| EFG1 | Filamentous growth protein | F: TATGCCCCAGCAAACAACTG |
| R: TTGTTGTCCTGCTGTCTGTC | ||
| EAP1 | Extracellular adherence protein | F: CTGCTCACTCAACTTCAATTGTCG |
| R: GAACACATCCACCTTCGGGA | ||
| SAP5 | Secreted aspartyl proteinase | F: CAGAATTTCCCGTCGATGAGA |
| R: CATTGTGCAAAGTAACTGCAACAG | ||
| ALS3 | Agglutinin-like protein | F: CTAATGCTGCTACGTATAATT |
| R: CCTGAAATTGACATGTAGCA | ||
| HWP1 | Hyphal cell wall protein | F: TGGTGCTATTACTATTCCGG |
| R: CAATAATAGCAGCACCGAAG | ||
| ECE1 | Extent of cell elongation protein | F: GCTGGTATCATTGCTGATAT |
| R: TTCGATGGATTGTTGAACAC |
Statistical Analysis
All assays were carried out in triplicate on at least three different occasions with independently grown cultures unless otherwise stated. All statistical analysis was performed using IBM SPSS Statistics 20.0 software program (IBM, Armonk, NY, USA). Statistical comparison between two groups was performed by Student’s t-test. Comparisons among multiple groups were performed by one-way ANOVA followed by LSD test. A P-value < 0.05 was considered statistically significant.
Results
Antimicrobial Activity and H2O2 Producing Ability of Lactobacilli
The antimicrobial activity of lactobacilli against C. albicans was evaluated using agar overlay method. The results showed that 15 of 18 (83.3%) Lactobacillus strains isolated from vaginal mucosa had inhibition activities on C. albicans growth (Table 2), among which more than half (8/15, 53.3%) had high inhibition activities ranked as +++.
Table 2.
Characteristics of 18 vaginal Lactobacillus strains.
| Strain | pH of CFS | H2O2 producing abilitya | GIAb |
|---|---|---|---|
| L. crispatus | |||
| A014 | 3.91 | ++ | +++ |
| A055 | 3.95 | ++ | ++ |
| B125 | 3.93 | +++ | ++ |
| B135 | 3.94 | +++ | +++ |
| B145 | 3.93 | +++ | +++ |
| B422 | 3.93 | ++ | +++ |
| B535 | 3.93 | +++ | ++ |
| L. gasseri | |||
| A054 | 3.92 | - | ++ |
| B062 | 3.86 | + | + |
| B351 | 3.93 | +++ | +++ |
| B451 | 3.95 | - | ++ |
| B542 | 3.97 | - | ++ |
| B554 | 3.93 | - | +++ |
| L. jensenii | |||
| A083 | 4.09 | +++ | - |
| B021 | 4.09 | +++ | - |
| B101 | 4.02 | +++ | +++ |
| B161 | 3.96 | ++ | - |
| B511 | 4.16 | +++ | +++ |
aThe H2O2 producing ability of Lactobacillus strains was graded as strongly positive (+++), positive (++), weakly positive (+), and negative (-) according the color intensity of colony on the TMB-MRS plate. bThe GIA was calculated by subtracting a CD of a Lactobacillus spot from the IZD and expressed as follows: GIA = (IZD – CD)/2. GIA < 0.5 mm was recorded as negative (-), [0.5, 2) mm as weak positive (+), [2, 3.5) mm as intermediate positive (++), and ≥ 3.5 mm as strong positive (+++).
The production of H2O2 by Lactobacillus strains was tested using the semi-qualitative TMB-HRP-MRS assay (Table 2). The strong-, moderate-, weak- and non-producers were 50.0% (9/18), 22.2% (4/18), 5.6% (1/18), and 22.2% (4/18), respectively.
Effect of CFS on C. albicans Growth
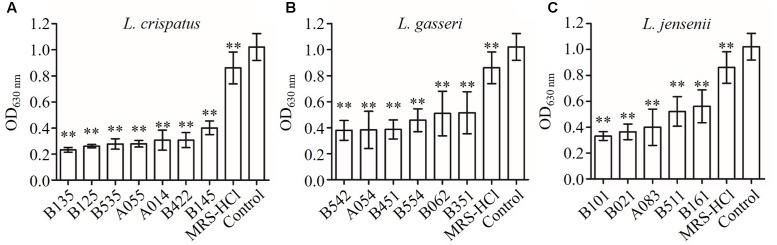
The strains of lactobacilli were assayed for their ability to produce inhibition compounds in CFS against the growth of C. albicans. The average pH value of CFS was 4.0 (in the range of 3.8–4.2), which was lower than fresh MRS broth (initial pH 6.5) (Table 2). To investigate whether low pH contributed to the growth inhibition effects of CFS, fresh MRS broth was adjusted to pH 4.0 with HCl (MRS-HCl). In the presence of CFS or MRS-HCl, the growth of C. albicans was significantly reduced when compared to the MRS control in general (Figure 1) (P < 0.01). In addition, while the MRS-HCl could significantly inhibit the growth of C. albicans (OD630nm MRS-HCl vs. control: 0.861 ± 0.122 vs. 1.021 ± 0.103, P = 0.008), each of the CFS could further inhibit C. albicans growth with significant difference compared to MRS-HCl (P < 0.01).
FIGURE 1.
Inhibition effects of CFS from different Lactobacillus strains on Candida albicans growth in YPD broth at 24 h after inoculation. (A) L. crispatus, (B) L. gasseri, (C) L. jensenii. Each bar is the mean ± SD from three independent experiments. ∗∗P < 0.01 compared to control by Student’s t-test.
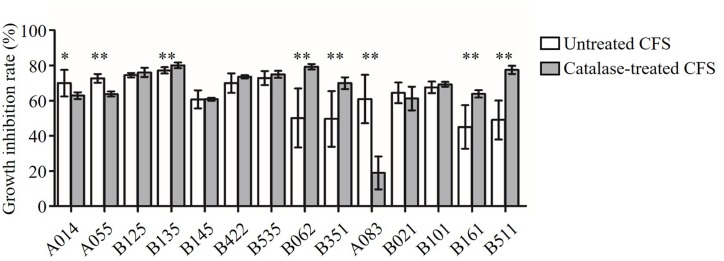
In order to test whether the antimicrobial activity was due to H2O2, the CFS was treated with catalase enzyme (1 mg/mL; Sigma, St. Louis, MO, USA) at 37°C for 1 h before approaching to the inhibition assay (Figure 2). The antimicrobial activity of six CFS (B125, B145, B422, B535, B021, and B101) was resistance to the catalase treatment (P > 0.05). These results suggested that these six strains might produce anti-Candida compounds with strong activities beyond the effect of H2O2. After catalase treatment, the growth inhibition rates of three CFS (A014, A055, and A083) were found to decrease, indicating that the observed anti-Candida effects of these three strains were partially attributed to the production of H2O2 (P < 0.05). However, it was strange that the growth inhibition abilities of five CFS (B135, B062, B351, B161, and B511) were found to significantly increase after catalase treatment.
FIGURE 2.
Inhibition of C. albicans growth by CFS and catalase-treated CFS. Growth inhibition rate (%) = (ODcontrol – ODCFS)/ODcontrol × 100. The statistical comparison between the catalase untreated and treated groups for each of the CFS was performed by Student’s t-test. ∗P < 0.05, ∗∗P < 0.01.
Effect of CFS on C. albicans Hyphal Formation
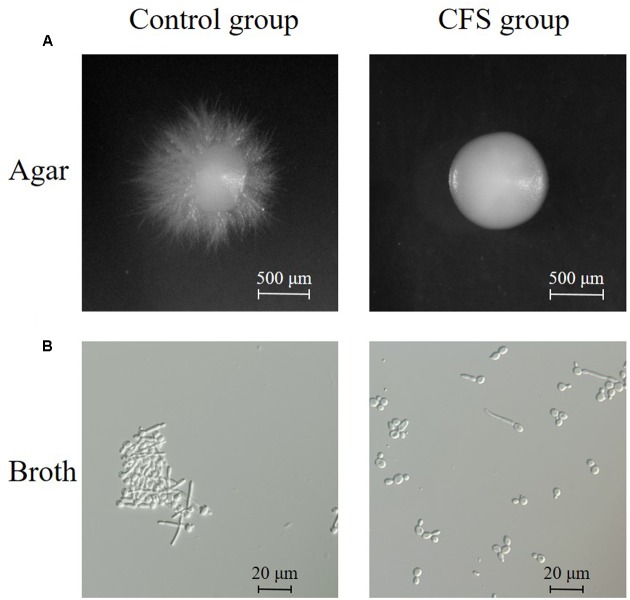
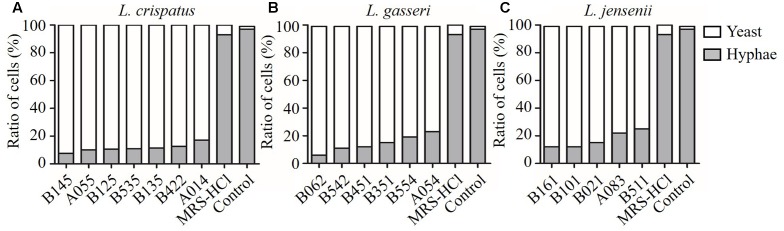
The effect of CFS on the hyphal formation was evaluated in RPMI 1640 agar medium containing 10% FBS (hypha-inducing condition). The yeast form and filamentous-proned colonies of C. albicans were counted. The results showed that the treatment with CFS was able to inhibit filamentation. Figure 3A was an example of C. albicans colony at absence or presence of CFS. In addition, the effect of CFS on the hyphal formation was evaluated in liquid medium supplemented with CFS. The results showed that most C. albicans cells grew as yeast (Figure 3B). As seen in Figure 4, about 98% of untreated cells formed hyphae and pseudohyphae over 4 h time course. When treated by MRS-HCl, there was a significant lower ratio of hyphal cells compared to that treated by MRS control (92.6 ± 8.0% vs. 97.8 ± 4.6%, P = 0.038). However, each of the CFS showed more significant inhibition effects on C. albicans hyphal formation compared to that of MRS-HCl (P < 0.01).
FIGURE 3.
Inhibition of hyphal formation by CFS. (A) Effect of CFS on C. albicans hyphal formation on solid RPMI 1640 + 10% FBS medium. Plates were incubated at 37°C for 5 days. Images of colony edges were obtained using a stereomicroscope, original magnification: 6.6×. (B) Effect of CFS on C. albicans hyphal formation in liquid RPMI 1640 + 10% FBS medium for 4 h at 37°C. Images of cellular morphology were obtained using a light microscopy, original magnification: 400×.
FIGURE 4.
Inhibition effects of CFS from different Lactobacillus strains on C. albicans hyphal formation. (A) L. crispatus, (B) L. gasseri, (C) L. jensenii. Effect of the addition of CFS on the yeast-to-hyphae conversion induced by liquid RPMI 1640 + 10% FBS medium at 37°C for 4 h in microtiter wells.
Comparison of Inhibitory Activities of Lactobacillus Species
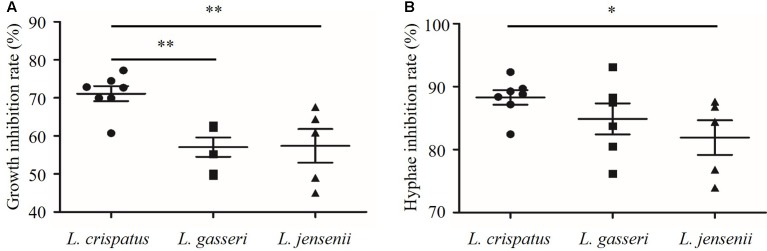
As shown in Figure 5, the growth and hyphal formation of C. albicans were inhibited by CFS from L. crispatus, L. gasseri, and L. jensenii. The growth inhibition rate of C. albicans treated by L. crispatus (71.1 ± 5.2%) was significantly higher than those treated by L. gasseri (57.1 ± 6.3 %) or L. jensenii (57.4 ± 9.9%), respectively (P < 0.01). The hyphae inhibition rate of C. albicans treated by L. crispatus (88.3 ± 3.02%) was significantly higher than that by L. jensenii (81.9 ± 6.2%) (P < 0.05). However, there was no significant difference between L. crispatus and L. gasseri treatments (88.3 ± 3.0% vs. 84.9 ± 6.0%, P > 0.05). Taking into consideration of growth and hyphal formation inhibition activities, we chose L. crispatus CFS to study the effects on the transcription of C. albicans virulence-related genes.
FIGURE 5.
Inhibition of growth and hyphal formation of C. albicans by CFS from L. crispatus, L. gasseri, and L. jensenii. (A) Growth inhibition rate (%) = (ODcontrol – ODCFS)/ODcontrol × 100, (B) Hyphae inhibition rate (%) = (Hyphae%control – Hyphae%CFS)/Hyphae%control × 100. Comparisons among three groups were performed by one-way ANOVA followed by LSD test. ∗P < 0.05, ∗∗P < 0.01.
Modulation of C. albicans Gene Expression by CFS
The expression levels of the hyphae-specific genes (ALS3, HWP1, ECE1, EAP1, and SAP5) and transcriptional regulatory genes (EFG1, TEC1, and NRG1) were quantified in C. albicans incubated with CFS by qRT-PCR. Figure 6 showed that CFS significantly down-regulated the expression of the hyphae-specific genes ALS3 (0.140-fold), HWP1 (0.075-fold), and ECE1 (0.045-fold), the differences were statistically significant compared to MRS control (P < 0.01). The gene expression level of SAP5 (1.283-fold) had no statistical difference compared to the control (P > 0.05). Strikingly, EAP1 gene, which involved in adhesion and biofilm formation, was found to be significantly up-regulated by CFS treatment (2.590-fold, P < 0.01). In addition, the expression level of negative transcriptional regulator NRG1 was also found to be significantly up-regulated (1.911-fold, P < 0.05), while two hyphae activator regulatory genes TEC1 (1.272-fold) and EFG1 (1.200-fold) involved in C. albicans morphogenesis were found to have no statistical difference compared to the control (P > 0.05).
FIGURE 6.
The expression levels (fold change) of virulence-related genes in C. albicans treated with CFS. ∗P < 0.05 and ∗∗P < 0.01 compared to control by Student’s t-test.
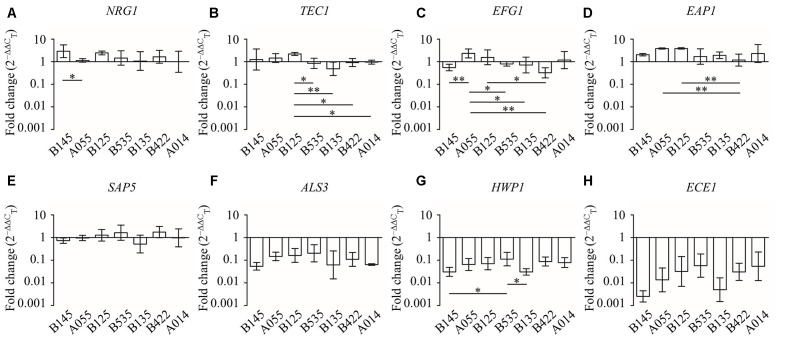
Comparing the different effects of CFS on C. albicans gene expression, we found that all seven L. crispatus CFS significantly down-regulated the expression of ALS3, HWP,1 and ECE1 genes (Figure 6), and there were no inter-strain difference except for gene HWP1 (by LSD test: B145 vs. B535, P = 0.026; B535 vs. B135, P = 0.024) (Figures 7F–H). In contrast, the up- and down-regulation phenomena seemed to exist for each of the other five C. albicans genes affected by CFS. According to the results of paired comparison, there were statistically significant inter-strain differences for NRG1, TEC1, EFG1, and EAP1 (Figures 7A–D) but not for SAP5 (Figure 7E). Collectively, the results suggested that each L. crispatus strains might have slightly different ability and mechanism to regulate C. albicans gene expression. Among seven tested L. crispatus strains, L. crispatus B145 displayed the strongest regulatory activities in terms of down-regulations of ALS3, HWP1, and ECE1 expressions, and up-regulation of NRG1 expression. Interestingly, this was in line with the finding that L. crispatus B145 had the strongest activity to inhibit yeast-to-hyphae transition (92.3 ± 9.2% hyphal formation inhibition).
FIGURE 7.
The expression levels (fold change) of virulence-related genes in C. albicans treated with seven different CFS from L. crispatus. (A) NRG1, (B) TEC1, (C) EFG1, (D) EAP1, (E) SAP5, (F) ALS3, (G) HWP1, (H) ECE1. Error bar represents SD from three biological replicates. Comparisons among multiple groups were performed by one-way ANOVA followed by LSD test. ∗P < 0.05, ∗∗P < 0.01.
Characterization of Anti-Candida-Related Compounds in CFS Derived from L. crispatus B145
In order to get some preliminary information about the chemical nature of the anti-Candida-related compounds secreted by L. crispatus, the strain B145 was used for further study. This strain was shown to have strong inhibition activities on C. albicans growth in the absence and presence of catalase. It also strongly inhibited the hyphal formation and some of the related gene expressions. The susceptibility to heat was evaluated by subjecting the B145 CFS to a number of thermal treatments: 60°C for 30 min; 100°C for 30 min; and 121°C for 15 min. The susceptibility to proteolytic enzymes was investigated by exposing CFS to proteinase-K and trypsin (1 mg/mL; KeyGen Biotech, Nanjing, China) for 3 h at 37°C (Sangmanee and Hongpattarakere, 2014).
The B145 CFS had about 60% growth inhibition activity on C. albicans after heat and protease treatments with no significant difference compared to that of the untreated CFS (P > 0.05) (Figure 8A). In addition, the anti-hyphal activities of CFS were significantly reduced after exposing to heat or protease treatments compared to that of the untreated CFS (P < 0.01) (Figure 8B). However, the residual activity of protease-treated CFS was significantly lower than those of heat treatments (P < 0.05).
FIGURE 8.
Inhibition of growth and hyphal formation of C. albicans by CFS from L. crispatus B145. CFS: untreated CFS from L. crispatus B145, CFS-1: CFS was heated at 80°C for 30 min; CFS-2: CFS was heated at 100°C for 30 min; CFS-3: CFS was heated at 121°C for 15 min; and CFS-4: CFS was treated with proteolytic enzymes. (A) Growth inhibition rate (%) = (ODcontrol – ODCFS)/ODcontrol × 100, (B) Hyphae inhibition rate (%) = (Hyphae%control – Hyphae%CFS)/ Hyphae%control × 100. ∗∗P < 0.01 compared to CFS by Student’s t-test.
These primarily results suggested that the growth and the hyphal inhibition activities of L. crispatus B145 CFS might be attributed to the antimicrobial compounds with different chemical nature in terms of heat stability and protease susceptibility.
Discussion
Vulvovaginal candidiasis, primarily caused by C. albicans, is one of the most common urogenital diseases affecting women of reproductive-age and associated to the disruption of the healthy vaginal microbiota (Achkar and Fries, 2010). Lactobacillus-dominated microbiota is thought to be a valuable biomarker for vaginal health since the Lactobacillus species can create a barrier against pathogen invasion (Petrova et al., 2015). L. crispatus, L. gasseri, and L. jensenii were the most frequently isolated species in the vagina of healthy women (Pendharkar et al., 2013; Al et al., 2014; Madhivanan et al., 2015), where C. albicans is often a minority co-habitant of vaginal microbiota. In this study, we investigated how these three Lactobacillus species affected C. albicans growth and yeast-to-hyphae transition, which are implicated in its pathogenesis.
We studied the antimicrobial activities of Lactobacillus strains against C. albicans using two assays, i.e., the agar overlay method and the microplate-based liquid medium assay. The results were similar in terms of having or not having inhibition activities except for three strains of L. jensenii (A083, B021, and B161) (Table 2 and Figure 1). The same phenomenon was also found by other researches. For example, Coman et al. (2014) evaluated the growth inhibition activities of L. rhamnosus IMC 501® and L. paracasei IMC 502® on different pathogens by several antimicrobial assays, such as agar well diffusion method and liquid co-culture assay. They found that the inhibition results were different within the different methods, and suggested that this might be due to the various antimicrobial mechanisms. In this study, the liquid medium assay using Lactobacillus CFS seemed to be more sensitive to assess the antimicrobial activities, where 100% (18/18) of the tested Lactobacillus strains showed anti-Candida activities rather than 83.3% (15/18) in agar overlay assay. We speculated that the solid agar might block the diffusion of antimicrobial compounds to reach the target C. albicans, which might affect those compounds with hydrophobic nature more significantly. This also suggests the necessity of using different methods in assessing the antimicrobial activities and comparing results from different studies in an objective way. Anyhow, our data suggests that the tested Lactobacillus strains had moderate to strong anti-Candida activities with inter-strain variations. Our results are consistent with some previous studies showing that several strains of lactobacilli were effective at inhibiting the growth of C. albicans (Parolin et al., 2015; Hütt et al., 2016).
Lactobacilli are known for their production of various antimicrobial compounds to prevent vaginal infection (Borges et al., 2014). H2O2 is one of the antimicrobial compounds. We found that Lactobacillus strains had different H2O2 production abilities, ranging from no evidence of H2O2 production to strong production (Table 2). However, all tested Lactobacillus strains showed growth inhibition effects on C. albicans regardless of H2O2 production, suggesting that they might produce some antimicrobial compounds in addition to H2O2. Organic acid production and consequent pH reduction by lactobacilli growth have also been suggested as one of the mechanisms for growth inhibition on Candida in previous studies (Jiang et al., 2015). However, we found that the anti-Candida activities of the tested CFS might not be largely attributed to low pH (Figure 1). Each of CFS from the tested Lactobacillus strains showed significantly higher antimicrobial activity than MRS-HCl liquid (pH 4.0) (P < 0.01). It suggests that other antimicrobial compounds might play roles in the growth inhibition effects of CFS on C. albicans. The antimicrobial compounds, such as bacteriocin-like substances and biosurfactants, have been suggested to contribute to the antagonistic effects of probiotic Lactobacillus strains against a variety of vaginal pathogens (Madhu and Prapulla, 2014; Maldonado-Barragán et al., 2016).
In this study, for the first time, the effects of CFS from three species of Lactobacillus (L. crispatus, L. gasseri, and L. jensenii) on yeast-to-hyphae transition of C. albicans were compared. The ability of yeast-to-hyphae transition is essential for C. albicans virulence (Mayer et al., 2013). The hyphae form, which can promote tissue penetration and escape from immune cell, is more prevalent in the infection process than yeast form (Lu et al., 2014). Blocking of virulence traits of pathogens, such as yeast-to-hyphae transition in C. albicans, has been recently considered as a new antifungal paradigm (Tsang et al., 2012). In this study, L. crispatus showed good inhibition activity on hyphal formation (Figures 4, 5). Longitudinal studies have also shown that the presence of L. crispatus promoted stability of the vaginal microbiota toward a healthy status (Verstraelen et al., 2009; Santiago et al., 2012). Our findings of L. crispatus strains with generally high inhibition activities on Candida growth and yeast-to-hyphae transition is very attractive. As far as we know, the clinical approved L. crispatus strains are very limited. Our results hint that L. crispatus strains are highly worthwhile for further investigation in developing the new antifungal strategies.
Our preliminary results revealed that the antimicrobial compounds from L. crispatus B145 against Candida growth were heat stable and protease resistance. But the same treatments would greatly reduce the residual activity against yeast-to-hyphae transition. This hints that the growth and yeast-to-hyphae transition inhibition activities would be attributed to different secretory antimicrobial compounds of L. crispatus B145. Tendulkar et al. (2007) have reported to identify a heat stable and protease resistance biosurfactant compound, which had anti-fungi activity. The compound was characterized to be lipopeptide in chemical nature. To further study whether L. crispatus B145 was able to inhibit different clinical relevant C. albicans strains, we tested the growth and hyphae inhibition effect of its CFS on five additional vaginal isolates of C. albicans. We found that the growth inhibition rates were from 50.7 to 68.6%, and the hyphae formation inhibition rates were from 77.9 to 91.7% (Supplementary Figure S1). These results confirmed our findings in C. albicans ATCC 10231. Further studies to purify the bioactive compounds from L. crispatus B145 and to distinguish different compounds in growth and hyphal inhibition would be interesting.
To elucidate the potential mechanisms involved in the inhibition of hyphal formation of C. albicans by the L. crispatus, we studied the expression levels of five hyphae-specific genes, i.e., ALS3, HWP1, ECE1, EAP1, and SAP5, in response to L. crispatus CFS under hyphae-inducing conditions. These genes encode proteins that are essential for hyphal formation and also play roles in Candida pathogenesis. ALS3 encodes Als3, which is a member of the agglutinin-like sequence gene family and contributes to the invasion of cells and subsequently cell damage (Liu and Filler, 2011). HWP1 encodes a cell wall mannose protein, which functions as an adhesion, required for hyphal formation and yeast adhesion to epithelial cells (Orsi et al., 2014a). ECE1 encodes a membrane protein, which is essential for cell elongation and biofilm formation (Fan et al., 2013). EAP1 gene encodes a glycosylphosphatidylinositol-anchored, glucan-cross-linked cell wall protein, in adhesion and biofilm formation (Li and Palecek, 2003). SAP5 encodes a member of the secreted aspartic proteases family, which are important for the pathogenesis of candidiasis (Sanglard et al., 1997). The results of qRT-PCR analysis revealed that all seven L. crispatus CFS significantly down-regulate the expression of hyphal genes ALS3, HWP1, and ECE1, associated with the inhibition of hyphal formation. Moreover, C. albicans hyphal maintenance requires specific transcriptional regulation mechanisms (Fan et al., 2013). We measured the expression of positive regulator genes of EFG1 and TEC1, and the negative regulator gene of NRG1 (Braun et al., 2001; Argimón et al., 2007; Daniels et al., 2015). Expression of the negative hyphal regulator gene NRG1 was found to be significantly up-regulated in C. albicans in the presence of L. crispatus CFS, implying that L. crispatus might repress the expression of hyphae-specific genes in a NRG1-dependent manner. To the best of our knowledge, this is the first study demonstrating the role of L. crispatus in inhibition of C. albicans virulence-related gene expressions. Interestingly, L. crispatus B145 was shown to strongly up-regulate NRG1 expression (3.398-fold) and down-regulate ALS3 (0.057-fold), HWP1 (0.033-fold), and ECE1 (0.003-fold) gene expressions. If the antibodies are available, experiments such as Western blot would further confirm this finding.
Conclusion
The present study demonstrated the in vitro inhibitory effects of vaginal L. crispatus, L. gasseri, and L. jensenii on C. albicans growth and hyphal formation. L. crispatus had a generally better ability to inhibit C. albicans and might down-regulate the expression of hyphae-specific genes ALS3, HWP1, and ECE1 in a NRG1-dependent manner. The effects might be due to their productions of some secretory antimicrobial compounds in addition to H2O2 and organic acids. The growth and hyphal inhibition compounds from L. crispatus B145 seemed to be different in chemical nature. Further investigations on L. crispatus B145 and other clinical isolates would facilitate the development of probiotic agents with great potentials against C. albicans.
Author Contributions
SW and TL designed experiments, analyzed data and wrote the paper; SW and QW carried out experiments; LY helped with assay set-up; HZ supervised the project and paper writing; EY gave critical review on the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (21672010).
Abbreviations
- ALS3
agglutinin-like protein 3
- CD
colony diameter
- CFS
cell-free supernatant
- EAP1
extracellular adherence protein 1
- ECE1
extent of cell elongation protein 1
- EFG1
enhanced filamentous growth 1
- FBS
fetal bovine serum
- GIA
growth inhibitory activity
- GSP1
GTP binding protein 1
- HWP1
hyphal wall protein 1
- IZD
inhibition zone diameter
- MRS
Man Rogosa and Sharpe
- NRG1
negative regulator of glucose-repressed 1
- OD
optical density
- PBS
phosphate buffered saline
- qRT-PCR
quantitative real-time reverse transcription PCR
- SAP5
secreted aspartyl proteinase 5
- SD
standard deviation
- TEC1
transposon enhancement control 1
- VVC
vulvovaginal candidiasis
- YPD
yeast extract-peptone-dextrose
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00564/full#supplementary-material
References
- Achkar J. M., Fries B. C. (2010). Candida infections of the genitourinary tract. Clin. Microbiol. Rev. 23 253–273. 10.1128/CMR.00076-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al K. I., Hamze M., Hober D., Chihib N. E., Drider D. (2014). Identification of vaginal lactobacilli with potential probiotic properties isolated from women in North Lebanon. Microb. Ecol. 67 722–734. 10.1007/s00248-014-0384-7 [DOI] [PubMed] [Google Scholar]
- Argimón S., Wishart J. A., Leng R., Macaskill S., Mavor A., Alexandris T., et al. (2007). Developmental regulation of an adhesin gene during cellular morphogenesis in the fungal pathogen Candida albicans. Eukaryot. Cell 6 682–692. 10.1128/EC.00340-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges S., Silva J., Teixeira P. (2014). The role of lactobacilli and probiotics in maintaining vaginal health. Arch. Gynecol. Obstet. 289 479–489. 10.1007/s00404-013-3064-9 [DOI] [PubMed] [Google Scholar]
- Braun B. R., Kadosh D., Johnson A. D. (2001). NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20 4753–4761. 10.1093/emboj/20.17.4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone A. (2015). Vulvovaginal Candida albicans infections: pathogenesis, immunity and vaccine prospects. BJOG 122 785–794. 10.1111/1471-0528.12994 [DOI] [PubMed] [Google Scholar]
- Coman M. M., Verdenelli M. C., Cecchini C., Silvi S., Orpianesi C., Boyko N., et al. (2014). In vitro evaluation of antimicrobial activity of Lactobacillus rhamnosus IMC 501, Lactobacillus paracasei® IMC 502® and SYNBIO® against pathogens. J. Appl. Microbiol. 117 518–527. 10.1111/jam.12544 [DOI] [PubMed] [Google Scholar]
- da Silva Dantas A., Lee K. K., Raziunaite I., Schaefer K., Wagener J., Yadav B., et al. (2016). Cell biology of Candida albicans-host interactions. Curr. Opin. Microbiol. 34 111–118. 10.1016/j.mib.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels K. J., Srikantha T., Pujol C., Park Y. N., Soll D. R. (2015). Role of Tec1 in the development, architecture, and integrity of sexual biofilms of Candida albicans. Eukaryot. Cell 14 228–240. 10.1128/EC.00224-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Carmo M. S., Noronha F. M., Arruda M. O., Costa E. P., Bomfim M. R., Monteiro A. S., et al. (2016). Lactobacillus fermentum ATCC 23271 displays in vitro inhibitory activities against Candida spp. Front. Microbiol. 7:1722 10.3389/fmicb.2016.01722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., He H., Dong Y., Pan H. (2013). Hyphae-specific genes HGC1, ALS3, HWP1, and ECE1 and relevant signaling pathways in Candida albicans. Mycopathologia 176 329–335. 10.1007/s11046-013-9684-6 [DOI] [PubMed] [Google Scholar]
- Förster T. M., Mogavero S., Dräger A., Graf K., Polke M., Jacobsen I. D., et al. (2016). Enemies and brothers in arms: Candida albicans and gram-positive bacteria. Cell. Microbiol. 18 1709–1715. 10.1111/cmi.12657 [DOI] [PubMed] [Google Scholar]
- Hütt P., Lapp E., Štšepetova J., Smidt I., Taelma H., Borovkova N., et al. (2016). Characterisation of probiotic properties in human vaginal lactobacilli strains. Microb. Ecol. Health Dis. 27:30484 10.3402/mehd.v27.30484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen I. D., Wilson D., Wachtler B., Brunke S., Naglik J. R., Hube B. (2012). Candida albicans dimorphism as a therapeutic target. Expert Rev. Anti Infect. Ther. 10 85–93. 10.1586/eri.11.152 [DOI] [PubMed] [Google Scholar]
- James K. M., MacDonald K. W., Chanyi R. M., Cadieux P. A., Burton J. P. (2016). Inhibition of Candida albicans biofilm formation and modulation of gene expression by probiotic cells and supernatant. J. Med. Microbiol. 65 328–336. 10.1099/jmm.0.000226 [DOI] [PubMed] [Google Scholar]
- Jiang Q., Stamatova I., Kari K., Meurman J. H. (2015). Inhibitory activity in vitro of probiotic lactobacilli against oral Candida under different fermentation conditions. Benef. Microbes 6 361–368. 10.3920/BM2014.0054 [DOI] [PubMed] [Google Scholar]
- Li F., Palecek S. P. (2003). EAP1, a Candida albicans gene involved in binding human epithelial cells. Eukaryot. Cell 2 1266–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Filler S. G. (2011). Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot. Cell 10 168–173. 10.1128/EC.00279-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu Y., Su C., Liu H. (2014). Candida albicans hyphal initiation and elongation. Trends Microbiol. 22 707–714. 10.1016/j.tim.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., Forney L. J., Ravel J. (2012). Vaginal microbiome: rethinking health and disease. Annu. Rev. Microbiol. 66 371–389. 10.1146/annurev-micro-092611-150157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhivanan P., Alleyn H. N., Raphael E., Krupp K., Ravi K., Nebhrajani R., et al. (2015). Identification of culturable vaginal Lactobacillus species among reproductive age women in Mysore. India. J. Med. Microbiol. 64 636–641. 10.1099/jmm.0.000070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhu A. N., Prapulla S. G. (2014). Evaluation and functional characterization of a biosurfactant produced by Lactobacillus plantarum CFR 2194. Appl. Biochem. Biotechnol. 172 1777–1789. 10.1007/s12010-013-0649-5 [DOI] [PubMed] [Google Scholar]
- Maldonado-Barragán A., Caballero-Guerrero B., Martín V., Ruiz-Barba J. L., Rodríguez J. M. (2016). Purification and genetic characterization of gassericin E, a novel co-culture inducible bacteriocin from Lactobacillus gasseri EV1461 isolated from the vagina of a healthy woman. BMC Microbiol. 16 37 10.1186/s12866-016-0663-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez R. C., Franceschini S. A., Patta M. C., Quintana S. M., Candido R. C., Ferreira J. C., et al. (2009). Improved treatment of vulvovaginal candidiasis with fluconazole plus probiotic Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14. Lett. Appl. Microbiol. 48 269–274. 10.1111/j.1472-765X.2008.02477.x [DOI] [PubMed] [Google Scholar]
- Matsubara V. H., Bandara H. M., Mayer M. P., Samaranayake L. P. (2016a). Probiotics as antifungals in mucosal candidiasis. Clin. Infect. Dis. 62 1143–1153. 10.1093/cid/ciw038 [DOI] [PubMed] [Google Scholar]
- Matsubara V. H., Wang Y., Bandara H. M., Mayer M. P., Samaranayake L. P. (2016b). Probiotic lactobacilli inhibit early stages of Candida albicans biofilm development by reducing their growth, cell adhesion, and filamentation. Appl. Microbiol. Biotechnol. 100 6415–6426. 10.1007/s00253-016-7527-3 [DOI] [PubMed] [Google Scholar]
- Mayer F. L., Wilson D., Hube B. (2013). Candida albicans pathogenicity mechanisms. Virulence 4 119–128. 10.4161/viru.22913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murina F., Graziottin A., Vicariotto F., De Seta F. (2014). Can Lactobacillus fermentum LF10 and Lactobacillus acidophilus LA02 in a slow-release vaginal product be useful for prevention of recurrent vulvovaginal candidiasis?: a clinical study. J. Clin. Gastroenterol. 48(Suppl. 1), S102–S105. 10.1097/MCG.0000000000000225 [DOI] [PubMed] [Google Scholar]
- Nobile C. J., Johnson A. D. (2015). Candida albicans biofilms and human disease. Annu. Rev. Microbiol. 69 71–92. 10.1146/annurev-micro-091014-104330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira V. M., Santos S. S., Silva C. R., Jorge A. O., Leão M. V. (2016). Lactobacillus is able to alter the virulence and the sensitivity profile of Candida albicans. J. Appl. Microbiol. 121 1737–1744. 10.1111/jam.13289 [DOI] [PubMed] [Google Scholar]
- Orsi C. F., Borghi E., Colombari B., Neglia R. G., Quaglino D., Ardizzoni A., et al. (2014a). Impact of Candida albicans hyphal wall protein 1 (HWP1) genotype on biofilm production and fungal susceptibility to microglial cells. Microb. Pathog. 6 20–27. 10.1016/j.micpath.2014.03.003 [DOI] [PubMed] [Google Scholar]
- Orsi C. F., Sabia C., Ardizzoni A., Colombari B., Neglia R. G., Peppoloni S., et al. (2014b). Inhibitory effects of different lactobacilli on Candida albicans hyphal formation and biofilm development. J. Biol. Regul. Homeost. Agents 28 743–752. [PubMed] [Google Scholar]
- Palacios S., Espadaler J., Fernández-Moya J. M., Prieto C., Salas N. (2016). Is it possible to prevent recurrent vulvovaginitis? The role of Lactobacillus plantarum I1001 (CECT7504). Eur. J. Clin. Microbiol. Infect. Dis. 35 1701–1708. 10.1007/s10096-016-2715-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palková Z., Váchová L. (2016). Yeast cell differentiation: lessons from pathogenic and non-pathogenic yeasts. Semin. Cell Dev. Biol. 57 110–119. 10.1016/j.semcdb.2016.04.006 [DOI] [PubMed] [Google Scholar]
- Parolin C., Marangoni A., Laghi L., Foschi C., Nahui P. R., Calonghi N., et al. (2015). Isolation of vaginal lactobacilli and characterization of anti-Candida activity. PLoS ONE 10:e131220 10.1371/journal.pone.0131220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual L. M., Daniele M. B., Giordano W., Pájaro M. C., Barberis I. L. (2008). Purification and partial characterization of novel bacteriocin L23 produced by Lactobacillus fermentum L23. Curr. Microbiol. 56 397–402. 10.1007/s00284-007-9094-4 [DOI] [PubMed] [Google Scholar]
- Pendharkar S., Magopane T., Larsson P. G., Bruyn G., Gray G. E., Hammarström L., et al. (2013). Identification and characterisation of vaginal lactobacilli from South African women. BMC Infect. Dis. 13:43 10.1186/1471-2334-13-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B. M., Yano J., Noverr M. C., Fidel P. J. (2014). Candida vaginitis: when opportunism knocks, the host responds. PLoS Pathog. 10:e1003965 10.1371/journal.ppat.1003965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova M. I., Lievens E., Malik S., Imholz N., Lebeer S. (2015). Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Front. Physiol. 6:81 10.3389/fphys.2015.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G., Charbonneau D., Erb J., Kochanowski B., Beuerman D., Poehner R., et al. (2003). Oral use of Lactobacillus rhamnosus GR-1 and L. fermentum RC-14 significantly alters vaginal flora: randomized, placebo-controlled trial in 64 healthy women. FEMS Immunol. Med. Microbiol. 35 131–134. [DOI] [PubMed] [Google Scholar]
- Sanglard D., Hube B., Monod M., Odds F. C., Gow N. A. (1997). A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect. Immun. 65 3539–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangmanee P., Hongpattarakere T. (2014). Inhibitory of multiple antifungal components produced by Lactobacillus plantarum K35 on growth, aflatoxin production and ultrastructure alterations of Aspergillus flavus and Aspergillus parasiticus. Food Control 40 224–233. 10.1016/j.foodcont.2013.12.005 [DOI] [Google Scholar]
- Santiago G. L., Tency I., Verstraelen H., Verhelst R., Trog M., Temmerman M., et al. (2012). Longitudinal qPCR study of the dynamics of L. crispatus, L. iners, A. vaginae, (sialidase positive) G. vaginalis, and P. bivia in the vagina. PLoS ONE 7:e45281 10.1371/journal.pone.0045281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R. S., Robbins N., Cowen L. E. (2011). Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol. Mol. Biol. Rev. 75 213–267. 10.1128/MMBR.00045-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P. E. (2011). Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9 737–748. 10.1038/nrmicro2636 [DOI] [PubMed] [Google Scholar]
- Sun L., Liao K., Wang D. (2015). Effects of magnolol and honokiol on adhesion, yeast-hyphal transition, and formation of biofilm by Candida albicans. PLoS ONE 10:e117695 10.1371/journal.pone.0117695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tendulkar S. R., Saikumari Y. K., Patel V., Raghotama S., Munshi T. K., Balaram P., et al. (2007). Isolation, purification and characterization of an antifungal molecule produced by Bacillus licheniformis BC98, and its effect on phytopathogen Magnaporthe grisea. J. Appl. Microbiol. 103 2331–2339. [DOI] [PubMed] [Google Scholar]
- Tsang P. W., Bandara H. M., Fong W. P. (2012). Purpurin suppresses Candida albicans biofilm formation and hyphal development. PLoS ONE 7:e50866 10.1371/journal.pone.0050866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraelen H., Verhelst R., Claeys G., De Backer E., Temmerman M., Vaneechoutte M. (2009). Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol. 9:116 10.1186/1471-2180-9-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila T., Romo J. A., Pierce C. G., McHardy S. F., Saville S. P., Lopez-Ribot J. L. (2016). Targeting Candida albicans filamentation for antifungal drug development. Virulence 7 1–9. 10.1080/21505594.2016.1197444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wang S., Dong L., Zhuang H., Li T. (2014). Identification of vaginal Lactobacillus isolates by 16S rDNA sequencing and characterization of the hydrogen peroxide production and autoaggregation activities of the predominant strains. Chin. J. Microecol. 10 1117–1122. [Google Scholar]
- Wang S., Wang Q., Xiao B., Zhang R., Wang B., Liao Q., et al. (2015). Characterization of vaginal Lactobacillus strains and their potential antagonistic effects on Candida albicans. Brit. Microbiol. Res. J. 6 185–195. 10.9734/BMRJ/2015/15116 [DOI] [Google Scholar]
- Xu H. Y., Tian W. H., Wan C. X., Jia L. J., Wang L. Y., Yuan J., et al. (2008). Antagonistic potential against pathogenic microorganisms and hydrogen peroxide production of indigenous lactobacilli isolated from vagina of Chinese pregnant women. Biomed. Environ. Sci. 21 365–371. 10.1016/S0895-3988(08)60056-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.