Abstract
Scope
The anticancer agent sulforaphane (SFN) acts via multiple mechanisms to modulate gene expression, including the induction of nuclear factor (erythroid-derived 2)-like 2 (Nrf2)-dependent signaling and the inhibition of histone deacetylase activity. Transcriptomics studies were performed in SFN-treated human colon cancer cells and in non-transformed colonic epithelial cells in order to pursue new mechanistic leads.
Methods and results
RNA-sequencing corroborated the expected changes in cancer-related pathways after SFN treatment. In addition to NAD(P)H quinone dehydrogenase 1 (NQO1) and other well-known Nrf2-dependent targets, SFN strongly induced the expression of Loc344887. This non-coding RNA was confirmed as a novel functional pseudogene for NmrA-like redox sensor 1 (NMRAL1), and was given the name NmrA-like redox sensor 2 pseudogene (NMRAL2P). Chromatin immunoprecipitation experiments corroborated the presence of Nrf2 interactions on the NMRAL2P genomic region, and interestingly, NMRAL2P also served as a co-regulator of NQO1 in human colon cancer cells. Silencing of NMRAL2P via CRISPR/Cas9 genome-editing protected against SFN-mediated inhibition of cancer cell growth, colony formation, and migration.
Conclusion
NMRAL2P is the first functional pseudogene to be identified both as a direct transcriptional target of Nrf2, and as a downstream regulator of Nrf2-dependent NQO1 induction. Further studies are warranted on NMRAL2P–Nrf2 crosstalk and the associated mechanisms of gene regulation.
Keywords: Colorectal cancer, CRISPR/Cas9, Loc344887, noncoding RNA
1 Introduction
Sulforaphane (SFN) is a dietary agent that exerts anticancer effects against various malignancies, including colorectal cancer [1, 2]. Chemopreventive outcomes of SFN have been attributed to multiple mechanisms [3–10]. SFN regulates antioxidant activity and the detoxification of carcinogens through induction of the nuclear factor (erythroid-derived 2)-like 2 (Nrf2) signaling pathway [11, 12]. Under normal conditions, Nrf2 is sequestered in the cytosol by protein partner Kelch-like ECH-associated protein 1 (Keap1). Upon treatment with SFN, Nrf2 dissociates from Keap1, translocates to the nucleus, and dimerizes with small musculoaponeurotic fibrosarcoma (MAF) proteins on the antioxidant response element (ARE) sequences of target genes [10, 13]. Target genes activated by Nrf2 include NAD(P)H quinone dehydrogenase 1 (NQO1), heme oxygenase 1 (HMOX1), and various glutathione S-transferases (GSTs). The response of these genes is influenced by nuclear/cytoplasmic trafficking of Nrf2, and by post-translational modifications that affect its interactions with small MAF proteins, chromatin remodeling factors, histone deacetylase (HDAC) enzymes, and other transcriptional regulators [14–18].
Among the epigenetic mechanisms implicated in human colon and prostate cancer cells, SFN has been shown to act via HDAC inhibition/turnover and changes in DNA methylation [19–22]. Acetylation of histone and non-histone proteins was linked to de-repression of tumor suppressor genes and the activation of apoptotic and G2/M cell arrest pathways, most notably in cancer cells as compared with the corresponding non-transformed cell lines [21–24].
Dietary isothiocyanates also alter the expression of various noncoding RNAs (ncRNAs), including microRNAs (miRNAs)[25] such as mir-155, mir-23b, and mir-27b in colonic epithelial cells [26], mir-155 in macrophages [27], miR-200c in bladder cancer cells [28], mir-21 in glioblastoma [29], and let-7 family members in pancreatic ductal adenocarcinoma cells [30]. Other ncRNAs, such as long noncoding RNAs (lncRNAs) and pseudogenes, also have been identified with roles in gene regulation, genome stability, cancer cell survival, and drug resistance [31–37]. There is a general lack of information on how these various ncRNAs might be impacted by diet and lifestyle factors.
While performing transcriptomics studies in SFN-treated colon cancer cells and in non-transformed colonic epithelial cells, we identified a ncRNA, Loc344887, that was directly regulated by Nrf2, and that served as a coactivator for NQO1. Localized on chromosome 3q27.2 and sharing 62% homology with the protein-coding gene NmrA-like redox sensor 1 (NMRAL1) on chromosome 16p13.3, the novel functional pseudogene was assigned the name NMRAL2P, and was pursued in mechanistic studies of cell viability, colony formation, and cell migration.
2 Materials and methods
2.1 Cell culture and treatments
Cell lines were obtained from ATCC and grown at 37°C in 5% CO2 with 1% penicillin/streptomycin. Human colon cancer cell lines HCT116 and HT29 were maintained in McCoy’s 5A media (Invitrogen) supplemented with 10% heat inactivated fetal bovine serum (FBS), whereas Caco2 cells and CCD841 non-transformed colonic epithelial cells were maintained in EMEM supplemented with 20% FBS. Treatments were performed when cells were ~70% confluent. Unless indicated otherwise, cells were incubated with 15 µM SFN or with the corresponding volume of dimethylsulfoxide (DMSO) vehicle. Allyl isothiocyanate (AITC), 6-methylsulfinylhexyl isothiocyanate (6-SFN), 9-methylsulfinylnonyl isothiocyanate (9-SFN), Oltipraz, tert-butyl hydroquinone (TBHQ), trichostatin A (TSA), suberoylanilide hydroxamic acid (SAHA), valproic acid (VPA), and sodium butyrate were used in some experiments. Concentrations were based on pre-determined IC50 values and prior reports [21, 22, 38].
2.2 RNA isolation and sequencing (RNA-seq)
RNA was isolated using the miRNeasy kit (Qiagen), according to the manufacturer’s protocol. For RNA-seq, RNA was isolated with Trizol (Life Technologies) and was purified and processed as reported [39]. In some experiments, the PARIS kit (ThermoFisher) was used to isolate nuclear and cytosolic RNA. Enrichment of nuclear RNA was confirmed by reference to metastasis associated lung adenocarcinoma transcript 1 (MALAT1). For a comprehensive list of the primers used, see Supplementary Table 1.
RNA-seq data quality was examined using Fastqc, and low quality reads (>50% bases with Q<30) were filtered out. Bowtie2 with default parameters was used to map reads to hg19 reference genome. Uniquely mapped genes were used to calculate the RPKM for each gene, and DESeq2 (R package) was used to identify significant differentially expressed genes with a threshold of fdr<0.05 and fold change >4. GOstats (R package) was used to perform KEGG pathway enrichment analysis, with a threshold of p<0.05. TCGA data for NMRAL2P were downloaded from the colon adenocarcinoma (COAD) dataset. Nrf2 target genes were selected from the ChIP-seq data of Chorley et al. [40], whereas Wnt signaling genes were identified from the gene list generated by Yu et al. [41].
2.3 Quantitative PCR (qPCR)
SuperScript III First-Strand Synthesis Master Mix (Invitrogen) was used on 1 µg of RNA to synthesize cDNA. qPCR was performed using SYBR Green I dye (Roche), cDNA, and gene-specific primers. Assays were run in a Light Cycler 96 or 480 (Roche) and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the 2−ΔΔCT method. Each experiment was repeated at least twice, and data were normalized to vehicle controls.
2.4 CRISPR-Cas9 genome editing
A small guide RNA (sgRNA NMRAL2P [upstream1], Supplementary Table 1), specific to the promoter region of NMRAL2P, was designed by CRISPR Design (crispr.mit.edu). Restriction cloning was used to insert the sgRNA into plasmid pSPCas9(BB)-2A–GFP (PX458), kindly provided by Dr. Feng Zhang (Addgene plasmid #48138) [42]. In brief, oligos were annealed by temperature ramp-down, phosphorylated, and ligated into the BbsI site of PX458 which contained sgRNA expression, Cas9 protein, and eGFP selection marker. PX458 plasmid (2.5 µg) was transfected into cells in a 6-well dish for 24 h. Green fluorescing cells were sorted individually into 96-well plates on a BD Biosciences FACSFusion Cell Sorter. Primers flanking exon 1 of NMRAL2P (see Supplementary Table 1) were used to screen genomic DNA of individual colonies, and PCR products were confirmed by sequencing.
2.5 siRNA transfection
Gene specific siRNAs (Sigma-Aldrich) or a Universal Control were transfected into cells using RNAiMax transfection reagent (Invitrogen), according to the manufacturer’s protocol. Unless stated otherwise, siRNA incubations were for 24–48 h. For siRNA primer sequences, see Supplementary Table 1.
2.6 MTT assays
Each treatment was performed in triplicate on NMRAL2P knockout cells, or the corresponding vector controls, plated at 1 × 104 cells per well. MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was added at 500 µg/ml and incubated at 37°C for 2 h. Resulting formazan dye was solubilized in DMSO and absorbance was measured at 562 nm (OD562).
2.7 Soft agar colony formation assays
Six-well plates were pre-coated with 0.6% agarose Type III-A (Sigma). Cells were mixed with 0.4% top agar and added to pre-coated plates at 3 × 104 cells/ well. After solidifying, 2 ml of liquid media containing SFN or DMSO was added to each well. Each treatment was performed in triplicate. Cells were incubated at 37°C for 2 weeks, and stained with 0.5% crystal violet in 6% formaldehyde. Two independent experiments were performed as biological replicates.
2.8 Transwell assays
To the upper chamber of a transwell insert was added 3 ×104 cells in serum-free media (Costar #3422), and serum-containing media without cells was added to the bottom chamber. After 24 h, inserts were fixed with 6% formaldehyde, the top of the membrane was swabbed, and then stained with 0.2% crystal violet. Membranes were washed and mounted onto glass slides. The number of cells that migrated through the insert was counted for ten 10X fields per treatment, for three wells per treatment. The experiment was repeated three times.
2.9 Immunoblotting
Cells were suspended in IP lysis buffer and lysed by freeze-thawing. Immunoblotting used the methodology described previously[21, 22], with primary antibodies to Keap1 (Cell Signaling #4617), Nrf2 (Cell Signaling #12721), NQO1(Cell Signaling #3187), and β-Actin (Sigma #A1978).
2.10 Chromatin immunoprecipitation (ChIP)
The ChIP-IT Express Sonication kit (Active Motif) was used according to the manufacturer’s protocol. Cells in 150 mm dishes were formaldehyde cross-linked, harvested, and then sonicated in a Bioruptor using 10-s intervals. Chromatin was immunoprecipitated with Nfr2 (Cell Signaling #3187) or Mafk antibodies (Abcam #ab50322) and pulled-down with Protein G magnetic beads (Active Motif). The DNA was reverse cross-linked and purified via the chromatin IP DNA purification kit (Active Motif). qPCR was performed using primers that flanked the AREs of HMOX1 and NQO1. Three putative AREs on the NMRAL2P locus were found by sequence analysis using the consensus sequence from Chorley et al [40]. For the primer sequences, see Supplementary Table 1.
3 Results
3.1 NMRAL2P is highly upregulated in SFN-treated colon cancer cells
RNA-seq was performed in human HCT116 colon cancer cells and CCD841 non-transformed colonic epithelial cells treated with vehicle or 15 µM SFN for 6 h, in triplicate. Principal component analysis confirmed that the two colonic epithelial cell lines had significantly different endogenous gene expression profiles, which became even more marked after SFN treatment (Supplementary Figure 1A). Approximately 50% of ~12,000 differentially expressed genes (DEGs) were upregulated and 50% were downregulated in HCT116 cells compared to CCD841 cells (Supplementary Figure. 1B). These DEGs likely reflect “cancer vs. non-cancer” differences, as well as genetic variation between the two cell lines. In HCT116 cells, 4846 genes were altered by SFN treatment (“SFN effect in cancer”), compared with 1691 genes in CCD841 cells (“SFN effect in non-transformed cells”, Supplementary Figure. 1B). The distribution and fold-changes of DEGs following incubation with SFN revealed a larger spread in HCT116 cells than in CCD841 cells (Supplementary Figure. 1C,D). Thus, not only were more genes altered in HCT116 cells, but the DEGs were changed by a larger fold-difference after SFN treatment.
Hierarchical clustering segregated between vehicle- and SFN-treated CCD841 and HCT116 cell lines (Fig. 1A). KEGG analysis showed enrichment of cancer-related pathways, including upregulation of p53 signaling and downregulation of Wnt/β-catenin signaling in SFN-treated HCT116 cells (Fig. 1B,C). Cell cycle targets included upregulated G2/M-related genes and downregulated G1/S-related genes, consistent with the reported role of SFN in G2/M arrest [21]. As anticipated, multiple Nrf2 target genes were upregulated in HCT116 cells following SFN treatment (Supplementary Fig 2). However, among the genes most highly reversed by SFN treatment, eleven that were under-expressed in HCT116 cells compared with CCD841 cells were strongly upregulated by SFN, and fifty-six constitutively overexpressed genes in HCT116 cells compared with CCD841 cells were markedly downregulated by SFN (Supplementary Fig 3). Notably, Loc344887 (NMRAL2P) was detected at low constitutive levels in HCT116 cells and was the most dramatically induced target of SFN (Supplementary Fig 3 and Figs. 1D,E).
Figure 1.
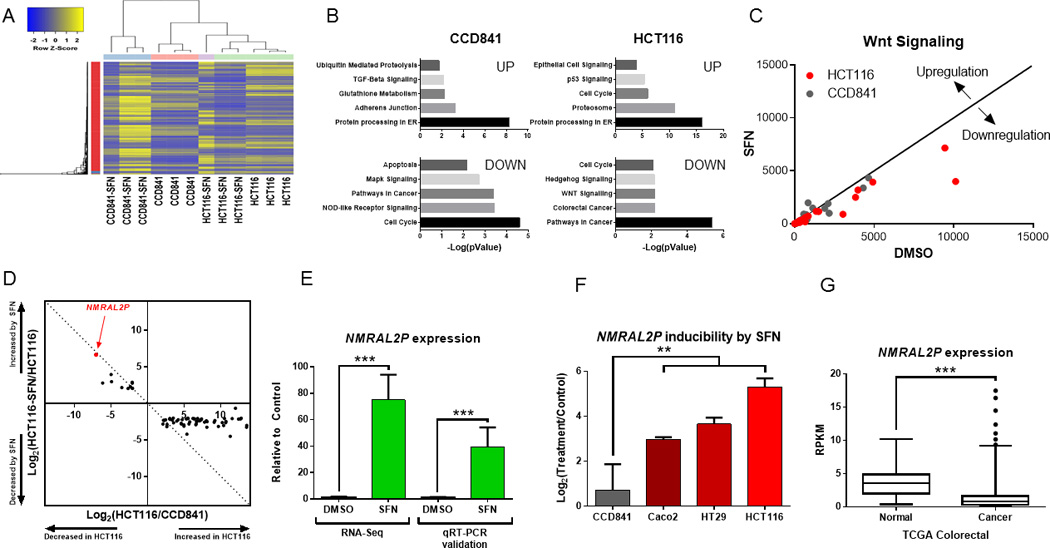
Identification of NMRAL2P as a novel target of SFN. (A) Heatmap of differentially expressed genes (DEGs) showed distinct clustering of HCT116 vs. CCD841 cells, in the presence and absence of SFN treatment. Each column represents a separate biological replicate for RNA-seq analysis. (B) Top five cancer-related pathways significantly upregulated and downregulated for each cell line (p<0.05). (C) DEGs associated with the Wnt signaling pathway, with each point designating the RPKM of a gene after treatment with SFN vs. vehicle control. The diagonal line represents a fold-change of zero. (D) Loc344887, renamed as NMRAL2P (HUGO Gene Nomenclature Committee, HGNC ID 52332), was identified as the most highly altered transcript in SFN-treated HCT116 cells (see also Supplementary Fig. 3). (E) qRT-PCR validation of NMRAL2P inducibility by SFN in HCT116 cells; mean±SD, n=3 (***p<0.001), representative data from an experiment repeated three times. (F) NMRAL2P induction was significantly greater in Caco2, HT29, and HCT116 colon cancer cells than in non-transformed CCD841 cells treated with SFN; mean±SD, n=3 (*p<0.05), from an experiment repeated three times. (G) Data from The Cancer Genome Atlas (TCGA) revealed significant NMRAL2P downregulation in human colorectal cancers (n=380) compared with normal colon (n=50, ***p<0.001).
Under identical SFN treatment conditions, NMRAL2P was induced more significantly in colon cancer cells than in non-transformed colonic epithelial cells (p<0.001), namely, 1.65-fold in CCD841 cells, 7.8-fold in Caco2 cells, 12.6-fold in HT29 cells, and 35-fold in HCT116 cells (Fig. 1F). Mining of The Cancer Genome Atlas (TCGA) (cancergenome.nih.gov) revealed that human colorectal cancers expressed significantly lower NMRAL2P levels than the corresponding normal tissues from patients, earmarking NMRAL2P as a potential new tumor suppressor biomarker (Fig. 1G, p<0.001).
3.2 NMRAL2P silencing protects colon cancer cells from SFN-mediated inhibition of cell growth, colony formation, and migration
An sgRNA with complementary sequence to the NMRAL2P promoter (see NMRAL2P[upstream1], Supplementary Table 1) was used to target Cas9 protein to the corresponding genomic region, seeking to disrupt transcription initiation. Screening of the genomic DNA of individual colonies, using PCR primers flanking exon 1 of NMRAL2P, identified a clone with a 390-bp deletion (Supplementary Fig 4, clone 5). Sequencing confirmed that the deletion was localized to the promoter region of NMRAL2P (Fig. 2A). The corresponding NMRAL2P knockout cells had similar overall growth and viability characteristics as the vector and mock controls (Supplementary Fig 5). No NMRAL2P expression was detected in the knockout cells before or after SFN treatment, in contrast to the parental HCT116 line under the same experimental conditions (Fig. 2B). In the MTT assay, NMRAL2P knockout cells were significantly less responsive than vector controls to low concentrations of SFN that attenuated cell viability (Fig. 2C). Inhibition in the colony formation assay at 7.5 µM SFN was partially rescued by NMRAL2P silencing (Fig. 2D), although this was not observed at 15 µM SFN, a concentration known to trigger autophagy and apoptosis in HCT116 cells [21,22]. A similar trend was noted in the transwell assay, with the inhibition of cell migration by SFN being partially rescued in NMRAL2P knockout cells incubated with 7.5 µM SFN (Fig. 2E, p<0.01).
Figure 2.
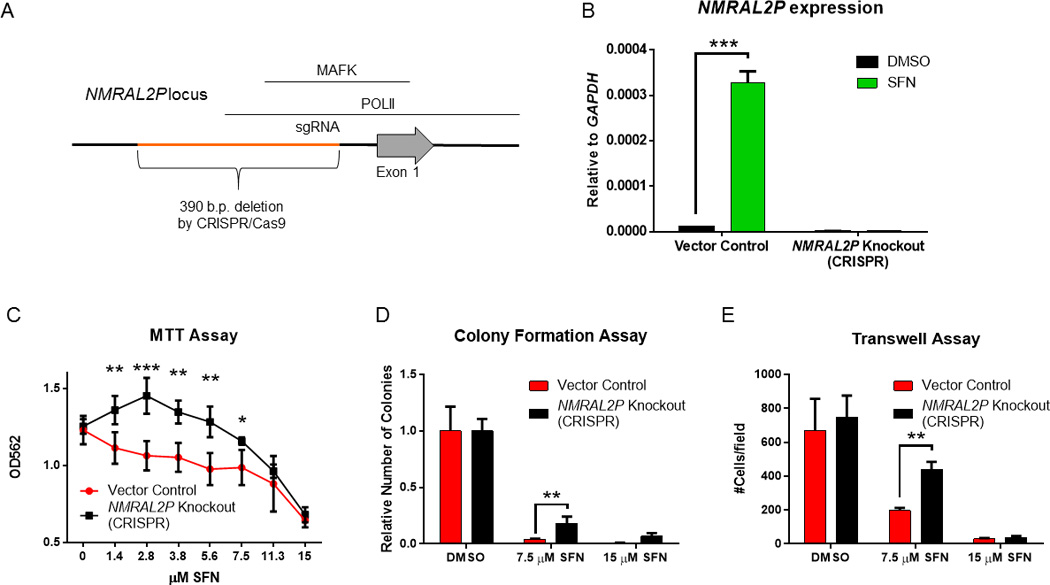
Phenotypic changes in colon cancer cells after NMRAL2P silencing. (A) In HCT116 cells, CRISPR/Cas9 genome-editing was used to delete a 390-base pair region in the NMRAL2P promoter (orange), interfering with RNA polymerase II and Mafk interactions, based on ENCODE Chip-seq data [66]. (B) Removal of the target sequence introduced the expected 390-bp deletion (see Supplementary Figure 4) and completely abrogated NMRAL2P inducibility by SFN; mean±SD, n=3 (***p<0.001), from an experiment repeated three times. (C) Enhanced viability of cells lacking NMRAL2P expression, 24 h after SFN treatment; actual absorbance readings at 562 nm in the MTT assay. (D) Colony formation and (E) transwell assays with NMRAL2P knockout and vector control cells. Data for colony formation are indicative of two independent experiments, whereas three separate experiments were conducted for MTT and transwell assays. In (C)-(E), data = mean±SD, n=3; *<0.05, **p<0.01, ***p<0.001.
3.3 NMRAL2P is regulated directly by Nrf2 in response to SFN treatment
In time-course experiments, NMRAL2P was upregulated within 1 h of SFN treatment and peaked at 8 h, similar to the well-known Nrf2 target gene HMOX1 and ahead of a second Nrf2-regulated gene, NQO1 (Fig. 3A). Several SFN analogs (AITC, 6-SFN, 9-SFN), Nrf2 activators (Oltipraz, TBHQ), and pan-HDAC inhibitors (TSA, SAHA, VPA, sodium butyrate) were compared as inducers of NMRAL2P expression. Surprisingly, pan-HDAC inhibitors either had no effect or reduced the expression of NMRAL2P at 6 and 24 h, whereas SFN analogs and Nrf2 activators induced the target gene, implicating a role for Nrf2 (Fig. 3B).
Figure 3.
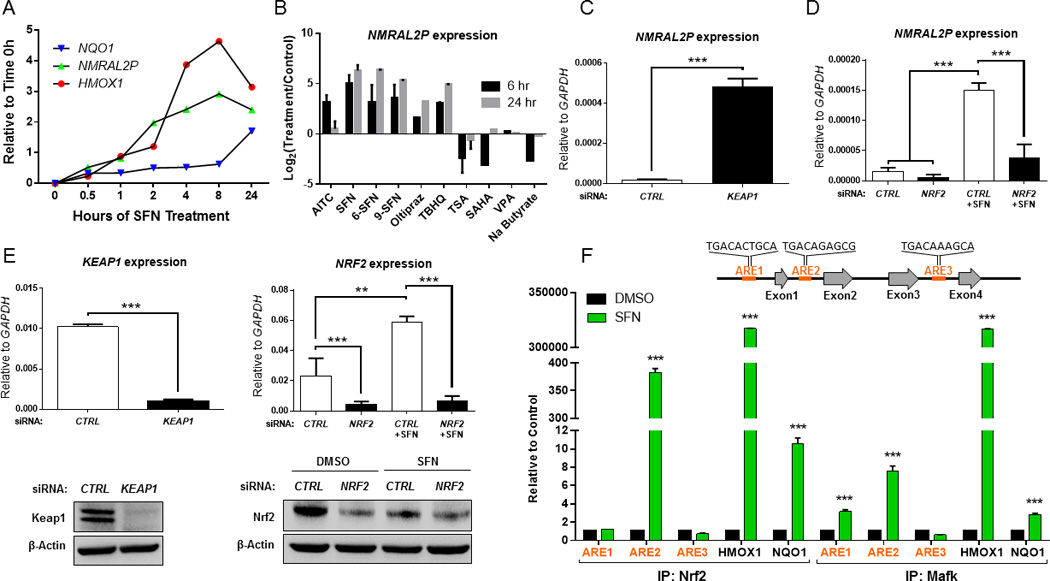
NMRAL2P is directly regulated by Nrf2. (A) Time-course for induction by SFN of NMRAL2P,HMOX1, and NQO1. (B) Changes in NMRAL2P expression in HCT116 cells treated with SFN and its analogs, Nrf2 activators, and pan-HDAC inhibitors at 6 and 24 h. (C) Induction of NMRAL2P following siRNA-mediated knockdown of Keap1, a negative regulator of Nrf2. (D) siRNA-mediated knockdown of Nrf2 interferes with NMRAL2P induction by SFN. (E) Confirmation by qRT-PCR and immunoblotting of siRNA targets from experiments shown in panels (C) and (D). (F) Chromatin immunoprecipitation (ChIP) assays using antibodies to Nrf2 and Mafk, with PCR primers recognizing known AREs in HMOX1 and NQO1 as positive controls. Three putative AREs were interrogated on NMRAL2P (ARE1, ARE2, ARE3). In panels (A)-(F), data = mean±SD from two or more independent experiments; **p<0.01, ***p<0.001.
The repressive partner of Nrf2, Keap1, was knocked down via siRNA-mediated inhibition, which resulted in highly significant induction of NMRAL2P, consistent with NMRAL2P activation by Nrf2 (Fig. 3C). In the reverse scenario, siRNA-mediated knockdown of Nrf2 interfered with the ability of SFN to activate NMRAL2P (Fig. 3D). Reduced expression of the corresponding siRNA targets, Keap1 and Nrf2, was confirmed both at the mRNA and protein level in these experiments (Fig. 3E).
In ChIP assays, three putative AREs were interrogated on the NMRAL2P locus (Fig. 3F). Upon SFN treatment, Nrf2 and its transcriptional coactivator partner, Mafk, were co-localized to ARE2, upstream of exon 2, and Mafk also interacted with ARE1, upstream of exon 1 (Fig. 3F). Neither Nrf2 nor Mafk were detected on ARE3, upstream of exon 4. As additional controls for the ChIP assays, Nrf2 and Mafk interactions were confirmed on HMOX1 and NQO1 (Fig. 3F). These findings supported the hypothesis that, like HMOX1 and NQO1, NMRAL2P was a direct transcriptional target of Nrf2.
3.4 NMRAL2P is a co-regulator of Nrf2-dependent NQO1 induction
In subcellular fractionation experiments, NMRAL2P was readily detected in the nucleus, but not in the cytosolic compartment of vehicle-treated HCT116 cells (Fig. 4A, black bars). After SFN treatment, there was an apparent increase in the abundance of NMRAL2P, both in the nuclear and cytosolic compartments (Fig. 4A, green bars). Future studies should seek to corroborate these findings, for example using fluorescence in situ hybridization, but we elected to focus on the nuclear aspects related to gene regulation.
Figure 4.
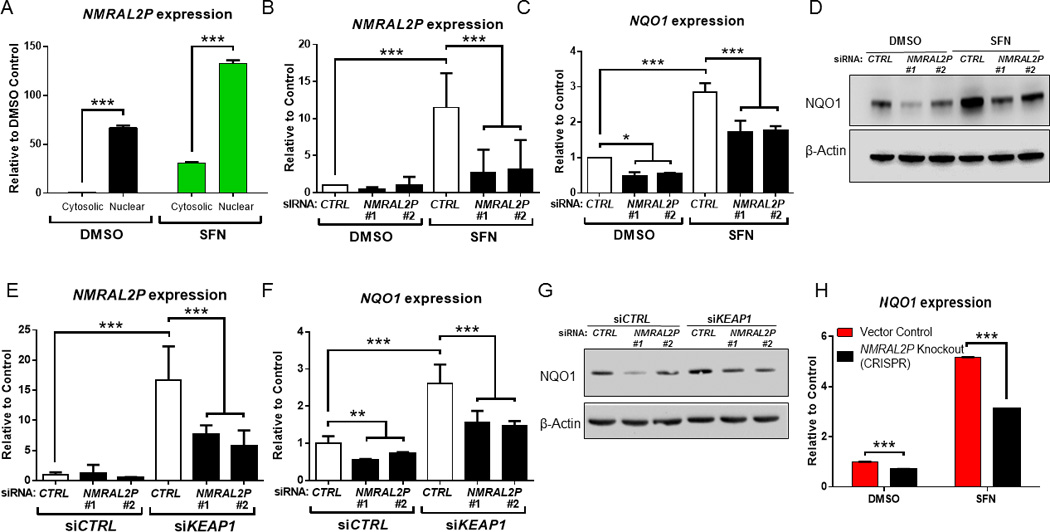
NMRAL2P is a non-coding RNA that influences NQO1 expression. (A) Higher expression of NMRAL2P in the nuclear vs. cytoplasmic compartment, before and after SFN treatment in HCT116 cells. (B,C) siRNAs targeting two different regions of NMRAL2P (siRNA #1 and #2) interfered with both NMRAL2P and NQO1 inducibility after SFN treatment. (D) Immunoblotting confirmed that NQO1 protein induction was abrogated in cells treated with NMRAL2P siRNAs. (E) NMRAL2P and (F) NQO1 expression following partial knockdown of both NMRAL2P and KEAP1. (G) Immunoblotting of NQO1 protein expression from the double knockdown experiments shown in panels (E) and (F). (H) NQO1 inducibility by SFN is attenuated in cells lacking NMRAL2P due to CRISPR/Cas9-mediated genome editing of the promoter region (see Fig 2A). In panels (A), (B), (C), (E), (F) and (H), data = mean±SD from two or more independent experiments; n=3; *p<0.05, **p<0.01, ***p<0.001.
From the time-course experiments (Fig. 3A), it was postulated that early induction of NMRAL2P might serve as an upstream regulator of NQO1. To test this hypothesis, siRNAs were targeted to two different regions of the NMRAL2P transcript (siRNAs #1 and #2), which resulted in significantly reduced NMRAL2P induction following SFN treatment (Fig. 4B). In these experiments, NQO1 mRNA and protein induction by SFN was attenuated significantly compared with the siRNA controls (Fig. 4C,D). Keap1 knockdown also was used to induce Nrf2; following siRNA-mediated silencing of NMRAL2P, a significant reduction was observed in the inducibility of both NMRAL2P (Fig. 4E) and NQO1 (Fig. 4F,G). Similar results were obtained in colon cancer cells lacking NMRAL2P expression due to CRISPR/Cas9 genome editing, with the inducibility of NQO1 being attenuated significantly after treatment with SFN (Fig. 4H).
Finally, no effect was seen on NMRAL1 after NMRAL2P knockdown or SFN treatment (Supplementary Fig 6), indicating that the siRNAs and PCR primers were specific for the functional pseudogene, rather than the protein-coding gene sharing 62% homology.
4 Discussion
Plasma SFN metabolites have been detected at 2 µM in people consuming broccoli sprouts [43], and chemopreventive outcomes in a mouse model of intestinal tumorigenesis were associated with tissues levels in the gastrointestinal tract of ~3–30 µM total SFN [44]. Although lower SFN concentrations might be considered in future RNA-seq experiments, minimizing apoptosis end-points [21, 22], we sought to parallel the prior transcriptome profiling in prostate cancer cells treated with 15 µM SFN, which implicated multiple cancer-related pathways [39]. In the current investigation of colon cancer cells treated with 15 µM SFN, multiple cancer-related pathways also were implicated, including cell cycle, hedgehog signaling, p53 signaling, Wnt signaling, colorectal cancer, and protein processing in ER (Figure 1B). Several Nrf2-dependent targets were upregulated by SFN in colon cancer cells (Supplementary Figure 2). However, our attention was drawn to Loc344887 (NMRAL2P) as a transcript with low constitutive levels in colon cancer cells that was dramatically induced by SFN (Supplementary Figure 3). Importantly, this ncRNA was identified as significantly downregulated in human colorectal cancer (Figure 1G), suggesting a possible tumor suppressor function in the colon, and the potential to serve as a clinical biomarker in patients at risk of developing malignancy of the large intestine.
We observed that NMRAL2P, but not the protein-coding gene NMRAL1, was strongly induced by SFN in colon cancer cells, and corroborated the presence of bona fide Nrf2/Mafk binding sites in the corresponding genomic region of NMRAL2P (Fig 3F). The ChIP assays focused on Nrf2 and Mafk, but we cannot rule out contributions from other MAF family members, or associated coactivators. Knockdown experiments that targeted either Keap1 or Nrf2 further supported the role of Nrf2 in regulating NMRAL2P, and loss of NMRAL2P implicated the ncRNA as a downstream activator of NQO1. We noted, however, that NQO1 inducibility was partially retained in SFN-treated colon cancer cells even after NMRAL2P silencing via siRNA treatment or CRISPR/Cas9 genome-editing (Figures 4C and 4H). This suggests that NMRAL2P probably cooperates with other factors in regulating NQO1 gene activity.
This is the first report to identify a functional pseudogene that is both a direct transcriptional target of Nrf2, and a downstream regulator of Nrf2-dependent NQO1 induction. Polymorphisms in NQO1 have been linked to increased risk for human colorectal cancer [45], and an anticancer role for NQO1 also has been identified in preclinical models. For example, in a rat colon carcinogenesis model, Oltipraz treatment resulted in reduced colonic aberrant crypt foci and tumor formation associated with NQO1 induction, whereas NQO1 knockout mice were more susceptible to radiation-induced myeloproliferative disease [46–48]. Thus, a change in NQO1 activity has potential implications for cancer susceptibility in the colon and in other tissues.
It is unlikely, however, that NMRAL2P serves as a master regulator of all Nrf2-dependent target genes. This point is perhaps best exemplified by the well-known Nrf2-dependent gene HMOX1. Thus, HMOX1 was induced rapidly by SFN (Figure 3A), but HMOX1, like NFE2L2 (the gene coding for Nrf2), was unaffected by NMRAL2P knockdown (Supplementary Figure 7). Prior reports noted that HMOX1 was upregulated at an earlier time-point than other Nrf2-dependent target genes, including NQO1 [49, 50], and implicated multiple factors in the dynamic regulation of HMOX1 [14, 51]. For example, Bach1 can inhibit HMOX1 induction by antagonizing Nrf2 binding [51], whereas the SWI/SNF chromatin remodeling factor BRG1 interacts with Nrf2 to selectively induce HMOX1 [14]. TET-dependent DNA methylation changes and post-translational modifications to Nrf2 also influence Keap1/Nrf2 interactions, and the extent of Nrf2 nuclear-cytoplasmic trafficking [17, 18, 52, 53]. These mechanisms would likely dictate the degree to which Nrf2, and perhaps Mafk, interact with NMRAL2P in the nuclear compartment.
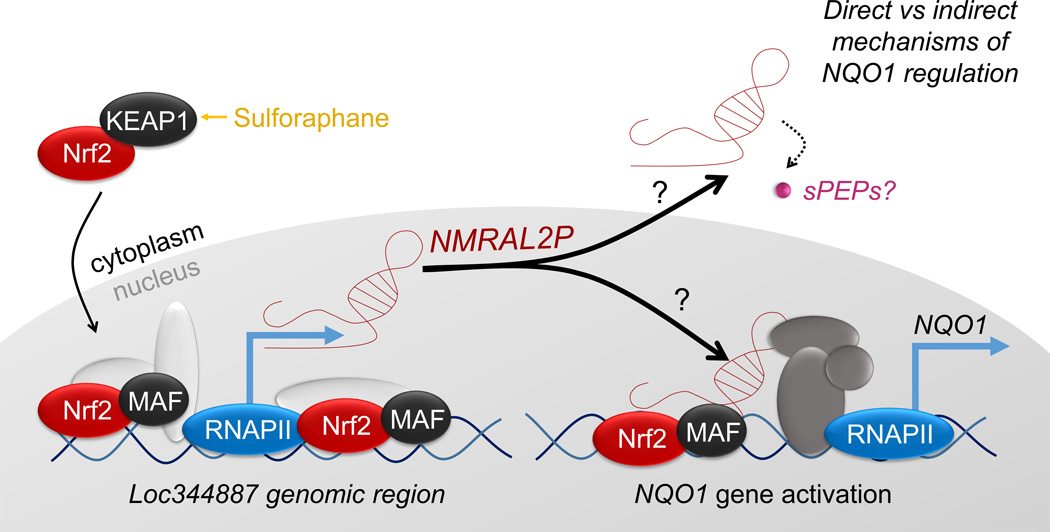
Several colon cancer-related lncRNAs have emerged as potential prognostic biomarkers [54–56]. Mechanisms that have been implicated include lncRNA-miRNA associations, lncRNA-protein interactions, and actions as miRNA precursors or pseudogenes [57, 58]. Pseudogene PTENP1 can serve as a miRNA “decoy” for the protein-coding gene phosphatase and tensin homolog (PTEN) [59], mutations in high-mobility group-1 pseudogenes (e.g., HMGA1P6 and HMGA1P7) alter their decoy functions that regulate HMGA1 [60], and pseudogene MYLKP1 regulates the mRNA stability of smooth muscle myosin light chain kinase, altering cell proliferation in cancer cells [61]. Considering the influence of SFN in regulating intracellular redox status, it is tempting to speculate on the role(s) of NMRAL2P as a functional pseudogene. One intriguing possibility centers on the emerging evidence for functional short peptides (sPEPs) encoded by minimal open reading frames [62]. Allowing for start codons besides ATG, the NMRAL2P transcript has several putative open reading frames for sPEPs; one of the hypothetical candidates based on conceptual translation, hCG2041270, was listed in prior studies with TBHQ [63]. Thus, we do not formally discriminate here between possible ncRNA and/or sPEP roles of NMRAL2P. Given the diverse actions of Nrf2 in cancer etiology [10, 12, 64, 65], we conclude that further studies are warranted on NMRAL2P/Nrf2 crosstalk and the associated direct versus indirect mechanisms of gene regulation (Figure 5).
Figure 5.
Working model for the induction of NMRAL2P, and its role as a downstream coactivator of NQO1 in SFN-treated colon cancer cells.
Supplementary Material
Acknowledgments
This research was supported in part by NIH grants CA090890, CA122959, ES00210, and ES023512, as well as the John S. Dunn Foundation, and a Chancellor’s Research Initiative.
G.S.J., P.R., D.E.W., E.H., and R.H.D. were responsible for the concept and design of the studies. L.B. and W.M.D. performed RNA-seq experiments. J.L. and D.S. conducted bioinformatics and other data analyses. Cell-based mechanistic experiments were performed by G.S.J., who with J.L. and R.H.D. were responsible for drafting of the manuscript. All authors read and approved the final iteration of the paper. We thank Drs. Margie Moczygemba and Sevinj Iskandarova of the Flow Cytometry Core for technical assistance.
Abbreviations
- AITC
allyl isothiocyanate
- ARE
antioxidant response element
- CRISPR
clustered regularly interspaced short palindromic repeats
- DMSO
dimethylsulfoxide
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HDAC
histone deacetylase
- HMOX1
heme oxygenase 1
- Keap1
kelch-like ECH-associated protein 1
- lncRNA
long noncoding RNA
- MAF
musculoaponeurotic fibrosarcoma
- MALAT1
metastasis associated lung adenocarcinoma transcript 1
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NMRAL1
NmrA-like redox sensor 1
- NMRAL2P
NmrA-like redox sensor 2 (functional pseudogene of NMRAL1)
- NQO1
NAD(P)H quinone dehydrogenase 1
- Nrf2
nuclear factor (erythroid-derived 2)-like 2
- SAHA
suberoylanilide hydroxamic acid
- SFN
sulforaphane
- 6-SFN
6-methylsulfinylhexyl isothiocyanate
- 9-SFN
9-methylsulfinylnonyl isothiocyanate
- sgRNA
small guide RNA
- sPEPS
short peptides encoded by minimal open reading frames
- TBHQ
tert-butylhydroquinone
- TSA
trichostatin A
- VPA
valproic acid
Footnotes
The authors have declared no conflicts of interest.
References
- 1.Chung FL, Conaway CC, Rao CV, Reddy BS. Chemoprevention of colonic aberrant crypt foci in Fischer rats by sulforaphane and phenethyl isothiocyanate. Carcinogenesis. 2000;21:2287–2291. doi: 10.1093/carcin/21.12.2287. [DOI] [PubMed] [Google Scholar]
- 2.Myzak MC, Dashwood WM, Orner GA, Ho E, et al. Sulforaphane inhibits histone deacetylase in vivo and suppresses tumorigenesis in Apc-minus mice. FASEB J. 2006;20:506–508. doi: 10.1096/fj.05-4785fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myzak MC, Dashwood RH. Chemoprotection by sulforaphane: keep one eye beyond Keap1. Cancer Lett. 2006;233:208–218. doi: 10.1016/j.canlet.2005.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho E, Beaver LM, Williams DE, Dashwood RH. Dietary Factors and Epigenetic Regulation for Prostate Cancer Prevention. Adv. Nutr. An Int. Rev. J. 2011;2:497–510. doi: 10.3945/an.111.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atwell LL, Beaver LM, Shannon J, Williams DE, et al. Epigenetic Regulation by Sulforaphane: Opportunities for Breast and Prostate Cancer Chemoprevention. Curr. Pharmacol. Reports. 2015;1:102–111. doi: 10.1007/s40495-014-0002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao Y, Tollefsbol TO. Impact of Epigenetic Dietary Components on Cancer through Histone Modifications. Curr. Med. Chem. 2015;22:2051–2064. doi: 10.2174/0929867322666150420102641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tortorella SM, Royce SG, Licciardi PV, Karagiannis TC. Dietary Sulforaphane in Cancer Chemoprevention: The Role of Epigenetic Regulation and HDAC Inhibition. Antioxid. Redox Signal. 2015;22:1382–1424. doi: 10.1089/ars.2014.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amjad AI, Parikh RA, Appleman LJ, Hahm E-R, et al. Broccoli-Derived Sulforaphane and Chemoprevention of Prostate Cancer: From Bench to Bedside. Curr. Pharmacol. Reports. 2015;1:382–390. doi: 10.1007/s40495-015-0034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuentes F, Paredes-Gonzalez X, Kong A-NT. Dietary Glucosinolates Sulforaphane, Phenethyl Isothiocyanate, Indole-3-Carbinol/3,3’-Diindolylmethane: Anti-Oxidative Stress/Inflammation, Nrf2, Epigenetics/Epigenomics and In Vivo Cancer Chemopreventive Efficacy. Curr. Pharmacol. Reports. 2015;1:179–196. doi: 10.1007/s40495-015-0017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L, Palliyaguru DL, Kensler TW, Colditz GA, et al. Frugal chemoprevention: targeting Nrf2 with foods rich in sulforaphane. Semin. Oncol. 2016;43:146–153. doi: 10.1053/j.seminoncol.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kensler TW, Wakabayashi N, Biswal S. Cell Survival Responses to Environmental Stresses Via the Keap1-Nrf2-ARE Pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 12.Qin S, Hou D-X. Multiple regulations of Keap1/Nrf2 system by dietary phytochemicals. Mol. Nutr. Food Res. 2016 doi: 10.1002/mnfr.201501017. [DOI] [PubMed] [Google Scholar]
- 13.Dinkova-Kostova AT, Talalay P. Direct and indirect antioxidant properties of inducers of cytoprotective proteins. Mol. Nutr. Food Res. 2008;(52 Suppl 1):S128–S138. doi: 10.1002/mnfr.200700195. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Ohta T, Maruyama A, Hosoya T, et al. BRG1 Interacts with Nrf2 To Selectively Mediate HO-1 Induction in Response to Oxidative Stress. Mol. Cell. Biol. 2006;26:7942–7952. doi: 10.1128/MCB.00700-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajendran P, Dashwood W-M, Li L, Kang Y, et al. Nrf2 status affects tumor growth, HDAC3 gene promoter associations, and the response to sulforaphane in the colon. Clin. Epigenetics. 2015;7:102. doi: 10.1186/s13148-015-0132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Li H, Liu L, Lu Y, et al. Methylation of arginine by PRMT1 regulates Nrf2 transcriptional activity during the antioxidative response. Biochim. Biophys. Acta - Mol. Cell Res. 2016;1863:2093–2103. doi: 10.1016/j.bbamcr.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Xu C, Yuan X, Pan Z, Shen G, et al. Mechanism of action of isothiocyanates: the induction of ARE-regulated genes is associated with activation of ERK and JNK and the phosphorylation and nuclear translocation of Nrf2. Mol. Cancer Ther. 2006;5:1918–1926. doi: 10.1158/1535-7163.MCT-05-0497. [DOI] [PubMed] [Google Scholar]
- 18.Keum Y-S, Yu S, Chang PP-J, Yuan X, et al. Mechanism of action of sulforaphane: inhibition of p38 mitogen-activated protein kinase isoforms contributing to the induction of antioxidant response element-mediated heme oxygenase-1 in human hepatoma HepG2 cells. Cancer Res. 2006;66:8804–8813. doi: 10.1158/0008-5472.CAN-05-3513. [DOI] [PubMed] [Google Scholar]
- 19.Ho E, Clarke JD, Dashwood RH. Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. J. Nutr. 2009;139:2393–2396. doi: 10.3945/jn.109.113332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu A, Wong CP, Yu Z, Williams DE, et al. Promoter de-methylation of cyclin D2 by sulforaphane in prostate cancer cells. Clin. Epigenetics. 2011;3:3. doi: 10.1186/1868-7083-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajendran P, Delage B, Dashwood WM, Yu T-W, et al. Histone deacetylase turnover and recovery in sulforaphane-treated colon cancer cells: competing actions of 14-3-3 and Pin1 in HDAC3/SMRT corepressor complex dissociation/reassembly. Mol. Cancer. 2011;10:68. doi: 10.1186/1476-4598-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajendran P, Kidane AI, Yu T-W, Dashwood W-M, et al. HDAC turnover, CtIP acetylation and dysregulated DNA damage signaling in colon cancer cells treated with sulforaphane and related dietary isothiocyanates. Epigenetics. 2013;8:612–623. doi: 10.4161/epi.24710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarke JD, Hsu A, Yu Z, Dashwood RH, et al. Differential effects of sulforaphane on histone deacetylases, cell cycle arrest and apoptosis in normal prostate cells versus hyperplastic and cancerous prostate cells. Mol. Nutr. Food Res. 2011;55:999–1009. doi: 10.1002/mnfr.201000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim E, Bisson WH, Löhr CV, Williams DE, et al. Histone and Non-Histone Targets of Dietary Deacetylase Inhibitors. Curr. Top. Med. Chem. 2016;16:714–731. doi: 10.2174/1568026615666150825125857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parasramka MA, Ho E, Williams DE, Dashwood RH. MicroRNAs, diet, and cancer: new mechanistic insights on the epigenetic actions of phytochemicals. Mol. Carcinog. 2012;51:213–230. doi: 10.1002/mc.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slaby O, Sachlova M, Brezkova V, Hezova R, et al. Identification of microRNAs regulated by isothiocyanates and association of polymorphisms inside their target sites with risk of sporadic colorectal cancer. Nutr. Cancer. 2013;65:247–254. doi: 10.1080/01635581.2013.756530. [DOI] [PubMed] [Google Scholar]
- 27.Wagner AE, Boesch-Saadatmandi C, Dose J, Schultheiss G, et al. Anti-inflammatory potential of allyl-isothiocyanate--role of Nrf2, NF-(κ) B and microRNA-155. J. Cell. Mol. Med. 2012;16:836–843. doi: 10.1111/j.1582-4934.2011.01367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan Y, Zhang L, Bao Y, Li B, et al. Epithelial-mesenchymal transition, a novel target of sulforaphane via COX-2/MMP2, 9/Snail, ZEB1 and miR-200c/ZEB1 pathways in human bladder cancer cells. J. Nutr. Biochem. 2013;24:1062–1069. doi: 10.1016/j.jnutbio.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Lan F, Pan Q, Yu H, Yue X. Sulforaphane enhances temozolomide-induced apoptosis because of down-regulation of miR-21 via Wnt/β-catenin signaling in glioblastoma. J. Neurochem. 2015;134:811–818. doi: 10.1111/jnc.13174. [DOI] [PubMed] [Google Scholar]
- 30.Appari M, Babu KR, Kaczorowski A, Gross W, et al. Sulforaphane, quercetin and catechins complement each other in elimination of advanced pancreatic cancer by miR-let-7 induction and K-ras inhibition. Int. J. Oncol. 2014;45:1391–1400. doi: 10.3892/ijo.2014.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serghiou S, Kyriakopoulou A, Ioannidis JPA. Long noncoding RNAs as novel predictors of survival in human cancer: a systematic review and meta-analysis. Mol. Cancer. 2016;15:50. doi: 10.1186/s12943-016-0535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng H, Zhang J, Shi J, Guo Z, et al. Role of long non-coding RNA in tumor drug resistance. Tumour Biol. 2016 doi: 10.1007/s13277-016-5125-8. [DOI] [PubMed] [Google Scholar]
- 33.Khanduja JS, Calvo IA, Joh RI, Hill IT, et al. Nuclear Noncoding RNAs and Genome Stability. Mol. Cell. 2016;63:7–20. doi: 10.1016/j.molcel.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majidinia M, Yousefi B. Long non-coding RNAs in cancer drug resistance development. DNA Repair (Amst) 2016;45:25–33. doi: 10.1016/j.dnarep.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Huang X, Zhi X, Gao Y, Ta N, et al. LncRNAs in pancreatic cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Xia J, Li Q, Yao Y, et al. NRF2/Long noncoding RNA ROR signaling regulates mammary stem cell expansion and protects against estrogen genotoxicity. J. Biol. Chem. 2014;289:31310–31318. doi: 10.1074/jbc.M114.604868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thai P, Statt S, Chen CH, Liang E, et al. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am. J. Respir. Cell Mol. Biol. 2013;49:204–211. doi: 10.1165/rcmb.2013-0159RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myzak MC, Karplus PA, Chung F-L, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64:5767–5774. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 39.Beaver LM, Buchanan A, Sokolowski EI, Riscoe AN, et al. Transcriptome analysis reveals a dynamic and differential transcriptional response to sulforaphane in normal and prostate cancer cells and suggests a role for Sp1 in chemoprevention. Mol. Nutr. Food Res. 2014;58:2001–2013. doi: 10.1002/mnfr.201400269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chorley BN, Campbell MR, Wang X, Karaca M, et al. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu EJ, Kim S-H, Kim HJ, Heo K, et al. Positive regulation of β-catenin-PROX1 signaling axis by DBC1 in colon cancer progression. Oncogene. 2015 doi: 10.1038/onc.2015.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ran FA, Hsu PD, Wright J, Agarwala V, et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atwell LL, Hsu A, Wong CP, Stevens JF, et al. Absorption and chemopreventive targets of sulforaphane in humans following consumption of broccoli sprouts or a myrosinase-treated broccoli sprout extract. Mol. Nutr. Food Res. 2015;59:424–433. doi: 10.1002/mnfr.201400674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu R, Khor TO, Shen G, Jeong W-S, et al. Cancer chemoprevention of intestinal polyposis in ApcMin/+ mice by sulforaphane, a natural product derived from cruciferous vegetable. Carcinogenesis. 2006;27:2038–2046. doi: 10.1093/carcin/bgl049. [DOI] [PubMed] [Google Scholar]
- 45.Zheng B, Wang Z, Chai R. NQO1 C609T polymorphism and colorectal cancer susceptibility: a meta-analysis. Arch. Med. Sci. 2014;10:651–660. doi: 10.5114/aoms.2014.44856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Begleiter A, Sivananthan K, Lefas GM, Maksymiuk AW, et al. Inhibition of colon carcinogenesis by post-initiation induction of NQO1 in Sprague-Dawley rats. Oncol. Rep. 2009;21:1559–1565. doi: 10.3892/or_00000388. [DOI] [PubMed] [Google Scholar]
- 47.Begleiter A, Sivananthan K, Curphey TJ, Bird RP. Induction of NAD(P)H quinone: oxidoreductase1 inhibits carcinogen-induced aberrant crypt foci in colons of Sprague-Dawley rats. Cancer Epidemiol. Biomarkers Prev. 2003;12:566–572. [PubMed] [Google Scholar]
- 48.Iskander K, Barrios RJ, Jaiswal AK. Disruption of NAD(P)H:quinone oxidoreductase 1 gene in mice leads to radiation-induced myeloproliferative disease. Cancer Res. 2008;68:7915–7922. doi: 10.1158/0008-5472.CAN-08-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xue M, Momiji H, Rabbani N, Barker G, et al. Frequency Modulated Translocational Oscillations of Nrf2 Mediate the Antioxidant Response Element Cytoprotective Transcriptional Response. Antioxid. Redox Signal. 2014;0:1–17. doi: 10.1089/ars.2014.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lo R, Matthews J. The aryl hydrocarbon receptor and estrogen receptor alpha differentially modulate nuclear factor erythroid-2-related factor 2 transactivation in MCF-7 breast cancer cells. Toxicol. Appl. Pharmacol. 2013;270:139–148. doi: 10.1016/j.taap.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 51.Sun J, Hoshino H, Takaku K, Nakajima O, et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002;21:5216–5224. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang H-C, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by Protein Kinase C Regulates Antioxidant Response Element-mediated Transcription. J. Biol. Chem. 2002;277:42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 53.Kang KA, Piao MJ, Kim KC, Kang HK, et al. Epigenetic modification of Nrf2 in 5-fluorouracil-resistant colon cancer cells: involvement of TET-dependent DNA demethylation. Cell Death Dis. 2014;5:e1183. doi: 10.1038/cddis.2014.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xin Y, Li Z, Shen J, Chan MTV, et al. CCAT1: a pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2016;49:255–260. doi: 10.1111/cpr.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology. 2015;149:1204.e12–1225.e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu M-D, Qi P, Du X. Long non-coding RNAs in colorectal cancer: implications for pathogenesis and clinical application. Mod. Pathol. 2014;27:1310–1320. doi: 10.1038/modpathol.2014.33. [DOI] [PubMed] [Google Scholar]
- 57.Xie X, Tang B, Xiao Y-F, Xie R, et al. Long non-coding RNAs in colorectal cancer. Oncotarget. 2016;7:5226–5239. doi: 10.18632/oncotarget.6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han D, Wang M, Ma N, Xu Y, et al. Long noncoding RNAs: novel players in colorectal cancer. Cancer Lett. 2015;361:13–21. doi: 10.1016/j.canlet.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 59.Rigoutsos I, Furnari F. Gene-expression forum: Decoy for microRNAs. Nature. 2010;465:1016–1017. doi: 10.1038/4651016a. [DOI] [PubMed] [Google Scholar]
- 60.Esposito F, De Martino M, Petti MG, Forzati F, et al. HMGA1 pseudogenes as candidate proto-oncogenic competitive endogenous RNAs. Oncotarget. 2014;5:8341–8354. doi: 10.18632/oncotarget.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han YJ, Ma SF, Yourek G, Park Y-D, et al. A transcribed pseudogene of MYLK promotes cell proliferation. FASEB J. 2011;25:2305–2312. doi: 10.1096/fj.10-177808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 2014;15:193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- 63.Yi YW, Oh S. Comparative analysis of NRF2-responsive gene expression in AcPC-1 pancreatic cancer cell line. Genes Genomics. 2015;37:97–109. doi: 10.1007/s13258-014-0253-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Menegon S, Columbano A, Giordano S. The Dual Roles of NRF2 in Cancer. Trends Mol. Med. 2016;22:578–593. doi: 10.1016/j.molmed.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 65.Kensler TW, Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis. 2010;31:90–99. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.ENCODE Project Consortium, T.E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.