Abstract
As the common birthplace of all human populations, modern humans have lived longer on the African continent than in any other geographical region of the world. This long history, along with the evolutionary need to adapt to environmental challenges such as exposure to infectious agents, has led to greater genetic variation in Africans. The vast genetic variation in Africans also extends to genes involved in the absorption, distribution, metabolism and excretion of pharmaceuticals. Ongoing cataloging of these clinically relevant variants reveals huge allele-frequency differences within and between African populations. Here, we examine Africa's large burden of infectious disease, discuss key examples of known genetic variation modulating disease risk, and provide examples of clinically relevant variants critical for establishing dosing guidelines. We propose that a more systematic characterization of the genetic diversity of African ancestry populations is required if the current benefits of precision medicine are to be extended to these populations.
Introduction
Modern humans evolved in Africa ~200 000 years ago and, although some groups first migrated out roughly 100 000 years ago to populate the world,1 others remained and eventually inhabited the entire continent.2 As groups migrated out of Africa they underwent bottlenecks leading to sharp reductions in population size and genetic diversity.3, 4, 5 To this day, African populations retain the most genetic diversity globally.6 In order to survive both within and out of Africa, early human populations had to adapt to their novel environments including new food resources, colder climates, higher altitudes and, especially, infectious diseases.7, 8 These adaptive requirements, facilitated by natural selection, led to an increased frequency of alleles that were beneficial in that environment. Owing to the fact that these adaptive requirements were driven by local environmental pressures, some of these evolutionarily advantageous alleles display geographic and ancestral specificity as observed in the genomes of present-day humans.8
Finding genetic signals of natural selection is becoming increasingly possible as high-throughput biotechnology and novel computational methods are developed.9, 10 The ability to perform genome-wide association studies and large-scale genetic-variation surveys have led to an exponential increase in the amount of publicly available genetic data,11 and an accompanying explosion in genomic comparisons both within and among populations.12, 13, 14 These data support the clinical significance of identifying genetic outcomes of natural selection such as pinpointing susceptibility loci for infectious diseases plaguing human populations15 or identifying responses to xenobiotic challenges.16 These advances have opened the field of genomic medicine, which takes an individual's genetic data into account when planning clinical care17, 18 and is indispensable in the push for personalized, or precision, medicine. In this capacity, genomics has the potential to inform clinicians and contribute to precision medicine on numerous fronts, such as disease prediction, treatment response and avoidance of adverse drug reactions (ADR) or off-target effects.19
Unfortunately, most clinical studies remain overwhelmingly European-centered19, 20, 21, 22, 23, 24 and the majority of current drug recommendations were established using data from clinical trials performed in Caucasian or Asian populations and may be inappropriate for African populations.19, 25 This sampling bias is not trivial, as estimates for both disease risk and allele frequencies vary significantly between worldwide populations, as well as between African populations.26, 27, 28, 29, 30 In fact, population-specific studies have established that Africa has the highest genetic diversity in the world,31 as is reflected culturally in the over 2000 languages spoken on the continent,32 and genetically in regional allele-frequency differences among African genomes29, 33 and in drug-metabolizing and -transporting genes in particular.34 As a result of this genetic heterogeneity, it is unsafe to extrapolate results from one African ancestral group to another, as a drug which is found to be in the correct plasma concentration in one group can cause reduced effectiveness or, more troublesome, complete drug toxicity in another.35, 36 The differential metabolism of codeine resulting from genetic variability in CYP2D6 provides an example. Owing to multiple gene-duplication events, some individuals have several copies of CYP2D6, leading to ultra-rapid metabolizing activity.37 These individuals quickly convert codeine into morphine, a dangerous outcome that can lead to severe ADR, including death. The global distribution of carriers varies widely, from very low rates reported in West Africa, to 3% of Northern Europeans, 5–10% of Southern Europeans, to a high of about 30% of Ethiopians.37, 38, 39, 40 These data, along with a strong warning from the US Food and Drug Administration against the use of codeine for the management of pain in children,41 and the current lack of infrastructure to genotype individuals, led the Ethiopian Food, Medicine and Health Care Administration and Control Authority to ban its use entirely.42
Genetic variations in genes involved in the ADME (absorption, distribution, metabolism and excretion) of pharmaceuticals influence drug safety and efficacy, and are the foundation of pharmacogenomic research and development.43, 44 Because genetic variation in ADME genes varies significantly between populations,29, 45 determining allele frequencies of genetic variants associated with known ADR in specific populations is critical for dosing guidelines and avoiding therapeutic failure. If the current benefits of personalized medicine are to be extended to African populations, a more thorough characterization of the diversity of African ancestries will be necessary33, 46, 47, 48, 49 and is the motivation for this review. In particular, we will examine Africa's burden of infectious disease, discuss known genetic adaptations to five selected diseases that are prevalent in Africa, and present examples of clinically relevant population-specific variants.
Malaria
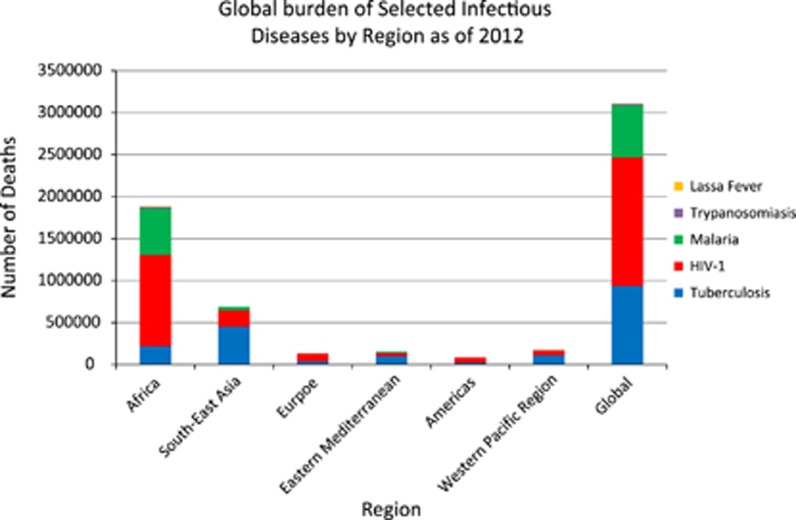
Transmitted to humans by infected female mosquitos, human malaria is caused by four species of the protozoan parasite Plasmodium: P. vivax, P. malariae, P. ovale and, the parasite that causes the most severe cases of malaria, P. falciparum.50 Malaria is endemic throughout most of the tropics, with P. falciparum producing the major burden of disease globally, followed by P. vivax.51 Owing to concerted global efforts, the number of deaths from malaria declined considerably between 2000 and 2015.50 Despite these significant gains, malaria still poses a significant worldwide public health problem, causing ~438 000 fatalities a year, with 292 000 deaths occurring in African children50 (Figure 1).
Figure 1.
The global burden of the diseases discussed in this review as of 2012. HAT, human African trypanosomiasis.
P. falciparum is the leading cause of malaria in Africa, whereas P. vivax is the dominant pathogen outside of Africa.52, 53 The Duffy antigen receptor for chemokines (ACKR1), commonly known as DARC, is the red blood cell receptor for the malaria-causing P. vivax.54 A Duffy-negative phenotype associated with rs2814778 abrogates DARC surface expression on red blood cells and confers resistance to P. vivax.55 This genetic variant is found extensively in malaria-endemic regions of sub-Saharan Africa, especially in West African populations (Supplementary Table 1).53, 54, 55, 56, 57 However, DARC-independent host cell invasion is apparently possible, as studies in Madagascar58 and Brazil59 have identified P. vivax parasites that successfully invaded Duffy-negative erythrocytes.
Upon infection, the parasite incubates in the liver and enters the blood stream infecting red blood cells and causing high fever, sweats, chills and frequently death.50, 60 However, the parasites do not thrive in the presence of sickle-shaped blood cells. Thus, one of the consequences of thousands of years of human co-existence with the malaria pathogen is that inherited red blood cell disorders known as sickle cell disease are maintained as a result of balancing selection.61 Sickle cell disease is a painful disease caused by abnormal hemoglobin alleles called hemoglobin S (HbS, ‘sickle') at rs334 (Supplementary Table 1). Individuals with two sickle alleles (HbSS) produce red blood cells which take the sickle shape, thus severely restricting their oxygen-carrying capacity and interrupting healthy blood flow. Despite the debilitating effects of the homozygous condition, HbS is maintained owing to protection against malaria, as P. falciparum cannot thrive in sickled red blood cells.61, 62, 63, 64 In fact, carriers of the sickle allele have 60% protection against overall mortality,65 resulting in a close geographical overlap of a high sickle allele frequency and malarial endemicity.66
G6PD (glucose-6-phosphate dehydrogenase) deficiency is an inherited enzyme abnormality and, similar to other red blood cell disorders, is prevalent in countries where malaria is historically endemic,67 signifying that the deficiency may confer some protection.68, 69, 70 Primaquine is the only drug licensed for the prevention and, in combination with other drugs, treatment of P. vivax. Unfortunately, the drug can cause severe hemolytic anemia in G6PD-deficient patients.71, 72
Malaria prophylaxis and treatment is challenging because the Plasmodium parasite is capable of generating drug resistance in a relatively short time.73 Therefore, an increasing number of African countries are heeding the recommendations of the World Health Organization for treating malaria by administering artemisinin-based combined treatments, which combines a form of artemisinin (either artemether or artesunate) with a longer-acting drug such as lumefantrine, amodiaquine, mefloquine or sulfadoxine-pyrimethamine.74, 75 Artemisinins work by quickly removing parasites from the blood, leaving fewer numbers upon which the partner drug must act.76 The implementation of combination therapy is intended to reduce parasite resistance, as it should be more difficult for P. falciparum to become resistant to two drugs with unrelated modes of action.77 However, resistance against the limited number of effective drugs is the foremost impediment to successfully treating malaria.78, 79
The artemisinin-based combined treatments presently recommended for treatment of uncomplicated malaria are substrates of CYP enzymes. These highly polymorphic enzymes metabolize >80% of the commonly used therapeutic drugs80 and influence individual variability in drug efficacy.34 It is well-known that allele frequencies in the corresponding CYP genes differ between African populations, translating into differences in drug response.81 For example, amodiaquine is regularly used in conjunction with artesunate, and, to a lesser extent, with sulfadoxine-pyrimethamine.82 The enzyme CYP2C8 metabolizes amodiaquine, as well as another malaria medication, chloroquine.83, 84 A variant of this enzyme, CYP2C8*2, results in the poor metabolizer phenotype, where individuals with at least one copy of CYP2C8*2 experience a longer drug half-life and increased adverse side effects.85 Allele frequencies of CYP2C8*2 differ between East and West Africa.84 Establishing an individual's pharmacogenetic profile with respect to these artemisinin-based combined treatment combinations will allow practitioners to better tailor treatment outcomes and improve rational drug use. A comprehensive list of the known CYP450 enzymes involved in the metabolism of antimalarial drugs includes: Artemether: CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP3A4 and CYP3A5; Lumefantrine: CYP2D6, CYP3A4 and CYP3A5; Amodiaquine; CYP1A1, CYP1B1 and CYP2C8; Mefloquine: CYP3A4 and CYP3A5; Chloroquine: CYP2C8, CYP2D6, CYP3A4 and CYP3A5; Sulfadoxine-pyrimethamine: CYP2C9 and CYP2D6; Primaquine: CYP1A1, CYP1A2, CYP3A4 and CYP3A5; and Quinine: CYP1A1, CYP1A2 and CYP3A5.81
Human african trypanosomiasis
Human African trypanosomiasis (HAT), also known as African sleeping sickness, is spread by blood-feeding tsetse flies infected with one of two protozoans, Trypanosoma brucei (TB) rhodesiense and T.b. gambiense.86, 87 T.b. rhodesiense causes East African (or Rhodesian) trypanosomiasis and accounts for 3% of cases annually, whereas two types of T.b. gambiense, termed groups 1 and 2, cause West African (or Gambian) trypanosomiasis and account for 97% of cases annually.87, 88 Primarily confined to remote rural areas of Sub-Saharan Africa,89, 90 incidences of the two illnesses generally map respectively to the east and west side of the African Rift Valley,91 although this historically defined boundary is shifting in Uganda as infected cattle move northwards, taking T.b. rhodesiense with them.92 Fortunately, disease prevalence has been steadily dropping since systematic global data collection began in 1940, reaching an all-time low of 3796 diagnosed cases in 2015,93 down from 30 000 diagnosed and an estimated 300 000 undiagnosed cases in 1995.94, 95
T.b. gambiense causes a prolonged chronic disease that can take decades before becoming fatal, whereas T.b. rhodesiense causes a more acute illness that progresses rapidly and can lead to death within weeks or months after infection.96, 97, 98 Regardless of the infecting agent, clinical progression of the illness is divided into two stages,96 (1) the early or hemolymphatic stage, characterized by nonspecific inflammatory symptoms similar to those associated with malaria or enteric fever, making diagnosis and treatment a challenge,99 and (2) the late meningoencephalitic stage, characterized by weight loss, behavioral changes, stupor and coma.96, 100 If left untreated, both can be fatal.98
There is an unequal incidence of HAT cases among people living in close geographical proximity to each other, suggesting a genetic component to HAT susceptibility.89 Thus far, genes associated with the susceptibility and severity of HAT include FAS and FASLG,101, 102 IL23R,103 SIGLEC6 and SIGLEC12,104 RNASEL, CXCR6 and IFIH1,105 APOL1,106 and OAS2/3.33
Although the burden of HAT on the African continent has plummeted in recent years, evolutionary signatures of the arms race between humans and trypanosomes are still evident in their genomes.86, 107, 108 For example, humans have a trypanosome lytic factor, which includes the key component APOL1. When trypanosome lytic factor is taken up by trypanosomes, APOL1 localizes to the lysosome and forms a pore that leads to osmotic swelling and rupture of the trypanosome. However, T.b. rhodesiense has evolved a serum resistance-associated virulence factor which, when expressed as a protein in trypanosome lytic factor-resistant lines, binds and inactivates APOL1.86 Variants in the APOL1 gene evade serum resistance-associated inactivation, restoring protection from HAT.109 In fact, the frequency of the HAT-protective G allele at rs73885319 is 26% among the 1000 Genomes AFR populations and as high as nearly 50% among the Esan in Nigeria (Table 1), whereas this variant is absent among non-African ancestry populations.6 Furthermore, using a different resistance mechanism, group 2 T.b. gambiense can also overcome APOL1, an adaptive strategy that reflects the ability of T.b. to continually evolve and infect new hosts.86 Curiously, the protective G allele at rs73885319 is common in Western Africa and rare to absent in Eastern Africa; the advantages conferred by APOL1 variants are not fully explained, although it has been suggested that protection may be conferred against other pathogens besides trypanosomes.109
Table 1. Frequency of G allele at rs73885319.
| Population | Sample size | Frequency | Reference | Population | Sample size | Frequency | Reference |
|---|---|---|---|---|---|---|---|
| Western Africa | Eastern Africa | ||||||
| Esan | 99 | 0.495 | 6 | Sena | 51 | 0.122 | 199 |
| Ibo | 411 | 0.488 | 200 | Malawi | 50 | 0.120 | 199 |
| Edo | 14 | 0.464 | 200 | Baganda | 100 | 0.080 | 33 |
| Akan | 361 | 0.436 | 200 | Barundi | 97 | 0.077 | 33 |
| Asante | 35 | 0.409 | 199 | Banyarwanda | 100 | 0.075 | 33 |
| Yoruba | 563 | 0.342 | 200 | Kikuyu | 55 | 0.064 | 200 |
| Ga-Adangbe | 158 | 0.247 | 200 | Luhya | 99 | 0.056 | 6 |
| Mandinka | 113 | 0.243 | 6 | Sandawe | 19 | 0.050 | 201 |
| Jola | 79 | 0.234 | 33 | Iraqw | 19 | 0.050 | 201 |
| Ewe | 45 | 0.189 | 200 | Hadza | 19 | 0.050 | 201 |
| Mende | 85 | 0.124 | 6 | Bantu Kenya | 12 | 0.045 | 112 |
| Wolof | 78 | 0.122 | 33 | Sudanese | 24 | 0.042 | 202 |
| Fula | 73 | 0.116 | 33 | Kalenjin | 99 | 0.020 | 200 |
| Bulsa | 22 | 0.114 | 199 | Anuak | 76 | 0.020 | 199 |
| Wolayta | 24 | 0 | 33 | ||||
| Southern Africa | Tygray | 21 | 0 | 202 | |||
| Zulu | 100 | 0.130 | 33 | Somali | 38 | 0 | 33 |
| Sotho | 86 | 0.081 | 33 | Sengwer | 19 | 0 | 201 |
| Bantu South Africa | 8 | 0.070 | 112 | Oromo | 26 | 0 | 33 |
| San | 7 | 0 | 112 | Maale | 76 | 0 | 199 |
| Gumuz | 24 | 0 | 33 | ||||
| Central Africa | Borana | 18 | 0 | 201 | |||
| Somie | 65 | 0.164 | 199 | Ari Cultivator | 24 | 0 | 202 |
| Congo | 55 | 0.109 | 199 | Ari Blacksmith | 17 | 0 | 202 |
| Bakola | 19 | 0.050 | 201 | Amhara | 42 | 0 | 33 |
| Biaka Pygmy | 36 | 0.042 | 112 | Afar | 76 | 0 | 199 |
| Mada | 19 | 0.030 | 201 | ||||
| Cameroon | 64 | 0.008 | 199 | Northern Africa | |||
| Mbuti Pygmy | 15 | 0 | 112 | Mozabite | 30 | 0.018 | 112 |
| Lemande | 18 | 0 | 201 | Kordofan | 30 | 0 | 199 |
| Fulani | 19 | 0 | 201 | ||||
Importantly, however, the protection from infectious disease that APOL1 variants confer appear to come at the cost of chronic disease risk. HAT-protective APOL1 variants have been strongly associated with kidney disease and, to a lesser degree, cardiovascular disease.110, 111 This association has been confirmed for nephropathies including focal segmental glomerulosclerosis,106, 112 HIV-associated nephropathy,112, 113 hypertension-attributed end-stage kidney disease,106 severe lupus nephritis114 and chronic kidney disease progression.115 Analyses of APOL1 HAT-protective/kidney disease risk variants have yielded some of the highest recorded odds ratios for a common variant: 29 and 89 for risk of HIV-associated nephropathy in African Americans112 and South African Blacks,113 respectively, and 17 for focal segmental glomerulosclerosis.112 The magnitude of effect suggested by these associations coupled with the relatively high prevalence of the risk alleles have been suggested to underlie the huge public health burden of kidney diseases among African ancestry individuals. The APOL1 story is a notable example of how the shaping of the genome in response to infectious disease can have an impact on chronic disease risk.
According to the CDC, four drugs are currently prescribed for the treatment of HAT, namely: pentamidine, suramin, melarsoprol and eflornithine,116 and some of the pharmacogenomics properties of some of these drugs have been described. For example, pentamidine is transported within the body by human OCT1, a broad selectivity transporter encoded by the highly polymorphic SCL22A1 gene,117, 118 variants of which affect the efficiency of uptake, distribution, and excretion of clinically relevant drugs.117, 119, 120 Furthermore, CYP450 enzymes with roles in the metabolism of pentamidine include CYP1A1, CYP1A2, CYP2C8, CYP2C19, CYP2B6, CYP3A4, CYP3A5 and CYP4A11.81
Human immunodeficiency virus
Sub-Saharan Africa accounts for 70% of the global burden of HIV, with nearly one out of every twenty-five adults infected with the virus121 (Figure 1). HIV is a retrovirus that causes the immune system to become weaker as infection progresses, eventually leading to the most advanced stage of infection, AIDS. There are two types of HIV, HIV-1 and HIV-2, of which the former is the predominant virus worldwide and is highly pathogenic.122 Because the virus affects the immune system, an HIV-positive individual is more susceptible to infections and complications from other illnesses, especially tuberculosis (TB), which are often the ultimate cause of death.123
Early research suggested that host genetic polymorphisms affect individual susceptibility to HIV-1, viral load (indicative of infectiousness), and the speed of disease progression.124, 125, 126 Although HIV-1 is not believed to have been in the human population long enough to have shaped the human genome, susceptibility to HIV-1 does depend in part on an individual's genetic architecture shaped by the evolutionary history of exposure to other infectious diseases.
A fully functioning receptor called CCR5 is required for HIV-1 virus entry.127, 128 A 32-base-pair deletion termed CCR5 Δ32 (rs333) leads to a frameshift in the coding sequence, resulting in a non-functional protein that is not expressed in the cell membrane.129, 130, 131 Individuals homozygous for CCR5 Δ32 do not express functional receptors and are resistant to HIV-1 infection.132, 133 In addition, heterozygosity for the deletion provides a reduction in functional CCR5, thereby conveying partial resistance to HIV-1 infection; when an infection does occur, the viral loads are lower, slowing the progression to AIDS.134
Data suggest that the CCR5 Δ32 deletion arose ~2900 years ago from a single mutation event135 and was subsequently subjected to positive selection,136, 137 resulting in a geographical distribution where it is more prevalent in Europeans and largely absent from Asian and African populations (Supplementary Table 1).129, 130, 136, 138, 139, 140 A twentieth century disease, HIV-1 has not been around long enough for selection pressure to drive CCR5 Δ32 from 0 to 10% in European populations.141 Another pathogen, Yersinia pestis, the cause of the bubonic plague, was proposed as an agent of positive selection for CCR5 Δ32.137 However, the variola virus, which causes smallpox, generates a stronger selective pressure than plague on pre-reproductive members of a population and therefore appears to be a more plausible candidate.142 In addition, both the variola virus and HIV-1 use chemokine receptors to enter white blood cells,143 whereas the plague bacterium has an entirely different mode of entry.
Selection pressure from two infectious agents, both flaviviruses, provides the best evidence thus far to explain why this deletion is not observed in Africa. First, individuals homozygous for CCR5 Δ32 are at higher risk for tick-borne encephalitis.144 Second, fully functional CCR5 reduces symptoms from infection with West Nile virus,145 named for the West Nile district of Uganda where it was first isolated in 1937.146 These data suggest that infectious agents had a role in shaping the genome. Whether CCR5 Δ32 is beneficial or deleterious in the context of a given infection (for example, HIV-1 infection versus West Nile virus) is contingent upon complex interactions between an infecting pathogen and the host immune system.
For those who have access to healthcare in Africa, treatment of HIV/AIDS with highly active antiretroviral therapy has drastically increased life expectancy for infected individuals,147, 148, 149 transforming a once certain fatal disease into a chronic condition. Because patients are now receiving continuing treatment, much of African pharmacogenetic research is focused on optimizing therapeutic responses and preventing ADRs150 by identifying clinically relevant genetic variants.151, 152, 153, 154, 155, 156
Drug efficacy disparities between African populations treated with same active antiretroviral therapy regimen have been reported. For example, efavirenz is metabolized to 8-hydroxyefavirenz mainly by CYP2B6.157 A specific variant of this enzyme, CYP2B6*6, is associated with a higher plasma efavirenz concentration. The frequency distribution of the CYP2B6*6 variant allele is significantly higher in Tanzanians (41.9%) than Ethiopians (31.4%)151 and has also been shown to vary between Zimbabweans (49%) and Ugandans (35%).158, 159
As we observed in the malaria discussion, many CYP450 enzymes are involved in the metabolism of drugs used to fight HIV. A comprehensive list includes: Efavirenz: CYP2B6, CYP3A4, and CYP3A5; Saquinavir: CYP3A4 and CYP3A5; Maraviroc: CYP3A4 and CYP3A5; Nevirapine: CYP2B6; Indinavir: CYP3A4 and CYP3A5; Nelfinavir: CYP3A4 and CYP3A5; Ritonavir: CYP3A4 and CYP3A5; and Lopinavir: CYP3A4 and CYP3A5.
Another example is the HLA-B*5701 allele, which is strongly associated with abacavir hypersensitivity syndrome.160 Abacavir causes serious ADR in 4–8% of patients.160 Screening for the HLA-B*5701 allele has been shown to greatly reduce the frequency of abacavir hypersensitivity syndrome.161, 162 There are clear frequency differences of the HLA-B*5701 allele between African populations, ranging from 0% in the Nigerian Yoruba to 3.3% in the Kenyan Luhya and 13.6% in the Kenyan Maasai.163 Such differences for a clinically relevant variant validates the importance of individual screening and demonstrates the inadequacy of group identity such as ‘African' in medical decision making at the individual level. In fact, the U.S. Food and Drug Administration currently recommends that screening for HLA-B*5701 be completed prior to administration of abacavir for all patients.163 Increasingly, genomic data can help predict immunological response to HIV/AIDS therapeutic medicine, as was observed in HIV infected women in Kenya164 and Zimbabwe165 taking nevirapine-based antiretroviral therapy.
Lassa fever
Lassa fever is a severe viral hemorrhagic fever caused by the Lassa virus. This RNA virus of the Arenaviridae family resides in the rodent vector Mastomys natalensis that lives in close contact with humans and sheds the virus in urine.166, 167 First described in 1969 in the town of Lassa, Nigeria,168, 169 Lassa fever is endemic in the West African countries of Guinea, Liberia, Nigeria and Sierra Leone,170, 171 although it does occur in other countries as well, such as Mali and Côte d'Ivoire.172, 173 The virus infects an estimated 300 000–500 000 people annually, resulting in thousands of deaths in the region.174, 175
The virus enters the cell via its cell-surface receptor, alpha-dystroglycan (DAG1), replicating in a wide variety of cell types.176 The glycosyltransferase LARGE is required for viral entry as it post-translationally modifies DAG1174 by producing a protein the virus needs to infect an individual.177 A genome-wide screen for recent selective sweeps identified a signal for positive selection at a 300 kb region exclusively within the LARGE gene in populations with West African ancestry.178 Further data supporting the hypothesis of this virus as a driver of selection was subsequently reported for LARGE as well for another gene biologically connected to Lassa fever, IL21.174, 179 For both genes, the signal has been localized to putative regulatory regions,174 a potentially important fact in the development of future therapies.
Although there are no approved vaccines for Lassa virus infection, the antiviral therapeutic ribavirin increases survival if given early in infection.180 The pharmacogenomics of ribavirin are well-known in relation to another infectious disease, as pegylated interferon-a and ribavirin-based regimens are the mainstay for treatment of patients with chronic hepatitis C virus genotype 1 infection. In this case in point, a patient's interleukin 28B (IL28B) genotype predicts drug response;181, 182, 183, 184 the same genotype is predicted to affect drug response when given to patients with Lassa Fever, although to date the literature is devoid of any Lassa Fever pharmacogenomic studies.
Tuberculosis
Africa carries the highest overall burden of TB, with 281 cases per 100 000 population in 2014, whereas the global average is 133 per 100 000 population.185 Approximately one-third of the global population has latent, asymptomatic TB,185 and although those individuals only have a 10% chance of developing active TB disease,186 risk is larger in individuals with compromised immune systems owing to conditions such as diabetes or HIV infection. Caused by the bacteria Mycobacterium tuberculosis, TB begins with fever, weight loss and coughing. Coughing progressively becomes worse with sputum and bloody coughs, chest pain and ultimately death.185, 187
Human genetic variation affects susceptibility to mycobacterial infections, and candidate gene-association studies have suggested roles for several genes and pathways.188 A recent genome-wide linkage study in Gambia and South Africa found suggestive linkage on 15q and Xq.189 Many studies have reported associations between susceptibility and resistance to TB and several HLA loci and/or alleles190, 191, 192, 193 and allele frequencies of these markers are known to vary between ethnic groups.194 One recent study from Uganda suggests the HLA-DQB1*03:03 allele may be associated with resistance to TB,186 but much work needs to be done between African populations to assess population-specific allele-frequency differences and/or corresponding pharmacogenomic outcomes. Moreover, the contributions of rare variants with potentially large effects or multiple genes of small effect warrant systematic investigation.188
Isoniazid, rifampicin and pyrazinamide are the most commonly used drugs to fight TB and, as we observed in previous examples, the highly polymorphic CYP450 enzymes are involved in metabolism of these drugs. For example, CYP1A2, CYP2C19 and CYP2E1 affect the metabolism of isoniazid. Metabolism of rifampin involves the actions of CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP3A4 and CYP3A5, CYP2C9, CYP2C19 and CYP2E1 affect the metabolism of ethambutol and the metabolism of pyrazinamide is affected by CYP1A2 and CYP3A4.81
Isoniazid, rifampicin and pyrazinamide can cause serious ADR in some individuals.195 The most serious is anti-TB drug-induced hepatotoxicity, which can lead to treatment failure and interruption, drug resistance, morbidity and mortality.196, 197 An increased risk of ADR is associated with SNPs rs1799929 and rs1495741 in NAT2.197 Ethiopian populations have significantly higher frequencies of these variants when compared with African ancestry 1000 Genomes populations (Supplementary Table 1),36 providing yet another example of the genetic diversity between different African ancestries. In a 2011 study,198 researchers examined the distribution of the SLCO1B1 rs4149032 polymorphism that is associated with low blood concentrations of rifampicin (Supplementary Table 1). They found that the variant allele occurred at a lower frequency in Caucasians or Asians than in African populations. The presence of rs4149032, and subsequent lower rifampicin concentration, means a higher dosage of the drug is required for African populations to obtain the target concentration.198 These two examples demonstrate again how African genetic diversity extends to clinically important genetic variants and supports the urgency of working to identify ancestry-specific pharmacogenomic variants.
Conclusion
This review details the complex history of infectious diseases in Africa and demonstrates how they have shaped African genomes. Documenting the vast genetic variation observed among African populations demonstrates the inadequacy of such group labels as ‘Blacks' or ‘Africans' in biomedical research, especially in the context of pharmacogenomics and medicine. It also calls for the need to engage more diverse populations across the continent to better document the scope and extent of genetic diversity in Africans to ensure they reap the benefits of the global efforts to use genomics to improve the precision of medicine for individuals. Given the extent of disease in Africa, combined with the monetary and human costs incurred from ADRs or ineffectiveness, the necessity for economical approaches to limit disease burden on the continent is evident. Although individualized screening prior to the selection of therapy is currently cost-prohibitive, comprehensive pharmacogenetic profiling of many African populations will produce data that will improve patient care by identifying populations at risk for developing drug toxicity or non-responsiveness until such time as truly personalized medicine can become a reality.
Acknowledgments
This research was supported by the Intramural Research Program of the Center for Research on Genomics and Global Health (CRGGH). The CRGGH is supported by the National Human Genome Research Institute, the National Institute of Diabetes and Digestive and Kidney Diseases, the Center for Information Technology, and the Office of the Director at the National Institutes of Health (1ZIAHG200362).
Footnotes
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
Disclaimer
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official view of the National Institutes of Health.
The authors declare no conflict of interest.
Supplementary Material
References
- Campbell MC, Tishkoff SA. The evolution of human genetic and phenotypic variation in Africa. Curr Biol 2010; 20: R166–R173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet 2008; 9: 403–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos W, Hoffman JI. Evidence that two main bottleneck events shaped modern human genetic diversity. Proc Biol Sci 2010; 277: 131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpending H, Rogers A. Genetic perspectives on human origins and differentiation. Annu Rev Genomics Hum Genet 2000; 1: 361–385. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA 2005; 102: 15942–15947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO et al. A global reference for human genetic variation. Nature 2015; 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaresque PL, Ballereau SJ, Jobling MA. Challenges in human genetic diversity: demographic history and adaptation. Hum Mol Genet 2007; 16 (Spec No. 2): R134–R139. [DOI] [PubMed] [Google Scholar]
- Fumagalli M, Sironi M, Pozzoli U, Ferrer-Admetlla A, Pattini L, Nielsen R. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet 2011; 7: e1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith C, Yang H, Landweber LF, Dunham I, Knoppers BM, Donnelly P et al. Where next for genetics and genomics? PLoS Biol 2015; 13: e1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The next-generation sequencing revolution and its impact on genomics. Cell 2013; 155: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. Next-generation sequencing platforms. Annu Rev Anal Chem 2013; 6: 287–303. [DOI] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science 2008; 319: 1100–1104. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA et al. Genetic structure of human populations. Science 2002; 298: 2381–2385. [DOI] [PubMed] [Google Scholar]
- Shriner D, Tekola-Ayele F, Adeyemo A, Rotimi CN. Genome-wide genotype and sequence-based reconstruction of the 140,000 year history of modern human ancestry. Sci Rep 2014; 4: 6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowell JL, Dowling NF, Yu W, Yesupriya A, Zhang L, Gwinn M. Trends in population-based studies of human genetics in infectious diseases. PLoS One 2012; 7: e25431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther 2011; 89: 464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenke M. Individualized genomics and the future of translational medicine. Mol Genet Genomic Med 2013; 1: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg GS, Willard HF. Genomic and personalized medicine: foundations and applications. Transl Res 2009; 154: 277–287. [DOI] [PubMed] [Google Scholar]
- Adeyemo A, Rotimi C. What does genomic medicine mean for diverse populations? Mol Genet Genomic Med 2014; 2: 3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Goldstein DB. Next generation disparities in human genomics: concerns and remedies. Trends Genet 2009; 25: 489–494. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA 2009; 106: 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MS Jr., Lara PN, Dang JH, Paterniti DA, Kelly K. Twenty years post-NIH Revitalization Act: enhancing minority participation in clinical trials (EMPaCT): laying the groundwork for improving minority clinical trial accrual: renewing the case for enhancing minority participation in cancer clinical trials. Cancer 2014; 120 (Suppl 7): 1091–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchard EG, Oh SS, Foreman MG, Celedon JC. Moving toward true inclusion of racial/ethnic minorities in federally funded studies. A key step for achieving respiratory health equality in the United States. Am J Respir Crit Care Med 2015; 191: 514–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SS, Galanter J, Thakur N, Pino-Yanes M, Barcelo NE, White MJ et al. Diversity in clinical and biomedical research: a promise yet to be fulfilled. PLoS Med 2015; 12: e1001918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai G, Davies G, Denti P, Steimer JL, McIlleron H, Zvada S et al. Pharmacometrics: opportunity for reducing disease burden in the developing world: the case of Africa. CPT Pharmacometrics Syst Pharmacol 2013; 2: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeyemo A, Rotimi C. Genetic variants associated with complex human diseases show wide variation across multiple populations. Public Health Genomics 2010; 13: 72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntzani EE, Liberopoulos G, Manolio TA, Ioannidis JP. Consistency of genome-wide associations across major ancestral groups. Hum Genet 2012; 131: 1057–1071. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Matise TC, North KE, Haiman CA, Fesinmeyer MD, Buyske S et al. Generalization and dilution of association results from European GWAS in populations of non-European ancestry: the PAGE study. PLoS Biol 2013; 11: e1001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos E, Doumatey A, Elkahloun AG, Shriner D, Huang H, Chen G et al. Pharmacogenomics, ancestry and clinical decision making for global populations. Pharmacogenomics J 2014; 14: 217–222. [DOI] [PubMed] [Google Scholar]
- Chen R, Corona E, Sikora M, Dudley JT, Morgan AA, Moreno-Estrada A et al. Type 2 diabetes risk alleles demonstrate extreme directional differentiation among human populations, compared to other diseases. PLoS Genet 2012; 8: e1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A et al. The genetic structure and history of Africans and African Americans. Science 2009; 324: 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashdan E. Ethnic diversity and its environmental determinants: effects of climate, pathogens, and habitat diversity. Am Anthropol 2001; 103: 968–991. [Google Scholar]
- Gurdasani D, Carstensen T, Tekola-Ayele F, Pagani L, Tachmazidou I, Hatzikotoulas K et al. The African Genome Variation Project shapes medical genetics in Africa. Nature 2015; 517: 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerb R, Fux R, Morike K, Kremsner PG, Gil JP, Gleiter CH et al. Pharmacogenetics of antimalarial drugs: effect on metabolism and transport. Lancet Infect Dis 2009; 9: 760–774. [DOI] [PubMed] [Google Scholar]
- Zanger UM, Turpeinen M, Klein K, Schwab M. Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal Bioanal Chem 2008; 392: 1093–1108. [DOI] [PubMed] [Google Scholar]
- Tekola-Ayele F, Adeyemo A, Aseffa A, Hailu E, Finan C, Davey G et al. Clinical and pharmacogenomic implications of genetic variation in a Southern Ethiopian population. Pharmacogenomics J 2015; 15: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aklillu E, Persson I, Bertilsson L, Johansson I, Rodrigues F, Ingelman-Sundberg M. Frequent distribution of ultrarapid metabolizers of debrisoquine in an Ethiopian population carrying duplicated and multiduplicated functional CYP2D6 alleles. J Pharmacol Exp Ther 1996; 278: 441–446. [PubMed] [Google Scholar]
- Bernal ML, Sinues B, Johansson I, McLellan RA, Wennerholm A, Dahl ML et al. Ten percent of North Spanish individuals carry duplicated or triplicated CYP2D6 genes associated with ultrarapid metabolism of debrisoquine. Pharmacogenetics 1999; 9: 657–660. [PubMed] [Google Scholar]
- Sachse C, Brockmoller J, Bauer S, Roots I. Cytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequences. Am J Hum Genet 1997; 60: 284–295. [PMC free article] [PubMed] [Google Scholar]
- Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics 2002; 3: 229–243. [DOI] [PubMed] [Google Scholar]
- FDA Drug Safety Communication. Available at http://wwwfdagov/Drugs/DrugSafety/ucm339112htm (accessed on 20 February 2013).
- Anberbir Y. Ethiopia: Authority Issues Red Alert On Codeine Drug. Available at: http://allafrica.com/stories/201511241318.html. (accessed on 21 November 2015).
- Hoehe MR, Kroslak T. Genetic variation and pharmacogenomics: concepts, facts, and challenges. Dialogues Clin Neurosci 2004; 6: 5–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Lu AY. Pharmacogenetics pharmacogenomics, and individualized medicine. Pharmacol Rev 2011; 63: 437–459. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang L, Zhou H, Stoneking M, Tang K. Global patterns of genetic diversity and signals of natural selection for human ADME genes. Hum Mol Genet 2011; 20: 528–540. [DOI] [PubMed] [Google Scholar]
- Rotimi C, Abayomi A, Abimiku A, Adabayeri VM, Adebamowo C, Adebiyi E et al. Research capacity. Enabling the genomic revolution in Africa. Science 2014; 344: 1346–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonkam A, Mayosi BM. Genomic medicine in Africa: promise, problems and prospects. Genome Med 2014; 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Tindana P, Littler K, Ramsay M, Rotimi C, Abayomi A et al. The H3Africa policy framework: negotiating fairness in genomics. Trends Genet 2015; 31: 117–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwo GH, Williams SM, Moore JH. The future of genomic medicine education in Africa. Genome Med 2015; 7: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. World Malaria Report 2015. Geneva, Switzerland 2015. Available at http://appswhoint/iris/bitstream/10665/200018/1/9789241565158_engpdf?ua=1 (accessed on 2 February 2016).
- Guerra CA, Gikandi PW, Tatem AJ, Noor AM, Smith DL, Hay SI et al. The limits and intensity of Plasmodium falciparum transmission: implications for malaria control and elimination worldwide. PLoS Med 2008; 5: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes RE, Reiner RC Jr., Battle KE, Longbottom J, Mappin B, Ordanovich D et al. Plasmodium vivax transmission in Africa. PLoS Negl Trop Dis 2015; 9: e0004222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg 2001; 64 (1-2 Suppl): 97–106. [DOI] [PubMed] [Google Scholar]
- Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med 1976; 295: 302–304. [DOI] [PubMed] [Google Scholar]
- Langhi DM Jr., Bordin JO. Duffy blood group and malaria. Hematology 2006; 11: 389–398. [DOI] [PubMed] [Google Scholar]
- Gething PW, Elyazar IR, Moyes CL, Smith DL, Battle KE, Guerra CA et al. A long neglected world malaria map: Plasmodium vivax endemicity in 2010. PLoS Negl Trop Dis 2012; 6: e1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes RE, Patil AP, Piel FB, Nyangiri OA, Kabaria CW, Gething PW et al. The global distribution of the Duffy blood group. Nat Commun 2011; 2: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci USA 2010; 107: 5967–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavasini CE, Mattos LC, Couto AA, Bonini-Domingos CR, Valencia SH, Neiras WC et al. Plasmodium vivax infection among Duffy antigen-negative individuals from the Brazilian Amazon region: an exception? Trans R Soc Trop Med Hyg 2007; 101: 1042–1044. [DOI] [PubMed] [Google Scholar]
- Wassmer SC, Taylor TE, Rathod PK, Mishra SK, Mohanty S, Arevalo-Herrera M et al. Investigating the pathogenesis of severe malaria: a multidisciplinary and cross-geographical approach. Am J Trop Med Hyg 2015; 93 (3 Suppl): 42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasvol G, Weatherall DJ, Wilson RJ. Cellular mechanism for the protective effect of haemoglobin S against P. falciparum malaria. Nature 1978; 274: 701–703. [DOI] [PubMed] [Google Scholar]
- Hill AV, Allsopp CE, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA et al. Common west African HLA antigens are associated with protection from severe malaria. Nature 1991; 352: 595–600. [DOI] [PubMed] [Google Scholar]
- Fumagalli M, Cagliani R, Pozzoli U, Riva S, Comi GP, Menozzi G et al. Widespread balancing selection and pathogen-driven selection at blood group antigen genes. Genome Res 2009; 19: 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison AC. Protection afforded by sickle-cell trait against subtertian malareal infection. Br Med J 1954; 1: 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aidoo M, Terlouw DJ, Kolczak MS, McElroy PD, ter Kuile FO, Kariuki S et al. Protective effects of the sickle cell gene against malaria morbidity and mortality. Lancet 2002; 359: 1311–1312. [DOI] [PubMed] [Google Scholar]
- Piel FB, Patil AP, Howes RE, Nyangiri OA, Gething PW, Williams TN et al. Global distribution of the sickle cell gene and geographical confirmation of the malaria hypothesis. Nat Commun 2010; 1: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes RE, Battle KE, Satyagraha AW, Baird JK, Hay SI. G6PD deficiency: global distribution, genetic variants and primaquine therapy. Adv Parasitol 2013; 81: 133–201. [DOI] [PubMed] [Google Scholar]
- Bienzle U, Ayeni O, Lucas AO, Luzzatto L. Glucose-6-phosphate dehydrogenase and malaria. Greater resistance of females heterozygous for enzyme deficiency and of males with non-deficient variant. Lancet 1972; 1: 107–110. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Varkonyi R, Cahinhinan N, Abbes S, Argyropoulos G, Destro-Bisol G et al. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science 2001; 293: 455–462. [DOI] [PubMed] [Google Scholar]
- Howes RE, Piel FB, Patil AP, Nyangiri OA, Gething PW, Dewi M et al. G6PD deficiency prevalence and estimates of affected populations in malaria endemic countries: a geostatistical model-based map. PLoS Med 2012; 9: e1001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alving AS, Carson PE, Flanagan CL, Ickes CE. Enzymatic deficiency in primaquine-sensitive erythrocytes. Science 1956; 124: 484–485. [DOI] [PubMed] [Google Scholar]
- Domingo GJ, Satyagraha AW, Anvikar A, Baird K, Bancone G, Bansil P et al. G6PD testing in support of treatment and elimination of malaria: recommendations for evaluation of G6PD tests. Malar J 2013; 12: 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahinas D, Folefoc A, Pillai DR. Targeting Plasmodium falciparum Hsp90: towards reversing antimalarial resistance. Pathogens 2013; 2: 33–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Guidelines for the treatment of malaria. 2015; Third edition. Available at: http://www.who.int/malaria/publications/atoz/9789241549127/en/ (accessed on 10 October 2016).
- Ogbonna A, Uneke CJ. Artemisinin-based combination therapy for uncomplicated malaria in sub-Saharan Africa: the efficacy, safety, resistance and policy implementation since Abuja 2000. Trans R Soc Trop Med Hyg 2008; 102: 621–627. [DOI] [PubMed] [Google Scholar]
- Roederer MW, McLeod H, Juliano JJ. Can pharmacogenomics improve malaria drug policy? Bull World Health Organ 2011; 89: 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung S, Pongtavornpinyo W, Hastings IM, Mills AJ, White NJ. Antimalarial drug resistance, artemisinin-based combination therapy, and the contribution of modeling to elucidating policy choices. Am J Trop Med Hyg 2004; 71 (2 Suppl): 179–186. [PubMed] [Google Scholar]
- Hyde JE. Drug-resistant malaria—an insight. FEBS J 2007; 274: 4688–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlotra RK, Henry-Halldin CN, Zimmerman PA. Application of pharmacogenomics to malaria: a holistic approach for successful chemotherapy. Pharmacogenomics 2009; 10: 435–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandara C, Swart M, Mpeta B, Wonkam A, Masimirembwa C. Cytochrome P450 pharmacogenetics in African populations: implications for public health. Expert Opin Drug Metab Toxicol 2014; 10: 769–785. [DOI] [PubMed] [Google Scholar]
- Bains RK. African variation at Cytochrome P450 genes: evolutionary aspects and the implications for the treatment of infectious diseases. Evol Med Public Health 2013; 20131: 118–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher JF, Aubouy A, Adeothy A, Cottrell G, Doritchamou J, Gourmel B et al. Comparison of sulfadoxine-pyrimethamine, unsupervised artemether-lumefantrine, and unsupervised artesunate-amodiaquine fixed-dose formulation for uncomplicated plasmodium falciparum malaria in Benin: a randomized effectiveness noninferiority trial. J Infect Dis 2009; 200: 57–65. [DOI] [PubMed] [Google Scholar]
- Li XQ, Bjorkman A, Andersson TB, Ridderstrom M, Masimirembwa CM. Amodiaquine clearance and its metabolism to N-desethylamodiaquine is mediated by CYP2C8: a new high affinity and turnover enzyme-specific probe substrate. J Pharmacol Exp Ther 2002; 300: 399–407. [DOI] [PubMed] [Google Scholar]
- Paganotti GM, Gramolelli S, Tabacchi F, Russo G, Modiano D, Coluzzi M et al. Distribution of human CYP2C8*2 allele in three different African populations. Malar J 2012; 11: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh S, Ouedraogo JB, Goldstein JA, Rosenthal PJ, Kroetz DL. Amodiaquine metabolism is impaired by common polymorphisms in CYP2C8: implications for malaria treatment in Africa. Clin Pharmacol Ther 2007; 82: 197–203. [DOI] [PubMed] [Google Scholar]
- Capewell P, Cooper A, Clucas C, Weir W, Macleod A. A co-evolutionary arms race: trypanosomes shaping the human genome, humans shaping the trypanosome genome. Parasitology 2015; 142 (Suppl 1): S108–S119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capewell P, Veitch NJ, Turner CM, Raper J, Berriman M, Hajduk SL et al. Differences between Trypanosoma brucei gambiense groups 1 and 2 in their resistance to killing by trypanolytic factor 1. PLoS Negl Trop Dis 2011; 5: e1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pays E, Vanhollebeke B, Uzureau P, Lecordier L, Perez-Morga D. The molecular arms race between African trypanosomes and humans. Nat Rev Microbiol 2014; 12: 575–584. [DOI] [PubMed] [Google Scholar]
- Welburn SC, Molyneux DH, Maudlin I. Beyond Tsetse—implications for research and control of human African trypanosomiasis epidemics. Trends Parasitol 2015; 32: 230–241. [DOI] [PubMed] [Google Scholar]
- Franco JR, Simarro PP, Diarra A, Jannin JG. Epidemiology of human African trypanosomiasis. Clin Epidemiol 2014; 6: 257–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn SC, Fevre EM, Coleman PG, Odiit M, Maudlin I. Sleeping sickness: a tale of two diseases. Trends Parasitol 2001; 17: 19–24. [DOI] [PubMed] [Google Scholar]
- Wardrop NA, Atkinson PM, Gething PW, Fevre EM, Picozzi K, Kakembo AS et al. Bayesian geostatistical analysis and prediction of Rhodesian human African trypanosomiasis. PLoS Negl Trop Dis 2010; 4: e914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health OrganizationCases of sleeping sickness drop to lowest level in 75 years. Geneva, Switzerland 2015. Available at http://www.who.int/trypanosomiasis_african/cases_drop_to_lowest_since_75_years/en/ (accessed on 2 February 2016).
- World Health OrganizationControl and surveillance of African trypanosomiasis. Report of a WHO Expert Committee. WHO technical report series 881: Geneva, Switzerland, 1998. [PubMed] [Google Scholar]
- Simarro PP, Diarra A, Ruiz Postigo JA, Franco JR, Jannin JG. The human African trypanosomiasis control and surveillance programme of the World Health Organization 2000-2009: the way forward. PLoS Negl Trop Dis 2011; 5: e1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checchi F, Filipe JA, Haydon DT, Chandramohan D, Chappuis F. Estimates of the duration of the early and late stage of gambiense sleeping sickness. BMC Infect Dis 2008; 8: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odiit M, Kansiime F, Enyaru JC. Duration of symptoms and case fatality of sleeping sickness caused by Trypanosoma brucei rhodesiense in Tororo, Uganda. East Afr Med J 1997; 74: 792–795. [PubMed] [Google Scholar]
- Lamour SD, Gomez-Romero M, Vorkas PA, Alibu VP, Saric J, Holmes E et al. Discovery of infection associated metabolic markers in human African trypanosomiasis. PLoS Negl Trop Dis 2015; 9: e0004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg JM. Human African trypanosomiasis: clinical presentation and immune response. Parasite Immunol 2004; 26: 469–476. [DOI] [PubMed] [Google Scholar]
- Checchi F, Funk S, Chandramohan D, Haydon DT, Chappuis F. Updated estimate of the duration of the meningo-encephalitic stage in gambiense human African trypanosomiasis. BMC Res Notes 2015; 8: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes MF, Nunes MP, Henriques-Pons A, Giese N, Morse HC 3rd, Davidson WF et al. Increased susceptibility of Fas ligand-deficient gld mice to Trypanosoma cruzi infection due to a Th2-biased host immune response. Eur J Immunol 1999; 29: 81–89. [DOI] [PubMed] [Google Scholar]
- Martins GA, Petkova SB, MacHado FS, Kitsis RN, Weiss LM, Wittner M et al. Fas-FasL interaction modulates nitric oxide production in Trypanosoma cruzi-infected mice. Immunology 2001; 103: 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro CM, Pontes MJ, Bird S, Chadzinska M, Scheer M, Verburg-van Kemenade BM et al. Trypanosomiasis-induced Th17-like immune responses in carp. PLoS One 2010; 5: e13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol 2007; 7: 255–266. [DOI] [PubMed] [Google Scholar]
- Fumagalli M, Pozzoli U, Cagliani R, Comi GP, Bresolin N, Clerici M et al. Genome-wide identification of susceptibility alleles for viral infections through a population genetics approach. PLoS Genet 2010; 6: e1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 2010; 329: 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean L, Chisi JE, Odiit M, Gibson WC, Ferris V, Picozzi K et al. Severity of human african trypanosomiasis in East Africa is associated with geographic location, parasite genotype, and host inflammatory cytokine response profile. Infect Immun 2004; 72: 7040–7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzureau P, Uzureau S, Lecordier L, Fontaine F, Tebabi P, Homble F et al. Mechanism of Trypanosoma brucei gambiense resistance to human serum. Nature 2013; 501: 430–434. [DOI] [PubMed] [Google Scholar]
- Thomson R, Genovese G, Canon C, Kovacsics D, Higgins MK, Carrington M et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci USA 2014; 111: E2130–E2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamal KJ, Tremaglio J, Friedman DJ, Ix JH, Kuller LH, Tracy RP et al. APOL1 genotype, kidney and cardiovascular disease, and death in older adults. Arterioscler Thromb Vasc Biol 2016; 36: 398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Bick AG, Flannick J, Friedman DJ, Genovese G, Parfenov MG et al. Increased burden of cardiovascular disease in carriers of APOL1 genetic variants. Circ Res 2014; 114: 845–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol 2011; 22: 2129–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasembeli AN, Duarte R, Ramsay M, Mosiane P, Dickens C, Dix-Peek T et al. APOL1 risk variants are strongly associated with HIV-associated nephropathy in Black South Africans. J Am Soc Nephrol 2015; 26: 2882–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman BI, Langefeld CD, Andringa KK, Croker JA, Williams AH, Garner NE et al. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol 2014; 66: 390–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsa A, Kao WH, Xie D, Astor BC, Li M, Hsu CY et al. APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med 2013; 369: 2183–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Parasites- African Trypanosomiasis (also known as Sleeping Sickness) 2016. Available at: http://www.cdc.gov/parasites/sleepingsickness/health_professionals/ (accessed on 10 October 2016).
- Arimany-Nardi C, Koepsell H, Pastor-Anglada M. Role of SLC22A1 polymorphic variants in drug disposition, therapeutic responses, and drug-drug interactions. Pharmacogenomics J 2015; 15: 473–487. [DOI] [PubMed] [Google Scholar]
- Shu Y, Leabman MK, Feng B, Mangravite LM, Huang CC, Stryke D et al. Evolutionary conservation predicts function of variants of the human organic cation transporter, OCT1. Proc Natl Acad Sci USA 2003; 100: 5902–5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorboulev V, Ulzheimer JC, Akhoundova A, Ulzheimer-Teuber I, Karbach U, Quester S et al. Cloning and characterization of two human polyspecific organic cation transporters. DNA Cell Biol 1997; 16: 871–881. [DOI] [PubMed] [Google Scholar]
- Zhang L, Dresser MJ, Gray AT, Yost SC, Terashita S, Giacomini KM. Cloning and functional expression of a human liver organic cation transporter. Mol Pharmacol 1997; 51: 913–921. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Global Health Observatory (GHO) data, HIV/AIDS 2015. Available at: http://www.who.int/gho/hiv/en/ (accessed on 10 October 2016).
- McCutchan FE. Global epidemiology of HIV. J Med Virol 2006; 78 (Suppl 1): S7–s12. [DOI] [PubMed] [Google Scholar]
- Worodria W, Massinga-Loembe M, Mazakpwe D, Luzinda K, Menten J, Van Leth F et al. Incidence and predictors of mortality and the effect of tuberculosis immune reconstitution inflammatory syndrome in a cohort of TB/HIV patients commencing antiretroviral therapy. J Acquir Immune Defic Syndr 2011; 58: 32–37. [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Bamshad M, Sato N, Mummidi S, Dhanda R, Catano G et al. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc Natl Acad Sci USA 1999; 96: 12004–12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliekelman P, Garner C, Slatkin M. Natural selection and resistance to HIV. Nature 2001; 411: 545–546. [DOI] [PubMed] [Google Scholar]
- Winkler C, An P, O'Brien SJ. Patterns of ethnic diversity among the genes that influence AIDS. Hum Mol Genet 2004; 13 (Spec No)1: R9–19. [DOI] [PubMed] [Google Scholar]
- Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 1996; 86: 367–377. [DOI] [PubMed] [Google Scholar]
- Le AQ, Taylor J, Dong W, McCloskey R, Woods C, Danroth R et al. Differential evolution of a CXCR4-using HIV-1 strain in CCR5wt/wt and CCR532/32 hosts revealed by longitudinal deep sequencing and phylogenetic reconstruction. Sci Rep 2015; 5: 17607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 1996; 382: 722–725. [DOI] [PubMed] [Google Scholar]
- Martinson JJ, Chapman NH, Rees DC, Liu YT, Clegg JB. Global distribution of the CCR5 gene 32-basepair deletion. Nat Genet 1997; 16: 100–103. [DOI] [PubMed] [Google Scholar]
- Hill CM, Littman DR. Natural resistance to HIV? Nature 1996; 382: 668–669. [DOI] [PubMed] [Google Scholar]
- Michael NL, Chang G, Louie LG, Mascola JR, Dondero D, Birx DL et al. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat Med 1997; 3: 338–340. [DOI] [PubMed] [Google Scholar]
- Huang Y, Paxton WA, Wolinsky SM, Neumann AU, Zhang L, He T et al. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med 1996; 2: 1240–1243. [DOI] [PubMed] [Google Scholar]
- Eugen-Olsen J, Iversen AK, Garred P, Koppelhus U, Pedersen C, Benfield TL et al. Heterozygosity for a deletion in the CKR-5 gene leads to prolonged AIDS-free survival and slower CD4 T-cell decline in a cohort of HIV-seropositive individuals. AIDS 1997; 11: 305–310. [DOI] [PubMed] [Google Scholar]
- Hummel S, Schmidt D, Kremeyer B, Herrmann B, Oppermann M. Detection of the CCR5-Delta32 HIV resistance gene in Bronze Age skeletons. Genes Immun 2005; 6: 371–374. [DOI] [PubMed] [Google Scholar]
- Libert F, Cochaux P, Beckman G, Samson M, Aksenova M, Cao A et al. The deltaccr5 mutation conferring protection against HIV-1 in Caucasian populations has a single and recent origin in Northeastern Europe. Hum Mol Genet 1998; 7: 399–406. [DOI] [PubMed] [Google Scholar]
- Stephens JC, Reich DE, Goldstein DB, Shin HD, Smith MW, Carrington M et al. Dating the origin of the CCR5-Delta32 AIDS-resistance allele by the coalescence of haplotypes. Am J Hum Genet 1998; 62: 1507–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem AH, Batzer MA. Distribution of the HIV resistance CCR5-Delta32 allele among Egyptians and Syrians. Mutat Res 2007; 616: 175–180. [DOI] [PubMed] [Google Scholar]
- Bharti D, Kumar A, Mahla RS, Kumar S, Ingle H, Yadav T et al. Low prevalence of CCR5-Delta32, CCR2-64I and SDF1-3'A alleles in the Baiga and Gond tribes of Central India. SpringerPlus 2015; 4: 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Nerurkar VR, Dashwood WM, Woodward CL, Ablan S, Shikuma CM et al. Genotype and allele frequency of a 32-base pair deletion mutation in the CCR5 gene in various ethnic groups: absence of mutation among Asians and Pacific Islanders. Int J Infect Dis 1999; 3: 186–191. [DOI] [PubMed] [Google Scholar]
- Novembre J, Galvani AP, Slatkin M. The geographic spread of the CCR5 Delta32 HIV-resistance allele. PLoS Biol 2005; 3: e339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani AP, Slatkin M. Evaluating plague and smallpox as historical selective pressures for the CCR5-Delta 32 HIV-resistance allele. Proc Natl Acad Sci USA 2003; 100: 15276–15279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani AS, Masters J, Zeng W, Barrett J, Pannu R, Everett H et al. Use of chemokine receptors by poxviruses. Science 1999; 286: 1968–1971. [DOI] [PubMed] [Google Scholar]
- Kindberg E, Mickiene A, Ax C, Akerlind B, Vene S, Lindquist L et al. A deletion in the chemokine receptor 5 (CCR5) gene is associated with tickborne encephalitis. J Infect Dis 2008; 197: 266–269. [DOI] [PubMed] [Google Scholar]
- Lim JK, Louie CY, Glaser C, Jean C, Johnson B, Johnson H et al. Genetic deficiency of chemokine receptor CCR5 is a strong risk factor for symptomatic West Nile virus infection: a meta-analysis of 4 cohorts in the US epidemic. J Infect Dis 2008; 197: 262–265. [DOI] [PubMed] [Google Scholar]
- Smithburn KHT, Burke A. A neurotropic virus isolated from the blood of a native of Uganda. Am J Trop Med Hyg 1940; 20: 471–492. [Google Scholar]
- Nsanzimana S, Remera E, Kanters S, Chan K, Forrest JI, Ford N et al. Life expectancy among HIV-positive patients in Rwanda: a retrospective observational cohort study. Lancet Global Health 2015; 3: e169–e177. [DOI] [PubMed] [Google Scholar]
- Ross EL, Weinstein MC, Schackman BR, Sax PE, Paltiel AD, Walensky RP et al. The clinical role and cost-effectiveness of long-acting antiretroviral therapy. Clin Infect Dis 2015; 60: 1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker LG, Venter F, Cohen K, Goemare E, Van Cutsem G, Boulle A et al. Provision of antiretroviral therapy in South Africa: the nuts and bolts. Antivir Ther 2014; 19 (Suppl 3): 105–116. [DOI] [PubMed] [Google Scholar]
- Aceti A, Gianserra L, Lambiase L, Pennica A, Teti E. Pharmacogenetics as a tool to tailor antiretroviral therapy: a review. World J Virol 2015; 4: 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngaimisi E, Habtewold A, Minzi O, Makonnen E, Mugusi S, Amogne W et al. Importance of ethnicity, CYP2B6 and ABCB1 genotype for efavirenz pharmacokinetics and treatment outcomes: a parallel-group prospective cohort study in two sub-Saharan Africa populations. PLoS One 2013; 8: e67946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart M, Whitehorn H, Ren Y, Smith P, Ramesar RS, Dandara C. PXR and CAR single nucleotide polymorphisms influence plasma efavirenz levels in South African HIV/AIDS patients. BMC Med Genet 2012; 13: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habtewold A, Amogne W, Makonnen E, Yimer G, Riedel KD, Ueda N et al. Long-term effect of efavirenz autoinduction on plasma/peripheral blood mononuclear cell drug exposure and CD4 count is influenced by UGT2B7 and CYP2B6 genotypes among HIV patients. J Antimicrob Chemother 2011; 66: 2350–2361. [DOI] [PubMed] [Google Scholar]
- Yimer G, Amogne W, Habtewold A, Makonnen E, Ueda N, Suda A et al. High plasma efavirenz level and CYP2B6*6 are associated with efavirenz-based HAART-induced liver injury in the treatment of naive HIV patients from Ethiopia: a prospective cohort study. Pharmacogenomics J 2012; 12: 499–506. [DOI] [PubMed] [Google Scholar]
- Mukonzo JK, Okwera A, Nakasujja N, Luzze H, Sebuwufu D, Ogwal-Okeng J et al. Influence of efavirenz pharmacokinetics and pharmacogenetics on neuropsychological disorders in Ugandan HIV-positive patients with or without tuberculosis: a prospective cohort study. BMC Infect Dis 2013; 13: 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade K, Geese WJ, Noor M, Flint O, Tebas P, Mulligan K et al. Genetic analysis implicates resistin in HIV lipodystrophy. AIDS 2008; 22: 1561–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward BA, Gorski JC, Jones DR, Hall SD, Flockhart DA, Desta Z. The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther 2003; 306: 287–300. [DOI] [PubMed] [Google Scholar]
- Mukonzo JK, Roshammar D, Waako P, Andersson M, Fukasawa T, Milani L et al. A novel polymorphism in ABCB1 gene, CYP2B6*6 and sex predict single-dose efavirenz population pharmacokinetics in Ugandans. Br J Clin Pharmacol 2009; 68: 690–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyakutira C, Roshammar D, Chigutsa E, Chonzi P, Ashton M, Nhachi C et al. High prevalence of the CYP2B6 516G→T(*6) variant and effect on the population pharmacokinetics of efavirenz in HIV/AIDS outpatients in Zimbabwe. Eur J Clin Pharmacol 2008; 64: 357–365. [DOI] [PubMed] [Google Scholar]
- Phillips E, Mallal S. Successful translation of pharmacogenetics into the clinic: the abacavir example. Mol Diagn Ther 2009; 13: 1–9. [DOI] [PubMed] [Google Scholar]
- Hetherington S, Hughes AR, Mosteller M, Shortino D, Baker KL, Spreen W et al. Genetic variations in HLA-B region and hypersensitivity reactions to abacavir. Lancet 2002; 359: 1121–1122. [DOI] [PubMed] [Google Scholar]
- Mallal S, Nolan D, Witt C, Masel G, Martin AM, Moore C et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet 2002; 359: 727–732. [DOI] [PubMed] [Google Scholar]
- Rotimi CN, Jorde LB. Ancestry and disease in the age of genomic medicine. N Engl J Med 2010; 363: 1551–1558. [DOI] [PubMed] [Google Scholar]
- Oluka MN, Okalebo FA, Guantai AN, McClelland RS, Graham SM. Cytochrome P450 2B6 genetic variants are associated with plasma nevirapine levels and clinical response in HIV-1 infected Kenyan women: a prospective cohort study. AIDS Res Ther 2015; 12: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhandire D, Lacerda M, Castel S, Mhandire K, Zhou D, Swart M et al. Effects of CYP2B6 and CYP1A2 genetic variation on nevirapine plasma concentration and pharmacodynamics as measured by CD4 cell count in Zimbabwean HIV-infected patients. Omics 2015; 19: 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monath TP, Newhouse VF, Kemp GE, Setzer HW, Cacciapuoti A. Lassa virus isolation from Mastomys natalensis rodents during an epidemic in Sierra Leone. Science 1974; 185: 263–265. [DOI] [PubMed] [Google Scholar]
- Mylne AQ, Pigott DM, Longbottom J, Shearer F, Duda KA, Messina JP et al. Mapping the zoonotic niche of Lassa fever in Africa. Trans R Soc Trop Med Hyg 2015; 109: 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame JD, Baldwin JM Jr., Gocke DJ, Troup JM. Lassa fever, a new virus disease of man from West Africa. I. Clinical description and pathological findings. Am J Trop Med Hyg 1970; 19: 670–676. [DOI] [PubMed] [Google Scholar]
- Buckley SM, Casals J. Lassa fever, a new virus disease of man from West Africa. 3. Isolation and characterization of the virus. Am J Trop Med Hyg 1970; 19: 680–691. [DOI] [PubMed] [Google Scholar]
- Bowen MD, Rollin PE, Ksiazek TG, Hustad HL, Bausch DG, Demby AH et al. Genetic diversity among Lassa virus strains. J Virol 2000; 74: 6992–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichet-Calvet E, Rogers DJ. Risk maps of Lassa fever in West Africa. PLoS Negl Trop Dis 2009; 3: e388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin S, Anaraki S, Gothard P, Walsh A, Brown D, Gopal R et al. The first case of Lassa fever imported from Mali to the United Kingdom, February 2009. Euro Surveil 2009; 14: pii. [PubMed] [Google Scholar]
- Gunther S, Emmerich P, Laue T, Kuhle O, Asper M, Jung A et al. Imported lassa fever in Germany: molecular characterization of a new lassa virus strain. Emerg Infect Dis 2000; 6: 466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen KG, Shylakhter I, Tabrizi S, Grossman SR, Happi CT, Sabeti PC. Genome-wide scans provide evidence for positive selection of genes implicated in Lassa fever. Philos Trans R Soc Lond B Biol Sci 2012; 367: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick JB. Clinical, epidemiologic, and therapeutic aspects of Lassa fever. Med Microbiol Immunol 1986; 175: 153–155. [DOI] [PubMed] [Google Scholar]
- Cao W, Henry MD, Borrow P, Yamada H, Elder JH, Ravkov EV et al. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 1998; 282: 2079–2081. [DOI] [PubMed] [Google Scholar]
- Kunz S, Rojek JM, Kanagawa M, Spiropoulou CF, Barresi R, Campbell KP et al. Posttranslational modification of alpha-dystroglycan, the cellular receptor for arenaviruses, by the glycosyltransferase LARGE is critical for virus binding. J Virol 2005; 79: 14282–14296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C et al. Genome-wide detection and characterization of positive selection in human populations. Nature 2007; 449: 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Cagliani R, Riva S, Pozzoli U, Biasin M, Piacentini L et al. Population genetics of IFIH1: ancient population structure, local selection, and implications for susceptibility to type 1 diabetes. Mol Biol Evol 2010; 27: 2555–2566. [DOI] [PubMed] [Google Scholar]
- McCormick JB, King IJ, Webb PA, Scribner CL, Craven RB, Johnson KM et al. Lassa fever. Effective therapy with ribavirin. N Engl J Med 1986; 314: 20–26. [DOI] [PubMed] [Google Scholar]
- Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 2009; 461: 399–401. [DOI] [PubMed] [Google Scholar]
- Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 2010; 138: 1338–1345. [DOI] [PubMed] [Google Scholar]
- Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet 2009; 41: 1100–1104. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet 2009; 41: 1105–1109. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Global tuberculosis report 2015. Available at: http://apps.who.int/iris/bitstream/10665/191102/1/9789241565059_eng.pdf (accessed on 10 October 2016).
- Wamala D, Buteme HK, Kirimunda S, Kallenius G, Joloba M. Association between human leukocyte antigen class II and pulmonary tuberculosis due to mycobacterium tuberculosis in Uganda. BMC Infect Dis 2016; 16: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev 2003; 16: 463–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller M, Hoal EG. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis 2010; 90: 71–83. [DOI] [PubMed] [Google Scholar]
- Bellamy R, Beyers N, McAdam KP, Ruwende C, Gie R, Samaai P et al. Genetic susceptibility to tuberculosis in Africans: a genome-wide scan. Proc Natl Acad Sci USA 2000; 97: 8005–8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettaneh A, Seng L, Tiev KP, Toledano C, Fabre B, Cabane J. Human leukocyte antigens and susceptibility to tuberculosis: a meta-analysis of case-control studies. Int J Tuberc Lung Dis 2006; 10: 717–725. [PubMed] [Google Scholar]
- Dubaniewicz A, Lewko B, Moszkowska G, Zamorska B, Stepinski J. Molecular subtypes of the HLA-DR antigens in pulmonary tuberculosis. Int J Infect Dis 2000; 4: 129–133. [DOI] [PubMed] [Google Scholar]
- Yuliwulandari R, Sachrowardi Q, Nakajima H, Kashiwase K, Hirayasu K, Mabuchi A et al. Association of HLA-A, -B, and -DRB1 with pulmonary tuberculosis in western Javanese Indonesia. Hum Immunol 2010; 71: 697–701. [DOI] [PubMed] [Google Scholar]
- Mishra G, Kumar N, Kaur G, Jain S, Tiwari PK, Mehra NK. Distribution of HLA-A, B and DRB1 alleles in Sahariya tribe of North Central India: an association with pulmonary tuberculosis. Infect Genet Evol 2014; 22: 175–182. [DOI] [PubMed] [Google Scholar]
- Lombard Z, Brune AE, Hoal EG, Babb C, Van Helden PD, Epplen JT et al. HLA class II disease associations in southern Africa. Tissue Antigens 2006; 67: 97–110. [DOI] [PubMed] [Google Scholar]
- Yew WW, Leung CC. Antituberculosis drugs and hepatotoxicity. Respirology 2006; 11: 699–707. [DOI] [PubMed] [Google Scholar]
- Tostmann A, Boeree MJ, Aarnoutse RE, de Lange WC, van der Ven AJ, Dekhuijzen R. Antituberculosis drug-induced hepatotoxicity: concise up-to-date review. J Gastroenterol Hepatol 2008; 23: 192–202. [DOI] [PubMed] [Google Scholar]
- Wang PY, Xie SY, Hao Q, Zhang C, Jiang BF. NAT2 polymorphisms and susceptibility to anti-tuberculosis drug-induced liver injury: a meta-analysis. Int J Tuberc Lung Dis 2012; 16: 589–595. [DOI] [PubMed] [Google Scholar]
- Chigutsa E, Visser ME, Swart EC, Denti P, Pushpakom S, Egan D et al. The SLCO1B1 rs4149032 polymorphism is highly prevalent in South Africans and is associated with reduced rifampin concentrations: dosing implications. Antimicrob Agents Chemother 2011; 55: 4122–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Kedem E, Rosset S, Haileselassie Y, Tzur S, Kra-Oz Z et al. Absence of APOL1 risk variants protects against HIV-associated nephropathy in the Ethiopian population. Am J Nephrol 2011; 34: 452–459. [DOI] [PubMed] [Google Scholar]
- Adeyemo AA, Tekola-Ayele F, Doumatey AP, Bentley AR, Chen G, Huang H et al. Evaluation of genome wide association study associated type 2 diabetes susceptibility loci in Sub Saharan Africans. Front Genet 2015; 6: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko WY, Rajan P, Gomez F, Scheinfeldt L, An P, Winkler CA et al. Identifying Darwinian selection acting on different human APOL1 variants among diverse African populations. Am J Hum Genet 2013; 93: 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani L, Kivisild T, Tarekegn A, Ekong R, Plaster C, Gallego Romero I et al. Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am J Hum Genet 2012; 91: 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.