Abstract
Acquired resistance to BRAFV600E inhibitors (BRAFi) in melanoma remains a common clinical obstacle, as is the case for any targeted drug therapy that can be developed given the plastic nature of cancers. While there has been significant focus on the cancer cell-intrinsic properties of BRAFi resistance, the impact of BRAFi resistance on host immunity has not been explored. Here we provide preclinical evidence that resistance to BRAFi in an autochthonous mouse model of melanoma is associated with restoration of myeloid-derived suppressor cells (MDSC) in the tumor microenvironment initially reduced by BRAFi treatment. In contrast to restoration of MDSC, levels of T regulatory cells remained reduced in BRAFi-resistant tumors. Accordingly, tumor gene expression signatures specific for myeloid cell chemotaxis and homeostasis reappeared in BRAFi-resistant tumors. Notably, MDSC restoration relied upon MAPK pathway reactivation and downstream production of the myeloid attractant CCL2 in BRAFi-resistant melanoma cells. Strikingly, while combination checkpoint blockade (anti-CTLA-4 + anti-PD-1) was ineffective against BRAFi-resistant melanomas, the addition of MDSC depletion/blockade (anti-Gr-1 + CCR2 antagonist) prevented outgrowth of BRAFi-resistant tumors. Our results illustrate how extrinsic pathways of immunosuppression elaborated by melanoma cells dominate the tumor microenvironment and highlight the need to target extrinsic as well as intrinsic mechanisms of drug resistance.
Keywords: vemurafenib, PLX4720, MDSC, regulatory T cell, CCL2
INTRODUCTION
Molecularly-targeted oncoprotein inhibitors are a powerful means for treating tumors with known oncogenic driver mutations. However, acquired resistance severely limits the clinical impact of these drugs. In patients with BRAFV600E-mutant melanoma, the FDA-approved BRAF-inhibitors vemurafenib and dabrafenib delay time to progression by a median of only 5–7 months (1,2). In many cases, resistance has been linked to tumor MAPK pathway reactivation through secondary activating mutations or overexpression of MAPK pathway-associated genes (3,4). Accordingly, MEK inhibitors are now commonly used in conjunction with BRAF inhibitors in patients (5). Nonetheless, acquired resistance still develops through MAPK pathway-related alterations, and typically within less than one year (6).
With oncogenic driver mutations as moving targets, a better approach to treating MAPKi-resistant cancers likely requires a better understanding of the tumor microenvironment (TME). While cancer cell-intrinsic properties of resistance have been extensively characterized, the immunological consequences of inhibitor resistance are not well understood. We and others have used mouse models to demonstrate that short-term BRAF-inhibition improves the ratio of CD8 T cells to myeloid derived suppressor cells (MDSCs), and to regulatory T cells (Tregs), in the TME (7–9). However it is not known for how long such favorable alterations are maintained. In one study of BRAFi-treated patients, a robust early infiltration of T cells into tumors was no longer observed at the time of progression (10). Further, BRAF/MEK-inhibitor-resistant patient melanomas were recently shown to have reduced CD8 T cell populations (11,12) and a more pronounced inflammatory monocyte/M2 macrophage gene expression profile (12). Despite these correlative studies, it is not yet known whether acquired inhibitor resistance directly promotes a more immunosuppressive tumor microenvironment, and which tumor-intrinsic factors might be required for this process.
Another goal in studying the immune constitution of MAPKi-resistant tumors is the rational design of effective immunotherapeutic strategies. While the first trial combining BRAFi with anti-CTLA-4 (ipiliumimab) was terminated due to hepatotoxicity (13), new trials combining BRAFi and MEKi, with anti-CTLA-4, anti-PD-1, and other immunotherapies, are currently underway (14). In preclinical studies, the efficacy of BRAF-inhibitors is enhanced by immunotherapies including anti-CSF-1R (15), anti-PD-1 (16), and adoptive T cell therapy (17). Nonetheless, how best to sequence the administration of oncoprotein inhibitors and immunotherapies remains unknown. Current best practice involves administering BRAFi as first-line therapy to alleviate symptoms in patients with advanced disease (18). This may be followed with checkpoint blockade immunotherapy or combination therapies, which are capable of generating more durable responses (19). However it is not known if inhibitor resistance might also impair tumor responsiveness to immunotherapy.
The present studies were aimed at determining how acquired inhibitor resistance alters tumor immune suppression, and using this knowledge to rationally administer immunotherapy in the drug resistance setting. We employed a mouse model of BRAFV600E-mutant autochthonous melanoma, in which resistance to the BRAF inhibitor PLX4720 evolves in the presence of an intact host immune system. We report that BRAFi-resistant tumors restore MDSCs and associated chemotactic gene signatures by melanoma cell-intrinsic MAPK-pathway-dependent mechanisms. Accordingly, MDSC elimination cooperates with dual checkpoint blockade immunotherapy to prevent the outgrowth of BRAFi-resistant variant tumors. These studies reveal a dynamic immunosuppressive response to a cancer during the course of prolonged oncoprotein inhibition, and show that BRAFi-resistant cancers depend on MDSCs for co-resistance to immunotherapy.
MATERIALS AND METHODS
Mice
Tyr::CreER+/−BrafCA/+Ptenlox/lox (Braf/Pten) mice (20) were a gift from Marcus Bosenberg (Yale). These mice were bred in-house onto a C57BL/6 background as previously described (7) with >98% purity obtained by congenic testing at the DartMouse™ Core Facility. C57BL/6 mice were obtained from both The Jackson Laboratory and Charles River. C57BL/6 Ccr2−/−, Rag1−/− and Cd8−/− mice were obtained from The Jackson Laboratory and bred in-house.
Mouse Model of BRAFi-Resistance
BRAFi resistance was induced in C57BL/6 mice bearing autochthonous Braf/Pten melanomas growing in skin grafts donated from Braf/Pten mice. This modified skin-graft tumor model extended the observable period of tumor growth by eliminating spontaneous tumor formation at distal sites. Mice were dorsally grafted with ~1 cm2 sections of Braf/Pten tail skin, and tumors were induced one week later by topical application of 4-hydroxy-tamoxifen (10ml of a 20 mM solution in DMSO) at the graft site. Mice bearing palpable melanomas (27–35 days post-induction) were fed PLX4720 (BRAFi) -containing diet ad libitum, for the designated period of time. PLX4720 (provided by Plexxikon Inc. under a Materials Transfer Agreement), was compounded in AIN-76A rodent diet (417 mg PLX4720/kg) by Research Diets, Inc. Tumors were deemed to have acquired resistance to BRAFi after demonstrating sustained growth (>10% increase in volume on 3 consecutive measurements) following initial growth stabilization for at least 3 weeks of treatment.
ERK protein detection
Whole Braf/Pten tumors were excised from untreated (0d) mice or mice treated with BRAFi for 4 days (4d), 55–60 days (60d), or 110–120 days (120d). Tumor lysates for Western blot were prepared with T-per buffer (ThermoFischer) supplemented with protease and phosphatase inhibitors (21). Protein was visualized as previously described (21) with anti-phospho-ERK (9101) and -total ERK (9102) antibodies (Cell Signaling, Beverly, MA).
Generation of BRAFi-resistant transplantable tumor line (BPR)
A tumor cell line, BPR, was derived from a single representative Braf/Pten tumor that had developed drug resistance following continual PLX4720 administration for 173 days. The cell line was generated using previously described methodology (21). A BRAFi-sensitive Braf/Pten cell line (termed BP), D4M.3A (21), was used as a control. BP and BPR cell lines were IMPACT tested (IDEXX BioResearch) for pathogens and were confirmed to be mycoplasma-free for the duration of the study using a TLR-2 activation-based assay (Mycoplasma Detection Kit, PlasmoTest). For transplantable tumor studies, mice were inoculated intradermally with 2.5×105 live BPR cells or BP cells.
EC50 determination
3×103 BPR or BP cells were cultured for 48h in complete DMEM-F12, and then switched to serum free media containing indicated concentrations of BRAFi in DMSO, or DMSO alone, for 48h. For in vitro experiments, PLX4032 (vemurafenib, Selleckchem) was used instead of PLX4720. Cell viability was assessed using alamarBlue (ThermoFischer) and fluorescence detected at 560nm excitation/590nm emission. Viability was normalized to DMSO controls, non-linear regression was used to generate a line of best fit, and EC50 values were calculated using Graphpad Prism software.
Flow cytometry
Tumors were harvested, weighed, minced, and digested for 45 minutes with gentle shaking at 37°C, in HBSS containing 7 mg/ml Collagenase D and 2 mg/ml DNase-I (Roche). Single cell suspensions were stained with antibodies including anti-CD45-APC-Cy7, anti-CD3-Vioblue, anti-CD8-PCP, anti-CD4-PCP, anti-Foxp3-FITC, anti-CD11b-APC, CD11b-PCP, anti-Gr-1-PE-Cy7, anti-Ly6C-PE, and anti-Ly6G-PE-Cy7 (BioLegend or eBioscience); anti-CCR2-APC or control Rat IgG2B-APC antibody were obtained from R&D. Cells were fixed/permeabilized using reagents from the Foxp3 staining kit (eBioscience). Flow cytometry was performed on a Miltenyi MACSQuant 10 Analyzer. Representative gating strategies are presented in Supplementary Fig. S1. To determine absolute cell number per gram of tumor, total numbers of cells in appropriate gates were multiplied by a correction factor for the proportion of the tumor sample run through the cytometer, and normalized for total tumor weight.
MDSC suppression assay
Tumors were digested as above, and CD11b+ cells were isolated magnetically by positive selection (Miltenyi). Purified CD11b+ cells were combined at indicated ratios with RBC-depleted C57BL/6 mouse splenocytes, and added to a 96 well plate pre-coated with anti-CD3 (5mg/ml, clone OKT3) and anti-CD28 (5mg/ml, clone PV-1) (BioXcell); to a final concentration of 3×105 splenocytes/200µl/well. Seventy-two hours later, supernatants were harvested and assayed for IFN-γ production by ELISA (R&D Systems).
Differential expression analysis
Gene expression analyses were performed on Agilent Whole Genome 8 × 60k DNA microarrays (see Supplemental material). Significance Analysis of Microarrays (SAM) (22) was used to identify significantly differentially expressed genes. For each comparison (untreated vs. treated samples at different time points), SAM version 4.0 was run as an Excel add-in using two class unpaired response type, logged (base 2) data and 500 permutations, with other parameters left unchanged. Genes with FDR q-value ≤10% were called significant. Expression data for significant genes were hierarchically clustered in Cluster 3.0 across genes and arrays using uncentered correlation similarity metric and average linkage clustering method and visualized in Java TreeView (23) version 1.1.6r4. Full gene expression data are available from NCBI GEO at GSE79206.
Functional enrichment analysis
Signatures of significant genes from SAM were analyzed via g:Profiler (24). Maximum size of functional category was set at 3500 genes and multiple testing correction was done using g:SCS, with other parameters left unchanged. In order to emphasize functional and pathway enrichment, regulatory motif and protein-protein interaction data were not included in the analyses.
Chemokine gene expression and protein detection
Total RNA was extracted (Qiagen RNeasy Kit) from whole tumors, or melanoma cells cultured in vitro with 300nM BRAFi (PLX4032, vemurafenib) or 300nM MEKi (PD0325901) for 48h, and translated to cDNA using a High Capacity RNA-to-cDNA kit (Applied Biosystems). Quantitative PCR was performed using prevalidated gene-specific primers (Ccl2, Cx3cl1, Cxcl3, Vegf) and master mix (Life Technologies) on a StepOne Plus Real-Time PCR System (Applied Biosystems). For protein detection, whole Braf/Pten tumors from untreated mice and mice treated for 5 days with BRAFi were excised, and protein extracts prepared as described for western blot. Total protein was quantified by Millipore Mouse 32-plex Cytokine/Chemokine Luminex, performed by the Immune Monitoring and Flow Cytometry Shared Resource (DartLab).
MDSC transwell recruitment assay
BPR cells were cultured in DMEM-F12 supplemented with FBS, then switched to serum-free media. Forty-eight hours later, BPR-conditioned medium (CM) was harvested. DMEM-F12 or CM (150 µl), with or without CCL2 blocking antibody (3µg/mL, Clone 123616, R&D Systems), was added to the bottom chamber of a 96-well transwell plate (Corning), and incubated overnight at 4°C. RBC-depleted splenocytes (5×105) resuspended in 50µL DMEM-F12 were placed in the top chamber of the transwell insert, and allowed to migrate for 5 hours at 37°C before cells from top and bottom chambers were harvested, stained for CD45, CD11b, Ly6C, and Ly6G, and populations were quantified by flow cytometry. To assess migration towards CCL2, 20ng/ml recombinant mouse CCL2 (Biolegend) was added to 0.5% FBS-enriched DMEM:F12 before incubating in the bottom transwell chamber overnight. RBC-depleted splenocytes resuspended in 0.5% FBS-enriched DMEM:F12 media were placed in the top chamber and allowed to migrate for 18 hours before being harvested and analyzed as described. Migration index was calculated as the number of cells with the indicated phenotype in the bottom chamber, divided by the total number of such cells in both chambers.
Experimental drug treatments and antibody depletions
All in vivo depletion or blockade antibodies were obtained from Bio-X-Cell. Anti-CCL2 (mAb clone 2H5) or hamster IgG isotype control was administered i.p. thrice weekly for a total of 6 doses (500 µg/dose for first 3 doses, 250 µg/dose for remaining doses). MEK1/2 inhibitor (MEKi) PD0325901 (Selleckchem) was compounded in aqueous vehicle (0.5% hydroxylpropyl cellulose, 0.2% Tween80), and administered at a dose of 25 mg/kg by oral gavage on the indicated days. Anti-PD-1 (250 µg/dose, mAb RMP1-14), and anti-CTLA-4 (100 µg/dose, mAb clone 9H10) were co-administered i.p. every 2–4 days for the first 5 doses beginning on indicated days, and then weekly for the remainder of the experiment. Anti-Gr-1 (mAb RB6-8C5, 200 µg/dose) antibody was administered i.p. twice weekly, beginning on indicated days. CCR2 antagonist (SC-202525, Santa Cruz Biotechnology) was administered i.p. at 90 µg/dose (in saline + 10% DMSO and 10% Tween80), every 2–3 days beginning on indicated days.
Statistical analyses
Statistical differences in cell subset abundances were determined using flow cytometry data and unpaired 2-tailed t-test, unless otherwise stated. Statistical differences in tumor sizes between groups, over time, were calculated by 2-way ANOVA with Tukey’s multiple comparisons test to assess differences at individual time points. Log-Rank (Mantel-Cox) analysis of Kaplan Meier data was used to assess differences in onset of resistance between experimental groups. Statistical analyses were performed using GraphPad Prism software.
RESULTS
Autochthonous Braf/Pten melanomas acquire resistance to the BRAF-inhibitor PLX4720
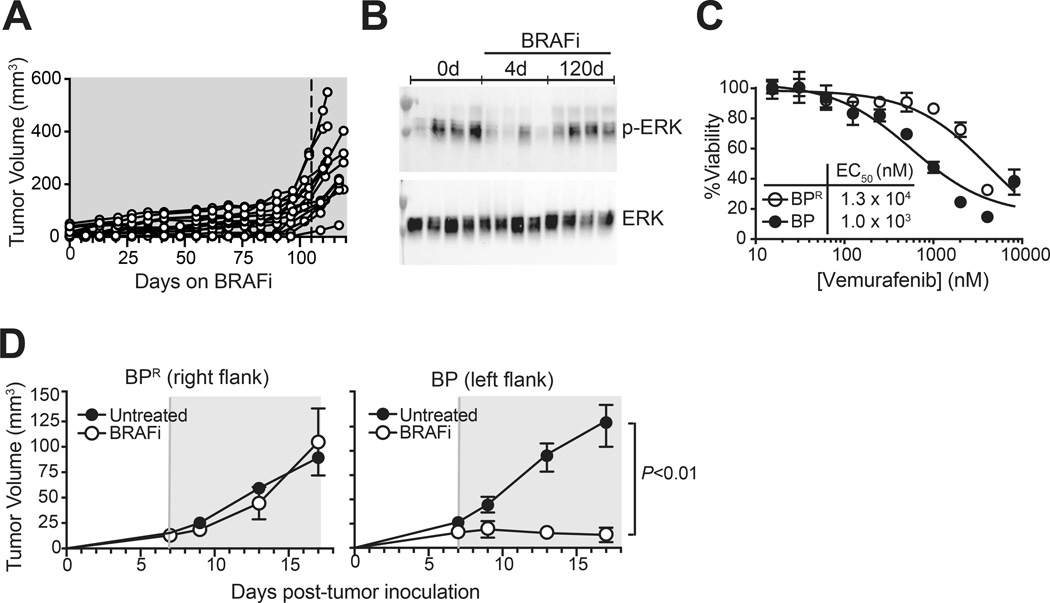
We previously documented that the vemurafenib analog PLX4720 (BRAFi) halts Braf/Pten tumor progression (7). After a prolonged period of growth arrest, Braf/Pten tumors almost universally escaped drug-mediated control (Fig. 1A). Defining the onset of resistance by the day at which tumor growth had increased by at least 10% on each of three prior consecutive measurements, resistance was attained in 45 out of 56 total mice observed for at least 110 days (mean onset 105 ± 23 days). Thus resistance was reproducible and occurred with predictable kinetics.
Figure 1. Braf/Pten tumors acquire resistance to BRAF-inhibitors.
(A) Mice bearing autochthonous Braf/Pten melanomas induced in syngeneic skin grafts (see Methods) received BRAFi (PLX4720) containing diet, and tumor growth was monitored. Data were pooled from 3 independent experiments each involving 4 or 5 mice (14 total). Dotted vertical line represents mean onset of resistance (105 days), calculated based an expanded cohort of 56 total mice. (B) Western blot was performed to detect phospho-ERK1/2 and total-ERK1/2 in lysates from tumors established and treated as in (A), for the indicated numbers of days. Lanes represent tumors from individual mice (4 per group). (C) Viability assay was used to determine EC50 of the BRAF-inhibitor vemurafenib against BPR versus BP melanoma cell lines (derived from BRAFi-resistant and sensitive Braf/Pten tumors, respectively). Symbols represent mean +/− standard deviation (SD) of 4 replicate wells per point. (D) BPR and BP tumors were grown on contralateral flanks of C57BL/6 mice, and BRAFi (PLX4720 in chow) was administered as indicated (treatment period shaded in grey). Symbols and error bars represent mean +/− SEM of 5 mice per group. Statistical significance was calculated by 2-way ANOVA. Data in B, C, and D are representative of at least 2 independent experiments with similar results.
After 120 days of continuous BRAFi treatment, resistant tumors were harvested for further characterization. Consistent with reactivation of MAPK pathway signaling, Western blot analysis showed restored pERK1/2 levels in resistant tumors, as compared to BRAFi-sensitive (4 day-treated) tumors (Fig. 1B). Further, a cell line (BPR) generated from a resistant tumor was one log-fold less sensitive to BRAFi in vitro as compared with the control BRAFi-sensitive BP cell line (21) (Fig. 1C). BPR cells also engrafted and grew as tumors in B6 mice, where their progression was unperturbed by BRAFi treatment, again in contrast to BP tumor controls (Fig. 1D). Thus prolonged BRAF-inhibition conferred resistance to Braf/Pten tumors that was cell-intrinsic, heritable, and associated with MAPK pathway reactivation.
The BRAF-inhibitor resistant tumor microenvironment is dominated by MDSCs
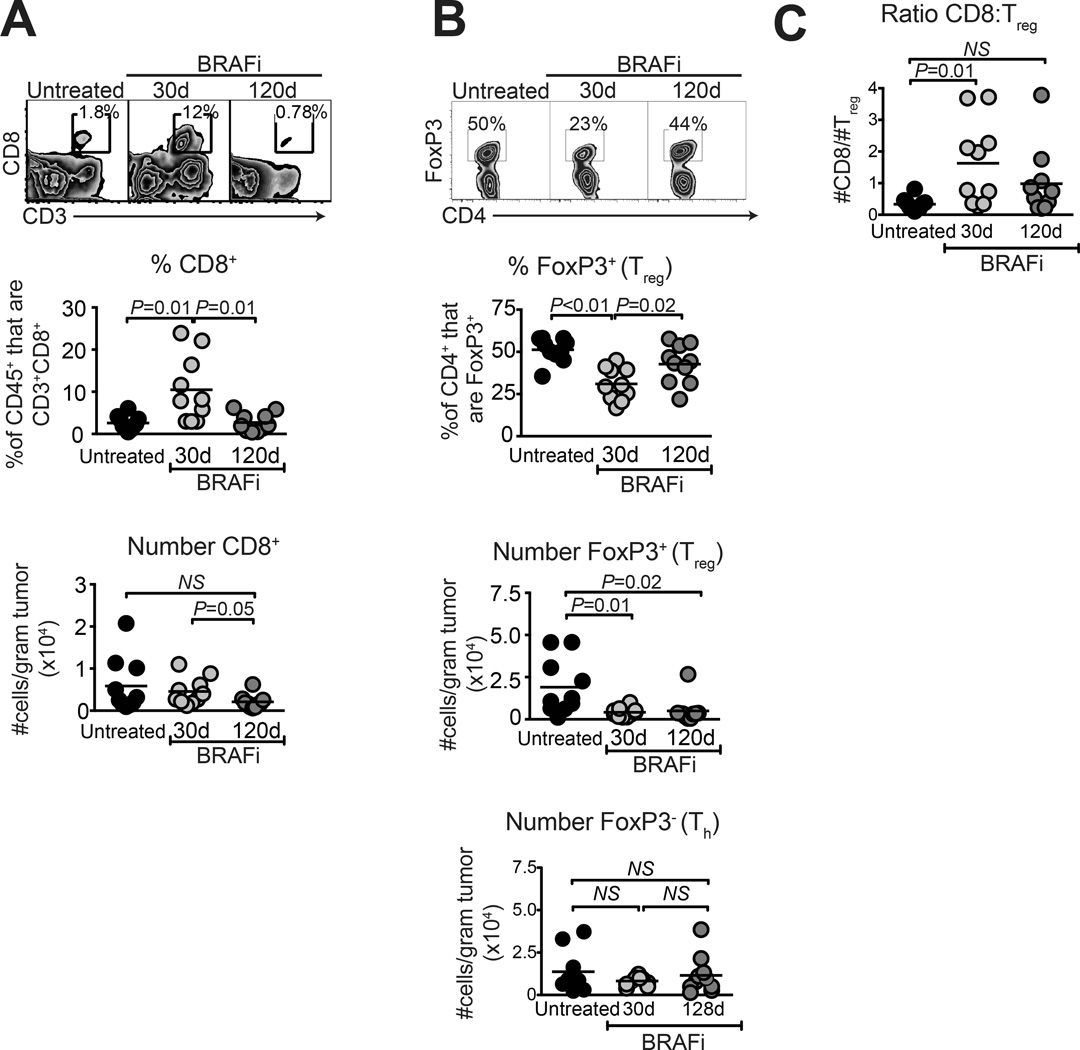
We previously published that 10-day BRAFi treatment leads to loss of intratumoral Tregs and MDSCs, and an increase in proportions of CD8 T cells (7). To ascertain long-term effects of therapy, mice underwent 30 or 120 days of treatment, and tumors were analyzed by flow cytometry. Tumors were size-matched (Supplementary Fig. S2) to eliminate effects of differential tumor size, as previously reported (7). On day 30, CD8 T cells were elevated by proportion out of total CD45+ cells, but not by absolute number (Fig. 2A). In contrast, CD4+Foxp3+ Tregs were decreased by proportion (of CD4+ cells) and by absolute number (Fig. 2B). Thus T cell alterations previously observed after 10d of BRAFi treatment (7) persisted during a full month of continued drug responsiveness. However, by day 120, proportions and absolute numbers of CD8 T cells were normalized to levels found in untreated tumors (Fig 2A). While Tregs significantly increased by proportion out of CD4 T cells, they were not restored by absolute number on day 120, consistent with a slight but insignificant increase in absolute numbers of Foxp3−CD4+ Th cells in the same tumors (Fig. 2B). Thus the overall CD8:Treg ratio at resistance was variable and not significantly different from untreated or day 30 treated tumors (Fig. 2C).
Figure 2. CD8 T cell and Treg proportions are restored to normal in BRAFi-resistant melanomas.
Braf/Pten melanomas were induced as in Fig. 1A, and mice received BRAFi-containing diet for the indicated times. Tumors were analyzed for proportion of (A) CD8+ T cells, pre-gated on live CD45+ cells and (B) FoxP3+ (Treg) and FoxP3− (Th) cells pre-gated on live CD45+CD3+CD4+ cells. Absolute numbers of each cell type (per gram tumor weight) are depicted in lower panels. (C) Ratios of intratumoral CD8 T cells to Treg cells were calculated for each sample. Data were pooled from two independent experiments each involving 4–5 mice per group; 30d samples taken on day 27 or 30, and 120d samples were taken on day 117 or 128, relative to starting BRAFi. All points represent individual mice and horizontal lines depict means; statistical significance was calculated by unpaired 2-tailed t-test.
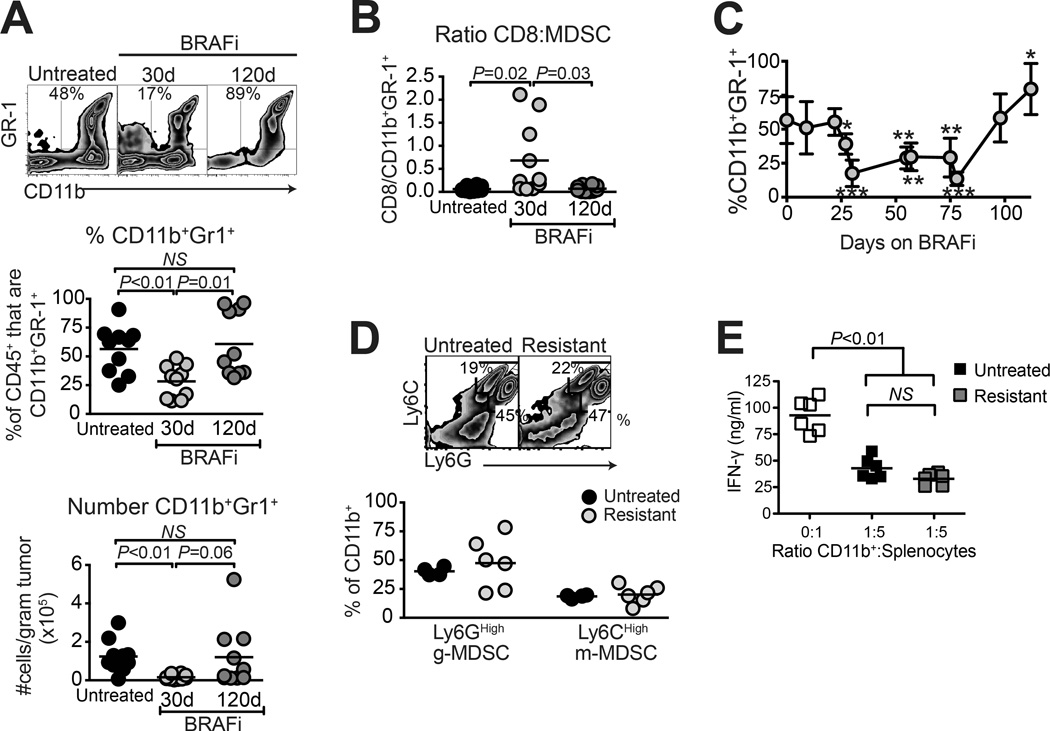
MDSCs (CD11b+Gr-1+) were also durably reduced after 30 days of BRAFi treatment, by proportion and absolute number (Fig. 3A), accounting for the apparent increase in CD8 T cell proportions within the CD45+ gate (Fig. 2A). However, in contrast to Tregs, MDSCs were entirely restored at resistance (Fig. 3A). As a result, favorable CD8:MDSC ratios were lost in resistant tumors (Fig. 3B). Analyses at intermediate time points showed that MDSC proportions were restored to untreated frequencies as early as day 98 (Fig. 3C), which slightly preceded the mean onset of rapid tumor growth (Fig. 1A). Furthermore, CD11b+ cells in resistant tumors were indistinguishable from those in untreated tumors with respect to Ly6CHigh monocytic and Ly6GHigh granulocytic subsets (Fig. 3D), and with regards to in vitro suppressive function (Fig. 3E). Thus, immunosuppressive MDSCs were efficiently restored in BRAFi-resistant melanomas.
Figure 3. Myeloid derived suppressor cell populations are restored in BRAFi-resistant melanomas.
Braf/Pten melanomas were induced as in Fig. 1A, and mice received BRAFi-containing diet for the indicated times. Tumors were analyzed for (A) Proportion and total number of CD11b+Gr-1+ cells, pre-gated on live CD45+ cells, and (B) ratio of CD8 T cells to CD11b+Gr-1+ cells. Samples were the same as those used in Fig. 2; significance was calculated by t-test. (C) Proportions of intratumoral CD11b+Gr-1+ cells were analyzed over time; data were pooled from 3 experiments, and significance was calculated by 1-way ANOVA (* P<0.05, ** P<0.01, ** P<0.001, *** P<0.0001). (D) Untreated and BRAFi-resistant tumors (129 days BRAFi) were analyzed for expression of Ly6G and Ly6C (gated on live CD45+CD11b+ cells) to differentiate g-MDSC and m-MDSC subsets. (A–D) Data points represent individual mice, and horizontal lines depict means. (E) In vitro assay was conducted using purified CD11b+ cells from untreated or BRAFi-resistant tumors (129 days BRAFi) as suppressors of IFN-γ production by activated splenocytes. Points represent individual wells; CD11b+ cells in each group were pooled from 2 tumors. Data in each panel were combined from two independent experiments each involving 4–5 mice/group; mice in A and B are the same as those shown in Fig. 2.
BRAFi-responsiveness and resistance reciprocally govern the tumor immune gene signature
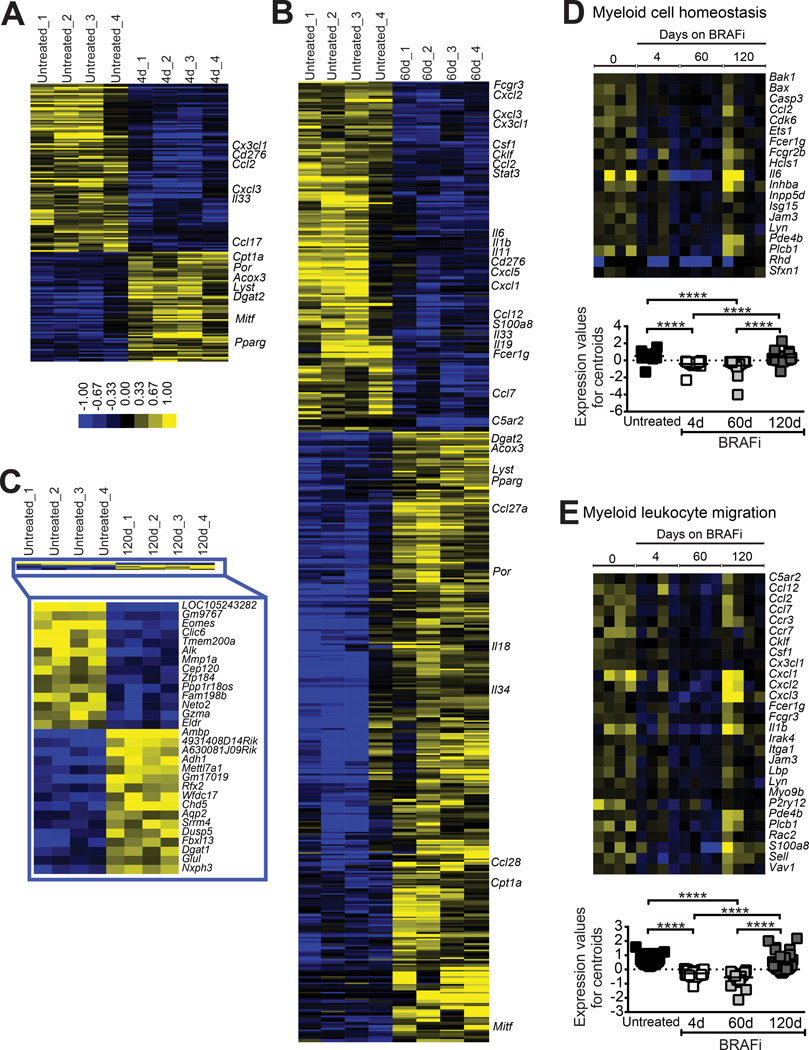
The above findings led to the hypothesis that tumor-derived factors that support MDSC occupancy in the tumor are decreased by BRAFi, but then restored at resistance. To identify candidate molecular mediators, DNA microarray analyses (Significance Analysis of Microarrays; SAM) were conducted on size-matched tumors (Fig. S1) from untreated mice (baseline), versus those treated with BRAFi for 4, 60, or 120 days.
On day 4 of treatment, a total of 885 transcripts were significantly differentially expressed (false discovery rate (FDR) < 10%) with a majority downregulated (537 transcripts down vs. 348 up) (Fig. 4A and Supplementary Fig. S3). Functional enrichment (g:Profiler) analysis indicated that the most significantly downregulated gene sets were involved in oncogenic processes (e.g. cell cycle and cell division). Regarding factors that could potentially recruit MDSCs, Ccl2, Cx3cl1, and Cxcl3, were among the few genes encoding secreted immune factors that were significantly downregulated (Fig. 4A), whereas other chemokine transcripts, including Ccl3, Ccl4, and Ccl5, were unaltered (Supplementary Fig. S3). Transcript for Cd276 (B7-H3), an inhibitor of T cell activation, was also reduced. No chemokine or cytokine transcripts were significantly upregulated on day 4, with enriched transcripts primarily involved in lipid metabolism and biosynthesis (e.g. Cpt1A, Por, Acox3, Pparγ; Fig. 4A). The melanocyte lineage transcription factor Mitf was also upregulated (Fig. 4A).
Figure 4. Evolution of the tumor immune gene signature during the acquisition of BRAFi resistance.
Braf/Pten tumors (generated as in Fig. 2) were analyzed by whole genome microarray, with SAM used to identify significantly differentially expressed genes (FDR ≤ 10%) comparing tumors from untreated mice to those that had been treated with BRAFi for 4 days (A), 60 days (B), or 120 days (C). Color bar refers to median-centered log2 fold change. In A and B, soluble immune mediators are highlighted, as well as candidate genes involved in tumor metabolic processes. In C, all significantly altered genes are listed. (D, E), g:Profiler tool was used for functional enrichment analysis to identify over-representation of Gene Ontology terms (biological processes) downregulated on d60 relative to untreated, and analyzed across all time points. Enriched gene sets included Myeloid Cell Homeostasis (D) and Myeloid Leukocyte Migration (E). Significance was calculated by Kruskal-Wallis test followed by Dunn’s multiple comparisons test, with **** denoting P<0.0001.
On day 60, transcripts for Ccl2, Cxcl3, and Cx3cl1 were still significantly decreased, with many additional changes apparent (1,121 down vs. 1,940 up; Fig. 4B and Supplementary Fig. S3). While cell cycle genes were still significantly downregulated on day 60, we also observed the presence of functional gene sets related to the immune response (e.g. defense response, inflammatory response and cytokine production). Transcripts for many additional chemokine/cytokine genes associated with myeloid cell recruitment and differentiation were downregulated on day 60 (e.g. S100a8, Cxcl2, Csf1, Cklf, and Cxcl5; Fig. 4B) and myeloid cell-associated transcripts (e.g. Fcgr3, Stat3, and Il6) were also decreased. Similar to day 4, g:Profiler analysis still indicated strongest increases in lipid metabolism and biosynthetic processes on day 60.
Strikingly, BRAFi-resistant (day 120) tumors exhibited very few significant differences as compared with baseline (14 transcripts down vs. 16 up; Fig. 4C). Both Eomes and GzA were significantly downregulated, potentially reflecting a decrease in immune cell effector function. Otherwise, both overall and immune-specific gene signatures were highly similar to untreated tumors. Accordingly, functional enrichment analysis indicated that global myeloid cell homeostasis and myeloid leukocyte migration gene signatures, which were significantly reduced on days 4 and 60, were restored to normal by day 120 (Fig. 4D and E). Thus while BRAF-inhibition induced remodeling of the tumor immune gene signature, baseline profiles favoring myeloid cell recruitment and homeostasis reappeared at resistance.
MAPK pathway reactivation drives the repopulation of MDSCs in BRAFi-resistant tumors
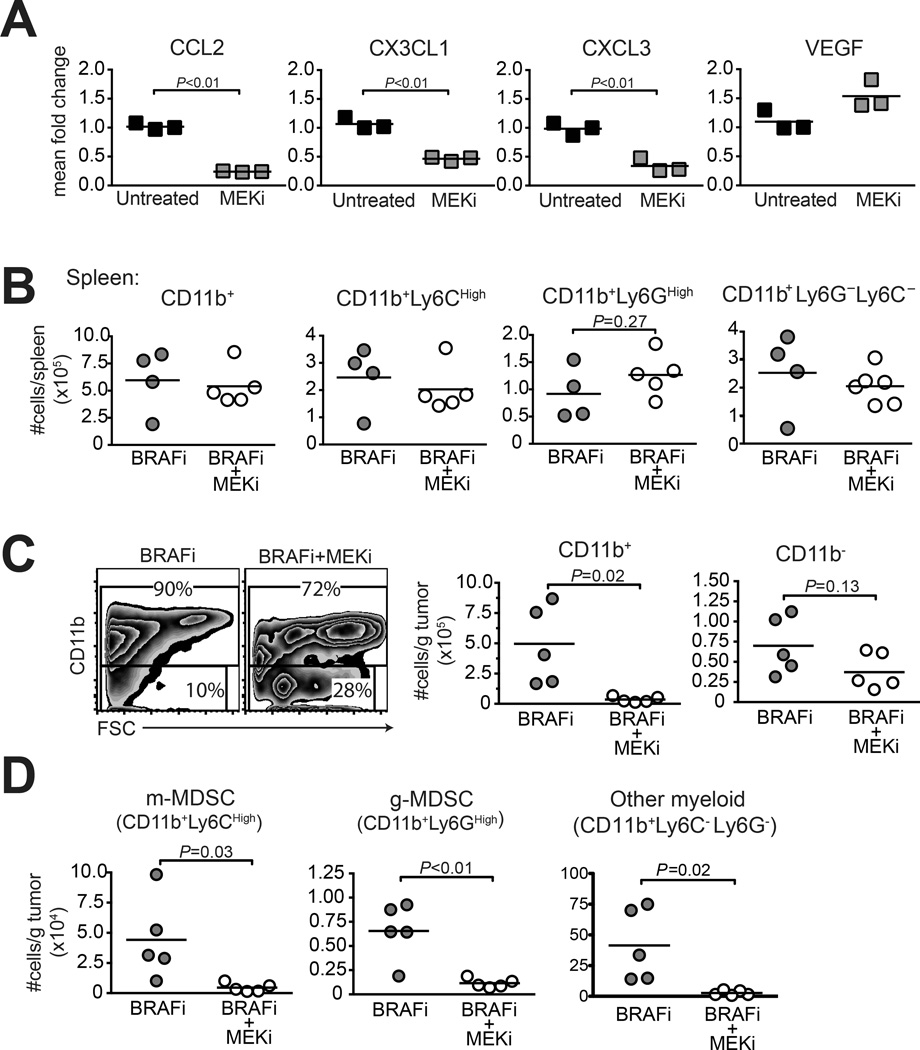
Microarray revealed the restoration of known mediators of myeloid cell recruitment (Fig. 4), concurrent with MAPK pathway reactivation as indicated by increased pERK1/2 (Fig. 1B). To determine if MAPK signaling restored myeloid cell-recruiting chemokine expression in BRAFi-resistant melanoma cells, BPR cells were treated with a MEK1/2 inhibitor (MEKi), and gene expression was analyzed. MEKi significantly reduced expression of Ccl2, Cx3cl1, and Cxcl3, but not Vegf (an irrelevant control which was also unchanged by microarray) (Fig. 5A). Accordingly, treatment of mice bearing established BPR tumors with MEKi, while having no effect on any CD11b+ cell subset in spleen (Fig. 5B), significantly and selectively reduced proportions and numbers of CD11b+ cells in tumors (Fig. 5C). Within this population, MEKi similarly reduced numbers of Ly6Chi monocytic (m)-MDSCs, Ly6Ghi granulocytic (g)-MDSC, and CD11b+ Ly6C−Ly6G− cells (Fig. 5D). Thus inhibition of MEK reduces chemokines associated with MDSC recruitment, and concurrently impairs global myeloid cell accumulation in BRAFi-resistant melanomas.
Figure 5. MEKi reduces intratumoral MDSC accumulation and suppresses the growth of BRAFi-resistant melanomas.
(A) qRT-PCR assessment of chemokine gene expression in BPR cells treated with 300 nM MEKi (PD0325901) or vehicle for 48h. Bars represent mean ±SEM of 3 wells. Statistical significance was calculated by unpaired t test. (B) BPR tumor-bearing mice receiving BRAFi treatment starting on d9 were co-treated with MEKi (versus vehicle) on days 16, 19, and 21. Spleen was analyzed by flow cytometry on day 22; absolute numbers of myeloid cells (CD11b+) and sub-populations (CD11b+Ly6CHigh, CD11b+Ly6GHigh, or CD11b+Ly6G−Ly6C−) were calculated and normalized per spleen. (C–D) Tumors taken from mice treated as in panel B were analyzed by flow cytometry for (C) total myeloid (CD11b+), and non-myeloid (CD11bneg) cells, and (D) total m-MDSCs (CD11b+Ly6CHigh) versus g-MDSCs (CD11b+Ly6GHigh); normalized per gram tumor weight. Points represent individual mice, horizontal lines depict means, and statistical significance was calculated by t test. Each panel depicts representative data of 2–3 independent experiments.
The CCL2/CCR2 axis restores monocytic MDSCs in resistant tumors
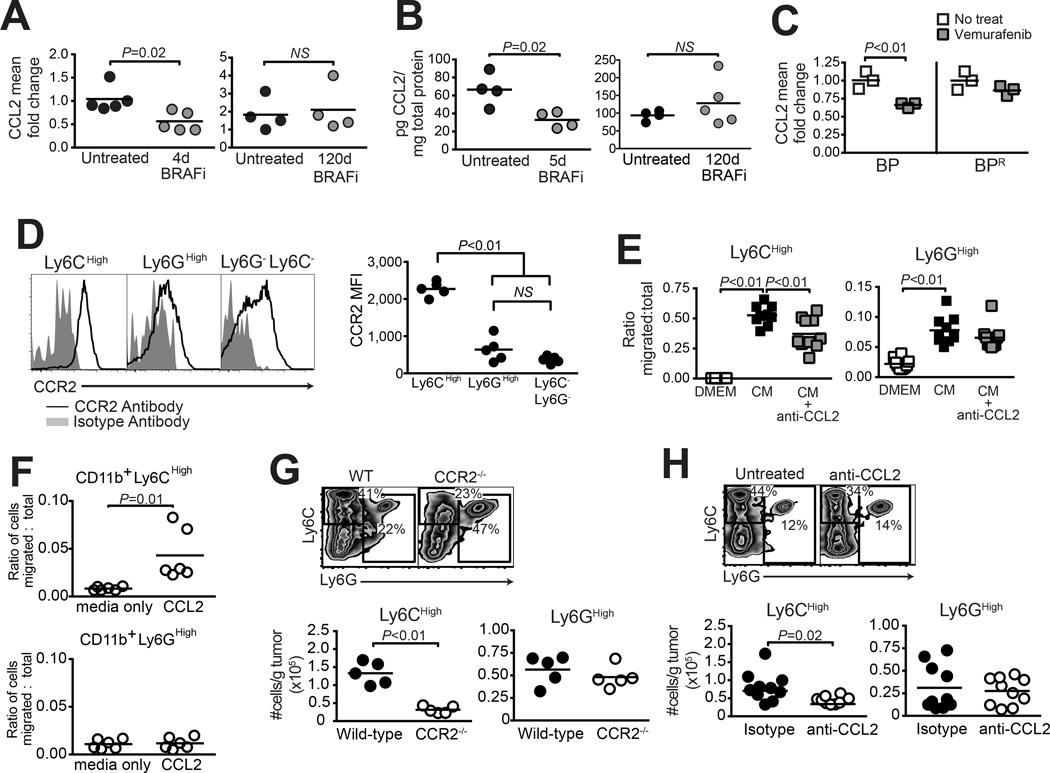
As Ccl2 gene expression was downregulated by BRAFi, and restored at resistance downstream of MEK, we next evaluated its role in MDSC recruitment to BRAFi-resistant tumors. To validate the CCL2 transcriptional signature, we confirmed the CCL2 reduction and restoration by real-time PCR (Fig. 6A), and by ELISA (Fig 6B). Additionally, BP melanoma cells, but not resistant BPR cells, downregulated CCL2 expression in response to BRAF-inhibition (Fig. 6C).
Figure 6. The CCL2/CCR2 axis mediates the accumulation of monocytic MDSCs in BRAFi-resistant melanomas.
(A–B) Braf/Pten tumors, induced as in Fig. 2 and treated with BRAFi for the indicated times, underwent (A) qRT-PCR analysis to detect whole tumor CCL2 gene expression, and (B) Luminex to determine CCL2 protein concentration. (C) qRT-PCR analysis of CCL2 expression in BP (sensitive) and BPR (resistant) melanoma cells in vitro +/− vemurafenib treatment for 48h. (D) Flow cytometric analysis of CCR2 expression on CD11b+ Ly6CHigh versus Ly6GHigh and Ly6G−Ly6C− subsets in transplanted BPR tumors. (E) Transwell assessment of migration of CD11b+Ly6CHigh (left) or CD11b+Ly6GHigh (right) MDSC subsets towards BPR tumor cell-conditioned medium (CM) +/− anti-CCL2-blocking antibody. (F) The migration of CD11b+Ly6CHigh (top) or CD11b+Ly6GHigh (bottom) MDSC subsets toward CCL2 was assessed by a transwell assay. Recombinant mouse CCL2 was added to 0.5% FBS-enriched DMEM:F12 in the bottom. Splenocytes derived from BPR-bearing mice were added to the top chamber and allowed to migrate for 18 hours before analyzing by flow cytometry. (G–H) Flow cytometric analysis of CD11b+ MDSC subsets in BPR tumors growing on (G) wild-type versus CCR2−/− mice, or (H) wild-type mice treated with anti-CCL2 blocking antibody (vs. isotype control) thrice weekly starting on day 0. Both experiments (G–H) were analyzed on day 11, with flow pre-gated on live CD45+ cells. Data in (H) are pooled from two identical experiments. In all experiments, symbols represent individual mice, squares represent individual wells, horizontal lines depict means, and statistical significance was calculated by unpaired t-test (except in D, which used paired t-test). Each experiment was conducted at least twice with similar results.
CCR2, the cognate receptor for CCL2, is a known marker of m-MDSCs (25). As expected, CCR2 was expressed at significantly higher levels on Ly6CHigh m-MDSCs, compared with Ly6GHigh g-MDSCs or Ly6C/Ly6G double negative myeloid cells, in BPR tumors (Fig. 6D). Furthermore, conditioned medium from BPR cells was sufficient to drive recruitment of both m-MDSCs and g-MDSCs in transwell assays. However only Ly6CHigh m-MDSC migration were blocked by neutralization of CCL2 (Fig. 6E), consistent with the finding that m-MDSCs, but not g-MDSCs, migrated in response to recombinant CCL2 (Fig. 6F). Finally, when BPR tumors were transplanted onto Ccr2−/− mice, intratumoral accumulation of m-MDSCs, but not g-MDSCs was significantly reduced as compared to wild type mice (Fig. 6G). A comparable reduction in m-MDSCs was observed in Ccr2−/− mice bearing BRAFi-sensitive autochthonous melanomas (Supplementary Fig. S4), indicating that similar mechanisms govern MDSC recruitment during initial tumor growth and at resistance. Importantly, antibody-mediated CCL2 blockade in BPR tumor-bearing wild type mice also led to a significant reduction in only the Ly6CHigh m-MDSC subset in tumors (Fig. 6H). Despite this, BPR tumor growth was not impaired in Ccr2−/− mice or mice receiving anti-CCL2 blocking antibody (Supplementary Fig. S4). Therefore, restored CCL2 production by BRAFi-resistant melanomas mediated the recruitment of CCR2-expressing monocytic MDSCs to the TME, although blocking this axis alone was insufficient to restrain tumor growth.
Checkpoint blockade cooperates with MDSC depletion + CCR2 antagonism to prevent BRAFi resistance
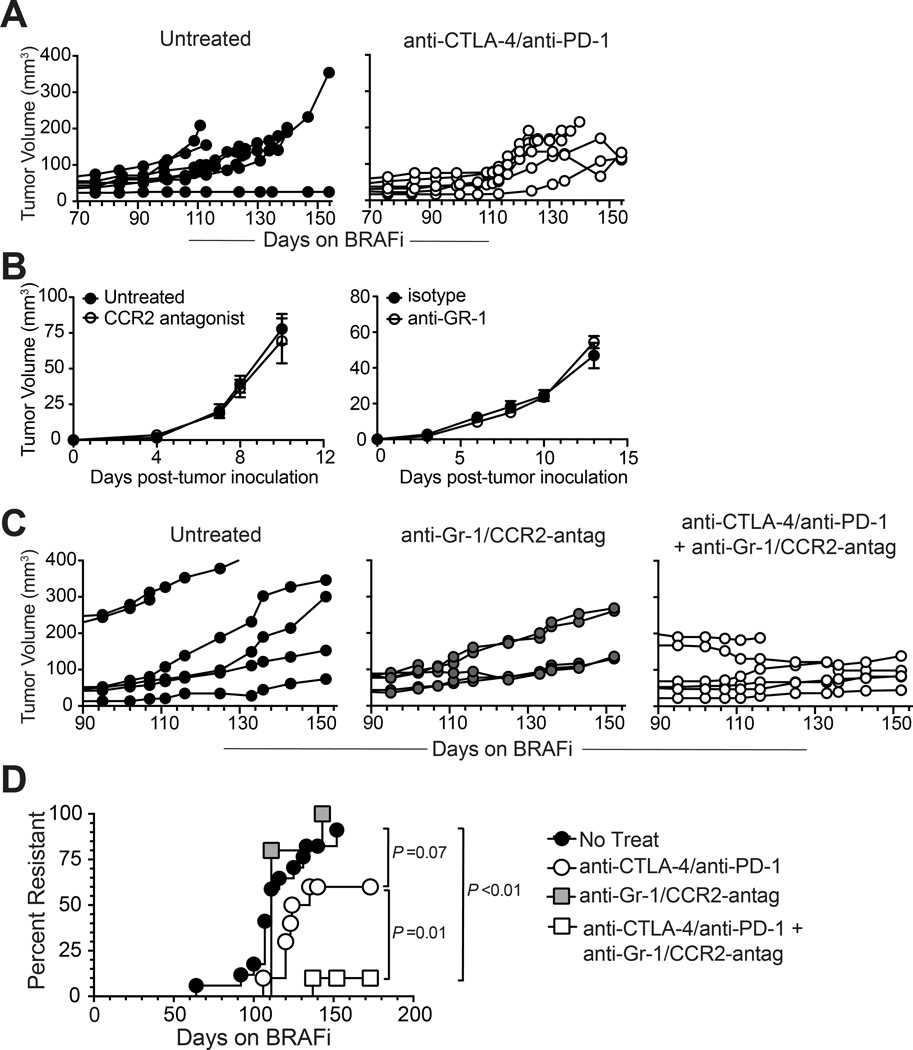
The re-establishment of immunosuppressive myeloid cells at resistance suggested that MDSCs could represent a barrier to effective immunotherapy of BRAFi-resistant tumors. Indeed, dual checkpoint blockade immunotherapy (anti-CTLA-4 + anti-PD-1), administered while tumors were still responding to BRAFi, did not reduce tumor size or delay the onset of BRAFi resistance (Fig. 7A), indicating that tumors were co-resistant to immunotherapy.
Figure 7. MDSC depletion/blockade synergizes with checkpoint blockade immunotherapy to prevent the development of BRAFi resistance.
Tumor growth in autochthonous Braf/Pten tumor-bearing mice receiving ongoing long-term treatment with BRAFi and various immunotherapeutic interventions; (A) Mice were treated +/− dual checkpoint blockade (anti-PD-1 + anti-CTLA-4 mAbs); data are pooled from two experiments each involving 3 mice/group. Checkpoint blockade was initiated on day 50 relative to BRAFi-initiation, and continued as described in Methods. (B) BPR tumor growth in wild-type mice treated with 90ug CCR2 antagonist every 2 days, beginning on day 0 (left), or treated with 200ug anti-GR-1 antibody on day 0 and day 3 versus isotype (right). (C) Mice were treated +/− MDSC depletion/blockade (anti-Gr-1 + CCR2 antagonist SC-202525), or dual checkpoint blockade + MDSC depletion/blockade. All treatments were initiated on day 85 and continued weekly as described in Methods. Dual checkpoint blockade (starting day 52) + MDSC depletion/blockade (starting day 89) was shown to be equally as effective in a second experiment. Tumor growth curves depict individual mice. (D) Onset of resistance in all mice studied receiving long-term BRAFi and immunotherapeutic interventions (n=18 mice in four combined experiments for no treatment, n=6 mice in a single experiment for MDSC depletion/blockade, n=10 mice in three experiments for dual checkpoint blockade, n=10 mice in two experiments for MDSC depletion/blockade + dual checkpoint blockade). Statistical significance was calculated by Mantel-Cox log rank test.
To separately address a role for MDSCs in promoting BRAFi resistance, MDSCs were depleted by a combination of anti-Gr-1 mAb, which eliminated g-MDSCs, and the CCR2-antagonist SC-202525, which selectively reduced intratumoral accumulation of m-MDSCs (Supplementary Fig. S5). However neither anti-Gr-1 mAb nor CCR2 antagonist treatment alone impaired the growth of BRAFi-resistant BPR tumors (Fig. 7B). Similarly, continuous administration of anti-Gr-1 in addition to CCR2-antagonist, beginning prior to drug resistance, did not impair the outgrowth of BRAFi-resistant tumors (Fig. 7C). MDSC depletion was also ineffective at regressing BRAFi-sensitive tumors that were responding to PLX (Supplementary Fig. S6). This apparent lack of anti-tumor immunity was consistent with the observation that BRAFi responsiveness and resistance occurred with similar kinetics in wild type versus Rag1−/− and Cd8−/− mice (Supplementary Fig. S7), indicating that host-endogenous immunity is insufficient to restrain the outgrowth of BRAFi-escape variant tumors. However, when anti-CTLA-4 + anti-PD-1 and anti-Gr-1 + CCR2-antagonist therapies were combined, the development of resistance to BRAFi was abrogated (Fig. 7C and D). Thus while melanoma cell-intrinsic BRAFi resistance mechanisms restored intratumoral MDSCs, targeting these cells eliminated co-resistance to immunotherapy.
DISCUSSION
Mechanisms of tumor resistance to oncoprotein inhibitors have been extensively characterized from the perspective of cancer cell-intrinsic properties (26). However, the immunology of drug resistance has remained poorly understood. The present studies establish that inhibitor resistance unfavorably alters the immune composition of the tumor microenvironment. Using a mouse model of in situ BRAFi resistance development, we show that resistance drives the restoration of immunosuppressive myeloid cells in tumors. This is accompanied by a universal shift in the transcriptional landscape of the tumor, and the reestablishment of myeloid cell chemotaxis and homeostasis signatures. Our data highlight the key roles for CCL2 as an intermediate, and MEK as a master regulator, of MDSC repopulation in BRAFi-resistant melanomas. These findings illustrate an intricate and evolving balance between cancer cells and immunosuppressive cells during long-term oncoprotein inhibition, and establish how rational manipulation of the tumor microenvironment can enable effective immunotherapy against aggressive, inhibitor-resistant cancers.
Our studies relied on an autochthonous mouse melanoma model that recapitulates several features of human disease. While these tumors do not regress as a result of treatment (7,27), tumor growth was stabilized by BRAFi for at least three months prior to the onset of resistance. This latency period is longer than reported for the aggressive inducible iBIP (BrafV600E Ink/Arf−/− Pten−/−) mouse model (28), but similar to that of a BRAFV600E transposon mutagenesis model (29), and within the range observed for melanoma patients (1,2). The synchronicity and reproducibility of resistance among inbred Braf/Pten mice is reminiscent of pan-resistance in patients, where multiple tumors escape BRAFi simultaneously (30). Restored p-ERK indicates a likely role for MAPK pathway reactivation as a mechanism of resistance, as is common in human tumors (3,4). Finally, the presence of immunosuppressive myeloid cells in resistant Braf/Pten tumors mirrors recent findings in MAPKi-resistant human melanomas, a fraction of which exhibit increased expression of monocyte and M2 macrophage-associated genes (12). Thus, resistance to BRAFi in Braf/Pten mice models multiple features of human disease.
Despite insignificant Braf/Pten tumor growth throughout more than 90 days of continuous BRAFi treatment, we detected significant alterations in the tumor immune microenvironment and immune gene signatures, predominantly with respect to MDSCs. Microarray analyses enabled us to resolve a small group of chemokines that we validated as having been produced by melanoma cells, regulated by MAPK signaling, and restored at resistance. Of these, we demonstrated CCL2 as a key mediator of monocytic MDSC recruitment to resistant tumors. While the CCL2/CCR2 axis was previously implicated as a determinant of acute responsiveness to BRAFi, in mice bearing transplantable melanoma, its role in recruiting MDSCs was not explored (8). Our finding that CCR2 is required for early m-MDSC entry into Braf/Pten tumor is consistent with the idea that CCL2 down regulation is responsible for m-MDSC loss following initial BRAFi-treatment. Interestingly, recent studies show that human melanoma cell lines increase production of CCL2 upon development of resistance to BRAFi, and that CCL2 is increased in serum of melanoma patients after extended vemurafenib treatment, where it is associated with poor clinical response (31). Roles for other MAPK-regulated chemokines identified herein have yet to be defined. However, there is precedence for CXCR2 (the cognate receptor for CXCL3) mediating granulocytic MDSC trafficking into tumors (32), and for CX3CR1 (the cognate receptor for CX3CL1) as a marker of some tumor-infiltrating MDSCs (33). Further investigation of these chemokines and other myeloid cell-supporting factors should provide greater insights regarding mechanisms of MDSC repopulation, as well as additional targets for immunotherapeutic intervention.
An important finding of the present work is the therapeutic cooperation between MDSC elimination and dual checkpoint blockade immunotherapy for the prevention of BRAFi escape. While MDSCs are known to serve various tumor-promoting roles (34), synergy with negative checkpoint blockade strongly suggests a role for T cell immunity in the present setting. Contributions of individual drugs need be further dissected, as a single checkpoint-blocking antibody combined with a single MDSC-targeting therapy could also yield therapeutic benefits. Our finding that BRAFi-resistant tumors did not respond to immune checkpoint blockade is consistent with the recent report that transcriptional signatures known to associate with MAPKi-resistance also associated with innate anti-PD-1 resistance (35). In fact, CCL2 restoration in BRAFi-resistant tumors may contribute to their non-responsiveness to dual checkpoint blockade, as CCL2 was reported to associate with innate anti-PD-1 resistance (35,36). This cross-resistance to anti-PD-1 therapy could hamper the efficacy of immune checkpoint blockade as second-line therapy in the absence of MDSC-targeting strategies. Our finding that CCL2 blockade impairs mMDSC recruitment without altering tumor growth does not necessarily implicate gMDSCs as dominant mediators of immune suppression. In fact co-depletion of both MDSC subsets was insufficient to impair growth of BRAFi-resistant tumors. Whether this implies the function of other less abundant cell types at resistance (e.g. Tregs) remains to be seen. More immunogenic tumor variants may also be needed to observe an effect of MDSC depletion, as our data from Rag1−/− and CD8−/− mice do not support host-endogenous immune surveillance against BRAFi-resistant tumors or even against tumors responding to initial BRAFi treatment, as we have previously shown (7). We do show that total MDSC depletion improves checkpoint blockade immunotherapy, although further studies will be required to determine if one MDSC subset dominates immune suppression in the immunotherapy setting. Finally, our data that combination immunotherapy (anti-CTLA-4 + anti-PD-1 + anti-Gr-1 + CCR2-antagonist) works uniquely by postponing resistance, but not by regressing tumors, lends support to the idea that BRAFi escape variants may be more susceptible to immunotherapy. Indeed checkpoint blockade immunotherapies function better in settings of higher tumor mutational load (37). Whether BRAFi resistance increases the number of immunogenic mutations in melanoma tumors remains to be seen.
In the clinic, melanoma patients with extensive, symptomatic disease are often started on first-line combination BRAFi + MEKi (18). The present studies do not directly address dual inhibitor resistance, however they provide a proof-of-principle understanding of how MDSC repopulation can occur in resistant tumors. Moreover, because frequent mechanisms of co-resistance to BRAFi+MEKi involve MAPK pathway reactivation (6), one would speculate that many tumors with resistance to dual inhibitors would rely on similar mechanisms of MDSC recruitment as demonstrated herein. Tumors with alternate resistance mechanisms will require further study to determine if MDSC recruitment is a general feature of resistance, or whether it applies only to resistance driven by MAPK pathway reactivation. As clinical studies are underway to determine how and when to optimally integrate immunotherapies for inhibitor treated patients (19,38), our findings indicate that MDSC depleting drugs may restore vulnerability to immune checkpoint blockade therapies. The efficacy of this approach might be altered in a setting of resistance to combined BRAFi and MEKi, as MEKi has been shown to improve immunotherapeutic effectiveness (17), but can also directly inhibit T cell function (39). The absence of reliable biomarkers to predict BRAFi resistance is a related clinical obstacle, although the present studies suggest that certain MAPK-driven chemokines, or MDSCs themselves, may serve as useful predictors of resistance. Despite these challenges, strategies to detect and eliminate MDSCs should better advance the complicated task of combining oncoprotein inhibitors and immunotherapies for cancer patients.
Supplementary Material
Acknowledgments
The authors thank M. Ernstoff, C. Brinckerhoff, B. Berwin, D. Mullins, and M. Jenkins for helpful discussions, technical guidance, and providing reagents. We thank Z. Fishman for critical review of this manuscript. PLX4720 was provided by Plexxikon Inc. under a Materials Transfer Agreement.
Financial support: NIH R21CA209375-01 (NCI), NIH R01CA120777-06 (NCI), ACS RSG LIB-121864, The Melanoma Research Alliance, and the Norris-Cotton Cancer Center (P30CA023108), to MJT. SMS and BTM were supported by T32GM00874-14. TBS was supported by NIH T32AI0073634. VM, TAW and MLW were supported by NIH P30AR061271. ZL and MLW received support from NIH R01AR061384-01 to MLW. TCC was supported by OCRF, NIH CA146122, NIH CA054174, and the Owens Foundation. Some analyses were carried out in DartLab, a shared resource supported by NIH P30CA023108-36 and NIH P30GM103415-14.
Footnotes
Potential Conflicts of Interest: None
REFERENCES
- 1.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380(9839):358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 2.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Eng J Med. 2012;366(8):707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman PB. Mechanisms of resistance to RAF inhibition in melanomas harboring a BRAF mutation. Am Soc Clin Oncol Educ Book. 2013:80–82. doi: 10.14694/EdBook_AM.2013.33.e80. [DOI] [PubMed] [Google Scholar]
- 4.Johnson DB, Menzies AM, Zimmer L, Eroglu Z, Ye F, Zhao S, et al. Acquired BRAF inhibitor resistance: A multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur J Cancer. 2015;51(18):2792–2799. doi: 10.1016/j.ejca.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eroglu Z, Ribas A. Combination therapy with BRAF and MEK inhibitors for melanoma: latest evidence and place in therapy. Ther Adv Med Oncol. 2016;8(1):48–56. doi: 10.1177/1758834015616934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Eng J Med. 2014;371(20):1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 7.Steinberg SM, Zhang P, Malik BT, Boni A, Shabaneh TB, Byrne KT, et al. BRAF Inhibition Alleviates Immune Suppression in Murine Autochthonous Melanoma. Cancer Immunol Res. 2014;2(11):1044–1050. doi: 10.1158/2326-6066.CIR-14-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knight DA, Ngiow SF, Li M, Parmenter T, Mok S, Cass A, et al. Host immunity contributes to the anti-melanoma activity of BRAF inhibitors. J Clin Invest. 2013;123(3):1371–1381. doi: 10.1172/JCI66236. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Ho PC, Meeth KM, Tsui YC, Srivastava B, Bosenberg MW, Kaech SM. Immune-based antitumor effects of BRAF inhibitors rely on signaling by CD40L and IFNgamma. Cancer Res. 2014;74(12):3205–3217. doi: 10.1158/0008-5472.CAN-13-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilmott JS, Long GV, Howle JR, Haydu LE, Sharma RN, Thompson JF, et al. Selective BRAF inhibitors induce marked T-cell infiltration into human metastatic melanoma. Clin Cancer Res. 2012;18(5):1386–1394. doi: 10.1158/1078-0432.CCR-11-2479. [DOI] [PubMed] [Google Scholar]
- 11.Cooper ZA, Reuben A, Spencer CN, Prieto PA, Austin-Breneman JL, Jiang H, et al. Distinct clinical patterns and immune infiltrates are observed at time of progression on targeted therapy versus immune checkpoint blockade for melanoma. Oncoimmunology. 2016;5(3):e1136044. doi: 10.1080/2162402X.2015.1136044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hugo W, Shi H, Sun L, Piva M, Song C, Kong X, et al. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell. 2015;162(6):1271–1285. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribas A, Hodi FS, Callahan M, Konto C, Wolchok J. Hepatotoxicity with combination of vemurafenib and ipilimumab. N Eng J Med. 2013;368(14):1365–1366. doi: 10.1056/NEJMc1302338. [DOI] [PubMed] [Google Scholar]
- 14.Aris M, Barrio MM. Combining immunotherapy with oncogene-targeted therapy: a new road for melanoma treatment. Front Immunol. 2015;6:46. doi: 10.3389/fimmu.2015.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mok S, Tsoi J, Koya RC, Hu-Lieskovan S, West BL, Bollag G, et al. Inhibition of colony stimulating factor-1 receptor improves antitumor efficacy of BRAF inhibition. BMC Cancer. 2015;15:356. doi: 10.1186/s12885-015-1377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper ZA, Juneja VR, Sage PT, Frederick DT, Piris A, Mitra D, et al. Response to BRAF inhibition in melanoma is enhanced when combined with immune checkpoint blockade. Cancer Immunol Res. 2014;2(7):643–654. doi: 10.1158/2326-6066.CIR-13-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu-Lieskovan S, Mok S, Homet Moreno B, Tsoi J, Robert L, Goedert L, et al. Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAF(V600E) melanoma. Sci Transl Med. 2015;7(279):279ra41. doi: 10.1126/scitranslmed.aaa4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibney GT, Atkins MB. Immunotherapy or molecularly targeted therapy: what is the best initial treatment for stage IV BRAF-mutant melanoma? Clin Adv Hematol Oncol. 2015;13(7):451–458. [PubMed] [Google Scholar]
- 19.Atkins MB, Larkin J. Immunotherapy Combined or Sequenced With Targeted Therapy in the Treatment of Solid Tumors: Current Perspectives. J Natl Cancer Inst. 2016;108(6):djv414. doi: 10.1093/jnci/djv414. [DOI] [PubMed] [Google Scholar]
- 20.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41(5):544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenkins MH, Steinberg SM, Alexander MP, Fisher JL, Ernstoff MS, Turk MJ, et al. Multiple murine BRaf(V600E) melanoma cell lines with sensitivity to PLX4032. Pigment Cell Melanoma Res. 2014;27(3):495–501. doi: 10.1111/pcmr.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 24.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler--a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesokhin AM, Hohl TM, Kitano S, Cortez C, Hirschhorn-Cymerman D, Avogadri F, et al. Monocytic CCR2(+) myeloid-derived suppressor cells promote immune escape by limiting activated CD8 T-cell infiltration into the tumor microenvironment. Cancer Res. 2012;72(4):876–886. doi: 10.1158/0008-5472.CAN-11-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das Thakur M, Stuart DD. Molecular pathways: response and resistance to BRAF and MEK inhibitors in BRAF(V600E) tumors. Clin Cancer Res. 2014;20(5):1074–1080. doi: 10.1158/1078-0432.CCR-13-0103. [DOI] [PubMed] [Google Scholar]
- 27.Hooijkaas A, Gadiot J, Morrow M, Stewart R, Schumacher T, Blank CU. Selective BRAF inhibition decreases tumor-resident lymphocyte frequencies in a mouse model of human melanoma. Oncoimmunology. 2012;1(5):609–617. doi: 10.4161/onci.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwong LN, Boland GM, Frederick DT, Helms TL, Akid AT, Miller JP, et al. Co-clinical assessment identifies patterns of BRAF inhibitor resistance in melanoma. J Clin Invest. 2015;125(4):1459–1470. doi: 10.1172/JCI78954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perna D, Karreth FA, Rust AG, Perez-Mancera PA, Rashid M, Iorio F, et al. BRAF inhibitor resistance mediated by the AKT pathway in an oncogenic BRAF mouse melanoma model. Proc Natl Acad Sci U S A. 2015;112(6):E536–E545. doi: 10.1073/pnas.1418163112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29(22):3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vergani E, Di Guardo L, Dugo M, Rigoletto S, Tragni G, Ruggeri R, et al. Overcoming melanoma resistance to vemurafenib by targeting CCL2-induced miR-34a, miR-100 and miR-125b. Oncotarget. 2016;7(4):4428–4441. doi: 10.18632/oncotarget.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Highfill SL, Cui Y, Giles AJ, Smith JP, Zhang H, Morse E, et al. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci Transl Med. 2014;6(237):237ra67. doi: 10.1126/scitranslmed.3007974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hart KM, Usherwood EJ, Berwin BL. CX3CR1 delineates temporally and functionally distinct subsets of myeloid-derived suppressor cells in a mouse model of ovarian cancer. Immunol Cell Biol. 2014;92(6):499–508. doi: 10.1038/icb.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ugel S, De Sanctis F, Mandruzzato S, Bronte V. Tumor-induced myeloid deviation: when myeloid-derived suppressor cells meet tumor-associated macrophages. J Clin Invest. 2015;125(9):3365–3376. doi: 10.1172/JCI80006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov. 2016;6(2):202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1462–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seifert H, Fisher R, Martin-Liberal J, Edmonds K, Hughes P, Khabra K, et al. Prognostic markers and tumour growth kinetics in melanoma patients progressing on vemurafenib. Melanoma Res. 2016;26(2):138–144. doi: 10.1097/CMR.0000000000000218. [DOI] [PubMed] [Google Scholar]
- 39.Boni A, Cogdill AP, Dang P, Udayakumar D, Njauw CN, Sloss CM, et al. Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Res. 2010;70(13):5213–5219. doi: 10.1158/0008-5472.CAN-10-0118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.