Abstract
Biodiesel production using microalgae would play a pivotal role in satisfying future global energy demands. Understanding of lipid metabolism in microalgae is important to isolate oleaginous strain capable of overproducing lipids. It has been reported that reducing starch biosynthesis can enhance lipid accumulation. However, the metabolic mechanism controlling carbon partitioning from starch to lipids in microalgae remains unclear, thus complicating the genetic engineering of algal strains. We here used “dynamic” metabolic profiling and essential transcription analysis of the oleaginous green alga Chlamydomonas sp. JSC4 for the first time to demonstrate the switching mechanisms from starch to lipid synthesis using salinity as a regulator, and identified the metabolic rate-limiting step for enhancing lipid accumulation (e.g., pyruvate-to-acetyl-CoA). These results, showing salinity-induced starch-to-lipid biosynthesis, will help increase our understanding of dynamic carbon partitioning in oleaginous microalgae. Moreover, we successfully determined the changes of several key lipid-synthesis-related genes (e.g., acetyl-CoA carboxylase, pyruvate decarboxylase, acetaldehyde dehydrogenase, acetyl-CoA synthetase and pyruvate ferredoxin oxidoreductase) and starch-degradation related genes (e.g., starch phosphorylases), which could provide a breakthrough in the marine microalgal production of biodiesel.
Increasing global energy demands have resulted in considerable effort to construct a recycling-oriented society utilizing novel sustainable energy sources1. Biomass has significant potential as an effective, environmentally benign renewable energy source that is biodegradable and non-toxic2. Microalgae are promising biodiesel producers due to their high growth rate, high CO2 fixation efficiency, high environmental stress tolerance, and the capacity to accumulate a large amount of lipid without competing for arable land and potable water2,3,4,5. However, the commercial production of microalgal biodiesel currently faces the serious challenges of low lipid productivity and high production costs2,6,7. Enhancing the economic feasibility of microalgal biodiesel will require critical engineering innovations in large-scale cultivation and breakthroughs for regulating lipid metabolism8.
Microalgae typically accumulates energy-rich compounds such as lipid and starch under environmental stress such as nitrogen depletion and high salinity by redirecting carbon flow towards these compounds for storage6,8,9. Synthetic pathway to produce lipid and starch by CO2 fixation have been determined in Chlamydomonas reinhardtii (C. reinhardtii) (Fig. 1); the genes encoding phosphoglucoisomerase (PGI), phosphoglucomutase (PGM), ADP-glucose phosphorylase (AGPP), α-amylase (AMY) and starch phosphorylase (SP) are related to starch biosynthesis and degradation10,11. Also, the genes encoding pyruvate dehydrogenase (PDH), pyruvate-ferredoxin oxidoreductase (PFOR), acetyl-CoA (AcCoA) carboxylase (ACC), pyruvate decarboxylase (PDC), acetaldehyde dehydrogenase (ALDH), AcCoA synthetase (ACS), and glycerol 3-phosphate dehydrogenase (GPDH) are related to lipid biosynthesis11,12,13,14,15,16,17. The compartmentalization of carbon metabolism in C. reinhardti was reviewed by Johnson et al. in detail9. Siaut et al. established that microalgae initially synthesize starch for a short-term energy reserve, then lipid is synthesized as the long-term energy storage compound18. Thus, previous studies have demonstrated that the lipid content of microalgae can be enhanced by switching carbon partitioning from starch to lipid synthesis using starch-related-gene-knockout approaches, while the cell growth of such engineered strains is inhibited18,19,20,21. Slower cell growth will result in a decreased lipid productivity even if the lipid content is improved.
Figure 1. Carbon partitioning from CO2 to starch and lipid accumulation and the key enzymes in Chlamydomonas spp.
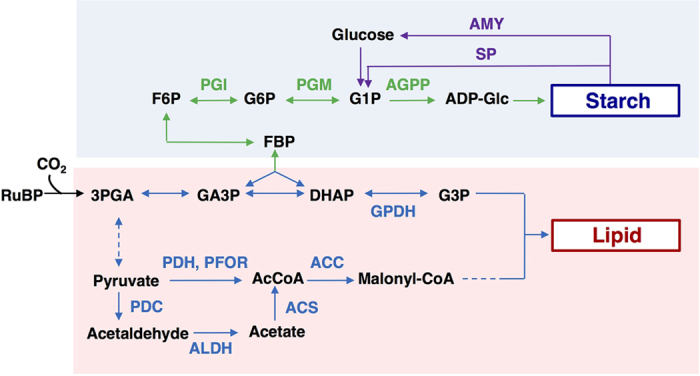
Metabolites: ribulose 1,5-bisphosphate carboxylase/oxygenase; RuBP, 3-phosphoglycerate; 3 PGA, glyceraldehyde 3- phosphate; GA3P, fructose 1,6-bisphosphate; FBP, dihydroxyacetone phosphate; DHAP, fructose 6-phosphate; F6P, glucose 6-phosphate; G6P, glucose 1-phosphate; G1P, ADP-Glucose; ADP-Glc, glycerol 3-phosphate; G3P, acetyl-CoA; AcCoA. Enzymes: phosphoglucoisomerase; PGI, phosphoglucomutase; PGM, ADP-Glc phosphorylase; AGPP, α-amylase; AMY, starch phosphorylase; SP, glycerol 3-phosphate dehydrogenase; GPDH, pyruvate dehydrogenase; PDH, pyruvate-ferredoxin oxidoreductase; PFOR, AcCoA carboxylase; ACC, pyruvate decarboxylase; PDC, acetaldehyde dehydrogenase; ALDH, AcCoA synthetase; ACS. Dotted lines represent multiple reaction steps.
Another current major limitation of microalgal biodiesel production is the shortage of freshwater resources22. This highlights the importance of developing microalgal strains that are highly tolerant of seawater. Moderate salinity stress may induce lipid accumulation in some microalgae, but also may cause growth inhibition8,23.
Metabolomics is a powerful tool for functional biology24,25,26 and allows the comprehensive profiling of metabolite accumulation in specific biosynthesis pathways21. Although several studies have focused on the pool size of key metabolites during partial metabolic profiling, there are no reports describing the regulation of “dynamic” metabolism in microalgae27,28. In addition, transcriptomic analysis is vital for providing comprehensive profiling of the mRNA levels of key enzymes related to environmental stress29,30.
We have developed feasible methods for lipid production using the oleaginous green alga Chlamydomonas sp. JSC4, which was isolated from the south Taiwan coast23,31,32. As a hopeful biomass producer, JSC4 possesses advantageous characteristics such as high growth potential and high starch/lipid content23,31. Importantly, lipid accumulation in JSC4 is effectively triggered by addition of sea salt (SS) in combination with nitrogen depletion, and the highest lipid productivity of 312 mg L−1 d−1 per unit dry cell weight (DCW) was attained previously under the photoautotrophic conditions31. Also, dynamic metabolic profiling using an in vivo13C labeling was carried out under different light intensities to reveal carbon metabolism after CO2 fixation31,33.
For efficient lipid production by microalgae, it is required to understand the regulatory networks that control carbon partition from starch to lipid9. However, the starch-to-lipid switching mechanisms in Chlamydomonas spp. under the salinity condition has yet to be elucidated. In this study, the switching mechanism was explored by monitoring in vivo dynamic metabolic profiles. Furthermore, mRNA regulation of the key genes involved in lipid biosynthesis and starch biosynthesis/biodegradation was assessed for the first time in JSC4. This study demonstrates that salinity is a powerful regulator for switching lipid/starch biosynthesis, and reveals the key mechanisms for maximizing the lipid production in JSC4.
Results
Feasibility of using JSC4 as a biodiesel producer
The growth performance of JSC4 under salinity stress was investigated by cultivating under 0%, 1%, or 2% (w/v) SS (Fig. 2). High biomass levels were obtained in medium containing 0% and 1% SS (5.6 g L−1 and 6.4 g L−1 after 9 days’ cultivation, respectively). JSC4 exhibits significant lipid accumulation more than 45% of DCW under 1% and 2% SS, which is obviously superior than the lipid content triggered by single stress of nitrogen depletion (Table 1). This result indicates that the salinity stress coupled with nitrogen depletion significantly improves the lipid content of JSC4. The highest lipid productivity of 358 mg L−1 d−1 was provided under 1% SS, which is superior than our previous results (in the range of 233–312 mg L−1 d−1)23,31 and previous studies (Table S1).
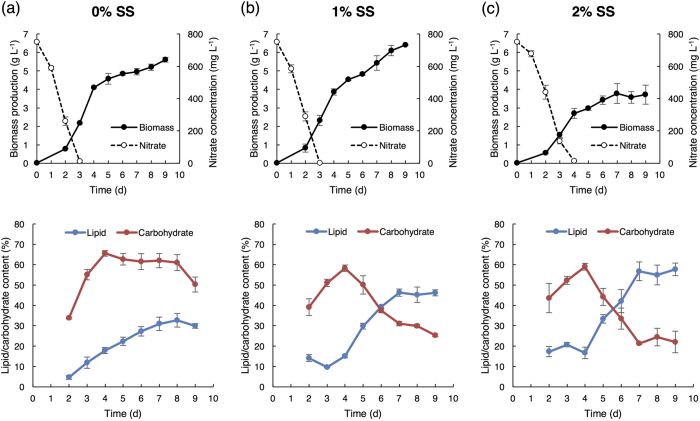
Figure 2. Energy storage compound switched from starch to lipid upon salinity stress.
Time-course profiles of biomass production, nitrate concentration, and lipid and carbohydrate content under (a) 0%, (b) 1%, and (c) 2% of sea salt (SS). Error bars indicate the standard deviation (SD) of three replicate experiments.
Table 1. Time-course profiles of biomass productivity under different SS conditions (0%, 1%, and 2%).
| SS (%) | Cultivation time (d) | Biomass productivity (mg-DCW L−1 d−1) | Lipid content (%) | Lipid productivity (mg L−1 d−1) |
|---|---|---|---|---|
| 0 | 3 | 724.3 ± 16.5 | 11.9 ± 2.8 | 86.2 ± 7.0 |
| 5 | 915.0 ± 58.3 | 22.4 ± 2.0 | 205.0 ± 5.8 | |
| 7 | 708.4 ± 24.6 | 31.0 ± 3.3 | 219.4 ± 22.4 | |
| 1 | 3 | 772.2 ± 92.0 | 9.7 ± 0.2 | 74.6 ± 5.3 |
| 5 | 907.3 ± 13.0 | 29.8 ± 1.4 | 270.7 ± 10.8 | |
| 7 | 775.2 ± 56.3 | 46.5 ± 1.8 | 358.9 ± 20.6 | |
| 2 | 3 | 510.3 ± 45.6 | 20.8 ± 1.0 | 105.9 ± 12.9 |
| 5 | 569.5 ± 37.9 | 33.4 ± 2.1 | 198.2 ± 20.1 | |
| 7 | 539.3 ± 76.6 | 56.9 ± 4.6 | 306.1 ± 19.2 |
Values are the averages of three replicated experiments, ±SD.
Energy storage compound switched from starch to lipid under salinity stress
Since most of the carbohydrate in Chlamydomonas spp. is starch19, we compared the time-course profiles of the total carbohydrate and total lipid content under different SS concentrations (Fig. 2 and Table 1). At 0% SS, the carbohydrate content sharply increased from 33.8% (Day 2) to 61.3% (Day 4) and remained constant thereafter, whereas the lipid content gradually increased and attained a maximum value of 32.8% (Day 8). The carbohydrate content in 1% or 2% SS also increased to around 60% during early-stage culture (Day 2 to Day 4), but then dramatically decreased after 4 days’ cultivation, in parallel with an increase in the lipid content. A maximum lipid content of 46.5% and 56.9% was obtained in 1% and 2% SS, respectively. These results illustrated that the dominant energy storage compound was rapidly switched from starch to lipid upon salinity stress. In addition, we observed salinity-induced lipid accumulation under a transmission electron microscope (TEM). The images depicted in Fig. S1 clearly show many large oil drops formed under salinity stress. Taken together, we demonstrate that energy-flow was shifted from carbohydrate (mainly starch) to lipid by salinity stress.
Time-course profiles of key metabolite in starch and lipid synthesis with/without salinity stress
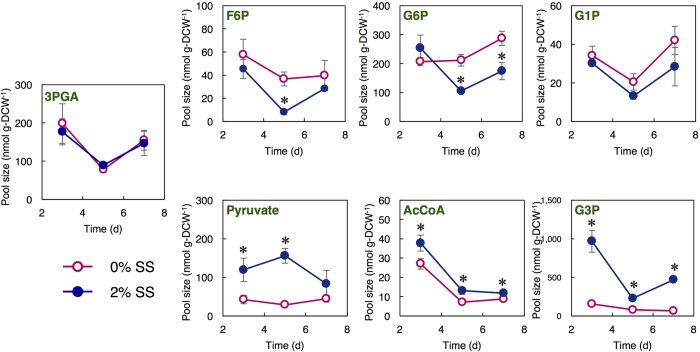
To investigate the metabolic mechanism at the lipid production stage, JSC4 cells were cultivated under 0% or 2% SS for 3, 5 and 7 days, and the intracellular metabolites were analyzed comprehensively. In the whole metabolites, we focused on the key metabolites related to starch and lipid synthesis and quantified the pool sizes (Fig. 3). The pool size of 3-phosphoglycerate (3PGA) was similar under the two conditions. Starch is synthesized in Chlamydomonas spp. cells by the polymerization of ADP-glucose (ADP-Glc)19. The pool sizes of the sugar phosphates (i.e., fructose 6-phosphate (F6P) and glucose 6-phosphate (G6P)) were significantly higher under 0% SS at Day 5 and 7. Taken these results with the carbohydrate content shown in Fig. 2, high pool sizes of these metabolites would be corresponding to high carbohydrate content under 0% SS.
Figure 3. Pool size of metabolic intermediates under different SS concentrations.
(0% and 2%) at Day 3, 5, and 7. ○ (pink) and • (blue) indicate 0% and 2% SS, respectively. Error bars indicate the SD of three replicate experiments (*p < 0.05 by Student’s t-test).
Lipid is biosynthesized from glycerol 3-phosphate (G3P) and malonyl-CoA9. The pool sizes of G3P, pyruvate, and AcCoA were higher under salinity stress (Fig. 3). During the lipid accumulation under 2% SS (Day 5 and 7), the pool sizes of G3P, pyruvate, and AcCoA were higher than under 0% SS. When combined the results obtained in Fig. 2, the pool sizes of these metabolites would be corresponding to the lipid content and vital for lipid accumulation.
Effects of salinity on 13C incorporation from CO2 into metabolic intermediates
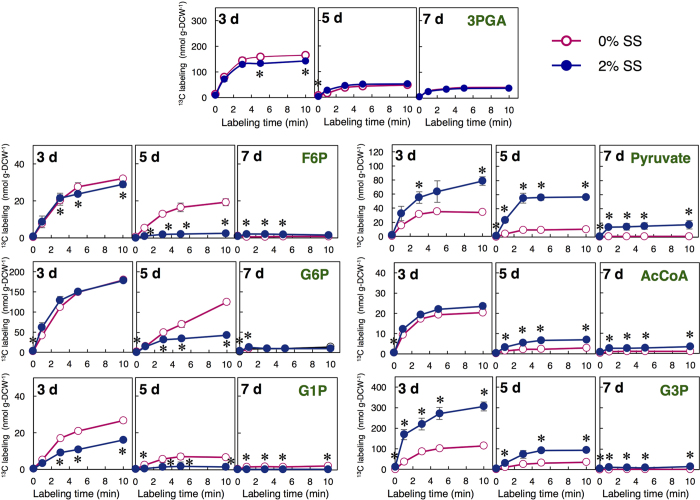
The newly accumulated level of each metabolic intermediate was determined by an in vivo13C-labeling assay31 using 13CO2. 13CO2 was transported and then fixed into 3PGA by ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBP). The assimilation rate of carbon into each metabolite is reflected by the 13C labeling amounts and thus can help elucidate the dynamics of starch and lipid biosynthesis. 13C labeling, in this study, is defined as the amount of carbon that is newly incorporated into metabolites. Figure 4 shows the 13C labeling for several metabolites in cells cultivated under 0% and 2% SS. All these13C-labeled metabolites were obviously decreased at from Day 3 to Day 7. Higher 13C labeling values for F6P, G6P, and glucose 1-phosphate (G1P) were observed under 0% SS than 2% SS on Day 5, whereas the 13C labeling of G3P was significantly increased at Day 3 and 5 under 2% SS. Moreover, the 13C labeling of pyruvate and AcCoA was higher under 2% SS at Day 5 and 7.
Figure 4. Time course of 13C labeling under different SS concentrations (0% and 2%) at Day 3, 5, and 7.
○ (pink) and • (blue) indicate 0% and 2% SS, respectively. Error bars indicate the SD of three replicate experiments (*p < 0.05 by Student’s t-test).
Relative quantification of mRNA levels related to lipid and starch synthesis with/without salinity stress
A better understanding of salinity-induced starch-to-lipid conversion in JSC4 requires time-course transcription analyses of the genes related to starch biosynthesis/degradation and lipid biosynthesis. Genome sequencing revealed that key genes in the lipid and starch metabolism pathway are conserved between C. reinhardtii and JSC4 (Fig. 1). The switching of starch/lipid synthesis induced by salinity stress described above was substantiated by quantifying mRNA levels by quantitative real-time PCR (qPCR) (Fig. 5).
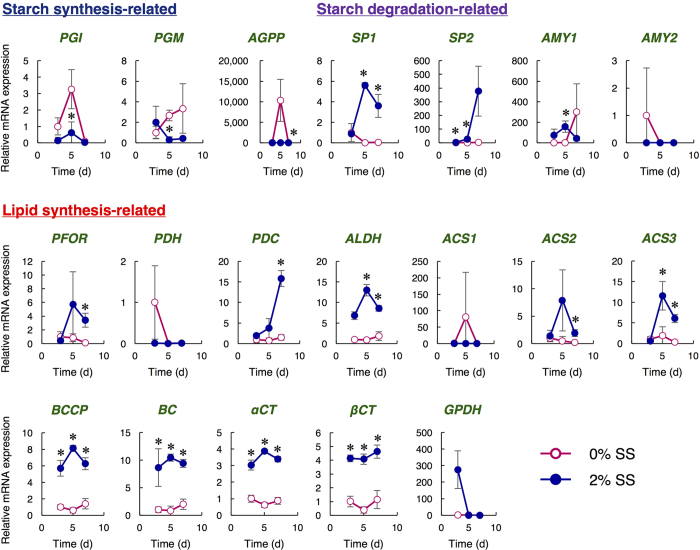
Figure 5. Relative quantification of mRNA under different SS concentrations (0% and 2%) at Day 3, 5, and 7.
○ (pink) and • (blue) indicate 0% and 2% SS, respectively. Biotin carboxyl carrier protein (BCCP), biotin carboxylase (BC), and carboxyltransferase α-subunit (αCT) and β-subunit (βCT) are components of ACC in Chlamydomonas sp. Shown are the relative mRNA levels normalized by the level of the RPL32 and by the level of each gene at the Day 3 under the 0% SS. Error bars indicate the SD of three replicate experiments (*p < 0.05 by Student’s t-test).
Transcription of genes involved in synthesis and degradation of starch10,11 was measured simultaneously. At Day 5, the mRNA levels of starch-synthesis-related genes such as PGI and PGM were significantly higher under 0% SS than under 2% SS. In particular, the level of AGPP drastically increased more than 10,000-fold from Day 3 to Day 5. The mRNA levels of enzymes involved in starch degradation, such as SP1 and SP2, were significantly higher under 2% SS than under 0% SS. The levels of SP1 and SP2 dramatically increased from Day 3 to Day 5, and the level of SP2 further increased 13.5-fold from Day 5 to Day 7. The level of AMY1 under salinity stress was higher than that under 0% SS at Day 3 and 5, while AMY2 expression was lower under 2% SS.
Next, transcription of genes involved in lipid synthesis11,12,13,14,15,16,17 was measured. Determination of the mRNA expression involved in lipid synthesis showed that PFOR mRNA under 2% SS increased approximately 10.5-fold from Day 3 to Day 5 and then remained constant; these levels are much higher than under 0% SS. The levels of PDC, ALDH, ACS2, and ACS3 under 2% SS were also significantly higher than under 0% SS. In particular, PDC showed 8.3-fold increase from Day 3 to Day 7 under 2% SS. ACS2 and ACS3 were enhanced from Day 3 to Day 5 under 2% SS (5.7- and 18.4-fold, respectively). ACC in Chlamydomonas spp. contains biotin carboxyl carrier protein (BCCP), biotin carboxylase (BC), and carboxyltransferase α-subunit (αCT) and β-subunit (βCT)16. The mRNA levels of BCCP, BC, αCT, and βCT was constantly higher under 2% SS. GPDH expression was higher under 2% SS at Day 3, but dramatically decreased thereafter, whereas extremely low signals of GPDH were detected under 0% SS throughout the cultivation period.
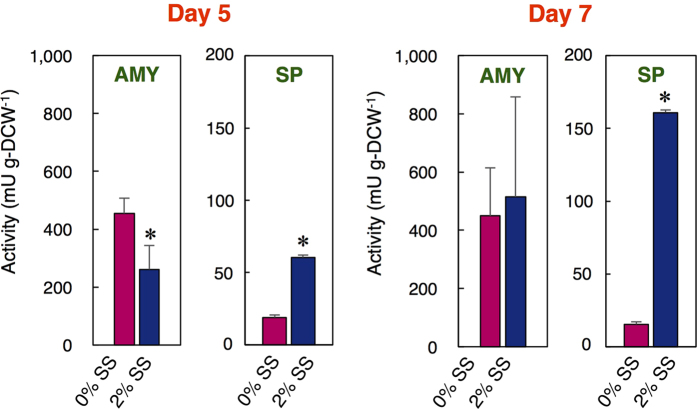
Determination of starch-degrading enzyme activity under salinity stress
AMY and SP activities in the cells were evaluated on Day 5 and 7 to investigate the starch degradation pathway affected by salinity. AMY activity was not largely affected by the addition of SS (Fig. 6). In contrast, SP activity was significantly increased under 2% SS (3.2- and 10.5-fold at Day 5 and 7, respectively).
Figure 6. Catalytic activity of AMY and SP at Day 5 and 7.
Error bars indicate the SD of three replicate experiments (*p < 0.05 by Student’s t-test).
Discussion
The economic feasibility of microalgal biodiesel can be enhanced by selecting/engineering a strain capable of maintaining high biomass production and rapid accumulation of lipid under environmental stress6. We previously reported that JSC4 is not only a robust producer of both biomass and lipid but a strain with high tolerances of irradiance and salt, which is quite suitable to apply for seawater-based outdoor cultivation23,31. Nitrogen stress is one of the key factors for enhancing lipid accumulation in many microalgal species3, however, the single stress of nitrogen depletion could not always induce the lipid accumulation of microalgae. In some microalgal strains, combining the dual stresses are required6. Salinity is also an important factor affecting the lipid content of some microalgae species, but their lipid productivity under high salinity stress is usually lower due to the concomitant strong inhibition of cell growth6. Metabolic profiling enables us to discover the metabolic mechanism for improving lipid accumulation in a microalga34. By utilizing knowledges of the lipid metabolism pathway, we can construct an oil-overproducing algal strain through genetic engineering20,34. However, the mechanism of salinity-induced lipid synthesis in JSC4 has not been elucidated in previous studies23,31,32. In this study, dynamic metabolic profiling together with transcription analysis under salinity stress were conducted for the first time, and the switching mechanisms from starch to lipid synthesis in JSC4 was demonstrated. We also successfully identified the metabolic rate-limiting step (e.g., pyruvate-to-AcCoA) and several key genes (e.g., ACC, PDC, ALDH, ACS, PFOR, and SP) for enhancing lipid accumulation in JSC4.
We show that salinity stress reduces starch and concomitantly enhances lipid accumulation (Fig. 2), and significantly increases the pool sizes of key lipid-synthesis-related metabolites (Fig. 3). Wang et al. indicated that salt causes osmotic stress, resulting in a dramatic accumulation of glycerol in Dunaliella tertiolecta and Dunaliella bardawil35. This is in agreement with the rapid accumulation of G3P under 2% SS shown in this study, since G3P is an important intermediate for glycerol synthesis. In Fig. 4, the 13C labeling of G1P was slightly higher at Day 5 under 0% SS, indicating that more G6P would be converted to G1P, and that the reaction from G6P to G1P might be activated to maintain a high starch content. In addition, a much higher 13C labeling of G6P was observed at Day 5 under 0% SS. Carbohydrate accumulation in Chlamydomonas spp. is mainly regulated by AGPP activity as the rate-limiting step19,21,36. We here propose that enhancing the reaction rate from G6P to G1P could also play an important role in improving starch accumulation in microalgae. In transgenic tobacco plants, overexpression of plastidial PGM increased starch content37. This research may support our proposal.
The 13C labeling of G3P was significantly higher under 2% SS, particularly at Day 3 and 5 (Fig. 4). The 13C labeling of AcCoA under 2% SS was also higher compared to under 0% SS. Thus, the higher 13C labeling of G3P and AcCoA under SS may correlate with higher lipid accumulation in the cell. On the other hand, the 13C labeling of pyruvate under salinity stress was clearly higher than that of AcCoA, likely due to not only an increase of carbon influx into pyruvate but also a limitation in the conversion to AcCoA. In Fig. 3, pyruvate was accumulated in the presence of SS, while the level of AcCoA was not largely increased. The results suggest that the lipid biosynthesis can be further improved by accelerating the metabolic reaction from pyruvate to AcCoA. By tracking the dynamic carbon flow, we found the likely rate-limiting steps in starch/lipid synthesis (e.g., starch: G6P-to-G1P; lipid: pyruvate-to-AcCoA), and proposed the strategy of starch-to-lipid conversion under salinity stress to effectively enhance lipid accumulation in microalgae. Decreased 13C labeling of these metabolites at Day 5 and 7 (Fig. 4) suggests that lipid production was performed not only by using newly incorporated CO2 but also by using intracellular carbon sources (i.e. carbohydrates).
Level of key gene transcripts is important for understanding the metabolic pathway. Although previous transcriptomic analyses in C. reinhardtii shed light on carbon partitioning between lipid and starch9, the levels of key transcripts related to starch and lipid biosynthesis and starch degradation under salinity stress were here evaluated simultaneously and dynamically for the first time in JSC4 (Fig. 5). Lower levels of the PGI and PGM transcript under 2% SS indicate that the conversion of F6P to G1P via G6P can be retarded to slow down starch biosynthesis. At Day 5, the drastically increased AGPP transcript under 0% SS may correlate to a high content of starch in the cells since the metabolic reaction by AGPP should be the main step for producing starch21. Interestingly, the significant enhancement of the SP1/SP2 transcripts under 2% SS at Day 5 and 7 indicates that SP1 and SP2 were immediately activated during salinity stress, thus aiding the switch from starch to lipid synthesis. With the results shown in Fig. 6, these findings clearly indicate that the upregulation of genes related to starch degradation by salinity stress could lead to the production of more SP, and the concomitant generation of more lipids. C. reinhardtii possesses functional AMY and SP as starch degradation enzymes38, however, it accumulates both lipid and starch under salinity stress18. Under salinity stress, the JSC4 cells produce lipid by degrading starch, and this would be the characteristic feature of this strain.
We focused on characterizing the mRNA levels of lipid biosynthesis-related genes (Fig. 5). The dramatic increase in PFOR transcript under 2% SS and the extremely low levels of PDH transcript was observed. These results raise the possibility that the metabolic reaction from pyruvate to AcCoA may be primarily catalyzed by PFOR, but not PDH. In the oleaginous green alga Chlorella desiccata, activation of the PDH-bypass, which consists of PDC, ALDH and ACS, is required for higher lipid biosynthesis under nitrogen deprivation39. In JSC4, expression levels of PDC, ALDH, and ACS were increased under SS. This result suggests that the PDH-bypass might also play important roles in lipid production under salinity condition. Notably, because GPDH is a critical enzyme in catalyzing the reversible conversion of dihydroxyacetone phosphate (DHAP) to G3P, the high level of the GPDH transcript at Day 3 under salinity stress indicates that large amounts of G3P would be produced and be available for lipid synthesis, which consistent with the results in Figs 3 and 4. Thus, by using transcription analysis combined with metabolic profiling, we suggest that carbon flow between starch and lipid biosynthesis in microalgae can be regulated by salinity stress. There is a possibility that some gene functions are also regulated at post-translational levels. Further examination of the specific enzymatic activity is required to strengthen the hypothesis obtained in this study.
The pool size of metabolites in Chlamydomonas spp. has been evaluated by metabolic profiling analysis. Bölling and Fiehn reported changes in the pool sizes of metabolites in C. reinhardtii following the depletion of various components of the growth medium27. However, changes in the pool size are not always reflected in the dynamic metabolism40. Accordingly, the dynamic metabolic turnover and carbon fraction of these metabolites must be investigated41,42. Recently, Kempa et al. reported the turnover of metabolites in C. reinhardtii by analyzing 13C isotopomer flow, however, it does not shed light on how to enhance a specific metabolic reaction41. Our study is the first report to provide comprehensive information regarding dynamic carbon flux and transcription related to the switching mechanisms between starch and lipid biosynthesis.
Increase in cellular biomass was observed after nitrate depletion (Fig. 2), likely due to the utilization of intracellular nitrogen source including protein. As previously reported, the JSC4 strain demonstrates protein reduction after nitrate depletion23. Some algae have shown cellular biomass increase along with intracellular protein reduction after the nitrogen depletion23,28. Further study is required to elucidate the biomass increment mechanism.
We suppose that the reason why JSC4 accumulates lipid under the salinity condition would be related to the fact that they originally live in the brackish-water region23. In the brackish water, nutrients required for photosynthesis might be sufficiently supplied from river. When JSC4 is carried to the open ocean by oceanic current, they might suffer from both starvation and salinity stress, which triggers accumulation of lipid as an energy source for long term survival. As the freshwater microalga C. reinhardtii, which was originally isolated from Amherst in the United States42, does not carry out the salinity-induced carbon flow switching18, it might be a brackish water-specific response to starvation.
Methods
Microorganism and growth conditions
Chlamydomonas sp. JSC423, isolated from a coastal area of southern Taiwan, was cultivated under phototrophic condition in Modified Bold (MB) 6N medium consisting of 8.8 mM NaNO3, 0.22 mM K2HPO4, 0.3 mM MgSO4, 0.17 mM CaCl2, 0.43 mM KH2PO4, 0.43 mM NaCl, and different concentrations of SS (Sigma-Aldrich Co., St. Louis, MO, USA). The levels of metals in the medium are described in the previous report43. After 3 days’ pre-culture, cells were inoculated into double-deck photobioreactor44, which has a first stage containing 2M NaHCO3/Na2CO3 to supply the desired concentration of CO2, and a second stage containing the culture broth, at an initial cell concentration of 20 mg L−1. The cells were cultured under the conditions as follows; light intensity, 250 μmol photons m−2 s−1 (white fluorescent lamps); CO2 aeration, 2% CO2; temperature, 30 °C.
Measurement of residual nitrate content
Nitrate concentration was measured using an optical method as previously reported45. The broth was centrifuged at 5,000 × g for 1 min, and the absorbance of the supernatant diluted 20-fold with distilled water was measured at 220 nm (i.e., Abs220) using UVmini-1240 UV-VIS spectrophotometer (Shimadzu, Kyoto, Japan). The residual nitrate content was evaluated using an appropriate calibration curve23.
Evaluation of lipid content
Cultivation of JSC4 was carried out under the same conditions as described above. Cells were collected by centrifugation at 5,000 × g for 1 min, washed with distilled water twice, and lyophilized. The dried cells were fractured with 0.5 mm glass beads using a multi-bead shocker (Yasui Kikai, Osaka, Japan) at 4 °C. The total lipids were extracted using the mixture of chloroform, methanol and water, and were esterified by Fatty Acid Methylation Kit (Nacalai Tesque, Kyoto, Japan) according to the previous method46. The fatty acid methyl esters (FAMEs) were identified and quantified by gas chromatography-mass spectrometry (GC-MS) on a GCMS-QP2010 Plus (Shimadzu) as described previously23. Samples were injected onto a DB-23 capillary column (60 m, 0.25 mm internal diameter, 0.15 μm film thickness; Agilent Technologies, Palo Alto, CA, USA). Helium was used as the carrier gas at a flow rate of 2.3 mL min−1. The injector, ion source, and interface source temperatures were set at 230, 230, and 250 °C, respectively. The oven temperature was initially set at 50 °C for 1 min, increased from 50 to 175 °C at a rate of 25 °C/min, increased from 175 to 230 °C at a rate of 4 °C/min, and held at 230 °C for 5 min. Supelco 37 Component FAME Mix (Sigma-Aldrich Co.) was utilized as a quantitative standard, and heptadecanoic acid (Sigma-Aldrich Co.) was used as an internal standard. Lipid productivity is calculated as overall lipid produced from Day 0.
Evaluation of carbohydrate content
The total carbohydrate content in the dried cells, prepared as above, was evaluated using a colorimetric method with an anthrone reagent47. The dried cells were incubated in anthrone solution (0.2% (w/v) anthrone in 75% (v/v) sulfuric acid) for 15 min at 100 °C. After cooling on ice, an aliquot was centrifuged at 5,000 × g for 1 min and the supernatant was measured at 620 nm (i.e., Abs620). The carbohydrate content was quantified using a standard curve generated using glucose.
Metabolic profiling
Cultivation of JSC4 was carried out under the same conditions as described above. Cell sampling was performed according to our previously reported method23. Cells were collected on 1-μm pore size polytetrafluoroethylene filter disks (Omnipore; Millipore, Billerica, MA, USA), washed with pre-chilled (4 °C) 20 mM ammonium carbonate, and immediately placed into 1 mL of pre-chilled (−30 °C) methanol containing 12.4 μM piperazine-1,4-bis(2-ethanesulfonic acid) as the internal standard. 300 μL of pre-chilled (4 °C) chloroform and 100 μL of pre-chilled (4 °C) water were added to provide a 10:3:1 (v/v/v) methanol:chloroform:water mixture. The cells were completely fractured with 300 μL of 0.5 mm glass beads using a multi-beads shocker (Yasui Kikai), then 396 μL of distilled water was added. The water layer was filtered through a Millipore 5 kDa cut-off filter and dried under vacuum using a FreeZone 2.5 Plus freeze dry system (Labconco, Kansas City, MO, USA).
The intermediate metabolites in lipid/starch synthesis pathway (e.g., 3 PGA, pyruvate, AcCoA, G3P, F6P, G6P, and G1P) were targeted and determined according to a previously described method28. Dried metabolites were dissolved in 20 μL of Milli-Q water and analyzed using a capillary electrophoresis-mass spectrometry (CE-MS) system comprising an Agilent G7100 CE system, an Agilent G6224AA LC/MSD time-of-flight system, and an Agilent 1200 series isocratic HPLC pump equipped with a 1:100 splitter for delivery of the sheath liquid. The CE separations were performed in a fused silica capillary (1 m × 50 μm i.d.) filled with 50 mM ammonium acetate (pH 9.0) for anionic metabolite analyses. The flow rate of the sheath liquid was set at 8 μL min−1. The electrospray ionization-mass spectrometry analyses were conducted in negative ion mode. Mass data were acquired at a rate of 1 spectra s−1 over the mass-to-charge ratio (m/z) range 70–1000.
In vivo13C-labeling
Using the same cell cultures as the metabolic profiling, in vivo13C-labeling was performed using sodium 13C-bicarbonate (NaH13CO3) as a carbon source as described previously31. Cells were harvested from culture broth at Day 3, 5, and 7 and resuspended in labeling medium (25 mM NaH13CO3) at the same cell density as in the culture broth. After time-course labeling for 1–10 min to monitor the accumulation rates, metabolites in approximately 10 mg of wet cells were analyzed using CE-MS as described above. The ratio of 13C to total carbon in each metabolite was calculated by searching for mass shifts between the 12C and 13C mass spectra31. 13C labeling, defined as “pool size (nmol g-DCW−1) × 13C fraction (%) (Fig. S2”), was used to evaluate the quantitative metabolic flux in the cells.
Transcript analysis
Cultivation of JSC4 was performed under the same conditions as described above. Cells were harvested from the broth by centrifugation at 5,000 × g for 1 min, frozen in liquid nitrogen, and milled using a mortar48. Total RNA was isolated from the frozen cell powder using an RNeasy Plus Universal Kit (QIAGEN, Tokyo, Japan). For qPCR experiments, complementary DNA was synthesized from approximately 100 ng of total RNA using a ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO, Osaka, Japan). qPCR was performed with THUNDERBIRD SYBR qPCR Mix (TOYOBO) using Mx qPCR Systems (Agilent). The average threshold cycle values were evaluated throughout the logarithmic amplification phase using triplicate samples, and were normalized by the level of the RPL32 (encoding ribosomal protein large subunit) and by the level of each gene at the Day 3 under 0% SS to evaluate relative levels of RNA transcription. The qPCR primers (Table S2) were designed based on each predicted gene sequence from the genome sequence, which was determined by GS FLX + (Roche) and MiSeq (Illumina) systems using genome DNA extracted by DNeasy Plant mini kit (QIAGEN) from the JSC4 cells.
Enzyme activity analysis
Cultivation of JSC4 was performed under the same conditions as described above. AMY and SP activities were analyzed from approximately 100 mg and 60 mg of wet cells, respectively. After harvesting by centrifugation at 5,000 × g for 1 min, cells were suspended in 1 mL of 50 mM Hepes-NaOH buffer (pH 7.0) containing 2 mM EDTA and 2 mM CaCl2, then were frozen and thawed with liquid nitrogen and 30 °C water four times38. The cells were broken by sonicating on ice for 30 s of 50% pulses; 30 s cooling period, five cycles. The suspension was centrifuged at 20,000 × g for 20 min at 4 °C, and the supernatant was purified on a PD-10 column system (GE-Healthcare Bio-Sciences KK, Tokyo, Japan). AMY activity of the cell extract was analyzed using an α-Amylase assay kit (Kikkoman Biochemifa Company, Tokyo, Japan)49. Hydrolysis of a model substrate, 2-chloro-4-nitrophenyl 65-azido-65-deoxy-β-maltopentaoside was detected by 2-chloro-4-nitrophenol (CNP) generation. 1 unit of the enzyme activity was defined as the amount of enzyme required to liberate 1 μmol of CNP per minute at 37 °C.
SP activity of the cell extract was evaluated according to a previous report50 with some minor modification. 50 μL of purified protein suspension was added to 395 μL of 50 mM HEPES-NaOH buffer (pH 7.0) containing 10 mM inorganic phosphoric acid and 10 mg mL−1 soluble starch (CAS number: 9005-84-9, Nacalai Tesque), incubated at 30 °C for 1 h, and boiled for 5 min to stop the catalytic reaction. Then 500 μL of 50 mM Tris-HCl buffer (pH 7.0) containing 120 mM MgCl2, 0.05 mM glucose 1,6-diphosphate, and 0.5 mM NADP was mixed with the suspension, 4 units of PGM and 2 units of G6P dehydrogenase were added, and reacted for 30 min at room temperature. The production of NADPH was monitored by absorbance at 365 nm using UVmini-1240 (Shimadzu). 1 unit of the enzyme activity was defined as the amount of enzyme required to liberate 1 μmol of G1P per minute at 30 °C.
Additional Information
How to cite this article: Ho, S.-H. et al. Dynamic metabolic profiling together with transcription analysis reveals salinity-induced starch-to-lipid biosynthesis in alga Chlamydomonas sp. JSC4. Sci. Rep. 7, 45471; doi: 10.1038/srep45471 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors would like to thank Ms. Mika Kakuda for technical assistance. We also thank Dr. Shinichiro Maruyama (Tohoku Univ.) for technical help in annotating the genome data of the strain JSC4. This work was supported by project P07015 of the New Energy and Industrial Technology Development Organization (NEDO). This work was also supported by State Key Laboratory of Urban Water Resource and Environment (Harbin Institute of Technology) (No. 2016TS07). The authors acknowledge the financial support of the National Science Council, Taiwan under grant no. 102-3113-P-006-016 and 101-3113-P-110-003.
Footnotes
The authors declare no competing financial interests.
Author Contributions S.H. designed the study, conducted the experiments, and drafted the manuscript. A.N. designed the study, conducted the experiments, interpreted the data and drafted the manuscript. Y.K. conducted the experiments of qPCR and commented on the experiment. H.Y. conducted the experiments of genome DNA extraction. J.C. provided strain JSC4 and commented on the manuscript. N.M. assisted in analysis of genome sequence. Y.H. designed the experiments of genome sequence analysis. J.M. participated in experimental design and revised the manuscript. T.H. designed the study, revised the manuscript and supervised the research. A.K. assisted with the laboratory management. All authors have read and approved the final manuscript.
References
- Georgianna D. R. & Mayfield S. P. Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 488, 329–335 (2012). [DOI] [PubMed] [Google Scholar]
- Moody J. W., McGinty C. M. & Quinn J. C. Global evaluation of biofuel potential from microalgae. Proc. Natl. Acad. Sci. USA 111, 8691–8696 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisti Y. Biodiesel from microalgae. Biotechnol. Adv. 25, 294–306 (2007). [DOI] [PubMed] [Google Scholar]
- Stephens E. et al. An economic and technical evaluation of microalgal biofuels. Nat. Biotechnol. 28, 126–128 (2010). [DOI] [PubMed] [Google Scholar]
- Larkum A. W., Ross I. L., Kruse O. & Hankamer B. Selection, breeding and engineering of microalgae for bioenergy and biofuel production. Trends Biotechnol. 30, 198–205 (2012). [DOI] [PubMed] [Google Scholar]
- Ho S. H. et al. Perspectives on engineering strategies for improving biofuel production from microalgae-A critical review. Biotechnol. Adv. 32, 1448–1459 (2014). [DOI] [PubMed] [Google Scholar]
- Wijffels R. H. & Barbosa M. J. An outlook on microalgal biofuels. Science (Washington) 329, 796–799 (2010). [DOI] [PubMed] [Google Scholar]
- Hu Q. et al. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54, 621–639 (2008). [DOI] [PubMed] [Google Scholar]
- Johnson X. & Alric J. Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints for carbon partitioning between oil and starch. Eukaryot. Cell 12, 776–793 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U. Compartmentation of glycolysis and of the oxidative pentose-phosphate pathway in Chlamydomonas reinhardii. Planta 167, 81–86 (1986). [DOI] [PubMed] [Google Scholar]
- Terashima M., Specht M. & Hippler M. The chloroplast proteome: a survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Curr. Genet. 57, 151–168 (2011). [DOI] [PubMed] [Google Scholar]
- Klöck G. & Kreuzberg K. Kinetic properties of a sn-glycerol-3-phosphate dehydrogenase purified from the unicellular alga Chlamydomonas reinhardtii. Biochim. Biophys. Acta. 991, 347–352 (1989). [DOI] [PubMed] [Google Scholar]
- Kreuzberg K., Klock G. & Grobheiser D. Subcellular distribution of pyruvate‐degrading enzymes in Chlamydomonas reinhardtii studied by an improved protoplast fractionation procedure. Physiol. Plant. 69, 481–488 (1987). [Google Scholar]
- Burgess S. J. et al. Identification of the elusive pyruvate reductase of Chlamydomonas reinhardtii chloroplasts. Plant Cell Physiol. 57, 82–94 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocker C. et al. Aldehyde dehydrogenase (ALDH) superfamily in plants: gene nomenclature and comparative genomics. Planta. 237, 189–210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalutskaya Z., Kharatyan N., Forchhammer K. & Ermilova E. Reduction of PII signaling protein enhances lipid body production in Chlamydomonas reinhardtii. Plant Sci. 240, 1–9 (2015). [DOI] [PubMed] [Google Scholar]
- Willeford K. O. & Gibbs M. Localization of the enzymes involved in the photoevolution of H2 from acetate in Chlamydomonas reinhardtii. Plant Physiol. 90, 788–791 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaut M. et al. Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 11, 7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. Inhibition of starch synthesis results in overproduction of lipids in Chlamydomonas reinhardtii. Biotechnol. Bioeng. 107, 258–268 (2010). [DOI] [PubMed] [Google Scholar]
- Trentacoste E. et al. Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc. Natl. Acad. Sci. USA 110, 19748–19753 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabawinski C. et al. Starchless mutants of Chlamydomonas reinhardtii lack the small subunit of a heterotetrameric ADP-glucose pyrophosphorylase. J. Bacteriol. 183, 1069–1077 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dismukes G. C. et al. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol. 19, 235–240 (2008). [DOI] [PubMed] [Google Scholar]
- Ho S. H. et al. Optimizing biodiesel production in marine Chlamydomonas sp. JSC4 through metabolic profiling and an innovative salinity-gradient strategy. Biotechnol. Biofuels 7, 97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran R., Reindl W. & Northen T. R. Mass spectrometry based metabolomics and enzymatic assays for functional genomics. Curr. Opin. Microbiol. 12, 547–552 (2009). [DOI] [PubMed] [Google Scholar]
- Diamond S., Jun D., Rubin B. E. & Golden S. S. The circadian oscillator in Synechococcus elongatus controls metabolite partitioning during diurnal growth. Proc. Natl. Acad. Sci. USA 112, E1916–E1925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link H., Kochanowski K. & Sauer U. Systematic identification of allosteric protein-metabolite interactions that control enzyme activity in vivo. Nat. Biotechnol. 31, 357–361 (2015). [DOI] [PubMed] [Google Scholar]
- Bölling C. & Fiehn O. Metabolite profiling of Chlamydomonas reinhardtii under nutrient deprivation. Plant Physiol. 139, 1995–2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasunuma T. et al. Dynamic metabolic profiling of cyanobacterial glycogen biosynthesis under conditions of nitrate depletion. J. Exp. Bot. 64, 2943–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby I. K. et al. Systems-level analysis of nitrogen starvation-induced modifications of carbon metabolism in a Chlamydomonas reinhardtii starchless mutant. Plant Cell. 25, 4305–4323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H. Q. et al. AlgaePath: comprehensive analysis of metabolic pathways using transcript abundance data from next-generation sequencing in green algae. BMC Genomics 15, 196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. H. et al. Dynamic metabolic profiling of the marine microalga Chlamydomonas sp. JSC4 and enhancing its oil production by optimizing light intensity. Biotechnol. Biofuels 8, 48 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi A. et al. Development of lipid productivities under different CO2 conditions of marine microalgae Chlamydomonas sp. JSC4. Bioresour. Technol. 152, 247–252 (2014). [DOI] [PubMed] [Google Scholar]
- Shinohara M. et al. Fixation of CO2 in Clostridium cellulovorans analyzed by 13C-isotopomer-based target metabolomics. AMB Express. 3, 61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radakovits R., Jinkerson R. E., Darzins A. & Posewitz M. C. Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell 9, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhuge J., Fang H. & Prior B. A. Glycerol production by microbial fermentation: a review. Biotechnol. Adv. 19, 201–223 (2001). [DOI] [PubMed] [Google Scholar]
- van den Koornhuyse N. et al. Control of starch composition and structure through substrate supply in the monocellular alga Chlamydomonas reinhardtii. J. Biol. Chem. 271, 16281–16287 (1996). [DOI] [PubMed] [Google Scholar]
- Uematsu K. et al. Expression of Arabidopsis plastidial phosphoglucomutase in tobacco stimulates photosynthetic carbon flow into starch synthesis. J. Plant Physiol. 169, 1454–1462 (2012). [DOI] [PubMed] [Google Scholar]
- Levi C. & Gibbs M. Starch degradation in synchronously grown Chlamydomonas reinhardtii and characterization of the amylase. Plant Physiol. 74, 459–463 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidan O. & Pick U. Acetyl-CoA synthetase is activated as part of the PDH-bypass in the oleaginous green alga Chlorella desiccate. J. Exp. Bot. 66, 7287–7298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondi M. & Liò P. Multi-omics and metabolic modelling pipelines: challenges and tools for systems microbiology. Microbiol. Res. 171, 52–64 (2015). [DOI] [PubMed] [Google Scholar]
- Kempa S. et al. An automated GCxGC-TOF-MS protocol for batch-wise extraction and alignment of mass isotopomer matrixes from differential 13C-labelling experiments: a case study for photoautotrophic-mixotrophic grown Chlamydomonas reinhardtii cells. J. Basic Microbiol. 49, 82–91 (2009). [DOI] [PubMed] [Google Scholar]
- Jang H. & Ehrenreich I. M. Genome-wide characterization of genetic variation in the unicellular, green alga Chlamydomonas reinhardtii. PLoS One 7, e41307 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges J. A., Franklin D. J. & Harrison P. J. Evolution of an artificial seawater medium: improvements in enriched seawater, artificial water over the last two decades. J. Phycol. 37, 1138–1145 (2001). [Google Scholar]
- Hasunuma T. et al. Overexpression of flv3 improves photosynthesis in the cyanobacterium Synechocystis sp. PCC6803 by enhancement of alternative electron flow. Biotechnol. Biofuels 7, 493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collos Y. et al. An optical method for the rapid measurement of micromolar concentrations of nitrate in marine phytoplankton cultures. J. Appl. Phycol. 11, 179–184 (1999). [Google Scholar]
- Bligh E. G. & Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
- Choi S. P., Nguyen M. T. & Sim S. J. Enzymatic pretreatment of Chlamydomonas reinhardtii biomass for ethanol production. Bioresour. Technol. 101, 5330–5336 (2010). [DOI] [PubMed] [Google Scholar]
- Liu C. et al. Validation of housekeeping genes for gene expression studies in an ice alga Chlamydomonas during freezing acclimation. Extremophiles 16, 419–425 (2012). [DOI] [PubMed] [Google Scholar]
- Inokuma K. et al. Efficient co-displaying and artificial ratio control of α-amylase and glucoamylase on the yeast cell surface by using combinations of different anchoring domains. Appl. Microbiol. Biotechnol. 99, 1655–1663 (2015). [DOI] [PubMed] [Google Scholar]
- Dauvillée D. et al. Plastidial phosphorylase is required for normal starch synthesis in Chlamydomonas reinhardtii. Plant J. 48, 274–285 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.