Abstract
Inositol polyphosphate multikinase (IPMK, ipk2, Arg82, ArgRIII) is an inositide kinase with unusually flexible substrate specificity and the capacity to partake in many functional protein–protein interactions (PPIs). By merging these two activities, IPMK is able to execute gene regulatory functions that are very unique and only now beginning to be recognized. In this short review, we present a brief history of IPMK, describe the structural biology of the enzyme and highlight a few recent discoveries that have shed more light on the role IPMK plays in inositide metabolism, nuclear signalling and transcriptional regulation.
Keywords: IPMK, ipk2, ArgR, inositide, multikinase, transcription, gene expression
Introduction
Inositides represent a large family of secondary messengers that are essential in the regulation of various cellular processes. This group of molecules may be classified either as water-soluble inositol polyphosphates (IPs) or inositol lipids [(phosphatidylinositols or phosphoinositides (PIPs)]. These ubiquitous molecules are descendants of myo-inositol, the most widely-occurring stereoisomer of inositol. As a six-carbon cyclitol harbouring one axial (C-2) and five equatorial hydroxy groups, myo-inositol provides a rich template for single and combinatorial phosphorylation at six positions, generating 63 unique inositol phosphates [1]. This number is magnified as inositol polyphosphates are either further phosphorylated to yield inositol pyrophosphates or conjugated to lipids to yield the phosphoinositides.
Besides their involvement in phospholipid metabolism to maintain cell membrane integrity, inositides are key mediators of diverse cellular signalling pathways. Elucidation of the physiological roles played by both IPs and PIPs in the cell have continued to receive increasing attention since the first definitive assignment of inositol 1,4,5-trisphosphate (IP3) as the calcium-releasing factor generated upon binding to IP3 receptors [2]. The signalling function of IP3 arose from the activation of phospholipase C (PLC) to hydrolyse membrane-bound phosphatidylinositol 4,5-bisphosphate (PIP2) into diacylglycerol (DAG) and IP3, which in turn activate protein kinase C (PKC) and allosterically stimulates IP3 receptors to release Ca2+ from intracellular stores respectively. These findings ushered in a decade and more of research in inositide metabolism, ranging from identification of more phosphorylated derivatives of lipid inositols and inositol phosphates [3–6], identification of inositide receptors and binding domains, characterization of their signalling roles [7], which may or may not be independent of each other, as well as the nuclear processes that are regulated by inositide–receptor interactions, to name a few. The reader is referred to these excellent reviews on these various themes [1,8–13].
Further investigations of the biochemical roles of inositol phosphates and lipid inositols in the cell have also raised important questions regarding the functional significance of higher inositol phosphate biosynthesis and other metabolic processes. These studies revealed the presence of a family of inositol phosphate kinases that catalyse the phosphorylation of inositol and its phosphorylated derivatives [14–16]. Interestingly, some of these enzymes have activities that are specific to the substrate [17,18] and can therefore be named in a more straightforward manner: inositol (1,4,5) trisphosphate 3-kinase (IP3K) specifically phosphorylates inositol (1,4,5) trisphosphate [(I(1,4,5)P3] at the 3-hydroxy position of the inositol ring to yield inositol (1,3,4,5) tetrakisphosphate [(I(1,3,4,5)P4]. Other enzymes however adopt a promiscuous catalytic activity towards several inositol phosphates. Inositol polyphosphate multikinase (IPMK, Figure 1A) is the most catalytically flexible member of this family and it is this enigmatic enzyme that shall be the focus of this short review.
Figure 1. Crystal structure of yeast IPMK reveals positions of amino acids involved in IPMK substrate specificity and PPIs.
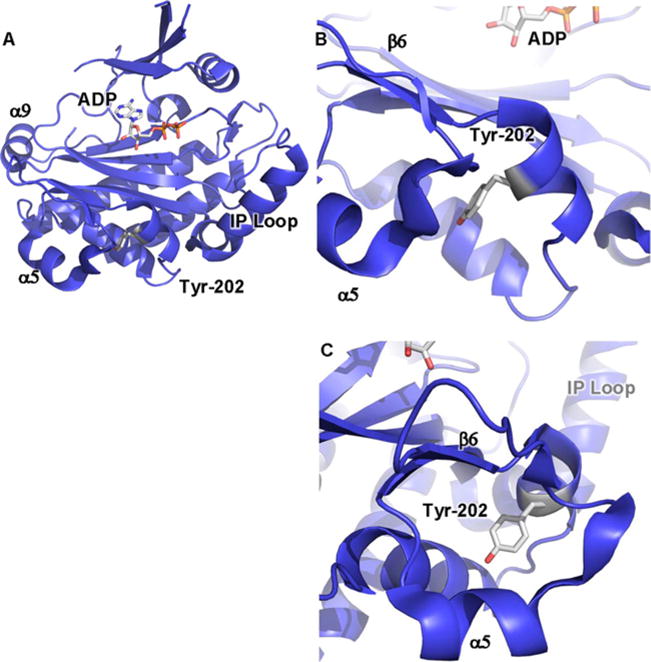
(A) Ribbon cartoon of the 2.0 Å crystal structure of S. cerevisiae IPMK [43] (PDB: 2IF8), one of only two IPMK structures solved to date, the other being the A. thaliana plant IPMK [30] (result not shown, PDB: 4FRF). The helix in the lower right labelled as ‘IP loop’ is the inositol binding helix, instilling much of the substrate specificity to the enzyme. ADP indicated as sticks. α9, α5 and β6 elements have been suggested to be a PPI surface. (B) Tyr174 of the mouse IPMK is phosphorylated in response to glucose and mediates IPMK interaction with AMPK, through an unknown mechanism. Tyr174 is conserved in human IPMK (Tyr191) and the depicted yeast IPMK (Tyr202), as indicated. (C) As in (B), rotated 90°.
IPMK is a versatile catalyst
IPMK assumed a number of aliases in the course of its history, perhaps, in part due to the variety of function it performs in the cell. It was first known as Arg82 (or ArgRIII) and was initially discovered as a transcription factor important in regulation of arginine metabolism in yeast [19–24]. However, the kinase activity of Arg82, detected as phosphorylation of IP3 at C-6 of the inositol ring, was first reported in pea extracts [25] and subsequently in budding yeast extracts [26]. Phosphorylation of IP3 to form inositol hexakisphosphate (IP6) via C-6, C-3 and C-2 kinase activities was also reported for this enzyme in Schizosaccharomyces pombe [27]. However, it was only in the late 1990s when independent work by Snyder and colleagues [28] and York and co-workers [29] showed that Arg82 displays dual C-3 and C-6 kinase activity that sequentially phosphorylates IP3 to IP4 to IP5 that this enzyme was pushed to the spotlight. Snyder and co-workers [28] aptly renamed the enzyme ‘inositol polyphosphate multikinase’ in honour of its catalytic flexibility. York and co-workers coined the yeast name ‘Ipk2’ for the same enzyme, but have now adopted the IPMK moniker as well [30]. Other investigators have showed that mammalian orthologues of IPMK catalyse pyrophosphorylation of IP5 to PPIP4 (diphosphorylated inositol phosphate) [31,32].
Besides being an inositol phosphate kinase, investigations showed that IPMK also possesses a nuclear phosphoinositide (phosphatidylinositol) kinase activity, catalysing phosphorylation of the phosphatidylinositol 4,5-bisphosphate (PIP2) to yield phosphatidyl 3,4,5-trisphosphate (PIP3) [33]. Interestingly, the related IP3-kinase shares the capacity to phosphorylate soluble inositol phosphate with IPMK, but phosphorylation of the lipid inositol PIP2 has only been observed with IPMK and not with IP3 3-kinase [10,34].
IPMK is a multi-functional regulatory protein
Whereas IPMK proves to be a flexible catalyst essential in inositide metabolism, recent studies show that the versatility of IPMK allows it to play critical roles in other nuclear functions such as transcriptional regulation, mRNA export and chromatin remodelling, among others [35–46]. The following is by no means an exhaustive list, but we present here some examples of the roles of IPMK in metabolic signalling networks.
The characterization of IPMK as an inositol phosphate kinase was preceded with nearly 20-year history as a transcriptional regulator of arginine metabolism [19–24,47]. IPMK is believed to be an important facilitator in the assembling of the transcriptional complex ArgR/Mcm1 that regulates the repression of arginine anabolic genes and the induction of catabolic genes in response to changes in the nitrogen sources in the cellular environment [48]. It remained controversial whether the kinase activity of IPMK is involved in its transcriptional role; however, it now seems clear that IPMK can function in transcriptional regulation simultaneously in both kinase-dependent [35] and kinase-independent [36] roles. The specifics of the kinase-dependence or independence of an isolated system are less important than the overall observations that IPMK can act both dependently and independently of its kinase activity.
The kinase-independent roles of IPMK have been highlighted with several recent discoveries of IPMK acting as an adaptor protein, stabilizing protein complexes that are of the highest biomedical importance. Kim et al. [37,38] identified IPMK as a novel cofactor of mTOR (mammalian target of rapamycin), a serine/threonine protein kinase important in the regulation of processes including cell growth, survival and proliferation, protein synthesis and transcription [39]. It exists as two distinct protein complexes, mTORC1 and mTORC2, each with different affinity toward a separate set of proteins. The mammalian IPMK genes contain N-terminal 60 residues that mediate direct binding of IPMK to mTORC1 and regulate mTORC1 signalling responses to amino acids [37,38].
IPMK has also been demonstrated to function in the physiological regulation of AMPK (AMP-activated protein kinase) [40–42], an enzyme that maintains cellular homoeostasis by triggering catabolic energy and antagonizing ATP-consuming anabolic pathways. Their findings show that IPMK is an AMPK-binding protein whose association with AMPK is controlled by glucose levels. Glucose induces phosphorylation of mouse IPMK at Tyr174 (yeast Tyr202 and human Tyr191), an amino acid that resides close to the catalytic core of IPMK (Figures 1B and 1C). Phosphorylation at IPMK Tyr174 enhances IPMK–AMPK interaction and AMPK signalling responses to glucose; however it remains unclear how phosphorylation of this tyrosine residue facilitates the IPMK–AMPK interaction, based on the yeast IPMK crystal structure [43].
Steroidogenic factor-1 (SF-1), a nuclear receptor that regulates genes essential for sexual development and production of steroids, has been shown to be an important target of IPMK [35,44]. IPMK interacts with the SF-1 protein and directly phosphorylates PIP2 bound to the SF-1 protein to yield PIP3 [35]. The crystal structures of SF-1 bound to PIP2 (Figure 2A, PDB: 4QK4) and PIP3 (Figure 2B, PDB: 4QJR) demonstrate high solvent accessibility of the inositide headgroup [44,45]. The interaction of IPMK with SF-1/PIP2 [35,46] raises interesting structural questions that may shed light on the importance of phosphoinositides in nuclear events such as transcription, as well as the molecular mechanisms by which IPMK can regulate lipid signalling within the nucleus.
Figure 2. Co-crystal structures of SF-1/PIP2 and SF-1/PIP3 demonstrate high solvent accessibility of the inositide headgroups.
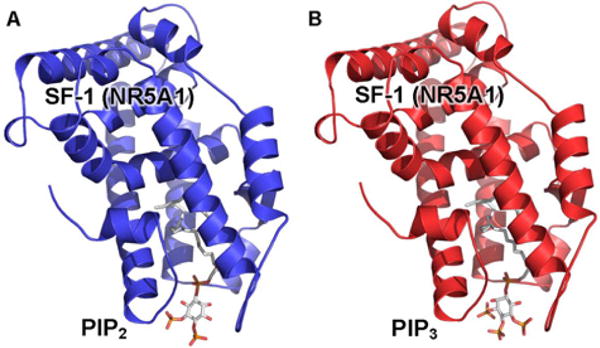
(A) Ribbon cartoon of the 2.8 Å co-crystal structure of the ligand-binding domain (LBD) of the nuclear receptor SF-1 bound to PIP2 (PDB: 4QK4), PIP2 indicated as sticks, revealing the solvent accessibility of the PIP2 headgroup. (B) Ribbon carton of the 2.4 Å crystal structure of the SF-1 LBD bound to PIP3 (PDB: 4QJR), PIP3 indicated as sticks. In both structures, the transcriptional co-activator peptide has been removed for clarity.
IPMK crystallography and catalysis
A glaring hole in our knowledge of IPMK is any structural information of the human orthologue, particularly given recent studies linking IPMK to intestinal carcinoids [49] and Huntington’s disease [50], both in studies from human patients. The only IPMK crystal structures that have been solved to date are the Saccharomyces cerevisiae (yeast) [43] and Arabidopsis thaliana (plant) [30] IPMKs. Despite low sequence homology (only approximately 17%) [30], these two orthologues exhibit high structural homology within their catalytic domains to the IP3-kinase family and both adopt the ATP-grasp fold [30,43]. Structural comparisons between the human IP3-kinase (PDB: 1W2D) [51], plant IPMK (PDB: 4FRF) [30] and yeast IPMK (PDB: 2IF8) [43] have shown how IPMKs achieve their hallmark substrate flexibility on several inositol phosphate species. This flexibility is contrasted with the high substrate specificity of IP3-kinases, which fall into the same family of ATP-grasp inositol kinases. In all these structures, it is the inositol phosphate-binding loop (IP loop, Figure 1A) that accounts for substrate selectivity by interacting with phosphorylated inositol species through the amino acid sequence on the ATP-facing surface of the IP loop helix.
The IP loop is the major structural mechanism that allows inositol kinases to achieve substrate specificity. Indeed, in an attempt to instil phosphatidylinositol lipid kinase activity in the human and rat IP3-kinases [34,51], we observed that removal of the IP loop from IP3-kinase produces a kinase with little specificity if any, as it will auto-phosphorylate IP3-kinase protein itself, as well as other proteins and phosphoinositides in the reaction (result not shown). More directed, modulatory experiments attempting to shift IPMK specificity have shown that the substrate specificity of the plant IPMK can be engineered by rational point mutations in the IP loop, dramatically reducing the plant IPMK 3-kinase activity, while not disturbing the 6-kinase activity [30]. Importantly, this uniquely engineered hypomorphic mutant of the plant IPMK can rescue the lethal phenotype of ipmk null Drosophila melanogaster flies [30], which may suggest that the 6-kinase activity is more important to very basic cellular or developmental IPMK functions, whereas the 3-kinase activity may participate more in fine-tuning cellular signalling. Regardless, the rational mutations afforded by the plant and yeast IPMK crystal structures have been invaluable in assessing how IPMK executes its catalytic functions.
IPMK as a kinase-independent adapter
Where crystallography has provided far less information is in how IPMK acts through catalytically independent mechanisms as an adapter protein in several transcriptional and signalling complexes, in addition to its promiscuous phosphorylated inositol and phosphatidylinositol kinase activities discussed above. Several recent studies have shown that the presence of IPMK stabilizes some very important protein signalling complexes, in a manner independent of the kinase activity of IPMK [37,38,52–55]. The laboratories performing these studies have also had the foresight to begin characterization of which structural domains in IPMK are important for those protein–protein interactions (PPIs). mTOR binds to IPMK via the N-terminal 60 amino acids of IPMK [37,38]. Serum response factor (SRF) requires the IPMK kinase domain itself for interaction (IPMK amino acids 93–182) [52], similar to what is required for the transcription factor and tumour suppressor p53 to interact with IPMK (IPMK amino acids 125–184) [53–55]. It is interesting that the catalytic domain of IPMK would mediate PPIs that do not require the IPMK kinase activity for regulation of the biological output from those complexes. A question that awaits exploration is if those important PPIs inhibit IPMK from executing IPMK kinase-dependent cellular functions in general inositide metabolism and how that may feedback into these signalling systems.
Although IPMK structures gave little insight into kinase-independent functions, the yeast IPMK crystal structure contains several helices (α9 and α5) and a β-sheet (β6) within the C-terminal domain that had been suspected to act as part of a PPI domain [43]. Primary structure analysis revealed that the human and yeast IPMKs contain large inserts immediately following the α9 helix, but that the sequences of those inserts are not conserved between IPMK orthologues in different species, are not ordered in any crystal structures to date and have no identified catalytic activity [30,43]. It remains to be seen if these disordered insert regions in the current IPMK crystal structures can become ordered if IPMK is complexed with proteins that interact with IPMK. Thus, it remains quite an open question as to what function the species-divergent and crystallographically disordered N- and C-terminal regions of IPMK may play in IPMK biology and which co-crystal structures may induce order in these regions?
Future trends in IPMK discovery
The discovery of direct IPMK phosphorylation of PIP2 bound to the nuclear receptor SF-1 [35,44], as well as the more recent discovery that IPMK effects the RNA-export activity of ALY through generation of nuclear PIP3 [56], suggests that IPMK may have an expansive, novel function specific to cellular nuclei, the phosphorylation of protein–inositide complexes. This function would unify IPMK catalytic flexibility with its ability to bind proteins, the inositide-binding proteins perhaps allowing the chemical inositide substrate to be optimally positioned for IPMK phosphorylation. Indeed, the enzyme kinetic parameters describing IPMK activity on PIP2 bound in micelles compared with PIP2 bound by the nuclear receptor SF-1 demonstrates that IPMK phosphorylates PIP2 in micelles with higher apparent velocity than PIP2 in SF-1, but with a ‘worse’ apparent KM (Figure 3) [35]. Those data are consistent with a model whereby IPMK PPIs with SF-1 enhance the KM, but slow down the enzyme when compared with PIP2 in micelles (perhaps due to slower substrate off rate).
Figure 3. Enzyme kinetic parameters of human IPMK activity on PIP2 in SF-1 or micelles.
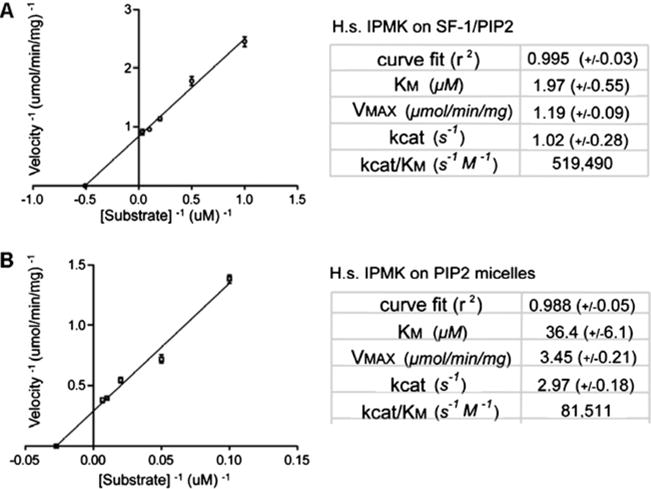
(A) Kinetic parameters of human IPMK activity on PIP2 bound by the nuclear receptor SF-1 [35]. (B) Kinetic parameters of human IPMK activity on PIP2 in micelles [35]. Although SF-1 slows the enzyme down, SF-1 improves the apparent KM, consistent with a PPI between IPMK and SF-1 facilitating IPMK phosphorylation of PIP2 bound by SF-1.
The details of the molecular interface between IPMK, PIP2 and SF-1 that enhance the overall activity of IPMK while still allowing substrate release will reveal how systems like this work in the nucleus and perhaps why they were selected for evolutionary reasons [46]. Further, the identification of novel protein–inositide complex substrates of IPMK will broaden the scope of IPMK influence in the nucleus, perhaps explaining why nuclear inositides are ubiquitous in all eukaryotic species examined to date, from yeast to plants to humans. The multiple roles IPMK plays in such central processes highlights the importance of this protein to eukaryotic cell physiology (Figure 4). Given that structure-based mutations introduced into the plant IPMK were able to decouple two of the most important enzyme activities of IPMK [30], a better understanding of IPMK structure and function will open the door for specific pharmacological targeting of these important IPMK activities.
Figure 4. IPMK has many cell physiological roles.
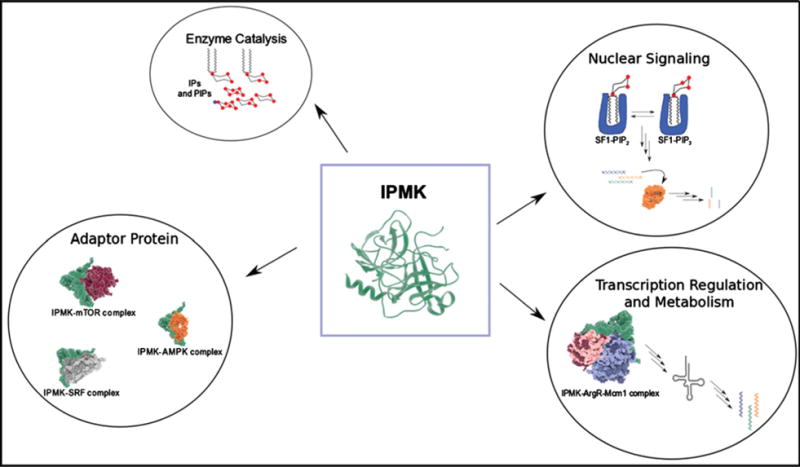
IPMK has well-established roles in inositol phosphate metabolism, in nuclear signalling through protein phosphoinositide complexes such as SF-1/PIP2, as a transcriptional co-regulator and in the cytoplasm as an adapter protein mediating several metabolic responses. The central role of IPMK in such a broad array of cellular processes highlights its importance to all eukaryotic cells.
Acknowledgments
Funding
This work was supported by the National Cancer Institute through grant 1K01CA172957 to R.D.B.
Abbreviations
- AMPK
AMP-activated protein kinase
- IP loop
inositol phosphate-binding loop
- IPMK
inositol polyphosphate multikinase
- mTOR
mammalian target of rapamycin
- PIP2
phosphatidylinositol 4,5-bisphosphate
- PIP3
phosphatidylinositol 3,4,5-trisphosphate
- PPI
protein–protein interaction
- PPIP4
diphosphorylated inositol phosphate
- SF-1
steroidogenic factor-1
References
- 1.York JD. Regulation of nuclear processes by inositol polyphosphates. Biochim Biophys Acta. 2006;1761:552–559. doi: 10.1016/j.bbalip.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 2.Streb H, Irvine RF, Berridge MJ, Schulz I. Release of calcium(2+) from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1,4,5-triphosphate. Nature. 1983;306:67–69. doi: 10.1038/306067a0. [DOI] [PubMed] [Google Scholar]
- 3.Batty IR, Nahorski SR, Irvine RF. Rapid formation of inositol 1,3,4,5-tetrakisphospate following muscarinic receptor stimulation of rat cerebral cortical slices. Biochem J. 1985;232:211–215. doi: 10.1042/bj2320211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irvine RF, Letcher AJ, Lander DJ, Downes CP. Inositol trisphosphates in carbachol-stimulated rat parotid glands. Biochem J. 1984;223:237–243. doi: 10.1042/bj2230237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitman M, Downes CP, Keeler M, Keller T, Cantley L. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature. 1988;332:644–646. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- 6.Traynor-Kaplan AE, Harris AL, Thompson BL, Taylor P, Sklar LA. An inositol tetrakisphosphate-containing phospholipid in activated neutrophils. Nature. 1988;334:353–356. doi: 10.1038/334353a0. [DOI] [PubMed] [Google Scholar]
- 7.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 8.Shears SB. How versatile are inositol phosphate kinases? Biochem J. 2004;377:265–280. doi: 10.1042/BJ20031428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seeds Andrew M, York John D. Inositol polyphosphate kinases: regulators of nuclear function. Biochem, Soc Symp. 2007:183–197. doi: 10.1042/BSS0740183. [DOI] [PubMed] [Google Scholar]
- 10.Resnick AC, Saiardi A. Inositol polyphosphate multikinase: metabolic architect of nuclear inositides. Front Biosci. 2008;13:856–866. doi: 10.2741/2726. [DOI] [PubMed] [Google Scholar]
- 11.Stevenson-Paulik J, Phillippy BQ. Inositol polyphosphates and kinases. Plant Cell Monogr. 2010;16:161–174. [Google Scholar]
- 12.Chakraborty A, Kim S, Snyder SH. Inositol pyrophosphates as mammalian cell signals. Sci Signal. 2011;4:re1, 11. doi: 10.1126/scisignal.2001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SJ, Lee HW, Baek JH, Cho YH, Kang HG, Jeong JS, Song J, Park HS, Chun KH. Activation of nuclear PTEN by inhibition of Notch signaling induces G2/M cell cycle arrest in gastric cancer. Oncogene. 2015 doi: 10.1038/onc.2015.80. [DOI] [PubMed] [Google Scholar]
- 14.Heslop JP, Irvine RF, Tashjian AH, Jr, Berridge MJ. Inositol tetrakis- and pentakisphosphates in GH4 cells. J Exp Biol. 1985;119:395–401. doi: 10.1242/jeb.119.1.395. [DOI] [PubMed] [Google Scholar]
- 15.Stephens LR. Preparation and separation of inositol tetrakisphosphates and inositol pentakisphosphates and the establishment of enantiomeric configurations by the use of L-iditol dehydrogenase. Methods Inositide Res. 1990:9–30. [Google Scholar]
- 16.Stephens L, Radenberg T, Thiel U, Vogel G, Khoo KH, Dell A, Jackson TR, Hawkins PT, Mayr GW. The detection, purification, structural characterization, and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s) J Biol Chem. 1993;268:4009–4015. [PubMed] [Google Scholar]
- 17.Irvine RF, Letcher AJ, Heslop JP, Berridge MJ. The inositol tris/tetrakisphosphate pathway - demonstration of ins(1,4,5)P3 3-kinase activity in animal tissues. Nature. 1986;320:631–634. doi: 10.1038/320631a0. [DOI] [PubMed] [Google Scholar]
- 18.Takazawa K, Perret J, Dumont JE, Erneux C. Molecular cloning and expression of a human brain inositol 1,4,5-trisphosphate 3-kinase. Biochem Biophys Res Commun. 1991;174:529–535. doi: 10.1016/0006-291x(91)91449-m. [DOI] [PubMed] [Google Scholar]
- 19.Bercy J, Dubois E, Messenguy F. Regulation of arginine metabolism in Saccharomyces cerevisiae: expression of the three ARGR regulatory genes and cellular localization of their products. Gene. 1987;55:277–285. doi: 10.1016/0378-1119(87)90287-3. [DOI] [PubMed] [Google Scholar]
- 20.Dubois E, Bercy J, Messenguy F. Characterization of two genes, ARGRI and ARGRIII required for specific regulation of arginine metabolism in yeast. Mol Gen Genet. 1987;207:142–148. doi: 10.1007/BF00331501. [DOI] [PubMed] [Google Scholar]
- 21.Messenguy F, Dubois E, Boonchird C. Determination of the DNA-binding sequences of ARGR proteins to arginine anabolic and catabolic promoters. Mol Cell Biol. 1991;11:2852–2863. doi: 10.1128/mcb.11.5.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu H, Dubois E, Broen P, Messenguy F. Functional analysis of ARGRI and ARGRIII regulatory proteins involved in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol Gen Genet. 1990;222:192–200. doi: 10.1007/BF00633817. [DOI] [PubMed] [Google Scholar]
- 23.Amar N, Messenguy F, El Bakkoury M, Dubois E. ArgRII, a component of the ArgR-Mcm1 complex involved in the control of arginine metabolism in Saccharomyces cerevisiae, is the sensor of arginine. Mol Cell Biol. 2000;20:2087–2097. doi: 10.1128/mcb.20.6.2087-2097.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubois E, Messenguy F. Pleiotropic function of ArgRIIIp (Arg82p), one of the regulators of arginine metabolism in Saccharomyces cerevisiae. Role in expression of cell-type-specific genes. Mol Gen Genet. 1994;243:315–324. doi: 10.1007/BF00301067. [DOI] [PubMed] [Google Scholar]
- 25.Chattaway JA, Drobak BK, Watkins PAC, Dawson AP, Letcher AJ, Stephens LR, Irvine RF. An inositol 1,4,5-trisphosphate-6-kinase activity in pea roots. Planta. 1992;187:542–545. doi: 10.1007/BF00199975. [DOI] [PubMed] [Google Scholar]
- 26.Estevez F, Pulford D, Stark MJR, Carter AN, Downes CP. Inositol trisphosphate metabolism in Saccharomyces cerevisiae: identification, purification and properties of inositol 1,4,5-trisphosphate 6-kinase. Biochem J. 1994;302:709–716. doi: 10.1042/bj3020709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ongusaha PP, Hughes PJ, Davey J, Michell RH. Inositol hexakisphosphate in Schizosaccharomyces pombe: synthesis from Ins(1,4,5)P3 and osmotic regulation. Biochem J. 1998;335(Pt 3):671–679. doi: 10.1042/bj3350671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- 29.Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 30.Endo-Streeter S, Tsui MK, Odom AR, Block J, York JD. Structural studies and protein engineering of inositol phosphate multikinase. J Biol Chem. 2012;287:35360–35369. doi: 10.1074/jbc.M112.365031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saiardi A, Nagata E, Luo HR, Sawa A, Luo X, Snowman AM, Snyder SH. Mammalian inositol polyphosphate multikinase synthesizes inositol 1,4,5-trisphosphate and an inositol pyrophosphate. Proc Natl Acad Sci USA. 2001;98:2306–2311. doi: 10.1073/pnas.041614598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang T, Caffrey JJ, Shears SB. The transcriptional regulator, Arg82, is a hybrid kinase with both monophosphoinositol and diphosphoinositol polyphosphate synthase activity. FEBS Lett. 2001;494:208–212. doi: 10.1016/s0014-5793(01)02351-1. [DOI] [PubMed] [Google Scholar]
- 33.Resnick AC, Snowman AM, Kang BN, Hurt KJ, Snyder SH, Saiardi A. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc Natl Acad Sci USA. 2005;102:12783–12788. doi: 10.1073/pnas.0506184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller GJ, Hurley JH. Crystal structure of the catalytic core of inositol 1,4,5-trisphosphate 3-kinase. Mol Cell. 2004;15:703–11. doi: 10.1016/j.molcel.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Blind RD, Suzawa M, Ingraham HA. Direct modification and activation of a nuclear receptor-PIP2 complex by the inositol lipid kinase IPMK. Sci Signal. 2012;5:ra44. doi: 10.1126/scisignal.2003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bosch D, Saiardi A. Arginine transcriptional response does not require inositol phosphate synthesis. J Biol Chem. 2012;287:38347–38355. doi: 10.1074/jbc.M112.384255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S, Snyder SH. Nutrient amino acids signal to mTOR via inositol polyphosphate multikinase. Cell Cycle. 2011;10:1708–1710. doi: 10.4161/cc.10.11.15559. [DOI] [PubMed] [Google Scholar]
- 38.Kim SY, Kim SWF, Maag D, Maxwell MJ, Resnick AC, Juluri KR, Chakraborty A, Koldobskiy MA, Cha SH, Barrow R, et al. Amino acid signaling to mTOR mediated by inositol polyphosphate multikinase. Cell Metab. 2011;13:215–221. doi: 10.1016/j.cmet.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bang S, Kim S, Dailey MJ, Chen Y, Moran TH, Snyder SH, Kim SF. AMP-activated protein kinase is physiologically regulated by inositol polyphosphate multikinase. Proc Natl Acad Sci USA. 2012;109:616–620. doi: 10.1073/pnas.1119751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bang S, Chen Y, Ahima RS, Kim SF. Convergence of IPMK and LKB1-AMPK signaling pathways on metformin action. Mol Endocrinol. 2014;28:1186–1193. doi: 10.1210/me.2014-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dailey MJ, Kim S. Inositol polyphosphate multikinase: an emerging player for the central action of AMP-activated protein kinase. Biochem Biophys Res Commun. 2012;421:1–3. doi: 10.1016/j.bbrc.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmes W, Jogl G. Crystal structure of inositol phosphate multikinase 2 and implications for substrate specificity. J Biol Chem. 2006;281:38109–38116. doi: 10.1074/jbc.M606883200. [DOI] [PubMed] [Google Scholar]
- 44.Blind RD, Sablin EP, Kuchenbecker KM, Chiu HJ, Deacon AM, Das D, Fletterick RJ, Ingraham HA. The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1. Proc Natl Acad Sci USA. 2014;111:15054–15059. doi: 10.1073/pnas.1416740111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sablin EP, Blind RD, Krylova IN, Ingraham JG, Cai F, Williams JD, Fletterick RJ, Ingraham HA. Structure of SF-1 bound by different phospholipids: evidence for regulatory ligands. Mol Endocrinol. 2009;23:25–34. doi: 10.1210/me.2007-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blind RD. Disentangling biological signaling networks by dynamic coupling of signaling lipids to modifying enzymes. Adv Biol Regul. 2014;54:25–38. doi: 10.1016/j.jbior.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu R, Paul BD, Smith DR, Tyagi R, Rao F, Khan AB, Blech DJ, Vandiver MS, Harraz MM, Guha P, et al. Inositol polyphosphate multikinase is a transcriptional coactivator required for immediate early gene induction. Proc Natl Acad Sci USA. 2013;110:16181–16186. doi: 10.1073/pnas.1315551110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dubois E, Dewaste V, Erneux C, Messenguy F. Inositol polyphosphate kinase activity of Arg82/ArgRIII is not required for the regulation of the arginine metabolism in yeast. FEBS Lett. 2000;486:300–304. doi: 10.1016/s0014-5793(00)02318-8. [DOI] [PubMed] [Google Scholar]
- 49.Sei Y, Zhao X, Forbes J, Szymczak S, Li Q, Trivedi A, Voellinger M, Joy G, Feng J, Whatley M, et al. A hereditary form of small intestinal carcinoid associated with a germline mutation in inositol polyphosphate multikinase. Gastroenterology. 2015;149:67–78. doi: 10.1053/j.gastro.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahmed I, Sbodio JI, Harraz MM, Tyagi R, Grima JC, Albacarys LK, Hubbi ME, Xu R, Kim S, Paul BD, Snyder SH. Huntington’s disease: neural dysfunction linked to inositol polyphosphate multikinase. Proc Natl Acad Sci USA. 2015 doi: 10.1073/pnas.1511810112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.González B, Schell MJ, Letcher AJ, Veprintsev DB, Irvine RF, Williams RL. Structure of a human inositol 1,4,5-trisphosphate 3-kinase: substrate binding reveals why it is not a phosphoinositide 3-kinase. Mol Cell. 2004;15:689–701. doi: 10.1016/j.molcel.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 52.Kim E, Tyagi R, Lee JY, Park J, Kim YR, Beon J, Chen PY, Cha JY, Snyder SH, Kim S. Inositol polyphosphate multikinase is a coactivator for serum response factor-dependent induction of immediate early genes. Proc Natl Acad Sci USA. 2013;110:19938–19943. doi: 10.1073/pnas.1320171110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu R, Sen N, Paul BD, Snowman AM, Rao F, Vandiver MS, Xu J, Snyder SH. Inositol polyphosphate multikinase is a coactivator of p53-mediated transcription and cell death. Sci Signal. 2013;6:ra22. doi: 10.1126/scisignal.2003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu R, Snyder SH. Gene transcription by p53 requires inositol polyphosphate multikinase as a co-activator. Cell Cycle. 2013;12:1819–1820. doi: 10.4161/cc.25119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li AG, Piluso LG, Cai X, Wei G, Sellers WR, Liu X. Mechanistic insights into maintenance of high p53 acetylation by PTEN. Mol Cell. 2006;23:575–587. doi: 10.1016/j.molcel.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 56.Wickramasinghe VO, Savill JM, Chavali S, Jonsdottir AB, Rajendra E, Gruner T, Laskey RA, Babu MM, Venkitaraman AR. Human inositol polyphosphate multikinase regulates transcript-selective nuclear mRNA export to preserve genome integrity. Mol Cell. 2013;51:737–750. doi: 10.1016/j.molcel.2013.08.031. [DOI] [PubMed] [Google Scholar]


