Li et al. show that OGT-mediated STAT3 O-GlcNAcylation, which is modulated by CUL3-Nrf2 signaling, negatively regulates STAT3 phosphorylation and IL-10 production in macrophages and exacerbates experimental colitis and colitis-associated cancer.
Abstract
Signal transducer and activator of transcription 3 (STAT3) is a key mediator of intestinal inflammation and tumorigenesis. However, the molecular mechanism that modulates STAT3 phosphorylation and activation is not fully understood. Here, we demonstrate that modification of STAT3 with O-linked β-N-acetylglucosamine (O-GlcNAc) on threonine 717 (T717) negatively regulates its phosphorylation and targets gene expression in macrophages. We further found that cullin 3 (CUL3), a cullin family E3 ubiquitin ligase, down-regulates the expression of the O-GlcNAc transferase (OGT) and inhibits STAT3 O-GlcNAcylation. The inhibitory effect of CUL3 on OGT expression is dependent on nuclear factor E2–related factor-2 (Nrf2), which binds to the Ogt promoter region and increases gene transcription. Myeloid deletion of Cul3 led to defective STAT3 phosphorylation in colon macrophages, which was accompanied by exacerbated colonic inflammation and inflammation-driven tumorigenesis. Thus, this study identifies a new form of posttranslational modification of STAT3, modulating its phosphorylation, and suggests the importance of immunometabolism on colonic inflammation and tumorigenesis.
Introduction
Colorectal cancer is a leading contributor to cancer-related deaths in the United States and is a major complication of inflammatory bowel disease (IBD; Clevers, 2004). Colorectal cancer and IBD are commonly associated with the exaggerated production of inflammatory cytokines, which are regulated by the activation of various cell signaling pathways (Karin and Greten, 2005; Yu et al., 2009; Saleh and Trinchieri, 2011; West et al., 2015). It has been well established that STAT3 and IL-10 play critical roles in the regulation of intestinal inflammation in both IBD patients and animals with experimental colitis. Genome-wide association studies indicated that STAT3 gene polymorphism is associated with increased susceptibility to IBD (Barrett et al., 2008; Jostins et al., 2012). Myeloid-derived STAT3 exerts a potent anti-inflammatory effect on chemically induced experimental colitis (Takeda et al., 1999). Meanwhile, STAT3 is important for survival and proliferation of intestinal epithelial cells (Bollrath et al., 2009; Grivennikov et al., 2009).
Recent studies of immune system metabolism, namely immunometabolism, have identified a tight link between metabolic reprogramming and hyperinflammation. It has been well documented that activation of immune cells is accompanied by metabolic changes toward increased glucose uptake, glycolysis, and pentose phosphate pathway activity (O’Neill and Hardie, 2013; Pearce et al., 2013). In addition to glycolysis and pentose phosphate pathway, a small portion of glucose metabolizes through the hexosamine biosynthesis pathway (HBP), which leads to the generation of its end product, UDP–N-acetylglucosamine (UDP-GlcNAc; Ruan et al., 2013). O-GlcNAc transferase (OGT) mediates the transfer of UDP-GlcNAc to serine or threonine residue of target proteins, which is called protein O-GlcNAcylation. Previous studies have discovered essential roles of protein O-GlcNAcylation in many fundamental biological activities, including signaling, transcription, and translation (Hart et al., 2011). A very recent study revealed a critical role of OGT in both T progenitor cell development in the thymus and mature T cell clonal expansion in the periphery (Swamy et al., 2016). Through metabolic profiling analysis, another study indicated that HBP is required for macrophage alternative activation (Jha et al., 2015), although a genetic animal model is further required to test this concept. The overall effect of OGT-mediated O-GlcNAcylation in innate immune function and its disease relevance remain elusive (Pearce and Everts, 2015).
Cullin-RING (really interesting new gene) ligases (CRLs) represent the largest E3 ubiquitin ligase family in eukaryotes (Genschik et al., 2013). Within the CRL family, CUL3-formed CRL3 has been reported to target more than 100 substrates for protein ubiquitination and degradation (Emanuele et al., 2011). Previous studies have established CUL3 as an essential regulator of a variety of important biological processes, including cell cycle, development, and cell stress response (Genschik et al., 2013). Recent studies have revealed a critical role of CUL3 in the adaptive immune system, including promoting germinal center B cell and NKT cell differentiation (Mathew et al., 2012) and inhibiting T follicular helper cell differentiation (Mathew et al., 2014). However, the role of CUL3 in the innate immune system and inflammation is not clear. In this study, we observed increased OGT expression and STAT3 O-GlcNAcylation in Cul3-deficient macrophages as a result of elevated Nrf2-mediated Ogt gene transcription. We also found that O-GlcNAcylation of STAT3 on T717 exerts an inhibitory effect on STAT3 phosphorylation and IL-10 production in macrophages and promotes disease severity in chemically induced colitis and a colitis-associated cancer (CAC) model. Our findings demonstrate a mechanistic link between O-GlcNAc signaling and STAT3 function during intestinal inflammation and tumorigenesis.
Results
Defective STAT3 phosphorylation and IL-10 production in Cul3-deficient macrophages
A previous study reported that whole-body deletion of CUL3 caused embryonic lethality (Singer et al., 1999). To examine the role of CUL3 in the innate immune system, we generated mice with a myeloid-conditional Cul3 deletion (Cul3Δmye) by crossing Cul3fl/fl mice (McEvoy et al., 2007) with lysosome M-Cre mice (Fig. S1 A). Cul3fl/fl mice were used as WT controls. Deletion of CUL3 protein in Cul3Δmye BM-derived macrophages (BMMs) was confirmed (Fig. S1 B). As a well-defined CRL3 target (Genschik et al., 2013), nuclear factor E2–related factor-2 (Nrf2) protein (Fig. S1 C) and its target gene Nqo1 transcripts (Fig. S1 D) were both dramatically increased in Cul3Δmye BMMs. These findings confirmed a successful CUL3 deletion in Cul3Δmye macrophages.
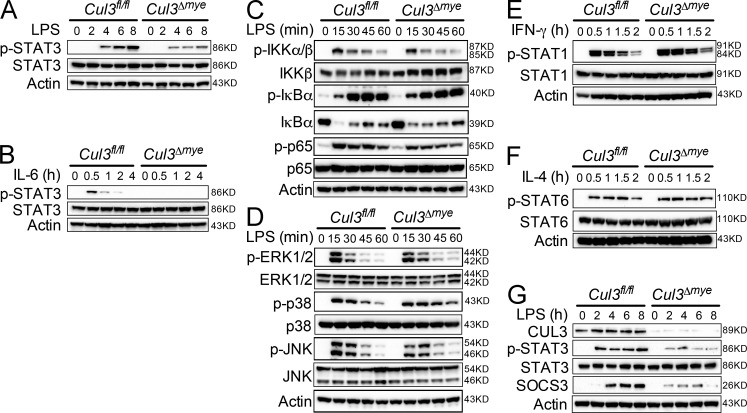
We tested the activation of various immune signaling pathways in Cul3Δmye macrophages. Cul3Δmye BMMs showed dramatically decreased STAT3 phosphorylation at Y705 in response to either LPS (Fig. 1 A) or IL-6 (Fig. 1 B). In contrast, Cul3Δmye BMMs exhibited slightly decreased NF-κB (Fig. 1 C), intact MAPK (Fig. 1 D) signaling upon LPS stimulation, intact STAT1 phosphorylation upon IFN-γ stimulation (Fig. 1 E), and intact STAT6 phosphorylation upon IL-4 stimulation (Fig. 1 F). LPS-induced STAT3 phosphorylation and up-regulation of suppressor of cytokine signaling 3 (SOCS3), a well-defined STAT3 transcriptional target, were also blunted in Cul3Δmye peritoneal macrophages (Fig. 1 G). These findings indicate a specific role of CUL3 in STAT3 phosphorylation in macrophages independent of stimuli.
Figure 1.
CUL3 is required for STAT3 phosphorylation in macrophages. (A and B) BMMs generated from Cul3fl/fl and Cul3Δmye mice were stimulated with 200 ng/ml LPS (A) or IL-6 (B) for the indicated periods. IKK, IκB kinase. Phosphorylation of STAT3 (Y705) was assayed with immunoblotting. (C and D) Immunoblotting of NF-κB (C) and MAPK (D) signaling molecules was performed in Cul3fl/fl and Cul3Δmye BMMs left untreated or treated with LPS for the indicated periods. ERK, extracellular signal–regulated kinase. (E and F) Immunoblotting of STAT1 phosphorylation (Y701) in response to 20 ng/ml IFN-γ (E) and STAT6 phosphorylation (Y641) in response to 20 ng/ml IL-4 (F) was performed in Cul3fl/fl and Cul3Δmye BMMs. (G) Immunoblotting of phosphorylated STAT3 (Y705) and its target protein suppressor of cytokine signaling 3 (SOCS3) was performed in peritoneal macrophages isolated from naive Cul3fl/fl and Cul3Δmye mice left untreated or treated with LPS for the indicated periods. The results are representative of three independent experiments.
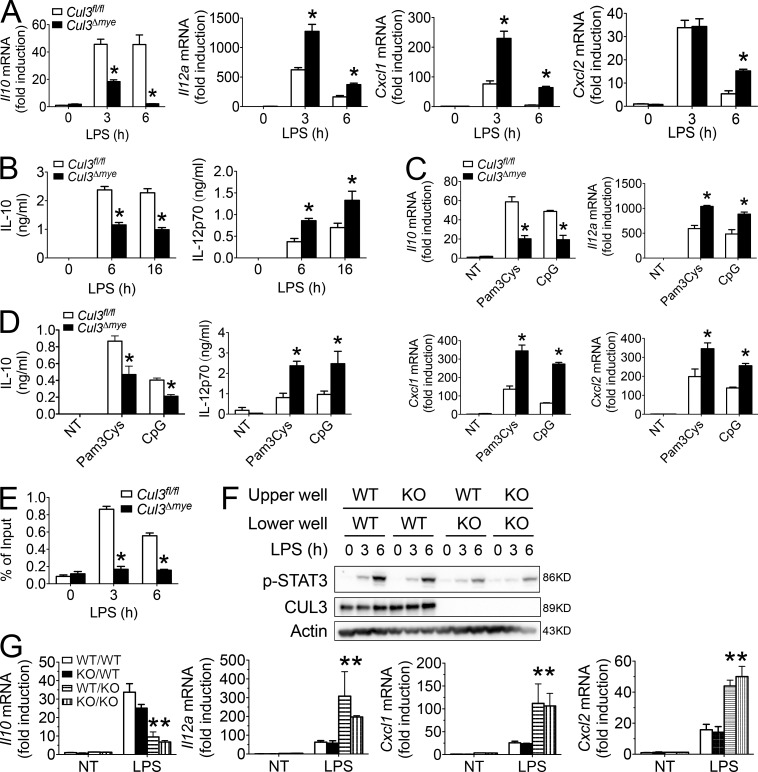
It has been shown that STAT3 is a key transcriptional factor mediating IL-10 production (Takeda et al., 1999). Cul3Δmye macrophages generated significantly lower levels of Il10 but higher levels of Il12a, Cxcl1, and Cxcl2 transcripts (Fig. 2 A), and a lower amount of IL-10 and higher amount of IL-12p70 protein upon LPS stimulation (Fig. 2 B). Macrophages activated by TLR2 (Pam3Cys) or TLR9 (CpG) agonists showed a similar phenotype (Fig. 2, C and D), suggesting a broad hyperinflammatory response in Cul3Δmye macrophages. Chromatin immunoprecipitation (ChIP) assays showed defective STAT3 binding at the promoter region of Il10 gene in LPS-stimulated Cul3Δmye macrophages, suggesting that CUL3 mediates TLR-induced STAT3 activation to cause chromosomal activation of Il10 promoter (Fig. 2 E).
Figure 2.
CUL3 mediates STAT3 phosphorylation and IL-10 production via a cell-intrinsic mechanism. (A–D) Cul3fl/fl and Cul3Δmye BMMs were left untreated or stimulated with 200 ng/ml LPS (A and B) for the indicated periods, 500 ng/ml PamCy3 (TLR2 agonist), or 5 µg/ml CpG oligonucleotide (TLR9 agonist; C and D) for 6 h. Cytokine transcripts including Il10, Il12a, Cxcl1, and Cxcl2 in the cells (A and C) and IL-10 and IL-12p70 proteins in the supernatants (B and D) were measured with RT-PCR and ELISA, respectively. NT, not treated. (E) ChIP assay of the binding of STAT3 at the promoter region of Il10 gene in Cul3fl/fl and Cul3Δmye BMMs left untreated or stimulated with LPS for 3 or 6 h. (F and G) Cul3fl/fl and Cul3Δmye BMMs were placed in either the upper or lower chamber of the Transwell co-culture system, followed by stimulation with LPS for the indicated periods. STAT3 phosphorylation (Y705; F) and cytokine transcripts including Il10, Il12a, Cxcl1, and Cxcl2 (G) in the cells placed in the lower chambers were assayed by immunoblotting and RT-PCR, respectively. The results shown are representative of three independent experiments and are expressed as mean ± SD. *, P < 0.05 versus controls.
STAT3 is a well-established signal transducer of the cytokine–cytokine receptor signaling pathway (O’Shea and Plenge, 2012). To examine whether defective STAT3 phosphorylation is caused by an altered extracellular cytokine environment for Cul3Δmye macrophages, we used a Transwell co-culture system in which Cul3f/fl and Cul3Δmye macrophages placed in distinct chambers shared an identical cytokine environment. Despite the genotypes of co-cultured cells in the upper chamber, Cul3Δmye BMMs in the lower chamber showed decreased STAT3 phosphorylation (Fig. 2 F), lower levels of Il10, and higher levels of Il12a, Cxcl1, and Cxcl2 transcripts (Fig. 2 G) after LPS stimulation compared with similarly treated WT BMMs (Fig. 2 F). These findings support a cell-intrinsic mechanism of CUL3 in mediating the STAT3–IL-10 signaling axis.
CUL3 negatively regulates OGT expression and protein O-GlcNAcylation
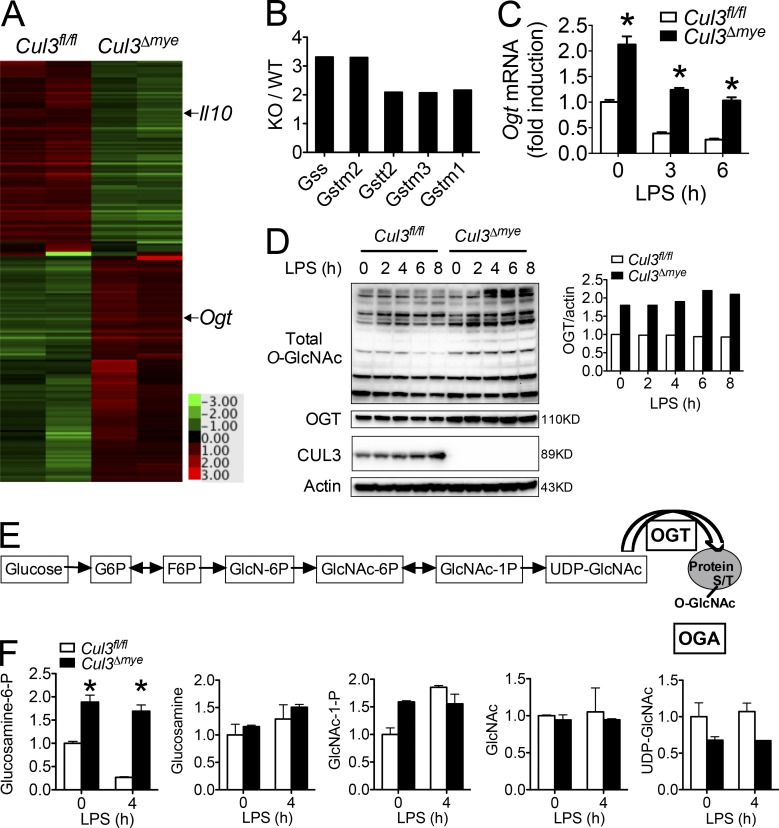
We next performed microarray analysis to explore the mechanism by which CUL3 mediates the STAT3–IL-10 signaling axis. After LPS stimulation, Cul3Δmye BMMs generated lower transcript levels of Il10 and several other STAT3 target genes, including Ahr (aryl hydrocarbon receptor), Klf4 (Kruppel-like factor 4), and Ccnd1 (cyclin D1; Fig. 3 A and Table S3; Yu et al., 2009). Several Nrf2 target genes, including Gss (glutathione synthetase), Gstm2 (GST Mu 2), Gstt2 (GST theta 2), Gstm3, and Gstm1 were expressed in Cul3Δmye BMMs at higher levels (Fig. 3 B), presumably because of elevated Nrf2 protein levels (Fig. S1 C). Interestingly, we observed higher expression of Ogt, which encodes a key enzyme for protein O-GlcNAcylation, in LPS-stimulated Cul3Δmye BMMs compared with similarly treated WT BMMs (Fig. 3 A and Table S3). RT-PCR analysis revealed that compared with WT BMMs, Cul3Δmye BMMs show a two- and threefold increase in Ogt transcript levels in naive status and upon LPS stimulation, respectively (Fig. 3 C). In line with increased Ogt transcript levels, Cul3Δmye BMMs contained markedly increased OGT protein levels (Fig. 3 D), as well as increased total O-GlcNAcylation (Fig. 3 D). It should be noted that despite reduced Ogt transcript levels, LPS stimulation up to 8 h failed to decrease OGT protein levels, indicating a prolonged half-life of OGT protein, as was recently reported (Yang et al., 2015a).
Figure 3.
CUL3 negatively regulates OGT expression and protein O-GlcNAcylation. (A and B) Gene profiling assay was performed with RNA samples extracted from Cul3fl/fl and Cul3Δmye BMMs stimulated with LPS for 4 h. A resultant heat map indicating Il10 and Ogt (A) and fold changes in Nrf2 target genes (B) are shown. (C and D) Cul3fl/fl and Cul3Δmye BMMs were left untreated or stimulated with 200 ng/ml LPS for the indicated periods. (C) Ogt transcript was measured with RT-PCR. (D) Immunoblotting (left) and densitometric analysis to quantify ratio of OGT to actin (right) in treated BMMs. (E and F) Cul3fl/fl and Cul3Δmye BMMs were left untreated or stimulated with 200 ng/ml LPS for 4 h. LC-MS/MS metabolomics analysis was performed to profile steady-state metabolites. A summary of HBP (E) and fold changes in HBP intermediate metabolites (F) are shown. In A, B, and F, the microarray and metabolomics experiment were performed once each, and the results in F are expressed as mean ± SD of duplicates. In C and D, the results are representative of three independent experiments and are expressed as mean ± SD. *, P < 0.05 versus controls.
To gain further insight into the effect of CUL3 on HBP activity, we performed targeted steady-state metabolomics profiling by mass spectrometry (MS; Yuan et al., 2012) in macrophages with or without LPS stimulation (Fig. 3 E). Compared with WT macrophages, we observed two- and fourfold increases in glucosamine-6-phosphate (GlcN-6P), the product of the committed step governing entry into HBP, in Cul3Δmye BMMs in naive status and upon LPS stimulation, respectively (Fig. 3 F and Table S4). These data suggest increased HBP activity in the absence of CUL3.
CUL3 inhibits STAT3 O-GlcNAcylation
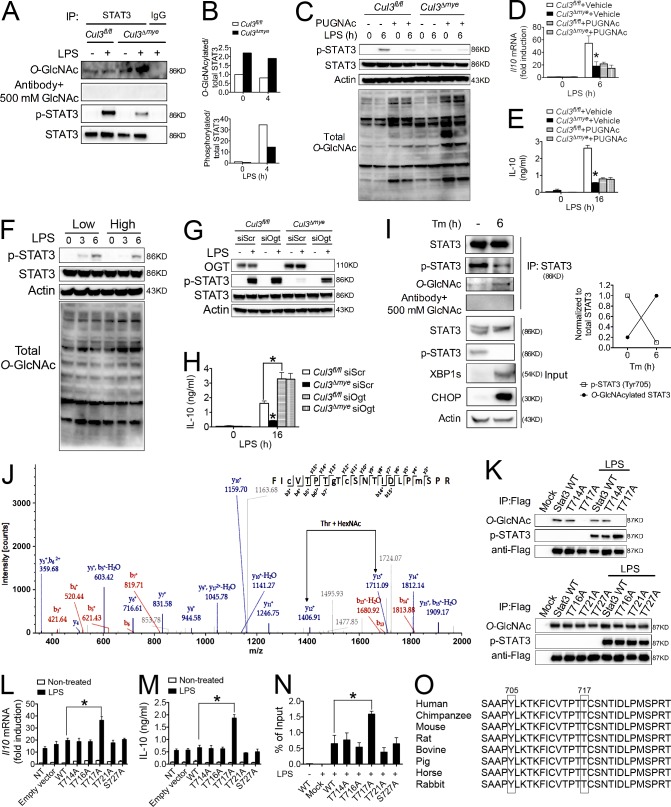
Previous studies suggested extensive cross-talk between O-GlcNAcylation and phosphorylation (Hart et al., 2011). The observation of defective STAT3 phosphorylation but increased total O-GlcNAcylation in Cul3Δmye macrophages prompted us to test the hypothesis that increased STAT3 O-GlcNAcylation inhibits its phosphorylation. We first measured STAT3 O-GlcNAcylation in Cul3fl/fl and Cul3Δmye macrophages by STAT3 immunoprecipitation, followed by immunoblotting with anti–O-GlcNAc antibody (because of the lack of specific antibody against O-GlcNAcylated STAT3). Cul3Δmye macrophages showed a twofold increase in O-GlcNAcylated STAT3 normalized to total STAT3 compared with WT cells (Fig. 4, A and B). The O-GlcNAc signal on STAT3 was abolished when anti–O-GlcNAc antibody was preincubated with 500 mM GlcNAc, indicating the specificity of the O-GlcNAc signal (Fig. 4 A). In contrast, LPS-induced STAT3 phosphorylation was dramatically attenuated in LPS-stimulated Cul3Δmye macrophages, suggesting a negative correlation between STAT3 O-GlcNAcylation and phosphorylation. We tested the relationship between STAT3 O-GlcNAcylation and phosphorylation using a pharmacological HBP activator. PUGNAc is a widely used inhibitor of O-GlcNAcase (OGA) that can potently enhance protein O-GlcNAcylation (Hart et al., 2011), and we confirmed its effect on promoting protein O-GlcNAcylation in macrophages (Fig. 4 C). We found that PUGNAc pretreatment dramatically attenuated LPS-induced STAT3 phosphorylation (Fig. 4 C) and IL-10 production (Fig. 4, D and E) in WT cells and abolished the differences between WT and Cul3Δmye macrophages. We next asked whether the inhibitory effect of STAT3 O-GlcNAcylation on its phosphorylation occurred in a physiological condition by lowering culture medium glucose level. Switching the glucose level from 25 to 5 mM in culture medium caused a decrease in total O-GlcNAcylation and an increase in STAT3 phosphorylation upon LPS stimulation (Fig. 4 F). These results indicate that glucose metabolism affects STAT3 phosphorylation through O-GlcNAc signaling.
Figure 4.
CUL3 inhibits STAT3 O-GlcNAcylation on T717. (A and B) Total STAT3 was immunoprecipitated (IP) from Cul3fl/fl and Cul3Δmye BMMs left untreated or stimulated with 200 ng/ml LPS for 4 h. Immunoblotting (A) and densitometric analysis to quantify ratio of either O-GlcNAcylated or phosphorylated STAT3 to total STAT3 (B) are shown. (C–E) Cul3fl/fl and Cul3Δmye BMMs were left untreated or stimulated with LPS for the indicated periods with or without pretreatment with 100 µM HBP activator PUGNAc. STAT3 phosphorylation (Y705) and total protein O-GlcNAcylation (C) and IL-10 transcript (D) and protein (E) were assayed by immunoblotting, RT-PCR, and ELISA, respectively. (F) STAT3 phosphorylation (Y705) was measured in WT BMMs left untreated or stimulated with LPS for 3 or 6 h in the presence of 5 or 25 mM glucose. (G and H) Cul3fl/fl and Cul3Δmye BMMs transfected with siScr or siOgt were left untreated or stimulated with LPS for 6 h. STAT3 phosphorylation (G) and IL-10 production (H) were assayed. (I and J) Total STAT3 was immunoprecipitated from liver tissue lysates of vehicle- or Tm-treated C57BL/6 mice. (I) Immunoblotting (left) and densitometric analysis (right) to quantify STAT3 O-GlcNAcylation and phosphorylation (Y705). (J) LC-MS/MS analysis was performed to identify T717 as a STAT3 O-GlcNAcylation site after Tm challenge. MS/MS spectrum of the 2+ ion at m/z 1,265.08667 corresponding to STAT3 peptide FIcVTPTgTcSNTIDLPmSPR. T177 is O-GlcNAcylated (gT). (K–N) A series of point mutations of STAT3 were generated with site-directed mutagenesis, as indicated. (K) Empty vector, FLAG-tagged WT, or mutant constructs of STAT3 were transfected into Cul3Δmye BMMs for 48 h, followed by LPS stimulation for 6 h. STAT3 O-GlcNAcylation was assayed as described in A. (L and M) IL-10 transcript (L) and protein (M) were measured by RT-PCR and ELISA, respectively. (N) ChIP assay was performed to measure the binding of WT or mutant constructs of STAT3 at the promoter region of Il10 gene. NT, not treated. (O) Cross-species sequence alignment of STAT3 revealed conserved O-GlcNAcylation site T717 and phosphorylation site Y705. In G, the LC-MS/MS experiment was performed once. In other experiments, the results are representative of three to five independent experiments and are expressed as mean ± SD. *, P < 0.05 versus controls.
Finally, we used siRNA-based Ogt gene knockdown to determine whether defective STAT3 phosphorylation and IL-10 production were dependent on elevated OGT expression in Cul3Δmye macrophages. Transfection with siOgt markedly rescued defective STAT3 phosphorylation (Fig. 4 G) and IL-10 production (Fig. 4 H) in Cul3Δmye macrophages compared with scrambled siRNA transfection. Collectively, these results indicate that CUL3 mediates STAT3 phosphorylation and IL-10 production through an O-GlcNAcylation–dependent mechanism.
STAT3 is O-GlcNAcylated on T717
We sought to identify the O-GlcNAcylation site or sites on STAT3 through MS analysis. Mouse livers were initially used as the model system because of their abundant STAT3 protein supply. A recent study suggested that the ER stress response promotes HBP activity and protein O-GlcNAcylation (Wang et al., 2014). We challenged C57BL/6 mice with vehicle or tunicamycin (Tm), a pharmacological inhibitor of protein glycosylation that is commonly used to induce ER stress (Ozcan et al., 2004). As expected, Tm challenge for 6 h caused the up-regulation of spliced X-box binding protein 1 (Xbp1s) and C/EBP homologous protein (CHOP), two ER stress target proteins (Fig. 4 I; Ozcan et al., 2004). Interestingly, Tm challenge dramatically enhanced STAT3 O-GlcNAcylation. In line with the negative correlation between STAT3 O-GlcNAcylation and phosphorylation observed in Cul3Δmye macrophages (Fig. 4 A), Tm challenge resulted in decreased STAT3 phosphorylation, a finding similar to that previously reported (Kimura et al., 2012). Therefore, mouse liver after ER stress induction serves as an ideal subject for proteomics analysis of STAT3 O-GlcNAcylation sites.
Immunoprecipitated STAT3 in total liver lysates prepared from vehicle- or Tm-treated mice was analyzed by liquid chromatography coupled to tandem MS (LC-MS/MS). A single peptide FICVTPTTCSNTIDLPMSPR (amino acid 710–729) was identified as carrying a HexNAc (+203.1 D) modification in liver sample from Tm-treated mouse, but not in that from vehicle-treated mouse (Fig. 4 J). Because of sufficient fragmentation by collision-induced dissociation (CID), the O-GlcNAc modification was localized to T717, whereas other potential O-GlcNAcylation sites (T714, T716, T721, and S727) in this peptide could not be confirmed. We also identified the phosphorylation of Y705 and S727, which was decreased in the Tm-treated mice (Fig. 4 J). These findings demonstrate a negative correlation between STAT3 O-GlcNAcylation and phosphorylation during ER stress. We further confirmed the aforementioned findings by analyzing Stat3 O-GlcNAcylation in mouse BMMs. In both nontreated and LPS-treated BMMs, the same peptide of Stat3 (amino acid 710–729), as observed in mouse liver Stat3, carried a HexNAc modification (Fig. S2). Therefore, the results from MS assay confirmed the biochemical analysis results of STAT3 O-GlcNAcylation (Fig. 4 A).
A series of mutant constructs of STAT3 were generated with those individual residues as potential O-GlcNAcylation sites mutated to alanine. To examine STAT3 O-GlcNAcylation sites, we transfected STAT3 WT or mutant constructs into Cul3Δmye macrophages, which represent a highly permissive environment for STAT3 O-GlcNAcylation (Fig. 4 A). STAT3 with T717A mutation showed a loss of O-GlcNAc signal, indicating that STAT3 is O-GlcNAcylated at T717 (Fig. 4 K). Macrophages transfected with STAT3 T717A mutant exhibited significantly increased STAT3 phosphorylation (Fig. 4 K) and IL-10 production at the transcript (Fig. 4 L) and protein (Fig. 4 M) levels upon LPS stimulation, compared with those transfected with WT STAT3. In agreement, ChIP assay revealed an increased binding of STAT3 T717A mutant to the Il10 promoter region, indicating an increased transcription activity in STAT3 upon the loss of O-GlcNAcylation (Fig. 4 N). Collectively, these data strongly indicate that STAT3 O-GlcNAcylation on T717 is an intrinsic inhibitory mechanism against its phosphorylation and transcriptional activity. Interestingly, STAT3 T717 and phosphorylation site Y705 are well conserved among metazoan species (Fig. 4 O), highlighting the cross-talk between O-GlcNAcylation on T717 and phosphorylation on Y705 as an evolutionarily conserved mechanism regulating STAT3 biological activity.
Previous studies suggest that OGT forms a functional protein complex with the serine/threonine phosphatase 1 (PP1; Wells et al., 2004). We therefore sought to determine whether PP1 is involved in the inhibitory effect of STAT3 O-GlcNAcylation on its phosphorylation. Pretreatment of cells with calyculin A, a widely used PP1 and PP2A inhibitor, increased LPS-induced Stat3 phosphorylation in both WT and Cul3Δmye cells (Fig. S3), which is consistent with previous study showing an antagonizing effect of protein phosphatase in STAT3 phosphorylation (Zgheib et al., 2012). However, decreased STAT3 phosphorylation was still observed in Cul3Δmye cells compared with WT cells in the presence of calyculin A, suggesting that PP1 or PP2A is not involved in the inhibitory effect of O-GlcNAcylation on phosphorylation of STAT3. Therefore, the mechanism whereby STAT3 O-GlcNAcylation inhibits its phosphorylation remains unidentified.
Elevated Nrf2 protein mediates increased Ogt transcription in Cul3Δmye macrophages
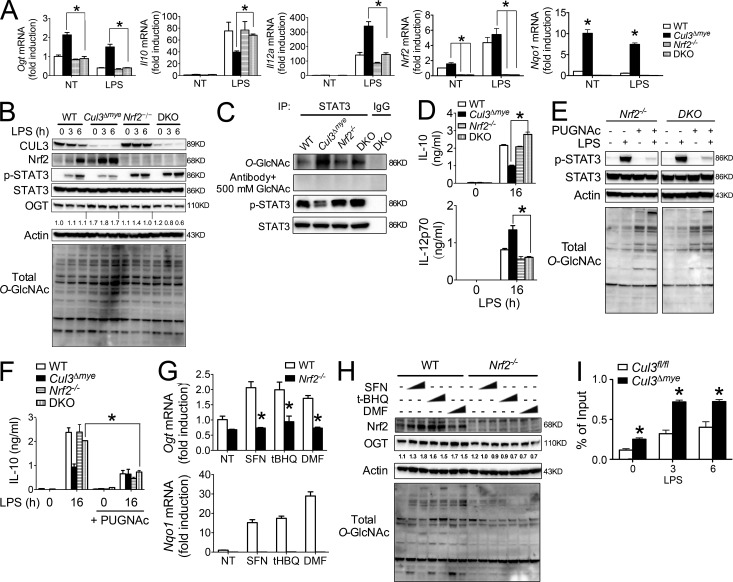
It is reasonable to hypothesize that certain CUL3-regulated transcriptional factors are responsible for elevated OGT levels in Cul3Δmye macrophages because the inhibitory effect of CUL3 on OGT occurs at the transcriptional level. Nrf2 has been firmly established as a target protein of CUL3-mediated protein ubiquitination and degradation (Emanuele et al., 2011), which has been confirmed in our hands (Fig. S1). We therefore tested whether elevated Nrf2 is an underlying mechanism for increased OGT transcription and decreased STAT3–IL-10 axis in Cul3Δmye macrophages by generating Cul3ΔmyeNrf2−/− double KO (DKO) mice. Although Nrf2−/− macrophages showed normal OGT expression compared with WT cells, deletion of Nrf2 abolished increased OGT expression at the transcript (Fig. 5 A) and protein (Fig. 5 B) levels, as well as elevated total O-GlcNAc signal, in Cul3Δmye macrophages. As a result, elevated STAT3 O-GlcNAcylation in Cul3Δmye macrophages was also abolished in DKO cells (Fig. 5 C). This result suggests that Nrf2 is not absolutely required for OGT expression, but elevated Nrf2 protein can increase OGT expression. Similar results were observed in autophagy-deficient macrophages showing increased MARCO (macrophage receptor with collagenous structure) expression caused by enhanced Nrf2 activation (Bonilla et al., 2013).
Figure 5.
Nrf2 promotes OGT expression in Cul3Δmye macrophages. (A–D) Cul3ΔmyeNrf2−/− DKO mice were generated. BMMs generated from mice with the indicated genotypes were left untreated or stimulated with 200 ng/ml LPS for the indicated periods. Transcript of Ogt, Il10, Il12a, Nrf2, and Nqo1 (A), STAT3 phosphorylation and total O-GlcNAc signal (B), STAT3 O-GlcNAcylation (C), and IL-10 and IL-12p70 proteins in the supernatants (D) were measured. NT, not treated. (E and F) BMMs of the indicated genotypes were left untreated or stimulated with LPS for 6 or 16 h in the absence or presence of 100 µM PUGNAc. STAT3 phosphorylation (Y705) and total protein O-GlcNAcylation (E) and IL-10 protein (F) were assayed by immunoblotting and ELISA, respectively. (G and H) WT and Nrf2−/− BMMs were treated with various Nrf2 activators for 16 h, including SFN (0.4 or 1 µM), tBHQ (2 or 5 µM), and DMF (10 or 40 µM). Transcript of Ogt and Nqo1 (G) and OGT, Nrf2, and total O-GlcNAc signal (H) were measured with RT-PCR and immunoblotting, respectively. (I) ChIP assay of the binding of Nrf2 at the promoter region of Ogt gene in Cul3fl/fl and Cul3Δmye BMMs left untreated or stimulated with LPS for 3 or 6 h. The results are representative of three independent experiments and are expressed as mean ± SD. *, P < 0.05 versus controls.
We took advantage of DKO macrophages to test the potential link between elevated Nrf2 and defective STAT3-IL-10 axis in Cul3Δmye macrophages. After LPS stimulation, defective STAT3 phosphorylation in Cul3Δmye macrophages was rescued in DKO macrophages (Fig. 5, B and C). In agreement, altered IL-10 and IL-12 production in Cul3Δmye macrophages were all normalized to the levels in WT cells by additional Nrf2 deletion (Fig. 5, A and D). These findings clearly state that elevated Nrf2 is responsible for increased OGT expression and STAT3 O-GlcNAcylation, as well as defective STAT3–IL-10 axis in Cul3Δmye macrophages. Normalization of OGT expression and total O-GlcNAc signal in DKO cells prompted us to test whether DKO cells were sensitive to PUGNAc-repressed STAT3–IL-10 axis, as observed in WT cells (Fig. 4 C). PUGNAc treatment caused a significant decrease in LPS-induced STAT3 phosphorylation (Fig. 5 E) and IL-10 production (Fig. 5 F) in Nrf2−/− or DKO macrophages, suggesting that Nrf2−/− or DKO macrophages show sensitivity to PUGNAc-inhibited STAT3 phosphorylation and IL-10 production similar to that of WT cells.
We further tested whether pharmacological activation of Nrf2 can increases OGT expression. Sulforaphane (SFN), tert-Butylhydroquinone (tBHQ), and dimethyl fumarate (DMF) are widely used Nrf2 activators (Jaramillo and Zhang, 2013). As expected, treatment of cells with the chemicals caused an increase in Nrf2 transcriptional activity (Fig. 5 G, bottom) and Nrf2 protein accumulation (Fig. 5 H). Interestingly, these chemicals also increased OGT at the transcript (Fig. 5 G, top) and protein (Fig. 5 H) levels, as well as total O-GlcNAc signals, in WT macrophages but not in Nrf2−/− macrophages, indicating Nrf2-mediated transcriptional activation of OGT and OGT-mediated protein O-GlcNAcylation. As an ultimate test of Nrf2-mediated OGT expression in Cul3Δmye macrophages, we performed ChIP analysis to examine whether Nrf2 directly enhances Ogt gene transcription by binding to the Ogt promoter region. Cul3Δmye macrophages showed enhanced Nrf2 binding at the Ogt promoter in the absence or presence of LPS stimulation (Fig. 5 F). Despite causing enhanced binding of Nrf2 at the Ogt promoter, short-term LPS challenge actually down-regulated Ogt in macrophages (Fig. 3 C), suggesting that additional inhibitory mechanisms exist to repress Ogt transcription. In sum, these results indicate that elevated Nrf2 caused by CUL3 deficiency promotes OGT expression and STAT3 O-GlcNAcylation, which subsequently affects STAT3 phosphorylation and IL-10 production (Fig. S2).
Cul3Δmye mice are more susceptible to experimental colitis and CAC
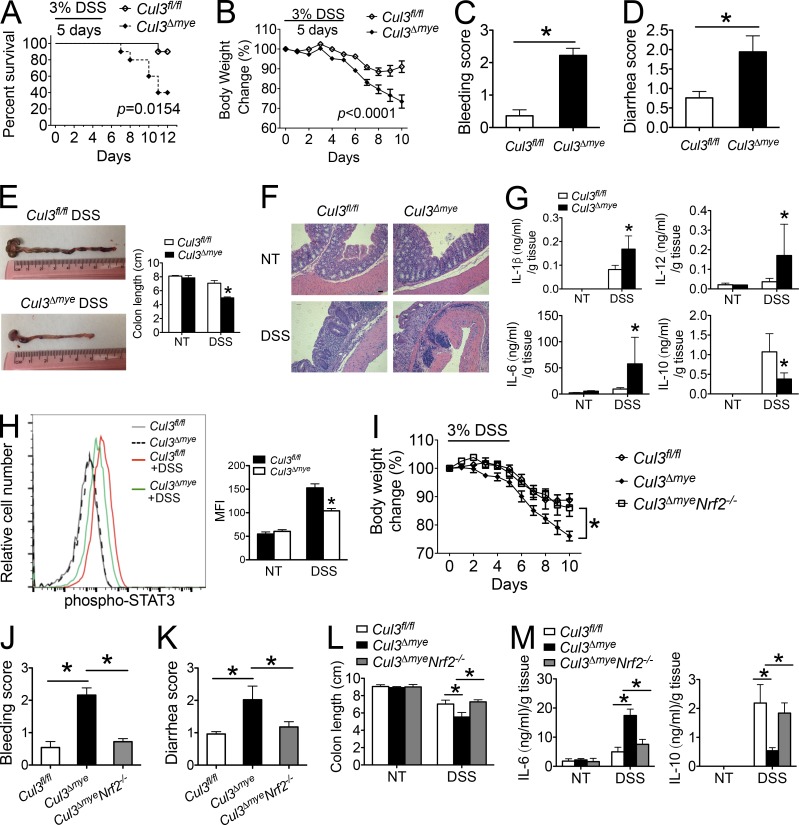
Numerous studies have established STAT3 and IL-10 as key mediators of intestinal inflammation and tumorigenesis (Yu et al., 2009; Grivennikov et al., 2010; O’Shea and Plenge, 2012; West et al., 2015). For example, myeloid deletion of Stat3 causes more severe intestinal inflammation during chemically induced colitis (Takeda et al., 1999). Il10-deficient mice develop spontaneous colitis (Moran et al., 2009). We studied the function of myeloid-derived CUL3 in intestinal inflammation during experimental colitis. Mice were challenged with 3% dextran sulfate sodium (DSS) for 5 d, and survival was assayed for 12 d (Wirtz et al., 2007). Cul3Δmye mice demonstrated significantly exacerbated mortality, weight loss, bleeding, diarrhea, and colon length shortening compared with WT mice (Fig. 6, A–E). Histological analysis revealed increased immune cell infiltration in DSS-treated Cul3Δmye mice (Fig. 6 F). To further evaluate the colonic inflammatory response induced by DSS treatment, colon explant cultures were established from untreated or DSS-treated mice. Production of inflammatory cytokines such as IL-1β, IL-6, and IL-12p70 were significantly enhanced in colon explant culture from Cul3Δmye mice, indicating exacerbated colonic inflammation (Fig. 6 G). In sharp contrast, IL-10 production in colon explant culture from Cul3Δmye mice was significantly decreased. In addition, STAT3 phosphorylation in colon macrophages was also significantly decreased in Cul3Δmye mice with colitis compared with those from WT mice with colitis, which was consistent with defective STAT3 phosphorylation in Cul3Δmye macrophages in vitro (Fig. 6 H).
Figure 6.
Cul3Δmye mice show increased susceptibility to chemically induced colitis. (A–H) Cul3fl/fl and Cul3Δmye mice were provided with drinking water containing 3% DSS for 5 d, followed by regular water for 5 d. (A–D) Kaplan–Meier plot of animal survival (A), body weight loss (B), bleeding score (C), and diarrhea score (D) in Cul3fl/fl (n = 35) and Cul3Δmye (n = 32) mice. (E) Representative colon photos (left) and quantification of colon length (right) of DSS-treated Cul3fl/fl and Cul3Δmye mice. (F) H&E staining of colons showing histopathology such as crypt loss and immune cell infiltration. Bar, 50 µm. (G) IL-1β, IL-6, IL-12p70, and IL-10 in colon explant cultures were measured by ELISA. (H) Histogram (left) and quantification of STAT3 phosphorylation (right) in colon macrophages (CD11b+F4/80+) was assayed by FACS analysis. NT, not treated. (I–M) Lethally irradiated C57BL/6 mice were reconstituted with BM cells from Cul3fl/fl, Cul3Δmye, or Cul3ΔmyeNrf2−/− donor mice (n = 10 per group), followed by DSS-colitis induction as described in A–H. (I–L) Body weight loss (I), bleeding score (J), diarrhea score (K), and quantification of colon length (L) were calculated. (M) IL-6 and IL-10 in colon explant cultures were measured by ELISA. The results shown are representative of three independent experiments and are expressed as mean ± SD. *, P < 0.05 versus controls.
We have shown that Nrf2 deletion rescued defective STAT3 phosphorylation and IL-10 production in Cul3Δmye macrophages (Fig. 5, A–D). To test whether Nrf2 deletion could abolish the DSS-colitis phenotype in Cul3Δmye mice, we generated BM chimeric mice in which lethally irradiated WT mice were reconstituted with BM cells isolated from Cul3fl/fl, Cul3Δmye, or Cul3ΔmyeNrf2−/− donor mice. Six weeks later, mice were subjected to the DSS-colitis model. We found that mice reconstituted with Cul3Δmye BM cells showed increased disease severity compared with the mice with WT BM cells, including exacerbated weight loss, bleeding, diarrhea, colon length shortening, inflammatory cytokine production, and decreased IL-10 production (Fig. 6, I–M). This phenotype was consistent with the increased disease severity in Cul3Δmye mice. In contrast, mice with Cul3ΔmyeNrf2−/− BM cells showed improved disease outcomes compared with mice with Cul3Δmye BM cells, indicating that Nrf2 mediates the in vivo function of myeloid-derived Cul3 in the DSS-colitis model.
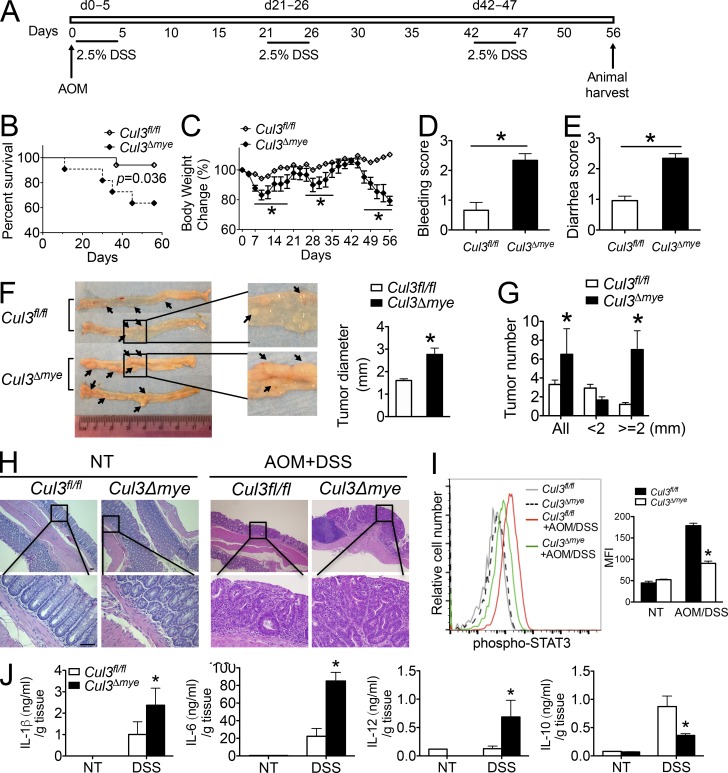
We next evaluated the function of myeloid-derived CUL3 in the development of an inflammation-driven CAC model induced with azoxymethane (AOM) plus three cycles of 2.5% DSS (Fig. 7 A; Wirtz et al., 2007). Over the entire course of the CAC model, AOM + DSS–treated Cul3Δmye mice showed a significant decrease in survival and exacerbated weight loss, bleeding, and diarrhea (Fig. 7, B–E). At the end of the CAC model, histological analyses revealed macroscopic colon polyps in the distal colons and rectums from the majority of AOM + DSS–treated mice. The maximal cross-sectional diameter (Fig. 7 F) and number (Fig. 7 G) of macroscopic polyps were significantly increased in Cul3Δmye mice compared with WT mice. Hematoxylin and eosin (H&E) staining of colon tissues revealed increased number and size of dysplastic lesions in AOM + DSS–treated Cul3Δmye mice, which was accompanied by enhanced immune cell infiltration (Fig. 7 H). In addition, STAT3 phosphorylation in colon macrophages was significantly decreased in Cul3Δmye mice (Fig. 7 I). In agreement, Cul3Δmye mice showed increased inflammatory cytokine production but decreased IL-10 production in colon tissue explant culture compared with WT mice (Fig. 7 J). In summary, these in vivo animal experiments suggest that myeloid deletion of CUL3 caused defective STAT3–IL-10 activation and exacerbated colonic inflammation and inflammation-driven tumors.
Figure 7.
Cul3Δmye mice are more susceptible to CAC model. (A) Schematic of AOM/DSS model of CAC. (B–E) Kaplan–Meier plot of animal survival (B), body weight loss (C), bleeding score (D), and diarrhea score (E) in Cul3fl/fl (n = 26) and Cul3Δmye (n = 22) mice after induction of CAC using AOM/DSS. (F) Macroscopic polyps (arrows) were identified in the distal and mid-colons isolated from Cul3fl/fl and Cul3Δmye mice that had completed the entire course of CAC model (left). The maximal cross-sectional diameter was quantified (right). (G) The number of macroscopic polyps was quantified. (H) H&E staining of colons showing histopathology such as crypt loss, immune cell infiltration, and polyp formation in Cul3fl/fl and Cul3Δmye mice that had completed the entire course of CAC model. Bars, 50 µm. (I) Histogram (left) and quantification of STAT3 phosphorylation (right) in colon macrophages (CD11b+F4/80+) assayed by FACS analysis (H). MFI, mean fluorescence intensity. (J) IL-1β, IL-6, IL-12p70, and IL-10 in colon explant cultures were measured by ELISA. The results shown are representative of two independent experiments and are expressed as mean ± SD. *, P < 0.05 versus controls.
Discussion
This study demonstrates an inhibitory effect of OGT-mediated STAT3 O-GlcNAcylation on macrophage STAT3 phosphorylation and IL-10 production (Fig. S4). STAT3 is a pleotropic transcription factor that mediates the transcription of numerous proteins involved in fundamental biological activities, including immune response, metabolism, cell cycle control, and development (Stark and Darnell, 2012). Upon stimulation with many cytokines, TLR agonists, and growth factors, STAT3 is activated by phosphorylation at T705 and S727. Recent studies have identified additional posttranslational modifications on STAT3 other than phosphorylation, including methylation on K140 and acetylation on K685 (Stark and Darnell, 2012). One previous study also reported O-GlcNAcylated STAT3 in 3T3-L1 adipocytes that was increased after insulin stimulation (Whelan et al., 2008). However, because of the lack of detailed information about the STAT3 O-GlcNAcylation site, it was difficult to test the causal relationship between STAT3 O-GlcNAcylation and its function. Our studies detected O-GlcNAcylated STAT3 in macrophages. We also identified T717 as the key O-GlcNAcylation site on STAT3 by unbiased LC-MS/MS analysis, followed by confirmatory studies of testing individual T→A or S→A mutants. Importantly, loss of O-GlcNAcylation by T717A mutation leads to enhanced STAT3 phosphorylation and transcriptional activity, which strongly argues that O-GlcNAcylation exerts an intrinsic inhibitory effect on STAT3 phosphorylation. These findings thus expand our current understanding of metabolic regulation of immune signaling pathways and highlight an essential role of immunometabolism in regulating immunity, inflammation, and inflammatory diseases.
Because of well-established cross-regulation between O-GlcNAcylation and phosphorylation (Hart et al., 2011), it is reasonable to predict that OGT-mediated O-GlcNAcylation plays a pivotal role in immune signaling and inflammation. Indeed, the studies of O-GlcNAc signaling in the immune system support the concept that O-GlcNAcylation enhances the activation of inflammatory immune signaling such as NF-κB (Yang et al., 2008). This notion is consistent with the observation that high glucose promotes inflammation (Yang et al., 2008). Several key molecules in TLR and NF-κB signaling pathways have been identified to be O-GlcNAcylated. For example, O-GlcNAcylation of IKKβ at S733 (Kawauchi et al., 2009), NF-κB p65 at T352 (Yang et al., 2008) and T305 (Allison et al., 2012), and c-Rel at S350 (Ramakrishnan et al., 2013) enhances their transcriptional activities. O-GlcNAcylation of TAB1 at S395, an important TLR signaling molecule, increased downstream NF-κB activation and cytokine production (Pathak et al., 2012). A recent study reported that mouse embryonic fibroblasts derived from a heterozygous O-GlcNAcase gene-deletion (Oga+/−) mouse strain contained elevated total O-GlcNAc signal, which led to increased promoter binding activity of p65 and cytokine production (Yang et al., 2015b). As a result, Oga+/− mice exhibited increased susceptibility to the DSS-colitis model. Our Cul3Δmye mice showed a phenotype similar to that of Oga+/− mice in the DSS-colitis model, indicating that O-GlcNAc signaling indeed promotes intestinal inflammation. Meanwhile, we found that elevated OGT protein and total O-GlcNAcylation caused defective activation of anti-inflammatory signaling pathway STAT3–IL-10 in Cul3Δmye macrophages. Therefore, it is reasonable to conclude that O-GlcNAc signaling affects inflammatory response through both pro- and anti-inflammatory signaling pathways. In addition to its classic enzymatic function, OGT also possesses enzymatic activity–independent function as a scaffold protein. For example, N-terminal truncated OGT, which lacks its enzymatic activity domain in C terminus, exhibited an inhibitory effect on gene transcription similar to that of full-length OGT in an overexpressing reporter system (Yang et al., 2002). This was caused by the requirement of N-terminal, but not C-terminal, portion of OGT binding to a critical transcriptional repressor, mSin3A (Yang et al., 2002). That study indicates that the enzymatic activity is not absolutely required for the function of OGT as transcriptional modulator. To fully understand enzymatic activity–dependent versus –independent effects of OGT on NF-κB signaling, a gene-deletion system is required that is further reconstituted with full-length versus N- or C-terminal OGT.
CUL3-formed E3 ubiquitin ligase has been indicated to target more than 100 proteins for ubiquitination and degradation including Nrf2, a master transcription factor for the antioxidative stress response (Genschik et al., 2013). We indeed observed that CUL3 deficiency results in increased Nrf2 protein accumulation, confirming a nonredundant function of CUL3 in maintaining Nrf2 protein at a low basal level. Although numerous previous studies have established that Nrf2 protects against oxidative stress–induced tissue damage through transcriptional activation of genes encoding a wide range of phase II detoxifying enzymes and antioxidants (Jaramillo and Zhang, 2013), some Nrf2-dependent effects could not be explained by its antioxidant function. Furthermore, the prolonged activation of Nrf2 has been shown to promote tissue damage, which again may be independent of its antioxidant function (Jaramillo and Zhang, 2013). For example, hyperactivated Nrf2 in Atg7-deficient liver has been shown to promote hepatocyte damage, but the underlying mechanism was not understood (Komatsu et al., 2010). It is highly possible that additional unknown Nrf2 target molecules and pathways exist to mediate pathological effect when Nrf2 is highly activated. Recent studies have shown that Nrf2 plays an essential role in mediating glucose and glutamine metabolic pathways (Mitsuishi et al., 2012; Singh et al., 2013). Using unbiased MS analysis, our study identified OGT as a novel transcriptional target of Nrf2, thus highlighting Nrf2-targeted metabolic pathways as important mechanisms modulating cell signaling. However, we also found that Nrf2 is not required for baseline OGT expression. Therefore, the molecular mechanism of OGT transcriptional control remains largely unknown. Interestingly, recent studies identified additional transcriptional factors other than Nrf2 as essential binding partners with CUL3, such as PLZF (promyelocytic leukemia zinc finger) and BCL6 (B cell lymphoma 6), in regulating NKT and B cell development (Mathew et al., 2012, 2014). Therefore, additional CUL3 target proteins other than Nrf2 may regulate baseline OGT transcription, which requires further investigation.
In addition to activation of the innate immune response, T cell activation and subtype skewing toward T helper 1 (Th1) and Th17 have also been indicated in the initiation and development of intestinal inflammation and inflammation-driven tumor (West et al., 2015). Genome-wide association studies of human IBD have identified a tight link between Th1- and Th17-related gene polymorphism and intestinal inflammation (Jostins et al., 2012). We detected significantly increased IL-6 and IL-12 production in colon tissues from Cul3Δmye mice in the DSS-colitis and CAC animal models, which was associated with exacerbated colonic inflammation and tumorigenesis. Because IL-6 and IL-12 are important inducing cytokines for Th17 and Th1 cell differentiation, respectively, further studies are warranted to explore the contribution of T cell–mediated inflammatory response to the disease severity affected by O-GlcNAc signaling.
In summary, our results provide an association between OGT-mediated glucose metabolism pathway and essential immune signaling in the innate immune system and expand our current understanding of metabolic regulation of the immune function and inflammation-associated diseases. Because of the complexity of established OGT target proteins, it is highly possible that OGT may affect macrophage function other than STAT3 and IL-10 production, requiring further investigation. Taking into account the other important roles of OGT in the adaptive immune system (Swamy et al., 2016) and tumorigenesis (Slawson and Hart, 2011), pharmacological modification of OGT activity presents a new therapeutic strategy for inflammation and inflammation-driven cancer.
Materials and methods
Mice
Cul3Δmye mice were generated by crossing Cul3fl/fl mice (McEvoy et al., 2007) with lysosome M-Cre mice. C57BL/6 mice, lysosome M-Cre mice, and Nrf2−/− mice were purchased from The Jackson Laboratory. Mice were treated in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and the Institutional Animal Care and Use Committee of the University of Nebraska Medical Center.
Reagents and antibodies
Ultrapure LPS and CpG oligonucleotide (ODN1826) were from InvivoGen. Recombinant murine IL-4, IL-6, and IFN-γ were from PeproTech. Pam3Cys was from EMC Microcollections. PUGNAc, Tm, GlcNAc, SFN, tBHQ, DMF, and AOM were from Sigma-Aldrich. DSS was from TDB Consultancy AB. Antibodies for immunoblotting included Phospho-Stat Antibody Sampler kit containing anti–p-STAT1 (Y701), anti–p-STAT3 (Y705), anti–p-STAT6 (Y641), anti–p-IKKα/β (S176/180), anti-IKKβ, anti–p-IκBα (S32), anti-IκBα, anti–p-p65 (S536), anti-p65, anti–p-ERK1/2 (T202/Y204), anti–p-JNK (T183/Y185), anti–p-p38 (T180/Y182), anti-OGT and anti–O-GlcNAc (Cell Signaling Technology), anti-STAT3, anti-SOCS3, anti-CHOP, HRP-conjugated anti–β-actin (Santa Cruz Biotechnology, Inc.), HRP-conjugated anti-FLAG (Sigma-Aldrich), anti-Xbp1s (BioLegend), and anti-CUL3 (BD). Anti-Nrf2 (Abcam) antibody was used for both immunoblotting and immunoprecipitation.
Cell culture and stimulation
BMMs were generated from Cul3fl/fl and Cul3Δmye mice in the presence of L-929 conditioned medium. Peritoneal macrophages were isolated by peritoneal lavage with 10 ml sterile DPBS containing 2% FBS. Macrophages were stimulated with LPS (200 ng/ml) for various time points as indicated in the figure legends. Cell culture supernatants were collected for ELISA. Cells were collected for RT-PCR or immunoblotting.
RT-PCR
Total RNA was extracted from in vitro cultured BMMs using Trisure (Bioline). cDNA synthesis was performed with Moloney murine leukemia virus reverse transcription (Invitrogen) at 38°C for 60 min. Real-time PCR was performed using SYBR Green PCR Master Mix in an Applied Biosystems StepOnePlus detection system. The fold difference in mRNA expression between treatment groups was determined by a standard ΔΔCt method. β-actin was analyzed as an internal control. The primer sequences of individual genes are listed in Table S1.
Immunoblotting
Electrophoresis of proteins was performed by using the NuPAGE system (Invitrogen) according to the manufacturer’s protocol. In brief, cultured BMMs were collected and lysed with RIPA buffer. Proteins were separated on a NuPAGE gel and transferred onto nitrocellulose membranes (Bio-Rad Laboratories). Appropriate primary antibodies and HRP-conjugated secondary antibodies were used, and proteins were detected using enhanced chemiluminescent (ECL) reagent (Thermo Fisher Scientific). Images were acquired with ChemiDoc MP System (Bio-Rad Laboratories).
ELISA
Cytokines generated by in vitro cultured BMMs and colon explants were quantified using the ELISA Set for mouse IL-1β, IL-6, IL-10, and IL-12p70 (BD) according to the manufacturer’s protocol.
Site-directed mutagenesis
pcDNA3 vector containing FLAG-tagged WT or S727A mouse Stat3 was a gift from A. Baldwin (University of North Carolina at Chapel Hill). Phusion Site-Directed mutagenesis kit (Thermo Fisher Scientific) was used to generate a series of STAT3 mutant constructs according to the manufacturer’s instructions. Primers used during mutagenesis PCR can be found in Table S2. The complete nucleotide sequences of all STAT3 mutants were double-checked by sequencing.
Cell transfection
BMMs were transfected with WT or mutant forms of STAT3 with X-tremeGENE HP DNA Transfection Reagent according to the manufacturer’s directions (Roche). The transfection efficiency was predetermined by FACS analysis of GFP by transfecting macrophages with pmaxGFP. For siRNA-based Ogt gene knockdown, BMMs were transfected with siScr or siOgt (Integrated DNA Technologies) using Lipofectamine RNAiMAX Transfection Reagent according to the manufacturer’s directions (Thermo Fisher Scientific).
ChIP assay
The ChIP procedure was performed using an assay kit (EMD Millipore) according to the manufacturer’s instructions. In brief, 3 × 106 BMMs were left untreated or stimulated with 200 ng/ml LPS for 3 or 6 h. DNA–protein structure was then cross-linked by 1% formaldehyde for 10 min at 37°C. Cells were collected and lysed in 400 µl SDS lysis buffer. The resulting lysate was sonicated to obtain DNA fragments ranging from 200 to 1,000 bp using a Branson Sonifier SLPe under the following condition: six times for periods of 15 s each. After centrifugation, the supernatant containing chromatin was diluted, and an aliquot (2% volume) was saved to indicate the input DNA in each sample. The remaining chromatin fractions were precleared with salmon sperm DNA/protein A agarose beads followed by immunoprecipitation with anti-STAT3 or anti-Nrf2 antibody overnight at 4°C with gentle rotation. Cross-linking was reversed for 4 h at 65°C and was followed by proteinase K digestion. DNA was purified by standard phenol/chloroform and ethanol precipitation and subjected to real-time PCR. Primers are as follows: for mouse Il10 promoter, forward 5′-CGACCAGTTCTTTAGCGCTTA-3′ and reverse 5′-TGTTCTTGGTCCCCCTTTTA-3′, which amplifies 172 bp from −180 to −9 relative to the transcription start site; for mouse Ogt promoter, forward 5′-GTACTTTAGCGCTTGCCAAA-3′ and reverse 5′-CATGACTGTAGAGAGCGCATG-3′, which amplifies 173 bp from −293 to −121 relative to the transcription start site.
Microarray
Total RNA was extracted from Cul3fl/fl and Cul3Δmye BMMs left untreated or stimulated with 200 ng/ml LPS for 4 h using RNeasy Mini kit (QIAGEN) according to the manufacturer’s instructions. The quality of total RNA was tested using Bioanalyzer 2000 (Agilent Technologies). RNA was digested with the RNase-Free DNase set and amplified into antisense RNA (aRNA). After one-color (Cy3) labeling, aRNA was loaded into a SurePrint G3 Mouse Gene Expression 8X60K Microarray (Agilent) and hybridized overnight in a hybridization oven. The hybridized array was washed and scanned, and data were extracted from the scanned image using Feature Extraction version 10.7 (Agilent Technologies).
Tm in vivo treatment
C57BL/6 mice were injected intraperitoneally with 1 mg/kg body weight Tm in sterile dextrose (150 mM) or vehicle control, as previously described (Ozcan et al., 2004). Liver tissues were collected 6 h after injection for biochemical analysis of STAT3 phosphorylation and O-GlcNAcylation and MS analysis of STAT3 O-GlcNAcylation sites.
STAT3 O-GlcNAcylation site mapping
Immunoprecipitated STAT3 from total liver lysates or BMMs was subjected to SDS-PAGE. The bands corresponding to STAT3 were excised, and the proteins were reduced, alkylated, and in-gel digested with trypsin. The peptides were extracted, lyophilized, resuspended in 2%ACN/0.1% formic acid, and analyzed by LC-MS/MS using a nanoAcquity (Waters Corp.) coupled to an LTQ Orbitrap Velos (Thermo Fisher Scientific). Samples were injected onto an in-house packed C18AQ (Michrom Biosciences) trap column (100 µm internal diameter × 2 cm, 5 µm particle size) and were washed with 1% ACN/0.1% formic acid for 20 min at 2 µl/min, then loaded onto an analytical column (C18AQ, 100 µm internal diameter × 22 cm, 5 µm particle size). The linear gradient for separation consisted of 5–36% mobile phase B over 120 min at a 250-nl/min flow rate, where mobile phase A was 0.1% formic acid in water and mobile phase B was 0.1% formic acid in ACN. The LTQ Orbitrap Velos was operated in data-dependent mode: the 10 most intense precursors were selected for subsequent collision-induced dissociation (CID) fragmentation. Resolution for the precursor scan (m/z 300–2,000) was set to 60,000 at m/z 400 with a target value of 106 ions. The MS/MS scans were acquired in the linear ion trap with a target value of 1,000. The normalized collision energy was set to 35% for CID. Precursors with unknown charge or a charge state of 1 and 4 or higher were excluded. Raw data files were processed using Proteome Discoverer version 2.0 (Thermo Fisher Scientific). Peak lists were searched against a Homo sapiens Uniprot database using Sequest. The following parameters were used to identify tryptic peptides for protein identification: 10 ppm precursor ion mass tolerance; 0.6 D product ion mass tolerance; up to two missed trypsin cleavage sites; carbamidomethylation of Cys set as a fixed modification; and hexNAc (+203.0794 D) of N/S/T, oxidation of M, and phosphorylation of S/T/Y set as variable modifications. The Percolator node was used to determine false discovery rates (FDRs), and a peptide FDR of 5% were used to filter all results.
Metabolomics
3 × 106 BMMs were left untreated or were stimulated with 200 ng/ml LPS for 4 h. Media from biological duplicates were fully aspirated, and 4 ml of 80% (vol/vol) methanol was added at dry ice temperatures. Cells were collected into conical tubes, and debris was removed through centrifugation at 2,000 g for 15 min. The resulting supernatant was evaporated using a refrigerated SpeedVac. Samples were resuspended using 20 µl HPLC-grade water for targeted LC-MS/MS. 10 µl was injected and analyzed using a 5500 QTRAP hybrid triple-quadrupole mass spectrometer (Sciex) coupled to a Prominence UFLC HPLC system (Shimadzu) via selected reaction monitoring (SRM) of a total of 261 endogenous water-soluble metabolites for steady-state analyses of samples. Some metabolites were targeted in both positive and negative ion mode for a total of 296 SRM transitions using positive/negative switching. ESI voltage was +4,900 V in positive ion mode and −4,500 V in negative ion mode. The dwell time was 4 ms per SRM transition, and the total cycle time was 1.89 s. Approximately 9–12 data points were acquired per detected metabolite. Samples were delivered to the MS via normal-phase chromatography using a 4.6-mm internal diameter × 10-cm Amide XBridge HILIC column (Waters Corp.) at 400 µl/min. Gradients were run starting from 85% buffer B (HPLC grade acetonitrile) to 42% B from 0 to 5 min; 42% B to 0% B from 5 to 16 min; 0% B held from 16 to 24 min; 0% B to 85% B from 24 to 25 min; and 85% B held for 7 min to re-equilibrate the column. Buffer A was comprised of 20 mM ammonium hydroxide/20 mM ammonium acetate, pH 9.0, in water/acetonitrile (95:5). Peak areas from the total ion current for each metabolite SRM transition were integrated using MultiQuant v2.1 software (Sciex). Data analysis was performed in Cluster3.0 and TreeViewer.
Colitis and CAC animal model
The induction of acute colitis by DSS (Wirtz et al., 2007) and CAC by AOM + DSS (Neufert et al., 2007) has been described previously. For the DSS-induced colitis model, Cul3fl/fl and Cul3Δmye mice were subjected to one cycle of 3% DSS exposure for 5 d. Mice were sacrificed, and inflammation assessments were made at day 7 after removal of DSS challenge. To evaluate tumorigenesis, mice received a single intraperitoneal injection (10 mg/kg body weight) of AOM followed by three cycles of DSS exposure. Mice were sacrificed, and tumor assessments were made 8 wk after AOM injection or when moribund. Colon lengths and histopathology were measured for each animal at the completion of each study. The bleeding score and diarrhea score were calculated as previously described (Allen et al., 2010; Salcedo et al., 2010).
BM chimera
Six-week old C57BL/6 mice were lethally irradiated (9.5 Gy). BM cells were prepared from femurs and tibiae of Cul3fl/fl, Cul3Δmye, or Cul3ΔmyeNrf2−/− donor mice. Irradiated recipient mice were reconstituted with 5 × 106 BM cells via the tail vein. After 6 wk, mice were subjected to one cycle of 3% DSS exposure for 5 d.
Statistics
Statistical analysis was performed with Prism 5.0 for Macintosh. All data are shown as mean ± SD. The mean values for biochemical data from each group were compared by Student’s t test. Comparisons between multiple time points were analyzed by repeated-measures ANOVA with Bonferroni post tests. Survival curves were generated by the product limit method of Kaplan and Meier, and significance was analyzed by log-rank test. In all tests, P values of less than 0.05 were considered statistically significant.
Online supplemental material
Fig. S1 shows the generation of myeloid conditional Cul3 gene deletion mice (Cul3Δmye). Fig. S2 shows the MS analysis of STAT3 O-GlcNAcylation in mouse BMMs. Fig. S3 shows no involvement of protein PP1 in the defective STAT3 phosphorylation in Cul3Δmye BMMs. Fig. S4 shows a schematic of CUL3-Nrf2 signaling-modulated OGT expression and STAT3 O-GlcNAcylation on STAT3 phosphorylation and IL-10 production in macrophages. Table S1 shows the list of RT-PCR primer sets for various mouse genes. Table S2 shows the list of primer sets used in the site-directed mutagenesis of STAT3. Table S3 shows the list of genes with increased and decreased expression levels in LPS-treated Cul3Δmye macrophages compared with similarly treated WT macrophages. Table S4 shows the list of metabolites in LPS-treated Cul3Δmye macrophages versus similarly treated WT macrophages.
Supplementary Material
Acknowledgments
We thank Hung-Ching Hsia from Dr. Albert Baldwin’s laboratory for STAT3 plasmids and Drs. Xiaochun Yu and Xiaoyong Yang for OGT plasmid. We thank Drs. Lee Graves and David Smalley from UNC Proteomics Core Facility for MS analysis, Dr. Yan Shi from the UNC Genomics Core Facility for microarray experiment, Dr. Joel Parker from the UNC Lineberger Cancer Center for microarray data analysis, and Min Yuan and Susanne Breitkopf for metabolomics experiments. We acknowledge Drs. Nat Moorman, Ben Major, Michael Emanuele, George Booz, and Lance Wells for helpful discussions.
This work was supported by National Institutes of Health grants K01DK098307 (to H. Wen), 5P01CA120964 (to J.M. Asara), and 5P30CA006516 (to J.M. Asara).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AOM
- azoxymethane
- BMM
- BM-derived macrophage
- CAC
- colitis-associated cancer
- ChIP
- chromatin immunoprecipitation
- CRL
- Cullin-RING (really interesting new gene) ligase
- DKO
- double KO
- DMF
- dimethyl fumarate
- DSS
- dextran sulfate sodium
- GlcNAc
- N-acetylglucosamine
- HBP
- hexosamine biosynthesis pathway
- IBD
- inflammatory bowel disease
- MS
- mass spectrometry
- O-GlcNAc
- O-linked β-GlcNAc
- OGT
- O-GlcNAc transferase
- SFN
- sulforaphane
- SRM
- selected reaction monitoring
- tBHQ
- tert-Butylhydroquinone
- Tm
- tunicamycin
References
- Allen I.C., TeKippe E.M., Woodford R.M., Uronis J.M., Holl E.K., Rogers A.B., Herfarth H.H., Jobin C., and Ting J.P.. 2010. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J. Exp. Med. 207:1045–1056. 10.1084/jem.20100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison D.F., Wamsley J.J., Kumar M., Li D., Gray L.G., Hart G.W., Jones D.R., and Mayo M.W.. 2012. Modification of RelA by O-linked N-acetylglucosamine links glucose metabolism to NF-κB acetylation and transcription. Proc. Natl. Acad. Sci. USA. 109:16888–16893. 10.1073/pnas.1208468109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., et al. Wellcome Trust Case Control Consortium . 2008. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 40:955–962. 10.1038/ng.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollrath J., Phesse T.J., von Burstin V.A., Putoczki T., Bennecke M., Bateman T., Nebelsiek T., Lundgren-May T., Canli O., Schwitalla S., et al. 2009. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 15:91–102. 10.1016/j.ccr.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Bonilla D.L., Bhattacharya A., Sha Y., Xu Y., Xiang Q., Kan A., Jagannath C., Komatsu M., and Eissa N.T.. 2013. Autophagy regulates phagocytosis by modulating the expression of scavenger receptors. Immunity. 39:537–547. 10.1016/j.immuni.2013.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. 2004. At the crossroads of inflammation and cancer. Cell. 118:671–674. 10.1016/j.cell.2004.09.005 [DOI] [PubMed] [Google Scholar]
- Emanuele M.J., Elia A.E., Xu Q., Thoma C.R., Izhar L., Leng Y., Guo A., Chen Y.N., Rush J., Hsu P.W., et al. 2011. Global identification of modular cullin-RING ligase substrates. Cell. 147:459–474. 10.1016/j.cell.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P., Sumara I., and Lechner E.. 2013. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): Cellular functions and disease implications. EMBO J. 32:2307–2320. 10.1038/emboj.2013.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S., Karin E., Terzic J., Mucida D., Yu G.Y., Vallabhapurapu S., Scheller J., Rose-John S., Cheroutre H., Eckmann L., and Karin M.. 2009. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 15:103–113. 10.1016/j.ccr.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S.I., Greten F.R., and Karin M.. 2010. Immunity, inflammation, and cancer. Cell. 140:883–899. 10.1016/j.cell.2010.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart G.W., Slawson C., Ramirez-Correa G., and Lagerlof O.. 2011. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80:825–858. 10.1146/annurev-biochem-060608-102511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo M.C., and Zhang D.D.. 2013. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 27:2179–2191. 10.1101/gad.225680.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha A.K., Huang S.C., Sergushichev A., Lampropoulou V., Ivanova Y., Loginicheva E., Chmielewski K., Stewart K.M., Ashall J., Everts B., et al. 2015. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 42:419–430. 10.1016/j.immuni.2015.02.005 [DOI] [PubMed] [Google Scholar]
- Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., et al. International IBD Genetics Consortium (IIBDGC) . 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 491:119–124. 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., and Greten F.R.. 2005. NF-kappaB: Linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 5:749–759. 10.1038/nri1703 [DOI] [PubMed] [Google Scholar]
- Kawauchi K., Araki K., Tobiume K., and Tanaka N.. 2009. Loss of p53 enhances catalytic activity of IKKβ through O-linked β-N-acetyl glucosamine modification. Proc. Natl. Acad. Sci. USA. 106:3431–3436. 10.1073/pnas.0813210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Yamada T., Matsumoto M., Kido Y., Hosooka T., Asahara S., Matsuda T., Ota T., Watanabe H., Sai Y., et al. 2012. Endoplasmic reticulum stress inhibits STAT3-dependent suppression of hepatic gluconeogenesis via dephosphorylation and deacetylation. Diabetes. 61:61–73. 10.2337/db10-1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., et al. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12:213–223. [DOI] [PubMed] [Google Scholar]
- Mathew R., Seiler M.P., Scanlon S.T., Mao A.P., Constantinides M.G., Bertozzi-Villa C., Singer J.D., and Bendelac A.. 2012. BTB-ZF factors recruit the E3 ligase cullin 3 to regulate lymphoid effector programs. Nature. 491:618–621. 10.1038/nature11548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R., Mao A.P., Chiang A.H., Bertozzi-Villa C., Bunker J.J., Scanlon S.T., McDonald B.D., Constantinides M.G., Hollister K., Singer J.D., et al. 2014. A negative feedback loop mediated by the Bcl6-cullin 3 complex limits Tfh cell differentiation. J. Exp. Med. 211:1137–1151. 10.1084/jem.20132267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy J.D., Kossatz U., Malek N., and Singer J.D.. 2007. Constitutive turnover of cyclin E by Cul3 maintains quiescence. Mol. Cell. Biol. 27:3651–3666. 10.1128/MCB.00720-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M., and Motohashi H.. 2012. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 22:66–79. 10.1016/j.ccr.2012.05.016 [DOI] [PubMed] [Google Scholar]
- Moran J.P., Walter J., Tannock G.W., Tonkonogy S.L., and Sartor R.B.. 2009. Bifidobacterium animalis causes extensive duodenitis and mild colonic inflammation in monoassociated interleukin-10-deficient mice. Inflamm. Bowel Dis. 15:1022–1031. 10.1002/ibd.20900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufert C., Becker C., and Neurath M.F.. 2007. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat. Protoc. 2:1998–2004. 10.1038/nprot.2007.279 [DOI] [PubMed] [Google Scholar]
- O’Neill L.A., and Hardie D.G.. 2013. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 493:346–355. 10.1038/nature11862 [DOI] [PubMed] [Google Scholar]
- O’Shea J.J., and Plenge R.. 2012. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity. 36:542–550. 10.1016/j.immuni.2012.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U., Cao Q., Yilmaz E., Lee A.H., Iwakoshi N.N., Ozdelen E., Tuncman G., Görgün C., Glimcher L.H., and Hotamisligil G.S.. 2004. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 306:457–461. 10.1126/science.1103160 [DOI] [PubMed] [Google Scholar]
- Pathak S., Borodkin V.S., Albarbarawi O., Campbell D.G., Ibrahim A., and van Aalten D.M.. 2012. O-GlcNAcylation of TAB1 modulates TAK1-mediated cytokine release. EMBO J. 31:1394–1404. 10.1038/emboj.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E.J., and Everts B.. 2015. Dendritic cell metabolism. Nat. Rev. Immunol. 15:18–29. 10.1038/nri3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E.L., Poffenberger M.C., Chang C.H., and Jones R.G.. 2013. Fueling immunity: Insights into metabolism and lymphocyte function. Science. 342:1242454 10.1126/science.1242454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan P., Clark P.M., Mason D.E., Peters E.C., Hsieh-Wilson L.C., and Baltimore D.. 2013. Activation of the transcriptional function of the NF-κB protein c-Rel by O-GlcNAc glycosylation. Sci. Signal. 6:ra75 10.1126/scisignal.2004097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H.B., Singh J.P., Li M.D., Wu J., and Yang X.. 2013. Cracking the O-GlcNAc code in metabolism. Trends Endocrinol. Metab. 24:301–309. 10.1016/j.tem.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo R., Worschech A., Cardone M., Jones Y., Gyulai Z., Dai R.M., Wang E., Ma W., Haines D., O’hUigin C., et al. 2010. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: Role of interleukin 18. J. Exp. Med. 207:1625–1636. 10.1084/jem.20100199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh M., and Trinchieri G.. 2011. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat. Rev. Immunol. 11:9–20. 10.1038/nri2891 [DOI] [PubMed] [Google Scholar]
- Singer J.D., Gurian-West M., Clurman B., and Roberts J.M.. 1999. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 13:2375–2387. 10.1101/gad.13.18.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Happel C., Manna S.K., Acquaah-Mensah G., Carrerero J., Kumar S., Nasipuri P., Krausz K.W., Wakabayashi N., Dewi R., et al. 2013. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J. Clin. Invest. 123:2921–2934. 10.1172/JCI66353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C., and Hart G.W.. 2011. O-GlcNAc signalling: Implications for cancer cell biology. Nat. Rev. Cancer. 11:678–684. 10.1038/nrc3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G.R., and Darnell J.E. Jr. 2012. The JAK-STAT pathway at twenty. Immunity. 36:503–514. 10.1016/j.immuni.2012.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamy M., Pathak S., Grzes K.M., Damerow S., Sinclair L.V., van Aalten D.M., and Cantrell D.A.. 2016. Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat. Immunol. 17:712–720. 10.1038/ni.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Clausen B.E., Kaisho T., Tsujimura T., Terada N., Förster I., and Akira S.. 1999. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 10:39–49. 10.1016/S1074-7613(00)80005-9 [DOI] [PubMed] [Google Scholar]
- Wang Z.V., Deng Y., Gao N., Pedrozo Z., Li D.L., Morales C.R., Criollo A., Luo X., Tan W., Jiang N., et al. 2014. Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell. 156:1179–1192. 10.1016/j.cell.2014.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells L., Kreppel L.K., Comer F.I., Wadzinski B.E., and Hart G.W.. 2004. O-GlcNAc transferase is in a functional complex with protein phosphatase 1 catalytic subunits. J. Biol. Chem. 279:38466–38470. 10.1074/jbc.M406481200 [DOI] [PubMed] [Google Scholar]
- West N.R., McCuaig S., Franchini F., and Powrie F.. 2015. Emerging cytokine networks in colorectal cancer. Nat. Rev. Immunol. 15:615–629. 10.1038/nri3896 [DOI] [PubMed] [Google Scholar]
- Whelan S.A., Lane M.D., and Hart G.W.. 2008. Regulation of the O-linked beta-N-acetylglucosamine transferase by insulin signaling. J. Biol. Chem. 283:21411–21417. 10.1074/jbc.M800677200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz S., Neufert C., Weigmann B., and Neurath M.F.. 2007. Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2:541–546. 10.1038/nprot.2007.41 [DOI] [PubMed] [Google Scholar]
- Yang W.H., Park S.Y., Nam H.W., Kim D.H., Kang J.G., Kang E.S., Kim Y.S., Lee H.C., Kim K.S., and Cho J.W.. 2008. NFκB activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc. Natl. Acad. Sci. USA. 105:17345–17350. 10.1073/pnas.0806198105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Zhang F., and Kudlow J.E.. 2002. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: Coupling protein O-GlcNAcylation to transcriptional repression. Cell. 110:69–80. 10.1016/S0092-8674(02)00810-3 [DOI] [PubMed] [Google Scholar]
- Yang Y.R., Kim D.H., Seo Y.K., Park D., Jang H.J., Choi S.Y., Lee Y.H., Lee G.H., Nakajima K., Taniguchi N., et al. 2015b Elevated O-GlcNAcylation promotes colonic inflammation and tumorigenesis by modulating NF-κB signaling. Oncotarget. 6:12529–12542. 10.18632/oncotarget.3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yin X., Yang H., and Xu Y.. 2015a Histone demethylase LSD2 acts as an E3 ubiquitin ligase and inhibits cancer cell growth through promoting proteasomal degradation of OGT. Mol. Cell. 58:47–59. 10.1016/j.molcel.2015.01.038 [DOI] [PubMed] [Google Scholar]
- Yu H., Pardoll D., and Jove R.. 2009. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer. 9:798–809. 10.1038/nrc2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Breitkopf S.B., Yang X., and Asara J.M.. 2012. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 7:872–881. 10.1038/nprot.2012.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgheib C., Zouein F.A., Chidiac R., Kurdi M., and Booz G.W.. 2012. Calyculin A reveals serine/threonine phosphatase protein phosphatase 1 as a regulatory nodal point in canonical signal transducer and activator of transcription 3 signaling of human microvascular endothelial cells. J. Interferon Cytokine Res. 32:87–94. 10.1089/jir.2011.0059 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.